Abstract
The mammalian skeleton presents an ideal system in which to study the genetic architecture of a set of related polygenic traits and the skeleton of the domestic dog (Canis familiaris) is arguably the best system in which to address the relationship between genes and anatomy. We have analyzed the genetic basis for skeletal variation in a population of >450 Portuguese Water Dogs. At this stage of this ongoing project, we have identified >40 putative quantitative trait loci (QTLs) for heritable skeletal phenotypes located on 22 different chromosomes, including the “X.” A striking aspect of these is the regulation of suites of traits representing bones located in different parts of the skeleton but related by function. Here we illustrate this by describing genetic variation in postcranial morphology. Two suites of traits are involved. One regulates the size of the pelvis relative to dimensions of the limb bones. The other regulates the shape of the pelvis. Both are examples of trade-offs that may be prototypical of different breeds. For the size of the pelvis relative to limb bones, we describe four QTLs located on autosome CFA 12, 30, 31, and X. For pelvic shape we describe QTLs on autosome CFA 2, 3, 22, and 36. The relation of these polygenic systems to musculoskeletal function is discussed.
Polygenic systems pose a problem for understanding disease etiology: Multiple phenotypes can arise from a single gene defect; and multiple genes can give rise to the same or similar symptom(s) (Moore 2003). We are analyzing the genetic basis for the morphology and development of the mammalian skeleton to obtain insight into the genetic architecture of a set of related polygenic traits. Skeletal metrics are quantitative traits that vary continuously depending on the genotype as well as the environment (nutrition, exercise). Such phenotypes are highly heritable (Mousseau and Roff 1987) and the assay conditions (X-rays) can be made precise.
Most genetic analyses of the mammalian skeleton have utilized the mouse. These have focused on size (see Vaughn et al. 1999; Liu et al. 2001; Brockmann et al. 2004) or the shape of particular parts of the murine skeleton, including aspects of the skull (Leamy et al. 1999; Workman et al. 2002) and limb bones (Beamer et al. 1999; Klingenberg et al. 2001; Leamy et al. 2002; Masinde et al. 2003; Lang et al. 2005). Most studies addressed specific questions such as the genetic relation between early and late development (Leamy et al. 1999; Vaughn et al. 1999) or relationships between skeletal size and shape to variation in organ size (Leamy et al. 2002; Rocha et al. 2004) or bone mineral density (Beamer et al. 1999). In most instances size and shape were regulated by a multitude of quantitative trait loci (QTLs) acting independently; however, a few instances of pleiotropy (Leamy et al. 2002) or antagonistic pleiotropy (Leamy et al. 1999) were reported. More recently, a number of pleiotropic effects have been identified in the analysis of murine mandibular morphology (Cheverud et al. 2004).
The skeleton of the domestic dog (Canis familiaris) represents an ideal system to address the relationship between genes and anatomy and to relate morphology to behavior (Stockard 1941). A large range of skeletal variation, both in size and shape, is encountered among dogs (Stockard 1941), where sizes range from 80 kg (Great Danes) to 0.5 kg (Chihuahuas), and shapes differ widely depending on the breed (e.g. greyhounds and pit bulls). The rapidity with which changes in morphology and behavior have been achieved by selective breeding suggests that among the loci controlling morphology some control large amounts of variation.
We have analyzed a population of Portuguese Water Dogs (PWDs), focusing on variation in both size and shape. For this analysis, we have employed principal component (PC) analysis to define heritable phenotypes that are independent aspects of skeletal variation. This approach uses a large number of skeletal metrics, thus increasing statistical power. However, it focuses on patterns of multiple metrics and will not capture independent variation of the metrics of individual bones. As such, we preferentially detect QTLs with pleiotropic effects.
The PWD population is ideal for such an analysis: Size and shape are segregating in this population despite its descent from relatively few founders (six ancestors account for ∼80% of the present gene pool) (see Molinari 1993; Chase et al. 1999). The wide range of values for the coefficient of inbreeding includes subpopulations that are highly inbred (homozygous, expressing recessive genes) as well as heterozygotes expressing phenotypes that result from interactions between haplotypes. Finally, accurate pedigree records coupled with outstanding owner cooperation facilitated the study.
