Abstract
Crossover (CO) is a key process for the accurate segregation of homologous chromosomes during the first meiotic division. In most eukaryotes, meiotic recombination is not homogeneous along the chromosomes, suggesting a tight control of the location of recombination events. We genotyped 71 single nucleotide polymorphisms (SNPs) covering the entire chromosome 4 of Arabidopsis thaliana on 702 F2 plants, representing 1404 meioses and allowing the detection of 1171 COs, to study CO localization in a higher plant. The genetic recombination rates varied along the chromosome from 0 cM/Mb near the centromere to 20 cM/Mb on the short arm next to the NOR region, with a chromosome average of 4.6 cM/Mb. Principal component analysis showed that CO rates negatively correlate with the G+C content (P =3×10-4), in contrast to that reported in other eukaryotes. COs also significantly correlate with the density of single repeats and the CpG ratio, but not with genes, pseudogenes, transposable elements, or dispersed repeats. Chromosome 4 has, on average, 1.6 COs per meiosis, and these COs are subjected to interference. A detailed analysis of several regions having high CO rates revealed “hot spots” of meiotic recombination contained in small fragments of a few kilobases. Both the intensity and the density of these hot spots explain the variation of CO rates along the chromosome.
Meiotic crossovers (COs) and sister chromatid cohesion provide physical links between homologous chromosomes ensuring proper chromosome segregation during the first meiotic division. In most eukaryotes, there is always at least one CO per pair of homologs (obligatory crossover) (Jones 1984, 1987). Cytological, genetic, and molecular studies in many organisms have demonstrated that COs are not evenly distributed along the chromosomes (Jones 1987; Carpenter 1988; Lynn et al. 2002). The tight control of the number and/or localization of COs is crucial. Mutations that reduce CO formation increase chromosome nondis-junction in organisms as diverse as Saccharomyces cerevisiae, Schizosaccharomyces pombe, Caenorhabditis elegans, Drosophila melanogaster (female), Arabidopsis thaliana, and the mouse (for review, see Lynn et al. 2004).
In yeast, the distribution of meiotic recombination events (COs and noncrossover gene conversions; NCOs) along chromosomes has been studied in detail by locating DNA double-strand breaks (DSBs), which initiate meiotic recombination (Baudat and Nicolas 1997; Gerton et al. 2000). These studies showed that DSBs tend to be clustered in chromosomal domains away from telomeres and centromeres (Gerton et al. 2000; Borde et al. 2004). In mammals, COs are also nonrandomly distributed along the chromosomes, with alternate domains having higher or lower levels of recombination (Kong et al. 2002; Nachman 2002). The CO rates tend to be low near the centromeres and increase toward the telomeres. In plants, the CO rates also vary along chromosomes (for review, see Anderson and Stack 2002). In general, centromeric regions have low CO rates compared to telomeric regions. However, in plants, there have been very few high-resolution studies in a single chromosome.
Many sequence parameters have been linked to the variation of CO rates in eukaryotes. In yeast and mammals, several studies have found a correlation between a high G+C content and a high rate of recombination in large domains (Gerton et al. 2000; Fullerton et al. 2001; Yu et al. 2001; Kong et al. 2002; Petes and Merker 2002; Jensen-Seaman et al. 2004). However, within 2-3 kb of the recombination initiation site no correlation between the G+C content and the distribution of COs in both yeast and humans was found (for review, see de Massy 2003) and, second, in human, rat, and mouse, when CpG ratio is included in a multiple regression analysis, correlation with the G+C content becomes negative (Kong et al. 2002; Jensen-Seaman et al. 2004). In wheat, barley, and maize, gene-rich regions are more recombinationally active than gene-poor regions (for review, see Schnable et al. 1998). In humans, female CO rates are not correlated with gene density on chromosome 21 (Lynn et al. 2000) whereas male CO rates are correlated, suggesting a different type of control. There are also conflicting results when correlating the density of transposable elements (TEs) and recombination rates (see Wright et al. 2003). Nevertheless, differences in meiotic CO rates between the sexes have been demonstrated in many higher eukaryotes (Lenormand and Dutheil 2005). Therefore, the primary DNA sequence itself cannot explain all of the variation of meiotic recombination.
In S. cerevisiae and S. pombe, hot spots have been defined as small DNA fragments of 1-2 kb, centered around meiotic DSBs that are repaired, using the homologous chromosome, to produce COs or NCOs (Keeney 2001). In mice, humans, and plants, several such regions have been studied in detail and have been found to share common features with hot spots described in yeast. These include high level of COs and NCOs clustered in small segments (1-2 kb) and a lack of clear consensus sequences (de Massy 2003; Kauppi et al. 2004; Rafalski and Morgante 2004). The distribution of meiotic hot spots along chromosomes is uneven, which suggests a local control of DSB formation. A lot of effort has been made recently to characterize this fine-scale variation of recombination rates mainly in humans but also in other eukaryotes for various reasons among which is to gain insight into the underlying mechanisms, to assist association studies, or to improve inferences from polymorphism data about selection and population history. However, except for S. cerevisiae, only a few regions have been characterized at the molecular level in other eukaryotes and more genome-wide studies are needed to unravel the determinants of hot spot activity.
The availability of the Arabidopsis genome sequence (The Arabidopsis Genome Initiative 2000) and the recent development of powerful high-throughput genotyping techniques (Gut 2001; Kwok 2001), allow us to determine precisely the location and rates of COs on one chromosome. Here, we show that CO rates are highly variable on chromosome 4 of Arabidopsis, with some regions having five times more COs than the chromosome average. The CO rates significantly negatively correlate with the G+C content and also significantly correlate with the density of single repeats and with CpG ratio. However, they do not correlate significantly either with genes, pseudogenes, transposable elements, or dispersed repeats. Our data also confirm that COs are subjected to interference on chromosome 4. Finally, we provide evidence of meiotic recombination hot spots and show that both their activity and density contribute to the variation of the CO rates.
Results
Chromosome 4 of A. thaliana is the smallest of its five chromosomes and presents several remarkable features (Fig. 1). It has an acrocentric architecture with a long arm 14.6 Mb long and short arm about 8 Mb long tipped by the nucleolar organizer region (NOR). This region is about 3.6-4 Mb long and is constituted of almost homogeneous ribosomal DNA repeats (Haberer et al. 1996). The available short arm sequence starts in the last proximal copy of the rDNA repeat (Mayer et al. 1999; The Arabidopsis Information Resource, http://www.arabidopsis.org/). In some accessions, including Columbia (Col) but not Landsberg (Ler), the short arm has a heterochomatic region, called the “knob,” identified cytologically (Fransz et al. 2000), primarily comprising transposable elements, in which a few genes are insulated (Mayer et al. 1999; Lippman et al. 2004). Moreover, an approximately 1.5-Mb-long region of the short arm, including the knob, is inverted between the two accessions, Col and Ler (Fransz et al. 2000).
