Abstract
Chronological life span (CLS) in Saccharomyces cerevisiae, defined as the time cells in a stationary phase culture remain viable, has been proposed as a model for the aging of post-mitotic tissues in mammals. We developed a high-throughput assay to determine CLS for ∼4800 single-gene deletion strains of yeast, and identified long-lived strains carrying mutations in the conserved TOR pathway. TOR signaling regulates multiple cellular processes in response to nutrients, especially amino acids, raising the possibility that decreased TOR signaling mediates life span extension by calorie restriction. In support of this possibility, removal of either asparagine or glutamate from the media significantly increased stationary phase survival. Pharmacological inhibition of TOR signaling by methionine sulfoximine or rapamycin also increased CLS. Decreased TOR activity also promoted increased accumulation of storage carbohydrates and enhanced stress resistance and nuclear relocalization of the stress-related transcription factor Msn2. We propose that up-regulation of a highly conserved response to starvation-induced stress is important for life span extension by decreased TOR signaling in yeast and higher eukaryotes.
Keywords: Saccharomyces cerevisiae, TOR, aging, life span, nutrients, yeast
Although aging and age-related diseases account for enormous social and economic costs, the mechanisms that underlie the gradual and progressive deterioration observed in humans remain poorly understood. However, research on model organisms such as mice, flies, worms, and yeast has provided insights into both genetic and environmental factors that can control aging. As with humans, each of these model organisms displays an exponential increase in mortality as it ages (Kaeberlein et al. 2001). Model organisms are useful for their similar basic biology and relatively short life span. Furthermore, the advent of reagents such as the genome-wide deletion collection of Saccharomyces cerevisiae strains (Winzeler et al. 1999) and high complexity Caenorhabditis elegans RNA interference (RNAi) libraries (Kamath et al. 2003; Rual et al. 2004) has made it feasible to conduct genome-wide screens for phenotypes, such as life span, that are difficult to screen by traditional methods. These tools have helped reveal genetic programs associated with increased longevity (Kenyon 1996, 2005; Kaeberlein 2004).
A growing body of data from studies on model systems indicates that aspects of aging have been conserved throughout evolution, because similar interventions can increase life span among evolutionarily divergent species. One such intervention is calorie restriction (CR), which can slow aging in virtually every biological system examined (Weindruch and Walford 1988; Masoro 2005). In addition to increasing life span, CR induces many similar physiological changes in diverse species (Longo and Finch 2003). For example, increased stress resistance, decreased ribosome biogenesis, and metabolic reprogramming in response to nutrient depletion are hallmarks of CR in yeast, worms, flies, and mammals.
Reduced activity of nutrient-responsive insulin-like signaling pathways is also associated with enhanced longevity in multicellular eukaryotes. For example, mutation of the C. elegans insulin/IGF-1 receptor homolog, Daf-2, dramatically increases life span and up-regulates stress response genes through the FOXO-like transcription factor Daf-16 (Kenyon et al. 1993; Kimura et al. 1997; Lin et al. 1997; Ogg et al. 1997). A similar pathway appears to regulate longevity in flies, as well (Hwangbo et al. 2004), and subsequent work in mice demonstrated that a variety of mutations conferring endocrine deficits in insulin-like signaling can extend mammalian life span (Brown-Borg et al. 1996; Coschigano et al. 2000; Flurkey et al. 2001, 2002; Bluher et al. 2003; Holzenberger et al. 2003). Down-regulation of insulin-like signaling pathway activity is likely to be responsible for a portion of the longevity benefit associated with CR, although evidence also exists that CR acts in a genetic pathway parallel to insulin/IGF-1 (Bartke et al. 2001; Hansen et al. 2005; Kaeberlein and Kennedy 2005).
The yeast S. cerevisiae can be used to model aging of multicellular eukaryotes in two distinct ways. The first, replicative life span (RLS), is a measure of the number of mitotic events an individual mother cell can undergo before senescence (Mortimer and Johnston 1959); the second, chronological life span (CLS), is a measure of the time a nondividing cell population can remain viable in liquid media (Fabrizio et al. 2003). RLS has been suggested to be a model for the aging of mitotic tissues, whereas CLS has been likened to the aging of post-mitotic tissues (MacLean et al. 2001; Longo 2003).
Most aging research conducted on yeast has measured RLS, and several dozen proteins are known to play a role in determining RLS (Kaeberlein et al. 2001). However, only a few genes have been implicated in the regulation of yeast chronological aging. Increased expression of genes coding for superoxide dismutase or catalase can increase CLS (Fabrizio et al. 2003), whereas deletion of these genes dramatically shortens CLS (Longo et al. 1996, 1999), suggesting that response to oxidative stress is of central importance in yeast CLS. Mutations that decrease the activity of the nutrient-responsive kinase Sch9 or adenylate cyclase (Cyr1), an activator of the cyclic AMP-dependent protein kinase (PKA), also increase CLS (Fabrizio et al. 2001, 2004). These proteins are normally activated in the presence of ample nutrients, especially glucose, and Sch9 and PKA are thought to signal in parallel pathways to increase glycolysis, ribosome biogenesis, growth, and cell cycle progression and to inhibit stress response pathways (Longo 2003; Jorgensen et al. 2004; Roosen et al. 2005).
The relationship between RLS and CLS in yeast has remained obscure. Many genetic interventions that increase RLS, including those that lead to activation of the histone deacetylase Sir2 (Kaeberlein et al. 1999), do not have a similar effect on CLS (Fabrizio et al. 2005; Kennedy 2005), and some mutations have been reported to have opposite effects in the two life span assays (Harris et al. 2001, 2003; Fabrizio et al. 2004). However, there is reason to believe that chronological and replicative aging are interconnected. First, chronologically aged cells display a reduced RLS (Ashrafi et al. 1999), and second, deletion of Sch9 or deletion of activating proteins of PKA signaling (including Cyr1) extends both RLS and CLS in yeast (Lin et al. 2000; Fabrizio et al. 2001, 2004). Sch9 has 47% identity to AKT, a key molecule involved in insulin-like signal transduction (Longo 2004), cellular senescence (Minamino et al. 2004; Miyauchi et al. 2004), and aging (Hertweck et al. 2004; Hamilton et al. 2005; Oh et al. 2005) in higher eukaryotes. Because decreased insulin-like peptide signaling has been linked to increased life span in worms, flies, and mice, the finding that Sch9 and PKA play a role in yeast RLS and CLS determination supports the idea that similar nutrient signaling pathways can influence life span in evolutionarily diverse species (Longo 2004).
We have employed the yeast deletion collection to conduct an unbiased and quantitative analysis of CLS on a genome-wide basis, with the goal of discovering deletions that confer enhanced longevity. Here we report that mutations resulting in decreased activity of the TOR signaling pathway increase CLS in yeast. TOR proteins are highly conserved throughout eukaryotes and have homology with phosphatidyl inositol kinases. Yeast has two different TOR proteins: Tor1, which can participate in the rapamycin-sensitive TORC1 complex, and the essential Tor2, which can participate in TORC1 and the rapamycin-insensitive TORC2 complex. TORC1 responds to nutrients and regulates a diverse set of cellular processes, including growth, metabolism, and stress response (Martin and Hall 2005). TORC2, which also responds to nutrients, regulates actin cytoskeletal remodeling (Martin and Hall 2005). Consistent with the phenotypes of the long-lived deletion strains we identify, inhibition of TOR signaling by amino acid depletion or by treatment with drugs that inhibit TOR activity also leads to enhanced CLS. In addition, reduction of TOR signaling improves resistance to heat and oxidative stress and induces the nuclear relocalization of the stress response transcription factor Msn2. These findings implicate the TOR pathway as an important regulator of aging in yeast. Combined with previous results in yeast and other model organisms, these data argue that conserved nutrient-sensing pathways play a key role in determining the rate of organismal aging.
