Abstract
Granulocyte colony-stimulating factor (G-CSF) mRNA contains two distinct types of cis-acting mRNA destabilizing elements in the 3′-untranslated region. In addition to several copies of the AU-rich element the G-CSF mRNA also contains a destabilizing region that includes several predicted stem-loop structures. We report here that the destabilizing activity resides in a single stem-loop structure within this region. A consensus sequence for the active structure has been derived by site-directed mutagenesis, revealing that a three-base loop of sequence YAU and unpaired bases either side of the stem contribute to the activity. The helical nature of the stem is essential and the stem must be less than 11 bp in length, but the destabilizing activity is relatively insensitive to the sequence within the helix. The stem-loop increases the rate of mRNA deadenylation, most likely by enhancing the processivity of the deadenylation reaction. A protein that binds the stem-loop, but not an inactive mutant form, has been detected in cytoplasmic lysates.
The cytokine granulocyte colony-stimulating factor (G-CSF) is secreted by fibroblasts and endothelial cells in response to the inflammatory mediators interleukin-1 (IL-1) and tumor necrosis factor alpha and by activated macrophages. It stimulates the production and maturation of neutrophils and is widely used to enhance patient recovery from the neutropenia that follows treatment with anticancer drugs.
In common with many other interleukins and hemopoietic cytokines, the synthesis of G-CSF is regulated both transcriptionally and through control of mRNA stability. In unstimulated cells G-CSF mRNA is unstable but becomes stabilized in response to IL-1 or tumor necrosis factor alpha, and also in the case of monocytes and macrophages, in response to lipopolysaccharide (11, 16). In monocytes the stabilization induced by lipopolysaccharide is reversed when the cells respond to the inhibitory cytokine IL-10 (6). At least two cis-acting mRNA destabilizing elements are present in the 3′-untranslated region (3′-UTR) of the G-CSF mRNA. In addition to several copies of the AU-rich element (ARE), G-CSF contains a structurally and functionally distinct element that has been called the stem-loop destabilizing element (SLDE) (5). The SLDE was identified as a result of its ability to destabilize mRNA in cells treated with calcium ionophore, which inhibits the function of the ARE but not the SLDE. It is likely that the presence of the SLDE in the G-CSF mRNA contributes to the specificity of regulation of G-CSF mRNA, which cannot be accounted for by AREs alone, since other cytokines with different patterns of expression also contain AREs. Examples of this are IL-10 mRNA, which unlike G-CSF is not rapidly destabilized in response to IL-10 receptor activation (6), and vascular endothelial growth factor (VEGF) mRNA, the stability of which is regulated in response to hypoxia (10, 20, 26).
The SLDE in G-CSF mRNA was previously localized to within a 184-nucleotide (nt) region of the 3′-UTR (5). This region contains at least two predicted stem-loop structures that are conserved in the human and mouse genes, at least one of which was found to be essential for destabilizing activity, although the extent to which other sequences or structures within the 184-nt segment contribute to the destabilizing function was not clear. We report here that the destabilizing element resides within a single stem-loop, describe features of the stem-loop that are essential for its activity, and show that the SLDE enhances the rate of shortening of the poly(A) tail.
MATERIALS AND METHODS
Plasmid construction.
Unless otherwise stated, all plasmids were constructed by insertion of the sequences described below between the KpnI and SacI sites of pfGH (18). SL11 contains the G-CSF 3′-UTR sequence shown in Fig. 1 which was generated by PCR amplification of the corresponding region of pfGH-SL2 (5) (equivalent to bases 941 to 1033 of GenBank entry X03438 [23]) using primers 176 (5′ GCTGGTACCTGAGGGTCCCCACCTGGG 3′) and 110 (5′ CGTGAGCTCGGGGAACACTGCTGTTTGAATATCAAACAGGGA 3′). The regions inserted to create SL13 and SL12 were generated by PCR amplification of the region of bases 984 to 1033 and 964 to 1033, respectively. The inserted sequences of all other SL11 variants except for SL20, SL25, and SL26 were generated by PCR amplification of the region of bases 941 to 1033 of SL11, with alterations in the primer oligonucleotides to generate the changed sequences shown in the respective figures. SL20 was constructed by ligation of three overlapping, chemically synthesized oligonucleotides with KpnI and SacI overhangs at the 5′ and 3′ termini, respectively. SL25 and SL26 were constructed by the insertion of a 43-bp chemically synthesized oligonucleotide linker (with KpnI overhangs) in forward and reverse orientation, respectively, into KpnI-digested SL13. Templates for preparation of probes for RNA electrophoretic mobility shift assay (REMSA) were prepared by inserting the KpnI-SphI fragment that contains the SLDE region from SL11 or SL11-4C into pGEM4Z that had been digested with KpnI and SphI, generating pGEM4Z-SL11 and pGEM4Z-SL11-4C, respectively. Template for preparation of poly(A)− marker RNA was prepared by inserting the 3′ UTR of SL11 with a BamHI site immediately after the polyadenylation site generated by PCR amplification of SL11 into pGEMTEasy, creating pGEMT3′GH.
