Abstract
Recognition of the domestic dog as a model for the comparative study of human genetic traits has led to major advances in canine genomics. The pathophysiological similarities shared between many human and dog diseases extend to a range of cancers. Human tumors frequently display recurrent chromosome aberrations, many of which are hallmarks of particular tumor subtypes. Using a range of molecular cytogenetic techniques we have generated evidence indicating that this is also true of canine tumors. Detailed knowledge of these genomic abnormalities has the potential to aid diagnosis, prognosis, and the selection of appropriate therapy in both species. We recently improved the efficiency and resolution of canine cancer cytogenetics studies by developing a small-scale genomic microarray comprising a panel of canine BAC clones representing subgenomic regions of particular interest. We have now extended these studies to generate a comprehensive canine comparative genomic hybridization (CGH) array that comprises 1158 canine BAC clones ordered throughout the genome with an average interval of 2 Mb. Most of the clones (84.3%) have been assigned to a precise cytogenetic location by fluorescence in situ hybridization (FISH), and 98.5% are also directly anchored within the current canine genome assembly, permitting direct translation from cytogenetic aberration to DNA sequence. We are now using this resource routinely for high-throughput array CGH and single-locus probe analysis of a range of canine cancers. Here we provide examples of the varied applications of this resource to tumor cytogenetics, in combination with other molecular cytogenetic techniques.
Cancers of both humans and domestic animals display considerable heterogeneity in their clinical behavior and response to therapy, a feature that extends even to tumor cases with comparable histopathology (Withrow and MacEwen 2001). These data suggest that existing modes of tumor subclassification will benefit from the introduction of new approaches to better define the spectrum of disease. The genomics revolution has provided valuable new tools and reagents that may be used to complement conventional histopathological classification of many malignancies. The introduction of molecular cytogenetic approaches to subclassify tumors according to the presence of key chromosome aberrations now plays an important role in the diagnosis, prognosis, and clinical management of many forms of human cancer (Mitelman Database of Chromosome Aberrations in Cancer 2005; http://cgap.nci.nih.gov/Chromosomes/Mitelman). We and others have demonstrated that canine tumors are also associated with recurrent chromosome aberrations (e.g., Hahn et al. 1994; Thomas et al. 2003a,b; Milne et al. 2004). Our studies of canine lymphoma and leukemia (Breen et al. 2003; Thomas et al. 2003a,b), in addition to ongoing studies of canine osteosarcoma and brain tumors, indicate that at least a proportion of these are evolutionarily related to chromosome aberrations in the corresponding human tumors, suggesting a conserved pathogenesis. These data suggest that the identification of recurrent, tumor-specific chromosome aberrations in canine tumors may provide a means by which to develop more sophisticated modes of diagnosis/prognosis for the benefit of our canine companions. Furthermore, this approach may also direct cancer research to key regions of the canine genome, and hence regions of the human genome that remain below the limits of detection in comparable studies of human tumors.
The power of a canine model for human diseases, including cancers, lies fundamentally in the unique demographic history of many dog breeds, which represent phenotypically distinct genetic isolates, characterized by unique constellations of morphology, behavior, and susceptibility to naturally occurring diseases. The restricted gene flow between breeds, combined with marked levels of inbreeding, has resulted in modern dog breeds with considerably reduced genetic heterogeneity (Parker et al. 2004; Sutter and Ostrander 2004; Sutter et al. 2004). Genetic investigations of canine diseases may thus be conducted among a reduced level of the background “noise” that is usually associated with investigations of more heterogeneous genomes such as our own. Identifying disease-associated genes in dog breeds is therefore likely to be simpler than in human populations.
One approach to identify genes that contribute to the origin and progression of tumors is to examine numerical and structural changes in genomic DNA isolated from malignant cells. Comparative genomic hybridization (CGH) analysis is a key research tool for the detection of such cytogenetic aberrations (Kallioniemi et al. 1994), in which the entire tumor genome is evaluated for regions of copy number imbalance in a single experiment. We previously developed canine metaphase-based CGH (mCGH) (Dunn et al. 2000; Thomas et al. 2001, 2003a) and subsequently generated a low-resolution microarray for the study of canine tumors by array-based CGH (aCGH) (Thomas et al. 2003b). We report here the extension of these studies to generate a comprehensive CGH array comprising 1158 canine BAC clones spanning the genome at intervals of 1-5 Mb with a mean interval of ∼2 Mb, providing up to fivefold greater resolution than mCGH. We anticipate that this resource will be key to gaining a greater understanding of both canine and human tumorigenesis. We are currently using the array for CGH analysis of a range of canine cancers including lymphoma, leukemia, osteosarcoma, and brain tumors. In this report we provide examples of the varied applications of this resource to tumor cytogenetics, in combination with other complementary molecular cytogenetic techniques.
