Abstract
A major goal in genomics is to understand how genes are regulated in different tissues, stages of development, diseases, and species. Mapping DNase I hypersensitive (HS) sites within nuclear chromatin is a powerful and well-established method of identifying many different types of regulatory elements, but in the past it has been limited to analysis of single loci. We have recently described a protocol to generate a genome-wide library of DNase HS sites. Here, we report high-throughput analysis, using massively parallel signature sequencing (MPSS), of 230,000 tags from a DNase library generated from quiescent human CD4+ T cells. Of the tags that uniquely map to the genome, we identified 14,190 clusters of sequences that group within close proximity to each other. By using a real-time PCR strategy, we determined that the majority of these clusters represent valid DNase HS sites. Approximately 80% of these DNase HS sites uniquely map within one or more annotated regions of the genome believed to contain regulatory elements, including regions 2 kb upstream of genes, CpG islands, and highly conserved sequences. Most DNase HS sites identified in CD4+ T cells are also HS in CD8+ T cells, B cells, hepatocytes, human umbilical vein endothelial cells (HUVECs), and HeLa cells. However, ∼10% of the DNase HS sites are lymphocyte specific, indicating that this procedure can identify gene regulatory elements that control cell type specificity. This strategy, which can be applied to any cell line or tissue, will enable a better understanding of how chromatin structure dictates cell function and fate.
Now that the genomes of many species have been sequenced, a major focus of genomics is to identify all gene regulatory elements within the noncoding DNA (Collins et al. 2003). This will be necessary to understand how gene expression is controlled in different cell types, stages of development, diseases, and species. A number of genome-wide technologies have been developed to identify the location of gene regulatory elements, such as sequence conservation, chromatin immunoprecipitation followed by microarray hybridization (ChIP-chip), and computational analyses. Since each method has its own sets of advantages and disadvantages, a combination of different techniques will likely be needed to successfully identify all gene regulatory elements. In addition, new methods will likely be needed to answer different questions not addressed by current technologies.
Historically, the mapping of DNase hypersensitive (HS) sites has been used to identify the location of regulatory regions, including enhancers, silencers, promoters, insulators, and locus control regions (Wu et al. 1979; Gross and Garrard 1988). Over the past 25 years since this method was developed, hundreds of DNase HS sites associated with specific loci have been described in the literature. Unfortunately, obtaining this information from the standard Southern blot approach is a tricky, time-consuming, and inaccurate task. While current estimates are that there are 25,000 human genes, researchers can only guess how many regulatory regions there are for every gene, as well as in which tissue they operate. Therefore, not only does the mapping of DNase HS sites across the genome in different cell types need to be scaled up, but these data also need to be made publicly available in a format that researchers can readily use and compare with other experimental data types.
Recently, we and others have described novel technologies to clone and sequence libraries of DNase HS sequences, which can be used to identify all active gene regulatory elements from a single cell type (Crawford et al. 2004; Sabo et al. 2004). In a pilot experiment (Crawford et al. 2004), we used conventional capillary sequencing to analyze clones from a DNase library and found that they are enriched for regions of the genome known to contain regulatory elements (e.g., CpG islands, regions immediately upstream of genes). However, despite extraordinary efforts to minimize random shearing of high-molecular-weight DNA, as well as reduce nonspecific DNase digestion, the background of nonspecific sequence tags was estimated to be ∼70%. To distinguish signal from noise, we determined that sequences from the DNase library that mapped within close proximity to each other are highly accurate at identifying valid DNase HS sites. Analysis of these “DNase clusters” allowed us to estimate that there are ∼100,000 DNase HS sites within nonactivated CD4+ T cells (Crawford et al. 2004).
To scale up and identify all valid DNase HS sites from background by using clustering analysis, we needed to significantly increase the sequencing throughput. Since only 20 bp of sequence is needed to uniquely map the position of most sequences within the genome, we determined that traditional sequencing methods that produce long sequence reads were poorly suited for this project, and methods capable of generating large numbers of short sequence tags would be advantageous.
Here, we describe high-throughput sequencing of a genomic CD4+ T-cell DNase library utilizing massively parallel signature sequencing (MPSS), which has been primarily used for sequencing EST tags from cDNA expression libraries (Brenner et al. 2000). Of the sequences that uniquely map to the genome, we identified 14,190 DNase clusters that correlate highly with valid DNase HS sites. While most of these sites were also HS in other cell types, ∼10% of these DNase HS sites are only present in lymphocytes, showing that we have identified both ubiquitous and cell type-specific gene regulatory elements.
Results
Sequence and DNase cluster analysis
A library of DNase HS sequences generated from CD4+ T cells from a human male donor was sequenced by using MPSS. Over 230,000 sequence tags (20 bases in length) were generated from five MPSS runs, of which 162,337 mapped to a single position in the human genome. The coordinates for the sequences that map to unique sites in the genome are publicly available (http://research.nhgri.nih.gov/DNaseHS/).