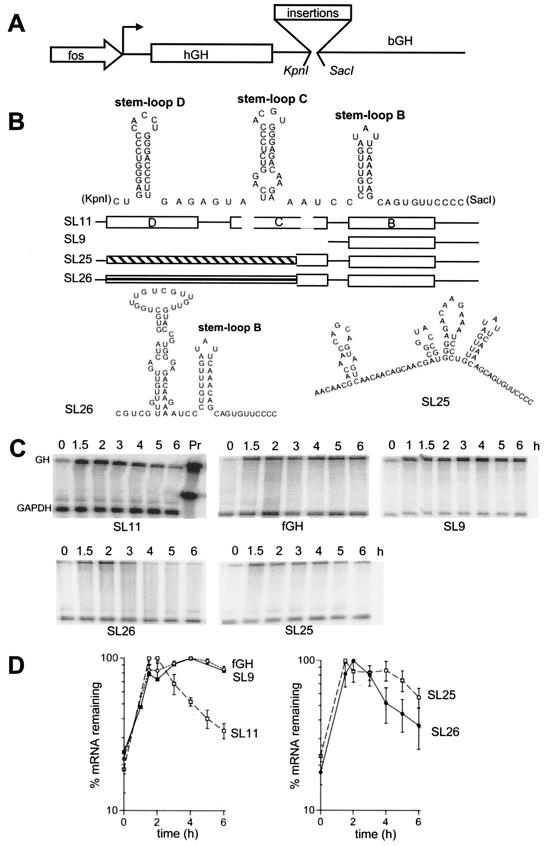
FIG. 1.
Predicted secondary structure of a region from the 3′-UTR of G-CSF that contains a potent destabilizing element and the effect of deletion or replacement of some stem-loops. (A) Schematic of the fGH reporter gene. The transcription start site is indicated by an arrow. The translated region, which is derived from human growth hormone (hGH), is boxed. (B) The predicted structure of the SLDE region of SL11 and schematics showing the region deleted or replaced in SL9, SL25, and SL26. Stem-loops in the schematic diagram are shown as open boxes, while the regions replaced in SL25 and SL26 are shown as hatched boxes. The predicted structure in SL26 is also shown. (C) RNase protection assays. RNA was isolated at the indicated times after serum stimulation of NIH 3T3 cells stably transfected with variants of the fGH gene containing the indicated sequences inserted in the 3′-UTR. The upper band is the protection product from probe to the growth hormone region of the fGH reporter; the lower band is the protection product from the GAPDH internal control. The SL11 gel also shows the migration of undigested probes. (D) Time courses of the RNase protection data. Each line on the graphs is labeled with the sequence name. The data shown are means and standard errors of the means from four (SL11), three (fGH), or two (SL25, SL26) experiments.
Cell culture and transfection.
NIH 3T3 cells were grown in Dulbecco's modified Eagle's medium with 7.5% fetal calf serum. Cells (2 × 106) were transfected with 20 μg of plasmid (linearized with ScaI) by CaPO4 precipitation, grown for 24 to 48 h, and selected in 400 μg of G418 per ml. After 10 to 12 days the resulting colonies (containing at least 100 independent clones) were pooled and maintained in 200 μg of G418 per ml. Serum stimulation of cells was performed essentially as described previously (18).
RNA isolation and analysis.
Total RNA was isolated by the guanidinium thiocyanate-acid phenol procedure (9) at various times after serum stimulation of cells. Specific transcripts were detected by RNase protection assay and quantified by PhosphorImager analysis (Molecular Dynamics, Sunnyvale, Calif.) as described previously (18). Data were normalized with respect to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as described previously (18) and plotted as means and standard errors of the means using data pooled from at least two time courses. Deadenylation assays were performed as described previously, except that the probe was prepared by in vitro transcription with SP6 polymerase of plasmid fGH that had been digested with SacI. This produces a probe complementary to 129 nt at the 3′ end of the SL11 and fGH mRNAs. Because the probe protects the 3′ end of the mRNA and RNase A (used for digestion) does not cleave at adenosine, the poly(A) tail remains attached to the duplex of probe and mRNA. The products of the RNase digestion were electrophoresed on nondenaturing polyacrylamide gels. Poly(A)− marker mRNA was prepared by in vitro transcription with T7 RNA polymerase from plasmid pGEMT3′GH digested with BamHI. This marker had the same migration as RNA prepared by in vitro deadenylation of total cellular RNA using oligo(dT) and RNase H as described previously (3). RNA secondary structure prediction was done using the mfold server (21) (http://bioinfo.math.rpi.edu/≈mfold/rna/form1.cgi).
REMSA.
Cytoplasmic extracts were prepared by hypotonic swelling of cells on ice in a solution containing 10 mM HEPES (pH 7.8), 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM dithiothreitol, and 0.5 mM phenylmethylsulfonyl fluoride for 15 min followed by the addition of NP-40 to a final concentration of 0.625%. Nuclei were removed by microcentrifugation for 30 s. RNA probe was prepared from SacI-digested pGEM4Z-SL11 or pGEM4Z-SL11-4C by in vitro transcription with SP6 RNA polymerase in the presence of 156 Ci of UTP per mmol. Binding reactions were performed in a volume of 10 μl using 1 μg of cytoplasmic protein and 20,000 cpm of probe in the presence of 25 mM Tris-Cl (pH 7.9), 100 mM KCl, 6.25 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol, 10 ng of tRNA per ml, and 100 ng of poly(dIdC) per ml for 15 min at 4°C followed by digestion with 2 U of RNase T1 (Worthington) per μl for 7 min. Samples were electrophoresed on 6% native polyacrylamide gels, after which the gels were dried and subjected to autoradiography.
RESULTS
Stem-loop B is the essential feature for destabilization.
Previous work has shown that a secondary structure-dependent destabilizing element resides within a 184-nt region of the 3′-UTR of the G-CSF mRNA (5). To localize the element we constructed several deletion and substitution mutants of the 184-nt region and assessed their destabilizing activity by insertion into the 3′-UTR of the reporter gene, fGH (Fig. 1). This reporter gene contains the growth hormone coding region and 3′-UTR but contains a linker region to allow insertions into the 3′-UTR. The promoter of the fGH gene is derived from the chicken FOS gene, allowing a pulse of transcription lasting for less than 1 h to be generated by serum stimulation. The fGH reporter has been used previously in studies of AU-destabilizing elements and in the initial identification of the SLDE.