Results
A high-throughput assay for CLS
The standard assay for yeast CLS requires growth of cells to stationary phase in liquid media, whereupon they cease division but remain metabolically active (Fabrizio et al. 2001). The percentage of viable cells is measured by periodically removing cells from the stationary phase culture and determining the fraction of these cells capable of forming a colony when plated onto rich media. Although this assay has been successful for the identification of several genes influencing CLS in yeast, the need for multiple serial dilution and plating steps makes this method poorly suited for high-throughput experiments.
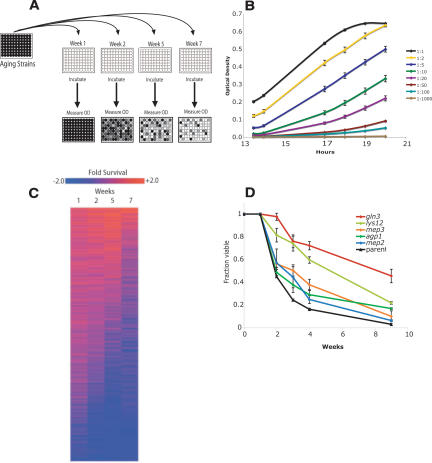
To conduct a genome-wide study of genetic factors influencing chronological aging in yeast, we developed a high-throughput method for measuring CLS (see Materials and Methods). This method involves first growing yeast cultures to stationary phase in individual wells of a 96-well plate. At various intervals (time points), 1 μL from each well is transferred into a corresponding well of a second 96-well plate containing 200 μL of fresh media. This second plate is incubated for 24 h at constant temperature, after which the optical density (OD) at 600 nm of each well is determined using a plate-reader (Fig. 1A). This OD corresponds to the number of viable cells in the inoculum (Fig. 1B), and we have validated this approach by direct comparison to the traditional colony-forming unit (CFU) assay (Supplementary Fig. 1).
Figure 1.
A genome-wide deletion screen for mutations that increase CLS. (A) A schematic diagram of high-throughput CLS measurement. Cells are aged in a 96-well plate and, at serial time points, aliquots are inoculated into a second 96-well plate containing rich media. After a fixed period of incubation, the OD of each well in the second plate is measured, and this OD corresponds to the number of viable cells originally pinned into the well. (B) Validation of the high-throughput screening method. Serial dilutions of yeast were inoculated into rich media, and OD measurements were taken at several time points. Observed OD is highly correlated to the number of viable cells inoculated into the media. (C) Relative survival of ∼4800 single-gene deletion strains over a 7-wk experiment. Ranked survival values (see Materials and Methods) were log2 normalized and projected as a heat map. Longer-lived strains appear red. (D) Deletion of TOR-regulated nitrogen acquisition genes extends life span. The integrals under the life span curves are gln3Δ = 6.55, lys12Δ = 5.39, mep3Δ = 3.95, agp1Δ = 3.65, and mep2Δ = 3.41, compared with the parental strain = 2.75 (P < 0.001 for all deletion strain life spans compared with the parental strain).
A genome-wide screen for longevity reveals deletion of components of the TOR pathway extends CLS
We measured the relative CLS for each deletion strain present in the yeast homozygous diploid deletion collection, an array of ∼4800 unique strains, each with a single homozygous gene deletion (Winzeler et al. 1999). For each strain, CLS was determined in triplicate (independent cultures), with viability at each time point calculated relative to the deletion collection mean viability for that time point (Supplemental Material). To determine relative CLS, we developed a ranking system (see Materials and Methods) that took into account the ability of each single-gene deletion mutant to maintain viability over the course of the experiment (Fig. 1C).
Our major focus was the identification of long-lived mutants rather than short-lived ones; although some of the short-lived mutants we identify are likely to represent models of accelerated aging, many deletions that shorten life span would be expected to cause a fitness deficit that might not have relevance to normal aging of the cells. Of the shortest-lived 300 deletion strains, 29% of these have deletions of genes associated with mitochondrial function, a significant enrichment relative to the entire genome (P =1.94×10-11) (Supplemental Material). This observation is consistent with previous work showing that respiratory-deficient yeast generate increased levels of reactive oxygen species (ROS) and have short CLS (Barros et al. 2004; Trancikova et al. 2004). In addition, we note that yeast lacking mitochondrial components essential for respiration cannot undergo the diauxic shift to use nonfermentable carbon sources, and do not enter stationary phase properly (Gray et al. 2004).
Of the 90 longest-lived deletion strains based on our genome-wide screen, 16 have mutations in genes that are implicated in TOR signaling and nutrient acquisition (Supplemental Material), the only apparent common Gene Ontology (Saccharomyces Genome Database, http://www.yeastgenome.org) function initially found. More recently, five other genes (SLM4, GTR1, GTR2, MEH1, and NVJ1) have been implicated in the regulation of autophagy, a TOR-regulated process (Roberts et al. 2003; Dubouloz et al. 2005). Additional deletions implicate genes for intracellular ion homeostasis, and cell wall organization and biogenesis. These and other genes may prove fruitful targets for further life span research (Supplemental Material).
Because TOR activity has been linked to yeast replicative aging (Kaeberlein et al. 2005), as well as to aging in both C. elegans and Drosophila melanogaster (Vellai et al. 2003; Kapahi et al. 2004; Meissner et al. 2004), we applied a more stringent CLS protocol (see Materials and Methods) to retest the 16 strains containing deletions with putative involvement in TOR signaling. In these more stringent tests, five of the TOR-related genes showed significantly increased life span in multiple CLS experiments (Fig. 1D), and we chose to focus on these for further study.
To quantify the magnitude of life span extension afforded by the deletions, we calculated the integral of the survival curve for each strain. This number serves as a useful statistic to describe survival differences over the course of an experiment. Deletion of the TOR-controlled transcription factor Gln3 conferred the largest magnitude of life span increase; the integral of the survival curve for the gln3Δ strain was 6.55 compared with 2.75 for the parental strain. Gln3 is a GATA transcription factor that transcriptionally up-regulates a large suite of genes encoding amino acid biosynthetic enzymes and ammonium and amino acid permeases (Shamji et al. 2000; Bertram et al. 2002). Deletion of GLN3 resulted in a doubling time ∼25% longer than wild-type cells (data not shown), suggesting that nutrient uptake and nitrogen assimilation become limiting for growth in this long-lived mutant.
In addition to GLN3, deletion of several Gln3-regulated genes also increased CLS. These included genes encoding Lys12 (integral of 5.39), which participates in lysine biosynthesis, the ammonium permeases Mep2 and Mep3 (integrals of 3.41 and 3.95, respectively), and the general amino acid permease Agp1 (integral of 3.65). These permeases are responsible for membrane transport of favored nitrogen sources and play a role in maintaining optimal nutrient status for cellular growth (Table 1).
Table 1.