Results
Integration of cytogenetic and genome assembly data
Of the 1158 clones represented on the array, 976 clones (84.3%) have been assigned to a precise cytogenetic location using multicolor fluorescence in situ hybridization (FISH) analysis (Breen et al. 2004). In addition, 1141 (98.5%) have been integrated within the 7.5× canine genome assembly (Lindblad-Toh et al. 2005), each at a location consistent with their cytogenetic assignment. Of the remaining 17 clones (1.5%), six clones were assigned to the Y chromosome and thus cannot be integrated into the female assembly, and 11 clones did not yield sequence data of sufficient quality to permit a conclusive in silico mapping assignment.
Array CGH analysis with a 2-Mb resolution canine BAC array
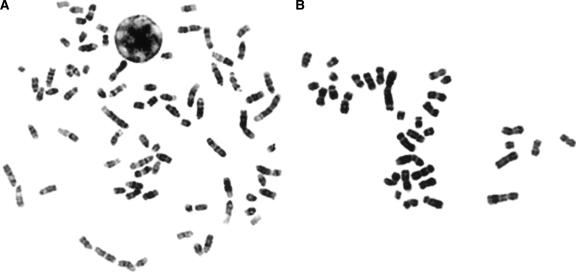
We selected a case of stage II appendicular osteosarcoma (OS-a) from our ongoing investigations of canine osteosarcoma (OS) to demonstrate the application of this array. Although cytogenetic studies of canine OS have thus far been limited to a single report (Mayr et al. 1991), our ongoing analyses indicate that, as with human OS, canine OS tumor karyotypes are typically complex and present with numerous genomic imbalances and structural rearrangements. Molecular cytogenetic techniques are therefore essential in order to determine conclusively the origins of the derivative chromosomes. Figure 1 shows a typical metaphase preparation from a normal canine cell alongside a cell from case OS-a. Conventional cytogenetic analysis revealed a chromosome number of 34 in all OS-a cells evaluated and a tumor karyotype comprising many metacentric chromosomes (Fig. 1B).
Figure 1.
(A) DAPI-banded metaphase spread from a clinically normal male dog. Note that the karyotype (2n = 78) comprises 38 pairs of acrocentric autosomes, a large submetacentric X chromosome, and a small metacentric Y chromosome. (B) Typical DAPI-banded metaphase preparation of a cell derived from canine osteosarcoma case OS-a. This tumor has a consistent chromosome number of 34 (30/30 cells) and presented with a complex karyotype comprising multiple metacentric chromosomes.
Reference versus reference control hybridization
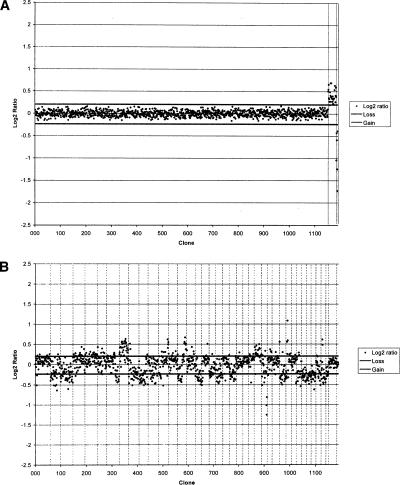
A series of self-self and sex-mismatch hybridizations were initially performed to assess array hybridization quality. Genomic DNA samples derived from clinically normal male and female dogs were differentially labeled and cohybridized onto the array under conditions described previously (Thomas et al. 2003b). All autosomal clones demonstrated fluorescence ratios approaching 1:1, representing the expected normal copy number in both reference individuals (Fig. 2A). The effects of the sex-mismatch hybridization are visible as increased representation of dog chromosome X (CFA X) DNA in the female (mean ratio 1.33:1), and of CFA Y DNA in the male (mean ratio 1:0.56).
Figure 2.
(A) Array CGH profile of a sex-mismatched hybridization performed with normal reference genomic DNA. Data are plotted as the mean, normalized, and background-subtracted log2 ratio of the replicate spots for each clone. Clones are plotted in genomic order from CFA 1qcen to CFA Yqtel. Clones derived from CFA X and CFA Y are shown with vertical broken lines on the right side of the profile. Log2 ratios representing genomic gain and loss are indicated by horizontal bars above and below the midline representing normal copy number. (B) Array CGH profile for case OS-a. Clones representing each chromosome are delineated by vertical bars. The profile demonstrates the range of genomic gains and losses present in the tumor, which are summarized in the text.