NIH 3T3 cells were transfected with the reporter gene constructs and the stability of the mRNA was determined in polyclonal pools of stably transfected cells following serum stimulation to induce a brief pulse of transcription. The 3′ half of the 184-nt region was found to be sufficient to destabilize the reporter mRNA (Fig. 1, SL11). This 93-nt region is predicted to contain three stem-loops, which we call B, C, and D. A deletion that completely removed stem-loops C and D eliminated the destabilizing effect (Fig. 1, SL9), but replacement of stem-loops C and D with a different sequence of similar length restored the destabilization (Fig. 1, SL26). This indicates that the sequences upstream of stem-loop B are not an essential part of the destabilizing element, but the destabilizing activity of stem-loop B can be influenced by neighboring sequences or structures. The dependence on the upstream region was made further apparent when another unrelated sequence of similar length was used to replace stem-loops C and D, producing SL25. In this case the substitution did not completely restore the destabilizing activity (Fig. 1).
Upstream sequences modulate the destabilization.
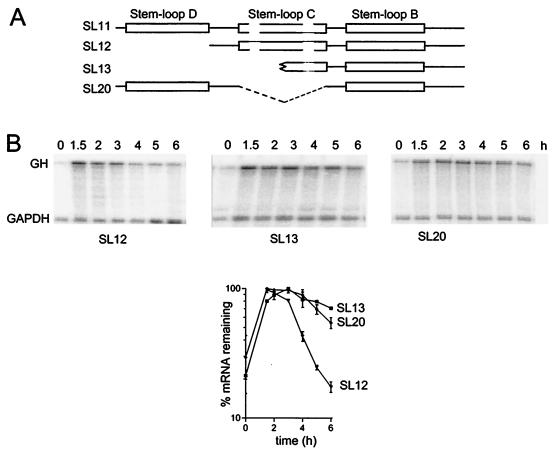
Further modifications of the region upstream of stem-loop B were made to investigate the way this region influences destabilization. Removal of stem-loop D had no effect on the destabilization (Fig. 2, SL12), but a larger deletion that also removed the sequences contributing to one strand of stem-loop C resulted in a reduced destabilizing activity (Fig. 2, SL13). The destabilizing activity was also reduced, but not completely eliminated, when stem-loop C was removed by an internal deletion that left stem-loop D intact (Fig. 2, SL20).
FIG.2.
The effect of deletion of individual stem-loops. (A) Schematic showing the truncated regions inserted into the fGH reporter. (B) RNase protection assays. RNA was isolated at the indicated times after serum stimulation of NIH 3T3 cells stably transfected with variants of the fGH gene containing the indicated sequences inserted in the 3′-UTR. The upper band is the protection product from probe for the growth hormone region of the fGH reporter; the lower band is the protection product from the GAPDH internal control. Representative RNase protection gels are shown for each construct and pooled data from two or more experiments are plotted below.
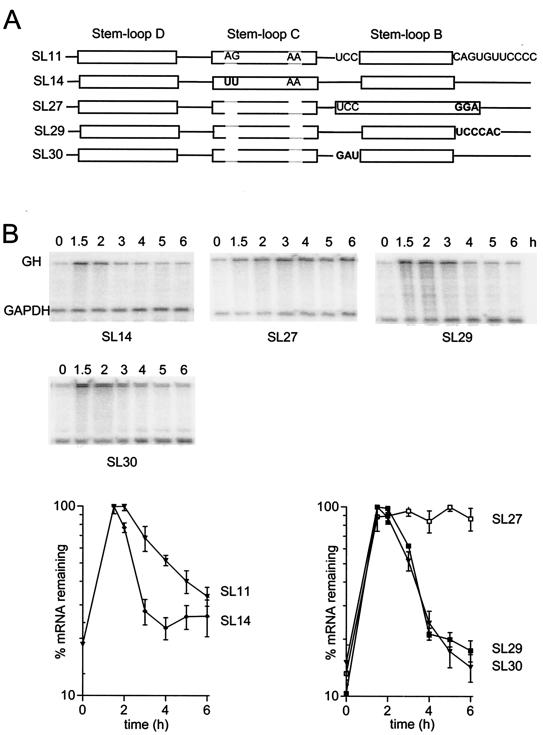
One possible explanation for the variable responses to deletion or substitution of sequences upstream of stem-loop B is that in some cases alternative secondary structures may form that prevent the formation of stem-loop B, either completely or in a proportion of RNA molecules. An alternative, but less likely, possibility is that some part of the stem-loop C region is actually part of the destabilizing element and by chance was recreated in the unrelated sequence that was substituted for stem-loop C in SL26. For example, stem-loop C and the substituted sequence in SL26 are both predicted to have stems with internal bulges containing two purines on each side of the bulge (Fig. 1). To investigate these possibilities we made two mutant constructs that do not have the purine bulges and that were designed to increase the likelihood that the mRNA folds to form stem-loops B and C. In one case, we mutated two bases in the internal bulge in stem-loop C to make them complementary to the opposing bases, thereby removing the bulge (Fig. 3, SL14). This would increase the stability of stem-loop C and hence its likelihood of formation. The SL14 mRNA was more unstable than SL11 mRNA, indicating that the bulge plays no part in the destabilizing element, but consistent with the possibility that a proportion of the SL11 mRNA might adopt an alternative, less active secondary structure which is avoided by lengthening the stem of stem-loop C. In SL27 we introduced a mutation designed to increase the length and hence the stability of stem-loop B. The three bases (CAG) immediately 3′ of stem-loop B were changed to GGA to allow pairing with the UCC that precedes the stem, thereby lengthening the stem by 3 bp. However, this mutation completely abolished the destabilizing activity (Fig. 3, SL27). To determine whether this loss of activity was due to a specific requirement for the single strand sequence either 5′ or 3′ of the stem, mutant forms were constructed with either the 5′ or the 3′ sequence changed, but in a way that avoided extending the length of stem-loop B. Replacing three bases 5′ of the stem (SL30) or six bases 3′ of the stem (SL29) had no effect on the destabilizing activity (Fig. 3), indicating that there is no specific sequence requirement at these positions. We conclude that the loss of destabilizing activity in SL27 was due to the increased length of the stem and thus the length of stem-loop B is an important determinant of its destabilizing activity.