Genes deleted in long-lived yeast strains
| Gene deletion | Protein function |
|---|---|
| GLN3 | TOR-regulated transcription of nitrogen acquisition genes |
| LYS12 | Lysine biosynthesis |
| MEP3 | Ammonium permease |
| AGP1 | Broad specificity amino acid permease |
| MEP2 | Ammonium permease |
Removal of preferred amino acids from the media extends life span
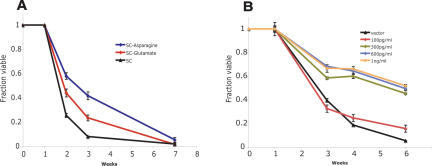
The TOR pathway is regulated by environmental amino acid quality, particularly intracellular glutamine levels (Martin and Hall 2005). Thus, deletion of genes for amino acid and nitrogen acquisition proteins might induce deficits in cellular amino acid levels and increase life span by diminishing TOR signaling. Therefore, we tested whether removal of preferred nitrogen sources from the media extends CLS. Asparagine (a high nitrogen quality amino acid) or glutamate (an intermediate nitrogen quality amino acid) (Cooper 2002; Crespo et al. 2002; Oliveira et al. 2005) was eliminated from the media while total amino acid concentration was held constant. Consistent with our hypothesis, we observed a life span extension in direct proportion to the quality of the amino acid removed from the media, with removal of the metabolically favorable asparagine conferring the larger life span extension (integral of 3.24 vs. 2.56 for glutamate removal and 1.99 for synthetic complete media) (Fig. 2A).
Figure 2.
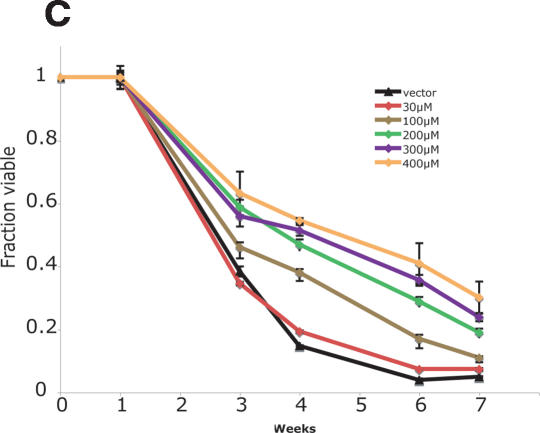
Multiple means of TOR pathway inhibition extend life span. (A) Removal of preferred amino acids from the media extends life span in proportion to the nutrient value of the eliminated amino acid, even though total nitrogen content is held constant. Removal of the preferred amino acid asparagine (integral = 3.24) or of the intermediately favored glutamate (integral = 2.56) confers increased survival compared with cells grown in synthetic complete media (integral = 1.99) (P < 0.001 for each amino acid drop out compared with synthetic complete). (B) Direct inhibition of the TOR pathway by low doses of rapamycin extends life span in a dose-responsive manner. The integrals under the life span curves are drug vehicle = 2.93, 100 pg/mL = 3.01, 300 pg/mL = 4.23, 600 pg/mL = 4.49, and 1 ng/mL = 4.51 (P < 0.001 for each dose >100 pg/mL vs. drug vehicle). (C) Inhibition of glutamine synthetase by the drug MSX extends life span by reducing intracellular glutamine, which reduces TOR signaling. The integrals under the life span curves are drug vehicle = 2.88, 30 μM = 2.95, 100 μM = 3.57, 200 μM = 4.11, 300 μM = 4.26, and 400 μM = 4.53 (P < 0.001 for each dose >30μM vs. drug vehicle).
Pharmacological inhibition of the TOR pathway extends CLS
Because abrogation of TOR-regulated nitrogen uptake or limitation of preferred nitrogen sources extended CLS, we wished to test directly if diminished TOR activity increased life span. We treated yeast cells with rapamycin, a potent and specific inhibitor of TORC1 signaling (Beck and Hall 1999; Cardenas et al. 1999; Shamji et al. 2000). At concentrations of rapamycin sufficient to slow, but not block, cell division, we observed a dose-dependent increase in CLS (Fig. 2B). The integral of the life span curve of drug vehicle-treated cells (2.93) increased to 4.23 at 300 pg/mL rapamycin and to 4.51 at 1 ng/mL rapamycin.
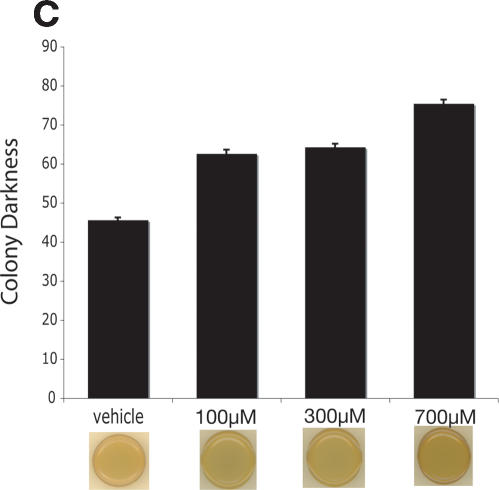
TOR activity can also be decreased by treating cells with methionine sulfoximine (MSX), an inhibitor of glutamine synthetase (Crespo et al. 2002). Because TOR activity is regulated by intracellular glutamine levels, inhibition of glutamine synthetase by MSX decreases both intracellular glutamine levels and TOR-dependent signaling (Crespo et al. 2002). Similar to treatment of cells with rapamycin, MSX increased CLS in a dose-dependent manner (Fig. 2C). The life span curve integral for vehicle-treated cells (2.88) increased to 3.57 at 100 μM MSX and to 4.53 at 400 μM MSX.
Long-lived mutants exhibit starvation phenotypes
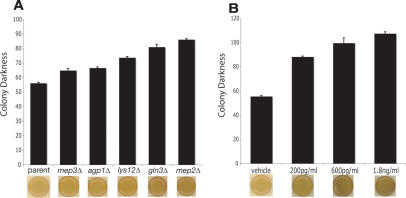
We speculated that long-lived mutants deficient in nitrogen acquisition might be responding to their environment as if they were grown under conditions of starvation. In all eukaryotic organisms studied to date, CR is characterized by an increase in gluconeogenesis (Hagopian et al. 2003) and the accumulation of energy storage molecules such as carbohydrates (Longo and Fabrizio 2002). Therefore, we examined the accumulation of the storage carbohydrate glycogen using an iodine vapor assay that stains yeast dark brown as they accumulate glycogen (Barbet et al. 1996; Enjalbert et al. 2000). Compared with the parental control, the long-lived gln3Δ, agp1Δ, mep2Δ, mep3Δ, and lys12Δ strains all exhibited significantly increased glycogen accumulation, even when grown on rich media (Fig. 3A). In addition, treatment with either rapamycin (Fig. 3B), as previously reported (Barbet et al. 1996), or MSX (Fig. 3C) increased glycogen accumulation in a dose-dependent manner. These observations are consistent with the induction of starvation-related genetic programs in the long-lived cells.
Figure 3.
Increased CLS correlates with increased starvation response. (A) Long-lived deletion strains accumulate glycogen on rich media as determined by an iodine vapor assay, which stains intracellular glycogen dark brown. (B) Rapamycin causes glycogen accumulation in a dose-dependent manner. (C) MSX treatment causes glycogen accumulation. For all groups, n = 9, bars are mean ± SEM; all test groups were significantly different than appropriate control (P < 0.001), and representative colonies are shown.
