Abstract
SINEs are retrotransposons that have enjoyed remarkable reproductive success during the course of mammalian evolution, and have played a major role in shaping mammalian genomes. Previously, an analysis of survey-sequence data from an individual dog (a poodle) indicated that canine genomes harbor a high frequency of alleles that differ only by the absence or presence of a SINEC_Cf repeat. Comparison of this survey-sequence data with a draft genome sequence of a distinct dog (a boxer) has confirmed this prediction, and revealed the chromosomal coordinates for >10,000 loci that are bimorphic for SINEC_Cf insertions. Analysis of SINE insertion sites from the genomes of nine additional dogs indicates that 3%-5% are absent from either the poodle or boxer genome sequences—suggesting that an additional 10,000 bimorphic loci could be readily identified in the general dog population. We describe a methodology that can be used to identify these loci, and could be adapted to exploit these bimorphic loci for genotyping purposes. Approximately half of all annotated canine genes contain SINEC_Cf repeats, and these elements are occasionally transcribed. When transcribed in the antisense orientation, they provide splice acceptor sites that can result in incorporation of novel exons. The high frequency of bimorphic SINE insertions in the dog population is predicted to provide numerous examples of allele-specific transcription patterns that will be valuable for the study of differential gene expression among multiple dog breeds.
Short interspersed elements (SINEs) are retrotransposons that have accumulated to very high copy numbers in many mammalian genomes. For example, at least 300 Mb (10%) of the human genome is composed of a single family of SINEs, known as Alus (Schmid 1996; Lander et al. 2001; Venter et al. 2001). SINEs accumulate by a “copy and paste” mechanism. Following transcription by RNA polymerase III, the transcripts can be reverse-transcribed and integrated into the genome at distinct locations (Eickbush 1992; Ohshima et al. 1996). There are no known mechanisms for specific removal of inserted SINEs.
SINEs must consume resources of their host for replication, expression, and amplification. In addition, novel transposition events can cause severe disruption of their host's cellular activities (see below). However, it is unclear whether SINEs are primarily intracellular parasites of defenseless host genomes, or if they are symbionts that are tolerated because of their occasional positive influences on genome evolution (Brosius and Gould 1992; Makalowski 2000). They have certainly been implicated in the dynamics of genome evolution, whereby new functional elements appear, and old ones become extinct. First, unequal homologous recombination between Alu elements has clearly contributed to human genomic diversity (Deininger and Batzer 1999). This process appears to underlie the diversification of specific genes (e.g., tropoelastin) (Szabo et al. 1999), or of large genomic regions that encompass multiple genes (e.g., segmental duplications) (Bailey et al. 2003). Second, during retrotransposition of a donor element, transcription past its normal cleavage site can lead to the transduction of 3′-sequences that flank the donor element. Previously this phenomenon has been described only for long interspersed elements (LINEs) (Goodier et al. 2000; Pickeral et al. 2000), although we also see evidence for SINE-mediated transduction of 3′-sequences in the dog genome (see Results and Discussion). Third, transcription of eukaryotic retrotransposons can interfere with expression of neighboring genes (Han et al. 2004). Transcripts of SINEs have also been reported to stimulate protein translation in a response to cellular stress (Schmid 1998; Rubin et al. 2002). Fourth, the insertion of SINEs within genes can have significant effects on mRNA splicing and protein expression. Approximately 75% of human genes contain Alus. There is now abundant evidence that retrotransposition of these elements into exons, or close to mRNA splicing signals, can have dramatic effects on the expression of cellular protein activities (Muratani et al. 1991; Wallace et al. 1991; Vidaud et al. 1993; Janicic et al. 1995; Halling et al. 1999; Mustajoki et al. 1999; Sukarova et al. 2001; Claverie-Martin et al. 2003; Ganguly et al. 2003). It is likely that they can also produce more subtle effects. Indeed, it has been estimated that at least 5% of alternatively spliced exons in the human transcriptome are derived from Alus (Sorek et al. 2002), and processes by which intronic Alus can become “exonized” have been described (Vervoort et al. 1998; Lev-Maor et al. 2003).
Although the human genome contains more than 1 million Alu elements, the vast majority were inserted prior to divergence of the human and ape lineages, and are therefore fixed in the genomes of current primate populations. However, among the ∼5000 young Alus that have integrated during the past 4-6 million years, ∼1200 elements have inserted so recently that they are bimorphic with respect to the presence or absence of insertion in different human genomes (Batzer and Deininger 2002). A genomic locus is considered bimorphic for a SINE insertion if it has two alleles in the general population that are distinguished by the absence or presence of a SINE insertion. A recent analysis of the dog genome, based on survey-sequencing, concluded that recent amplification of canine SINEs has led to a much higher frequency of bimorphic SINE insertions (Kirkness et al. 2003). However, in the absence of a draft genome sequence, the genomic context of these sequence variations was generally unclear. Here, we have extended the initial observations by identifying genomic coordinates and context for more than 10,000 bimorphic SINE insertions. We consider the potential phenotypic consequences of this genomic variability, and the potential utility of SINEs as abundant, evenly distributed polymorphisms that can help us better understand the ancestral relationships between the diverse dog populations of today.
Results and Discussion
Approaches to identify loci that are bimorphic for SINE insertions
For this study, we compared an assembly of survey-sequence data from a poodle genome (derived from 1.5× coverage) (Kirkness et al. 2003), and a draft sequence of a boxer genome (derived from 7.5× coverage) (http://www.genome.gov/12511476). These have been termed CanSS and CanFam1, respectively. The focus of the comparison was a family of SINE elements, termed SINEC_Cf (RepBase release 7.11). The SINEC_Cf repeats comprise a major subfamily of canine-specific SINEs that are likely derived from a tRNA, and contain internal control elements for transcription by RNA polymerase III (Minnick et al. 1992; Bentolila et al. 1999). A SINEC_Cf repeat (∼200 bp) can be distinguished from related SINEs by a two-base insertion (RG) at position 91. Homologous SINEs have been described in a variety of carnivore species (Vassetzky and Kramerov 2002). Previously, we reported that CanSS contains ∼233,000 fragments of SINEC_Cf repeats, with a combined length of 33.8 Mb (Kirkness et al. 2003). As expected for a more complete assembly, RepeatMasker analysis of CanFam1 (http://genome.ucsc.edu/cgi-bin/hgTables) identified fewer SINEC_Cf fragments (∼170,000), but a similar combined length (29.3 Mb). Both analyses indicate that SINEC_Cf sequences represent ∼15% of the total length of all SINEs in the dog genome.
We surveyed a large selection of SINEC_Cf sequences for their distribution between the two sequenced genomes. Using an approach described previously (Kirkness et al. 2003), segments of CanSS that contain full-length SINEC_Cf repeats with flanking sequences were masked for common repeats, and aligned with CanFam1. Loci were considered as potentially bimorphic for SINEC_Cf insertions when both flanks of a CanSS SINE were contiguous on CanFam1 (i.e., the SINE is absent from CanFam1), and the match was unique. The requirement for a unique match is a more stringent criterion than was applied previously, and, as a consequence, heterozygous SINE insertions in the boxer genome were not scored. Examples of such heterozygosity were implied when the flanks of a CanSS SINE could be aligned to two regions of CanFam1 (one on a defined chromosome, and one on the 91 Mb of sequence that has not been assigned to a specific chromosome). However, the requirement for a unique alignment ensured that SINE insertions within any recently duplicated regions of the dog genome were not mistakenly considered as bimorphic insertions. Alignment of 50,500 CanSS segments with CanFam1 revealed 3987 loci (7.9%) that are predicted to be bimorphic for SINEC_Cf insertions (Table 1; Supplemental Table S1). Using the same approach, 92,580 SINE-containing segments of CanFam1 were aligned with CanSS to identify 6575 loci (7.1%) where the SINEC_Cf repeat is absent from the homologous region of CanSS (Table 1; Supplemental Table S1). Again, it should be noted that this is a minimum estimate, as it discounts the 7% of SINEC_Cf insertions that are predicted to be heterozygous in the sequenced poodle genome (Kirkness et al. 2003). In combination, these analyses revealed 10,562 loci for which there is strong evidence of bimorphism for SINE insertions between the two sequenced genomes.
Table 1.
