Abstract
Novel transcriptional units (TUs) are EST-supported transcribed features not corresponding to known genes. Unconventional gene pairs (UGPs) are pairs of genes and/or TUs sharing exon-to-exon cis-antisense overlaps or putative bidirectional promoters. Computational TU and UGP discovery followed by manual curation was performed in the entire published 34.9-Mb human chromosome 22 euchromatic sequence. Novel TUs (n = 517) were as abundant as known genes (n = 492) and typically did not have nonprimate DNA and protein homologies. One hundred seventy-one (33%) of TUs, but only 13 (3%) of genes, both lacked nonprimate conservation and localized to gaps in the human–mouse BLASTZ alignment. Novel TUs were richer in exonic primate-specific interspersed repetitive elements (P = 0.001) and were more likely to rely on splice junctions provided by them, than were known genes: 19% of spliced TUs, versus 5% of spliced genes, had a splice site within a primate-specific repeat. Hence, novel TUs and known genes may represent different portions of the transcriptome. Two hundred nine (21%) of chromosome 22 transcripts participated in 77 cis-antisense and 42 promoter-sharing UGPs. Transcripts involved simultaneously in both UGP types were more common than was expected (P = 0.01). UGPs were nonrandomly distributed along the sequence: 89 (75%) clustered in distinct regions, the sum of which equaled 4.4 Mb (<13% of the chromosome). Eighty (67%) of the UGPs possessed significant locus structure differences between primates and rodents. Since some TUs may be functional noncoding transcripts and since the cis-regulatory potential of UGPs is well recognized, TUs and UGPs specific to the primate lineage may contribute to the genomic basis for primate-specific phenotypes.
Despite the publication of a highly accurate human genome sequence (International Human Genome Sequencing Consortium [IHGSC] 2004) with comprehensive annotation (Hubbard et al. 2002; Kent et al. 2002), the functional definition of a mammalian gene remains in flux (Okazaki and Hume 2003), in part because more of the genome is transcribed than is accounted for by reference protein-coding gene sets. Sequencing random clones from normalized and subtracted high-quality cDNA libraries continues to uncover previously uncharacterized transcripts, many of which do not correspond to mRNAs of conserved protein-coding genes (Carninci et al. 2003). The continued growth of dbEST has resulted in the availability of >2 million sequences not matching any annotated genes (Larsson et al 2005) and has facilitated definition of ∼25,000 transcript models distinct from known genes (Shklar et al 2005). Since the loci encoding such transcripts may not fit conventional definitions of a gene, the term transcriptional unit (TU) has been introduced (Carninci et al. 2003).
Transcriptome hybridization to genomic tiling arrays suggests that noncoding TUs are surprisingly widespread and that existing annotations greatly underestimate the number of transcribed features (Shoemaker et al. 2001; Kapranov et al. 2002; Rinn et al. 2003). Functions have been identified for some non-coding RNAs. They can serve as hosts of intron-encoded snoRNAs (Numata et al. 2003) and as host genes or direct precursors of small trans-acting microRNAs, which themselves constitute an extremely important functional class of noncoding RNAs (Rodriguez et al. 2004). They can also be involved in mRNA processing, transcription factor recruitment, and chromatin remodeling, suggesting that an extensive RNA-based regulatory network may exist in higher organisms (Mattick 2003). In addition, ORFless functional RNAs encoding telomerase and endoribonuclease components have been reported (Topper and Clayton 1990; Feng et al. 1995), emphasizing the significance of noncoding TUs.
A second intriguing feature of genome organization emerging from cDNA discovery projects is the abundance of unconventional gene pairs (UGPs), each comprised of two transcripts that overlap or are in close proximity to one another, in a manner suggesting coordinated regulation of gene expression. These UGPs include naturally occurring cis-antisense pairs, as well as genes and/or TUs that share a putative bidirectional promoter. Noncoding TUs and UGPs may be functionally linked in that many noncoding RNAs are antisense to coding genes and have been shown to regulate gene expression in many species (Hildebrandt and Nellen 1992; Delihas and Forst 2001; Lee and Ambros 2001; Misra et al. 2002; Kramer et al. 2003; Numata et al. 2003). In humans, naturally occurring cis-antisense transcripts are relevant to diverse aspects of genome dynamics, including imprinting (Sleutels et al. 2002); pathogenesis of neurodegenerative disorders (Andres et al. 2003); methylation in inherited anemia (Tufarelli et al. 2003); gene vestigialization (Millar et al. 1999); splicing regulation, X-inactivation, and RNA editing (Shendure and Church 2002); and retrotransposition (Ejima and Yang 2003) leading to de novo generation of primate-specific genes expressed in the brain (Courseaux and Nahon 2001).
We define a TU as one or more flcDNA-supported and/or EST-supported transcripts mapping to the same locus and sharing exonic sequence on the same strand. In this report, we refer to TUs identical to known genes as “known genes,” or simply “genes,” and to TUs identified by our analysis but devoid of known-gene identities and public-database annotations as “novel TUs,” or simply “TUs.” The goal of the present study was to annotate TUs and UGPs on chromosome 22 (chr22) and to characterize their incidence, evolutionary conservation, and distribution along the genomic landscape of the chromosome.
Results
Characterization of known genes and novel TUs
To catalog known genes and novel TUs, all genomic clones comprising the chr22 tiling path were subjected to a Perl-based analysis pipeline (see Methods). For every clone, matching ESTs and cDNAs were identified and their exon–intron structures defined. Transcripts with better scoring matches at a genomic locale other than the query clone were excluded. EST-supported transcribed features without full-length cDNA evidence were operationally defined as putative novel TUs. These TUs were manually analyzed to eliminate ESTs with ambiguous orientation and those likely originating from pre-mRNA and genomic contaminants. Remaining TUs were further curated to minimize artifactual fragmentation of genes with long UTRs into multiple transcript models; EST clusters within 10 kb of known-gene boundaries with expression profiles complementary to the known genes were generally considered UTR extensions and not standalone TUs.
Chr22 yielded 1009 transcript models: 492 genes and 517 TUs (Table 1). Most known genes were supported by the Sanger chr22 reference gene catalog (Collins et al. 2003), whereas most novel TUs were not previously annotated (Table 2).
Table 1.
Known genes and novel TUs on chr22
| Splicing and support | Known genes | Novel TUs | Total |
|---|---|---|---|
| Spliced, supported by multiple GenBank accessions | 379 | 53 | 434 |
| Spliced, supported by a single GenBank accession | 55 | 102 | 157 |
| Unspliced, supported by multiple GenBank accessions | 38 | 198 | 234 |
| Unspliced, single-accession, with 3′ AATAAA or ATTAAA | 20 | 164 | 184 |
| Total | 492 | 517 | 1009 |
Table 2.
Known genes and novel TUs on chr22 identified by our analysis compared with those annotated by the Sanger Centre
| Known genes | Novel TUs | |
|---|---|---|
| Matches a Sanger partial or complete coding gene | 404 | 47 |
| Matches a Sanger noncoding gene | 9 | 8 |
| Matches a Sanger pseudogene | 16 | 11 |
| Without a Sanger equivalent | 69 | 451 |
| Total | 498 | 517 |
The total in this table is 1015 because seven genes in our data set matched two Sanger genes each and in six of the seven cases the matched Sanger genes were not homologous to anything in our data set other than the gene that merged them.
