Abstract
Overexpression of the human multidrug resistance gene 1 (MDR1) is a negative prognostic factor in leukemia. Despite intense efforts to characterize the gene at the molecular level, little is known about the genetic events that switch on gene expression in P-glycoprotein-negative cells. Recent studies have shown that the transcriptional competence of MDR1 is often closely associated with DNA methylation. Chromatin remodeling and modification targeted by the recognition of methylated DNA provide a dominant mechanism for transcriptional repression. Consistent with this epigenetic model, interference with DNA methyltransferase and histone deacetylase activity alone or in combination can reactivate silent genes. In the present study, we used chromatin immunoprecipitation to monitor the molecular events involved in the activation and repression of MDR1. Inhibitors of DNA methyltransferase (5-azacytidine [5aC]) and histone deacetylase (trichostatin A [TSA]) were used to examine gene transcription, promoter methylation status, and the chromatin determinants associated with the MDR1 promoter. We have established that methyl-CpG binding protein 2 (MeCP2) is involved in methylation-dependent silencing of human MDR1 in cells that lack the known transcriptional repressors MBD2 and MBD3. In the repressed state the MDR1 promoter is methylated and assembled into chromatin enriched with MeCP2 and deacetylated histone. TSA induced significant acetylation of histones H3 and H4 but did not activate transcription. 5aC induced DNA demethylation, leading to the release of MeCP2, promoter acetylation, and partial relief of repression. MDR1 expression was significantly increased following combined 5aC and TSA treatments. Inhibition of histone deacetylase is not an overriding mechanism in the reactivation of methylated MDR1. Our results provide us with a clearer understanding of the molecular mechanism necessary for repression of MDR1.
Tumors become resistant to chemotherapy by a variety of mechanisms, including altered DNA repair, alterations in scavenging enzymes, and increased drug efflux. Although clinical drug resistance may be multifactorial, understanding these various mechanisms is important and may afford a means of improving treatment regimens. One of the most extensively studied mechanisms of multidrug resistance is associated with the overexpression of the MDR1 product, P glycoprotein (Pgp) (for reviews see references 27 and 55). A transmembrane protein, Pgp acts as an efflux pump, reducing intracellular drug levels and thus cytotoxic activity. We and others have demonstrated that the human MDR1 promoter is activated by changes in CpG methylation in B-cell chronic lymphocytic leukemia (26), acute myeloid leukemia (38), and tumor cell lines (12).
DNA hypermethylation is associated with transcriptional silencing and is accompanied by the accumulation of deacetylated histones on heterochromatin (13, 21). Methylation is often associated with the pathological silencing of tumor suppressor genes in human cancer and neurodevelopmental syndrome (20, 23). In contrast, transcriptionally active chromatin is characterized by the enrichment of hyperacetylated histone and is generally associated with hypomethylated chromatin (2, 34). The mechanisms underlying the correlation between DNA methylation and histone deacetylation in the control of gene expression have been established by recent biochemical experiments (24, 42). Evidence has emerged that a family of methyl-CpG binding proteins binds heterochromatin to stably repress transcription (10, 18). The transcriptional repressor methyl-CpG binding protein 2 (MeCP2 ) is the best-characterized family member (18, 41). MeCP2 is typically associated on heterochromatin in a methylation-dependent manner by its methyl-binding domain (MBD) and can displace histone H1 for nucleosome binding, indicating that it can dynamically interact with assembled chromatin (30, 40). The transcriptional-repression domain of MeCP2 recruits the corepressors mSin3 and histone deacetylases (HDACs) (24, 42), causing transcriptional repression (41). Moreover, repression by MeCP2 is partially overcome by the incubation of cells with the HDAC inhibitor trichostatin A (TSA).
Four other methyl-CpG binding proteins have been identified and are related to MeCP2 by virtue of their MBD motif (18). MBD1 is an integral chromosomal protein (45) and contrary to previous reports is not part of the MeCP1 repressor complex (10). MBD1 is a methylation-dependent transcriptional repressor that forms a complex with HDAC (45). MBD2 is associated with HDAC1 in the MeCP1 corepressor complex (4) and induces transcriptional repression dependent upon histone deacetylation (43). MBD3 also associates with HDAC and is a subunit of the Mi-2 complex (59), which also include Mta1-like, p66, Rpd3, and RbA p48/p46 (57, 58). These findings are consistent with the concept that CpG methylation represses gene activity by the formation of specialized chromatin. Although MeCP2, MBD1, MBD2/MeCP1, and MBD3 recruit HDAC corepressor complex, there are a number of features that distinguish each MBD protein (18, 35, 39, 45). Sparsely methylated genes do not provide strong ligands for MBD2/MeCP1 and therefore cannot fully repress transcription (5). MeCP2, on the other hand, can bind to a single symmetrical methyl-CpG, and repression is most striking around the density of 1 methyl-CpG pair per 100 bp (39, 41). These differences taken together with the distinct HDAC complexes associated with MBD1, MBD2, and MBD3 suggest that the mechanism by which these proteins silence transcription is different from that elucidated for MeCP2 (43, 45, 59).
The finding that CpG methylation of the MDR1 promoter results in transcriptional silencing supports a model in which methyl-CpG binding proteins might be involved in transcriptional control. To define the mechanism of silencing, we used chromatin immunoprecipitation (ChIP) to monitor the determinants of remodeling of MDR1 chromatin concomitant with activation using inhibitors of DNA methyltransferase and HDAC. One biological consequence of CpG methylation is the silencing of MDR1, which correlates with binding of MeCP2 to the promoter. Although the inhibition of the HDAC activity by TSA results in the accumulation of hyperacetylated core histones, this does not activate transcription unless chromatin is also demethylated and MeCP2 is released. These experiments provide a definitive demonstration of the hierarchy of DNA methylation relative to histone deacetylation. Transcriptional control is managed by two mechanisms; when densely methylated, MDR1 is transcriptionally silent by a mechanism that is TSA independent. Upon demethylation, activation of MDR1 is mediated by HDAC. Our cell line system provides a valuable model for understanding the molecular mechanisms involved in MDR1 transcriptional regulation.
MATERIALS AND METHODS
Cell culture and drug treatments.
CEM cells were grown in RPMI 1640 medium (Sigma) supplemented with 10% fetal calf serum (Trace), l-glutamine (2 mM), and 1× gentamicin at 37°C in a 5% CO2 atmosphere. CEM-A7R was established from the CEM-A7 cell line in the absence of doxorubicin (66). Cells were split to low density 18 h before drug insult. Cells were treated with TSA (Upstate Biotechnology; 10 μg in ethanol) or 5-azacytidine (5aC) (Sigma; 100 mM in complete culture medium) for approximately 3 days. For analysis of MDR1 transcription and bisulfite sequencing, TSA was added to cells at 50 ng/ml and 5aC was added at 1 μM for 32 h followed by two additional doses for 16 h, respectively. For the combination 5aC-TSA cocktail, cells were treated with a single dose of 5aC for 32 h followed by two additional doses of 1 μM 5aC and 50 ng of TSA per ml for 16 h. For the analysis of acetyl-H3, acetyl-H4, and HDAC-1 ChIPs, CEM-CCRF cells were treated with TSA (100 ng/ml), 5aC (1 μM), or a combination cocktail of 5aC (1 μM) and TSA (100 ng/ml) for 8 h or 3 days. For ChIP with methyl-CpG binding proteins, CEM-CCRF cells were treated with TSA (50 ng/ml with two additional doses of 50 ng/ml for 16 h) or 5aC (1 μM for 32 h with two additional doses of 0.5 μM for 16 h). For the combination 5aC-TSA treatment analysis, cells were incubated with 5aC (1 μM) for 32 h with two additional doses of 5aC (0.5 μM) and TSA (50 ng/ml) for 16 h.
Antibodies and Western blot analysis.
Antibodies against MBD1, MBD2, MBD3, and MBD4 were purchased from Santa Cruz. Anti-MeCP2 and -MBD2b antibodies have been described (24, 59). Antibodies against HDAC-1 and acetylated histones H3 and H4 were purchased from Upstate Biotechnology. Protein samples were size fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nylon membrane, and Western blotting was performed by using standard techniques.
Bisulfite genomic sequencing.
Genomic DNA was prepared from CEM cells and bisulfite conversion of genomic DNA was carried out following the protocol developed and described by Clark and coworkers (7). Bisulfite-treated DNA was then desalted, eluted in water, and desulfonated in the presence of 0.3 M NaOH at 37°C for 45 min. The DNA solution was neutralized in the presence of ammonium acetate. The bisulfite-treated DNA was prepared in water for hot-start PCR amplification. Experimental PCR protocols and DNA primer sequences are available upon request. Amplification products were cloned into pGEM-T vector as recommended by the manufacturer (Promega). Plasmid clones were grown and DNA was isolated with a plasmid purification kit (Qiagen) and cycle sequenced by using dye terminator chemistry. The cycling parameters were 1 cycle of 95°C for 30 s followed by 25 cycles of 95°C for 30 s, 50°C for 15 s, and 60°C for 4 min. Microspin G-50 columns (Amersham Pharmacia Biotech) were used in the removal of dye-labeled nucleotides, and the samples were air dried and analyzed by automated sequencing (Automated DNA Analysis Facility, School of Biochemistry and Molecular Biology, University of New South Wales, Sydney, Australia).
RNA and RT-PCR.
Cells were washed twice in 1× phosphate-buffered saline, and cell numbers were normalized. All manipulations were performed on ice. RNA was prepared with guanidine isothiocyanate solution followed by silica gel membrane purification following the protocol recommended by the manufacturer (Qiagen). RNA was eluted with RNase-free water, quantified at an optical density at 260/280 nm, and normalized for first-strand cDNA synthesis. Reverse transcription (RT) was performed with a first-strand cDNA kit following the protocol recommended by the manufacturer (Life Technologies). For analysis of MDR1 in CEM cells, RT-PCR was performed with H3.3 primers (yielding a 215-bp product and corresponding to exon 2 [positions 282 to 301] and exon 3 [476 to 495]) and MDR1 (yielding a 157-bp product and corresponding to exon 21 [2596 to 2615] and exon 22 [2733 to 2752]) and have been described (61). The cycling parameters consisted of 95°C for 60 s, followed by 32 cycles at 95°C for 60 s, 58°C for 60 s, and 72°C for 60 s and 1 cycle at 72°C for 10 min. RT-PCR products were then combined size fractionated by PAGE, and visualized by autoradiography.