We have found that skeletal variation in PWD shape is regulated by suites of loci that control variation along a trade-off continuum between different functional morphological states. These trade-offs often parallel transitions encountered in the ontogenetic changes between newborns and the adult state (heterochrony), suggesting that changes in functional anatomy of the adult arise from the genetic system that controls development. Here we illustrate these concepts with a description of the genetic basis for trade-offs involving the pelvis.
Results
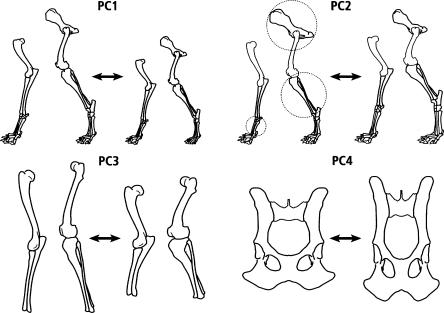
Figure 1 presents schematic versions of the axes of variation of the first four PCs in the analysis of the post-cranial skeleton of the PWD. The first PC (Fig. 1) describes the variation in skeletal size. Subsequent components describe aspects of variation in shape. PC2 (Fig. 1) represents a trade-off (inverse correlation) between the various metrics of the pelvis versus the width of limb bones and the size of the limb joints and feet; PC3 (Fig. 1) represents a trade-off between various aspects of skeletal length versus width (represented here by the major limb bones); and PC4 (Fig. 1) is a trade-off between dimensions of pelvic bones, leading to change in the shape of the pelvis. These four PCs explain 70% of the total variation in the postcranial skeleton: PC1, 54%; PC2, 10%; PC3, 7%; and PC4, 3%. Heritabilities (see Methods) of these four components ranged from 67% for PC4 up to 73% for PC3.
Figure 1.
Axes of variation of principal components 1 through 4. PC1 indicates size; PC2 is a trade-off between large pelvis, thin distal limbs, and short feet (metatarsal and metacarpal) vs. smaller pelvis, thicker distal limbs, and longer feet; PC3 is a trade-off between long thin bones vs. short thick bones; PC4, pelvic shape, is a trade-off between a longer narrower pelvis vs. shorter wider pelvis (see radiographs in Fig. 2). PC2 skeletal traits relevant to Figure 5 are circled.
The main skeletal shape axes of variation involving the pelvis are described by the loadings of PC2 and PC4 (Table 1). PC2 involves a trade-off between skeletal components that are structurally separate but functionally related (the size of the pelvis and the dimensions of distal limb bones). On the other hand, PC4 is primarily composed of hip metrics and represents a trade-off between the length and width of the pelvis that also changes the ratio of the upper to the lower parts (lengths of the ilium vs. ischium). This can be seen in radiographs of animals with extreme values of PC4 (Fig. 2), where the inverse correlation between the variation in the ischium, oscoxa, and hip width, on the one hand, and the ilium, ilium span, and ischial tuberosity, on the other, result in a shorter wider pelvis (Fig. 2, left) as opposed to a longer narrower pelvis (Fig. 2, right). (This difference is presented schematically in Fig. 1.) The loadings for PC2 (the trade-off between a large pelvis, thin limb bones, and shorter feet, on the one hand, and smaller pelvis, thicker limb bones, and longer distal limb elements, on the other) define differences that distinguish some breeds of dog. For example, greyhounds have longer relatively thinner legs than do pit bulls. We compared pelvic metrics of these two breeds (Table 2) and found that they supported the trade-off for PC2 in Table 1. In general, greyhounds have a significantly larger pelvis than do pit bulls. The pelvic shapes of these two breeds also resemble, to some extent, the trade-off described above for PC4. The breed with the relatively larger ilium (ilium length/ischium length) also has the larger ischial tuberosity. However, the ratio of the ischium length to the ischial tuberosity is not significantly different.
Table 1.
