Abstract
Recent large-scale analyses of mainly full-length cDNA libraries generated from a variety of mouse tissues indicated that almost half of all representative cloned sequences did not contain an apparent protein-coding sequence, and were putatively derived from non-protein-coding RNA (ncRNA) genes. However, many of these clones were singletons and the majority were unspliced, raising the possibility that they may be derived from genomic DNA or unprocessed pre-mRNA contamination during library construction, or alternatively represent nonspecific “transcriptional noise.” Here we show, using reverse transcriptase-dependent PCR, microarray, and Northern blot analyses, that many of these clones were derived from genuine transcripts of unknown function whose expression appears to be regulated. The ncRNA transcripts have larger exons and fewer introns than protein-coding transcripts. Analysis of the genomic landscape around these sequences indicates that some cDNA clones were produced not from terminal poly(A) tracts but internal priming sites within longer transcripts, only a minority of which is encompassed by known genes. A significant proportion of these transcripts exhibit tissue-specific expression patterns, as well as dynamic changes in their expression in macrophages following lipopolysaccharide stimulation. Taken together, the data provide strong support for the conclusion that ncRNAs are an important, regulated component of the mammalian transcriptome.
In recent years there have been increasing reports of functional non-protein-coding RNAs (ncRNAs) that are involved or implicated in developmental, tissue-specific, and disease processes, including X-chromosome dosage compensation, germ cell development and embryogenesis, neural and immune cell development, kidney and testis development, B-cell neoplasia, lung cancer, prostate cancer, cartilage-hair hypoplasia, spinocerebellar ataxia type 8, DiGeorge syndrome, autism, and schizophrenia (see Pang et al. 2005). Many putative ncRNAs are alternatively spliced and/or polyadenylated (Sutherland et al. 1996; Tam et al. 1997; Bussemakers et al. 1999; Raho et al. 2000; Charlier et al. 2001; Wolf et al. 2001). Smaller ncRNAs, termed microRNAs, have also been shown to be involved in developmental processes in both plants and animals, as well as implicated in disease (Carrington and Ambros 2003; Mattick and Makunin 2005). Recent evidence suggests that these microRNAs are derived from the introns of capped and polyadenylated protein-coding transcripts as well as the exons and introns of non-protein-coding transcripts, many of which are derived from “intergenic” regions (Cai et al. 2004; Rodriguez et al. 2004; Seitz et al. 2004; Mattick and Makunin 2005; Ying and Lin 2005). In addition, many complex genetic phenomena, including cosuppression, imprinting, methylation, and gene silencing (see Mattick and Gagen 2001; Mattick 2003; Kawasaki and Taira 2004; Ting et al. 2005), as well as the heterochromatization of centromeres and other aspects of chromosome dynamics (Mochizuki et al. 2002; Hall et al. 2003; Volpe et al. 2003), are now known or are strongly implied to be directed or mediated by RNA signaling.
Broad insight into the repertoire of transcripts expressed in animals has primarily been obtained by systematic sequencing of full-length cDNA libraries (Okazaki et al. 2002; Ota et al. 2004; Stolc et al. 2004; Carninci et al. 2005) (see below), and by transcript profiling using whole-chromosome oligonucleotide arrays (Kapranov et al. 2002, 2005; Bertone et al. 2004; Cawley et al. 2004; Kampa et al. 2004; Schadt et al. 2004; Stolc et al. 2004; Cheng et al. 2005), both of which indicate that non-protein-coding transcripts are abundant, and in the case of mammals may account for at least half of all transcripts. Moreover, RNA expression analyses of well-studied genomic regions, such as β-globin (Ashe et al. 1997), bithorax-abdominal A/B (Lipshitz et al. 1987; Sanchez-Herrero and Akam 1989; Drewell et al. 2002), and various imprinted loci (Sleutels et al. 2002; Georges et al. 2003; Holmes et al. 2003), all show a consistent picture—that is, that the majority of these regions is transcribed and that the transcripts can be derived from “intergenic” regions and from both strands.
Many candidate ncRNAs in the mouse have emerged from the RIKEN Mouse Gene Encyclopedia project (Okazaki et al. 2002; Numata et al. 2003). This project was based on construction of comprehensive full-length cDNA libraries using oligo(dT) priming and advanced techniques for trapping 5′-caps of mature RNAs, with the aim of fully characterizing the mouse transcriptome, as well as obtaining full-length protein-coding sequences and reference information about transcriptional start sites. The success of this approach was confirmed by the fact that a high percentage of the obtained protein coding sequences are, indeed, full length (Okazaki et al. 2002; Furuno et al. 2003).
RIKEN's cDNA libraries were made from a large variety of mouse cells, tissues, and developmental stages, using aggressive normalization procedures to remove abundant sequences and improve the coverage of the transcriptome (Carninci et al. 2003). More than 2 million clones obtained from these libraries were sample sequenced at their 5′- and 3′-ends, which were then binned on this basis. Many of these bins contained multiple clones (representing repetitive sampling of abundant transcripts), including splice variants (Zavolan et al. 2003), whereas others contained only singletons. Full-length sequencing and analysis by the FANTOM2 consortium of >60,000 putative full-length transcripts revealed that these were derived from ∼33,400 distinct loci (transcription units with one or more promoters and polyadenylation sites), of which 47% (almost 16,000) lack an apparent protein-coding region as judged by manual annotation (Okazaki et al. 2002). Some of these clones have since been shown to encode small peptides (Grimmond et al. 2003). A minority of these transcripts could be confidently aligned with the human genome (Numata et al. 2003). Lack of sequence conservation, however, does not imply lack of function, as several known functional ncRNAs (such as Xist) are rapidly evolving (Pang et al. 2006). Most of these sequences were unspliced, and a high proportion were singletons, which raises the possibility that the putative ncRNA set contained significant genomic or pre-mRNA contamination, or spurious transcripts, the risks of which are increased by the procedures used to enrich for rarer sequences. In order, therefore, to establish whether or not these sequences are genuine transcripts, a matter that could ultimately have a profound impact on our understanding of mammalian biology, we have undertaken a series of structured studies on the putative ncRNAs identified by FANTOM2.
Results
Bioinformatics analysis of the non-coding transcript set
For the purposes of this analysis, we divided the clones in the FANTOM2 representative transcript set (RTS), which have unique identifier numbers, into those that are known or predicted to be protein coding (17,594), which have corresponding RPS (representative protein transcript) identifier numbers, and those that do not have such designation (15,815), which are potential non-coding RNAs (Okazaki et al. 2002; Furuno et al. 2003; Numata et al. 2003). These transcripts will henceforth be referred to as “mRNA” (protein coding and flanking exonic sequences) or “putative ncRNA” (putative non-protein-coding RNA).
Given that the 15,815 putative ncRNA sequences were manually curated by different individuals, it is possible that a proportion of them may have been inconsistently assigned. We therefore sought to identify likely contaminating coding transcripts by imposing additional filters based on (1) a synonymous/nonsynonymous mutation analysis using the CRITICA coding region identification tool (Badger and Olsen 1999) and (2) a longest open reading frame search (see Supplemental Methods for a full description of the methods used in the bioinformatics analysis). CRITICA predicts 33,303 out of 60,770 Fantom 2 cDNAs (55%) to be protein coding, but only 384 (2%) of the 15,815 putative ncRNA sequences. Since the 15,815 set was identified without using comparative analysis, this result provides independent evidence that nearly all of them are non-coding. Of the 15,815 putative ncRNAs, 2120 (13%) had complete or incomplete ORFs >100 codons, and 191 of these were also predicted as protein coding by CRITICA. The remaining set of 13,502 (85%) putative ncRNAs that passed both filters will henceforth be referred to as the “filtered” ncRNA set. We cannot, of course, rule out the possibility that some of the sequences in these sets encode small peptides or constitute parts of 5′- or 3′-UTRs linked to protein-coding exons, although it should also be noted that the average distance of the ncRNA transcripts to an annotated protein-coding locus in the same orientation is 74 kb (see below).