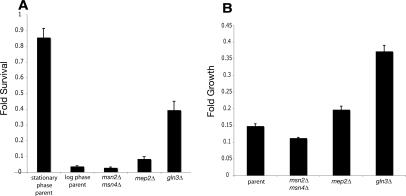
Another response to starvation is an up-regulation of certain stress responses that enable cells to withstand a variety of challenges, including heat and oxidative stress. Thus, we tested the long-lived mep2Δ and gln3Δ deletion strains for their ability to survive acute exposure to high heat or to grow in the presence of the drug paraquat, which generates the oxidative radical superoxide. As a positive control for heat stress resistance, we used wild-type cells that had already entered stationary phase. Cells in stationary phase are heat-shock resistant due to the up-regulation of stress resistance programs, including those regulated by Msn2 and its partially redundant partner Msn4 (Garreau et al. 2000). We compared the heat stress resistance of the stationary phase cells to that of logarithmically growing wild-type, gln3Δ, mep2Δ, and msn2Δ msn4Δ cells. Both the mep2Δ and gln3Δ strains showed significantly improved ability to recover from heat shock relative to logarithmically growing wild-type cells (P = 0.0159 and P < 0.0001, respectively) (Fig. 4A) and enhanced ability to grow in 1 mM paraquat (P = 0.002 and P < 0.0001, respectively) (Fig. 4B). These observations are consistent with a chronically up-regulated stress response as a consequence of cells sensing a nutrient-poor environment.
Figure 4.
Long-lived strains are resistant to lethal heat-stress or oxidative insult. (A) Mid-log cells were treated with a 2-min, 55°C heat shock, and survival was calculated relative to untreated isogenic strains. The mep2Δ and gln3Δ strains were significantly protected compared with the parental strain (P < 0.0159 for mep2Δ and P < 0.0001 for gln3Δ). (B) Mid-log cells were inoculated into media containing 1 mM of the free-radical-generating drug paraquat, and growth was compared with untreated isogenic strains after 15 h. Deletion of MEP2 or GLN3 affords significant protection from paraquat (P = 0.002 for mep2Δ and P < 0.0001 for gln3Δ). For all groups, n = 6, and bars are mean ± SEM.
Msn2 localizes to the nucleus in TOR-deficient cells
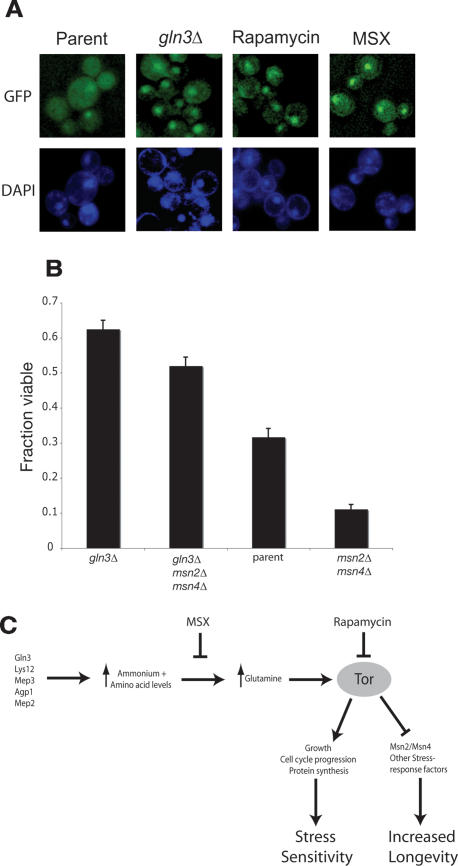
As yeast cells begin to sense depletion of nutrients in their environment, the stress-responsive transcription factors Msn2 and Msn4 relocalize from the cytoplasm to the nucleus and orchestrate the preparation for starvation conditions in a TOR-dependent manner (Beck and Hall 1999; Crespo et al. 2002). Because TOR is known to be a sensor of amino acid quality and Msn2 is maintained in the cytoplasm by active TOR signaling (Schmelzle et al. 2004), we tested whether Msn2 relocalized to the nucleus in the long-lived gln3Δ strain, or upon treatment of cells with the drugs rapamycin or MSX. We used fluorescence microscopy to track the subcellular localization of an Msn2-GFP fusion protein expressed from the endogenous MSN2 promoter at the chromosomal MSN2 locus. Prior to entry into stationary phase, we observed increased nuclear localization of this fusion protein in the gln3Δ strain, or in wild-type cells treated with 600 pg/mL rapamycin or 200 μM MSX, compared with the untreated wild-type cells (Fig. 5A). Relocalization of Msn2-GFP to the nucleus occurred at ∼8 h post-inoculation in the case of these life-extending interventions, but not until ∼24 h in vector-treated cells. This observation is indicative of a precocious starvation response in the long-lived gln3Δ or drug-treated cells.
Figure 5.
Partial inhibition of TORC1 activity causes precocious nuclear localization of Msn2. (A) Eight hours after strains were inoculated into SC media, an Msn2-GFP fusion protein accumulates in the nucleus of gln3Δ cells and cells treated with 600 pg/mL rapamycin or 200 μM MSX, but not in untreated parental cells. The frequency of Msn2-GFP nuclear localization is 56% of gln3Δ cells, 52% of rapamycin-treated cells, and 57% of MSX-treated cells, but only 12% of untreated parental control cells. Approximately 100 cells were examined per group. (B) Msn2 and Msn4 are required for the full life span extension of the gln3Δ strain. We measured viability 4 wk post-inoculation and observed that transcriptional activity of Msn2 and Msn4 is crucial for maximal CLS. However, the gln3Δ mutation activates an Msn2- and Msn4-independent pathway that can extend life span relative to wild type cells. (C) An integrated model linking long-lived deletion strains, reduced amino acid levels, and rapamycin treatment to increased stress response and longevity.
Msn2 and Msn4 are required for the full life span extension of gln3Δ cells
We wished to test if the function of the partially redundant transcription factors Msn2 and Msn4 was required for the life span extension of gln3Δ cells. We compared the viability of cells 4 wk after inoculation into synthetic complete media and found that the presence of Msn2 and Msn4 is required for the full life span extension benefit of the gln3Δ mutation (P = 0.005 gln3Δ vs. gln3Δ msn2Δ msn4Δ) (Fig. 5B). However the gln3Δ mutation can extend viability (relative to the parental strain) even when combined with deletion of both MSN2 and MSN4 (P = 0.0001 gln3Δ msn2Δ msn4Δ vs. parental strain).
In summary, we find that reduced TOR signaling, either by diminished nitrogen source acquisition or biosynthesis or by direct inhibition of TOR signaling, leads to an increased ability to respond to stress. This increase in stress tolerance is at least in part due to an Msn2/Msn4-dependent transcriptional response, likely leading to an increased life span.
Discussion
From a screen for long-lived single-gene deletion mutants using the yeast ORF deletion collection, we have identified the nutrient-responsive TOR kinase as a primary determinant of yeast CLS. We describe five gene deletions and two pharmacological interventions that decrease TOR activity and increase CLS. In addition, we show that altering the growth medium in a manner known to decrease TOR activity also increases CLS. We provide evidence that reduced TOR signaling can extend the longevity of quiescent yeast cells, at least partly by activating the stress-responsive transcription factor Msn2.
TOR activity determines life span in response to amino acids
Yeast can grow on a variety of nitrogen sources, but not all nitrogen sources support optimal growth. Glutamine, asparagine, and ammonium are all preferred nitrogen sources because they are easily converted to a variety of amino acids, and glutamine is known to be a key determinant of TOR signaling (Crespo et al. 2002). One mechanism by which intracellular glutamine (and TOR activity) can be decreased is through reduction of amino acids used as preferred nitrogen sources in the growth medium (Crespo et al. 2002). We show that removal of either asparagine or glutamate from the media significantly increases CLS. Removal of nonessential amino acids has also been reported to increase RLS (Jiang et al. 2000), possibly through a TOR-dependent mechanism. Consistent with this hypothesis, mutations resulting in decreased TOR activity have recently been shown to also increase RLS (Kaeberlein et al. 2005).
