Abstract
Based mostly on the results of in vitro experiments, ADF (actin-depolymerizing factor) proteins are thought to be key modulators of the dynamic organization of the actin cytoskeleton. The few studies concerned with the in vivo function of ADF proteins that have been reported to date were performed almost exclusively using single-cell systems and have failed to produce consistent results. To investigate ADF functions in vivo and during the development of multicellular organs, we generated transgenic Arabidopsis plants that express a cDNA encoding an ADF protein (AtADF1) in the sense or the antisense orientation under the control of a strong constitutively active promoter. Selected lines with significantly altered levels of AtADF protein expression were characterized phenotypically. Overexpression of AtADF1 resulted in the disappearance of thick actin cables in different cell types, caused irregular cellular and tissue morphogenesis, and reduced the growth of cells and organs. In contrast, reduced AtADF expression promoted the formation of actin cables, resulted in a delay in flowering, and stimulated cell expansion as well as organ growth. These results are consistent with the molecular functions of ADF as predicted by in vitro studies, support the global roles of ADF proteins during the development of a multicellular organism, and demonstrate that these proteins are key regulators of F-actin organization, flowering, and cell and organ expansion in Arabidopsis.
INTRODUCTION
The plant actin cytoskeleton is known to play key roles in the morphogenesis and function of highly specialized cell types, such as pollen tubes, root hairs, trichomes (leaf hairs), and stomatal guard cells. In addition, as demonstrated using single-cell model systems, F-actin structures also are required for a number of essential cellular processes, including cell division and cell expansion, which are thought to be crucially important for the morphogenesis of multicellular plant organs (reviewed in Kost et al., 1999b). However, few studies have addressed directly the functions of the actin cytoskeleton during plant development (Ramachandran et al., 2000; Baluska et al., 2001).
A substantial body of work, performed mostly using animal or yeast systems, has shown that the organization and function of the actin cytoskeleton are controlled by a large and heterogenous family of actin binding proteins (Ayscough, 1998). Among these, proteins of the actin-depolymerizing factor (ADF/cofilin) class stand out as potential key regulators of F-actin organization (Carlier et al., 1997; reviewed in Staiger et al., 1997; Bamburg, 1999). Genes encoding ADF/cofilin proteins have been identified and cloned from a variety of organisms, including Saccharomyces cerevisiae (Iida et al., 1993; Moon et al., 1993), Dictyostelium discoideum (Aizawa et al., 1995), and Drosophila melanogaster (Gunsalus et al., 1995). In higher plants, cDNAs encoding ADF proteins have been cloned from Lilium longiflorum (Kim et al., 1993), Brassica napus (Kim et al., 1993), maize (Rozycka et al., 1995; Lopez et al., 1996), and Arabidopsis (Dong et al., 2001). Whereas the genomes of most nonplant organisms appear to contain only one or two genes encoding ADF/cofilin proteins, ADF genes were shown to form a large family in Arabidopsis that consists of at least nine members (AtADF1 to AtADF9; Dong et al., 2001). Surprisingly large families of cytoskeletal genes typically are found in the genomes of a variety of plant species (Meagher et al., 1999).
The different ADF/cofilin proteins identified in various organisms show high sequence homology. Extensive in vitro studies have indicated that they are functionally well conserved as well. ADF/cofilin proteins were shown to bind to monomeric (G-) and filamentous (F-) actin in a pH-dependent manner (Yonezawa et al., 1985; Hawkins et al., 1993; Hayden et al., 1993). It has been suggested that they promote F-actin disassembly by severing actin filaments (Mabuchi, 1983; Moriyama and Yahara, 1999; Pope et al., 2000) and by significantly enhancing the rate of monomer dissociation from the “pointed” (unstable) ends of actin filaments (Carlier et al., 1997; Maciver et al., 1998). The ability of ADF/cofilin proteins to stimulate monomer dissociation from actin filaments was discovered in part through in vitro experiments using recombinant AtADF1 (Carlier et al., 1997), the protein whose in vivo functions we are investigating in this study. It has been demonstrated that ADF/cofilin proteins destabilize the association of monomers with the pointed ends of actin filaments by inducing conformational changes upon binding to F-actin polymers (McGough et al., 1997).
Not surprisingly, considering the fact that ADF/cofilin proteins are thought to be key modulators of F-actin organization, the activity of these proteins was found to be controlled by different signaling molecules. Phosphatidylinositol 4,5-bisphosphate, a well-known actin regulator, was demonstrated to inhibit the activity of ADF/cofilin proteins by preventing them from binding to actin (Yonezawa et al., 1990). More recently, phosphorylation of a conserved serine residue at the N terminus was found to inhibit the actin binding activity of ADF/cofilin proteins (Agnew et al., 1995; Moriyama et al., 1996; Smertenko et al., 1998). Kinases were identified that mediate specific phosphorylation of this site (Arber et al., 1998; Smertenko et al., 1998; Yang et al., 1998; Y. Hong and N.-H. Chua, unpublished data).
Despite the extensive in vitro characterization of ADF/cofilin proteins, little has been done to investigate the functions of these proteins in living cells. Cofilin was found to be essential for the vegetative growth of yeast cells (Iida et al., 1993; Moon et al., 1993) and for the development of Drosophila. Disruption of an ADF/cofilin gene in the Drosophila twinstar mutant was shown to inhibit centrosome migration and cytokinesis, which leads to the early arrest of larval development (Gunsalus et al., 1995). To our knowledge, the description of the twinstar phenotype is the only report available in the literature that addresses the function of ADF/cofilin proteins in the context of the development of a multicellular organism.
In a few studies, the effects of experimentally increasing ADF/cofilin protein levels in living cells have been investigated. All of these studies were performed using single-cell systems, and their results are controversial. Microinjection of cofilin into muscle cells induced the formation of short F-actin–cofilin rods (Nagaoka et al., 1995). In contrast, overexpression of cofilin in Dictyostelium was found to stimulate F-actin bundling, membrane ruffling, and cell movement (Aizawa et al., 1996, 1997). In Tradescantia stamen hair cells, microinjected maize ADF1 (ZmADF1) transformed longitudinal actin cables into thick, transverse arrays (Hussey et al., 1998). We demonstrated recently that transiently expressed AtADF proteins, including AtADF1, bind to F-actin, reduce the number and length of actin filaments, and may promote F-actin bundling in living onion and tobacco cells (Dong et al., 2001).
To initiate the investigation of the functions of the large AtADF gene family in vivo and during plant development, we generated transgenic Arabidopsis lines that express the cDNA encoding one member (AtADF1) in the sense or the antisense orientation under the control of the strong and constitutively expressed cauliflower mosaic virus (CaMV) 35S promoter. This approach allowed us to analyze the phenotypes of plants that either produced 30 to 40 times more AtADF1 than wild-type plants or showed significantly reduced expression of AtADF1 and other AtADF isoforms. Altered levels of AtADF expression in Arabidopsis plants were found to affect F-actin organization, cell expansion, organ growth, and flowering time.
RESULTS
Generation and Molecular Characterization of Transgenic Arabidopsis Lines with Altered Levels of AtADF Expression
To explore the functions of ADF proteins during plant growth and development, we cloned the AtADF1 cDNA downstream of a CaMV 35S promoter in both the sense and the antisense orientations. The CaMV 35S promoter was chosen because it is known to be constitutively active in Arabidopsis plants and to confer particularly high expression levels in the vascular tissue, an expression pattern that matches that of the AtADF1 promoter (Dong et al., 2001). The resulting sense and antisense constructs were transformed into Arabidopsis (ecotype C24) using Agrobacterium tumefaciens–mediated gene transfer. The growth and development of transgenic Arabidopsis plants with altered levels of ADF expression were analyzed.