Sensitivity and specificity of known gene identification
We defined the sensitivity of our method as the percentage of Sanger genes we successfully detected and annotated. Of the 577 Sanger genes that were neither pseudogenes nor immunoglobulins, 468 (Table 2, rows 1, 2) were identified by our approach, for a sensitivity of 81%. To determine the reason for this potentially subpar sensitivity, we analyzed the 109 Sanger genes that lacked equivalents in our data set (Table 3). Only 11 of these Sanger genes were missing due to problems with our algorithm. The rest were undetected because they did not meet our criteria for a gene or TU: that a genomic sequence be transcribed, that the transcript be represented by a feature other than a single unspliced nonpolyadenylated cDNA or EST, and that the sequence not contain any immunoglobulin homology. Therefore, the discrepancy between our and Sanger catalogs is due primarily to differences in operational definitions of transcribed features, with our definition being more rigorous.
Table 3.
Categorization of the 109 Sanger chr22 genes without equivalents in our data set
| Reason for lack of a known gene or TU corresponding to a Sanger gene | No. of Sanger genes |
|---|---|
| Unknown; Sanger gene passes GSPS and LOCUS criteria | 11 |
| Sanger gene is transcriptionally silent,a but not in a recent duplication | 34 |
| Sanger gene is putatively transcriptionally silent,b and in a recent duplication | 35 |
| Sanger gene is homologous to immunoglobulin gene segments | 9 |
| Sanger gene is transcribed, but as an unspliced nonpolyadenylated singleton | 19 |
| Special case | 1 |
Transcriptionally silent: no public ESTs or flcDNAs overlap any exons of the Sanger gene model on the sense strand of that model
Putatively transcriptionally silent: the Sanger gene model is in a recent paralogous segmental duplication. Some public ESTs and/or flcDNAs have high sense-strand homologies to the Sanger gene model. However, these ESTs/cDNAs match another copy of the duplicated region better than they match the copy containing the Sanger gene model being considered. Therefore, the Sanger model is most likely transcriptionally silent
We defined the specificity of our approach as the fraction of Sanger pseudogenes that our algorithm examined and excluded, rather than mistakenly including them among genes or TUs. Of the 234 Sanger pseudogenes, 207 did not match any of our genes or TUs, for a specificity of 88%. The other 27 Sanger pseudogenes all had cDNA or EST evidence for sense-strand transcription and thus were included in our analysis.
Quality assessment of novel TUs
Our automated characterization of transcribed features did not consider sequence at putative splice junctions. Therefore, one assessment of the quality of the TUs was to check whether their splice sites were canonical (GT-AG). We subjected randomly selected subsets of 25 spliced TUs with multiple EST support and 50 spliced TUs with single EST support, comprising a total of 126 introns, to manual splice-junction analysis by using Spidey (Wheelan et al. 2001). Nearly all splice sites (107 of 126) were canonical. Virtually all others differed from GT-AG by only one nucleotide, with GC-AG being the most common (six of 19). No U12 (AT-AC) splice sites were seen, consistent with the observation that GC-AG is the second most common mammalian intron type while AT-AC is extremely rare (Burset et al. 2000; Chong et al. 2004). Based on this sample, we conclude that the majority of spliced TUs represent real transcripts.
To test the quality of unspliced, singleton-EST TUs, we checked for perfect identity of ESTs to genomic sequence at canonical AATAAA or ATTAAA polyadenylation signals present within the 3′-most 40 bp of the ESTs. Only four of 100 randomly selected singleton-EST TUs had sequencing errors. This indicates that the majority of those TUs which are defined solely by singleton ESTs probably originate from biologically real, canonically polyadenylated transcripts.
Our splice-based and polyadenylation-based estimates of the fraction of novel TUs representing biologically real transcripts (85% and 96%, respectively) are likely conservative, because completely noncanonical splice sites and polyadenylation signals have been reported in mammals (Caffrey et al. 2000; Chong et al. 2004). Hence, some TUs without consensus splice junction or polyadenylation signal sequences might reflect the existence of even more transcribed features.
Nonprimate homologies and protein-coding potential of known genes and novel TUs
We inferred nonprimate DNA homologies from BLASTN sequence similarities between the human query and any nonprimate sequence in GenBank, and BLASTZ sequence similarities between the human query and the mouse genome (see Methods). A major distinction between novel TUs and known genes was that only 108 of 517 TUs (21%), in contrast to 423 of 492 genes (86%), had nonprimate homologies detectable by BLASTN. Examined by using the more sensitive BLASTZ alignment, 345 of 517 TUs (67%) had homologies in the mouse genome (Supplemental Tables 1, 2). The 172 TUs lacking homologies were located in gaps of the global human/mouse BLASTZ alignment, suggesting a recent evolutionary origin. Thirty-eight of those 172 TUs resided in genomic sequences present in human but not in mouse.
We then evaluated whether some of these human genes and TUs might be protein-coding despite high nucleotide-level divergence (see Methods). However, BLASTX alignments indicated that ORFs of only 17 (25%) of the genes and 13 (3%) of the TUs primate-specific by BLASTN had homology to nonprimate proteins (Supplemental Table 3). Therefore, the majority of genes and TUs apparently specific to primates are unlikely to represent highly diverged coding transcripts.
Finally, we compared ORF lengths of genes and TUs on chr22 (Supplemental Table 4). Gene ORF lengths significantly exceeded TU ORF lengths (P = 0.0001 by Wilcoxon rank-sum test), suggesting that TUs, to a greater extent than genes, are representative of the noncoding portion of the transcriptome.
Our chr22 results parallel a comparative analysis of human chromosome 21 (chr21) by Gardiner et al. (2003), in which numerous species-specific spliced transcripts equivalent to our nonconserved TUs were reported in both human and mouse. While lacking interspecies BLAST homologies, nearly all of those transcripts could be verified by RT-PCR. Thus, nonconserved TUs are not merely EST-database artifacts and may define a novel class of primate-specific genes (Gardiner et al. 2003).
Primate-specific exonic sequences in known genes and novel TUs
We hypothesized that some TUs are evolutionarily young transcribed features that are primate-specific rather than mammalian-wide. We used Alu and Mer1 interspersed repeats as markers of primate specificity (Kawashima et al. 1992) of putatively exonic sequences. Novel TUs were significantly enriched in expressed primate-specific repeats relative to known genes: 3.5% of an average known gene's reference transcript, versus 9.5% of an average TU's reference transcript, consisted of such repeats (P = 0.001, Wilcoxon rank sum test). In total, 71 kb of known gene and novel TU exonic sequences consisted of primate-specific repeats.
Thirty of 155 novel TUs (19%) versus 21 of 434 spliced known genes (5%) had at least one splice junction within a primate-specific repetitive element (P < 0.0001, two-sample binomial z-test), suggesting that engagement of novel intrarepeat splice sites during primate evolution may have been more frequent in the TUs than in the known genes (Supplemental Table 5).
Characterization of cis-antisense UGPs
We identified 77 cis-antisense UGPs (Fig. 1A; Supplemental Table 6). Twenty-three pairs were tail-to-tail and 13 were head-to-head, consistent with the observation that in mammals tail-to-tail antisense overlaps are more common than are head-to-head antisense overlaps (Edgar 2003).
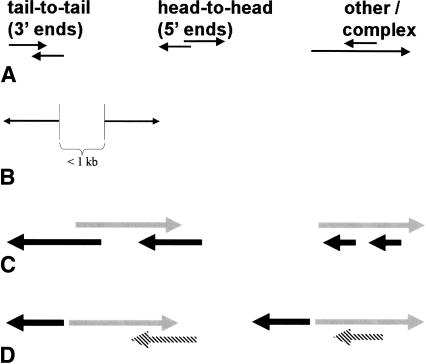
Figure 1.