Figure 1.
Variation of the CO rates on chromosome 4 of A. thaliana. The numbers refer to the intervals given in Supplemental Table 2. The dotted line represents the average CO rate on chromosome 4 (4.6 cM/Mb). A schematic representation of chromosome 4 of A. thaliana is aligned with the diagram. (Black box) NOR (nucleolar organizer region), (gray box) heterochromatic knob, (diamond-shaped box) centromere.
We genotyped a population of 736 F2 plants resulting from a cross between Col and Ler (see Methods) with 71 SNPs (Supplemental Table 1) chosen from the Monsanto database (Jander et al. 2002) to be evenly spaced on the Arabidopsis chromosome 4. The average interval between two SNPs was 204 kb on the long arm (60 SNPs) and 239 kb on the short arm (11 SNPs).
Variation of CO rates across chromosome 4
After SNP genotyping, we analyzed the variation in CO rates in 702 plants (34 plants had missing data for more than 24 markers and were thus discarded). On average, we genotyped 666 plants (thus representing 1332 meioses because in an F2 plant each chromosome comes from an independent meiosis) per interval. We verified that there was no bias in the segregation of each marker. The cumulated genetic distance of the chromosome was estimated to be 83.9 cM, of which 69 cM corresponded to the long arm (Supplemental Table 2).
As the intervals were small, the genetic length of each interval can be simply calculated by dividing the number of recombinant chromosomes by the number of meioses analyzed. Genetic recombination varied greatly along the chromosome, from 0 cM/Mb next to the centromere, to 20.2 cM/Mb next to the NOR (Supplemental Table 2; Fig. 1). The frequencies of COs in different intervals could not be directly compared because of both the variation in interval length and the number of analyzed chromosomes. Therefore, we developed a statistical approach to unambiguously identify intervals that were significantly either “colder” or “hotter” than the chromosome average. The approach is based on a simply binomial model of the number of COs in each interval, so that the “temperature” of an interval is determined by the probability that the number of COs in it exceeds the expected one, under the assumption that the recombination rate is constant along the chromosome. We implemented a statistical program (TETRA) to compute both the average number of COs per nucleotide, and the significance of the observed values from the binomial model (see Methods).
TETRA calculated an average of 4.6 × 10–8 COs/nucleotide, which is, on average, 1 cM for 217 kb for chromosome 4. Among the 70 intervals tested, TETRA identified 30 intervals with a significant deviation from the average rate of COs; 12 intervals had a significantly lower rate (cold) and 18 had a significantly higher rate (hot) (P > 0.95 and P < 0.05 for the cold and hot intervals, respectively; Supplemental Table 2). The hot intervals were not randomly distributed: four (intervals 67-70) were clustered on the short arm next to the NOR and eight (intervals 43-56) were clustered in a 3-Mb region on the long arm next to the centromere (Fig. 1). There was almost no genetic recombination in the centromeric and inverted region (intervals 58-63) and no clustering of the cold intervals was observed outside the centromeric region. In the middle of the long arm, there were alternate hot and cold intervals, although the “temperature” of most of these intervals was not significantly different from the chromosome average. In summary, the COs were unevenly distributed along chromosome 4 with alternating hot and mildly cold regions.
Correlation of CO rates with primary sequence features
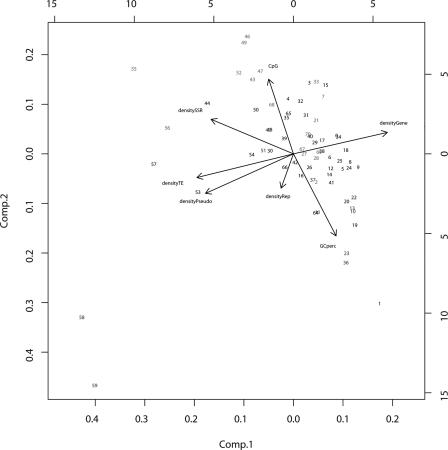
We performed a principal component analysis to determine the most relevant genome features that correlated with the observed CO frequencies. The genes, pseudogenes, G+C content, and CpG log ratio, as well as repeated sequences, such as transposable elements (TEs) and single repeats (SSR), were carefully listed from both publicly available data and in-house computed analysis (see Methods). In each interval, we took the G+C content and the CpG ratio (see Methods) and calculated the density of each of the other features. We analyzed the whole chromosome, excluding the intervals 60-63 contained in the inverted region. On the first principal component axis, accounting for 46.4% of the variation, we found that gene, pseudogene, and TE densities contribute the most to the composition diversity of the intervals (Fig. 2). However, this axis shows that these features do not occur randomly along the chromosome, but follow two opposite gradients: The gene density is low in the pericentromeric and subtelomeric regions and high in the middle of chromosome arms, whereas the opposite is true for pseudogene and TE density. On the second principal component axis, adding 22.6% to the explained variation, the GC content and the CpG ratio appear to be more relevant to CO rates' variation. The intervals showing a significantly higher rate of COs tend to cluster regions of low G+C content where the CpG ratio is high. Conversely, the intervals with a low CO rate cluster in regions of high G+C content where the CpG ratio is low (Fig. 2).
Figure 2.
Principal component analysis of chromosome 4 of A. thaliana. Numbers refer to the intervals given in Figure 1 and Supplemental Table 2. Hot intervals are indicated in red; cold intervals are indicated in blue.
A regression analysis carried out between the CO rate and the G+C content or CpG ratio confirmed these trends with R2 of 0.18 (P = 3 × 10–4) for G+C content and an R2 of 0.20 (P = 1.3 × 10–4) for the CpG ratio. The regression was stronger when analyzing only the long arm of the chromosome, with R2 = 0.36 (P = 4 × 10–7) for G+C content and R2 = 0.22 (P = 1.7 × 10–4) for the CpG ratio. Of the other regressions tested (gene density, pseudogenes, etc.), only the SSR density had a significant correlation with CO rates (R2 = 0.13; P = 3 × 10–3). Therefore, unlike the results obtained in several other eukaryotes, in which a high CO rate tends to correlate with a high G+C content, we suggest that on chromosome 4 of A. thaliana a high CO rate correlates with a low G+C content. The CpG ratio and SSR density also weakly correlate with CO rates.