ChIP.
CEM-CCRF and CEM-A7R cells were treated with TSA, 5aC, or 5aC-TSA as described earlier. ChIP analysis was performed following the instructions recommended by the supplier of acetyl-H3, acetyl-H4, and HDAC-1 antibodies (Upstate Biotechnology) with some modifications. In brief, 2.4 × 106 cells were used in each ChIP. Cells were pelleted by centrifugation for 5 min at 2,000 rpm and resuspended in 1× phosphate-buffered saline for a total of two washes. Cells were harvested, and proteins were cross-linked to DNA by adding formaldehyde to a final concentration of 1.0% for 10 min at room temperature. Samples were pelleted and resuspended in 0.6 ml of SDS lysis buffer with the addition of complete protease inhibitor cocktail tablets (Roche Molecular Biochemicals). Formaldehyde-fixed cells were allowed to settle on ice for 10 min. Cells were spun down by brief centrifugation, and the supernatant was carefully aspirated. Cross-linked cells were resuspended in 0.4 ml of SDS lysis buffer including protease inhibitors and sonicated to shear chromatin. Before the addition of antibody, samples were precleared with 60 μl of salmon sperm DNA-protein A agarose for 30 min at 4°C with agitation. The soluble chromatin fraction was collected, and 5 μl of anti-acetyl-H3 (residues 1 to 21), anti-acetyl-H4 (2 to 19), or anti-HDAC-1 (53 to 482) antibodies was added as recommended by the manufacturer. ChIPs for methyl-CpG binding proteins were performed with 10 μl of antibody to the transcriptional-repression domain of MeCP2 or 20 μl of MBD1, MBD2, MBD3, or MBD4 antibody, added to the respective samples and incubated overnight at 4°C with rotation. Immune complexes were collected with salmon sperm DNA-protein A-agarose or protein A/G-agarose (Santa Cruz), washed, and eluted with buffer (1% SDS, 0.1 M NaHCO3). Protein-DNA cross-links were reversed at 65°C overnight. Respective samples were treated with proteinase K, recovered by phenol-chloroform extraction, and ethanol precipitated. DNA pellets were washed in 70% ethanol, resuspended in water, and saved for competitive PCR amplification as previously described (14). Heterologous DNA competitors were designed based on MDR1 for competitive PCR and used as an internal reference for quantitation. Amplification reaction mixtures included equimolar amounts of DNA standards for competitive analysis of MDR1 intron 1 (296 to 595 relative to the transcription initiation site) with the primers 5′-TCAGGAGCTCCTGGAGCAGC-3′ (sense) and 5′-GAGCGCCCGCCGTTGATGCC-3′ (antisense). The MDR1 DNA target is 300 bp long, and its competitor is 240 bp long. Primers were added at 50 pmol/reaction. The cycling parameters consisted of an initial denaturation at 95°C for 1 min, followed by 32 cycles of 95°C for 60 s, 58°C for 60 s, and 72°C for 60 s, with a final extension at 72°C for 10 min, on a GeneAmp PCR system 9700 version 2.25 (PE Applied Biosystems). Samples (30%, or 16 μl) of the PCR mixtures were removed, size fractionated by PAGE, and stained with 0.6 μg of ethidium bromide per ml. Quantitative analyses were performed with a MultiImage light cabinet and quantitated with ChemiImager 4400 low-light imaging system version 5.1 software (Alpha Innotech Corp).
MDR1 transcription and MeCP2 ChIP analyses in Xenopus laevis oocytes.
MDR1 plasmid was methylated in vitro with SssI methylase according to the manufacturer's instructions (New England Biolabs) and injected into Xenopus oocytes as described previously (24). For analysis of MDR1 transcription, a titration series (13.8 nl of DNA containing 0.25, 0.5, 1, and 2 ng/oocyte) of DNA was prepared and injected into 40 Xenopus oocyte nuclei. For TSA treatment, oocytes were pretreated with 100 ng of the HDAC inhibitor per ml for 6 h, collected, and injected with either unmethylated or methylated DNA. Oocytes were collected 16 h after injection to allow for chromatin assembly, homogenized and used for RNA isolation with RNAzol (Cinna Scientific). The RNA was treated with RQ1 RNase-free DNase following the instructions recommended by the supplier (Promega) and then redissolved in an appropriate volume of water for first-strand cDNA synthesis. Transcription was assayed using competitive RT-PCR. Primers used to amplify MDR1 correspond to exon 1 of MDR1 sense (10 to 30) and antisense (110 to 120) sequences: 5′-AGTAGCGGCTCTTCCAAGCTC-3′ and 5′-CGCTCCTTGGAACGGCCACCA-3′. The MDR1 cDNA target is 121 bp long, and its competitor is 160 bp long. Xenopus histone H4 RNA was used as an internal reference for cDNA synthesis; primers 5′-GGTCTGGGCAAAGGAGGCGC-3′ (sense) and 5′-GTCCCGGATAACGTTCTCCA-3′ (antisense) yield a 180-bp cDNA product. Competitive RT-PCR products were separated on an 8% polyacrylamide gel stained with ethidium bromide. For ChIP analysis, 40 oocytes were injected with 1.0 ng of unmethylated or SssI-methylated MDR1 plasmid. Oocytes were collected 16 h after injection and cross-linked with 1% formaldehyde (final concentration) for 10 min at room temperature. ChIPs were performed with 8 μl of full-length anti-MeCP2 antibody, immune complexes were collected, and DNA was quantified by competitive PCR.
RESULTS
5aC but not TSA changes the CpG methylation pattern of the MDR1 CpG island.
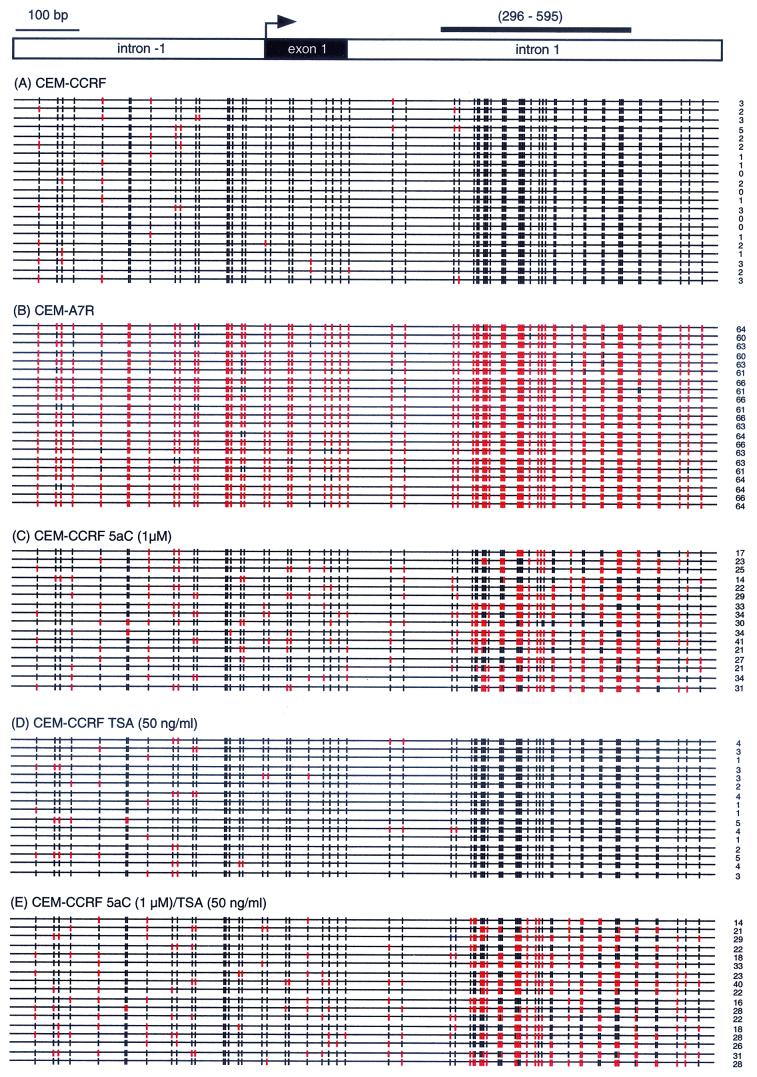
Transcriptional inactivity of MDR1 can be associated with histone deacetylation (22, 27) and DNA methylation in both cancer cell lines and clinical samples (26, 38). The CpG methylation status of the MDR1 promoter in the T-cell leukemia cell lines CEM-CCRF (drug sensitive) and CEM-A7R (drug resistant) was examined by bisulfite genomic sequencing. The MDR1 promoter (1.15 kb with 66 CpG dinucleotides) was hypermethylated in CEM-CCRF cells (Fig. 1A), in which MDR1 is transcriptionally silent. In contrast, the active MDR1 gene in Pgp-expressing CEM-A7R cells was hypomethylated (Fig. 1B). These results correlate well with our earlier observations that MDR1 gene expression is associated with differences in the methylation status of the promoter (26). Treatment of the CEM-CCRF cells with 5aC significantly reduced the level of CpG methylation in the MDR1 promoter (Fig. 1C) (P < 0.001, Student's paired t test), whereas TSA did not change the extent of CpG methylation (Fig. 1D). Furthermore, the combination of 5aC and TSA treatment in the CEM-CCRF cell line did not significantly extend the degree of CpG demethylation of the MDR1 promoter (Fig. 1E) (P < 0.26). Taken together, these results reveal that the core promoter of MDR1 in CEM-CCRF cells is hypermethylated, whereas in CEM-A7R the CpG island is hypomethylated. The results also show that only the demethylating agent 5aC and not the HDAC inhibitor TSA changes the CpG methylation pattern of the MDR1 promoter.
FIG. 1.
