Abstract
Gene regulation by AP-1 transcription factors in response to Jun N-terminal kinase (JNK) signaling controls essential cellular processes during development and in pathological situations. Here, we report genetic and molecular evidence that the histone acetyltransferase (HAT) Chameau and the histone deacetylase DRpd3 act as antagonistic cofactors of DJun and DFos to modulate JNK-dependent transcription during thorax metamorphosis and JNK-induced apoptosis in Drosophila. We demonstrate in cultured cells that DFos phosphorylation mediated by JNK signaling plays a central role in coordinating the dynamics of Chameau and DRpd3 recruitment and function at AP-1-responsive promoters. Activating the pathway stimulates the HAT function of Chameau, promoting histone H4 acetylation and target gene transcription. Conversely, in response to JNK signaling inactivation, DRpd3 is recruited and suppresses histone acetylation and transcription. This study establishes a direct link among JNK signaling, DFos phosphorylation, chromatin modification, and AP-1-dependent transcription and its importance in a developing organism.
Keywords: JNK signaling, Fos, histone acetylation, epigenetic control of transcription, Drosophila, development
The transcriptional response to signal transduction via the evolutionary conserved Jun N-terminal kinase (JNK) pathway controls essential cellular processes, including morphogenesis, differentiation, and apoptosis during development and in physiopathological situations (Eferl and Wagner 2003; Xia and Karin 2004). A large body of research supports the model that in response to extracellular stimulation, JNK activates the transcriptional effector AP-1 by phosphorylation, and thereby reprograms target gene expression (Noselli and Agnes 1999; Kockel et al. 2001; Weston and Davis 2002). AP-1 mainly consists of Fos and Jun family proteins that can form homodimers or heterodimers and bind DNA through conserved bZIP domains. Given the importance of chromatin dynamics in the control of gene expression, recent work has focused on AP-1 interaction partners capable of chromatin modification and remodeling, notably enzymes able to reversibly modify histone tails by acetylation. The nuclear receptor corepressor (NCoR)/histone deacetylase 3 (HDAC3) complex was thus found to inhibit the JNK pathway (Zhang et al. 2002). The checkpoint function of the NCoR complex is relieved by c-Jun phosphorylation, which directs its removal from the promoter (Ogawa et al. 2004) and promotes the recruitment of the TAF7 RNA polymerase II (RNA pol II) subunit (Munz et al. 2003). AP-1 recruits DRpd3/HDAC1 to reverse histone acetylation at the promoter of the attacin-A gene and its activation by the NF-κB transcription factor Relish associated with the histone acetyltransferase (HAT) Pcaf (Kim et al. 2005). Other reports on stimulation of Fos or Jun activities by CBP (Bannister and Kouzarides 1995; Benkoussa et al. 2002) suggest that the recruitment of HAT coactivator complexes at target promoters mediate nucleosome acetylation and stimulate transcription. These findings, gained from experiments performed in vitro and in cultured cells, indicate that chromatin dynamics plays a central role in the cellular response to JNK signaling. However, the epigenetic mechanisms that control the transcriptional response in intact organisms remain unclear.
JNK signaling regulates a number of different processes during Drosophila development, including pupal thorax closure and apoptotic cell death in imaginal discs. The role of JNK signaling is well established in these two morphogenetic events. In the former, it drives the migration of wing discs toward the midline and their fusion into a continuous epidermal structure; in the latter, it apoptotically eliminates cells exposed to inappropriate proximodistal patterning cues (for review, see Kockel et al. 2001). The present study provides genetic and molecular evidence that Chameau (Chm), a MYST domain HAT previously reported to act in epigenetic mechanisms of transcriptional control (Grienenberger et al. 2000) and to support histone acetylation at replication origins (Aggarwal and Calvi 2004), and the HDAC DRpd3 modulate the transactivation potential of AP-1 during thorax closure and JNK-induced apoptosis in an antagonistic manner. Furthermore, by deciphering the mode of action of Chm and DRpd3 in vitro and in cultured cells, we provide mechanistic insights into the regulation of AP-1 function by chromatin.
Results
Chm enhances JNK signaling during thorax closure
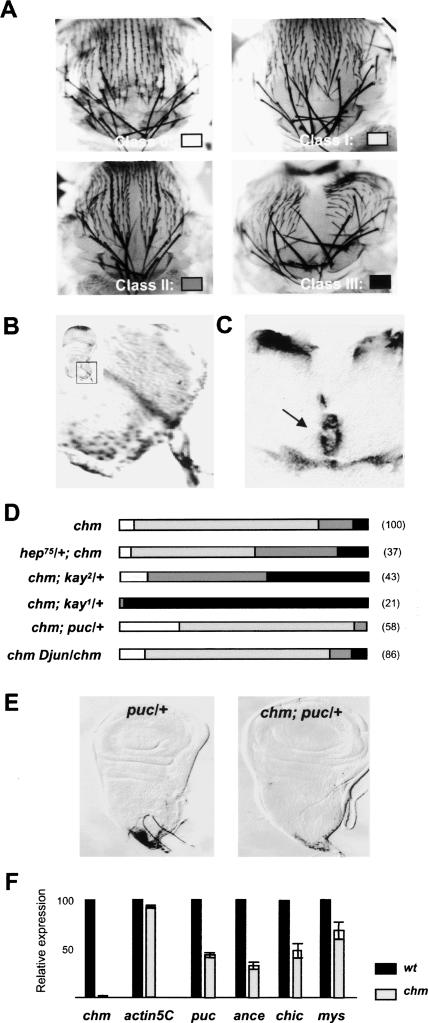
Animals homozygous for the amorphous allele chm14 lack zygotic chm function and die during late pupal stages. Pharate adults display morphological aberrations, especially a cleft of variable depth and width along the thorax midline. To quantitatively assess the strength of this phenotype, we adopted a system of four phenotypic classes, ranging from the absence to a mild, pronounced, or severe cleft (Fig. 1A). chm is expressed in the two epithelia of wing discs, the columnar epithelium (data not shown), and the peripodial membrane (Fig. 1B), and by 8 h after puparium formation (APF), when the two contralateral discs meet at the dorsal midline, transcription proceeds in fusion regions only (Fig. 1C). Together, the mutant phenotype and the expression pattern of chm support a function in migration and/or fusion of wing discs during metamorphosis.
Figure 1.
Chm enhances JNK signaling during thorax closure. (A) Variable penetrance of chm thoracic cleft phenotype. (B) chm expression in peripodial cells as revealed by β-Gal activity in the large peripodial nuclei of wing discs from the chm7lacZ enhancer trap line. The inset shows the region of the disc that has been magnified. (C) chm expression at sites of imaginal disc fusion revealed by in situ hybridization on dissected pupae 8 h APF. Arrow indicates the dorsal midline. (D) Genetic interactions between chm and JNK pathway components encoding genes hep, kay, puc, and Djun, as revealed by changes in the strength of the cleft phenotype. For the color code representing the different phenotypic classes refer to A. The numbers of examined pupae are shown in parentheses. (E) puc expression in wild-type (left) and chm mutant (right) discs, revealed by β-Gal activity in the enhancer trap line pucE69. (F) Transcription levels of chm; of actin5C, which is not an AP-1 target; and of the target genes puc, mys, ance, and chic in wild-type and chm wing discs 1 h APF.