PC2 and PC4 loadings
| PC2 | Loading | PC4 | Loading |
|---|---|---|---|
| Pelvic inlet | – 0.23 | Ischium | – 0.45 |
| Pelvic outlet | – 0.21 | Radius ID | – 0.20 |
| Oscoxa | – 0.17 | Tibia ID | – 0.17 |
| Trochanter | – 0.15 | Oscoxa | – 0.12 |
| Ischium | – 0.15 | Olecranon | 0.12 |
| Ilium | – 0.15 | Anklein-lever | 0.15 |
| Vertebral height | – 0.14 | Hip width | 0.15 |
| Hip width | – 0.10 | Sacrum | 0.16 |
| Ischial tuberosity | – 0.10 | Ilium | 0.17 |
| Ilium span | – 0.10 | Ilium span | 0.20 |
| Olecranon | 0.10 | Ischial tuberosity | 0.26 |
| Anklein-lever | 0.10 | ||
| Foot | 0.10 | ||
| Humerus head | 0.14 | ||
| Glenoid | 0.14 | ||
| Acetabulum | 0.14 | ||
| Calcaneus | 0.15 | ||
| Metacarpal | 0.15 | ||
| Tarsaljoint | 0.15 | ||
| Tibia OD | 0.20 | ||
| Radius OD | 0.23 | ||
| Femur OD | 0.25 | ||
| Humerus OD | 0.26 |
Principal component loadings (eigenvectors) for PC2 and PC4 are listed. Only those values with a bootstrap significance of ≤0.05 are shown. Reference points for selected metrics of PC4 are displayed in Figure 1. Additional reference landmarks for PC2 are described in Chase et al. 2002: Table 2; (Fig. 3B,D,E). OD = outer diameter; ID = internal diameter.
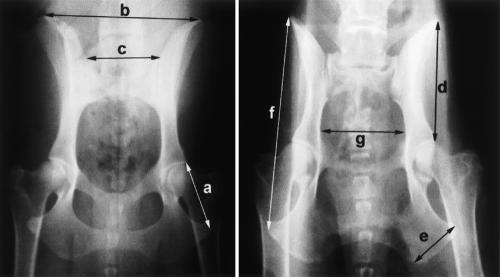
Figure 2.
Hip X-rays from two size-matched dogs at opposite extremes of the PC4 axis of variation. Individual metrics are indicated: a, ischium; b, ilium span; c, sacrum; d, ilium; e, ischial tuberosity; f, oscoxa; and g, pelvic inlet. The differences in pelvic shape are presented schematicallyin Figure 1.
Table 2.
Mean values of mass-specific lengths (length × mass-0.33) and length ratios for two breeds of dog greyhound (GH) and pit bull (PB)
| Trait | GH | PB |
|---|---|---|
| Ilium length/ischium lengtha | 1.598 | 1.461 |
| Pelvis lengtha | 6.307 | 5.457 |
| Ilium lengtha | 3.939 | 3.266 |
| Ischium length/pelvic lengtha | 0.391 | 0.410 |
| Ischium widtha | 4.536 | 3.933 |
| Acetabular breadtha | 0.831 | 0.753 |
| Ischial tuberositya | 1.770 | 1.559 |
| Ischium lengtha | 2.469 | 2.234 |
| Pelvis length vs. ischium width | 1.395 | 1.392 |
| Ilium span | 3.673 | 3.606 |
| Ischium length/ischial tuberosity | 1.395 | 1.433 |
Sample size was four dogs for each breed.
Significant differences: two tail t-test corrected by using sequential Bonferroni
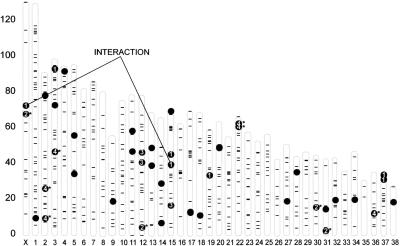
Because each dog has a value for each of the PCs, the PCs are phenotypes for which we can identify QTLs. We have identified QTLs regulating variation of each of the PCs. Their location on the canine physical map is presented in Figure 3. We identified QTLs by their significant association with simple sequence repeat (SSR) markers distributed over the genome. QTLs regulating variation of PC1, PC2, PC3, and PC4 are denoted by black dots with the numerals 1, 2, 3, or 4. Other QTLs for additional PCs with significant heritabilities are represented by dots without numbers. As with the mouse, these QTLs suggest a genetic architecture in which the genetic basis of post-cranial skeletal traits are distributed throughout the genome (>35 QTLs on 26 chromosomes). In addition, an interaction between two PC1 QTLs that contributes to size sexual dimorphism is shown (see Chase et al. 2005b).
Figure 3.