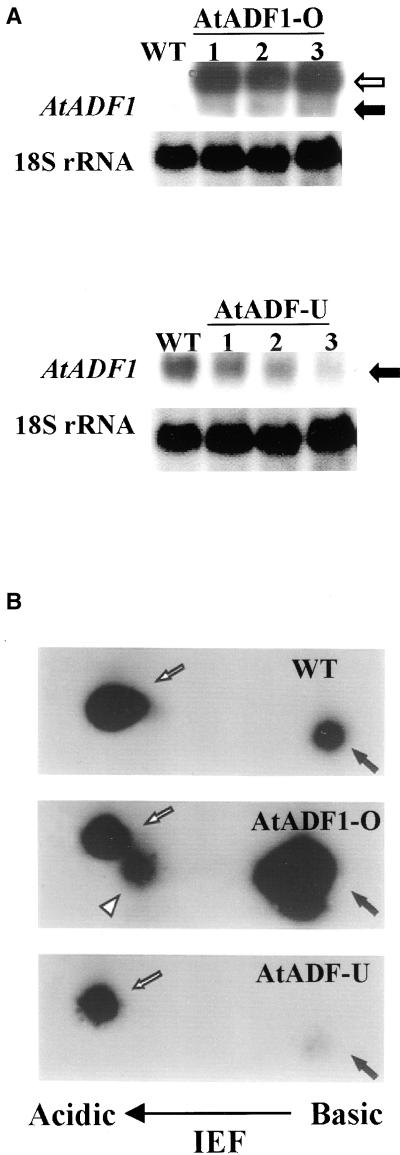
In total, 10 lines transformed with the sense construct (AtADF1-O lines) and nine lines transformed with the antisense construct (AtADF-U lines) were obtained. Of these, six AtADF1-O lines and four AtADF-U lines showed phenotypes that were similar within each of the two groups of lines. Three independent lines of each group with distinct transgene integration sites, as determined by DNA gel blot analysis (data not shown), and with characteristic phenotypes were selected for further analysis. RNA gel blot experiments revealed that the selected AtADF1-O lines contained 30 to 50 times more AtADF1 transcript than did wild-type plants (Figure 1A, top). In contrast, in all AtADF-U lines analyzed, the amount of transcript detected with a probe corresponding to a fragment of the AtADF1 5′ untranslated region was reduced two to three times (Figure 1A, bottom). Difficulties with the identification of probes that allow the detection of gene-specific transcripts of particular AtADF genes (Dong et al., 2001) prevented us from analyzing in detail the expression levels of each of the AtADF genes in AtADF-U plants.
Figure 1.
Molecular Characterization of Selected AtADF1-O and AtADF-U Lines.
(A) RNA gel blot analysis. A total of 10 μg (top) or 30 μg (bottom) of total RNA prepared from 10-day-old seedlings was loaded in each lane. Blots were hybridized with antisense RNA transcribed from the AtADF1 5′ untranslated region and with a probe that recognizes 18S rRNA to control for loading. Closed arrows, endogenous AtADF1 mRNA; open arrow, transcript of the AtADF1 transgene (200 to 300 bp longer than the endogenous transcript because of the addition of the E9 poly[A] signal [Leu et al., 1996]). WT, wild type.
(B) Two-dimensional immunoblotting. For each panel, 80 μg of total protein was separated by SDS-PAGE followed by isoelectric focusing (IEF). A polyclonal antibody raised against AtADF1 was used to detect ADF proteins. Closed arrows, AtADF1; open arrows, other ADF isoform; arrowhead, spot detected exclusively when AtADF1-O protein preparations were blotted, likely representing phosphorylated AtADF1 (same molecular mass but different charge; AtADF proteins are known to be regulated by phosphorylation [Y. Hong and N.-H. Chua, unpublished data]).
AtADF1 protein levels in the AtADF1-O lines were increased 30 to 40 times, as detected by one-dimensional (data not shown) and two-dimensional immunoblotting (data for one AtADF1-O line are shown in Figure 1B, middle; similar data were obtained for the other two lines), using a polyclonal antibody that recognizes several AtADF isoforms, including AtADF1. In the AtADF-U lines, AtADF1 protein levels were reduced dramatically and the expression of other ADF isoforms was decreased at least threefold (data for one AtADF-U line are shown in Figure 1B, bottom; similar data were obtained for the other two lines). Because isoform-specific AtADF antibodies were not available, it was not possible to determine exactly which AtADF isoforms displayed reduced expression in AtADF-U plants.
Altered ADF Expression Levels Affect Flowering Time and Plant Growth
AtADF1-O and AtADF-U plants were fully fertile (data not shown) and indistinguishable from wild-type plants in terms of most aspects of their development and morphology. However, AtADF-U plants were considerably delayed in flowering and produced ∼50% more rosette leaves than did wild-type and AtADF1-O plants. In addition, throughout ontogenesis, AtADF1-O and AtADF-U plants were clearly distinct in size from wild-type plants.
Under standard growth conditions in our greenhouse, wild-type Arabidopsis plants produced a mean of 17 rosette leaves before they formed an inflorescence, and the first flowers opened on average 35 days after germination (Table 1). Although AtADF1-O plants behaved very similarly, AtADF-U plants began flowering ∼2 weeks later, after they had formed an average of 26 rosette leaves (Table 1).
Table 1.
Flowering Time (Days after Germination) and Rosette Leaf Number of Wild-Type, AtADF1-O, and AtADF-U Plants
| Type of Plant | Flowering Time | Leaf Number |
|---|---|---|
| Wild typea | 35.33 (±1.377) | 16.57 (±0.952) |
| AtADF1-Oa | 37.07 (±1.093) | 16.46 (±1.098) |
| AtADF-Ua | 49.33 (±2.786) | 25.61 (±1.903) |
a Data represent three independent experiments conducted with at least 15 plants of each genotype. The 95% confidence intervals are given in parentheses.
The most striking phenotype displayed by AtADF1-O and AtADF-U plants was their abnormal size. The length of adult inflorescences, as well as of cotyledons, hypocotyls, and roots at the seedling stage, was decreased notably in the case of AtADF1-O plants and increased significantly in the case of AtADF-U plants (Table 2 and Figure 2), indicating that ADF proteins are key regulators of tissue expansion in developing Arabidopsis organs.
Table 2.
Average Length (in millimeters) of Different Organs of 10-Day-Old Light-Grown Seedlings and of the Inflorescences of 68-Day-Old Adult Plants
| Type of Plant | Cotyledon | Hypocotyl | Root | Inflorescence |
|---|---|---|---|---|
| Wild typea | 3.90 (±0.096) | 2.59 (±0.096) | 25.10 (±0.929) | 288.5 (±11.08) |
| AtADF1-Oa | 2.13 (±0.088) | 2.42 (±0.123) | 21.45 (±0.386) | 279.0 (±15.69) |
| AtADF-Ua | 4.28 (±0.123) | 2.80 (±0.114) | 27.75 (±0.815) | 364.1 (±29.99) |
a The 95% confidence intervals are given in parentheses.
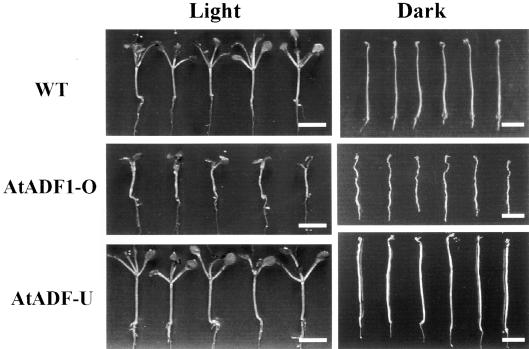
Figure 2.
Phenotypes of Light- and Dark-Grown Wild-Type, AtADF1-O, and AtADF-U Seedlings.
Seedlings grown in vitro for 10 days under a regular light regimen (left) or in complete darkness (right) are shown. Top row, wild-type (WT) seedlings; middle row, AtADF1-O seedlings; bottom row, AtADF-U seedlings.  .
.
The hypocotyl of etiolated (dark-grown) seedlings is an extremely rapidly expanding organ that is quite amenable to experimental analysis. Therefore, we decided to focus on this organ for a more detailed analysis of the function of AtADF proteins in the control of tissue growth.