FIG.3.
Lengthening stem-loop C enhances instability but lengthening stem-loop B abrogates destabilization. (A) Schematic showing the mutations introduced into SL11. (B) RNase protection assays. RNA was isolated at the indicated times after serum stimulation of NIH 3T3 cells stably transfected with variants of the fGH gene containing the indicated sequences inserted in the 3′-UTR. The upper band is the protection product from probe for the growth hormone region of the fGH reporter; the lower band is the protection product from the GAPDH internal control. Representative RNase protection gels are shown for each construct and pooled data from two or more experiments are plotted below.
Essential features of the stem-loop destabilizing element.
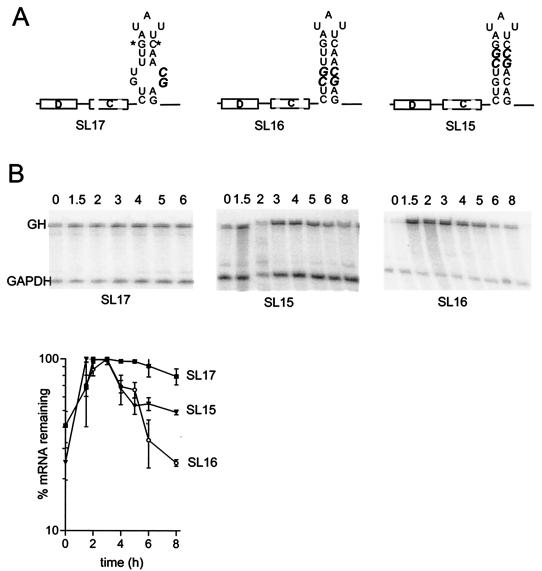
Various changes were made to the stem or loop of stem-loop B to further assess how dependent the destabilizing effect is on the sequence or structure of this stem-loop. Partial disruption of the stem by mutation of two consecutive bases (SL17) eliminated the destabilizing effect, but restoration of base pairing by introducing compensating mutations in the other strand of the stem largely restored the destabilizing effect (SL16) (Fig. 4). This result is consistent with stem-loop B being the functional element. It also suggests that the sequence within the stem is not critical for function so long as base pairing is maintained, consistent with the previous finding that an AU base pair can be replaced by a GC base pair (shown by asterisks in Fig. 4) without loss of destabilizing activity (5). However, when compensated changes were introduced at another location (near the middle of the stem) there was a partial loss of destabilizing activity, indicating that the destabilizing activity is not completely insensitive to the sequence within the helical region (SL15, Fig. 4).
FIG.4.
The destabilizing effect requires base pairing in the stem but is not highly dependent on the sequence. (A) Schematic showing changes introduced into the stem of stem-loop B. Changed bases are shown in bold. Asterisks indicate the GC base pair that replaces the AU base pair that is present in the wild-type G-CSF. This substitution was previously shown to maintain destabilizing activity (5). (B) RNase protection assay gels and quantification. RNA was isolated at the indicated times after serum stimulation of NIH 3T3 cells stably transfected with variants of the fGH gene containing the indicated sequences inserted in the 3′-UTR. The upper band is the protection product from probe for the growth hormone region of the fGH reporter; the lower band is the protection product from the GAPDH internal control. Representative RNase protection gels are shown for each construct and pooled data from two or more experiments are plotted below.
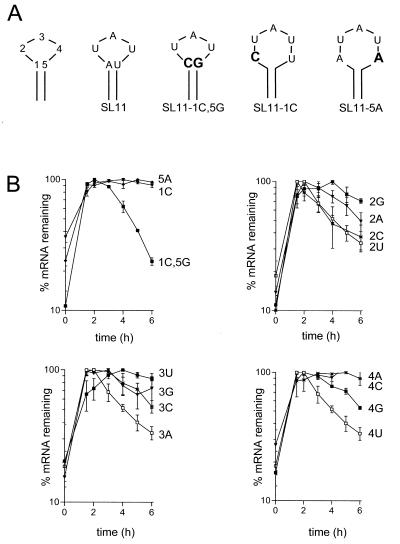
The mfold algorithm for predicting RNA secondary structure (21) gives two possible structures for the loop of stem-loop B, one with a three-base loop (as shown in Fig. 1 and 4) and the other with a five-base loop in which the AU pair at the top of the stem is not base paired. The predicted free energies for the two structures are −7.2 and −7.5 kcal/mol, respectively, slightly favoring the five-base loop. To determine whether the element functions best as a three-base loop or a five-base loop we changed the potential AU closing base pair to a GC base pair, creating SL11-1C,5G (loop positions are numbered 1 to 5 as for a five-base loop; see Fig. 5). This change, which is predicted to strongly favor a three-base loop (ΔG = −8.7 kcal/mol), had no effect on the destabilizing activity (Fig. 5). On the other hand, forcing the structure to have a five-base loop, either by mutation of the A to a C (SL11-1C) or the U to an A (SL11-5A), abolished the destabilization (Fig. 5). Thus, the functional destabilizing element has a three-base loop.