As a further quality control measure, we tested for the presence of misoriented transcripts among the filtered set by checking for CT-AC versus GT-AG splice junctions. Of the 13,502 sequences, 13,410 were successfully mapped to the genome, and 3350 of these are spliced according to standard criteria (see Supplemental Methods). Of the spliced clones, 21 have CT-AC junctions and 2675 have GT-AG. The remaining 654 include some infrequent splices (33 GC-AG and 12 AT-AC), but mostly correspond to imperfections in the transcript-genome alignment, often caused by gaps in the genome sequence. In any case, among cases in which the orientation can be determined from splice signals, the misorientation rate is extremely low.
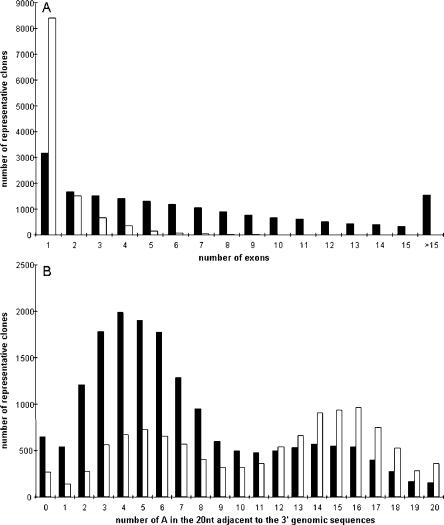
We compared the features of the 13,502 putative ncRNA sequences in the filtered set with those of mRNAs (Table 1). Comparing the cDNA sequences against the mouse genome showed that 72% of the mapped ncRNA sequences were uninterrupted by an intron, while only 18% of the mRNAs were unspliced. In addition, while they have similar length, most spliced ncRNAs consist of only one or very few exons, in contrast to mRNAs (Fig. 1A). The ncRNA exons are correspondingly larger than those of mRNAs, which provides further evidence that these sequences are different from protein-coding sequences. We also examined the number of clones in the mRNA and putative ncRNA sets in relation to mouse EST data. Almost half (48%) of the 13,502 putative ncRNAs were singletons, in contrast to only 9% of the mRNAs, suggesting that ncRNA sequences are expressed at lower levels than protein-coding sequences, a result confirmed by microarray analyses (see below). This result was also supported by the average incidence of mRNA and putative ncRNA clones generated during the high-throughput phase of the RIKEN cDNA cloning project, as judged by end-sequence EST data (Table 1).
Table 1.
Distribution of the numbers and exon structures of mRNA and filtered ncRNA sequences in the FANTOM 2 representative transcript set
| Total | Unspliced | Average number of exons | Number of singleton clones | Average number of EST hits | |
|---|---|---|---|---|---|
| mRNA | 17,594 | 3158 | 6.81 | 1508 | 48.5 |
| ncRNA | 13,502 | 9777 | 1.49 | 6496 | 5.1 |
Figure 1.
(A) Distribution of exon number in mRNAs and putative ncRNAs. (B) Distribution of the number of As within the following 20 nt of the adjacent 3′ genomic sequence. The black bars are mRNAs (protein coding and flanking exonic sequences), and the white bars are putative ncRNAs.
The average size of the mRNA-derived reverse transcripts was 2.1 kb, whereas that of the filtered ncRNA-derived reverse transcripts was 1.8 kb (Table 2). We also looked for the presence of the major polyadenylation signal, AATAAA and ATTAAA, as well as minor polyadenylation signals, in the mRNA and filtered ncRNA sets (Supplemental Table 1). We found that many ncRNAs lacked recognizable polyadenylation signals. Moreover, evidence from documented ncRNAs (Pang et al. 2005) and from Northern analyses (see below) suggests that some ncRNAs are relatively long, which would create substantial difficulties for cDNA cloning protocols, for a variety of well-established technical reasons. We were interested, therefore, in examining how many cDNAs encoding putative ncRNA transcripts might have been derived from polyadenylated transcripts, and what percentage might have been derived by internal oligo(dT) priming from internal A-rich sequences in longer transcripts (Nam et al. 2002), compared to mRNA-derived cDNAs.
Table 2.
Average length and incidence of the major polyadenylation signals in mRNAs and filtered ncRNAs in the FANTOM 2 representative transcript set
|
Number of representative clones
|
Average length (nt)
|
|||||
|---|---|---|---|---|---|---|
| pA+ | pA– | Total | pA+ | pA– | All | |
| mRNA | 8646 | 8948 | 17,594 | 2085 | 2210 | 2149 |
| ncRNA | 4111 | 9391 | 13,502 | 1543 | 1914 | 1801 |
| Total | 12,757 | 18,339 | 31,096 | |||
The designation pA+ indicates the presence of the major polyadenylation signal (AATAAA or ATTAAA) within 30 nt of the 3′-end sequence of the transcript and pA– indicates the absence of this signal. Minor polyadenylation signals were observed in 2567 mRNAs and 2103 ncRNAs, but there was no significant bias in the distribution of these signals between the two sets (Supplemental Table 1).
We analyzed the genomic sequence immediately 3′ to the cDNA sequences for both the filtered ncRNA and mRNA clones, excluding the terminal poly(A) sequences in the clones that are derived from oligo(dT) priming, plotting the number of As in the following 20 bases versus the number of clones at each point (Fig. 1B). The results show that the majority of mRNAs had a normal distribution less than the mean (8.7 A) in the adjacent 3′ genomic sequence, indicating that most were primed from post-transcriptionally added poly(A) stretches, as would be expected. The presence of a small second peak around a higher A content suggests that a small fraction may have derived from internal priming of longer primary transcripts. In contrast, the ncRNA transcripts showed a strong bimodal distribution, with one peak similar to that found with mRNA sequences (containing ∼40% of the ncRNA sequences, average size 1.5 kb), and a second at a higher A content (containing the other ncRNA sequences, average size 2.0 kb), indicating that ∼60% of the ncRNA sequences were likely to be derived from false priming from A-rich sequences within longer transcripts, which was subsequently verified by PCR analyses (see below). In agreement with this prediction, several fragments of the very long imprinted ncRNA Air (Sleutels et al. 2002) upstream of A-rich sequences were found in the cDNA libraries (data not shown).
