Abstract
Eukaryotic initiation factor 4E (eIF4E) binding proteins (4E-BPs) regulate the assembly of initiation complexes required for cap-dependent mRNA translation. 4E-BP1 undergoes insulin-stimulated phosphorylation, resulting in its release from eIF4E, allowing initiation complex assembly. 4E-BP1 undergoes caspase-dependent cleavage in cells undergoing apoptosis. Here we show that cleavage occurs after Asp24, giving rise to the N-terminally truncated polypeptide Δ4E-BP1, which possesses the eIF4E-binding site and all the known phosphorylation sites. Δ4E-BP1 binds to eIF4E and fails to become sufficiently phosphorylated upon insulin stimulation to bring about its release from eIF4E. Therefore, Δ4E-BP1 acts as a potent inhibitor of cap-dependent translation. Using a mutagenesis approach, we identify a novel regulatory motif of four amino acids (RAIP) which lies within the first 24 residues of 4E-BP1 and which is necessary for efficient phosphorylation of 4E-BP1. This motif is conserved among sequences of 4E-BP1 and 4E-BP2 but is absent from 4E-BP3. Insulin increased the phosphorylation of 4E-BP3 but not sufficiently to cause its release from eIF4E. However, a chimeric protein that was generated by replacing the N terminus of 4E-BP3 with the N-terminal sequence of 4E-BP1 (containing this RAIP motif) underwent a higher degree of phosphorylation and was released from eIF4E. This suggests that the N-terminal sequence of 4E-BP1 is required for optimal regulation of 4E-BPs by insulin.
Control of mRNA translation plays a pivotal role in regulating gene expression under a wide variety of conditions in mammalian cells (reviewed in reference 27). The principal control point in mRNA translation is the initiation process, which is largely regulated through changes in the states of phosphorylation of proteins (eukaryotic initiation factors [eIFs] and other components) involved in this process (for reviews see references 11, 31, 34). One of the main regulatory steps of translation initiation involves the formation of eIF4F complexes, which are required to recruit ribosomal subunits to the mRNA during cap-dependent initiation (reviewed in reference 32). The cap contains a 7-methylguanosine triphosphate moiety linked through a 5′-5′-phosphodiester bond to the 5′ end of the mRNA. eIF4E, one component of eIF4F, binds directly to the cap. eIF4E also interacts with the scaffold protein eIF4G, which in turn binds RNA helicase eIF4A to form the eIF4F complex (32). eIF4G also interacts with the poly(A)-binding protein and the eIF4E kinases, Mnk1 and Mnk2 (reviewed in reference 11; see also reference 35).
The interaction between eIF4E and eIF4G is modulated by the association of eIF4E-binding proteins (4E-BPs) with eIF4E. 4E-BPs and eIF4G interact with eIF4E at overlapping binding sites (13, 23). 4E-BPs therefore compete with eIF4G for binding to eIF4E and act as inhibitors of eIF4F complex formation. Of the three known 4E-BPs (4E-BP1, -2, and -3), 4E-BP1 is the best characterized. 4E-BP1 undergoes phosphorylation at five well-characterized S/T-P sites (Thr36, -45, and -69 and Ser64 and -82 [numbering according to the rat sequence]) in response to agents that activate mRNA translation, e.g., insulin (reviewed in references 11, 21). Phosphorylation of these sites leads to the release of 4E-BP1 from eIF4E. However, phosphorylation of sites proximal to the eIF4E-binding motif are considered to be of particular importance in regulating this dissociation of 4E-BP1 (i.e., Thr45 and Ser64) (10, 29, 30). 4E-BP1 phosphorylation in vivo is blocked by the immunosuppressant drug rapamycin, indicating that it requires the activity of mTOR (mammalian target of rapamycin). Although mTOR can phosphorylate 4E-BP1 in vitro, it is not clear that it is responsible for this in vivo (2, 3, 29, 40). 4E-BP2 and 4E-BP3 each contain homologous T/S-P sites that correspond to the phosphorylation sites in 4E-BP1. The available data suggest that 4E-BP2 phosphorylation may be regulated in a manner similar to that of 4E-BP1 and that it is also dependent on mTOR (22). The regulation of 4E-BP3 has not so far been reported.
During apoptosis protein synthesis is inhibited and several translation factors are cleaved in a caspase-dependent manner (4-8, 25, 26, 28, 38). In some cases, cleavage is known to impair or otherwise alter the functions of the factors (5, 25). We have recently shown that prior to apoptosis, in response, e.g., to DNA damage, 4E-BP1 and other targets of mTOR signaling undergo dephosphorylation. This leads to the increased binding of 4E-BP1 to eIF4E and loss of eIF4F complexes (38, 39). This is thought to inhibit cap-dependent translation while allowing the translation of mRNAs containing internal ribosome entry sites (IRESs), whose translation is cap independent. The translation of mRNAs encoding a number of proteins involved during apoptosis is thought to be regulated through an IRES (9, 15, 17, 18, 37), allowing the ongoing synthesis of these proteins during apoptosis.
Following caspase activation, 4E-BP1 undergoes cleavage to generate two fragments, one of which retains the ability to sequester eIF4E (6, 38). Here we identify the site of cleavage and show that the major cleavage product potently inhibits the upregulation of the cap-dependent translation that normally occurs during insulin stimulation. Although this fragment contains all the sites of phosphorylation, it undergoes a lower level of phosphorylation than full-length 4E-BP1 in response to insulin. This suggests that the cleaved N-terminal fragment contains features important for the phosphorylation of 4E-BP1. We localize this regulatory element to a sequence of four amino acids (RAIP motif) which is conserved among the sequences of 4E-BP1 and 4E-BP2 but which is absent from 4E-BP3. Insulin fails to induce a sufficient level of 4E-BP3 phosphorylation to facilitate its release from eIF4E. However, a 4E-BP1/4E-BP3 chimera which has the N-terminal end of 4E-BP3 replaced by the N terminus of 4E-BP1, containing the RAIP motif, became more highly phosphorylated, causing its dissociation from eIF4E upon insulin stimulation. This indicates that the extreme N-terminal sequence of 4E-BP1 containing the RAIP element plays an important role in the regulation of the 4E-BPs by insulin.
MATERIALS AND METHODS
Chemicals and biochemicals.
Staurosporine was obtained from Sigma, and oligonucleotides were synthesized by MWG Biotech Inc. [32P]orthophosphate was purchased from Amersham Corp., and Microcystin-LR was supplied by Calbiochem. All other reagents were purchased as previously described (38).
Plasmids.
PCR-directed fragments encoding rat 4E-BP1 and human 4E-BP3 were digested with HindIII and BamHI and cloned into pcDNA3.1myc/3his− (from Invitrogen) to generate pcDNA3.1 expressing 4E-BP1 and 4E-BP3, respectively, containing C-terminal Myc and His tags. In a similar manner, pcDNA3.1 plasmids encoding 8Δ/4E-BP1, 16Δ/4E-BP1, and 24Δ/4E-BP1 were made. Site-directed mutagenesis was carried out according to the manufacturer's instructions (by using QuikChange; Stratagene) to make an array of 4E-BP1 and 4E-BP3 point mutants.
A KpnI restriction site was introduced into the coding sequencefor 4E-BP1 between nucleotides 115 and 120 and into the 4E-BP3 coding sequence between nucleotides 75 and 80 without altering the sequence of the encoded protein to generate pcDNA3.1(4E-BP1 KpnI) and pcDNA(4E-BP3 KpnI), respectively, by site-directed mutagenesis (see above). pcDNA3.1(4E-BP3 KpnI) was digested with KpnI and HindIII to obtain a cDNA encoding the C terminus of 4E-BP3 (185 nucleotides) and the linearized pcDNA3.1myc/3his− vector containing the coding sequence for the N terminus of 4E-BP3. Both restriction digest products were subcloned into pcDNA3.1(4E-BP1 KpnI) digested with KpnI and HindIII to obtain pcDNA3.1 expressing either nBP1/cBP3 or nBP3/cBP1.
For bacterial expression, cDNAs for both 4E-BP1 and 16Δ/4E-BP1 were subcloned into pET21a+ (from Novagene) digested with NdeI and XhoI to generate pET21a+ expressing 4E-BP1 and 16Δ/4E-BP1, respectively, with His tags at the C termini.
Cell culture and cytotoxic treatment.
