Abstract
Higher plants use photoperiodic cues to regulate many aspects of development, including the transition from vegetative to floral development. The EARLY FLOWERING3 (ELF3) gene is required for photoperiodic flowering and normal circadian regulation in Arabidopsis. We have cloned ELF3 by positional methods and found that it encodes a novel 695–amino acid protein that may function as a transcriptional regulator. ELF3 transcript level is regulated in a circadian manner, as is expected of a zeitnehmer input pathway component. Overexpression of the LATE ELONGATED HYPOCOTYL gene, which has been proposed to function as a clock component, did not abolish circadian regulation of ELF3 transcription, providing further evidence that ELF3 is a circadian clock input pathway component.
INTRODUCTION
Both plants and animals use seasonal cues to synchronize their reproductive development with the external environment. The primary seasonal cue used by organisms is daylength, or photoperiod. Higher plants use photoperiodic cues to regulate many aspects of development, including the transition from vegetative to floral development. Arabidopsis is a facultative long-day (LD) plant, flowering earlier in LD conditions than in short-day (SD) conditions.
Mutational analysis of Arabidopsis has led to the discovery of a number of genes that are required for the photoperiodic regulation of flowering. Mutations in the CONSTANS (CO) and GIGANTEA (GI) genes result in late flowering in LD conditions but have little or no effect in SD conditions, suggesting that the wild-type genes are required for the promotion of flowering in inductive (LD) conditions. CO encodes a zinc finger protein that likely functions as a transcription factor, and GI encodes a novel putative membrane protein (Putterill et al., 1995; Fowler et al., 1999). In contrast to co and gi mutants, flowering locus t (ft) mutants flower late in both LD and SD conditions (Ruiz-García et al., 1997). FT belongs to a family of putative membrane-associated proteins that can bind hydrophobic ligands (Kardailsky et al., 1999; Kobayashi et al., 1999). One proposed molecular function of FT is the generation of peptide molecules that could act as transmissible signals (Kobayashi et al., 1999). FT appears to function partially downstream of CO. Although the identification of genes that control flowering has begun to reveal a great deal about the regulation of cell fate during plant development, these genes have not revealed how circadian clock function influences the vegetative-to-floral transition.
Circadian clocks appear to be involved in photoperiodic responses in cyanobacteria, fungi, plants, and animals. Although a molecular feedback loop required for circadian regulation in Neurospora and animals has been described in detail, the molecular mechanisms of plant circadian clocks remain largely unknown. Genetic approaches have resulted in the isolation of a number of intriguing recessive mutations that alter the free-running period of the Arabidopsis circadian clock, including TIMING OF CAB (TOC) 1, CIRCADIAN CLOCK ASSOCIATED (CCA) 1, and ZEITLUPE (ZTL) (Somers et al., 1998, 2000; Green and Tobin, 1999). ZTL encodes a novel F-box–containing protein that may be involved in light-dependent proteolysis. TOC1 encodes a histidine kinase similar to those of bacterial two-component signaling systems (Strayer et al., 2000). CCA1 encodes a single MYB domain–containing transcription factor whose expression is regulated in a circadian manner. The overexpression of CCA1, or of the closely related gene LATE ELONGATED HYPOCOTYL (LHY), results in photoperiod-insensitive early flowering and arrhythmicity of circadian clock–associated gene expression (Schaffer et al., 1998; Wang and Tobin, 1998). In addition, both CO and GI are regulated in a circadian manner.
Mutations in the EARLY FLOWERING3 (ELF3) locus also result in the loss of both photoperiod sensitivity and circadian regulation, making ELF3 a candidate for linking circadian clock function with the photoperiodic induction of flowering. elf3 mutant plants flower early and at the same developmental time in both LD and SD light conditions (Zagotta et al., 1996). The long hypocotyl phenotype of elf3 mutant plants suggests a defect in light reception or the transduction of light signals (Zagotta et al., 1996). In addition, leaf movements and circadian clock–regulated gene expression are arrhythmic in elf3 mutants in constant light conditions but not in constant dark conditions, suggesting that a circadian clock remains functional in the absence of wild-type ELF3 function (Hicks et al., 1996). On the basis of these results, the ELF3 gene product was proposed to function in a light input pathway to the circadian oscillator. The absence of ELF3 was hypothesized to alter the coordination of light and circadian regulatory pathways, resulting in the altered flowering time and photoperiodic insensitivity observed in elf3 mutants. This model is supported by recent results showing that ELF3 is required to gate light input to the circadian oscillator, altering the sensitivity of the central oscillator to light at a particular point in the circadian cycle (McWatters et al., 2000).
To elucidate the molecular mechanism of ELF3 function, we have isolated the ELF3 gene by positional cloning methods. ELF3 encodes a novel protein of 695 amino acids that may function as a transcriptional regulator. ELF3 mRNA level is regulated in a cyclic manner, peaking at ∼14 to 16 hr after sunrise regardless of daylength. Continuation of cyclic expression in constant conditions shows that ELF3 is regulated by a circadian clock. Constitutive expression of LHY does not abolish the circadian rhythm of ELF3 gene expression, suggesting that additional genes, and potentially additional feedback loops, are involved in the regulation of ELF3.
