Abstract
Activating transcription factor 1 (ATF1), CREB, and the cyclic AMP (cAMP) response element modulatory protein (CREM), which constitute a subfamily of the basic leucine zipper transcription factors, activate gene expression by binding as homo- or heterodimers to the cAMP response element in regulatory regions of target genes. To investigate the function of ATF1 in vivo, we inactivated the corresponding gene by homologous recombination. In contrast to CREB-deficient mice, which suffer from perinatal lethality, mice lacking ATF1 do not exhibit any discernible phenotypic abnormalities. Since ATF1 and CREB but not CREM are strongly coexpressed during early mouse development, we generated mice deficient for both CREB and ATF1. ATF1−/− CREB−/− embryos die before implantation due to developmental arrest. ATF1+/− CREB−/− embryos display a phenotype of embryonic lethality around embryonic day 9.5 due to massive apoptosis. These results indicate that CREB and ATF1 act in concert to mediate signals essential for maintaining cell viability during early embryonic development.
The activating transcription factor 1 (ATF1), CREB, and the cyclic AMP response element modulatory protein (CREM) share high sequence homology and mediate the transcriptional response to various extracellular signals, including peptide hormones (15, 27), growth factors (10, 11, 26), neurotransmitters, and Ca2+ (17, 23). Activation of the CREB/CREM/ATF1 proteins is mediated via different signaling pathways which converge to phosphorylate a distinct serine residue (22). Phosphorylation of CREB by mitogen-activated protein kinase and Akt/protein kinase B has been implicated as important for cellular survival in cultured cells (4, 9). CREB is also thought to act as an antiapoptotic factor in sympathetic neurons (19). In human clear cell sarcoma, the ATF1 gene is fused to the genes encoding Ewing's sarcoma protein and seems to be responsible for maintaining tumor viability (5). It was also suggested that ATF1 is upregulated in human metastatic melanoma cells. Disruption of ATF1 activity in these cells by using an inhibitory anti-ATF1 antibody fragment suppressed their tumorigenicity and metastatic potential in nude mice (14).
As previously reported, the corresponding genes of CREB and CREM have been inactivated by gene targeting in mice. Mice lacking the CREM gene exhibit an arrest in spermatogenesis (3, 16), whereas CREB-deficient mice die perinatally due to atelectasis of the lung (21). We show here that loss of ATF1 function after inactivation of the ATF1 gene in mice does not cause any obvious phenotypic abnormalities. Our expression studies showed that ATF1 and CREB but not CREM are strongly coexpressed during early mouse development. To identify the role of both proteins during early development and to circumvent possible compensatory effects, we therefore generated mice deficient for both ATF1 and CREB. Interestingly, complete inactivation of both proteins, ATF1 and CREB, results in embryonic death before implantation. Embryos with only one functional ATF1 allele in the absence of CREB develop further but die around embryonic day 9.5 (E9.5). Therefore, ATF1 and CREB proteins play a crucial role during early mouse development. They can compensate for each other's function, although they are not equivalent, implying that the influence of each factor differs during early development.
MATERIALS AND METHODS
Immunohistochemistry.
Mouse embryos were fixed, embedded, and sectioned as described for in situ hybridization (29). Sections were deparaffinated and rehydrated through an ethanol series. Endogenous peroxidases were blocked by incubation with 3% H2O2 in distilled H2O. After microwave treatment with Antigen Retrieval Citra Solution (BioGenex), sections were washed with phosphate-buffered saline (PBS), blocked in PBS containing 1.5% normal goat serum (Sigma), and incubated with the first antibody overnight. Detection was performed with Vecta Stain Elite Kit (Serva) followed by diaminobenzidine substrate (Roche) incubation. The following first antibodies were used: mouse ATF1, amino acids 10 to 36; and mouse CREB, amino acids 136 to 150.
Generation and genotyping of ATF1−/− mice.
The mouse ATF1 gene was disrupted in embryonic stem (ES) cells using a deletion vector strategy (see Fig. 1). This deletion disrupts the gene and removes the coding information for both the protein kinase A domain and the leucine zipper domain (residues 200 to 1380 of the cDNA sequence). Six out of 100 neomycin-resistant clones were identified by Southern blotting. Three independent ES cell clones were used for injection into C57BL/6 blastocysts to generate chimeric mice. Chimeras were then mated with both C57BL/6 and 129/SvJ mice to generate outbred and isogenic colonies, respectively. Heterozygous mice were intercrossed to obtain ATF1−/− mice, which were genotyped by PCR analysis of tail DNA using the following primers: 1′ (5′-GCAGGTGATGGAAGACAGATCATTCC-3′), 2′ (5′-AGACCTGCCTCCT-CACCTAACTGCC-3′), and 3′ (5′-AAGCGCCATTCGCCATTCAGGC-3′). ATF1- as well as CREB-deficient animals were bred in a C57BL/6 × 129/SvEv hybrid genetic background. The offspring of ATF1+/− CREB+/− heterozygous matings were recovered from the mothers at E18.5 via caesarean section. At different days of gestation, litters from intercrosses were serially sectioned and genotyped via PCR.
FIG. 1.
Immunohistochemical analysis revealed overlapping expression of ATF1 and CREB during early embryogenesis. At E3.5 ATF1 could be detected in the whole blastocyst, whereas CREB was mainly located in the ICM cells. During further embryogenesis at E6.0 and E7.5, ATF1 is expressed in all embryonic, extraembryonic, and trophectoderm-derived cells. CREB was found only in epiblast cells and lineages deriving thereof. A clear border can be seen between extraembryonic parts and the epiblast (arrowheads). In adult testis, the expression pattern of both proteins is very distinct. ATF1 is highly expressed in spermatocytes of the pachytene stage, whereas CREB protein is found only in Sertoli cells (arrows).
Western blot analysis.