The Size and Morphology of Hypocotyl Cells in Etiolated Seedlings Are Controlled by ADF Expression Levels
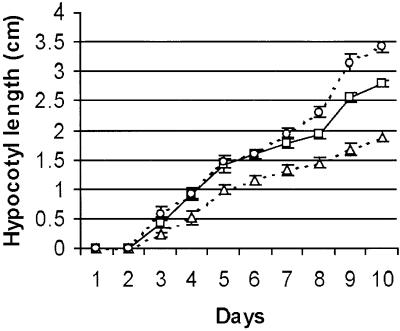
The average hypocotyl lengths of wild-type, AtADF1-O, and AtADF-U seedlings growing on culture plates in the dark were determined at daily intervals during the first 10 days after germination. Significant differences in the lengths of wild-type and transgenic hypocotyls were detected after 4 days in the case of AtADF1-O seedlings and after 8 days in the case of AtADF-U seedlings (Figure 3). Wild-type seedlings grew to a mean length of 2.8 cm in 10 days. At the same time, AtADF1-O and AtADF-U hypocotyls had reached an average of 67.5 and 122% of that length, respectively (Figures 2 and 3). Wild-type and AtADF-U hypocotyls were straight, whereas AtADF1-O hypocotyls developed a wavy morphology (Figure 2).
Figure 3.
Elongation Kinetics of Etiolated Wild-Type, AtADF1-O, and AtADF-U Hypocotyls.
Seedlings were grown in vitro for 10 days in complete darkness. Each time point represents 15 seedlings. Open squares, wild type; open triangles, AtADF1-O; open circles, AtADF-U. Error bars indicate ±sd.
Measurement of the mean cell length in hypocotyls of 10-day-old etiolated seedlings showed that AtADF1-overexpressing cells had reached only 66% (329.5 ± 12 μm [95% confidence interval]) of the length of wild-type cells (501.5 ± 18 μm), whereas cells with reduced expression of AtADF proteins were 24% longer (622.5 ± 29 μm) than such cells. These results demonstrate that alterations in the extent of cell expansion accounted for all of the observed differences in hypocotyl length between wild-type and transgenic seedlings.
Not only the length but also the radial diameter and the morphology of hypocotyl cells were affected by altered AtADF protein levels, as observed using scanning electron microscopy and histology. AtADF1-O hypocotyl cells were irregularly shaped and often displayed a clearly increased radial diameter (Figures 4F and 4G). As a consequence of this, the circumference of AtADF1-O hypocotyls was enlarged (Figure 4F), and tissue organization was disrupted significantly (Figures 4B, 4F, and 4G). Cells in the epidermis were particularly affected, which resulted in an uneven appearance of this cell layer in surface views (Figure 4B). It is likely that these defects were responsible for the wavy morphology of etiolated AtADF1-O hypocotyls described above. Although the radial diameter of cells in AtADF-U hypocotyls also was increased, these cells were as regularly shaped as wild-type cells. Correspondingly, we observed an enlargement of the circumference of AtADF-U hypocotyls but no disruption of tissue organization (Figures 4C, 4H, and 4I).
Figure 4.
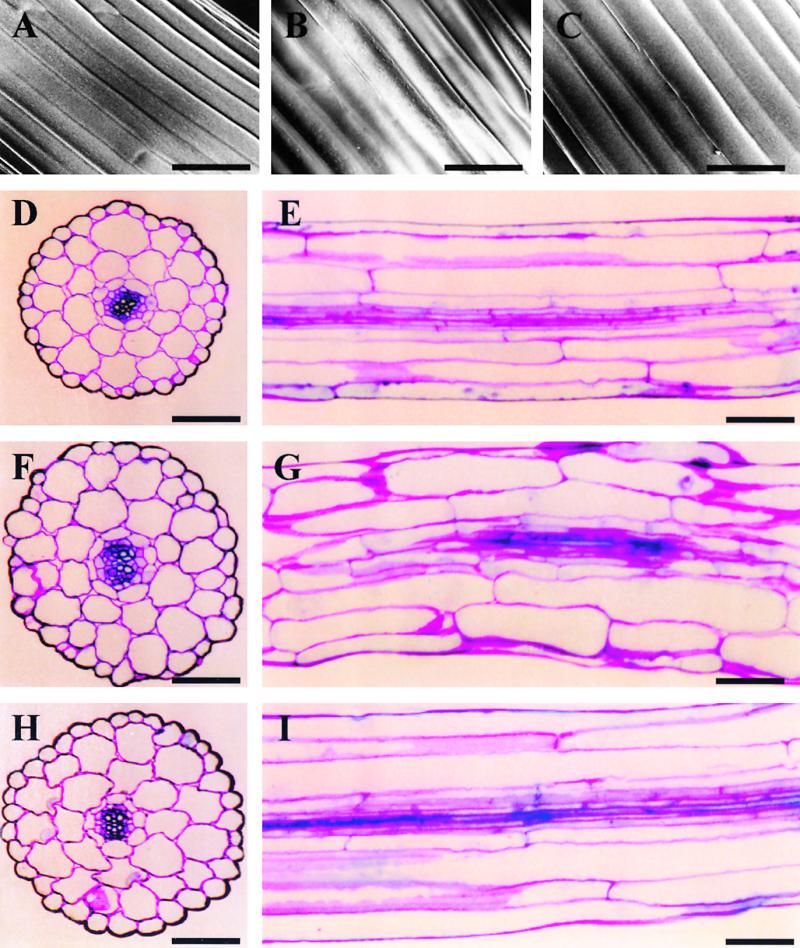
Scanning Electron Microscopy and Histological Analysis of Etiolated Hypocotyls.
Specimens were taken from the central region of the hypocotyls of 10-day-old dark-grown seedlings. (A) to (C) show surface views generated using scanning electron microscopy, and (D) to (I) show histological sections of cell walls stained with toluidine blue O. (D), (F), and (H) are cross-sections, and (E), (G), and (I) are longitudinal sections.
(A), (D), and (E) Wild type.
(B), (F), and (G) AtADF1-O.
(C), (H), and (I) AtADF-U.
Because of the wavy morphology of AtADF1-O hypocotyls, it was impossible to obtain longitudinal sections that showed extended stretches of vascular tissue. The vascular tissue in (G) therefore appears incomplete.  .
.
Altered Levels of ADF Expression Affect F-Actin Organization in Hypocotyl Cells
To investigate the effects of altered levels of ADF expression on the organization of the actin cytoskeleton in hypocotyl cells, we introduced into AtADF1-O and AtADF-U lines by genetic crosses a cDNA encoding a green fluorescent protein (GFP)–mouse talin fusion protein (GFP-mTn), which was shown to allow noninvasive observation of F-actin structures in living plant cells and tissues (Kost et al., 1998), under the control of a CaMV 35S promoter. Expression of the GFP-mTn fusion protein in AtADF1-O and AtADF-U plants did not affect their phenotypes (data not shown).
Confocal imaging of GFP-mTn fluorescence emitted from cells of dark- and light-grown wild-type hypocotyls revealed the presence in these cells of an F-actin network consisting of thick, often longitudinally oriented cables and finer, randomly arranged filaments (Figures 5A and 5D). Thick actin cables were particularly abundant in cells of etiolated hypocotyls, indicating that their presence is associated with rapid cell elongation (Figure 5A).
Figure 5.
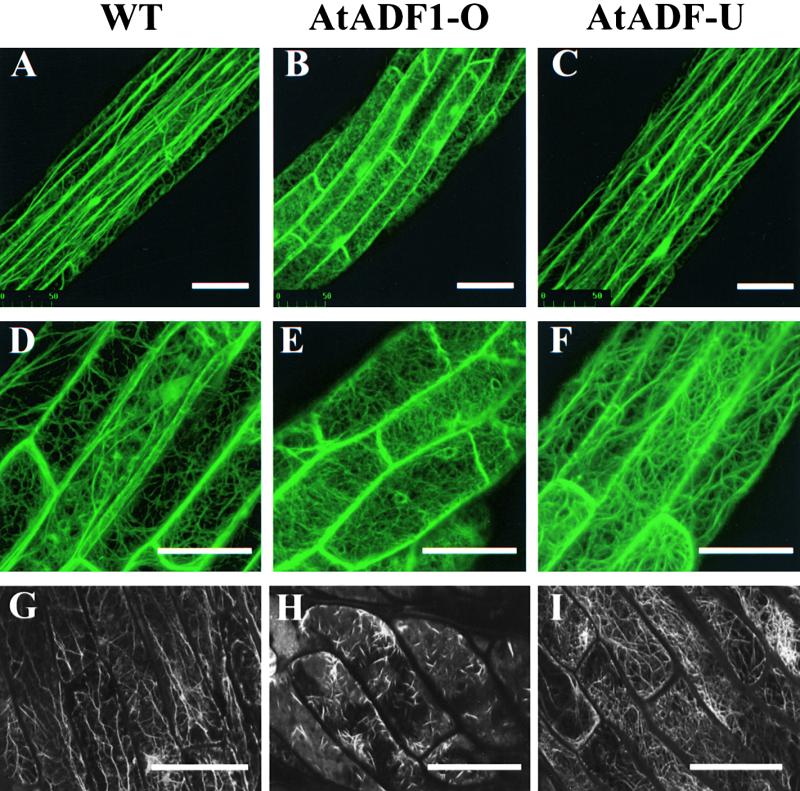
F-Actin Organization in Epidermal Cells of Hypocotyls and Petioles.