As flies defective in JNK signaling display thorax defects similarly to chm mutants, we asked whether Chm might play a role in JNK signaling during thorax closure. Thus, we tested genetic interactions between chm and genes encoding components of the JNK pathway (Fig. 1D). Reducing the gene dosage of positive effectors, such as DJNKK/hemipterous (hep) and Dfos/kayak (kay), does not per se affect thorax closure but exacerbates the chm phenotype: Heterozygosity for hep raises the proportion of class II phenotypes from 14% to 39%; reducing Dfos activity has a more pronounced effect, as class III rises from 6% to 97% in the case of the null allele kay1 and to 40% with the hypomorphic allele kay2. Conversely, heterozygosity for puckered (puc), a repressor of JNK activity, dominantly suppresses the thoracic cleft of chm mutants, as no chm; puc/+ pharate adult presents a class III phenotype. Moreover, this genotype produces viable adult escapers, an effect already observed with lethal combinations of hep and Dfos alleles rescued by puc heterozygosity (Agnes et al. 1999; Zeitlinger and Bohmann 1999). These positive and negative genetic interactions indicate that Chm cooperates with JNK signaling during thorax closure. We noticed, however, that heterozygosity for Djun does not exacerbate the chm phenotype (Fig. 1D). The observation that Dfos, but not Djun, alleles dominantly interact with chm suggests that DFos acts independently of DJun in the context of Chm-dependent thorax closure processes.
Next, we examined AP-1 target gene transcription. The puc gene is activated by JNK signaling in proximal peripodial cells (Agnes et al. 1999; Zeitlinger and Bohmann 1999; Pastor-Pareja et al. 2004). In chm mutant discs, puc expression is strongly reduced but can still be detected, consistent with a stimulatory rather than essential function of Chm for JNK signaling in these cells (Fig. 1E). To confirm such a function, we compared transcription levels of puc and three additional bona fide JNK targets—ance, chickadee (chic), and myospheroid (mys) (Jasper et al. 2001)—in wild-type and chm mutant wing discs. Results from quantitative RT–PCR show that the transcription of the four target genes is significantly reduced in a chm background (Fig. 1F). We conclude that Chm supports JNK-dependent gene activation during wing disc development and thorax closure.
Chm acts positively in the JNK-dependent apoptotic pathway
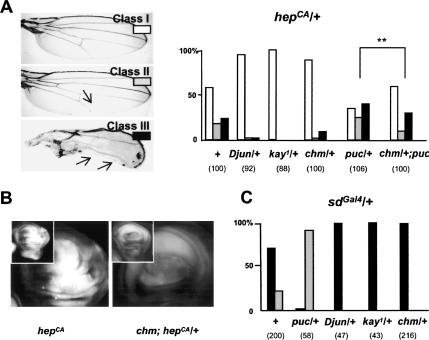
To determine whether chm contributes to JNK-dependent processes in addition to thorax closure, we examined JNK-induced apoptotic cell death. In the insertion trap line hepCA, JNK signaling is ectopically activated in the wing blade primordium, which causes cell death and induces notches of variable extent in the adult wing (Adachi-Yamada et al. 1999). Eliminating one copy of Djun, Dfos, or chm rescues the hepCA wing phenotype. Furthermore, loss of one copy of chm partially reverts the enhancement of the hepCA notching phenotype that is observed as a consequence of puc heterozygosity (Fig. 2A). Further supporting the notion of cooperation between JNK and Chm, cell death as detected by acridine orange staining in hepCA wing discs is abrogated in chm homozygous conditions (Fig. 2B). JNK-dependent apoptosis is also induced in response to morphogen gradient distortion caused by misexpression of scalloped (sd) (Liu et al. 2000) or optomotor blind (omb) (Adachi-Yamada et al. 1999). We observed that heterozygosity for puc aggravates—and for Djun, Dfos, or chm dominantly rescues—the notched wing phenotype of flies heterozygous for sd (Fig. 2C) or omb (data not shown). Thus, Chm acts as a positive effector in the JNK-regulated apoptotic pathway. Interestingly, in this process chm cooperates with both Dfos and Djun genes.
Figure 2.
chm is required for JNK-induced apoptosis. (A) Variable severity of the notched wing phenotype induced by hepCA (left, arrows show wing notches) and changes induced in the relative abundance of the phenotypic classes by heterozygosis for Djun, kay, puc, chm, and chm plus puc (right). (B) Acridine orange staining of hepCA and chm; hepCA/+ wing discs. (C) Changes in the sdGal4/+ notched wing phenotype induced by puc, Djun, kay, or chm heterozygosis. The numbers of examined wings are shown in parentheses.
Chm acts as a coactivator of AP-1
Based on the genetic interactions described above, we proceeded to analyze the epistatic relationship between Chm and AP-1 factors. To this end, we used UAS transgenes and the MZ980Gal4 driver that promotes expression specifically in the proximal region of the wing disc and later along the prospective junction of the contralateral discs (Martin-Blanco et al. 2000). Driving UASchm in these cells fully rescues the thoracic cleft of chm mutants, while Dfos, Djun, or both Dfos and Djun expression in the MZ980 domain do not (Fig. 3A). Thus, chm is not epistatic to Dfos or Djun, which indicates that Chm is not upstream of AP-1 in the pathway and does not control the expression of Dfos and Djun. RT–PCR from RNA samples of wild-type and chm mutant wing discs confirmed that Dfos and Djun transcription is not affected by chm loss of function (data not shown). This finding is consistent with Chm cooperating with JNK signaling at the level of the transcriptional effectors of the pathway.
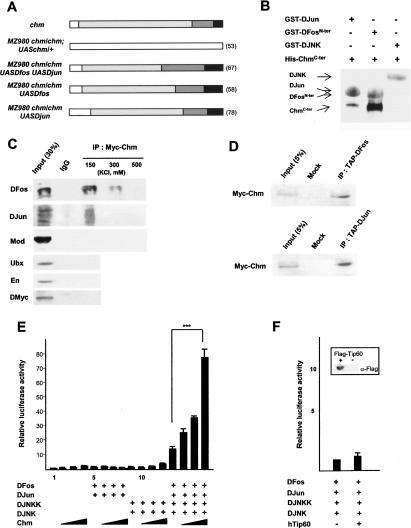
Figure 3.
Chm acts as a nuclear coactivator of DJun and DFos. (A) Rescue of chm thoracic cleft phenotype by Chm, DFos, DJun, or DJun plus DFos expression in the MZ980 domain. Refer to Figure 1A for the color code of phenotypic classes. (B) GST pull-down assays. Binding of His-ChmCter to GST fusions of DJun, DFosN-ter or DJNK was revealed by Western blot probed by anti-His and anti-GST antibodies, and secondary antibodies coupled to peroxydase (brown) and alkaline phosphatase (blue). (C) Immunoprecipitation of nuclear extracts from chm; 69BGal4/UASMyc-chm third instar larvae using IgG or anti-Myc. Immunoprecipitates were washed by increasing salt concentration and analyzed by Western blot using antibodies to DFos, DJun, Mod, En, Ubx, and DMyc as a negative control. (D) Western blots probed with anti-Myc of immunoprecipitates obtained by anti-TAP from nuclear extracts of larvae ubiquitously expressing Myc-Chm and TAP-DFos or Myc-Chm and TAP-DJun, or of wild-type larvae (Mock). (E) Luciferase assays of HEK293 cells transfected with the reporter construct and combinations of expression vectors for DJNKK, DJNK, DFos, DJun, and Chm (50, 100, and 250 ng/well). Histograms represent luciferase activity relative to a value of 1 for transfection with reporter alone (error bars show standard deviations, five independent assays). (F) Luciferase assays with DJNKK, DJNK, DFos, DJun, and hTip60 (250 ng/well). Reference value of 1 in the absence of hTip60; four independent assays. (Inset) Western blot probed with anti-Flag of cell extracts transfected with a Flag-hTip60-producing or empty vector.