Thirty micrograms of nuclear extract of ATF1−/− and control tissues was loaded per lane, resolved on a sodium dodecyl sulfate-10% polyacrylamide gel, and transferred to a nitrocellulose membrane by semidry electroblotting. The membrane was blocked in Tris-buffered saline containing 0.1% Tween 20 and 5% dried defatted milk, incubated with an immunoglobulin A (IgA)-type monoclonal antibody which specifically recognizes ATF1 (MAb5, a generous gift that we received from Steven Hinrichs). The blot was then treated with a peroxidase-coupled anti-mouse antibody (Vector) diluted in Tris-buffered saline containing 0.1% Tween 20 and 1/1,000 normal goat serum. Subsequently, detection of the signal was performed with an enhanced chemiluminescence kit (Vector) and the membrane was exposed on Fuji film.
β-Galactosidase staining.
E3.5 and E7.5 embryos were collected, washed twice in PBS, and fixed for 10 min at 4°C in 4% paraformaldehyde (PFA). After fxation, embryos were again washed in PBS and incubated in staining solution: 4 mM K3(Fe[CN]6); 4 mM K4(Fe[CN]6); 2 mM MgCl2; 0.02% NP-40; 0.01% Na-deoxycholate; 5 mM EGTA, and 0.4 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (Biomol)/ml in PBS. They were then washed and postfixed for 2 h in 4% PFA before being stored in 70% ethanol.
In vitro culture of embryos.
Embryos were collected at E3.5, transferred to ES cell medium, and incubated in a humidified atmosphere at 37°C and 5% CO2 for 7 days. ES cell medium was made up as follows: 1× Earle's balanced salt solution (Gibco BRL), 0.2% sodium bicarbonate (Gibco BRL), 0.33 mM sodium pyruvate (Gibco BRL), 100 U of penicillin-100 mg of streptomycin (Gibco BRL), 0.1 mM EDTA, and 0.4% bovine serum albumin (Sigma), sterile filtered. Differentiation of the embryos was monitored by taking photographs every 24 h.
In situ hybridization and histology.
Embryos were collected at different developmental stages, fixed in 4% PFA (pH 7.2) overnight, dehydrated through an ethanol series, cleared in toluene, embedded in paraffin, and sectioned (thickness, 7 μm). In situ prehybridizations, hybridizations, and probe synthesis were carried out as described previously (29). Probes for hybridization were hydrolyzed to 200-bp fragments and dissolved at a concentration of 60 ng/ml in hybridization solution. Slides were dipped in Kodak NTB2 emulsion diluted 1:1 with water, exposed at 4°C for 5 to 10 days, and developed using Kodak D19 developing solution and Kodakfix at 15°C for 4 min. Sections were stained with hematoxylin and eosin and visualized using a Zeiss Axiophot microscope.
Hoechst 33258 staining of blastocysts.
Embryos were collected at E3.5. The zona pellucida was removed by pronase treatment, and the embryos were fixed overnight in 4% PFA in M2 medium (12). They were incubated individually on slides for 15 min in 20-μl drops of 50 mM Tris HCl, pH 7.5, 150 mM NaCl, and 1% NP-40; dried; and washed with PBS and were then stained 10 min with Hoechst 33258 (Sigma), washed in PBS, and mounted. A Zeiss axiophot microscope was used for fluorescence reading.
In situ detection of apoptotic cells.
Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) analysis of the E3.5 embryos was performed as previously described (18). Paraffin sections of embryos were deparaffinated and rehydrated through an ethanol series. Following a rinse in H2O, they were incubated in 20 μg of proteinase K/ml and for 5 min in 2% H2O2 at room temperature. The following treatment was performed according to the in situ cell detection kit (Roche). Positive cells were detected with the Vecta Stain Elite Kit (Serva) and with diaminobenzidine substrate (Roche) as already described for immunohistochemistry.
RESULTS
Expression pattern of ATF1.
Immunohistochemical analysis of ATF1 expression showed high levels of ATF1 during early development that decreased during further maturation. At E3.5 ATF1 was expressed in trophectoderm and inner cell mass (ICM) cells (Fig. 1). At E6.0 and E7.5, a uniform level of ATF1 protein was detected in embryonic and extraembryonic tissue and trophectoderm-derived cells (Fig. 1). In contrast, CREB at E3.5 was expressed only in the ICM and during later stages in the epiblast and cells derived thereof (Fig. 1). CREM could not be detected at all (data not shown). During later development and in adult mice, ATF1 expression was, contrary to the rather ubiquitously expressed CREB, highest in testis (data not shown). In testis the two proteins were differentially expressed: CREB staining was located in the Sertoli cells (Fig. 1, arrows), whereas ATF1 was found specifically in spermatocytes of the pachytene stage (Fig. 1). In adults ATF1 protein could also be detected in muscle, fat, and chorioid plexus but to a lesser extent (data not shown).
Generation of mice lacking functional ATF1.
The mouse ATF1 gene was disrupted in ES cells by homologous recombination. Part of exon 3 and of exons 4 to 7 encoding the kinase-inducible domain, the basic region, and the leucine zipper domain was replaced by a β-galactosidase/neomycin resistance cassette inserted in frame (Fig. 2A). The mutant ES cells were used to generate heterozygous mutant mice, which were intercrossed to obtain mice homozygous for the ATF1 mutation. Genotyping of the offspring at 4 weeks showed a Mendelian distribution of the three expected genotypes (Fig. 2B). Western blot analysis revealed that the ATF1 protein is clearly absent in ATF1−/− mice. In addition, β-galactosidase staining of ATF1+/− and CREB+/− embryos showed that, as expected, the LacZ gene is expressed in accordance with the immunohistochemical data during early embryogenesis (Fig. 2C). ATF1−/− mice were fertile and did not exhibit any apparently abnormal phenotype. Histological examination of all major tissues and organs revealed no differences between mutant animals and wild-type littermates (data not shown). The absence of obvious early developmental defects in ATF1−/− or CREB−/− mice suggests that each factor can compensate for the absence of the other during early mouse development.
FIG. 2.
