Abstract
Recent studies have shown that casein kinase I ɛ (CKIɛ) is an essential regulator of the mammalian circadian clock. However, the detailed mechanisms by which CKIɛ regulates each component of the circadian negative-feedback loop have not been fully defined. We show here that mPer proteins, negative limbs of the autoregulatory loop, are specific substrates for CKIɛ and CKIδ. The CKI phosphorylation of mPer1 and mPer3 proteins results in their rapid degradation, which is dependent on the ubiquitin-proteasome pathway. Moreover, CKIɛ and CKIδ are able to induce nuclear translocation of mPer3, which requires its nuclear localization signal. The mutation in potential phosphorylation sites on mPer3 decreased the extent of both nuclear translocation and degradation of mPer3 that are stimulated by CKIɛ. CKIɛ and CKIδ affected the inhibitory effect of mPer proteins on the transcriptional activity of BMAL1-CLOCK, but the inhibitory effect of mCry proteins on the activity of BMAL1-CLOCK was unaffected. These results suggest that CKIɛ and CKIδ regulate the mammalian circadian autoregulatory loop by controlling both protein turnover and subcellular localization of mPer proteins.
In most living organisms, behavioral and physiological processes display ∼24-h rhythms that are controlled by circadian pacemakers (6, 15, 22, 37). Although these rhythms persist in constant conditions, fluctuations of the natural environment entrain rhythms to precisely 24-h periods. The circadian clock is made up of three components: an input pathway adjusting the time, a central oscillator generating the circadian signal, and an output pathway manifesting itself in circadian physiology and behavior (10, 12, 23, 25). In mammals, the master circadian pacemaker is located in the suprachiasmatic nucleus, which controls neural and humoral signals that either drive output rhythms or synchronize peripheral oscillators with the day-night cycle (3, 38).
Autoregulatory feedback loops of gene expression are believed to provide the rhythm-generating mechanisms. Since several clock genes are conserved in flies and mammals, the fundamental mechanism may be evolutionarily conserved (40). In mammals, the positive limb of the feedback loop is composed of CLOCK and BMAL1 (2, 4, 8, 16), while the negative limb is composed of cryptochrome proteins (Cry1 and Cry2) and period proteins (Per1, Per2, and Per3) (1, 5, 27, 29, 31, 32, 33, 35, 41).
In Drosophila melanogaster, doubletime (dbt) was identified as a kinase, which is thought to phosphorylate dPer (Drosophila Period) (17, 21). Missense mutations in dbt result in an altered circadian rhythm. Null alleles of dbt result in hypophosphorylation of dPer and arrhythmia. Dbt-mediated phosphorylation destabilizes dPer so that the level of dPer increases only when dTim (Drosophila Timeless) is increasing (17, 21, 24). In the Syrian hamster, the tau mutation is a spontaneous, semidominant mutation causing a short-period phenotype. Using a positional syntenic cloning strategy, the tau locus is revealed to encode casein kinase I ɛ (CKIɛ), a mammalian homolog of Dbt, and the mutation causes substitution of a cysteine for a conserved arginine at residue 178 (19). The ability of the mutant CKIɛ to phosphorylate the mPer proteins is reduced compared to that of wild-type CKIɛ. Moreover, recent studies suggest that deficiency of hPer phosphorylation by CKIɛ may be implicated in human sleep disorders. Familial advanced sleep phase syndrome (FASPS) can be attributed to a missense mutation in hPer2, which causes hypophosphorylation of the hPer2 protein by CKIɛ in vitro (34). The polymorphism in a region of the hPer3 gene, a presumable CKIɛ binding domain, is suggested to be associated with delayed sleep phase syndrome (DSPS) (7). Thus, it is generally believed that CKIɛ is an essential regulator of the mammalian circadian clock, but the mechanisms by which CKIɛ regulates each components of the circadian negative-feedback loop have not been fully defined.
In this study, we examined interactions of CKIɛ and CKIδ with each component of the feedback loop and found that CKIɛ and CKIδ bind to and phosphorylate mPer proteins specifically. Recent reports suggested that CKIɛ could regulate subcellular localization of mPer proteins (30, 36) and affect the stability of hPer1 and mPer1 (14, 36). We have here identified potential CKIɛ phosphorylation sites on mPer3 and shown that CKIɛ and CKIδ induce nuclear translocation of mPer3 by phosphorylating it. This nuclear import is shown to require a nuclear localization signal (NLS) in mPer3. We have also shown that the ubiquitin-proteasome system operates in the CKIɛ-mediated degradation of mPer proteins. Finally, our results have revealed that CKIɛ and CKIδ reduce the inhibitory effect of mPer2 on the transcriptional activity of BMAL1-CLOCK but enhance the effect of mPer3. These results suggest that CKIɛ and CKIδ control each function of mPer1, mPer2, and mPer3 differently by specifically interacting with and phosphorylating them and thus regulate the mammalian circadian autoregulatory loop.
MATERIALS AND METHODS
Reagents and plasmids.
Anti-Myc monoclonal antibody (9E10) and rabbit anti-Myc polyclonal antibody (A-14) were purchased from Santa Cruz Biotechnology, Inc. Rabbit anti-hemagglutinin (HA) polyclonal antibody was from Clontech. The monoclonal antibody directed against the CKIɛ carboxyl-terminal region from Transduction Laboratories. The open reading frames of mouse clock proteins and CKI were subcloned into pcDNA3, pcDNA3(HA), or pcDNA3(Myc), and these constructs were utilized for transfections and in vitro translations.
In vitro translation and coimmunoprecipitation.
Each of the clock proteins or the CKIɛ was separately synthesized in vitro by using TNT T7 Coupled Reticulocyte Lysate System (Promega) according to the manufacturer's instructions. For the coimmunoprecipitation experiments, 10-μl aliquots of each specific TNT reaction mixture were mixed and incubated for 30 min at 30°C. Incubation buffer (50 mM Tris-Cl, 50 mM NaCl, 100 mM NaF, 10% glycerol, 2 mM EDTA, 2 mM sodium orthovanadate, 10 mM sodium pyrophosphate, 1 mM dithiothreitol [DTT], 1% Nonidet P-40 [pH 8.0]; supplemented with 0.1 TIU of aprotinin/ml, 10 μg of leupeptin/ml, and 1 mM phenylmethylsulfonyl fluoride) was then added, and each of these mixtures was incubated with anti-HA antibody and protein G-Sepharose beads (Pharmacia, Uppsala, Sweden) at 4°C for 2 h. These beads were washed three times with 500 μl of the incubation buffer. After resolution by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), the immunoprecipitates were detected by a Bio-Rad phosphorimager. The data were analyzed by using Molecular Analyst software (Bio-Rad).
Bacterial expression and purification of recombinant CKIɛ.
For bacterial expression, the open reading frame of CKIɛ or CKIɛ(KR) was subcloned into the expression vector pGEX-5T (Pharmacia Biotech). Escherichia coli cells (BL21) transformed with pGEX-CKIɛ or pGEX-CKIɛ(KR) were grown overnight to saturation in 10 ml of Luria-Bertani (LB) medium containing 50 μg of ampicillin/ml. The cells were grown in 2 liters of LB medium containing 50 μg of ampicillin/ml at 37°C to reach an A600 of 0.6. After the temperature shift to 25°C, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 1 mM, and the cells were cultured for 9 h. Glutathione S-transferase (GST)-CKIɛ or GST-CKIɛ(KR) was purified by affinity chromatography on glutathione-Sepharose 4B (Pharmacia Biotech).
GST pull-down assay.
Each of the Myc-tagged clock proteins was expressed in COS7 cells, and the cells were lysed in the incubation buffer (see above). The lysates precleared by centrifugation were incubated with GST (100 ng), GST-CKIɛ (20 ng), or GST-CKIɛ(KR) (20 ng), as well as glutathione-Sepharose 4B, for 16 h at 4°C. The beads were then washed twice with the incubation buffer. After they were resolved by SDS-PAGE, the precipitates were analyzed by immunoblotting.
In vitro kinase assay.
Each of the Myc-tagged clock proteins was expressed in COS7 cells, and the cells were lysed in the incubation buffer. Myc-tagged proteins were immunoprecipitated by incubating the cell lysates with anti-Myc antibody (9E10) and protein G-Sepharose beads for 2 h at 4°C. The beads were washed twice with the incubation buffer and once with a kinase reaction buffer (30 mM HEPES [pH 7.5], 7 mM MgCl2 50 μg of bovine serum albumin [BSA]/ml, 50 μM ATP, 0.5 mM DTT). The washed beads were mixed with a kinase reaction buffer supplemented with either GST-CKIɛ (20 ng) or GST-CKIɛ(KR) (20 ng), plus 5 μCi of [γ-32P]ATP, and incubated for 30 min at 30°C. The reaction was stopped by the addition of Laemmli sample buffer. After resolution by SDS-PAGE, substrate phosphorylation was detected with a Bio-Rad phosphorimager. The data were analyzed by using Molecular Analyst software (Bio-Rad).