RESULTS
Positional Cloning of ELF3
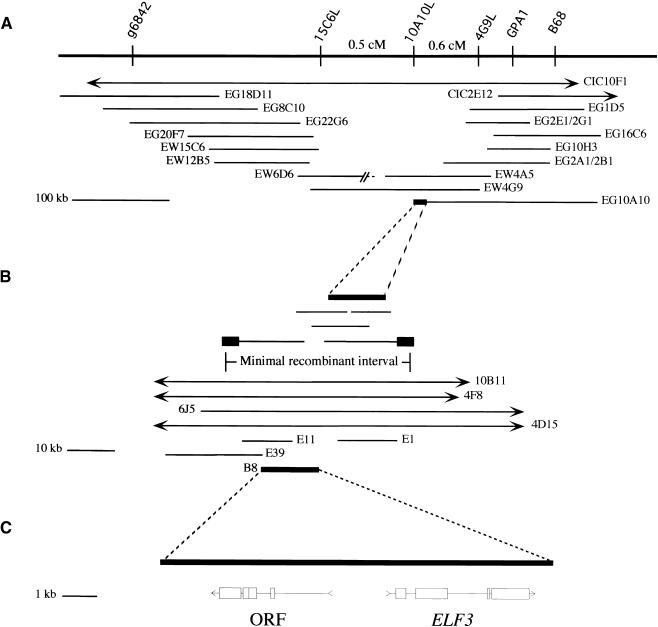
The ELF3 gene was isolated on the basis of its chromosomal map position. ELF3 was mapped initially to the middle of chromosome 2, between molecular markers g6842 and GPA1 (Zagotta et al., 1996). A yeast artificial chromosome (YAC) contig covering this region (H. Goodman, personal communication) was refined, and several restriction fragment length polymorphisms (RFLPs) in the region were identified using YAC end clones as probes (Figure 1A). Genomic cosmid clones (Figure 1B) were isolated from this region and used to identify RFLPs distinguishable from ELF3 by recombination. This defined an ∼60-kb region of chromosome 2 that contained the ELF3 gene.
Figure 1.
Map-Based Cloning of ELF3.
(A) Physical mapping of ELF3. Molecular markers are shown above the line with relevant recombination distances, and the YAC contig of ∼500 kb of Arabidopsis genomic DNA is shown below the line. YAC 6D6 is chimeric, with the unlinked region represented by a dashed line. cM, centimorgan.
(B) YAC end clone 10A10L was used as a starting point for identifying λ and cosmid clones. RFLP analysis localized ELF3 to the 60-kb minimal recombinant interval shown. The closed boxes indicate the RFLPs. BAC clones spanning this region were identified, and cosmid subclones of BAC 4D15 were used for transformation rescue. Cosmid B8 complemented elf3-1 and elf3-3 mutant phenotypes (see Figure 2).
(C) The positions and structures of two transcription units within cosmid B8 are shown. ORF, open reading frame.
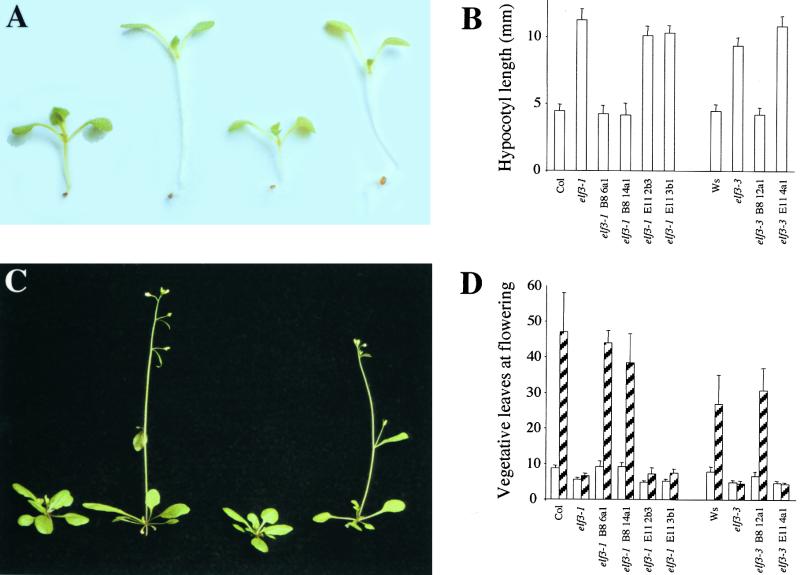
Bacterial artificial chromosomes (BACs) that spanned this minimal recombinant interval were identified from the Texas A&M University BAC library (Figure 1B). A cosmid mini-library of BAC 4D15 was constructed, and members of a group of cosmid clones that spanned the region of interest were transformed individually into elf3 mutant plants. Transformation with an 11-kb cosmid clone, B8, rescued all elf3 mutant phenotypes assayed (Figures 1B and 2). Cosmids E1, E11, and E39 did not rescue the elf3 mutant (Figure 2 and data not shown).
Figure 2.
Complementation of elf3 Mutants.
(A) Ten-day-old seedlings grown in SD conditions. Left to right: wild-type Columbia, elf3-1, transgenic elf3-1 containing cosmid B8, and transgenic elf3-1 containing cosmid E11.
(B) Hypocotyl lengths of 10-day-old seedlings grown in SD conditions. Col, wild-type Columbia; Ws, Wassilewskija. Error bars indicate ±sd (n = 20).
(C) Twenty-five-day-old plants grown in LD conditions. Left to right: wild-type Columbia, elf3-1, transgenic elf3-1 containing cosmid B8, and transgenic elf3-1 containing cosmid E11.
(D) Flowering time expressed as vegetative leaves produced before flowering. Plants were grown in either SD or LD conditions. Error bars indicate ±sd (n ≥ 16).