Inactivation of the mouse ATF1 gene by gene targeting. (A) Shown is part of the ATF1 locus with exons 3 to 7 (black boxes) encoding portions of the kinase-inducible domain (exon 3) and all of the leucine zipper domain (exon 7) (top). Targeting construct containing a 3.8-kb 5′ homology and a 1.6-kb 3′ homology fragment inserted into the pHM3 vector is shown (middle). The targeted ATF1 gene is shown at bottom. lacZ-neo, β-galactosidase/neomycin-resistance cassette; Bg, BglII; E, EcoRI. (B) Top, PCR genotyping of genomic DNA isolated from embryos of heterozygous intercrosses. Detection of the wild-type allele corresponds to a 260-bp fragment, and the mutant allele corresponds to a 420-bp fragment. Bottom, Western blot analysis of nucleus extracts of kidney, liver, and testis shows clear loss of ATF1 in tissues of ATF1−/− mice. (C) Whole-mount β-galactosidase stainings of ATF1+/− and CREB+/− embryos. LacZ staining confirms the expression pattern of ATF1 and CREB revealed by immunohistochemistry (Fig. 1).
Generation of mice deficient for ATF1 and CREB.
To rule out compensatory effects, mice harboring the mutated ATF1 allele were crossed with our previously derived mice which carried a disrupted CREB allele (21). Postnatal analysis of progeny from ATF1+/− CREB+/− intercrosses revealed that, as expected, mice carrying a homozygous disruption of the ATF1 gene did not show any abnormalities. CREB−/− mice suffered from perinatal lethality, as was reported beforehand (21). At all developmental stages examined (adult, E18.5, E9.5, E7.5, and E6.0), double-heterozygous animals as well as ATF1−/− CREB+/− animals showed no phenotypic or histological abnormalities, compared to wild-type littermates (Table 1). However, ATF1+/− CREB−/− animals as well as ATF1−/− CREB−/− animals were absent postnatally, indicating an phenotype of embryonic lethality (Table 1). ATF1−/− CREB−/− embryos were recovered only from the preimplantational stage at E3.5. ATF1+/− CREB−/− embryos were not detected later than E9.5, even though at all earlier stages examined (E7.5, E6.0, and E3.5), embryos of this genotype were isolated at the expected Mendelian ratio (Table 1). Interestingly, ATF1−/− CREB+/− embryos developed completely normally during embryogenesis and after birth.
TABLE 1.
Numbers and percentages of dissected embryos
| Embryonic day | Embryo no. in total | No. of embryos isolated in % (expected %) for different genotypes
|
|||||
|---|---|---|---|---|---|---|---|
| ATF1+/+CREB−/− | ATF1−/−CREB+/+ | ATF1−/−CREB+/− | ATF1+/−CREB−/− | ATF1−/−CREB−/− | Genotype not determined | ||
| 18.5a | 147 | 6.1 (6.3) | 7.6 (6.3) | 12.2 (12.5) | 0 (12.5) | 0 (6.3) | |
| 9.5a | 161 | 4.4 (6.3) | 6.2 (6.3) | 8.1 (12.5) | 14.3 (12.5) | 0 (6.3) | |
| 7.5a | 68 | 7.4 (6.3) | 4.4 (6.3) | 14.7 (12.5) | 14.7 (12.5) | 0 (6.3) | |
| 3.5b | 37 | 10.8 (12.5) | 35.1 (25.0) | 13.5 (12.5) | 5.4 (12.5) | 5.4 | |
| 3.5c | 82 | 26.8 (25.0) | 53.7 (50.0) | 15.9 (25.0) | 3.7 | ||
Offspring of ATF1+/− CREB+/− intercrosses.\
Offspring of ATF1−/− CREB+/− × ATF1+/− CREB+/− matings.\
Offspring of ATF1−/− CREB+/− intercrosses.
Defective development of preimplantation embryos lacking CREB and ATF1.
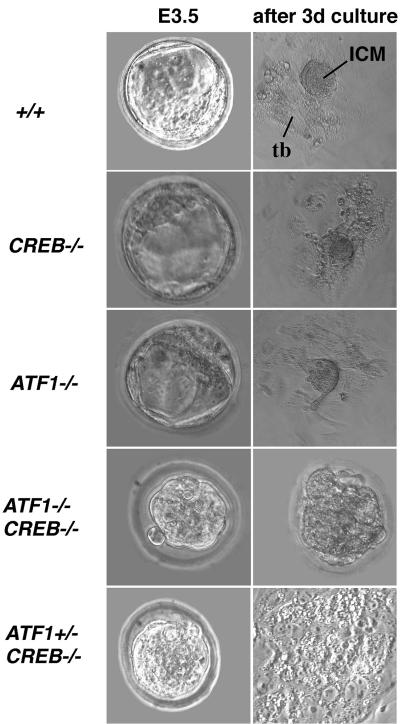
To examine the early lethality of ATF1−/− CREB−/− and ATF1+/− CREB−/− embryos, we isolated preimplantation embryos. In contrast to the wild type (ATF1−/− and CREB−/− blastocysts), embryos of both genotypes, ATF1+/− CREB−/− and ATF1−/− CREB−/−, exhibited a morula-like appearance without blastocoel or ICM cells present (Fig. 3). To determine whether the observed phenotype of ATF1+/− CREB−/− and ATF1−/− CREB−/− embryos was caused by a delay or an arrest in development, embryos were collected at E3.5 and individually cultured in vitro. Wild-type embryos proliferated, hatched from the zona pellucida, and differentiated into a trophoblast monolayer with the ICM apparent as an overlying compact cell aggregate (Fig. 3). Embryos carrying a disruption of only one of the two genes (ATF1−/− or CREB−/−) behaved like wild-type embryos and exhibited no altered outgrowth development (Fig. 3). In contrast, ATF1−/− CREB−/− did not develop to the blastocyst stage and did not hatch from their zona pellucida. Cells degenerated and died within the first 4 days of culture (Fig. 3). ATF1+/− CREB−/− embryos showed a milder phenotype with a delayed hatching from the zona pellucida. They formed a trophoblast monolayer, however, without developing a proper ICM on top (Fig. 3).