Array CGH and targeted FISH analysis of a canine osteosarcoma case
Using our 2-Mb array, aCGH analysis of case OS-a demonstrated a vast range of DNA copy number aberrations throughout the canine genome (Fig. 2B), a detailed description of which lies outside the scope of the present study. In summarized form, the aberrations observed by aCGH included overrepresentation of regions of CFA 4, 5, 8, 12, 14q dist., 25, and X, and underrepresentation of regions of CFA 2, 3, 7qdist., 9, 10, 11, 14qprox., 16, 18qprox, 19, 21, 26, 29, 33, 34, 35, 36, 37, 38, and Y.
To compare and correlate aCGH data with information generated by other means, we selected 16 clones represented on the array with which to perform multicolor single-locus probe (SLP) FISH analysis on fixed tumor cell preparations of OS-a. These represented clones distributed throughout the genome that displayed a range of normal and abnormal aCGH ratios. All probes were first hybridized onto metaphase chromosome preparations from clinically normal dogs and produced reliable hybridization signals that confirmed a normal copy number (2n = 2) in all cells analyzed (data not shown). Probes were then applied to OS-a tumor cell preparations, and images were acquired from 30 representative metaphase spreads and interphase nuclei.
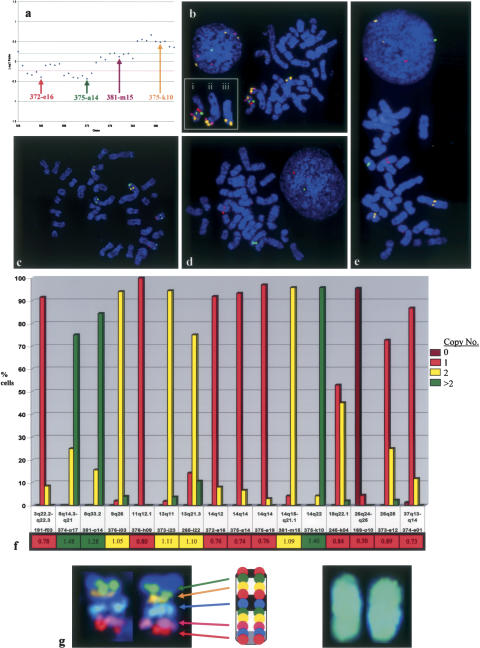
There was a close correlation between the copy number indicated by the aCGH analysis and that revealed by the targeted SLP analysis. For example, the aCGH data indicated a copy number loss of the proximal end of CFA 14, a normal copy number of a small segment in the distal half, and a copy number gain of the distal end of this chromosome (Fig. 3a). These findings were supported by FISH analysis of four spatially separated SLPs from these regions (Fig. 3b), which also revealed structural rearrangements involving CFA 14. One largely intact CFA 14 homolog was translocated onto another chromosome by an apparent centric fusion. The proximal region of the other CFA 14 homolog was absent, whereas the distal end was present within two additional aberrant chromosome structures, indicating a genomic amplification event. aCGH also showed overrepresentation of regions of CFA 8. This was supported by SLP analysis of two BAC clones from these regions, with 75%-85% of cells analyzed showing more than two copies. Figure 3c shows a metaphase spread in which FISH analysis showed that the copy number of the CFA 8 BACs was normal, but revealed distinct structural rearrangements. Both the aCGH and SLP analyses detected underrepresentation of regions of CFA 3, 18, and 37, and a homozygous deletion of a region including CFA 26q24-q25 (Fig. 3d). aCGH showed a normal copy number of CFA 9q25-q26; however SLP analysis indicated that both homologs of CFA 9 were grossly abnormal (Fig. 3e). A summary of the concordance between SLP and aCGH analyses for each of the 16 loci is shown in Figure 3f.
Figure 3.