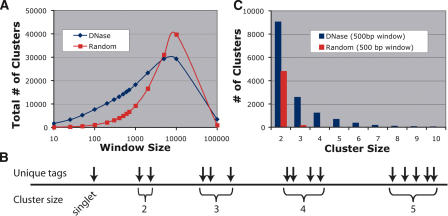
We expected sequence tags that map in close proximity to other tags would represent true DNase HS sites, while tags that map in isolation would not. We defined DNase clusters as sequence tags that map within a certain number of base pairs (window size) to one another. DNase clusters occur more frequently in this DNase library than in in silico libraries generated from random regions of the genome (Fig. 1A). The largest difference in the number of clusters between the DNase and random libraries occurred at a 500-bp window. Figure 1B shows the representative cluster sizes (the number of sequences from the DNase library that mapped within each cluster). Compared with the random libraries, the DNase library has twice as many cluster sizes of two, and significantly more cluster sizes of three and greater for the 500-bp window size (Fig. 1C). Only clusters identified with a 500-bp window size were used for subsequent analyses.
Figure 1.
Clustering analysis of sequences from DNase and random libraries. (A) The total number of clusters was determined for 15 different window sizes. For window sizes <5000 bp, there are larger numbers of clusters for the DNase library. For very large window sizes (100,000 bp), the majority of the DNase and random libraries cluster within only a few regions of the genome. (B) Example of how unique tags cluster into different sizes. The greatest spread between DNase and random libraries occurs at a 500-bp window. (C) Within this optimal window size, there are twice as many DNase clusters of two compared with random. Clusters of three or more are rarely found in random libraries.
Validation of DNase HS sites by real-time PCR
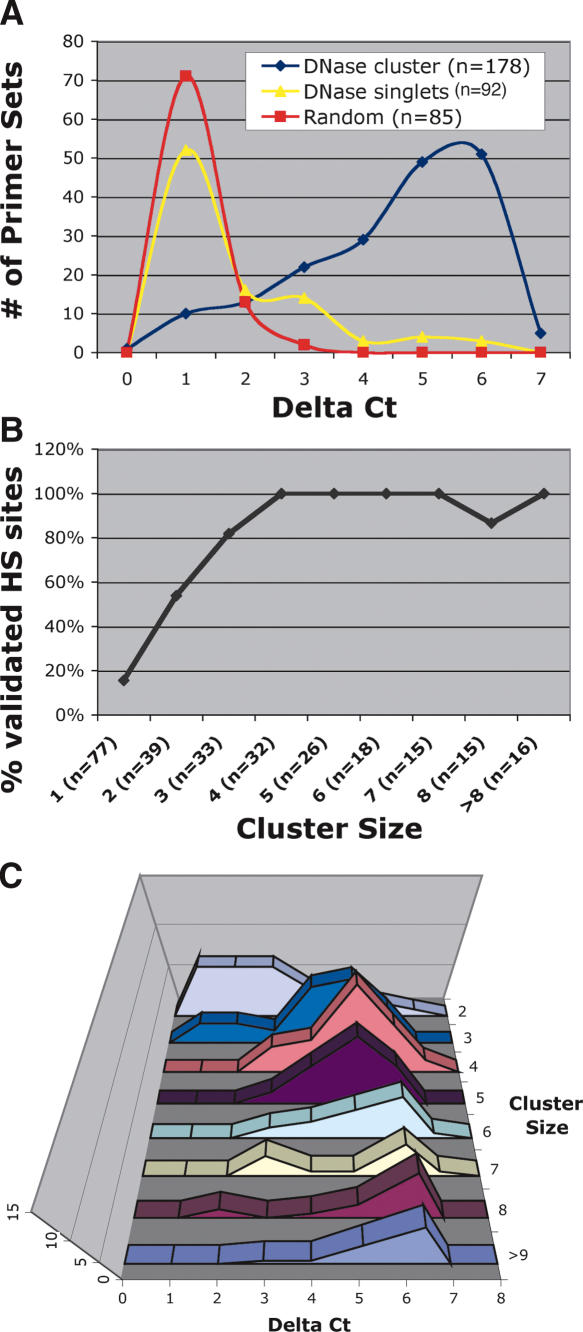
Real-time PCR (McArthur et al. 2001) was used to test whether sequence tags from the DNase library represented valid DNase HS sites (Fig. 2A). Delta (Δ) Ct (the difference in threshold cycles) values display the relative amount of DNase sensitivity between primer sets by comparing amplification from DNase digested and nondigested DNA. Higher Δ Ct values mark regions that are more sensitive to DNase digestion than are regions with lower Δ Ct values. To determine the precision by which clustering predicts valid DNase HS sites, primer sets were designed to flank both DNase singlets and DNase clusters. The overall background level of DNase digestion was determined by using primer sets that flanked randomly chosen, but unique, regions of the genome.
Figure 2.