FIG. 3.
Functional disruption of both CREB and ATF1 results in preimplantation defects. Embryos of different genotypes were isolated at E3.5. ATF1+/− CREB−/− and ATF1−/− CREB−/− embryos show a morula-like structure, while wild-type and single-knockout embryos are comprised of trophectoderm surrounding ICM and blastocoel. Outgrowths after 4 days of in vitro culture of wild-type, ATF1−/−, and CREB−/− blastocysts develop a monolayer of trophoblast cells with the ICM cells on top. No outgrowth of ATF1−/− CREB−/− embryos occurs after 4 days in culture. Embryos seem to arrest in development; cells appear necrotic and are still surrounded by the zona pellucida. ATF1+/− CREB−/− embryos hatch from the zona pellucida; a trophoblast monolayer is present, but no obvious ICM can be seen. tb, trophoblast monolayer. 3d, 3 days.
ATF1+/− CREB−/− embryos lack normal epiblasts.
Later postimplantational development of ATF1+/− CREB−/− embryos at E6.0, E7.5 and E9.5 was analyzed histologically. This revealed that they were smaller than their littermates and developed severe phenotypic abnormalities. They exhibited a variable phenotype, with some containing, instead of a proper egg cylinder, an accumulation of cells similar to epiblast cells at the distal end of the conceptus. Others were composed only of extraembryonic tissue (Fig. 4). At E6.0, ATF1+/− CREB−/− embryos fail to develop a pseudostratified columnar epithelial organization of the epiblast (Fig. 4). At E7.5, when embryonic mesoderm, amnion, chorion, and allantois are clearly distinguishable in wild-type embryos, ATF1+/− CREB−/− embryos lack more differentiated structures. The extraembryonic ectoderm appears rather normal; however, in the embryonic part the normal organization of the three germ layers, amnion, and allantois is absent. Despite the failure of proper epiblast formation, embryonic cells of the ATF1+/− CREB−/− embryos seem to differentiate at least in part. At E9.5, some embryos show cells morphologically similar to mesodermal tissue at their distal end; others are composed of only blood islands surrounded by extraembryonic ectoderm.
FIG. 4.
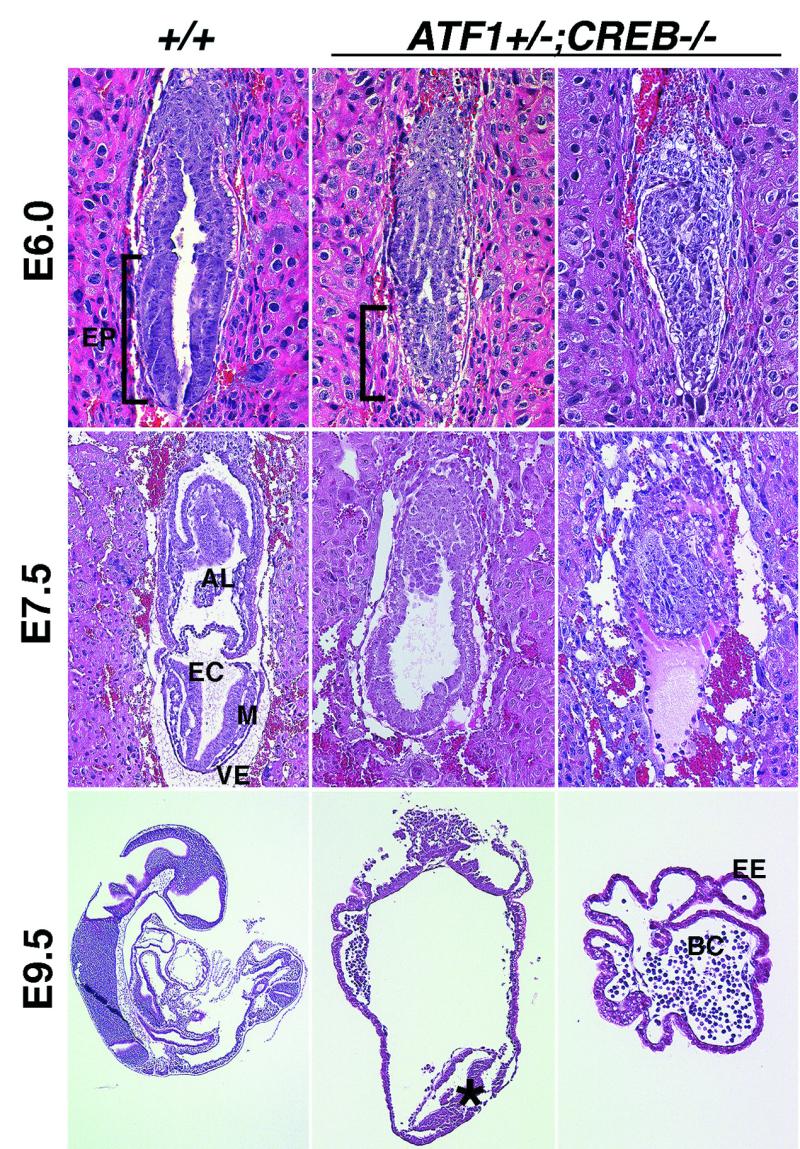
ATF1+/− CREB−/− embryos exhibit severe embryonic disorganization. Sections of hematoxylin-and-eosin-stained wild-type and ATF1+/− CREB−/− embryos at embryonic stages E6.0, E7.5, and E9.5 are shown. Two types of ATF1+/− CREB−/− embryos could be isolated. One group showed a strong reduction and malformation of the embryonic region, whereas the other one was comprised only of extraembryonic ectoderm lacking the embryonic part completely. At E6.0 ATF1+/− CREB−/− embryos fail to form the pseudostratified columnar epithelium of the epiblast. At E7.5, gastrulation is almost complete and the three germ layers have developed from the epiblast. In ATF1+/− CREB−/− embryos, germ layers cannot be distinguished and extraembryonic mesoderm-derived structures like amnion, allantois, and chorion are missing. E9.5 embryos of this genotype fail to develop any structural organization compared to wild-type embryos of the same age. Some of the ATF1+/− CREB−/− embryos develop, instead of a structured embryo, a group of mesoderm-like cells (star) at the distal end of the conceptus. Others comprise only blood cells surrounded by extraembryonic ectoderm. AL, allantois; EC, embryonic ectoderm; EP, epiblast; M, mesoderm; VE, visceral endoderm; EE, extraembryonic ectoderm; and BC, blood cells.
