Abstract
Sequence-based analyses have predicted that ∼35% of mammalian alternative splicing (AS) events produce premature termination codon (PTC)-containing splice variants that are targeted by the process of nonsense-mediated mRNA decay (NMD). This led to speculation that AS may often regulate gene expression by activating NMD. Using AS microarrays, we show that PTC-containing splice variants are generally produced at uniformly low levels across diverse mammalian cells and tissues, independently of the action of NMD. Our results suggest that most PTC-introducing AS events are not under positive selection pressure and therefore may not contribute important functional roles.
Keywords: Alternative splicing, microarray analysis, nonsense-mediated mRNA decay, premature termination codon
Alternative splicing (AS), the process by which exons in transcripts are joined in different combinations to generate multiple mRNA variants, represents an important mechanism for the expression of structurally and functionally distinct proteins from a limited number of genes (Graveley 2001; Black 2003). Regulation of AS plays critical roles in cell growth, differentiation, and cell death, and aberrant AS has been implicated in many human diseases (Smith and Valcarcel 2000; Caceres and Kornblihtt 2002; Cartegni et al. 2002). It has been estimated that at least 74% of human genes contain one or more alternative exons (Johnson et al. 2003), yet it is not known to what extent the resulting splice variants specify functionally relevant transcripts and proteins.
Previous studies have shown that the introduction of premature termination codons (PTCs) in spliced transcripts can activate transcript degradation via the process of nonsense-mediated mRNA decay (NMD) (Hillman et al. 2004; Maquat 2004; Alonso 2005). NMD is important for the removal of nonfunctional PTC-containing transcripts (Mendell et al. 2004; Mitrovich and Anderson 2005). It has also been shown to function in the autoregulation of transcript levels of several RNA-binding proteins, including splicing factors (Morrison et al. 1997; Sureau et al. 2001; Wollerton et al. 2004; for review, see Lareau et al. 2004). In addition to a PTC, NMD requires a set of upstream frameshift (UPF) protein factors that associate with the spliced transcripts via interactions with a post-splicing exon junction complex (Lykke-Andersen et al. 2000; Kim et al. 2001; Lejeune et al. 2002; Gehring et al. 2003). One or more of the UPF proteins, including the essential UPF1 protein (Medghalchi et al. 2001), subsequently recruit RNA nucleases that initiate degradation of the PTC-containing transcripts (Lejeune et al. 2003; for review, see Baker and Parker 2004).
Recent bioinformatics analyses of expressed sequence tag (EST) and cDNA sequence data have predicted that ∼35% of AS events have the potential to introduce PTCs that could elicit NMD of transcripts (Green et al. 2003; Lewis et al. 2003). This finding led to speculation that NMD activated by AS may represent a widely used mechanism by which gene expression is down-regulated (Hillman et al. 2004; Neu-Yilik et al. 2004; Alonso 2005; Lejeune and Maquat 2005), and it has also been proposed that the introduction of PTCs by AS serves as a mechanism for the tissue-specific regulation of gene expression (Hillman et al. 2004; Holbrook et al. 2004; Raes and Van de Peer 2005). However, no study has yet examined experimentally the actual levels of PTC-containing mRNA splice variants in normal cells and tissues, and it is not known to what extent AS functions globally to regulate transcript levels via NMD. Moreover, although it has been reported that short interfering duplex RNA (siRNA)-mediated depletion of UPF1 in cultured cells results in changes in the levels (both up and down) of transcripts for ∼9% of genes (Mendell et al. 2004), it was not determined to what extent these effects might be coupled to AS events that introduce PTCs.
In the present study, we used a recently described quantitative microarray platform (Pan et al. 2004) to determine the relative levels of PTC-containing versus non-PTC-containing splice variants in mammalian cells and tissues of diverse origin. The majority of PTC-containing splice variants were found to be present at low steady-state abundance and only very rarely appeared to be subject to tissue-specific regulation. Efficient depletion of UPF1 resulted in increased levels of PTC-containing splice variants for some genes, but only a small proportion of the total number of genes that encode PTC-introducing AS events displayed pronounced UPF1-dependent changes in alternative splicing levels, and these effects were not significantly correlated with the effects of UPF1 depletion on transcripts levels. These experimental data, together with supporting computational analyses, suggest that approximately one-third or more of AS events located within the open reading frames (ORFs) of mammalian genes produce PTC-containing splice variant mRNAs that are not under significant selection pressure, and therefore may not play important functional roles.