A 20-kb genomic region including cosmid B8 was subcloned and sequenced. Sequence analysis revealed two potential transcription units contained within cosmid B8, neither of which showed significant similarity to genes of known function. The molecular nature of elf3 mutant alleles was used to determine that the more distal gene contained on cosmid B8 was ELF3. The EcoRI fragment from the distal end of B8 identified a 1- to 2-kb deletion in elf3-2, which was generated by fast neutron mutagenesis (data not shown). Sequence analysis of eight additional elf3 alleles identified further mutations within this transcription unit (Figure 3A).
Figure 3.
DNA Coding and Amino Acid Sequence of ELF3 and Sequence Comparison with Putative Orthologs.
(A) The DNA sequence corresponding to ELF3 cDNA clone 8.2 is shown, along with the predicted amino acid sequence of the longest open reading frame. Amino acid numbering is shown at right, and intron positions are marked with inverted triangles. Molecular changes in eight elf3 alleles are shown above the DNA sequence. The closed inverted triangle is a cryptic splice site used in some elf3-7 transcripts (see text). A potential nuclear targeting signal is shown in boldface. Translation stop codon is indicated by an asterisk.
(B) Black boxes indicate identical residues, and gray boxes indicate conserved residues between ELF3 and ELF3-like predicted proteins in other plant species. Dashes indicate gaps in the sequences. Tomato, soybean, and maize ELF3-like proteins were predicted from partial sequences from expressed sequence tag clones. The rice ELF3-like protein was predicted from genomic sequence.
ELF3 Encodes a Novel Protein
The ELF3 gene has four exons and three introns and is predicted to encode a novel soluble protein of 695 amino acids that is particularly rich in serine, proline, and glutamine (Figure 3A). Database searches revealed strong sequence similarity between ELF3 and predicted proteins from tomato, soybean, rice, and maize (Figure 3B) and weaker similarity between ELF3 and an Arabidopsis protein of unknown function on chromosome 3 (GenBank accession number BAB01726).
ELF3 contains a proline-rich region between amino acids 440 and 540 that consists of ∼25% proline residues. This region is shared by both the tomato expressed sequence tag and the Arabidopsis ELF3-like protein from chromosome 3. An acidic region is located from amino acids 206 to 320, and part of this region is shared by ELF3-like proteins from tomato and rice. A threonine-rich stretch of amino acids is located near the C-terminal end of the protein from amino acids 636 to 652, and three short runs of glutamine residues are found between amino acids 544 and 585. The length of the first glutamine repeat is polymorphic between the Columbia and Wassilewskija ecotypes: the Wassilewskija gene contains 16 glutamine codons, whereas the Columbia gene contains seven (data not shown). The proline-rich region, the acidic region, and the threonine/glutamine–rich region could play a role in transcriptional activation. A potential nuclear targeting signal beginning at amino acid 591 was predicted by analysis with PROSORT2 (Figure 3A), which is consistent with a role for ELF3 in transcriptional regulation. Potential nuclear targeting signals also were predicted in the ELF3-like proteins from tomato and rice. Although a nuclear targeting signal was not identified in the partial sequences available from soybean and maize ELF3-like proteins, these were predicted to be nuclear proteins on the basis of their generally basic nature. Finally, ELF3 contains a large number of potential phosphorylation sites, suggesting that phosphorylation may be involved in ELF3 regulation.
The elf3-1, elf3-3, elf3-4, elf3-5, and elf3-9 alleles all contain single base changes that result in premature stop codons (Figure 3A). elf3-6 and elf3-8 are independently derived alleles that contain identical single base changes at the exon 4 splice acceptor site. This mutation leads to the inclusion of 28 amino acids from a different reading frame before a stop codon is encountered. All of these alleles result in a strong mutant phenotype (Hicks et al., 1996; Zagotta et al., 1996) and are likely to be either null alleles or strong reduced-function alleles.
elf3-7 is a weak allele that causes early flowering and long hypocotyl phenotypes. However, elf3-7 homozygous mutants remain sensitive to photoperiod, flowering earlier in LD than in SD (J. Reed, personal communication; K.A. Hicks, unpublished results). The elf3-7 allele contains a single base change at the exon 1 splice donor site (Figure 3A). Analysis of elf3-7 transcripts by reverse transcriptase–polymerase chain reaction (PCR) (data not shown) indicated the use of several cryptic splice sites that resulted in several different transcripts, all but one of which contain premature stop codons. On the basis of similarity to the strong alleles described above, we deduce that transcripts containing a premature stop codon are unlikely to confer the weak phenotype observed in the elf3-7 allele. However, the use of one in-frame cryptic splice site was observed. The use of this splice site is predicted to result in the loss of eight amino acids, a molecular change that could be consistent with the observed partial function phenotype.