ATF1+/− CREB−/− embryos are reduced in the number of pluripotent cells but differentiate.
To define cell identities of the accumulated cells at the distal end of ATF1+/− CREB−/− embryos, we examined the expression of a panel of molecular marker genes showing regionalized expression in embryos at stages E6.0 and E7.5 by in situ hybridization. The expression analysis in the ATF1+/− CREB−/− embryos showed that a number of markers (oct-4 for pluripotent epiblast cells [20], brachyury and fgf-8 for primitive streak mesoderm [8, 28], bmp-4 for extraembryonic mesoderm [30], and hnf-3β and lim-1 as early organizer genes [1, 2]) were present in distally localized cells (Fig. 5A and data not shown). The initiation of gastrulation was confirmed by the presence of fgf-8 and brachyury. However, the mesoderm-like cells appear to differentiate into mainly posterior, extraembryonic mesoderm, which was shown by the high levels of bmp-4 in some embryos and the resulting blood cells at E9.5 (Fig. 4 and data not shown). Expression of oct-4 showed that in ATF1+/− CREB−/− embryos, cells committed to the embryonic lineage were markedly reduced in number or completely absent (Fig. 5A). At E3.5, Hoechst staining of ATF1−/− CREB−/− embryos revealed that they also contained fewer blastomeres than did their wild-type littermates (Fig. 5B).
FIG. 5.
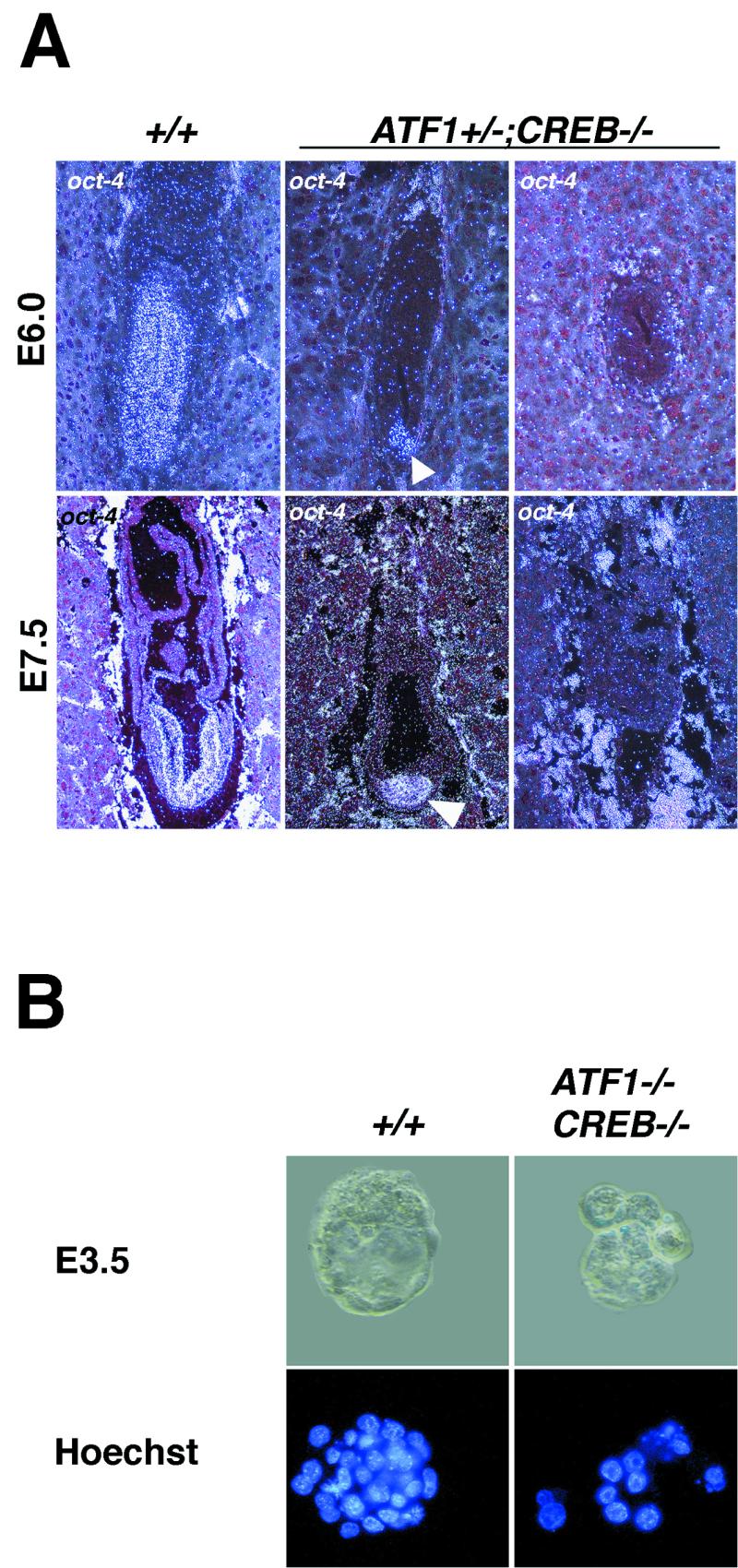
Number of totipotent cells in ATF1−/− CREB−/− embryos and of pluripotent cells in ATF1+/− CREB−/− embryos is strongly reduced. (A) oct-4 expression in wild-type and ATF1+/− CREB−/− embryos at E6.0 and E7.5 detected by in situ hybridization. Pluripotent cells of the epiblast appear positive for Oct-4. In contrast to the wild type, ATF1+/− CREB−/− embryos develop a strongly reduced embryonic part with fewer epiblast cells or lack pluripotent cells completely. (B) Hoechst staining reveals that ATF1−/− CREB−/− embryos are comprised of fewer blastomeres than are their wild-type littermates.