Cell culture and transfections.
COS7 cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum and antibiotics (100 U of penicillin and 0.2 mg of kanamycin/ml). The cells were seeded in 35-mm dishes for 24 h before transfection at a density of 5 × 105 cells per dish. In cotransfection, the total amount of DNA per well was adjusted to 1 μg (or 1.1 μg for luciferase assays) by adding pcDNA3 vector as carrier. The plasmids were mixed with Lipofectamine Plus (Life Technologies) and in serum-free Opti-MEM (Life Technologies). After transfection, the cells were incubated in DMEM supplemented with 10% fetal calf serum for an additional 36 h before extraction. Cells were scraped into 100 μl of incubation buffer (see above). The cell lysates were centrifuged at 12,000 × g for 30 min. For the immunoprecipitation experiments, the supernatant was then mixed and incubated with an antibody and protein G-Sepharose.
Phosphatase treatment.
Each of Myc-tagged mPer proteins expressed in COS7 cells was immunoprecipitated with anti-Myc antibody (9E10) and protein G-Sepharose. The beads were washed and resuspended in phosphatase buffer (50 mM Tris-HCl, 0.1 mM EDTA, 5 mM DTT, 0.01% Brij 35, 2 mM MnCl2; pH 7.0) and then incubated with 40 U of purified lambda phosphatase (New England Biolabs) for 30 min at 30°C. The phosphatase reaction was stopped by the addition of Laemmli sample buffer.
Antibody.
Polyclonal antisera were raised in rabbits against two synthetic peptides from the deduced amino acid sequences of mPer1. An N-terminal peptide (SGPLEGADGGGD) corresponding to amino acids 2 to 13 and a C-terminal peptide (DGLGLEPMEEGGGE) corresponding to amino acids 1223 to 1236 were synthesized and conjugated to BSA. Each antibody was affinity purified with the peptide-conjugated cellulose and termed anti-mPer1(N) antibody and anti-mPer1(C) antibody, respectively. For immunoprecipitation of endogenous mPer1 protein, NIH 3T3 cells were incubated in hypotonic lysis buffer (20 mM Tris-HCl [pH 7.4], 5 mM MgCl2, 8 mM KCl, 1 mM dithiothreitol, 1 μg of leupeptin/ml, 1 μg of pepstatin/ml, and 1 mM phenylmethylsulfonyl fluoride) and homogenized. After centrifugation at 12,000 × g for 10 min, the resulting supernatant was mixed with each of antibodies and protein G-Sepharose beads.
Mutagenesis.
The mutants used were constructed by PCR-based mutagenesis. PCR was performed by using KOD plus DNA polymerase (Toyobo), a thermostable DNA polymerase isolated from Pyrococcus kodakaraensis KOD1. DpnI restriction enzyme-treated PCR products were transformed into E. coli. Positive clones were picked up, and mutagenesis was verified by sequencing.
Ubiquitination.
To show ubiquitination of mPer proteins, each of Myc-tagged mPer proteins was coexpressed with HA-tagged ubiquitin. The cells were lysed in 1% SDS-Tris buffer (1% SDS, 20 mM Tris-HCl [pH 6.8], 5 mM EDTA) and boiled for 10 min. The lysates were diluted 10-fold into the incubation buffer (see above) and then subjected to anti-Myc immunoprecipitation.
Pulse-chase experiments.
To study the metabolic stability of mPer proteins expressed in COS7 cells, the cells were incubated for 1 h in methionine-cysteine-deficient medium and then for 30 min in 300 μCi of [35S]-labeled methionine-cysteine/ml. The cells were washed with DMEM and incubated for various lengths of time in DMEM-10% fetal calf serum with or without MG-132. The cells were lysed in the incubation buffer (see above) containing protease inhibitors, and the cell lysates were centrifuged at 12,000 × g for 15 min. For the immunoprecipitation, the supernanant was then mixed and incubated with anti-Myc antibody (9E10) and protein G-Sepharose.
Cell staining.
Cells were fixed by the direct addition of 3.7% formaldehyde (final concentration) to the cell culture medium and then permeabilized with 0.5% Triton X-100 in phosphate-buffered saline (PBS) for 5 min. The coverslips were incubated with 3% BSA-PBS dissolving primary antibodies at 4°C overnight and then with the appropriate secondary antibodies at room temperature for 1 h. The cells were finally mounted in Mowiol and examined by using a Zeiss Axiophoto.
Luciferase assays.
Each transfection contained the arginine vasopressin promoter cloned into pGL3-Basic (50 ng; Promega) and cytomegalovirus β-galactosidase (50 ng). Amounts of the constructs transfected varied depending on the experiment (see the figure legends for details). The total amount of DNA per well was adjusted by adding pcDNA3 vector as carrier. At 36 h after transfection, COS7 cells were scraped into 150 μl of reporter lysis buffer (Promega) and centrifuged at 12,000 × g for 5 min after being vortex mixed for 15 s. The supernatant was used as a cell extract to detect the luciferase activity. The luciferase assay was carried out by using a luciferase assay system (Promega). In brief, 20 μl of the room temperature cell extract was mixed with 100 μl of room temperature luciferase assay reagent containing the substrate. The reaction was performed and measured in a luminometer (LB9507; Berthold). To determine the transfection efficiency in each assay, the β-galactosidase activity was measured according to the protocol of Promega, and the data were normalized for β-galactosidase activity.
RESULTS
CKIɛ and CKIδ bind to and phosphorylate mPer proteins.
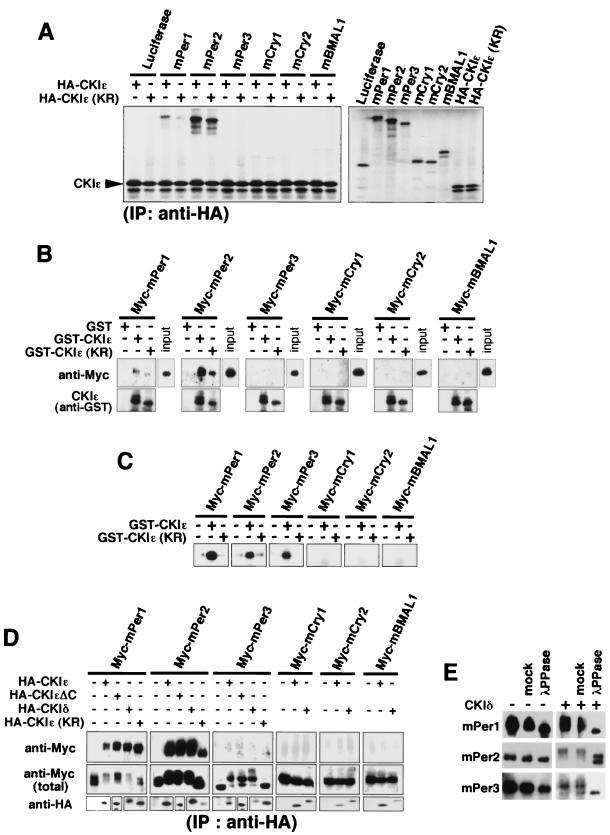
We first examined, by using various biochemical methods, the possible physical interactions of CKIɛ with each component of the mammalian circadian feedback loop in order to search for specific targets of CKIɛ. mPer1, mPer2, mPer3, mCry1, mCry2, and mBMAL1 were separately synthesized in vitro and incubated with in vitro-synthesized HA-tagged CKIɛ. By immunoprecipitation with anti-HA antibody, mPer1 and mPer2 were coimmunoprecipitated with CKIɛ (Fig. 1A). A kinase-dead form of CKIɛ [i.e., CKIɛ(KR)] also coimmunoprecipitated with mPer1 and mPer2 (Fig. 1A). A GST pulldown assay with GST-fused CKIɛ or CKIɛ(KR) toward these clock proteins expressed in COS7 cells also showed the interaction of CKIɛ with mPer1 and mPer2 (Fig. 1B). In both assays, mPer2 bound to CKIɛ more tightly than did mPer1 (Fig. 1A and B). In kinase assays, all of the three mPer proteins were readily phosphorylated by recombinant CKIɛ, whereas mCry1, mCry2, or mBMAL1 was not significantly phosphorylated by CKIɛ under the same conditions (Fig. 1C). Physical interaction of CKIɛ with mPer1, mPer2, and mPer3 and CKIɛ phosphorylation of the mPer proteins were recently reported (30). We then examined the association of CKIδ or CKIɛ with these clock proteins in cultured cells, since CKIδ is closest to CKIɛ (97% identity in the kinase domain) of the seven identified members of CKI (9). Coexpression and coimmunoprecipitation assays showed that both CKIɛ and δ bound to mPer1 and mPer2 specifically and that both kinases, but not CKIɛ(KR), were able to cause a significant electrophoretic mobility shift in mPer1, mPer2, and mPer3 (Fig. 1D). Especially, the same results were obtained by using a C-terminus-deleted mutant of CKIɛ (CKIɛΔC, Fig. 1D), which lacks a C-terminal region outside the catalytic domain. Therefore, it may be that CKIɛ and CKIδ recognize and bind to mPer proteins through their kinase domain in their primary sequence. The electrophoretic retardation of the mPer protein bands resulted from phosphorylation, since it was reversed by the phosphatase treatment (Fig. 1E). These results, taken together, suggest that CKIɛ and CKIδ phosphorylate mPer1, mPer2, and mPer3 specifically and bind to mPer1 and mPer2 tightly and that only mPer proteins among clock components can be specific targets of CKI. We were able to detect binding of mPer3 to CKI under milder wash conditions in which other proteins did not bind to CKI (data not shown). Thus, the binding affinity of mPer3 for CKI appears to be lower than that of mPer1 or mPer2 but is still higher than that of other proteins.