Results and Discussion
Quantitative profiling of PTC-containing splice variants in normal mammalian tissues
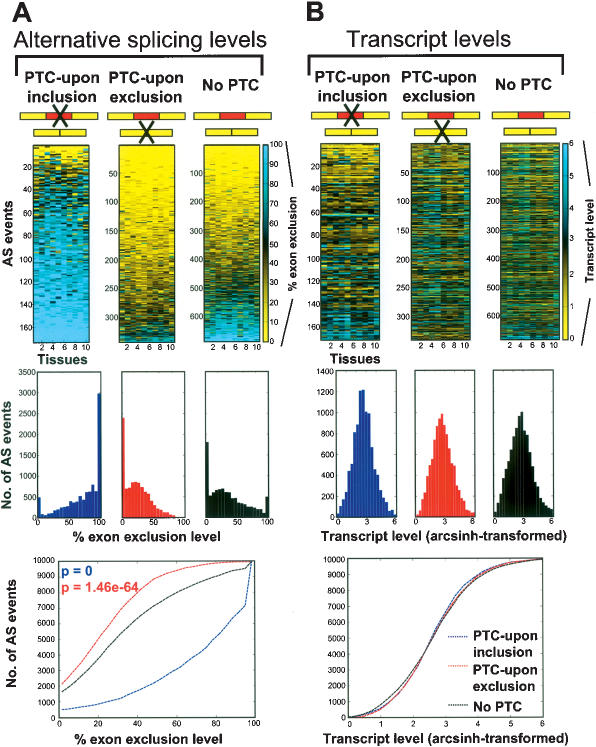
A recently described quantitative AS microarray platform (Pan et al. 2004) was employed to survey the levels of splice variants produced from genes that do or do not encode PTC-introducing AS events. This survey was initially conducted across 10 diverse adult mouse tissues. The AS microarray contains sets of six oligonucleotide probes (specific for exon and splice junction sequences) to monitor the inclusion/exclusion levels of 3126 sequence-verified cassette-type alternative exons. A computer algorithm, referred to as the “Generative model for the Alternative Splicing Array Platform” (GenASAP), was used to infer the percent exclusion levels of each alternatively spliced exon (i.e., the percentage of the total transcripts from a gene with a skipped alternative exon) (Pan et al. 2004). GenASAP also generates a confidence rank for each percent exclusion value. Percent exclusion levels in the top half of the ranks correlate well with independent RT-PCR measurements and were used for most of the analyses in the present study (refer to Supplementary Tables S1-S3 for data). Using this system, we compared the percent exclusion levels of alternative exons represented on the microarray that have the potential to introduce PTCs that can activate NMD, with the percent exclusion levels of alternative exons that do not have the potential to introduce PTCs that can activate NMD (Fig. 1A).
Figure 1.
Microarray comparison of AS and transcript levels of genes containing AS events predicted, based on sequence analysis, to introduce a PTC upon exon inclusion (“PTC-upon inclusion”), introduce a premature termination codon upon exon exclusion (“PTC-upon exclusion”), or not to introduce a PTC (“No PTC”) (refer to Supplementary Tables S1, S2 for more information). Alternative splicing and corresponding transcript levels of genes were profiled in 10 diverse mouse tissues, arranged from left to right in each panel (lanes 1-10: brain, heart, intestine, kidney, liver, lung, muscle, salivary, spleen, and testis). (A) Alternative splicing levels are represented by percent exon exclusion (i.e., the percent of the total transcripts from a gene with a skipped exon, indicated by the color scale). The AS events are ordered from low to high percent exon exclusion (based on the average value for the 10 tissues) down the Y-axis. (B) Transcript levels are represented by the averages of arcsinh-transformed intensity measurements of the probes specific for the flanking constant exons (indicated by the color scale). The order of AS events down the Y-axis in B is the same as in A, allowing a direct comparison. Histograms below the microarray clustergrams in A represent the distribution of AS events with the percent exon exclusion level range indicated. Histograms in B represent the distribution of AS events with the transcript levels indicated. The percent exon exclusion and transcript levels measured in the 10 tissues in each of the PTC-upon inclusion, PTC-upon exclusion, and No PTC categories were resampled 10,000 times to normalize for the different numbers of AS events in each category. The line curves (bottom row) show cumulative distributions of percent exon exclusion or transcript levels of the AS events, using the same data as used to plot the histograms. The distributions of percent exon exclusion levels between the PTC-upon inclusion (blue text)/exclusion (red text) and No PTC categories are significantly different (Wilcoxon rank sum test). In contrast, differences in the distributions of transcript levels between the PTC-upon inclusion/exclusion and No PTC categories are not statistically significant.