Physical map of SSR markers used in QTL identification: Location of skeletal QTLs on the dog genome. For each autosome and the X chromosome, the physical positions of the markers used in the genome scan are indicated (–). Markers that show a significant association with a phenotype are indicated with black dots; those for PC1—4 are numbered. Other QTLs for PCs with significant heritabilities are indicated by additional black dots. An interaction of PC1 QTLs between the X chromosome and chromosome 15 is indicated (Chase et al. 2005b).
Four QTLs for PC2 were located on the X chromosome and on autosomes CFA 12, 30, and 31. Five QTLs associated with PC 4 were located on CFA 2, 3, 22, and 36 (two QTLs were located on CFA 3). The two QTLs shown on CFA 22 are probably the same locus associated with two different markers. Table 3 presents details of the location of each QTL as well as the amount of variation within the PC that they explain. In all, the PC2 QTLs explain ∼12.5% of the total variation or 18% of the heritable variation. The PC4 QTLs explain ∼50% of the heritable variation. (Syntenic comparison with the human, mouse, and rat genomes suggests a number of candidate genes that might be involved. These are listed as Supplemental material available online.)
Table 3.
QTL characterization
| PC | Marker associated with QTL | Chromosome | Location bp | BLUP R2 | P-value |
|---|---|---|---|---|---|
| 2 | FH2656 | X | 60,748,537 | 0.028 | 0.0013 |
| 2 | FH3324 | CFA12 | 3,999,454 | 0.055 | 0.0027 |
| 2 | FH2269 | CFA30 | 14,820,034 | 0.015 | 0.0025 |
| 2 | FH2189 | CFA31 | 2,000,000 | 0.027 | 0.0337 |
| 4 | FH2431 | CFA2 | 8,860,467 | 0.069 | 0.0128 |
| 4 | FH3395 | CFA2 | 24,878,538 | 0.104 | 0.0162 |
| 4 | FH2388 | CFA3 | 44,745,542 | 0.048 | 0.0022 |
| 4 | FH2320 | CFA3 | 44,880,174 | 0.049 | 0.0094 |
| 4 | FH3337 | CFA22 | 58,232,711 | 0.051 | 0.0036 |
| 4 | FH2538 | CFA22 | 60,047,493 | 0.043 | 0.0065 |
| 4 | FH2323 | CFA36 | 11,380,453 | 0.040 | 0.0001 |
All markers significantly associated with PC2 and PC4 are listed along with the physical position of the marker, the amount of variation explained by the marker, and the P-value for each association. P-values are based on allele sharing tests (Methods) and adjusted for the number of trials in the genome scan (e.g., 700). Possible candidate genes are listed in a supplemental table (Table 3b) online.
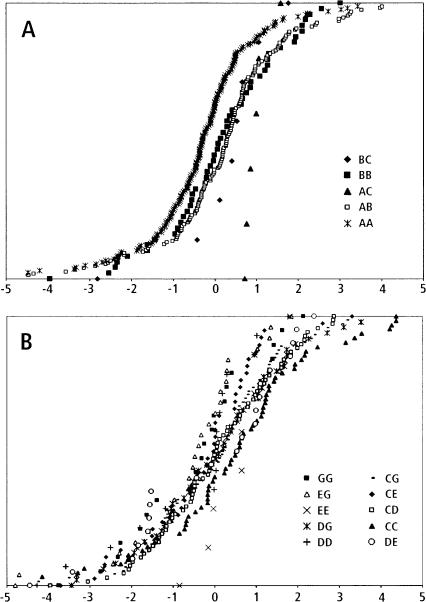
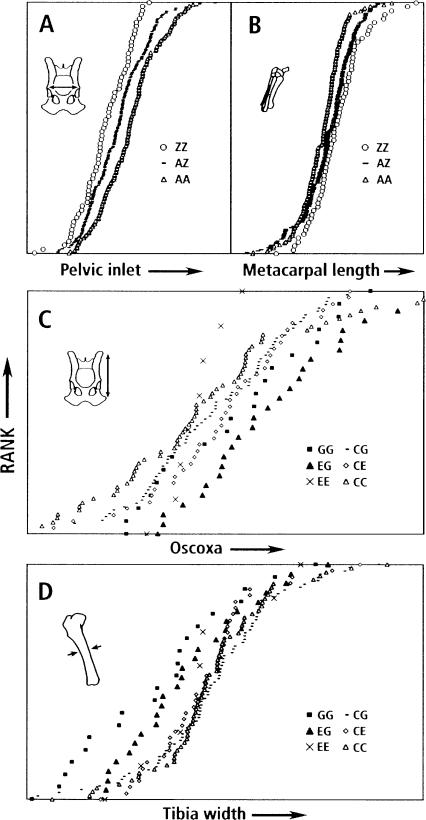
Figure 4 contrasts the complexity of two different QTLs for PC2. One, associated with SSR marker FH2269, is relatively simple, involving one minor and two major alleles of the marker. Note that the B haplotype is dominant to the A (AB = BB > AA, a significant relationship). The other QTL, associated with marker FH3324, is complex with >15 marker genotypes derived from seven alleles (only 10 genotypes are presented in Fig. 4). This also presents different examples of haplotype relationships— e.g., G dominant to E (GG = GE < EE), or C and G additively related (GG < CG < CC)—however, the smaller number of dogs with particular genotypes make these relationships less significant and they can only be regarded as suggestive. The trade-off between size of the pelvis and the dimensions of distal bones (PC2) can be seen in the effects of QTL genotypes on particular bones. In order to avoid effects of size and shape components other than PC2, we compared residuals of bone metrics obtained after removing the