Genomic coordinates of bimorphic SINE insertions
| Dog breed | Sequence ID | Start coordinate | End coordinate | Poodle sequence ID | Expected insertion | Boxer chromosome | Coordinate | Orientation |
|---|---|---|---|---|---|---|---|---|
| Boxer | chr01 | 3320493 | 3320616 | AACN010667811 | 557 | chr01 | 3320493 | + |
| Poodle | CE563274 | 497 | 374 | chr01 | 3903727 | — | ||
| Boxer | chr01 | 4033961 | 4034084 | AACN010457626 | 424 | chr01 | 4033961 | + |
| Boxer | chr01 | 4263197 | 4263074 | AACN010384528 | 799 | chr01 | 4263197 | — |
| Boxer | chr01 | 4674003 | 4673880 | AACN010148521 | 1453 | chr01 | 4674003 | — |
| Poodle | CE666043 | 360 | 237 | chr01 | 7738326 | + | ||
| Poodle | AACN010508306 | 188 | 311 | chr01 | 8008980 | — | ||
| Poodle | AACN010763349 | 556 | 433 | chr01 | 8380849 | — | ||
| English shepherd | TI389899990 | 168 | 291 | chr01 | 8480134 | + | ||
| Boxer | chr01 | 8688840 | 8688963 | CE514608 | 246 | chr01 | 8688840 | + |
| Boxer | chr01 | 8883852 | 8883975 | AACN010090863 | 287 | chr01 | 8883852 | + |
| Poodle | AACN010699373 | 517 | 640 | chr01 | 9062009 | + | ||
| Boxer | chr01 | 9483194 | 9483317 | CE826798 | 292 | chr01 | 9483194 | + |
| Boxer | chr01 | 9813098 | 9813221 | AACN010538407 | 224 | chr01 | 9813098 | + |
| Poodle | CE366308 | 330 | 207 | chr01 | 10844127 | + | ||
| Boxer | chr01 | 11362455 | 11362578 | AACN010063024 | 1426 | chr01 | 11362455 | + |
| Boxer | chr01 | 11748564 | 11748441 | AACN010413170 | 664 | chr01 | 11748564 | — |
| Boxer | chr01 | 11931616 | 11931493 | AACN010029257 | 3387 | chr01 | 11931616 | — |
| Poodle | AACN010775895 | 173 | 296 | chr01 | 12079479 | + | ||
| Boxer | chr01 | 12104226 | 12104103 | AACN010165887 | 1149 | chr01 | 12104226 | — |
| Boxer | chr01 | 12141439 | 12141562 | AACN010415886 | 845 | chr01 | 12141439 | + |
| Poodle | CE792917 | 540 | 417 | chr01 | 12168215 | + | ||
| Boxer | chr01 | 12285426 | 12285549 | AACN010039890 | 320 | chr01 | 12285426 | + |
| Boxer | chr01 | 12591205 | 12591328 | CE574347 | 179 | chr01 | 12591205 | + |
| Poodle | AACN010707012 | 641 | 764 | chr01 | 12615665 | + | ||
| Poodle | AACN010435963 | 443 | 566 | chr01 | 13122073 | — | ||
| Boxer | chr01 | 13391958 | 13391835 | AACN010271007 | 572 | chr01 | 13391958 | — |
| Poodle | AACN010604073 | 291 | 168 | chr01 | 13465651 | + | ||
| Poodle | CE571271 | 203 | 326 | chr01 | 14175158 | — | ||
| Boxer | chr01 | 14417857 | 14417980 | AACN010113778 | 158 | chr01 | 14417857 | + |
| Poodle | AACN010184489 | 1793 | 1670 | chr01 | 14507734 | — | ||
| Poodle | AACN010749955 | 477 | 600 | chr01 | 14517253 | + | ||
| Boxer | chr01 | 14564268 | 14564391 | AACN010271540 | 329 | chr01 | 14564268 | + |
| Boxer | chr01 | 14929470 | 14929593 | AACN010717696 | 548 | chr01 | 14929470 | + |
| Boxer | chr01 | 15150723 | 15150600 | AACN010916377 | 44 | chr01 | 15150723 | — |
| Boxer | chr01 | 15151989 | 15152112 | AACN010425836 | 1089 | chr01 | 15151989 | + |
| Poodle | CE412993 | 68 | 191 | chr01 | 15183132 | — | ||
| Poodle | CE056395 | 581 | 458 | chr01 | 15200679 | — | ||
| Poodle | AACN010373145 | 322 | 199 | chr01 | 15623445 | + | ||
| Poodle | AACN011003395 | 69 | 192 | chr01 | 15849317 | + | ||
| Poodle | AACN010870015 | 437 | 314 | chr01 | 15885701 | + | ||
| Boxer | chr01 | 16017574 | 16017697 | AACN010108721 | 892 | chr01 | 16017574 | + |
| Poodle | AACN010615570 | 227 | 104 | chr01 | 16441116 | + | ||
| Boxer | chr01 | 16771014 | 16771137 | AACN010816174 | 568 | chr01 | 16771014 | + |
| Boxer | chr01 | 16948663 | 16948786 | AACN010808689 | 337 | chr01 | 16948663 | + |
| Boxer | chr01 | 17165450 | 17165327 | AACN010826518 | 634 | chr01 | 17165450 | — |
| Bedlington terrier | TI356819018 | 473 | 350 | chr01 | 17632332 | — | ||
| Poodle | AACN010370372 | 423 | 546 | chr01 | 21284948 | — |
Each SINE is defined by a dog breed, a sequence ID, and the start and end coordinates of the core SINE consensus sequence within the sequence ID. For boxer, the sequence IDs and coordinates refer to CanFam1 (http://genome.ucsc.edu). For poodle, they refer to GenBank accessions (http://www.ncbi.nih.gov/Genbank/); for other breeds, they refer to sequences that are available from the NCBI Trace Archive (http://www.ncbi.nlm.nih.gov/Traces/). For boxer SINEs that are absent from the poodle genome, the table lists a homologous poodle sequence ID, and the coordinate where the SINE insertion would be expected. For all SINEs, the coordinates of the insertion and the orientation of the SINE are mapped relative to CanFam1. The complete table of 11,265 SINEs is provided as Supplemental Table S1.
It was of interest to determine how many additional bimorphic loci may exist in the general dog population that are not revealed by the large collections of sequence data from two specific dogs. In order to address this question, we used ∼1 million random genomic sequence reads that have been generated from nine dogs of different breeds, four wolves, and a coyote (http://www.genome.gov/12511476). Reads that contained SINEC_Cf repeats, and sufficient flanking sequence, were processed and aligned with both CanSS and CanFam1 as described above. For the nine domesticated dogs, analysis of ∼1000 SINEC_Cf repeats per dog indicated that 7.0%-10.9% were absent from CanSS, 6.7%-11.5% were absent from CanFam1, and 2.8%-5.0% were absent from both genomes (Table 2). The corresponding values for novel SINE insertions in the genomes of wild canids overlapped the range for domesticated dogs (4.1%-4.9%; Chinese and Spanish wolves), or were substantially larger (7.9%-9.2%; Californian coyote, Alaskan wolf, and Indian wolf). However, the sample sizes for bimorphic SINE insertions were relatively small for each of the wild canids (15-36), and the differences between them cannot yet be considered as significant.
Table 2.
Survey of bimorphic SINE insertions from random genomic sequence of multiple dogs
| Breed | Sequence reads | SINEC_Cf with unique flanks | SINEC_Cf absent from CanSS (%) | SINEC_Cf absent from CanFam1 (%) | SINEC_Cf absent from both (%) |
|---|---|---|---|---|---|
| Beagle | 99,648 | 990 | 69 (7.0) | 66 (6.7) | 28 (2.8) |
| Labrador Retriever | 99,744 | 942 | 78 (8.3) | 69 (7.3) | 28 (3.0) |
| German Shepherd | 100,743 | 987 | 72 (7.3) | 75 (7.6) | 30 (3.0) |
| Italian Greyhound | 98,208 | 889 | 79 (8.9) | 72 (8.1) | 33 (3.7) |
| English Shepherd | 99,648 | 954 | 88 (9.2) | 81 (8.5) | 38 (4.0) |
| Bedlington Terrier | 102,240 | 969 | 106 (10.9) | 92 (9.5) | 40 (4.1) |
| Portugese Water Dog | 97,728 | 1013 | 95 (9.4) | 94 (9.3) | 42 (4.1) |
| Alaskan Malamute | 100,704 | 932 | 96 (10.3) | 87 (9.3) | 45 (4.8) |
| Rottweiler | 102,143 | 1040 | 106 (10.2) | 120 (11.5) | 52 (5.0) |
| Chinese Gray Wolf | 23,423 | 169 | 16 (9.5) | 17 (10.1) | 7 (4.1) |
| Spanish Gray Wolf | 22,176 | 185 | 18 (9.7) | 15 (8.1) | 9 (4.9) |
| Californian Coyote | 23,790 | 240 | 29 (12.1) | 28 (11.7) | 19 (7.9) |
| Alaskan Gray Wolf | 21,696 | 220 | 34 (15.4) | 36 (16.4) | 20 (9.1) |
| Indian Gray Wolf | 22,560 | 227 | 32 (14.1) | 29 (12.8) | 21 (9.2) |
Whole-genome shotgun reads from the NCBI Trace Archive were derived from nine dogs of different breeds, four wolves, and a coyote. Reads that contained SINEC_Cf sequences with nonrepetitive flanks were searched against CanSS and CanFam1, as described in the Methods section.
The preceding analysis indicates that many thousands of SINEC_Cf insertions within canine genomes are not represented in either CanSS or CanFam1. However, the analysis also demonstrates that random genomic sequencing is an inefficient means to identify these novel loci. Only ∼1% of the sequence reads contained sufficient SINE and flanking sequences to permit comparison with the two reference genomes. Consequently, we have developed a methodology that specifically targets SINEC_Cf repeats and flanking sequence for amplification and sequencing. The methodology is centered on a simple inverse-PCR that exploits both the high level of sequence conservation between SINEC_Cf repeats, and the sequence variation between SINEC_Cf repeats and related canine SINEs (Fig. 1). The consensus sequence of SINEC_Cf repeats contains a single, conserved CATG sequence that can be cleaved by the frequently cutting restriction enzyme, NlaIII. After self-ligation of digested genomic fragments, sequences upstream of SINEC_Cf repeats are amplified selectively with primers corresponding to segments of the SINEC_Cf repeat that differ from related SINEs. Cloning of the PCR products yields libraries of SINEC_Cf flanking sequences that can be sequenced readily. Limited sequencing of seven trial libraries demonstrated that >88% of inserts contain SINEC_Cf repeats (with >100 bp of flanking sequence). Importantly, after alignment of these flanking sequences with CanSS and CanFam1, 2.7%-6.8% identified the loci of novel SINEC_Cf insertions. In order to validate these data, 29 of the loci were amplified from the genomic DNA of 8-15 dogs (three to five breeds). Of these, 26 were confirmed as sites of variable SINEC_Cf content, and three were monomorphic for the absence of a SINE in all of the tested dogs (Fig. 2). When scaled up, this simple assay should identify several thousands of novel bimorphic SINEC_Cf insertions, and complement subtractive hybridization approaches that were recently reported to identify bimorphic Alu insertions in human populations (Mamedov et al. 2005).