(A) to (F) show GFP fluorescence emitted from the central regions of the hypocotyls of 10-day-old dark-grown (top row) and light-grown (middle row) GFP-mTn–expressing seedlings. (G) to (I) show fluorescein phalloidin fluorescence emitted from cotyledon petioles of 10-day-old dark-grown fixed seedlings. Projections of serial confocal optical sections are shown.  .
.
(A), (D), and (G) Wild type (WT).
(B), (E), and (H) AtADF1-O.
(C), (F), and (I) AtADF-U.
Interestingly, altered AtADF protein levels were found to cause significant changes in F-actin organization in hypocotyl cells. Cells of both light- and dark-grown AtADF1-O hypocotyls contained no thick actin cables, whereas fine actin filaments appeared less affected (Figures 5B and 5E). In contrast, AtADF-U hypocotyl cells consistently showed a slight but clear increase in the number of actin cables compared with wild-type cells (Figures 5C and 5F).
Because GFP-mTn binding may interfere with the interaction between actin filaments and AtADF proteins, we confirmed these observations using fluorescein phalloidin staining of actin filaments in wild-type, AtADF1-O, and AtADF-U seedlings in the absence of the GFP-mTn fusion protein. Although this technique did not yield reproducible results when applied to hypocotyl cells, it allowed the visualization in cotyledon petiole cells of essentially the same effects of altered AtADF protein levels on F-actin organization described above (Figures 5G, 5H, and 5I). AtADF1 overexpression resulted in the disruption of actin cables present in wild-type cells (Figures 5G and 5H), whereas reduced AtADF expression stimulated the formation of actin cables (Figure 5I). The difference in the appearance of the remaining F-actin structures in AtADF1-O cells, as visualized using GFP-mTn expression (Figures 5B and 5E) and fluorescein phalloidin staining (Figure 5H), may result either from a stabilization of actin filaments by bound GFP-mTn or from fluorescein phalloidin staining artifacts.
F-Actin Reorganization Caused by Altered AtADF Expression Levels Affects the Morphology and Function of Specialized Cell Types
The actin cytoskeleton has been shown to be essential for the morphogenesis and function of a number of highly specialized cell types, including root hairs (Miller et al., 1998), trichomes (Mathur et al., 1999; Szymanski et al., 1999), and stomatal guard cells (Eun and Lee, 1997). Using scanning electron microscopy and stable expression of the GFP-mTn fusion protein, we investigated the morphology and the actin cytoskeleton, respectively, of these three cell types in wild- type plants and plants with altered ADF expression levels.
Severe disruption of F-actin organization and morphological aberrations were detected in all three cell types when AtADF1-O plants were examined. Careful analysis of AtADF-U plants revealed no abnormalities in trichomes and stomatal guard cells but showed a clear increase in the average length of root hairs, which was accompanied by changes in F-actin organization (Figure 6 and Table 3).
Figure 6.
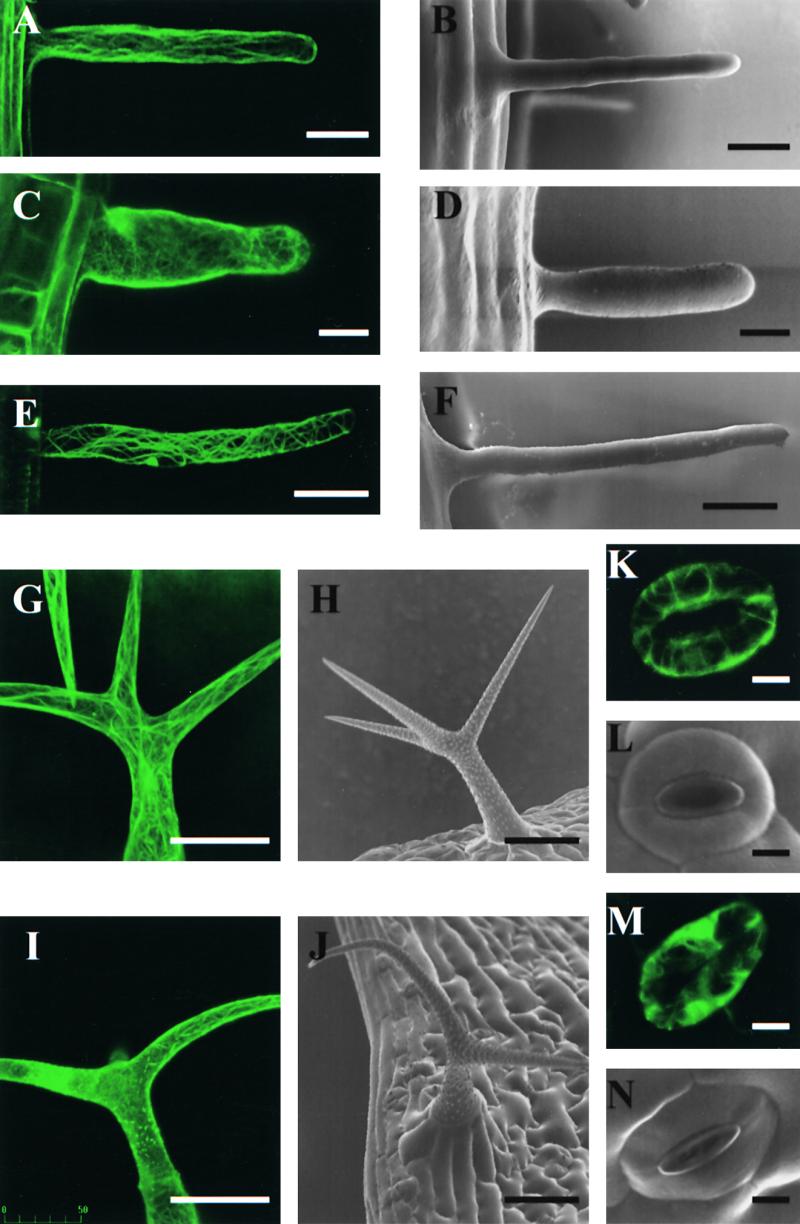
Morphology and Actin Cytoskeleton of Specialized Cell Types.
(B), (D), (F), (H), (J), (L), and (N) show cell morphology as observed using scanning electron microscopy; (A), (C), (E), (G), (I), (K), and (M) show F-actin organization as visualized using stable expression of GFP-mTn. Projections of serial confocal optical sections are shown. All images depict selected cells with representative phenotypes. Wild-type trichomes display a variable degree of branching (Mathur and Chua, 2000), which was not affected significantly by AtADF1 overexpression (data not shown).
(A) and (B) Wild-type root hair.
(C) and (D) AtADF1-O root hair.
(E) and (F) AtADF-U root hair.
(G) and (H) Wild-type trichome.
(I) and (J) AtADF1-O trichome.
(K) and (L) Wild-type stomata.
(M) and (N) AtADF1-O stomata.
 in (A) to (N)
in (A) to (N)  .
.
Table 3.
Average Length of Wild-Type, AtADF1-O, and AtADF-U Root Hairs
| Type of Plant | Root Hair Length (μm) | Percentage of Wild Type |
|---|---|---|
| Wild type | 173.6 (±8.48) | 100.0 |
| AtADF1-Oa | 128.3 (±4.97) | 73.9 |
| AtADF-Ua | 225.0 (±9.45) | 129.6 |
a Because the percentage of abnormal root hairs on individual AtADF1-O and AtADF-U plants was quite variable (see Discussion), transgenic plants with a high percentage (>70%) of affected root hairs were selected for this analysis. The 95% confidence interval is given in parentheses.