ELF3 Transcript Level Cycles Daily
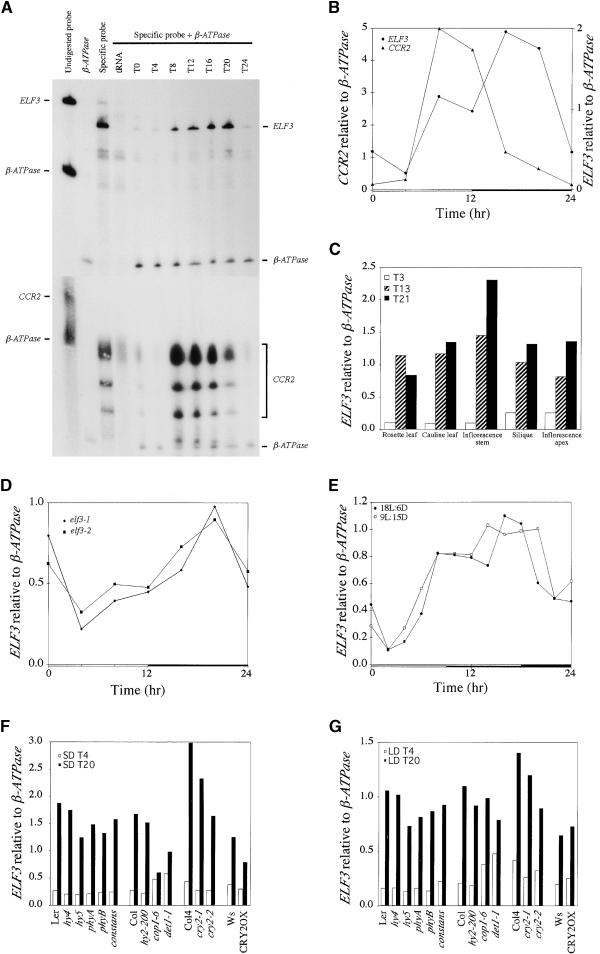
As shown in Figures 4A and 4B, ELF3 transcript level varied during a 24-hr period, with a trough in the early day and a peak in the early night. The maximal transcript level was observed ∼16 hr after dawn in 12-hr-light/12-hr-dark cycles. In the same RNA samples, expression of the circadian clock–regulated gene CCR2 was highest in the late day, as has been reported (Kreps and Simon, 1997). Diurnal cycling of the ELF3 transcript was observed in a variety of adult shoot tissues (Figure 4C). A very low level of ELF3 expression also was detected in seedling root tissue and followed a similar temporal pattern of expression (data not shown). Diurnal cycling of ELF3 transcript was observed in elf3 mutant alleles (Figure 4D and data not shown).
Figure 4.
ELF3 Transcript Levels in Wild-Type and Mutant Backgrounds.
Ten micrograms of total RNA was hybridized with antisense riboprobes for ELF3 or CCR2 and then subjected to RNase digestion, as described in Methods. β-ATPase served as an internal control, and tRNA was used as a negative control. Time 0 (T0) indicates dawn. Quantitative representation of expression data normalized to β-ATPase is shown in all panels except (A). Open and closed bars along x-axis represent light and dark photoperiods, respectively.
(A) Wild-type seedlings were grown in 12-hr-light/12-hr-dark conditions for ∼1 week, and samples were collected every 4 hr. The specific probe was complementary to ELF3 at top and to CCR2 at bottom.
(B) Quantification of ELF3 and CCR2 mRNA levels shown in (A) after normalization to β-ATPase mRNA. The experiment was repeated at least three times.
(C) ELF3 transcript level in mature wild-type plants grown in 9-hr-light/15-hr-dark SD conditions. Tissue samples were collected at T3, T13, and T21.
(D) ELF3 transcript level in elf3 mutant seedlings grown in 12-hr-light/12-hr-dark conditions. Samples were collected every 4 hr. The experiment was repeated five times for elf3-1.
(E) ELF3 transcript level in wild-type seedlings grown in either 9-hr-light/15-hr-dark SD conditions (9L:15D) or 18-hr-light/6-hr-dark LD conditions (18L:6D). Samples were collected every 2 hr. The experiment was repeated twice.
(F) and (G) ELF3 transcript level in wild-type and mutant seedlings grown in 9-hr-light/15-hr-dark SD conditions (F) or 18-hr-light/6-hr-dark LD conditions (G). Samples were collected at T4 and T20. Col, Columbia; Ler, Landsberg; phy, phytochrome; Ws, Wassilewskija.
Phenotypic analysis suggested that ELF3 plays a role in light signal transduction in addition to its role in regulating flowering time. In an effort to determine the potential upstream regulators of ELF3, we measured the level of ELF3 transcript in known mutants that show defects in light signal transduction and/or floral induction. None of the mutations tested had a profound effect on the level or gross temporal pattern of ELF3 expression. Expression of ELF3 in the cop1-6 background was somewhat altered from that of the wild type in SD conditions, with higher expression than in the wild type observed at T4 and lower expression observed at T20; this effect on ELF3 expression by the cop1-6 mutation was less pronounced in LD conditions.