Swiss 3T3 cells were treated and maintained as described before (38), and HEK293 cells were handled similarly. Transient transfections on HEK293 cells (8 μg of DNA per 10-cm2 plate) were performed as described previously (14) with the following modifications: air was bubbled into 2× HEPES-buffered saline by using a drawn-out glass Pasteur pipette during the slow addition of the CaCl2-DNA mixture. Cells were extracted as previously described (38).
Assays to determine translation factor phosphorylation and association.
Phosphorylation of 4E-BP1 was assessed as previously described (38) (using 20 μg of protein). Anti-4E-BP1 antiserum or 4E-BP1 phospho-specific antibodies (detecting phosphorylation at Thr36 and/or Thr45 [Thr36/45], Ser64, or Thr69; New England Biolabs) were used. To assess the phosphorylation state of p70 S6 kinase, mobility shift gel assays were performed by resolving samples on a 10 acrylamide-0.1% bisacrylamide gel, and Western blots were developed as described earlier (38). Affinity chromatography on m7GTP-Sepharose was used to isolate eIF4E and its associated proteins from cell lysates (200 μg of protein). The proteins bound to eIF4E were analyzed as described earlier (38). 4E-BP3 phosphorylation was determined by immunoprecipitating Myc-tagged 4E-BP3 from transiently transfected HEK293 cell lysates (400 μg of protein) with an anti-Myc antibody (clone 9E10; Sigma). These samples were subjected to the Immobilized pH Gradients (IPGphor) isoelectric focusing system from Amersham Pharmacia Biotech using the IPGphor 7-cm strip with a pH range of 4 to 7, according to the manufacturer's protocol. Precast Novex (pH 4 to 10) ZOOM gels were used for the second dimension. 4E-BP3 was transferred to Immobilon using the Novex transfer system. In vivo 32P radiolabeling was carried out by serum starving transiently transfected HEK293 cells overnight in medium lacking phosphate. These cells were treated and pulsed with 1 mCi of [32P]orthophosphate for 2 h. His-tagged 4E-BP1 (or mutants) was purified from cell extracts on Ni-agarose and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography. Quantification of incorporation of the 32P label was carried out by measuring the radioactivity associated with the overexpressed protein after excision from the SDS-PAGE gel.
Protein purification.
BL21DE3plysS cells were transformed with pET4E-BP1 or pETΔ16/BP1 individually. Two hundred fifty milliliters of culture was grown until the optical density at 600 nM reached 0.6, and then expression was induced with 0.5 mM IPTG for 3 h at 30°C. The culture was pelleted and resuspended in lysis buffer containing 20 mM Tris, pH 7.5, 1 M NaCl, 5% (vol/vol) glycerol, 10 mM imidazole, 5 mM β-mercaptoethanol, 0.1% (vol/vol) Triton X-100, and protease inhibitors (complete; Roche) diluted to 1×. The culture was lysed by one freeze-thaw cycle and sonicated four times for 30-s intervals. The debris was removed by centrifugation at 12,000 rpm for 15 min (using a Beckman Centrifuge JA25.50 rotor). The carboxy-terminal His-tagged 4E-BP1 and Δ16/BP1 proteins were purified on Ni-nitrilotriacetic acid (NTA) agarose beads, which were washed three times in lysis buffer containing 20 mM imidazole and then eluted with lysis buffer containing 500 mM imidazole to a final volume of 0.5 ml. The eluted proteins were dialyzed overnight against the mTOR assay buffer (see below). The in vitro mTOR kinase assay was carried out as previously described (19) on 1 μg of either recombinant 4E-BP1his or Δ16/BP1his.
Cap-dependent translation assay.
HEK293 cells were cotransfected with an array of pcDNA3.1myc/his vectors and a bicistronic construct containing the green fluorescent protein (GFP) and chloramphenicol acetyltransferase (CAT) reporter genes (provided by A. A. M. Thomas, University of Utrecht). Cells were serum starved overnight in medium lacking l-methionine and pulsed with 20 μCi of [35S]methionine for 2 h after stimulation. GFP and CAT proteins were immunoprecipitated from the cell extracts (300 μg of protein) by using anti-GFP (from Sigma) and anti-CAT (from 5-Prime-3-Prime). The samples were resolved on an SDS-polyacrylamide gel containing 12.5% acrylamide, which was stained, fixed, washed with Amplify (Amersham Pharmacia Biotech) and then subjected to fluorography to detect the incorporation of [35S]methionine into both GFP and CAT.
RESULTS
Identification of the caspase-dependent cleavage site of 4E-BP1.
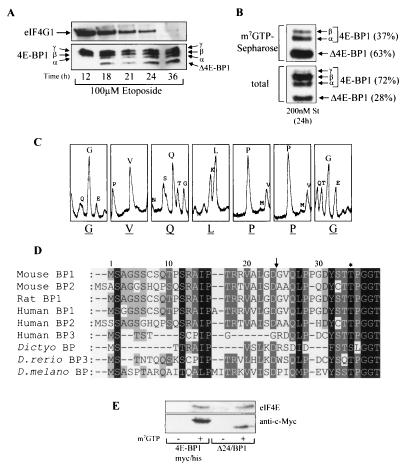
Our previous studies have shown that the treatment of cells with potent inducers of apoptosis, either DNA-damaging agents (38) or staurosporine (39), leads to the cleavage of 4E-BP1 to produce a product termed Δ4E-BP1 and that this is blocked by a broad-spectrum caspase inhibitor, Z-VAD.FMK. It has also been shown that eIF4G1 is cleaved in a caspase-dependent manner during apoptosis (4-6, 8, 26). The time courses of both eIF4G1 and 4E-BP1 cleavage in Swiss 3T3 cells undergoing apoptosis induced by etoposide, a DNA-damaging agent, were analyzed (Fig. 1A). The amount of full-length eIF4G1, measured in total-cell lysates, gradually decreased during apoptosis. However, eIF4G1 was still present in cells after 24 h of etoposide treatment, albeit at reduced levels compared to the level at 12 h. Complete loss of intact eIF4G1 was apparent by 36 h. 4E-BP1 underwent cleavage to yield a product termed Δ4E-BP1 after 18 h of etoposide treatment.
FIG. 1.
Identification of the caspase-dependent cleavage of 4E-BP1. (A) Swiss 3T3 cells were treated with etoposide for the times indicated. The samples were analyzed for total eIF4G1 (top) and 4E-BP1 (bottom). The α-, β-, and γ-species of 4E-BP1 and cleavage product Δ4E-BP1 are labeled. (B) Swiss 3T3 cells were treated with staurosporine (St) for 24 h, and extracts were subjected to affinity chromatography on m7GTP-Sepharose (see Materials and Methods). The percentages of uncleaved 4E-BP1 (γ, β, and α) and cleaved (Δ4E-BP1) 4E-BP1 bound to eIF4E (top) and total levels present within cell extracts (bottom) were quantified by densitometry (National Institutes of Health Image, version 1.61). (C) Δ4E-BP1 (isolated as described for panel B) was excised from the membrane and subjected to Edman degradation, yielding N-terminal sequence GVQLPPG. (D) N-terminal alignment of 4E-BPs. Arrow, cleavage site; ∗, phosphorylation site at Thr36 in mouse and rat 4E-BP1 and at Thr37 in human 4E-BP1. 4E-BP1 homologues from D. discoideum (Dictyo), D. rerio, and D. melanogaster (D. melano) are also shown. (E) Cell lysates prepared from serum-starved HEK293 cells overexpressing either 4E-BP1myc/his or Δ24/BP1 (see Fig. 2A for details of these mutants) were subjected to affinity chromatography using either m7GTP-Sepharose (+) or Sepharose (CL-4B; −). The levels of purified recombinant 4E-BP1 and eIF4E were analyzed.
Δ4E-BP1 has previously been shown to bind to eIF4E in apoptotic cells (6, 38). To study this further, eIF4E was purified by affinity chromatography on m7GTP-Sepharose from extracts of Swiss 3T3 cells that had been treated with staurosporine (Fig. 1B, top). The majority of the 4E-BP1 bound to eIF4E was in the cleaved state (63% of the total; Fig. 1B, top). Analysis of 4E-BP1 within these cells revealed that Δ4E-BP1 was 28% of the total (Fig. 1B, bottom). These data suggest that Δ4E-BP1 bound preferentially to eIF4E compared to the uncleaved 4E-BP1 within apoptotic cells.