Analysis of transcript sequence data corresponding to the AS events represented on the mouse AS microarray indicated that 514 (42.7%) of the 1204 AS events located in genes with known ORFs have the potential to result in the introduction of a PTC and trigger NMD (Supplementary Table S1); 172 (14.3%) of the AS events introduce a PTC upon inclusion of the alternative exon (“PTC-upon inclusion” category), and 342 (28.4%) of the AS events introduce a PTC upon skipping of the alternative exon (“PTC-upon exclusion” category). The remaining 690 (57.3%) AS events are not expected to introduce a PTC or activate NMD (“No PTC” category).
Global suppression of PTC-containing splice variants in mammalian tissues
A comparison of the percent exon exclusion levels among the three AS event categories across the 10 mouse tissues revealed striking differences that are consistent with the widespread suppression of splice variants that contain PTCs introduced by AS: 84.9% of the AS events in the PTC-upon inclusion category have an average percent exon exclusion level >50% (i.e., overall high alternative exon exclusion), while, in contrast, 92.1% of the AS events in the PTC-upon exclusion category have an average percent exon exclusion level <50% (i.e., overall high alternative exon inclusion) (Fig. 1A). On average, the percent exon exclusion levels of AS events in the No PTC category are intermediate between the PTC-upon inclusion and PTC-upon exclusion categories. Consistent with our previous result indicating that the majority of alternative exons in mammalian tissues do not undergo pronounced skipping (Pan et al. 2004), the majority of AS events in the No PTC category are highly included.
We also analyzed steady-state transcript levels of the same genes that were analyzed for percent exon exclusion levels in the three AS event categories in Figure 1A (Fig. 1B). Transcript levels were determined using averages of the signals from flanking constitutive exon probes on the microarray. Remarkably, although there are pronounced differences in the overall percent exon exclusion levels among the three categories, the overall steady-state transcript levels of the genes in each category are not significantly different (Fig. 1B). We obtained similar results when analyzing transcript profiling data (Zhang et al. 2004) for an overlapping set of genes in 45 additional mouse cell and tissue types (Supplementary Fig. S1). These results suggest that NMD does not function on a frequent basis to reduce overall steady-state levels of transcripts of genes that encode PTC-introducing AS events.
The data in Figure 1A demonstrate that PTC-containing transcripts generated by AS are generally present at low abundance levels in diverse adult mammalian tissues, compared with transcripts that do not contain PTCs. This observation was confirmed by performing RT-PCR assays on nine representative examples where the PTC-containing splice variant was predicted from the microarray data to represent the lower abundance variant in all 10 tissues. In all nine cases (90 independent RT-PCR reactions), the expected pattern was observed and the PTC-containing splice variant was generally found to be uniformly low in abundance in the 10 tissues (Supplementary Fig. S2). In addition, in a small fraction of the microarray data (Fig. 1A), the predicted PTC-containing splice variants appeared to be more abundant in one or more tissues than the splice variants predicted not to contain a PTC. However, out of 12 representative cases examined across the 10 tissues (120 independent RT-PCR reactions), only three were validated by RT-PCR assays (Supplementary Fig. S2). This low validation rate can be attributed to the observation that many of the latter predictions are from low-ranking GenASAP data (refer to the legend for Supplementary Fig. S2 for more information). These data suggest that possible regulatory mechanisms resulting in the increased ratio of PTC-containing to non-PTC-containing splice variants in mammalian tissues are extremely rare.
The results described above are consistent with previous proposals that NMD may act efficiently and on a frequent basis to reduce the levels of PTC-containing transcripts. However, it is also possible that many of the PTC-containing splice variants are initially produced at low levels, independently of the action of NMD. In order to investigate this possibility, we next analyzed the global effects of depletion of the critical NMD factor UPF1 on the percent exon exclusion levels of alternative exons that do or do not have the potential to introduce PTCs.
Global analysis of the role of UPF1 in reducing PTC-containing splice variants
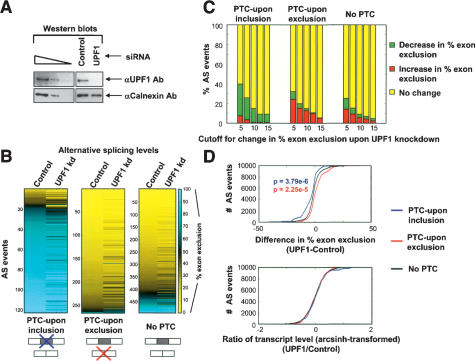
Using a new microarray designed for the quantitative profiling of 3055 sequence-verified human cassette-type AS events (Fig. 2; refer to Supplementary Tables S1, S3), the percent exon exclusion levels in the three AS event categories (PTC-upon inclusion/exclusion and No PTC) were compared in HeLa cells, following treatment of the cells with either a siRNA shown previously to specifically and efficiently knock down the expression of UPF1, or with a control, nonspecific siRNA (Fig. 2A; Kim et al. 2005). Western blotting showed that the UPF1 siRNA reduced UPF1 to ∼5% of its normal endogenous level, whereas the control siRNA did not significantly alter the level of UPF1 (Fig. 2A). In cells treated with the control siRNA, the overall distributions of percent exon exclusion levels in the three AS event categories were remarkably similar to those observed in each of the 10 mouse tissues using the mouse AS microarray (cf. Figs. 1A and 2B, Control). This demonstrates that cell types of diverse origin and from different mammalian species maintain overall low levels of PTC-containing splice variants.
