Abstract
The PLZF (promyelocytic leukemia zinc finger) transcriptional repressor, when fused to retinoic acid receptor alpha (RARα), causes a refractory form of acute promyelocytic leukemia. The highly conserved N-terminal BTB (bric a brac, tramtrack, broad complex)/POZ domain of PLZF plays a critical role in this disease, since it is required for transcriptional repression by the PLZF-RARα fusion protein. The crystal structure of the PLZF BTB domain revealed an obligate homodimer with a highly conserved charged pocket formed by apposition of the two monomers. An extensive structure-function analysis showed that the charged pocket motif plays a major role in transcriptional repression by PLZF. We found that mutations of the BTB domain that neutralize key charged pocket residues did not disrupt dimerization, yet abrogated the ability of PLZF to repress transcription and led to the loss of interaction with N-CoR, SMRT, and histone deacetylases (HDACs). We extended these studies to the Bcl-6 protein, which is linked to the pathogenesis of non-Hodgkin's lymphomas. In this case, neutralizing the charged pocket also resulted in loss of repression and corepressor binding. Experiments with purified protein showed that corepressor-BTB interactions were direct. A comparison of the PLZF, Bcl-6, and the FAZF (Fanconi anemia zinc finger)/ROG protein shows that variations in the BTB pocket result in differential affinity for corepressors, which predicts the potency of transcriptional repression. Thus, the BTB pocket represents a molecular structure involved in recruitment of transcriptional repression complexes to target promoters.
The BTB (bric a brac, tramtrack, broad complex) domain is found in a large and diverse family of proteins. BTB domains are highly conserved from Drosophila to Homo sapiens and are present in 5 to 10% of all zinc finger proteins (1, 54). In the annotation of the human genome sequence, BTB domains were found in 113 proteins, representing the 28th most common motif in the human proteome (29). BTB proteins play critical roles in development, homeostasis, and neoplasia (3, 9, 22, 28, 47, 48).
We are interested in a subset of BTB proteins involved in silencing gene expression. Among these, the PLZF (promyelocytic leukemia zinc finger) protein was identified as the translocation partner of retinoic acid receptor alpha (RARα) in t(11;17)(q23;q21) retinoid-resistant acute promyelocytic leukemia (6, 32). PLZF is a growth suppressor that blocks proliferation and myeloid differentiation through silencing of target genes, including cell cycle regulators (2, 46, 52). Another BTB protein, Bcl-6 (B-cell lymphoma 6), is rearranged in at least 35% of diffuse large cell lymphomas, as well as in follicular and AIDS-related lymphomas, with aberrant expression usually driven by heterologous promoter regions. (14, 35, 51). Bcl-6 is normally expressed in germinal center B cells, as well as intrafollicular CD4+ cells (4, 10). In lymphomas, persistent expression of Bcl-6 is believed to block lymphocyte differentiation, leaving these cells in a state in which they are competent to continue proliferating and vulnerable to further mutagenesis (45). Bcl-6 directly represses promoters of gene products such as CD69, cyclin D2, and MIP1α (45).
Initial studies of the transcriptional function of PLZF, Bcl-6, and PLZF-RARα led to the conclusion that the BTB domain of these proteins was essential for dimerization, transcriptional repression, and nuclear microspeckled localization (10, 13). Furthermore, the BTB domains can be autonomous transcriptional repression motifs. In transient transfection assays, BTB domains fused to the GAL4 DNA binding domain (DBD) repressed reporter genes containing GAL4 binding sites (5, 30).
The discovery of the role of histone deacetylases (HDACs) and corepressor proteins such as N-CoR and SMRT in transcriptional repression led a number of groups to ask whether PLZF, PLZF-RARα, and Bcl-6 worked through interaction with such factors (7, 11, 12, 16, 18, 19, 21, 23, 34, 50). Some of these reports suggested a key role for the BTB domain. For example, Guidez et al. found that the BTB domain was important for the ability of PLZF-RARα to repress transcriptional repression and bind N-CoR (18). Others were unable to detect an interaction between the PLZF BTB domain and N-CoR by using glutathione S-transferase (GST) protein affinity chromatography (16). None of these studies fully addressed the critical question of whether the BTB domains could directly touch transcriptional corepressors. The protein interaction studies employed included yeast and mammalian two-hybrid models, coimmunoprecipitations, and GST pulldowns with in vitro-transcribed and -translated proteins (7, 11, 12, 16, 18, 19, 21, 23, 34, 50). In both eukaryotic cells as well as in reticulocyte lysates, other proteins such as Sin3A are present and could mediate bridging interactions between the BTB domain and other corepressors.
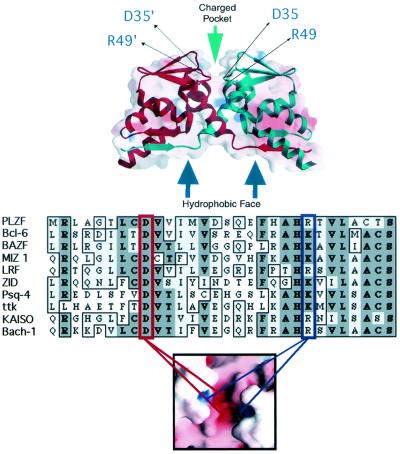
Given the frequent representation of the BTB domain in the transcription factor repertoire, its highly conserved nature and its role in hematologic malignancies, we wished to understand the molecular basis of its structure and functions. A crystallographic analysis of the PLZF BTB dimer at a 1.9-Å resolution revealed an intertwined, obligate homodimer with an extensive hydrophobic interface (Fig. 1) (1). A striking surface feature was a charged pocket formed by apposition of the two monomers (1, 31).
FIG. 1.
Structural motifs of the BTB dimer. (Top panel) Superimposed ribbon and charge-mapping views of the PLZF BTB dimer (red, negative charge; blue, positive charge). The BTB monomer ribbons are shown in blue and red, respectively. The green arrow indicates the highly conserved charged pocket, and the blue arrows indicate the hydrophobic oligomerization surface region. The thin black arrows indicate the location of key, conserved charged pocket residues in both monomers. (Middle panel) Sequence alignment of the N terminus of several different BTB domains. This region of the BTB corresponds mainly to the charged pocket. The red box indicates the conserved aspartate residue, which in PLZF is at position 35. The blue box indicates the conserved basic pocket residue, which in PLZF is an arginine at position 49. The location of these two residues in the charged pocket is shown in the lower panel, which is a charge mapping viewed from the “top” of the dimer.
To test the function of the pocket and other dimer features, we performed a battery of biochemical, transcriptional, and biological assays on an extensive set of mutants (36). We examined the ability of isolated BTB domain mutants and full-length PLZF mutants to properly fold, dimerize, repress transcription, form high-molecular-weight complexes, localize to nuclear microspeckles, and suppress cell proliferation. We found that mutations that resulted in misfolding led to proteins that could not dimerize and were nonfunctional. However, the most prominent finding of these studies was that proper charge alignment in the BTB pocket was required for its transcriptional repression function, even when the overall structural integrity and dimerization ability of the domain were preserved (36).
Given these results, we hypothesized that the PLZF BTB charged pocket represents a novel site for recruitment of corepressor-HDAC complexes. In our present studies, we find that the proper charge alignment in the BTB pocket is required not only for transcriptional repression, but also for binding of corepressors. We show for the first time that the BTB-corepressor interaction is direct and is mediated by discreet regions of N-CoR and SMRT. Furthermore, these results were perfectly reproducible in the case of Bcl-6 repression and its interaction with N-CoR, SMRT, and the Bcl-6-specific corepressor B-CoR (24). Finally, natural variations in key residues of the charged pocket between PLZF, Bcl-6, and the FAZF (Fanconi anemia zinc finger) proteins demonstrate that changes in affinity for corepressors can in part predict the ability of different BTB domains to efficiently repress transcription. Thus, the BTB pocket may represent a novel repression complex recruitment motif and is a major transcriptional effector site for PLZF and Bcl-6.
MATERIALS AND METHODS
Plasmids and mutant BTB domains.
The DNA segment encoding residues 1 to 137 of PLZF comprising the BTB region was previously described (36). The R49D, D33N, D41R, R49Q, and D35N/R49Q PLZF BTB mutants were described in reference 36. Additional PLZF mutants used herein were constructed by PCR amplification from human PLZF cDNA by using a 5′ primer containing a BamHI site (5′-TATGGATCCATGGATCTGACAAAA-3′) and a 3′ primer containing an XbaI site 5′-TCACTCTAGAGCGGCCATGGTGGCCTCCGTGTCATT-3′). BTB domain mutations were created by PCR-mediated mutagenesis with the following specific oligonucleotides: for R49K, 5′-CACGCCCACAAGACGGTGCTG-3′ and 5′-CAGCACCGTGCTGTGGGCGTG-3′; for R49S, 5′-CACGCCCACAGCACGGTGCTG-3′ and 5′-CAGCACCGTGCTGTGGGCGTG-3′; and for D35N, 5′-ACTTTGTGCAATGTGGTCATC and 5′-GATGACCACATTGCACAAAGT-3′. A DNA segment containing residues 5 to 129 comprising the BTB domain of Bcl-6 was PCR amplified with a 5′ BamHI site-containing primer (5′-TATGGATCCGCTGACAGCCAGATC-3′) and a 3′ HindIII-XbaI site-containing primer (5′-GCTCTAGAAAGCTTTTCACTGGCCTTAATAAA-3′). To introduce PCR-generated point mutations, the following oligonucleotides were used: for D33N, 5′-ATCTTGACTAATGTTGTCATT-3′; for K47Q, 5′-AGAGCCCATCAAACGGTCCTC-3′ and 5′-GAGGACCGTTTGATGGGCTCT-3′; for K47D, 5′-AGAGCCCATGACACGGTCCTC-3′ and 5′-GAGGACCGTGTCATGGGCTCT-3′; for K47R, 5′-AGAGCCCATAGAACGGTCCTC-3′ and 5′-GAGGACCGTTCTATGGGCTCT-3′; and for K47S, 5′-AGAGCCCATAGTACGGTCCTC-3′ and 5′-GAGGACCGTACTATGGGCTCT-3′. A construct encoding the FAZF BTB domain (amino acids 1 to 125) was amplified from the FAFZF cDNA by using the BamHI site-containing primer 5′-TCGGATCCGTGATGTCCCTGCCCCCC-3′ and the 3′ XbaI-HindIII site-containing primer 5′-CGTCTAGAAAGCTCCAGGCCTGGATCTGGCTT-3′. The following point mutations were introduced: D29N (5′-GCACTCTGTAATACTCTGATC-3′ and 5′-GATCAGAGTATTACAGAGTGC-3′), S44R (5′-CCCGCCCACAGACTGGTGCTA-3′ and 5′-TAGCACCAGCAGTCTGTGGGCGGG-3′), and S44K (5′-CCCGCCCACAAACTGGTGCTA-3′ and 5′-TAGCACCAGTTTGTGGGCGGG-3′). Amplified fragments were gel purified (Qiagen, Valencia, Calif.) and digested for insertion into the pBXG1 GAL4 DBD mammalian expression vector (36) and the pSP73 vector for in vitro transcription and translation (Promega, Madison, Wis.). Sequences encoding the Bcl-6 and PLZF BTB domains were cloned into pET-32(a) (Novagen, Madison, Wis.), to yield an Escherichia coli thioredoxin protein, followed by a 56-amino-acid linker containing a six-His affinity tag and ending with the BTB domains of PLZF or Bcl-6. PLZFΔBTB, lacking the first 120 N-terminal amino acids of PLZF, was described previously (13). Full-length PLZF was cloned into the EcoRI site of pCDNA3.1myc/his+A (Invitrogen, Carlsbad, Calif.). This plasmid was partially digested with EcoRV and SfiI to remove sequences encoding the wild-type BTB domain and replaced with EcoRV-SfiI fragments derived from pSP73 vectors harboring the mutant BTB domains. The correct reading frame and sequence were confirmed for all plasmids by automated DNA sequencing (Utah State University Biotechnology Center, Logan, and the OCI Sequencing Facility at the Princess Margaret Hospital, Toronto, Ontario, Canada). Expression vectors for SMRT and N-CoR were a gift of Mitch Lazar (University of Pennsylvania). The B-CoR expression vector and the VP16-N-CoR, -SMRT, or -B-CoR fusions for mammal two-hybrid assays were previously described (23, 24). The coding region of mouse N-CoR (amino acids 1351 to 1616) was cloned into the T7 polymerase-based expression vector pET-16-b(+) (Novagen).