ELF3 Transcript Level Shows a Circadian Rhythm in Constant Conditions
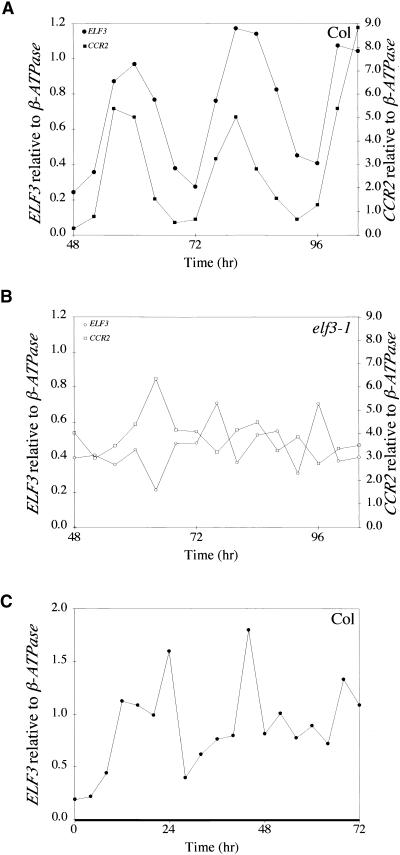
The observation of a diurnal rhythm of ELF3 transcript level suggested that an endogenous circadian clock might regulate ELF3 expression. One hallmark of circadian rhythms is that they continue in constant conditions. Therefore, ELF3 expression was analyzed in wild-type seedlings entrained to 12-hr-light/12-hr-dark cycles and transferred to constant conditions. Figure 5A shows that ELF3 transcript level continued to cycle for at least 4 days after transfer to constant light conditions, indicating that ELF3 is regulated by a circadian clock. The rhythmic expression of a previously described circadian clock–regulated gene, CCR2, also is shown in Figure 5A. Although ELF3 and CCR2 expression was not in phase in 12-hr-light/12-hr-dark cycles (Figures 4A and 4B), expression of these genes appeared to be relatively in phase after 48 hr in constant light conditions (Figure 5A). This altered phasing of gene expression was consistently observed. A similar experiment was conducted in which wild-type seedlings were transferred to constant dark after entrainment in light/dark cycles, and ELF3 transcript level was measured. Cycling of ELF3 transcript level was observed during the first day of constant dark conditions; however, rhythmic expression of ELF3 was not maintained at detectable levels (Figure 5C). Damping of circadian oscillations also has been observed for a number of other circadian clock–regulated genes, including CAB, CAT2, CCA1, and CCR2 (Millar and Kay, 1991; Kreps and Simon, 1997; Zhong et al., 1997; Wang and Tobin, 1998).
Figure 5.
ELF3 Transcript Level Continues to Cycle in Constant Conditions.
Wild-type ([A] and [C]) and elf3 mutant (B) seedlings were entrained under 12-hr-light/12-hr-dark conditions and then shifted to either constant light ([A] and [B]) or constant dark (C) at time 0, which corresponds to normal dawn. Samples were taken every 4 hr and analyzed by RNase protection. ELF3 and CCR2 transcript levels were normalized to β-ATPase. Experiments were repeated at least three times. Col, Columbia.
ELF3 Transcript Cycling Requires ELF3
Previously, we showed that ELF3 is required for the circadian regulation of the cab2 promoter and of leaf sleep movements in constant light (Hicks et al., 1996). To determine whether ELF3 function is required for the circadian regulation of its own expression, we analyzed ELF3 gene expression in elf3 mutant seedlings that were first entrained to 12-hr-light/12-hr-dark cycles and then transferred to constant light. Figure 5B shows that ELF3 transcript levels did not cycle in the elf3-1 mutant after transfer to constant light conditions, indicating that ELF3 is required for its own circadian regulation. The loss of rhythmicity in CCR2 expression in the elf3-1 mutant is shown as well. Similar results were observed for two additional mutant alleles, elf3-2 and elf3-3 (data not shown).
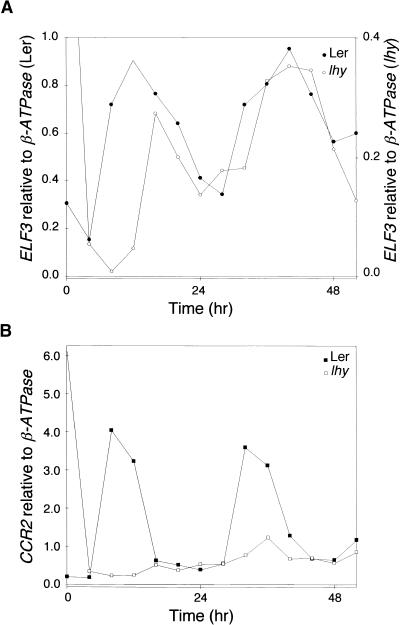
Constitutive Expression of LHY Does Not Abolish the Circadian Rhythm of ELF3 Gene Expression
The rhythmic expression of two MYB-related transcription factors, CCA1 and LHY, has been shown to be required for circadian regulation in Arabidopsis. Constitutive overexpression of either of these genes disrupts the circadian regulation of a number of genes and results in arrhythmic leaf sleep movements (Schaffer et al., 1998; Wang and Tobin, 1998). Therefore, we anticipated that overexpression of LHY also would lead to a loss of rhythmic ELF3 gene expression. However, as shown in Figure 6A, the constitutive overexpression of LHY did not abolish the circadian rhythm of ELF3 gene expression. Although the overexpression of LHY resulted in a reduction in ELF3 transcript levels, gene expression was rhythmic for at least 2 days in constant light. In contrast, Figure 6B shows that the overexpression of LHY resulted in low levels of CCR2 transcript that did not show a circadian rhythm, as reported previously (Schaffer et al., 1998).
Figure 6.
ELF3 Transcript Level Continues to Cycle in lhy Mutant Seedlings.
Wild-type Landsberg (Ler) and lhy mutant seedlings were entrained under 12-hr-light/12-hr-dark conditions and then shifted to constant light at time 0, which corresponds to normal dawn. Samples were taken every 4 hr and analyzed by RNase protection. ELF3 (A) and CCR2 (B) transcript levels were normalized to β-ATPase. Scales for ELF3 (A) differ between wild type and lhy. The experiment was repeated twice.