FIG.5.
The structure and sequence of the B loop are important for its function as a destabilizing element. (A) Schematic showing changes to the closing base pair of stem-loop B. The structure at left shows the numbering of bases used in naming of constructs. Changed bases are shown in bold. (B) RNA degradation time courses. Variant forms of SL11 with changes either to the closing base pair of stem-loop B as shown in panel A or point mutations in the three-base loop as indicated by the label on the time course were transfected and analyzed by RNase protection assay as described in Materials and Methods and the legend for Fig. 1.
To determine whether any variation in the sequence of the three-base loop is tolerated we measured the effect of substitution at each of the three positions (Fig. 5). (Note that the bases of the UAU loop are numbered 2, 3, and 4.) Changing U2 to a purine reduced the destabilizing effect, but C at this position was as effective as the wild-type U (Fig. 5B, upper right graph). Changing A3 to any other base reduced the destabilizing effect. Changing U4 to C or A eliminated the destabilizing effect, while converting this base to G significantly reduced the destabilizing activity. Thus, the activity of the SLDE is highly dependent on the sequence of the loop, with the most active structure being a three-base loop with the sequence PyAU.
The stem-loop destabilizing element enhances the rate of deadenylation.
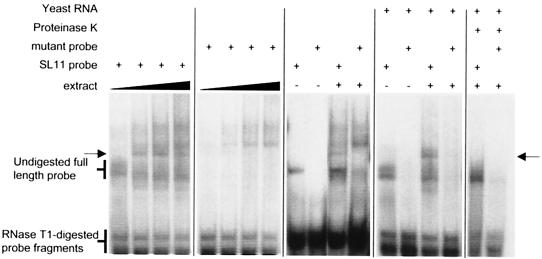
The SLDE could function in an analogous way to the ARE through binding one or more proteins that interact with other components of the RNA degradation machinery. Alternatively, it could function as the cleavage site for a specific endoribonuclease. We used RNase protection assays with probes spanning the SLDE region in an attempt to detect possible endonucleolytic cleavage products but were unable to detect any such products (data not shown). However, when we performed a REMSA on cytoplasmic extract using either an RNA probe that contains the SLDE (SL11 probe) or a probe containing a point mutation in the loop that eliminates destabilizing activity (SL11-4C probe), a complex was observed to form on the functional RNA but not on the mutant RNA even at the highest protein concentration used (Fig. 6). This complex was sensitive to digestion by proteinase K and was insensitive to competition by excess heterologous RNA (whereas slower migrating complexes that formed on both the wild-type and mutant probe were sensitive to competition by the heterologous RNA). Although we cannot rule out the possibility that endonucleolytic cleavage at the SLDE generates products that are too unstable to be detectable by RNase protection assay, the detection of a complex in the REMSA that is sensitive to an inactivating point mutation suggests that the destabilization may be mediated through interaction with a binding protein.
FIG.6.
Electrophoretic mobility shift detection of a complex on the SL11 RNA. SL11 probe corresponding to the sequence shown in Fig. 1 or a mutant probe containing a single inactivating point mutation in the SLDE loop (Fig. 5, SL11-4C) was incubated with cytoplasmic extract from BALB/c 3T3 cells and subjected to digestion with T1 nuclease, and the products were electrophoresed on a native 6% polyacrylamide gel. The first pair of panels show complexes resulting from incubation of SL11 and SL11-4C probes, respectively, with 0.625, 1.25, 2.5 and 5 μg of cytoplasmic protein extract. A complex that forms on the SLDE but not the mutant probe is indicated with arrows. Other panels show the complexes formed on SL11 or SL11-4C probe with or without 1 μg of protein extract, in the presence or absence of 1.6 μg of yeast RNA per ml plus 20 μg of tRNA per ml, and with or without digestion for 5 min by 10 μg of proteinase K per ml.
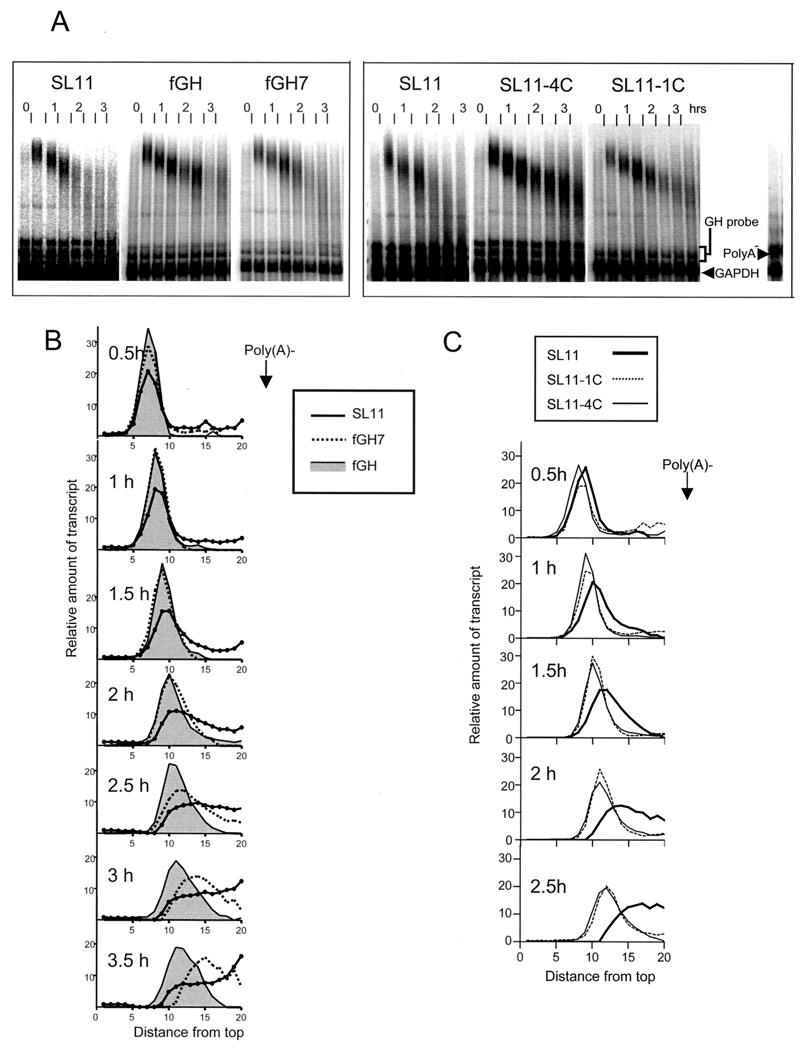