The MDR1 promoter is differentially methylated in MDR1− and MDR1+ CEM cells. The demethylating agent 5aC and not the HDAC inhibitor TSA changes the CpG methylation pattern of the MDR1 allele. The MDR1 CpG island was analyzed for DNA methylation status by bisulfite sequencing. Each row represents a single cloned allele, and the positions of methylated and unmethylated CpG sites are shown as black and red vertical lines, respectively. The number of unmethylated CpG sites are represented on the right of each sequenced allele. In total there are 66 CpG dinucleotides within the 1.15-kb region of the MDR1 promoter. (A) The MDR1 promoter is hypermethylated in CEM-CCRF cells. (B) The transcriptionally active promoter in CEM-A7R cells is hypomethylated. (C) 5aC treatment significantly reduces the number of methylated CpG sites in CEM-CCRF cells (P < 0.001). (D) In contrast, TSA treatment does not significantly change the number of methylated CpG sites. (E) Combined 5aC-TSA treatment does not further extend 5aC-induced demethylation pattern (P < 0.26). (Top) The thick black line corresponds to the region in intron 1 (296 to 595 relative to the transcription initiation site [→]) used in ChIP experiments (Fig. 5 to 7).
CpG methylation and HDAC operate in synergy to maintain the transcriptional silence of MDR1.
We have previously shown MDR1 can be reactivated, albeit at low levels, following DNA demethylation with 5aC and 5-aza-2′deoxycytidine (26). This allowed us to examine the link between DNA methylation and histone deacetylation in silencing MDR1 transcription. The Pgp-negative cell line CEM-CCRF (Fig. 2, lane 1) was treated with 5aC and/or TSA. In three separate experiments, TSA (50 ng/ml) treatment alone was insufficient to reactivate the silent allele (Fig. 2, lane 2) and MDR1 was not reactivated with any tested dose (10, 100, and 300 ng/ml) of TSA up to 5 days (data not shown). The HDAC inhibitor sodium butyrate also failed to reactivate MDR1 (data not shown). Treatment of cells with 5aC alone induced a small yet reproducible increase in MDR1 mRNA levels (Fig. 2, lane 3). However, treatment of CEM-CCRF cells with TSA following 5aC (1 μM) resulted in robust MDR1 gene expression (Fig. 2, lane 4) with a 10-fold induction of gene expression relative to 5aC alone. Kinetic studies demonstrated that transcriptional activation of MDR1 was first observed at 60 h after 5aC treatment and peaked at 72 h (data not shown).
FIG. 2.
CpG methylation is dominant in keeping MDR1 inactive. HDAC inhibition is insufficient to reactivate the normally silent hypermethylated MDR1 gene in drug-sensitive CEM-CCRF cells. Quantitative RT-PCR analysis shows that TSA (50 ng/ml) treatment of CEM-CCRF cells does not activate MDR1 transcription. Treatment with 5aC (1 μM) induces low-level reactivation of MDR1 compared to untreated CEM-CCRF cells. However, coincubation of 5aC-treated CEM-CCRF cells with TSA induces high levels of gene reactivation. For simplicity, the difference in MDR1 and H3.3 amplification yields after 32 PCR cycles is shown. In this example, amplification rates between MDR1 and H3.3 were determined to be equivalent in a series of PCR cycles. Results are averages of three experiments with standard deviations.
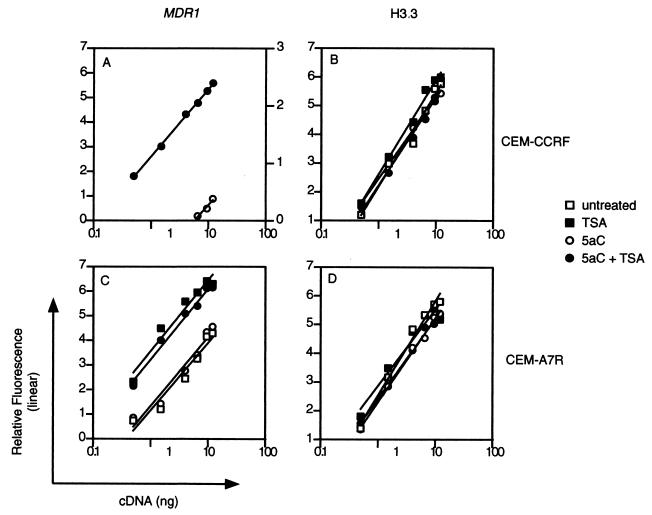
To accurately measure the level of MDR1 gene activation in a more quantitative fashion, we employed a quantitative PCR approach. The amplification kinetics of MDR1 was compared at a fixed number of PCR cycles to confirm linear phase amplification (Fig. 3A and C). The housekeeping gene encoding histone 3.3 (H3.3) was used as an internal reference (Fig. 3B and D). Figure 3A compares the levels of MDR1 mRNA by RT-PCR following 5aC and 5aC-TSA treatments in CEM-CCRF cells by examining amplification efficiencies on titrated cDNA. Regression analysis shows that the significance values (r2) in the amplification gradient were 0.95 and 0.99 for 5aC- and 5aC-TSA-treated cells, respectively. TSA alone did not activate MDR1 gene transcription and thus is not shown in Fig. 3A. These results indicate that in CEM-CCRF cells, the inhibition of CpG methylation and HDAC operate in synergy to maintain the silent state of MDR1 when hypermethylated.
FIG. 3.
Demethylating agent 5aC does not change endogenous MDR1 levels in drug-resistant CEM-A7R cells, but HDAC inhibition can upregulate gene transcription. A comparative kinetic analysis of H3.3 and MDR1 was done by cDNA titration. To test the linearity of amplification, 12 ng of cDNA was serially diluted to 0.5 ng and amplified for 32 cycles. Amplification kinetics were compared for MDR1 and H3.3 in CEM-CCRF and CEM-A7R cells. Amplification yields were plotted as relative fluorescence versus cDNA titration for untreated, TSA-treated, 5aC-treated, and 5aC-TSA-treated CEM-CCRF and CEM-A7R cells. Similar results were obtained in three independent experiments.
We believe that a condition of MDR1 transcriptional fitness is dependent on the CpG methylation status of the gene. If this explanation is correct, we reasoned that treatment of the MDR1-positive CEM-A7R cell line with HDAC inhibitors would further upregulate transcription from a hypomethylated promoter. This hypothesis follows from the observation that the MDR1 promoter in drug-resistant CEM-A7R cells is generally undermethylated (Fig. 1). We therefore quantitated MDR1 mRNA levels in Pgp-expressing CEM-A7R cells treated with TSA and/or 5aC (Fig. 3C). In three separate experiments, treatment of drug-resistant CEM-A7R cells with TSA increased MDR1 transcription measured by quantitative RT-PCR (Fig. 3C) (r2 = 0.94). Treatment with 5aC over 3 days failed to significantly upregulate transcription above that of untreated CEM-A7R cells (r2 = 0.95). Interestingly, combined treatments of 5aC and TSA did not increase MDR1 transcription above the levels determined for TSA treatment alone (r2 = 0.98), suggesting that histone deacetylation inhibition is sufficient to raise transcription levels when the MDR1 promoter is hypomethylated in CEM-A7R but insufficient when the promoter is hypermethylated in CEM-CCRF cells. These results suggest that the mechanism of MDR1 gene repression is methylation dependent and HDAC independent when the promoter is hypermethylated and HDAC dependent when the promoter is hypomethylated.
Expression of the family of methyl-CpG binding proteins.
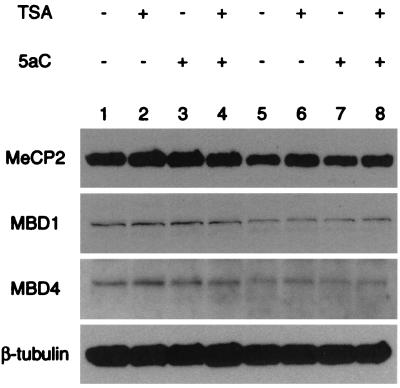
Previous work demonstrated that members of the methyl-CpG binding protein family, MBD1, MBD2 (MeCP1), MBD3, and MeCP2, mediate transcriptional repression and are present in distinct HDAC-containing complexes (24, 42, 43, 45). MBD4 is the exception and has been described as an endonuclease that forms a complex with the DNA mismatch-repair protein MLH1 (19, 48). In order to determine which MBD protein(s) might be bound to the methylated MDR1 promoter, we examined the expression of these proteins in CEM-CCRF and CEM-A7R cells. MeCP2 was strongly expressed in these cells (Fig. 4). MBD1 and MBD4 were also expressed, and MBD2 and MBD3 were undetectable (Fig. 4). Treatment of CEM-CCRF and CEM-A7R cells with TSA and/or 5aC for 3 to 5 days did not alter MeCP2, MBD1, or MBD4 protein expression levels. Polyclonal antisera raised against MBD2b (59) also failed to detect endogenous protein by Western blotting (data not shown).
FIG. 4.
The profile of methyl-CpG binding proteins MeCP2, MBD1, and MBD4 is not changed in cells treated with 5aC and/or TSA. CEM-CCRF and CEM-A7R cells lack the known methyl-CpG binding proteins MBD2 and MBD3. Shown are immunoblots of CEM-CCRF (lanes 1 to 4) and CEM-A7R (lanes 5 to 8) nuclear proteins probed with polyclonal antibodies against MeCP2, MBD1, and MBD4.
Based on the binding properties of MeCP2 (36) and MBD1 (18) we hypothesized that either could be associated with the MDR1 promoter. If this explanation was correct, we reasoned that demethylation with 5aC should result in the release of methyl-CpG binding proteins from MDR1. To test this hypothesis, we used a ChIP strategy on formaldehyde-fixed cells to quantitate the occupancy of endogenous methyl-CpG binding complexes on the MDR1 promoter.
Association with transcriptionally silent chromatin: MeCP2 but not MBD1 is released from the MDR1 promoter.
