Abstract
Phosphatidylinositol transfer proteins (PITPs) modulate signal transduction pathways and membrane-trafficking functions in eukaryotes. Here, we describe the characterization of a gene family from Lotus japonicus that encodes a novel class of plant PITP-like proteins (LjPLPs) and that is regulated in an unusual nodule-specific manner. Members of this gene family were identified based on their nucleotide sequence homology with a previously described cDNA, LjNOD16, which encodes the L. japonicus late nodulin Nlj16. Nlj16 or highly related amino acid sequences are shown to constitute C-terminal domains of LjPLPs and are suggested to function as specific plasma membrane targeting modules. The expression patterns of one member of this gene family (LjPLP-IV) revealed that LjNOD16 mRNA synthesis in nodules is the result of the transcriptional activity of a nodule-specific promoter located in an intron of the LjPLP-IV gene. This intron-borne bidirectional promoter also generates nodule-specific antisense transcripts derived from the N-terminal PITP domain coding region of the LjPLP-IV gene. We propose that Nlj16 protein synthesis and LjPLP-IV antisense transcript generation are components of an elaborate mechanism designed to control LjPLP synthesis and/or functioning in nodules.
INTRODUCTION
Phosphatidylinositol transfer proteins (PITPs) are defined operationally by their ability to transfer phosphatidylinositol (PtdIns) or phosphatidylcholine (PtdCho) monomers between membrane bilayers in vitro (Cleves et al., 1991; Wirtz, 1991). Studies in a number of eukaryotic systems have demonstrated that PITPs function to regulate various aspects of lipid metabolism (reviewed in Cleves et al., 1991; B.G. Kearns et al., 1998; Li et al., 2000), but the first clue about their significance in vivo emerged from studies of Saccharomyces cerevisiae (Novick et al., 1980; Bankaitis et al., 1989, 1990; Aitken et al., 1990) showing their essential role in the formation of secretory vesicles and in protein transport from the Golgi complex. In vitro studies in mammalian systems have suggested that PITPs play important roles in promoting the activities of various inositol lipid-signaling pathways by regulating the production of certain phosphoinositides (Hay and Martin, 1993; Hay et al., 1995; Ohashi et al., 1995; Cunningham et al., 1996; Jones et al., 1998; Simon et al., 1998). These inositol-containing phospholipids have been shown to belong to a large and heterogeneous group of lipids with both structural and key regulatory roles in a wide range of important cellular signaling events (Drøbak et al., 1999).
Although PITPs generally are considered to promote signal transduction, physiological studies in Drosophila rdgB mutants have suggested a signal adaptation function for the RdgB PITP, the loss of which results in neurodegenerative disease of the fly retina (Milligan et al., 1997). Thus, PITPs may downregulate, as well as promote, signal transduction pathways. The mechanisms by which PITPs perform these various functions are not known, and present data are not easily reconciled by a simple model that PITPs act exclusively to regulate phosphoinositide synthesis (B.G. Kearns et al., 1998).
Plant proteins similar to yeast PITPs also have been identified and characterized. These proteins include the Ssh1p and Ssh2p polypeptides from soybean and AtSEC14 from Arabidopsis (Jouannic et al., 1998; M.A. Kearns et al., 1998). Plant PITPs share ∼25% amino acid identity (36 to 50% similarity) with the S. cerevisiae Sec14p protein but do not have any sequence similarity with mammalian PITPs and appear to have different biochemical properties compared with the corresponding yeast and metazoan PITPs (Jouannic et al., 1998; M.A. Kearns et al., 1998). Interestingly, Ssh1p has been found to be subjected to phosphorylation in response to various environmental stress conditions (including hyperosmotic stress), suggesting a physiological role in plant osmoprotection (M.A. Kearns et al., 1998).
As part of a search for expressed sequence tags correlated with late stages of symbiotic root nodule development in the model legume plant Lotus japonicus, we previously identified and characterized a novel nodule-specific cDNA, LjNOD16, that corresponds to a highly abundant mRNA species present in nitrogen-fixing root nodules (Kapranov et al., 1997; Szczyglowski et al., 1997). LjNOD16 mRNA was localized to infected, bacteroid-containing cells of L. japonicus nodules, and the mRNA was shown to encode a 15.5-kD protein termed nodulin16 (Nlj16; Kapranov et al., 1997). Analysis of the predicted Nlj16 amino acid sequence revealed several interesting features, including the presence of two α-helical regions with a high propensity to form coiled-coil structures and a positively charged C terminus (Kapranov et al., 1997).
In an attempt to generate further insights into the possible function of the Nlj16 protein in nodules, and in the context of earlier observations suggesting that Nlj16 may represent a truncated version of a much larger protein(s) (Kapranov et al., 1997), we initiated a detailed characterization of L. japonicus cDNA and genomic sequences that are related to LjNOD16.
Here, we report an unanticipated relationship between Nlj16 and the predicted protein products of four members of a previously undescribed gene family that encode novel PITP-like proteins (LjPLPs). All members of this novel gene family share a two-domain structure consisting of an N-terminal PITP-like domain joined to a C-terminal domain composed of amino acid sequences identical to, or highly related to, nodulin Nlj16. The latter domain is suggested to function as a specific plasma membrane targeting module. We further demonstrate that LjNOD16 expression in nodule tissues results from an unusual transcriptional program that is controlled by a nodule-specific promoter located within an intron of the L. japonicus LjPLP gene (LjPLP-IV). A striking aspect of this transcriptional program is the ability of the same promoter sequence to drive, in reverse orientation, the synthesis of unusual nodule-specific antisense transcripts corresponding to the PITP domain coding region of LjPLP-IV. These data indicate the presence of a novel family of developmentally regulated genes encoding Sec14p-like proteins and suggest a role for Nlj16 in exerting a dominant negative effect apparently directed at inactivating the expression of specific LjPLPs in nodules.
RESULTS
LjNOD16 Shares a Region of Similarity with a Novel Class of PITP-like Genes
We suggested previously that Nlj16 represents a domain module that is used more generally in higher plant proteins (Kapranov et al., 1997). Therefore, we initiated studies to determine whether the Nlj16 domain was present in other L. japonicus proteins. We initially focused on the isolation of LjNOD16-related cDNA species from L. japonicus nodules and characterized a single class of cDNAs in detail.
The longest cDNA clone of this class (pCR5h-24) consisted of 2453 bp predicted to encode a protein composed of two distinct domains (Figure 1). The C-terminal domain of this polypeptide shared significant similarity with the putative coiled-coil domain of Nlj16 (75 and 87% primary sequence identity and similarity, respectively). Interestingly, the N-terminal domain shared significant primary sequence homology with the major S. cerevisiae PITP Sec14p (39% identity and 59% similarity; Bankaitis et al., 1989). We designated this cDNA, and its corresponding gene, the L. japonicus PITP-like protein gene (LjPLP-I; Figure 1).
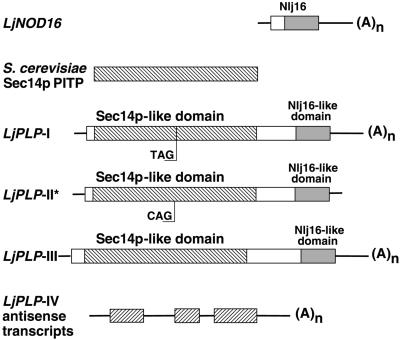
Figure 1.
Scheme of LjPLP cDNAs.
The boxes represent coding regions, and the lines correspond to the 5′ and 3′ untranslated regions (UTRs) of the cDNAs. The hatched boxes represent either Sec14p or Sec14p-like domains, the shaded boxes represent Nlj16-like domains, and the open boxes correspond to regions sharing no apparent sequence similarity with the other cDNAs. The positions of the stop TAG222 codon in LjPLP-I and the corresponding CAG222 codon in LjPLP-II are indicated. The asterisk indicates that the LjPLP-II cDNA sequence was deduced from the nucleotide sequence of the corresponding L. japonicus genomic region. The ORFs present in the LjPLP-IV antisense transcripts, which encode Sec14p-like domains, are represented by inversely hatched boxes, and the intron sequences are represented by lines. The nucleotide sequences for LjPLP-I, LjPLP-II, and LjPLP-III have GenBank accession numbers AF366900, AF366901, and AF367433, respectively. The nucleotide sequence of the LjPLP-IV gene, from which the LjPLP-IV antisense transcripts originate, has GenBank accession number AF367434.