Expression, purification, and interaction of PLZF, N-CoR, and SMRT fragments.
The preparation of PLZF BTB protein was described previously (1), and Bcl-6 BTB was expressed and purified by a similar protocol. N-CoR fragment 1351 to 1616 containing an N-terminal six-His tag was expressed in E. coli BL21(DE3) cells grown in Luria-Bertani (LB) broth with induction with 0.2 mM isopropyl β-d-thiogalactopyranoside (IPTG). Cells were harvested and lysed by cavitation in an Emulsiflex-C5 (Avestin, Ottawa, Ontario, Canada). The resulting lysate was cleared by centrifugation, and the soluble supernatant was purified by metal chelation chromatography on a nickel-nitrilotriacetic acid (Ni-NTA) column (Qiagen). The peak fractions containing the His-tagged protein were pooled, concentrated, and further purified by size-exclusion chromatography on a Superdex-75 column (Pharmacia Biotech; 16 by 600 mm) equilibrated in a mixture containing 250 mM NaCl and 20 mM Tris-HCl (pH 8.0). Equimolar amounts of 10-His-tagged N-CoR(1351-1616) or 6-His-tagged thioredoxin and Bcl-6 BTB or PLZF BTB were mixed overnight at 4°C. The mixture was then loaded onto a Ni-NTA spin column (Qiagen) equilibrated in buffer A (200 mM NaCl, 20 mM Tris-HCl [pH 8.0], 10 mM imidazole, 5 mM β-mercaptoethanol) and washed twice with 600 μl of buffer A. Bound protein was then eluted with 400 μl of buffer A containing 300 mM imidazole.
Reporter assays.
Reporter constructs used in these experiments included (GAL4)5-tk-luciferase (Luc) and (interleukin-3 receptor [IL-3R])4-tk-Luc, the latter containing PLZF binding sites (36). A tk-Luc construct lacking specific binding sites was used as a negative control for the above reporters and a tk-Renilla luciferase plasmid was included as an internal control. 293T cells were plated in 12-well tissue culture dishes at a density of 2 × 105 per well and transfected with the Superfect lipid reagent (Qiagen). Dual luciferase assays were performed (Promega), and luciferase activity was measured with an MLX microtiter plate luminometer (Dynex Technologies; Chantilly, Va.). All transfection experiments were performed in duplicate or quadruplicate 3 to 10 times. The fold repression of transcription was calculated relative to transcription of the reporters in the presence of the relevant empty expression vector and normalized to the internal control. Immunoblotting with the appropriate antibodies confirmed expression of all of the transfected proteins. The amount of transfected plasmids is indicated in the figure legends.
Mammalian two-hybrid assays.
For the B-CoR, N-CoR, and SMRT experiments, 293T cells were grown in 12-well tissue culture plates at a density of 2 × 105 cells per well. The B-CoR experiments were also performed with HeLa cells transfected with TFX20 lipid reagent (Promega). The bait plasmids were the same GAL4-BTB fusion proteins described above, at 25 ng per well. The prey plasmids consisted of the VP-16 corepressor fusions as described above. Activation of (GAL)5-tk-Luc (for N-CoR and SMRT) or simian virus 40 (SV40)-(GAL4)4-Luc (for B-CoR) compared to GAL1-147 was normalized to that of the internal control and tabulated as fold activation of the hybrid formed between the interacting proteins.
In vitro translation immunoprecipitations.
Interaction of BTB domains with N-CoR, SMRT, and B-CoR was determined by precipitating the corepressors with the antibodies listed below and visualizing the BTB domains by 35S autoradiography of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. Five hundred nanograms of DNA was used for each reaction. Reactions were performed as per the TnT coupled in vitro transcription-translation system manufacturer's protocol (Promega). Aliquots of 5 μl of each sample were run by PAGE as controls of expression. The remaining 45 μl of the sample was combined with the in vitro-translated potential protein partner and incubated for at least 2 h with continuous rotating. The cocktail of in vitro translates was then split between two tubes. Protein A beads with the specific antibody of interest preattached (see below) were added to one of the tubes. Protein A beads with a nonspecific antibody of the correct isotype was added to the other tube. In both cases, the volume was brought up with Tris-buffered saline (TBS) buffer containing protease inhibitors (Roche Molecular Biochemicals, Indianapolis, Ind.). These tubes were incubated for at least 4 h at 4°C. The beads were then spun and washed six times with repeated rotation in TBS plus 0.5% Tween 20. The beads were then placed in Laemmli buffer for running on SDS-PAGE. The antibodies were rabbit polyclonal N-CoR (H-303) antibodies, goat polyclonal SMRT (N-20) antibodies, c-myc (9E10) mouse monoclonal antibodies (Santa Cruz, Santa Cruz, Calif.), and control preimmune rabbit and goat polyclonal immunoglobulin G (IgG) (Jackson Immuno-Research; West Grove, Pa.).
Transient transfection immunoprecipitation.
293T cells were plated at a density of 106 cells per well and transfected with the indicated plasmids at a final concentration of 1 μg of DNA per well of a six-well plate. Transfections were performed with the Superfect reagent as directed (Qiagen). The cells were harvested 48 h after transfection for immunoblotting. Twenty microliters of protein A-Sepharose beads (Roche Molecular Biochemicals Indianapolis, Ind.) was washed twice in 0.2 M sodium borate (pH 9.0). The following antibodies were used for immunoprecipitations and/or immunoblotting: PLZF mouse monoclonal antibody (IgG2a isotype) (33), rabbit polyclonal GAL (DBD), HDAC1 (H-51), HDAC2 (H-54), and N-CoR (H-303) antibodies, goat polyclonal SMRT (N-20) antibodies (Santa Cruz), isotype control preimmune mouse monoclonal IgG2a, and control preimmune rabbit and goat polyclonal IgG (Jackson ImmunoResearch). To prepare antibody-coated beads, 10 μl of 50% protein A-agarose beads slurry was washed with 0.2 M boric acid. The antibodies were added at a concentration of 1 mg/ml and mixed for 1 h at room temperature. The beads were washed again at pH 9.0 at room temperature in sodium borate followed by washing in 0.2 M triethanolamine (pH 8.5) and freshly measured dimethylpimelidate at a final concentration of 20 mM and mixed for 1 h at room temperature. The beads were resuspended in 0.2 M ethanolamine for 5 min, before being transferred into phosphate-buffered saline (PBS). (Unless stated otherwise, all chemicals were obtained from Sigma, St. Louis, Mo.). Cells were harvested with PBS at 4°C and exposed to lysis buffer (150 mM NaCl, 20 mM Tris-Cl, Tween 20 [pH 7.4], plus protease inhibitors) on ice for 15 min. This suspension was centrifuged at 2,000 × g for 5 min at 4°C. The supernatant was precleared by exposure to beads bound to a nonspecific rabbit IgG (Zymed, San Francisco, Calif.) and mixed for approximately 1 h at 4°C. The beads were then pelleted and exposed to specific antibodies of interest bound to protein A-Sepharose or their isotype controls and mixed on a rotator overnight. The pellets were washed in fresh cold lysis buffer six times—the last three times with NP-40 instead of Tween 20 in the lysis buffer. The beads were then placed in Laemmli buffer, and the precipitated proteins were released by boiling. This was followed by electrophoresis through an SDS-12% polyacrylamide gel and transfer to an Immobilon P membrane (Millipore, Bedford, Mass.). Immunoblotting was performed with a 1:1,000 dilution of monoclonal PLZF antibodies (100 mg/ml) or a 1:500 dilution of SMRT, HDAC1, HDAC2, or N-CoR primary antibodies followed by a 1:3,000 dilution of horseradish peroxidase-conjugated anti-mouse, anti-rabbit, or anti-goat secondary antibody (Roche Molecular Biochemicals). Autoradiographs were preformed with the ECL enhanced chemiluminescence kit (Amersham Pharmacia, Buckinghamshire, United Kingdom). Direct immunoblotting was performed with 20 μl of lysates from transfected 293T cells.
RESULTS
Mutating the BTB domain.
A highly conserved charged pocket is a critical functional motif of the PLZF BTB dimer (36). The most conserved charged residues in the PLZF pocket are an aspartate at position 35 and an arginine at position 49 (Fig. 1). Each monomer of the BTB dimer contributes a wall to the pocket in which the D35 and R49 residues are present, leading to the coordinately charged pocket containing the two positive and two negative charges (see bottom panel of Fig. 1). Aspartate 35 is nearly invariant throughout the BTB domain family, while position 49 is most often an arginine or a lysine, with very few exceptions (see Fig. 1 alignment). The introduction of point mutations into these residues is highly informative in studying the function of the BTB pocket. Point mutations that result in loss of these two charges functionally inactivate BTB-dependent repression while maintaining the ability to dimerize and oligomerize (36) (described below).