Interference on chromosome 4
We obtained 1171 COs for 1404 analyzed meioses. This corresponded to an average of 0.8 events per chromatid and per meiosis, corresponding to 1.6 COs per pair of homologous chromosomes (bivalents) per meiosis. There were, on average, 1.3 events on the long arm and 0.3 events on the short arm. However, if we take into account the 1.5 Mb that are inverted between the two parental lines, and therefore “forbidden” from forming and/or recovering COs, the ratio of COs per megabase on the short arm was double that of the long arm (0.18 vs. 0.09).
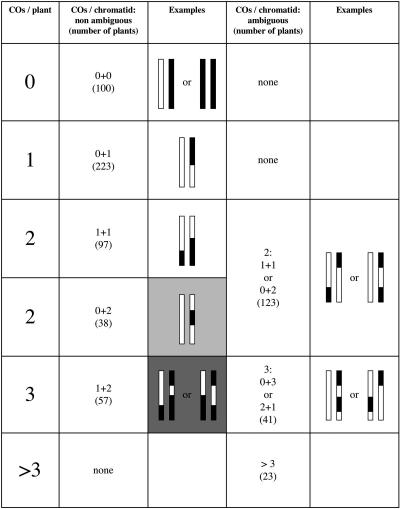
For 515 pairs of chromosomes, we were able to determine unequivocally the number of exchanges that each chromatid had undergone (0, 1, or 2 COs) during meiosis (Fig. 3). For 123 pairs of chromosomes harboring two exchanges, we could not unambiguously attribute the recombination events to one or the other chromatid. We reassigned them either to the “1 + 1” or the “2 + 0” class (see Fig. 3) on the basis of the prorata between the sizes of these latter classes in the nonambiguous class with two exchanges. The 41 pairs that exhibit three exchanges that could not be credited to one or the other chromatid were considered to fall in the “2 + 1” class (that is, we assumed “3 + 0” pairs to be very rare). For the remaining pairs (23), which display four or more CO, we could not attribute CO unambiguously to parental chromosomes, so we discarded them. As expected, one exchange event was the most common occurrence (692 chromatids). Furthermore, we compared the observed distribution of the number of COs to what is expected under a Poisson distribution (Supplemental Table 3), Test of χ2 goodness-of-fit shows that the null Poisson hypothesis can be strongly rejected (χ2 = 121.8, P < 5 × 10–4). Hence, we can conclude that multiple COs do not occur on chromosome 4 independently one from each other.
Figure 3.
Number of CO events per chromatid deduced from the genotype of an F2 plant. When an F2 plant displays two COs, either the extremities of both chromosomes are homozygous, and the COs are on one chromatid or both, or the extremities are heterozygous and there is ambiguity between one event on each chromatid or two events on the same chromatid. A similar approach has been used to analyse F2 plants that displays three COs. When an F2 plant displays more than three COs, the recombination history cannot be inferred from the genotype. In the columns “COs/chromatid,” the numbers in parentheses represent the number of plants in each category. (Light gray box) plants used in the interference analysis. (Dark gray box) plants containing a chromatid with two COs but that could not be used in the interference analysis.
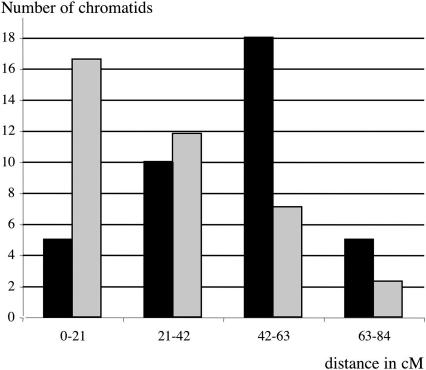
For each of the 38 plants having two precisely located COs on the same chromatid (Fig. 3, light gray box), we calculated the genetic distance between the two COs. The distance varied from 1.17 to 62.8 cM with a mean distance of 44.1 cM. The mean expected value for randomly distributed double COs was one-third of the chromosome, being 27.9 cM (see Methods). We then classified the 38 plants into four groups: group 1, with events separated by less than 25% of the chromosome (0-21 cM); group 2, with events separated by more than 25% but less than 50% of the chromosome (21-42 cM); group 3, with events separated by more than 50% but less than 75% of the chromosome (42-63 cM); and group 4, with events separated by more than 75% of the chromosome (63-83.9 cM) (Fig. 4). We compared the observed distribution with the expected distribution if COs were located independently of each other. We found a very strong probability (χ2 = 27.9, P < 5 × 10–3) that double COs were not located independently of each other. The same analysis on only the long arm also showed that the observed distribution and the observed mean distance (36 cM) were very different from the theoretical values (23 cM; data not shown). We then looked at the effect of the centromere on interference. For the 12 chromosomes having one CO on the short arm and the other on the long arm, the mean distance was 59.6 cM, that is, 70% of the genetic length of the chromosome, while the mean distance between two COs occurring on the long arm represents 52% of the genetic length of the long arm. These results confirm that CO location on chromosome 4 is affected by interference and that the centromere is not a barrier to interference.
Figure 4.
Distribution of the distances in centiMorgans between double COs. (Histogram in black) observed distribution of double COs in our F2 (see text); (histogram in gray) theoretical distribution of double COs if the position of one CO is independent of the second (Methods).