Then, we investigated whether Chm directly binds to DFos and/or DJun using GST pull-down assays. The C-terminal half of Chm (amino acids 494–812), which contains the MYST domain, displays strong in vitro affinity for an N-terminal fragment of DFos (including the N terminus and the basic DNA-binding domain), and binds DJun as well, although less efficiently. Conversely, the cytoplasmic kinase Basket (Bsk)/DJNK does not bind to the His-Chm fusion protein (Fig. 3B). The Chm N terminus (amino acids 20–400) does not associate with DFos or DJun (data not shown). Similar experiments with GST-fused DFos deletion mutants identified the basic region of DFos as the predominant Chm-interacting domain, although significant association with the C-terminal part of DFos was also observed (see Supplementary Fig. S1). Immunoprecipation assays followed by Western analyses confirmed that these interactions occur in vivo. Both DFos and DJun coprecipitate with Myc-Chm from nuclear extracts of larvae expressing a Myc-tagged version of the protein (Fig. 3C). DFos is eluted from the immunoprecipitate at higher salt concentrations than DJun, which indicates a more stable association with Chm and that Chm to DFos interaction can occur in the absence of DJun. Confirming the specificity of the assay, nuclear proteins unrelated to JNK signaling, the chromatin-associated protein Modulo (Mod), the homeodomain transcription factors Ultrabithorax (Ubx) and Engrailed (En), as well as the basic helix–loop–helix (bHLH) factor DMyc are not precipitated by Myc-Chm (Fig. 3C). In reciprocal experiments, an anti-TAP antibody coprecipitates Myc-Chm from nuclear extracts of larvae ubiquitously expressing Myc-Chm and TAP-DFos or TAP-DJun (Fig. 3D). The results of the in vitro and the in vivo experiments, taken together, show that Chm, DFos and DJun can directly interact and form multimeric protein complexes in larvae.
To further characterize the cooperation between DFos, DJun and Chm, we performed reporter gene assays in cultured cells. HEK293 cells were transiently transfected with an AP-1-dependent luciferase reporter in the presence or absence of Chm, DJun and DFos, DJNKK and DJNK. Chm either alone, or in the presence of DFos and DJun or of DJNKK and DJNK, had no significant effect (Fig. 3E). However, when DJNKK, DJNK, DFos, and DJun were supplied together, Chm stimulated AP-1-dependent transcription in a dose-dependent manner. Thus, Chm promotes the transactivation potential of AP-1 only when the JNK pathway is active. This effect seems to be specific for Chm, as expression of hTip60, another MYST HAT (Hilfiker et al. 1997), does not change luciferase activity (Fig. 3F). These data identify Chm as a transcriptional coactivator of DFos and DJun.
DFos recruits Chm to chromatin to promote transcriptional activity
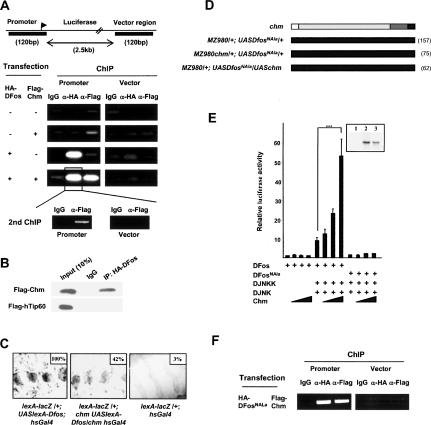
Next, we focused on the functional relevance of the Chm/DFos interaction. To investigate whether this interaction occurs on AP-1 target DNA, we performed chromatin immunoprecipitation (ChIP) assays in HEK293 cells transfected with an AP-1 reporter and constructs for HA-DFos and Flag-Chm. DFos efficiently and specifically binds the reporter promoter. Chm does so only when expressed together with DFos (Fig. 4A). No significant binding was observed on the vector DNA. To ascertain that Chm and DFos simultaneously associated with promoter sequences, we performed a sequential ChIP experiment. After a first ChIP with an antibody to HA (DFos) and dissociation of the antibodies from the immunoprecipitated complexes, a second ChIP was performed using anti-Flag (Chm) antibody. PCR detection then showed that the complex eventually pulled-down contains both DFos and Chm associated with the promoter (Fig. 4A, bottom). As a control, Chm but not hTip60 was coprecipitated by DFos from cells transfected with HA-DFos and either Flag-Chm or Flag-hTip60 (Fig. 4B). We conclude that Chm is recruited to chromatin in a sequence specific manner by DFos to AP-1-binding sites.
Figure 4.
Cooperation between Chm and DFos in transcriptional activation. (A) ChIP with IgG, anti-HA, and anti-Flag of chromatin extracts from HEK293 cells transfected with vectors producing Flag-Chm, HA-DFos, or both. (Top) Occupancy was assayed by real-time PCR using primers for regions (shown in black) encompassing AP-1-binding sites (left) or a vector region (right). (Bottom) Second ChIP with anti-Flag from chromatin complexes precipitated by anti-HA. (B) Western blots probed with anti-Flag of immunoprecipitates obtained by anti-HA or IgG from nuclear extracts of HEK293 cells transfected with vectors producing HA-DFos and Flag-Chm (top) or HA-DFos and Flag-hTip60 (bottom). (C) Magnified views of larval epidermis stained for β-galactosidase upon lexA–lacZ reporter induction by LexA–DFos chimera in an otherwise wild-type (left) or chm mutant (middle) background, and of lexA–lacZ animals as a control (right). Levels of β-galactosidase expression are given as the percentage of the value obtained for lexA–lacZ/+; UASLexA–Dfos; hsGal4 larvae. (D) Lack of genetic interactions of chm with the DfosNAla variant during thorax closure. Refer to Figure 1A for the color code of phenotypic classes. (E) Luciferase assays of HEK293 cells transfected with DFos and Chm (50, 100, and 250 ng/well) in the presence or absence of JNK and DJNKK, and with DFosNAla and Chm in the presence of JNK and DJNKK. Reference value of 1 for transfection with reporter alone, four independent assays. (Inset) Western blot, probed with anti-DFos antibody, of cell extracts transfected with an empty vector (lane 1) and with vectors producing DFosNAla (lane 2) or DFos (lane 3). (F) ChIP with IgG, anti-HA, and anti-Flag of chromatin extracts from HEK293 cells transfected with vectors producing Flag-Chm and HA-DFosNAla. Occupancy was assayed by real-time PCR using primers for promoter and vector regions.
Next, we asked whether Chm acts by promoting DFos DNA-binding or transactivating potential. This was addressed in an in vivo assay, using a fly line that expresses a chimera between the DNA-binding LexA protein and DFos, and permits monitoring transactivation by DFos independent of its DNA-binding activity (Szuts and Bienz 2000). We observed that a lacZ reporter cloned downstream of LexA-binding sites is strongly activated in the larval epidermis by DFos–LexA and that chm mutation reduces lacZ transcription (Fig. 4C). This result suggests that Chm supports gene activation in the animal by a mechanism improving DFos transactivation capacity.