Figure 2.
Microarray analysis of the role of the essential NMD factor UPF1 in controlling AS and transcript levels. (A) Western blot of total cellular protein isolated from HeLa cells 3 d after transfection with either an siRNA that reduces expression of UPF1 (UPF1 kd), or a nonspecific control siRNA (Control) (Kim et al. 2005). The protein lysates were probed with an antibody specific for UPF1 (Lykke-Andersen et al. 2000), and with an anti-Calnexin antibody as a loading control. Serial threefold dilutions of protein in the left three lanes indicate that the Western blot is semiquantitative and permitted an estimation of the UPF1 depletion efficiency at ∼95%. (B) Microarray comparison of percent exon exclusion levels profiled in Control and UPF1-depleted HeLa cells. Genes are ordered down the Y-axis from low to high percent exon exclusion in the Control columns, and the same gene orders are preserved in the UPF1 kd columns. Data for percent exon exclusion levels are shown for the three AS event categories (PTC-upon inclusion/exclusion and No PTC). The color scale is as shown in Figure 1A. (C) The bar graphs indicate the percent of AS events (Y-axis) in the three AS event categories that show an increase, decrease, or no change in percent exon exclusion level, measured in each case as the difference in the percent exon exclusion between the UPF1 kd and Control kd. Different cutoffs (ranging from at least 5% to at least 15%) for the difference in percent exon exclusion levels were used, as indicated on the X-axis. The enrichment for percent exon exclusion changes in the PTC-upon inclusion/exclusion categories that result in increased levels of the PTC-containing variant upon UPF1 knockdown is statistically significant (p < 10-4 for 5%-10% difference and p < 0.05 for 15% difference, Fisher's exact test). (D) The difference in percent exon exclusion values (upper panel) and ratios of transcript levels (lower panel) between the UPF1-depleted and Control samples for the three AS event categories are displayed in cumulative distribution plots. These plots were generated using data for the same AS genes as shown in B, which represent all AS events with GenASAP percent exon exclusion values ranking in the top half of the data (refer to the text). The microarray measurements in the PTC-upon inclusion/exclusion and No PTC were resampled 10,000 times to normalize for different numbers of AS events in each category. The changes in percent exon exclusion levels between the PTC-upon inclusion/exclusion and No PTC categories, upon UPF1 knockdown, are significantly different (Wilcoxon rank sum test). The differences in ratios of transcript levels between the PTC-upon inclusion/exclusion and No PTC categories, upon UPF1 knockdown, are not statistically significant.
Consistent with a critical role for NMD in the removal of PTC-containing transcripts, increased ratios of PTC-containing to non-PTC-containing splice variants were observed for 80%-90% of AS events that displayed a change in exon exclusion level in cells depleted of UPF1 (Fig. 2C). However, only a relatively small proportion of the total genes in the PTC-upon inclusion/exclusion categories displayed substantial changes in percent exon exclusion levels upon depletion of UPF1. Whereas 30%-40% of the genes in the PTC-upon inclusion/exclusion categories displayed a 5% or greater difference in percent exon exclusion level between the control and UPF1-depeleted cells, <10% of the genes displayed a percent exon exclusion level difference of 15% or higher (Fig. 2C).
At a significantly lower frequency (0%-10% of PTC-introducing AS events that display a 15% or greater change in exon exclusion level), depletion of UPF1 also resulted in increased ratios of predicted non-PTC-containing to PTC-containing splice variants (Fig. 2C). Moreover, a somewhat smaller number of genes in the No PTC category also displayed changes in percent exon exclusion levels upon UPF1 depletion, but these changes involved similar frequencies of increased exon inclusion or exclusion (Fig. 2C). Both of these effects could be attributed to separate activities of UPF1 on transcript levels operating in a direct manner, for example, by acting on PTC-introducing events not accounted for by our sequence analysis, or could be due to possible indirect effects. In summary, the results described above indicate that NMD may reduce the levels of many PTC-containing splice variants to a small extent, whereas only a relatively small fraction of PTC-introducing AS events appear to be subject to more pronounced regulation by NMD.