To study further the properties of Δ4E-BP1, it was necessary to identify the caspase-dependent cleavage site. To do this, Swiss 3T3 cells were treated with staurosporine and Δ4E-BP1 was copurified with eIF4E on m7GTP-Sepharose and then subjected to N-terminal sequence analysis (Fig. 1C). This revealed that 4E-BP1 was cleaved after Asp24 at an atypical caspase cleavage motif, ALGD. The scissile bond (Asp-Gly) and adjacent residues are fully conserved in rat and mouse 4E-BP1 sequences and partially conserved in human 4E-BP1 (VLGD) but absent from either 4E-BP2 or 4E-BP3 (Fig. 1D). Given that Δ4E-BP1 is recognized by our antiserum raised against the peptide ESQFEMDI, which corresponds to the extreme C terminus of 4E-BP1, Δ4E-BP1 must contain an intact C terminus. Δ4E-BP1 therefore lacks the first 24 residues of the full-length protein. As a control to confirm that the interaction of Δ4E-BP1 with eIF4E (Fig. 1B) was specific and not due to its binding to the Sepharose resin, affinity chromatography was carried out using either m7GTP-Sepharose or unmodified Sepharose beads (Fig. 1E). Δ24/BP1 (a truncation mutant that mimics Δ4E-BP1; for more detail see Fig. 2A) was not retained on Sepharose itself, showing that the binding of Δ24/BP1 to m7GTP-Sepharose is specific and presumably facilitated through its interaction with eIF4E.
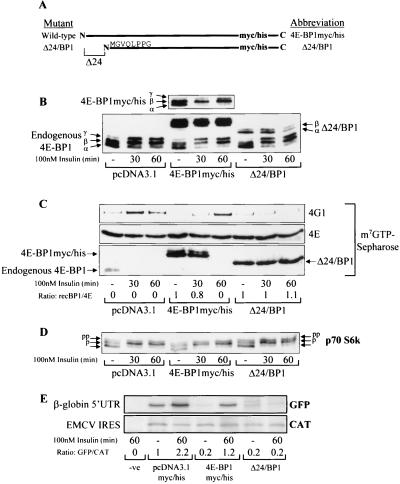
FIG. 2.
The cleaved fragment of 4E-BP1 (Δ24/BP1) acts as a dominant inhibitor of eIF4F complex assembly and cap-dependent translation. (A) Diagrammatic representation of mutants 4E-BP1myc/his and Δ24/BP1. The amino acid sequence indicates the N-terminus of Δ24/BP1. (B) HEK293 cells were transiently transfected with the empty vector or pcDNA3.1myc/his− expressing either 4E-BP1myc/his or Δ24/BP1, as indicated. These cells were serum starved and treated for 30 or 60 min (as indicated) with insulin. Cell extracts were analyzed to assess 4E-BP1 phosphorylation. 4E-BP1, endogenous 4E-BP1 phosphorylation; 4E-BP1myc/his (top), shorter exposure of the overexpressed 4E-BP1myc/his. (C) The cell extracts from panel B were subjected to affinity chromatography on m7GTP-Sepharose (see Materials and Methods). The amounts of eIF4G1 (4G1), eIF4E (4E), endogenous 4E-BP1 (4E-BP1), 4E-BP1myc/his, and Δ24/BP1 were analyzed. The ratios of recombinant 4E-BP1 bound to eIF4E (recBP1/4E) were assessed by densitometry (National Institutes of Health [NIH] Image, version 1.61). (D) p70 S6 kinase phosphorylation was analyzed. Arrows, differently phosphorylated species (pp, most-phosphorylated species; lowest arrow, least-phosphorylated species). (E) A bicistronic vector containing both a GFP reporter gene downstream from the β-globin 5′ untranslated region (UTR) and a CAT reporter gene downstream from the encephalomyocarditis virus (EMCV) IRES was cotransfected into HEK293 cells as indicated. Lane −ve, no-DNA control. Cell extracts were analyzed for cap-dependent translation as described in Materials and Methods. The ratios of the amounts of [35S]methionine incorporated into GFP and CAT were determined by densitometry (NIH Image, version 1.61); these ratios were normalized against the empty-vector serum-starved control (value set at 1).
Δ4E-BP1 potently inhibits eIF4F complex assembly.
A 4E-BP1 mutant lacking residues 1 to 24 and with C-terminal Myc and His tags (Δ24/BP1; see Materials and Methods) was generated to mimic Δ4E-BP1. Full-length 4E-BP1 with C-terminal Myc and His tags (4E-BP1myc/his) was also generated for use as a control. 4E-BP1myc/his and Δ24/BP1 were overexpressed in HEK293 cells, and these cells were treated as indicated in the legend for Fig. 2. As judged by immunoblotting, the level of expression of Δ24/BP1 was considerably lower than that of 4E-BP1myc/his and slightly lower than that of endogenous 4E-BP1 (Fig. 2B). Both endogenous 4E-BP1 and 4E-BP1myc/his became more highly phosphorylated after insulin treatment, as indicated by increased proportions of the upper hyperphosphorylated γ-isoform and the loss of the least-phosphorylated α-species. Δ24/BP1 underwent only partial phosphorylation, with a modest increase in the more slowly migrating form. The levels of 4E-BP1 and eIF4G1 bound to eIF4E were then compared (Fig. 2C). 4E-BP1myc/his competed with the endogenous 4E-BP1 for binding to eIF4E. Insulin treatment, as expected, caused a time-dependent dissociation of 4E-BP1myc/his from eIF4E, allowing consequent association of eIF4G1 with eIF4E. This indicates that the response of 4E-BP1myc/his to insulin is similar to that of endogenous 4E-BP1. 4E-BP1myc/his completely dissociated from eIF4E by 60 min. However, this was slower than the dissociation of endogenous 4E-BP1, probably reflecting the greater total amount of 4E-BP1 within these cells that must be phosphorylated for this to occur. In contrast, Δ24/BP1 was not released from eIF4E, i.e., the ratio of Δ24/BP1 to eIF4E did not decrease with insulin. As a consequence, Δ24/BP1 inhibited eIF4F complex assembly even though its expression levels are lower than those for 4E-BP1myc/his.
Since the endogenous 4E-BP1 still became phosphorylated in these cells (Fig. 2B), it is unlikely that Δ24/BP1 inhibits signaling through mTOR. To confirm this, the regulation of p70 S6 kinase (another target of mTOR signaling) was also examined. The activity of p70 S6 kinase is regulated by phosphorylation: more highly phosphorylated forms exhibit higher kinase activity and migrate more slowly during SDS-PAGE. Insulin treatment caused similar shifts to the more highly phosphorylated isoforms of p70 S6 kinase in cells expressing Δ24/BP1 and in cells transiently transfected with the empty vector (Fig. 2D), confirming that Δ24/BP1 does not inhibit mTOR signaling.
Δ24/BP1 inhibits cap-dependent translation.
Since Δ24/BP1 blocked insulin-stimulated eIF4F assembly, it was anticipated that, in insulin-stimulated cells, it would specifically inhibit cap-dependent translation, while 4E-BP1myc/his would not. To study this, HEK293 cells were cotransfected with a bicistronic vector and the 4E-BP1 constructs. The bicistronic vector contained both a GFP reporter gene downstream from the β-globin gene 5′ untranslated region, to monitor cap-dependent mRNA translation, and a CAT reporter gene downstream from a picornavirus (encephalomyocarditis virus) IRES. The latter monitors cap-independent mRNA translation (Fig. 2E). Overexpression of either 4E-BP1myc/his or Δ24/BP1 inhibited the basal level of cap-dependent mRNA translation, compared to that in cells transiently transfected with the empty vector (observed as a fivefold decrease in [35S]methionine incorporation into GFP). Incorporation into GFP increased significantly and by similar amounts after insulin treatment in cells transfected with either the empty vector or 4E-BP1myc/his, demonstrating an increase in cap-dependent mRNA translation in both cases. In contrast, no such increase was observed in cells overexpressing Δ24/BP1 (ratio of 0.2, sixfold less than that observed with 4E-BP1myc/his). The levels of incorporation into CAT for cells expressing 4E-BP1myc/his and those expressing Δ24/BP1 did not differ significantly, suggesting that these two proteins do not interfere with IRES-driven (cap-independent) mRNA translation, as expected, and reported earlier for 4E-BP1 (1).
Δ24/BP1 shows reduced phosphorylation upon insulin stimulation.