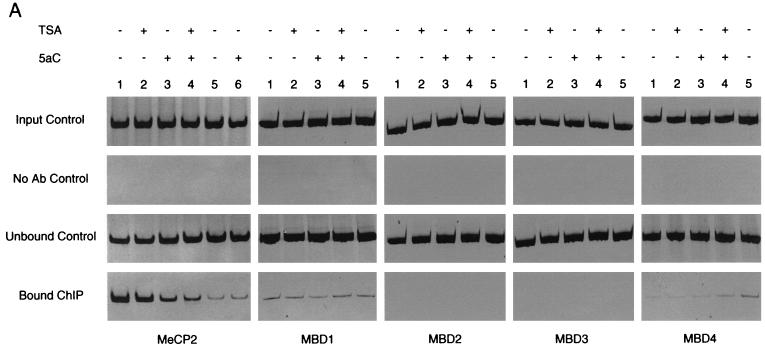
In order to compare enrichment of chromatin-associated proteins we employed a competitive PCR strategy to quantitate the ChIP assay. Competitive PCR was performed with heterologous DNA standards (240 bp) that differ in size from endogenous or target DNA (300 bp). This is the method of choice for PCR quantitation because heterologous competitors satisfy linear-phase kinetics, and as a result quantitation is independent of exponential-phase determination (14). Taking into account the number of cells used in the ChIP (before and after TSA and/or 5aC exposure) and the efficiency of chromatin sonication after cross-linking, we quantified protein association on the MDR1 promoter. Cleared chromatin solution (sonicated fragments were approximately 500 bp) were immunoprecipitated with a panel of antibodies specific for MeCP2, MBD1, MBD2, MBD3, and MBD4 (Fig. 5A) and quantified by competitive PCR (Fig. 5B).
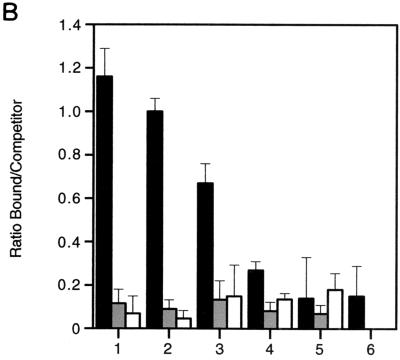
FIG. 5.
Profile of methyl-CpG binding proteins on MDR1 CpG island chromatin by ChIP. (A) Precipitous release of MeCP2 from the MDR1 promoter following treatment of CEM-CCRF cells with the demethylating agent 5aC and the HDAC inhibitor TSA. MBD2 and MBD3 are not engaged on methylated MDR1 chromatin. CEM-CCRF cells were treated with TSA (50 ng/ml), 5aC (1 μM for 32 h followed by two additional doses of 0.5 μM for 16 h), or a combination of 5aC and TSA (1 μM 5aC for 32 h with two additional doses of 0.5 μM 5aC and 50-ng/ml TSA for 16 h). CEM-A7R cells were treated with 5aC alone, as described above for CEM-CCRF cells. Controls show input genomic DNA before addition of antibody and unbound DNA after elution. Soluble chromatin used in immunoprecipitations had a typical size of ≤0.5 kb visualized by gel electrophoresis. Sonicated preparations above this threshold were not used for ChIP analysis. (B) Quantitation of MeCP2-, MBD1-, and MBD4-bound MDR1-immunoprecipitated DNA analyzed by competitive PCR. ChIPs were performed with CEM-CCRF (bars 1 to 4) or CEM-A7R (bars 5 to 6) cells as described for panel A. MeCP2 (black bar), MBD1 (grey bar), and MBD4 (white bar) localization was quantified by competitive PCR. MBD2 and MBD3 are not shown. Data were plotted as the ratio of bound MeCP2 to competitor DNA and expressed as relative fluorescence. Bars 1 and 5, untreated cells; bar 2, TSA-treated cells; bars 3 and 6, 5aC-treated cells; bar 4, 5aC-TSA-treated cells. Quantitation of MBD1 and MBD4 in 5aC-treated CEM-A7R cells was not performed (bar 6). Error bars show standard deviations of three independent tests.
In the first set of experiments, we analyzed MeCP2 association with the MDR1 promoter. Treatment of CEM-CCRF cells with 5aC alone leads to a modest yet consistent reduction in MeCP2 localization (Fig. 5). Surprisingly, the combination of TSA and 5aC treatment in CEM-CCRF cells resulted in a further reduction in MeCP2 binding. Treatment with TSA alone did not significantly reduce the degree of MeCP2 occupancy on the MDR1 promoter. There was little association of MeCP2 with MDR1 in CEM-A7R cells, and this is consistent with the hypomethylation status of the allele (Fig. 1). These data demonstrate that demethylation is associated with the release of MeCP2 from the MDR1 promoter. To test the specificity of MeCP2 binding on an unrelated gene, we analyzed MeCP2 localization on the deoxycytidine kinase (dCK) promoter, known to be unmethylated in CEM-CCRF cells (29). In untreated cells, MeCP2 does not localize on exon 1 of the dCK promoter (unpublished observation).
ChIP analysis shows a dramatic difference in the level of MBD1 and MBD4 chromatin association compared with that of MeCP2 (Fig. 5). Interestingly, levels of bound MBD1 and MBD4 on MDR1 chromatin are comparable to quantified MeCP2 levels in untreated CEM-A7R cells, and this might reflect the global binding nature of these proteins. Surprisingly, exposure of cells to 5aC did not affect the level of MBD1 association, in striking contrast to the measured level of MeCP2 enrichment on the endogenous promoter. PCR analysis did not detect MBD2 or MBD3 occupancy on the CpG island (Fig. 5A), and this is in agreement with our earlier observation that CEM-CCRF and CEM-A7R cells lack these methyl-CpG binding proteins. Neither MBD1 nor MBD4 was found to be associated with the unmethylated dCK promoter (unpublished observation). Taken together, these results demonstrate that MeCP2 association with MDR1 is dramatically reduced when the gene is demethylated. There is a differential pattern in methyl-CpG binding proteins on the promoter, and MeCP2 release appears to be independent of MBD1 and/or MBD4 loading.
Relationship between the release of MeCP2 and histone acetylation.
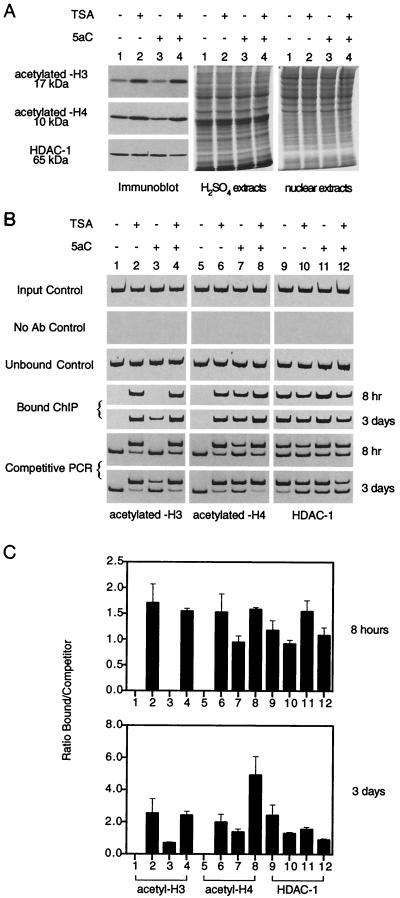
Next, we investigated whether treatment with 5aC might affect histone acetylation. Global histone acetylation was assessed with antibodies specific for the acetylated forms of these proteins. Figure 6A shows that histones H3 and H4 were hyperacetylated in CEM-CCRF cells treated with TSA but not with 5aC. Treatment of cells with the demethylating agent has been demonstrated to restore histone acetylation on methylation-mediated transcriptionally silent promoters (8), so we examined histone acetylation on the MDR1 promoter. To test this proposition, ChIPs with antibodies against acetyl-H3 and acetyl-H4 were performed in CEM-CCRF cells treated with TSA, 5aC, or 5aC-TSA for either 8 h or 3 days (Fig. 6B and 6C). TSA induced hyperacetylation of histones H3 (Fig. 6B, lanes 1 to 4) and H4 (lanes 5 to 8) compared to their respective untreated controls. 5aC treatment partially restored acetyl-H4-bound chromatin. 5aC-TSA treatment completely restored the amount of associated acetylated H3 and H4 on the MDR1 promoter. These results show that even in the absence of transcriptional activation, gene demethylation can induce local promoter-associated histone hyperacetylation.
FIG. 6.
MDR1 chromatin is enriched in acetylated histone following gene demethylation and HDAC inhibition. (A) Western blot analysis of acetyl-H3, acetyl-H4, and HDAC-1. HDAC inhibition by TSA results in hyperacetylation of H3 and H4. 5aC treatment does not alter the levels of global core histone acetylation. Acid and nuclear extracts were prepared from CEM-CCRF cells for acetylated-histone (acetyl-H3 and -H4, 17 and 10 kDa) and HDAC-1 (65 kDa) detection, respectively. (B) Histone acetylation enrichment on MDR1 chromatin. ChIPs were performed with CEM-CCRF cells treated with TSA (100 ng/ml), 5aC (1 μM), or a combination for 8 h or 3 days. Associated acetylated H3 and H4 histones analyzed by competitive PCR are also shown. Controls show input genomic DNA before addition of antibodies and unbound DNA after elution. Soluble chromatin used in immunoprecipitations had a typical size of ≤0.5 kb visualized by gel electrophoresis. Sonicated preparations above this threshold were not used for ChIP analysis. (C) ChIP profile of acetylated/deacetylated histones on MDR1 chromatin. Competitive PCR was used to aid in quantitation. ChIPs were performed with CEM-CCRF cells as described above (Fig. 5), and relative localization was quantified. Quantitation was plotted as a mean ratio of bound antibody DNA to competitor DNA after 8 h or 3 days of treatment and expressed as relative fluorescence. Bars 1, 5, and 9, untreated cells; bars 2, 6, and 10, TSA-treated cells; bars 3, 7, and 11, 5aC-treated cells; bars 4, 8, and 12, 5aC-TSA-treated cells. Error bars show the standard deviations of three independent tests.