JNK-mediated phosphorylation of DFos regulates the functional interaction with Chm
So far, we have seen that DFos and Chm physically interact and functionally cooperate in the execution of JNK signals to mediate apoptosis, morphogenesis, and gene activation. Next, we addressed how JNK influences the function of DFos and Chm at the molecular level. DFos is a substrate for JNK and several phosphorylation sites have been reported (Ciapponi et al. 2001). The relevance of JNK-mediated phosphorylation of DFos in thorax closure is illustrated by the strong cleft phenotype induced by the expression in the MZ980 domain of a variant, DFosNAla, in which the two DJNK threonine residues that represent DJNK targets were mutated into alanine (Fig. 4D). Interestingly, this effect cannot be reversed by Chm overexpression, indicating that the protein cannot promote gene activation by DFosNAla. To further validate this interpretation, we performed reporter gene assays in HEK293 cells. The luciferase reporter is activated by DFos, DJNKK and DJNK, and in this context Chm expression boosts transcription in a concentration-dependent manner whereas no such effect is observed using DFosNAla instead of the wild-type protein (Fig. 4E). These data strongly suggest that the cooperation between DFos and Chm in gene activation is regulated by a mechanism that involves DFos phosphorylation. Interestingly, phosphorylation by DJNK does not seem to be required for Chm to DFos binding as DFosNala recruits Chm to chromatin as efficiently as the wild-type protein (Fig. 4F).
Chm promotes H4 acetylation at AP-1 target sites
The recruitment of Chm to AP-1-binding sites by DFos suggests a mechanism for gene activation that involves histone modification by the HAT activity of Chm. As the bZIP domain of DFos is sufficient to recruit Chm, and to avoid the additional effects of the transactivation and phosphorylation domains of DFos, we used a truncated version of DFos that essentially comprises the bZIP domain. Thus, we devised a simple experimental system relying on DFosbZIP expression in HEK293 cells to explore the mode of action of Chm.
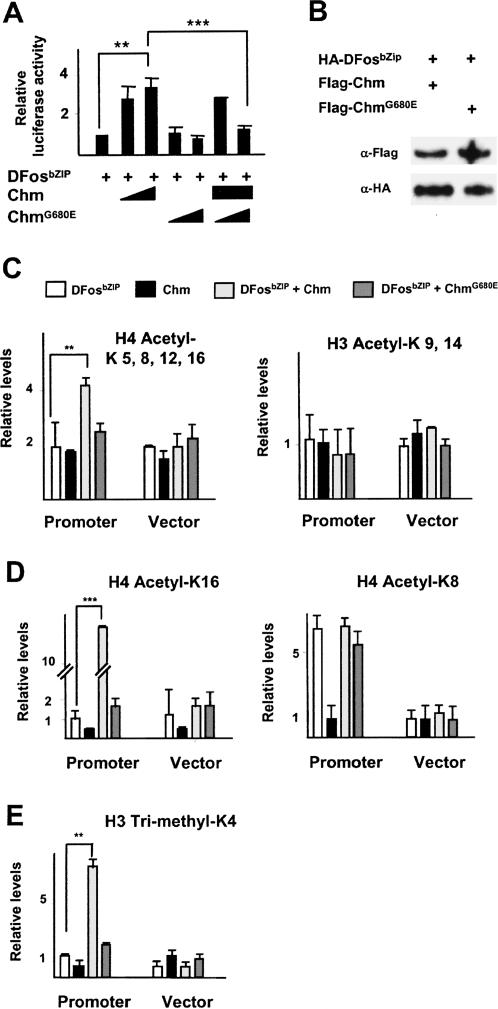
We first tested the requirement of Chm HAT activity for DFos-mediated transcription. Coexpression of DFosbZIP with Chm results in a significant stimulation of reporter activity. This is dependent on the enzymatic activity of Chm, because a point mutation in the acetyl-CoA-binding site (G680E) that abolishes HAT activity (Grienenberger et al. 2002) abrogates reporter activation (Fig. 5A). The effect of ChmG680E is dominant as indicated by luciferase inhibition when the level of the mutant protein is raised leaving that of the wild-type constant. In these experiments, similar levels of mutant and wild-type Chm proteins are expressed (data not shown) and bind chromatin in transfected HEK293 cells (Fig. 5B). The dominant negative effect of ChmG680E was also observed using the full-length DFos protein (data not shown).
Figure 5.
Chm promotes H4 acetylation at AP-1-responsive sequence. (A) Luciferase assays of HEK293 cells transfected with DFosbZIP, Chm, and/or ChmG680E (100 or 250 ng/well). Reference value of 1 for transfection with DFosbZIP alone; three independent assays. (B) Western blots, probed with antibody to Flag and HA tags, of chromatin preparations from cells transfected with vectors producing HA-DFosbZIP and Flag-Chm or Flag-ChmG680E. (C–E) ChIP of chromatin extracts from HEK293 cells transfected with DFosbZIP, Chm, DFosbZIP and Chm, or DFosbZIP and ChmG680E. Immunoprecipitations were performed by antibodies to tetra-acetylated H4 (C, left), acetylated H3K9/K14 (C, right), acetylated H4K16 (D, left), acetylated H4K8 (D, right), and trimethylated H3K4 (E). Data of real-time PCR are given as relative levels compared with cells transfected with reporter alone (average of four independent assays).
To assess whether HAT-dependent AP-1 reporter activation correlated with changes in histone modification, we next examined the acetylation pattern at the reporter gene promoter and its dependence on Chm enzymatic activity by performing ChIP experiments with acetylation site-specific antibodies. Chm recruitment by DFosbZIP results in an increase of H4 tetra-acetylation but does not change K9/K14 acetylation of H3, while ChmG680E has no significant effect (Fig. 5C). Thus, H4 is a likely target of DFos-directed acetylation by Chm. In ChIP assays with antibodies to mono-acetylated H4 isoforms, Chm was found to enhance the H4K16 acetylation over levels observed in the presence of DFosbZIP alone, an effect that is not supported by ChmG680E (Fig. 5D). Thus, H4K16 is a target lysine of Chm HAT activity. Interestingly, DFosbZIP promotes H4K8 acetylation but Chm addition does not raise the level further. This indicates that H4K8 is not a Chm target, and therefore suggests that DFosbZIP recruits another HAT to modify this lysine.
Then, we examined Chm function in transcription activation. H3K4 trimethylation constitutes a chromatin mark for transcriptionally active loci (Lachner and Jenuwein 2002). Consistent with the coactivator function of Chm, the level of H3K4 trimethylation at the promoter is increased in the presence of both Chm and DFosbZIP. Again, Chm HAT activity is required in this process, as using the ChmG680E variant has no effect (Fig. 5E). This suggests that H4 acetylation by Chm is a prerequisite for the definition of a histone modification pattern instructive for improved transcription.
DRpd3 interacts with DFos and counteracts Chm function
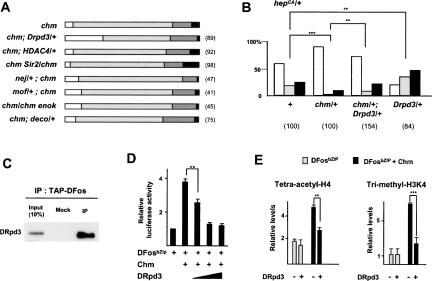
The identification of HAT activities as mediators of DFos-dependent histone acetylation and gene activation raises the question of which HDACs might function to counter these processes. A hypomorphic mutation of the gene encoding the Drosophila homolog of HDAC1, Drpd3, dominantly suppresses the thoracic cleft of chm mutants, whereas available mutations in other HDAC genes, including DHDAC4 and Dsir2, have no effect (Fig. 6A). Furthermore, heterozygosity for Drpd3 enhances the hepCA notched wing phenotype and counteracts the phenotypic rescue caused by the mutation of one copy of chm (Fig. 6B). Thus, Drpd3 and chm antagonistically control JNK signaling during thorax closure and the JNK-dependent apoptotic pathway. As H4K8 is modified by another HAT than Chm (Fig. 5D), we also tested genetic interactions with available mutations in HAT genes. Reducing the gene dosage of nejire/CBP, mof, enok, or deco neither aggravated nor rescued the chm thorax phenotype (Fig. 6A).
Figure 6.