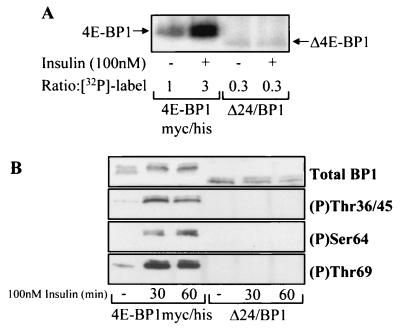
The data in Fig. 2 suggested that all or part of the N-terminal region removed from Δ4E-BP1 is necessary for the full regulation of phosphorylation of 4E-BP1 in response to insulin. To verify this, we compared the levels of incorporation of radiolabel into 4E-BP1myc/his and Δ24/BP1 in insulin-treated cells. HEK293 cells expressing either 4E-BP1myc/his or Δ24/BP1 were metabolically labeled with [32P]orthophosphate and treated with insulin. Insulin caused a threefold increase in incorporation of the 32P radiolabel into 4E-BP1myc/his (Fig. 3A). In contrast, insulin failed to increase the extent of incorporation of the 32P radiolabel into Δ24/BP1.
FIG. 3.
The cleaved form of 4E-BP1 does not undergo efficient insulin-stimulated phosphorylation. (A) HEK293 cells overexpressing either 4E-BP1myc/his or Δ24/BP1. In vivo radiolabeling was carried out as described in Materials and Methods and 32P label incorporation into 4E-BP1 was quantified ([32P]-label; the 4E-BP1myc/his serum-starved-only sample is standardized to 1). (B) HEK293 cells overexpressing either 4E-BP1myc/his or Δ24/BP1 at comparable levels were treated as indicated. The extracts were analyzed for total 4E-BP1 levels and phosphorylation of 4E-BP1 at Thr36 and 45, Ser64, and Thr69 as indicated.
4E-BP1 undergoes phosphorylation on at least five sites (11) in vivo. To investigate the phosphorylation of individual sites in 4E-BP1myc/his and Δ24/BP1, we employed the available phospho-specific antibodies that detect 4E-BP1 when phosphorylated at Thr36/45, Ser64, or Thr69 (29). When the same cell extracts used in Fig. 2B were analyzed, the phosphorylation of Ser64 coincided with the release of 4E-BP1myc/his from eIF4E, which became maximal at 1 h (data not shown). This indicates that phosphorylation at this site is important for the release of 4E-BP1 from eIF4E. It is interesting that Ser64 phosphorylation is only observed for the γ-species, which is not able to bind eIF4E. This is consistent with recent data indicating that phosphorylation of Ser64 weakens the binding of 4E-BP1 to eIF4E (20). Because the expression levels of Δ24/BP1 were much lower than those of 4E-BP1myc/his, the experiment was repeated under altered conditions. A reduced amount of the 4E-BP1myc/his expression vector was used during the transient transfections so that both 4E-BP1myc/his and Δ24/BP1 were expressed in HEK293 cells to similar degrees for better comparison of their phosphorylation (Fig. 3B). In all cases, there was no detectable phosphorylation of Δ24/BP1 at Thr36 and 45, Ser64, or Thr69, while this was clearly observed for 4E-BP1myc/his. Longer exposure of the Western blot, when using the Thr69 phospho-specific antibody, revealed a trace of Thr69 phosphorylation (data not shown), indicating that the phosphorylation of Δ24/BP1 in response to insulin is substantially, but not completely, impaired.
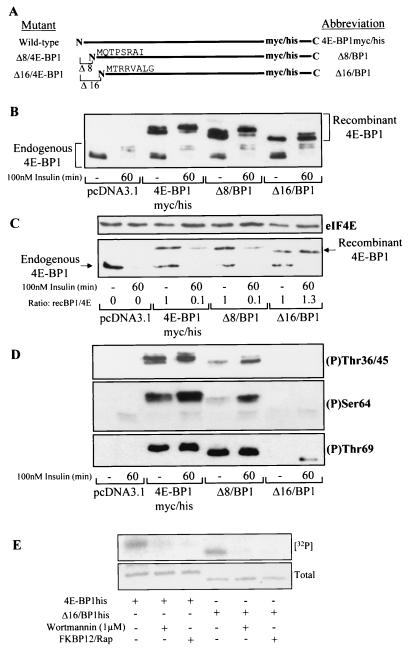
The region between residues 9 and 16 in 4E-BP1 is required for efficient phosphorylation.
The N terminus of 4E-BP1 thus appears to be essential for the regulation of phosphorylation at Thr36/45, Ser64, and Thr69 in vivo. To identify the region necessary for these phosphorylation events, thus allowing release from eIF4E, two progressively shorter truncations were generated (Δ8/BP1, lacking residues 1 to 8, and Δ16/BP1, lacking residues 1 to 16; Fig. 4A). The total levels of these overexpressed 4E-BP1 peptides were similar (Fig. 4B). Δ16/BP1 was phosphorylated less well in response to insulin treatment (Fig. 4B) than 4E-BP1myc/his and Δ8/BP1, as manifested by the decreased proportion undergoing a mobility shift from the faster-migrating hypophosphorylated isoforms (α) to the slower-migrating hyperphosphorylated isoforms (β and γ). Association of the truncated 4E-BP1 polypeptides with eIF4E was analyzed (Fig. 4C). In all cases, the overexpressed 4E-BP1 proteins competed with endogenous 4E-BP1 for binding to eIF4E. 4E-BP1myc/his and Δ8/BP1 each dissociated from eIF4E after insulin treatment; in contrast Δ16/BP1 did not, again showing defective regulation.
FIG. 4.
Residues between 8 and 16 are required for efficient phosphorylation of 4E-BP1. (A) Diagrammatic representation of mutants 4E-BP1myc/his, Δ8/BP1, and Δ16/BP1. The amino acid sequences indicate the N termini of these truncated proteins. (B) HEK293 cells were transiently transfected with plasmids expressing either 4E-BP1myc/his, Δ8/BP1, or Δ16/BP1 as indicated. The pcDNA3.1myc/his vector was used as a control. Cells were serum starved and treated with insulin as indicated. The extracts were analyzed by using the anti-4E-BP1 antibody. (C) The cell extracts from panel B were subjected to affinity chromatography on m7GTP-Sepharose as described for Fig. 2C. The ratios of recombinant 4E-BP1 bound to eIF4E (recBP1/4E) were assessed by densitometry (National Institutes of Health Image, version 1.61), where the ratio of recombinant 4E-BP1 to eIF4E under serum-starved conditions is normalized to unity. (D) Cell extracts prepared for panel B were analyzed for 4E-BP1 phosphorylated at Thr36/45 (top), Ser64 (middle), and Thr69 (bottom). (E) An in vitro assay of mTOR kinase activity against recombinant 4E-BP1his and Δ16/BP1his was carried out as described in Materials and Methods. mTOR kinase activity was measured in the presence and absence of wortmannin or an FKBP12-rapamycin complex (FKBP12/Rap). Incorporation of the 32P label into the substrates ([32P]) was assessed by autoradiography. The total levels were determined by staining the gel with Coomassie brilliant blue. A photograph of the stained gel is shown (Total).
The levels of phosphorylation at Thr36/45, Ser64, and Thr69 (Fig. 4D) were also compared (Fig. 4B shows a loading control). Phosphorylation of Δ16/BP1 at all sites was markedly decreased compared with that of 4E-BP1myc/his and Δ8/BP1. At longer exposures, a faint band appeared when Δ16/BP1 was probed with the Thr36/45 phospho-specific antibody, suggesting that there was some phosphorylation at either Thr36 or Thr45 after insulin treatment, albeit much less than for 4E-BP1myc/his or Δ8/BP1. Δ16/BP1 was phosphorylated at Thr69 upon insulin stimulation, although to a significantly lesser degree than 4E-BP1myc/his and Δ8/BP1. The partial mobility shift of Δ16/BP1 from the α- to the β-species upon stimulation with insulin (Fig. 4B) is likely due to the phosphorylation of this protein at Thr69 (Fig. 4D), as this event causes a shift to the β-form, as previously reported by Mothe-Satney et al. (30). These data therefore reveal that a region between residues 9 and 16 of 4E-BP1 is required for efficient phosphorylation of 4E-BP1 and therefore for its release from eIF4E (to allow eIF4F complex formation). Considering that the loss of this region of 4E-BP1 does not fully impair the phosphorylation of 4E-BP1 observed upon insulin treatment (especially at Thr69), it appears that other regions within 4E-BP1 still permit some insulin-responsive phosphorylation events.