The PLZF BTB domain is physically and functionally required for interactions with corepressors.
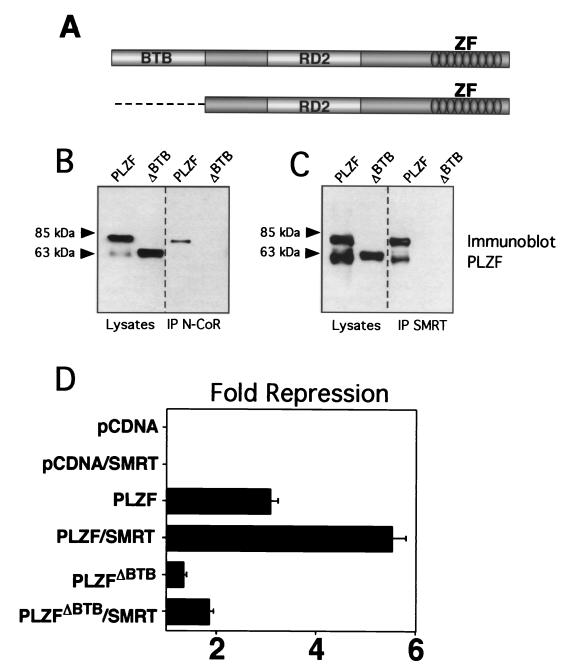
We hypothesized that the BTB charged pocket mediates transcriptional repression through a pivotal role in recruiting corepressors to the BTB domain of PLZF. However, PLZF has two other characterized functional motifs: a C-terminal array of nine zinc fingers that bind to specific DNA targets as well as other proteins and a second repression domain (RD2) that interacts with the ETO corepressor (Fig. 2A) (27, 30, 37). Thus, we first analyzed the impact of BTB loss of function by deleting the entire domain and studying the functional and physical effects on interactions with the N-CoR and SMRT corepressors. We transfected 293T cells with PLZF or PLZFΔBTB and N-CoR or SMRT and confirmed expression of all proteins by immunoblotting. Coimmunoprecipitations were performed with SMRT or N-CoR antibodies. In both cases, the full-length form of PLZF could be coprecipitated with the corepressors, while the form deleted for the BTB domain could not. We believe that the second band is a C-terminally truncated form of PLZF that is occasionally seen in these experiments. Thus, the BTB domain was required for in vivo interaction with the corepressors (Fig. 2B). To assess PLZF repression, we employed a construct containing four multimerized PLZF binding sites (2, 36). PLZF typically represses this reporter threefold, and SMRT approximately doubles the repression to approximately sixfold (Fig. 2C). In contrast, PLZFΔBTB represses less than 1.5-fold, and this effect was not significantly enhanced by SMRT (enhanced to 1.8-fold). In our experience, SMRT can variably enhance repression of several reporter constructs, although this effect is small compared to the enhancement of the BTB proteins under study. Enhancement by N-CoR is more difficult to show in these cells, possibly because they have plentiful endogenous N-CoR (data not shown). We conclude that the BTB domain is required for effective physical interactions with SMRT and N-CoR and functional interaction with SMRT and is required for the transcriptional effects of PLZF.
FIG. 2.
The BTB domain is physically and functionally critical for interactions with corepressors. (A) PLZF contains discrete functional domains, including the N-terminal BTB domain, a less-well-characterized second repression domain (RD2), and nine C-terminal Zn fingers (ZF). The PLZFΔBTB construct used in these experiments is also shown. (B) Coimmunoprecipitations (IP) performed in 293T cells. Cells (4 × 105) were transfected with 1 μg of PLZF or PLZFΔBTB expression vectors. Endogenous N-CoR was precipitated, and the resulting fraction was immunoblotted to detect PLZF. Expression of both proteins is confirmed by direct Western blotting of the cell lysates as shown in the left two lanes. The lower band in the PLZF lanes is an alternate PLZF product commonly seen in cells transfected with the PLZF expression vector. (C) Coimmunoprecipitations with endogenous SMRT similar to panel B. (D) 293T cells (2 × 105) were transfected with 200 ng of pCDNA, pCDNA-PLZF, or pCDNA-PLZFΔBTB, with or without 150 ng of CMX-SMRT. The cells were cotransfected with 50 ng of a luciferase reporter containing four PLZF binding sites from the IL-3Rα chain promoter and a tk-renilla internal control. Results are expressed as fold repression of luciferase normalized to the internal control.
The PLZF BTB charged pocket is required for transcriptional repression and corepressor interaction.
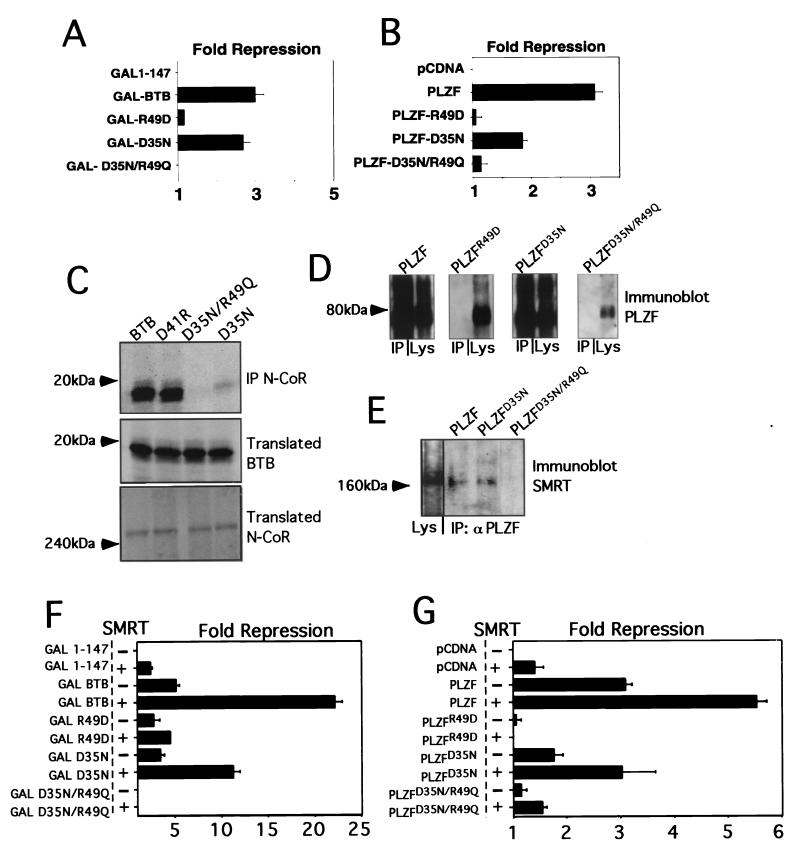
Having confirmed the central role of the PLZF BTB domain in corepressor recruitment and repression, we next tested the role of the BTB pocket in these processes. Although the pocket in the wild-type protein has a high charge density due to the two arginines and two aspartates, it carries a formal net charge of zero. These residues are in close proximity to each other, but do not interact through salt bridging. We used three mutant BTB domains we had previously characterized (36). These are as follows. (i) The first is an R49D point mutation, which if present in the folded dimer, would present a net charge of −4 in the pocket. This mutation results in an unfolded nonfunctional BTB domain (36). (ii) The second is a D35N mutation, which would create a +2 charge in the pocket. Such BTB domains partially preserve dimerization and repression (36). (iii) The third is a double point mutant, D35N/R49Q, which produces a pocket with no net charge, as with the wild-type protein, but which lacks the high charge density. This mutant domain has an improved stability profile relative to the two other mutants, but has lost all repression activity when tested in the context of full-length PLZF. In fact, when fused to heterologous DBDs, this BTBD35N/R49Q mutant is a transcriptional activator (36). All of these mutants were tested as fusions to the GAL4 DBD (amino acids 1 to 147) or were inserted into full-length PLZF.
GAL-BTB fusions and full-length PLZF species with mutant BTB domains were cotransfected with the (GAL)5-tk-Luc or (IL-3Rα)4-tk-Luc reporters, respectively. Expression of the BTB proteins was verified by immunoblotting (data not shown). Consistent with our previous results, in both cases the R49D misfolding mutant was nonfunctional, the D35N mutant was partially functional, and the dimerization-competent D35N/R49Q double mutant was unable to repress (Fig. 3A and B).
FIG. 3.
The PLZF BTB charged pocket is required for transcriptional repression and corepressor interaction. (A) 293T cells (2 × 105) were cotransfected with 200 ng of the indicated GAL-fusion effectors, 50 ng of (GAL)5-tk-Luc reporter, and 5 ng of tk-renilla. Results are expressed as fold repression of luciferase normalized to the internal control. (B) 293T cells (2 × 105) were cotransfected with 200 ng of the indicated wild-type and mutant PLZF species, 50 ng of (IL-3Rα)4-tk-Luc reporter, and 5 ng of tk-renilla. (C) The ability of N-CoR to precipitate BTB domains was tested by allowing in vitro-translated N-CoR to interact with in vitro-translated BTB domains. Both proteins were 35S labeled and visualized by fluorography. The top gel shows BTB domains after precipitation with N-CoR antibodies; the middle and bottom gels show the presence of the indicated BTB species N-CoR in the input lysates. IP, immunoprecipitation. (D) Western blots of transfected full-length PLZF proteins with wild-type or point mutant BTB domains after immunoprecipitation with N-CoR antibodies. The lysate input and corresponding immunoprecipitation reactions are paired in each of the boxes shown. (E) PLZF monoclonal antibody was used to immunoprecipitate lysates from 293T cells transfected with full-length wild-type PLZF and PLZF harboring mutations in the BTB domain. Western blots were performed with the resolved proteins to determine the presence of the SMRT corepressor. Lane 1 shows the presence of SMRT in the cell lysates. (F) Similar to panel A, 293T cells were cotransfected with GAL-BTB fusions and the GAL reporter, with or without 150 ng of SMRT, as indicated. (G) 293T cells were transfected as shown in panel B with full-length PLZF or the indicated mutants along with the IL-3Rα reporter, with or without 150 ng of SMRT.