The possibility exists that some or many of the ncRNA sequences were derived from internal priming of unprocessed pre-mRNAs (Okazaki et al. 2002; Carninci et al. 2003). To address this, we examined the number of unspliced ncRNA transcripts lacking canonical polyadenylation sites that lay within the introns of larger transcription units in the same orientation, acknowledging that bona fide separate transcripts can and have been documented to originate within introns of larger genes (see, e.g., Levinson et al. 1990; Mount and Henikoff 1993; Reisman et al. 1996; Polesskaya et al. 2003) as is clearly evident from the Ensembl and UCSC genome browsers. We found that 30% of filtered ncRNA transcripts fulfill this criterion, and are therefore potential fragments of pre-mRNAs. Another 1% are same-orientation spliced transcripts within introns of larger transcription units, but unless both exons are previously unrecognized alternative exons of the larger transcript, these transcripts must be independent. The majority lies outside of, or in the opposite orientation to, known transcription units and is therefore either genuine ncRNA transcripts (complete or partial) or fragments of unidentified protein-coding genes or unidentified introns at the extremities of known protein-coding genes. The average distance of the putative ncRNA transcripts in the filtered set to an annotated protein-coding locus in the same orientation is 74 kb.
The ncRNA sequences were not derived from genomic contamination
Given the aggressive normalization procedures used to obtain clones from low-abundance transcripts, there is also a risk some of the putative ncRNA clones were derived from genomic contamination. To address this, we undertook an RT-PCR analysis directed at 74 filtered clones selected at random from a cDNA library constructed from testis (Supplemental Table 2), which contained a large proportion of putative ncRNA sequences, consistent with previous observations that developing sperm express large amounts of RNA (Miller et al. 1999). Of these clones, 53 were singletons, 27 were unspliced, and 19 were both. Total RNA was extracted from mouse testis and brain cortex by the same methodology as used for cDNA library construction, and subjected to RT-PCR using primers specifically designed against the 74 selected sequences (Supplemental Table 3). In testis, almost all (71/74) amplifications produced a product of the predicted size with threshold cycle values (Ct) values <30 in the presence of reverse transcriptase, but not when reverse transcriptase was omitted from the reactions (Supplemental Fig. 1; Supplemental Table 2). Of the predicted products, 17 were also detected in brain cortex with Ct values <30, with no correlation between the Ct values observed for these sequences in the two tissues, indicating that the expression of many of these ncRNAs is tissue-specific. This was confirmed by microarray analyses (see below).
We also examined whether longer transcripts could be detected in those cases suspected of having been internally primed from A-rich sequences, using RT-PCR with primer pairs that spanned the end of the cDNA transcript, that is, with the one primer in the cloned transcript sequence and the other in the downstream genomic sequence. Thirteen clones from the set of transcripts showing 13 or more As in the 20-bp downstream genomic sequence were selected from testis, and almost three-quarters (9/13) were amplified by PCR with Ct values <30, in a reverse transcriptase-dependent manner (see Supplemental Table 4), indicating that these were, indeed, longer transcripts, and providing additional evidence that the cloned sequences were derived from genuine transcripts.
Tissue profiling of putative ncRNAs
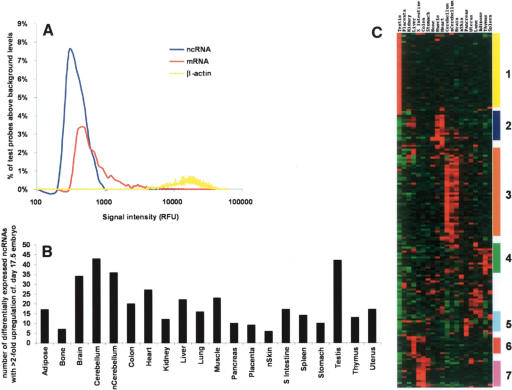
To determine whether the ncRNAs exhibit tissue-specific expression and how their expression levels and patterns compare to those of mRNAs, we examined the expression profile of 1602 filtered ncRNAs across 20 tissues using cDNA microarrays. We also examined the expression of 5179 protein-coding mRNAs in a wide range of different tissues (see Methods). Figure 2A shows that overall expression of the putative ncRNAs was lower than that of their protein-coding counterparts, in keeping with previous reports that ncRNAs are often found in low abundance (Cawley et al. 2004; Kampa et al. 2004), a feature that is not surprising if, as expected, their function is mainly regulatory (Mattick 2004). We used stringent criteria to look for tissue-specific expression. To reduce the likelihood of false positives, we filtered out any clones whose expression levels were close to the background range. Approximately 11% (178) of filtered ncRNAs and 32% (1649) of mRNAs were found to be not only up-regulated by at least twofold compared to day 17.5 whole embryo reference tissue but also adjudged as differentially expressed using analysis of variance (ANOVA) with Bonferroni multiple testing correction (see Methods). Figure 2B shows that the differentially expressed ncRNAs were up-regulated across a range of tissues, and that certain tissues (e.g., central nervous system and testis) appeared enriched for ncRNAs. Notably, many of the up-regulated ncRNAs (110/178; 62%) were overexpressed in multiple, usually developmentally related tissues, and hierarchical clustering clearly demonstrated tissue-specific groups of ncRNAs (Fig. 2C; Supplemental Table 5). The additional observation that 50 of 178 (28%) of these up-regulated ncRNAs were derived from singleton clones, only 26% of which were intragenic to a longer transcription unit in the same orientation, further strengthens the likelihood that singleton clones represent distinct transcripts that are biologically relevant.
Figure 2.
Tissue profiling of putative ncRNAs. RNA was isolated from 20 mainly adult tissues, and corresponding cDNA probes were prepared and hybridized to RIKEN 20K-2 microarrays as described in Methods. (A) Relative frequency histogram of signal intensity of mRNAs and filtered ncRNAs in the microarrays. Mean signal intensity values for each sequence were calculated for individual tissues relative to negative control cDNAs from Arabidopsis (Accession nos. X98108, X13611, X90769, Z99707, AF004393, Z49777, Q03943, U58284) and a positive control (β-actin). To compare the distribution of signals, the fluorescence values for mRNAs and ncRNAs were grouped into bins of every 100 relative fluorescence units (RFU). The number of signals within each bin was then converted to a percentage of the total number of signals across all tissues for each category. To distinguish real from background levels, the background frequency for each bin (as estimated by the Arabidopsis controls) was subtracted from each of the remaining three groups, and the resulting relative frequency values were plotted. (B) Tissue distribution of up-regulated ncRNAs. Here 178 differentially expressed ncRNAs with >2-fold up-regulation relative to the day 17.5 whole embryo reference tissue were identified using Welch ANOVA with Bonferroni multiple testing correction (P = 0.01). To determine tissue distribution, the number of up-regulated ncRNAs within each tissue was calculated and plotted. (C) Hierarchical clustering of up-regulated ncRNAs. Here 178 differentially expressed ncRNAs showing greater than twofold up-regulation in expression were hierarchically clustered according to tissue expression using the Cluster tool (Eisen et al. 1998). Groups of clones showing tissue-specific expression patterns are indicated: (1) testis; (2) muscle; (3) central nervous system; (4) thymus; (5) pancreas; (6) liver; (7) enteric tract. Tissues labeled with “n” are neonatal tissues. The primary data used for the cluster analysis, including the GenBank ID numbers and the relative intensity ratios of the differentially expressed ncRNA sequences, are given in Supplemental Table 5.