We also examined the effects of UPF1 knockdown on transcript levels (using averages of the signals from flanking constitutive exon probes on the microarray), both for the subset of genes that display a significant change in percent exon exclusion level, and also for all genes corresponding to the top half-ranking GenASAP AS events analyzed in the PTC-upon inclusion/exclusion and No PTC categories in Figure 2A (Fig. 2D; Supplementary Fig. S3). Surprisingly, in either case, although UPF1 results in overall increases in the ratios of PTC-containing to non-PTC-containing splice variants, the overall distributions of transcript levels in the PTC-upon inclusion/exclusion and No PTC AS event categories were not significantly different. This is evident from a comparison of cumulative distribution plots showing the differences in percent exclusion levels of the AS events in UPF1-depleted versus control cells (Fig. 2D, upper panel), and the differences between the transcript levels for the same sets of genes (Fig. 2D, lower panel). Similar results were obtained when comparing the overall transcript levels for the subset of genes in each AS event category that show a 10% or greater change in exon exclusion level between the UPF1-depleted and control cells (Supplementary Fig. S3).
In order to investigate the effects of UPF1 depletion on transcript levels further, we performed RT-PCR assays on sets of AS events that display a range of UPF1-dependent changes in percent exon exclusion in the PTC-upon inclusion/exclusion and No PTC categories. Representative RT-PCR reactions from each AS event category are shown in Supplementary Figure S4. Interestingly, it is apparent from the RT-PCR data that depletion of UPF1, while generally resulting in increased levels of PTC-containing splice variants, in many cases also results in the simultaneous decrease in level of non-PTC-containing splice variants, such that the overall transcript levels are not significantly altered. In other cases analyzed, the change in splice variant ratio detected in the microarray data and by RT-PCR assay involved only very minor increases in PTC-containing variants, without detectable decreases in the non-PTC-containing splice variant from the same genes, but the resulting total transcript level changes may be below the limit of the sensitivity of detection on the microarray. These results therefore show that UPF1-dependent changes in percent exon exclusion levels on average do not detectably affect transcript levels of the corresponding genes. Moreover, consistent with the results of a recent microarray profiling study showing that the transcript levels for ∼9% of human genes show at least a 1.9-fold increase or decrease upon depletion of UPF1 (Mendell et al. 2004), we find that the transcript levels for ∼6% of the genes measured on the human AS microarray are affected to the same extent by depletion of UPF1 (data not shown). However, consistent with the results in Figure 2 and Supplementary Figure S4, these genes are not significantly associated with PTC-introducing AS events (Supplementary Fig. S5).
The results described above provide evidence that NMD may only play a limited role in the suppression of PTC-containing splice variants. The majority of PTC-containing transcripts produced by AS do not appear to undergo pronounced UPF1-dependent suppression but instead are already present at low abundance. Thus, taken together with the results in Figure 1, the data in Figure 2 and Supplementary Figures S3-S5 support the view that ∼80% of PTC-introducing AS events, which comprise as many as one-third of all cassette exon AS events that overlap known ORFs, produce low levels of splice variants independently of NMD.
Global selection pressure acting against PTC-introducing AS events
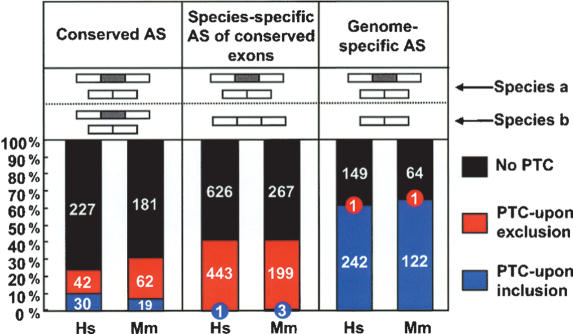
To further assess the possible functional significance of the majority of PTC-containing splice variants, which, based on our microarray data, are generally found to be present at low abundance levels, we next determined what proportion of these are mouse-specific versus conserved between human and mouse (Fig. 3). Conserved AS events are those that can be detected in both human and mouse transcripts. Two different types of species-specific AS events have been defined by comparisons of human and mouse genome and transcript sequences (Modrek and Lee 2003; Pan et al. 2005; Yeo et al. 2005). One type, referred to as “species-specific AS of conserved exons,” is represented by a conserved exon that is alternatively spliced in mouse but constitutively spliced in orthologous human transcripts. The other type, referred to as “genome-specific AS,” is represented by an alternative exon in mouse transcripts that is not detected in the orthologous human transcripts. We find that both types of species-specific AS events are more often represented by PTC-introducing AS events than are conserved AS events (p < 0.005, Fisher's exact test) (Fig. 3). Very similar results were obtained when comparing a larger data set of sequence-validated human AS events (p < 10-8, Fisher's exact test) (Fig. 3; Supplementary Table S4). These results are consistent with previous observations showing that conserved AS events more often are frame-preserving than species-specific AS events (Philipps et al. 2004; Resch et al. 2004; Sorek et al. 2004; Yeo et al. 2005). Indeed, a very recent report has provided evidence, also from the comparative analysis of human and mouse sequences, that only ∼5% of all cassette alternative exons are both conserved and have the potential to introduce PTCs (Baek and Green 2005). Given the likelihood that species-specific AS events, relative to conserved AS events, more often generate nonfunctional transcripts, the observations summarized above support the conclusion from the microarray comparisons of AS and transcript levels that PTC-containing transcripts generated by AS are not expected to often represent functionally relevant splice variants.