Schematic illustrations of principal UGP genomic structures. Each arrow represents one transcribed feature. Only the orientation of the transcribed features is shown. Individual exons of the features are not shown. (A) Three types of exon-to-exon cis-antisense pairs. (Left) A tail-to-tail cis-antisense pair of transcribed features (genes, TUs, or one gene and one TU) on opposite strands in the same locus, with overlap of the last exons (“tails”) of the two features. Only the last exons are involved in the overlap. (Center) A head-to-head cis-antisense pair of transcribed features on opposite strands in the same locus, with overlap of the first exons (“heads”) of the two features. Only the first exons are involved in the overlap. (Right) cis-antisense pairs that are neither tail-to-tail nor head-to-head are collectively referred to as “other” or “complex” in this article. Only one of the many possible configurations of cis-antisense pairs is shown (B) A pair of transcribed features with a putative bidirectional promoter. (C) A gene (gray) that is antisense to multiple other genes (black). (D) A gene (gray) that shares a bidirectional promoter with another gene (solid black) and harbors an antisense transcript (hatched black).
Surprisingly, the remaining 41 cis-antisense UGPs did not fit either category. Eight structural types of complex antisense pairs could be distinguished by manual annotation (Supplemental Table 7). In the most common structure, an unspliced antisense transcript overlapped one internal exon of a spliced transcript. The unspliced transcript is a novel TU in 10 of 11 of those cases. In other structures, multiple categories of terminal–terminal, terminal–internal, and internal–internal exon overlap were seen, revealing a substantial diversity and complexity of antisense-overlap structures.
A gene-only approach to annotation would miss more than half of the cis-antisense pairs on chromosome 22. Our results confirm those of Yelin et al. (2003) that gene–TU cis-antisense pairs are most common, followed by gene–gene and TU–TU cis-antisense pairs.
For both gene–gene and gene–TU pairs, complex pairs were the prevalent type, followed by tail-to-tail and finally head-to-head pairs. Therefore, the complexity of genomic structure of a given antisense pair does not appear to depend on whether or not the pair includes a TU.
Characterization of putative bidirectionally promoted UGPs
We identified 42 putative bidirectionally promoted gene pairs on chr22: 21 were gene–gene, 18 gene–TU, and 3 TU–TU (Fig. 1B; Supplemental Table 8). As above, annotation limited to genes with full-length cDNA support would miss approximately half of these pairs. Putative bidirectional promoter sizes ranged from 10–919 bp, with the median of 256 bp—consistent with the distribution observed by Trinklein et al. (2004).
Most (81%) of the putative bidirectional promoters on chr22, and all bidirectional promoters of gene–gene pairs, overlapped CpG islands (Supplemental Table 8). This is consistent with evidence that the majority of RNA polymerase II–transcribed genes initiating at bidirectional promoters have a CpG island between them (Adachi and Lieber 2002).
Anecdotal reports in the literature suggested that mammalian CpG-island bidirectional promoters are frequently devoid of TATA boxes (Smith et al. 1990; Qvist et al. 1998; Seki et al. 2002). Conversely, CpG islands appear less frequently in promoters that contain both TATA boxes and initiator regions (Suzuki et al. 2001). We searched for instances of the relaxed TATA-box consensus (Kutach and Kadonaga 2000) on both strands of the 42 putative bidirectional promoters. The number of putative bidirectional promoters with and without TATA-box consensus sequences were approximately equal, although TATA-less promoters comprise only a minority of RNAPolII promoters (Lewin 2000). The proportion of TATA-less putative bidirectional promoters for gene–gene pairs in our data set (12 of 21; 57%) was approximately equal to that for gene–TU pairs (10 of 18; 56%), although all gene–gene pairs included CpG islands.
Simultaneous involvement of some transcribed features in multiple UGPs
Seven genes participated in cis-antisense overlaps with two other genes or TUs (Fig. 1C), while one (UNC84B) participated in three independent cis-antisense overlaps (Supplemental Table 9).
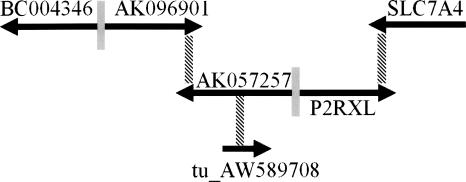
We also identified all transcript models on chr22 that shared a putative bidirectional promoter with a second model while also participating in a cis-antisense pair with a third (Fig. 1D). Sixteen genes and four TUs were in this category (Supplemental Table 10). Their counts are summarized in Table 4. Significantly more genes and TUs are involved in both cis-antisense pairs and putative bidirectional promoter pairs than predicted by the frequencies of the two types of independent events (20 observed vs. 11.76 expected, P = 0.01). Therefore, for a given transcript model, presence of one UGP type increases the probability of the other. A remarkable chain (group of genes and TUs connected by multiple UGPs)—six genes and TUs linked by three cis-antisense pairs and two putative bidirectional promoters—is shown in Figure 2.
Table 4.
Observed numbers of transcript models involved in UGPs on chr22
| With cis-antisense | |||
|---|---|---|---|
| With bidirectional promoter | Yes | No | |
| Yes | 20 | 64 | |
| No | 125 | 800 | |
Figure 2.
A contiguous chain of five genes and one TU linked by five UGPs on chr22 (BC004346-SLC7A4). Arrows indicate the direction of transcription of the labeled genes and TU. Cis-antisense overlaps are represented by hatched black vertical rectangles. Putative bidirectional promoters are represented by solid gray vertical rectangles. For clarity, individual exons are not shown. Drawing is not to scale.
Distribution of UGPs along the genomic sequence
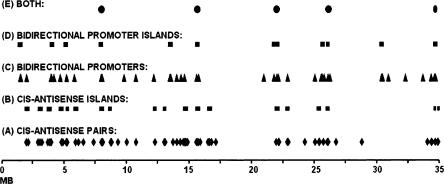
The distribution of UGPs on chr22 is illustrated in Figure 3. Most UGPs mapped closely to one another within several UGP clusters. We refer to these clusters as UGP islands, operationally defined as regions with at least two UGPs ≤250 kb from each other.
Figure 3.
Clustering of UGPs along 35 Mb of chr22q. UGPs cluster near one another more frequently than expected by chance on the 35 Mb of human chr22q. Cis-antisense gene pairs (A) are defined as forming an island (B) if two pairs lie within 250 kb of one another. Similarly, pairs of genes that may share a putative bidirectional promoter (C) are defined as forming an island (D) if they lie within 250 kb of one another. Sixty-three of 77 (82%) cis-antisense gene pairs and 26 of 42 (62%) of gene pairs sharing a putative bidirectional promoter lie in islands. Ten thousand simulations of distributions of these features into 35 Mb of genomic space did not yield any distribution with clustering as great as that observed (see text for details). Five regions of 22q harbor both cis-antisense islands and putative-bidirectional promoter islands (E).
To determine the proportion of human chr22 sequence within UGP islands, we first measured the length of each genomic region corresponding to a cis-antisense UGP island (for coordinates, see Supplemental Table 6). The combined length of the cis-antisense UGP islands on Figure 3 was 3.4 Mb. We emphasize that this was the sum of lengths of UGP islands, rather than of the extremely small exon-to-exon cis-antisense overlaps themselves. The sum of UGP islands enriched in cis-antisense UGPs represented a small fraction of the chr22 sequence, and the majority of the cis-antisense UGPs (63 of 77; 82%) resided in that small fraction (3.4 Mb; 10%) of the total sequence.