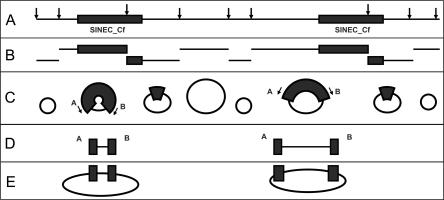
Figure 1.
Construction of libraries that are enriched for SINEC_Cf elements and flanking sequence. (A) Genomic DNA is cleaved with the frequently cutting restriction enzyme, NlaIII. (B) The cleaved fragments are self-ligated. (C) The circularized products are subjected to PCR using SINEC_Cf-specific primers. (D) The linear products are size-selected and cloned in a plasmid vector. (E) Inserts are sequenced with a vector-specific primer.
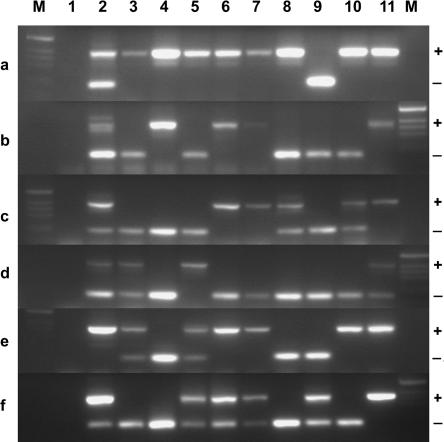
Figure 2.
Validation of putative bimorphic SINE insertions. Six representative examples (a-f) of PCR products from loci that were predicted to contain bimorphic SINE insertions after analysis of SINEC_Cf libraries (see Fig. 1). Primers were designed to the flanks of the predicted insertion site, and templates were genomic DNA from a combination of mixed-breed dogs (lane 2), three Bernese Mountain dogs (lanes 3-5), three pugs (lanes 6-8), and three dachshunds (lanes 9-11). Product lengths were derived from DNA markers (lanes M), and were consistent with the presence (+) or absence (–) of a SINE insertion.
The current collections of canine genomic sequence data have therefore revealed >11,000 loci that are bimorphic for SINEC_Cf insertions (Supplemental Table S1). An additional 86,000 loci that contain a SINEC_Cf repeat on at least one allele of CanFam1 and CanSS can also be examined for variability in a wider selection of dogs. Furthermore, the inverse-PCR approach described above permits novel SINEC_Cf insertions to be identified with ∼90-fold higher efficiency than from random genomic sequence reads. Together, these approaches should permit the identification of at least 10,000 additional bimorphic loci in the general dog population. This genomic variability will be a very useful resource for the study of ancestral relationships between different canine lineages. Specifically, bimorphic SINE elements offer two advantages over other types of common polymorphisms. First, the presence of a SINE element represents identity by descent, since the probability that two different young SINE repeats would integrate independently at the same chromosomal location is small. Although there have been a few reports of parallel insertion events (Kass et al. 2000; Cantrell et al. 2001), these are considered to be very rare, at least for primate Alus (Roy-Engel et al. 2002; Salem et al. 2005). Second, the ancestral state of each SINE insertion polymorphism is known to be the absence of the SINE element, and this can be used to root trees of population relationships. In contrast, other types of genetic polymorphism, such as VNTRs and SNPs, can be identical by state if they have arisen from independent parallel mutations at different times and have not been inherited from a common ancestor. Bimorphic Alu-insertion polymorphisms have been used to study human origins, ancestral relationships, and demography (for review, see Batzer and Deininger 2002; Watkins et al. 2003). In some respects, dog breeds resemble geographically isolated human populations, but with a higher degree of isolation, and narrower bottlenecks. In addition, the complex genomic structure of modern dog populations presents specific challenges, owing to the recent origin of most dog breeds (<300 yr), and their derivation from multiple ancestral types (Parker et al. 2004).
Previously, evolutionary studies of canine lineages have focused mainly on variations of mitochondrial DNA or VNTRs (Vila et al. 1997; Savolainen et al. 2002; Koskinen 2003; Parker et al. 2004). These approaches indicate that modern dog breeds were first domesticated from wolves, possibly in East Asia, and that many dog breeds that share morphologies, behaviors, and geographical origins can be segregated by genotype. However, in common with most types of marker, variations of mitochondrial DNA and VNTRs have limitations for evolutionary analyses (Ellegren 2000; Sigurgardottir et al. 2000). Bimorphic SINE insertions offer the advantages of identity-by-descent, and easy typing methodologies, that make these abundant variations a valuable additional resource for identifying the ancestral relationships between different dog breeds, and between domesticated dogs and wild canids.
It is also relevant to note that recent observations of extensive linkage disequilibrium in the dog indicate that association studies to find genes that contribute to diseases and traits could be conducted using as few as 30,000 evenly distributed genomic markers (Sutter et al. 2004). It is conceivable that bimorphic SINEs could provide many of these markers if the throughput for SINE-typing could be scaled up to that currently used for SNPs. One approach for high-throughput typing could use the total amplification of SINEC_Cf flanks (Fig. 1D). The products would be labeled and hybridized to microarrays of oligonucleotides that represent known SINE flanks. By this means, genomes could be scored for the presence or absence of many thousands of SINEs in a single hybridization assay.
Recent LINE activity in the canine genome
Active long interspersed elements (LINEs) are autonomous retrotransposons that likely provide the enzymic activities that are required by SINEs for their propagation (Jurka 1997; Dewannieux et al. 2003). The high frequency of bimorphic SINE insertions in the dog genome may therefore be indicative of highly active LINEs. Identification of active LINEs can pose a major problem for genome assemblies that are based on the whole-genome shotgun approach. Owing to their abundance, similarity, and the fact that they cannot be spanned by individual sequence reads, LINEs are often imprecisely assembled as collapsed contigs. For example, the draft mouse genome contained only 12 full-length LINEs with intact open reading frames (ORFs), although at least 3000 were predicted to exist (Waterston et al. 2002). Similarly, our analysis of CanFam1 revealed only four LINEs with two intact ORFs among the 3226 LINEs that are long enough to be functional (>4.5 kb). The vast majority of these elements contain frameshift mutations or in-frame stop codons. However, at present, we cannot readily distinguish assembly errors from genuine mutations that disrupt the ORFs. Consequently, the number of canine LINEs that are potentially active cannot be estimated reliably.
Another approach to compare recent LINE activity in the dog and human genomes is an analysis of 3′-truncated LINEs. Recent differences in LINE activity should be reflected by detectable differences between the numbers of recent LINE insertions. Owing to the fact that most LINE insertions are 3′-truncations, this analysis is not dependent on precise assembly of full-length elements. We considered the 3′-terminal 500 bases of the youngest known dog and human LINEs (L1_Y_Cf, and L1HS respectively; RepBase Update 9.1). These were aligned with the dog and human genomes, and alignments that spanned at least 98% of the query sequence were categorized by percentage nucleotide identity. For L1_Y_Cf, there were 2700 genomic segments that shared at least 98% identity. For L1HS, the value was 792. That is, among the youngest elements, there are approximately threefold more L1_Y_Cf - like elements in the dog genome, than L1HS-like elements in the human genome. This is consistent with a recent higher level of LINE activity in the dog lineage.
Evidence of recent LINE activity, in the form of bimorphic LINE insertions, is a difficult problem to represent in genome assemblies because the shorter allele (lacking the LINE) is preferentially selected for the final assembly. This is exemplified in the CanFam1 assembly. A BAC clone (GenBank accession no. AC147784.3) from the same dog that was sequenced for CanFam1, contains a full-length L1_Y_Cf (bases 83,981-90,278) with two uninterrupted ORFs. However, although the BAC sequence is represented in CanFam1 (chr29, bases 36,992,169-37,155,933), the LINE is absent. Analysis of the raw sequence reads (from the Trace Archive) that cover the insertion site reveals two alleles in which the element is either absent (e.g., accession nos. 294,160,392 and 285,880,943) or present (e.g., accession nos. 290,601,686 and 237,812,340). This is clearly an example of a bimorphic LINE insertion. We have begun to examine this phenomenon in more detail by using the 3′-ends of LINEs (plus unique flanking sequence) from the poodle genome sequence. When searched against CanFam1, alignments that span the flanking sequence, but not the LINE sequence, indicate potential bimorphic insertions of LINEs. Our preliminary analysis suggests that there are more than a hundred of such candidate regions that can now be tested by amplification of the regions from multiple dog genomes.
The analysis of LINEs in the dog genome has revealed examples of intact elements that are potentially active, and evidence for a higher level of recent activity than in the human genome. It is therefore possible that increased LINE activity has contributed to the relatively high frequency of bimorphic SINE insertions in the canine genome.