An unusual feature of the LjPLP-I cDNA is the presence of an in-frame TAG stop codon within the Sec14p-like domain at codon 222 (TAG222). To rule out the possibility that this TAG222 codon was the product of a cloning artifact, the genomic DNA region encompassing TAG222 was amplified from L. japonicus DNA by polymerase chain reaction (PCR), and the nucleotide sequences of four independently generated PCR products were determined. Two of these PCR products were found to be identical to the corresponding region of the LjPLP-I cDNA, containing TAG stop codons at equivalent nucleotide positions. Thus, the in-frame TAG222 codon appears to be genuine, and the LjPLP-I cDNA is unlikely to encode a functional protein product. Unexpectedly, the two other genomic PCR products were found to be highly similar to, but not identical with, the LjPLP-I cDNA. These genomic PCR products contained a CAG codon in place of the TAG222 codon present in LjPLP-I. These data suggest the presence of at least one other LjPLP gene in the L. japonicus genome, which we designated LjPLP-II (Figure 1).
To characterize the LjPLP-II gene and its potential protein product, a 5.5-kb genomic DNA fragment containing the entire LjPLP-II coding region was cloned, and its DNA sequence was determined. The corresponding region was found to be 98% identical to the LjPLP-I cDNA at the nucleotide level and to contain an uninterrupted open reading frame (ORF) encoding 550 amino acids comprising both Sec14p- and Nlj16-like domains (Figure 1).
Screening of a L. japonicus nodule-specific cDNA library with a DNA probe corresponding to the Sec14p-like domain of the LjPLP-I cDNA was used to identify additional members of the LjPLP gene family. Two new classes of cDNAs were isolated (LjPLP-III and LjPLP-IV; Figure 1). The longest of these cDNAs (LjPLP-III) was 2256 bp in length and encoded a protein of 625 amino acids, initiating with an ATG codon at nucleotide position +77 relative to the cDNA 5′ end. The predicted structure of the deduced protein product of this ORF was found to be the same as the two-domain Sec14p-Nlj16 structure of the PLPs described above (Figure 1). The second class of cDNAs recovered in this screen was unusual. These cDNAs were found to correspond to endogenous antisense transcripts derived from yet another member of the LjPLP gene family (LjPLP-IV; Figure 1). In LjPLP-IV, the Sec14p-Nlj16 domain structure that typifies the LjPLP-I to LjPLP-III gene products is conserved.
The initial indication of the presence of antisense LjPLP-IV transcripts in nodules was obtained in a search for ORFs in LjPLP-IV cDNAs. This revealed multiple short ORFs on the noncoding strand interrupted by intron sequences (see below). Conceptual translation of these ORFs revealed amino acid sequences sharing a high degree of similarity with the Sec14p domains of LjPLPs (Figure 1) as well as Sec14p-like proteins from other plant species (see Discussion). LjPLP-IV cDNAs differed both in size (0.5 to 1.5 kb) and in the positions of their 5′ and 3′ ends, but all of these cDNAs contained poly(A)+ sequences at their 3′ ends, indicating that they were derived from mRNA transcripts (Figure 1). As described in detail below, these antisense transcripts appear to arise from unusual transcriptional events driven by a nodule-specific bidirectional promoter located within an intron of the LjPLP-IV gene.
Differential Expression of LjPLP Genes
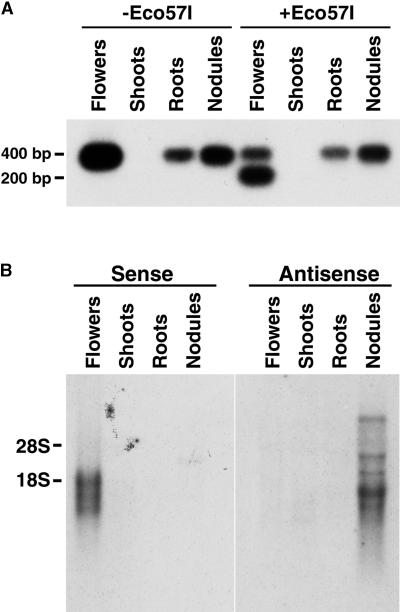
cDNAs derived from LjPLP-I mRNA, containing the UAG222 stop codon, could be distinguished from those derived from LjPLP-II mRNA, containing a CAG codon at the equivalent nucleotide position (CAG222), by digestion with restriction endonuclease Eco57I. To examine the expression of the corresponding genes, we performed reverse transcription (RT)–PCR amplification using total RNA from different L. japonicus tissues. A pair of primers designed specifically to anneal to LjPLP-I or LjPLP-II mRNAs, but not to the LjPLP-III or LjPLP-IV mRNAs, was used. These primers were designed to amplify a 400-bp region encoding the TAG222 and CAG222 codons of the LjPLP-I and LjPLP-II mRNAs, respectively. Eco57I digestion was expected to cleave the RT-PCR product derived from the LjPLP-II mRNA essentially in half, generating a doublet of 200 bp. RT-PCR products of 400 bp were identified from mRNA samples derived from L. japonicus flowers, roots, and nodules but not from shoot tissues (Figure 2A). However, only flower-derived RT-PCR products were found to be cleaved by Eco57I, generating products of 400 and 200 bp. These results demonstrate that although the production of LjPLP-I mRNA occurs in a variety of L. japonicus tissues, LjPLP-II transcription appears to be limited to flowers. RNA gel blot analysis demonstrated the presence of LjPLP-III–derived transcripts in L. japonicus flowers, uninfected roots, and nodules but not in shoot tissues (data not shown).
Figure 2.
Tissue-Specific Expression of the LjPLP-I, LjPLP-II, and LjPLP-IV Genes.
(A) Fragments (400-bp), encompassing either the UAG222 stop codon or the CAG222 triplet-containing regions of LjPLP-I and LjPLP-II mRNAs, respectively, were amplified using RT-PCR. The 400- and 200-bp fragments observed in the samples digested with Eco57I correspond to the LjPLP-I and LjPLP-II transcripts, respectively.
(B) Tissue-specific expression of the LjPLP-IV transcripts. Ten micrograms of total RNA was hybridized with radiolabeled, strand-specific, RNA probes complementary to the presumed sense or antisense transcripts of the LjPLP-IV gene. The positions of 28S and 18S rRNAs are indicated.
LjPLP-IV gene expression was investigated using radiolabeled, strand-specific RNA probes complementary to derived antisense or sense transcripts. Interestingly, LjPLP-IV antisense transcripts were found to predominate in L. japonicus nodules (Figure 2B). The broad size distribution of the antisense transcripts found to hybridize was consistent with the size heterogeneity of the corresponding cDNAs (see above). Moreover, LjPLP-IV sense transcripts were found to be present in L. japonicus flowers and to constitute a family of closely migrating bands (Figure 2B). These data suggest the presence of a functional LjPLP-IV gene with diverse mRNA transcripts.
LjPLP-IV Antisense Transcripts and LjNOD16 mRNA Are Derived from the Same Gene
Because both LjPLP-IV antisense transcripts and LjNOD16 transcripts appear to be expressed only in mature nitrogen-fixing nodules, and because the predicted protein products of the short ORFs present on the minus strands of the antisense transcripts and the Nlj16 protein correspond to the two domains that characterize the PLP proteins, we postulated that antisense LjPLP-IV transcripts and the LjNOD16 mRNA could be produced by divergent transcription from a promoter within the LjPLP-IV gene.