FIG. 1.
CKIɛ and CKIδ bind to and phosphorylate mPer proteins specifically. (A) Luciferase (control), mPer1, mPer2, mPer3, mCry1, mCry2, and mBMAL1 were separately synthesized in vitro by using TNT T7 Coupled Reticulocyte Lysate System (right panel) and then mixed with in vitro-synthesized HA-tagged CKIɛ or CKIɛ(KR) and incubated. Each of these mixtures was subjected to immunoprecipitation with anti-HA antibody (12CA5). After resolution by SDS-PAGE, immunoprecipitates were detected by a Bio-Rad phosphorimager (left panel). (B) Myc-tagged mPer1, mPer2, mPer3, mCry1, mCry2, or mBMAL1 was expressed in COS7 cells. Each of the cell extracts was incubated with GST, GST-fused CKIɛ, or GST-fused CKIɛ(KR) (expressed in E. coli, and purified) and glutathione-Sepharose 4B. The beads were then washed with the incubation buffer. After resolution by SDS-PAGE, proteins were analyzed by immunoblotting with anti-Myc antibody (A-14). (C) Myc-tagged mPer1, mPer2, mPer3, mCry1, mCry2, or mBMAL1 was expressed in COS7 cells. The cell extracts were subjected to immunoprecipitation with anti-Myc antibody (9E10). The washed beads were mixed with a kinase reaction buffer supplemented with GST-fused CKIɛ or GST-fused CKIɛ(KR), as well as [γ-32P]ATP, and incubated for 30 min. After resolution by SDS-PAGE, substrate phosphorylation was detected by a Bio-Rad phosphorimager. (D) Myc-tagged mPer1, mPer2, mPer3, mCry1, mCry2, or mBMAL1 was expressed in COS7 cells, together with HA-tagged CKIɛ, CKIɛΔC, CKIδ, or CKIɛ(KR). The cell lysates were subjected to immunoprecipitation with anti-HA antibody (12CA5). After resolution by SDS-PAGE, immunoprecipitates were analyzed by immunoblotting with anti-Myc antibody (A-14). (E) Myc-tagged mPer1, mPer2, or mPer3 was expressed in COS7 cells, together with or without CKIδ. The cell lysates were subjected to immunoprecipitation with anti-Myc antibody (9E10), and the washed beads were resuspended in phosphatase buffer and incubated with or without (mock) 40 U of purified lambda phosphatase for 30 min. After resolution by SDS-PAGE, the immunoprecipitates were analyzed by immunoblotting with anti-Myc antibody (A-14).
In vivo association between mPer1 and CKIɛ.
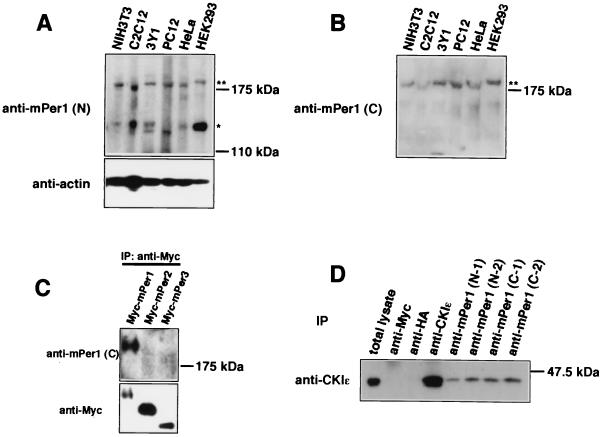
We produced two kinds of anti-mPer1 antibodies by using an N-terminal peptide sequence (residues 2 to 13) and a C-terminal peptide sequence (residues 1223 to 1236), respectively, as antigens. The antisera were affinity purified on each of the peptides and were termed anti-mPer1(N) antibody and anti-mPer1(C) antibody, respectively. The anti-mPer1(N) antibody recognized two major bands with relative molecular masses of ca. 200 and 140 kDa, respectively, in various mammalian cells (Fig. 2A). The 140-kDa band may correspond to a 140-kDa band which was previously revealed by using a similar anti-N-terminal peptide antibody (11). Multiple bands may be attributed to alternative splicing (29) and/or to some posttranslational modification. Anti-mPer1(C) antibody, however, recognized only the 200-kDa band in the same cells (Fig. 2B). It specifically reacted with Myc-mPer1 among Myc-tagged versions of mPer1, mPer2, and mPer3 (Fig. 2C). When extracts from NIH 3T3 cells were subjected to immunoprecipitation with each of these antibodies against mPer1 [anti-mPer1(C) antibodies produced in two rabbits, C-1 and C-2, and anti-mPer1(N) antibodies produced in two rabbits, N-1 and N-2], CKIɛ was coimmunoprecipitated in every case (Fig. 2D). Control immunoprecipitation with anti-Myc antibody or anti-HA antibody did not precipitate CKIɛ (Fig. 2D). Therefore, we can conclude that endogenous mPer1 binds to endogenous CKIɛ in NIH 3T3 cells.
FIG. 2.
In vivo association between mPer1 and CKIɛ. (A and B) Various mammalian cells were lysed in the incubation buffer. After the cell extracts were resolved by SDS-PAGE, they were analyzed by immunoblotting with anti-mPer1(N) antibody or anti-actin antibody (loading control) (A) or with anti-mPer1(C) antibody (B). (C) Myc-tagged mPer1, mPer2, or mPer3 was expressed in COS7 cells, and the cell lysates were subjected to immunoprecipitation with anti-Myc antibody (9E10). These immunoprecipitates were analyzed by immunoblotting with anti-mPer1(C) antibody (upper panel) or anti-Myc antibody (A-14) (lower panel). (D) For coimmunoprecipitation experiments, NIH 3T3 cells were incubated in hypotonic lysis buffer and homogenized. Each of antibodies and protein G-Sepharose beads were added to the resulting supernatant. The beads were then washed with the hypotonic lysis buffer. The immunoprecipitates were analyzed by immunoblotting with anti-CKIɛ antibody. Anti-HA antibody and anti-Myc antibody were used as a nonrelevant antibody control.
mPer, mCry, and CKIɛ form a ternary complex.