Validation of DNase clusters by real-time PCR. Delta (Δ) Ct values represent the number of additional cycles to achieve threshold amplification from nuclear DNA treated with DNase I compared with nuclear DNA not treated with DNase I. (A) Most primer sets that flank random regions of the genome display Δ Ct values less than two. Approximately 20% of primer sets that flank sequences from the DNase library that do not cluster with other sequences display Δ Ct values greater than two. Eighty percent of primer sets that flank DNase clusters of two or more display Δ Ct values greater than two. (B) The percentage of primer sets that have Δ Ct values greater than two was determined for each cluster size (% validated sites). Clusters of two were ∼50% accurate at identifying valid HS sites, while clusters of three or more were highly accurate at identifying valid HS sites. (C) The distribution of Δ Ct values was determined for different cluster sizes. Note that the highest cluster sizes have the highest Δ Ct values.
Since 95% of the primer sets surrounding random regions of the genome displayed Δ Ct values less than two, we defined this value to be our threshold for hypersensitivity. Approximately 16% of the DNase singlets have a Δ Ct value greater than two. However, 85% of DNase cluster sizes of two or more display Δ Ct values of greater than two, indicating that DNase clusters accurately identify valid DNase HS sites from background. Primer sets designed outside of the immediate HS region from 14 DNase clusters displayed marked reduction in sensitivity to DNase, indicating that DNase clusters are enriched for the most HS regions of the genome (data not shown).
To determine the minimum cluster size required to identify a valid HS site, each primer set that flanks a DNase cluster was separated into different cluster sizes (Fig. 2B). With a threshold for hypersensitivity set at a Δ Ct value of two, ∼50% of cluster sizes of two represented valid HS sites, 80% of cluster sizes of three represented valid HS sites, and cluster sizes of four or more represented valid HS sites ∼100% of the time. The small dip in clusters of eight likely represents two regions where the PCR primers did not directly flank the HS region. Not all DNase clusters are equally HS to DNase digestion (Fig. 2C). The largest cluster sizes show mean Δ Ct values around six, whereas the smaller clusters tend to have a lower mean. There is an intriguing suggestion of bimodality in the data (most prominent for the cluster of seven). The peaks of this bimodal distribution differ by approximately three cycles, corresponding to a 23, eightfold, difference in hypersensitivity.
Predicting the total number of DNase HS sites throughout the genome
To determine the total number of DNase HS sites within the genome of CD4+ T cells, the corrected number of clusters based on the real-time PCR validation results was determined (Table 1). Looking at the distribution of true sites that are represented by singletons and by clusters of two through nine, we observed that this distribution deviates markedly from a Poisson distribution (χ2 > 15,000). This strongly suggests that not all HS sites are equally accessible to DNase digestion.
Table 1.
Statistical modeling of DNase HS data to determine best fit
|
No. of DNase clusters
|
Best
|
||||
|---|---|---|---|---|---|
| Cluster size | % valida | Normalized (observed)b | 1 statec | 2 stated | |
| Singlet | 123,000 | 16% | 19,680 | 13,380 | 19,587 |
| 2 | 9039 | 54% | 4881 | 11,708 | 5161 |
| 3 | 2565 | 80% | 2078 | 6829 | 1863 |
| 4 | 1216 | 100% | 1216 | 2987 | 1160 |
| 5 | 674 | 100% | 674 | 1045 | 753 |
| 6 | 348 | 100% | 348 | 305 | 424 |
| 7 | 165 | 100% | 165 | 76 | 205 |
| 8 | 90 | 100% | 90 | 16 | 87 |
| >9 | 118 | 100% | 118 | 3 | 49 |
| Χ2 | 15,115 | 170 | |||
Percentage validated values were derived from real-time PCR results for each cluster size
Normalized (observed) values are the number of DNase clusters multiplied by percentage valid
Best 1 state represents a single Poisson distribution that best fits observed data
Best 2 state represents two classes of Poisson distributions that best fit observed data. One class assumes 8% of the DNase HS sites are eight times more hypersensitive to DNase digestion
By considering also the real-time PCR data in Figure 2C, we propose a model in which there are two classes of sites: one with hypersensitivity 1×, and the other 8×. By fitting the data in Table 1 to just one variable (the proportion in the more HS class), we get a dramatically improved fit to the observed data (χ2 = 170). This model suggests that there are ∼74,000 true DNase HS sites in CD4+ T cells, but that ∼5900 (8%) of these are considerably more sensitive than are the rest. The model further suggests that the majority of the most sensitive sites have already been identified in clusters of two or more. But to identify 95% of all true DNase HS sites across the genome with a cluster of two or more, ∼1.75 million sequence reads would be needed. To identify these sites with a cluster of three or more would require ∼2.3 million sequence reads.