In addition to responding to the composition of the growth media, TOR activity can also be altered by mutations that affect intracellular nitrogen levels. We identified five long-lived single-gene deletion strains (gln3Δ, agp1Δ, mep2Δ, mep3Δ, and lys12Δ), each lacking a gene important for glutamine metabolism (Fig. 5B). For example, GLN3 codes for a transcription factor that up-regulates GLN1, the essential gene that codes for glutamine synthetase. Deletion of GLN3 results in decreased transcription of Gln1, which is predicted to decrease intracellular glutamine levels (and hence TOR activity) (Stanbrough et al. 1995; Cunningham et al. 1996). Consistent with this effect, treatment of cells with MSX, an inhibitor of Gln1, decreases intracellular glutamine levels and thus TOR activity (Crespo et al. 2002). We find that MSX also increases CLS (Fig. 2C). Agp1 is an amino acid transporter of broad substrate specificity capable of taking up the preferred amino acids asparagine and glutamine (Schreve et al. 1998), Mep2 and Mep3 are permeases that import ammonium, and Lys12 catalyzes an important step in lysine biosynthesis; all of these deletions are also anticipated to limit intracellular amino acid pools and, thus, decrease TOR activity (Fig. 5B).
Evidence from higher eukaryotes suggests that a decrease in amino acid levels can increase life span in organisms other than yeast. In rats or mice, reduction of dietary methionine levels results in a significant life span extension, comparable to that of CR (Orentreich et al. 1993; Miller et al. 2005). In flies, limitation of yeast extract, the dietary source of amino acids, provides a greater longevity enhancement than reduction of sucrose (Piper et al. 2005). Mutation of INDY, which encodes a carboxylate nutrient transporter localized to the gut and fat body of flies, increases life span (Rogina et al. 2000; Inoue et al. 2002; Knauf et al. 2002; Marden et al. 2003). The INDY protein is a high-affinity transporter for several metabolites of the TCA cycle, including α-ketoglutarate (Knauf et al. 2002), an important precursor in glutamine synthesis, and we speculate that reduced INDY expression might increase life span through a reduction in TOR signaling.
In C. elegans, deletion of the intestinal di- and tripeptide transporter PEP-2 causes an increase in stress resistance and synergizes with reduced insulin signaling to increase life span (Meissner et al. 2004). PEP-2 mutation dramatically enhances the life span increase due to a weak Tor RNAi allele but does not confer additional life span increase in the context of a strong Tor RNAi allele (Meissner et al. 2004), suggesting that PEP-2 mutation reduces the uptake of amino acids and thereby inhibits TOR signaling.
Nutrient signaling and stress response
How does decreased TOR activity result in increased CLS? Reduced TOR signaling leads to many characteristics of preparation for stationary phase, and this early preparation seems to be critical for the survival benefit. We observe maximal life span increase by rapamycin or MSX treatment only if the drug is administered before the cells enter stationary phase (our unpublished results). Thus, advanced accumulation of storage carbohydrates and up-regulation of stress-responsive proteins before entry into stationary phase may be essential to increased life span.
Although we cannot rule out the possibility that decreased translation rate plays a role in the observed life span increase for the various interventions we report, extension of CLS is more likely principally due to the induction of stress-responsive processes repressed by TOR. These processes include transcriptional up-regulation of genes for enzymes capable of detoxifying free radicals, degrading damaged proteins through autophagy, and protecting other vital cellular functions. Similar transcriptional mechanisms can extend life span in other organisms and are negatively regulated by nutrient signaling (Lee et al. 2003; Murphy et al. 2003; Giannakou et al. 2004; Hwangbo et al. 2004). These nutrient-sensing pathways may be the product of a common theme conserved throughout evolution, allowing organisms to shunt their energy investments into processes of self-maintenance and repair in the face of scarce nutrients. For example, in worms, flies, and mice, caloric restriction or reduced insulin signaling activates members of the FOXO fork head transcription factor family (Longo and Finch 2003). These transcription factors are important for the increased expression of a variety of genes involved in stress response and are functional homologs of yeast Msn2 and Msn4.
Conclusion
Although the detailed molecular mechanism by which CR extends life span remains unknown, results from experimental organisms suggest that cellular responses to nutrients are mediated through broadly conserved nutrient-sensing pathways that can extend life span when signaling through these pathways is reduced. Accumulating evidence from yeast, worms, and flies indicates a central role for the TOR kinase in this process. Genetic and environmental interventions that decrease TOR activity are promising avenues for further exploration in mammalian systems and may be useful for improving human health.
Materials and methods
Yeast strains and media
All CLS experiments were conducted in the BY4743 diploid background (Winzeler et al. 1999). The yeast ORF deletion collection (Winzeler et al. 1999) was obtained from Open Biosystems. Genomic GFP fusion to MSN2 (Huh et al. 2003) was generated in a gln3Δ background by crossing and sporulation. A control Msn2-GFP strain was generated in the same manner and had identical auxotrophic markers and mating type as the gln3Δ MSN2-GFP strain. A complete list of strains is provided in the Supplemental Material.
Unless otherwise noted, CLS experiments were carried out in 2% glucose synthetic complete media (Ausubel et al. 2003) supplemented with a fourfold excess of the amino acids for which the strains were auxotrophic. For amino acid drop-out experiments, bulk amino acid content was held constant by addition of mass equivalent excess of all other amino acids (Ausubel et al. 2003). Glutamine is not present in SC media, and therefore, asparagine was withheld instead. For OD viability assays, cells were grown in standard yeast extract peptone media with 2% glucose (Ausubel et al. 2003). We verified that sporulation was not taking place during diploid chronological aging experiments by the absence of tetrad asci in aged cultures examined using a microscope.
OD assay for determining viability
As a surrogate for determining the number of viable cells in aging cultures by counting CFUs on solid agar media, a method was developed whereby the number of viable yeast cells inoculated into a known volume of YPD media was determined based on OD of the culture after a fixed period of outgrowth. This method was validated by inoculating serial dilutions of yeast cells into 200 μL YPD media contained in wells of a 96-well plate. OD measurements were taken at serial time points and in the linear range of our measurement (0.05-0.6). The OD of each well after outgrowth was highly correlated with the number of viable cells inoculated into the liquid, as determined by CFU survival assays. OD measurements were also found to correlate well with CFU-based survival measurements on aging populations of cells or with heat-killed cells (Supplemental Material).
High-throughput CLS assay of the deletion collection
The entire deletion set (fifty-six 96-well plates) was inoculated into fresh YPD media in 96-well plates (∼1 μL per well) using a high-density Biomek FX replica pinning robot (Beckman-Coulter). Cells were cultured at 30°C for 3 d. CLS assays were initiated by transferring ∼1 μL per well from 3-d-old YPD cultures into individual wells of 96-well plates containing 150 μL SC media in each well. Aging cultures were maintained at 30°C in sealed plastic bags under high humidity to prevent loss of culture volume due to evaporation. At various time points, ∼1 μL from each well was transferred to a second 96-well plate that contained 200 μL YPD. These inoculated YPD plates were incubated for 24 h at 25°C, before gentle shaking to evenly suspend the yeast culture, after which the optical density of each well was measured using a Wallace plate reader (Perkin Elmer). In this manner, the entire deletion collection was measured in triplicate at 1, 2, 5, and 7 wk after inoculation into SC media. To calculate survival, the average OD for each strain was divided by the mean OD of the entire deletion collection for each time point. This calculation represents a relative survival value. For each strain, this value was summed for time points 2, 5, and 7 wk and was used to rank the deletion strains for further testing (Supplemental Material).