Figures 6A and 6B depict the actin cytoskeleton and the morphology, respectively, of wild-type Arabidopsis root hairs. Similar images of the actin cytoskeleton and the shape of Arabidopsis root hairs can be found in the literature (Miller et al., 1998; Baluska et al., 2000). It has been demonstrated that the longitudinally oriented actin cables in root hairs are essential for the elongation of these cells. Many of the root hairs formed by AtADF1-O plants displayed highly irregular F-actin organization and lacked longitudinally oriented actin cables (Figure 6C). Such root hairs clearly were shorter than their wild-type counterparts and showed an increased radial diameter (Figures 6C and 6D). On AtADF-U roots, abnormally elongated root hairs often were observed that contained an increased number of longitudinally oriented actin cables (Figure 6E). Statistical analysis revealed that the average length of root hairs was affected to a similar extent as the average length of cells in hypocotyls of etiolated seedlings (see above and Table 3).
We and others have analyzed in detail the organization and functions of the actin cytoskeleton in Arabidopsis trichomes (Mathur et al., 1999; Szymanski et al., 1999; Mathur and Chua, 2000). These studies have shown that an intact actin cytoskeleton is required for the normal morphogenesis and elongation of trichome branches. Compared with wild-type trichomes (Figures 6G and 6H), AtADF1-O trichomes frequently were found to contain aberrant F-actin structures (Figure 6I) and to show distorted (curved or shortened) branches (Figure 6J).
Stomatal guard cells contain radial arrays of actin cables whose disintegration was shown to be associated with stomatal closure (Eun and Lee, 1997). We observed such actin cables in GFP-mTn–expressing wild-type guard cells (Figure 6K). In contrast, many of the stomatal guard cells found in the epidermis of AtADF1-O leaves displayed a completely disrupted actin cytoskeleton (Figure 6M). Such cells generally were irregularly shaped, and affected stomata were partially or entirely closed (Figure 6N).
DISCUSSION
Evaluation of the Approach Used in This Study
Complete knockout of the ADF/cofilin genes in Saccharomyces and Drosophila was found to inhibit vegetative growth and arrest early development, respectively, indicating that these genes have essential functions in both organisms. In contrast to these organisms, Arabidopsis contains a large family of AtADF genes (Dong et al., 2001). Knockout of a single member of this gene family may cause severe defects in early embryogenesis, if the disrupted gene encodes an isoform with unique and essential functions, or may result in no detectable phenotype at all, if the functions of the product of the mutated gene can be supplanted by other isoforms. In both cases, limited information can be gained on the function of AtADF proteins during the development of Arabidopsis plants. Therefore, we took a different approach. By expressing the AtADF1 cDNA in the sense or antisense orientation under the control of the CaMV 35S promoter in transgenic Arabidopsis plants, we generated lines that showed a significant increase in the expression of AtADF1 or a clear decrease in the expression of AtADF1 and at least one other AtADF isoform. Altering the expression of AtADF proteins using this technique made it possible to obtain plants with moderate phenotypes that were able to go through ontogenesis and reproduce. Under standard growth conditions, these plants were indistinguishable from wild-type plants in many aspects of their morphology and development, but they displayed a number of clear abnormalities that allowed interesting conclusions to be drawn concerning the functions of AtADF proteins in vivo and during plant ontogenesis. AtADF1-O or AtADF-U plants growing under stress conditions are expected to display additional defects.
Although in AtADF-U lines the expression of at least one isoform other than AtADF1 also was reduced, we focused specifically on the effects of altered AtADF1 protein levels. It is likely, therefore, that our results reveal only some of the functions that AtADF proteins perform during Arabidopsis development. Also inherent in the approach we have followed, the development of phenotypes in specific cell types and tissues depends on the activity of the 35S promoter. We chose this promoter because its activity pattern in Arabidopsis plants largely overlaps that of the AtADF1 promoter (Dong et al., 2001). Although the 35S promoter generally is considered to be constitutively active in all plant organs, it has been shown to confer variable expression levels in different tissues (Benfey et al., 1990) and to express at low levels during early embryogenesis (C.-H. Dong and N.-H. Chua, unpublished observation) and pollen tube growth (Twell et al., 1989). Therefore, it is not surprising that all investigated AtADF1-O and AtADF-U lines showed normal fertility, although the elongation of pollen tubes is known to be highly dependent on a functional actin cytoskeleton (Taylor and Hepler, 1997; Kost et al., 1999a). In addition, the activity of ADF proteins in living cells is believed to be controlled by complex regulatory mechanisms (Yonezawa et al., 1985, 1990; Hawkins et al., 1993; Hayden et al., 1993; Agnew et al., 1995; Moriyama et al., 1996; Smertenko et al., 1998) that may affect the development of phenotypes in certain cell types or tissues, particularly in AtADF1-O lines.
Most of the described effects of altered AtADF gene expression levels were observed consistently in independent transgenic lines and in all plants analyzed. However, only a fraction of the root hairs, trichomes, and stomata of individual transgenic plants was typically found to display abnormalities. Furthermore, the percentage of abnormal root hairs, trichomes, and stomata was variable among different transgenic lines and also among individual plants of the same line (ranging from a low percentage to >70% of the cells). Apart from positional effects resulting from different transgene integration sites, two additional factors may have contributed to the phenotypic variability in these single cells. We have observed that marker genes (GFP, GUS, GFP-mTn) under the control of the CaMV 35S promoter often are expressed only in a fraction of the trichomes, root hairs, and stomata of individual transgenic Arabidopsis plants, indicating low level or inconsistent activity of this promoter in these cell types (C.-H. Dong, J. Mathur, and N.-H. Chua, unpublished observation). In addition, it is possible that regulatory mechanisms that control AtADF1 activity, which may be highly responsive to slight changes in environmental conditions, were partially responsible for the observed variability in the percentage of abnormal root hairs, trichomes, and stomata on AtADF1-O plants.
AtADF1 Proteins Appear to Control the Disassembly of Actin Cables in Different Cell Types
AtADF1-O plants displayed very clear defects in the organization of the actin cytoskeleton in a variety of cell types. In hypocotyl cells of light- and dark-grown seedlings, as well as in root hairs and in a number of other cell types we have examined (root and cotyledon cells; C.-H. Dong and N.-H. Chua, unpublished data), overexpression of AtADF1 caused the complete disappearance of thick actin cables. In some trichomes and stomatal guard cells, we observed complete disruption of the actin cytoskeleton as a consequence of AtADF1 overexpression. These findings are consistent with the ability of ADF proteins to stimulate F-actin depolymerization, which was predicted based on in vitro studies (Carlier et al., 1997; Maciver et al., 1998; Moriyama and Yahara, 1999), and also with the results of experiments in which the levels of ADF proteins were increased in single cells by microinjection (Nagaoka et al., 1995; Hussey et al., 1998) or transient transformation (Aizawa et al., 1996; Dong et al., 2001).
The effects of significantly reduced expression of AtADF1 and other AtADF isoforms on the organization of the actin cytoskeleton were more moderate than those of AtADF1 overexpression, but they were clearly detectable. Hypocotyl cells, root hairs, and other cell types (root and cotyledon cells; C.-H. Dong and N.-H. Chua, unpublished data) of AtADF-U plants generally were found to contain normal F-actin structures, but they displayed a clear increase in the number of actin cables.
Together, our data indicate that the main function of AtADF1, and possibly of the entire AtADF protein family, in Arabidopsis plants may be the developmentally or environmentally controlled breakdown of thick actin cables.
Actin Cables Have Essential Functions during Cell Elongation and Cellular Morphogenesis
Thick actin cables that appear to be the target of AtADF protein activity were found be present in particularly high numbers in rapidly elongating cells, such as cells in etiolated hypocotyls and root hairs, indicating that they may have a function in polar cell growth. In fact, our results demonstrate that the disruption of these cables by AtADF1 overexpression significantly inhibited cell elongation, whereas the stimulation of their formation by reduced AtADF expression promoted this process. Consistent with the former finding, the elongation of root hairs (Miller et al., 1998), the expansion of other types of root cells (Baskin and Bivens, 1995), and the growth of Arabidopsis seedlings (Baluska et al., 2001) were reported to be inhibited by F-actin–depolymerizing drugs. Root hairs elongate by tip growth, during which cell expansion occurs exclusively at the tip (Kropf et al., 1998). The longitudinally oriented actin cables in these cells are believed to serve as tracks for the myosin-mediated movement of secretory vesicles containing newly synthesized cell membrane and cell wall material toward the growing tip (reviewed in Taylor and Hepler, 1997). Although hypocotyl cells, like most other types of plant cells, expand by diffuse polar growth, during which cells expand in all directions to a certain degree (Gendreau et al., 1997; Kropf et al., 1998), it is conceivable that actin cables also are essential for the elongation of these cells because of their ability to facilitate the delivery of new cell wall and membrane material to growth sites. Misdelivery of such material resulting from the disruption of intracellular trafficking could account for the irregular cellular morphology observed in AtADF1-O hypocotyls.