effects of PC1, PC3, and PC4. In Figure 5, the QTL phenotypes “width of the pelvic inlet” and “length of a metacarpal bone” display an antagonistic pleiotropy in their association with different marker genotypes of FH2269. Whereas this trade-off is highly significant, a similar trade-off seen with the FH3324-associated QTL (also shown in Fig. 5) can only be regarded as suggestive. Here the trade-off is between the length of the pelvis (oscoxa) and the inner diameter of the tibia. This trade-off involves phenotypes associated with two particular marker genotypes, EG and GG. Because of the large number of other genotypes associated with this QTL and, consequently, the lower number of dogs with any particular genotype (Fig. 4), the statistical significance of this trade-off is greatly reduced.
Figure 4.
Simple and complex examples of PC 2 QTLs. (A) All FH2269 marker genotypes. (B) Ten of >15 FH3324 genotypes are shown. QTLs associated with FH2269 and FH3324 are graphed as cumulative distributions. For each marker genotype, all individuals are ranked on the y-axis according to the pedigree-corrected PC2 value on the x-axis. Pedigree effects were removed by using jackknife techniques.
Figure 5.
Trade-offs: Effects of genotypes on pelvis and limb bone metrics for PC2 QTLs associated with FH2269 and FH3324. The pelvic inlet (A; see Fig. 2, g) is compared to the length of the metacarpal bone (B); the oscoxa (C; see Fig. 2, f) is compared to the width of the tibia (D). In A and B, the genotypes were simplified by combining haplotypes (B + C = Z). In C and D, only six genotypes are shown: GG and EG are compared with EE, CG, CE, and CC. In each graph, dogs are ranked on the y-axis in order of increasing phenotype (x-axis), resulting in cumulative distributions of phenotypes for each genotype. Pedigree effects were removed by using jackknife techniques. Effects of PCs 1, 3, and 4 also were removed (see Chase et al. 2004). Thus, x-axis values are residuals.
Discussion
Suites of positively and negatively correlated traits govern variation in canine skeletal size and shape (Chase et al. 2002). The metrics of the limbs and pelvis define a PC for size and three for shape, of which two involve the pelvis: PC2 is a trade-off between the overall size of the pelvis and the weight and strength of the limbs (thickness of the main limb bones and joints as well as length of the feet). Functionally, this represents a trade-off between high-speed, energy-efficient running (speed) versus limb strength. This trade-off is prototypical of contrasting breeds such as the greyhound and pit bull. A large pelvic girdle associated with relatively narrow limb bones is consistent with high-speed running and locomotor economy. Mammals specialized for running exhibit large pelvic muscles and relatively slender limbs (Hildebrand and Goslow 2001; Pasi and Carrier 2003). The alternative pattern of small pelvis and relatively thick limb bones appears consistent with a more generalized locomotor anatomy in which more of the body is carried by the forelimbs, leading to a less massive pelvic girdle, and the diameter of the limb bones need not be constrained by the energetics of high-speed running. PWDs do not resemble greyhounds or pit bulls (see Chase et al. 2002: Fig. 1), but this axis of skeletal variation has allowed breeders to vary their shape and activity to promote terrestrial speed as opposed to prowess in water activities such as pulling objects while swimming.