Validation of long-lived deletion strains
Verification of putative long-lived deletion strains identified from the genome-wide screen was carried out by independent retesting of CLS. Strains were removed from frozen stock and streaked onto solid YPD agar media. CLS cultures were obtained by lightly inoculating each strain into 150 μL SC media in 96-well plates. Cultures were maintained and viability was determined by OD after outgrowth as described above, except transfer from aging SC cultures into YPD was carried out manually using micropipettors. One week after inoculation into aging plates, when cells reach maximal density, the initial time point was taken, and this OD corresponds to 100% viability for that strain. Subsequent weekly OD measurements were divided by the initial OD measurement for that well to yield fraction viable. By comparing each strain back to itself at the first time point, growth rate differences are taken into account. All life span experiments reported were conducted at least three times, with multiple replicates of each strain. Integrals of the life span curves were calculated by summing the trapezoids created by the viability time points. Treatments were assigned a P value by calculating the variance of integrals between biological replicates for a given treatment, and then comparing this to the integrals for wild-type or drug vehicle-treated cells using a t-test.
Pharmacological inhibition of the TOR pathway
Rapamycin (Sigma) was suspended in ethanol at a stock concentration of 1 mg/mL and stored at -20°C. MSX (Sigma) was dissolved in sterile water at a concentration of 100 mM and stored at 4°C. Drugs were added at the indicated concentration at the time of inoculation of the aging culture.
Glycogen accumulation, heat shock, and paraquat resistance
Glycogen accumulation was measured essentially as described previously (Enjalbert et al. 2000). In brief, strains were spotted on solid YPD media and allowed to grow for 48 h at 30°C before a 2-min incubation above a monolayer of solid iodine crystals. Images were scanned and colony darkness was quantified by using ImageJ software (Wayne Rasband, National Institutes of Health, Bethesda, MD). Statistics were calculated using the Student's t-test.
Heat shock resistance assays were conducted on exponentially growing cells in SC media. Strains were exposed to 55°C or 25°C (as a control) for 2 min and placed on ice for 1 min, and then 5 μL aliquots from the heat-treated or control cells were transferred to YPD media. After 16 h of growth at 25°C, OD measurements were taken for the heat-stressed and the control cultures, and fold survival was calculated by dividing the OD of the heat-stressed cultures by the control cultures. Statistics were calculated using the Student's t-test.
Paraquat resistance assays were conducted by inoculating 5 μL of exponentially growing yeast culture into 200 μL of YPD or 200 μL of YPD with 1 mM paraquat (methyl viologen, Sigma). After 16 h of growth, the OD of the cultures was measured, and fold survival was calculated by dividing the OD of the heat-stressed cultures by the control cultures. Statistics were calculated using the Student's t-test.
Fluorescence microscopy
Cells were incubated overnight in tubes at 30°C in YPD media. One hundred microliters of these cultures was then inoculated into 2 mL of SC media with drug vector, 600 pg/mL rapamycin or 200 μM MSX, and incubated at 30°C. At time points 2, 8, 12, and 24 h post-inoculation into SC media, small aliquots of cells were removed from culture, incubated for 10 min with 5 μg/mL DAPI, and examined using a Ziess Delta Vision fluorescence microscope.
Strains
The parent strain used was BY4743 (Mata/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ0 met15Δ0/MET15 lys2Δ0/LYS2). The homozygous deletions gln3, mep2, mep3, agp1, and lys12, and the heterozygous GFP strain (MSN2-GFP/MSN2) were in this background.
Acknowledgments
We thank George Martin and the members of the Fields laboratory for advice and helpful discussion. R.W.P. III thanks L. Rhode and the Powers family for support. This work was funded in part by NIH training grant P30 AG013280, and an award to R.W.P. III from the University of Washington Nathan Shock Center of Excellence for the Basic Biology of Aging. S.F. is an investigator of the Howard Hughes Medical Institute.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1381406.
References
- Ashrafi, K., Sinclair, D., Gordon, J.I., and Guarente, L. 1999. Passage through stationary phase advances replicative aging in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 96: 9100-9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. 2003. Current protocols in molecular biology. John Wiley and Sons, New York.
- Barbet, N.C., Schneider, U., Helliwell, S.B., Stansfield, I., Tuite, M.F., and Hall, M.N. 1996. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 7: 25-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros, M.H., Bandy, B., Tahara, E.B., and Kowaltowski, A.J. 2004. Higher respiratory activity decreases mitochondrial reactive oxygen release and increases life span in Saccharomyces cerevisiae. J. Biol. Chem. 279: 49883-49888. [DOI] [PubMed] [Google Scholar]
- Bartke, A., Wright, J.C., Mattison, J.A., Ingram, D.K., Miller, R.A., and Roth, G.S. 2001. Extending the lifespan of long-lived mice. Nature 414: 412. [DOI] [PubMed] [Google Scholar]
- Beck, T. and Hall, M.N. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402: 689-692. [DOI] [PubMed] [Google Scholar]
- Bertram, P.G., Choi, J.H., Carvalho, J., Chan, T.F., Ai, W., and Zheng, X.F. 2002. Convergence of TOR-nitrogen and Snf1-glucose signaling pathways onto Gln3. Mol. Cell. Biol. 22: 1246-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluher, M., Kahn, B.B., and Kahn, C.R. 2003. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science 299: 572-574. [DOI] [PubMed] [Google Scholar]
- Brown-Borg, H.M., Borg, K.E., Meliska, C.J., and Bartke, A. 1996. Dwarf mice and the ageing process. Nature 384: 33. [DOI] [PubMed] [Google Scholar]
- Cardenas, M.E., Cutler, N.S., Lorenz, M.C., Di Como, C.J., and Heitman, J. 1999. The TOR signaling cascade regulates gene expression in response to nutrients. Genes & Dev. 13: 3271-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, T.G. 2002. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: Connecting the dots. FEMS Microbiol. Rev. 26: 223-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coschigano, K.T., Clemmons, D., Bellush, L.L., and Kopchick, J.J. 2000. Assessment of growth parameters and life span of GHR/BP gene-disrupted mice. Endocrinology 141: 2608-2613. [DOI] [PubMed] [Google Scholar]
- Crespo, J.L., Powers, T., Fowler, B., and Hall, M.N. 2002. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc. Natl. Acad. Sci. 99: 6784-6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham, T.S., Svetlov, V.V., Rai, R., Smart, W., and Cooper, T.G. 1996. G1n3p is capable of binding to UAS(NTR) elements and activating transcription in Saccharomyces cerevisiae. J. Bacteriol. 178: 3470-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouloz, F., Deloche, O., Wanke, V., Cameroni, E., and De Virgilio, C. 2005. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol. Cell 19: 15-26. [DOI] [PubMed] [Google Scholar]