Location of validated DNase HS sites within the annotated genome
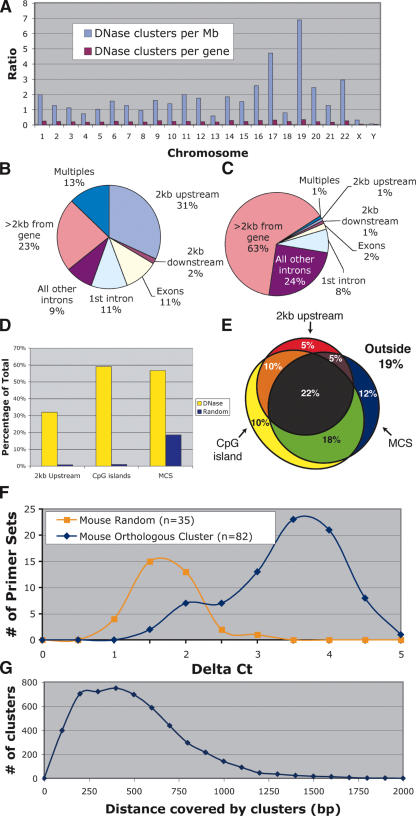
The location of the 5159 DNase HS sites that represented cluster sizes of three or more were mapped relative to chromosomes, genes, CpG islands, and highly conserved sequences. A higher proportional number of DNase HS sites were identified on chromosomes 17 and 19, which are known to be particularly gene rich. When DNase HS sites were normalized for the number of genes on each chromosome, a relatively equal number of DNase HS sites per autosome were detected (Fig. 3A). The X and Y chromosomes, however, displayed an unusually low number of DNase HS sites relative to genes, even after compensating for the presence of only one of each sex chromosome within the male genome. This indicates that the sex chromosomes in T cells may contain fewer active regulatory elements.
Figure 3.
Location of DNase clusters of three or more relative to the annotated genome. (A) DNase clusters were mapped to each chromosome, and the density of sites per Mb was determined (blue bars). DNase clusters are significantly overrepresented on chromosomes 17 and 19, which are known to be especially gene rich. No differences were detected when the density of DNase clusters per gene was determined for each chromosome (red bars). (B) The location of DNase clusters relative to genes was determined. Multiples represent DNase clusters that were <2 kb from more than one gene. (C) For comparison, a library of randomly chosen coordinates was also mapped relative to genes. (D) The percentage of DNase and random sites that map to annotated regions of the genome often used to search for gene regulatory elements; regions <2 kb upstream of genes, within CpG islands, and within multispecies conserved sequences (MCS). (E) A Venn diagram shows the percentage of DNase clusters that map within one or more annotated regions of the genome (each region is represented by a circle or oval). “Outside” represents the percentage of DNase HS sites that do not map to any of the three categories. (F) Most human DNase HS sites are also hypersensitive at orthologous regions (mouse cluster) in mouse. These regions displayed higher Δ Ct values than do randomly selected controls. (G) Size of DNase HS sites (in base pairs) was calculated by subtracting the start and stop positions of each DNase cluster.
The locations of DNase HS sites were mapped relative to genes. We found that 31% of DNase HS sites map to regions within 2 kb upstream of genes, while only 2% map within 2 kb downstream of genes (Fig. 3B). Of the 31% of DNase HS sites that map within the transcribed regions of genes, about one-third map to the first intron. Interestingly, 23% of DNase HS sites map >2 kb from any gene, indicating these mark the location of long distance regulatory elements or the presence of previously unknown transcripts. This is different than a random distribution (Fig. 3C). About 60% of DNase HS sites map within CpG islands, which are regions that often contain promoters of housekeeping genes (Fig. 3D). In addition, 55% map nearby to multispecies conserved sequences (MCSs) (Fig. 3D). These percentages are significantly higher than those in the random data set.
Since many of these annotated regions overlap, these findings are not completely independent. For example, many CpG islands map to regions upstream of genes, and these promoter regions are often conserved between multiple species. Thus, we analyzed the amount of overlap between regions of the genome that are within 2 kb upstream of a gene, within a CpG island, and within sequences that are highly conserved (Fig. 3E). Approximately 80% of DNase HS sites map to one or more of these annotated regions. Only 18% of random control sequences map to one or more of these regions (data not shown).
To test whether human DNase HS sites are functionally conserved in mouse, we designed primer sets to flank orthologous regions in the mouse genome. Real-time PCR was performed on mouse CD4+ T-cell DNA that was treated with and without DNase. To determine the background level of DNase hypersensitivity, primer sets were designed around random regions of the mouse genome. Of the mouse regions that are orthologous to human DNase HS sites, most were HS in mouse compared with random controls (Fig. 3F). The small bump at a Δ Ct value of two may represent orthologous regions of chromatin that are not HS in mouse, since four of these regions have high Δ Ct (>2.5) values in human.
To determine the approximate size of each DNase HS site, the distance between the start and stop coordinates of each cluster of three or more was calculated (Fig. 3G). On average, most DNase HS sites spanned ∼100-1000 bp.
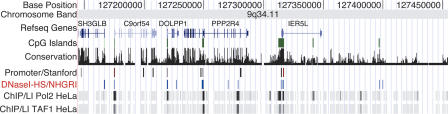
We are participating in the ENCODE (Encyclopedia of DNA Elements) Consortium, a group whose goal is to identify all gene regulatory elements throughout the genome (The Encode Consortium 2004). A carefully selected 1% of the genome is being analyzed with different methods, including sequence conservation, ChIP-chip, promoter/enhancer identification, origins of replication, and others. These data are being made publicly available on the UCSC (University of California, Santa Cruz) genome browser (http://genome.ucsc.edu/ENCODE/). For the data that have already been deposited, there is a high degree of correlation between the DNase HS sites reported here and other data types, indicating that these different experimental methods are identifying similar functional regions of the genome (Fig. 4).