Interestingly, we have found that reduced expression in Arabidopsis of genes encoding profilin, an actin binding protein with complex and not very well characterized in vivo functions, dramatically reduced plant growth and hypocotyl elongation (Ramachandran et al., 2000). These observations also suggest crucial functions of the actin cytoskeleton in the elongation of plant cells, although plants with reduced profilin protein levels did not show obvious defects in F-actin organization. In contrast to AtADF proteins, which apparently depolymerize actin cables, Arabidopsis profilins may control dynamic properties of the actin cytoskeleton that are required for cell elongation.
Abnormal Cell Expansion Causes Corresponding Defects in Organ Growth
Plant cells in developing tissues are constrained within rigid cell walls and are unable to change shape rapidly or to move. Defects in cell elongation or cellular morphogenesis therefore are expected to translate directly into corresponding abnormalities at the tissue and organ level (Kost et al., 1999b). In accordance with this, we found that altered levels of AtADF expression affected the elongation of root hairs, hypocotyl cells, and intact hypocotyls to a similar extent. Also, we found that irregular cellular morphogenesis in etiolated AtADF1-O hypocotyls was accompanied by abnormal tissue organization and by a wavy morphology of the entire organ.
Actin-Dependent Cellular Processes Other Than Cell Expansion Are Unaffected in AtADF1-O and AtADF-U Plants
Drosophila ADF/cofilin null mutants display severely defective centrosome separation and cytokinesis (Gunsalus et al., 1995). The mechanisms that mediate nuclear division and particularly cytokinesis in plants are very different from those used by Drosophila. However, although the exact functions of actin filaments during these processes in plants are not well understood, they are believed to depend on the actin cytoskeleton. The same is true for a number of other cellular processes that are similarly important for organogenesis and development, such as cell-to-cell communication through plasmodesmata and the differentiation of vascular cells (Ding et al., 1996; Fukuda, 1997). Interestingly, we detected no abnormalities in AtADF1-O or AtADF-U plants that would indicate defects in one of the processes listed above (with the exception of a flowering delay in AtADF-U plants, which may result from defects in actin-dependent signaling processes, as discussed below). Apparently, the F-actin structures in Arabidopsis plants that are involved in these processes are not essential, are not regulated by ADF proteins at all, or are controlled by an ADF isoform other than AtADF1.
AtADF1 Overexpression Disturbs Trichome Morphogenesis
Long actin cables have been shown to be required for the normal morphogenesis of Arabidopsis trichomes, particularly for the extension of branches. Pharmacological disruption of these cables resulted in severely distorted trichomes with twisted stems and short, curved branches (Mathur et al., 1999; Szymanski et al., 1999). Some of the trichomes produced by AtADF1-O plants displayed a completely disassembled actin cytoskeleton and showed similar phenotypes. However, although the branches of these trichomes were curved and/or shortened significantly, stem growth was never affected. In developing trichomes, GFP-mTn expression under the control of the CaMV 35S promoter was always low during early stages and reached higher levels only close to maturity (J. Mathur and N.-H. Chua, unpublished observation). It is reasonable to assume that the expression of the AtADF1 transgene in AtADF1-O plants, which was controlled by the same promoter, followed a similar pattern. Apparently, stems of AtADF1-O trichomes developed normally before the expression of the AtADF1 transgene reached inhibitory levels.
AtADF1 May Have an Important Function in the Regulation of Stomatal Closure
The disassembly of radially arranged actin cables in stomatal guard cells induced by abscisic acid is believed to result in changes in the morphology of these cells and in stomatal closure (Eun and Lee, 1997). We found that the disruption of F-actin cables in guard cells caused by AtADF1 overexpression produced exactly the same effects. These observations suggest the interesting possibility that ADF proteins may act as effectors downstream of abscisic acid signals in the regulation of stomatal closure.
Reduced AtADF1 Expression Delays Flowering
Although the genetic and environmental control of flowering induction in Arabidopsis has been studied extensively at the molecular level (reviewed in Koornneef et al., 1998; Simpson et al., 1999), little evidence for the involvement of the actin cytoskeleton in this process has been reported. It is believed that the switch from vegetative to reproductive development occurs when certain cells in the shoot meristem change their identity under the control of developmentally and environmentally regulated signals. We recently observed early flowering of Arabidopsis plants that displayed reduced growth as a consequence of lower levels of profilin gene expression (Ramachandran et al., 2000). In the study presented here, we demonstrate a significant delay in the induction of flowering in AtADF-U lines, which grew larger than wild-type plants. Both observations indicate the possible involvement of the actin cytoskeleton in the signaling processes underlying flower induction. The actin cytoskeleton has been implicated in the transport of developmental signals between cells through the plasmodesmata (McLean et al., 1997). Intercellular signaling processes, which may be sensitive to changes in the actin cytoskeleton resulting from the reduced expression of actin binding proteins, could be essential for the induction of flowering in the Arabidopsis shoot meristem. Alternately, it is possible that lower levels of actin binding proteins altered flowering time indirectly by affecting plant growth and not directly via effects on F-actin organization. The elucidation of the exact nature of this interesting link between the expression levels of actin binding proteins and the induction of flowering in Arabidopsis clearly requires further investigation.
Conclusions
In conclusion, our results demonstrate that the main function of AtADF1 in Arabidopsis plants is the controlled breakdown of actin cables that are essential for cell and organ expansion as well as for stomatal closure. In addition, we present evidence that actin structures controlled by AtADF proteins may be required for signaling processes involved in the induction of flowering. These findings contribute significantly to an improved understanding of the functions of AtADF proteins and of the actin cytoskeleton in living plant cells and during plant development. Analysis of the effects of altered expression levels of other AtADF isoforms is required to complement our work and to reveal possible additional functions of the AtADF gene family in Arabidopsis plants.
METHODS
Plant Growth and Transformation
Wild-type and transgenic Arabidopsis thaliana (ecotype C24) seed were sterilized and then sowed on plates containing Murashige and Skoog medium (Sigma) with 3% sucrose. Plant growth conditions were as described previously (Dong et al., 2001). The AtADF1 cDNA, which was obtained from the Arabidopsis Biological Resource Center (Columbus, OH), was cloned in both orientations downstream of the 35S promoter into the binary vector pVIP96 (Leu et al., 1995), and the resulting constructs were transferred into Arabidopsis according to Valvekens et al. (1988). AtADF1-O and AtADF-U lines were crossed with transgenic plants containing a 35S::green fluorescent protein–mouse talin (GFP-mTn) construct (the same transgenic line described by Kost et al., 1998).
RNA Gel Blot Analysis
RNA prepared from 10-day-old seedlings was separated on 1.2% agarose–formaldehyde gels and transferred onto nylon membranes. Antisense RNA fragments transcribed from the 5′ untranslated region of the AtADF1 cDNA were used as hybridization probes. 18S rRNA served as the loading control.
Two-Dimensional Gel Electrophoresis and Protein Gel Blot Analysis
AtADF1–glutathione S-transferase fusion proteins expressed in Escherichia coli were purified on glutathione–sepharose affinity columns (Pharmacia). AtADF1 was released from the fusion protein bound to the resin by thrombin treatment. Purified AtADF1 was used to immunize rabbits according to a standard protocol (Hong et al., 1996). Specific AtADF antibodies were obtained by affinity purification.