Evidence for the existence of hot spots of recombination
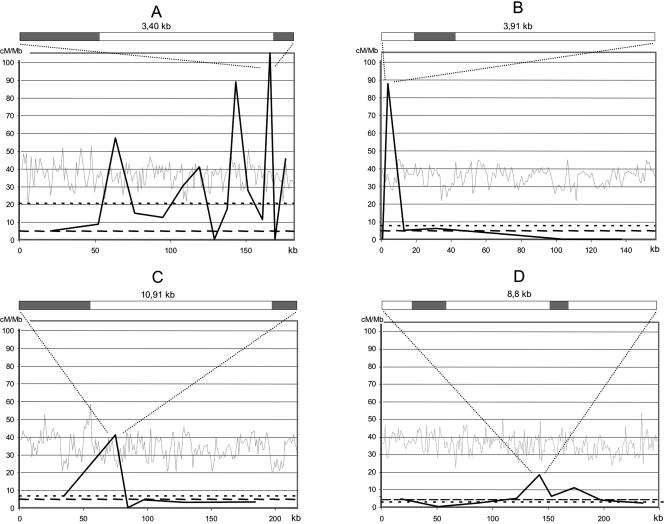
We further investigated several of the 14 intervals having the highest CO rates together with one interval with a slightly above average CO rate and one cold interval. For each interval, we genotyped the corresponding recombinant plants using a set of SNP or indel markers, giving precise locations of the exchange points. We divided the hottest interval (interval 70; Fig. 1) into 15 parts to map the COs at a precision of a few kilobases (Fig. 5A). We found a clearly nonhomogeneous distribution of exchange events. Two very small fragments (3.4 and 3.2 kb) 20 kb apart exhibited a very high rate of COs (>85 cM/Mb), being 15 times higher than the chromosome average (4.6 cM/Mb) and four times higher than the interval average (20.2 cM/Mb). We found that two other fragments in interval 70 had moderately high rates of genetic recombination (40 and 55 cM/Mb, 8 to 10 times the chromosome average). We also analyzed another hot interval (interval 21, Fig. 5B) in the middle of the long arm (Fig. 1). We found one DNA fragment displaying a large increase of genetic recombination in this interval. The recombination rates in the remainder of this interval were mostly lower than the chromosome average. We also observed the same type of “spotty” CO distribution in the other hot intervals that we investigated (7, 55, 56 and 68, 69; data not shown).
Figure 5.
Fine-scale analysis of the distribution of CO breakpoints in four intervals. (A) interval 70: 44.5kb-3; 16.4kb-2; 6.2kb-5; 19.5kb-4; 17.5kb-3; 9.4kb-4; 12.3kb-7; 8.1kb-0; 8.3kb-2; 3.2kb-4; 13.0kb-5; 6.4kb-1; 3.4kb-5; 3.3kb-0; 11.0kb-7. (B) interval 21: 2.5kb-0; 3.9kb-5; 13.8kb-1; 23.6kb-2; 45.7kb-2; 29.2kb-0; 38.6kb-0. (C) interval 57: 69.5kb-6; 12.2kb-7; 7.1kb-0; 17.4kb-1; 46.5kb-2; 65.0kb-3. (D) interval 37: 35.2kb-2; 32.7kb-0; 38.1kb-1; 8.8kb-2; 12.6kb-1; 28.3kb-1; 20.2kb-1; 49.2kb-2; 38.7kb-1. For each interval the size of each DNA fragment is given and the number of recombinant plants. (Light gray line) G+C content calculated in 1-kb windows. (Black line) CO rates in centiMorgans per megabase. (Small dotted line) interval COs rate average. (Large dotted line) chromosome 4 COs rate average. Above each major peak is the gene organization of the fragment. (Gray box) gene.
We then analyzed interval 57 (Fig. 1), which did not appear to have a significantly high rate of genetic recombination when analyzed by TETRA (6.6 cM/Mb; P = 0.08). We found one DNA fragment of 12 kb displaying a high rate of genetic recombination (40 cM/Mb) whereas the remainder of the interval displayed CO rates below the chromosome average (Fig. 5C). Interval 37 was found to be significantly cold when analyzed by TETRA (2.7 cM/Mb; P > 0.98). We performed the same kind of analysis as for the other intervals. We found a dispatch of the 12 CO exchanges in 9 of the 10 fragments studied (Fig. 5D) with a maximum of two events in an 8.8-kb fragment. This small fragment seems to exhibit a slightly higher CO rate than the genome average (Fig. 5D). However, more plants would be needed to confirm this difference. For the four regions analyzed, hot spots did not seem to correlate with G+C content or gene organization (Fig. 5A-D).
Discussion
We obtained a very detailed genetic map of chromosome 4 of A. thaliana by genotyping a series of 71 SNP markers on 702 F2 plants issued from an F1 Col/Ler hybrid. The total size of the genetic map was estimated at 83.9 cM, which is consistent with other maps obtained from crosses of the same accessions: the classical map (76 cM; Meinke et al. 1998), the RFLP map (74.4 cM; Schmidt et al. 1995; Liu et al. 1996), tetrad analysis using the quartet mutation (85 cM; Copenhaver et al. 1998; Lam et al. 2005), and first versions of the RIL genetic map (76 cM; Lister and Dean 1993).
We found that, on average, a chromosome 4 bivalent undergoes 1.6 crossovers per meiosis. Copenhaver et al. found an average of 1.5 COs on chromosome 4 in male meiosis in a Col/Ler cross (Copenhaver et al. 1998; Lam et al. 2005). Meiotic recombination has also been assessed using cytology by recording the numbers and locations of chiasmata on metaphase I bivalents in pollen mother cells of several accessions, including Col and Ler (Sanchez-Moran et al. 2002). Both the genetic and cytological methods gave consistent results, with the mean chiasma frequency being 1.6 for chromosome 4. Therefore, CO frequency on chromosome 4 during meiosis of a Col/Ler F1 hybrid is not greatly different from that in the parents.
In most eukaryotes, “positive interference” (i.e., the probability of COs occurring next to each other is lower than expected) affects the distribution of multiple COs on a single chromosome (see Zickler and Kleckner 1999). However, not all the COs seem to interfere, and recent data suggest two pathways for crossovers in S. cerevisiae, in humans, and in A. thaliana: one pathway being sensitive to interference (class I) and the other insensitive (class II) (Copenhaver et al. 2002; Housworth and Stahl 2003; Higgins et al. 2004; Hollingsworth and Brill 2004; Stahl et al. 2004; Lam et al. 2005; Mercier et al. 2005). We show also that COs are subjected to interference on chromosome 4. Double COs on the same chromatid are significantly further than one-third of the chromosome length apart, contrary to what is expected for randomly distributed COs. In addition, our results suggest that interference is insensitive to centromere as previously proposed by Colombo and Jones (1997) and that the centromere may increase the strength of interference on chromosome 4. However, there is not complete interference, as we observed double COs only a few centiMorgans apart. We could assume, as suggested by previous studies (Copenhaver et al. 2002; Lam et al. 2005) that these close double COs are insensitive to interference and the distant double COs are sensitive to interference. In yeast, the level of interference has been shown to depend on the size of the chromosome with the short chromosomes harboring less interference (Kabback et al. 1999). However, the disparity in size of the chromosomes is less pronounced in Arabidopsis, with the shortest chromosome being more than two-thirds of the size of the longest chromosome, while in yeast there is a fourfold difference in size. Moreover, Lam et al. (2005) recently provided evidences that in Arabidopsis NOR-bearing chromosomes (i.e., chromosomes 2 and 4) exhibit more interference than the others and suggested that the NOR region itself rather than the size of the chromosome could influence interference. Further analyses are needed to determine whether interference varies along the chromosome, as recently suggested in a study on rice (Esch 2005).