MeCP2 has been proposed to interact with the transcriptional corepressors mSin3A and HDAC to direct a large component of gene repression in model systems (24, 42). Several lines of evidence suggest that MeCP2 is involved in methylation-dependent, dominant silencing of MDR1. First, there is an inverse correlation between MeCP2 localization and transcriptional activity of MDR1 in CEM-CCRF and CEM-A7R cells. Second, 5aC reduces DNA methylation and the association with MeCP2 but not MBD1. Third, TSA treatment induces significant acetylation of H3 and H4 histones but does not activate MDR1 transcription. This suggests that histone deacetylation is not an overriding mechanism, a phenomenon previously reported for FMR1 transcriptional silencing in fragile X cells (8). Therefore, we addressed the question of whether direct HDAC-1 association was affected after demethylation and acetylation of MDR1 chromatin. Cells treated with TSA consistently show reduced HDAC-1 binding on the promoter (Fig. 6C, compare lanes 9 and 10). Treatment with TSA alone for 3 days has a greater effect on HDAC-1 binding than MeCP2 (Fig. 5). These results are consistent with the conclusion that HDAC-1 is brought to the promoter through association with MeCP2 on hypermethylated chromatin (24, 42).
MeCP2 is enriched on methylated MDR1 chromatin in X. laevis oocytes.
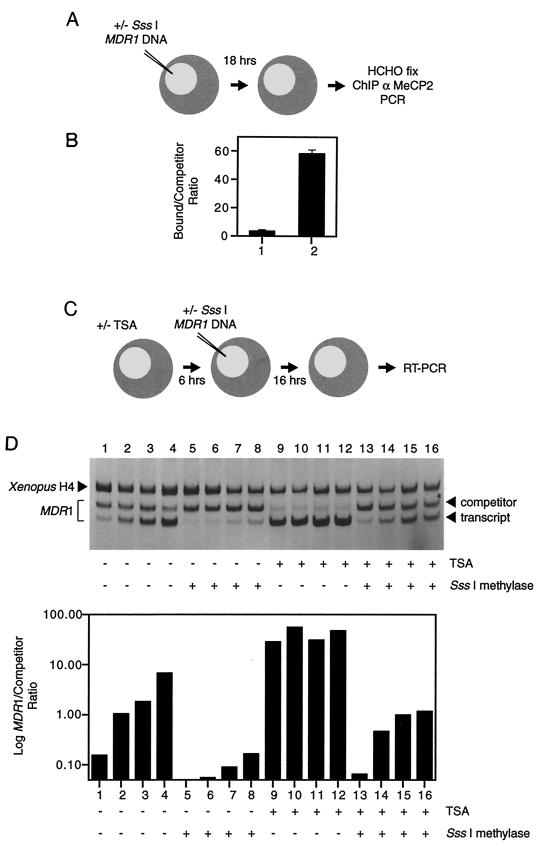
We next examined the molecular mechanisms of transcriptional silencing on the methylated MDR1 promoter using a X. laevis oocyte model system in order to investigate the generality of our observations. Sequence comparison of Xenopus MeCP2 reveals that it is very similar to mammalian MeCP2 proteins, exists as a complex with HDAC, and stably associates with methylated nucleosomal DNA to repress transcription (24). We microinjected SssI-methylated and unmethylated MDR1 DNA templates into frog oocytes and examined MeCP2 association and transcription from the MDR1 promoter. Following a ChIP assay using anti-MeCP2 antibody, immunopurified DNA was analyzed by competitive PCR. The association of MeCP2 with MDR1 was enhanced more than 11-fold following DNA methylation compared to unmethylated DNA (Fig. 7A). In addition, treatment with TSA did not release MeCP2 from methylated chromatin (unpublished observation). We have not detected Xenopus MBD1 in association with methylated templates under these conditions (data not shown). The differential association of MeCP2 with methylated versus unmethylated MDR1 constructs in Xenopus allowed a direct analysis of the consequence of MeCP2 association for chromatin remodeling and transcriptional repression.
FIG. 7.
ChIP of MeCP2 and transcription of MDR1 in X. laevis oocytes. (A) Strategy to immunoprecipitate chromatin-associated MeCP2 onto the MDR1 promoter in X. laevis oocytes. SssI-methylated and unmethylated MDR1 DNAs were injected into oocyte nuclei and allowed to assemble chromatin. Oocytes were collected for ChIP 16 h after DNA injection. (B) Binding affinity of MeCP2 is dependent on the CpG methylation status of MDR1 chromatin. Xenopus oocytes were microinjected (1 ng of template/oocyte) with unmethylated (lane 1) or SssI-methylated (lane 2) plasmid. Oocytes were collected 16 h after injection, and proteins were cross-linked with formaldehyde. MeCP2 was immunoprecipitated from soluble chromatin following sonication. Competitive PCR analysis was used to detect and quantify MeCP2-immunoprecipitated DNA. Error bars show the standard deviations of competitive PCR quantitation of three independent tests. (C) Strategy to investigate the role of methylation-mediated transcriptional repression of MDR1 by inhibiting HDAC. Oocytes were pretreated with TSA for 6 h. Transcription of the MDR1 promoter was analyzed 16 h after injection by RT-PCR. (D) CpG methylation has the capacity to silence MDR1 transcription, and TSA can partially alleviate this repression. A titrated series (0.25, 0.5, 1, and 2 ng/oocyte) of unmethylated and SssI-methylated MDR1 DNA was injected into Xenopus oocytes. Transcription was assayed by competitive RT-PCR, and Xenopus histone H4 served as an internal control. Similar results were obtained in three independent experiments.
CpG methylation is dominant in silencing MDR1 transcription in X. laevis oocytes.
Methylation-mediated transcriptional repression was assayed in frog oocytes with the HDAC inhibitor TSA. Microinjection of a titrated (1:2) series of unmethylated MDR1 DNA into Xenopus oocyte nuclei results in strong transcription after a 16-h incubation period to allow chromatin assembly. Transcription of MDR1 is repressed more than 10-fold when completely methylated templates are injected, relative to Xenopus histone H4 (Fig. 7B, compare lanes 1 to 4 with lanes 5 to 8). TSA (100 ng/ml) enhanced MDR1 gene transcription of the unmethylated templates more than 50-fold (Fig. 7B, lanes 9 to 12) compared to untreated oocytes. Methylation-dependent transcriptional repression was only partially relieved when oocytes were incubated with TSA in a dose-dependent manner (Fig. 7B, compare lanes 5 to 8 with lanes 13 to 16; the difference in transcriptional activity is between two- and fourfold), suggesting that transcriptional competence of MDR1 is managed by CpG methylation and silencing cannot be alleviated by the inhibition of HDAC. These results are consistent with our in vivo transcriptional work in the CEM-CCRF cell line model (Fig. 2). Under similar conditions, addition of 5aC does not lead to DNA demethylation in oocytes because templates do not replicate and has very little effect on transcription either alone or in combination with TSA (unpublished observation). Thus, CpG methylation is dominant in silencing MDR1 transcription through recruitment of MeCP2, and whereas the inhibition of HDAC by TSA allows the remodeling of chromatin when the template is unmethylated to facilitate transcription, the efficiency of the process is significantly reduced when chromatin is methylated. These results provide strong in vivo evidence in a distinct system for the repressive functions of MDR1 promoter methylation independent of the recruitment of HDAC. Figure 8 summarizes the molecular mechanisms involved in this phenomenon.
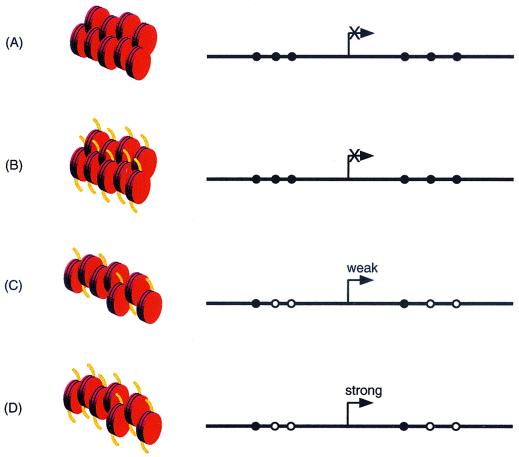
FIG. 8.
Molecular mechanisms by which methylation might facilitate transcriptional silencing of MDR1 by MeCP2 repressor. (A) Methylation-dependent silencing of MDR1 transcription. ChIP analysis shows that MeCP2 repressor complex is associated with the promoter (closed circles, methylated CpGs; open circles, unmethylated CpG sites). MeCP2 assembly on the MDR1 promoter forms a more stable repressive chromatin structure. Basal transcriptional machinery and RNA polymerase are unable to gain access to the promoter, and the consequence is strong repression. Although not shown, chromatin fragments containing MBD1 and MBD4 immunoprecipitates show less abundant association with the methylated MDR1 promoter. Organization of nucleosome particles is shown in red. (B) The inability of TSA to unlock repression caused by methylation is confirmed by MeCP2 localization. The HDAC inhibitor TSA cannot relieve transcriptional silencing, suggesting that when the MDR1 promoter is hypermethylated, repression is independent of HDAC activity (acetylated histones tails are shown in yellow). (C) Demethylation of the MDR1 promoter by 5aC reduces the silencing power of methylation by a two-step process. MeCP2 repressor complex is quantitatively released from the MDR1 promoter, and this is accompanied by incomplete acetylation of histones H3 and H4. Transcription of MDR1 is activated, albeit weakly. (D) Transcription of MDR1 is strongest in cells treated with TSA-5aC. Effective histone acetylation complexes and stable transcription complexes replace the MeCP2 corepressor. The assembly of nucleosomes is less compact, allowing transcription factor access. The promoter appears to function similarly to TSA-inducible, unmethylated promoter, such as that in MDR1-positive CEM-A7R cells.
DISCUSSION
We originally observed that methylation of MDR1 was inversely correlated with transcription; however, the molecular events that switch on gene expression in MDR1-negative cells remained unclear. We now extend these results and establish that the biological consequence of CpG methylation is the silencing of MDR1 through the formation of repressive chromatin. We find that the combination of 5aC and TSA leads to the precipitous release of MeCP2 and the staged enrichment of acetyl-H3 and acetyl-H4 concomitant with transcriptional activation.
Model of MDR1 transcriptional regulation.