We next tested whether these mutations affected the interaction of the BTB domain with corepressors. We first performed coimmunoprecipitations with in vitro-translated GAL-BTB fusions and corepressors. The proteins were labeled with [35S]methionine and allowed to interact prior to precipitation with N-CoR antibodies. The BTB domains were then visualized by fluorography (Fig. 3C). Wild-type BTB and BTBD41R (a mutation outside of the pocket which represses in a manner similar to wild type) both interacted with N-CoR. BTBD35N/R49Q did not interact with N-Cor, while BTBD35N maintained some interaction. BTBR49D was not tested, since it does not properly fold (36). We next cotransfected full-length PLZF species with N-CoR or SMRT and performed coimmunoprecipitations. In Fig. 3D, lysates were precipitated with N-CoR antibodies and immunoblotted for PLZF. Wild-type PLZF and PLZFD35N interacted with N-CoR, while PLZFR49D and PLZFD35N/R49Q did not. In Fig. 3E, lysates were precipitated with PLZF antibodies and immunoblotted for SMRT. PLZF and PLZFD35N interacted with SMRT, while PLZFD35N/R49Q did not.
Functional interaction was similarly affected. In reporter assays with (GAL)5-tk-Luc, cotransfection of SMRT enhanced GAL-BTB and GAL-D35N, but did not enhance GAL-R49D or GAL-D35N/R49Q (Fig. 3F). Similarly, in assays using the (IL-3Rα)4-tk-Luc reporter, PLZF and PLZFD35N were enhanced by SMRT. In contrast, PLZFR49D was not enhanced and PLZFD35N/R49Q was only minimally enhanced (Fig. 3G). From these experiments, we conclude that the D35N/R49Q loss-of-pocket-charge mutant is unable to bind to corepressors and to mediate repression. This suggests that correct charge alignment within the BTB pocket is required for stable interaction of corepressors. Interestingly, the single-pocket mutant D35N seems to partially reduce BTB-dependent repression and corepressor interaction. This was more evident in the context of full-length protein. The fact that the D35N/R49Q mutant severely affects full-length PLZF repression and corepressor association underscores the critical nature of the BTB domain for transcriptional silencing.
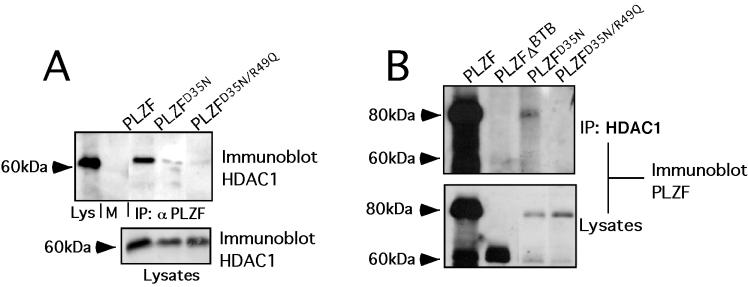
The PLZF BTB charged pocket is required for efficient recruitment of HDACs.
The ability of PLZF to repress transcriptional repression has been linked to its capacity to recruit HDACs, probably through the bridging effects of corepressors (7, 18). We tested the ability of full-length PLZF with BTB mutations to interact with HDAC1 in cotransfection and/or coimmunoprecipitation experiments When PLZF monoclonal antibodies were used to precipitate the cell lysates, PLZFD35N was partially and PLZFD35N/R49Q was almost completely defective in forming a complex with HDAC1 (Fig. 4A). Conversely, when the lysates were precipitated with HDAC1 antibodies, PLZF was efficiently coprecipitated, but PLZFΔBTB and PLZFD35N/R49Q were not. As before, PLZFD35N was partially defective for association with HDAC1 (Fig. 4B). Similar results were seen in coimmunoprecipitations with HDAC2 (not shown). Therefore, the BTB pocket is required for efficient association of PLZF with HDACs, consistent with a major role of this motif in mediating the HDAC-dependent repression effects of PLZF.
FIG. 4.
The PLZF BTB charged pocket is required for efficient recruitment of HDACs. 293T cells (106) were transfected with 1 μg of full-length PLZF or mutants and 1 μg of HDAC1 expression plasmid. (A) Lysates were subjected to immunoprecipitation (IP) with PLZF antibodies and then immunoblotted with HDAC1 antibodies. The first lane shows lysate input (Lys), followed by lysates after precipitation with preimmune rabbit sera and then respective coprecipitations with wild-type and mutant forms of PLZF. The lower gel shows expression of HDAC- 1 in the input lysates. (B) Precipitations were performed with HDAC1-specific antibodies, and the resulting proteins were immunoblotted with PLZF antisera. The PLZF species used in each case is indicated. Lysate controls are shown below.
The charged pocket is required for Bcl-6 BTB transcriptional repression and interaction with corepressors.
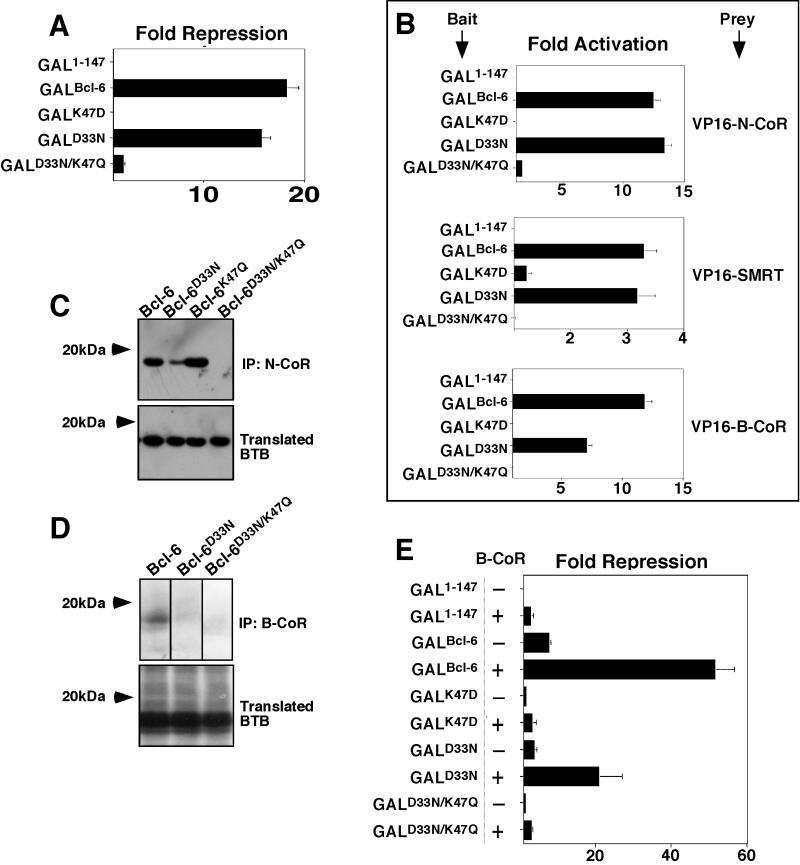
We next wished to determine whether the charged pocket was equally important for another BTB repressor. Bcl-6 is a transcriptional repressor that is aberrantly expressed in many cases of non-Hodgkin's lymphoma and is known to interact with corepressors and HDACs. We created mutations in the Bcl-6 charged residues equivalent to those of PLZF, predicted to form the pocket of its BTB domain. Specifically these included K47D, D33N, and D33N/K47Q. (The fact that Bcl-6 has a lysine instead of arginine in the pocket will be discussed below.) These BTB domains were fused to GAL1-147 and tested for their ability to repress the (GAL)5-tk-Luc reporter. As before, all proteins were detected by immunoblotting (data not shown). As in the case of PLZF, the K47D mutant did not repress, the partially defective D33N mutant was able to repress to almost the same extent as the wild type, and the D33N/K47Q loss-of-charge mutant also failed to repress transcription (Fig. 5A). To determine whether loss of repression correlates with loss of corepressor binding, we first performed mammalian two-hybrid experiments with the Bcl-6 GAL-BTB fusions as bait and VP16-N-CoR, -SMRT, and -B-CoR as prey. B-CoR is a Bcl-6-specific corepressor, which interacts with Bcl-6, but not PLZF. The interaction likely requires the same corepressor binding site on the BTB domain, since the interaction of B-CoR or N-CoR and SMRT with Bcl-6 was shown to be mutually exclusive (24). Interaction between Bcl-6 and each corepressor was confirmed in vivo by activation of the (GAL)5 reporter. As in the case of PLZF, Bcl-6D33N was still capable of interacting with each corepressor, while Bcl-6K47D mutants did not. None of the corepressors could interact in vivo with the Bcl-6D33N/R49Q mutant, indicating that the correct charge alignment of residues in the pocket was required to recruit the corepressors (Fig. 5B). Furthermore, wild-type Bcl-6 BTB could be coprecipitated with N-CoR as well as the single-charge-neutralization mutant Bcl-6D33N and the additional control single-charge mutant Bcl-6K47Q. However, the double-charge-neutralization mutant Bcl-6D33N/K47Q did not interact with the corepressor (Fig. 5C). In similar assays, B-CoR precipitated wild-type but not double mutant BTB domains. The interaction with Bcl-6D33N in this case was present, although considerably weakened (Fig. 5D). B-CoR was previously shown to enhance transcriptional repression by Bcl-6 (24). We tested whether B-CoR could enhance repression by the point mutants in reporter assays. B-CoR efficiently enhanced transcriptional repression by GAL-Bcl-6BTB and was able to enhance Bcl-6D33N as well. The Bcl-6D33N/R49Q mutant was not enhanced by B-CoR, nor was the misfolding mutant Bcl-6K47D mutant (Fig. 5E). Therefore, the model developed for PLZF could be applied to Bcl-6, indicating that the integrity of the charged pocket motif of the Bcl-6 BTB domain is critical for recruitment of corepressors and allows BTB-dependent transcriptional repression.
FIG. 5.
The charged pocket is required for Bcl-6 BTB transcriptional repression and interaction with corepressors. Wild-type and mutant Bcl-6 BTB domains were analyzed in several assays. (A) 293T cells (2 × 105) were transfected with 100 ng of GAL-Bcl-6 expression vectors and 50 ng of the (GAL)5-tk-Luc reporter and 5 ng of tk-renilla as an internal control. The results are expressed as fold repression of luciferase normalized to the internal control. (B) Mammalian two-hybrid assays performed in 293T cells transfected with either 25 ng of GAL-Bcl-6BTB bait plasmids and 150 ng of VP16-N-CoR or -SMRT prey. HeLa cells were transfected with 5 ng of GAL-Bcl-6BTB bait and 25 ng of VP16-B-CoR prey. Results are expressed as fold activation of the (GAL)5-tk-Luc reporter normalized to the internal control (tk-renilla in N-CoR and SMRT experiments and a cytomegalovirus-β-galactosidase [CMV-β-Gal] construct in B-CoR experiments in HeLa cells). B-CoR interaction was also tested in 293T cells with similar results (not shown). (C) In vitro-translated 35S-labeled Bcl-6 wild-type and mutant BTB domains were mixed with in vitro-translated N-CoR. Lysates were precipitated with N-CoR antibodies and resolved by fluorography. The top gel shows the presence of BTB domains after coimmunoprecipitation (IP), and the bottom gel verifies the presence of the BTB domains in the lysates prior to precipitation. (D) In vitro-translated Bcl-6 BTB species were allowed to interact with myc-tagged B-CoR and immunoprecipitated with myc epitope antibodies. The top gel shows lysates after coimmunoprecipitation, and the bottom gel shows the lysates prior to precipitation. (E) Reporter assays were performed as in panel A. The ability of B-CoR to enhance repression by the wild type or mutant Bcl-6BTB was determined by transfecting 100 ng of the B-CoR expression plasmid to the indicated cell cultures.