- Enjalbert, B., Parrou, J.L., Vincent, O., and Francois, J. 2000. Mitochondrial respiratory mutants of Saccharomyces cerevisiae accumulate glycogen and readily mobilize it in a glucose-depleted medium. Microbiology 146: 2685-2694. [DOI] [PubMed] [Google Scholar]
- Fabrizio, P., Pozza, F., Pletcher, S.D., Gendron, C.M., and Longo, V.D. 2001. Regulation of longevity and stress resistance by Sch9 in yeast. Science 292: 288-290. [DOI] [PubMed] [Google Scholar]
- Fabrizio, P., Liou, L.L., Moy, V.N., Diaspro, A., Selverstone Valentine, J., Gralla, E.B., and Longo, V.D. 2003. SOD2 functions downstream of Sch9 to extend longevity in yeast. Genetics 163: 35-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio, P., Pletcher, S.D., Minois, N., Vaupel, J.W., and Longo, V.D. 2004. Chronological aging-independent replicative life span regulation by Msn2/Msn4 and Sod2 in Saccharomyces cerevisiae. FEBS Lett. 557: 136-142. [DOI] [PubMed] [Google Scholar]
- Fabrizio, P., Gattazzo, C., Battistella, L., Wei, M., Cheng, C., McGrew, K., and Longo, V.D. 2005. Sir2 blocks extreme lifespan extension. Cell 123: 655-667. [DOI] [PubMed] [Google Scholar]
- Flurkey, K., Papaconstantinou, J., Miller, R.A., and Harrison, D.E. 2001. Lifespan extension and delayed immune and collagen aging in mutant mice with defects in growth hormone production. Proc. Natl. Acad. Sci. 98: 6736-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flurkey, K., Papaconstantinou, J., and Harrison, D.E. 2002. The Snell dwarf mutation Pit1(dw) can increase life span in mice. Mech. Ageing Dev. 123: 121-130. [DOI] [PubMed] [Google Scholar]
- Garreau, H., Hasan, R.N., Renault, G., Estruch, F., Boy-Marcotte, E., and Jacquet, M. 2000. Hyperphosphorylation of Msn2p and Msn4p in response to heat shock and the diauxic shift is inhibited by cAMP in Saccharomyces cerevisiae. Microbiology 146: 2113-2120. [DOI] [PubMed] [Google Scholar]
- Giannakou, M.E., Goss, M., Junger, M.A., Hafen, E., Leevers, S.J., and Partridge, L. 2004. Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science 305: 361. [DOI] [PubMed] [Google Scholar]
- Gray, J.V., Petsko, G.A., Johnston, G.C., Ringe, D., Singer, R.A., and Werner-Washburne, M. 2004. `Sleeping beauty': Quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 68: 187-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagopian, K., Ramsey, J.J., and Weindruch, R. 2003. Caloric restriction increases gluconeogenic and transaminase enzyme activities in mouse liver. Exp. Gerontol 38: 267-278. [DOI] [PubMed] [Google Scholar]
- Hamilton, B., Dong, Y., Shindo, M., Liu, W., Odell, I., Ruvkun, G., and Lee, S.S. 2005. A systematic RNAi screen for longevity genes in C. elegans. Genes & Dev. 19: 1544-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, M., Hsu, A.L., Dillin, A., and Kenyon, C. 2005. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS. Genet. 1: e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, N., MacLean, M., Hatzianthis, K., Panaretou, B., and Piper, P.W. 2001. Increasing Saccharomyces cerevisiae stress resistance, through the overactivation of the heat shock response resulting from defects in the Hsp90 chaperone, does not extend replicative life span but can be associated with slower chronological ageing of nondividing cells. Mol. Genet. Genomics 265: 258-263. [DOI] [PubMed] [Google Scholar]
- Harris, N., Costa, V., MacLean, M., Mollapour, M., Moradas-Ferreira, P., and Piper, P.W. 2003. Mnsod overexpression extends the yeast chronological (G(0)) life span but acts independently of Sir2p histone deacetylase to shorten the replicative life span of dividing cells. Free Radic. Biol. Med. 34: 1599-1606. [DOI] [PubMed] [Google Scholar]
- Hertweck, M., Gobel, C., and Baumeister, R. 2004. C. elegans SGK-1 is the critical component in the Akt/PKB kinase complex to control stress response and life span. Dev. Cell 6: 577-588. [DOI] [PubMed] [Google Scholar]
- Holzenberger, M., Dupont, J., Ducos, B., Leneuve, P., Geloen, A., Even, P.C., Cervera, P., and Le Bouc, Y. 2003. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature 421: 182-187. [DOI] [PubMed] [Google Scholar]
- Huh, W.K., Falvo, J.V., Gerke, L.C., Carroll, A.S., Howson, R.W., Weissman, J.S., and O'Shea, E.K. 2003. Global analysis of protein localization in budding yeast. Nature 425: 686-691. [DOI] [PubMed] [Google Scholar]
- Hwangbo, D.S., Gershman, B., Tu, M.P., Palmer, M., and Tatar, M. 2004. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature 429: 562-566. [DOI] [PubMed] [Google Scholar]
- Inoue, K., Fei, Y.J., Huang, W., Zhuang, L., Chen, Z., and Ganapathy, V. 2002. Functional identity of Drosophila melanogaster Indy as a cation-independent, electroneutral transporter for tricarboxylic acid-cycle intermediates. Biochem. J. 367: 313-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, J.C., Jaruga, E., Repnevskaya, M.V., and Jazwinski, S.M. 2000. An intervention resembling caloric restriction prolongs life span and retards aging in yeast. FASEB J. 14: 2135-2137. [DOI] [PubMed] [Google Scholar]
- Jorgensen, P., Rupes, I., Sharom, J.R., Schneper, L., Broach, J.R., and Tyers, M. 2004. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes & Dev. 18: 2491-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein, M. 2004. Aging-related research in the `-omics' age. Sci. Aging Knowledge Environ. 2004: pe39. [DOI] [PubMed] [Google Scholar]
- Kaeberlein, M. and Kennedy, B.K. 2005. Large-scale identification in yeast of conserved ageing genes. Mech Ageing Dev 126: 17-21. [DOI] [PubMed] [Google Scholar]
- Kaeberlein, M., McVey, M., and Guarente, L. 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes & Dev. 13: 2570-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2001. Using yeast to discover the fountain of youth. Sci. Aging Knowledge Environ. 2001: pe1. [DOI] [PubMed] [Google Scholar]
- Kaeberlein, M., Powers III, R.W., Steffen, K.K., Westman, E.A., Hu, D., Dang, N., Kerr, E.O., Kirkland, K.T., Fields, S., and Kennedy, B.K. 2005. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310: 1193-1196. [DOI] [PubMed] [Google Scholar]
- Kamath, R.S., Fraser, A.G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231-237. [DOI] [PubMed] [Google Scholar]
- Kapahi, P., Zid, B.M., Harper, T., Koslover, D., Sapin, V., and Benzer, S. 2004. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 14: 885-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy, B.K. 2005. The enigmatic role of Sir2 in aging. Cell 123: 548-550. [DOI] [PubMed] [Google Scholar]
- Kenyon, C. 1996. Ponce d'elegans: Genetic quest for the fountain of youth. Cell 84: 501-504. [DOI] [PubMed] [Google Scholar]
- ____. 2005. The plasticity of aging: Insights from long-lived mutants. Cell 120: 449-460. [DOI] [PubMed] [Google Scholar]
- Kenyon, C., Chang, J., Gensch, E., Rudner, A., and Tabtiang, R. 1993. A C. elegans mutant that lives twice as long as wild type. Nature 366: 461-464. [DOI] [PubMed] [Google Scholar]
- Kimura, K.D., Tissenbaum, H.A., Liu, Y., and Ruvkun, G. 1997. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277: 942-946. [DOI] [PubMed] [Google Scholar]
- Knauf, F., Rogina, B., Jiang, Z., Aronson, P.S., and Helfand, S.L. 2002. Functional characterization and immunolocalization of the transporter encoded by the life-extending gene Indy. Proc. Natl. Acad. Sci. 99: 14315-14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.S., Lee, R.Y., Fraser, A.G., Kamath, R.S., Ahringer, J., and Ruvkun, G. 2003. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat. Genet. 33: 40-48. [DOI] [PubMed] [Google Scholar]