(a) Enlarged partial aCGH profile from Fig. 2B, showing 36 BAC clones distributed along the length of CFA 14. The pattern of fluorescence ratios indicates a copy number loss of the proximal region of CFA 14, interrupted by a small region with a normal copy number. A copy number gain is observed for clones within CFA 14qdist. (b) SLP analysis of four clones from CFA 14 supports these findings. Two proximal clones, 372-e16 (14q12, labeled red) and 375-a14 (14q14, green), are both present in a single copy on one arm of a novel metacentric chromosome in >90% of cells analyzed (shown as “i” in the inset). Two distal clones on CFA 14, 381-m15 (14q15-q21.1, labeled purple) and 375-k10 (14q22, yellow), are also present on the same derivative chromosome arm, suggesting a centric fusion event involving a grossly intact copy of CFA 14. Both distal clones are also present within a second novel chromosome structure (shown as “ii” in the inset). Clone 381-m15 has a normal copy number in >95% of cells, whereas chromosome duplication and translocation have generated a third copy of clone 375-k10 in >96% of cells (shown as “iii” in the inset). Inset: the three derivative chromosomes from this spread that contain segments of CFA 14. (c) SLP analysis of two clones from each of CFA 8 (374-o17, labeled yellow; 381-014, green) and CFA 13 (373-i23, red; 265-l22, blue). This figure shows one of the 15%-25% of cells in this tumor with two copies of both CFA 8 clones. Although all four probes show a normal copy number, there is clear indication of structural rearrangements involving both CFA 8 and CFA 13. (d) SLP analysis of clones from CFA 3 (191-f03, red), CFA 18 (245-k04, green) and CFA 37 (374-e01, purple) demonstrated a single copy number, correlating with observations from aCGH. A fourth probe, clone 169-o10 (CFA 26, labeled yellow) indicated a homozygous deletion (i.e., no visible signal) in > 95% of cells studied, which is also apparent from aCGH analysis. (e) SLP analysis of clones from CFA 11 (376-h09, red), CFA 14 (375-e19, purple), and CFA 26 (373-e12, green) demonstrated a single copy number. Clone 372-i12 (CFA 9q26, yellow) is present as two copies, correlating with the normal aCGH profile; however, it is clear from the position of the probe signals that CFA 9 has undergone structural rearrangement. (f) Combined summary of the aCGH and SLP analyses for these 16 loci. The frequency of probe signals (n = 0, 1, 2, or >2) in the cell population is plotted against the corresponding locus. Beneath each BAC address the aCGH ratio of each locus is indicated and color-coded according to the threshold of copy number: loss (red), normal (yellow), gain (green). In each instance there is agreement between the copy number data derived from both approaches. (g) Application of a chromosome-specific tiling panel in FISH analysis. The central feature shows a CFA 38 ideogram and the cytogenetic location of 10 BAC clones that have been mapped to this chromosome. When all 10 of these clones are labeled with the same fluorochrome, cohybridization results in painting of the chromosome as shown on the right. Conversely, if individual clones are labeled with different fluorochromes, the pool of fluorescently labeled BAC clones (in this instance, five clones) may be used to generate a multicolored chromosome-specific “tiling-set” for the study structural rearrangements (left).
Discussion
Canine genome analysis has advanced significantly in recent months with the generation of a fully integrated cytogenetic/radiation hybrid map (Breen et al. 2004), a 1.5× genome sequence (Kirkness et al. 2003), and a physically anchored 7.5× genome sequence assembly (Lindblad-Toh et al. 2005). The availability of these key resources empowers the dog as a model system by making available a comprehensive genomics `tool-box' with which to perform critical experiments to further our understanding of the genetics of both human and companion animal health.
Towards this aim we have produced a genome-wide CGH microarray for the dog, with an average clone spacing of 2 Mb, using canine BAC clones that have been integrated into the 7.5× genome assembly and also assigned to the canine karyotype by FISH analysis. To demonstrate the application of this resource we have included aCGH data from the analysis of a canine osteosarcoma case that showed a wide range of karyotypic abnormalities. This case served to demonstrate the approach by which these data may be validated and interpreted in combination with the use of complementary molecular cytogenetic techniques. The development of genomic microarrays for CGH analysis of tumors is both labor-intensive and costly, and as such is likely to remain limited to those species for which the value of such a resource can be justified. A range of genomic CGH arrays exist for the human and murine genomes, currently with effective resolving power of up to 0.08 Mb (Ishkanian et al. 2004) and 1 Mb (Chung et al. 2004), respectively. With the development of this canine genomic microarray, the dog therefore continues to represent one of the best equipped mammalian systems in which to perform genomic investigations.