Total protein was extracted from 10-day-old seedlings in PBS containing 1 mM phenylmethylsulfonyl fluoride. Protein concentrations in the supernatant were determined using the Bradford method (Bradford, 1976). Two-dimensional gel electrophoresis was performed according to the O'Farrell method (Hochstrasser et al., 1988) with some modifications. A total of 80 μg of protein was precipitated with 8 volumes of cold acetone. The protein pellet was redissolved subsequently in iso-urea solution E (0.1 mg of DTT, 0.4 g of cholamidopropyldimethylhydroxy propanesulfonate [Bio-Rad], 5.4 g of urea, and 500 μL of BioLyte 3/10 ampholyte [Bio-Rad] in 6 mL of deionized water). Twelve percent SDS-PAGE and isoelectric focusing were performed in a Protein II chamber (Bio-Rad).
For protein gel blot analysis, proteins were transferred onto a nitrocellulose membrane. The membrane was blocked with 3% nonfat milk in PBS containing 0.05% Tween-20 for 1 hr at room temperature before incubation with affinity-purified anti-ADF IgG. Alkaline phosphatase–conjugated anti–rabbit IgG (Promega) was used as the secondary antibody.
Measurement of Hypocotyl and Cortical Cell Length
Wild-type and transgenic seedlings were grown in darkness on vertical square plates. The lengths of the hypocotyls of 20 randomly selected seedlings were determined. Longitudinal sections of these hypocotyls were then prepared, and cell length was measured using a transmission light microscope (Olympus, Tokyo, Japan). At least 20 cells were measured for each sample.
Flowering Time and Leaf Number
The flowering times of wild-type, AtADF1-O, and AtADF-U plants were measured as number of days from seed germination to the opening of the first flower. Rosette leaf numbers were counted when the first flower was formed on the main inflorescence.
Observation of F-Actin Structures
Ten-day-old etiolated seedlings were placed in a tube containing a slurry of an abrasive substance called EZE-LAP (a lapping compound; number 700 WF; Scour Pads, Sydney, Australia) in phosphate buffer, pH 7.2, and vortexed for 1 min. After washing three times with phosphate buffer, fragments of petioles and hypocotyls were excised and incubated in fluorescein phalloidin (Molecular Probes, Eugene, OR) in phosphate buffer for 2 hr at room temperature. The specimens were mounted on a glass slide with the same buffer, and the epidermal cells were visualized with a confocal microscope (model MRC 600 or MRC 1024; Bio-Rad).
GFP-mTn–expressing Arabidopsis seedlings were mounted in water, and F-actin organization in living hypocotyl cells was imaged using the same confocal microscope with the pinhole/iris opened relatively wide.
Acknowledgments
We thank Yangsan Chan, Caoming Wen, and Weiping Tang for technical assistance. Ann L. Cleary's (Plant Cell Biology Group, Research School of Biological Sciences, Australian National University, Canberra) assistance with the fluorescein phalloidin staining is greatly appreciated. We also are grateful to Qi Xie and Yiqun Bao for helpful discussions. This work was supported in part by United States Department of Energy Grant No. DOE94ER20143 to N.-H.C.
References
- Agnew, B.J., Minamide, L.S., and Bamburg, J.R. (1995). Reactivation of phosphorylated actin depolymerizing factor and identification of the regulatory site. J. Biol. Chem. 270, 17582–17587. [DOI] [PubMed] [Google Scholar]
- Aizawa, H., Sutoh, K., Tsubuki, S., Kawashima, S., Ishii, A., and Yahara, I. (1995). Identification, characterization, and intracellular distribution of cofilin in Dictyostelium discoideum. J. Biol. Chem. 270, 10923–10932. [DOI] [PubMed] [Google Scholar]
- Aizawa, H., Sutoh, K., and Yahara, I. (1996). Overexpression of cofilin stimulates bundling of actin filaments, membrane ruffling, and cell movement in Dictyostelium. J. Cell Biol. 132, 335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa, H., Fukui, Y., and Yahara, I. (1997). Live dynamics of Dictyostelium cofilin suggests a role in remodeling actin latticework into bundles. J. Cell Sci. 110, 2333–2344. [DOI] [PubMed] [Google Scholar]
- Arber, S., Barbayyannis, F.A., Hanser, H., Schneider, C., Stanyon, C.A., Bernard, O., and Caroni, P. (1998). Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature 393, 805–809. [DOI] [PubMed] [Google Scholar]
- Ayscough, K.R. (1998). In vivo functions of actin-binding proteins. Curr. Opin. Cell Biol. 10, 102–111. [DOI] [PubMed] [Google Scholar]
- Baluska, F., Salaj, J., Mathur, J., Braun, M., Jasper, F., Samaj, J., Chua, N.-H., Barlow, P.W., and Volkmann, D. (2000). Root hair formation: F-actin–dependent tip growth is initiated by local assembly of profilin-supported F-actin meshworks accumulated within expansin-enriched bulges. Dev. Biol. 227, 618–632. [DOI] [PubMed] [Google Scholar]
- Baluska, F., Jasik, J., Edelmann, H.G., Salajova, T., and Volkmann, D. (2001). Latrunculin B–induced plant dwarfism: Plant cell elongation is F-actin dependent. Dev. Biol. 231, 113–124. [DOI] [PubMed] [Google Scholar]
- Bamburg, J.R. (1999). Proteins of the ADF/cofilin family: Essential regulators of actin dynamics. Annu. Rev. Cell Biol. 15, 185–230. [DOI] [PubMed] [Google Scholar]
- Baskin, T.I., and Bivens, N.J. (1995). Stimulation of radial expansion in Arabidopsis roots by inhibitors of actomyosin and vesicle secretion but not by various inhibitors of metabolism. Planta 197, 514–521. [DOI] [PubMed] [Google Scholar]
- Benfey, P.N., Ren, L., and Chua, N.-H. (1990). Combinatorial and synergistic properties of CaMV 35S enhancer subdomains. EMBO J. 9, 1685–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Carlier, M.F., Santolini, J., Lanrent, V., Melki, R., Didry, D., Hong, Y., Xia, G.-X., Chua, N.-H., and Pantolni, D. (1997). Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: Implication in actin-based motility. J. Cell Biol. 136, 1307–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, B., Kwon, M.O., and Warnberg, L. (1996). Evidence that actin filaments are involved in controlling the permeability of plasmodesmata in tobacco mesophyll. Plant J. 10, 157–164. [Google Scholar]
- Dong, C.-H., Kost, B., Xia, G.-X., and Chua, N.-H. (2001). Molecular identification and characterization of Arabidopsis AtADF1, AtADF5 and AtADF6 genes. Plant Mol. Biol., in press. [DOI] [PubMed]
- Eun, S.-O., and Lee, Y. (1997). Actin filaments of guard cells are reorganized in response to light and abscisic acid. Plant Physiol. 115, 1491–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda, H. (1997). Tracheary element differentiation. Plant Cell 9, 1147–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau, E., Traas, J., Desnos, T., Grandjean, O., Caboche, M., and Hofte, H. (1997). Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 114, 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus, K.C., Bonaccorsi, S., Williams, E., Verni, F., Gatti, M., and Goldberg, M.L. (1995). Mutations in twinstar, a Drosophila gene encoding a cofilin/ADF homologue, result in defects in centrosome migration and cytokinesis. J. Cell Biol. 131, 1243–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins, M., Pope, B., Maciver, S.K., and Weeds, A.G. (1993). Human actin depolymerizing factor mediates a pH-sensitive destruction of actin filaments. Biochemistry 32, 9985–9993. [DOI] [PubMed] [Google Scholar]
- Hayden, S.D.M., Miller, P.S., Brauweiler, A., and Bamburg, J.R. (1993). Analysis of the interactions of actin depolymerizing factor with G- and F-actin. Biochemistry 32, 9994–10004. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, D.F., Harrington, M.G., Hochstrasser, A.C., Miller, M.J., and Merril, C.R. (1988). Methods for increasing the resolution of two-dimensional protein electrophoresis. Anal. Biochem. 173, 424–435. [DOI] [PubMed] [Google Scholar]
- Hong, Y., Takano, M., Liu, C.-M., Gasch, A., Chye, M.-L., and Chua, N.-H. (1996). Expression of three members of the calcium-dependent protein kinase gene family in Arabidopsis thaliana. Plant Mol. Biol. 30, 1259–1275. [DOI] [PubMed] [Google Scholar]