The ARE mRNA destabilizing element as well as the recently described c-fos coding region determinant accelerate the rate of mRNA deadenylation (8, 12). To examine whether the SLDE influences mRNA deadenylation we compared the deadenylation rate of SL11 to that of fGH (which is stable), fGH7, which contains an ARE that was previously shown to accelerate the deadenylation rate about twofold (18), or two stable mutant forms of SL11. The SL11 and fGH7 mRNAs were more rapidly deadenylated than the fGH mRNA or the stable mutants, SL-1C and SL-4C (Fig. 7). During the first hour following serum stimulation, while the transcription pulse was occurring, all the mRNAs accumulated with poly(A) tails of similar lengths. The stable mRNA transcripts (fGH, SL11-1C, and SL11-4C) subsequently shortened in a relatively homogeneous manner, with the band on the gel broadening only gradually over time, indicative of a distributive mechanism of deadenylation, as previously observed for the fGH mRNA (18). In contrast, for the fGH7 mRNA, and even more so for the SL11 mRNA, the majority of transcripts migrated more rapidly once the transcription pulse (which takes place during the first hour) had ceased. Furthermore, the band on the gel rapidly broadened, indicating a more rapid, processive deadenylation. This indicates that the SLDE, like the ARE, destabilizes at least in part by enhancing the rate of mRNA deadenylation and that this enhancement involves an increase in the processivity of the deadenylation reaction.
FIG. 7.
The SLDE enhances deadenylation. (A) To monitor poly(A) tail lengths, RNase A-resistant hybrids formed between the mRNA and a probe that spans the 3′ end of the mRNA were electrophoresed on nondenaturing gels. RNA was isolated at half-hour intervals after serum stimulation of NIH 3T3 cells stably transfected with SL11, fGH (a stable mRNA that deadenylates slowly), or fGH7 (which contains an ARE that enhances the deadenylation rate) (left panel) or with SL11 or two stable mutants of SL11 as shown (right panel). A GAPDH coding region probe was included as an internal standard. The lane on the right shows the migration from in vitro-prepared poly(A)− RNA. Note that the two bands immediately above the GAPDH band are residual degradation products from the GH probe and are not due to accumulation of deadenylated mRNA in vivo. (B) To compare deadenylation rates, individual lanes from the left panel were scanned by phosphorimager and the signal intensity from the top of the gel to near the expected location of deadenylated RNA was plotted for each time point. The SL11 trace is shown as a solid line, the fGH7 trace is shown as a dotted line, and the fGH trace is shaded. (C) Plots of individual lanes from the right panel were scanned by phosphorimager and the signal intensity was plotted. The SL11 trace is shown as a thick line, the SL11-4C trace is shown as a thin line, and the SL11-1C trace is shown as a dashed line.
DISCUSSION
Structure of the destabilizing element.
Our data indicate that the destabilizing activity of the G-CSF SLDE resides primarily in a single stem-loop. We found that the degree of destabilization can be influenced by the upstream region, but the observation that the upstream region can be replaced by a different sequence with little loss of effect argues against any sequence specificity requirement of this upstream region. The variable effect of changing the upstream sequence could result from the requirement for a single-stranded region to either side of the essential stem-loop. We have noticed that the constructs with changes to the upstream region that resulted in reduced destabilizing activity are predicted by the mfold program to have some of the five bases preceding the stem involved in base pairing, whereas these bases are predicted to be single stranded in the constructs that have destabilizing activity similar to or greater than that of SL11. However, this correlation has not been tested experimentally.
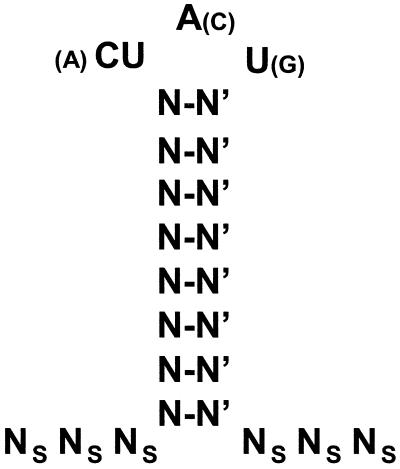
The other features of the SLDE that are important for its function are the size and sequence of the loop and the length of the stem. These features are summarized in Fig. 8, which presents a consensus structure for the SLDE based on the analysis of function of the various mutations we constructed. Briefly, the key features are a loop sequence of YAU, a stem that is stable but less than 11 bp in length, and the presence of unpaired bases flanking the stem. The optimal stem length has not been determined, but we presume that the naturally occurring length of 8 bp is close to the optimum. We are presently using the consensus to search for other genes that may also contain an SLDE.
FIG. 8.