It has been reported that mTOR phosphorylates 4E-BP1 (at least in vitro) at Thr36 and -45 (see, e.g., reference 29). It was thus possible that residues 9 to 16 of 4E-BP1 played a role in the efficiency with which mTOR could phosphorylate 4E-BP1. To examine this, 4E-BP1 and Δ16/BP1 were tested as substrates for mTOR (Fig. 4E). Phosphorylation was inhibited by two agents which block mTOR kinase activity, FKBP12-rapamycin and wortmannin, confirming that the phosphorylation observed was mTOR dependent. mTOR phosphorylated both proteins with similar efficiencies in vitro. Given that phosphorylation of Δ16/BP1 in vivo was greatly impaired, compared to that of 4E-BP1, these data suggest that mTOR may not be the major kinase involved in vivo.
Identification of the RAIP sequence as an important regulatory element in 4E-BP1.
The analysis above strongly implies that residues 9 to 18 are critical for the regulation of 4E-BP1 phosphorylation in vivo. This region is highly conserved between 4E-BP1 and 4E-BP2 homologues from mammalian species (Fig. 5A). However, human 4E-BP3 has a shorter N-terminal region, which differs markedly from that of 4E-BP1 or 4E-BP2.
FIG. 5.
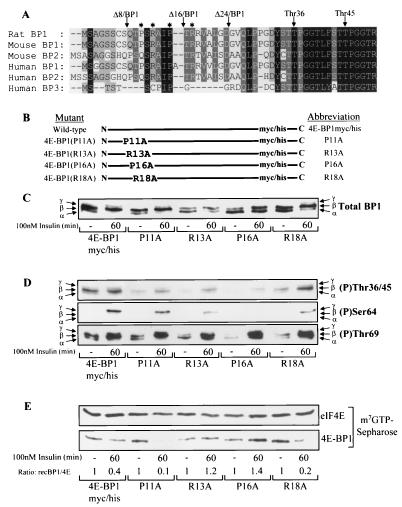
4E-BP1 R13A and 4E-BP1 P16A mutants show reduced basal and insulin-stimulated phosphorylation, preventing dissociation from eIF4E. (A) N-terminal sequence homology of 4E-BPs. The Thr36 phosphorylation site and truncations Δ8/BP1, Δ16/BP1, and Δ24/BP1 are shown. ∗, point mutations P11A, R13A, P16A, and R18A. (B) Diagrammatic representation of mutants used. (C) HEK293 cells were transiently transfected with plasmids expressing the mutants shown above, serum starved, and stimulated with insulin where indicated. 4E-BP1 mutants were analyzed using the anti-Myc antibody. (D and E) The cell extracts from panel C were analyzed for 4E-BP1 phosphorylated at Thr36/45 (top), Ser64 (middle), and Thr69 (bottom) (D) or subjected to affinity chromatography on m7GTP-Sepharose as described for Fig. 2C (E), prior to SDS-PAGE and Western blotting as indicated.
To investigate further the potential importance of this region, four point mutations were made within this section of full-length 4E-BP1 (Fig. 5B). When each of these polypeptides was expressed in HEK293 cells, the total levels of overexpressed 4E-BP1 were similar (Fig. 5C). The R13A and P16A mutations each caused a significant defect in the basal phosphorylation of 4E-BP1, resulting in a higher proportion of the protein migrating as the least-phosphorylated α-species. The wild-type, P11A, and R18A proteins each showed similar shifts in migration to more highly phosphorylated species upon insulin stimulation, where the β-species is the main form, with a smaller amount resolving as the γ-isoform. In contrast, insulin apparently elicited a smaller increase in phosphorylation of either R13A or P16A mutants than of the wild type, as manifested by an equal distribution between α- and β-isoforms and no appearance of the γ-isoform. To analyze the extent of phosphorylation more thoroughly, the degrees of phosphorylation at Thr36/45, Ser64, and Thr69 were also compared (Fig. 5D), with Fig. 5C showing the loading control. For the R13A and P16A mutants, the basal phosphorylation of all sites was less than that for the wild type. For Thr69, basal phosphorylation was reduced for the R13A, P16A, and R18A mutants, although insulin increased this to a level comparable to that for the wild type in each case. For Ser64, insulin was only able to induce a small degree of phosphorylation of the R13A mutant and no signal at all was observed for the P16A mutant. The R18A mutant also showed a reduced degree of Ser64 phosphorylation upon insulin stimulation compared to the wild-type 4E-BP1 protein. The abilities of insulin to bring about the release of these 4E-BP1 mutants from eIF4E were also compared (Fig. 5E). Wild-type 4E-BP1 and the P11A and R18A mutants each substantially dissociated from eIF4E upon insulin stimulation. In contrast, the R13A and P16A point mutants did not.
The above data suggest that the conserved sequence that includes Arg13 and Pro16 is important for normal 4E-BP1 regulation. To explore the role of this RAIP sequence more thoroughly, additional point mutants were made (summarized in Fig. 6A). The phosphorylation of these mutants was analyzed (Fig. 6B and C). In all cases, mutants containing the Ile15-to-Ala mutation showed the greatest defects in phosphorylation compared to the corresponding mutants where Ile15 was retained. For example, the basal phosphorylation of Thr36/45, Ser64, and Thr69 was almost undetectable in the AAAP, RAAA, and AAAA proteins. Insulin increased the signal with all the phospho-specific antisera for R13A, P16A, and AAIA mutants, while little or no phosphorylation of Thr36/45 or Ser64 was observed in the corresponding point mutants having Ala at position 15 (Fig. 6C). Insulin caused increased phosphorylation of Thr69 in these proteins, but this was again much weaker than in the mutants where Ile15 was retained (Fig. 6C). This suggests that Ile15 is one of the critical determinants for regulating 4E-BP1 phosphorylation in vivo.
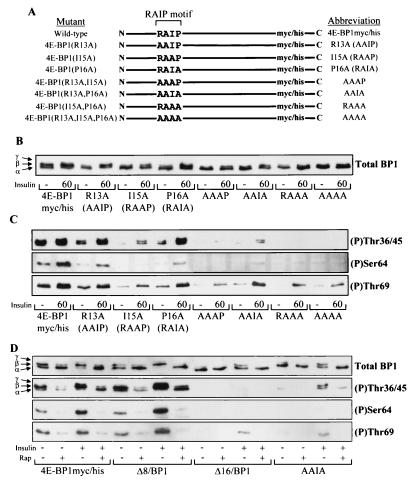
FIG. 6.
Residues within the RAIP motif (13 to 16) are necessary for efficient phosphorylation of 4E-BP1. (A) Diagrammatic representation of 4E-BP1myc/his, R13A, I15A, P16A, AAAP, AAIA, RAAA, and AAAA mutants. (B and C) HEK293 cells were transiently transfected with plasmids expressing the mutants shown above where indicated. The cells were serum starved and stimulated (where indicated) with insulin. 4E-BP1 was analyzed as described for Fig. 5C (B) or analyzed for 4E-BP1 phosphorylated at Thr36/45 (top), Ser64 (middle), or Thr69 (bottom) (C). (D) HEK293 cells overexpressing 4E-BP1myc/his, Δ8/BP1, Δ16/BP1, and the AAIA mutant to similar protein levels were serum starved. These cells were then pretreated with 100 nM rapamycin (Rap) for 30 min prior to insulin stimulation (100 nM insulin for 60 min, where indicated). The cell lysates were subjected to SDS-PAGE and Western blotting using the anti-Myc antibody (Total BP1) or the phospho-specific 4E-BP1 antibodies as indicated.
To assess whether the phosphorylation of the N-terminal mutant 4E-BP1 polypeptides was dependent on mTOR, as for wild-type 4E-BP1, HEK293 cells expressing these proteins were pretreated with rapamycin for 30 min prior to being stimulated with insulin. The phosphorylation of truncation mutants Δ8/BP1 and Δ16/BP1, as well as that of the AAIA mutant, was compared to that of the wild-type 4E-BP1myc/his (Fig. 6D). The AAIA mutant was chosen for this study since it showed only an intermediate defect in insulin-induced phosphorylation, i.e., it still showed some regulation by insulin (Fig. 6C). The basal and insulin-induced phosphorylation of both 4E-BP1myc/his and Δ8/BP1 was reduced by rapamycin treatment but not completely blocked, i.e., some of the β-species and a signal with the Thr36/45 phospho-specific antibody were still observed. Rapamycin also blocked the phosphorylation of the AAIA mutant induced by insulin (Fig. 6C). The faint Thr69 signal observed with the Δ16/BP1 and AAIA proteins after insulin stimulation was abolished with rapamycin treatment, showing that this residual phosphorylation, like that of wild-type 4E-BP1, is dependent on mTOR signaling.