Evidence for SINE-mediated transduction of 3′-flanking sequences
When transcription of a retrotransposon fails to terminate at the end of the element, the additional downstream genomic sequence that is transcribed can be mobilized to a new genomic location along with the element. This mechanism of transduction is thought to occur during retrotransposition of LINE-1 elements when the normal polyadenylation signal is bypassed in favor of a second, downstream signal (Goodier et al. 2000; Pickeral et al. 2000). However, transduction of 3′-flanking sequences by SINEs has not been described previously. Analysis of flanking sequences and target site duplications for SINEC_Cf elements in the dog genome revealed several examples of short genomic segments (60-120 bp) that appear to have been transduced during retrotransposition of SINEC_Cfs (Supplemental Table S2). These examples include a short segment of Chromosome 8 that is replicated downstream of a SINEC_Cf at eight other genomic locations. They also include a bimorphic insertion, where a SINE appears to have transposed a short segment from Chromosome 13 to Chromosome 1 of the boxer genome, although the latter locus lacks both elements in the sequenced poodle genome. The genomic variability that results from bimorphic insertions of SINEs may therefore extend to additional flanking sequences for some active SINEC_Cfs. Although the mechanism for these putative transduction events remains to be explored, the sequences are consistent with transcription of active SINEC_Cf repeats through their 3′-flanking sequences, with polyadenylation at downstream cleavage sites.
Distribution of bimorphic SINE insertions across the dog genome
The 92,580 SINEC_Cf repeats from CanFam1, and the 11,265 SINEC_Cf repeats that are bimorphic between CanFam1, CanSS, and the Trace Archive reads, are distributed among all chromosomes with frequencies of 29.7-43.8 per Mb and 2.8-6.0 per Mb, respectively (Table 3). The local GC content for 1 kb upstream (median = 38.2%) and 1 kb downstream (38.0%) of SINEC_Cf insertion sites is only slightly lower than the genome average for 1-kb nonoverlapping windows of CanFam1 (39.5%). In addition, there is no indication that SINEC_Cf repeats are preferentially located within genes. The Ensembl annotation of CanFam1 (http://www.ensembl.org/Canis_familiaris; release 27.1.1) identifies 18,201 genes. These span 34% of the CanFam1 sequence, and contain 36% of the CanFam1 SINEC_Cf repeats (and 33% of the bimorphic SINEC_Cf repeats) that were identified in this study. Across the whole genome, 48% of annotated genes contain at least one CanFam1 SINEC_Cf (mean; 3.8 per gene), and 14% contain at least one bimorphic SINEC_Cf (mean; 1.5 per gene).
Table 3.
Distribution of SINEC_Cf repeats and bimorphic SINEC_Cf insertions (bSINEC_Cf) among dog chromosomes and annotated genes
|
SINEC_Cf
|
bSINEC_Cf
|
Annotated genes
|
||||||
|---|---|---|---|---|---|---|---|---|
| Chromosome | Length (Mb) | No. | per Mb | No. | per Mb | No. | % with SINEC_Cf | % with bSINEC_Cf |
| 1 | 124.898 | 4520 | 36.2 | 516 | 4.1 | 1058 | 45.4 | 11.5 |
| 2 | 87.725 | 3125 | 35.6 | 373 | 4.3 | 672 | 52.7 | 14.9 |
| 3 | 95.080 | 3116 | 32.8 | 398 | 4.2 | 457 | 55.6 | 16.2 |
| 4 | 91.326 | 3183 | 34.9 | 383 | 4.2 | 521 | 49.3 | 14.2 |
| 5 | 92.065 | 3103 | 33.7 | 360 | 3.9 | 916 | 43.7 | 9.9 |
| 6 | 79.106 | 2938 | 37.1 | 332 | 4.2 | 805 | 48.1 | 12.3 |
| 7 | 83.037 | 3054 | 36.8 | 338 | 4.1 | 586 | 58.0 | 15.4 |
| 8 | 77.375 | 2931 | 37.9 | 452 | 5.8 | 567 | 51.1 | 16.6 |
| 9 | 53.643 | 2347 | 43.8 | 312 | 5.8 | 846 | 44.8 | 13.4 |
| 10 | 72.717 | 2720 | 37.4 | 341 | 4.7 | 600 | 53.5 | 16.0 |
| 11 | 75.770 | 2856 | 37.7 | 316 | 4.2 | 480 | 49.2 | 11.9 |
| 12 | 75.456 | 3048 | 40.4 | 399 | 5.3 | 539 | 46.6 | 16.0 |
| 13 | 66.160 | 2124 | 32.1 | 257 | 3.9 | 348 | 48.6 | 12.6 |
| 14 | 63.550 | 2261 | 35.6 | 269 | 4.2 | 360 | 52.8 | 15.8 |
| 15 | 67.238 | 2626 | 39.1 | 264 | 3.9 | 461 | 52.5 | 12.1 |
| 16 | 60.308 | 2066 | 34.3 | 251 | 4.2 | 388 | 47.9 | 14.9 |
| 17 | 66.886 | 2521 | 37.7 | 326 | 4.9 | 545 | 49.2 | 13.4 |
| 18 | 66.174 | 2208 | 33.4 | 278 | 4.2 | 851 | 33.5 | 7.3 |
| 19 | 56.914 | 1978 | 34.8 | 272 | 4.8 | 176 | 58.0 | 18.2 |
| 20 | 61.173 | 2381 | 38.9 | 282 | 4.6 | 884 | 39.6 | 9.3 |
| 21 | 53.028 | 1896 | 35.8 | 268 | 5.1 | 408 | 44.9 | 15.4 |
| 22 | 64.236 | 2350 | 36.6 | 324 | 5.0 | 205 | 52.7 | 20.5 |
| 23 | 55.586 | 2000 | 36.0 | 273 | 4.9 | 290 | 64.5 | 20.7 |
| 24 | 50.732 | 1810 | 35.7 | 267 | 5.3 | 487 | 45.2 | 14.2 |
| 25 | 54.437 | 1922 | 35.3 | 271 | 5.0 | 379 | 49.3 | 17.2 |
| 26 | 41.010 | 1668 | 40.7 | 244 | 5.9 | 425 | 47.5 | 15.8 |
| 27 | 49.087 | 2089 | 42.6 | 267 | 5.4 | 471 | 48.2 | 16.3 |
| 28 | 42.450 | 1479 | 34.8 | 166 | 3.9 | 310 | 57.1 | 14.2 |
| 29 | 44.860 | 1528 | 34.1 | 180 | 4.0 | 185 | 57.8 | 14.6 |
| 30 | 43.165 | 1708 | 39.6 | 199 | 4.6 | 361 | 59.8 | 19.1 |
| 31 | 41.216 | 1350 | 32.8 | 204 | 4.9 | 173 | 48.6 | 19.7 |
| 32 | 41.834 | 1834 | 43.8 | 252 | 6.0 | 204 | 60.3 | 27.0 |
| 33 | 34.471 | 1328 | 38.5 | 198 | 5.7 | 205 | 65.4 | 25.9 |
| 34 | 45.233 | 1389 | 30.7 | 216 | 4.8 | 211 | 48.8 | 19.4 |
| 35 | 29.507 | 875 | 29.7 | 146 | 4.9 | 168 | 44.0 | 18.5 |
| 36 | 33.961 | 1172 | 34.5 | 146 | 4.3 | 163 | 58.3 | 18.4 |
| 37 | 33.897 | 1118 | 33.0 | 164 | 4.8 | 207 | 56.0 | 16.4 |
| 38 | 26.492 | 863 | 32.6 | 146 | 5.5 | 193 | 44.0 | 14.5 |
| X | 126.913 | 5429 | 42.8 | 356 | 2.8 | 735 | 44.6 | 8.2 |
| Un | 91.063 | 3666 | 40.3 | 259 | 2.8 | 361 | 21.6 | 5.0 |
Insertion of SINEs close to exons has been shown to cause aberrant splicing of transcripts in both human (Wallace et al. 1991; Ganguly et al. 2003) and dog (Lin et al. 1999). Insertion of SINEs within exons has also been reported to disrupt gene expression and cause diseases in both human and dog. For example, an Alu insertion within exon 11 of CLCN5 is associated with Dent's disease in human (Claverie-Martin et al. 2003), while a SINEC_Cf insertion within exon 2 of the PLPTA gene is associated with centronuclear myopathy in Labrador retrievers (Pele et al. 2005). The Ensembl annotation of CanFam1 lists 163 exon junctions that are within 100 bases of a SINEC_Cf repeat. However, none of the 92,580 SINEC_Cf repeats from CanFam1 are annotated as residing within an exon. This is likely to be misleading for at least two reasons. First, there are examples of introns within annotated genes that consist entirely of SINEC_Cf sequence (Ensembl genes, ENSCAFG00000000578, ENSCAFG00000000879, ENSCAFG00000001129, ENSCAFG00000004064, ENSCAFG00000008336, ENSCAFG00000017490, ENSCAFG00000017848). If these are, indeed, transcribed genes, the SINEC_Cf insertion would be expected to affect gene expression. However, it is possible that at least some of these examples are processed pseudogenes that have no functional significance whether or not they have acquired SINE insertions. The second reason for an absence of SINEC_Cf sequences in annotated exons relates to our limited knowledge of canine gene structures. Relative to human and mouse, the dog is not well represented in GenBank by cDNA sequences. For example, dbEST (http://www.ncbi.nlm.nih.gov/dbEST; release 100104) contains 6.0 million expressed sequence tags (ESTs) from human, 4.2 million from mouse, but only ∼155,000 from dog. Consequently, the gene annotation of CanFam1 relies more heavily on sequence comparisons with genes from other species that have been validated by ESTs and full-length cDNAs. Such comparisons would not be expected to reveal dog-specific repeats within predicted transcripts.