Consensus sequence for activity of the stem-loop destabilizing element. A schematic representation of the functional element summarizing the results of the mutagenesis data are shown. N-N′, an essential base pair whose sequence can vary; Ns, bases that must be single stranded. Optimal bases in the loop are shown in large print and suboptimal bases are shown in parentheses.
The structure of the destabilizing element is conserved across species.
With the sequence of the G-CSF 3′-UTR known for seven species (human, mouse, rat, cat, cow, pig, and chicken), it is interesting to compare the different sequences. The 3′-UTR sequences are rather divergent across the different species, but two sequence segments are conserved in all the species. These are the ARE near the 5′end of the 3′-UTR and the SLDE. Figure 9 shows an alignment of the first 450 nt of the 3′-UTRs from the seven species. The 3′-UTR sequences are either incomplete or quite divergent beyond this region. The conserved ARE region is located approximately 40 to 80 nt downstream of the stop codon in all seven species. The location of the conserved SLDE region is different in chickens than in the mammalian species (about 350 nt from the stop codon in mammals versus 160 nt in the chicken), but it is the longest contiguous segment of sequence conservation across all species anywhere in the 3′-UTR. This conservation of sequence across species is consistent with the AREs and the SLDE having a function and suggests that the function is conserved from birds to mammals. The stem of the SLDE in the chicken mRNA is predicted to be 1 bp shorter than in the other species but is otherwise identical. The nonessential stem-loop (stem-loop C) that precedes the SLDE is conserved in the mammalian species but not in the chicken, adding further weight to the conclusion that stem-loop C is not essential for function.
FIG. 9.
Comparison of part of the 3′-UTR sequences of G-CSF mRNAs from different species. Sequences were aligned using the GCG programs CLUSTAL and GAP. Regions conserved across all seven species are boxed. The lower box includes the SLDE. The sequences (and their GenBank accession numbers) are from cow (AF092533 [15]), cat (AB042552 [27]), human (X03438 [23]), mouse (M13926 [25]), pig (Y10494 [17]), rat (U37101 [13]), and chicken (X14477 [19]). The numbering shown above the sequences is with respect to the start of the human 3′-UTR and below is with respect to the start of the chicken 3′-UTR.
Mechanism of destabilization.
Destabilizing elements located in the 3′-UTRs of some mRNAs have been found to be sites of endonucleolytic cleavage (1, 2, 4, 14, 24). The local RNA secondary structure is implicated in the activity of several such elements, either because the cleavage site is located within a loop region or because the secondary structure appears to play a role in targeting the endonuclease to the RNA (2, 14, 24). However, we found that the destabilizing effect of the SLDE is more likely to be analogous to that of the ARE, which functions by promoting deadenylation of the mRNA. We observed in transcription pulse time courses that both elements enhance the deadenylation rate and rapidly produce heterogeneity of poly(A) tail length, indicative of a processive mechanism of shortening, whereas stable mutants of SL11 did not enhance deadenylation. However, it is unlikely that the activities of the SLDE and the ARE are mediated through binding the same cellular factor, because the SLDE is structurally different from the ARE.
It is interesting that two distinct destabilizing elements that target the same step in degradation coexist in a single transcript. This situation has recently been described for the c-fos mRNA, which has a coding region determinant as well as AREs in the 3′-UTR, both of which promote mRNA deadenylation (12). The c-fos coding region determinant may provide a means of coupling mRNA degradation to translation that is not provided by the 3′ AREs. In the case of G-CSF, the SLDE may provide a response to a regulatory pathway that is not afforded by the AREs or may allow rapid degradation of the mRNA under circumstances that normally lead to stabilization of ARE-containing transcripts. For example, the IL-2 and IL-3 mRNAs, both of which contain 3′ AREs, are stabilized in response to signaling by the stress-activated protein kinase pathway (7, 22). Perhaps the rapid decay afforded by the ARE and the SLDE is subject to regulation by different pathways, so that the G-CSF mRNA, which contains both types of element, requires activation of two signaling pathways for the mRNA to be stabilized, whereas some other ARE-containing mRNAs might respond to the stress-activated protein kinase (or JNK) pathway alone. This will be an interesting question to investigate further.
Acknowledgments
We are grateful to all members of the lab for advice and discussion during the course of this work and to Michael Zuker for helpful advice on RNA secondary structure, Tom Gonda for advice and critical reading of the manuscript, and Cathy Lagnado for assistance with analysis and presentation of deadenylation data.
This work was supported by program grant #973204 from the NH&MRC of Australia.