Numerous studies have attempted to understand the factors responsible for genetic recombination variations and to identify primary sequence features that may correlate with this variability. In many sexual organisms, such as mammals, birds, yeast, drosophila, and nematodes, positive correlations between the CO rates and G+C content have been observed at the scale of several hundred of kilobases (Hurst et al. 1999; Gerton et al. 2000; Fullerton et al. 2001; Marais et al. 2001; Takano-Shimizu 2001; Yu et al. 2001; Birdsell 2002; Kong et al. 2002; Jensen-Seaman et al. 2004). Gerton et al. (2000) suggested that regions of high G+C content stimulate recombination. Alternatively, several recent studies have proposed that high levels of recombination may create regions with high G+C content, probably through a biased gene conversion (BGC) toward G+C. In other words, meiotic recombination modifies the base composition through the average density of recombination hot spots (see below; Galtier et al. 2001; Birdsell 2002; Montoya-Burgos et al. 2003; Meunier and Duret 2004). However, in humans, rats, and mice, when CpG ratio is included in a multiple regression model, the correlation with the G+C content becomes negative (Kong et al. 2002; Jensen-Seaman et al. 2004).
In contrast, we found that regions of low G+C content and high CpG ratio on chromosome 4 of A. thaliana tend to have higher rates of genetic recombination. Therefore, the BGC hypothesis suggested to explain the correlation found in other eukaryotes may not apply in Arabidopis. However, homologs of genes believed to participate in G+C-biased mismatch repair in other organisms exist in the genome of Arabidopsis (Birdsell 2002). It is also possible that in Arabidopsis BGC cannot affect the nucleotide content due to the high level of inbreeding of the plant that does not favor the formation of heteroduplex DNAs. However, this would explain an absence of correlation but not a negative correlation. In contrast to our results, a study recently reported no correlation between the G+C content and CO rates in Arabidopsis (Marais et al. 2004). We suggest that our observation is due to the higher precision of our recombination map because we studied 702 plants compared to the 101 RILs used in the study of Marais et al. (2004). Therefore, our study in Arabidopsis questions the assumptions made for G+C correlation and so, the problem of causation remains an open query. Data from more species are needed and may reveal a species-specific lineage in the evolution of recombination.
A fine-scale analysis of several intervals showed peaks in crossover activity. For example, in the hottest interval (interval 70; Fig. 1), CO breakpoints are found in 12 of the 14 fragments tested, even though there is clustering in two small regions 20 kb apart (Fig. 5A). In other intervals, including one not having a significantly high CO rate, one small DNA fragment accounts for most of the genetic recombination of the interval. In the genome of A. thaliana, this punctuate distribution of CO activity strongly suggests recombination hot spots where recombination events group around an initiation site. In plants, several hot spots of CO activity have been described. The 140-kb a1-sh2 region in maize has peaks of CO activity (three to six times the genome average) in three small intervals (1.7-3.4 kb) (Yao et al. 2002). Other loci in maize or in rice, such as bronze or waxy, show some properties of hot spots (Dooner and Martinez-Ferez 1997; Okagaki and Weil 1997; Inukai et al. 2000). For the bronze locus, unlike for yeast and mammals, it has been suggested that recombination is initiated uniformly along the gene and not at a preferential site. Therefore, although existence of hot spots seems to be the rule rather than the exception in plants and other higher eukaryotes, there may be some differences. However, all studies on higher eukaryotes looked at no more than one or two intervals that were often selected for a phenotype associated with the recombination event. Therefore, it is difficult to determine whether the observed hot spot patterns in these intervals can be applied to the whole genome. Here, we show that a punctuate distribution of hot spots is a general feature of the chromosome that is not restricted to significantly recombinogenic regions. Our results also strongly suggest that recombination is initiated at preferential sites all along the chromosome. However, both the intensity and the density of the recombination sites influence the variation of recombination, as hot regions contain one or several very hot spots whereas a mildly warm interval would contain only one mild hot spot. Cold regions may contain few spots with a higher rate of recombination than the genome average, but it remains to be demonstrated. A similar result has recently been obtained in the human genome, where strong hot spots have been detected in narrow regions of strong LD and weak hot spots in regions of strong marker association (Jeffreys et al. 2005). We have identified more than 10 small DNA fragments that may behave as hot spots on chromosome 4 of A. thaliana. Further experiments are needed to confirm the strength and the precise location of the initiation site of these hot spots. Their fine characterization and analysis in other genetic backgrounds is needed to determine the factors that govern their activity and distribution.
Methods
F2 recombinant population construction, genomic DNA extraction
The two Arabidopsis accessions, Columbia and Landsberg erecta, were crossed to obtain an F1 hybrid. Self-fertilization from a single F1 was carried out to obtain F2 seeds. Seeds were grown in soil in long-day conditions in the greenhouse. At the rosette stage, the whole material of 736 F2 plants together with plant material from the two parental accessions was collected. DNA was extracted as described (Loudet et al. 2002).
Selection of SNPs
Most of the SNPs were chosen from the Monsanto database (Jander et al. 2002). When convenient SNPs were not found in the database, DNA fragments were amplified at the desired position on the genomic DNA of the two parental accessions and sequenced to identify a SNP suitable for genotyping. A list of the SNPs used in this study is given in Supplemental Table 1. A couple of primers were designed for each SNP to obtain a PCR fragment containing the predicted SNP. A list of the PCR primers used in this study is given in Supplemental Table 1. The PCRs were carried out on the parental accession DNAs using standard conditions: 94°C 4 min, (94°C 45 sec, 52°C 45 sec, 72°C 1 min) × 35 cycles, 72°C, with Eurobio 1× reaction buffer and Taq polymerase. PCR fragments were sequenced (Genome Express) to check the presence and position of the SNPs.
SNP genotyping
At each of the SNP sites, DNA extracted from the F2 plants was genotyped either by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry, as described by Sauer et al. (2000) or by fluorescence based techniques: the Amplifluor technology (Serological Corporation) or the TaqMan technology (Applied Biosystems). For a number of SNPs, the results obtained with one technique (usually mass spectrometry) were confirmed with one of the two other methods. The techniques used for each SNP are given in Supplemental Table 1.