Similarly, we measured each region corresponding to an island of putatively bidirectionally promoted UGPs (Supplemental Table 8). The combined length of the putative bidirectional promoter UGP islands was 1.5 Mb. This was the sum of lengths of several extensive genomic regions enriched in putatively bidirectionally promoted UGPs, not of the extremely small putative bidirectional promoters themselves. The sum of UGP islands enriched in putative bidirectional promoters represented a small fraction of the chr22 sequence, and the majority of putative bidirectional promoters (26 of 42; 62%) resided in that small fraction (1.5 Mb; 4%) of the total genomic sequence.
Visual examination of UGP island map locations revealed five areas of substantial overlap between cis-antisense islands and putative bidirectional promoter islands. These regions were simultaneously enriched in both types of UGPs. They are represented by ovals at the top of Figure 3 and are found approximately at Mb 8, 16, 22, 26, and 35 on the map.
Islands of putative bidirectional promoters were weakly correlated with locally high CpG island density. Qualitative comparison of our UGP island distribution with Sanger Institute's SuperMap22 did not demonstrate any correlation of UGP islands of either class with GC content, SINE or LINE density, recombination hotspots, human–mouse synteny breakpoints, or recent segmental duplications.
The recent genomewide assessment of cis-antisense pairs in the mouse (Kiyosawa et al. 2003) may corroborate our UGP island findings. Figure 3 of that study portrays visually apparent clustering of exon-to-exon cis-antisense pairs on almost every autosome, highly similar to that on human chr22 in our Figure 3. Nevertheless, Kiyosawa et al. did not comment on the fact that many of their antisense pairs mapped onto the genome in close proximity to one another, or on large genomic regions enriched in, or depleted in, antisense pairs. Those investigators only noted the scarcity of overlapping gene pairs on the X-chromosome and the extent of coverage of known imprinted regions in their study.
To assess significance of UGP clustering, we nonparametrically derived four chromosome-wide P-values expressing the likelihood that the observed incidence of antisense pairs, antisense pairs within antisense UGP islands, putative bidirectional promoters, and putative bidirectional promoters within bidirectional-promoter UGP islands can occur by chance (see Methods). For each of the four characteristics, we searched 10,000 gene distribution simulations for instances where the simulated incidence of the UGPs or islands under consideration exceeded the actual incidence. No such instances were found. Therefore, all four P-values were <10-4.
For simulations, we divided chr22 into 20 intervals with different genomic sizes but approximately equal numbers of transcribed features and thus different gene densities. Intervals with similar gene densities had widely varying interval-specific P-values (probabilities that the observed complexity can be matched by chance). Therefore, our analysis does not support the hypothesis that the visually apparent clustering of UGPs along chr22 into UGP islands depends entirely on gene density. This result is consistent with earlier observations that incidence of bidirectional promoters does not correlate with gene density (Adachi and Lieber 2002).
In silico expression profiling of UGPs
We compiled expression profiles for all chr22 UGPs in silico by using cellular and tissue origins indicated in the GenBank entries for all cDNAs and ESTs representing members of those UGPs.
For 35 (45%) of the 77 antisense pairs, human ESTs suggested expression of both members of the pair in the same tissue or cell type, allowing the possibility of post-transcriptional regulation by dsRNA mechanisms. This is less than the 67% seen in a small-scale (n = 39) experimental test of expression profile complementarity in cis-antisense pairs identified in silico (Shendure and Church 2002). Therefore, strand-specific microarray-based transcript detection (Kumar et al. 2002) targeted toward tissues from which the antisense ESTs originated may uncover a greater extent of expression profile complementarity than do in silico surveys alone.
Of those 35 pairs, 18 were gene–gene, 16 were gene–TU, and one was TU–TU. These proportions approximately mirrored the proportions of the three pair types in the total chr22 antisense data set. These proportions support the nonartifactual nature of TUs: If TUs cis-antisense to genes were likely to be derived from artifactually misoriented ESTs of those genes, then gene–TU cis-antisense pairs with expression profile complementarity would be disproportionately common, rather than occur at a frequency corresponding to their frequency in the total data set.
Thirty-four (81%) of the 42 putative bidirectionally promoted transcript model pairs on chr22 had expression profile complementarity. This is significantly greater than the 45% seen for antisense pairs. Therefore, antisense pairs may be less likely to have expression profile complementarity than do putative bidirectionally promoted pairs. This extent of expression profile complementarity in putative bidirectionally promoted pairs is consistent with the finding that the majority of human bidirectional promoters are coregulatory and contain cis-regulatory elements affecting both genes at once (Trinklein et al. 2004).
Human–mouse comparative analysis of UGPs
In 19 of the 35 chr22 antisense pairs with expression profile complementarity, genomic organization of the locus was conserved between human and mouse. In the other 16, one or both members of each human pair lacked a transcribed ortholog or positional equivalent, based on mouse public flcDNA and EST data. Five of the 16 cases of human–mouse genomic structure differences at cis-antisense loci were characterized by the expression of both members of the antisense pair in human brain. This proportion (31%) is markedly greater than the proportion of all chr22 cis-antisense pairs in which both members are expressed in human brain (10 of 77; 13%). This raises the intriguing possibility that some antisense-mediated gene expression regulatory mechanisms in human brain are specific to the human lineage and occur at loci harboring genomic structure distinctions relative to mouse. It has been previously shown that species-specific gene expression patterns in humans are most evident in the brain (Enard et al. 2002).
Of the 84 putatively bidirectionally promoted transcripts on chr22 (60 genes and 24 TUs), 25 (30%) had no BLASTN-detectable homologies to any mouse transcribed or genomic sequences in the NR, EST, and MGSCv3 divisions of GenBank. They included seven genes and 18 TUs. Therefore, 12% (7/60) of putatively bidirectionally promoted genes and 75% (18/24) of putatively bidirectionally promoted TUs lacked mouse homology. In addition, only 10 (24%) of the 42 putative bidirectionally promoted pairs had their genomic structure completely conserved in the mouse. The present analysis suggests that putative bidirectionally promoted as well as cis-antisense UGPs frequently have lineage-specific genomic structures and on occasion harbor lineage-specific transcripts.
Discussion
Parallels to previous TU and UGP analyses
UGPs and novel EST-supported TUs have been identified in the human genome in previous studies (including Shendure and Church 2002; Gardiner et al. 2003; Trinklein et al. 2004). However, four aspects of our analysis are nonredundant relative to those studies.
Automated identification of EST-derived TUs was combined with manual curation on a whole-chromosome scale. As a result, while our known gene set and those in previous analyses overlapped to a great extent, our novel TUs are mostly devoid of existing annotations. Although many ESTs comprising our TUs are displayed by the UCSC and Ensembl genome browsers, those portals retain other ESTs from low-quality data sets, may erroneously map ESTs in recent segmental duplications, harbor EST orientation ambiguities, and do not derive TU genomic structures from individual EST/genome alignments.
Irregularities in UGP incidence along the genomic landscape have not been previously reported. In contrast, we emphasize the distribution of UGPs along chr22. Most UGPs reside in distinct regions that together constitute just a small portion of the chromosome's total sequence. By using a nonparametric simulation approach, we rejected the hypothesis that UGP clustering is directly proportional to transcribed feature density. The absence of a correlation between UGP clustering and the genomic parameters we examined raises the possibility that the proximity of multiple UGPs at UGP islands is important for the function or regulation of UGP components or perhaps for chromosome structure, and may thus be maintained through selection.