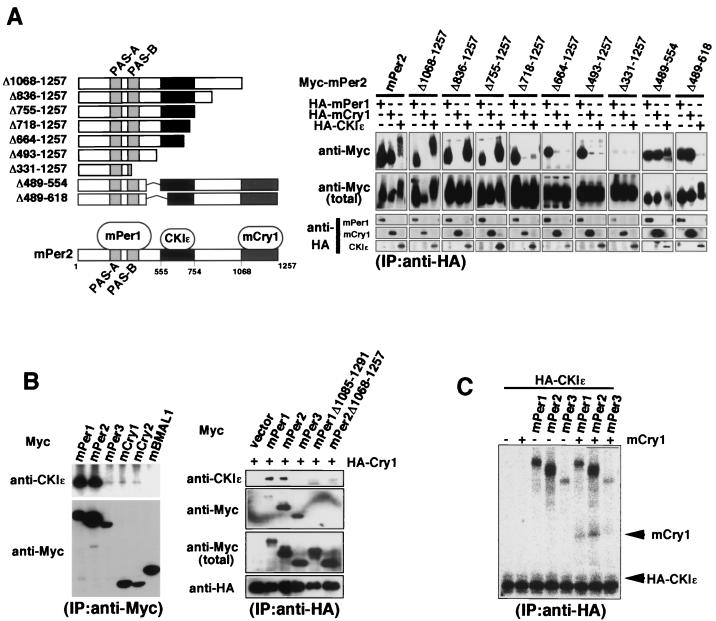
To determine a CKIɛ binding domain on mPer proteins, various deletion mutants of Myc-tagged mPer2 (see Fig. 3A, left) were expressed with HA-tagged CKIɛ in COS7 cells, and coimmunoprecipitation assays were performed with anti-HA antibodies. The results shown in Fig. 3A (right) suggested that the CKIɛ binding domain of mPer2 is located between residues 555 and 754 (Fig. 3A). This result is essentially the same as the recently reported result for a CKIɛ binding domain in mPer2 (34). Our result concerning an mPer1 binding site on mPer2 (Fig. 3A) was also consistent with the previous results indicating that mPer proteins homo- or heterodimerize through their PER-ARNT-SIM (PAS) domains (39). Our assay further identified an mCry1 binding domain on mPer2 as a C-terminal region (residues 1068 to 1257; Fig. 3A), the region being highly conserved among three mPer proteins. Deletion of the corresponding region in both mPer1 and mPer3 also resulted in deficiency of their interaction with mCry1 (data not shown). Since the CKIɛ binding domain and the mCry1 binding domain on mPer2 are not overlapping, mPer2, mCry1, and CKIɛ could form a ternary complex. Actually, endogenous CKIɛ was coimmunoprecipitated with HA-mCry1 only when full-length mPer1 or full-length mPer2 but not mPer3 or the C-terminal (i.e., the mCry binding site) deleted forms of mPer1 or mPer2 (mPer1Δ1085-1291 or mPer2Δ1068-1257) was coexpressed (Fig. 3B, right panel). Also, in vitro-synthesized mCry1 was coimmunoprecipitated with HA-CKIɛ only in the presence of mPer1 or mPer2 (Fig. 3C). These results suggest that CKIɛ, mCry1, and either mPer1 or mPer2 form a ternary complex.
FIG. 3.
mPer, mCry, and CKIɛ form a ternary complex. (A) In the left panel is a diagram of the deletion constructs of mPer2 and a summary of the results of the coimmunoprecipitation experiments (a CKIɛ binding domain and an mCry1 binding domain are indicated by dark gray and light gray boxes, respectively). In the right panel a CKIɛ binding domain and an mCry1 binding domain on mPer2 are identified. For coimmunoprecipitation experiments, Myc-tagged mPer2 or each of various fragments of mPer2 was expressed in COS7 cells, together with HA-tagged mPer1, mCry1, or CKIɛ. HA-tagged mPer1, mCry1, or CKIɛ was immunoprecipitated with anti-HA antibody (12CA5). The immunoprecipitates were analyzed by immunoblotting with anti-Myc antibody (A-14). (B, left panel) For the coimmunoprecipitation of endogenous CKIɛ, Myc-tagged mPer1, mPer2, mPer3, mCry1, mCry2, or mBMAL1 was expressed in COS7 cells. The cell lysates were subjected to immunoprecipitation with anti-Myc antibody (A-14). After resolution by SDS-PAGE, the immunoprecipitates were analyzed by immunoblotting with anti-CKIɛ antibody. (B, right panel) mPer, mCry, and CKIɛ form a ternary complex in cultured cells. Myc-tagged mPer1, mPer2, mPer3, mPer1Δ1085-1291, or mPer2Δ1068-1257 was expressed in COS7 cells, together with HA-tagged mCry1. The cell lysates were subjected to immunoprecipitation with anti-HA polyclonal antibody. After resolution by SDS-PAGE, the immunoprecipitates were analyzed by immunoblotting with anti-CKIɛ antibody. (C) mPer, mCry, and CKIɛ form a ternary complex in vitro. mPer1, mPer2, mPer3, mCry1, or HA-tagged CKIɛ was separately synthesized in vitro by using TNT T7 Coupled Reticulocyte Lysate System. mPer1, mPer2, or mPer3 was then mixed with HA-tagged CKIɛ, together with or without mCry1, and incubated. Each of these mixtures was then immunoprecipitated with anti-HA antibody (12CA5). The immunoprecipitates were detected with a Bio-Rad phosphorimager.
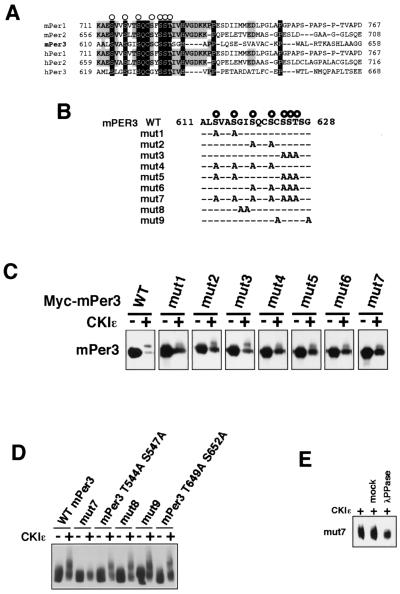
Identification of potential CKIɛ phosphorylation sites on mPer3.
CKIɛ phosphorylation sites on mPer proteins may lie around and/or in the CKIɛ binding domain. Sequences corresponding to a consensus motif for CKIɛ phosphorylation (20) are present in the middle region of the CKIɛ binding domain on mPer2 and conserved in all Per proteins (Fig. 4A and B). A recent report suggested that these residues in Per2 are a subset of CKI phosphorylation sites (34). Then, various mutants of mPer3 in which part or all of the seven conserved serine and threonine residues are replaced by alanines (Fig. 4B, mut1 to mut7) were produced and expressed with or without CKIɛ in COS7 cells. Wild-type mPer3 showed a marked mobility shift in SDS-PAGE in the presence of CKIɛ; ca. 70% of wild-type mPer3 was in the phosphorylated (i.e., the mobility-shifted) form in the presence of CKIɛ (Fig. 4C). In contrast, the ratio of the mobility-shifted band in the presence of CKIɛ was reduced in all of the mutants of mPer3; only ca. 10 to 40% of the molecules were in the phosphorylated form. The pattern of the CKI-induced electrophoretic retardation of mutant proteins was slightly but reproducibly different from that of wild-type proteins. The mutations in each sites do not produce completely the same results. For example, mut1 differed slightly but reproducibly from mut2 or mut3. We speculate that, as reported in hPer2 (34), phosphorylation of mPER3 S613 (the first serine in potential phosphorylation sites) would create CKIɛ recognition sites for phosphorylation of downstream serine or threonine residues. In mut7 in which all of the conserved serine and threonine residues in this region were mutated, the ratio of the shifted band was greatly reduced reproducibly (Fig. 4C). We then made mutants of mPer3 in which nonphosphorylated residues in the region were replaced by alanines (Fig. 4B, mut8 and mut9). In addition, we also generated two mutants in which other serine and threonine residues in the CKIɛ binding domain were replaced by alanines (mPer3 T544A S547A and mPer3 T649A S652A in Fig. 4D). The results showed that all of these mutants exhibited essentially the same CKI-mediated electrophoretic retardation as that of wild-type mPer3 (Fig. 4D). Furthermore, we confirmed that the newly made mPer2 mutant corresponding to the mPer3 mut7 was also able to bind to CKIɛ (data not shown). Therefore, our data indicate that the reduction of the mobility-shifted bands in mut1 to mut7 results from the mutation of phosphorylation sites. It should be noted, however, that even in mut7, the CKIɛ-dependent mobility shift was detected (Fig. 4E). These results suggest that these serine and threonine residues are potential phosphorylation sites for CKIɛ, as shown previously in Per2 (34), and that there are other phosphorylation sites.
FIG. 4.