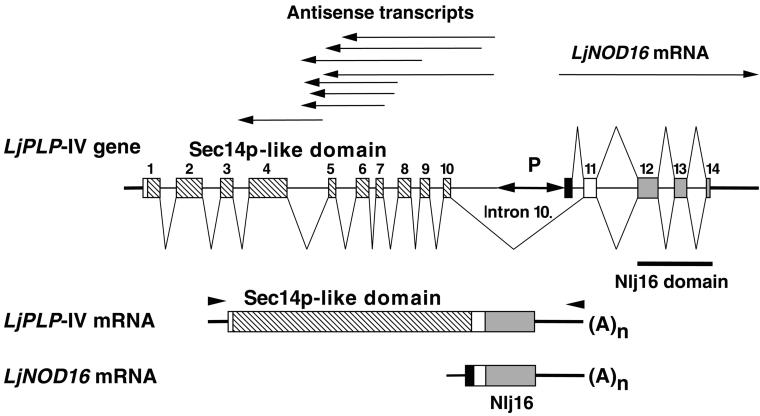
To test this hypothesis, two complementary approaches were used. First, a phage λ clone containing LjNOD16 was isolated from a L. japonicus genomic DNA library. DNA sequence analysis of this clone revealed the presence of a region encoding a Sec14p-like domain, with a nucleotide sequence of the noncoding strand identical to the LjPLP-IV antisense transcripts, immediately upstream of the LjNOD16 gene (Figure 3). This result confirmed the linkage between the DNA regions encoding the Sec14p-like domain of LjPLP-IV and Nlj16 in the L. japonicus genome. To determine whether these two domains were present in a single transcriptional unit, a RT-PCR primer was designed that hybridized 77 bp upstream from the presumed ATG initiation codon of the PITP-like domain. A suitable downstream primer was derived from the nucleotide sequence of the 3′ untranslated region (UTR) of the LjNOD16 mRNA (Figure 3). Indeed, a 1903-bp RT-PCR product was generated by these primers from total RNA of L. japonicus nodule and flower tissues and was found to contain a 482-residue ORF that was identical to the LjPLP-IV protein sequences inferred from the conceptual translation of LjPLP-IV–derived antisense cDNAs. The presence of a stop codon (TAA) 36 bp upstream from the putative ATG initiation codon suggested that the RT-PCR product encompassed the entire LjPLP-IV coding region. The primary amino acid sequence of the LjPLP-IV C-terminal Nlj16 domain was found to be identical to amino acids 16 to 141 of nodulin Nlj16 (Figures 3 and 4; see also below). The LjNOD16 mRNA region that contains the 5′ UTR and codons 1 to 15 of the Nlj16 protein was found to derive from intron 10 of the LjPLP-IV gene and to be absent from the mRNA encoding the full-length LjPLP-IV protein (Figure 3; see also below).
Figure 3.
Schemes of the LjPLP-IV Genomic Region and the LjPLP-IV Sense Transcripts and the LjNOD16 Transcript.
Exons are numbered and represented by boxes, introns are symbolized by thin lines, and thick lines indicate the 5′ and 3′ UTRs. Differently shaded boxes correspond to either Sec14p-like or Nlj16 domains, as described in the legend to Figure 1. The solid box represents the first exon of the LjNOD16 transcript. The positions of various cDNA clones corresponding to either LjPLP-IV antisense transcripts or the LjNOD16 transcript are indicated by arrows above the diagram of the LjPLP-IV gene. The localization of the bidirectional promoter (P) within intron 10 is indicated. The positions of primers used to amplify the LjPLP-IV mRNA are indicated by arrowheads.
Figure 4.
Amino Acid Sequence Alignment between L. japonicus LjPLP-IV, Nodulin Nlj16, AtPLP (Hypothetical Arabidopsis Protein CAB16843.1), and Yeast Sec14p Proteins.
The alignment was generated using the PileUp function of the Genetics Computer Group package (Madison, WI) and SeqVu software version 1.0.1 (Garvan Institute of Medical Research, Sydney, Australia). Similar residues are boxed, and identical amino acids are boxed and shaded. Dashes represent gaps in the sequences, and asterisks indicate the conserved amino acid residues required for PtdIns binding activity of the yeast Sec14p protein (Sha et al., 1998).
A Bidirectional, Tissue-Specific Promoter Is Present within Intron 10 of LjPLP-IV
The intron–exon structure of the coding region of LjPLP-IV was determined by comparing the corresponding nucleotide sequences of the genomic clone and the product of the RT-PCR amplification reaction. The LjPLP-IV gene was found to contain at least 14 exons and 13 introns (Figure 3). Intron 10 was the largest intron, defining the boundary between the N-terminal PITP-like domain and the C-terminal Nlj16 domain coding regions of LjPLP-IV. Because transcription of antisense LjPLP-IV RNAs and the LjNOD16 mRNA appeared to be initiated within intron 10, this region was postulated as a location for a bidirectional nodule-specific promoter.
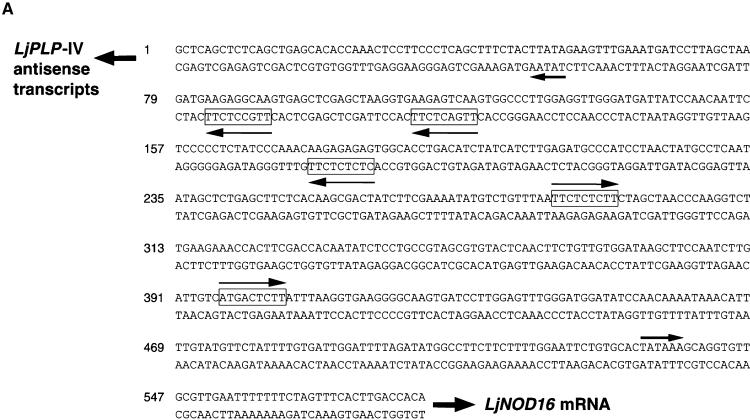
This promoter region of the intron was proposed to reside within a 581-bp region of intron 10 bounded by the 5′ ends of the longest antisense LjPLP-IV transcript and the LjNOD16 transcript (Figure 3). Two TATA box–like sequences were identified ∼40 bp upstream of the 5′ termini of the longest antisense and LjNOD16 cDNAs within this region. Moreover, a number of DNA sequence motifs exhibiting a high degree of similarity with nodulin box consensus sequence (5′-TTGTCTCTT-3′) were found in this region as well (Figure 5A) (Ramlov et al., 1993; Szczyglowski et al., 1994). These observations are consistent with the existence of a nodule-specific bidirectional promoter region in this intron (Figure 5A).
Figure 5.
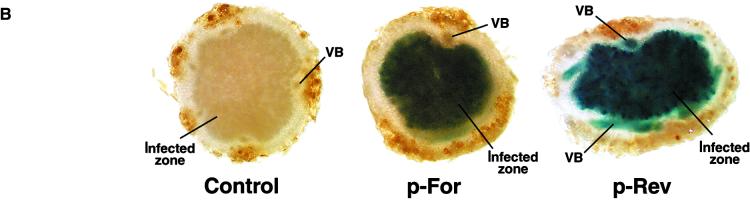
The Internal Bidirectional Promoter of the LjPLP-IV Gene Directs the Expression of the β-Glucuronidase Reporter Gene to the Central Infected Zone of L. corniculatus Nodules.
(A) Nucleotide sequence of the intron-localized bidirectional promoter of the LjPLP-IV gene. The directions of transcription of the LjPLP-IV antisense transcripts and the LjNOD16 transcript are indicated by arrows. The positions and presumed orientations of TATA box–like sequences are indicated by arrows and nodulin gene conserved promoter motifs are indicated by boxed sequences.
(B) Histochemical localization of GUS activity in nodule hand sections derived from transgenic L. corniculatus plants harboring the promoter–GUS fusions. VB, nodule vascular bundle (for further details see text).
The functionality of a nodule-specific promoter within LjPLP-IV intron 10 was demonstrated using transgenic plants. The putative 581-bp promoter region was fused in both orientations to a uidA reporter gene generating the forward construct (p-For) in the direction of LjNOD16 expression and a reverse construct (p-Rev) corresponding to LjPLP-IV antisense expression. Staining of hand-cut nodule sections clearly revealed that the putative LjPLP-IV intron 10 promoter directed β-glucuronidase (GUS) expression only in the central, infected zone of the nodule (Figure 5B). Other L. corniculatus tissues (roots, leaves, and flowers) did not reveal any significant cytological staining for GUS activity (data not shown). The p-Rev construct displayed strong GUS expression in vascular bundles of the nodule in addition to the infected zone (Figure 5B). A promoterless uidA construct was used as a negative control in these experiments. As expected, this control construct failed to generate GUS expression in the central infected zone of the nodules (Figure 5B). Although the level of resolution of these experiments did not allow the assignment of reporter gene activity to an infected versus uninfected cell-specific level, it established that there is a tissue-specific pattern directed by the LjPLP-IV intron 10 promoter that confers nodule-infected zone expression highly similar to that reported for the late nodulin Srglb3 promoter (Szczyglowski et al., 1994) and consistent with the localization of LjNOD16 mRNA to the infected cells of L. japonicus nodules (Kapranov et al., 1997). Therefore, we conclude that the LjPLP-IV intron 10 region contains a unique, bidirectional promoter activity involved in tissue (nodule)-specific gene expression.