We uncovered human–mouse structural differences putatively relevant to the regulation of transcripts in UGPs. Just one-third of the chr22 UGPs did not possess such differences. Shendure and Church (2002) found that an even smaller proportion of human cis-antisense pairs in their set (5%) had conserved genomic organization in mouse, although their methodology did not rely on manual curation to the extent that ours did. Nevertheless, their conclusion that certain antisense overlaps may be lineage-specific rather than mammalian-wide agrees with ours. While our manuscript was in preparation, Veeramachaneni et al. (2004), in addition to describing numerous chains similar to that on our Figure 2, reported that only a small fraction of gene overlaps have identical structures in human and mouse. Yet, as they only considered known genes and not EST-derived primate-specific TUs, they likely overestimated the extent of human–mouse conservation in gene overlaps.
Our cis-antisense findings extend upon previous studies. Kiyosawa et al. (2003) performed comprehensive analysis of mouse cis-antisense based on full-length cDNA data and did not tabulate chains; in contrast, we focused on a single human chromosome, incorporated ESTs, and catalogued transcript models involved in multiple UGPs. The proportion of cis-antisense pairs with coexpression evidence on human chr22 (45%) was greater than that reported by Kiyosawa et al. in their Figure 1 ([480 + 274]/[1252 + 1229] = 30%), perhaps due to our use of ESTs. Yelin et al. (2003) utilized ESTs in their genomewide study of cis-antisense in human. While we lacked their means for experimental validation of the antisense pairs, we treated ESTs less conservatively, allowing singletons and doubletons if splice site and polyadenylation signal criteria were met. Only 34 (44%) of our 77 chr22 antisense pairs were also reported by Yelin et al., suggesting that our approach can identify pairs missed by their procedure. A set of mammalian cis-antisense pair examples was discovered by Shendure and Church (2002) when public EST collections were still quite limited. Because of limited sequences available at the time and a UniGene-based strategy, just 144 pairs were identified in the human genome by those investigators.
Cis-regulation, nonconservation, and the bimodal transcriptome
Highly significant differences in nonprimate conservation, protein-coding potential, and exonic primate-specific sequence content were observed between known genes and novel TUs, as there is minimal overlap between the ranges of these genomic parameters in genes and TUs. With respect to these parameters, the human transcriptome may be bimodal, with known conserved coding genes and novel lineage-specific noncoding TUs defining its two major fractions.
The most striking feature of novel TUs relative to known genes is the near-absence of nonhuman homologies. We infer that some TUs are lineage specific to primates and perhaps solely to humans. If some of these TUs are functional, then their lineage specificity can provide part of the genomic basis for primate- and human-specific phenotypes.
The potentially large number of lineage-specific transcripts in humans lends new credence to the assertion that our ability to model human biological processes in nonhuman models must be critically reexamined (Margolin 2001). It is commonly stated that, once the few known lineage-specific gene family expansions in humans and mice are taken into account, human genes without mouse orthologs are rare to nonexistent. Our results call for a reexamination of this assumption as well.
This is the first analysis to tabulate demonstrably primate-specific sequences in exons on human chr22, which add up to 71 kb (Supplemental Table 5). Although 71 kb of exonic sequence is not a lot, it is a highly conservative estimate due to its omission of primate-specific sequences other than Alu and Mer1 elements and its failure to account for primate-specific repeats in alternatively spliced and polyadenylated regions that are not parts of our reference transcripts. Even this small amount of sequence, however, affords an interesting glimpse into how much of a human chromosome can become newly recruited into transcribed structures specifically in the course of primate evolution.
One of the most noteworthy properties of our chr22 UGP set was the frequent incidence of genes and TUs participating in multiple types and instances of UGPs. This challenges the accepted view that clusters of closely spaced but functionally unrelated genes in mammals are rare (Angiolillo et al. 2002), because practically all chr22 UGPs are pairs of genes and/or TUs without sequence homology to one another outside of the cis-antisense overlap, and because most gene–gene pairs lack evidence for involvement of the two products in common pathways. Clusters of more than three apparently functionally unrelated transcript models joined by a combination of UGPs have been observed in this study (Fig. 2).
Together, such genes and TUs signify that regulatory relationships specified by the genomic proximity or overlap of expressed features may be more complex than is simple coregulation or antiregulation of bidirectionally promoted pairs or the downregulation of a sense gene by an antisense TU. We propose that clusters of apparently functionally unrelated genes and TUs linked by combinations of UGPs are analogous to the sentences of a new sequence-based regulatory language. The words of this language are the transcribed features themselves. The exon-to-exon cis-antisense overlaps in which they are involved, and the bidirectional promoters that some of them share, are the punctuation marks. The sentences are to be deciphered along the genomic sequence.
It is therefore distressing that the majority of transcripts involved in multiple UGPs do not have a known function. Transcriptome-wide studies should move beyond large-scale cDNA sequencing and toward large-scale functional investigations of the sequenced transcripts. If they do not, any sequence-based regulatory language will be as mystifying as a language with a non-Latin alphabet is to a monolingual English speaker.
Implications for gene birth and primate-specific phenotypes
The abundance of TUs and UGPs in human genomic sequence and the putative primate-lineage specificity of a subset of TU sequences and UGP genomic structures raise at least three questions. First, which TUs and UGPs are functionally important? TUs that participate in UGPs might be better candidates for functional, versus stochastic, transcription, especially if the UGP is an antisense pair in which both members are expressed in the same tissue. Even when cis-antisense pairs characterized by spatiotemporally mutually exclusive expression profiles of the pair members are considered, it is possible that one member's expression profile was altered in the course of evolution because of the appearance of an antisense counterpart, leading to lineage-specific modification of function even in the absence of any in vivo hybridization between the UGP-encoded transcripts.
Second, which TUs are evolutionarily young genes? The origin of new genes is recognized as a fundamental biological process that is essential for the appearance of novel biological functions and makes a major contribution to genetic diversity. However, the exact mechanisms giving rise to new genes remain to be elucidated, although one mechanism by which new genes are created is the shuffling of existing coding-gene exons, which generates both coding and noncoding new genes and is often facilitated by retrotransposition (Long et al. 2003). The potentially large number of novel TUs lacking coding-gene homologies in humans, however, may mean that additional mechanisms are involved. While mouse sequences are useful in evaluating whether a TU arose before or after the mammalian radiation, determining which TUs represent young genes requires more nonhuman primate genomic and cDNA sequences than are presently available. Putative primate orthologs represented in cDNA/EST libraries would be useful for establishing that TUs are in fact young, although conserved and nonartifactual, transcribed features. Absence of primate orthologs may signal bona fide human-specific genes but could also indicate that the TUs are artifactual and produced by transcription initiation inefficiency or stochastic initiation from weak promoters. Such transcripts are not necessarily functionally irrelevant, however, because the expression of any transcript can theoretically fall under selective constraint if the initial instance of expression confers an advantage (Ohlsson et al. 2001).
The genomic structures of certain human TUs and UGPs strongly suggest that the existence of those TUs and UGPs is made possible by primate-specific sequences, primate-specific genomic structures, or both. Therefore, the third question is whether primate-specific, and possibly human-specific, TUs and UGPs comprise an essential part of the genomic basis of primate-specific phenotypic characteristics and of the phenotypes that so strikingly differentiate humans from other primates, respectively.
Methods
Definitions
We define known genes as those represented by at least one experimentally based, full-length cDNA in the NT division of GenBank, regardless of coding potential. We define novel TUs as transcribed features in the genome other than known genes. TUs are predicted in silico from EST-to-genomic DNA alignments in which the ESTs do not correspond to exons of known genes. ESTs comprising a TU must be canonically spliced (GT-AG introns) and/or canonically polyadenylated (AATAAA or ATTAAA polyadenylation signal within 40 bp of the submitter-indicated 3′ end). Since all EST-to-genomic alignments were manually curated, the presence of the polyadenylation signals in high-quality EST and genomic sequence was verified. Combined with the requirement for splicing and/or canonical polyadenylation, this effectively eliminated ESTs primed from genomic (A)n stretches. We excluded ESTs from the ORESTES (Strausberg et al. 2002) and RAGE (Harrington et al. 2001) data sets because ORESTES includes many unspliced, singleton, and chimeric ESTs indicative of overall low data quality, and RAGE is derived from cell lines with artificial promoter insertions and therefore is not representative of naturally occurring transcription.