DRpd3 interacts with DFos and counteracts Chm function. (A) Genetic interactions of chm with HDAC- and HAT-encoding genes during thorax closure, revealed by the strength of the thoracic cleft phenotype. The numbers of examined pupae are given in parentheses. Refer to Figure 1A for the color code of phenotypic classes. (B) Changes in the distribution of the wing phenotype classes associated with hepCA (see Fig. 2A) by heterozygosis for Drpd304556 allele and suppression of the rescuing effect of chm heterozygosis. Refer to Figure 2A for the color code of phenotypic classes. (C) Western blots, revealed by anti-DRpd3, of immunoprecipitates obtained by anti-TAP from nuclear extracts of wild-type (Mock) and armGal4 UASTAP-Dfos/UASMyc-chm third instar larvae. (D) Luciferase assays of HEK293 cells transfected with DFosbZIP, Chm (250 ng/well), and increasing amounts of DRpd3 (50, 100, or 250 ng/well). Reference value of 1 for cells transfected with DFosbZIP alone; four independent assays. (E) ChIP of chromatin extracts from HEK293 cells transfected with DFosbZIP or DFosbZIP and Chm, in the presence or absence of DRpd3. Immunoprecipitations were performed with antibodies to tetra-acetylated H4 (left) and to trimethylated H3K4 (right). Data are represented as a relative levels compared with transfection with the reporter alone (average of three independent assays).
To examine the effect of DRpd3 on AP-1-dependent transcription at the molecular level, we asked whether DRpd3 could directly interact with DFos, as Chm does. DRpd3 coprecipitates with TAP-DFos from nuclear extracts of larvae expressing UASTAP-Dfos under the control of armGal4 (Fig. 6C). GST pull-down assays reveal that DFos physically interacts with DRpd3 and that the only DFos fragments that precipitate DRpd3 share the ZIP domain (Supplementary Fig. S1). Thus, DRpd3 and Chm physically interact with the ZIP and the basic domains of DFos, respectively. Finally, to assess the functional relevance of these interactions, we tested the effect of DRpd3 expression on DFosbZIP/Chm-induced transcription and histone modifications in HEK293 cells. Increasing DRpd3 levels while leaving Chm constant results in reporter repression (Fig. 6D). Consistent with this inhibitory effect, DRpd3 reduces H4 tetra-acetylation and H3K4 trimethylation mediated by DFosbZIP/Chm at the promoter (Fig. 6E), indicating that DRpd3 restores a repressive transcriptional environment. Thus, Chm and DRpd3 behave as antagonistic cofactors of DFosbZIP in HEK293 cells. Considered together, these data strongly suggest that the nuclear response to JNK signaling is determined by the opposing activities of this HAT/HDAC pair.
Regulation of Chm and DRpd3 recruitment and function by JNK signaling
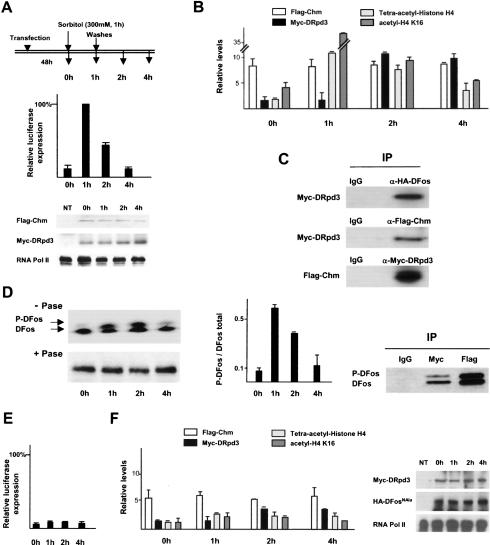
To investigate the roles of Chm and DRpd3 in JNK phosphorylation-dependent transactivation, we performed experiments with the full-length version of DFos. To modulate JNK activity we employed sorbitol treatment, which can reversibly activate the pathway in cultured cells (Bagowski et al. 2003). In HEK293 cells transfected with Chm, DRpd3 and DFos vectors, luciferase transcription rapidly increases after sorbitol addition and is progressively shut-down after sorbitol removal (Fig. 7A). Therefore, reversible stimulation by sorbitol constitutes an appropriate system to correlate molecular changes at AP-1-responsive promoter with JNK-dependent transcription.
Figure 7.
Chm and DRpd3 recruitment and function upon JNK signaling activation and deactivation. (A–C) Compilation of data obtained from RT–PCR, ChIP, and Western blot analyses during JNK signaling activation and inhibition upon sorbitol addition (300 mM for 1 h) and removal in HEK293 cells transfected with HA-DFos, Flag-Chm, and Myc-DRpd3. (A, top) At each time point described, total RNA was harvested for RT–PCR to access luciferase transcription, and chromatin extract was prepared for ChIP analyses. (Middle) luciferase transcription. (Bottom) Control Western blots of cell extracts for Chm and DRpd3 expression, and RNA pol II as a loading control. (NT) Nontransfected cells. (B) ChIP with anti-Flag, anti-Myc, anti-tetra-acetylated H4, and anti-acetyled H4K16 antibodies. RT–PCR data are normalized relative to GAPDH expression. ChIP data are depicted as relative levels compared with transfection with the reporter alone (average of three independent assays). (C) Western blots, probed for proteins indicated to the left, of proteins associated with chromatin fragments extracted 1 h after sorbitol removal and immunoprecipitated by antibodies to Flag (Chm), Myc (DRpd3), or HA (DFos). (D, left) Phosphorylation status of chromatin-bound HA-DFos was assessed by Western blot of chromatin samples probed by anti-HA. +Pase and –Pase refer to chromatin samples digested or not with calf intestinal alkaline phosphatase. (Middle) Time course upon sorbitol addition and removal of P-DFos/total DFos ratio obtained by phosphoimager gel scan. (Right) Western blot for DFos and P-DFos of chromatin fragments extracted 1 h after sorbitol removal and immunoprecipitated by Myc-DRpd3 and Flag-Chm. (E,F) Compilation of data obtained from similar experiments as in A and B, but using HA-DFosNAla instead of HA-DFos (average of four independent assays). (Right) Western blots of cell extracts for DRpd3, DFosNAla, and RNA pol II.
We performed ChIP assays to assess changes in reporter promoter occupancy throughout the experiment (Fig. 7B). We found that Chm is always and at invariant levels associated with the promoter, indicating that its recruitment by DFos does not depend on JNK signaling. H4 acetylation levels at the promoter, as monitored by the relative abundance of H4K16 and tetra-acetylated H4, dynamically changed throughout the time course of induction and correlated with transcriptional activity (Fig. 7B). Because DFos-directed acetylation by Chm is responsible for these modifications (Fig. 5D), this result indicates that the protein, while present at the promoter, is unable to acetylate H4 in nonstimulated cells, and therefore that JNK signaling stimulates the latent HAT activity of Chm and permits subsequent transcriptional activation.
Whereas Chm is found constitutively associated, DRpd3 recruitment to the promoter depends on JNK signaling. Before sorbitol addition, which we could consider as the inactive steady state, DRpd3 does not associate the promoter (1.35-fold over background). DRpd3 thus appears dispensable for maintenance of the inactive steady state. While activating the pathway upon sorbitol addition has no significant effect on DRpd3 tethering, the HDAC starts to be recruited to the promoter after sorbitol elimination (14-fold over background 1 h after sorbitol removal) and the levels of H4 acetylation and transcription then decrease simultaneously and progressively. Thus, the down-regulation of the transcriptional response to JNK activation after dissipation of the signal is at least in part mediated by recruitment of DRpd3 to the promoter already occupied by Chm. Supporting this notion, DRpd3 is found on chromatin fragments immunoprecipitated by Chm and conversely Chm is associated with fragments containing DRpd3 (Fig. 7C). Together, these results suggest that DRpd3 assumes a transient function to reduce histone modification and transcription levels previously stimulated by Chm.