A discrete domain of N-CoR and SMRT interacts directly with the BTB domains of PLZF and Bcl-6.
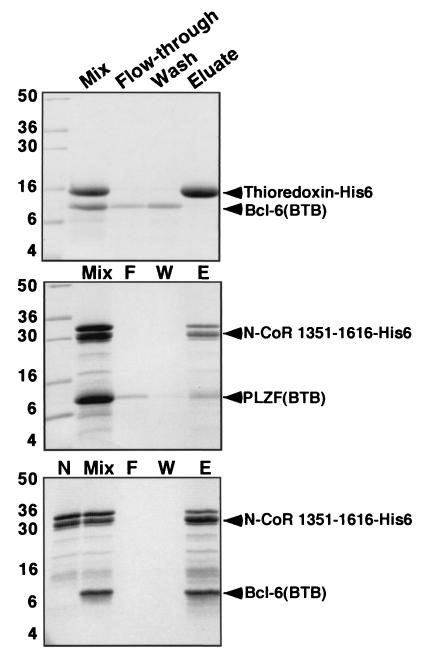
Although it is clear from our experiments and those of others that the BTB domain is required for interaction with corepressors, it was not certain whether this interaction was direct. To answer this question, as well as to map the interacting region of the corepressors, we purified six-His-tagged N-CoR and SMRT fragments expressed in E. coli. These proteins were allowed to interact with bacterially expressed and purified PLZF and Bcl-6 BTB domains. The complex was then affinity purified over Ni-NTA columns, eluted by imidazole washing, and resolved by SDS-PAGE (Fig. 6). Bcl-6 was captured to a greater extent than PLZF, suggesting a higher affinity for corepressors. The fragment of N-CoR interacting with the BTB domain was mapped to residues 1351 to 1616 of N-CoR. Representative experiments are shown in Fig. 6. Our preliminary results demonstrate that a fragment of SMRT between positions 1413 to 1497 also binds directly to these BTB domains with similar affinity. Thus, we show for the first time that both PLZF and Bcl-6 BTB domains interact directly with N-CoR and SMRT and have mapped the direct interaction to a restricted region of the corepressor proteins.
FIG. 6.
A discrete domain of N-CoR and SMRT interacts directly with the BTB domains of PLZF and Bcl-6. In vitro interaction of Bcl-6BTB and PLZFBTB with a fragment of N-CoR. Purified BTB domain was mixed in solution with the indicated histidine-tagged protein, followed by Ni-NTA affinity purification. The samples were resolved on SDS-PAGE gel (14% polyacrylamide) followed by Coomassie staining. Molecular weight markers are indicated. Lanes: Mix, mixture before loading on the Ni-NTA column; F, column flowthrough (unbound protein); W, column wash; E, imidazole eluate (releases His-tagged proteins); N (bottom gel), purified residues 1351 to 1616 before mixing with BTB domain. The N-CoR fragment is a doublet; the smaller band corresponds to a partial proteolysis product that retains the His tag. Bcl-6BTB interacts strongly with the N-CoR fragment, while the PLZFBTB domains interact, but to a lesser extent. Neither interacts with His-tagged thioredoxin.
Transcriptional repression and corepressor binding correlate with the identity of critical pocket residues in PLZF, Bcl-6, and FAZF.
Given the difference in affinity between PLZF and Bcl-6 for corepressor binding, we decided to reexamine the exact BTB domain sequences of these proteins. One obvious difference was the presence of lysine in the pocket of Bcl-6 rather than the arginine found in the pocket of PLZF (Fig. 7A). We further noticed that another BTB protein under active study in our laboratory, the PLZF-like FAZF protein, contains a polar serine instead of a positively charged residue at the equivalent position (Fig. 7A). In previous studies, we showed that FAZF binds to the same DNA sequences as PLZF, but is not as effective a repressor (20). In light of these findings, we compared the abilities of the BTB domains of FAZF, Bcl-6, and PLZF to repress transcription when fused to the GAL4 DBD. An equal amount of each expression vector was transfected into 293T cells, and similar levels of expression were verified by immunoblotting. In these assays, the Bcl-6 BTB domain repressed transcription over 200-fold, while PLZF was much weaker, inhibiting transcription only 10-fold (Fig. 7B). The FAZF BTB domain did not detectably repress transcription. When the ability of these BTB domains to interact in vivo with N-CoR or SMRT was tested by mammalian two-hybrid analysis, only Bcl-6 interacted strongly enough to activate the reporter (Fig. 7C). The differential affinity for corepressors could not be accounted for by differential expression levels of GAL4-BTB fusions (Fig. 7D). Thus, these BTB proteins have differences in their charged pockets, correlating with a differential ability to both repress transcription and interact with corepressors.
FIG. 7.
Transcriptional repression and corepressor binding correlate with the identity of the critical charged pocket residues in PLZF, Bcl-6, and FAZF. (A) Sequence alignment of the pocket region of the BTB domains of PLZF, Bcl-6, and FAZF. The arrowheads indicate the critical charged residues: D35 in PLZF (this is D33 in Bcl-6 and D29 in FAZF) and R49 in PLZF (this is K47 in Bcl-6 and S44 in FAZF). The S44 residue of FAZF is circled. (B) 293T cells were transfected with 400 ng of the GAL1-147 or respective GAL-BTB fusions from PLZF, Bcl-6, or FAZF along with 100 ng of (GAL)5-tk-Luc reporter and 5 ng of tk-renilla control plasmids. (C) Mammalian two-hybrid experiments were performed with 25 ng of the indicated GAL constructs as bait and 150 ng of VP16-N-CoR or -SMRT as prey. The results are expressed as fold activation of the luciferase reporter normalized to the internal control. (D) GAL4 DBD immunoblots corresponding to the mammalian two-hybrid assay described in panel C were performed in 293T cells transfected with equivalent amounts of GAL-BTB fusions and GAL-VP16 prey plasmids as indicated. Lanes: N, VP16-N-CoR cotransfection; lane S: VP16-SMRT cotransfection.
Swapping critical Bcl-6 charged pocket residues alters transcriptional repression and corepressor binding properties of BTB domains.
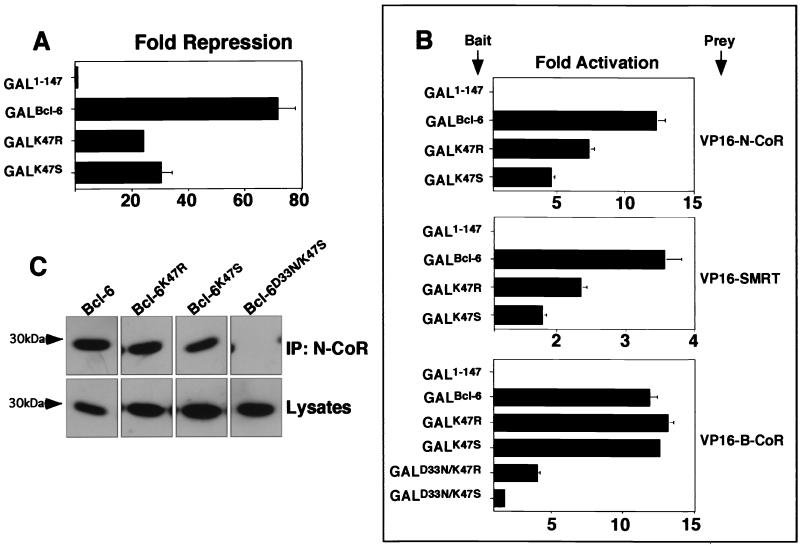
We postulated that the differences in the charged pocket among Bcl-6, PLZF, and FAZF could account, in part, for the differences in potency of repression and extent of corepressor binding. To test this notion, we introduced the arginine residue from PLZF or the serine from FAZF into position 47 of Bcl-6 usually occupied by lysine (Fig. 7A). The resulting mutants, Bcl-6K47R and Bcl-6K47S, were partially impaired for repression compared to the wild type (Fig. 8A), suggesting that this single-residue change could alter interaction with corepressors. Consistent with this notion, Bcl-6K47R and Bcl-6K47S exhibited impaired association to N-CoR and SMRT as measured by the mammalian two-hybrid assay (Fig. 8B). In contrast, in the case of the Bcl-6-specific B-CoR corepressor, we did not detect a difference in interaction by changing residue 47. However, when the Bcl-6K47R and Bcl-6K47S mutations were combined with the Bcl-6D33N mutation to make Bcl-6D33N/K47R and Bcl-6D33N/K47S, a severe loss of B-CoR binding was uncovered (Fig. 8B). This indicates that residues 33 and 47 are critical for B-CoR interaction, yet other, perhaps more subtle differences in the pocket or other portions of the Bcl-6 BTB domain may play a critical role for the selective binding of B-CoR to Bcl-6. Finally, consistent with reporter assays and mammalian two-hybrid results, N-CoR antibodies coimmunoprecipitated slightly less Bcl-6K47R than wild-type Bcl-6 and still less Bcl-6K47S than Bcl-6K47R (Fig. 8C). Bcl-6K47S/D33N, which, similar to Bcl-6D33N/K47Q, results in loss of pocket charge, did not interact with N-CoR at all.
FIG. 8.
Swapping critical Bcl-6 charged pocket residues alters transcriptional repression and corepressor binding properties. Mutant Bcl-6 BTB domains were generated by swapping the K47 residue for arginine (as in PLZF) or serine (as in FAZF). (A) Reporter assays in 293T cells transfected with 25 ng of GAL1-147 or GAL-Bcl-6BTB mutants as indicated along with 100 ng of reporter and 5 ng of the internal control. Results are tabulated as fold repression compared to that of GAL1-147 normalized to the internal control. (B) Mammalian two-hybrid assays in 293T cells transfected with 25 ng of GAL-Bcl-6 bait and 150 ng of VP-16-N-CoR or -SMRT prey or 5 ng of GAL-Bcl-6 bait with 25 ng of VP16-B-CoR prey in HeLa cells. Results are reported as fold activation of the (GAL)5-tk-Luc reporter construct. B-CoR interaction was also tested in 293 T cells with similar results (not shown). (C) Coimmunoprecipitations (IP) were performed with in vitro-translated BTB domains and N-CoR as described previously. The top section shows fluorography of BTB domains precipitated with N-CoR antibodies, and the bottom section shows the appropriate lysates prior to immunoprecipitation.