Figure 4.
An example of multiple genome-wide technologies used to identify gene regulatory elements. This is a screen shot from the UCSC genome browser ENCODE region Enr232 (chr9: 127,144,681-127,454,484). Shown in the DNase I-HS/NHGRI track are the locations of DNase clusters of two or more, as well as other data tracks. Names and location of exons and introns are indicated in RefSeq Gene track. The conservation track measures the degree of sequence conservation among human, chimp, mouse, rat, and chicken. The Promoter/Stanford track displays relative activity of predicted promoters in luciferase reporter assays (Trinklein et al. 2003). ChIP/L1 displays ChIP-chip data for DNA Polymerase II (Pol2) and transcription initiation factor TFIID subunit 1 (TAF1) from HeLa cells, as determined by the University of California at San Diego (Kim et al. 2005). Note the overlap of many experimental data types.
Expression analysis of genes near DNase HS sites
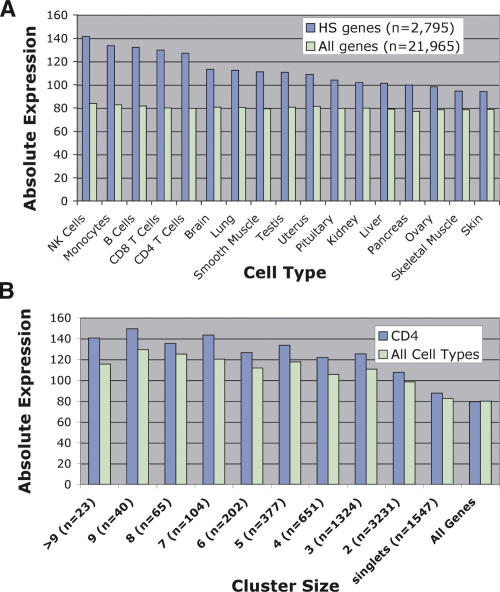
To determine if DNase HS sites are associated with elevated levels of nearby gene expression, we determined the average expression value of genes that had a nearby DNase cluster of three or more. This data set included genes with DNase clusters located either within the gene or within 2 kb upstream or downstream. The average expression value from this set of 2795 genes was determined from multiple human tissues and was compared to average gene expression values from all genes (Fig. 5A). Compared with the expression value of all genes, those genes with a nearby DNase HS site have higher average expression values in all cell types. In addition, genes with nearby DNase HS sites have the highest average expression values in peripheral blood cell types, including CD4+ T cells. This suggests not only that DNase HS sites are present near genes that are highly expressed but also that the locations of these sites mark genes with the highest average expression within the cell type that the DNase library was derived from.
Figure 5.
DNase HS sites identify genes that have higher levels of expression using microarray analyses. (A) Average expression values of genes that had a DNase HS site nearby were compared to average expression value of all genes. Genes that had a DNase HS site nearby had higher levels of gene expression in all primary tissues. In addition, the highest levels of gene expression were from peripheral blood cell types, including natural killer (NK), monocytes, B cells, and CD8+ and CD4+ T cells. (B) Average expression values of genes that are associated with different cluster sizes were determined from CD4+ T cells as well as the averaged gene expression from all primary tissues (all cell types). “All genes” represents the average expression value of all genes on the Affymetrix U133A expression array.
Since we had identified DNase clusters of all sizes, we next wanted to determine if larger cluster sizes were associated with higher average gene expression than were smaller cluster sizes. When we compared expression of genes nearby clusters of three or more, we detected a similar increase in average gene expression for each of these cluster sizes. A smaller increase in average gene expression was detected for genes associated with clusters of two, which is likely attributed to the increased false-positive rate within this cluster size. Genes associated with DNase singlets, which had the highest false-positive rate, had only a minor increase in average gene expression, and all genes showed no difference in gene expression (Fig. 5B).