The N terminus of 4E-BP1 facilitates insulin-stimulated phosphorylation of 4E-BP3 and its release from eIF4E.
The above data suggest that the RAIP motif is necessary for the mTOR-dependent regulation of 4E-BP1 phosphorylation upon insulin stimulation. 4E-BP3 lacks this motif (Fig. 5A). We, therefore, asked whether insulin stimulation could also regulate the phosphorylation of 4E-BP3. Following insulin stimulation, 4E-BP3 did not dissociate from eIF4E and eIF4F complex formation did not occur, i.e., no eIF4G1 was copurified with eIF4E (Fig. 7B). This indicates that the binding of 4E-BP3 to eIF4E is not modulated in response to insulin and that its regulation thus differs from that of 4E-BP1. We tested whether the observed difference between 4E-BP1 and 4E-BP3 might be due to their differing N termini by analyzing two chimeric 4E-BPs. In one chimera (nBP1/cBP3), residues 1 to 19 of 4E-BP3 were replaced by residues 1 to 32 of 4E-BP1. The other chimera (nBP3/cBP1) had the converse replacement. The constructs are summarized in Fig. 7A. Insulin caused a shift in the migration of 4E-BP1 toward the more slowly migrating species, while no significant shift was apparent with either 4E-BP3myc/his or nBP1/cBP3 (Fig. 7C). Samples of the same cell extracts used in Fig. 7C were subjected to m7GTP-Sepharose chromatography (Fig. 7D). Insulin stimulation caused 50% of nBP1/cBP3 to dissociate from eIF4E (Fig. 7D) but did not elicit significant release of nBP3/cBP1. It thus appears that the N-terminal regions of these proteins are indeed important in the regulation of their functions by insulin. In particular, the N terminus of 4E-BP1 is required for insulin to bring about the phosphorylation events necessary for the release of 4E-BPs from eIF4E. In contrast, the N terminus of 4E-BP3 is unable to promote a sufficient degree of phosphorylation of 4E-BP3myc/his and nBP3/cBP1 during insulin stimulation to cause their release from eIF4E (Fig. 7D). It seemed probable that the anti-Thr36/45 antibody would cross-react with 4E-BP3 at residue Thr23 (which has the same flanking sequence as Thr36 in 4E-BP1; Fig. 7A). Insulin stimulation increased the signal observed with this antibody for 4E-BP3, showing that it can bring about some degree of phosphorylation of 4E-BP3, albeit not a sufficient degree to cause its release from eIF4E. The basal signal for the nBP1/cBP3 chimera was stronger than that for 4E-BP3myc/his, suggesting a higher basal level of phosphorylation, although this could reflect a higher affinity of this antibody for the former protein.
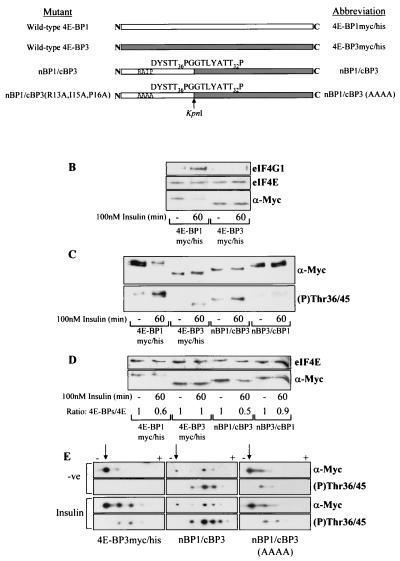
FIG. 7.
Insulin-induced phosphorylation of 4E-BP3 is increased when 4E-BP3 contains the N terminus of 4E-BP1. (A) Diagrammatic representation of mutants used. White and grey boxes, 4E-BP1 and 4E-BP3 proteins, respectively. These mutants each possess C-terminal Myc and His tags (not shown; see Fig. 2A). Arrow, KpnI site used in creating the chimeras. The amino acid sequence expressed adjacent to this site within each chimera is indicated. The phosphorylation sites in 4E-BP1 (Thr36 and -45) and the corresponding sites in 4E-BP3 (Thr23 and -32) are indicated. (B) Cell samples from HEK293 cells overexpressing tagged 4E-BP1 or 4E-BP3 were subjected to affinity chromatography on m7GTP-Sepharose as described for Fig. 2C. (C) HEK293 cells expressing either 4E-BP1myc/his, 4E-BP3myc/his, nBP1/cBP3, or nBP3/cBP1 were stimulated with insulin as indicated. The extracts were analyzed using anti-Myc antiserum (α-Myc) to detect the levels of expression and using 4E-BP1 phospho-Thr36/45 antiserum to analyze phosphorylation of 4E-BP3 at Thr23 and 4E-BP1 at Thr36/45. (D) The cell extracts from panel C were subjected to affinity chromatography with m7GTP-Sepharose. The ratios of recombinant 4E-BP protein bound to eIF4E (4E-BPs/4E) were determined by densitometric analysis of the blots, where the ratios during serum-starved conditions are normalized to 1. (E) HEK293 cells overexpressing either 4E-BP3myc/his, nBP1/cBP3, or nBP1/cBP3(AAAA) were treated with insulin as indicated. The cell extracts were analyzed by isoelectric focusing as described in Materials and Methods. The overexpressed protein was analyzed using anti-Myc antiserum and the 4E-BP1 phospho-Thr36/45 antiserum (to analyze phosphorylation of 4E-BP3 at Thr23 and of 4E-BP1 at Thr36/45). Arrows, positions of the most basic, least phosphorylated form of each protein that lies closest to the cathode (−). Upon phosphorylation the 4E-BP3 protein migrates further toward the anode (+), and so the observed shift toward the anode from the point of origin is representative of increasing phosphorylation.
Due to sequence differences between 4E-BP1 and 4E-BP3, it was considered that the 4E-BP1 Ser64 and Thr69 phospho-specific antibodies would probably not cross-react with 4E-BP3. It is also interesting that neither the insulin-induced phosphorylation of 4E-BP3 at Thr23 nor the phosphorylation of nBP1/cBP3 was sufficient to alter significantly the mobility of these polypeptides during SDS-PAGE (Fig. 7C). We therefore employed an isoelectric focusing technique to resolve differentially phosphorylated species of 4E-BP3 and nBP1/cBP3 (Fig. 7E). One main species of 4E-BP3 was apparent when using the anti-Myc antiserum, and no signal was observed with the anti-phospho-Thr36/45 serum. Insulin caused a shift in the migration of 4E-BP3, yielding two more-acidic forms, which both reacted with the phospho-specific antiserum for Thr36/45. The picture for nBP1/cBP3 was quite different, as it showed a much higher basal level of phosphorylation (Fig. 7E) based on its migration on isoelectric focusing. Furthermore, these species also reacted strongly with the anti-phospho-Thr36/45 antibody. Insulin induced a further shift toward the anode, with the appearance of an additional acidic form probably containing five phosphate groups. This indicates that the N terminus of 4E-BP1 confers a much higher basal level of phosphorylation on 4E-BP3. To examine whether the RAIP sequence element was necessary for the high degree of phosphorylation observed in nBP1/cBP3, we generated an nBP1/cBP3 triple-alanine substitution mutant in which Arg13, Ile15, and Pro16 were replaced [designated nBP1/cBP3(AAAA)]. This nBP1/cBP3(AAAA) mutant showed only a limited degree of basal phosphorylation such that the pattern was similar to that for 4E-BP3myc/his. Its phosphorylation was not increased further upon insulin treatment (Fig. 7E). This experiment reveals that the RAIP sequence element is indeed required for insulin to elicit the extent of phosphorylation observed for nBP1/cBP3.