Identification of potential CKIɛ phosphorylation sites on mPer3. (A) Amino acid sequences of the mammalian Per family. Amino acid identities and similarities are indicated by dark gray and light gray boxes, respectively. The conserved serine-threonine residues are indicated by open circles. (B) PCR-generated alanine mutations were made in the mPer3 conserved serine-threonine cluster and compared with the wild-type (WT) mPer3 sequence. (C and D) Wild-type (WT) mPer3 or each of various mutants of mPer3 (0.7 μg of DNA) was expressed in COS7 cells, together with or without CKIɛ (0.3 μg of DNA). The cell extracts were subjected to immunoblotting with anti-Myc antibody (A-14). Equal amounts of proteins were loaded. The electrophoretic retardation of the mPer3 protein bands results from phosphorylation. (E) Myc-tagged mut7 was expressed in COS7 cells, together with or without CKIɛ. The cell lysates were subjected to immunoprecipitation with anti-Myc antibody (9E10) and incubated with or without (mock) purified lambda phosphatase. The immunoprecipitates were analyzed by immunoblotting with anti-Myc antibody (A-14).
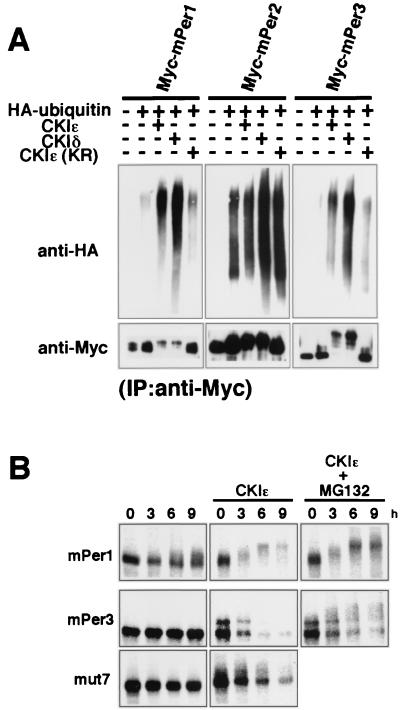
CKI promotes ubiquitination of mPer proteins.
We noticed that the protein levels of mPer1 and mPer3 were significantly reduced when coexpressed with CKIɛ or CKIδ (see Fig. 1D and Fig. 4C) and that the protein levels of the potential phosphorylation site mutants of mPer3 were relatively increased (see Fig. 4C). So, we hypothesized that CKIɛ and CKIδ promote the ubiquitin-proteasome pathway-dependent degradation of mPer proteins. First, we examined ubiquitination of mPer proteins. Each of Myc-tagged mPer proteins was coexpressed with HA-ubiquitin in the presence or absence of CKIɛ or CKIδ. Then, immunoprecipitates with anti-Myc antibody were immunoblotted with anti-HA antibody. In the case of Myc-mPer1 and Myc-mPer3, CKIɛ or CKIδ, but not CKIɛ(KR), promoted the accumulation of high-molecular-mass species recognized by the anti-HA antibody (Fig. 5A), indicating that mPer1 and mPer3 become multiply ubiquitinated in response to phosphorylation by CKIɛ or CKIδ. Since mPer2 protein seemed to be highly ubiquitinated even in the absence of CKI, the effect of CKI was not clearly detected. We then examined the metabolic stability of mPer proteins. To this end, we expressed Myc-mPer proteins in COS7 cells and pulsed the cells with 35S-labeled methionine-cysteine. In the presence of CKIɛ, shifts in electrophoretic mobility of mPer3 occurred (Fig. 5B), and the amount of the labeled mPer3 protein decreased rapidly (Fig. 5B). A similar result was previously reported for hPer1 and mPer1 (14, 36) (Fig. 5B). The addition of MG-132, an inhibitor of the 26S proteasome, blocked significantly this rapid decline of the protein level (Fig. 5B). The potential phosphorylation site mutant of mPer3, mut7, showed a longer half-life than wild-type mPer3 in the presence of CKIɛ (Fig. 5B). The degradation of mPer2 also appeared to be accelerated by CKIɛ-mediated phosphorylation (14; data not shown). These results taken together suggest that the ubiquitin-proteasome system operates in the CKI-mediated enhancement of degradation of mPer proteins.
FIG. 5.
CKI promotes ubiquitination of mPer proteins. (A) To show ubiquitination of mPer proteins, each of Myc-tagged mPer proteins was coexpressed with HA-tagged ubiquitin in the presence or absence of CKIɛ, CKIδ, or CKIɛ(KR). The cell lysates were then subjected to immunoprecipitation with anti-Myc antibody (9E10). After resolution by SDS-PAGE, immunoprecipitates were analyzed by immunoblotting with anti-HA polyclonal antibody. The accumulation of high-molecular-mass species recognized by the anti-HA antibody indicates that Myc-tagged proteins become multiply ubiquitinated. (B) To study the metabolic stability of mPer proteins, each of Myc-tagged mPer proteins was expressed in COS7 cells, together with or without CKIɛ. The cells were incubated in methionine-cysteine-deficient medium and then pulsed with 35S-labeled methionine-cysteine. The cells were incubated for the indicated lengths of time in DMEM-10% fetal calf serum with or without MG-132. The cell lysates were subjected to immunoprecipitation with anti-Myc antibody (9E10). The immunoprecipitates were detected with a Bio-Rad phosphorimager.
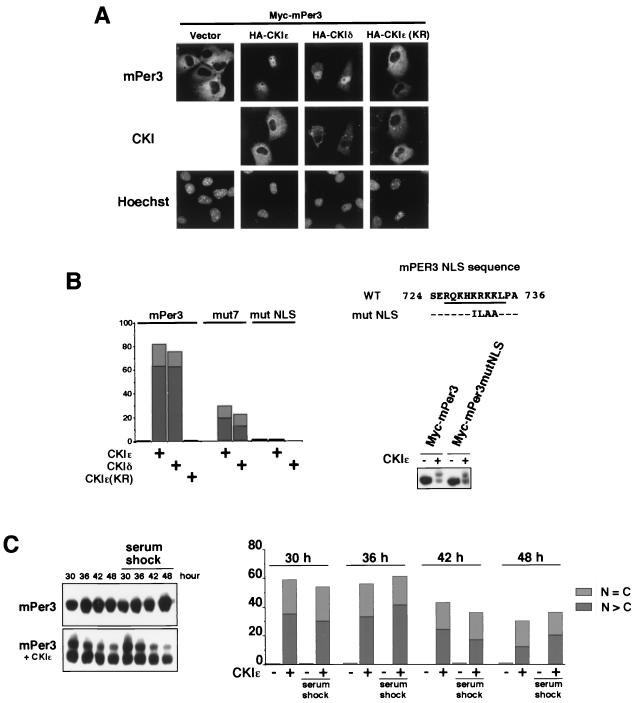
CKI-induced nuclear translocation of mPer3 depends on CKI-mediated phosphorylation and NLS of mPer3.
We examined the effect of expression of CKIɛ or CKIδ on subcellular localization of mPer proteins in COS7 cells. Subcellular distribution of mPer1 and mPer2 was not significantly altered by CKIɛ or CKIδ (data not shown). In contrast, a marked change in subcellular localization of mPer3 was induced by CKIɛ and CKIδ (Fig. 6A and B). Thus, mPer3 translocated from the cytoplasm to the nucleus when coexpressed with CKIɛ or CKIδ. The expressed CKIɛ and CKIδ were mainly cytoplasmic (Fig. 6A). We could detect the CKI binding to mPer3 under milder wash conditions. However, the binding affinity is not so high, probably explaining the dissociation of mPer3 from CKI after phosphorylation; thus, CKI remains in the cytoplasm. Although these results are not totally in good agreement with the recent reports (30, 36), our data are consistent with the previous finding of the CKIɛ-induced nuclear translocation of mPer3 (30). The differences might result from the differences in experimental conditions, the cells used, and the species of the molecules used.
FIG. 6.
