Abstract
Recent evidence points to considerable transcription occurring in non-protein-coding regions of eukaryote genomes. However, their lack of conservation and demonstrated function have created controversy over whether these transcripts are functional. Applying a novel cloning strategy, we have cloned 100 novel and 61 known or predicted Caenorhabditis elegans full-length ncRNAs. Studying the genomic environment and transcriptional characteristics have shown that two-thirds of all ncRNAs, including many intronic snoRNAs, are independently transcribed under the control of ncRNA-specific upstream promoter elements. Furthermore, the transcription levels of at least 60% of the ncRNAs vary with developmental stages. We identified two new classes of ncRNAs, stem-bulge RNAs (sbRNAs) and snRNA-like RNAs (snlRNAs), both featuring distinct internal motifs, secondary structures, upstream elements, and high and developmentally variable expression. Most of the novel ncRNAs are conserved in Caenorhabditis briggsae, but only one homolog was found outside the nematodes. Preliminary estimates indicate that the C. elegans transcriptome contains ∼2700 small non-coding RNAs, potentially acting as regulatory elements in nematode development.
Small non-protein-coding RNAs (ncRNAs) such as microRNAs (miRNAs) have received increasing attention as regulatory factors shaping cellular and organismal life. A catalog of reported miRNAs from various eukaryotes now covers 400 different RNA species (Liu et al. 2005). In Caenorhabditis elegans, 120 miRNAs have been identified experimentally and computationally. However, the remainder of the C. elegans small non-coding transcriptome has received far less attention (Stricklin et al. 2005). So far the C. elegans spliceosomal snRNAs and a small number of box C/D snoRNAs (Thomas et al. 1990; Higa et al. 2002; Wachi et al. 2004) have been experimentally verified, whereas other common eukaryotic ncRNAs (RNase P RNA, Y RNA, SRP RNAs) have only been identified as putative loci in the genomic sequence (Harris et al. 2003; Stricklin et al. 2005). Investigations into the small non-coding transcriptome of several eukaryote model organisms have invariably revealed several ncRNAs that could not be assigned to any known functional class (Huttenhofer et al. 2001; Marker et al. 2002; Yuan et al. 2003). A recent study of the protist Disctyostelium discoideum found 16 sequences (out of a total of 36 novel ncRNAs) that, based on conserved internal and upstream motifs, could be assigned to one of two novel classes of small non-coding RNAs (Aspegren et al. 2004). Together, these studies clearly indicate the presence of a still unexplored segment of the eukaryote small non-coding transcriptome.
One particularly intriguing aspect of the non-coding transcriptome is its potential to fill the regulatory gap created by the surprisingly low number of protein-coding genes in higher organisms. Between one-celled yeast, thousand-celled nematodes, and trillion-celled mammals, there is a difference of a mere 6000 to 19,000 to 25,000 in protein-coding gene numbers; regulation by non-coding RNA has been suggested to account for this discrepancy (Mattick 2003, 2004). The recent census of the known and verified ncRNA population stands at around 5300 different species (Liu et al. 2005), and may be as high as 20,000 if mRNA-like transcripts are included (Pang et al. 2005), of which a considerable fraction has been ascribed regulatory roles. High-density microarray analyses indicate a large quantity of transcriptionally active DNA outside annotated or predicted protein-coding regions (Okazaki et al. 2002; Yamada et al. 2003; Bertone et al. 2004; Cawley et al. 2004; Kampa et al. 2004; Stolc et al. 2005). This would bring the number of non-coding transcripts into several tens of thousands, if a substantial fraction of the transcriptionally active DNA actually codes for functionally active transcripts. The significance of these mostly non-conserved transcripts, however, still remains highly contentious (Wang et al. 2004; Huttenhofer et al. 2005).
We have attempted to map out part of the small non-coding transcriptome of C. elegans. Except for miRNAs, we have retrieved transcripts of nearly all predicted and previously verified small ncRNAs in C. elegans in addition to 100 novel ncRNAs. Of these novel ncRNAs, one-third do not belong to any known functional class of ncRNAs, and at least two groups of transcripts display conserved features indicative of two novel functional classes. The genomic organization of the C. elegans small ncRNAs is also peculiar in two aspects. Contrary to other metazoans, several of the snoRNA genes appear to be transcribed from independent promoters, of which at least one type bears little sequence resemblance to previously described ncRNA promoters (Thomas et al. 1990). Moreover, a considerable fraction of apparently independently transcribed ncRNA genes (of various functional classes) is located in introns of protein-coding genes, a type of genomic organization seldom observed in other organisms.
Results
ncRNA-specific library
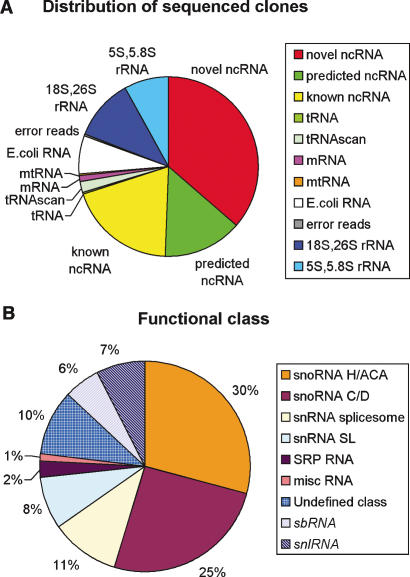
We used a new strategy to construct an ncRNA-specific full-length library of C. elegans from mixed-stage worms and eggs (see Methods). Our procedure ensured a substantial fraction of full-length clones with defined 5′- and 3′-termini, while allowing us to distinguish between 5′-capped and uncapped transcripts. The number of fragments of unwanted RNAs (rRNAs, tRNAs, and mRNAs) was also significantly reduced compared to previous efforts (see Discussion). Full-length cDNA libraries of both capped and uncapped transcripts were established, and altogether 2178 clones were sequenced (Fig. 1A). Thirty-six percent of the clones in our library represented 100 novel transcripts with confirmed expression (Northern blot). Another 34% contained 61 different representatives of all known and predicted families of short ncRNAs in C. elegans, indicating that the cloning strategy had very high sensitivity. Only four potential novel RNAs were discarded because of lack of detectable expression or defined 5′-and 3′-ends. Northern blot analyses of different developmental stages and environmental condition revealed substantial variation in expression of at least 60% of the novel transcripts.
Figure 1.
Clonal and functional distributions. (A) Distribution of sequenced library clones on different RNA species and categories. The E. coli RNAs are contaminants from food bacteria ingested by C. elegans. (tRNAscan) tRNAs detected by tRNAscan (Lowe and Eddy 1997); (mtRNA) mitochondrial RNA. (B) Functional distribution of all novel and known ncRNAs detected in this study. Sectors representing RNAs of un defined and novel functional classes are hatched or gridded. misc RNA includes RNase P RNA and Y RNA.