The dynamics of Chm activation and DRpd3 recruitment to the promoter was next considered with regard with DFos phosphorylation. Sorbitol addition increases the ratio of phosphorylated DFos (P-DFos) to nonphosphorylated DFos in the chromatin-bound fraction, and sorbitol removal decreases the ratio (Fig. 7D). Thus, DFos phosphorylation correlates with Chm-mediated transcriptional activation and DFos dephosphorylation with down-regulation by DRpd3. Furthermore, Western analysis of immunoprecipitated chromatin revealed that 1 h after sorbitol removal Chm, DFos and P-DFos are present on the same chromatin fragments, whereas DRpd3 coprecipitates predominantly with fragments containing unphosphorylated DFos (Fig. 7D, right). Pull-down assays indicate that DFos interacts in vitro with both Chm and DRpd3, and P-DFos only with Chm (see Supplementary Fig. S2). Together, these data suggest that the DFos phosphorylation state is the likely connection between sorbitol stimulus, Chm HAT activity unmasking and DRpd3 recruitment. To investigate this point further, we performed sorbitol experiments using the DFosNAla variant, which cannot be phosphorylated, instead of the wild-type protein. Consistent with our earlier finding (Fig. 4E), Chm still is tethered to the promoter. However, sorbitol addition no longer stimulates H4 acetylation and target gene transcription (Fig. 7E,F). This lack of JNK responsiveness correlates with a significantly reduced change in promoter occupancy by DRpd3 in response to JNK signaling. Thus, the DFos phosphorylation status controls Chm HAT activity and DRpd3 recruitment to the promoter.
Discussion
Chm/DRpd3 partnership in the developmental response to JNK signaling
This study establishes a direct link between JNK signaling and chromatin modification via AP-1 transcription factors in an intact developing organism, an effect that rests on the HAT coactivator Chm and the HDAC corepressor DRpd3. This conclusion holds for thoracic closure and JNK-induced apoptosis but not for another JNK-dependent morphogenetic event, the embryonic dorsal closure. Although this process shares many similarities with thorax closure (Kockel et al. 2001), it is not affected in embryos deprived of maternal and zygotic chm contribution (data not shown). Thus, Chm is not required for all JNK signaling-dependent mechanisms. Furthermore, when it is involved, Chm is not essential for JNK signaling: The incomplete penetrance of chm phenotypes and the decreased but remaining expression of AP-1 target genes in mutant discs illustrate that chm loss-of-function compromises but does not block JNK signaling. Thus, Chm is not an obligatory component in the pathway but its recruitment supports the transcriptional efficiency of AP-1 during imaginal development. DRpd3 counteracts this function, indicating that the balanced activity of the two antagonistic factors constitutes a new regulatory mechanism for JNK-dependent transcription.
The functional partnership between Chm and DRpd3 in the control of JNK signaling seems specific to this HAT/HDAC pair, as mutations in other HDAC or HAT genes do not modify the chm thorax closure phenotype. Particularly illustrative is the absence of genetic interactions with mutant alleles of mof, since Mof is a MYST HAT of the same substrate specificity as Chm—H4K16 (Akhtar and Becker 2000)—and DFos interacts with Chm through the conserved MYST domain. Interestingly, a recent report has connected Chm and DRpd3 functions during Drosophila oogenesis (Aggarwal and Calvi 2004). The two proteins exhibit opposite effects on H4 acetylation and activity of replication origins, indicating that the functional antagonism of Chm and DRpd3 regulates several chromatin-dependent processes, including transcription and replication.
Although genetic experiments emphasize the specificity of Chm/DRpd3 partnership, our data do not imply that DRpd3's only role in JNK signaling is in balancing Chm function. Neither do they exclude that other HDACs nor HATs participate in the JNK response. For example, in cultured cells DRpd3 recruitment by AP-1 changes histone acetylation levels established earlier by the HAT Pcaf at the attacin promoter from a permissive to a repressive status (Kim et al. 2005), and a HDAC3 repressor complex maintains c-Jun transcriptionally inactive (Weiss et al. 2003). It is therefore likely that different HATs and HDACs control AP-1-regulated transcription, depending on the physiological context. What specifies HAT and HDAC commitment in a given process remains to be explored.
Mechanisms of Chm/DRpd3-mediated control of AP-1-dependent transcription
Insights into the mechanism by which Chm supports JNK target gene transcription were initially obtained from observations made in the developing animal. First, Chm improves the activation of a lexA–lacZ reporter by the LexA–DFos fusion protein, revealing that Chm stimulates the transactivating potential of DFos when it is tethered to a promoter. Second, the thorax cleft phenotype caused by DFosNAla, a protein deficient for the JNK phosophorylation sites, is not rescued upon simultaneous overexpression of Chm. As the DFosNAla variant recruits Chm as efficiently as the wild-type protein, this suggests that transcriptional improvement by Chm requires DFos phosphorylation, providing an attractive link between Chm function and JNK signaling.
Our study of the mode of action of the antagonistic cofactors Chm and DRpd3 in cultured cells has provided further insights into a chromatin-based mechanism that executes a modulation of the transcriptional response to JNK signaling. We propose the following model, based on the dynamics of cofactor recruitment and activity, chromatin modification and transcriptional status related to reversible activation of the pathway. In response to JNK signaling, Chm HAT activity sets up a histone modification pattern that is instructive for transcriptional enhancement. Consistent with this notion Chm acetylates H4, with a marked preference for K16, and facilitates H3K4 trimethylation. However, AP-1 likely also engages HATs of different substrate specificity, since H4K8 acetylation, a modification required for the recruitment of the SWI/SNF-activating complex (Agalioti et al. 2002), is directed by DFos in the absence of Chm. After JNK signaling has ceased, DRpd3 gets recruited to promoters occupied by unphosphorylated DFos and counteracts the effects of Chm by reversing histone modifications, which results in transcriptional down-regulation. Strikingly, the recruitment of DRpd3 seems not to result from the displacement of Chm from the promoter, since invariant levels of Chm are associated with the promoter in sorbitol experiments, whereas DRpd3 starts to be recruited only once the signal has been eliminated. Thus, as opposed to an exchange of a HAT coactivator complex for an HDAC corepressor complex, which occurs for instance between Pcaf/NF-κb and DRpd3/AP-1 complexes at the attacin promoter (Kim et al. 2005), a complex containing both Chm and DRpd3 could then form at the target promoter whose activity changes the histone modification pattern back to a pattern less permissive to transcription. Thus, DRpd3 most likely functions during a transient phase from a transcriptionally active to silent status. Its absence from the promoter at the inactive steady state in nonstimulated cells, suggests that unphosphorylated DFos then lies in a conformational environment that prevents DRpd3 recruitment by the ZIP domain.