Statistical analysis of CO rates: TETRA
We define P as the probability of having a CO in a specific position of the chromosome, and assume that P ≪ 1. The probability Pi of observing one CO in the ith interval on a chromosome is approximated as PLi, where Li stands for the length of the ith interval. If the number of informative chromosomes for the interval i (i.e., the number of chromosomes for which both SNPs delimiting the interval are available) is written as Vi, then the number, Ni, of COs in the ith interval is distributed according to a binomial B(Vi,PLi) under the null hypothesis that the COs rate is constant along the chromosome. More explicitly, we have for all
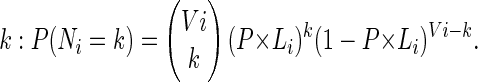 |
According to the observed values, ni, of the number of COs in the different intervals, TETRA computes the average CO rate, P, along the whole chromosome. It then computes the P-value, Ti, of the observed number of COs under the above binomial model, that is:
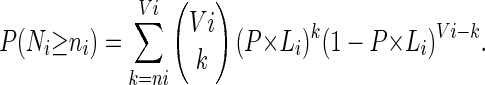 |
This P-value can be interpreted as the probability that the number of COs in the ith interval exceeds its observed value under the model of homogeneous CO rate along the chromosome.
Statistical analysis of COs interference
We derive the probability distribution function of the distribution of distances between the two COs by assuming that the locations of the two COs are independently uniformly distributed random variables. L is the length of the chromosome, and x and y are locations of the two COs. x and y are uniformly distributed in [0, L]. The distance r = (x–y). The distribution of x and y is symmetric under the exchange of x and y; we can condition on x > y. The probability distribution function P(r) is proportional to the length of the segment, in the x, y plane, between the points of coordinates (r, 0) and (L, L–r), which is itself proportional to L–r. Imposing the normalization of the probability distribution function, we obtain P(r) = 2(1–r/L). The expectation value of r is thus 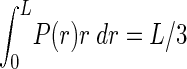 . We can deduce the probabilities P1, P2, P3, P4 of r being in each of the four bins [0, L/4], [L/4, L/2], [L/2, 3L/4], [3L/4, L]. For instance
. We can deduce the probabilities P1, P2, P3, P4 of r being in each of the four bins [0, L/4], [L/4, L/2], [L/2, 3L/4], [3L/4, L]. For instance 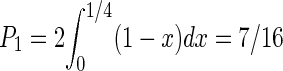 . Similarly, we find P2 = 5/16, P3 = 3/16, P4 = 1/16. These probabilities are used in the analysis of Figure 4.
. Similarly, we find P2 = 5/16, P3 = 3/16, P4 = 1/16. These probabilities are used in the analysis of Figure 4.
Correlation studies
The A. thaliana genomic sequence and its annotation were down-loaded from the TIGR Web site (http://ftp.tigr.org/pub/data/a_thaliana/ath1/). Gene and pseudogene annotations have been extracted from TIGR-XML files from release 5 of the genome annotation. Transposable elements have been re-annotated using the RMBLR procedure from the TE annotation pipeline described by Quesneville et al. (2005). The TE reference set used is derived from the A. thaliana RepeatMasker repeat library (March 6, 2004). The same TE family consecutive TE fragments (on both the genome and the reference TE) have been automatically joined if separated by a sequence composed of more than 80% of other TE insertions (in this case we have a nested TE). Otherwise they are joined if a gap of 5000 nucleotides or a region of mismatches 500 nucleotides long separate them.
Single repeats (SSR) were found using the Tandem Repeat Finder program (Benson 1999), and repeats by a BLASTN all-by-all using BLASTER and GROUPER (Quesneville et al. 2005) without using any simple link clustering coverage constraint. G+C content and CpG were counted with in-house python scripts. CpG ratios were computed by taking the log10 of the C+G dinucleotide frequency divided by the product of G and C frequencies. All data and analysis results were stored in a MySQL database and retrieved by SQL queries.
Statistical analyses were carried out using the R software environment (http://cran.r-project.org)
Detection of hot spots
We halved each interval by choosing a convenient SNP (or indel) either from the Monsanto database or by DNA sequencing (see above). We then sequenced the DNA fragment containing the SNP in the recombinant plants and finally distributed the plants according to their genotype within one or the other half interval. This was iteratively repeated until the location of CO breakpoints was obtained within a few kilobases. A list of the SNPs, indels, and corresponding primers is given in Supplemental Table 1. Genomic DNA from plants was amplified by PCR in standard conditions (see above) with an annealing temperature adapted for each set of primers.
Acknowledgments
We thank Mathilde Grelon, Raphaël Mercier, Eric Jenczewski, and Valérie Borde for critical reading of the manuscript and Sylvie Jolivet for technical assistance. All the members of the “Méiose et Recombinaison” group provided helpful comments and participated in stimulating discussions. Marc Mézard kindly provided the statistical analysis of interference. This work was supported by grants from the Institut National de la Recherche Agronomique (to C.M.) and the European Union (Epigenome Network of Excellence to V.C.).
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4319006.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Anderson, L.K. and Stack, S.M. 2002. Meiotic recombination in plants. Curr. Genomics 3 507-525. [Google Scholar]
- The Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796-815. [DOI] [PubMed] [Google Scholar]
- Baudat, F. and Nicolas, A. 1997. Clustering of meiotic double-strand breaks on yeast chromosome III. Proc. Natl. Acad. Sci. 94 5213-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, G. 1999. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 27 573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birdsell, J.A. 2002. Integrating genomics, bioinformatics, and classical genetics to study the effects of recombination on genome evolution. Mol. Biol. Evol. 19 1181-1197. [DOI] [PubMed] [Google Scholar]
- Borde, V., Lin, W., Novikov, E., Petrini, J.H., Lichten, M., and Nicolas, A. 2004. Association of Mre11p with double-strand break sites during yeast meiosis. Mol. Cell 13 389-401. [DOI] [PubMed] [Google Scholar]
- Carpenter, A.T.C. 1988. Thoughts on recombination nodules, meotic recombination, and chiasmata. In Genetic recombination (eds. E.R. Kucherlapati and G.R. Smith), pp. 529-548. American Society for Microbiology, Washington, DC.