Swapping critical charged pocket residues in FAZF and PLZF alters transcriptional repression and corepressor interactions.
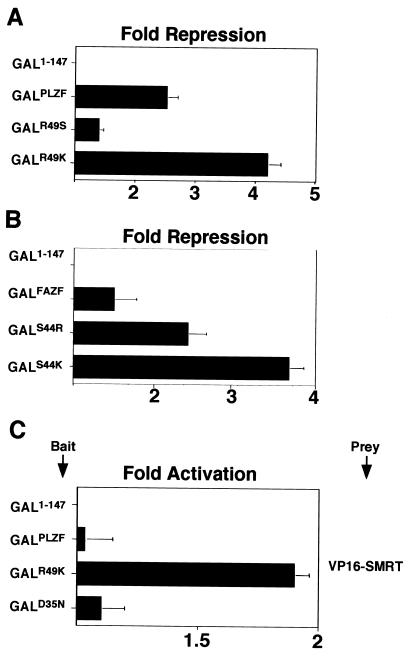
In experiments reciprocal to those described above, mutants of PLZF were constructed by inserting the lysine characteristic of Bcl-6 or serine of FAZF at position 49 of the BTB domain of PLZF. Likewise, a FAZF BTB domain was constructed containing lysine (Bcl-6) or arginine (PLZF) at position 44. In reporter assays with GAL-BTB fusions, PLZFR49K was a more powerful repressor than the wild type, simulating Bcl-6; although mirroring the situation for FAZF, PLZFR49S was a weaker repressor than wild-type PLZF (Fig. 9A). Conversely, changing FAZF toward PLZF (FAZFS44R) yielded a stronger repressor than wild-type FAZF, and mutation of FAZF toward Bcl-6 (FAZFS44K) was more powerful still (Fig. 9B). As noted above, under our experimental conditions, PLZF and FAZF were not able to generate a mammalian two-hybrid signal with N-CoR and SMRT. However, PLZFR49K, which repressed more powerfully than wild-type PLZF, was able to yield a signal in the two-hybrid assay with SMRT. Thus, changing a single residue in the pocket enhanced the affinity of PLZF in vivo for SMRT (Fig. 9C). In conclusion, differences in critical pocket residues among different BTB proteins can predict the transcriptional potency of the BTB domain and the strength of its interaction with corepressors.
FIG. 9.
Swapping critical charged pocket residues in FAZF and PLZF alters transcriptional repression and corepressor interactions. Mutant PLZF BTB domains were generated by swapping the R49 residue for lysine (as in Bcl-6) or serine (as in FAZF). FAZF mutants were also made by swapping the S44 for lysine (as in Bcl-6) or arginine (as in PLZF). (A and B) Reporter assays performed with 2 × 105 293T cells cotransfected with 400 ng of GAL1-147 or GAL-BTB fusions as indicated with 100 ng of reporter and 5 ng of internal control. (C) Mammalian two-hybrid assay performed in 293T cells with 25 ng of GAL1-147 or GAL-PLZFBTB bait and 150 ng of VP16-SMRT prey as indicated.
DISCUSSION
BTB domain-containing proteins comprise a large and diverse family of factors involved in multiple cellular processes. The common property of BTB domains is their ability to mediate protein-protein interactions, including homodimerization (1). These interactions often result in higher-order oligomer formation shown to be of functional relevance for the Bach1, GAGA, and PLZF proteins among others (2, 25, 36, 53). The highly conserved nature of the BTB domain sequence predicts that most if not all such domains fold into the obligate dimer structure seen in the 1.9-Å resolution crystal structure of the PLZF BTB dimer (1). The importance of this architectural function is highlighted by experiments where the Bcl-6 BTB domain was fused to the Drosophila ttk69 protein in place of its own BTB domain. This Bcl-6-ttk69 hybrid could block retinal photoreceptor cell differentiation in a manner similar to wild-type ttk69 (49). Similarly, the mod(mdg4) BTB domain was able to substitute for the GAGA BTB domain in reporter assays of transcriptional activation by GAGA (42). The interchangeability of these domains suggests that the common dimerization and oligomerization function of the BTB motif is necessary for many if not all activities of this class of proteins.
The BTB domain of PLZF is of critical interest in the pathophysiology of the t(11;17)(q23;q21) retinoid refractory variant of acute promyelocytic leukemia. In this disease, the PLZF BTB domain is required for transcriptional repression by the PLZF-RARα fusion product, leading to dominant-negative silencing of genes involved in myeloid differentiation (13, 16, 18, 34). The BTB domain is also required for transcriptional repression by wild-type PLZF, resulting in target gene silencing characterized by growth suppression, cell cycle arrest, and differentiation arrest in myeloid cells (2, 36, 46, 52). Numerous other BTB domains, such as those of Bcl-6, BAZF, KAISO, Hic-1, and Bach1, are required for transcriptional repression as well (5, 8, 10, 39, 41, 44, 53). Several BTB domains, including those of PLZF and Bcl-6, autonomously repress transcription when fused to the GAL4 DBD (5, 30, 44). In contrast, BTB domains such as that of the MAZR BTB/ZF protein are not repressors (26). In fact, both the MAZR and Miz BTB domains are required for transcriptional activation by their respective proteins (26, 40). Therefore, while the basic BTB domain structure mediates an architectural function, subtle variations among different BTB domains might play a role in determining distinctive transcriptional activities.
In our previous studies, we found that the alignment of charged residues within the BTB pocket was required for autonomous transcriptional repression by the PLZF BTB dimer. When the two conserved charged residues of the pocket, D35 and R49, were switched to polar amino acids, the BTB domain no longer repressed and in fact activated a reporter construct (36). The importance of the charged pocket effect for overall function of PLZF was underscored by the fact that full-length PLZF harboring the double mutant BTB domain was severely impaired for transcriptional repression and growth suppression, yet could still oligomerize and localize to characteristic nuclear speckles. In contrast, PLZFΔBTB was completely nonfunctional. This allowed a distinction to be made for PLZF between general BTB architectural effects and BTB-dependent transcriptional repression (36).
The PLZF BTB domain associates with the N-CoR and SMRT corepressors as well as HDAC1 (18, 50). In the current study, we found that the PLZF BTB domain is required for stable in vivo interaction with N-CoR or SMRT, as well as for the ability of SMRT to enhance repression by PLZF. Given our previous results, we hypothesized that the charged pocket is required for recruitment of these corepressors. The only previously characterized recruitment interaction between a DNA binding transcription factor and the N-CoR-SMRT complexes at the structural level was that of nuclear hormone receptors such as RARα. In this case the N-CoR or SMRT LXXI/HIXXXI/L helix (or CoRNR box) binds to a hydrophobic pocket in the AF2 domain of the nuclear receptor (15). In the current report, we find that the PLZF BTB domain D35N/R49Q double pocket mutant, which fails to repress transcription, did not physically or functionally interact with N-CoR and SMRT. Full-length PLZFD35N/R49Q was previously shown to bind to DNA as a low-mobility gel shift complex and form nuclear speckles, both indicative of intact dimerization or oligomerization by the BTB domain. Yet herein, we show that PLZFD35N/R49Q does not repress and does not bind to corepressors. Therefore, these results suggest that the charged pocket of the PLZF BTB is specifically required for N-CoR-SMRT recruitment and transcriptional repression and could represent a docking site for corepressors with PLZF. Interestingly, HDAC1 also required the intact pocket in order to complex with PLZF. A number of studies identified N-CoR and SMRT in the context of different protein complexes, some of which contain class I HDACs and some of which contain class II HDACs (43). Such interactions may depend on cell type and differentiation stage, as well as on the specific transcription factor that recruits N-CoR and SMRT (43). Thus, HDAC1 is possibly recruited to PLZF indirectly through N-Cor or SMRT. The nature of the entire repression complex that PLZF assembles on target genes is yet unknown. Our preliminary data show that PLZF fractionates from cells both as an ∼600-kDa complex as well as a 2-MDa complex 2. Identification of the subunits of these complexes and their interactions will clarify the relationship among N-CoR, SMRT, HDACs, and PLZF.
Due to its importance in lymphoid differentiation and lymphomagenesis, we also analyzed Bcl-6, another member of the BTB-zinc finger repressor family that binds to N-CoR and SMRT. Bcl-6 also interacts with B-CoR, a specific corepressor not homologous to SMRT and N-CoR, that does not interact with PLZF (23). As in the case of PLZF, the BTB domain is the main corepressor interaction site for Bcl-6 (23, 24). The interaction of the Bcl-6 BTB domain with each of these corepressors is mutually exclusive, suggesting that the interaction of B-CoR, SMRT, and N-CoR occurs through the same unique binding site (24). The equivalent conserved pocket residues of Bcl-6 correspond to positions D33 and K47. When these residues were mutated in the Bcl-6 BTB domain, the mutant domain could neither repress transcription nor interact with N-CoR, SMRT, or B-CoR, consistent with the idea that the pocket is the binding site or that the integrity of the pocket is required for proper presentation of the corepressor binding site. In addition, these experiments indicated that our model for the PLZF BTB domain could be extended to other proteins.
While our data suggest that an interaction site of corepressors to PLZF and Bcl-6 is the BTB charged pocket, it was unknown whether this interaction was direct. Previous studies that examined BTB-corepressor interactions employed GST affinity chromatography with in vitro-translated proteins. However, these assays are hampered by the fact the reticulocyte lysates do not represent a pure source of protein and contaminants such as Sin3A, and others could act as bridging factors (18, 23, 50). Using proteins expressed and purified from E. coli, we showed for the first time that PLZF and Bcl-6 directly bound to N-CoR or SMRT and localized this interaction to discrete regions of these corepressors. In addition, the Bcl-6 BTB domain has a higher affinity for corepressors than PLZF, consistent with previous data generated by GST pulldown assays (23). Given our data suggesting that the charged pocket may be an interaction site for corepressors, one might predict that subtle variations between PLZF and Bcl-6 could account for these differences. Furthermore, if BTB-dependent repression is contingent on corepressor recruitment, a higher-affinity interaction would most likely translate into more potent repression by Bcl-6 than PLZF.