Genomic location of the novel ncRNAs
The genetic loci of the novel transcripts show no particular chromosomal distribution, but a major fraction (55%) is oriented in the sense direction within the transcribed region of a known or predicted gene, intronic loci constituting the larger part of these. Among functionally annotated genes hosting one or more intronic ncRNA loci, 30% code for a ribosomal protein, and a considerable fraction (24% of all novel loci) is also located within a C. elegans operon (see Supplemental material). Three loci are found in UTRs of coding genes, and one locus (cen42) overlaps the junction between a coding exon and the following intron. Ten loci (10%) are located antisense to introns of protein-coding genes, and 36% of the novel loci have intergenic positions.
The data reveal two aspects of the genomic organization of small ncRNA genes specific to C. elegans. The first is that the large fraction of intronic ncRNA loci does not only include snoRNAs, but also snRNA, SL2 RNA, and SRP RNA loci. Secondly, a considerable fraction of the snoRNAs detected has intergenic loci. Of the 89 snoRNA loci in our material, 30 are intergenic, and with a few exceptions, these 30 loci also display conserved upstream motifs (see below).
Functional distribution of the ncRNAs
Of the 161 known and novel ncRNAs detected, roughly one-half correspond to snoRNA-like transcripts, one-fourth have other known ncRNA functions, and the remaining one-fourth have no obvious cellular function (Fig. 1B). The 100 novel ncRNAs fall in two major categories, novel snoRNA-like transcripts (69) and transcripts not belonging to any previously known functional class (31). Among the snoRNA-like transcripts, 42 have conserved sequence elements and secondary structure (data not shown) resembling the box H/ACA subclass, whereas 27 transcripts have been classified as box C/D snoRNAs. One box H/ACA snoRNA was found to contain conserved elements common to the vertebrate snR30 snoRNA involved in rRNA processing (Atzorn et al. 2004).
C. elegans ncRNAs are conserved in Caenorhabditis briggsae
Of our 100 novel ncRNAs, 70 have recognizable counterparts in C. briggsae with >40% sequence identity. The average identity of the transcribed novel ncRNAs sequences (60%) is around thrice that of the immediate flanking 5′ and 3′ sequences (16.1% and 21.5%, respectively). Nearly one-third of the novel, functionally unknown ncRNAs (10 out of 31), along with 22 novel snoRNA-like transcripts, are >85% identical in C. briggsae, a conservation level on par with that of the spliceosomal snRNAs. One transcript (CeN96) was identified by two short conserved sequence motifs as a C. elegans homolog of the yeast/vertebrate snR30/U17 snoRNA required for processing of 18S rRNA (Atzorn et al. 2004). Aside from CeN96, no sequence homologs of the novel ncRNAs could be found in other species.
A majority of the C. elegans ncRNAs show developmentally variable expression
Using Northern blots, we observed transcription levels of 20 known or predicted and 86 novel ncRNA families (comprising 44 and 87 different transcripts, respectively). Total RNA was extracted at 12 different developmental stages from egg to mature adult, in dauer worms and after heat-shock treatment. Hybridization signals from Northern blots were recorded to obtain quantitative estimates of the ncRNA expression levels. Sixty of the ncRNA families (covering 61 ncRNA species) showed variations in expression exceeding two standard deviations at certain developmental stages or environmental conditions. An additional 40 families (57 species) showed similar tendencies of regulated expression at a lower level of variation. Only six ncRNA families were devoid of variation in expression over all conditions tested. Among these were four of the five spliceosomal snRNA families tested (the fifth, snRNA U5, displayed a slight [I = 1.08] tendency to elevated transcription in dauer worms) (see Supplemental material for details).
Clustering the ncRNAs according to similarities in expression produced six distinct profiles comprising altogether 72 transcripts (Supplemental Fig. S-1). Eight ncRNA families, the majority of which are novel ncRNAs of unknown function, displayed significantly elevated expression toward the later worm stages. While only one ncRNA showed significantly elevated expression at early development stages, at least 11 others showed slight increases. Several ncRNAs, mostly snoRNA-like transcripts expressed from the UM2 containing loci, showed highest expression during middle stages of development (see below for details).
The dauer worm cluster was particularly important because 28 different snoRNAs showed significantly elevated expression at this stage. Conversely, all snRNAs showed unaltered expression in dauer, indicating that the data reflect physiologically relevant ncRNA levels. Because most of the dauer-expressed transcripts are snoRNA genes, probably transcribed with and processed from their host gene transcripts, it is also possible that the elevated expression levels of the ncRNAs in the dauer state represent altered activation of their host genes, or differential processing of pre-mRNAs to mRNAs or to snoRNAs (de Turris et al. 2004). Six ncRNAs with significantly elevated expression in starved L1 (L1s) larvae also showed high expression in dauer worms, possibly indicating a relation to hunger stress. Heat-shock treatment (30°C for 3 h) increased expression above twofold in three ncRNAs, two of which also displayed high dauer expression, suggesting a role in stress-related regulatory processes.
Two novel functional classes, sbRNAs and snlRNAs
Analysis of the 31 novel ncRNAs that could not be assigned to any known functional class identified two new functional classes of ncRNAs. According to secondary structure features and internal motifs, we labeled the respective groups stem-bulge RNAs (sbRNAs) and snRNA-like RNAs (snlRNAs).
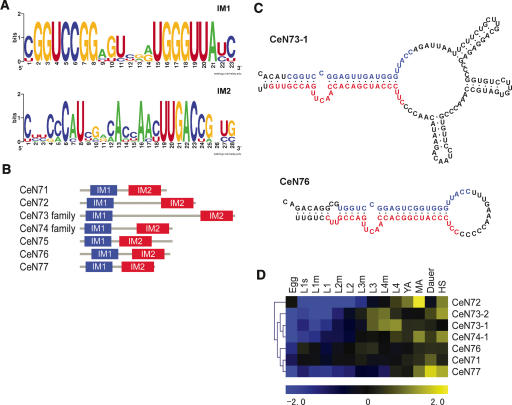
The stem-bulge RNA class (sbRNAs), consisting of nine novel transcripts, is characterized by two distinct motifs (IM1 and IM2) located at the 5′- and 3′-termini (Fig. 2A,B). These conserved motifs have the potential of forming a double-stranded helix featuring a bulge with a conserved sequence (AACUU) separated by a single-stranded RNA loop of varying length (Fig. 2C). The construct could serve as a binding site for common protein/protein complexes. Searches for combinations of the IM1 and IM2 sequence revealed four additional high confidence hits in the C. elegans genome, and 11 in the C. briggsae genome (data not shown). All sbRNAs share a common upstream motif (UM3, see below) including a TATA-box. A few of the sbRNA genes (e.g., cen73 and cbp9) are obvious sequence and structural homologs. But for most of the C. briggsae sbRNAs, only IM1 and IM2 are conserved, whereas the intervening loop shows little sequence conservation. No sequence homologs of sbRNAs were found in any other organism; however, the recently discovered Class I ncRNAs in Dictyostelium shows a similar pattern of internal stem-forming conserved 5′- and 3′-end motifs (Aspegren et al. 2004), possibly indicating some relationship to the nematode sbRNAs. An additional aspect of the sbRNA loci is that they frequently occur in clusters of two or three closely located genes, reflecting perhaps a dependence on a common DNA environment. The expression levels of most sbRNAs vary considerably during the course of C. elegans development, with a tendency toward higher expression in later stages of worm development (Fig. 2D).