REFERENCES
- 1.Binder, R., J. A. Horowitz, J. P. Basilion, D. M. Koeller, R. D. Klausner, and J. B. Harford. 1994. Evidence that the pathway of transferrin receptor mRNA degradation involves an endonucleolytic cleavage within the 3′ UTR and does not involve poly(A) tail shortening. EMBO J. 13:1969-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Binder, R., S.-P. L. Hwang, R. Ratnasabapathy, and D. L. Williams. 1989. Degradation of apolipoprotein II mRNA occurs via endonucleolytic cleavage at 5′-AAU-3′/5/-UAA-3′ elements in single-stranded loop domains of the 3′-noncoding region. J. Biol. Chem. 264:16910-16918. [PubMed] [Google Scholar]
- 3.Brewer, G., and J. Ross. 1988. Poly(A) shortening and degradation of the 3′ A+U-rich sequences of human c -myc mRNA in a cell-free system. Mol. Cell. Biol. 8:1697-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, B. D., I. D. Zipkin, and R. M. Harland. 1993. Sequence-specific endonucleolytic cleavage and protection of mRNA in Xenopus and Drosophila. Genes Dev. 7:1620-1631. [DOI] [PubMed] [Google Scholar]
- 5.Brown, C. Y., C. A. Lagnado, and G. J. Goodall. 1996. A cytokine mRNA-destabilizing element that is structurally and functionally distinct from A+U-rich elements. Proc. Natl. Acad. Sci. USA 93:13721-13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, C. Y., C. A. Lagnado, M. A. Vadas, and G. J. Goodall. 1996. Differential regulation of the stability of cytokine mRNAs in lipopolysaccharide-activated blood monocytes in response to interleukin-10. J. Biol. Chem. 271:20108-20112. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C. Y., F. Del Gatto-Konczak, Z. Wu, and M. Karin. 1998. Stabilization of interleukin-2 mRNA by the c-Jun NH2-terminal kinase pathway. Science 280:1945-1949. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C. Y., N. Xu, and A. B. Shyu. 1995. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol. Cell. Biol. 15:5777-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 10.Dibbens, J. A., D. L. Miller, A. Damert, W. Risau, M. A. Vadas, and G. J. Goodall. 1999. Hypoxic regulation of vascular endothelial growth factor mRNA stability requires the cooperation of multiple RNA elements. Mol. Biol. Cell 10:907-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ernst, T. J., A. R. Ritchie, G. D. Demetri, and J. D. Griffin. 1989. Regulation of granulocyte- and monocyte-colony stimulating factor mRNA levels in human blood monocytes is mediated primarily at a post-transcriptional level. J. Biol. Chem. 264:5700-5703. [PubMed] [Google Scholar]
- 12.Grosset, C., C. Y. Chen, N. Xu, N. Sonenberg, H. Jacquemin-Sablon, and A. B. Shyu. 2000. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell 103:29-40. [DOI] [PubMed] [Google Scholar]
- 13.Han, S. W., N. Ramesh, and W. R. Osborne. 1996. Cloning and expression of the cDNA encoding rat granulocyte colony-stimulating factor. Gene 175:101-104. [DOI] [PubMed] [Google Scholar]
- 14.Hanson, M. N., and D. R. Schoenberg. 2001. Identification of in vivo mRNA decay intermediates corresponding to sites of in vitro cleavage by polysomal ribonuclease 1. J. Biol. Chem. 276:12331-12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heidari, M., and M. E. Kehrli, Jr. 2000. Cloning, sequencing, and analysis of cDNA encoding bovine granulocyte-colony stimulating factor. Vet. Immunol. Immunopathol. 73:183-191. [DOI] [PubMed] [Google Scholar]
- 16.Koeffler, H. P., J. Gasson, and A. Tobler. 1988. Transcriptional and posttranscriptional modulation of myeloid colony-stimulating factor expression by tumor necrosis factor and other agents. Mol. Cell. Biol. 8:3432-3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kulmburg, P., M. Radke, B. Mezes, R. Mertelsmann, and F. M. Rosenthal. 1997. Cloning and sequence analysis of the immediate promoter region and cDNA of porcine granulocyte colony-stimulating factor. Gene 197:361-365. [DOI] [PubMed] [Google Scholar]
- 18.Lagnado, C. A., C. Y. Brown, and G. J. Goodall. 1994. AUUUA is not sufficient to promote poly(A) shortening and degradation of an mRNA: the functional sequence within AU-rich elements may be UUAUUUA(U/A)(U/A). Mol. Cell. Biol. 14:7984-7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leutz, A., K. Damm, E. Sterneck, E. Kowenz, S. Ness, R. Frank, H. Gausepohl, Y. C. Pan, J. Smart, M. Hayman, et al. 1989. Molecular cloning of the chicken myelomonocytic growth factor (cMGF) reveals relationship to interleukin 6 and granulocyte colony stimulating factor. EMBO J. 8:175-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levy, A. P., N. S. Levy, and M. A. Goldberg. 1996. Post-transcriptional regulation of vascular endothelial growth factor by hypoxia. J. Biol. Chem. 271:2746-2753. [DOI] [PubMed] [Google Scholar]
- 21.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 22.Ming, X. F., M. Kaiser, and C. Moroni. 1998. c-jun N-terminal kinase is involved in AUUUA-mediated interleukin-3 mRNA turnover in mast cells. EMBO J. 17:6039-6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagata, S., M. Tsuchiya, S. Asano, Y. Kaziro, T. Yamazaki, O. Yamamoto, Y. Hirata, N. Kubota, M. Oheda, H. Nomura, et al. 1986. Molecular cloning and expression of cDNA for human granulocyte colony-stimulating factor. Nature 319:415-418. [DOI] [PubMed] [Google Scholar]
- 24.Scheper, W., D. Meinsma, P. E. Holthuizen, and J. S. Sussenbach. 1995. Long-range RNA interaction of two sequence elements required for endonucleolytic cleavage of human insulin-like growth factor II mRNAs. Mol. Cell. Biol. 15:235-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsuchiya, M., S. Asano, Y. Kaziro, and S. Nagata. 1986. Isolation and characterization of the cDNA for murine granulocyte colony-stimulating factor. Proc. Natl. Acad. Sci. USA 83:7633-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White, F. C., S. M. Carrol, and M. P. Kamps. 1995. VEGF mRNA is reversibly stabilized by hypoxia and persistently stabilised in VEGF-overexpressing human tumor cell lines. Growth Factors 12:289-301. [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto, A., A. Iwata, K. Tuchiya, A. Katsumata, K. Oishi, T. Saito, H. Tsujimoto, A. Hasegawa, and S. Ueda. 2001. Molecular cloning and expression of the cDNA encoding feline granulocyte colony-stimulating factor. Gene 274:263-269. [DOI] [PubMed] [Google Scholar]