- Lin, K., Dorman, J.B., Rodan, A., and Kenyon, C. 1997. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278: 1319-1322. [DOI] [PubMed] [Google Scholar]
- Lin, S.J., Defossez, P.A., and Guarente, L. 2000. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289: 2126-2128. [DOI] [PubMed] [Google Scholar]
- Longo, V.D. 2003. The Ras and Sch9 pathways regulate stress resistance and longevity. Exp. Gerontol. 38: 807-811. [DOI] [PubMed] [Google Scholar]
- ____. 2004. Ras: The other pro-aging pathway. Sci. Aging Knowledge Environ. 2004: pe36. [DOI] [PubMed] [Google Scholar]
- Longo, V.D. and Fabrizio, P. 2002. Regulation of longevity and stress resistance: A molecular strategy conserved from yeast to humans? Cell Mol. Life Sci. 59: 903-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo, V.D. and Finch, C.E. 2003. Evolutionary medicine: From dwarf model systems to healthy centenarians? Science 299: 1342-1346. [DOI] [PubMed] [Google Scholar]
- Longo, V.D., Gralla, E.B., and Valentine, J.S. 1996. Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae: Mitochondrial production of toxic oxygen species in vivo. J. Biol. Chem. 271: 12275-12280. [DOI] [PubMed] [Google Scholar]
- Longo, V.D., Liou, L.L., Valentine, J.S., and Gralla, E.B. 1999. Mitochondrial superoxide decreases yeast survival in stationary phase. Arch. Biochem. Biophys. 365: 131-142. [DOI] [PubMed] [Google Scholar]
- MacLean, M., Harris, N., and Piper, P.W. 2001. Chronological lifespan of stationary phase yeast cells: A model for investigating the factors that might influence the ageing of postmitotic tissues in higher organisms. Yeast 18: 499-509. [DOI] [PubMed] [Google Scholar]
- Marden, J.H., Rogina, B., Montooth, K.L., and Helfand, S.L. 2003. Conditional tradeoffs between aging and organismal performance of Indy long-lived mutant flies. Proc. Natl. Acad. Sci. 100: 3369-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, D.E. and Hall, M.N. 2005. The expanding TOR signaling network. Curr. Opin. Cell Biol. 17: 158-166. [DOI] [PubMed] [Google Scholar]
- Masoro, E.J. 2005. Overview of caloric restriction and ageing. Mech. Ageing Dev. 126: 913-922. [DOI] [PubMed] [Google Scholar]
- Meissner, B., Boll, M., Daniel, H., and Baumeister, R. 2004. Deletion of the intestinal peptide transporter affects insulin and TOR signaling in Caenorhabditis elegans. J. Biol. Chem. 279: 36739-36745. [DOI] [PubMed] [Google Scholar]
- Miller, R.A., Buehner, G., Chang, Y., Harper, J.M., Sigler, R., and Smith-Wheelock, M. 2005. Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell 4: 119-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino, T., Miyauchi, H., Tateno, K., Kunieda, T., and Komuro, I. 2004. Akt-induced cellular senescence: Implication for human disease. Cell Cycle 3: 449-451. [PubMed] [Google Scholar]
- Miyauchi, H., Minamino, T., Tateno, K., Kunieda, T., Toko, H., and Komuro, I. 2004. Akt negatively regulates the in vitro lifespan of human endothelial cells via a p53/p21-dependent pathway. EMBO J. 23: 212-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer, R.K. and Johnston, J.R. 1959. Life span of individual yeast cells. Nature 183: 1751-1752. [DOI] [PubMed] [Google Scholar]
- Murphy, C.T., McCarroll, S.A., Bargmann, C.I., Fraser, A., Kamath, R.S., Ahringer, J., Li, H., and Kenyon, C. 2003. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424: 277-283. [DOI] [PubMed] [Google Scholar]
- Ogg, S., Paradis, S., Gottlieb, S., Patterson, G.I., Lee, L., Tissenbaum, H.A., and Ruvkun, G. 1997. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389: 994-999. [DOI] [PubMed] [Google Scholar]
- Oh, S.W., Mukhopadhyay, A., Svrzikapa, N., Jiang, F., Davis, R.J., and Tissenbaum, H.A. 2005. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc. Natl. Acad. Sci. 102: 4494-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira, E.M., Mansure, J.J., and Bon, E.P. 2005. Gln3p and Nil1p regulation of invertase activity and SUC2 expression in Saccharomyces cerevisiae. FEMS Yeast Res. 5: 605-609. [DOI] [PubMed] [Google Scholar]
- Orentreich, N., Matias, J.R., DeFelice, A., and Zimmerman, J.A. 1993. Low methionine ingestion by rats extends life span. J. Nutr. 123: 269-274. [DOI] [PubMed] [Google Scholar]
- Piper, M.D., Mair, W., and Partridge, L. 2005. Counting the calories: The role of specific nutrients in extension of life span by food restriction. J. Gerontol. A Biol. Sci. Med. Sci. 60: 549-555. [DOI] [PubMed] [Google Scholar]
- Roberts, P., Moshitch-Moshkovitz, S., Kvam, E., O'Toole, E., Winey, M., and Goldfarb, D.S. 2003. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell 14: 129-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogina, B., Reenan, R.A., Nilsen, S.P., and Helfand, S.L. 2000. Extended life-span conferred by cotransporter gene mutations in Drosophila. Science 290: 2137-2140. [DOI] [PubMed] [Google Scholar]
- Roosen, J., Engelen, K., Marchal, K., Mathys, J., Griffioen, G., Cameroni, E., Thevelein, J.M., De Virgilio, C., De Moor, B., and Winderickx, J. 2005. PKA and Sch9 control a molecular switch important for the proper adaptation to nutrient availability. Mol. Microbiol. 55: 862-880. [DOI] [PubMed] [Google Scholar]
- Rual, J.F., Ceron, J., Koreth, J., Hao, T., Nicot, A.S., Hirozane-Kishikawa, T., Vandenhaute, J., Orkin, S.H., Hill, D.E., van den Heuvel, S., et al. 2004. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14: 2162-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzle, T., Beck, T., Martin, D.E., and Hall, M.N. 2004. Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol. Cell Biol. 24: 338-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreve, J.L., Sin, J.K., and Garrett, J.M. 1998. The Saccharomyces cerevisiae YCC5 (YCL025c) gene encodes an amino acid permease, Agp1, which transports asparagine and glutamine. J. Bacteriol. 180: 2556-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamji, A.F., Kuruvilla, F.G., and Schreiber, S.L. 2000. Partitioning the transcriptional program induced by rapamycin among the effectors of the Tor proteins. Curr. Biol. 10: 1574-1581. [DOI] [PubMed] [Google Scholar]
- Stanbrough, M., Rowen, D.W., and Magasanik, B. 1995. Role of the GATA factors Gln3p and Nil1p of Saccharomyces cerevisiae in the expression of nitrogen-regulated genes. Proc. Natl. Acad. Sci. 92: 9450-9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trancikova, A., Weisova, P., Kissova, I., Zeman, I., and Kolarov, J. 2004. Production of reactive oxygen species and loss of viability in yeast mitochondrial mutants: Protective effect of Bcl-xL. FEMS Yeast Res. 5: 149-156. [DOI] [PubMed] [Google Scholar]
- Vellai, T., Takacs-Vellai, K., Zhang, Y., Kovacs, A.L., Orosz, L., and Muller, F. 2003. Genetics: Influence of TOR kinase on lifespan in C. elegans. Nature 426: 620. [DOI] [PubMed] [Google Scholar]
- Weindruch, R.H. and Walford, R.L. 1988. The retardation of aging and disease by dietary restriction. Thomas, Springfield, IL.
- Winzeler, E.A., Shoemaker, D.D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J.D., Bussey, H., et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901-906. [DOI] [PubMed] [Google Scholar]