Figure 2.
The stem-bulge RNAs of C. elegans. (A) Sequential composition of the 5′-end (IM1; E = 1.2 × 10–19) and 3′-end (IM2; E = 1.0 × 10–20) motifs of the sbRNAs (see Supplemental material for details on the E-value). (B) Relative positions of IM1 and IM2 within each of the verified sbRNAs. (C) Predicted (Mfold) secondary structure of sbRNAs CeN73-1 and CeN76. (D) Relative expression of seven of the sbRNAs at heatshock (HS) and different developmental stages (Egg through Dauer; see complete list of abbreviations in Supplemental material).
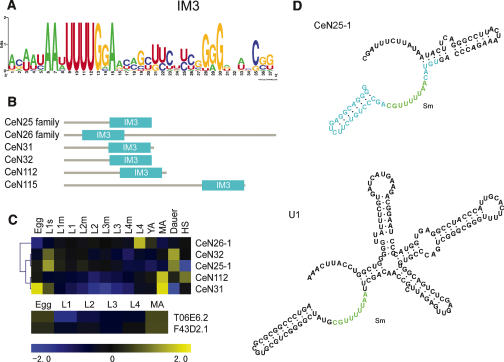
The second novel class, snlRNAs, consisting of eight novel transcripts, is characterized by the presence of internal motif 3 (IM3) (Fig. 3A; Supplemental Table S-2). The IM3 sequence includes the conserved sequence AARUUUUGGA reported as the Sm protein-binding site in spliceosomal snRNAs (Riedel et al. 1987), but is at least 20 nt longer than a traditional Sm protein-binding site, and may thus contain binding sites for additional proteins (Fig. 3D). The IM3 sequence is mostly located in the 3′-terminal half of the transcripts (Fig. 3B), and is also found in all SL2 snRNAs and the predicted U3 snoRNAs. The presence of the Sm binding site within IM3 possibly indicates that snlRNAs have a role in splicing or other Sm related functions. Similar to snRNAs, several of the snlRNAs appear as multi-copy gene families consisting of two or more ncRNAs with minor sequence differences. With a few exceptions, the snlRNA loci share the same upstream motif (UM1, see below) with C. elegans spliceosomal snRNAs.
Figure 3.
The snlRNAs of C. elegans. (A) Sequence of the IM3 motif of snlRNAs (E = 1.8 × 10–37). (B) Internal position of IM3 in the snlRNAs. (C) Relative expression of five snlRNAs and two C. elegans cyclin mRNAs (T06E6.2 and F43D2.1; Wang and Kim 2003). (D) Comparison of the secondary structures of snlRNA CeN25-1 and snRNA U1. In addition to the Sm-binding site, both RNAs show a similar 3′-tail stem-loop structure, but the remainder of the IM3 motif is absent in U1 snRNA.
Like spliceosomal snRNAs, most snlRNAs appear to have high expression levels, which tend to increase even more toward later stages of worm development. The snlRNA CeN31 is particularly intriguing, showing high expression both in the egg/embryo stage and in the mature adult, whereas intervening stages all have low expression (Fig. 3C). This class of ncRNAs correlates strongly with several mRNAs in C. elegans (Wang and Kim 2003), including two of the seven cyclin genes (Fig. 3C), suggesting involvement of these ncRNAs in cell division.
ncRNA-specific upstream motifs
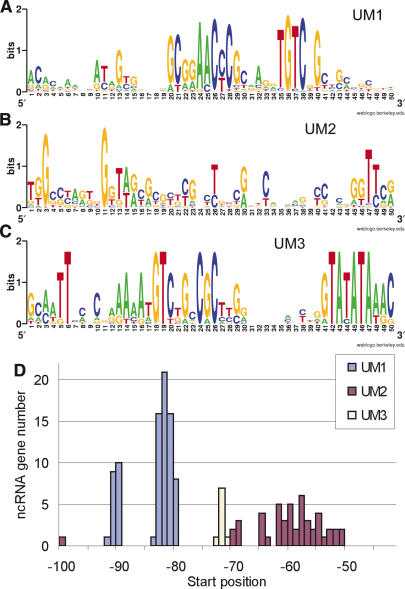
Analysis of the 100-bp 5′-end flanking sequences of the novel ncRNAs using MEME (Bailey and Elkan 1995) yielded three distinct 50-bp upstream motifs (UM1-3) (Fig. 4). The first of these, upstream motif 1 (UM1), is found at 84 loci, including all but two spliceosomal snRNA loci, and contains a highly conserved core sequence that overlaps the snRNA proximal sequence element (PSE) previously identified in C. elegans (Thomas et al. 1990). UM1 was also found at six new box C/D snoRNAs loci and at a majority of the spliced leader (SL) RNA loci.
Figure 4.
Upstream motifs discovered at ncRNA loci. (A) Upstream motif 1 (UM1; E = 4.0 × 10–521). (B) Upstream motif 2 (UM2; E = 7.3 × 10–179). (C) Upstream motif 3 (UM3; E = 1.1 × 10–38). (For explanation of E, see Fig. 2.) (D) Distribution of distances from motif position 1 of UM1, UM2, and UM3, respectively, to 5′-end of the ncRNA transcripts. UM1 and UM3 have defined distances from start of the motif to the 5′-end of transcript. The two peaks for UM1 represent distances for loci with (smaller peak) and without (larger peak) an additional TATA-box. The distances between UM2 and transcript 5′-ends are more variable, possibly indicative of post-transcriptional 5′-end processing for this group of ncRNAs.
Upstream motif 2 (UM2) was found at the loci of 47 ncRNAs, of which 40 (31 novel) had snoRNA-like features. The remaining seven transcripts could not be assigned to any known class of ncRNAs. UM2 is on average located 20 bp closer to the transcript 5′-end than UM1, and differs substantially from UM1 in base composition (Fig. 4D). UM2 contains no highly conserved core motif. Rather, the most conserved bases are concentrated toward both ends of the motif. An interesting feature of these two conserved ends is that they show considerable resemblance to the box A and box B components of the internal tRNA promoter. There are no tRNA annotations upstream of any of the 198 loci of our data set. However, four pseudo-tRNA annotations occur upstream of four UM2-containing loci (see Supplemental material for details).
Upstream motif 3 (UM3) is shared by all C. elegans verified and predicted sbRNA loci, and was also found at all predicted C. briggsae sbRNA loci. The motif is composed of a highly conserved central core sequence followed invariably by a canonical TATA-box located at approximately -30 bp (Figs. 4C and 5D). The UM3 core shares a small submotif (TGTCNG) with the UM1 core sequence, but otherwise the two motifs show little sequence similarity. Based on their upstream motifs, their genomic locations, and several other factors, we suggest that the 161 known and novel ncRNAs detected in our study can be divided into several different biogenesis groups (Fig. 5 and Supplemental materials).
Figure 5.