Analysis of DNAse HS sites within multiple cell types
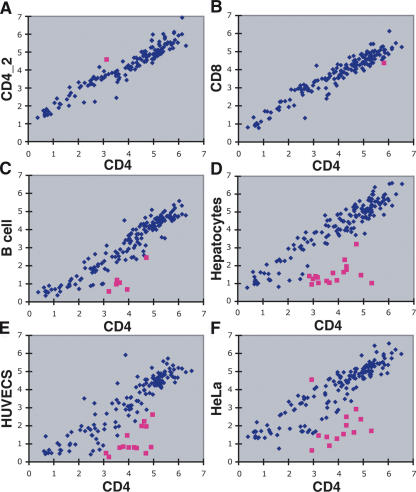
We were interested in determining the number of DNase HS sites that were CD4+ T-cell specific, and whether the presence or absence of DNase HS sites in different cell types correlated with changes in nearby gene expression. Real-time PCR, using 180 primer sets that flank CD4+ T-cell DNase sequences (representing both singlets and clusters), was used to determine relative DNase hypersensitivity between different cell types. To show this procedure produces highly reproducible data, real-time PCR was performed on DNase-treated DNA from two independent CD4+ T-cell preparations (Fig. 6A). To identify DNase HS sites that were CD4+ T-cell specific, real-time PCR was performed on DNase-treated DNA from a number of different human cell types, including CD8+ T cells, B cells, hepatocytes, human umbilical vein endothelial cells (HUVECs), and HeLa cells (Fig. 6B-F). No significant differences were detected between CD4+ and CD8+ T cells (except for the rare outlier), which was expected due to their similarity. Six CD4+ HS sites were found not to be HS in B cells, while a larger number were found not to be HS in hepatocytes (17), HUVECs (14), and HeLa (12) cells. The presence or absence of these outlier DNase HS sites was variable in non-T-cell types (Table 2).
Figure 6.
Cell type specificity of CD4+ T cell-specific DNase HS sites using real-time PCR. Red squares identify “outlier” DNase clusters that were hypersensitive in CD4+ T cells, but not hypersensitive in other cell types. Δ Ct values (both x- and y-axes) mark the relative hypersensitivity of each DNase cluster for each cell type. (A) Two independent CD4+ T cell preparations display that this method is highly reproducible. The single outlier represents rare data points that are >3 SD from the mean. (B) Only one outlier was detected between CD4+ and CD8+ T cells. (C) Five DNase clusters were identified as not hypersensitive in B cells. (D,E,F) Additional DNase clusters were identified as not hypersensitive in hepatocytes, human umbilical vein endothelial cells (HUVEC), and HeLa cells.
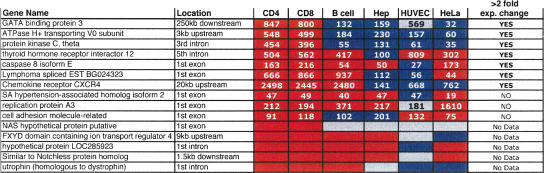
Table 2.
Comparisons of DNase clusters that vary in hypersensitivity in different cell types
The location of each DNase cluster relative to the nearest gene is displayed. Red boxes identify regions that are hypersensitive (Δ Ct > 2) and are not statistical outliers. Blue boxes identify regions that are not hypersensitive (Δ Ct > 2) and are statistical outliers. Gray boxes identify regions that are hypersensitive (Δ Ct > 2), but are statistical outliers. The relative expression values of each gene, as determined by Affymetrix U133A expression arrays, are displayed within each box (Su et al. 2002). Genes that display a greater than twofold decrease in gene expression, when a DNase HS site is not present in non-T cells, are indicated at the right. “No Data” represent genes that were not analyzed on the Affymetrix U133A microarra.
To determine if the presence or absence of each outlier DNase HS site is associated with changes in nearby gene expression, expression values of the closest genes were compared from different tissues. When a DNase HS site was absent in non-T-cell types (outlier), most of the nearest genes displayed a more than twofold decrease in gene expression (Table 2). When a HS site was present in all cell types (nonoutlier; n = 145), only 10% showed a more than twofold difference in gene expression (data not shown).
Luciferase reporter assays
To characterize whether DNase HS sites displayed enhancer activity, we cloned 77 regions that flanked DNase clusters of three or more downstream of a luciferase reporter gene. This reporter construct contained a SV40 promoter. In addition, 40 randomly selected regions were also cloned downstream of the luciferase gene. Each clone was cotransfected with a Renilla luciferase control plasmid into Jurkat and HeLa cell lines, and firefly luciferase-to-Renilla luciferase ratios were determined. In Jurkat and HeLa cells, a positive control plasmid that contained a SV40 enhancer displayed 1.5- and 40-fold higher levels of basal level transcription, respectively. We were unable to detect enhancer activity from any of the DNase HS or random clones when transfected into HeLa cells (data not shown). However, when transfected into Jurkat cells, we identified one DNase HS site that increased basal level of transcription by sixfold. This region, which was 20 kb upstream of the CXCR4 gene, was only HS in lymphocytes (Table 1).
Discussion
Encouraged by an initial low-throughput pilot study to identify DNase HS sites (Crawford et al. 2004), we have greatly expanded this work and now report a detailed analysis of 40 times as many sequence tags from a DNAse HS library. We have found that the MPSS method is capable of generating large numbers of short sequence tags from a genomic DNase HS library, and the resulting clusters of sites provide a rich data set for understanding genome-wide gene regulation. To achieve a nearly comprehensive view of DNase HS sites from a single cell type, however, it may be necessary to obtain 2 million genomic sequence tags. However, since a larger number of sequences might increase the cluster sizes required to identify valid DNase HS sites from background, the actual number of required sequences might be substantially higher. In addition, it may also be necessary to sequence DNase libraries that are digested with different concentrations of DNase. While MPSS regularly achieves these high numbers of EST tags from expression libraries, it has not been possible thus far to do so for DNase HS sites. This likely relates to complexity issues; the DNase HS method is attempting to recover one site from several hundred kilobases of lightly digested DNA, whereas the EST application aims to tag one site per mRNA molecule, of average size 2 kb. To achieve another order of magnitude in sequence tag density, additional improvements in the MPSS method will be needed. Next-generation sequencing technology that is based on in situ amplified DNA clusters and on sequencing by synthesis may provide the necessary increase in throughput over the MPSS process. An alternative method would be to hybridize DNase-treated end-labeled genomic DNA to tiled microarrays covering the entire genome.