TUs represent genomic segments capable of generating transcripts, regardless of the coding capacity of those transcripts. They are inferred solely from EST evidence, with a single clone sufficient to define a TU in some cases, although a TU may not be supported by a singleton unspliced EST without a canonical polyadenylation signal. For every TU supported by multiple ESTs, we used the 5′-most EST-supported putative transcription start site to define the 5′ boundary, and the 3′-most EST-supported polyadenylation site to define the 3′ boundary. We defined TUs in a strand-specific fashion, as did Okazaki et al. (2002) and Carninci et al. (2003).
UGPs are of two types. An exon-to-exon cis-antisense gene pair is a pair of overlapping genes or TUs, transcribed from the opposite strands of the same locus (Fig. 1A). A putative bidirectionally promoted pair is a pair of divergently transcribed features whose transcription start sites are separated by <1 kb of genomic sequence (Fig. 1B).
Perl-based TU discovery and UGP analysis pipeline
We developed a three-stage Perl-based high-throughput automated annotation pipeline. We modified the bioperl.org open-source BLAST parsers (Stajich et al. 2002) to make them aware of the transcriptional orientation of cDNAs and ESTs matching genomic sequences. The first stage utilized these parsers to analyze all cDNA and EST BLAST matches against RepeatMasker-processed query genomic sequence and determined which nongenic EST matches completely and precisely satisfied our operational definition of a TU. Only primary EST and cDNA evidence was used. We did not use third-party annotations or any transcripts bearing NM and XM designations. In the second stage, all cDNA matches and all nongenic EST matches remaining after the first stage were subjected to BLASTN against the entire human genomic sequence in the NR and HTGS databases. Matches with homologies to genomic regions other than the region in which they were originally identified, and with equal or higher BLAST scores associated with homologies to those other regions, were automatically eliminated due to their putatively segmentally duplicated or pseudogenic nature. The third stage automatically compiled complete exon–intron structures for every gene and TU, quoting exact coordinates of every element of the structure on the genomic sequence, accession numbers of cDNAs or ESTs supporting each exon of each gene and TU, and the extent of apparent involvement of the gene or TU in UGPs in a table suitable for manual curation.
Nonprimate conservation analysis of gene and TU DNA and protein sequences
The criteria for BLASTN were as follows: masked input, Expect = 10, word size = 11. Queries were subjected to RepeatMasker (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker) prior to BLASTN processing. BLASTN outputs were manually curated to exclude low-complexity hits. Only sequences directly supported by wet-laboratory evidence, rather than models built by National Center for Biotechnology Information (NCBI) curators, were deemed acceptable. Therefore, BLAST hits beginning with NM and XM (DNA) and NP and XP (protein) were excluded during the curation of BLAST outputs. For sequences with no nonprimate hits reported after BLASTN searches of the NR and HTGS databases, a BLASTN search of the whole-genome mouse shotgun assembly (mm4) with identical criteria was performed to test for the possibility that the sequences had mouse homologies absent from the mouse sequences in NR and HTGS due to the fragmentary nature of mouse genome coverage by those two databases.
BLASTZ alignments to mouse were visualized from the July 2003 human assembly at the UCSC Genome Browser. Only exons of the human genes and TUs were considered when reporting whether a BLASTZ alignment existed. No distinction was made between partial and complete overlaps of an exon with a sequence block alignable to mouse by BLASTZ. Genes and TUs whose exons at least partially corresponded to blocks on the “Chained BLASTZ mouse/human alignment,” but not to blocks on the “BLASTZ mouse, tight subset of best alignments,” were reported as “aligned but not tight.” For TUs involved in antisense pairs, we excluded the region of cis-antisense overlap in all BLASTN and BLASTZ analyses, focusing instead on sequence conservation in exonic regions unique to the TU and not shared with potentially conserved genes in the same locus.
BLASTX criteria were as follows: low complexity filtering enabled, Expect = 10, word size = 3, matrix BLOSUM62. Only sense-strand protein homologies outside of masked repetitive and low-complexity sequence were considered. Homologies to low-complexity protein sequences corresponding to unmasked DNA sequences, including but not limited to proline-rich and glycine-rich tracts, were disregarded during the manual analysis. BLASTX-suggested putative orthologs were considered only if their ORF direction corresponded to the correct direction of transcription of the query.
Nonparametric assessment of UGP distribution
We conducted a nonparametric assessment of the probability that the observed incidence and clustering of UGPs were due to chance, by performing a simulation. In each run of the simulation, we retained the genomic size, number of exons, and orientation of genes and TUs. However, we used a random number generator to generate a simulated position along the genomic sequence for each gene, so that the same gene would have a different position in every simulation. We kept track of the number of simulated UGPs that arose in every simulation. The percentage of simulation runs in which both the total number of simulated UGPs and the number of simulated UGPs clustered into UGP islands met or exceeded the actual observed number and the actual observed UGP-island clustering of UGPs in the corresponding chromosomal interval served as the nonparametric P-value. Ten thousand distinct simulation runs were performed for each of the 20 intervals into which we divided chr22. The intervals were selected without regard to genomic size but with the criterion that each interval must have ∼50 consecutive gene models (known genes and/or novel TUs).
Expression profile complementarity and interspecies comparative analysis of UGPs
To test whether two members of an antisense pair are expressed in the same tissue or cell type in human, we used bl2seq to extract the longest region of transcript overlap. We used this region as a query in a BLASTN search of the human EST database. After eliminating ORESTES and RAGE ESTs, we examined BLAST output for evidence that the query sequence is transcribed in both directions. The presence of 5′ EST reads matching the query in a “Plus/Plus” orientation as well as other 5′ EST reads with a “Plus/Minus” match, and/or the presence of 3′ EST reads with “Plus/Minus” HSPs (indicating transcription of the query's sense strand) as well as other 3′ EST reads with “Plus/Plus” HSPs (indicating transcription of the query's antisense strand), constituted such evidence. We obtained library origin information from the FASTA descriptor line of every EST matching the query and used the cDNA Library Finder (http://cgap.nci.nih.gov/Tissues/LibraryFinder) to identify the corresponding tissue or cell type of origin, creating two lists of tissues of origin: one for sense-strand and one for antisense-strand ESTs. If the same tissue or cell type of origin appeared in both lists, we considered that as evidence for potential expression of both members of the antisense pair in the same tissue or cell type. ESTs from unclear-origin and pooled-tissue libraries were excluded. Normal and tumor specimens of the same tissue were nonequivalent. For example, sense-strand transcription in normal brain and antisense transcription in a brain tumor would not count as a complementary expression profile, whereas sense and antisense transcription in brain tumors (regardless of whether or not they were observed in the same cDNA library) would count as a complementary expression profile.
To determine if a human antisense pair was conserved at the orthologous mouse locus, we submitted the region of overlap to BLASTN against the MGSCv3 mouse genome database at NCBI and used the highest-scoring hit on the mouse genome as a BLASTN query against the mouse subset of dbEST. We searched for evidence that the mouse query is transcribed in both directions, with an approach identical to that used in human. In addition, if one or both members of the human pair had mouse orthologs, and if the human configuration was either tail-to-tail or head-to-head, we searched for positional equivalents of antisense transcripts in the mouse by analyzing the directionality of mouse EST matches to the last or first exon, respectively, of the mouse transcripts. We did not perform expression profile complementarity testing in mouse.