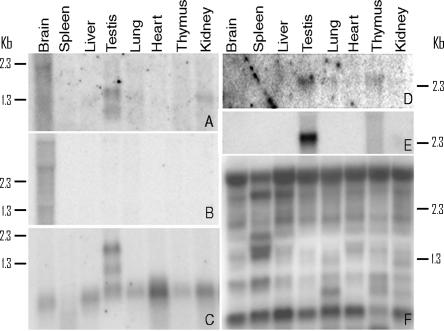
PCR amplification does not give an insight into the transcript size or complexity. To further validate the tissue specificity, and to identify the actual transcripts, eight candidate tissue-specific ncRNAs selected from the filtered set to reflect a range of tissue-specific patterns (see Supplemental Table 6) were analyzed using Northern blots. Six of the eight target ncRNAs were detectable on Northern blots, and exhibited tissue specificity that correlated with the pattern observed in the RIKEN microarray tissues expression profile (Fig. 3; cf. Supplemental Tables 6 and 7). The ncRNAs varied in their level of expression and in their size, ranging from <1 kb to >5 kb. Comparison of the sizes of the cDNAs with their RNA targets (see Fig. 3) suggests that some are full length and some are partial copies, consistent with bioinformatics and PCR analyses (Fig. 1; Supplemental Table 4). One of the probes (GenBank ID AK014924) detected multiple bands (Fig. 3F). Genomic analysis of the probe sequence showed that it contains a stretch of 45 bp that occurs with 98% identity at 11 different positions throughout the mouse genome, none of which corresponds to known protein-coding genes. Thus, this probe may be detecting a family of non-coding RNAs that share a common sequence.
Figure 3.
Northern blot analysis of the size and tissue distribution of selected ncRNAs. Details of the methods can be found in the Supplemental material. The full-length cDNA probes of the eight ncRNAs used in this analysis were obtained from the DNABook (Hayashizaki and Kawai 2004). The corresponding GenBank IDs (and sizes) are: (A) AK035433 (1318 bp); (B) AK028310 (2905 bp); (C) AK017092 (765 bp); (D) AK040014 (139 bp); (E) AK040058 (1141 bp); (F) AK014924 (1799 bp). Total RNA was used, and blots were exposed in a PhosphorImager cassette for 24 h at 4°C. The positions and sizes (in kilobases) of molecular mass markers are shown.
The Northern analyses also demonstrated the presence of tissue-specific alternatively spliced variants of these ncRNAs (Fig. 3, see, e.g., testis samples in A and C), in keeping with previous reports (Sutherland et al. 1996; Tam et al. 1997; Bussemakers et al. 1999; Raho et al. 2000; Charlier et al. 2001; Wolf et al. 2001). This indicates that there may be an additional level of complexity in non-coding RNAs, since (if the observed alternative splicing is meaningful and not some type of tissue-specific noise) it suggests that not only are different ncRNA isoforms relevant in different tissues, but also that it may be important which sequences reside in exons and in introns, possibly because of different subsequent processing pathways and destinations (Rodriguez et al. 2004).
Having demonstrated tissue-specific expression of ncRNAs, we were interested to examine whether there was any evidence of correlation between the expression levels of ncRNAs and mRNAs. We therefore clustered the 178 ncRNAs and 1649 mRNAs detected as being differentially regulated on the cDNA microarrays and specifically looked for clusters in which ncRNAs were highly correlated (R > 0.9) to mRNAs (see Methods). We found 25 groups, containing altogether 65 ncRNAs and 148 mRNAs (Supplemental Table 8). The largest of these contained 23 ncRNAs and 96 mRNAs, all of which were highly expressed in testis (Supplemental Fig. 2A). Included in this cluster were numerous mRNAs implicated in testicular and/or sperm function, including Theg (Nayernia et al. 1999; Yanaka et al. 2000), Tcp10 (Schimenti et al. 1988), Man2a2 (Akama et al. 2002), Adam1b/Ftna (Cho et al. 1997), Adam24 (Zhu et al. 1999), Clgn (Ikawa et al. 1997), and Dnahc8 (Samant et al. 2002). The next largest cluster contained 12 ncRNAs and 19 mRNAs, all of which were highly expressed in the central nervous system (Supplemental Fig. 2B). Several of these mRNAs are known to be have roles in brain function, including Ntng1 (Nakashiba et al. 2000), Dlgh2/PSD93 (Tao et al. 2003; Parker et al. 2004), Nell2 (Matsuyama et al. 2004), and Gabrg2 (Gunther et al. 1995). Clearly, it will be interesting to follow up in future studies whether the ncRNAs identified here play any role in the biology of the mRNAs whose expression patterns they share.
Dynamic regulation of ncRNA transcription
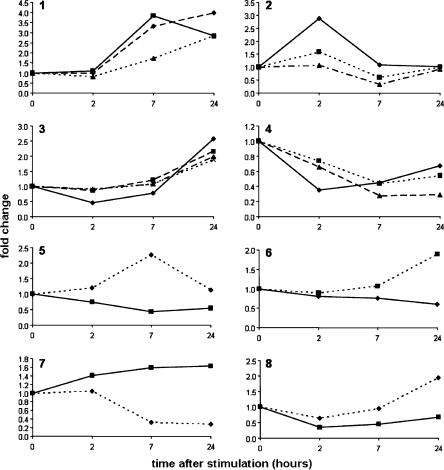
It has previously been reported that mammalian macrophages, a key component of the innate immune response, are among the most complex sources of mRNA (Wells et al. 2003a,b). The expression profile of mouse bone-marrow-derived macrophages changes radically in response to activation by microorganism-derived products such as lipopolysaccharide (LPS). To determine whether ncRNAs exhibit similar dynamic alterations in expression, we examined the time course of macrophage activation in response to LPS. Our array profiling in this system revealed that 70 (1.4%) of 5171 filtered ncRNAs were regulated significantly across the time course of LPS response in C57Bl/6 mouse cells, 53 up-regulated and 17 down-regulated. For the purpose of validation, the dynamics of ncRNA expression were measured directly by quantitative real-time PCR using RNA extracted from mouse macrophages at different time points after LPS activation, and primers specifically designed against 13 selected sequences from the filtered ncRNA set (Supplemental Tables 3 and 7). The results (Fig. 4, Panels 1-4) confirmed that the putative ncRNAs are detectable in macrophages and are, indeed, tightly regulated, confirming what had been previously observed in the microarray profiles. Furthermore, K-means analysis revealed clusters of ncRNAs that share coregulated expression (Fig. 4, Panels 1-4), similar to the coregulated groups of protein-encoding transcripts observed previously in the macrophage mRNA population upon LPS activation (Wells et al. 2003a).
Figure 4.
Dynamics of the expression of putative ncRNAs during macrophage activation by LPS. Panels 1-4 show quantitative real-time PCR profiles for 13 selected ncRNAs, as follows: (1) AK008526 (- ♦ -), AK017043 (—▪—), and AK007024 (- ▴ - -); (2) AK019555 (- - ▴ - -), AV079268 (—♦—) and AK017039 (- ▪ - -); (3) AK009126 (- ▪ -), AK033985 (—♦—), AK017432 (x - -), and AK007998 (- - ▴ - -); (4) AK008218 (- ▴ -), AK018521 (—♦—), and AK035433 (- - ▪ -). Panels 5-8 show examples of expression patterns of selected sense-antisense pairs of ncRNAs with their cognate protein-coding transcripts in macrophages. The sense-antisense pairs are mRNAs (- - ♦ - -) and ncRNAs (—▪—), as follows: (5) Cacnb4/AK035433 (correlation coefficient = -0.77, P-value = 0.23); (6) Ubxd6 (Rep-8)/AK017432 (correlation coefficient = -0.83, P-value = 0.17); (7) Atp5l/AK017092 (correlation coefficient = -0.79, P-value = 0.21); (8) Slc22a6/AK018521 (correlation coefficient = 0.54, P-value = 0.46). RNA was isolated at time 0, and at 2, 7, and 24 h after LPS stimulation. The abscissa indicates the average fold change in ncRNA/mRNA expression compared to time 0.