DISCUSSION
ELF3 Encodes a Novel Protein
The ELF3 gene is predicted to encode a novel protein. The ELF3 sequence was previously identified as pyk20 in an unrelated promoter tagging approach (Puzio et al., 1999). Searches of current databases revealed significant sequence similarity to ELF3 only in other higher plants. ELF3 may function as a transcription factor on the basis of the presence of sequence features that are commonly found in transcriptional regulators: a proline-rich region, an acidic region, and a threonine/glutamine–rich region (Figure 3). ELF3-like proteins from tomato and rice share these sequence features in part, strengthening the possibility that ELF3 encodes a novel transcription factor. Although none of these proteins appears to contain a DNA binding domain, it is possible that ELF3 acts to regulate transcription in concert with other factors. Given the pleiotropic nature of the elf3 mutant phenotypes, many of which are related directly to defects in light perception, it is likely that ELF3 functions in photoreceptor-mediated signal transduction pathways. It is now clear that some higher plant photoreceptors, such as the phytochromes, function in the nucleus; thus, the ELF3 protein may interact with these photoreceptors to regulate gene transcription.
There are a number of examples of altered gene expression in elf3 mutants that suggest potential downstream targets of transcriptional regulation by ELF3. The rhythmic expression of GI, LHY, CAB, and CCR2 is abolished in constant light conditions in elf3 null mutants (Hicks et al., 1996; Schaffer et al., 1998; Fowler et al., 1999) (Figure 5). In addition, the expression of GI was higher in elf3 null mutants in LD cycles, particularly during the later half of the cycle, when ELF3 expression normally peaks (Fowler et al., 1999). The question remains whether ELF3 functions to regulate the transcription of some or all of these genes directly, or in contrast, whether ELF3 affects their expression indirectly by way of the circadian clock.
ELF3 Transcript Is Regulated in a Circadian Manner
On the basis of the early flowering phenotype of elf3 null alleles, ELF3 likely functions as a repressor of flowering. In wild-type Arabidopsis, flowering is more delayed in SD conditions than in LD conditions, presumably because of the increased level or activity of a floral repressor in SD conditions. However, no difference in ELF3 transcript level was observed between LD and SD conditions (Figure 4E). Diurnal cycling of ELF3 transcript occurred in both conditions, with the peak level of transcript occurring ∼16 to 18 hr after dawn. Although this corresponds to the middle of the dark period in 9-hr-light/15-hr-dark SD conditions, maximal transcript levels were observed at the light-to-dark transition in 18-hr-light/6-hr-dark LD conditions.
Significantly, elf3-1 mutants display a semidominant early flowering phenotype in LD conditions but a fully recessive early flowering phenotype in SD conditions (data not shown). This observation, together with the observation of the partial mutant phenotype in the elf3-7 splice site mutant, suggests that the ELF3 protein itself functions in a quantitative manner. Thus, the relative abundance of functional ELF3 protein in the nucleus may serve as a mechanism to measure the length of day and to transmit this information to the circadian clock, with a high level of active ELF3 protein being found during the long night in SD-grown plants.
We propose that although the level of ELF3 transcript was similar in LD and SD conditions, the activity of ELF3 protein may be decreased by LD conditions. This decrease could be caused at the molecular level by ELF3 interaction with, or modification by, another protein or ligand that is strictly regulated by light. This interaction or modification would occur only in the presence of light. Thus, maximum ELF3 activity would be observed in SD conditions, when the peak level of ELF3 transcript overlaps with the dark period. One candidate for directing such interactions with the ELF3 protein is phytochrome B, which was found to interact with ELF3 in yeast two-hybrid experiments and in vitro (Liu et al., 2001). This proposed interaction mechanism could account for the quantitative aspect of photoperiodism in Arabidopsis.
Our results on the nature of ELF3 transcriptional regulation, namely, that ELF3 transcripts accumulate in a circadian manner and that this accumulation is dependent on the presence of functional ELF3 protein in constant light conditions (Figure 5), strengthen the conclusion that ELF3 functions within a zeitnehmer (“time-taker”) feedback loop (McWatters et al., 2000) and suggest that the input pathway is regulated rhythmically by feedback from the central oscillator to ELF3 transcription.
Constitutive Expression of LHY Does Not Abolish the Circadian Rhythm of ELF3 Gene Expression
The rhythmic expression of central oscillator components is required for circadian function in systems such as Neurospora and animals, in which the molecular feedback loop has been well characterized (Dunlap, 1999). Therefore, the constitutive expression of an Arabidopsis oscillator component is expected to result in a loss of rhythmicity. Two MYB-related transcription factors, CCA1 and LHY, have been proposed as oscillator components on the basis of the disruption of circadian regulation when either factor is expressed constitutively (Schaffer et al., 1998; Wang and Tobin, 1998). However, the continued rhythmic expression of ELF3 in the presence of constitutive LHY expression suggests that LHY is not a component of the ELF3-regulating central oscillator. It remains possible that multiple clocks function within Arabidopsis and that the ELF3-related clock is separate from the clock that requires LHY function.
ELF3 Gene Function in Flowering Plants
Database searches for gene sequences related to ELF3 have revealed significant homology only with sequences from other higher plant species. This leaves open the possibility that the ELF3 protein, and the homologous proteins of other plant species, function in a regulatory hierarchy that has evolved specifically in the plant kingdom. If this is the case, then plants may have developed novel molecular mechanisms for the regulation of input signals to the circadian clock, or there may exist a type of circadian clock that is specific to plants.