We tested for expression profile complementarity in each putative bidirectionally promoted pair with the same protocol as that used for antisense analysis. To determine the structure of the orthologous mouse locus, we used the reference transcripts (flcDNAs, ESTs, or hand-constructed contigs bearing the most characteristic exon–intron footprints) of the two members of each human pair as BLASTN queries against the mouse subsets of the NT and EST databases. The top-scoring mouse hit, if any, was the putative mouse ortholog. When both human pair members had putative mouse orthologs, the mouse orthologs were submitted to BLASTN against the MGSCv3 mouse genome database at NCBI (Expect = 10, default filter, no MegaBLAST). The coordinates of their 5′ ends on the mouse genomic sequence were used to determine locus structure in the mouse. Whenever only one of the two human pair members had a putative mouse ortholog, we manually interpreted BLAST output for 1 kb of genomic sequence upstream of the 5′-most known end of the mouse ortholog, searching for divergently transcribed ESTs indicative of positionally equivalent TUs. When curating the outputs of BLAST searches against the mouse EST database, we eliminated all ESTs with “RIKEN” in their FASTA descriptor, due to well-known misorientation problems in the RIKEN mouse EST set.
Acknowledgments
We thank Phil Green and Debbie Nickerson for helpful discussions and guidance, as well as Ming K. Lee for assistance with SeqHelp and advice on Perl programming. This work was supported by NIH training grants HG-00035 and CA-09437, as well as by NIH R01 grant CA-27632.
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.3883606.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Adachi, N. and Lieber, M.R. 2002. Bidirectional gene organization: A common architectural feature of the human genome. Cell 109 807-809. [DOI] [PubMed] [Google Scholar]
- Andres, A.M., Soldevila, M., Saitou, N., Volpini, V., Calafell, F., and Bertranpetit, J. 2003. Understanding the dynamics of Spinocerebellar ataxia 8 (SCA8) locus through a comparative genetic approach in humans and apes. Neurosci. Lett. 336 143-146. [DOI] [PubMed] [Google Scholar]
- Angiolillo, A., Russo, G., Porcellini, A., Smaldone, S., D'Alessandro, F., and Pietropaolo, C. 2002. The human homologue of the mouse Surf5 gene encodes multiple alternatively spliced transcripts. Gene 284 169-178. [DOI] [PubMed] [Google Scholar]
- Burset, M., Seledtsov, I.A., and Solovyev, V.V. 2000. Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res. 28 4364-4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey, J.J., Safrany, S.T., Yang, X., and Shears, S.B. 2000. Discovery of molecular and catalytic diversity among human diphosphoinositol-polyphosphate phosphohydrolases: An expanding Nudt family. J. Biol. Chem. 275 12730-12736. [DOI] [PubMed] [Google Scholar]
- Carninci, P., Waki, K., Shiraki, T., Konno, H., Shibata, K., Itoh, M., Aizawa, K., Arakawa, T., Ishii, Y., Sasaki, D., et al. 2003. Targeting a complex transcriptome: The construction of the mouse full-length cDNA encyclopedia. Genome Res. 13 1273-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, A., Zhang, G., and Bajic, V.B. 2004. Information for the Coordinates of Exons (ICE): A human splice sites database. Genomics 84 762-766. [DOI] [PubMed] [Google Scholar]
- Collins, J.E., Goward, M.E., Cole, C.G., Smink, L.J., Huckle, E.J., Knowles, S., Bye, J.M., Beare, D.M., and Dunham, I. 2003. Reevaluating human gene annotation: A second-generation analysis of chromosome 22. Genome Res. 13 27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courseaux, A. and Nahon, J.L. 2001. Birth of two chimeric genes in the Hominidae lineage. Science 291 1293-1297. [DOI] [PubMed] [Google Scholar]
- Delihas, N. and Forst, S. 2001. MicF: An antisense RNA gene involved in response of Escherichia coli to global stress factors. J. Mol. Biol. 313 1-12. [DOI] [PubMed] [Google Scholar]
- Edgar, A.J. 2003. The gene structure and expression of human ABHD1: Overlapping polyadenylation signal sequence with Sec12. BMC Genomics 4 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejima, Y. and Yang, L. 2003. Trans mobilization of genomic DNA as a mechanism for retrotransposon-mediated exon shuffling. Hum. Mol. Genet. 12 1321-1328. [DOI] [PubMed] [Google Scholar]
- Enard, W., Khaitovich, P., Klose, J., Zollner, S., Heissig, F., Giavalisco, P., Nieselt-Struwe, K., Muchmore, E., Varki, A., Ravid, R., et al. 2002. Intra- and interspecific variation in primate gene expression patterns. Science 296 340-343. [DOI] [PubMed] [Google Scholar]
- Feng, J., Funk, W.D., Wang, S.S., Weinrich, S.L., Avilion, A.A., Chiu, C.P., Adams, R.R., Chang, E., Allsopp, R.C., and Yu, J. 1995. The RNA component of human telomerase. Science 269 1236-1241. [DOI] [PubMed] [Google Scholar]
- Gardiner, K., Fortna, A., Bechtel, L., and Davisson, M.T. 2003. Mouse models of Down syndrome: How useful can they be? Comparison of the gene content of human chromosome 21 with orthologous mouse genomic regions. Gene 318 137-147. [DOI] [PubMed] [Google Scholar]
- Harrington, J.J., Sherf, B., Rundlett, S., Jackson, P.D., Perry, R., Cain, S., Leventhal, C., Thornton, M., Ramachandran, R., Whittington, J., et al. 2001. Creation of genome-wide protein expression libraries using random activation of gene expression. Nat. Biotechnol. 19 440-445. [DOI] [PubMed] [Google Scholar]
- Hildebrandt, M. and Nellen, W. 1992. Differential antisense transcription from the Dictyostelium EB4 gene locus: Implications on antisense-mediated regulation of mRNA stability. Cell 69 197-204. [DOI] [PubMed] [Google Scholar]
- Hubbard, T., Barker, D., Birney, E., Cameron, G., Chen, Y., Clark, L., Cox, T., Cuff, J., Curwen, V., Down, T., et al. 2002. The Ensembl genome database project. Nucleic Acids Res. 30 38-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (IHGSC). 2004. Finishing the euchromatic sequence of the human genome. Nature 431 931-945. [DOI] [PubMed] [Google Scholar]
- Kapranov, P., Cawley, S.E., Drenkow, J., Bekiranov, S., Strausberg, R.L., Fodor, S.P., and Gingeras, T.R. 2002. Large-scale transcriptional activity in chromosomes 21 and 22. Science 296 916-919. [DOI] [PubMed] [Google Scholar]
- Kawashima, I., Mita-Honjo, K., and Takiguchi, Y. 1992. Characterization of the primate-specific repetitive DNA element MER1. DNA Seq. 2 313-318. [DOI] [PubMed] [Google Scholar]
- Kent, W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M., and Haussler, D. 2002. The human genome browser at UCSC. Genome Res. 12 996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosawa, H., Yamanaka, I., Osato, N., Kondo, S., and Hayashizaki, Y. 2003. Antisense transcripts with FANTOM2 clone set and their implications for gene regulation. Genome Res. 13 1324-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, C., Loros, J.J., Dunlap, J.C., and Crosthwaite, S.K. 2003. Role for antisense RNA in regulating circadian clock function in Neurospora crassa. Nature 421 948-952. [DOI] [PubMed] [Google Scholar]
- Kumar, A., Harrison, P.M., Cheung, K.H., Lan, N., Echols, N., Bertone, P., Miller, P., Gerstein, M.B., and Snyder, M. 2002. An integrated approach for finding overlooked genes in yeast. Nat. Biotechnol. 20 58-63. [DOI] [PubMed] [Google Scholar]
- Kutach, A.K. and Kadonaga, J.T. 2000. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol. Cell. Biol. 20 4754-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson, T.P., Murray, C.G., Hill, T., Fredriksson, R., and Schioth, H.B. 2005. Comparison of the current RefSeq, Ensembl and EST databases for counting genes and gene discovery. FEBS Lett. 579 690-698. [DOI] [PubMed] [Google Scholar]
- Lee, R.C. and Ambros, V. 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294 862-864. [DOI] [PubMed] [Google Scholar]
- Lewin, B. 2000. Genes VII. Oxford University Press, New York.