Several of the ncRNAs map to the opposite (antisense) strand of protein-coding genes. Figure 4 (Panels 5-8) shows the expression of four such overlapping sense/antisense transcript pairs following LPS stimulation of macrophages. The results show that there is no consistent pattern of coordinate expression of these overlapping transcripts, suggesting that, if these ncRNAs are part of local cis-antisense regulatory loops controlling the transcription, stability, or the translation of the overlapping mRNAs, the relationship between the two is not straightforward. However, this does not deny the possibility that some or many sense-antisense transcripts may be coordinately regulated in different ways (Cawley et al. 2004; Katayama et al. 2005).
Subclassification of the experimentally validated ncRNAs
We then further characterized those ncRNAs identified as being differentially expressed on the tissue and macrophage arrays. These 242 transcripts (172 from the tissue arrays, 64 from the macrophage arrays, and six from both) had already passed the bioinformatics filter described earlier, but the possibility remained that some of them may still have represented non-genuine ncRNAs either because they: (1) may contain some protein-coding capacity that was missed by previous filters because of frameshift or other sequencing errors; (2) may be internally primed from pre-mRNA of a known protein-coding transcript; (3) may represent a known or possible unannotated UTR fragment of a protein-coding transcript; or (4) were internally primed from a longer unknown transcript whose coding status remains undetermined. To assess how many of them were high-confidence ncRNA candidates, we therefore manually assessed each for the above four possibilities (see Supplemental Methods). We found that 25 (10%) were in category 1, 47 (19%) were in category 2, 34 (14%) were in category 3, and 21 (9%) were in category 4. Thus, the remainder (115/242, of which almost half were spliced) of the experimentally validated set appear unequivocally genuine. These sequences were also mapped to the human genome using the alignment net files available at UCSC, and 174 human syntenic regions were identified, of which 54 overlapped with transcripts on the same strand.
Discussion
The results presented here show that the cloned non-protein-coding sequences in the RIKEN cDNA collection are not, in the main, derived from genomic or pre-mRNA contamination. Most of the putative ncRNA sequences examined, using reverse-transcriptase-dependent PCR analysis of testis, brain, and stimulated macrophage samples, and microarray profiling or Northern blots with RNA isolated from various tissues, have been shown to be detectable as transcripts that are not related to known or predicted protein-coding genes. Their level of expression is on average lower than that of mRNAs, but without knowing their function we can make no inference about the significance of that fact, although it might be expected if these RNAs had mainly regulatory functions. Low levels of expression are equally true of mRNAs encoding developmental transcriptional regulators.
Like mRNAs, a proportion of the identified ncRNA transcripts exhibit tissue-specific expression. While it might be argued that these ncRNA transcripts partly or largely reflect a background of “transcriptional noise” that varies according to the differential accessibility of chromatin to transcription factors in different types of cells, the existence of such noise, in the sense of illegitimate transcripts from promiscuous promoters as opposed to the stochastic (noisy) firing of bona fide promoters (see, e.g., Elowitz et al. 2002; Ozbudak et al. 2002; Blake et al. 2003), has not been demonstrated, although it may occur. However, the fact that at least some of these ncRNAs exhibit dynamical alteration in their level of expression in response to an acute physiological stimulus (Fig. 4), in patterns that parallel the behavior of mRNAs, suggests that these transcripts are intentional and that their expression is controlled by similar mechanisms.
A similar conclusion was reached by Cawley et al. (2004), who showed that a significant number of non-coding RNAs are regulated in response to retinoic acid, and concluded that protein-coding and non-coding genes are bound by common transcription factors and regulated by common environmental signals. In addition, genome-wide transcriptome analysis has identified large numbers of developmentally coordinated non-coding transcripts in Drosophila that are more conserved than non-expressed non-coding sequences (Stolc et al. 2004). In well-characterized loci, such as the bithorax-abdominal A/B locus, insertions or deletions in sequences encoding ncRNAs (which are themselves developmentally regulated) cause developmental phenotypes, suggesting that these RNAs are involved in gene regulation (Lipshitz et al. 1987; Sanchez-Herrero and Akam 1989; Drewell et al. 2002). Whether these ncRNAs are genetically active in trans (either locally or at a distance), or are produced simply as a consequence of transcriptional interference to activate or repress cis-acting controls, as has been suggested from recent studies in Drosophila (Bender and Fitzgerald 2002; Drewell et al. 2002; Hogga and Karch 2002; Rank et al. 2002) and yeast (Martens et al. 2004), remains to be determined.
The numbers of identified ncRNAs are almost certainly an underestimate of the real numbers, for several reasons. Their relatively low abundance, the majority only identified thus far as singletons in very large and aggressively normalized cDNA libraries containing some 2 × 106 primary clones, suggests that most ncRNAs remain to be identified. The source of RNA for these libraries is primarily RNAs that have 5′-caps and 3′-poly(A) tails, which excludes all RNAs that do not stably possess these features, including those (such as the precursors of miRNAs) that are post-transcriptionally processed to smaller species. In addition, since 60% of the cloned ncRNA sequences appear to be internally primed from poly(A)-rich segments, the likelihood is that many transcripts that lack such segments within a reasonable distance of the 5′-end may have been missed because of the difficulties in cloning longer transcripts. Indeed, there are several fragments of the ncRNA transcript Air, which is >100 kb in length and is clearly functional as a cis-acting transcriptional regulator (Sleutels et al. 2002), present in the FANTOM2 clone set, all of which have a genome-encoded A-rich region following their 3′-ends.
The RIKEN cDNA collection has revealed very approximately equal numbers of mRNA and ncRNA transcripts, which correlates with estimates made from whole-genome transcript analysis using tiled chips, querying limited numbers of cell types (Cawley et al. 2004; Kampa et al. 2004). Evidence summarized elsewhere suggests that ncRNAs may play an important role in the control of differentiation and development, by a variety of mechanisms (Mattick 2003, 2004). Although the RIKEN project included developing embryonic tissues in library construction, embryonic RNAs were not used as test tissues on the microarrays, and it is unlikely that the full spectrum of non-coding transcripts expressed in specific developmental sites and temporal windows was fully sampled. By extension from the limited number of loci that have been intensively studied to date, and ongoing efforts at identifying full-length cDNAs, we would suggest that at least half of all transcripts in mammals do not encode protein (Mattick 2003). A sizeable proportion appear to arise from genuine promoters (Cawley et al. 2004). Detailed computational analysis of ncRNA sequence relationships within genomic and transcriptomic networks, their evolution in different species, and the use of a range of experimental approaches including siRNA knock-down, targeted deletion, and ectopic expression with appropriate phenotypic assays, will be required to establish the roles of ncRNAs in mammalian and metazoan biology.
Methods
Details of the bioinformatics analyses, PCR amplifications, and Northern blots are given in the Supplemental material.