One key feature of the canine array described here is that each locus represented is also available as a BAC probe for defined FISH analysis, of which 84.3% have been assigned to a precise, unique cytogenetic location by FISH. A subset of 804 clones has been used to anchor the most recent iteration of the canine RH-map to the dog karyotype (Breen et al. 2004), and 98.5% of the clones have been integrated into the 7.5× genome sequence assembly (Lindblad-Toh et al. 2005). This provides two key advantages: (1) chromosome aberrations detected by aCGH analysis can be confirmed and quantified by subsequent multicolor FISH analysis of the corresponding BAC clone onto tumor chromosome preparations and/or interphase nuclei, and (2) cytogenetic aberrations may be translated directly into genome sequence without the need for additional resources or experimental work. This in turn allows for detailed examination of known and putative genes residing within regions of genomic imbalance for their potential role in the disease process or other phenotypes of interest.
The combination of aCGH and SLP techniques clearly provides a synergistic approach towards a clearer understanding of tumor genome organization. A normal aCGH ratio in isolation indicates solely a balanced representation of that specific locus in the global tumor cell population. As can be seen for clones 376-i03 (CFA 9q26, Fig. 3e) and 381-m15 (CFA 14q15-q21.1, Fig. 3b) in case OS-a, the chromosome on which a locus resides in the tumor cell may be structurally aberrant despite its normal copy number. A locus that is overrepresented in one subset of the cell population but underrepresented in other cells within the same tumor specimen may balance out to generate a normal aCGH profile. Direct SLP analysis, however, allows for detailed evaluation of individual cells within a population and thus permits a comprehensive assessment of copy number. SLP analysis can also detect tumor heterogeneity and/or contamination of the specimen with normal tissue. Accurate and representative enumeration of probe signals in an SLP analysis can, however, be time-consuming when performed at a genome-wide level. This is particularly prohibitive in the absence of prior information regarding chromosome aberrations within the tumor, where the selection of appropriate probes itself represents a significant challenge. This challenge is exacerbated for tumors with particularly extensive genomic aberrations, exemplified by the canine osteosarcoma case described here. Our ongoing strategy is therefore to use aCGH to generate a genome-wide assessment of chromosome imbalances in tumors, and to guide in the selection of SLP probes for a more detailed analysis of these findings.
The primary application of the array described here is likely to be the detection and characterization of chromosome aberrations in canine tumors. With the advent of this resource, large-scale, genome-wide analysis of genomic imbalances in canine tumors now becomes a feasible approach. The use of an array in which all clones have a defined cytogenetic location overcomes the requirement to be skilled in chromosome identification, an aspect of canine cytogenetics that remains challenging. The integration of BAC mapping data with the canine genome assembly also allows for the generation of higher-resolution arrays, which may either be genome-wide or comprise a targeted selection of additional clones selected from the assembly for the construction of chromosome-specific microarrays. These will be particularly useful for higher-resolution studies of small genomic regions. Additional applications will include the study of nontumor-related genomic imbalances such as congenital and developmental disorders, which, as with tumor studies, may also generate data of potential relevance to human medicine. BACs can also be used as probe combinations in FISH analysis. For example, the resolution limits of fluorescence optics allow us to pool chromosome-specific panels of BAC clones spaced at intervals <3-4 Mb to form a “pseudo-paint” probe for studying entire chromosomes (Fig. 3g). DNA isolated from individual, aberrant chromosomes (isolated by flow-sorting or microdissection) may be amplified, fluorescently labeled, and hybridized to the array in order to establish the genomic origin of novel chromosome structures present in malignant cells (e.g., Fiegler et al. 2003). The evaluation of structural chromosome rearrangements is often intractable to conventional cytogenetic analyses, and can be particularly difficult in the dog due to its challenging karyotype. This resource offers potential for studying patterns of chromosome conservation between related species by crossspecies array painting analysis. The array may also help to determine whether individual dog populations demonstrate the short regions of natural genomic polymorphism that were recently identified in humans (Iafrate et al. 2004; Sebat et al. 2004). We currently breed-match tumor samples to obviate this possibility, and our studies thus far have not identified such polymorphic regions in dog breeds. As more genomic analyses are undertaken it seems likely that such polymorphic regions will be encountered, and the very nature of the dog as a collection of diverse breeds would make such a study particularly fascinating. This array represents a valuable and integrative addition to the high-quality resources that have resulted from the success of the collaborative and expanding network of the canine genome mapping community. Collectively, these resources provide the necessary foundations to support efficient canine and comparative genome studies with benefit to both man and his best friend.