Figure 3.
AS events with the potential to trigger NMD are more often represented by species-specific alternative exons than conserved alternative exons. Percentages of total AS events with or without the potential to activate NMD that are conserved or species-specific in human and mouse are shown. Alternative exons were identified in human and mouse ortholog gene pairs using EST and cDNA sequence data, as described previously (Pan et al. 2005) (see also Materials and Methods), and also scored for their potential to introduce a PTC and elicit NMD. “Conserved AS” indicates detection of sequence-conserved alternative and flanking exons in a human and mouse ortholog pair; “Species-specific AS of conserved exons” indicates a conserved exon that is alternatively spliced in one species and constitutively spliced in the other; and “Genome-specific AS” indicates detection of an AS event in one species and sequence evidence for only the spliced constitutive exons in the other species.
Conclusions
Ongoing analyses of human and mouse genome and transcript sequence data, as well as microarray studies, are documenting a vast repertoire of alternative spliced mRNAs. A major question of the post-genomic era, therefore, is the extent to which these alternatively spliced transcripts are functionally significant. Combined with information from sequence analyses on the conservation of these splice variants, as well as computational predictions as to which AS events have the potential to introduce PTCs that can activate NMD, our results address this question using an experimental, microarray-based approach that allows the levels of thousands of these splice variants to be simultaneously monitored in individual cell and tissue types. Importantly, our observation stemming from the use of this system indicating that the majority of PTC-containing transcripts are present at uniformly low abundance levels in normal mammalian tissues, and that only a relatively small proportion of PTC-containing splice variants appear to be substantially regulated by NMD, suggests that functionally important AS events are concentrated among the remaining two-thirds of AS events that do not introduce PTCs.
Materials and methods
Identification of AS events in human and mouse transcripts
Detection and filtering of AS events were performed essentially as described previously (Pan et al. 2004, 2005). To identify cassette and mutually exclusive AS events in human transcripts, we used cDNA/EST sequences data from UniGene (ftp://ftp.ncbi.nih.gov/repository/UniGene; Build #158) and human genome sequence data (ftp://ftp.ncbi.nih.gov/genomes/H_sapiens).
Cell culture, transfection, RNA purification, and protein analysis
Human HeLa cells (2 × 108) were propagated in DMEM medium (GIBCO-BRL) containing 10% fetal bovine serum (GIBCO-BRL) and transiently transfected with 100 nM in vitro synthesized siRNA (Dharmacon) using Oligofectamine (Invitrogen). Protein and total RNA were isolated 3 d later. Control and UPF1 siRNAs and conditions for Western blotting were described previously (Kim et al. 2005). Additional information is provided in the Supplemental Material.
Microarray hybridization, image processing, and data analysis
Microarray design, hybridization, and data analysis for 3055 human AS events (represented on a single 22K Agilent microarray) were performed essentially as described previously (Pan et al. 2004). Information on AS events represented on the human microarray is provided in Supplementary Table S3. Analysis of percent exon exclusion levels for 3126 AS events in 10 mouse tissues was performed using data described in our previous study (Pan et al. 2004). The microarray analysis of AS levels in HeLa cells treated with siRNAs (Fig. 2), used percent exon exclusion values generated in the top half (higher confidence)-ranking portion of the data. Percent exon exclusion values and associated rankings for these values were generated using the GenASAP algorithm, as described previously (Pan et al. 2004). Details of the methods for analyzing transcript levels of alternatively spliced genes are provided in the Supplemental Material.
Detection of PTC-containing splice variants and categorization of conserved and species-specific alternative exons
Available information on ORFs for genes represented in the human and mouse AS microarrays (Figs. 1, 2; Supplementary Tables S2, S3), as well as in a larger data set of human AS events (Fig. 3; Supplementary Table S4), was obtained from the header-line of the corresponding human and mouse cDNA sequences in the UniGene database. Termination codons were mapped to the ORF sequences in relation to the splice junctions. A termination codon was considered premature (PTC) and potentially NMD-triggering if it was found 50 or more nucleotides upstream of the last exon-exon junction (Nagy and Maquat 1998). After applying the “50-nucleotide rule,” it was determined that 42.7% of the 1204 mouse AS events and 41.8% of the 2270 human AS events have the potential to elicit NMD (Supplementary Table S1). Additional details of methods for detection of PTC-introducing AS events, as well as methods for detection of conserved and species-specific AS events, is provided in the Supplemental Material.