CKI-induced nuclear translocation of mPer3 depends on CKI-mediated phosphorylation and NLS of mPer3. (A) Myc-tagged mPer3 was expressed in COS7 cells, together with or without HA-tagged CKIɛ, CKIδ or CKIɛ(KR). The cells were fixed and incubated with primary antibodies (anti-Myc antibody [9E10] and anti-HA polyclonal antibody) and then with the appropriate secondary antibodies. The cells were examined by using a Zeiss Axiophoto. (B, left panel) Myc-tagged mPer3, mut7, or mutNLS (see the right panel) (0.9 μg of DNA) was expressed in COS7 cells, together with or without HA-tagged CKIɛ, CKIδ, or CKIɛ(KR) (0.1 μg of DNA). After being stained, the cells were classified into three categories in terms of location of Myc-tagged mPer proteins as nucleus (N > C), cytoplasm (N < C), or both nucleus and cytoplasm (N = C). The classifications “N > C” and “N = C” are indicated by dark gray and light gray boxes, respectively. More than 150 cells were examined, and the percentages of N > C and N = C are shown. Experiments were performed twice with similar results. (B, right panel) A PCR-generated mutation was made in a presumable mPer3 NLS (mutNLS) and compared to the wild-type (WT) sequence. WT mPer3 or mPer3mutNLS was expressed in COS7 cells, together with or without CKIɛ. The cell extracts were subjected to immunoblotting with anti-Myc antibody (A-14). (C) The cells were subjected to transfection and kept in a medium containing 1% serum, after the serum shock (50% for 2 h). Myc-tagged mPer3 was expressed in COS7 cells, together with or without CKIɛ. The cells were then lysed (or fixed for staining) 30, 36, 42, and 48 h after the serum shock (time = 0 h). In the left panel, cell extracts were subjected to immunoblotting with anti-Myc antibody (A-14). The electrophoretic retardation of the mPer3 protein bands results from phosphorylation. In the right panel, cells were classified after being stained into three categories with regard to the location of Myc-tagged mPer3: N > C, N < C, or N = C. The N > C and N = C classifications are indicated by dark gray and light gray boxes, respectively. More than 250 cells were examined, and the percentages of the N > C and N = C groups are shown.
The CKI-induced nuclear translocation of the potential CKI phosphorylation site mutant of mPer3, mut7, was suppressed compared to that of wild-type mPer3 (Fig. 6B). Moreover, CKIɛ(KR) did not induce nuclear translocation of mPer3 (Fig. 6A and B). These data suggest that CKI-induced nuclear translocation of mPer3 depends on the phosphorylation. The incomplete suppression in the case of mut7 may also indicate the existence of other phosphorylation sites. We then examined whether nuclear translocation of mPer3 is dependent on its putative NLS sequence, the existence of which was suggested by Yagita et al. (39). We made a mutant mPer3 in which four consecutive basic amino acids in this region were replaced by nonpolar amino acids (Fig. 6B). This mutant mPer3 remained in the cytoplasm even when CKIɛ or CKIδ was coexpressed (Fig. 6B). The mutation of this region does not affect CKI binding (Fig. 6B, right panel). Thus, this sequence is in fact an NLS sequence, and CKI-induced nuclear translocation of mPer3 requires this NLS of mPer3. It is possible that CKIɛ- or CKIδ-mediated phosphorylation of mPer3 leads to unmasking of somehow buried NLS and thus induces nuclear translocation of mPer3. We performed a series of experiments in cells after a serum shock and thus examined CKIɛ-mediated phosphorylation and nuclear translocation of mPer3 in cells oscillating in a circadian fashion. At all time points, CKI-mediated phosphorylation and nuclear translocation of mPer3 occurred (Fig. 6C), as in the case of nonsynchronized cells. Therefore, these results suggest that phosphorylation and changes in subcellular localization of exogenously expressed Per proteins by exogenously expressed CKI may not be significantly affected by the endogenous circadian clock.
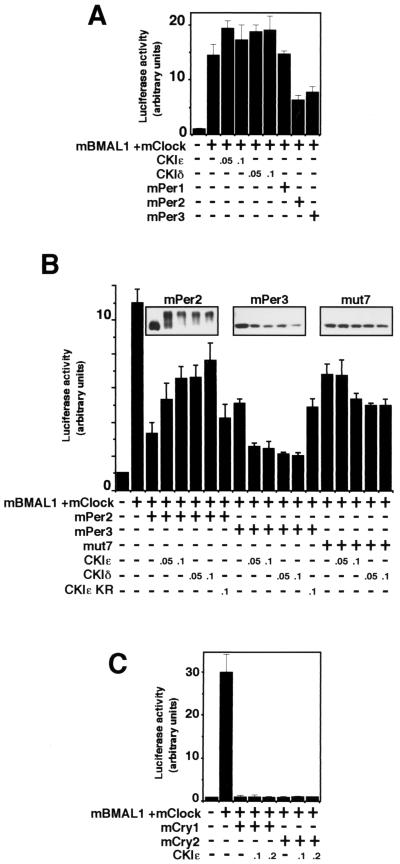
CKIɛ and CKIδ reduce the inhibitory effect of mPer2 on the transcriptional activity of BMAL1-CLOCK but enhance the inhibitory effect of mPer3.
BMAL1-CLOCK binds to E box elements in the mPer1 gene promoter to activate transcription (2, 4, 8, 16). The same machinery functions in expression of the clock output genes such as arginine vasopressin (13). It has been reported that each of the three mPer proteins can negatively regulate BMAL1-CLOCK-mediated transcription (4). In our experiments, mPer2 and mPer3, but not mPer1, inhibited significantly BMAL1-CLOCK-mediated transcription (Fig. 7A). CKIɛ and CKIδ elevated the transcriptional activity of BMAL1-CLOCK 1.2- to 1.3-fold (Fig. 7A). Interestingly, CKIɛ or CKIδ reduced the inhibitory effect of mPer2 on the transcriptional activity of BMAL1-CLOCK (Fig. 7B). This might result, at least in part, from the CKI-dependent enhancement of protein turnover of mPer2 (Fig. 7B, upper panel), although the steady-state amount of mPer2 was not markedly decreased in the presence of CKIɛ or CKIδ. Remarkably, CKIɛ or CKIδ enhanced the inhibitory effect of mPer3 on the activity of BMAL1-CLOCK (Fig. 7B), even though the amount of mPer3 was decreased significantly (Fig. 7B). This may be attributed, at least in part, to CKIɛ- or CKIδ-dependent nuclear translocation of mPer3 (Fig. 6). In fact, the inhibitory activity of the potential phosphorylation site mutant of mPer3, mut7, was not significantly enhanced by CKI, although the CKI-induced decrease of the protein level was less significant (Fig. 7B). CKIɛ or CKIδ did not affect the inhibitory effect of mCry1 and mCry2 on the transcriptional activity of BMAL1-CLOCK (Fig. 7C). These results suggest that the effect of CKI on the E-box-mediated transcription is clearly seen only in the presence of mPer proteins and that the inhibitory effects of mPer2 and mPer3 are differently affected by CKI.
FIG. 7.
The effect of CKI on the E-box-mediated transcription is seen only in the presence of mPer proteins, and the inhibitory effects of mPer2 and mPer3 are differently affected by CKI. The amounts of the constructs transfected varied depending on the experiment: mBMAL1 (100 ng), mClock (100 ng), and mPer (700 ng) (A and B) and mBMAL1 (250 ng), mClock (250 ng), and mCry (400 ng) (C). The amount of CKI expression vector (in micrograms) is shown in each panel. The total amount of DNA per well was adjusted to 1.1 μg by adding pcDNA3 vector as the carrier. At 36 h after transfection, COS7 cells were scraped into 150 μl of reporter lysis buffer and then centrifuged. The supernatant was used to detect the luciferase activity. The data were normalized for β-galactosidase activity. The relative luciferase activities (means ± the standard deviations; n = 3) are presented. These experiments were repeated three times, and the data shown are representative. (A) The effect of CKIɛ, CKIδ, mPer1, mPer2, or mPer3 on mBMAL1-mCLOCK-mediated transcription. (B) CKIɛ and CKIδ reduce the inhibitory effect of mPer2 on the transcriptional activity of BMAL1-CLOCK but enhance the inhibitory effect of mPer3. The amounts of the mPer proteins are shown (upper panel). mut7 is one of the potential phosphorylation site mutants of mPer3 (see Fig. 4B). The data for the effects of CKIɛ(KR) on the inhibitory effect of mPer proteins were normalized, since CKIɛ(KR) suppressed the transcriptional activity of BMAL1-CLOCK to some extent (data not shown). (C) CKIɛ or CKIδ does not affect the inhibitory effect of mCry1 and mCry2 on the transcriptional activity of BMAL1-CLOCK.
DISCUSSION
We have presented here several lines of evidence suggesting that CKIɛ and CKIδ regulate the mammalian circadian feedback loop by interacting with and phosphorylating mPer proteins. First, we examined interaction of CKIɛ and CKIδ with each component of the feedback loop and found that both CKIɛ and CKIδ bind to and phosphorylate mPer proteins specifically. Evidence was also presented for in vivo association of mPer1 with CKIɛ in NIH 3T3 cells. Second, CKIɛ and CKIδ were shown to promote the rapid degradation of mPer proteins. Third, CKIɛ and CKIδ were found to induce nuclear translocation of mPer3. Subcellular localization of mCry1, mCry2, and mBMAL1 was not affected at all by CKIɛ or CKIδ (data not shown). Finally, the effects of CKIɛ and CKIδ on E-box-mediated transcription were observed only in the presence of mPer proteins. These results, together with the recently reported results concerning the interaction of CKIɛ with mPer proteins (14, 30, 36), suggest that CKIɛ and CKIδ play a role in regulation of the circadian rhythm by controlling the functions of mPer proteins. The shortened period length of the circadian rhythm in tau mutation (19) may be attributed to hypophosphorylation of mPer proteins.