Methods
Selection of BAC clones for microarray generation
All clones were derived from the RPCI-81 canine BAC library (Li et al. 1999). Of these, 87 were present on our first generation canine genomic microarray, among which are 26 clones containing canine orthologs of human cancer-related genes (Thomas et al. 2003c). Three CFA Y clones have been reported elsewhere (Bannasch et al. 2005). The remaining clones were selected from the current integrated cytogenetic/radiation hybrid map of the dog (Breen et al. 2004) in order to span each chromosome at an average interval of ∼2 Mb, assuming a genome size of 2.4 Gb (Lindblad-Toh et al. 2005).
Clones were placed within the Dog 1.0 whole-genome shotgun assembly (Lindblad-Toh et al. 2005) by aligning BAC end sequence data to the genome using BLAST (Altschul et al. 1997). BAC alignments were defined as either a unique, high-quality alignment of one BAC end or a high-quality alignment of both ends with accurate relative positioning. Clone addresses and their chromosomal locations (as determined by cytogenetic analysis and/or integration into the canine genome assembly) are provided in the Supplemental material, located at http://www.cvm.ncsu.edu/mbs/breen_matthew.htm. Cytogenetic mapping was performed according to our routine multicolor FISH protocols (Breen et al. 2004).
DOP-PCR amplification of DNA templates and microarray generation
BAC DNA was extracted using the Qiagen R.E.A.L. Prep 96 Plasmid kit and amplified by degenerate oligonucleotide primer (DOP)-PCR using three different degenerate primers in separate reactions as described (Fiegler et al. 2003; Thomas et al. 2003b). Two μL of each DOP-PCR product were pooled and used as the template in a second round of amplification using a 5′ amino-linked PCR primer as described (Fiegler et al. 2003; Thomas et al. 2003b). PCR products were arrayed onto amine-binding slides (3D link-activated slides, Motorola) using a MicroGrid II arrayer (BioRobotics) as described (Fiegler et al. 2003). Arrays were then subjected to a series of validation procedures described elsewhere (Thomas et al. 2003b), including self-self and sex-mismatch hybridizations with germline DNA isolated from clinically normal dogs. Eight clones generated anomalous results in these procedures, predominantly representing putative autosomal clones with apparent sex chromosome ratios, suggesting errors in mapping or clone selection. These clones were excluded from further analysis, resulting in a total of 1158 clones on the array.
Initiation of canine osteosarcoma cell line OS-a
Tumor tissue was obtained from a family-owned Rottweiler (male, age 9) with stage II appendicular osteosarcoma, using an approved protocol. Grossly visible tumor was dissected from adjacent normal tissue, rinsed in sterile phosphate buffered saline solution, and divided into three portions that were (1) fixed in 10% neutral buffered formalin, (2) snap frozen, and (3) disaggregated into single-cell suspensions through two cycles of 45 sec each, 15 sec apart in a MediMachine (Becton Dickinson Immunocytometry Systems). Cells were sequentially passaged until they reached ∼80% confluence. By the second passage, the cultures were homogeneous and consisted of large polygonal to plump spindloid or slightly rounded cells, with no evidence of stromal cells. Histologic diagnosis of OS was verified from formalin-fixed tissues (Idexx Veterinary Services). Osteoblastic origin of the isolated cells was confirmed at the third passage by expression of alkaline phosphatase and osteocalcin (IHC Services). High-molecular-weight DNA and metaphase/interphase preparations were generated from low-passage (n = 3) cell line material using conventional techniques.
Array CGH and SLP analyses of canine osteosarcoma OS-a
The reference individuals used in this study were shown previously to demonstrate a normal karyotype (Dunn et al. 2000; Thomas et al. 2001, 2003a). Test (tumor) and reference (normal) DNA probes were labeled with Cyanine3-dCTP or Cyanine5-dCTP (Perkin Elmer) as required, using a BioPrime Array CGH Labeling System (Invitrogen) following the manufacturer's recommendations. Hybridization was performed as described (Thomas et al. 2003b). Arrays were scanned using a ScanArray 4000 at 10-μm resolution and analyzed with ScanArray Express version 3.0 (Perkin Elmer). Each spot position was automatically located, and manual adjustments were made as necessary. Spots with poor morphology and those impinged by fluorescent debris were excluded from further analysis. In each instance a minimum of 98% of clones passed these exclusion criteria. Fluorescence intensities were calculated for each spot after local background subtraction, and normalized to a mean 1:1 ratio on the autosomal clones; ratios of normalized values were than established. The mean fluorescence ratio of each duplicate was then converted to a log2 ratio in order to weight genomic gains and losses equally, and plotted graphically. Following standard conventions for CGH analysis, clones demonstrating a test:reference fluorescence ratio greater than 1.15:1 (gain) or less than 0.85:1 (loss) were classed as aberrant. The subsequent FISH analysis of single-locus probes was performed as described (Breen et al. 2004), and results were scored by two independent investigators with no prior knowledge of the expected copy number.