Nlj16 Contains a Functional Plasma Membrane Targeting Domain
The data presented above suggest the presence of a new family of L. japonicus proteins that exhibit a two-domain arrangement, with a Sec14p-like domain at the N terminus and an Nlj16-like domain at the C terminus. This type of association of Sec14p and Nlj16 domains appears to be conserved in analogous genes in higher plants (see Discussion). To gain further insight into the functional properties of this domain arrangement, we investigated the functional properties of the Sec14p and Nlj16 domains of LjPLP-IV.
The Nlj16 domain of the LjPLP-IV protein contains two cysteine residues at the extreme C terminus that are preceded by a polybasic region (Figures 4 and 6F). These two motifs have been reported to cooperate in membrane targeting of prenylated proteins (Hancock et al., 1991; Zhang and Casey, 1996; Rodriguez-Concepcion et al., 1999). Therefore, we examined whether Nlj16 may be membrane associated. Differential centrifugation experiments in conjunction with anti-Nlj16 antibody revealed that the Nlj16 protein cosedimented with fractions that pelleted at 10,000 and 100,000g (data not shown), suggesting its association with the membrane compartments. To further investigate the subcellular localization of the Nlj16 and LjPLP-IV proteins, we constructed mGFP5–Nlj16 and mGFP5–LjPLP-IV fusion proteins and examined their expression in bombarded onion epidermal cells. Control cells, which expressed mGFP5 alone from the cauliflower mosaic virus 35S promoter, were found to display a broad distribution of mGFP5 fluorescence in the cytoplasm, nucleus, and transvacuolar strands (Figure 6A). In contrast, the C-terminal Nlj16 extension targeted the mGFP5–Nlj16 chimeric protein exclusively to the cell periphery (Figure 6B). Additional evidence supporting this notion was derived from the localization of fluorescence in a small subset of transformed onion cells transiently expressing mGFP5–Nlj16 and undergoing plasmolysis. In such cells, mGFP5 fluorescence always was detected at the protoplast periphery, and no fluorescence was associated with cell walls or vacuolar membranes (Figures 6D and 6E). These data suggest that the Njl16 domain is capable of directing mGFP5 to the plant plasma membrane. Similarly, the mGFP5–LjPLP-IV fusion protein also localized to the plasma membrane (data not shown).
Figure 6.
The Conserved Nlj16 Domain Mediates the Plasma Membrane Targeting of the LjPLP-IV Protein.
(A) to (E) Subcellular localization of the mGFP5–Nlj16 fusion proteins in onion epidermal cells. Fluorescent images of onion cells expressing mGFP5 alone (A), the mGFP5–Nlj16 fusion ([B] and [D]), and the mGFP5–Nlj16ΔCC fusion (C) are shown. (E) is a bright-field image of the cell shown in (D). The onion cell shown in (D) and (E) has undergone plasmolysis, and mGFP5 fluorescence can be seen on the periphery of the protoplast. P, protoplast; V, vacuole; CW, cell wall.
(F) Alignment of the C-terminal Nlj16-like domains of LjPLP-II, LjPLP-III, LjPLP-IV, and Arabidopsis AtPLP. Similar residues are boxed, and identical amino acids are boxed and shaded. Dashes represent gaps in the sequences.
Structural Requirements for Nlj16 Domain–Mediated Plasma Membrane Targeting
To further investigate this targeting phenomenon, we analyzed the individual roles of the Nlj16 polybasic domain and the CC motif in mGFP5–Nlj16 targeting to the plasma membrane. The Nlj16 CC motif was found to be necessary for targeting, as revealed by the observed diffuse distribution of mGFP5–Nlj16ΔCC within all accessible intracellular compartments; this localization resembled that of mGFP5 alone (Figure 6C). Although required for the localization of the chimeric protein to the plasma membrane, the CC motif was found to be insufficient for targeting by itself. Rather, mGFP5 localization to the plasma membrane required both the polybasic region and the CC motif (Nlj16 residues KKKQKKKTFFCC; data not shown).
As indicated above, the LjPLP-IV Nlj16 domain is very similar to the C-terminal domains of other PLP proteins from L. japonicus and other plant species (Figure 6F). Notable differences include variations in the C-terminal amino acids of these proteins; for example, the residues corresponding to the LjPLP-IV CC motif are CW and WA in LjPLP-III and LjPLP-II, respectively (Figure 6F). Further transient expression experiments demonstrated that the Nlj16-like domain of LjPLP-III (Nlj16-III) also is able to function as a plasma membrane targeting domain and that the CW motif is essential for this function (data not shown). In contrast to Nlj16, however, the C-terminal basic region of Nlj16-III (RQAEAKLRKKRFCW) was found to be insufficient to direct mGFP5 to the plasma membrane (data not shown). These results indicate that other, as yet unidentified, signals may be required for the plasma membrane targeting activity of Nlj16-III. In this regard, the likelihood that Nlj16 or Nlj16-like domains form coiled-coil structures suggests that, in addition to their potential to interact electrostatically (via charged C-terminal domains) with plasma membrane phospholipids, the Nlj16 domains also may interact with as yet unidentified protein partners (Kapranov et al., 1997).
The LjPLP-IV Sec14p-like Domain Complements a Yeast sec14 Mutant Phenotype
Because amino acid sequence homology has been found to be insufficient to conclude whether a Sec14p-like protein is a PITP (Salama et al., 1990; M.A. Kearns et al., 1998; Li et al., 2000), we further investigated the relationship between the Sec14 domain of LjPLP-IV and the yeast Sec14 protein on a functional level.
Constitutive expression of LjPLP-IV driven by the yeast phosphoglycerate kinase (PGK) promoter failed to rescue the growth of a temperature-sensitive sec14-1 yeast strain at the restrictive temperature (Figure 7A, construct pGK-IV). However, expression of the LjPLP-IVΔCC protein, which lacks the two C-terminal cysteine residues apparently required for plasma membrane targeting of LjPLP-IV in plant cells, was found to rescue the growth of the temperature-sensitive sec14-1 yeast mutant at 37°C (Figure 7A). Similar results were obtained when a truncated form of LjPLP-III lacking the entire Nlj16-like domain (LjPLP-IIIΔ) was used (Figure 7A, construct pGK-IIIΔ). In addition, the expression of truncated LjPLP-IIIΔ and LjPLP-IV proteins lacking the C-terminal Nlj16 domains was found to complement the lethal phenotype associated with the null yeast sec14Δ1 allele present in strain CTY558 in plasmid shuffle experiments (data not shown). These data indicate that the Sec14p-like domains of LjPLP-III and LjPLP-IV share functional properties with yeast Sec14p and may represent PITP modules. These results also demonstrate that functional Nlj16-like domains interfere with the ability of LjPLP Sec14p domains to substitute for Sec14p function in yeast, probably because they direct targeting of LjPLP Sec14p domains to the plasma membrane.
Figure 7.