Transcription of SINEC_Cf elements
Although current annotation of CanFam1 fails to identify SINEC_Cf repeats within predicted exons, an analysis of dog ESTs identified 120 examples of cDNA clones that contain complete or partial SINEC_Cf sequences (Table 4). Approximately half of these are located in the “sense” orientation at the 3′-ends of cDNAs. This common location may be an experimental artifact, caused by oligo(dT) priming of cDNA synthesis at internal, A-rich regions of the primary transcript that are provided by the SINE. However, it is also possible that at least some of these examples arise from the use of the known polyadenylation signals within most tRNA-related SINEs, including SINEC_Cf (Borodulina and Kramerov 2001). Depending on the context of these signals, they could cause premature cleavage and polyadenylation of Pol II-derived dog mRNAs. The cDNAs that terminate with a SINEC_Cf sequence include four examples in which the SINEC_Cf repeats are absent from CanFam1, and are therefore likely to represent bimorphic insertions (Table 5). Notably, three of these four examples are located within annotated genes that are transcribed in the same orientation as the SINEC_Cf repeat that terminates the cDNA. It will be of interest to know if such SINE insertions cause a significant level of premature polyadenylation for the transcripts of the genes in which they have inserted. This should be relatively easy to determine using tissues from dogs that are heterozygous for the SINE insertions, as these tissues should also express normal transcripts from one allele.
Table 4.
Dog ESTs that contain SINEC_Cf sequences
| EST ID | Start | End | Orientation | Type | EST ID | Start | End | Orientation | Type | EST ID | Start | End | Orientation | Type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BI395282 | 100 | 292 | (+) | 1 | CK999028 | 9 | 191 | (–) | 1 | CF411877 | 509 | 701 | (+) | 2 |
| BI396013 | 247 | 428 | (+) | 1 | CN000189 | 1 | 200 | (–) | 1 | CK997835 | 373 | 553 | (–) | 2 |
| BI817000 | 122 | 307 | (+) | 1 | CN000259 | 1 | 185 | (–) | 1 | CN000198 | 540 | 735 | (+) | 2 |
| BM536771 | 134 | 316 | (–) | 1 | CN000862 | 1 | 198 | (–) | 1 | CN001701 | 354 | 549 | (–) | 2 |
| BM538738 | 29 | 206 | (–) | 1 | CN003480 | 9 | 177 | (–) | 1 | CN003009 | 132 | 323 | (+) | 2 |
| BM540896 | 1 | 178 | (–) | 1 | CN003481 | 52 | 232 | (+) | 1 | CN003100 | 198 | 370 | (–) | 2 |
| BQ091505 | 75 | 251 | (+) | 1 | CN004223 | 4 | 186 | (–) | 1 | CN003272 | 372 | 553 | (+) | 2 |
| BQ091808 | 34 | 230 | (–) | 1 | CO615070 | 1 | 177 | (–) | 1 | CN003734 | 467 | 653 | (+) | 2 |
| BQ233944 | 258 | 445 | (+) | 1 | CO618415 | 1 | 170 | (–) | 1 | CN003833 | 466 | 649 | (+) | 2 |
| BQ233951 | 80 | 248 | (+) | 1 | CO622584 | 1 | 192 | (–) | 1 | CN004818 | 224 | 406 | (–) | 2 |
| BQ234215 | 272 | 443 | (+) | 1 | CO622645 | 1 | 184 | (–) | 1 | CN005854 | 337 | 520 | (+) | 2 |
| BQ234327 | 32 | 200 | (+) | 1 | CO624201 | 14 | 199 | (–) | 1 | CO620822 | 82 | 253 | (–) | 2 |
| BQ234548 | 80 | 248 | (+) | 1 | CO631931 | 1 | 181 | (–) | 1 | CO630670 | 115 | 296 | (+) | 2 |
| BQ235019 | 404 | 574 | (+) | 1 | CO633032 | 16 | 212 | (–) | 1 | CO688822 | 181 | 367 | (+) | 2 |
| BQ235154 | 401 | 585 | (+) | 1 | CO633878 | 19 | 187 | (–) | 1 | CO708098 | 261 | 444 | (+) | 2 |
| BQ235636 | 335 | 521 | (+) | 1 | CO634532 | 1 | 173 | (–) | 1 | BM536886 | 187 | 337 | (–) | 3 |
| BQ235876 | 129 | 321 | (+) | 1 | CO635086 | 1 | 173 | (–) | 1 | BM539513 | 338 | 459 | (–) | 3 |
| BQ788275 | 291 | 460 | (+) | 1 | CO677843 | 1 | 171 | (–) | 1 | BM540124 | 47 | 198 | (–) | 3 |
| BQ788475 | 291 | 465 | (+) | 1 | Z97735 | 3 | 192 | (–) | 1 | BQ234114 | 244 | 395 | (–) | 3 |
| BU744447 | 60 | 271 | (–) | 1 | Z97747 | 1 | 180 | (–) | 1 | BU749657 | 397 | 532 | (–) | 3 |
| BU744716 | 70 | 232 | (–) | 1 | Z97810 | 30 | 213 | (–) | 1 | BU751410 | 37 | 167 | (–) | 3 |
| BU744952 | 61 | 227 | (–) | 1 | BI389234 | 124 | 312 | (–) | 2 | CK996159 | 35 | 156 | (–) | 3 |
| BU745150 | 87 | 273 | (–) | 1 | BM538367 | 123 | 312 | (–) | 2 | CK996423 | 275 | 436 | (+) | 3 |
| BU749508 | 66 | 248 | (–) | 1 | BM541044 | 246 | 428 | (+) | 2 | CN002508 | 157 | 318 | (–) | 3 |
| BU751148 | 61 | 243 | (–) | 1 | BM735692 | 178 | 358 | (–) | 2 | CO629118 | 58 | 205 | (+) | 3 |
| BU751489 | 114 | 297 | (–) | 1 | BQ234408 | 316 | 500 | (+) | 2 | CO668671 | 394 | 545 | (+) | 3 |
| CF406750 | 65 | 260 | (–) | 1 | BQ235081 | 316 | 500 | (+) | 2 | CO675560 | 265 | 414 | (+) | 3 |
| CF407503 | 69 | 250 | (–) | 1 | BQ235579 | 204 | 335 | (–) | 2 | CO675838 | 117 | 270 | (+) | 3 |
| CF408296 | 58 | 245 | (–) | 1 | BU750849 | 192 | 398 | (+) | 2 | CO684930 | 118 | 267 | (+) | 3 |
| CF408332 | 59 | 246 | (–) | 1 | BU751135 | 148 | 345 | (+) | 2 | BF228953 | 25 | 193 | (+) | 4 |
| CF409934 | 107 | 310 | (–) | 1 | BU751296 | 557 | 726 | (–) | 2 | BF228988 | 18 | 194 | (+) | 4 |
| CF410986 | 145 | 327 | (–) | 1 | BU751297 | 324 | 492 | (+) | 2 | BQ234445 | 5 | 187 | (+) | 4 |
| CF411174 | 218 | 411 | (–) | 1 | CF406935 | 280 | 463 | (+) | 2 | BQ234448 | 10 | 146 | (+) | 4 |
| CF413125 | 130 | 324 | (–) | 1 | CF406941 | 595 | 755 | (–) | 2 | BQ235047 | 9 | 143 | (+) | 4 |
| CK996008 | 4 | 199 | (–) | 1 | CF408437 | 286 | 452 | (+) | 2 | BQ235092 | 10 | 150 | (+) | 4 |
| CK996233 | 93 | 277 | (–) | 1 | CF409696 | 259 | 439 | (–) | 2 | BQ235470 | 9 | 144 | (+) | 4 |
| CK997330 | 1 | 181 | (–) | 1 | CF409992 | 367 | 556 | (+) | 2 | BQ235629 | 9 | 191 | (+) | 4 |
| CK997507 | 3 | 174 | (–) | 1 | CF410165 | 274 | 451 | (–) | 2 | BQ235644 | 9 | 169 | (+) | 4 |
| CK997864 | 1 | 172 | (–) | 1 | CF410277 | 296 | 489 | (–) | 2 | BQ290082 | 9 | 136 | (+) | 4 |
| CK998336 | 23 | 191 | (–) | 1 | CF411091 | 230 | 412 | (+) | 2 | CK995704 | 1 | 162 | (+) | 4 |
Each EST is represented by its GenBank accession number, the start and end coordinates of the SINEC_Cf sequence within the EST, the orientation of the SINEC_Cf sequence (+/–), and the type of insertion (1–4). Four types of insertion were distinguished. A complete SINEC_Cf sequence was located at either the 3′-end of the cDNA (type 1) or within the cDNA sequence (type 2). Partial SINEC_Cf sequences that have arisen from the use of splice acceptor sites within the element were termed type 3. Finally, there were ESTs that consist entirely of SINEC_Cf sequence (type 4).
Table 5.