RT-PCR assays
RT-PCR reactions (Supplementary Figs. S2, S4) were carried out as described previously (Pan et al. 2004, 2005), with modifications as described in the Supplemental Material.
Acknowledgments
We thank Leo Lee for help with data analysis; and John Calarco, Timothy Hughes, Jim Ingles, Joanna Ip, May Khanna, Susan McCracken, and Miles Wilkinson for helpful discussions and comments on the manuscript. We are also grateful to Jens Lykke-Andersen for the anti-UPF1 antibody. Y.K.K. and L.E.M. are supported by GM059614. O.S. and A.L.S. are sup-ported by NSERC graduate scholarships. This work was supported by operating grants from the CIHR and NCIC to B.J.B.
Supplemental material is available at http://www.genesdev.org and http://www.utoronto.ca/intron/AS.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1382806.
References
- Alonso, C.R. 2005. Nonsense-mediated RNA decay: A molecular system micromanaging individual gene activities and suppressing genomic noise. Bioessays 27: 463-466. [DOI] [PubMed] [Google Scholar]
- Baek, D. and Green, P. 2005. Sequence conservation, relative isoform frequencies, and nonsense-mediated decay in evolutionarily conserved alternative splicing. Proc. Natl. Acad. Sci. 102: 12813-12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, K.E. and Parker, R. 2004. Nonsense-mediated mRNA decay: Terminating erroneous gene expression. Curr. Opin. Cell Biol. 16: 293-299. [DOI] [PubMed] [Google Scholar]
- Black, D.L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72: 291-336. [DOI] [PubMed] [Google Scholar]
- Caceres, J.F. and Kornblihtt, A.R. 2002. Alternative splicing: Multiple control mechanisms and involvement in human disease. Trends Genet. 18: 186-193. [DOI] [PubMed] [Google Scholar]
- Cartegni, L., Chew, S.L., and Krainer, A.R. 2002. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 3: 285-298. [DOI] [PubMed] [Google Scholar]
- Gehring, N.H., Neu-Yilik, G., Schell, T., Hentze, M.W., and Kulozik, A.E. 2003. Y14 and hUpf3b form an NMD-activating complex. Mol. Cell 11: 939-949. [DOI] [PubMed] [Google Scholar]
- Graveley, B.R. 2001. Alternative splicing: Increasing diversity in the proteomic world. Trends Genet. 17: 100-107. [DOI] [PubMed] [Google Scholar]
- Green, R.E., Lewis, B.P., Hillman, R.T., Blanchette, M., Lareau, L.F., Garnett, A.T., Rio, D.C., and Brenner, S.E. 2003. Widespread predicted nonsense-mediated mRNA decay of alternatively-spliced transcripts of human normal and disease genes. Bioinformatics 19 Suppl. 1: i118-i121. [DOI] [PubMed] [Google Scholar]
- Hillman, R.T., Green, R.E., and Brenner, S.E. 2004. An unappreciated role for RNA surveillance. Genome Biol. 5: R8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook, J.A., Neu-Yilik, G., Hentze, M.W., and Kulozik, A.E. 2004. Nonsense-mediated decay approaches the clinic. Nat. Genet. 36: 801-808. [DOI] [PubMed] [Google Scholar]
- Johnson, J.M., Castle, J., Garrett-Engele, P., Kan, Z., Loerch, P.M., Armour, C.D., Santos, R., Schadt, E.E., Stoughton, R., and Shoemaker, D.D. 2003. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302: 2141-2144. [DOI] [PubMed] [Google Scholar]
- Kim, V.N., Kataoka, N., and Dreyfuss, G. 2001. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science 293: 1832-1836. [DOI] [PubMed] [Google Scholar]
- Kim, Y.K., Furic, L., Desgroseillers, L., and Maquat, L.E. 2005. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell 120: 195-208. [DOI] [PubMed] [Google Scholar]
- Lareau, L.F., Green, R.E., Bhatnagar, R.S., and Brenner, S.E. 2004. The evolving roles of alternative splicing. Curr. Opin. Struct. Biol. 14: 273-282. [DOI] [PubMed] [Google Scholar]
- Lejeune, F. and Maquat, L.E. 2005. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr. Opin. Cell Biol. 17: 309-315. [DOI] [PubMed] [Google Scholar]
- Lejeune, F., Ishigaki, Y., Li, X., and Maquat, L.E. 2002. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: Dynamics of mRNP remodeling. EMBO J. 21: 3536-3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune, F., Li, X., and Maquat, L.E. 2003. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell 12: 675-687. [DOI] [PubMed] [Google Scholar]