- Colombo, P.C. and Jones, G.H. 1997. Chiasma interference is blind to centromeres. Heredity 79 214-227. [DOI] [PubMed] [Google Scholar]
- Copenhaver, G.P., Browne, W.E., and Preuss, D. 1998. Assaying genome-wide recombination and centromere functions with Arabidopsis tetrads. Proc. Natl. Acad. Sci. 95 247-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver, G.P., Housworth, E.A., and Stahl, F.W. 2002. Crossover interference in Arabidopsis. Genetics 160 1631-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy, B. 2003. Distribution of meiotic recombination sites. Trends Genet. 19 514-522. [DOI] [PubMed] [Google Scholar]
- Dooner, H.K. and Martinez-Ferez, I.M. 1997. Recombination occurs uniformly within the bronze gene, a meiotic recombination hotspot in the maize genome. Plant Cell 9 1633-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch, E. 2005. Estimation of gametic frequencies from F2 populations using the EM algorithm and its application in the analysis of crossover interference in rice. Theor. Appl. Genet. 111 100-109. [DOI] [PubMed] [Google Scholar]
- Fransz, P.F., Armstrong, S., de Jong, J.H., Parnell, L.D., van Drunen, C., Dean, C., Zabel, P., Bisseling, T., and Jones, G.H. 2000. Integrated cytogenetic map of chromosome arm 4S of A. thaliana: Structural organization of heterochromatic knob and centromere region. Cell 100 367-376. [DOI] [PubMed] [Google Scholar]
- Fullerton, S.M., Carvalho, A.B., and Clark, A.G. 2001. Local rates of recombination are positively correlated with GC content in the human genome. Mol. Biol. Evol. 18 1139-1142. [DOI] [PubMed] [Google Scholar]
- Galtier, N., Piganeau, G., Mouchiroud, D., and Duret, L. 2001. GC-content evolution in mammalian genomes: The biased gene conversion hypothesis. Genetics 159 907-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton, J.L., DeRisi, J., Shroff, R., Lichten, M., Brown, P.O., and Petes, T.D. 2000. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 97 11383-11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gut, I.G. 2001. Automation in genotyping of single nucleotide polymorphisms. Hum. Mutat. 17 475-492. [DOI] [PubMed] [Google Scholar]
- Haberer, G., Fischer, T.C., and Torres-Ruiz. R.A. 1996. Mapping of the nucleolus organizer region on chromosome 4 in Arabidopsis thaliana. Mol. Gen. Genet. 250 123-128. [DOI] [PubMed] [Google Scholar]
- Higgins, J.D., Armstrong, S.J., Franklin, F.C., and Jones, G.H. 2004. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: Evidence for two classes of recombination in Arabidopsis. Genes & Dev. 18 2557-2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth, N.M. and Brill, S.J. 2004. The Mus81 solution to resolution: Generating meiotic crossovers without Holliday junctions. Genes & Dev. 18 117-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housworth, E.A. and Stahl, F.W. 2003. Crossover interference in humans. Am. J. Hum. Genet. 73 188-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst, L.D., Brunton, C.F., and Smith, N.G. 1999. Small introns tend to occur in GC-rich regions in some but not all vertebrates. Trends Genet. 15 437-439. [DOI] [PubMed] [Google Scholar]
- Inukai, T., Sako, A., Hirano, H.Y., and Sano, Y. 2000. Analysis of intragenic recombination at wx in rice: Correlation between the molecular and genetic maps within the locus. Genome 43 589-596. [DOI] [PubMed] [Google Scholar]
- Jander, G., Norris, S.R., Rounsley, S.D., Bush, D.F., Levin, I.M., and Last, R.L. 2002. Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129 440-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys, A.J., Neumann, R., Panayi, M., Myers, S., and Donnelly, P. 2005. Human recombination hot spots hidden in regions of strong marker association. Nat. Genet. 37 601-606. [DOI] [PubMed] [Google Scholar]
- Jensen-Seaman, M.I., Furey, T.S., Payseur, B.A., Lu, Y., Roskin, K.M., Chen, C.F., Thomas, M.A., Haussler, D., and Jacob, H.J. 2004. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 14 528-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, G.H. 1984. The control of chiasma distribution. Symp. Soc. Exp. Biol. 38 293-320. [PubMed] [Google Scholar]
- Jones, G.H. 1987. Chiasmata. In Meiosis (ed. P.B. Moens), pp. 213-244. Academic Press, London.
- Kaback, D.B., Barber, D., Mahon, J., Lamb, J., and You, J. 1999. Chromosome size-dependent control of meiotic reciprocal recombination in Saccharomyces cerevisiae: The role of crossover interference. Genetics 152 1475-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi, L., Jeffreys, A.J., and Keeney, S. 2004. Where the crossovers are: Recombination distributions in mammals. Nat. Rev. Genet. 5 413-424. [DOI] [PubMed] [Google Scholar]
- Keeney, S. 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52 1-53. [DOI] [PubMed] [Google Scholar]
- Kong, A., Gudbjartsson, D.F., Sainz, J., Jonsdottir, G.M., Gudjonsson, S.A., Richardsson, B., Sigurdardottir, S., Barnard, J., Hallbeck, B., Masson, G., et al. 2002. A high-resolution recombination map of the human genome. Nat. Genet. 31 241-247. [DOI] [PubMed] [Google Scholar]
- Kwok, P.Y. 2001. Methods for genotyping single nucleotide polymorphisms. Annu. Rev. Genomics Hum. Genet. 2 235-258. [DOI] [PubMed] [Google Scholar]
- Lam, S., Horn, S.R., Radford, S.J., Housworth, E.A., Stahl, F.W., and Copenhaver, G.P. 2005. Crossover interference on NOR-bearing chromosomes in Arabidopsis. Genetics 170 807-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand, T. and Dutheil, J. 2005. Recombination difference between sexes: A role for haploid selection. PLoS Biol. 3 e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman, Z., Gendrel, A.V., Black, M., Vaughn, M.W., Dedhia, N., McCombie, W.R., Lavine, K., Mittal, V., May, B., Kasschau, K.D., et al. 2004. Role of transposable elements in heterochromatin and epigenetic control. Nature 430 471-476. [DOI] [PubMed] [Google Scholar]
- Lister, C. and Dean, C. 1993. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 4 745-750. [DOI] [PubMed] [Google Scholar]