Examples of dog cDNAs that terminate with a bimorphic SINEC_Cf (+) sequence
| SINEC_Cf(+) | GGGATCC..167..AAAAA | Ensembl gene prediction | |
|---|---|---|---|
| 1 | BI817000(+) | 1-101-CCATTAGAGGATATAAAGAAGGGATCC..174..AAAAA-307 | ENSCAFG00000016640 (+) |
| Cfa6 | 18637451-18637551-CCATTAGAGGATATAAAGAAgattgct | ||
| 2 | BQ234215(+) | 9-251-CAGAGGGTTAAAATGCTTCAGGGATCC..160..AATAA-443 | ENSCAFG00000010152 (+) |
| Cfa21 | 49587244-49587487-CAGAGGGTTAAAATGCTTCAaattgtc | ||
| 3 | CK997864(–) | 1-372-TCAGCTGGTAGGAATGAATTGGGATCC..156..TAAAA-564 | – |
| Cfa31 | 20121516-20121868-TCAGCTGGTAGGAATGAATatggatat | ||
| 4 | BQ233944(+) | 10-237-GTGTTAGAACTCCATCAACAGGGATCC..176..AAAAA-445 | ENSCAFG00000013709 (+) |
| Cfa17 | 66653098-66653326-GTGTTAGAACTCCATCAACAttttccc |
The termini of the consensus SINEC_Cf (+) sequence is included for comparison. For examples 1–4, the GenBank accession numbers (and orientations) refer to dog ESTs [e.g., BI817000 (+)] that can be aligned with specific chromosomal regions of CanFam1. The nucleotide identity between the EST and genomic sequences is terminated by a SINEC_Cf sequence at the 3′-end of the cDNA sequence. These are all examples of transcribed SINEC_Cf insertions that are bimorphic in the dog population. Three of the four examples map within introns of putative genes in the (+) orientation (Ensembl annotation).
Among the ESTs of Table 4, there are also examples of dog cDNAs that have acquired additional exons (relative to their human orthologs) owing to the splicing of transcribed SINEC_Cf elements. An example is illustrated by the canine Tipin gene (Fig. 3). Relative to human, the dog Tipin transcript has acquired an additional exon owing to the insertion of a SINEC_Cf repeat downstream of the first exon. When transcribed in the (–) orientation, a SINEC_Cf repeat provides characteristic sequence motifs that permit it to act as a 3′-splice acceptor site, resulting in activation of a cryptic 5′-splice site downstream of the element. A similar mechanism has been described for exonization of an Alu within the RPE gene in primates (Krull et al. 2005). In the case of canine Tipin, the novel exon is predicted to be untranslated although its effect on Tipin protein expression is currently unknown. Table 6 provides details of 10 distinct cDNAs where SINEC_Cf repeats are spliced into transcripts by using precisely the same splice acceptor site. For most of these, alignment of the cDNA sequences with CanFam1 confirms the predicted splicing pattern. However, there are also two examples (9 and 10) in which the SINEC_Cf repeat is absent from CanFam1. These likely represent additional examples of bimorphic SINEC_Cf insertions that are transcribed. Four of the cDNAs appear to represent dog orthologs of known human genes, and the SINE insertion disrupts the homologous open reading frame for three of these (see legend to Table 6).
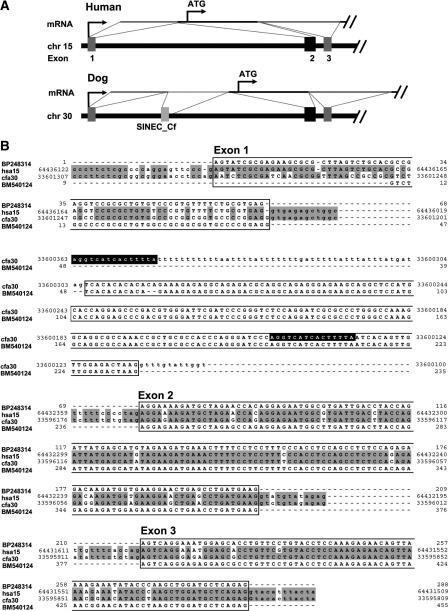
Figure 3.
Splicing of a SINEC_Cf sequence into the canine mRNA for Tipin. (A) The 5′-end of the canine Tipin gene differs from that of human by inclusion of an additional untranslated exon that is derived from a SINEC_Cf. (B) Alignment of sequences representing the 5′-ends of Tipin mRNA from human (GenBank accession no. BP248314) and dog (BM540124) with genomic sequences from human (NCBI build 34) and dog (CanFam1). Nucleotide identity between the human and dog genomic sequences is shaded, exons are boxed, and intronic sequences are in lower case. Between exons 1 and 2, the dog genome contains a SINEC_Cf sequence that is flanked by a characteristic 15-bp duplication of the insertion site (white lettering on black). This element provides a 3′-splice acceptor site, and causes activation of a cryptic 5′-splice site downstream of the element, resulting in the incorporation of the SINEC_Cf sequence within the dog Tipin mRNA.
Table 6.
Examples of dog cDNAs that have incorporated a SINEC_Cf (–) sequence via a splice acceptor site within the element
| SINEC_Cf(–) | TTTTTATTTATTTATGATAGTCACACAGAG.. 141.. GGATCCC | |
|---|---|---|
| 1 | BM540124(+) | 38-GCCCCGGAGG TCACACACAC.. 134.. GGATCCC AGGTCATCAC-208 |
| Cfa30 | 33601222-GCCCCGGAGGgtgagagctg... tttatgatagTCACACACAC.. 136.. GGATCCC AGGTCATCAC-33600139 | |
| 2 | CN002508(+) | 154-CACTGATGAG TCACACAGAG.. 138.. GGATCCC CATTTCATTT-328 |
| Cfa2 | 68067110-CACTGATGAGgtatgaattg... tttatgatagTCACACAGAG.. 138.. GGATCCC CATTTCATTT-68064412 | |
| 3 | CO675838(–) | 306-GCACACTCTG TCACACAGAG.. 138.. GGATCCC AAGCCAAGCA-478 |
| Cfa6 | 11279927-GCACACTCTGgtcagttcca... tttatgatagTCACACAGAG.. 141.. GGATCCC AAGCCAAGCA-11279642 | |
| 4 | BM536886(+) | 177-CAGCCCACAG ACATAGAGAG.. 134.. GGATCCC CTCATCTGAC-347 |
| Cfa9 | 26197334-CAGCCCACAGgtaaagtatt... ttcatgatagACATAGAGAG.. 134.. GGATCCC CTCATCTGAC-26192495 | |
| 5 | CO668671(–) | 109-CATCCCAGAG GCNCNCAGTG.. 136.. GGATCCC CTACATTGCT-281 |
| Cfa14 | 47081174-CATCCCAGAGgtaagagtat... tttatgatagGCACACAGTG.. 138.. GGATCCC CTACATTGCT-47047028 | |
| 6 | BU749657(+) | 322-GCAATCACGG AGAGAGAGAG.. 114.. GGATCCC CGGGTTAAGG-472 |
| Cfa15 | 52463523-GCAATCACGGgtaaggtctt... tttattatagGCACACAGTG.. 114.. GGATCCC CGGGTTAAGG-52469700 | |
| 7 | BQ234114(+) | 233-CCCACCACAG TCACAGAGAG.. 136.. GGATCCC AGATCCATCT-405 |
| Cfa26 | 24551559-CCCACCACAGgtataaacaa... tcagtcacagTCACAGAGAG.. 136.. GGATCCC AGATCCATCT-24550169 | |
| 8 | CO684930(–) | 328-ATCAAAGCTG TCACACAGAG.. 132.. GGATCCC CATGTGTGTG-496 |
| Cfa27 | 34794751-ATCAAAGCTGgtgagataca... tctatgatagTCACACAGAG.. 138.. GGATCCC CATGTGTGTG-34795736 | |
| 9 | CO675560(–) | 183-TCTTCAAGCT TCACAGAGAG.. 134.. GGATCCC CCGTTAGTTG-353 |
| Cfa22 | 49073614-TCTTCAAGCTgtgagtgcgg... TTAGTTG-49088654 | |
| 10 | CO629118(–) | 320-ACCTCTAAAG TCNCAGAGAG.. 134.. GGATCCC TATGCTCACT-493 |
| Cfa35 | 28868224-ACCTCTAAAGgtcagtcaca... TATGCTCACT-28867050 |
The location of the splice site (↓) follows a canonical “AG” dinucleotide, and is illustrated on the consensus SINEC_Cf (–) sequence. For examples 1–8, the GenBank accession numbers (and orientations) refer to dog ESTs [e.g., BM540124 (+)] that can be aligned with specific chromosomal regions of the dog genome sequence. The alignments of EST and genomic sequences include exons (upper case), introns (lower case), and coordinates of the aligned EST and genomic sequences. All introns are flanked by canonical “gt” and “ag” dinucleotides. For examples 9 and 10, sequences that flank the SINEC_Cf of the EST can be aligned with genomic sequence, but the SINEC_Cf sequence is absent. These are examples of bimorphic SINEC_Cf insertions that have become incorporated within cDNAs as alternative exons. For three of the dog ESTs (CN002508, BM536886, CO668671), the SINE insertion disrupts the open reading frame of the homologous human cDNA (phosphorylase kinase, β subunit, NM_000293; TRIM37 mRNA, NM_015294; phosphodiesterase 1C mRNA, NM_005020).