The cell line model system described here allowed a coordinated approach to examine transcriptional regulation of MDR1 using inhibitors of HDAC and DNA methylation. We have demonstrated that methylation exerts a repressive influence on MDR1 chromatin. These results are consistent with the observations that histone deacetylation can reactivate gene expression only after demethylation (6). Interestingly, Coffee and colleagues showed that TSA treatment was unable to fully restore H3 acetylation on the methylated FMR1 promoter by ChIP (8). We clearly demonstrate that hyperacetylation of both H3 and H4 histone is not an overriding mechanism in transcriptional reactivation. Repressor complex association is most likely gene specific and would be influenced by methylation status, histone acetylation-deacetylation patterns, and transcription factor requirements for each promoter. For example, MBD2 is selectively associated with transcriptionally silent p14/16 promoters in human colon carcinoma cells, and this is independent of MeCP2 chromatin association (33), whereas MeCP2 is reported to be involved in repression of the imprinted genes Snrpn and U2af1-rs1 (17) and proviral silencing (31).
We provide evidence that 5aC will cause dynamic H3 and H4 acetylation on the MDR1 promoter (presumably a consequence of MeCP2-HDAC release following demethylation), although accumulation of these histones is lower than with TSA treatment. It is not surprising that 5aC alone causes minor changes in transcription (26), because hyperacetylation is necessary to achieve expression. Genome-wide location analysis will provide valuable information on how widespread acetylation occurs following HDAC inhibition.
The most direct mechanism by which MeCP2 represses MDR1 transcription would be to directly prevent the binding and assembly of basal transcriptional machinery to the promoter (25). In our model system, transcriptional competence is correlated with MeCP2 release from the promoter, and under the same conditions this does not significantly activate transcription unless an effective histone acetylation complex is introduced. MeCP2 could interfere with transcription complex assembly or utilization by RNA polymerase through contact with the basal transcriptional machinery, as seen with Sin3 (37, 65). Considering that methylated DNA has the capacity to direct the assembly of repressive chromatin (24), we also believe that MeCP2 could sequester methylation specific and transcriptionally repressive chromatin to an inactive nuclear matrix (64).
The methyl-CpG density is an important parameter in gene silencing (5). Experiments using MeCP2 show that transcription is dependent on methylation but not linearly related to methyl-CpG density. Transcriptional repression is most striking around a density of 1 methyl-CpG pair per 100 bp, while methylation below this threshold has little result (41). In the model we present, a fully methylated MDR1 promoter has on average 5 to 6 methyl-CpG pairs per 100 bp. This methylation density would presumably ensure enough MeCP2 binding sites to assemble repressive chromatin. Interestingly, cells treated with 5aC show an approximately twofold reduction in methyl-CpG frequency over the promoter, yet this is correlated with a small increase in MDR1 gene transcription (Fig. 1 and 2). This encourages the view that release of MeCP2 from specific regions of the promoter may be necessary for transcriptional reactivation. Using our cell model system and ChIP assay we can begin to address these pressing questions.
We have discussed methylation-mediated transcriptional repression; however, we must emphasize there are examples that suggest that MDR1 can be reactivated solely by HDAC inhibitors in SW620 colon cancer cells (22). In light of our results, how do we explain these data? It is worth considering the endogenous methylation status of MDR1 in this model rather than focusing on sequence-specific DNA-binding proteins as an endpoint in negotiating transcription. Bisulfite genomic sequencing performed in our laboratory with SW620 cells has revealed the MDR1 promoter is naturally undermethylated and associated with acetylated histones. Inhibition of HDAC by TSA is sufficient in upregulating MDR1 transcription, a phenomenon similar to that described for the hypomethylated gene in CEM-A7R cells (unpublished data).
Inhibition of HDAC activity is necessary but insufficient to activate transcription.
With the ever-growing number of HDACs in the class II family, it seems likely that other HDAC complexes may operate either independently of or in association with the MeCP2 repressor. Further work is needed to discriminate whether other HDACs are involved in silencing or able to complement the inhibition of deacetylated histones by TSA. In our experiments, TSA alone has no functional effect on MDR1 gene transcription (Fig. 2). Does this mean that the silent chromatin state is solely dependent on promoter methylation, or could patterns of histone hypoacetylation contribute? The stability of methylation-mediated silencing is greatly enhanced on the inactive X chromosome by a mechanism defined by histone underacetylation (60). It is well established that histone hypoacetylation of Xi chromatin is correlated with silencing (11). Once again, TSA alone was completely without effect, whereas 5aC induced demethylation and the combined 5aC-TSA treatment could alleviate silencing on the inactive X chromosome (11).
It is conceivable, then, that methylation-dependent repressors operate in an already-hypoacetylated chromatin context. In such a model, methylation could reinforce the silencing strength already established by HDAC. Although we predict that HDAC is guided to transcriptionally repressed chromatin by genomic methylation patterns, the realization that MBD proteins exist with HDACs (large pools of HDAC activity can be immunoprecipitated with MeCP2 antibodies [24, 42]) support this view. However, we cannot dismiss the possibility that MDR1 chromatin is naturally underacetylated. Patterns of histone hypoacetylation once laid down may no longer require the ongoing activity of histone acetylases and deacetylases to maintain them, thereby creating a default deacetylated state. Indeed, if promoter acetylation is required to generate accessible chromatin and/or stably recruit basal transcriptional machinery to the promoter, might promoter hypoacetylation provide a signal for DNA methylation? It remains a possibility that the primary mechanism of repression is histone underacetylation and that methylation of the promoter is a secondary consequence of transcriptional arrest. Patterns of deacetylated histones on the genome could augment DNA methylation by recruitment of de novo methyltransferases (44). As a consequence, the binding of methyl-CpG binding proteins to methylated DNA would complete transcriptional silencing.
This view is also complicated by recent studies demonstrating that underacetylated chromatin is not restricted to inactive promoters (15). Surprisingly large hypoacetylated regions have been identified within the active and hyperacetylated chromatin domain of the β-globulin gene. These results would then suggest that spatial histone acetylation profiles need not always correlate with transcriptional activity, but more importantly, patterns of histone hyper- and hypoacetylation coexist within chromatin domains, and this could expand the dynamic range of transcriptional regulation. The model of gene silencing we present would need to take account of temporal and spatial organization of chromatin determinants in directing transcriptional regulation (9, 49). For example, are local regions of MDR1 responsible for silencing? Conversely, what chromosomal region(s) is acetylated to drive transcription? It is apparent that transcriptional silencing involves the complex interaction of several different components. Among the activities that are dependent on methylation and histone deacetylation examined in this study are ATPase remodeling complexes that alter chromatin accessibility and transcriptional competence (47). The prototype of this remodeling machine is the evolutionary conserved human SWI/SNF, a multisubunit complex composed of Brm and Brg1 subunits (62, 63). Recent, experimental data showed that these constituents are associated with transcriptional repressors like Sin (46, 53). Chromatin remodeling by Mi-2, a member of the SWI/SNF superfamily, has also been purified with HDAC activity and shown to repress transcription in a methylation-dependent manner (59). These experiments lend support to the idea that the MeCP2-HDAC complex is not the sole connection to methylation-mediated silencing. These results provide direct links to epigenetic control and silencing with chromatin remodeling and histone modification. Experiments are under way to examine these possibilities.
Interestingly, inactive X chromatin of differentiated cells displays a low rate of histone acetate turnover and consequently shows little or no histone acetylase accumulation following exposure to inhibitors of HDAC (21, 28). The idea that cells used in this study might display low turnover rates of histone acetylase-deacetylase is possible although unlikely, given that the accumulation of hyperacetylated histones can be detected as soon as 30 min after TSA exposure (A. El-Osta and A. P. Wolffe, unpublished results). Our experiments show that TSA alone could not reactivate gene expression; however, it is not completely without effect. We observe targeted association of acetyl-H3 and acetyl-H4 with methylated MDR1 chromatin in the absence of active transcription, and this is correlated with changes in chromatin conformation (A. El-Osta and A. P. Wolffe, unpublished findings). The formaldehyde fixation and ChIP assay offers the ability to demonstrate endogenous binding of chromatin-associated determinants on the MDR1 promoter. The assay measures the steady-state binding of histone acetylation and deacetylation, and this would not reflect functional enzymatic turnover. Despite this limitation, the ChIP assay provides a direct path to the study of structure and function of chromatin and how these factors are organized and regulated on promoters in vivo.
Alternative methylation dependent mechanisms repressing MDR1.
The imprinted genes Snrpn and U2af1-rs1 show unique histone hypoacetylation patterns between paternal and maternal alleles (17). A feature of the nonexpressed maternal allele is correlated with CpG methylation and the in vivo association of MeCP2 with U2af1-rs1 and H3 hypoacetylation of lysines K14, K9, and K18. In contrast, methylation was not associated with histone H4 deacetylation, suggesting that histones H3 and H4 are independently regulated. Taken together, these data suggest that while patterns of CpG methylation are important in recruiting HDAC, other mechanisms are likely to be involved in silencing, of which methylation-mediated histone deacetylation is just one.
We have already discussed the idea that MeCP2 may not be the sole determinant by which transcriptional repression is achieved. DNMT1 itself associates with HDAC activity to bring about transcriptional silencing (16, 50). ChIP experiments performed in our laboratory with DNMT1 antibody (50) suggest that it is not associated with the MDR1 promoter (A. El-Osta, K. D. Robertson, and A. P. Wolffe, unpublished data). These results preclude a direct DNMT1 association with MDR1 in the 300-bp region of intron 1. However, we cannot formally exclude the possibility that DNMT1 resides elsewhere on chromatin and escape sequence detection.
The silencing properties of MBD1 are interesting, since TSA does overcome transcriptional repression but MBD1 antibodies do not deplete HDAC1 (45). Therefore, the mode of repression is different from that described for MeCP2. The mechanism of repression remains uncharacterized, since HDAC1, Mi-2, and Sin3 do not associate with MBD1. We had anticipated that demethylation would reduce MBD1 promoter association. This discordance in MeCP2 and MBD1 release might reflect differences in spatial localization and/or sensitivities to 5aC. Whether the two proteins randomly localize to methyl-CpG sites or have differing promoter binding specificities remains to be determined. However, we do know that overexpressed MBD1 can bind to DNA in methylation-deficient cells that fail to localize MeCP2 or MBD2 (18), suggesting that the protein may bind indiscriminately to heterochromatin. Demethylation of the MDR1 promoter following 5aC treatment is not complete (on average, 34 of a total of 66 methyl-CpG sites are demethylated), and this could explain the differences in protein segregation. Alternatively, it is possible that MeCP2 outcompetes MBD1 for methyl-CpG sites.