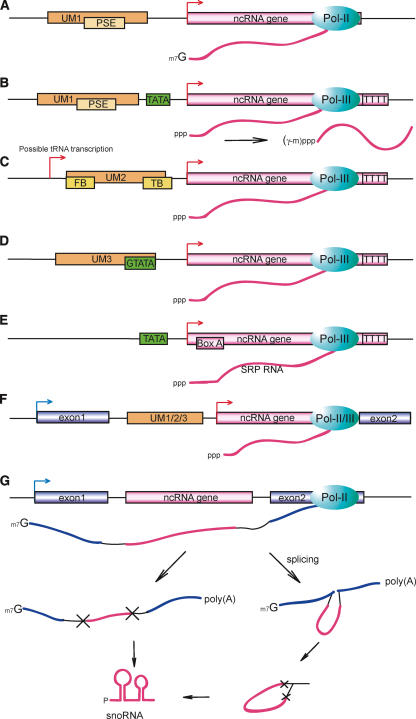
Arrangements of transcriptional elements and genomic locations of small non-coding ncRNA loci, as inferred from genomic and experimental data. (A) TATA-less loci with UM1. This type of locus is characterized by the Upstream Motif 1 and is found both intergenically and intronically. Transcripts from TATA-less UM1 loci generally carry a 5′-end cap, most likely transcribed by RNA polymerase II, and make up biogenesis group I-A, which comprises most spliceosomal snRNAs, a fraction of the SL RNAs, most snlRNAs, and a few C/D snoRNAs along with some unclassified transcripts. (B) Loci with UM1 and a TATA-box. This type of locus combines the UM1 with a TATA-box, and most often a tract of four or more Ts is found within 10 bp of the transcript 3′-terminus. Known RNA polymerase III transcripts like U6 snRNA and RNase P RNA are found at this type of locus. The transcripts may have a single methyl group added at the γ-phosphate post-transcriptionally, as is commonly found in U6 and 7SK snRNAs (Gupta et al. 1990). (C) Loci with UM2. This type of locus comprises a number of both intergenic and intronic snoRNA-like transcripts, along with a few uncharacterized ncRNAs, and makes up biogenesis group II. Transcripts are generally uncapped, and an oligo-T tract is found close to the 3′-terminus, indicating transcription by RNA polymerase III. FB (Front Box) and TB (Tail Box) are the most conserved 15-bp motifs within the 100-bp upstream sequence of these loci, and show strong resemblance to Box A and Box B of the tRNA promoter. A “possible tRNA transcription” initiation site has been indicated to account for the possibility that UM2 is transcribed as a part of the primary transcript (see Supplemental material for details). (D) Loci with UM3. This type of locus has only been found in sbRNAs, and is characterized by UM3, which contains a TATA-box preceded by a strongly conserved G residue. The loci are terminated by an oligo-T tract, and most transcripts are uncapped, suggesting transcription by RNA polymerase III. (E) SRP RNA loci. The C. elegans SRP RNA loci are characterized by a rudimentary TATA-box and a Box A element at ∼10-20 bp downstream of the transcription start, and are terminated by an oligo-T tract. (F) Independently transcribed intronic loci. This type of locus represents subgroups of locus types A-E, in which both the transcribed sequence and the corresponding control elements (promoter, terminator) are found within the intron of a protein-coding gene. This type of locus is found for all the above promoter elements, but is most common for UM1 and UM2 type loci. (G) Motif-less intronic loci. These loci are exclusively made up of snoRNA-like genes, and are often found within an intron of a ribosomal gene. The distance between the ncRNA locus and the preceding exon is generally short (<50 bp) and AT-rich. Transcription is initiated from the host gene promoter, and the snoRNA is processed either directly from the pre-mRNA, or from a spliced intron lariat.
ncRNA cap structure correlates to upstream motifs
Our ncRNA library construction strategy was designed to discriminate between 5′-end capped and non-capped transcripts. Of the 161 known and novel ncRNAs detected, 52 had more than a 95% probability of carrying a 5′-end cap, and an additional 11 were found to have more than an 80% probability of being capped (Supplemental Table S-1). Most capped transcripts originated from the TATA-less UM1 loci, or from intergenic loci without a discernible upstream motif.
The 50 TATA-less UM1 ncRNAs had either positive (42) or undetermined (8) cap status, suggesting a strong relationship between cap structure and this upstream motif. In our data, U6 snRNA also showed strong indications of a 5′-end cap, probably implying post-transcriptional processing of a γ-monomethyl-GTP cap previously found on human U6 snRNA (Gupta et al. 1990). Ninety-one transcripts had more than an 80% probability (62 had a >95% probability) of not carrying a cap. These were derived mainly from intronic loci without upstream motifs, or from both intergenic and intronic loci with upstream motifs other than UM1.
Intronic loci show signs of independent transcription
A feature of the C. elegans small non-coding transcriptome not found in other organisms (Dominski et al. 2003) is the high frequency of conserved motif elements upstream of intronic ncRNA loci. Of the 198 different chromosomal loci corresponding to 161 known and novel ncRNAs, 88 are located in the sense direction within an intron of a verified or predicted protein-coding gene. More than 50% of these have conserved upstream sequences (mainly UM1 and UM2) (Fig. 5F). Comparing these intronic “motif loci” to intronic ncRNA loci without a discernible upstream motif (“non-motif loci”), we found several striking differences that further suggest independent transcription of intronic motif loci from host genes. Most transcripts from intronic UM1 loci appear to carry 5′-end caps, indicative of independent RNA polymerase II transcription. The intervening sequence between the ncRNA 5′-end and the preceding exon is also generally more A/T-rich and its median size much shorter at non-motif loci (35 nt) than at UM1 (253 nt) and UM2 (152 nt) loci (Supplemental Fig. S-2). More than 60% of the intronic non-motif loci (all snoRNAs) reside in ribosomal or other translational related genes (Supplemental Table S-1). In other organisms, such genes are known to have a high frequency of the TOP-type promoters, known to control the processing of cotranscribed, intronic snoRNAs (de Turris et al. 2004). Only 5% of the host genes with intronic UM2 loci belong to this category of genes. Also, when comparing the cellular levels of ncRNAs (represented by library clones) and their host gene mRNAs (represented by public EST data), we observe a correlation between expression levels of motif-free ncRNAs and host gene mRNAs. A similar trend was not found for motif-containing loci, suggesting that ncRNA expression is unrelated to their host genes' transcription (Supplemental Fig. S-3).
High frequency of independently transcribed snoRNA loci
The data also indicate a high degree of independently transcribed snoRNAs in C. elegans. Whereas independent transcription of sno/scaRNAs is common in plants (Brown et al. 2003), most metazoan sno/scaRNA genes are intronic, and only a few intergenic loci have been reported to exhibit independent transcription (Tycowski et al. 2004). The snoRNAs in our material map to 89 loci, of which 30 are intergenic and (with two exceptions) associated with either UM1 or UM2. Of the remaining 59 intronic snoRNA loci, four contain UM1 and 19 contain UM2, suggesting that close to 60% of all snoRNAs in C. elegans are, in fact, independently transcribed.