PC4 describes variation in the shape of the pelvis. This axis of variation also is contrasted in breeds such as the greyhound and pit bull. In this case the breed difference parallels the PC with respect to the relative lengths of the ischium and ilium. Although the variation in the ischial tuberosity is suggestive, it is not significant. The function of hip shape is less obvious, although it will again be important for running, since the lengths of the ilium and ischial tuberosity constrain the size of the muscles— gluteal and hamstring muscles, respectively—that extend the hind limbs.
Variation in particular skeletal metric can be regulated independently in conjunction with different suites of traits. Thus, in the PWD, the thickness of the limb bones varies with the size of the dog and with three different PCs defining trade-offs in shape: skull versus limb diameter (Chase et al. 2002), pelvis size versus limb thickness (PC2), and limb length versus width (PC3) (data not shown). We have found that different, unlinked, genetic loci (Fig. 3) govern these independent aspects of limb variation.
As with the skull and post-cranial anatomy, we have found that individual QTLs can regulate metrics of different bones that are related by function. Thus, the size of the pelvic inlet and the length of the metacarpal bone are regulated by a common QTL such that they are inversely correlated (a larger pelvis being associated with a smaller fore foot). Again, a QTL appears to regulate both variation in pelvis length and the width of the tibia. Either example could result from a single gene that coregulates morphology of multiple bones or from separate linked genes, each regulating the structure of an individual bone. Both explanations would arise as a consequence of selection for function.
The trade-offs that we have discussed may be relevant to the evolution of humans. A trade-off between pelvis size and metrics of limb robustness is seen in the transition from Australopithecus (large pelvis less robust limb bones) to Homo (smaller pelvis more robust limbs), albeit for different functional purposes (Wolpoff 1999). It will be interesting to see if hominids and canids have used the same genes to implement such trade-offs.
Finally, we note that QTLs related to pelvis shape may be relevant to disease. Recently, we have found that a particular haplotype of the QTL associated with marker FH2388 is associated with osteoarthritis in the coxofemoral joint. This appears to result directly from the action of the QTL haplotype rather than indirectly from changes in pelvic shape produced by the QTL (Chase et al. 2005a).
Methods
X-rays for phenotypes as well as blood for DNA genotypes were obtained from owners of PWDs through the Georgie Project (K. Miller, director; http://www.georgieproject.com). We have increased the number of dogs in the data set previously used for analysis of the full skeleton (Chase et al. 2002) and focused on metrics of the hip and limb as described in Chase et al. (2002: Fig. 3B,D,E; Table 2). Phenotypes (skeletal metrics from radiographs) and DNA genotypes (alleles of SSR markers) were obtained from 463 PWDs. Methods for analyzing the radiographs and for genotyping DNA with SSR markers have been described in detail (Chase et al. 2002, 2004). Specific, easily recognized, landmarks were used to define bone dimensions. These landmarks were accurately recognized by three different people. All points were recorded by using the paths function of Adobe Photoshop and exported to an Excel spreadsheet. Path length data were adjusted for measurement error (removal of extreme outliers) and checked for systematic bias of any particular individual scoring of radiographs. Missing values of particular bones were imputed by using multiple regression of all other metrics. Finally, path lengths were adjusted for sex. A table of the adjusted path lengths used is appended as Supplemental online material.
Narrow sense heritabilities (the percentage of additive genetic variation in the total variation) were calculated by using the polygenic function of SOLAR (Almasy and Blangero 1998). Few programs are available that are capable of processing a pedigree as complex as the PWD pedigree. Those programs designed to handle general pedigrees take too long to complete a computational analysis of the entire genome. This precludes the use of simulations/permutations to establish the frequency of false positives. In light of these limitations, we have chosen single marker analysis instead of interval mapping as a more robust, albeit conservative, test of association. Methods for identifying QTLs and for estimating the genotypic means of markers near a QTL in an unstructured population have been published (Lynch and Walsh 1998; Chase et al. 2002, 2004).
Briefly, QTLs are identified by using an allele sharing method. This method compares pedigree-corrected allele sharing values at a marker with phenotypic similarity to establish an association between marker and phenotype. Monte Carlo simulations of a random trait with heritability equal to the target phenotype are used to estimate the NULL distribution of the marker-phenotype association. This NULL distribution is used to estimate the significance of the association between the marker and the target phenotype. P-values were adjusted for ∼700 markers tested. Marker genotypic means are estimated by using a mixed model in which the marker genotypes are treated as fixed effects and the additive genetic background and error deviations are treated as random effects.