Other mechanisms of methylation-dependent repression have been shown to function independently of HDAC activity. Baylin and colleagues recently demonstrated that DMAP1 (DNMT1-associated protein) partners with DNMT1 and HDAC2 to repress transcription (51). Experiments from their laboratory show DMAP1 mediated repression was not relieved by TSA. Might DMAP1 indirectly participate with MeCP2 to maintain gene repression? DNMT1 has been shown to associate with MBD2 and MBD3 (54). Interestingly, a mechanism dependent on methylation of histone H3 was recently revealed by Bannister and colleagues (1). Heterochromatin protein 1 recognizes methylation on H3 to reestablish new heterochromatin protein 1-coated chromatin and maintenance of transcriptional repression. These recent studies highlight the rapid progress in understanding how DNA and protein methylation can contribute to transcriptional silencing. It is clear that methylation-dependent silencing acts through several complexes that include methyl-CpG binding proteins, HDAC, and the enzyme that maintains DNA methylation.
Difference in the ability to relieve silencing by inhibiting HDAC.
A number of studies now show distinct differences in the ability of TSA to relieve repression by using in vitro DNA constructs (nonchromosomal), such as transfected plasmids (13, 52), compared to naturally methylated endogenous (chromosomal) genes (6, 8). TSA exposure does not completely relieve transcriptional repression in transfected cell lines (42) or DNA microinjection systems (24). How can these results be validated with our observations regarding the discordance in the relief of methylation dependent silencing by TSA? We find it useful to consider the overall chromosomal network in the different model systems. We propose that the difference in capacity of DNA methylation to silence gene activity is strengthened when operating within the natural nuclear matrix. Methylated chromosomal DNA is dominant in maintaining transcriptional repression, and examples have been reported (6). Nonchromosomal DNA may not accurately mimic endogenous chromatin and thus will not reflect the organizational complexity of DNA methylation with reference to DNA-protein association, direct binding partners, and chromatin assembly. These changes in model systems would influence the ability of DNA methylation to direct chromatin structure and function.
In this report we provide evidence that TSA can upregulate MDR1 transcription of unmethylated DNA templates in Xenopus oocytes. Interestingly, unlike in our cell line model, TSA can in part alleviate repression of in vitro methylated constructs in Xenopus. While methylated DNA injected into oocyte nuclei demonstrates several epigenetic hallmarks typical of transcriptionally repressed chromatin (namely, promoter methylation, histone deacetylation and reduced DNase I nuclear accessibility) (24), this approach fundamentally focuses on the core MDR1 promoter region (excluding other sequence components) and thus may not mirror all remodeling activities and chromatin recruiting elements in the chromosome. We would expect then that in a chromosomal context, the repressor complex associated with MDR1 has a greater capacity to target large transcriptionally inactive chromatin domains.
Assays that monitor chromatin by nuclease accessibility reveal that acetylation is critical in decondensing methylated chromatin (DNase I insensitive) to a more transcriptionally active and less compact arrangement (3, 13; A. El-Osta, unpublished data). This raises the following question: why does 5aC-TSA displace more MeCP2 than 5aC alone? We propose that this is a change in promoter accessibility to MeCP2 cross-links formed by formaldehyde. The structural changes observed by nucleosomes mediated by deacetylation would compact chromatin, thereby facilitating nucleosome-nucleosome cooperation, whereas acetylation of specific lysine residues at the N-terminal domain of histones destabilize nucleosome interactions (32, 56). Changes in local chromatin structure generated by TSA exposure can also modulate methylated DNA conformation (13). In order to determine whether MeCP2 repositioning is a function of changes in chromatin organization by inhibitors of DNA methyltransferase and HDAC (3), we intend to continue our in vivo cross-linking studies to analyze the spatial MeCP2 binding profile on specific sequences of the gene. The availability of DNA microarray technology will allow us to explore these questions on a larger genomic scale with greater resolution.
In conclusion, we used ChIP technology to determine effects of treatment with the demethylation agent 5aC and the HDAC inhibitor TSA to study MDR1 gene regulation. These experiments show that the silencing mechanism mediated by MeCP2 is independent of HDAC activity when the promoter is heavily methylated. Demethylation and release of MeCP2 from the MDR1 promoter prepare chromatin to reestablish histone acetylation and thus transcriptional activation. We have identified some of the chromatin determinants involved in silencing and suggest that the release of MeCP2 provides one gateway to the transcriptional control of MDR1.
Acknowledgments
We thank P. L. Jones for anti-MeCP2, P. Wade for MBD2b, and K. D. Robertson for DNMT1 antibodies and continued support of the work. We thank S. Baylin, F. Magdinier, R. Johnstone, and S. Russell for critical reading of the manuscript.
A. El-Osta is a Senior Research Fellow with the Fragile X (FRAXA) Research Foundation of the United States of America and was supported by an Anti-Cancer Council of Victoria Post-Doctoral Research Fellowship (Australia). This work was supported in part by the Human Frontiers Science Program Organization (HFSP) and UICC International Cancer Technology Transfer Fellowship (ICRETT) awards.
Footnotes
A.E.-O. dedicates this work to the living memory of his good friend and mentor Alan P. Wolffe.
REFERENCES
- 1.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 2.Bauer, W. R., J. J. Hayes, J. H. White, and A. P. Wolffe. 1994. Nucleosomal structural changes due to acetylation. J. Mol. Biol. 236:685-690. [DOI] [PubMed] [Google Scholar]
- 3.Bestor, T. H. 1998. Gene silencing. Methylation meets acetylation. Nature 393:311-312. [DOI] [PubMed] [Google Scholar]
- 4.Boyes, J., and A. Bird. 1991. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell 64:1123-1134. [DOI] [PubMed] [Google Scholar]
- 5.Boyes, J., and A. Bird. 1992. Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO J. 11:327-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cameron, E. E., K. E. Bachman, S. Myohanen, J. G. Herman, and S. B. Baylin. 1999. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 21:103-107. [DOI] [PubMed] [Google Scholar]
- 7.Clark, S. J., J. Harrison, C. L. Paul, and M. Frommer. 1994. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 22:2990-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coffee, B., F. Zhang, S. T. Warren, and D. Reines. 1999. Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nat. Genet. 22:98-101. [DOI] [PubMed] [Google Scholar]
- 9.Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97:299-311. [DOI] [PubMed] [Google Scholar]
- 10.Cross, S. H., R. R. Meehan, X. Nan, and A. Bird. 1997. A component of the transcriptional repressor MeCP1 shares a motif with DNA methyltransferase and HRX proteins. Nat. Genet. 16:256-259. [DOI] [PubMed] [Google Scholar]
- 11.Csankovszki, G., A. Nagy, and R. Jaenisch. 2001. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J. Cell Biol. 153:773-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desiderato, L., M. W. Davey, and A. A. Piper. 1997. Demethylation of the human MDR1 5′ region accompanies activation of P-glycoprotein expression in a HL60 multidrug resistant subline. Somat. Cell Mol. Genet. 23:391-400. [DOI] [PubMed] [Google Scholar]
- 13.Eden, S., T. Hashimshony, I. Keshet, H. Cedar, and A. W. Thorne. 1998. DNA methylation models histone acetylation. Nature 394:842.. [DOI] [PubMed] [Google Scholar]
- 14.El-Osta, A., P. Kantharidis, and J. Zalcberg. 1999. Absolute quantitation of MDR1 transcripts using heterologous DNA standards—validation of the competitive RT-PCR (CRT-PCR) approach. BioTechniques 26:1114-1124. [DOI] [PubMed] [Google Scholar]
- 15.Forsberg, E. C., K. M. Downs, H. M. Christensen, H. Im, P. A. Nuzzi, and E. H. Bresnick. 2000. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. USA 97:14494-14499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuks, F., W. A. Burgers, A. Brehm, L. Hughes-Davies, and T. Kouzarides. 2000. DNA methyltransferase dnmt1 associates with histone deacetylase activity. Nat. Genet. 1:88-91. [DOI] [PubMed] [Google Scholar]
- 17.Gregory, R. I., T. E., Randall, C. A., Johnson, S. Khosla, I. Hatada, L. P. O'Neill, B. M. Turner, and R. Feil. 2001. DNA methylation is linked to deacetylation of histone H3, but not H4, on the imprinted genes Snrpn and U2af1-rs1. Mol. Cell. Biol. 21:5426-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendrich, B., and A. Bird. 1998. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 18:6538-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hendrich, B., U. Hardeland, H. H. Ng, J. Jiricny, and A. Bird. 1999. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature 401:301-304. [DOI] [PubMed] [Google Scholar]
- 20.Herman, J. G., J. Jen, A. Merlo, and S. B. Baylin. 1996. Hypermethylation-associated inactivation indicates a tumor suppressor role for p15INK4B. Cancer Res. 56:722-727. [PubMed] [Google Scholar]
- 21.Jeppesen, P., and B. M. Turner. 1993. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell 74:281-289. [DOI] [PubMed] [Google Scholar]