Expression profiling
The 20 tissues selected for analysis were mainly from the major adult organs and included spleen, thymus, kidney, heart, lung, liver, brain, cerebellum, 10-day neonatal cerebellum, placenta, testis, uterus, pancreas, small intestine, stomach, colon, 10-day neonatal skin, bone, muscle, and adipose tissue. Day 17.5 whole embryo was used as the common reference tissue. RNA extraction and cDNA preparation and hybridization to the RIKEN 20K-2 cDNA arrays were performed as previously described (Bono et al. 2003), and experiments were conducted a minimum of four times. Non-coding and protein-coding probes were identified based on manual annotation of the FANTOM2 collection (Okazaki et al. 2002), and data analysis was performed using the Genespring 6.0 software package (Silicon Genetics). In brief, Cy3/Cy5 ratio data were log-transformed, then normalized using a Lowess procedure and median centering, and Welch ANOVA with Bonferroni multiple testing correction (P = 0.01) was used to find differentially expressed genes. To reduce the likelihood of false-positive discovery arising from variation of signals in the background range, a further filter was applied by removing any clones whose raw expression was below median background levels (as estimated from 3× SSC controls) in either the Cy3 or Cy5 channels in a majority of samples. Hierarchical clustering of clones based on tissue expression patterns was performed with the Cluster tool (Eisen et al. 1998) using average linkage clustering on log-transformed Cy3/Cy5 ratio data. To identify groups of correlated ncRNAs and mRNAs, QT clustering (Heyer et al. 1999) was used in the first instance to group any transcripts that were differentially expressed and correlated with a minimum R value of 0.9 (minimum cluster size = 2). Those groups that contained at least one ncRNA were then subject to hierarchical clustering (as described above), and any two or more groups that were sufficiently similar (R > 0.9) were then merged to produce the final clusters.
Acknowledgments
This study was supported by research grants for the National Project on Genome Network Analysis and the RIKEN Genome Exploration Research Project from the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government to Y.H. It was also supported by grants from the Australian Research Council to J.S.M. and D.A.H. The authors thank Takeya Kasukawa and Pär Engstrom for assistance with the bioinformatics analyses and Khairina Tajul Arifin for assistance with the microarray analyses.
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4200206.
Footnotes
[Supplemental material is available online at www.genome.org. The microarray data from this study have been submitted to the Gene Expression Omnibus under accession nos. GSD275 and GSE3098.]
References
- Akama, T.O., Nakagawa, H., Sugihara, K., Narisawa, S., Ohyama, C., Nishimura, S., O'Brien, D.A., Moremen, K.W., Millan, J.L., and Fukuda, M.N. 2002. Germ cell survival through carbohydrate-mediated interaction with Sertoli cells. Science 295 124-127. [DOI] [PubMed] [Google Scholar]
- Ashe, H.L., Monks, J., Wijgerde, M., Fraser, P., and Proudfoot, N.J. 1997. Intergenic transcription and transinduction of the human β-globin locus. Genes & Dev. 11 2494-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger, J.H. and Olsen, G.J. 1999. CRITICA: Coding region identification tool invoking comparative analysis. Mol. Biol. Evol. 16 512-524. [DOI] [PubMed] [Google Scholar]
- Bender, W. and Fitzgerald, D.P. 2002. Transcription activates repressed domains in the Drosophila bithorax complex. Development 129 4923-4930. [DOI] [PubMed] [Google Scholar]
- Bertone, P., Stolc, V., Royce, T.E., Rozowsky, J.S., Urban, A.E., Zhu, X., Rinn, J.L., Tongprasit, W., Samanta, M., Weissman, S., et al. 2004. Global identification of human transcribed sequences with genome tiling arrays. Science 306 2242-2246. [DOI] [PubMed] [Google Scholar]
- Blake, W.J., Kaern, M., Cantor, C.R., and Collins, J.J. 2003. Noise in eukaryotic gene expression. Nature 422 633-637. [DOI] [PubMed] [Google Scholar]
- Bono, H., Yagi, K., Kasukawa, T., Nikaido, I., Tominaga, N., Miki, R., Mizuno, Y., Tomaru, Y., Goto, H., Nitanda, H., et al. 2003. Systematic expression profiling of the mouse transcriptome using RIKEN cDNA microarrays. Genome Res. 13 1318-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussemakers, M.J., van Bokhoven, A., Verhaegh, G.W., Smit, F.P., Karthaus, H.F., Schalken, J.A., Debruyne, F.M., Ru, N., and Isaacs, W.B. 1999. DD3: A new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 59 5975-5979. [PubMed] [Google Scholar]
- Cai, X., Hagedorn, C.H., and Cullen, B.R. 2004. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10 1957-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci, P., Waki, K., Shiraki, T., Konno, H., Shibata, K., Itoh, M., Aizawa, K., Arakawa, T., Ishii, Y., Sasaki, D., et al. 2003. Targeting a complex transcriptome: The construction of the mouse full-length cDNA encyclopedia. Genome Res. 13 1273-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci, P., Kasukawa, T., Katayama, S., Gough, J., Frith, M.C., Maeda, N., Oyama, R., Ravasi, T., Lenhard, B., Wells, C., et al. 2005. The transcriptional landscape of the mammalian genome. Science 309 1559-1563. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C. and Ambros, V. 2003. Role of microRNAs in plant and animal development. Science 301 336-338. [DOI] [PubMed] [Google Scholar]
- Cawley, S., Bekiranov, S., Ng, H.H., Kapranov, P., Sekinger, E.A., Kampa, D., Piccolboni, A., Sementchenko, V., Cheng, J., Williams, A.J., et al. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116 499-509. [DOI] [PubMed] [Google Scholar]
- Charlier, C., Segers, K., Wagenaar, D., Karim, L., Berghmans, S., Jaillon, O., Shay, T., Weissenbach, J., Cockett, N., Gyapay, G., et al. 2001. Human-ovine comparative sequencing of a 250-kb imprinted domain encompassing the callipyge (clpg) locus and identification of six imprinted transcripts: DLK1, DAT, GTL2, PEG11, antiPEG11, and MEG8. Genome Res. 11 850-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, J., Kapranov, P., Drenkow, J., Dike, S., Brubaker, S., Patel, S., Long, J., Stern, D., Tammana, H., Helt, G., et al. 2005. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308 1149-1154. [DOI] [PubMed] [Google Scholar]
- Cho, C., Turner, L., Primakoff, P., and Myles, D.G. 1997. Genomic organization of the mouse fertilin β gene that encodes an ADAM family protein active in sperm-egg fusion. Dev. Genet. 20 320-328. [DOI] [PubMed] [Google Scholar]
- Drewell, R.A., Bae, E., Burr, J., and Lewis, E.B. 2002. Transcription defines the embryonic domains of cis-regulatory activity at the Drosophila bithorax complex. Proc. Natl. Acad. Sci. 99 16853-16858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. 95 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz, M.B., Levine, A.J., Siggia, E.D., and Swain, P.S. 2002. Stochastic gene expression in a single cell. Science 297 1183-1186. [DOI] [PubMed] [Google Scholar]