Loss of CREB and ATF1 leads to increased programmed cell death.
To determine whether cell loss occurred due to programmed cell death, we performed TUNEL assays. Remarkably, after 1 day of in vitro culture of E3.5 ATF1−/− CREB−/− embryos, most of the blastomeres showed intense TUNEL labeling, whereas, in control embryos, no signal for dying cells could be detected (Fig. 6 A). At E3.5, TUNEL stainings of ATF1+/− CREB−/− embryos did not differ from those for control littermates (data not shown). But during later development at E7.5, a massive number of apoptotic cells could be detected in distal parts of ATF1+/− CREB−/− embryos overlapping the region of oct-4 expression, whereas, in wild-type littermates, only few cells underwent programmed cell death (Fig. 5 and 6B). Therefore, cellular suvival in both ATF1−/− CREB−/− and ATF1+/− CREB−/− embryos is severely affected but at different developmental stages.
FIG. 6.
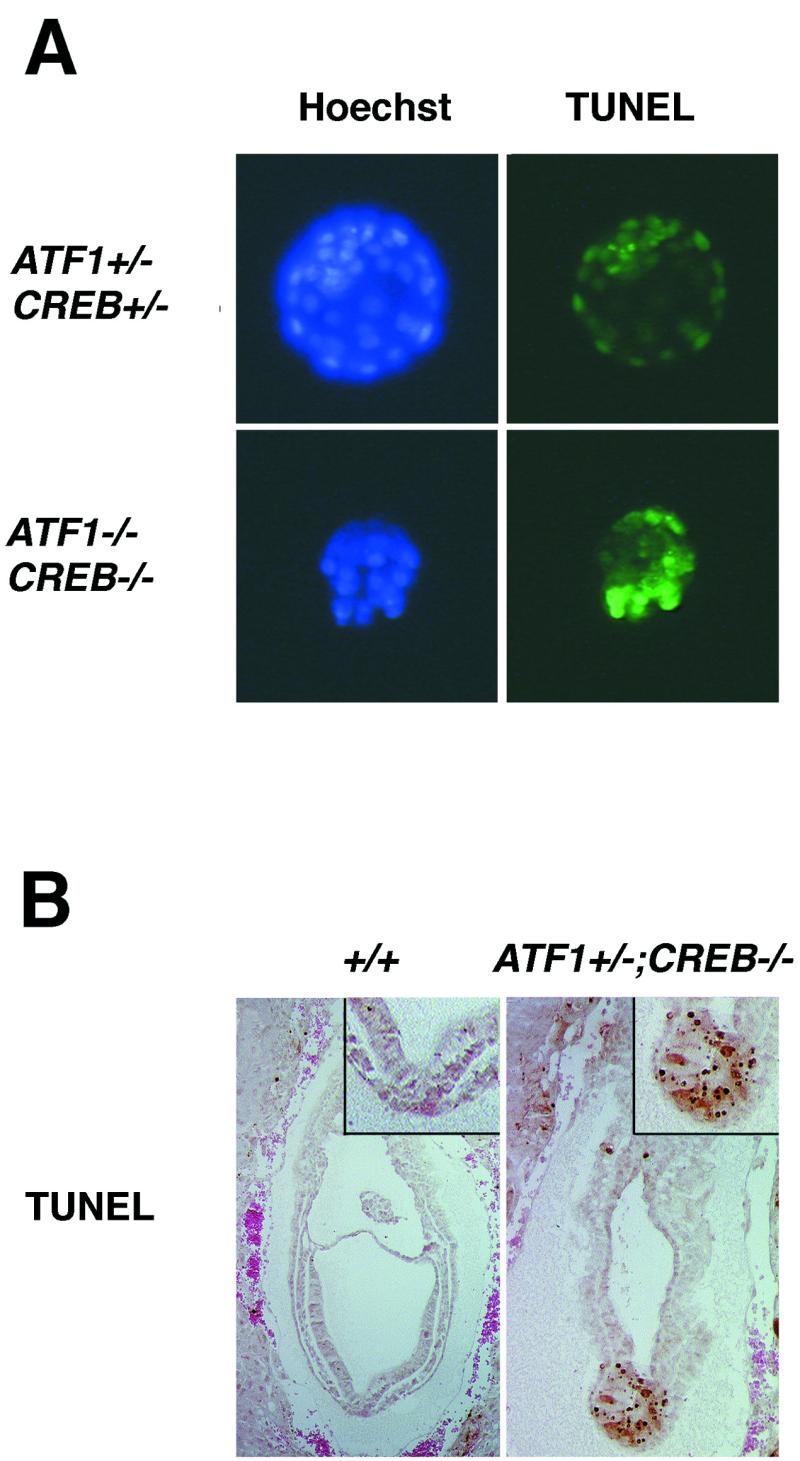
Both ATF1 and CREB are important for cellular survival during early development. In situ cell death detection shows an increasing number of dying cells in ATF1−/− CREB−/− as well as in ATF1+/− CREB−/− embryos but at different developmental stages. (A) After 1 day of culture, control blastocysts do not show any TUNEL-labeled cells, whereas almost all cells of ATF1−/− CREB−/− embryos, isolated at E3.5, seem to be dying. (B) At E7.5, ATF1+/− CREB−/− mutant embryos show, compared to the wild types, intense TUNEL staining in the distal part of the embryo overlapping the oct-4-expressing region.
DISCUSSION
ATF1 can compensate for CREB function in its absence.