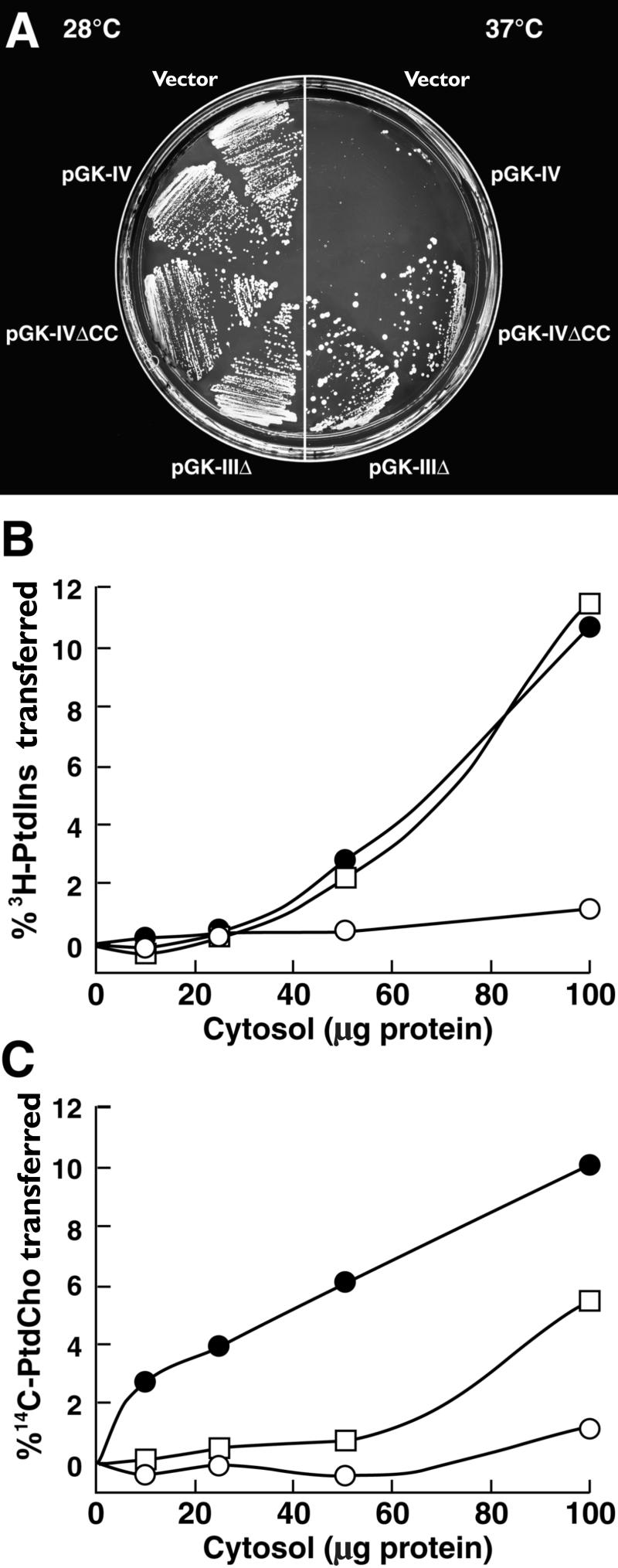
The LjPLP-IV Protein Complements the Temperature-Sensitive Phenotype of the Yeast sec14 Mutant and Exhibits PtdIns and PtdCho Transfer Activity.
(A) The growth phenotype of yeast strain CTY1079 (sec14-1ts Δspo14) harboring the indicated constructs (see Methods). Individual transformants were streaked on selective (−Ura) media and grown at permissive (28°C) and nonpermissive (37°C) temperatures.
(B) and (C) Phospholipid transfer activities. The ability of the recombinant LjPLP-IV Sec14p domain (LjPLP-IV–Sec14p) to transfer 3H-PtdIns (B) or 14C-PtdCho (C) was measured in cytosol prepared from E. coli. Activities are represented as the percentage of total input 3H-PtdIns or 14C-PtdCho transferred from donor membranes to unlabeled acceptor membranes during a 30-min incubation at 37°C. Cytosol values are presented as amounts of protein added to the assay. Closed circles, Sec14p; open squares, LjPLP-IV–Sec14p; open circles, E. coli cytosol.
Assay blanks represented the addition of buffer alone to the transfer assay reactions. These reactions defined assay background, and background values were subtracted from each assay to yield transfer values. Ranges of 12,695 to 15,121 cpm and 22,697 to 28,560 cpm of input 3H-PtdIns and 14C-PtdCho, respectively, were used in each assay. Background values for these assays ranged from 237 to 374 for the PtdIns transfer assays and from 238 to 391 for the PtdCho transfer assays. The data shown are from a representative experiment of at least three independent assays.
LjPLP-IV Exhibits Intrinsic PtdIns and PtdCho Transfer Activities in Vitro
To obtain more direct evidence that LjPLP-IV is a classic PITP, we expressed a recombinant LjPLP-IV Sec14p domain (LjPLP-IV–Sec14p) in Escherichia coli and measured PtdIns and PtdCho transfer activities in crude E. coli cytosolic fractions. In control experiments, recombinant Sec14p cytosolic fractions were able to transfer 10.6% of the total input 3H-PtdIns at the highest concentration tested. Cytosolic fractions prepared from bacteria expressing LjPLP-IV–Sec14p were able to catalyze the in vitro transfer of 11.6% of input 3H-PtdIns from donor to acceptor membranes (Figure 7B).
Similar results were obtained when PtdCho transfer activities were measured (Figure 7C). Bacterial cytosolic fractions harboring recombinant Sec14p catalyzed vigorous rates of PtdCho transfer activities (10% of total input substrate transferred). Recombinant LjPLP-IV–Sec14p cytosolic fractions also exhibited significant PtdCho transfer activities (5.6% of total input 14C-PtdCho transferred at the highest cytosol concentration tested; Figure 7C). These data demonstrate that LjPLP-IV is a PITP that affects both PtdIns and PtdCho transfer. Thus, the Sec14p domain of LjPLP-IV harbors both intrinsic PtdIns and PtdCho transfer activity. These biochemical properties identify the LjPLP-IV Sec14p module as a classic PITP (Li et al., 2000).
DISCUSSION
The LjPLP Family Defines a Novel Two-Domain Structure
We have identified four LjPLP genes in the L. japonicus genome whose structures are characterized by a two-domain arrangement in which an N-terminal Sec14p coding region is linked to a C-terminal Nlj16 coding region. The Nlj16 and Nlj16-like C-terminal domains of LjPLPs constitute a signature of this new class of plant Sec14p-like proteins. The presence of a large C-terminal extension alone is an unusual feature for PITPs (Lopez et al., 1994; B.G. Kearns et al., 1998; Li et al., 2000). Yet, LjPLP structure appears to be conserved widely in higher plants. In database searches using LjNOD16, we were able to identify a number of plant genes encoding Sec14p-like proteins with C-terminal domains very similar to the putative coiled-coil domain of Nlj16. With the exception of one maize expressed sequence tag (GenBank accession number AJ006545), all of these genes are derived from the Arabidopsis genome sequencing project (e.g., GenBank accession numbers Z99708.1, AC007212.6, AL023094.2, AC006841_29, and T08565; Figure 4).
Interestingly, the two Arabidopsis expressed sequence tags (168K8 and 110G16) found previously to share similarity with the Nlj16 protein (Kapranov et al., 1997) actually represent Sec14p-like proteins with Nlj16 domains (GenBank accession numbers AC006841_29 and T08565, respectively). Thus, the Nlj16-type coiled-coil domain is linked intimately with a novel class of Sec14p-like proteins in both leguminous and nonleguminous plants. We designate this novel class of higher plant Sec14p-like proteins as plant PITP-like proteins (PLPs). We emphasize, however, that this Sec14p–Nlj16 module is not a feature of all higher plant PITPs. The Sec14p-like Ssh1p and Ssh2p proteins of soybean (M.A. Kearns et al., 1998) and the AtSec14p from Arabidopsis (Jouannic et al., 1998) consist of only the Sec14p domain.
Potential Roles for LjPLPs in Promoting Signaling
Our data suggest that the LjPLP-III and LjPLP-IV proteins are plasma membrane–localized PITPs whose expression is subject to developmental regulation. This conclusion derives from the findings that the Nlj16 domains of both proteins likely represent plasma membrane targeting motifs and that the Sec14p domain of LjPLP-IV (and probably also that of LjPLP-III) is a classic PITP domain with intrinsic PtdIns and PtdCho transfer activities. LjPLP-II also may represent a membrane-associated PITP.
By analogy with mammalian PITPs, LjPLPs may promote phosphoinositide-driven signaling cascades initiated at the plant cell plasma membrane. Because LjPLPs appear not to be cytoplasmic proteins, their mode of action could be restricted to a PtdIns presentation mode. Because LjPLPs harbor Sec14p domains, the activities of these proteins may resemble the functions of Sec14p-like PITPs more closely than those of metazoan PITPs. In this regard, a family of nonclassic PITPs, yeast Sec14p homologs (SFH proteins), have been described that resemble LjPLPs in their membrane association properties (Li et al., 2000).