PC analysis
PC analysis (Jackson 1991) defines independent factors (eigenvectors or loadings) that describe a decreasing amount of the total variation. Forty metrics taken from the pelvis and fore- and hind-limbs (Chase et al. 2002; http://www.georgieproject.com) of ∼450 PWDs formed the matrix used for PC analysis. PC analysis on age- and sex-corrected data used the prcomp function in R environment for statistical computing (http://www.r-project.org/). The data were centered (mean = 0) and scaled (variance = 1). The prcomp function uses singular value decomposition to calculate the PCs on this normalized data and returns a matrix of variable loadings (e.g., the eigenvectors). Each eigenvector defines a linear transformation of the original input data (a PC score). The resulting PC score is not correlated with any other PC scores (i.e., PCs defined by other eigenvectors). Thus, the original input data were transformed to a set of independent scores that explain a decreasing amount of the total variation (Jackson 1991). The interpretation of these eigenvectors is important in exploring the biology underlying these patterns of variation. The magnitude of the loadings and their relative signs (correlations or inverse correlation) describes the influence of different traits on PCs determining size or shape. We used bootstrap analysis to place confidence intervals about the individual trait loadings for each eigenvector. Important trait loadings should remain significant; i.e., confidence intervals should not include zero.
Supplementary Material
Acknowledgments
This research was supported by the National Science Foundation grant NSF IBN-0212141 to D.C. and the National Institutes of Health/NIGMS grant 63056 to K.G.L., by a gift from the Judith L. Chiara Foundation, and gifts from >100 PWD owners. We thank Makiko Uemera, Kerry Matz, and Tyler Jarvik for technical assistance. Materials (blood and X-rays) were obtained from owners by Deborah Broughton through the Georgie Project (Karen Miller, director).
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3800005.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Almasy, L. and Blangero, J. 1998. Multipoint quantitative trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 62: 1198-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beamer, W.G., Shultz, K.L., Churchill, G.A., Frankel, W.N., Baylink, D.J., Rosen, C.J., and Donahue, L.R. 1999. Quantitative trait loci for bone density in C57BL/6J and CAST/EiJ inbred mice. Mamm. Genome 10: 1043-1049. [DOI] [PubMed] [Google Scholar]
- Brockmann, G.A., Karatayli, E., Haley, C.S., Renne, U., Rottmann, O.J., and Karle, S. 2004. QTLs for pre- and post-weaning body weight and body composition in selected mice. Mamm. Genome 15: 593-609. [DOI] [PubMed] [Google Scholar]
- Chase, K., Adler, F.R., Miller-Stebbings, K., and Lark, K.G. 1999. Teaching a new dog old tricks: Identifying quantitative trait loci (in dogs) using lessons from plants. J. Hered. 90: 43-51. [DOI] [PubMed] [Google Scholar]
- Chase, K., Carrier, D.R., Adler, F.R., Jarvik, T., Ostrander, E.A., Lorentzen, T.D., and Lark, K.G. 2002. Genetic basis for systems of skeletal quantitative traits: Principal component analysis of the canid skeleton. Proc. Natl. Acad. Sci. 99: 9930-9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase, K., Lawler, D.F., Adler, F.R., Ostrander, E.A., and Lark, K.G. 2004. Bilaterally asymmetric effects of quantitative trait loci (QTLs): QTLs that affect laxity in the right versus left coxofemoral (hip) joints of the dog (Canis familiaris). Am. J. Med. Genet. 124A: 239-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase, K., Lawler, D.F., Carrier, D.R., and Lark, K.G. 2005a. Genetic regulation of osteoarthritis: A QTL regulating cranial and caudal acetabular osteophyte formation in the hip joint of the dog (Canis familiaris). Am. J. Med. Genet. 135A: 334-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase, K., Carrier, D.R., Adler, F.R., Ostrander, E.A., and Lark, K.G. 2005b. Interaction between the X chromosome and an autosome regulates size sexual dimorphism in Portuguese Water Dogs. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Cheverud, J.M., Ehrich, T.H., Vaughn, T.T., Koreishi, S.F., Linsey, R.B., and Pletscher, L.S. 2004. Pleiotropic effects on mandibular morphology, II: Differential epistasis and genetic variation in morphological integration. J. Exp. Zoolog. B Mol. Dev. Evol. 302: 424-435. [DOI] [PubMed] [Google Scholar]
- Hildebrand, M. and Goslow, G.E. 2001. Analysis of vertebrate structure. John Wiley, New York.