AP-1 phosphorylation plays a pivotal role in regulating the epigenetic response to JNK signaling. An “activation by derepression” model has been recently proposed following which JNK-dependent phosphorylation induces the release of an HDAC3 repressor complex from the N terminus of c-Jun (Weiss et al. 2003; Ogawa et al. 2004). We have seen that DFosbZIP, which lacks JNK phosphorylation sites and domains other than the bZIP that could recruit HDAC complexes, can promote transcription in nonstimulated cells. This suggests that a phosphorylation-dependent process similar to the activation by derepression model mediates transactivation by full-length DFos when JNK signaling is active. Our work furthermore supports additional regulatory roles for AP-1 phosphorylation as indicated by the dependence of Chm HAT activity and DRpd3 recruitment on the DFos phosphorylation state. We propose that phosphorylation not only releases an HDAC corepressor complex (other than DRpd3) and leads to activation by derepression but also unmasks Chm HAT function and results in increased transcriptional efficiency, whereas dephosphorylation promotes the recruitment of DRpd3 to reverse transcriptional activation. Considering the opposite functions of chm and Drpd3 in JNK-dependent thorax closure and apoptosis during fly development, it is tempting to speculate that the balanced activities of the Chm HAT and the DRpd3 HDAC allow transient JNK target gene activation as well as fine-tuning of JNK transcriptional output in vivo.
Materials and methods
Fly stocks and manipulations
The following fly lines were used: UASLexA–Dfos and LL lexA–lacZ reporter construct (M. Bienz, Cambridge, UK); kay1, kay2, DjunIA109, UASDjun, UASDfos, UASDfosNAla, UASTAP-Djun, and UASTAP-Dfos (D. Bohmann); deco1 (M.L. Goldberg, Ithaca, NY); mof1 (J.C. Lucchesi, Atlanta, GA); enok1 (L. Luo, Stanford, CA); MZ980Gal4 (E. Martin-Blanco, Barcelona, Spain); hepCA and ombbi (K. Matsumoto, Nagoya, Japan); hep1 (S. Noselli, Nice, France); DSir205327 (S.M. Parkhurst, Nice, France); the null chm14 allele (Grienenberger et al. 2002) and chm7lacZ (J. Pradel); DHDAC4KG05226, Drpd304556, nej3, pucE69, sdGal4, armGal4, 69BGal4, and hsGal4 (Bloomington). UASMyc-chm was generated using a Myc-tag/chm cDNA fusion cloned in pUAS-T, and UAS-Myc-chmG680E by replacing an Age1/Xba1 fragment of pUAS-T-Myc-chm by the homologous fragment with G680E point mutation. In situ hybridizations and immunodetections were performed according to standard procedures. Digoxigenin RNA-labeled probes were generated according to manufacturer's protocol (Boehringer-Mannheim). All primer sequences used in this work are available on request.
Larval nuclear extract preparation, immunoprecipitation, and β-galactosidase activity quantification
Nuclei were isolated from third instar larvae using a slightly modified version of the protocol of Lehmann and Koolman (1988). Nuclear preparations (10 mM Tris; 0.1 mM EDTA; 150 mM KCl; 10% glycerol; 1 mM DTT, leupeptin, aprotinin, and pepstatin protease inhibitors at pH 7.5) were immunoprecipitated by anti-Myc or rabbit-IgG coupled to agarose beads (Sigma) for 2 h at 4°C. Beads were washed five times with increasing salt concentrations (150–500 mM KCl), boiled in Laemmli buffer and eluted proteins subjected to 10% SDS-PAGE, transferred onto nitrocellulose membrane, and probed with monoclonal antibodies to the Myc Tag (Santa Cruz), Mod (Garzino et al. 1992), En (4D9, Developmental Studies Hybridoma Bank), Ubx (FP3.38), and DMyc (Prober and Edgar 2000) or polyclonal antibodies to DFos (Zeitlinger and Bohmann 1999), DJun (Santa Cruz), or HDAC1/DRpd3 (Abcam). Quantitative assays of β-galactosidase activity were performed as described in Simon and Lis (1987), and the values obtained from triplicate assays on batches of 20 larvae of each genotype are given as a relative percentage, setting to 100% the value obtained for lexA–lacZ/+; UASLexA–Dfos; hsGal4 animals.
Cell culture transfection and luciferase assays
HEK293 cells were grown in DMEM with 10% FBS and transiently transfected using the calcium phosphate precipitation method. Expression vectors pAP-1-Luciferase, pCEFL-HA-DFos, pCEFL-HA-DJun, pCEFL-HA-DJNK, and pCEFL-HA-DJNKK were kindly provided by S.X. Hou (Frederick, MD), and pCMV2-hTip60 was provided by K.V. Kandror (Boston, MA). Vectors for Flag-Chm and Flag-ChmG680E expression were generated from PCR-amplified chm and chmG680E cDNAs cloned in frame in pXJ-Flag. pDEST26-HA-FosbZIP, pDEST26-HA-FosNAla, and pDEST26-Myc-DRpd3 expression vectors were obtained by PCR amplification of the corresponding coding sequences. All constructs were sequence-verified. Reporter plasmid (1 μg) and expression vectors (50 ng, except when stated otherwise) were transfected with 100 ng of CMV-β-Gal plasmid, and total DNA was scaled up to 10 μg with pBluescript. For luciferase assays, cells were lysed in 300 μL of lysis buffer (25 mM Tris, 2 mM EDTA, 1 mM DTT, 10% Glycerol, 1% Triton X-100 at pH 7.8) by three cycles of freeze-thawing, and extracts were analyzed for β-Gal (30 μL) or luciferase (50 μL) activities. For immunoprecipitations, nuclear extracts from transfected HEK293 cells were treated as described above.
GST pull-down assays
GST fusions DFosA, DFosD, DJun, and DJNK are described in Ciapponi et al. (2001). His-ChmNter and His-ChmCter were generated by subcloning BamH1/StuI (Nter) or StuI/HindIII (Cter) fragments of chm cDNAs in pQE31. DFosB, DFosC, and DFosE deletions were generated by PCR amplification and Gateway recombination. Recombinant proteins were expressed in Escherichia coli BL21. 35S-methionine-labeled DRpd3 was produced by in vitro transcription/translation according to manufacturer's instructions (Promega). Pull-down assays were adapted from Ciapponi et al. (2001), using 300 mM NaCl and 1.2 μg/mL of ethidium bromide in the binding buffer.
ChIP analyses
Assays were performed with soluble chromatin from 2 × 108 HEK293 cells. Antibodies to modified histones were from Upstate Biotechnology and from Abcam. Anti-HA-agarose, anti-Myc-agarose, IgG-agarose, and anti-Flag-Sepharose were from Sigma. ChIP assays were performed as in Cawley et al. (2004). To ascertain that variations in histone modification levels were not due to nucleosome displacement, removal, or enrichment, controls by ChIP with anti-H4 were systematically performed. No significant changes in nucleosome occupancy were observed.
RT–PCR assays
RNA from wing imaginal discs (1 h APF) or transfected HEK293 cells was extracted by Trizol (GIBCO-BRL) and purified by RNeasy column (Qiagen). One microgram of total RNA was used for first-strand cDNA reaction using oligo d(T) and SuperScript II reverse transcriptase (Invitrogen) for 50 min at 42°C. RNA quantification by real-time PCR was performed as described above, and normalized with reference to rp49 RNA for wing imaginal discs and GAPDH for HEK293 cells.
Statistical analyses
Statistical analyses were performed with the Student's t-test: ns indicates no statistical difference, P < 0.6; *, P < 0.1; **, P < 0.01; and ***, P < 0.001.
Acknowledgments
We thank A.J. Saurin for critical reading of the manuscript, P. Ramain for initial help in cell culture, and colleagues and the Bloomington centre for fly lines. This work was supported by the CNRS, grants from “l'Association pour la Recherche contre le Cancer” (ARC) and “la Ligue Nationale Contre Le Cancer” (LNCC), and fellowships from “le Ministère de la Recherche et de la Technologie” (MRT) and l'ARC.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.359506.
Corresponding authors.