Downregulation of LjPLP-IV Function in Nodules
The L. japonicus LjPLP-IV gene is a particularly interesting member of the novel two-domain LjPLPs because it encodes at least three different mRNA species that are expressed differentially in L. japonicus nodules and/or flowers. A full-length 1.9-kb sense LjPLP-IV transcript, encoding the LjPLP-IV protein, accumulates predominantly in L. japonicus flowers. The same transcript also is present in nodules, albeit at a significantly diminished level. In contrast, two distinct classes of mRNA species derived from the same LjPLP-IV gene, antisense LjPLP-IV transcripts and LjNOD16, accumulate to high levels in L. japonicus nodules. These transcripts appear to originate as a result of tissue-specific activation of an internal bidirectional promoter that is localized within intron 10 of the LjPLP-IV gene (i.e., the intron marking the boundary between the Sec14p and Nlj16 domain coding regions of LjPLP-IV).
The developmentally regulated expression of antisense RNAs that derive from the LjPLP-IV Sec14p domain coding region and the expression of nodulin Nlj16 suggest that LjPLP-IV function is suppressed in nodules. Antisense RNA expression would be expected to reduce LjPLP-IV expression, and levels of LjPLP-IV sense mRNA are much lower in nodules than in flowers. Furthermore, nodulin Nlj16 expression itself could involve a developmental reprogramming of transcription within the LjPLP structural gene to direct the synthesis of a protein that represents a truncated LjPLP-IV lacking the Sec14p domain. Thus, Nlj16 might act in synergy with the antisense RNAs in blocking LjPLP-IV activity in nodules. Because nodulin Nlj16 likely is targeted to the plasma membrane, it may act as a dominant negative form of LjPLP-IV. Thus, Nlj16 might compete for a specific binding site or partner that normally interacts with LjPLP-IV or other LjPLP proteins, thereby preventing LjPLP-related biological function(s) in this plant organ.
The putative downregulation of LjPLP function in nodules may be relevant to the role of plant phospholipid metabolism in promoting the symbiosis between legumes and their endosymbionts. The activation of lipid signaling pathways in association with Nod factor–mediated root hair deformation in Vicia sativa subsp nigra has been postulated (den Hartog et al., 2001). Expression of a root- and nodule-specific isoform of soybean PtdIns 3-kinase in association with the proliferation of peribacteroid membranes in infected cells of the nodule also has been documented (Hong and Verma, 1994). The latter observation lends support to the hypothesis that plant PtdIns 3-phosphate may play an important role in membrane biogenesis and the development of endosymbiotic compartments in nodules (Hong and Verma, 1994; Drøbak et al., 1999). If LjPLPs function to downregulate such a signaling pathway, then their activity would have to be reduced to sustain nodule development.
Finally, because sense and antisense LjPLP-IV transcripts likely coexist in L. japonicus nodules, double-stranded RNA species could be generated. Such double-stranded RNA molecules, in addition to their presumed inhibitory role in LjPLP gene expression in nodules, could serve as signaling molecules involved in the long-distance coordination of plant developmental processes related to symbiotic nitrogen fixation (e.g., signaling between nodules and flowers). This signaling could occur in a manner similar to the phenomenon of post-transcriptional gene silencing (Waterhouse et al., 1998). A detailed developmental analysis of LjPLP-IV gene expression in nodules and flowers is required to test this hypothesis.
METHODS
Plant Material and Growth Conditions
Lotus japonicus ecotype B-129-S9 Gifu plants were germinated and grown as described (Kapranov et al., 1997; Szczyglowski et al., 1997). Nodules, leaves, and stems of L. japonicus plants inoculated with Mezorhizobium loti strain NZP2235 (Jarvis et al., 1982) were harvested 35 days after inoculation; control uninoculated roots were collected from axenically grown L. japonicus plants of the same age. L. japonicus flowers were obtained from 2- to 3-month-old plants.
Transgenic L. corniculatus plants were grown as described (Szczyglowski et al., 1994). Fully mature nodules, leaves, flowers, and root segments were harvested from transgenic plants 42 to 45 days after inoculation and used directly for histochemical analyses.
Screening of L. japonicus Genomic DNA and Nodule-Specific cDNA Libraries
A L. japonicus genomic DNA library and a cDNA library from mature nodules of the same plant species were kindly provided by Dr. Jens Stougaard (Århus University, Århus, Denmark). To isolate LjNOD16-related genes, we screened L. japonicus genomic and cDNA libraries under low-stringency conditions with a 530-bp EcoRI-HindIII fragment of the LjNOD16 cDNA, representing the entire coding region of Nlj16, as a probe. In addition, to isolate LjPLP-III and LjPLP-IV cDNAs, the L. japonicus cDNA library was screened with the phosphatidylinositol transfer protein (PITP)–like domain-containing fragment of the LjPLP-I cDNA (base pairs 44 to 1282) under low-stringency conditions.
Nucleic Acid Isolation and RNA Gel Blot Analyses
Genomic DNA and total RNA from various L. japonicus tissues were isolated as described by Kapranov et al. (1997) and Szczyglowski et al. (1997). RNA gel blot analyses were performed essentially as described (Kapranov et al., 1997; Szczyglowski et al., 1997). For hybridization with strand-specific RNA probes, the filters were prehybridized in 100 mM potassium phosphate buffer, pH 6.8, 5 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate), 1 × Denhardt's solution (1 × Denhardt's solution is 0.02% Ficoll, 0.02% polyvinylpyrrolidone, and 0.02% BSA), 0.1% SDS, and 100 μg/mL denatured salmon sperm DNA at 50°C for 4 hr. Hybridization was performed in 70 mM potassium phosphate buffer, pH 6.8, 3.6 × SSC, 0.7 × Denhardt's solution, 7.0% dextran sulfate, 71 μg/mL denatured salmon sperm DNA, and 50% deionized formamide at 65°C. The filters were washed for 15 min in 2 × SSC and 0.1% SDS, for 15 min in 1 × SSC and 0.1% SDS, and for 15 min in 0.1 × SSC and 0.1% SDS at 65°C.
Radiolabeled RNA probes were prepared as follows. Template DNA (0.5 to 1 μg) was linearized and incubated in a buffer containing 40 mM Tris, pH 7.5, 8 mM MgCl2, 2 mM spermidine, 25 mM NaCl, 10 mM DTT, 40 units of placental RNase inhibitor (Boehringer Mannheim Biochemicals, Indianapolis, IN), 0.5 mM ATP, 0.5 mM GTP, 0.5 mM CTP, 15 μM UTP, 50 μCi of α-32P-UTP, and 20 units of T3 or T7 RNA polymerase (Boehringer Mannheim Biochemicals) in a total volume of 20 μL. The labeling reactions were performed for 1 hr at 37°C. The DNA template was removed from the reaction mixture by adding 10 units of RNase-free DNase I (Boehringer Mannheim Biochemicals) and incubating at 37°C for 15 min. Radiolabeled RNA probes were purified on Bio-Spin 6 chromatography columns (Bio-Rad) according to the manufacturer's instructions.
Polymerase Chain Reaction Amplification and DNA Sequencing
The 400-bp genomic polymerase chain reaction (PCR) fragments encompassing the regions of the LjPLP-I or LjPLP-II gene containing the TAG or CAG codon, respectively, were amplified with PCR from L. japonicus genomic DNA using a pair of common primers, DB588 and DB544. PCR was performed for 30 cycles with the following cycle profile: a 1-min DNA denaturation step at 94°C, a 1-min annealing step at 55°C, and a 1-min extension step at 72°C.
A 5.5-kb genomic DNA fragment corresponding to the entire coding region of the LjPLP-II gene was amplified by PCR from the corresponding phage lysate by using forward (DB562) and reverse (DB561) primers. These primers were designed based on the nucleotide sequences of the 5′ and 3′ untranslated regions (UTRs), respectively, of LjPLP-I cDNA. PCR was performed for 30 cycles using Pfu DNA polymerase (Promega) under the following cycling conditions: a 1-min denaturation step at 94°C, a 1-min annealing step at 55°C, and a 7-min extension step at 72°C. The PCR product was cloned and sequenced. Reverse transcription (RT)–PCR was performed essentially as described (Szczyglowski et al., 1998), except that RT was performed using 200 units of Superscript II reverse transcriptase (GIBCO). Sequences of the primers are available from the authors upon request.
Chimeric Gene Constructs and Generation of Transgenic L. corniculatus Plants
Chimeric gene constructs were prepared using standard molecular techniques. Briefly, the 581-bp DNA fragment derived from intron 10 of the LjPLP-IV gene was amplified by PCR and cloned in both orientations into the unique BamHI restriction site of the pBI101-derived binary vector (Clontech, Palo Alto, CA). This resulted in the construction of the forward (p-For) and reverse (p-Rev) binary vectors, which are named with respect to the direction of the β-glucuronidase (GUS) coding region.