- Long, M., Deutsch, M., Wang, W., Betran, E., Brunet, F.G., and Zhang, J. 2003. Origin of new genes: Evidence from experimental and computational analyses. Genetica 118 171-182. [PubMed] [Google Scholar]
- Margolin, J. 2001. From comparative and functional genomics to practical decisions in the clinic: A view from the trenches. Genome Res. 11 923-925. [DOI] [PubMed] [Google Scholar]
- Mattick, J.S. 2003. Challenging the dogma: The hidden layer of non-protein-coding RNAs in complex organisms. Bioessays 25 930-939. [DOI] [PubMed] [Google Scholar]
- Millar, R., Conklin, D., Lofton-Day, C., Hutchinson, E., Troskie, B., Illing, N., Sealfon, S.C., Hapgood, J. 1999. A novel human GnRH receptor homolog gene: Abundant and wide tissue distribution of the antisense transcript. J. Endocrinol. 162 117-126. [DOI] [PubMed] [Google Scholar]
- Misra, S., Crosby, M.A., Mungall, C.J., Matthews, B.B., Campbell, K.S., Hradecky, P., Huang, Y., Kaminker, J.S., Millburn, G.H., Prochnik, S.E., et al. 2002. Annotation of the Drosophila melanogaster euchromatic genome: A systematic review. Genome Biol. 3 research0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numata, K., Kanai, A., Saito, R., Kondo, S., Adachi, J., Wilming, L.G., Hume, D.A., Hayashizaki, Y., and Tomita, M. 2003. Identification of putative noncoding RNAs among the RIKEN mouse full-length cDNA collection. Genome Res. 13 1301-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson, R., Paldi, A., and Graves, J.A. 2001. Did genomic imprinting and X chromosome inactivation arise from stochastic expression? Trends Genet. 17 136-141. [DOI] [PubMed] [Google Scholar]
- Okazaki, Y. and Hume, D.A. 2003. A guide to the mammalian genome. Genome Res. 13 1267-1272. [Google Scholar]
- Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., Nikaido, I., Osato, N., Saito, R., Suzuki, H., et al. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420 563-573. [DOI] [PubMed] [Google Scholar]
- Qvist, H., Sjostrom, H., and Noren, O. 1998. The TATA-less, GC-rich porcine dipeptidylpeptidase IV (DPPIV) promoter shows bidirectional activity. Biol. Chem. 379 75-81. [PubMed] [Google Scholar]
- Rinn, J.L., Euskirchen, G., Bertone, P., Martone, R., Luscombe, N.M., Hartman, S., Harrison, P.M., Nelson, F.K., Miller, P., Gerstein, M., et al. 2003. The transcriptional activity of human chromosome 22. Genes & Dev. 17 529-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, A., Griffiths-Jones, S., Ashurst, J.L., and Bradley, A. 2004. Identification of mammalian microRNA host genes and transcription units. Genome Res. 14 1902-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, Y., Ikeda, S., Kiyohara, H., Ayabe, H., Seki, T., and Matsui, H. 2002. Sequencing analysis of a putative human O-sialoglycoprotein endopeptidase gene (OSGEP) and analysis of a bidirectional promoter between the OSGEP and APEX genes. Gene 285 101-108. [DOI] [PubMed] [Google Scholar]
- Shendure, J. and Church, G.M. 2002. Computational discovery of sense-antisense transcription in the human and mouse genomes. Genome Biol. 3 research0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shklar, M., Strichman-Almashanu, L., Shmueli, O., Shmoish, M., Safran, M., and Lancet, D. 2005. GeneTide—Terra Incognita Discovery Endeavor: A new transcriptome focused member of the GeneCards/GeneNote suite of databases. Nucleic Acids Res. 33 D556-D561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker, D.D., Schadt, E.E., Armour, C.D., He, Y.D., Garrett-Engele, P., McDonagh, P.D., Loerch, P.M., Leonardson, A., Lum, P.Y., Cavet, G., et al. 2001. Experimental annotation of the human genome using microarray technology. Nature 409 922-927. [DOI] [PubMed] [Google Scholar]
- Sleutels, F., Zwart, R., and Barlow, D.P. 2002. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415 810-813. [DOI] [PubMed] [Google Scholar]
- Smith, M.L., Mitchell, P.J., and Crouse, G.F. 1990. Analysis of the mouse Dhfr/Rep-3 major promoter region by using linker-scanning and internal deletion mutations and DNase I footprinting. Mol. Cell. Biol. 10 6003-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich, J.E., Block, D., Boulez, K., Brenner, S.E., Chervitz, S.A., Dagdigian, C., Fuellen, G., Gilbert, J.G., Korf, I., Lapp, H., et al. 2002. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 12 1611-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausberg, R.L., Feingold, E.A., Grouse, L.H., Derge, J.G., Klausner, R.D., Collins, F.S., Wagner, L., Shenmen, C.M., Schuler, G.D., Altschul, S.F., et al. 2002. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. 99 16899-16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, Y., Tsunoda, T., Sese, J., Taira, H., Mizushima-Sugano, J., Hata, H., Ota, T., Isogai, T., Tanaka, T., Nakamura, Y., et al. 2001. Identification and characterization of the potential promoter regions of 1031 kinds of human genes. Genome Res. 11 677-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topper, J.N. and Clayton, D.A. 1990. Characterization of human MRP/Th RNA and its nuclear gene: Full length MRP/Th RNA is an active endoribonuclease when assembled as an RNP. Nucleic Acids Res. 18 793-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein, N.D., Aldred, S.F., Hartman, S.J., Schroeder, D.I., Otillar, R.P., and Myers, R.M. 2004. An abundance of bidirectional promoters in the human genome. Genome Res. 14 62-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufarelli, C., Stanley, J.A., Garrick, D., Sharpe, J.A., Ayyub, H., Wood, W.G., and Higgs, D.R. 2003. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat. Genet. 34 157-165. [DOI] [PubMed] [Google Scholar]
- Veeramachaneni, V., Makalowski, W., Galdzicki, M., Sood, R., and Makalowska, I. 2004. Mammalian overlapping genes: The comparative perspective. Genome Res. 14 280-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheelan, S.J., Church, D.M., and Ostell, J.M. 2001. Spidey: A tool for mRNA-to-genomic alignments. Genome Res. 11 1952-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelin, R., Dahary, D., Sorek, R., Levanon, E.Y., Goldstein, O., Shoshan, A., Diber, A., Biton, S., Tamir, Y., Khosravi, R., et al. 2003. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 21 379-386. [DOI] [PubMed] [Google Scholar]
Web site references
- http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker; RepeatMasker.
- http://cgap.nci.nih.gov/Tissues/LibraryFinder; cDNA Library Finder.