Mouse embryos lacking ATF1 and CREB suffer embryonic lethality at the morula stage, whereas ATF1+/− CREB−/− embryos show a delayed development, with affected survival of pluripotent cells, resulting in embryonic death around E9.5. Since embryos homozygous for null alleles of either ATF1 or CREB are reported to develop normally, it appears that neither gene alone is required for normal pre- and peri-implantation development. From our results we deduce that functional redundancy between ATF1 and CREB during the pre- and peri-implantation period accounts for the lack of an abnormal phenotype in single-mutant ATF1−/− or CREB−/− embryos. The presence of a single CREB allele is sufficient to compensate for the absence of ATF1, whereas a single, intact allele of ATF1 in the absence of functional CREB leads to early embryonic lethality. The resulting severity of defects in embryos carrying a mutation in both genes is therefore dependent on gene dosage and is sensitive to the allele present.
ATF1 and CREB mediate signals essential for survival.
Arrest of ATF1−/− CREB−/− embryos at the morula stage coincides with the end of cleavage and the need for fibroblast growth factor (FGF) input for further differentiation. Blocking FGF signaling results in prevention of attachment and outgrowth of blastocysts in vitro (6). Previous data show that FGF can regulate CREB and ATF1 via the mitogen-activated protein kinase pathway, indicating that they might be necessary to mediate those signals important for early differentiation (26). ATF1+/− CREB−/− embryos also exhibit a differentiation defect. At E6.0 their ICM fails to develop into the pseudostratified columnar epithelium of the epiblast, which might affect the normal interaction of embryonic ectoderm and visceral endoderm. The process of cavitation depends on signals from the visceral endoderm and is a result of both programmed cell death and selective cell survival (7). Therefore, the interplay of death and survival signals might be disturbed in ATF1+/− CREB−/− embryos, leading to excessive apoptosis.
CREB, CREM, and ATF1 exhibit similar functions in different cellular systems.
The results presented here reveal the importance of the CREB/CREM/ATF1 family of transcription factors in the maintenance of cell viability in vivo, which is in line with studies using dominant-negative CREB (13, 24, 25, 31). It seems that cell survival in a variety of tissues and at various stages of development is dependent on which proteins of the CREB/CREM/ATF1 family are present. Here we show that, during early mouse development, only CREB and ATF1 play a major role in cell survival, since CREM is not expressed. CREM alone, on the other hand, was shown to be involved in the survival specifically of male germ cells (3, 16), where it is the only member of the family expressed in postmitotic spermatids. In addition, the notion that CREB, CREM, and ATF1 are important for survival is strongly supported by the findings which show that CREB and CREM together are crucial for cell survival in neurons of the central nervous system, where ATF1 is not expressed (T. Mantamadiotis and T. Lemberger, unpublished data). Whether the involved signaling pathways are the same in the different cellular systems has to be elucidated. The exact signaling cascades leading to activation of ATF1 and CREB and the mechanisms by which CREB/CREM/ATF1 family members support cell survival are also not known. Analysis of target genes that are significantly affected in our animal model will possibly reveal some of the survival factors which CREB and ATF1 control during early mouse development.
Acknowledgments
We are grateful to Sergei Arsenian for supplying the protocol for the PCR analysis of materials from sections, to Hans Schöler for the support with a plasmid containing oct-4 cDNA, and to Steven Hinrichs for providing us the MAb5 antibody. We also thank Dagmar Bock for help with in situ hybridization experiments; Annette Klewe-Nebenius for ES cell injection; and Erich Greiner, Theo Mantamadiotis, and Stefan Berger for critically reading the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft through SFB 601, the Fonds der Chemischen Industrie, the Alexander von Humboldt-Stiftung through the Max-Planck-Forschungspreis für Internationale Kooperation 1998, and the Volkswagen-Stiftung.
REFERENCES
- 1.Ang, S. L., and J. Rossant. 1994. HNF-3 beta is essential for node and notochord formation in mouse development. Cell 78:561-574. [DOI] [PubMed] [Google Scholar]
- 2.Barnes, J. D., J. L. Crosby, C. M. Jones, C. V. Wright, and B. L. Hogan. 1994. Embryonic expression of Lim-1, the mouse homolog of Xenopus Xlim-1, suggests a role in lateral mesoderm differentiation and neurogenesis. Dev. Biol. 161:168-178. [DOI] [PubMed] [Google Scholar]
- 3.Blendy, J. A., K. H. Kaestner, G. F. Weinbauer, E. Nieschlag, and G. Schutz. 1996. Severe impairment of spermatogenesis in mice lacking the CREM gene. Nature 380:162-165. [DOI] [PubMed] [Google Scholar]
- 4.Bonni, A., A. Brunet, A. E. West, S. R. Datta, M. A. Takasu, and M. E. Greenberg. 1999. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science 286:1358-1362. [DOI] [PubMed] [Google Scholar]
- 5.Bosilevac, J. M., R. J. Olsen, J. A. Bridge, and S. H. Hinrichs. 1999. Tumor cell viability in clear cell sarcoma requires DNA binding activity of the EWS/ATF1 fusion protein. J. Biol. Chem. 274:34811-34818. [DOI] [PubMed] [Google Scholar]
- 6.Chai, N., Y. Patel, K. Jacobson, J. McMahon, A. McMahon, and D. A. Rappolee. 1998. FGF is an essential regulator of the fifth cell division in preimplantation mouse embryos. Dev. Biol. 198:105-115. [DOI] [PubMed] [Google Scholar]