The binary vectors described above were transferred independently into Agrobacterium rhizogenes A4 (Tempe and Casse-Delbart, 1989) using the freeze–thaw method of Hofgen and Willmitzer (1988). Transgenic L. corniculatus cv Rodeo plants were generated as described previously (Szabados et al., 1990; Szczyglowski et al., 1994). GUS activity in the nodule hand sections and other L. corniculatus tissues was analyzed histochemically (Jefferson et al., 1987; Szczyglowski et al., 1994) as described by Malamy and Benfey (1997). Stained tissues were examined using a Wild Heerburgg M420 stereoscope (Wild Leitz Ltd., Heerbrugg, Switzerland). The images of stained nodule sections were generated using a Kodak DC120 digital camera and processed using Adobe (Mountain View, CA) Photoshop 5.02 software.
Subcellular Localization of the mGFP5–Nlj16 Fusion Protein in Onion Epidermal Cells
All fusion proteins used in subcellular localization studies were constructed in a derivative of pAVA319 vector (von Arnim et al., 1998) in which the gfp cDNA was replaced with the cDNA encoding mGFP5 (pAVA393; kindly provided by A.G. von Arnim, University of Tennessee, Knoxville) (Siemering et al., 1996). Plasmid DNA from each construct was delivered into the onion epidermal cells using 1.6-μm gold particles and the Biolistic PDS-1000 gene transformation system (Dupont), essentially as described by Varagona et al. (1992). The bombarded onion epidermal explants were incubated for 18 to 24 hr in the light on solid Murashige and Skoog (1962) medium (GIBCO) containing 30 g/L sucrose and 180 mg/L KH2PO4. The fluorescence of mGFP5 fusion proteins was analyzed using a Zeiss (Jena, Germany) Axiophot epifluorescence microscope with the following parameters: emission at 470 ± 20 nm, beam splitter at 510 nm, and excitation at 540 ± 20 nm. Images were obtained using a Kodak DC120 digital camera and Adobe Photoshop 5.02 software. Each transfection experiment was repeated at least two times.
Yeast Complementation Experiments
The yeast strains CTY1079 (MATα, ura3-52, lys2-801, Δhis3-200, sec14-1ts Δspo14::HIS3; Xie et al., 1998; Phillips et al., 1999) and CTY558 (MATα, ade2, ade3, leu2, Δhis3-200, ura3-52, sec14Δ1:: HIS3/pCTY11; Lopez et al., 1994) were used for the complementation experiments. The entire LjPLP-IV cDNA was cloned into the yeast expression vector YePlac195PGK under the control of the phosphoglycerate kinase (PGK) promoter to generate construct pGK-IV. The constructs pGK-IVΔCC and pGK-IIIΔ express the truncated LjPLP-IV and LjPLP-III proteins that lack the CC residues or the entire Nlj16-like domain, respectively.
Plasmids YEplac195PGK, pGK-IV, pGK-IVΔCC, and pGK-IIIΔ were introduced into the yeast strains according to the procedure of Elble (1992). CTY1079 transformants were selected initially at 28°C on defined yeast medium lacking uracil. Complementation of the temperature-sensitive phenotype of the sec14-1 allele in strain CTY1079 was evaluated after 4 days of growth on selective medium at 37°C. Five independent CTY1079 transformants containing each plasmid were evaluated. Complementation of the null sec14Δ1 allele was performed essentially as described (Lopez et al., 1994).
Phosphatidylinositol and Phosphatidylcholine Transfer Assays
The Sec14p domain of LjPLP-IV (amino acids 1 to 365) was fused to the His6 tag in the pET30a vector and expressed in Escherichia coli strain BL21De3 (Novagen, Madison, WI). Freshly transformed E. coli were grown in Luria-Bertani broth supplemented with kanamycin (25 μg/mL) at 37°C. Protein expression was induced with isopropylthio-β-galactoside (1 mM), and cultures were shifted to 28°C for 3 hr. Total E. coli lysate, prepared either from control strain BL21De3 or from derivative strains individually expressing LjPLP-IV–Sec14p or His6–Sec14p, was clarified by centrifugation at 100,000g (M.A. Kearns et al., 1998). This clarified fraction represented total cytosol and was assayed for phosphatidylinositol (PtdIns) and phosphatidylcholine (PtdCho) transfer, as described previously (Aitken et al., 1990). Recombinant His6-tagged SEC14 was expressed from pRE526, as described by Skinner et al. (1995).
Acknowledgments
We thank Ryan Bushey for excellent technical assistance with generation of transgenic plants and Drs. Mark Johnson and Jim Kastenmayer for their helpful advice during the course of the work. We also thank Marlene Cameron and Kurt Stepnitz for their expert help with preparation of the figures. P.K., F.J.d.B., and K.S. were supported by United States Department of Energy Grants DE-FG02-91ER20021 and 98-35305-6551 from the United States Department of Agriculture. S.M.R. and V.A.B. were supported by Grant GM44530 from the National Institutes of Health.
References
- Aitken, J.F., van Heusden, G.P.H., Temkin, M., and Dowhan, W. (1990). The gene encoding the phosphatidylinositol transfer protein is essential for cell growth. J. Biol. Chem. 265, 4711–4717. [PubMed] [Google Scholar]
- Bankaitis, V.A., Malehorn, D.E., Emr, S.D., and Greene, R. (1989). The Saccharomyces cerevisiae SEC14 gene encodes a cytosolic factor that is required for transport of secretory proteins from the yeast Golgi complex. J. Cell Biol. 108, 1271–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis, V.A., Aitken, J.F., Cleves, A.E., and Dowhan, W. (1990). An essential role for a phospholipid transfer protein in yeast Golgi function. Nature 347, 561–562. [DOI] [PubMed] [Google Scholar]
- Cleves, A.E., McGee, T.P., and Bankaitis, V.A. (1991). Phospholipid transfer proteins: A biological debut. Trends Cell Biol. 1, 30–34. [DOI] [PubMed] [Google Scholar]
- Cunningham, E., Tan, S.W., Swigart, P., Hsuan, J., Bankaitis, V., and Cockcroft, S. (1996). The yeast and mammalian isoforms of phosphatidylinositol transfer protein can all restore phospholipase C–mediated inositol lipid signaling in cytosol-depleted RBL-2H3 and HL60 cells. Proc. Natl. Acad. Sci. USA 93, 6589–6593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hartog, M., Musgrave, A., and Munnik, T. (2001). Nod factor–induced phosphatidic acid and diacylglycerol pyrophosphate formation: A role for phospholipase C and D in root hair deformation. Plant J. 25, 55–65. [DOI] [PubMed] [Google Scholar]
- Drøbak, K.K., Dewey, R.E., and Boss, W.F. (1999). Phosphoinositide kinases and the synthesis of polyphosphoinositides in higher plant cells. Int. Rev. Cytol. 189, 95–130. [DOI] [PubMed] [Google Scholar]
- Elble, R. (1992). A simple and efficient procedure for transformation of yeasts. BioTechniques 13, 18–20. [PubMed] [Google Scholar]
- Hancock, J.F., Cadwallader, K., Paterson, H., and Marshall, C.J. (1991). A CAAX or a CAAL motif and a second signal are sufficient for plasma membrane targeting of ras proteins. EMBO J. 10, 4033–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, J.C., and Martin, T.F.J. (1993). Phosphatidylinositol transfer protein required for ATP-dependent priming of Ca2+-activated secretion. Nature 366, 572–575. [DOI] [PubMed] [Google Scholar]
- Hay, J.C., Fisette, P.L., Jenkins, G.H., Fukami, K., Takenawa, T., Anderson, R.E., and Martin, T.F.J. (1995). ATP-dependent inositide phosphorylation required for Ca2+-activated secretion. Nature 372, 173–177. [DOI] [PubMed] [Google Scholar]
- Hofgen, R., and Willmitzer, L. (1988). Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 16, 9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Z., and Verma, D.P.S. (1994). A phosphatidylinositol 3-kinase is induced during soybean nodule organogenesis and is associated with membrane proliferation. Proc. Natl. Acad. Sci. USA 91, 9617–9621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis, B.D.W., Pankhurst, C.E., and Patel, J.J. (1982). Rhizobium loti, a new species of legume root nodule bacteria. Int. J. Syst. Bacteriol. 32, 378–380. [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, S.M., Alb, J.G., Jr., Phillips, S.E., Bankaitis, V.A., and Howell, K.E. (1998). A phosphatidylinositol-3-kinase and phosphatidylinositol transfer protein cooperate to drive phosphatidylinositol-3-phosphate–dependent formation of constitutive transport vesicles from the trans-Golgi network. J. Biol. Chem. 273, 10349–10354. [DOI] [PubMed] [Google Scholar]