Estimates of 2700 ncRNA genes
Given established and novel ncRNA data, we tried estimating the size of the C. elegans non-coding transcriptome in three ways. The first estimate was based on the finding that, with respect to conservation in C. briggsae, the ncRNA-containing introns show a radically different distribution from other introns (see Supplemental material for details). Assuming that the distribution of known ncRNA-containing introns is representative of all introns hosting an ncRNA locus, we took introns ranging from 50 to 130 bp to represent introns without an ncRNA locus (no ncRNA in C. elegans was found in an intron shorter than 135 bp). Using linear regression analysis, we inferred the number of ncRNA-containing introns to be between 0.9% and 2.3% of the total intron population, somewhat depending on the set of introns used for the non-ncRNA-containing set. When extrapolated to accommodate also for possible intergenic ncRNAs, a C. elegans non-coding transcriptome of 1600-4100 different species is obtained, with 2385 as the most likely estimate (Table 1).
Table 1.
Estimates of the C. elegans ncRNA transcriptome
| Model | Estimated no. of ncRNA species |
|---|---|
| 1. Intron conservation | 2385 |
| 2. Conserved upstream motifs | 2757 |
| 3. Clone no. versus expression level | 2936 |
| Average | 2693 |
Model 1 is based on the difference between conservation between ncRNA-containing introns and the total intron population when comparing Caenorhabditis elegans to Caenorhabditis briggsae. Basically, the introns shorter than 130 bp are taken to represent non-ncRNA introns, and the fraction of ncRNA-containing introns in the total intron population is inferred by linear regression analysis. Correcting for the fraction of intergenic ncRNA loci yields the estimated number of ncRNAs. Model 2 is based on the occurrence of upstream motif 1 (UM1) in the C. elegans genome, corrected for one highly repetitive sequence, and adjusted for nonmotif loci. Model 3 is a statistical calculation based on the correlation between the frequency of identical clones in the ncRNA library, and the concentration (expression level) of the corresponding ncRNA as estimated from Northern blots. (All three models are explained in detail in the Supplemental material.)
A second approach used the observation that a considerable fraction of both known and novel ncRNA loci harbors conserved upstream elements. Performing Meta-MEME (Grundy et al. 1997) searches for the conserved small ncRNA upstream motifs across the entire C. elegans genome yielded 1404, 527, and 65 sites (cutoff: E < 0.1) for UM1, UM2, and UM3, respectively. However, because a relatively large fraction of both UM2 and UM3 “genomic hits” overlapped with exons of protein-coding genes, and many additional UM2 sites overlapped with a considerable number of tRNA and pseudo-tRNA genes (see above and Supplemental material), we based the estimate on the occurrences of UM1 only. Accounting for the fraction of ncRNA genes in our library not having UM1, we obtained an estimate of 2757 different ncRNA species in the C. elegans small non-coding transcriptome (Table 1).
A third approach, used the correlation between the number of sequenced clones of each ncRNA and their cellular concentrations as observed from the Northern blots to establish a multinominal model (see Supplemental materials for details), and arrived at an estimate of 2936 different small ncRNAs. Taken together, these three estimates all point to an ncRNA transcriptome of a few thousand species, with a figure close to 2700 as the most likely estimate (Table 1).
Discussion
Our data show that the eukaryotic non-coding transcriptome still harbors plenty of novelty. Using a novel cloning strategy, we have identified 100 novel non-coding transcripts with verified expression. Three elements of our cloning approach contributed significantly to our result. Fractioning of total RNA on an anion resin instead of through PAGE reduced contamination from fragmented high-molecular weight RNAs (Eddy 2001; Huttenhofer et al. 2001). Specific targeting of the most important RNA contaminants through oligo-coated magnetic beads prior to cloning enabled us to reach a pre-screen ncRNA detection efficiency of 36% on novel ncRNAs (70% for all small ncRNAs) (Fig. 1A), compared to the 3%∼7% obtained in previous studies (Huttenhofer et al. 2001). Finally, ligating adaptors to both 3′- and 5′-ends protected terminal nucleotides from exonuclease degradation and guaranteed that incompletely reverse-transcribed RNAs would not be cloned. This yielded full-length sequences and allowed determination of the transcript 5′-end structure, which, in turn, enabled us to establish correlations between upstream motifs, 5′-cap status, and 3′-end termination signals indicative of the ncRNA mode of biogenesis.
A large contingent of snoRNA-like transcripts was to be expected, as this fraction of the C. elegans transcriptome has been less studied than miRNAs (Higa et al. 2002; Wachi et al. 2004; Stricklin et al. 2005). However, our data also include 31 transcripts for which no sequential or structural homolog outside the nematodes could be found. Several of these carry unique motifs both upstream and internally, and may well represent novel functional classes of ncRNAs. RNomics efforts in other model organisms have all revealed 25%-50% clones that could not be ascribed to any known functional category of ncRNAs (Huttenhofer et al. 2001; Marker et al. 2002; Yuan et al. 2003; Wang et al. 2004), but only in Dictyostelium could these transcripts be grouped into potentially novel classes of ncRNAs based on common internal and upstream motifs (Aspegren et al. 2004). Of the 31 novel clones in our data, we could discern two groups with common features, comprising 17 different transcripts. Among these, nine stem-bulge RNAs (sbRNAs) display characteristics slightly resembling Dictyostelium Class I RNAs (Aspegren et al. 2004), in that the defining sequence motifs are found at the RNA 5′- and 3′-termini, and have the potential to form a stem-loop structure containing a bulge with a conserved sequence. The loci of both classes also contain upstream motifs common to all genes within each group. However, the length and composition of the sequence motifs defining the respective classes are not conserved between Caenorhabditis and Dictyostelium, and there is no further evidence to support any functional relationship between the two ncRNA categories. The snRNA-like RNAs (snlRNAs) clearly share an Sm protein-binding site with both spliceosomal snRNAs and SL RNAs (Zeiner et al. 2004). However, the conserved sequence element in this group extends beyond the Sm-binding site, and may allow for binding of additional proteins not participating in the general spliceosomal processes. With a few exceptions, the snlRNAs also share the UM1/PSE upstream motif with both snRNAs and several other ncRNAs, possibly also pointing to involvement in similar processes. The C. elegans protein-coding genes contain a large fraction of very short introns that may require additional RNA factors for correct splicing, and a preliminary suggestion would be that the snlRNAs are somehow involved in splicing related activities.
The lack of conservation beyond C. briggsae for all but one of the novel transcripts is conspicuous, but not entirely unprecedented. Previous attempts to detect small ncRNAs (Huttenhofer et al. 2001; Marker et al. 2002; Tang et al. 2002; Yuan et al. 2003) have resulted in the identification of a group of small non-coding transcripts not assignable to any known class of small ncRNAs. Furthermore, finding a conserved homolog for these transcripts even in related species has been notoriously difficult. Moreover, as we have regarded as “known” or “predicted” all ncRNAs that have been annotated in the C. elegans genome based on sequence homology in other species, it is not altogether unreasonable that our novel ncRNAs should lack non-nematode homologs. The recent observation that even ultraconserved vertebrate non-coding sequences are not found in nematodes or flies (Bejerano et al. 2004; Woolfe et al. 2005) may indicate a general lack of conservation of non-coding regulatory elements between vertebrates and non-vertebrates.