Next, we determined potential CKIɛ phosphorylation sites on mPer3. The effect of CKI on the degradation and change in subcellular localization was most marked in mPer3. Therefore, we selected mPer3. Our results demonstrated that the highly conserved serine-threonine cluster in the middle portion of the CKI-interacting region of mPer proteins is a subset of potential CKI phosphorylation sites of mPer3. A recent report suggested that the S662G mutation causes hypophosphorylation of Per2 by CKI in vitro (34). The analyses with the potential phosphorylation site mutants have shown that the identified potential phosphorylation sites are required for the optimal CKIɛ-mediated nuclear translocation and rapid degradation of mPer3. In locomotor activity rhythms, the circadian cycle length was significantly (0.5 h) shorter in mPer3-deficient mice than that in controls (26). Thus, mPer3 may play some role in determining the normal circadian period. Moreover, alterations of CKI-dependent regulation of subcellular localization and protein stability of Per proteins may cause sleep disorders such as FASPS and DSPS (7, 34).
Interestingly, although both the CKIɛ-interacting domain and the phosphorylation sites on three mPer proteins seem to be located in nearly the same region of the molecules (36) (Fig. 3A), the effects of CKIɛ-mediated phosphorylation on each mPer protein are not the same. It has previously been reported that the coexpression of CKIɛ leads to masking of the NLS of mPer1 and thus its cytoplasmic retention (36). On the other hand, our results have shown that CKIɛ and CKIδ are able to induce the nuclear translocation of mPer3, which requires its NLS. The phosphorylation in this case may lead to unmasking of the mPer3 NLS. Therefore, masking or unmasking of signal sequences such as NLS by the phosphorylation-mediated conformational change may determine the subcellular localization of mPer proteins. Moreover, while CKIɛ and CKIδ reduce the inhibitory effect of mPer2 on the transcriptional activity of BMAL1-CLOCK, they enhance the inhibitory effect of mPer3. The enhancement of the inhibitory effect of mPer3 by CKIɛ and CKIδ appears to result from nuclear translocation of mPer3. We examined whether the CKIɛ affected the inhibitory effect of mPer3mutNLS on the transcriptional activity of BMAL1-CLOCK and found that CKIɛ did not affect the inhibitory effect of mPer3mutNLS (data not shown). Thus, the CKI-mediated phosphorylation could have different effects on regulation of the function of each mPer protein. It is thought that each of three mPer proteins may have specific roles in the mammalian circadian feedback loop. Actually, in the suprachiasmatic nucleus, expression of mPer1 and mPer2 is rapidly induced by light exposure, whereas that of mPer3 is not induced at any phase of the clock (31, 41). Moreover, the detailed analyses in mutant mice suggest a model in which mPer2 rhythmically stimulates the expression of BMAL1, whereas mPer3 may function as an “output” protein, transducing the oscillation of the central circadian clock to effector genes (28). Our result that the CKI-mediated phosphorylation induces specific changes in each of mPer proteins suggests that CKIɛ and CKIδ may function to ensure that each mPer proteins should have different and specific roles in the mammalian circadian autoregulatory loop.
In Drosophila, Dbt-mediated phosphorylation destabilizes dPer so that the level of dPer increases only when dTim increases (17, 21, 24). Thus, one way in which phosphorylation may contribute to circadian timing is to destabilize mPer proteins. In this study, we have shown that CKIɛ and CKIδ promote degradation of mPer proteins, which is dependent on the ubiquitin-proteasome pathway. Although it has not yet been examined whether the activity of CKIɛ or CKIδ shows a circadian oscillation, the mRNA level of doubletime in Drosophila or the protein level of CKIɛ in hamster brain has been shown to be nearly constant (17, 19). Per proteins are expressed in a circadian fashion, and thus the expressed Per proteins become phosphorylated by CKI, which appears to be constitutively active. Therefore, it is possible that the CKI-mediated phosphorylation and degradation of Per proteins would apparently show a circadian oscillation. In Drosophila, the dPer-dTim interaction is necessary for both stabilization and nuclear translocation of dPer. Only the dPer monomer is thought to be a substrate for Dbt. In mammalian cells, the Per-Tim interaction is not so apparent, and Per-Per and Per-Cry interactions appear to be dominant. Per proteins in these dimers might be poorer substrates for CKI than monomeric forms of Per proteins. These points should be investigated in more detail in future experiments.
Cry proteins have previously been shown to be strong candidates for negative regulators in the autoregulatory loop (18). Recent studies on tau, FASPS and DSPS have shown that the CKI-mediated phosphorylation of mPer proteins also plays an important role in the robust circadian rhythm (7, 19, 34). Although the effect of CKI on the BMAL1-CLOCK-mediated transcription was not very large, CKI may regulate the circadian period length by controlling both protein turnover and subcellular localization of Per proteins. Since clock proteins bind to one another in a complex manner, the changes of subcellular localization and protein stability in mPer proteins may evoke complex changes in the fundamental machinery of circadian clock. Our results have shown that mPer, mCry, and CKI form a ternary complex and therefore CKI might affect the functions of Cry proteins through Per proteins. Further studies on posttranscriptional regulation of mPer proteins would uncover molecular mechanisms by which the robust circadian rhythm is maintained and thus will lead to molecular understanding of human sleep disorders such as FASPS and DSPS.
Acknowledgments
We thank K. Terasawa for the preparation of HA-ubiquitin pcDNA3, pcDNA3(HA), and pcDNA3(Myc). We also thank members of our laboratory for helpful comments and suggestions.
This work was supported by grants-in-aid from the Ministry of Education, Science, and Culture of Japan (to E.N.).