The recent expansion of SINEs in the dog genome, reflected by a high frequency of bimorphic SINE insertions, provides a unique opportunity to explore the influence of SINEs on the evolution of a mammalian genome. For many thousands of genes, an individual dog will carry two alleles that differ by their content of SINEs. It is therefore possible to assess the impact of SINEs on gene expression patterns within individuals (or even within individual cells) rather than requiring a comparison between multiple individuals or between multiple species. The high frequency of bimorphic SINE insertions in the dog is predicted to provide numerous examples of allele-specific splicing patterns that can be studied further by correlating their potential functional effects with their distribution between dog breeds. Consequently, it is likely that canine bimorphic SINE insertions will provide us with evidence of how insertion elements can mold a mammalian genome, as well as the means to identify genetic relationships between the diverse lineages of current canine populations.
Methods
Loci that are bimorphic for SINEC_Cf insertions between CanSS and CanFam1
The unmasked assembly of a draft boxer genome sequence (CanFam1) was downloaded from the UCSC Genome Bioinformatics site (http://hgdownload.cse.ucsc.edu/downloads.html#dog). The unmasked assembly of a survey-sequenced poodle genome sequence (CanSS) has been described previously (Kirkness et al. 2003). A sequence consisting of bases 1-124 of the SINEC_Cf consensus (RepBase release 7.11) was searched against CanSS using NCBI BLAST (version 2.2.4). The output was filtered for alignments that included at least bases 5-120 of the SINEC_Cf consensus, had no gaps, and had fewer than 11 mismatches. Aligned segments of the survey sequence, together with 50 bases of 5′-sequence, and 175 bases of 3′-sequence, were extracted from the contigs, and masked for canine SINEs and low-complexity sequences using RepeatMasker (version 07/02). The output was filtered for sequence fragments that retained at least 30 consecutive unmasked bases on both flanks of the masked SINEC_Cf sequence. These sequences were then searched against CanFam1 using NCBI BLAST (-W 15, -v 5, -b 5, -F F). In order for a SINEC_Cf to be scored as potentially bimorphic, it was necessary for the flanks of the query to align with only a single fragment of CanFam1, and for these flanks to be contiguous on the homologous CanFam1 fragment. The same approach was used to identify SINEC_Cf repeats in CanFam1 that are absent from CanSS.
Loci that are bimorphic for SINEC_Cf insertions from multiple breeds of dog
Approximately 1 million whole-genome shotgun reads that were derived from nine dogs of different breeds, four wolves, and a coyote were downloaded from the NCBI Trace Archive (ftp://ftp.ncbi.nih.gov/pub/TraceDB). The correlation between “center-_project” IDs (S229-S245) and specific dog breeds was provided by Kerstin Lindblad-Toh (The Broad Institute). Selection of SINEC_Cf repeats and flanking sequences was performed as for CanSS and CanFam1 segments (see above), except that they were restricted to bases 25-675 of each shotgun read (in order to avoid low-quality bases). The filtered sequences were then searched against both CanSS and CanFam1, and scored as described above.
Evidence for SINE-mediate transduction of 3′-flanking sequences
For each of the 92,580 SINEC_Cf-containing segments of CanFam1, 100 bases that flank the 3′-end of the element were masked (RepeatMasker) and searched against the complete collection of 143,080 SINEC_Cf-containing segments from CanFam1 and CanSS using NCBI BLAST. Distinct segments that shared nucleotide identity for >50 consecutive bases were subject to further manual alignment, and annotated for SINEC_Cf sequences, target-site duplications, and transposed 3′-sequences. Potential transduction events were indicated when a target-site duplication (plus 3′-flanking sequence) of one element was contained within the target site duplications of another.
Characterization of SINEC_Cf sequences within dog ESTs
Approximately 155,000 dog ESTs in dbEST (http://www.ncbi.nlm.nih.gov/dbEST/; release 100104) were searched with bases 1-124 of the SINEC_Cf consensus sequence as described above for CanSS and CanFam1. Those ESTs that contained SINEC_Cf sequences were downloaded from dbEST and aligned to CanFam1 using BLAT (http://www.genome.ucsc.edu/cgi-bin/hgBlat) and NCBI BLAST.
Assay for identification of novel loci that are bimorphic for SINEC_Cf insertions
Dog genomic DNA (100 ng; Novagen) was digested with NlaIII in 20 μL, heat-inactivated (65°C, 20 min), and ligated overnight at 20°C in 500 μL with 10,000 U of DNA ligase (New England Biolabs). After phenol-extraction and ethanol-precipitation, ∼20 ng were amplified in 50 μL with 0.5 U of Platinum Taq DNA Polymerase, 0.2 mM each dNTP, 1× buffer, 1.5 mM MgCl2, and 1.2 μM of the following primers: 5′-GGTATCAACGCAGAGTGGCC GCCTCGGCCCTGGGCCAAAGGCAGG, 5′-GGTATCAACG CAGAGTGGCCGCCT, 5′-ATTCTAGAGGCCATTACGGCCTC GAATCCCACRTCRGGCTCCYRG, 5′-ATTCTAGAGGCCATTAC GGCCTCG.
The PCR-amplification was 30 cycles of 95°C (45 sec), 60°C (1 min), and 72°C (2 min). Products of >300 bp were purified from agarose gels using the QIAquick Gel Extraction system (Qiagen), and cloned using the TOPO TA Cloning system (Invitrogen). After transformation, plasmid templates were prepared from white colonies and sequenced using the M13F primer. Seven independent libraries were constructed, and 772 clones were sequenced. High-quality sequence data were obtained for 81%-92% of the clones from each library.
Supplementary Material
Acknowledgments
We thank Jeremy Peterson for assisting with the comparison of GC contents among different genomic regions. We also thank Kristen Gansberger and Shruti Goal for assistance with construction of SINE-enriched libraries of canine genomic fragments.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3765505.
Footnotes
[Supplemental material is available online at www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: Kerstin Lindblad-Toh.]
References
- Bailey, J.A., Liu, G., and Eichler, E.E. 2003. An Alu transposition model for the origin and expansion of human segmental duplications. Am. J. Hum. Genet. 73: 823-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer, M.A. and Deininger, P.L. 2002. Alu repeats and human genomic diversity. Nat. Rev. Genet. 3: 370-379. [DOI] [PubMed] [Google Scholar]
- Bentolila, S., Bach, J.M., Kessler, J.L., Bordelais, I., Cruaud, C., Weissenbach, J., and Panthier, J.J. 1999. Analysis of major repetitive DNA sequences in the dog (Canis familiaris) genome. Mamm. Genome 10: 699-705. [DOI] [PubMed] [Google Scholar]
- Borodulina, O.R. and Kramerov, D.A. 2001. Short interspersed elements (SINEs) from insectivores. Two classes of mammalian SINEs distinguished by A-rich tail structure. Mamm. Genome 12: 779-786. [DOI] [PubMed] [Google Scholar]
- Brosius, J. and Gould, S.J. 1992. On “genomenclature”: A comprehensive (and respectful) taxonomy for pseudogenes and other “junk DNA.” Proc. Natl. Acad. Sci. 89: 10706-10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantrell, M.A., Filanoski, B.J., Ingermann, A.R., Olsson, K., DiLuglio, N., Lister, Z., and Wichman, H.A. 2001. An ancient retrovirus-like element contains hot spots for SINE insertion. Genetics 158: 769-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie-Martin, F., Gonzalez-Acosta, H., Flores, C., Anton-Gamero, M., and Garcia-Nieto, V. 2003. De novo insertion of an Alu sequence in the coding region of the CLCN5 gene results in Dent's disease. Hum. Genet. 113: 480-485. [DOI] [PubMed] [Google Scholar]
- Deininger, P.L. and Batzer, M.A. 1999. Alu repeats and human disease. Mol. Genet. Metab. 67: 183-193. [DOI] [PubMed] [Google Scholar]
- Dewannieux, M., Esnault, C., and Heidmann, T. 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35: 41-48. [DOI] [PubMed] [Google Scholar]
- Eickbush, T.H. 1992. Transposing without ends: The non-LTR retrotransposable elements. New Biol. 4: 430-440. [PubMed] [Google Scholar]
- Ellegren, H. 2000. Microsatellite mutations in the germline: Implications for evolutionary inference. Trends Genet. 16: 551-558. [DOI] [PubMed] [Google Scholar]
- Ganguly, A., Dunbar, T., Chen, P., Godmilow, L., and Ganguly, T. 2003. Exon skipping caused by an intronic insertion of a young Alu Yb9 element leads to severe hemophilia A. Hum. Genet. 113: 348-352. [DOI] [PubMed] [Google Scholar]
- Goodier, J.L., Ostertag, E.M., and Kazazian Jr., H.H. 2000. Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum. Mol. Genet. 9: 653-657. [DOI] [PubMed] [Google Scholar]
- Halling, K.C., Lazzaro, C.R., Honchel, R., Bufill, J.A., Powell, S.M., Arndt, C.A., and Lindor, N.M. 1999. Hereditary desmoid disease in a family with a germline Alu I repeat mutation of the APC gene. Hum. Hered. 49: 97-102. [DOI] [PubMed] [Google Scholar]