- Lewis, B.P., Green, R.E., and Brenner, S.E. 2003. Evidence for the wide-spread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. 7: 189-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen, J., Shu, M.D., and Steitz, J.A. 2000. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103: 1121-1131. [DOI] [PubMed] [Google Scholar]
- Maquat, L.E. 2004. Nonsense-mediated mRNA decay: Splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol. 5: 89-99. [DOI] [PubMed] [Google Scholar]
- Medghalchi, S.M., Frischmeyer, P.A., Mendell, J.T., Kelly, A.G., Lawler, A.M., and Dietz, H.C. 2001. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum. Mol. Genet. 10: 99-105. [DOI] [PubMed] [Google Scholar]
- Mendell, J.T., Sharifi, N.A., Meyers, J.L., Martinez-Murillo, F., and Dietz, H.C. 2004. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 36: 1073-1078. [DOI] [PubMed] [Google Scholar]
- Mitrovich, Q.M. and Anderson, P. 2005. mRNA surveillance of expressed pseudogenes in C. elegans. Curr. Biol. 15: 963-967. [DOI] [PubMed] [Google Scholar]
- Modrek, B. and Lee, C.J. 2003. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat. Genet. 34: 177-180. [DOI] [PubMed] [Google Scholar]
- Morrison, M., Harris, K.S., and Roth, M.B. 1997. smg mutants affect the expression of alternatively spliced SR protein mRNAs in Caenorhabditis elegans. Proc. Natl. Acad. Sci. 94: 9782-9785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, E. and Maquat, L.E. 1998. A rule for termination-codon position within intron-containing genes: When nonsense affects RNA abundance. Trends Biochem. Sci. 23: 198-199. [DOI] [PubMed] [Google Scholar]
- Neu-Yilik, G., Gehring, N.H., Hentze, M.W., and Kulozik, A.E. 2004. Nonsense-mediated mRNA decay: From vacuum cleaner to Swiss army knife. Genome Biol. 5: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Q., Shai, O., Misquitta, C., Zhang, W., Saltzman, A.L., Mohammad, N., Babak, T., Siu, H., Hughes, T.R., Morris, Q.D., et al. 2004. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell 16: 929-941. [DOI] [PubMed] [Google Scholar]
- Pan, Q., Bakowski, M.A., Morris, Q., Zhang, W., Frey, B.J., Hughes, T.R., and Blencowe, B.J. 2005. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet. 21: 73-77. [DOI] [PubMed] [Google Scholar]
- Philipps, D.L., Park, J.W., and Graveley, B.R. 2004. A computational and experimental approach toward a priori identification of alternatively spliced exons. RNA 10: 1838-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes, J. and Van de Peer, Y. 2005. Functional divergence of proteins through frameshift mutations. Trends Genet. 21: 428-431. [DOI] [PubMed] [Google Scholar]
- Resch, A., Xing, Y., Alekseyenko, A., Modrek, B., and Lee, C. 2004. Evidence for a subpopulation of conserved alternative splicing events under selection pressure for protein reading frame preservation. Nucleic Acids Res. 32: 1261-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C.W. and Valcarcel, J. 2000. Alternative pre-mRNA splicing: The logic of combinatorial control. Trends Biochem. Sci. 25: 381-388. [DOI] [PubMed] [Google Scholar]
- Sorek, R., Shamir, R., and Ast, G. 2004. How prevalent is functional alternative splicing in the human genome? Trends Genet. 20: 68-71. [DOI] [PubMed] [Google Scholar]
- Sureau, A., Gattoni, R., Dooghe, Y., Stevenin, J., and Soret, J. 2001. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 20: 1785-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollerton, M.C., Gooding, C., Wagner, E.J., Garcia-Blanco, M.A., and Smith, C.W. 2004. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell 13: 91-100. [DOI] [PubMed] [Google Scholar]
- Yeo, G.W., Van Nostrand, E., Holste, D., Poggio, T., and Burge, C.B. 2005. Identification and analysis of alternative splicing events conserved in human and mouse. Proc. Natl. Acad. Sci. 102: 2850-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., Morris, Q.D., Chang, R., Shai, O., Bakowski, M.A., Mitsakakis, N., Mohammad, N., Robinson, M.D., Zirngibl, R., Somogyi, E., et al. 2004. The functional landscape of mouse gene expression. J. Biol. 3: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]