- 7.Coucouvanis, E., and G. R. Martin. 1995. Signals for death and survival: a two-step mechanism for cavitation in the vertebrate embryo. Cell 83:279-287. [DOI] [PubMed] [Google Scholar]
- 8.Crossley, P. H., and G. R. Martin. 1995. The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development 121:439-451. [DOI] [PubMed] [Google Scholar]
- 9.Du, K., and M. Montminy. 1998. CREB is a regulatory target for the protein kinase Akt/PKB. J. Biol. Chem. 273:32377-32379. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh, A., and M. E. Greenberg. 1995. Distinct roles for bFGF and NT-3 in the regulation of cortical neurogenesis. Neuron 15:89-103. [DOI] [PubMed] [Google Scholar]
- 11.Ginty, D. D., A. Bonni, and M. E. Greenberg. 1994. Nerve growth factor activates a Ras-dependent protein kinase that stimulates c-fos transcription via phosphorylation of CREB. Cell 77:713-725. [DOI] [PubMed] [Google Scholar]
- 12.Hogan, B. L. M., R. S. P. Beddington, F. Constantini, and E. Lacy. 1994. Manipulating the mouse embryo. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 13.Jean, D., M. Harbison, D. J. McConkey, Z. Ronai, and M. Bar-Eli. 1998. CREB and its associated proteins act as survival factors for human melanoma cells. J. Biol. Chem. 273:24884-24890. [DOI] [PubMed] [Google Scholar]
- 14.Jean, D., C. Tellez, S. Huang, D. W. Davis, C. J. Bruns, D. J. McConkey, S. H. Hinrichs, and M. Bar-Eli. 2000. Inhibition of tumor growth and metastasis of human melanoma by intracellular anti-ATF-1 single chain Fv fragment. Oncogene 19:2721-2730. [DOI] [PubMed] [Google Scholar]
- 15.Klemm, D. J., W. J. Roesler, T. Boras, L. A. Colton, K. Felder, and J. E. Reusch. 1998. Insulin stimulates cAMP-response element binding protein activity in HepG2 and 3T3-L1 cell lines. J. Biol. Chem. 273:917-923. [DOI] [PubMed] [Google Scholar]
- 16.Nantel, F., L. Monaco, N. S. Foulkes, D. Masquilier, M. LeMeur, K. Henriksen, A. Dierich, M. Parvinen, and P. Sassone-Corsi. 1996. Spermiogenesis deficiency and germ-cell apoptosis in CREM-mutant mice. Nature 380:159-162. [DOI] [PubMed] [Google Scholar]
- 17.Pende, M., T. L. Fisher, P. B. Simpson, J. T. Russell, J. Blenis, and V. Gallo. 1997. Neurotransmitter- and growth factor-induced cAMP response element binding protein phosphorylation in glial cell progenitors: role of calcium ions, protein kinase C, and mitogen-activated protein kinase/ribosomal S6 kinase pathway. J. Neurosci. 17:1291-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rassoulzadegan, M., Y. Yang, and F. Cuzin. 1998. APLP2, a member of the Alzheimer precursor protein family, is required for correct genomic segregation in dividing mouse cells. EMBO J. 17:4647-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riccio, A., S. Ahn, C. M. Davenport, J. A. Blendy, and D. D. Ginty. 1999. Mediation by a CREB family transcription factor of NGF-dependent survival of sympathetic neurons. Science 286:2358-2361. [DOI] [PubMed] [Google Scholar]
- 20.Rosner, M. H., M. A. Vigano, K. Ozato, P. M. Timmons, F. Poirier, P. W. Rigby, and L. M. Staudt. 1990. A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature 345:686-692. [DOI] [PubMed] [Google Scholar]
- 21.Rudolph, D., A. Tafuri, P. Gass, G. J. Hammerling, B. Arnold, and G. Schutz. 1998. Impaired fetal T cell development and perinatal lethality in mice lacking the cAMP response element binding protein. Proc. Natl. Acad. Sci. USA 95:4481-4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaywitz, A. J., and M. E. Greenberg. 1999. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu. Rev. Biochem. 68:821-861. [DOI] [PubMed] [Google Scholar]
- 23.Sheng, M., M. A. Thompson, and M. E. Greenberg. 1991. CREB: a Ca(2+)-regulated transcription factor phosphorylated by calmodulin-dependent kinases. Science 252:1427-1430. [DOI] [PubMed] [Google Scholar]
- 24.Somers, J. P., J. A. DeLoia, and A. J. Zeleznik. 1999. Adenovirus-directed expression of a nonphosphorylatable mutant of CREB (cAMP response element-binding protein) adversely affects the survival, but not the differentiation, of rat granulosa cells. Mol. Endocrinol. 13:1364-1372. [DOI] [PubMed] [Google Scholar]
- 25.Struthers, R. S., W. W. Vale, C. Arias, P. E. Sawchenko, and M. R. Montminy. 1991. Somatotroph hypoplasia and dwarfism in transgenic mice expressing a non-phosphorylatable CREB mutant. Nature 350:622-624. [DOI] [PubMed] [Google Scholar]
- 26.Tan, Y., J. Rouse, A. Zhang, S. Cariati, P. Cohen, and M. J. Comb. 1996. FGF and stress regulate CREB and ATF-1 via a pathway involving p38 MAP kinase and MAPKAP kinase-2. EMBO J. 15:4629-4642. [PMC free article] [PubMed] [Google Scholar]
- 27.Walker, W. H., L. Fucci, and J. F. Habener. 1995. Expression of the gene encoding transcription factor cyclic adenosine 3′,5′-monophosphate (cAMP) response element-binding protein (CREB): regulation by follicle-stimulating hormone-induced cAMP signaling in primary rat Sertoli cells. Endocrinology 136:3534-3545. [DOI] [PubMed] [Google Scholar]
- 28.Wilkinson, D. G., S. Bhatt, and B. G. Herrmann. 1990. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature 343:657-659. [DOI] [PubMed] [Google Scholar]
- 29.Wilkinson, D. G., and M. A. Nieto. 1993. Detection of mRNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 225:361-373. [DOI] [PubMed] [Google Scholar]
- 30.Winnier, G., M. Blessing, P. A. Labosky, and B. L. Hogan. 1995. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 9:2105-2116. [DOI] [PubMed] [Google Scholar]
- 31.Xie, S., J. E. Price, M. Luca, D. Jean, Z. Ronai, and M. Bar-Eli. 1997. Dominant-negative CREB inhibits tumor growth and metastasis of human melanoma cells. Oncogene 15:2069-2075. [DOI] [PubMed] [Google Scholar]