- Furuno, M., Kasukawa, T., Saito, R., Adachi, J., Suzuki, H., Baldarelli, R., Hayashizaki, Y., and Okazaki, Y. 2003. CDS annotation in full-length cDNA sequence. Genome Res. 13 1478-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges, M., Charlier, C., and Cockett, N. 2003. The callipyge locus: Evidence for the trans interaction of reciprocally imprinted genes. Trends Genet. 19 248-252. [DOI] [PubMed] [Google Scholar]
- Grimmond, S.M., Miranda, K.C., Yuan, Z., Davis, M.J., Hume, D.A., Yagi, K., Tominaga, N., Bono, H., Hayashizaki, Y., Okazaki, Y., et al. 2003. The mouse secretome: Functional classification of the proteins secreted into the extracellular environment. Genome Res. 13 1350-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther, U., Benson, J., Benke, D., Fritschy, J.M., Reyes, G., Knoflach, F., Crestani, F., Aguzzi, A., Arigoni, M., Lang, Y., et al. 1995. Benzodiazepine-insensitive mice generated by targeted disruption of the γ 2 subunit gene of γ-aminobutyric acid type A receptors. Proc. Natl. Acad. Sci. 92 7749-7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, I.M., Noma, K., and Grewal, S.I. 2003. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc. Natl. Acad. Sci. 100 193-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashizaki, Y. and Kawai, J. 2004. A new approach to the distribution and storage of genetic resources. Nat. Rev. Genet. 5 223-228. [DOI] [PubMed] [Google Scholar]
- Heyer, L.J., Kruglyak, S., and Yooseph, S. 1999. Exploring expression data: Identification and analysis of coexpressed genes. Genome Res. 9 1106-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogga, I. and Karch, F. 2002. Transcription through the iab-7 cis-regulatory domain of the bithorax complex interferes with maintenance of Polycomb-mediated silencing. Development 129 4915-4922. [DOI] [PubMed] [Google Scholar]
- Holmes, R., Williamson, C., Peters, J., Denny, P., and Wells, C. 2003. A comprehensive transcript map of the mouse Gnas imprinted complex. Genome Res. 13 1410-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawa, M., Wada, I., Kominami, K., Watanabe, D., Toshimori, K., Nishimune, Y., and Okabe, M. 1997. The putative chaperone calmegin is required for sperm fertility. Nature 387 607-611. [DOI] [PubMed] [Google Scholar]
- Kampa, D., Cheng, J., Kapranov, P., Yamanaka, M., Brubaker, S., Cawley, S., Drenkow, J., Piccolboni, A., Bekiranov, S., Helt, G., et al. 2004. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 14 331-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov, P., Cawley, S.E., Drenkow, J., Bekiranov, S., Strausberg, R.L., Fodor, S.P., and Gingeras, T.R. 2002. Large-scale transcriptional activity in chromosomes 21 and 22. Science 296 916-919. [DOI] [PubMed] [Google Scholar]
- Kapranov, P., Drenkow, J., Cheng, J., Long, J., Helt, G., Dike, S., and Gingeras, T.R. 2005. Examples of the complex architecture of the human transcriptome revealed by RACE and high density tiling arrays. Genome Res. 15 987-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama, S., Tomaru, Y., Kasukawa, T., Waki, K., Nakanishi, M., Nakamura, M., Nishida, H., Yap, C.C., Suzuki, M., Kawai, J., et al. 2005. Antisense transcription in the mammalian transcriptome. Science 309 1564-1566. [DOI] [PubMed] [Google Scholar]
- Kawasaki, H. and Taira, K. 2004. Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature 431 211-217. [DOI] [PubMed] [Google Scholar]
- Levinson, B., Kenwrick, S., Lakich, D., Hammonds Jr., G., and Gitschier, J. 1990. A transcribed gene in an intron of the human factor VIII gene. Genomics 7 1-11. [DOI] [PubMed] [Google Scholar]
- Lipshitz, H.D., Peattie, D.A., and Hogness, D.S. 1987. Novel transcripts from the ultrabithorax domain of the bithorax complex. Genes & Dev. 1 307-322. [DOI] [PubMed] [Google Scholar]
- Martens, J.A., Laprade, L., and Winston, F. 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429 571-574. [DOI] [PubMed] [Google Scholar]
- Matsuyama, S., Aihara, K., Nishino, N., Takeda, S., Tanizawa, K., Kuroda, S., and Horie, M. 2004. Enhanced long-term potentiation in vivo in dentate gyrus of NELL2-deficient mice. Neuroreport 15 417-420. [DOI] [PubMed] [Google Scholar]
- Mattick, J.S. 2003. Challenging the dogma: The hidden layer of non-protein-coding RNAs in complex organisms. Bioessays 25 930-939. [DOI] [PubMed] [Google Scholar]
- ———. 2004. RNA regulation: a new genetics? Nat. Rev. Genet. 5 316-323. [DOI] [PubMed] [Google Scholar]
- Mattick, J.S. and Gagen, M.J. 2001. The evolution of controlled multitasked gene networks: The role of introns and other noncoding RNAs in the development of complex organisms. Mol. Biol. Evol. 18 1611-1630. [DOI] [PubMed] [Google Scholar]
- Mattick, J.S. and Makunin, I.V. 2005. Small regulatory RNAs in mammals. Hum. Mol. Genet. 14 R121-R132. [DOI] [PubMed] [Google Scholar]
- Miller, D., Briggs, D., Snowden, H., Hamlington, J., Rollinson, S., Lilford, R., and Krawetz, S.A. 1999. A complex population of RNAs exists in human ejaculate spermatozoa: Implications for understanding molecular aspects of spermiogenesis. Gene 237 385-392. [DOI] [PubMed] [Google Scholar]
- Mochizuki, K., Fine, N.A., Fujisawa, T., and Gorovsky, M.A. 2002. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell 110 689-699. [DOI] [PubMed] [Google Scholar]
- Mount, S. and Henikoff, S. 1993. Nested genes take flight. Curr. Biol. 3 372-374. [DOI] [PubMed] [Google Scholar]
- Nakashiba, T., Ikeda, T., Nishimura, S., Tashiro, K., Honjo, T., Culotti, J.G., and Itohara, S. 2000. Netrin-G1: A novel glycosyl phosphatidylinositol-linked mammalian netrin that is functionally divergent from classical netrins. J. Neurosci. 20 6540-6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam, D.K., Lee, S., Zhou, G., Cao, X., Wang, C., Clark, T., Chen, J., Rowley, J.D., and Wang, S.M. 2002. Oligo(dT) primer generates a high frequency of truncated cDNAs through internal poly(A) priming during reverse transcription. Proc. Natl. Acad. Sci. 99 6152-6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayernia, K., von Mering, M.H., Kraszucka, K., Burfeind, P., Wehrend, A., Kohler, M., Schmid, M., and Engel, W. 1999. A novel testicular haploid expressed gene (THEG) involved in mouse spermatid-Sertoli cell interaction. Biol. Reprod. 60 1488-1495. [DOI] [PubMed] [Google Scholar]
- Numata, K., Kanai, A., Saito, R., Kondo, S., Adachi, J., Wilming, L.G., Hume, D.A., Hayashizaki, Y., and Tomita, M. 2003. Identification of putative noncoding RNAs among the RIKEN mouse full-length cDNA collection. Genome Res. 13 1301-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., Nikaido, I., Osato, N., Saito, R., Suzuki, H., et al. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420 563-573. [DOI] [PubMed] [Google Scholar]
- Ota, T., Suzuki, Y., Nishikawa, T., Otsuki, T., Sugiyama, T., Irie, R., Wakamatsu, A., Hayashi, K., Sato, H., Nagai, K., et al. 2004. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 36 40-45. [DOI] [PubMed] [Google Scholar]