Supplementary Material
Acknowledgments
Supported by grants from the American Kennel Club Canine Health Foundation to M.B./E.O. (CHF 2214) and M.B./J.M. (CHF 2254). The dog BAC library was gridded by the Human Genome Mapping Resource Centre, Hinxton, UK, supported by a project grant (052908/Z/97) from the Wellcome Trust. E.A.O. and F.G. gratefully acknowledge support from the AKC Canine Health Foundation and U.S. Army grant BAA DAAD 19-01-1-0658. We thank Shengdar Tsai for advice regarding data analysis, Oliver Dovey for performing microarray printing, and Dannika Bannasch for allowing us to use canine Y-chromosome BACs prior to publication.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3825705.
Footnotes
[Supplemental material is available online at www.genome.org and http://www.cvm.ncsu.edu/mbs/breen_matthew.htm.]
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannasch, D.L., Bannasch, M.J., Ryun, J.R., Famula, T.R., and Pedersen, N.C. 2005. Y chromosome haplotype analysis in purebred dogs. Mamm. Genome 16: 273-280. [DOI] [PubMed] [Google Scholar]
- Breen, M., Thomas, R., Faircloth, S., Mahoney, J., Pettengill, M., Scott, A., Thomson, S., Hudson, R., Bianco, S., Fosmire, S., et al. 2003. Molecular cytogenetics of canine lymphoma and leukaemia. In Genes, dogs and cancer: 3rd Annual Canine Cancer Conference. (ed. J. Modiano), pp. P3021.0903. International Veterinary Information Service, Ithaca NY (www.ivis.org), Seattle, WA.
- Breen, M., Hitte, C., Lorentzen, T.D., Thomas, R., Cadieu, E., Sabacan, L., Scott, A., Evanno, G., Parker, H.G., Kirkness, E.F., et al. 2004. An integrated 4249 marker FISH/RH map of the canine genome. BMC Genomics 5: 65-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, Y.J., Jonkers, J., Kitson, H., Fiegler, H., Humphray, S., Scott, C., Hunt, S., Yu, Y., Nishijima, I., Velds, A., et al. 2004. A whole-genome mouse BAC microarray with 1-Mb resolution for analysis of DNA copy number changes by array comparative genomic hybridization. Genome Res. 14: 188-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn, K.A., Thomas, R., Binns, M.M., and Breen, M. 2000. Comparative genomic hybridization (CGH) in dogs—Application to the study of a canine glial tumour cell line. Vet. J. 160: 77-82. [DOI] [PubMed] [Google Scholar]
- Fiegler, H., Carr, P., Douglas, E.J., Burford, D.C., Hunt, S., Scott, C.E., Smith, J., Vetrie, D., Gorman, P., Tomlinson, I.P., et al. 2003. DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer 36: 361-374. [DOI] [PubMed] [Google Scholar]
- Hahn, K.A., Richardson, R.C., Hahn, E.A., and Chrisman, C.L. 1994. Diagnostic and prognostic importance of chromosomal aberrations identified in 61 dogs with lymphosarcoma. Vet. Pathol. 31: 528-540. [DOI] [PubMed] [Google Scholar]
- Iafrate, A.J., Feuk, L., Rivera, M.N., Listewnik, M.L., Donahoe, P.K., Qi, Y., Scherer, S.W., and Lee, C. 2004. Detection of large-scale variation in the human genome. Nat. Genet. 36: 949-951. [DOI] [PubMed] [Google Scholar]
- Ishkanian, A.S., Malloff, C.A., Watson, S.K., DeLeeuw, R.J., Chi, B., Coe, B.P., Snijders, A., Albertson, D.G., Pinkel, D., Marra, M.A., et al. 2004. A tiling resolution DNA microarray with complete coverage of the human genome. Nat. Genet. 36: 299-303. [DOI] [PubMed] [Google Scholar]
- Kallioniemi, O.P., Kallioniemi, A., Piper, J., Isola, J., Waldman, F.M., Gray, J.W., and Pinkel, D. 1994. Optimizing comparative genomic hybridization for analysis of DNA sequence copy number changes in solid tumors. Genes Chromosomes Cancer 10: 231-243. [DOI] [PubMed] [Google Scholar]