Our data show that the clustering approach can accurately distinguish true DNase HS sites from background noise. Clusters of three or more tags within a 500-bp window almost always prove to be valid. About 80% of such DNase HS sites map to regions of the genome that are expected to contain gene regulatory elements (regions 2 kb upstream of genes, CpG islands, or highly conserved sequences). However, 20% of all DNase HS sites fall outside of these categories, indicating that current annotation of the human genome does not yet include all regulatory elements or that DNase sites may also mark the location of non-regulatory structural elements. Our data also suggest that chromatin structure, in addition to primary sequence, has been conserved throughout evolution. Of the human DNase HS sites where an orthologous region in mouse could be identified, most of these regions were also HS in mouse.
Analyzing DNase HS sites between different cell types should identify regulatory domains that control housekeeping versus cell type-specific functions. We were surprised to find that ∼90% of DNase HS sites in CD4+ T cells are present in all cell types tested, indicating that only ∼10% of gene regulatory regions in this tissue control cell type specificity. Of this 10%, a number of sites were HS in every cell type except for one, suggesting that cells may have combinatorial control over the expression of certain genes. In general, when DNase HS sites are not universally present in all cell types, the level of expression of the nearest gene correlates with the presence of the DNase HS sites. The exceptions (e.g., BG024323 in HeLa cells) (Table 1) may represent gene regulatory elements that have open chromatin but lack the full complement of transcription factors necessary to initiate transcription. A number of these transcriptionally “poised” regions have been described for the globin genes (Gross and Garrard 1988; Hebbes et al. 1992; Schneider et al. 2004).
We postulated that many of these DNase HS sites would mark the location of enhancers. By cloning a number of these regions into a luciferase reporter vector, however, we identified only one out of 77 DNase HS sites that display enhancer-like activity. This result may indicate that most DNase HS sites have other functions (promoters, silencers, insulators) or that other cis- or trans-regulatory elements, or other epigenetic signals and large-scale chromatin architecture, are absent in our test system but are required for enhancer activity.
We believe that generating DNAse HS site libraries from primary tissues will ultimately be necessary to understand how genes are regulated in vivo. However, since many organs are composed of heterogenous cell types, it may be some time before large numbers of homogenous cell types can be teased away from these tissues. In the meantime, this protocol may need to be optimized to work with established cell lines. Since newly replicated DNA is more susceptible to DNase digestion than bulk chromatin, it may be necessary to use cell lines that are either synchronized or blocked in a certain part of the cell cycle to reduce levels of background (Hewish 1977).
How many DNase HS sites do we believe we have identified? Of the 5159 clusters of three or greater, we predict that most are true HS sites. In addition, ∼50% of the 9000 clusters of two are estimated to be true HS sites, and 16% of the remaining 123,000 singlets are estimated to be true HS sites. Therefore, we believe that we have identified ∼30,000 DNase HS sites in CD4+ T cells. While it is difficult to know which singlets are authentic, those that map to promoters, CpG islands, or highly conserved sequences are more likely to represent true HS sites.
All of the data described here are publicly available (http://research.nhgri.nih.gov/DNaseHS/), and should provide a rich resource for investigating genome function. In addition, this Web site identifies all singlets and clusters that map to promoters, CpG islands, or highly conserved elements. Another Web site developed by the ENCODE Consortium allows for comparisons of DNase HS sites to other data types, including sequence conservation, ChIP-chip, origins of replication, etc. (http://genome.ucsc.edu/ENCODE/). Together these data will provide important insights into the vast complexity of how genes, chromatin structure, regulatory signals, and transcription factors function together on a genome-wide scale.
Methods
Preparation of DNase I-treated DNA
Intact nuclei were prepared and digested with DNase I as previously described (Crawford et al. 2004). Briefly, cells were lysed with 0.1% NP40 and nuclei were collected by centrifugation. Intact nuclei were treated with different concentrations (0-12 U) of DNase I for 10 min, and reactions were stopped with 0.1 M EDTA. Digested high-molecular-weight DNA was embedded in 1.0% InCert (BioWhittaker) low-melt gel agarose. To remove protein, DNA plugs were washed with LIDS buffer (1% lauryl sulfate, 10 mM Tris-Cl, 100 mM EDTA) overnight at 37°C and were subsequently washed three times in 0.2× NDS (0.5 M EDTA at pH 8.0, 10 mM Tris base, 1% N-lauroylsarcosine sodium salt), followed by three washes in 50 mM EDTA. Optimal concentrations of DNase generated a smear of high-molecular-weight fragments (>100 kb) when analyzed by pulsed field gel electrophoresis. DNased ends were blunt ended in gel with T4 DNA Polymerase, melted at 65°C, and purified by phenol extraction and ethanol precipitation.