- 22.Jin, S., and K. W. Scotto. 1998. Transcriptional regulation of the MDR1 gene by histone acetyltransferase and deacetylase is mediated by NF-Y. Mol. Cell. Biol. 18:4377-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones, P. A., and P. W. Laird. 1999. Cancer epigenetics comes of age. Nat. Genet. 21:163-167. [DOI] [PubMed] [Google Scholar]
- 24.Jones, P. L., G. J. Veenstra, P. A. Wade, D. Vermaak, S. U. Kass, N. Landsberger, J. Strouboulis, and A. P. Wolffe. 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19:187-191. [DOI] [PubMed] [Google Scholar]
- 25.Kaludov, N. K., and A. P. Wolffe. 2000. MeCP2 driven transcriptional repression in vitro: selectivity for methylated DNA, action at a distance and contacts with the basal transcription machinery. Nucleic Acids Res. 28:1921-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kantharidis, P., A. El-Osta, M. deSilva, D. M. Wall, X. F. Hu, A. Slater, G. Nadalin, J. D. Parkin, and J. R. Zalcberg. 1997. Altered methylation of the human MDR1 promoter is associated with acquired multidrug resistance. Clin. Cancer Res. 3:2025-2032. [PubMed] [Google Scholar]
- 27.Kantharidis, P., S. El-Osta, M. de Silva, G. Lee, X. F. Hu, and J. Zalcberg. 2000. Regulation of MDR1 gene expression: emerging concepts. Drug Res. Updates 3:99-108. [DOI] [PubMed] [Google Scholar]
- 28.Keohane, A. M., J. S. Lavender, L. P. O'Neill, and B. M. Turner. 1998. Histone acetylation and X inactivation. Dev. Genet. 22:65-73. [DOI] [PubMed] [Google Scholar]
- 29.Leegwater, P. A., R. R. De Abreu, and F. Albertioni. 1998. Analysis of DNA methylation of the 5′ region of the deoxycytidine kinase gene in CCRF-CEM-sensitive and cladribine (CdA)- and 2-chloro-2′-arabino-fluoro-2′-deoxyadenosine (CAFdA)-resistant cells. Cancer Lett. 130:169-173. [DOI] [PubMed] [Google Scholar]
- 30.Lewis, J. D., R. R. Meehan, W. J. Henzel, I. Maurer-Fogy, P. Jeppesen, F. Klein, and A. Bird. 1992. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 69:905-914. [DOI] [PubMed] [Google Scholar]
- 31.Lorincz, M. C., D. Schubeler, and M. Groudine. 2001. Methylation-mediated proviral silencing is associated with MeCP2 recruitment and localized histone H3 deacetylation. Mol. Cell. Biol. 21:7913-7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 33.Magdinier, F., and A. P. Wolffe. 2001. Selective association of the methyl-CpG binding protein MBD2 with the silent p14/p16 locus in human neoplasia. Proc. Natl. Acad. Sci. USA 98:4990-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McQueen, H. A., G. Siriaco, and A. P. Bird. 1998. Chicken minichromosomes are hyperacetylated, early replicating and gene rich. Genome Res. 8:621-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meehan, R. R., J. D. Lewis, S. McKay, E. L. Kleiner, and A. P. Bird. 1989. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell 58:499-507. [DOI] [PubMed] [Google Scholar]
- 36.Meehan, R. R., J. D. Lewis, and A. P. Bird. 1992. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 20:5085-5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muscat, G. E., L. J. Burke, and M. Downes. 1998. The corepressor N-CoR and its variants RIP13a and RIP13Delta1 directly interact with the basal transcription factors TFIIB, TAFII32 and TAFII70. Nucleic Acids Res. 26:2899-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakayama, M., M. Wada, T. Harada, J. Nagayama, H. Kusaba, K. Ohshima, M. Kozuru, H. Komatsu, R. Ueda, and M. Kuwano. 1998. Hypomethylation status of CpG sites at the promoter region and overexpression of the human MDR1 gene in acute myeloid leukemias. Blood 92:4296-4307. [PubMed] [Google Scholar]
- 39.Nan, X., R. R. Meehan, and A. Bird. 1993. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 21:4886-4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nan, X., P. Tate, and A. Bird. 1996. DNA methylation specifies chromosomal localization of MeCP2. Mol. Cell. Biol. 16:414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nan, X., J. Campoy, and A. Bird. 1997. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell 88:471-481. [DOI] [PubMed] [Google Scholar]
- 42.Nan, X., H. H. Ng, C. A. Johnson, C. D. Laherty, B. M. Turner, R. N. Eisenman, and A. Bird. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393:386-389. [DOI] [PubMed] [Google Scholar]
- 43.Ng, H. H., Y. Zhang, B. Hendrich, C. A. Johnson, B. M. Turner, H. Erdjument-Bromage, P. Tempst, D. Reinberg, and A. Bird. 1999. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet. 23:58-61. [DOI] [PubMed] [Google Scholar]
- 44.Ng, H. H., and A. Bird. 1999. DNA methylation and chromatin modification. Curr. Opin. Genet. Dev. 9:158-163. [DOI] [PubMed] [Google Scholar]
- 45.Ng, H. H., P. Jeppesen, and A. Bird. 2000. Active repression of methylated genes by the chromosomal protein MBD1. Mol. Cell. Biol. 20:1394-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perez-Martin, J., and A. D. Johnson. 1998. The C-terminal domain of Sin1 interacts with the SWI-SNF complex in yeast. Mol. Cell. Biol. 18:4157-4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peterson, C. L. 2000. ATP-dependent chromatin remodeling: going mobile. FEBS Lett. 476:68-72. [DOI] [PubMed] [Google Scholar]
- 48.Petronzelli, F., A. Riccio, G. D. Markham, S. H. Seeholzer, M. Genuardi, M. Karbowski, A. T. Yeung, Y. Matsumoto, and A. Bellacosa. 2000. Investigation of the substrate spectrum of the human mismatch-specific DNA N-glycosylase MED1 (MBD4): fundamental role of the catalytic domain. J. Cell Physiol. 185:473-480. [DOI] [PubMed] [Google Scholar]
- 49.Reinke, H., P. D. Gregory, and W. Horz. 2001. A transient histone hyperacetylation signal marks nucleosomes for remodeling at the PHO8 promoter in vivo. Mol. Cell 7:529-538. [DOI] [PubMed] [Google Scholar]
- 50.Robertson, K. D., S. Ait-Si-Ali, T. Yokochi, P. A. Wade, P. L. Jones, and A. P. Wolffe. 2000. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 25:338-342. [DOI] [PubMed] [Google Scholar]
- 51.Rountree, M. R., K. E. Bachman, and S. B. Baylin. 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 25:269-277. [DOI] [PubMed] [Google Scholar]
- 52.Schubeler, D., M. C. Lorincz, D. M. Cimbora, A. Telling, Y. Q. Feng, E. E. Bouhassira, and M. Groudine. 2000. Genomic targeting of methylated DNA: influence of methylation on transcription, replication, chromatin structure, and histone acetylation. Mol. Cell. Biol. 20:9103-9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sif, S., A. J. Saurin, A. N. Imbalzano, and R. E. Kingston. 2001. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 15:603-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tatematsu, K., Yamazaki, T., and F. Ishikawa. 2000. MBD2-MBD3 complex binds to hemi-methylated DNA and forms a complex containing DNMT1 at the replication foci in late S phase. Genes Cells 5:677-688. [DOI] [PubMed] [Google Scholar]
- 55.Ueda, K., A. Yoshida, and T. Amachi. 1999. Recent progress in P-glycoprotein research. Anticancer Drug Des. 14:115-121. [PubMed] [Google Scholar]
- 56.Ura, K., H. Kurumizaka, S. Dimitrov, G. Almouzni, and A. P. Wolffe. 1997. Histone acetylation: influence on transcription, nucleosome mobility and positioning, and linker histone-dependent transcriptional repression. EMBO J. 16:2096-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wade, P. A., P. L. Jones, D. Vermaak, G. J. Veenstra, A. Imhof, T. Sera, C. Tse, H. Ge, Y. B. Shi, J. C. Hansen, and A. P. Wolffe. 1998. Histone deacetylase directs the dominant silencing of transcription in chromatin: association with MeCP2 and the Mi-2 chromodomain SWI/SNF ATPase. Cold Spring Harbor Symp. Quant. Biol. 63:435-445. [DOI] [PubMed] [Google Scholar]
- 58.Wade, P. A., P. L. Jones, D. Vermaak, and A. P. Wolffe. 1998. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr. Biol. 8:843-846. [DOI] [PubMed] [Google Scholar]
- 59.Wade, P. A., A. Gegonne, P. L. Jones, E. Ballestar, F. Aubry, and A. P. Wolffe. 1999. Mi-2 complex couples DNA methylation to chromatin remodeling and histone deacetylation. Nat. Genet. 1:62-66. [DOI] [PubMed] [Google Scholar]
- 60.Wakefield, M. J., A. M. Keohane, B. M. Turner, and J. A. Graves. 1997. Histone underacetylation is an ancient component of mammalian X chromosome inactivation. Proc. Natl. Acad. Sci. USA 94:9665-9668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wall, D. M., S. El-Osta, D. Tzelepis, I. Bertoncello, P. Kantharidis, S. T. Chou, J. R. Zalcberg, and J. D. Parkin. 1997. Expression of MDR1 and mrp in the normal B-cell homologue of B-cell chronic lymphocytic leukaemia. Br. J. Haematol. 96:697-707. [DOI] [PubMed] [Google Scholar]
- 62.Wang, W., Y. Xue, S. Zhou, A. Kuo, B. R. Cairns, and G. R. Crabtree. 1996. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 10:2117-2130. [DOI] [PubMed] [Google Scholar]
- 63.Wang, W., J. Cote, Y. Xue, S. Zhou, P. A. Khavari, S. R. Biggar, C. Muchardt, G. V. Kalpana, S. P. Goff, M. Yaniv, J. L. Workman, and G. R. Crabtree. 1996. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15:5370-5382. [PMC free article] [PubMed] [Google Scholar]
- 64.Weitzel, J. M., H. Buhrmester, and W. H. Stratling. 1997. Chicken MAR-binding protein ARBP is homologous to rat methyl-CpG-binding protein MeCP2. Mol. Cell. Biol. 17:5656-5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wong, C. W., and M. L. Privalsky. 1998. Transcriptional repression by the SMRT-mSin3 corepressor: multiple interactions, multiple mechanisms, and a potential role for TFIIB. Mol. Cell. Biol. 18:5500-5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zalcberg, J. R., X. F. Hu, D. M. Wall, S. Mirski, S. Cole, G. Nadalin, M. De Luise, J. D. Parkin, V. Vrazas, L. Campbell, and P. Kantharidis. 1994. Cellular and karyotypic characterization of two doxorubicin resistant cell lines isolated from the same parental human leukemia cell line. Int. J. Cancer 57:522-528. [DOI] [PubMed] [Google Scholar]