- Jackson, J.E. 1991. A user's guide to principal components. Wiley, New York.
- Klingenberg, C.P., Leamy, L.J., Routman, E.J., and Cheverud, J.M. 2001. Genetic architecture of mandible shape in mice: Effects of quantitative trait loci analyzed by geometric morphometrics. Genetics 157: 785-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, D.H., Sharkey, N.A., Mack, H.A., Vogler, G.P., Vandenbergh, D.J., Blizard, D.A., Stout, J.T., and McClearn, G.E. 2005. Quantitative trait loci analysis of structural and material skeletal phenotypes in C57BL/6J and DBA/2 second-generation and recombinant inbred mice. J. Bone Miner. Res. 20: 88-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leamy, L.J., Routman, E.J., and Cheverud, J.M. 1999. Quantitative trait loci for early- and late-developing skull characters in mice: A test of the genetic independence model of morphological integration. Am. Nat. 153: 201-214. [DOI] [PubMed] [Google Scholar]
- Leamy, L.J., Pomp, D., Eisen, E.J., and Cheverud, J.M. 2002. Pleiotropy of quantitative trait loci for organ weights and limb bone lengths in mice. Physiol. Genomics 10: 21-29. [DOI] [PubMed] [Google Scholar]
- Liu, X., Bunger, L., and Keightley, P.D. 2001. Characterization of a major X-linked quantitative trait locus influencing body weight of mice. J. Hered. 92: 355-357. [DOI] [PubMed] [Google Scholar]
- Lynch, M. and Walsh, B. 1998. Genetics and analysis of quantitative traits. Sinauer Associates, Boston, MA.
- Masinde, G.L., Wergedal, J., Davidson, H., Mohan, S., Li, R., Li, X., and Baylink, D.J. 2003. Quantitative trait loci for periosteal circumference (PC): Identification of single loci and epistatic effects in F2 MRL/SJL mice. Bone 32: 554-560. [DOI] [PubMed] [Google Scholar]
- Molinari, C. 1993. The Portuguese Water Dog. ELO-Publicidade, Portugal.
- Moore, J.H. 2003. The ubiquitous nature of epistasis in determining susceptibility to common human diseases. Hum. Hered. 56: 73-82. [DOI] [PubMed] [Google Scholar]
- Mousseau, T.A. and Roff, D.A. 1987. Natural selection and the heritability of fitness components. Heredity 59: 181-197. [DOI] [PubMed] [Google Scholar]
- Pasi, B.M.C. and Carrier, D.R. 2003. Functional tradeoffs in the limb muscles of dogs selected for running versus fighting. J. Evol. Biol. 16: 324-332. [DOI] [PubMed] [Google Scholar]
- Rocha, J.L., Eisen, E.J., Van Vleck, L.D., and Pomp, D. 2004. A large-sample QTL study in mice, II: Body composition. Mamm. Genome 15: 100-113. [DOI] [PubMed] [Google Scholar]
- Stockard, C. 1941. The genetic and endocrinic basis for differences in form and behavior. Wistar Institute of Anatomy and Biology, Philadelphia, PA.
- Vaughn, T.T., Pletscher, L.S., Peripato, A., King-Ellison, K., Adams, E., Erikson, C., and Cheverud, J.M. 1999. Mapping quantitative trait loci for murine growth: A closer look at genetic architecture. Genet. Res. 74: 313-322. [DOI] [PubMed] [Google Scholar]
- Wolpoff, M.H. 1999. Paleoanthropology. McGraw-Hill, Boston, MA.
- Workman, M.S., Leamy, L.J., Routman, E.J., and Cheverud, J.M. 2002. Analysis of quantitative trait locus effects on the size and shape of mandibular molars in mice. Genetics 160: 1573-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web site references
- http://www.georgieproject.com; Georgie Project home page.
- http://www.r-project.org/; the R Project for Statisticical Computing home page.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.