DISCUSSION
In this report we show that 4E-BP1 is cleaved during apoptosis to generate a product (Δ4E-BP1) which possesses the eIF4E-binding site and all the known sites of phosphorylation but which does not undergo a sufficient level of insulin-stimulated phosphorylation to facilitate its dissociation from eIF4E. Its basal level of phosphorylation is also low compared to that of full-length 4E-BP1. The unphosphorylated cleavage product (Fig. 2B and 3A and B) functions as a potent inhibitor of eIF4F complex formation in apoptotic cells, as cleavage of even a small proportion of the total 4E-BP1 leads to substantial binding of eIF4E to this protein (Fig. 1B). As expected from its ability to block the formation of eIF4F complexes, Δ4E-BP1 also prevented the stimulation of cap-dependent translation by insulin (Fig. 2E). eIF4G1 (4-6, 8, 26) and eIF4G2 (24) are also cleaved by caspases, and these effects are likely to contribute to the inhibition of translation during apoptosis. It was proposed that the cleavage of eIF4G1 would impair cap-dependent translation but that the C-terminal fragment of eIF4G1 might mediate cap-independent translation (7, 18). Our data indicate that early in apoptosis, while cells still contain significant amounts of intact eIF4G1, Δ4E-BP1 may serve to inhibit eIF4F complex formation (Fig. 1A). Indeed Δ4E-BP1 within apoptotic cells was observed to be sequestered to eIF4E to a greater extent than the full-length 4E-BP1 (Fig. 1B). This suggests that during the early stages of apoptosis, when a small proportion of 4E-BP1 is cleaved, the ability of eIF4E to form initiation complexes would likely be blocked, thus causing the impairment of cap-dependent translation. eIF4F complexes are not required for IRES-driven translation, and, consistent with this, Δ4E-BP1 did not affect the translation of an IRES-driven reporter (Fig. 2E). This implies that IRES-driven translation may be maintained, or even favored, in apoptotic cells containing Δ4E-BP1, perhaps thereby potentially allowing the increased expression of apoptotic mediators such as c-Myc, APAF-1, and DAP5/p97, whose mRNAs contain IRESs (reviewed in references 7 and 18).
4E-BP1 is cleaved at the Asp24-Gly25 bond, and this was blocked by Z-VAD.FMK, which implies that 4E-BP1 is cleaved by caspase(s). This study defines a novel feature of 4E-BP1, a sequence near the N terminus which is required for the regulation of its phosphorylation in vivo. Our initial experiments indicated that the region between residues 9 and 16 of 4E-BP1 was critical for its phosphorylation upon insulin stimulation. Mutational analysis revealed that the RAIP motif within this region is a key regulatory feature of 4E-BP. This is perhaps surprising given that it is not adjacent to any of the phosphorylation sites. This motif may be required for the association of kinase(s) responsible for phosphorylating 4E-BP1. It is interesting that a motif in the N-terminal half of c-Jun is required for binding to the c-Jun N-terminal kinase (JNK) (16). This motif is again distal to the phosphorylation sites but facilitates the phosphorylation of c-Jun by JNK. In a manner analogous to this, and given that the phosphorylation of 4E-BP1 is thought to be ordered (10, 29, 30), the RAIP motif may be involved in the “docking” of kinases responsible for phosphorylating the initial sites in 4E-BP1 (especially those acting on Thr36, Thr45, and perhaps Thr69). The phosphorylation of these initial sites may be necessary for the phosphorylation of Ser64, which leads to the release of 4E-BP1 from eIF4E. To explore this analogy further, wild-type 4E-BP1 and the 4E-BP1 R13A, I15A, and P16A mutants (used as negative controls) were purified on Ni-NTA agarose from HEK293 cells to find any potential kinases interacting with this motif. This analysis did not reveal any obviously associated candidate (data not shown), perhaps because the association of the kinase(s) with 4E-BP1 may be transient. Significant (but reduced) phosphorylation of Thr69 was detected for several mutants which showed no detectable phosphorylation of Thr36 or -45. This suggests, in agreement with the results of Mothe-Satney et al. (29), that phosphorylation of Thr69 may not be completely dependent on phosphorylation of Thr36 or -45. Although the work shown here reveals that the RAIP motif within the N terminus is critical for the extent to which 4E-BP1 is phosphorylated upon insulin stimulation, other residues that flank this RAIP motif are also likely to contribute to the regulation of 4E-BP1. This notion is supported by the modest decrease in the extent of phosphorylation of the R18A mutant (Fig. 5D), although the effect of this point mutation was not as severe as those of the mutations within the RAIP sequence.
The RAIP motif is conserved between 4E-BP1 and 4E-BP2 (Fig. 1D). Little work has been done on 4E-BP2, but the available data suggest that it is regulated in a manner similar to that for 4E-BP1 (22). In contrast, 4E-BP3 does not contain this motif and possesses only the IP (it is hard to align these sequences as the N terminus of 4E-BP3 is much shorter than that of 4E-BP1 and quite divergent; Fig. 5A). Our data show that 4E-BP3 is not sufficiently phosphorylated to cause its dissociation from eIF4E in response to insulin and so differs from 4E-BP1 (Fig. 7D). It is tempting to speculate that this lack of regulation might reflect the absence of the RAIP motif from 4E-BP3. The observation that 4E-BP3 does not dissociate from eIF4E upon insulin stimulation suggests that its in vivo function is not to regulate eIF4F assembly in response to signals such as insulin but to modulate it under other conditions. The generation of a chimera containing the N terminus of 4E-BP1 greatly increased the level of phosphorylation of the 4E-BP3 protein, and this effect was completely dependent on the presence of the RAIP motif within the portion of 4E-BP1 present in the chimera (Fig. 7E). This again underlines the importance of this motif for 4E-BP phosphorylation.
It is interesting that the insect (Drosophila melanogaster) homologue THOR entirely lacks the RAIP sequence, and the putative homologue from the zebrafish Danio rerio also does not possess the whole motif, although like 4E-BP3 it contains related sequence CPIP (Fig. 1D). This could be interpreted as indicating that this motif may not be required for regulation of 4E-BP in these animal species. However, the putative homologue from slime mold Dictyostelium discoideum does possess this sequence. Mammalian targets for regulation by mTOR signaling such as p70 S6 kinase (12, 36) and elongation factor 2 kinase (33) do not possess RAIP sequences. These data suggest that the role of this sequence motif may be restricted to 4E-BP1 and 4E-BP2. A key question for future work is how this motif functions to promote insulin-stimulated phosphorylation of 4E-BPs.
Acknowledgments
We thank A. A. M. Thomas (University of Utrecht) and R. M. Denton (University of Bristol) for supplying antibodies. The cDNA encoding human 4E-BP3 was a kind gift from F. Poulin and N. Sonenberg (McGill University, Montreal). We thank N. Morrice (University of Dundee) for his help in sequencing Δ4E-BP1. We also thank J. E. Harthill (University of Dundee) for technical assistance in isoelectric focusing, P. R. Shepherd (University College, London) for providing the FLAG-tagged mTOR construct, P. H. Scott (University of Glasgow) for supplying the recombinant FKBP12 protein, and both G. C. Scheper and T. P. Herbert (University of Dundee) for critical reading of the manuscript.
This work was supported by the Wellcome Trust through the award of a Prize Studentship to A. R. Tee.