- Kirkness, E.F., Bafna, V., Halpern, A.L., Levy, S., Remington, K., Rusch, D.B., Delcher, A.L., Pop, M., Wang, W., Fraser, C.M., et al. 2003. The dog genome: Survey sequencing and comparative analysis. Science 301: 1898-1903. [DOI] [PubMed] [Google Scholar]
- Li, R., Mignot, E., Faraco, J., Kadotani, H., Cantanese, J., Zhao, B., Lin, X., Hinton, L., Ostrander, E.A., Patterson, D.F., et al. 1999. Construction and characterization of an eightfold redundant dog genomic bacterial artificial chromosome library. Genomics 58: 9-17. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh, K., Wade, C.M., Mikkelsen, T.S., Karlsson, E.K., Jaffe, D.B., Kamal, M., Clamp, M., Chang, J.L., Kulbokas III, E.J., Zody, M.C., et al. 2005. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature (in press). [DOI] [PubMed]
- Mayr, B., Eschborn, U., Loupal, G., and Schleger, W. 1991. Characterisation of complex karyotype changes in two canine bone tumours. Res. Vet. Sci. 51: 341-343. [DOI] [PubMed] [Google Scholar]
- Milne, B.S., Hoather, T., O'Brien, P.C.M., Yang, F., Ferguson-Smith, M.A., Dobson, J., and Sargan, D. 2004. Karyotype of soft tissue sarcomas: A multi-colour, multi-species approach to canine chromosome painting. Chromosome Res. 12: 825-835. [DOI] [PubMed] [Google Scholar]
- Parker, H., Kim, L., Sutter, N., Carlson, S., Lorentzen, T., Malek, T., Johnson, G., DeFrance, H., Ostrander, E.A., and Kruglyak, L. 2004. Genetic structure of the purebred domestic dog. Science 304: 1160-1164. [DOI] [PubMed] [Google Scholar]
- Sebat, J., Lakshmi, B., Troge, J., Alexander, J., Young, J., Lundin, P., Maner, S., Massa, H., Walker, M., Chi, M., et al. 2004. Large-scale copy number polymorphism in the human genome. Science 305: 525-528. [DOI] [PubMed] [Google Scholar]
- Sutter, N.B. and Ostrander, E.A. 2004. Dog star rising: The canine genetic system. Nat. Rev. Genet. 5: 900-910. [DOI] [PubMed] [Google Scholar]
- Sutter, N.B., Eberle, M.A., Parker, H.G., Pullar, B.J., Kirkness, E.F., Kruglyak, L., and Ostrander, E.A. 2004. Extensive and breed-specific linkage disequilibrium in Canis familiaris. Genome Res. 14: 2388-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, R., Smith, K.C., Gould, R., Gower, S.M., Binns, M.M., and Breen, M. 2001. Molecular cytogenetic analysis of a novel high-grade canine T-lymphoblastic lymphoma demonstrating co-expression of CD3 and CD79a cell markers. Chromosome Res. 9: 649-657. [DOI] [PubMed] [Google Scholar]
- Thomas, R., Smith, K.C., Ostrander, E.A., Galibert, F., and Breen, M. 2003a. Chromosome aberrations in canine multicentric lymphomas detected with comparative genomic hybridisation and a panel of single locus probes. Br. J. Cancer 89: 1530-1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, R., Fiegler, H., Ostrander, E.A., Galibert, F., Carter, N.P., and Breen, M. 2003b. A canine cancer-gene microarray for CGH analysis of tumors. Cytogenet. Genome Res. 102: 254-260. [DOI] [PubMed] [Google Scholar]
- Thomas, R., Bridge, W., Benke, K., and Breen, M. 2003c. Isolation and chromosomal assignment of canine genomic BAC clones representing 25 cancer-related genes. Cytogenet. Genome Res. 102: 249-253. [DOI] [PubMed] [Google Scholar]
- Withrow, S.J. and MacEwen, E.G. 2001. Small animal clinical oncology. W.B. Saunders, Philadelphia, PA.
Web site references
- http://www.cvm.ncsu.edu/mbs/breen_matthew.htm; Primary authors' laboratory Web site, with details of all clones used on this array.
- http://cgap.nci.nih.gov/Chromosomes/Mitelman; Mitelman Database of Chromosome Aberrations in Cancer 2005. Mitelman, F., Johansson, B., and Mertens, F. (eds).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.