- Han, J.S., Szak, S.T., and Boeke, J.D. 2004. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature 429: 268-274. [DOI] [PubMed] [Google Scholar]
- Janicic, N., Pausova, Z., Cole, D.E., and Hendy, G.N. 1995. Insertion of an Alu sequence in the Ca2+-sensing receptor gene in familial hypocalciuric hypercalcemia and neonatal severe hyperparathyroidism. Am. J. Hum. Genet. 56: 880-886. [PMC free article] [PubMed] [Google Scholar]
- Jurka, J. 1997. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc. Natl. Acad. Sci. 94: 1872-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass, D.H., Raynor, M.E., and Williams, T.M. 2000. Evolutionary history of B1 retroposons in the genus Mus. J. Mol. Evol. 51: 256-264. [DOI] [PubMed] [Google Scholar]
- Kirkness, E.F., Bafna, V., Halpern, A.L., Levy, S., Remington, K., Rusch, D.B., Delcher, A.L., Pop, M., Wang, W., Fraser, C.M., et al. 2003. The dog genome: Survey sequencing and comparative analysis. Science 301: 1898-1903. [DOI] [PubMed] [Google Scholar]
- Koskinen, M.T. 2003. Individual assignment using microsatellite DNA reveals unambiguous breed identification in the domestic dog. Anim. Genet. 34: 297-301. [DOI] [PubMed] [Google Scholar]
- Krull, M., Brosius, J., and Schmitz, J. 2005. Alu-SINE exonization: En route to protein-coding function. Mol. Biol. Evol. 22: 1702-1711. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Lev-Maor, G., Sorek, R., Shomron, N., and Ast, G. 2003. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science 300: 1288-1291. [DOI] [PubMed] [Google Scholar]
- Lin, L., Faraco, J., Li, R., Kadotani, H., Rogers, W., Lin, X., Qiu, X., de Jong, P.J., Nishino, S., and Mignot, E. 1999. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin orexin receptor 2 gene. Cell 98: 365-376. [DOI] [PubMed] [Google Scholar]
- Makalowski, W. 2000. Genomic scrap yard: How genomes utilize all that junk. Gene 259: 61-67.11163962 [Google Scholar]
- Mamedov, I.Z., Arzumanyan, E.S., Amosova, A.L., Lebedev, Y.B., and Sverdlov, E.D. 2005. Whole-genome experimental identification of insertion/deletion polymorphisms of interspersed repeats by a new general approach. Nucleic Acids Res. 33: e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnick, M.F., Stillwell, L.C., Heineman, J.M., and Stiegler, G.L. 1992. A highly repetitive DNA sequence possibly unique to canids. Gene 110: 235-238. [DOI] [PubMed] [Google Scholar]
- Muratani, K., Hada, T., Yamamoto, Y., Kaneko, T., Shigeto, Y., Ohue, T., Furuyama, J., and Higashino, K. 1991. Inactivation of the cholinesterase gene by Alu insertion: Possible mechanism for human gene transposition. Proc. Natl. Acad. Sci. 88: 11315-11319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustajoki, S., Ahola, H., Mustajoki, P., and Kauppinen, R. 1999. Insertion of Alu element responsible for acute intermittent porphyria. Hum. Mutat. 13: 431-438. [DOI] [PubMed] [Google Scholar]
- Ohshima, K., Hamada, M., Terai, Y., and Okada, N. 1996. The 3′ ends of tRNA-derived short interspersed repetitive elements are derived from the 3′ ends of long interspersed repetitive elements. Mol. Cell. Biol. 16: 3756-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, H.G., Kim, L.V., Sutter, N.B., Carlson, S., Lorentzen, T.D., Malek, T.B., Johnson, G.S., DeFrance, H.B., Ostrander, E.A., and Kruglyak, L. 2004. Genetic structure of the purebred domestic dog. Science 304: 1160-1164. [DOI] [PubMed] [Google Scholar]
- Pele, M., Tiret, L., Kessler, J.L., Blot, S., and Panthier, J.J. 2005. SINE exonic insertion in the PTPLA gene leads to multiple splicing defects and segregates with the autosomal recessive centronuclear myopathy in dogs. Hum. Mol. Genet. 14: 1417-1427. [DOI] [PubMed] [Google Scholar]
- Pickeral, O.K., Makalowski, W., Boguski, M.S., and Boeke, J.D. 2000. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 10: 411-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy-Engel, A.M., Carroll, M.L., El-Sawy, M., Salem, A.H., Garber, R.K., Nguyen, S.V., Deininger, P.L., and Batzer, M.A. 2002. Non-traditional Alu evolution and primate genomic diversity. J. Mol. Biol. 316: 1033-1040. [DOI] [PubMed] [Google Scholar]
- Rubin, C.M., Kimura, R.H., and Schmid, C.W. 2002. Selective stimulation of translational expression by Alu RNA. Nucleic Acids Res. 30: 3253-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem, A.H., Ray, D.A., and Batzer, M.A. 2005. Identity by descent and DNA sequence variation of human SINE and LINE elements. Cytogenet. Genome Res. 108: 63-72. [DOI] [PubMed] [Google Scholar]
- Savolainen, P., Zhang, Y.P., Luo, J., Lundeberg, J., and Leitner, T. 2002. Genetic evidence for an East Asian origin of domestic dogs. Science 298: 1610-1613. [DOI] [PubMed] [Google Scholar]
- Schmid, C.W. 1996. Alu: Structure, origin, evolution, significance and function of one-tenth of human DNA. Prog. Nucleic Acid Res. Mol. Biol. 53: 283-319. [DOI] [PubMed] [Google Scholar]
- ———. 1998. Does SINE evolution preclude Alu function? Nucleic Acids Res. 26: 4541-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurgardottir, S., Helgason, A., Gulcher, J.R., Stefansson, K., and Donnelly, P. 2000. The mutation rate in the human mtDNA control region. Am. J. Hum. Genet. 66: 1599-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek, R., Ast, G., and Graur, D. 2002. Alu-containing exons are alternatively spliced. Genome Res. 12: 1060-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukarova, E., Dimovski, A.J., Tchacarova, P., Petkov, G.H., and Efremov, G.D. 2001. An Alu insert as the cause of a severe form of hemophilia A. Acta Haematol. 106: 126-129. [DOI] [PubMed] [Google Scholar]
- Sutter, N.B., Eberle, M.A., Parker, H.G., Pullar, B.J., Kirkness, E.F., Kruglyak, L., and Ostrander, E.A. 2004. Extensive and breed-specific linkage disequilibrium in Canis familiaris. Genome Res. 14: 2388-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo, Z., Levi-Minzi, S.A., Christiano, A.M., Struminger, C., Stoneking, M., Batzer, M.A., and Boyd, C.D. 1999. Sequential loss of two neighboring exons of the tropoelastin gene during primate evolution. J. Mol. Evol. 49: 664-671. [DOI] [PubMed] [Google Scholar]
- Vassetzky, N.S. and Kramerov, D.A. 2002. CAN—A pan-carnivore SINE family. Mamm. Genome 13: 50-57. [DOI] [PubMed] [Google Scholar]
- Venter, J., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- Vervoort, R., Gitzelmann, R., Lissens, W., and Liebaers, I. 1998. A mutation IVS8+0.6kbdelTC creating a new donor splice site activates a cryptic exon in an Alu-element in intron 8 of the human β-glucuronidase gene. Hum. Genet. 103: 686-693. [DOI] [PubMed] [Google Scholar]
- Vidaud, D., Vidaud, M., Bahnak, B.R., Siguret, V., Gispert Sanchez, S., Laurian, Y., Meyer, D., Goossens, M., and Lavergne, J.M. 1993. Haemophilia B due to a de novo insertion of a human-specific Alu subfamily member within the coding region of the factor IX gene. Eur. J. Hum. Genet. 1: 30-36. [DOI] [PubMed] [Google Scholar]
- Vila, C., Savolainen, P., Maldonado, J.E., Amorim, I.R., Rice, J.E., Honeycutt, R.L., Crandall, K.A., Lundeberg, J., and Wayne, R.K. 1997. Multiple and ancient origins of the domestic dog. Science 276: 1687-1689. [DOI] [PubMed] [Google Scholar]
- Wallace, M.R., Andersen, L.B., Saulino, A.M., Gregory, P.E., Glover, T.W., and Collins, F.S. 1991. A de novo Alu insertion results in neurofibromatosis type 1. Nature 353: 864-866. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Watkins, W.S., Rogers, A.R., Ostler, C.T., Wooding, S., Bamshad, M.J., Brassington, A.M., Carroll, M.L., Nguyen, S.V., Walker, J.A., Prasad, B.V., et al. 2003. Genetic variation among world populations: Inferences from 100 Alu insertion polymorphisms. Genome Res. 13: 1607-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web site references
- ftp://ftp.ncbi.nih.gov/pub/TraceDB; NCBI site for downloading traces.
- http://genome.ucsc.edu/cgi-bin/hgTables; UCSC site for querying genome sequences.
- http://hgdownload.cse.ucsc.edu/downloads.html#dog; UCSC site for downloading sequence data.
- http://www.ensembl.org/Canis_familiaris; Ensembl site for annotation of dog genome.
- http://www.genome.gov/12511476; NHGRI links to draft genome sequence.
- http://www.ncbi.nlm.nih.gov/dbEST; NCBI site for ESTs.
- http://www.genome.ucsc.edu/cgi-bin/hgBlat; UCSC site for rapid identification of DNA sequence similarity.
- http://genome.ucsc.edu; University of California, Santa Cruz site contains the reference sequence and working draft assemblies for a large collection of genomes.
- http://www.ncbi.nih.gov/Genbank/; National Institutes of Health site contains an annotated collection of all publicly available DNA sequences.
- http://www.ncbi.nlm.nih.gov/Traces/; National Center for Biotechnology Information site contains an archive of DNA sequence traces.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.