REFERENCES
- 1.Beretta, L., A.-C. Gingras, Y. V. Svitkin, M. N. Hall, and N. Sonenberg. 1996. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent translation. EMBO J. 15:658-664. [PMC free article] [PubMed] [Google Scholar]
- 2.Brunn, G. J., C. C. Hudson, A. Sekulic, J. M. Williams, H. Hosoi, P. J. Houghton, J. C. Lawrence, and R. T. Abraham. 1997. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science 277:99-101. [DOI] [PubMed] [Google Scholar]
- 3.Burnett, P. E., R. K. Barrow, N. A. Cohen, S. H. Snyder, and D. M. Sabatini. 1998. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl. Acad. Sci. USA 95:1432-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bushell, M., L. McKendrick, R. U. Jaenicke, M. J. Clemens, and S. J. Morley. 1999. Caspase-3 is necessary and sufficient for cleavage of protein synthesis eukaryotic initiation factor 4G during apoptosis. FEBS Lett. 451:332-336. [DOI] [PubMed] [Google Scholar]
- 5.Bushell, M., D. Poncet, W. E. Marissen, H. Flotow, R. E. Lloyd, M. J. Clemens, and S. J. Morley. 2000. Cleavage of polypeptide chain initiation factor eIF4GI during apoptosis in lymphoma cells: characterisation of an internal fragment by caspase-3-mediated cleavage. Cell Death Differ. 7:628-636. [DOI] [PubMed] [Google Scholar]
- 6.Bushell, M., W. Wood, M. J. Clemens, and S. J. Morley. 2000. Changes in integrity and association of eukaryotic protein synthesis initiation factors during apoptosis. Eur. J. Biochem. 267:1083-1091. [DOI] [PubMed] [Google Scholar]
- 7.Clemens, M. J., M. Bushell, I. W. Jeffrey, V. M. Pain, and S. J. Morley. 2000. Translation initiation factor modifications and the regulation of protein synthesis in apoptotic cells. Cell Death Differ. 7:603-615. [DOI] [PubMed] [Google Scholar]
- 8.Clemens, M. J., M. Bushell, and S. J. Morley. 1998. Degradation of eukaryotic polypeptide chain initiation factor (eIF) 4G in response to induction of apoptosis in human lymphoma cell lines. Oncogene 17:2921-2931. [DOI] [PubMed] [Google Scholar]
- 9.Coldwell, M. J., S. A. Mitchell, M. Stoneley, M. MacFarlane, and A. E. Willis. 2000. Initiation of APAF-1 translation by internal ribosome entry. Oncogene 19:899-905. [DOI] [PubMed] [Google Scholar]
- 10.Gingras, A.-C., S. P. Gygi, B. Raught, R. D. Polakiewicz, R. T. Abraham, M. F. Hoekstra, R. Aebersold, and N. Sonenberg. 1999. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 13:1422-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gingras, A.-C., B. Raught, and N. Sonenberg. 1999. eIF4 translation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 12.Gout, I., T. Minami, K. Hara, Y. Tsujishita, V. Filonenko, M. D. Waterfield, and K. Yonezawa. 1998. Molecular cloning and characterization of a novel p70 S6 kinase, p70 S6 kinase β containing a proline-rich region. J. Biol. Chem. 273:30061-30064. [DOI] [PubMed] [Google Scholar]
- 13.Haghighat, A., S. Mader, A. Pause, and N. Sonenberg. 1995. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 14:5701-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall-Jackson, C. A., D. A. Cross, N. Morrice, and C. Smythe. 1999. ATR is a caffeine-sensitive, DNA-activated protein kinase with a substrate specificity distinct from DNA-PK. Oncogene 18:6707-6713. [DOI] [PubMed] [Google Scholar]
- 15.Henis-Korenblit, S., N. Levy-Strumpf, D. Goldstaub, and A. Kimchi. 2000. A novel form of DAP5 protein accumulates in apoptotic cells as a result of caspase cleavage and internal ribosome entry site-mediated translation. Mol. Cell. Biol. 20:496-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hibi, M., A. Lin, T. Smeal, A. Minden, and M. Karin. 1993. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 7:2135-2148. [DOI] [PubMed] [Google Scholar]
- 17.Holcik, M., C. Lefebvre, C. Yeh, T. Chow, and R. G. Korneluk. 1999. A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat. Cell Biol. 1:190-192. [DOI] [PubMed] [Google Scholar]
- 18.Holcik, M., N. Sonenberg, and R. G. Korneluk. 2000. Internal ribosome initiation of translation and the control of cell death. Trends Genet. 16:469-473. [DOI] [PubMed] [Google Scholar]
- 19.Huang, T. S., M. L. Kuo, J. Y. Shew, Y. W. Chou, and W. K. Yang. 1996. Distinct p53-mediated G1/S checkpoint responses in two NIH3T3 subclone cells following treatment with DNA-damaging agents. Oncogene 13:625-632. [PubMed] [Google Scholar]
- 20.Karim, M. M., J. M. X. Hughes, J. Warricker, G. C. Scheper, C. G. Proud, and J. E. G. McCarthy. 2001. A quantitative molecular model for modulation of mammalian translation by the eIF4E-binding protein 1. J. Biol. Chem. 276:20750-20757. [DOI] [PubMed] [Google Scholar]
- 21.Lawrence, J. C., and R. T. Abraham. 1997. PHAS/4E-BPs as regulators of mRNA translation and cell proliferation. Trends Biochem. Sci. 22:345-349. [DOI] [PubMed] [Google Scholar]
- 22.Lin, T. A., and J. C. Lawrence. 1996. Control of the translational regulators PHAS-I and PHAS-II by insulin and cAMP in 3T3-L1 adipocytes. J. Biol. Chem. 271:30199-30204. [DOI] [PubMed] [Google Scholar]
- 23.Mader, S., H. Lee, A. Pause, and N. Sonenberg. 1995. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4γ and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 15:4990-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marissen, W. E., A. Gradi, N. Sonenberg, and R. E. Lloyd. 2000. Cleavage of eukaryotic initiation factor 4GII correlates with translation inhibition during apoptosis. Cell Death Differ. 7:1234-1243. [DOI] [PubMed] [Google Scholar]
- 25.Marissen, W. E., Y. Guo, A. A. M. Thomas, R. L. Matts, and R. E. Lloyd. 2000. Identification of caspase 3-mediated cleavage and functional alteration of eukaryotic initiation factor 2α in apoptosis. J. Biol. Chem. 275:9314-9323. [DOI] [PubMed] [Google Scholar]
- 26.Marissen, W. E., and R. E. Lloyd. 1998. Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase-3 during inhibition of translation in apoptotic cells. Mol. Cell. Biol. 18:7565-7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathews, M. B., N. Sonenberg, and J. W. B. Hershey. 1997. Origins and targets of translational control, p. 1-30. In J. W. B. Hershey, M. B. Mathews, and N. Sonenberg (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Morley, S. J., L. McKendrick, and M. Bushell. 1998. Cleavage of translation initiation factor 4G (eIF4G) during antiFas IgM-induced apoptosis does not require signalling through the p38 mitogen-activated protein (MAP) kinase. FEBS Lett. 438:41-48. [DOI] [PubMed] [Google Scholar]
- 29.Mothe-Satney, I., G. J. Brunn, L. P. McMahon, C. T. Capaldo, R. T. Abraham, and J. C. Lawrence. 2000. Mammalian target of rapamycin-dependent phosphorylation of PHAS-1 in four (S/T)P sites detected by phospho-specific antibodies. J. Biol. Chem. 275:33836-33843. [DOI] [PubMed] [Google Scholar]
- 30.Mothe-Satney, I., D. Yang, P. Fadden, T. A. J. Haystead, and J. C. Lawrence. 2000. Multiple mechanisms control phosphorylation of PHAS-I in five (S/T)P sites that govern translational repression. Mol. Cell. Biol. 20:3558-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Proud, C. G. 1992. Protein phosphorylation in translational control. Curr. Top. Cell Regul. 32:243-369. [DOI] [PubMed] [Google Scholar]
- 32.Raught, B., A.-C. Gingras, and N. Sonenberg. 2000. Regulation of ribosome recruitment in eukaryotes, p. 245-293. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Redpath, N. T., N. T. Price, and C. G. Proud. 1996. Cloning and expression of cDNA encoding protein synthesis elongation factor-2 kinase. J. Biol. Chem. 271:17547-17554. [PubMed] [Google Scholar]
- 34.Rhoads, R. E. 1999. Signal transduction pathways that regulate eukaryotic protein synthesis. J. Biol. Chem. 274:30337-30340. [DOI] [PubMed] [Google Scholar]
- 35.Scheper, G. C., N. A. Morrice, and C. G. Proud. 2001. The MAP kinase signal-integrating kinase Mnk2 is an eIF4E kinase with high basal activity in mammalian cells. Mol. Cell. Biol. 21:743-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shima, H., M. Pende, Y. Chen, S. Fumagalli, G. Thomas, and S. C. Kozma. 1998. Disruption of the p70S6k/p85S6k gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 17:6649-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stoneley, M., S. A. Chappell, C. L. Jopling, M. Dickens, M. MacFarlane, and A. E. Willis. 2000. c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol. Cell. Biol. 20:1162-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tee, A. R., and C. G. Proud. 2000. DNA damage causes inactivation of translational regulators linked to mTOR signalling. Oncogene 19:3021-3031. [DOI] [PubMed] [Google Scholar]
- 39.Tee, A. R., and C. G. Proud. 2001. Staurosporine inhibits phosphorylation of translational regulators linked to mTOR. Cell Death Differ. 8:841-849. [DOI] [PubMed] [Google Scholar]
- 40.Yang, D., G. J. Brunn, and J. C. Lawrence. 1999. Mutational analysis of sites in the translational regulator, PHAS-I, that are selectively phosphorylated by mTOR. FEBS Lett. 453:387-390. [DOI] [PubMed] [Google Scholar]