One possible explanation for such a novel system in multicellular plants is that higher plants are uniquely constrained within their environment. For example, some species of flowering plants quite accurately measure slight changes in daylength to promote or repress critical developmental events such as the initiation of flowering. The precise seasonal timing of such events is crucial for plant reproduction and population dispersal. It will be of great interest to determine whether the putative ELF3 homologs identified in other flowering plant species play similar roles in regulating circadian clock function and aspects of plant development such as the initiation of flowering. Although there is clear evidence that ELF3 functions to repress floral initiation in the LD plant Arabidopsis, what function does the tomato homolog of ELF3 have in day-neutral varieties of that species? To investigate questions such as this, it will be essential to elucidate the molecular function of the ELF3 protein in Arabidopsis and to determine if the homologous proteins in species such as tomato have equivalent functions. In addition, it will be critical to determine whether the components of the circadian clock and the pathways that regulate the initiation of flowering are conserved among these higher plant species. It remains a possibility that not only are circadian components and regulatory mechanisms divergent among bacteria, fungi, plants, and animals but that even within the plant kingdom multiple molecular mechanisms exist that control circadian clock function and, in turn, related developmental processes.
METHODS
Plant Material
elf3-1, elf3-2, elf3-3, and elf3-4 have been described (Hicks et al., 1996; Zagotta et al., 1996). elf3-5, elf3-7, elf3-8, and elf3-9 were isolated in the Arabidopsis thaliana Columbia ecotype by using ethyl methanesulfonate mutagenesis. elf3-7 and elf3-9 were kindly provided by Jason Reed (University of North Carolina, Chapel Hill), and elf3-8 was kindly provided by Andrew Millar (University of Warwick, Coventry, UK). elf3-6 was isolated in the Wassilewskija ecotype from the publicly available Feldmann T-DNA insertion populations (Arabidopsis Biological Resource Center, Columbus, OH).
pif was kindly provided by José Martinez-Zapater (Instituto Valenciano de Investigaciones Agrarias, Valencia, Spain). aux1, hy4 (2.23N), hy5 (Ci88), co, det1-1, and cop1-6 were obtained from the Arabidopsis Biological Resource Center. phyA-201 and phyB-5 were kindly provided by Joanne Chory (Salk Institute for Biological Studies, La Jolla, CA) (Nagatani et al., 1993; Reed et al., 1994). cry2-1, cry2-2, and CRY2OX were kindly provided by Chentao Lin (University of California, Los Angeles) (Guo et al., 1998).
Cloning and Sequencing of ELF3
Meiotic recombination events were generated near ELF3 by screening for recombination events between elf3-1 and the flanking markers erecta and pif. Crosses were analyzed between elf3-1 (Columbia ecotype) and erecta aux1 (Landsberg ecotype) or erecta pif1 (Landsberg ecotype). Restriction fragment length polymorphism (RFLP) mapping was performed by DNA gel blot analysis according to standard protocols.
The yeast artificial chromosome (YAC) contig spanning the region between g6842 and GPA1 was provided by Howard Goodman (Massachusetts General Hospital, Boston). YAC end clones were obtained by plasmid rescue or inverse polymerase chain reaction (PCR). End clones were used to refine the contig by DNA gel blot hybridization and to map RFLPs between the Columbia and Landsberg ecotypes.
Genomic clones were isolated from the Mulligan and Davis λ genomic library, the Olszewski cosmid library (Olszewski et al., 1988), the Texas A&M University (TAMU) bacterial artificial chromosome (BAC) library (Choi et al., 1995), and a cosmid library constructed from TAMU BAC 4D15 by using the pOCA28 vector. Cosmid clones were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation, and elf3-1 and elf3-3 were transformed by vacuum infiltration (Bechtold et al., 1993; Bent et al., 1994).
DNA fragments were subcloned into the pBluescript SK− vector (Stratagene) for sequence analysis. The ELF3 gene was amplified by PCR from genomic DNA of mutant alleles and subcloned into pBluescript SK− or pCR2.1 (Invitrogen, Carlsbad, CA) for sequence analysis. Mutations were confirmed by direct sequencing of a pool of at least 10 PCR products. elf3-7 transcripts were isolated with reverse transcriptase–PCR by using the cDNA cycle kit (Invitrogen), and PCR products were subcloned as described above.
RNA Analysis
Tissue samples were harvested as described above, and total RNA was extracted essentially as described by Nagy et al. (1988). RNA used for the experiment shown in Figure 6 was kindly provided by Isabelle Carré (University of Warwick). RNase protection assays were performed according to standard protocols. Templates were prepared with PCR amplification from DNA subclones by using one primer complementary to the vector sequence and a second primer complementary to the insert. The ELF3 primer sequence was 5′-TGGCACCTAGCTCTCATC-3′, the CCR2 primer sequence was 5′-CGCTTGTTATGCTTCTACTTGG-3′, and the β-ATPase primer sequence was 5′-TTCCTTGAGAGCTACGAGATG-3′. The CCR2 plasmid was provided by J. Kreps (Torrey Mesa Research Institute, La Jolla, CA). β-ATPase was obtained as an expressed sequence tag clone (GenBank accession number N96685) from the Arabidopsis Biological Resource Center. The intensity of protected fragments was measured by PhosphorImager analysis (Molecular Dynamics, Sunnyvale, CA).