Developmentally variable expression of the C. elegans non-coding transcriptome is also intriguing, particularly considering that the constitutively expressed spliceosomal snRNAs displayed very stable expression. We therefore assume these variations in expression are not artifactual but physiologically relevant. Few small ncRNAs are likely to themselves have catalytic or purely structural functions; instead, developmentally regulated ncRNAs may regulate the activity of other gene products. Also, snoRNAs show a remarkably high frequency of variably expressed transcripts, possibly indicating that methylation and Ψ-uridylation of rRNA may actually modify ribosomal properties. Several mammalian snoRNAs have been shown to have tissue-specific expression (Cavaille et al. 2000). In Archaea, rRNA methylation has been shown to depend on culturing temperature (Noon et al. 1998). Adaptive rRNA modifications in response to varying environment conditions have also been suggested as an explanation to the extended repertoire of snoRNAs in plants (Brown et al. 2003). As most of the variably expressed snoRNAs in our data showed increased levels at the dauer stage, this might indicate a role for differential modification of rRNA nucleotides between normal and hunger-stressed worms.
Transcriptional analysis of the data gave three major results. The first was an extension of previous observations (Hernandez 2001; Ohler et al. 2004) that small ncRNAs have unique promoter structures. Particularly interesting is the strong resemblance between UM2 and the tRNA internal promoter, perhaps suggesting that TFIIIC (or a TFIIIC-like factor) binds UM2 and recruits RNA polymerase III. In Arabidopsis an arrangement of a tRNAGly gene preceding a snoRNA43 gene has been found at several loci, and a similar tRNAMet-snoRNA locus was also observed in rice (Kruszka et al. 2003). One of the Arabidopsis loci was demonstrated to give rise to a dicistronic primary transcript that was cleaved by the tRNA 3′-end-processing enzyme RNase Z, releasing the snoRNA from the tRNA. We have no evidence for a dicistronic primary transcript, possibly because it is very short-lived. An alternative possibility is that an originally internal tRNA promoter has been transformed to act as an (non-transcribed) upstream core promoter. Whatever the case, this arrangement of tRNA (or tRNA-like) and snoRNA genes in both C. elegans and plants possibly points to a very old evolutionary solution to the transcription of snoRNA genes that predates the divergence of plants and animals.
The second major result from the transcriptional analysis was the high number of snoRNA-like transcripts with apparently independent transcription. Whereas independent transcription of sno/scaRNAs is common in plants (Brown et al. 2003), it has been reported for very few sno/scaRNAs in animals (Tycowski et al. 2004). In C. elegans, 52 of 88 known and novel snoRNA-like transcripts appear to have an upstream promoter element and/or an intergenic location. The third result was the extent to which intronic C. elegans ncRNAs are independently transcribed. Whereas independent transcription from intronic loci has been reported (Dominski et al. 2003), in C. elegans >50% of all intronic ncRNA loci show strong signs of independent transcription.
Recent studies have indicated that the total number of non-protein-coding transcripts in both human (Bertone et al. 2004; Cawley et al. 2004; Kampa et al. 2004) and mouse (Okazaki et al. 2002) genomes may be far higher than previous estimates. Assuming that ncRNAs play a major role in specifying eukaryotic multicellular complexity (Mattick 2003, 2004), the complexity and number of cell fates in a nematode (of the order 103) should imply a higher number of non-coding transcripts than in a unicellular organism, but lower than in more complex organisms like insects or vertebrates. Present estimates for ncRNAs in Escherichia coli stand at some hundreds (Vogel et al. 2003; Zhang et al. 2004; Saetrom et al. 2005), whereas data on human Chromosomes 21 and 22 (Cawley et al. 2004; Kampa et al. 2004; Cheng et al. 2005) would indicate a number at least two orders of magnitude higher for mammals. An estimate of ∼2700 small non-coding RNAs in C. elegans compares well with the 3000-4000 predicted ncRNA loci reported in the most recent computational survey of this organism (Missal et al. 2005). A number of ncRNAs in the lower thousands would amount to a class of possible regulatory molecules in C. elegans as large as the complete set of protein signal transduction factors (Chervitz et al. 1998), and might fit the bill of this complex an organism.
Methods
ncRNA-specific library
Total RNA was isolated from mixed-stage worms and eggs according to the Trizol (Invitrogen) protocol, then loaded on a Qiagentip (Qiagen) and eluted with a 0.6 ∼ 1.0 M NaCl gradient of QRW2 buffer (Qiagen RNA/DNA Handbook) at 50°C. Fractions corresponding to tRNAs, small RNAs (80-500 nt), and high-molecular-weight mRNA/rRNA were collected. The Ambion MicrobExpress kit was adapted to remove remaining mRNAs and rRNAs: The small RNA fraction was hybridized to a mixture of specifically designed oligonucleotides in binding buffer, then unwanted RNA molecules were targeted and removed from ncRNA by a magnetic beads-based process as per protocol (Ambion). The enriched ncRNA pool was cloned using an adaptor-mediated library construction protocol (modified from Elbashir et al. 2001): RNAs were dephosphorylated with calf intestine alkaline phosphatase (Fermentas), then ligated to the 3′-adaptor oligonucleotide by T4 RNA ligase (Fermentas). The ligation product was split into two aliquots; one was treated with PolyNucleotide Kinase (PNK; Fermentas) to phosphorylate uncapped RNA, and the other was treated with Tobacco Acid Pyrophosphatase (TAP; Epicentre) to remove 5′-end methyl-guanosine caps from capped RNA. Thereafter, both were ligated to the 5′-adaptor oligonucleotide. Small molecules and excessive adaptors were removed from the ligation products with the mirVana miRNA isolation kit (Ambion) and reverse-transcribed (RT) with Thermoscript (Invitrogen) at 50°C, using oligo 3RT as the RT primer. Library normalization (when applied) was carried out by adding mRNA and rRNA from the above removal procedure to the RT products, then denaturing at 98°C in hybridization buffer, followed by reannealing at 70°C and treatment with duplex-specific nuclease (DSN; Evrogen) for 25 min. The cDNA was PCR-amplified by using Platinum Taq (Invitrogen) with 3RT and 5CD primers for 15 cycles, digested with PaeI and SacI (Fermentas) and cloned in pGEM-4Z (Promega; see Supplemental material for the oligonucleotide sequences used in this study).
5′- and 3′-RACE
RACE was performed by PCR amplification of the RT products (see above), with one primer designed specific to the ncRNA sequence and another primer being either 5CD or 3RT for 5′- and 3′-RACE, respectively.
Northern blot
RNA probes were synthesized and labeled by in vitro transcription of plasmids with T7 RNA polymerase (Fermentas) and Dig-11-UTP (Roche). Total RNA extracted from 12 different developmental stages and two environmental conditions was analyzed. Northern blotting was performed per standard and manufacturers' protocols. Blots were hybridized in ULTRAhyb (Ambion) at 68°C overnight, then treated with Blocking and Washing Buffer (Roche) and detected by CDP-star (Roche). Chemiluminescent signals were recorded in an image system ChemiCapt 3000 (Vilber). See Supplemental material for definitions of developmental stages and details of the Northern blot analysis.