REFERENCES
- 1.Albrecht, U., Z. S. Sun, G. Eichele, and C. C. Lee. 1997. A differential response of two putative mammalian circadian regulators, mPer1 and mPer2, to light. Cell 91:1055-1064. [DOI] [PubMed]
- 2.Antoch, M. P., E. J. Song, A. M. Chang, M. H. Vitaterna, Y. Zhao, L. D. Wilsbacher, A. M. Sangoram, D. P. King, L. H. Pinto, and J. S. Takahashi. 1997. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell 89:655-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, S. A., and U. Schibler. 1999. The ins and outs of circadian timekeeping. Curr. Opin. Genet. Dev. 9:588-594. [DOI] [PubMed] [Google Scholar]
- 4.Darlington, T. K., K. Wager-Smith, M. F. Ceriani, D. Staknis, N. Gekakis, T. D. Steeves, C. J. Weitz, J. S. Takahashi, and S. A. Kay. 1998. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science 280:1599-1603. [DOI] [PubMed] [Google Scholar]
- 5.Devlin, P. F., and S. A. Kay. 1999. Cryptochromes—bringing the blues to circadian rhythms. Trends Cell Biol. 9:295-298. [DOI] [PubMed] [Google Scholar]
- 6.Dunlap, J. C. 1999. Molecular bases for circadian clocks. Cell 96:271-290. [DOI] [PubMed] [Google Scholar]
- 7.Ebisawa, T., M. Uchiyama, N. Kajimura, K. Mishima, Y. Kamei, M. Katoh, T. Watanabe, M. Sekimoto, K. Shibui, K. Kim, Y. Kudo, Y. Ozeki, M. Sugishita, R. Toyoshima, Y. Inoue, N. Yamada, T. Nagase, N. Ozaki, O. Ohara, N. Ishida, M. Okawa, K. Takahashi, and T. Yamauchi. 2001. Association of structural polymorphisms in the human period3 gene with delayed sleep phase syndrome. EMBO Rep. 2:342-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gekakis, N., D. Staknis, H. B. Nguyen, F. C. Davis, L. D. Wilsbacher, D. P. King, J. S. Takahashi, and C. J. Weitz. 1998. Role of the CLOCK protein in the mammalian circadian mechanism. Science 280:1564-1569. [DOI] [PubMed] [Google Scholar]
- 9.Gietzen, K. F., and D. M. Virshup. 1999. Identification of inhibitory autophosphorylation sites in casein kinase I epsilon. J. Biol. Chem. 274:32063-32070. [DOI] [PubMed]
- 10.Hall, J. C. 1998. Genetics of biological rhythms in Drosophila. Adv. Genet. 38:135-184. [DOI] [PubMed] [Google Scholar]
- 11.Hastings, M. H., M. D. Field, E. S. Maywood, D. R. Weaver, and S. M. Reppert. 1999. Differential regulation of mPer1 and mTIM proteins in the mouse suprachiasmatic nuclei: new insights into a core clock mechanism. J. Neurosci. 19:RC11. [DOI] [PMC free article] [PubMed]
- 12.Iwasaki, H., and J. C. Dunlap. 2000. Microbial circadian oscillatory systems in Neurospora and Synechococcus: models for cellular clocks. Curr. Opin. Microbiol. 3:189-196. [DOI] [PubMed] [Google Scholar]
- 13.Jin, X., L. P. Shearman, D. R. Weaver, M. J. Zylka, G. J. de Vries, and S. M. Reppert. 1999. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell 96:57-68. [DOI] [PubMed] [Google Scholar]
- 14.Keesler, G. A., F. Camacho, Y. Guo, D. Virshup, C. Mondadori, and Z. Yao. 2000. Phosphorylation and destabilization of human Period I clock protein by human casein kinase I epsilon. Neuroreport 11:951-955. [DOI] [PubMed] [Google Scholar]
- 15.King, D. P., and J. S. Takahash. 2000. Molecular genetics of circadian rhythms in mammals. Annu. Rev. Neurosci. 23:713-742. [DOI] [PubMed] [Google Scholar]
- 16.King, D. P., Y. Zhao, A. M. Sangoram, L. D. Wilsbacher, M. Tanaka, M. P. Antoch, T. D. Steeves, M. H. Vitaterna, J. M. Kornhauser, P. L. Lowrey, F. W. Turek, and J. S. Takahashi. 1997. Positional cloning of the mouse circadian clock gene. Cell 89:641-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kloss, B., J. L. Price, L. Saez, J. Blau, A. Rothenfluh, C. S. Wesley, and M. W. Young. 1998. The Drosophila clock gene doubletime encodes a protein closely related to human casein kinase Iɛ. Cell 94:97-107. [DOI] [PubMed] [Google Scholar]
- 18.Kume, K., M. J. Zylka, S. Sriram, L. P. Shearman, D. R. Weaver, X. Jin, E. S. Maywood, M. H. Hastings, and S. M. Reppert. 1999. mCry1 and mCry2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98:193-205. [DOI] [PubMed] [Google Scholar]
- 19.Lowrey, P. L., K. Shimomura, M. P. Antoch, S. Yamazaki, P. D. Zemenides, M. R. Ralph, M. Menaker, and J. S. Takahashi. 2000. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 288:483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morgan, D. O., and H. L. De Bondt. 1994. Protein kinase regulation: insights from crystal structure analysis. Curr. Opin. Cell Biol. 6:239-246. [DOI] [PubMed] [Google Scholar]
- 21.Price, J. L., J. Blau, A. Rothenfluh, M. Abodeely, B. Kloss, and M. W. Young. 1998. doubletime is a novel Drosophila clock gene that regulates Period protein accumulation. Cell 94:83-95. [DOI] [PubMed] [Google Scholar]
- 22.Reppert, S. M. 1998. A clockwork explosion! Neuron 21:1-4. [DOI] [PubMed] [Google Scholar]
- 23.Rosbash, M. 1995. Molecular control of circadian rhythms. Curr. Opin. Genet. Dev. 5:662-668. [DOI] [PubMed] [Google Scholar]
- 24.Saez, L., and M. W. Young. 1996. Regulation of nuclear entry of the Drosophila clock proteins Period and Timeless. Neuron 17:911-920. [DOI] [PubMed] [Google Scholar]
- 25.Scully, A. L., and S. A. Kay. 2000. Time flies for Drosophila. Cell 100:297-300. [DOI] [PubMed] [Google Scholar]
- 26.Shearman, L. P., X. Jin, C. Lee, S. M. Reppert, and D. R. Weaver. 2000. Targeted disruption of the mPer3 gene: subtle effects on circadian clock function. Mol. Cell. Biol. 20:6269-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shearman, L. P., M. J. Zylka, D. R. Weaver, L. F. Kolakowski, and S. M. Reppert. 1997. Two Period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron 19:1261-1269. [DOI] [PubMed] [Google Scholar]
- 28.Shearman, L. P., S. Sriram, D. R. Weaver, E. S. Maywood, I. Chaves, B. Zheng, K. Kume, C. C. Lee, G. T. van der Horst, M. H. Hastings, and S. M. Reppert. 2000. Interacting molecular loops in the mammalian circadian clock. Science 288:1013-1019. [DOI] [PubMed] [Google Scholar]
- 29.Sun, Z. S., U. Albrecht, O. Zhuchenko, J. Bailey, G. Eichele, and C. C. Lee. 1997. RIGUI, a putative mammalian ortholog of the Drosophila Period gene. Cell 90:1003-1011. [DOI] [PubMed] [Google Scholar]
- 30.Takano, A., K. Shimizu, S. Kani, R. M. Buijs, M. Okada, and K. Nagai. 2000. Cloning and characterization of rat casein kinase 1ɛ. FEBS Lett. 477:106-112. [DOI] [PubMed] [Google Scholar]
- 31.Takumi, T., K. Taguchi, S. Miyake, Y. Sakakida, N. Takashima, C. Matsubara, Y. Maebayashi, K. Okumura, S. Takekida, S. Yamamoto, K. Yagita, L. Yan, M. W. Young, and H. Okamura. 1998. A light-independent oscillatory gene mPer3 in mouse SCN and OVLT. EMBO J. 17:4753-4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tei, H., H. Okamura, Y. Shigeyoshi, C. Fukuhara, R. Ozawa, M. Hirose, and Y. Sakaki. 1997. Circadian oscillation of a mammalian homologue of the Drosophila Period gene. Nature 389:512-516. [DOI] [PubMed] [Google Scholar]
- 33.Thresher, R. J., M. H. Vitaterna, Y. Miyamoto, A. Kazantsev, D. S. Hsu, C. Petit, C. P. Selby, L. Dawut, O. Smithies, J. S. Takahashi, and A. Sancar. 1998. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science 282:1490-1494. [DOI] [PubMed] [Google Scholar]
- 34.Toh, K. L., C. R. Jones, Y. He, E. J. Eide, W. A. Hinz, D. M. Virshup, L. J. Ptacek, and Y. H. Fu. 2001. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 291:1040-1043. [DOI] [PubMed] [Google Scholar]
- 35.van der Horst, G. T., M. Muijtjens, K. Kobayashi, R. Takano, S. Kanno, M. Takao, J. de Wit, A. Verkerk, A. P. Eker, D. van Leenen, R. Buijs, D. Bootsma, J. H. Hoeijmakers, and A. Yasui. 1999. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398:627-630. [DOI] [PubMed] [Google Scholar]
- 36.Vielhaber, E., E. Eide, A. Rivers, Z. H. Gao, and D. M. Virshup. 2000. Nuclear entry of the circadian regulator mPer1 is controlled by mammalian casein kinase I epsilon. Mol. Cell. Biol. 20:4888-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wager-Smith, K., and S. A. Kay. 2000. Circadian rhythm genetics: from flies to mice to humans. Nat. Genet. 26:23-27. [DOI] [PubMed] [Google Scholar]
- 38.Whitmore, D., N. Cermakian, C. Crosio, N. S. Foulkes, M. P. Pando, Z. Travnickova, and P. Sassone-Corsi. 2000. A clockwork organ. Biol. Chem. 381:793-800. [DOI] [PubMed] [Google Scholar]
- 39.Yagita, K., S. Yamaguchi, F. Tamanini, G. T. van Der Horst, J. H. Hoeijmakers, A. Yasui, J. J. Loros, J. C. Dunlap, and H. Okamura. 2000. Dimerization and nuclear entry of mPer proteins in mammalian cells. Genes Dev. 14:1353-1363. [PMC free article] [PubMed] [Google Scholar]
- 40.Young, M. W. 2000. Life's 24-hour clock: molecular control of circadian rhythms in animal cells. Trends Biochem. Sci. 25:601-606. [DOI] [PubMed] [Google Scholar]
- 41.Zylka, M. J., L. P. Shearman, D. R. Weaver, and S. M. Reppert. 1998. Three Period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron 20:1103-1110. [DOI] [PubMed] [Google Scholar]