- Hussey, P.J., Yuan, M., Calder, G., Khan, S., and Lloyd, C.W. (1998). Microinjection of pollen-specific actin-depolymerizing factor, ZmADF1, reorientates F-actin strands in Tradescantia stamen hair cells. Plant J. 14, 353–357. [Google Scholar]
- Iida, K., Moriyama, K., Matsumoto, S., Kawasaki, H., Nishida, E., and Yahara, I. (1993). Isolation of a yeast essential gene, COF1, that encodes a homologue of mammalian cofilin, a low-Mr actin-binding and depolymerizing protein. Gene 124, 115–120. [DOI] [PubMed] [Google Scholar]
- Kim, S.-R., Kim, Y., and An, G. (1993). Molecular cloning and characterization of anther-preferential cDNA encoding a putative actin-depolymerizing factor. Plant Mol. Biol. 21, 39–45. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Alonso-Blanco, C., Peeters, A.J.M., and Soppe, W. (1998). Genetic control of flowering time in Arabidopsis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 345–370. [DOI] [PubMed] [Google Scholar]
- Kost, B., Spielhofer, P., and Chua, N.-H. (1998). A GFP–mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin. Plant J. 16, 393–401. [DOI] [PubMed] [Google Scholar]
- Kost, B., Lemichez, E., Spielhofer, P., Hong, Y., Tolias, K., Carpenter, C., and Chua, N.-H. (1999. a). Rac homologues and compartmentalized phosphatidylinositol 4,5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J. Cell Biol. 145, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost, B., Mathur, J., and Chua, N.-H. (1999. b). Cytoskeleton in plant development. Curr. Opin. Plant Biol. 2, 462–470. [DOI] [PubMed] [Google Scholar]
- Kropf, D.L., Bisgrove, S.R., and Hable, W.E. (1998). Cytoskeletal control of polar growth in plant cells. Curr. Opin. Cell Biol. 10, 117–122. [DOI] [PubMed] [Google Scholar]
- Leu, W.-M., Cao, X.-L., Wilson, T.J., Snustad, D.P., and Chua, N.-H. (1995). Phytochrome A and phytochrome B mediate the hypocotyl-specific downregulation of TUB1 by light in Arabidopsis. Plant Cell 7, 2187–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez, I., Anthony, R.G., Maciver, S.K., Jiang, C.-J., Khan, S., Weeds, A.G., and Hussey, P.J. (1996). Pollen specific expression of maize genes encoding actin depolymerizing factor-like proteins. Proc. Natl. Acad. Sci. USA 93, 7415–7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabuchi, I. (1983). An actin-depolymerizing protein (depactin) from starfish oocytes: Properties and interaction with actin. J. Cell Biol. 97, 1612–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver, S.K., Pope, B.J., Whytock, S., and Weeds, A.G. (1998). The effect of two actin depolymerizing factors (ADF/cofilins) on actin filament turnover: pH sensitivity of F-actin binding by human ADF, but not of Acanthamoeba actophorin. Eur. J. Biochem. 256, 388–397. [DOI] [PubMed] [Google Scholar]
- Mathur, J., and Chua, N.-H. (2000). Microtubule stabilization leads to growth reorientation in Arabidopsis trichomes. Plant Cell 12, 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur, J., Spielhofer, P., Kost, B., and Chua, N.-H. (1999). The actin cytoskeleton is required to elaborate and maintain spatial patterning during trichome cell morphogenesis in Arabidopsis thaliana. Development 126, 5559–5568. [DOI] [PubMed] [Google Scholar]
- McGough, A., Pope, B., Chiu, W., and Weeds, A. (1997). Cofilin changes the twist of F-actin: Implications for actin filament dynamics and cellular function. J. Cell Biol. 138, 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean, B.G., Hempel, F.D., and Zambryski, P.C. (1997). Plant intercellular communication via plasmodesmata. Plant Cell 9, 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher, R.B., McKinney, E.C., and Kandasmy, M.K. (1999). Isovariant dynamics expand and buffer the responses of complex systems: The diverse plant actin gene family. Plant Cell 11, 995–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, D.D., Ruijter, N.C.A., Bisseling, T., and Emons, A.M.C. (1998). The role of actin in root hair morphogenesis: Studies with lipochito-oligosaccharide as a growth stimulator and cytochalasin as an actin perturbing drug. Plant J. 17, 141–154. [Google Scholar]
- Moon, A., Janmey, P.A., Louie, K.A., and Drubin, D.G. (1993). Cofilin is an essential component of the yeast cortical cytoskeleton. J. Cell Biol. 120, 421–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama, K., and Yahara, I. (1999). Two activities of cofilin, severing and accelerating directional depolymerization of actin filaments, are affected differentially by mutations around the actin-binding helix. EMBO J. 18, 6752–6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama, K., Iida, K., and Yaharra, I. (1996). Phosphorylation of Ser-3 of cofilin regulates its essential function on actin. Genes Cells 1, 73–86. [DOI] [PubMed] [Google Scholar]
- Nagaoka, R., Kusano, K.I., Abe, H., and Obinata, T. (1995). Effects of cofilin on actin filamentous structures in cultured muscle cells. J. Cell Sci. 108, 581–593. [DOI] [PubMed] [Google Scholar]
- Pope, B.J., Gonsior, S.M., Yeoh, S., McGough, A., and Weeds, A.G. (2000). Uncoupling actin filament fragmentation by cofilin from increased subunit turnover. J. Mol. Biol. 298, 649–661. [DOI] [PubMed] [Google Scholar]
- Ramachandran, S., Christensen, H., Ishimaru, Y., Dong, C.-H., Wen, C.-M., Cleary, A.L., and Chua, N.-H. (2000). Profilin plays a role in cell elongation, cell shape maintenance, and flowering in Arabidopsis. Plant Physiol. 124, 1637–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozycka, M., Khan, S., Lopez, I., Greenland, A.J., and Hussey, P.J. (1995). A Zea mays pollen cDNA encoding a putative actin-depolymerizing factor. Plant Physiol. 107, 1011–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, G.G., Gendall, A.R., and Dean, C. (1999). When to switch to flowering. Annu. Rev. Cell Dev. Biol. 15, 519–550. [DOI] [PubMed] [Google Scholar]
- Smertenko, A.P., Jiang, C.J., Simmons, N.J., Weeds, A.G., Davies, D.R., and Hussey, P.J. (1998). Ser6 in the maize actin-depolymerizing factor, ZmADF3, is phosphorylated by a calcium-stimulated protein kinase and is essential for the control of functional activity. Plant J. 14, 187–193. [DOI] [PubMed] [Google Scholar]
- Staiger, C.J., Gibbon, B.C., Kovar, D.R., and Zonia, L.E. (1997). Profilin and actin-depolymerizing factor: Modulators of actin organization in plants. Trends Plant Sci. 2, 275–281. [Google Scholar]
- Szymanski, D.B., Marks, M.D., and Wick, S.M. (1999). Organized F-actin is essential for normal trichome morphogenesis in Arabidopsis. Plant Cell 11, 2331–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, L.P., and Hepler, P.K. (1997). Pollen germination and tube growth. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 461–491. [DOI] [PubMed] [Google Scholar]
- Twell, D., Klein, T.M., Fromm, M.E., and McCormick, S. (1989). Transient expression of chimeric genes delivered into pollen by microprojectile bombardment. Plant Physiol. 91, 1270–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens, D., Van Montagu, M., and Lijsebettens, M. (1988). Agrobacterium tumefaciens–mediated transformation of Arabidopsis thaliana root explants using kanamycin selection. Proc. Natl. Acad. Sci. USA 85, 5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, N., Higuchi, O., Ohashi, K., Nagata, K., Wada, A., Kangawa, K., Nishida, E., and Mizuno, K. (1998). Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature 393, 809–812. [DOI] [PubMed] [Google Scholar]
- Yonezawa, N., Nishida, E., and Sakai, H. (1985). pH control of actin polymerization by cofilin. J. Biol. Chem. 260, 14410–14412. [PubMed] [Google Scholar]
- Yonezawa, N., Nishida, E., Iida, K., Yahara, I., and Sakai, H. (1990). Inhibition of the interactions of cofilin, destrin, and deoxyribonuclease I with actin by phosphoinositides. J. Biol. Chem. 265, 8382–8386. [PubMed] [Google Scholar]