- Jouannic, N., Lepetit, M., Vergnolle, C., Cantrel, C., Gardies, A.-M., Kader, J.-C., and Arondel, V. (1998). Isolation of a cDNA from Arabidopsis thaliana that complements the sec14 mutant of yeast. Eur. J. Biochem. 258, 402–410. [DOI] [PubMed] [Google Scholar]
- Kapranov, P., de Bruijn, F.J., and Szczyglowski, K. (1997). Novel, highly expressed late nodulin gene (LjNOD16) from Lotus japonicus. Plant Physiol. 113, 1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns, B.G., Alb, J.G., Jr., and Bankaitis, V.A. (1998). Phosphatidylinositol transfer proteins: The long and winding road to physiological function. Trends Cell Biol. 8, 276–282. [DOI] [PubMed] [Google Scholar]
- Kearns, M.A., Monks, D.E., Fang, M., Rivas, M.P., Courtney, P.D., Chen, J., Prestwich, G.D., Theibert, A.B., Dewey, R.E., and Bankaitis, V.A. (1998). Novel developmentally regulated phosphoinositide binding proteins from soybean whose expression bypasses the requirements for an essential phosphatidylinositol transfer protein in yeast. EMBO J. 17, 4004–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Routt, S., Xie, Z., Cui, X., Fang, M., Kearns, M.A., Bard, M., Kirsch, D., and Bankaitis, V.A. (2000). Identification of a novel family of nonclassical yeast PITPs whose function modulates activation of phospholipase D and Sec14p-independent cell growth. Mol. Biol. Cell 11, 1989–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez, M.C., Nicaud, J.-M., Skinner, H.B., Vergnolle, C., Kader, J.C., Bankaitis, V.A., and Gaillardin, C. (1994). A phosphatidylinositol/phosphatidylcholine transfer protein is required for differentiation of the dimorphic yeast Yarrowia lipolytica from the yeast to the mycelial form. J. Cell Biol. 125, 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy, J.E., and Benfey, P.N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124, 33–44. [DOI] [PubMed] [Google Scholar]
- Milligan, S.E., Alb, J.G., Jr., Elagina, R., Bankaitis, V.A., and Hyde, D.R. (1997). The phosphatidylinositol transfer protein domain of Drosophila retinal degeneration protein B is required for photoreceptor cell survival and recovery from light stimulation. J. Cell Biol. 139, 351–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Novick, P., Field, C., and Schekman, R. (1980). Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell 21, 205–215. [DOI] [PubMed] [Google Scholar]
- Ohashi, M., de Vries, J.K., Frank, R., Snoek, G., Bankaitis, V.A., Wirtz, K., and Huttner, W.B. (1995). A role for phosphatidylinositol transfer protein in secretory vesicle formation. Nature 377, 544–547. [DOI] [PubMed] [Google Scholar]
- Phillips, S.E., Sha, B., Topalof, L., Xie, Z., Alb, J.G., Klenchin, V.A., Swigart, P., Cockcroft, S., Martin, T.F.J., Luo, M., and Bankaitis, V.A. (1999). Yeast sec14p deficient in phosphatidylinositol transfer activity is functional in vivo. Mol. Cell 4, 187–197. [DOI] [PubMed] [Google Scholar]
- Ramlov, K.B., Laursen, N.B., Stougaard, J., and Marcker, K.A. (1993). Site-directed mutagenesis of the organ-specific element in the soybean leghemoglobin lbc3 gene promoter. Plant J. 4, 577–580. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Concepcion, M., Yalovsky, S., Zik, M., Fromm, H., and Gruissem, W. (1999). The prenylation status of a novel plant calmodulin directs plasma membrane or nuclear localization of the protein. EMBO J. 18, 1996–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama, S.R., Cleves, A.E., Malehorn, D.E., Whitters, E.A., and Bankaitis, V.A. (1990). Cloning and characterization of Kluyveromyces lactis SEC14, a gene whose product stimulates Golgi secretory function in Saccharomyces cerevisiae. J. Bacteriol. 172, 4510–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha, B., Phillips, S.E., Bankaitis, V.A., and Luo, M. (1998). Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol-transfer protein. Nature 391, 506–510. [DOI] [PubMed] [Google Scholar]
- Siemering, K.R., Golbik, R., Sever, R., and Haseloff, J. (1996). Mutations that suppress the thermosensitivity of green fluorescent protein. Curr. Biol. 6, 1653–1663. [DOI] [PubMed] [Google Scholar]
- Simon, J.-P., Morimoto, T., Bankaitis, V.A., Gottlieb, T.A., Ivanov, I.E., Adesnik, M., and Sabatini, D.D. (1998). An essential role for phosphatidylinositol transfer protein in the scission of COPI-coated vesicles from the trans-Golgi network. Proc. Natl. Acad. Sci. USA 95, 11181–11186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner, H.B., McGee, T.P., McMaster, C.R., Fry, M.R., Bell, R.M., and Bankaitis, V.A. (1995). Phosphatidylinositol transfer protein stimulates yeast Golgi function by inhibiting choline-phosphate cytidyltransferase activity. Proc. Natl. Acad. Sci. USA 92, 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabados, L., Ratet, P., Grunenberg, B., and de Bruijn, F.J. (1990). Functional analysis of the Sesbania rostrata leghemoglobin glb3 gene 5′-upstream region in transgenic Lotus corniculatus and Nicotiana tabacum plants. Plant Cell 2, 973–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczyglowski, K., Szabados, L., Fujimoto, S.Y., Silver, D., and de Bruijn, F.J. (1994). Site-specific mutagenesis of the nodule-infected cell expression (NICE) element and the AT-rich element ATRE-BS2* of the Sesbania rostrata leghemoglobin glb3 promoter. Plant Cell 6, 317–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczyglowski, K., Hamburger, D., Kapranov, P., and de Bruijn, F.J. (1997). Construction of a Lotus japonicus late nodulin expressed sequence tag library and identification of novel nodule-specific genes. Plant Physiol. 114, 1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczyglowski, K., Kapranov, P., Hamburger, D., and de Bruijn, F.J. (1998). The Lotus japonicus LjNOD70 nodulin gene encodes a protein with similarities to transporters. Plant Mol. Biol. 37, 651–661. [DOI] [PubMed] [Google Scholar]
- Tempe, J., and Casse-Delbart, F. (1989). Plant gene vectors and genetic transformation: Agrobacterium Ri plasmids. In Cell Culture and Somatic Cell Genetics of Plants, Vol. 6, J. Schell and I.K. Vasil, eds (San Diego, CA: Academic Press), pp. 25–49.
- Varagona, M.J., Schmidt, R.J., and Raikhel, N.V. (1992). Nuclear localization signal(s) required for nuclear targeting of the maize regulatory protein Opaque-2. Plant Cell 4, 1213–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim, A.G., Deng, X.-W., and Stacey, M.G. (1998). Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221, 35–43. [DOI] [PubMed] [Google Scholar]
- Waterhouse, P.M., Graham, M., and Wang, M.-B. (1998). Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl. Acad. Sci. USA 95, 13959–13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz, K.W.A. (1991). Phospholipid transfer proteins. Annu. Rev. Bio-chem. 60, 73–99. [DOI] [PubMed] [Google Scholar]
- Xie, Z., Fang, M., Rivas, M.P., Faulkner, A., Sternweis, P.C., Engebrecht, J., and Bankaitis, V.A. (1998). Phospholipase D activity is required for suppression of yeast phosphatidylinositol transfer protein defects. Proc. Natl. Acad. Sci. USA 95, 12346–12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F.L., and Casey, P.J. (1996). Protein prenylation: Molecular mechanisms and functional consequences. Annu. Rev. Biochem. 65, 241–269. [DOI] [PubMed] [Google Scholar]