Determination of ncRNA sequences and 5′-structure
The 161 ncRNA sequences were either determined as a consensus of aligned sequencing reads belonging to the same ncRNA species, when these agreed with size estimates from Northern blots (most cases), by joining two adjacent ncRNAs inferred from genomic and Northern blots' information (four cases), or from elongation of consensus sequences based on RACE and Northern data (10 cases). The probability of an ncRNA being 5′-capped or not was determined from a statistic model based on the distribution of known capped and uncapped small RNAs in TAP or PNK libraries (see Supplemental material, “Capping Probability”).
Computational analysis
C. elegans genome annotation and sequence data, and the C. briggsae genome data, were downloaded from WormBase (version WS123) (Harris et al. 2003). The 161 ncRNA sequences were mapped to 198 C. elegans genome loci by BLASTN. These loci, as well as their equal-sized 5′- and 3′-flanking sequences, were searched by BLASTN for homologs in the C. briggsae genome, with default parameters except for having the low-complexity filter switched off. The conservation score of a sequence used here is defined as the identical residues count, divided by the length of the sequence, in the alignment of the best High-Scoring Segment Pair.
The MEME motif discovery tool (version 3.0.13) (Bailey and Elkan 1995) was used to search for conserved motifs in all the ncRNA sequences and 100-bp upstream sequences of all ncRNA gene loci, respectively. This discovered three clearly discernible upstream motifs: UM1, UM2, and UM3; and three internal motifs: IM1, IM2, and IM3. The Meta-MEME software (Grundy et al. 1997) was used to search for further UM1, UM2, and UM3 sites in the C. elegans genome, using Hidden Markov Models (HMM) for each upstream motif that had been generated from MEME-produced weight matrices. To avoid the high scores of tandem repeat regions when using a Meta-MEME scan on a whole-genome scale, repeats were masked prior to the HMM search. An E-value threshold (0.1) was chosen such that most (>90%) of the upstream motifs associated with ncRNAs in our material could be identified.
Acknowledgments
We thank Haitao Guo, Jinzhao Li, and Jun Liu for early experiment discussion; Zhihua Zhang for Perl programming; and Muhammad Nauman Aftab and Jimmy Ye for careful reading of the manuscript. The C. elegans strain N2 used in this work was provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources. This work was supported by the Chinese Academy of Sciences Grant Nos. KSCX2-2, KJCX1-08, and KSCXZ-SW-223; National Sciences Foundation of China Grant No. 39890070; the National High Technology Development Program of China under Grant No. 2002AA231031; National Key Basic Research & Development Program 973 under Grant Nos. 2002CB713805 and 2003CB715900; and Beijing Science and Technology Commission Grant Nos. H020220030130 and H010210010113.
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4139206.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession nos. AY948555-AY948719.]
References
- Aspegren, A., Hinas, A., Larsson, P., Larsson, A., and Soderbom, F. 2004. Novel non-coding RNAs in Dictyostelium discoideum and their expression during development. Nucleic Acids Res. 32 4646-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atzorn, V., Fragapane, P., and Kiss, T. 2004. U17/snR30 is a ubiquitous snoRNA with two conserved sequence motifs essential for 18S rRNA production. Mol. Cell. Biol. 24 1769-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, T.L. and Elkan, C. 1995. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 3 21-29. [PubMed] [Google Scholar]
- Bejerano, G., Pheasant, M., Makunin, I., Stephen, S., Kent, W.J., Mattick, J.S., and Haussler, D. 2004. Ultraconserved elements in the human genome. Science 304 1321-1325. [DOI] [PubMed] [Google Scholar]
- Bertone, P., Stolc, V., Royce, T.E., Rozowsky, J.S., Urban, A.E., Zhu, X., Rinn, J.L., Tongprasit, W., Samanta, M., Weissman, S., et al. 2004. Global identification of human transcribed sequences with genome tiling arrays. Science 306 2242-2246. [DOI] [PubMed] [Google Scholar]
- Brown, J.W.S., Echeverria, M., and Qu, L.-H. 2003. Plant snoRNAs: Functional evolution and new modes of gene expression. Trends Plant Sci. 8 42-49. [DOI] [PubMed] [Google Scholar]
- Cavaille, J., Buiting, K., Kiefmann, M., Lalande, M., Brannan, C.I., Horsthemke, B., Bachellerie, J.P., Brosius, J., and Huttenhofer, A. 2000. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. 97 14311-14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawley, S., Bekiranov, S., Ng, H.H., Kapranov, P., Sekinger, E.A., Kampa, D., Piccolboni, A., Sementchenko, V., Cheng, J., Williams, A.J., et al. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116 499-509. [DOI] [PubMed] [Google Scholar]
- Cheng, J., Kapranov, P., Drenkow, J., Dike, S., Brubaker, S., Patel, S., Long, J., Stern, D., Tammana, H., Helt, G., et al. 2005. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308 1149-1154. [DOI] [PubMed] [Google Scholar]
- Chervitz, S.A., Aravind, L., Sherlock, G., Ball, C.A., Koonin, E.V., Dwight, S.S., Harris, M.A., Dolinski, K., Mohr, S., Smith, T., et al. 1998. Comparison of the complete protein sets of worm and yeast: Orthology and divergence. Science 282 2022-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Turris, V., Di Leva, G., Caldarola, S., Loreni, F., Amaldi, F., and Bozzoni, I. 2004. TOP promoter elements control the relative ratio of intron-encoded snoRNA versus spliced mRNA biosynthesis. J. Mol. Biol. 344 383-394. [DOI] [PubMed] [Google Scholar]
- Dominski, Z., Yang, X.-C., Purdy, M., and Marzluff, W.F. 2003. Cloning and characterization of the Drosophila U7 small nuclear RNA. Proc. Natl. Acad. Sci. 100 9422-9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy, S.R. 2001. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2 919-929. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Lendeckel, W., and Tuschl, T. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 15 188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy, W.N., Bailey, T.L., Elkan, C.P., and Baker, M.E. 1997. Meta-MEME: Motif-based hidden Markov models of protein families. Comput. Appl. Biosci. 13 397-406. [DOI] [PubMed] [Google Scholar]
- Gupta, S., Busch, R., Singh, R., and Reddy, R. 1990. Characterization of U6 small nuclear RNA cap-specific antibodies. Identification of γ-monomethyl-GTP cap structure in 7SK and several other human small RNAs. J. Biol. Chem. 265 19137-19142. [PubMed] [Google Scholar]
- Harris, T.W., Lee, R., Schwarz, E., Bradnam, K., Lawson, D., Chen, W., Blasier, D., Kenny, E., Cunningham, F., Kishore, R., et al. 2003. WormBase: A cross-species database for comparative genomics. Nucleic Acids Res. 31 133-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, N. 2001. Small nuclear RNA genes: A model system to study fundamental mechanisms of transcription. J. Biol. Chem. 276 26733-26736. [DOI] [PubMed] [Google Scholar]