- Liu, Y.G., Mitsukawa, N., Lister, C., Dean, C., and Whittier, R.F. 1996. Isolation and mapping of a new set of 129 RFLP markers in Arabidopsis thaliana using recombinant inbred lines. Plant J. 10 733-736. [DOI] [PubMed] [Google Scholar]
- Loudet, O., Chaillou, S., Camilleri, C., Bouchez, D., and Daniel-Vedele, F. 2002. Bay-0 x Shahdara recombinant inbred line population: A powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor. Appl. Genet. 104 1173-1184. [DOI] [PubMed] [Google Scholar]
- Lynn, A., Kashuk, C., Petersen, M.B., Bailey, J.A., Cox, D.R., Antonarakis, S.E., and Chakravarti, A. 2000. Patterns of meiotic recombination on the long arm of human chromosome 21. Genome Res 10 1319-1332. [DOI] [PubMed] [Google Scholar]
- Lynn, A., Koehler, K.E., Judis, L., Chan, E.R., Cherry, J.P., Schwartz, S., Seftel, A., Hunt, P.A., and Hassold, T.J. 2002. Covariation of synaptonemal complex length and mammalian meiotic exchange rates. Science 296 2222-2225. [DOI] [PubMed] [Google Scholar]
- Lynn, A., Ashley, T., and Hassold, T. 2004. Variation in human meiotic recombination. Annu. Rev. Genomics Hum. Genet. 5 317-349. [DOI] [PubMed] [Google Scholar]
- Marais, G., Mouchiroud, D., and Duret, L. 2001. Does recombination improve selection on codon usage? Lessons from nematode and fly complete genomes. Proc. Natl. Acad. Sci. 98 5688-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais, G., Charlesworth, B., and Wright, S.I. 2004. Recombination and base composition: The case of the highly self-fertilizing plant Arabidopsis thaliana. Genome Biol. 5 R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, K. Schuller, C., Wambutt, R., Murphy, G., Volckaert, G., Pohl, T., Dusterhoft, A., Stiekema, W., Entian, K.D., Terryn, N., et al. 1999. Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. Nature 402 769-777. [DOI] [PubMed] [Google Scholar]
- Meinke, D.W., Cherry, J.M., Dean, C., Rounsley, S.D., and Koornneef, M. 1998. Arabidopsis thaliana: A model plant for genome analysis. Science 282 662, 679-682. [DOI] [PubMed] [Google Scholar]
- Mercier, R., Jolivet, S., Vezon, D., Huppe, E., Chelysheva, L., Giovanni, M., Nogue, F., Doutriaux, M.P., Horlow, C., Grelon, M., et al. 2005. Two meiotic crossover classes cohabit in Arabidopsis: One is dependent on MER3, whereas the other one is not. Curr. Biol. 15 692-701. [DOI] [PubMed] [Google Scholar]
- Meunier, J. and Duret, L. 2004. Recombination drives the evolution of GC-content in the human genome. Mol. Biol. Evol. 21 984-990. [DOI] [PubMed] [Google Scholar]
- Montoya-Burgos, J.I., Boursot, P., and Galtier, N. 2003. Recombination explains isochores in mammalian genomes. Trends Genet. 19 128-130. [DOI] [PubMed] [Google Scholar]
- Nachman, M.W. 2002. Variation in recombination rate across the genome: Evidence and implications. Curr. Opin. Genet. Dev. 12 657-663. [DOI] [PubMed] [Google Scholar]
- Okagaki, R.J. and Weil, C.F. 1997. Analysis of recombination sites within the maize waxy locus. Genetics 147 815-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes, T.D. and Merker, J.D. 2002. Context dependence of meiotic recombination hotspots in yeast: The relationship between recombination activity of a reporter construct and base composition. Genetics 162 2049-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesneville, H., Bergman, C.M., Andrieu, O., Autard, D., Nouaud, D., Ashburner, M., and Anxolabehere, D. 2005. Combined evidence annotation of transposable elements in genome sequences. PLoS Comput. Biol. 1 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski, A. and Morgante, M. 2004. Corn and humans: Recombination and linkage disequilibrium in two genomes of similar size. Trends Genet. 20 103-111. [DOI] [PubMed] [Google Scholar]
- Sauer, S., Lechner, D., Berlin, K., Lehrach, H., Escary, J.L., Fox, N., Gut, I.G. 2000. A novel procedure for efficient genotyping of single nucleotide polymorphisms. Nucleic Acids Res. 28 E13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Moran, E., Armstrong, S.J., Santos, J.L., Franklin, F.C., and Jones, G.H. 2002. Variation in chiasma frequency among eight accessions of Arabidopsis thaliana. Genetics 162 1415-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, R., West, J., Love, K., Lenehan, Z., Lister, C., Thompson, H., Bouchez, D., and Dean, C. 1995. Physical map and organization of Arabidopsis thaliana chromosome 4. Science 270 480-483. [DOI] [PubMed] [Google Scholar]
- Schnable, P.S., Hsia, A.P., and Nikolau, B.J. 1998. Genetic recombination in plants. Curr. Opin. Plant Biol. 1 123-129. [DOI] [PubMed] [Google Scholar]
- Stahl, F.W., Foss, H.M., Young, L.S., Borts, R.H., Abdullah, M.F., and Copenhaver, G.P. 2004. Does crossover interference count in Saccharomyces cerevisiae? Genetics 168 35-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano-Shimizu, T. 2001. Local changes in GC/AT substitution biases and in crossover frequencies on Drosophila chromosomes. Mol. Biol. Evol. 18 606-619. [DOI] [PubMed] [Google Scholar]
- Wright, S.I., Agrawal, N., and Bureau, T.E. 2003. Effects of recombination rate and gene density on transposable element distributions in Arabidopsis thaliana. Genome Res. 13 1897-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, H., Zhou, Q., Li, J., Smith, H., Yandeau, M., Nikolau, B.J., and Schnable, P.S. 2002. Molecular characterization of meiotic recombination across the 140-kb multigenic a1-sh2 interval of maize. Proc. Natl. Acad. Sci. 16 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, A., Zhao, C.F., Fan, Y., Jang, W.H., Mungall, A.J., Deloukas, P., Olsen, A., Doggett, N.A., Ghebranious, N., Broman, K.W., et al. 2001. Comparison of human genetic and sequence-based physical maps. Nature 409 951-953. [DOI] [PubMed] [Google Scholar]
- Zickler, D. and Kleckner, N. 1999. Meiotic chromosomes: Integrating structure and function. Annu. Rev. Genet. 33 603-754. [DOI] [PubMed] [Google Scholar]
Web site references
- http://www.arabidopsis.org/; The Arabidopsis information resource.
- http://ftp.tigr.org/pub/data/a_thaliana/ath1/; Arabidopsis sequence database.
- http://cran.r-project.org; R software.