Acknowledgments
We are grateful to I. Carré for providing RNA samples and to R. Formosa and H. Foss for technical assistance. This research was supported by National Science Foundation Grant MCB-9808208 and by U.S.–Israel Binational Agricultural Research and Development Agency Grant US-2964-97 to D.R.W. and by a National Institutes of Health postdoctoral fellowship to K.A.H.
References
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 316, 1194–1199. [Google Scholar]
- Bent, A.F., Kunkel, B.N., Dahlbeck, D., Brown, K.L., Schmidt, R., Giraudat, J., Leung, J., and Staskawicz, B.J. (1994). RPS2 of Arabidopsis thaliana: A leucine-rich repeat class of plant disease resistance genes. Science 265, 1856–1860. [DOI] [PubMed] [Google Scholar]
- Choi, S., Creelman, R.A., Mullet, J.E., and Wing, R.A. (1995). Construction and characterization of a bacterial artificial chromosome library of Arabidopsis thaliana. Weeds World 2, 17–20. [Google Scholar]
- Dunlap, J.C. (1999). Molecular bases for circadian clocks. Cell 96, 271–290. [DOI] [PubMed] [Google Scholar]
- Fowler, S., Lee, K., Onouchi, H., Samach, A., Richardson, K., Morris, B., Coupland, G., and Putterill, J. (1999). GIGANTEA: A circadian clock–controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18, 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, R.M., and Tobin, E.M. (1999). Loss of the circadian clock–associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc. Natl. Acad. Sci. USA 96, 4176–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., Yang, H., Mockler, T.C., and Lin, C. (1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 279, 1360–1363. [DOI] [PubMed] [Google Scholar]
- Hicks, K.A., Millar, A.J., Carré, I.A., Somers, D.E., Straume, M., Meeks-Wagner, D.R., and Kay, S.A. (1996). Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274, 790–792. [DOI] [PubMed] [Google Scholar]
- Kardailsky, I., Shukla, V.K., Ahn, J.H., Dagenais, N., Christensen, S.K., Nguyen, J.T., Chory, J., Harrison, M.J., and Weigel, D. (1999). Activation tagging of the floral inducer FT. Science 286, 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kobayashi, Y., Kaya, H., Goto, K., and Iwabuchi, M. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. [DOI] [PubMed] [Google Scholar]
- Kreps, J.A., and Simon, A.E. (1997). Environmental and genetic effects on circadian clock–regulated gene expression in Arabidopsis. Plant Cell 9, 297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X.L., Covington, M.F., Fankhauser, C., Chory, J., and Wagner, D.R. (2001). ELF3 encodes a circadian clock–regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13, 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWatters, H.G., Bastow, R.M., Hall, A., and Millar, A.J. (2000). The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature 408, 716–720. [DOI] [PubMed] [Google Scholar]
- Millar, A.J., and Kay, S.A. (1991). Circadian control of cab gene transcription and mRNA accumulation in Arabidopsis. Plant Cell 3, 541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani, A., Reed, J.W., and Chory, J. (1993). Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol. 102, 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, F., Kay, S.A., and Chua, N.-H. (1988). Analysis of gene expression in transgenic plants. In Plant Molecular Biology Manual, S. Gelvin and R. Schilperoort, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 1–29.
- Olszewski, N., Martin, F., and Ausubel, F. (1988). Specialized binary vector for plant transformation: Expression of the Arabidopsis thaliana AHAS gene in Nicotiana tabacum. Nucleic Acids Res. 16, 10765–10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill, J., Robson, F., Lee, K., Simon, R., and Coupland, G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857. [DOI] [PubMed] [Google Scholar]
- Puzio, P.S., Lausen, J., Almeida-Engler, J., Cai, D., Gheysen, G., and Grundler, F.M.W. (1999). Isolation of a gene from Arabidopsis thaliana related to nematode feeding structures. Gene 239, 163–172. [DOI] [PubMed] [Google Scholar]
- Reed, J.W., Nagatani, A., Elich, T.D., Fagan, M., and Chory, J. (1994). Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104, 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-García, L., Madueno, F., Wilkinson, M., Haughn, G., Salinas, J., and Martinez-Zapater, J.M. (1997). Different roles of flowering-time genes in the activation of floral initiation genes in Arabidopsis. Plant Cell 9, 1921–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer, R., Ramsay, N., Samach, A., Corden, S., Putterill, J., Carré, I.A., and Coupland, G. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93, 1219–1229. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Webb, A.A.R., Pearson, M., and Kay, S.A. (1998). The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 125, 485–494. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Schultz, T.F., Milnamow, M., and Kay, S.A. (2000). ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101, 319–329. [DOI] [PubMed] [Google Scholar]
- Strayer, C., Oyama, T., Schultz, T.F., Raman, R., Somers, D.E., Mas, P., Panda, S., Kreps, J.A., and Kay, S.A. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289, 768–771. [DOI] [PubMed] [Google Scholar]
- Wang, Z.-Y., and Tobin, E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207–1217. [DOI] [PubMed] [Google Scholar]
- Zagotta, M.T., Hicks, K.A., Jacobs, C.I., Young, J.C., Hangarter, R.P., and Meeks-Wagner, D.R. (1996). The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J. 10, 691–702. [DOI] [PubMed] [Google Scholar]
- Zhong, H., Resnick, A., Straume, M., and McClung, C.R. (1997). Effects of synergistic signaling by phytochrome A and cryptochrome1 on circadian clock–regulated catalase expression. Plant Cell 9, 947–955. [DOI] [PMC free article] [PubMed] [Google Scholar]