- Higa, S., Maeda, N., Kenmochi, N., and Tanaka, T. 2002. Location of 2′-O-methyl nucleotides in 26S rRNA and methylation guide snoRNAs in Caenorhabditis elegans. Biochem. Biophys. Res. Comm. 297 1344-1349. [DOI] [PubMed] [Google Scholar]
- Huttenhofer, A., Kiefmann, M., Meier-Ewert, S., O'Brien, J., Lehrach, H., Bachellerie, J.P., and Brosius, J. 2001. RNomics: An experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J. 20 2943-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhofer, A., Schattner, P., and Polacek, N. 2005. Non-coding RNAs: Hope or hype? Trends Genet. 21 289-297. [DOI] [PubMed] [Google Scholar]
- Kampa, D., Cheng, J., Kapranov, P., Yamanaka, M., Brubaker, S., Cawley, S., Drenkow, J., Piccolboni, A., Bekiranov, S., Helt, G., et al. 2004. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 14 331-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruszka, K., Barneche, F., Guyot, R., Ailhas, J., Meneau, I., Schiffer, S., Marchfelder, A., and Echeverria, M. 2003. Plant dicistronic tRNA-snoRNA genes: A new mode of expression of the small nucleolar RNAs processed by RNase Z. EMBO J. 22 621-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C., Bai, B., Skogerbo, G., Cai, L., Deng, W., Zhang, Y., Bu, D., Zhao, Y., and Chen, R. 2005. NONCODE: An integrated knowledge database of non-coding RNAs. Nucleic Acids Res. 33 D112-D115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, T. and Eddy, S. 1997. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25 955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marker, C., Zemann, A., Terhorst, T., Kiefmann, M., Kastenmayer, J.P., Green, P., Bachellerie, J.P., Brosius, J., and Huttenhofer, A. 2002. Experimental RNomics: Identification of 140 candidates for small non-messenger RNAs in the plant Arabidopsis thaliana. Curr. Biol. 12 2002-2013. [DOI] [PubMed] [Google Scholar]
- Mattick, J.S. 2003. Challenging the dogma: The hidden layer of non-protein-coding RNAs in complex organisms. Bioessays 25 930-939. [DOI] [PubMed] [Google Scholar]
- ———. 2004. RNA regulation: A new genetics? Nat. Rev. Genet. 5 316-323. [DOI] [PubMed] [Google Scholar]
- Missal, K., Zhu, X., Rose, D., Deng, W., Skogerboe, G., Chen, R., and Stadler, P.F. 2005. Prediction of structured noncoding RNAs in the genome of the nematode Caenorhabitis elegans. J. Exp. Zoolog. B Mol. Dev. Evol. (in press). [DOI] [PubMed]
- Noon, K.R., Bruenger, E., and McCloskey, J.A. 1998. Posttranscriptional modifications in 16S and 23S rRNAs of the Archaeal hyperthermophile Sulfolobus solfataricus. J. Bacteriol. 180 2883-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler, U., Yekta, S., Lim, L.P., Bartel, D.P., and Burge, C.B. 2004. Patterns of flanking sequence conservation and a characteristic upstream motif for microRNA gene identification. RNA 10 1309-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., Nikaido, I., Osato, N., Saito, R., Suzuki, H., et al. FANTOM Consortium. RIKEN Genome Exploration Research Group Phase I & II Team. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420 563-573. [DOI] [PubMed] [Google Scholar]
- Pang, K.C., Stephen, S., Engstrom, P.G., Tajul-Arifin, K., Chen, W., Wahlestedt, C., Lenhard, B., Hayashizaki, Y., and Mattick, J.S. 2005. RNAdb—A comprehensive mammalian noncoding RNA database. Nucleic Acids Res. 33 D125-D130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel, N., Wolin, S., and Guthrie, C. 1987. A subset of yeast snRNAs contains functional binding sites for the highly conserved Sm antigen. Science 235 328-331. [DOI] [PubMed] [Google Scholar]
- Saetrom, P., Sneve, R., Kristiansen, K.I., Snove Jr., O., Grunfeld, T., Rognes, T., and Seeberg, E. 2005. Predicting non-coding RNA genes in Escherichia coli with boosted genetic programming. Nucleic Acids Res. 33 3263-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolc, V., Samanta, M.P., Tongprasit, W., Sethi, H., Liang, S., Nelson, D.C., Hegeman, A., Nelson, C., Rancour, D., Bednarek, S., et al. 2005. Identification of transcribed sequences in Arabidopsis thaliana by using high-resolution genome tiling arrays. Proc. Natl. Acad. Sci. 102 4453-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricklin, S.L., Griffiths-Jones, S., and Eddy, S.R. 2005. C. elegans noncoding RNA genes. In WormBook (ed. The C. elegans Research Community), p. 7, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Tang, T.H., Bachellerie, J.P., Rozhdestvensky, T., Bortolin, M.L., Huber, H., Drungowski, M., Elge, T., Brosius, J., and Huttenhofer, A. 2002. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. 99 7536-7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, J., Lea, K., Zucker-Aprison, E., and Blumenthal, T. 1990. The spliceosomal snRNAs of Caenorhabditis elegans. Nucleic Acids Res. 18 2633-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski, K.T., Aab, A., and Steitz, J.A. 2004. Guide RNAs with 5′ caps and novel box C/D snoRNA-like domains for modification of snRNAs in Metazoa. Curr. Biol. 14 1985-1995. [DOI] [PubMed] [Google Scholar]
- Vogel, J., Bartels, V., Tang, T.H., Churakov, G., Slagter-Jager, J.G., Huttenhofer, A., and Wagner, E.G.H. 2003. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 31 6435-6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachi, M., Ogawa, T., Yokoyama, K., Hokii, Y., Shimoyama, M., Muto, A., and Ushida, C. 2004. Isolation of eight novel Caenorhabditis elegans small RNAs. Gene 335 47-56. [DOI] [PubMed] [Google Scholar]
- Wang, J. and Kim, S.K. 2003. Global analysis of dauer gene expression in Caenorhabditis elegans. Development 130 1621-1634. [DOI] [PubMed] [Google Scholar]
- Wang, J., Zhang, J., Zheng, H., Li, J., Liu, D., Li, H., Samudrala, R., Yu, J., and Wong, G.K.-S. 2004. Mouse transcriptome: Neutral evolution of `non-coding' complementary DNAs. Nature 431 758. [PubMed] [Google Scholar]
- Woolfe, A., Goodson, M., Goode, D.K., Snell, P., McEwen, G.K., Vavouri, T., Smith, S.F., North, P., Callaway, H., Kelly, K., et al. 2005. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 3 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, K., Lim, J., Dale, J.M., Chen, H., Shinn, P., Palm, C.J., Southwick, A.M., Wu, H.C., Kim, C., Nguyen, M., et al. 2003. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302 842-846. [DOI] [PubMed] [Google Scholar]
- Yuan, G., Klambt, C., Bachellerie, J.P., Brosius, J., and Huttenhofer, A. 2003. RNomics in Drosophila melanogaster: Identification of 66 candidates for novel non-messenger RNAs. Nucleic Acids Res. 31 2495-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiner, G.M., Foldynova, S., Sturm, N.R., Lukes, J., and Campbell, D.A. 2004. SmD1 is required for spliced leader RNA biogenesis. Eukaryotic Cell 3 241-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Zhang, Z., Ling, L., Shi, B., and Chen, R. 2004. Conservation analysis of small RNA genes in Escherichia coli. Bioinformatics 20 599-603. [DOI] [PubMed] [Google Scholar]