References
- Adachi-Yamada T., Fujimura-Kamada, K., Nishida, Y., and Matsumoto, K. 1999. Distortion of proximodistal information causes JNK-dependent apoptosis in Drosophila wing. Nature 400: 166–169. [DOI] [PubMed] [Google Scholar]
- Agalioti T., Chen, G., and Thanos, D. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111: 381–392. [DOI] [PubMed] [Google Scholar]
- Aggarwal B.D. and Calvi, B.R. 2004. Chromatin regulates origin activity in Drosophila follicle cells. Nature 430: 372–376. [DOI] [PubMed] [Google Scholar]
- Agnes F., Suzanne, M., and Noselli, S. 1999. The Drosophila JNK pathway controls the morphogenesis of imaginal discs during metamorphosis. Development 126: 5453–5462. [DOI] [PubMed] [Google Scholar]
- Akhtar A. and Becker, P.B. 2000. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell 5: 367–375. [DOI] [PubMed] [Google Scholar]
- Bagowski C.P., Besser, J., Frey, C.R., and Ferrell Jr., J.E. 2003. The JNK cascade as a biochemical switch in mammalian cells: Ultrasensitive and all-or-none responses. Curr. Biol. 13: 315–320. [DOI] [PubMed] [Google Scholar]
- Bannister A.J. and Kouzarides, T. 1995. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J. 14: 4758–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkoussa M., Brand, C., Delmotte, M.H., Formstecher, P., and Lefebvre, P. 2002. Retinoic acid receptors inhibit AP-1 activation by regulating extracellular signal-regulated kinase and CBP recruitment to an AP-1-responsive promoter. Mol. Cell Biol. 22: 4522–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawley S., Bekiranov, S., Ng, H.H., Kapranov, P., Sekinger, E.A., Kampa, D., Piccolboni, A., Sementchenko, V., Cheng, J., Williams, A.J., et al. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116: 499–509. [DOI] [PubMed] [Google Scholar]
- Ciapponi L., Jackson, D.B., Mlodzik, M., and Bohmann, D. 2001. Drosophila Fos mediates ERK and JNK signals via distinct phosphorylation sites. Genes & Dev. 15: 1540–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eferl R. and Wagner, E.F. 2003. AP-1: A double-edged sword in tumorigenesis. Nat. Rev. Cancer 3: 859–868. [DOI] [PubMed] [Google Scholar]
- Garzino V., Pereira, A., Laurenti, P., Graba, Y., Levis, R.W., Le Parco, Y., and Pradel, J. 1992. Cell lineage-specific expression of modulo, a dose-dependent modifier of variegation in Drosophila. EMBO J. 11: 4471–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienenberger A., Miotto, B., Sagnier, T., Cavalli, G., Schramke, V., Geli, V., Mariol, M.C., Berenger, H., Graba, Y., and Pradel, J. 2002. The MYST domain acetyltransferase Chameau functions in epigenetic mechanisms of transcriptional repression. Curr. Biol. 12: 762–766. [DOI] [PubMed] [Google Scholar]
- Hilfiker A., Hilfiker-Kleiner, D., Pannuti, A., and Lucchesi, J.C. 1997. mof, a putative acetyltransferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J. 16: 2054–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasper H., Benes, V., Schwager, C., Sauer, S., Clauder-Munster, S., Ansorge, W., and Bohmann, D. 2001. The genomic response of the Drosophila embryo to JNK signaling. Dev. Cell 1: 579–586. [DOI] [PubMed] [Google Scholar]
- Kim T., Yoon, Y., Cho, H., Lee, W-B., Kim, J., Song, Y-H., Kim, S.N., Yoon, J.H., Kim-Ha, J., and Kim, Y-J. 2005. Downregulation of lipopolysaccharide response in Drosophila by negative crosstalk between the AP-1 and the NF-B signaling modules. Nat. Immunol. 6: 211–218. [DOI] [PubMed] [Google Scholar]
- Kockel L., Homsy, J.G., and Bohmann, D. 2001. Drosophila AP-1: Lessons from an invertebrate. Oncogene 20: 2347–2364. [DOI] [PubMed] [Google Scholar]
- Lachner M. and Jenuwein, T. 2002. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 14: 286–298. [DOI] [PubMed] [Google Scholar]
- Lehmann M. and Koolman, J. 1988. Ecdysteroid receptors of the blowfly Calliphora vicina: Partial purification and characterization of ecdysteroid binding. Mol. Cell Endocrinol. 57: 239–249. [DOI] [PubMed] [Google Scholar]
- Liu X., Grammont, M., and Irvine, K.D. 2000. Roles for scalloped and vestigial in regulating cell affinity and interaction between the wing blade and wing hinge. Dev. Biol. 228: 287–303. [DOI] [PubMed] [Google Scholar]
- Martin-Blanco E., Pastor-Pareja, J.C., and Garcia-Bellido, A. 2000. JNK and decapentaplegic signaling control adhesiveness and cytoskeleton dynamics during thorax closure in Drosophila. Proc. Natl. Acad. Sci. 97: 7888–7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munz C., Psichari, E., Mandilis, D., Lavigne, A.C., Spiliotaki, M., Oehler, T., Davidson, I., Tora, L., Angel, P., and Pintzas, A. 2003. TAF7 (TAFII55) plays a role in the transcription activation by c-Jun. J. Biol. Chem. 278: 21510–21516. [DOI] [PubMed] [Google Scholar]
- Noselli S. and Agnes, F. 1999. Roles of the JNK signaling pathway in Drosophila morphogenesis. Curr. Opin. Genet. Dev. 9: 466–472. [DOI] [PubMed] [Google Scholar]
- Ogawa S., Lozach, J., Jepsen, K., Sawka-Verhelle, D., Perissi, V., Sasik, R., Rose, D.W., Johnson, R.S., Rosenfeld, M.G., and Glass, C.K. 2004. A nuclear receptor corepressor transcriptional checkpoint controlling activator protein 1-dependent gene networks required for macrophage activation. Proc. Natl. Acad. Sci. 101: 14461–14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor-Pareja J.C., Grawe, F., Martin-Blanco, E., and Garcia-Bellido, A. 2004. Invasive cell behavior during Drosophila imaginal disc eversion is mediated by the JNK signaling cascade. Dev. Cell 7: 387–399. [DOI] [PubMed] [Google Scholar]
- Prober D.A. and Edgar, B.A. 2000. Ras1 promotes cellular growth in the Drosophila wing. Cell 100: 435–446. [DOI] [PubMed] [Google Scholar]
- Simon J.A. and Lis, J.T. 1987. A germline transformation analysis reveals flexibility in the organization of heat shock consensus elements. Nucleic Acid Res. 15: 2971–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szuts D. and Bienz, M. 2000. LexA chimeras reveal the function of Drosophila Fos as a context-dependent transcriptional activator. Proc. Natl. Acad. Sci. 97: 5351–5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss C., Schneider, S., Wagner, E.F., Zhang, X., Seto, E., and Bohmann, D. 2003. JNK phosphorylation relieves HDAC3-dependent suppression of the transcriptional activity of c-Jun. EMBO J. 22: 3686–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston C.R. and Davis, R.J. 2002. The JNK signal transduction pathway. Curr. Opin. Genet. Dev. 12: 14–21. [DOI] [PubMed] [Google Scholar]
- Xia Y. and Karin, M. 2004. The control of cell motility and epithelial morphogenesis by Jun kinases. Trends Cell. Biol. 14: 94–101. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J. and Bohmann, D. 1999. Thorax closure in Drosophila: Involvement of Fos and the JNK pathway. Development 126: 3947–3956. [DOI] [PubMed] [Google Scholar]
- Zhang J., Kalkum, M., Chait, B.T., and Roeder, R.G. 2002. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell 9: 611–623. [DOI] [PubMed] [Google Scholar]