We showed that both of these predictions are accurate. First, we compared and contrasted the protein sequences of PLZF, Bcl-6, and FAZF. These proteins did differ in a conserved positive residue in the charged pocket, arginine in PLZF, lysine in Bcl-6, and unique among BTB proteins, serine in FAZF. Second, in reporter assays, the BTB domain of Bcl-6 was a far more powerful repressor than that of PLZF, while the FAZF BTB was transcriptionally inert. These differences were reflected in the relative affinities of these BTB domains for corepressors. Furthermore, the differences among these proteins in repression activity and affinity for corepressors could be partially transferred by swapping this residue among the three different domains. The fact that these residues, located deep within the pocket of the BTB domain, are so important for corepressor binding, again supports a central role for this structure in formation of the BTB-dependent repression complex.
Most BTB domains possess the conserved charged pocket; therefore, it is not surprising that many other BTB transcription factors bind to corepressors, including Bach-2, GAGA, and ZID (23, 38). Since our residue-swapping experiments did not fully transfer repression functions between Bcl-6, PLZF, and FAZF BTB domains, other structural elements of the BTB domain must be important for its function as well. Furthermore, B-CoR, while requiring the charged pocket of Bcl-6 to form a complex with the BTB fails to bind other BTB domains. Bcl-6 has a small sequence proximal to the pocket between residues 63 and 68, which could play a role in this specific interaction. The HIC-1 protein has a charged pocket, but fails to bind to N-CoR or SMRT (8). This might be explained by a unique 13-residue insertion in its BTB domain potentially altering the three-dimensional structure of the pocket and/or other sites of the BTB domain usually required for corepressor interaction (17). Ongoing comparative studies of BTB proteins should indicate what other features of the BTB domain might be required for its potency as a repression domain and ability to selectively bind cofactors.
Finally, while the BTB domain is required for PLZF and Bcl-6 transcriptional repression, both of these proteins have second repression domains. In PLZF, this domain, located between residues 200 and 300, binds to the ETO protein (37) and can function autonomously when tethered to GAL4. However, PLZF deleted for the BTB domain or mutated in the charged pocket could not repress transcription. This indicates that there must be close cooperation between the two domains, with the second repression domain offering additional contact sites for corepressors, and the BTB domain contributing both binding sites for N-CoR or SMRT and dimerization or oligomerization activity.
In conclusion, in the course of our efforts to understand the molecular pathogenesis of specific hematologic malignancies, we identified critical residues in the BTB domains of PLZF and Bcl-6 required for corepressor binding and transcriptional repression. We also show that while BTB domains have a common function in mediating dimerization or oligomerization, variations between BTB domains exist that modulate transcriptional potency and corepressor binding. The groove of the PLZF and Bcl-6 charged pocket offers a potential site for interaction with specific residues of corepressors. Confirmation of this model may allow the design of molecules to specifically prevent the interaction of the repression machinery with the BTB transcription factors. These experiments will allow dissection of BTB-dependent and BTB-independent mechanisms in the biological actions of PLZF and other factors. Finally, inhibitors of the pocket could directly or indirectly prevent interaction with corepressors and offer a novel transcriptional therapy approach to the treatment of Bcl-6-associated non-Hodgkin's lymphoma, t(11;17) acute promyelocytic leukemia, and other disorders.
Acknowledgments
J.D.L. is supported by NIH R01 CA 59936 and American Cancer Society Award DHP 160. V.B. is supported by NIH R29 CA 71540. G.G.P. is supported by National Cancer Institute of Canada grant 012103. K.F.A. is supported by a Canadian Institutes of Health Research Doctoral Research Award. A.M. is supported by NIH K08 CA73762.
We thank Samuel Waxman and Kathy Borden for continued support.
REFERENCES
- 1.Ahmad, K. F., C. K. Engel, and G. G. Prive. 1998. Crystal structure of the BTB domain from PLZF. Proc. Natl. Acad. Sci. USA 95:12123-12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball, H. J., A. Melnick, R. Shaknovich, R. A. Kohanski, and J. D. Licht. 1999. The promyelocytic leukemia zinc finger (PLZF) protein binds DNA in a high molecular weight complex associated with cdc2 kinase. Nucleic Acids Res. 27:4106-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barna, M., N. Hawe, L. Niswander, and P. P. Pandolfi. 2000. Plzf regulates limb and axial skeletal patterning. Nat. Genet. 25:166-172. [DOI] [PubMed] [Google Scholar]
- 4.Cattoretti, G., C. C. Chang, K. Cechova, J. Zhang, B. H. Ye, B. Falini, D. C. Louie, K. Offit, R. S. Chaganti, and R. Dalla-Favera. 1995. BCL-6 protein is expressed in germinal-center B cells. Blood 86:45-53. [PubMed] [Google Scholar]
- 5.Chang, C. C., B. H. Ye, R. S. Chaganti, and R. Dalla-Favera. 1996. BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc. Natl. Acad. Sci. USA 93:6947-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, Z., N. J. Brand, A. Chen, S. J. Chen, J. H. Tong, Z. Y. Wang, S. Waxman, and A. Zelent. 1993. Fusion between a novel Kruppel-like zinc finger gene and the retinoic acid receptor-alpha locus due to a variant t(11;17) translocation associated with acute promyelocytic leukaemia. EMBO J. 12:1161-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.David, G., L. Alland, S. H. Hong, C. W. Wong, R. A. DePinho, and A. Dejean. 1998. Histone deacetylase associated with mSin3A mediates repression by the acute promyelocytic leukemia-associated PLZF protein. Oncogene 16:2549-2556. [DOI] [PubMed] [Google Scholar]
- 8.Deltour, S., C. Guerardel, and D. Leprince. 1999. Recruitment of SMRT/N-CoR-mSin3A-HDAC-repressing complexes is not a general mechanism for BTB/POZ transcriptional repressors: the case of HIC-1 and gammaFBP-B. Proc. Natl. Acad. Sci. USA 96:14831-14836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dent, A. L., A. L. Shaffer, X. Yu, D. Allman, and L. M. Staudt. 1997. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science 276:589-592. [DOI] [PubMed] [Google Scholar]
- 10.Dhordain, P., O. Albagli, S. Ansieau, M. H. Koken, C. Deweindt, S. Quief, D. Lantoine, A. Leutz, J. P. Kerckaert, and D. Leprince. 1995. The BTB/POZ domain targets the LAZ3/BCL6 oncoprotein to nuclear dots and mediates homomerisation in vivo. Oncogene 11:2689-2697. [PubMed] [Google Scholar]
- 11.Dhordain, P., O. Albagli, R. J. Lin, S. Ansieau, S. Quief, A. Leutz, J. P. Kerckaert, R. M. Evans, and D. Leprince. 1997. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc. Natl. Acad. Sci. USA 94:10762-10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhordain, P., R. J. Lin, S. Quief, D. Lantoine, J. P. Kerckaert, R. M. Evans, and O. Albagli. 1998. The LAZ3(BCL-6) oncoprotein recruits a SMRT/mSIN3A/histone deacetylase containing complex to mediate transcriptional repression. Nucleic Acids Res. 26:4645-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong, S., J. Zhu, A. Reid, P. Strutt, F. Guidez, H. J. Zhong, Z. Y. Wang, J. Licht, S. Waxman, C. Chomienne et al. 1996. Amino-terminal protein-protein interaction motif (POZ-domain) is responsible for activities of the promyelocytic leukemia zinc finger-retinoic acid receptor-alpha fusion protein. Proc. Natl. Acad. Sci. USA 93:3624-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaidano, G., F. Lo Coco, B. H. Ye, D. Shibata, A. M. Levine, D. M. Knowles, and R. Dalla-Favera. 1994. Rearrangements of the BCL-6 gene in acquired immunodeficiency syndrome-associated non-Hodgkin's lymphoma: association with diffuse large-cell subtype. Blood 84:397-402. [PubMed] [Google Scholar]
- 15.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 16.Grignani, F., S. De Matteis, C. Nervi, L. Tomassoni, V. Gelmetti, M. Cioce, M. Fanelli, M. Ruthardt, F. F. Ferrara, I. Zamir, C. Seiser, M. A. Lazar, S. Minucci, and P. G. Pelicci. 1998. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature 391:815-818. [DOI] [PubMed] [Google Scholar]
- 17.Guerardel, C., S. Deltour, and D. Leprince. 1999. Evolutionary divergence in the broad complex, tramtrack and bric a brac/poxviruses and zinc finger domain from the candidate tumor suppressor gene hypermethylated in cancer. FEBS Lett. 451:253-256. [DOI] [PubMed] [Google Scholar]
- 18.Guidez, F., S. Ivins, J. Zhu, M. Soderstrom, S. Waxman, and A. Zelent. 1998. Reduced retinoic acid-sensitivities of nuclear receptor corepressor binding to PML- and PLZF-RARalpha underlie molecular pathogenesis and treatment of acute promyelocytic leukemia. Blood 91:2634-2642. [PubMed] [Google Scholar]
- 19.He, L. Z., F. Guidez, C. Tribioli, D. Peruzzi, M. Ruthardt, A. Zelent, and P. P. Pandolfi. 1998. Distinct interactions of PML-RARalpha and PLZF-RARalpha with co-repressors determine differential responses to RA in APL. Nat. Genet. 18:126-135. [DOI] [PubMed] [Google Scholar]
- 20.Hoatlin, M. E., Y. Zhi, H. Ball, K. Silvey, A. Melnick, S. Stone, S. Arai, N. Hawe, G. Owen, A. Zelent, and J. D. Licht. 1999. A novel BTB/POZ transcriptional repressor protein interacts with the Fanconi anemia group C protein and PLZF. Blood 94:3737-3747. [PubMed] [Google Scholar]