- Ozbudak, E.M., Thattai, M., Kurtser, I., Grossman, A.D., and van Oudenaarden, A. 2002. Regulation of noise in the expression of a single gene. Nat. Genet. 31 69-73. [DOI] [PubMed] [Google Scholar]
- Pang, K.C., Stephen, S., Engström, P.G., Tajul-Arifin, K., Chen, W., Wahlestedt, C., Lenhard, B., Hayashizaki, Y., and Mattick, J.S. 2005. RNAdb—A comprehensive mammalian noncoding RNA database. Nucleic Acids Res. 33 D125-D130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, K.C., Frith, M.C., and Mattick, J.S. 2006. Rapid evolution of noncoding RNAs: Lack of conservation does not mean lack of function. Trends Genet. 22 (in press). [DOI] [PubMed]
- Parker, M.J., Zhao, S., Bredt, D.S., Sanes, J.R., and Feng, G. 2004. PSD93 regulates synaptic stability at neuronal cholinergic synapses. J. Neurosci. 24 378-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesskaya, O.O., Haroutunian, V., Davis, K.L., Hernandez, I., and Sokolov, B.P. 2003. Novel putative nonprotein-coding RNA gene from 11q14 displays decreased expression in brains of patients with schizophrenia. J. Neurosci. Res. 74 111-122. [DOI] [PubMed] [Google Scholar]
- Raho, G., Barone, V., Rossi, D., Philipson, L., and Sorrentino, V. 2000. The gas 5 gene shows four alternative splicing patterns without coding for a protein. Gene 256 13-17. [DOI] [PubMed] [Google Scholar]
- Rank, G., Prestel, M., and Paro, R. 2002. Transcription through intergenic chromosomal memory elements of the Drosophila bithorax complex correlates with an epigenetic switch. Mol. Cell Biol. 22 8026-8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisman, D., Balint, E., Loging, W.T., Rotter, V., and Almon, E. 1996. A novel transcript encoded within the 10-kb first intron of the human p53 tumor suppressor gene (D17S2179E) is induced during differentiation of myeloid leukemia cells. Genomics 38 364-370. [DOI] [PubMed] [Google Scholar]
- Rodriguez, A., Griffiths-Jones, S., Ashurst, J.L., and Bradley, A. 2004. Identification of mammalian microRNA host genes and transcription units. Genome Res. 14 1902-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samant, S.A., Ogunkua, O., Hui, L., Fossella, J., and Pilder, S.H. 2002. The T complex distorter 2 candidate gene, Dnahc8, encodes at least two testis-specific axonemal dynein heavy chains that differ extensively at their amino and carboxyl termini. Dev. Biol. 250 24-43. [DOI] [PubMed] [Google Scholar]
- Sanchez-Herrero, E. and Akam, M. 1989. Spatially ordered transcription of regulatory DNA in the bithorax complex of Drosophila. Development 107 321-329. [DOI] [PubMed] [Google Scholar]
- Schadt, E.E., Edwards, S.W., GuhaThakurta, D., Holder, D., Ying, L., Svetnik, V., Leonardson, A., Hart, K.W., Russell, A., Li, G., et al. 2004. A comprehensive transcript index of the human genome generated using microarrays and computational approaches. Genome Biol. 5 R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimenti, J., Cebra-Thomas, J.A., Decker, C.L., Islam, S.D., Pilder, S.H., and Silver, L.M. 1988. A candidate gene family for the mouse t complex responder (Tcr) locus responsible for haploid effects on sperm function. Cell 55 71-78. [DOI] [PubMed] [Google Scholar]
- Seitz, H., Royo, H., Bortolin, M.L., Lin, S.P., Ferguson-Smith, A.C., and Cavaille, J. 2004. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res. 14 1741-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels, F., Zwart, R., and Barlow, D.P. 2002. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415 810-813. [DOI] [PubMed] [Google Scholar]
- Stolc, V., Gauhar, Z., Mason, C., Halasz, G., van Batenburg, M.F., Rifkin, S.A., Hua, S., Herreman, T., Tongprasit, W., Barbano, P.E., et al. 2004. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science 306 655-660. [DOI] [PubMed] [Google Scholar]
- Sutherland, H.F., Wadey, R., McKie, J.M., Taylor, C., Atif, U., Johnstone, K.A., Halford, S., Kim, U.J., Goodship, J., Baldini, A., et al. 1996. Identification of a novel transcript disrupted by a balanced translocation associated with DiGeorge syndrome. Am. J. Hum. Genet. 59 23-31. [PMC free article] [PubMed] [Google Scholar]
- Tam, W., Ben-Yehuda, D., and Hayward, W.S. 1997. bic, a novel gene activated by proviral insertions in avian leukosis virus-induced lymphomas, is likely to function through its noncoding RNA. Mol. Cell Biol. 17 1490-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Y.X., Rumbaugh, G., Wang, G.D., Petralia, R.S., Zhao, C., Kauer, F.W., Tao, F., Zhuo, M., Wenthold, R.J., Raja, S.N., et al. 2003. Impaired NMDA receptor-mediated postsynaptic function and blunted NMDA receptor-dependent persistent pain in mice lacking postsynaptic density-93 protein. J. Neurosci. 23 6703-6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting, A.H., Schuebel, K.E., Herman, J.G., and Baylin, S.B. 2005. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat. Genet. 37 906-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe, T., Schramke, V., Hamilton, G.L., White, S.A., Teng, G., Martienssen, R.A., and Allshire, R.C. 2003. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 11 137-146. [DOI] [PubMed] [Google Scholar]
- Wells, C.A., Ravasi, T., Faulkner, G.J., Carninci, P., Okazaki, Y., Hayashizaki, Y., Sweet, M., Wainwright, B.J., and Hume, D.A. 2003a. Genetic control of the innate immune response. BMC Immunol. 4 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells, C.A., Ravasi, T., Sultana, R., Yagi, K., Carninci, P., Bono, H., Faulkner, G., Okazaki, Y., Quackenbush, J., Hume, D.A., et al. 2003b. Continued discovery of transcriptional units expressed in cells of the mouse mononuclear phagocyte lineage. Genome Res. 13 1360-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, S., Mertens, D., Schaffner, C., Korz, C., Dohner, H., Stilgenbauer, S., and Lichter, P. 2001. B-cell neoplasia associated gene with multiple splicing (BCMS): The candidate B-CLL gene on 13q14 comprises more than 560 kb covering all critical regions. Hum. Mol. Genet. 10 1275-1285. [DOI] [PubMed] [Google Scholar]
- Yanaka, N., Kobayashi, K., Wakimoto, K., Yamada, E., Imahie, H., Imai, Y., and Mori, C. 2000. Insertional mutation of the murine kisimo locus caused a defect in spermatogenesis. J. Biol. Chem. 275 14791-14794. [DOI] [PubMed] [Google Scholar]
- Ying, S.Y. and Lin, S.L. 2005. Intronic microRNAs. Biochem. Biophys. Res. Commun. 326 515-520. [DOI] [PubMed] [Google Scholar]
- Zavolan, M., Kondo, S., Schonbach, C., Adachi, J., Hume, D.A., Hayashizaki, Y., and Gaasterland, T. 2003. Impact of alternative initiation, splicing, and termination on the diversity of the mRNA transcripts encoded by the mouse transcriptome. Genome Res. 13 1290-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, G.Z., Lin, Y., Myles, D.G., and Primakoff, P. 1999. Identification of four novel ADAMs with potential roles in spermatogenesis and fertilization. Gene 234 227-237. [DOI] [PubMed] [Google Scholar]