- 21.Hong, S. H., G. David, C. W. Wong, A. Dejean, and M. L. Privalsky. 1997. SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor alpha (RARalpha) and PLZF-RARalpha oncoproteins associated with acute promyelocytic leukemia. Proc. Natl. Acad. Sci. USA 94:9028-9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horowitz, H., and C. A. Berg. 1996. The Drosophila pipsqueak gene encodes a nuclear BTB-domain-containing protein required early in oogenesis. Development 122:1859-1871. [DOI] [PubMed] [Google Scholar]
- 23.Huynh, K. D., and V. J. Bardwell. 1998. The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT. Oncogene 17:2473-2484. [DOI] [PubMed] [Google Scholar]
- 24.Huynh, K. D., W. Fischle, E. Verdin, and V. J. Bardwell. 2000. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 14:1810-1823. [PMC free article] [PubMed] [Google Scholar]
- 25.Katsani, K. R., M. A. Hajibagheri, and C. P. Verrijzer. 1999. Co-operative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. EMBO J. 18:698-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi, A., H. Yamagiwa, H. Hoshino, A. Muto, K. Sato, M. Morita, N. Hayashi, M. Yamamoto, and K. Igarashi. 2000. A combinatorial code for gene expression generated by transcription factor Bach2 and MAZR (MAZ-related factor) through the BTB/POZ domain. Mol. Cell. Biol. 20:1733-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koken, M. H., A. Reid, F. Quignon, M. K. Chelbi-Alix, J. M. Davies, J. H. Kabarowski, J. Zhu, S. Dong, S. Chen, Z. Chen, C. C. Tan, J. Licht, S. Waxman, H. de The, and A. Zelent. 1997. Leukemia-associated retinoic acid receptor alpha fusion partners, PML and PLZF, heterodimerize and colocalize to nuclear bodies. Proc. Natl. Acad. Sci. USA 94:10255-10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai, Z. C., and Y. Li. 1999. Tramtrack69 is positively and autonomously required for Drosophila photoreceptor development. Genetics 152:299-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, R. Funke, D. Gage, K. Harris, A. Heaford, J. Howland, L. Kann, J. Lehoczky, R. LeVine, P. McEwan, K. McKernan, J. Meldrim, J. P. Mesirov, C. Miranda, W. Morris, J. Naylor, C. Raymond, M. Rosetti, R. Santos, A. Sheridan, C. Sougnez, N. Stange-Thomann, N. Stojanovic, A. Subramanian, D. Wyman, J. Rogers, J. Sulston, R. Ainscough, S. Beck, D. Bentley, J. Burton, C. Clee, N. Carter, A. Coulson, R. Deadman, P. Deloukas, A. Dunham, I. Dunham, R. Durbin, L. French, D. Grafham, S. Gregory, T. Hubbard, S. Humphray, A. Hunt, M. Jones, C. Lloyd, A. McMurray, L. Matthews, S. Mercer, S. Milne, J. C. Mullikin, A. Mungall, R. Plumb, M. Ross, R. Shownkeen, S. Sims, R. H. Waterston, R. K. Wilson, L. W. Hillier, J. D. McPherson, M. A. Marra, E. R. Mardis, L. A. Fulton, A. T. Chinwalla, K. H. Pepin, W. R. Gish, S. L. Chissoe, M. C. Wendl, K. D. Delehaunty, T. L. Miner, A. Delehaunty, J. B. Kramer, L. L. Cook, R. S. Fulton, D. L. Johnson, P. J. Minx, S. W. Clifton, T. Hawkins, E. Branscomb, P. Predki, P. Richardson, S. Wenning, T. Slezak, N. Doggett, J. F. Cheng, A. Olsen, S. Lucas, C. Elkin, E. Uberbacher, M. Frazier et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 30.Li, J. Y., M. A. English, H. J. Ball, P. L. Yeyati, S. Waxman, and J. D. Licht. 1997. Sequence-specific DNA binding and transcriptional regulation by the promyelocytic leukemia zinc finger protein. J. Biol. Chem. 272:22447-22455. [DOI] [PubMed] [Google Scholar]
- 31.Li, X., H. Peng, D. C. Schultz, J. M. Lopez-Guisa, F. J. Rauscher III, and R. Marmorstein. 1999. Structure-function studies of the BTB/POZ transcriptional repression domain from the promyelocytic leukemia zinc finger oncoprotein. Cancer Res. 59:5275-5282. [PubMed] [Google Scholar]
- 32.Licht, J. D., C. Chomienne, A. Goy, A. Chen, A. A. Scott, D. R. Head, J. L. Michaux, Y. Wu, A. DeBlasio, W. H. Miller, Jr., et al. 1995. Clinical and molecular characterization of a rare syndrome of acute promyelocytic leukemia associated with translocation (11;17). Blood 85:1083-1094. [PubMed] [Google Scholar]
- 33.Licht, J. D., R. Shaknovich, M. A. English, A. Melnick, J. Y. Li, J. C. Reddy, S. Dong, S. J. Chen, A. Zelent, and S. Waxman. 1996. Reduced and altered DNA-binding and transcriptional properties of the PLZF-retinoic acid receptor-alpha chimera generated in t(11;17)-associated acute promyelocytic leukemia. Oncogene 12:323-336. [PubMed] [Google Scholar]
- 34.Lin, R. J., L. Nagy, S. Inoue, W. Shao, W. H. Miller, Jr., and R. M. Evans. 1998. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature 391:811-814. [DOI] [PubMed] [Google Scholar]
- 35.Lo Coco, F., B. H. Ye, F. Lista, P. Corradini, K. Offit, D. M. Knowles, R. S. Chaganti, and R. Dalla-Favera. 1994. Rearrangements of the BCL6 gene in diffuse large cell non-Hodgkin's lymphoma. Blood 83:1757-1759. [PubMed] [Google Scholar]
- 36.Melnick, A., K. F. Ahmad, S. Arai, A. Polinger, H. Ball, K. L. Borden, G. W. Carlile, G. G. Prive, and J. D. Licht. 2000. In-depth mutational analysis of the promyelocytic leukemia zinc finger BTB/POZ domain reveals motifs and residues required for biological and transcriptional functions. Mol. Cell. Biol. 20:6550-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melnick, A. M., J. J. Westendorf, A. Polinger, G. W. Carlile, S. Arai, H. J. Ball, B. Lutterbach, S. W. Hiebert, and J. D. Licht. 2000. The ETO protein disrupted in t(8;21)-associated acute myeloid leukemia is a corepressor for the promyelocytic leukemia zinc finger protein. Mol. Cell. Biol. 20:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muto, A., H. Hoshino, L. Madisen, N. Yanai, M. Obinata, H. Karasuyama, N. Hayashi, H. Nakauchi, M. Yamamoto, M. Groudine, and K. Igarashi. 1998. Identification of Bach2 as a B-cell-specific partner for small maf proteins that negatively regulate the immunoglobulin heavy chain gene 3′ enhancer. EMBO J. 17:5734-5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okabe, S., T. Fukuda, K. Ishibashi, S. Kojima, S. Okada, M. Hatano, M. Ebara, H. Saisho, and T. Tokuhisa. 1998. BAZF, a novel Bcl6 homolog, functions as a transcriptional repressor. Mol. Cell. Biol. 18:4235-4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peukert, K., P. Staller, A. Schneider, G. Carmichael, F. Hanel, and M. Eilers. 1997. An alternative pathway for gene regulation by Myc. EMBO J. 16:5672-5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prokhortchouk, A., B. Hendrich, H. Jorgensen, A. Ruzov, M. Wilm, G. Georgiev, A. Bird, and E. Prokhortchouk. 2001. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 15:1613-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Read, D., M. J. Butte, A. F. Dernburg, M. Frasch, and T. B. Kornberg. 2000. Functional studies of the BTB domain in the Drosophila GAGA and Mod(mdg4) proteins. Nucleic Acids Res. 28:3864-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rietveld, L. E., E. Caldenhoven, and H. G. Stunnenberg. 2001. Avian erythroleukemia: a model for corepressor function in cancer. Oncogene 20:3100-3109. [DOI] [PubMed] [Google Scholar]
- 44.Seyfert, V. L., D. Allman, Y. He, and L. M. Staudt. 1996. Transcriptional repression by the proto-oncogene BCL-6. Oncogene 12:2331-2342. [PubMed] [Google Scholar]
- 45.Shaffer, A. L., X. Yu, Y. He, J. Boldrick, E. P. Chan, and L. M. Staudt. 2000. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity 13:199-212. [DOI] [PubMed] [Google Scholar]
- 46.Shaknovich, R., P. L. Yeyati, S. Ivins, A. Melnick, C. Lempert, S. Waxman, A. Zelent, and J. D. Licht. 1998. The promyelocytic leukemia zinc finger protein affects myeloid cell growth, differentiation, and apoptosis. Mol. Cell. Biol. 18:5533-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Staudt, L. M., A. L. Dent, A. L. Shaffer, and X. Yu. 1999. Regulation of lymphocyte cell fate decisions and lymphomagenesis by BCL-6. Int. Rev. Immunol. 18:381-403. [DOI] [PubMed] [Google Scholar]
- 48.Usui-Aoki, K., H. Ito, K. Ui-Tei, K. Takahashi, T. Lukacsovich, W. Awano, H. Nakata, Z. F. Piao, E. E. Nilsson, J. Tomida, and D. Yamamoto. 2000. Formation of the male-specific muscle in female Drosophila by ectopic fruitless expression. Nat. Cell Biol. 2:500-506. [DOI] [PubMed] [Google Scholar]
- 49.Wen, Y., D. Nguyen, Y. Li, and Z. C. Lai. 2000. The N-terminal BTB/POZ domain and C-terminal sequences are essential for Tramtrack69 to specify cell fate in the developing Drosophila eye. Genetics 156:195-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong, C. W., and M. L. Privalsky. 1998. Components of the SMRT corepressor complex exhibit distinctive interactions with the POZ domain oncoproteins PLZF, PLZF-RARalpha, and BCL-6. J. Biol. Chem. 273:27695-27702. [DOI] [PubMed] [Google Scholar]
- 51.Ye, B. H., F. Lista, F. Lo Coco, D. M. Knowles, K. Offit, R. S. Chaganti, and R. Dalla-Favera. 1993. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science 262:747-750. [DOI] [PubMed] [Google Scholar]
- 52.Yeyati, P. L., R. Shaknovich, S. Boterashvili, J. Li, H. J. Ball, S. Waxman, K. Nason-Burchenal, E. Dmitrovsky, A. Zelent, and J. D. Licht. 1999.Leukemia translocation protein PLZF inhibits cell growth and expressionof cyclin A. Oncogene 18:925-934. [DOI] [PubMed] [Google Scholar]
- 53.Yoshida, C., F. Tokumasu, K. I. Hohmura, J. Bungert, N. Hayashi, T. Nagasawa, J. D. Engel, M. Yamamoto, K. Takeyasu, and K. Igarashi. 1999. Long range interaction of cis-DNA elements mediated by architectural transcription factor Bach1. Genes Cells 4:643-655. [DOI] [PubMed] [Google Scholar]
- 54.Zollman, S., D. Godt, G. G. Prive, J. L. Couderc, and F. A. Laski. 1994. The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc. Natl. Acad. Sci. USA 91:10717-10721. [DOI] [PMC free article] [PubMed] [Google Scholar]