Massively parallel signature sequencing
Biotinylated linkers (annealed product of 5′ bio-CTGGTCGTAGCATCTTGTAGCATAGTCCGAC3′ and 3′CAGCATCGTAGAACATCGTATCAGGCTG-P) containing a MmeI restriction site (TCCGAC) at the 3′ end were attached to the blunt-ended DNased ends. Digestion with MmeI cuts 20 bp into the sequence adjacent to the DNase HS site, leaving a 2-bp overhang. After purification of DNased ends on streptavidin beads, a second set of linkers (annealed product of 5′TATCACTAAGATGCTGACGGCTGTT and 3′NNATAGTGATTCTACGACTGCCGA5′) containing a two-base degenerate overhang is ligated to the opposite end. Inserts, along with the linkers, are PCR amplified from the beads (primers: 5′ FAM-CTGGTCGTAGCATCTTGTAGCA and 3′ATAGTGATTCTACGACTGCCGA-FAM5′), and the inserts are PAGE-selected to minimize the adaptor contaminants. After digesting with SfaNI, a single nucleotide (dTTP) is added to the 3′ restriction site. The remaining steps are identical to the previously described MPSS protocol (Brenner et al. 2000). Briefly, these products are cloned into a vector that contains 107 different 32-mer oligonucleotide tags. Products are amplified, hybridized to immobilized beads that contain complementary sequences to each tag, and sequenced by using successive rounds of digestion followed by hybridization to sequence specific fluorescently tagged decoder linkers.
Alignment, clustering analysis, and random library generation
The 20-bp MPSS sequence tags were aligned to the National Center for Biotechnology Information (NCBI) human genome build 34 by using megaBLAST optimized for short sequences (-W 16 -q -20 -a 2 -FF). Only perfect matches that had a single unique alignment within the genome were used for further analysis. To identify clusters of sequence tags, the distance between each mapped tag was determined. Starting from one end of each chromosome, a sequence tag within a certain distance (window size) of another tag was marked as a cluster. If the next subsequent tag was also within the window size, it was also grouped within that cluster. The number of clusters were determined by using 15 different window sizes. Random coordinate libraries were generated as previously described (Crawford et al. 2004). Briefly, random coordinates were chosen throughout the genome, and 20 bases of adjacent sequence were extracted from each coordinate. The sequences from the random coordinates were aligned to the genome using megaBLAST, and only perfect matches that had a single unique alignment were used for further analysis.
Statistical analyses
The program for finding the best fit was written in C (available upon request). The distribution of singletons and DNase clusters that represented valid DNase HS sites was estimated from real-time PCR data. We generated expected hit distributions for comparison under models with a single mean hit rate and with two mean hit rates eightfold apart by using Poisson expectations and searching over a dense grid of possible hit rates and underlying the true number of DNase HS sites. In the case of the second model (corresponding to two DNase hypersensitivity levels), an additional parameter (proportion of sites in the more HS category) was optimized. The fit was dramatically improved by adding this one degree of freedom. By using the best fit model, we generated expected outcomes under larger numbers of sequence reads to estimate the number of sequences required to identify 95% of all DNase HS sites.
Comparison to genome annotation and gene expression data
The location of DNase clusters and random coordinates were compared to RefSeq genes and CpG islands, which were downloaded as tracks from the UCSC Genome Browser (http://genome.ucsc.edu/) (Karolchik et al. 2003). In addition, we also analyzed their location relative to MCSs, which were generated from a genome-wide multisequence alignment of human, chimpanzee, mouse, and rat (available at UCSC) by using a previously described approach (Margulies et al. 2003). The percentage of DNase clusters that contain at least one MCS within 100 bp from the center of each DNase cluster was determined. These data were compared to the percentage of randomly chosen unique regions of the genome (±100 bp) that contain a MCS.
Expression data comprising the GNF normal tissue database (on Affymetrix U133A microarrays) were obtained in raw .CEL form from relevant cell types from Novartis (Su et al. 2002). Expression data from HUVEC cells were downloaded (again in raw .CEL format) from the Gene Expression Omnibus Web site (http://www.ncbi.nlm.nih.gov/geo/). RNA from HeLa cells used in this study was purified by using RNeasy (Qiagen) and hybridized to Affymetrix U133Aplus arrays. All expression data were normalized together by using RMA (Irizarry et al. 2003) via the BioConductor project's Affymetrix package (http://www.bioconductor.org).
Isolation of cell types
Primary human CD4+ T cells, CD8+ T cells, and B cells were purified from apheresed blood samples from anonymous donors (National Institutes of Health Blood Bank, Institutional Review Board exemption issued by National Institutes of Health Office of Human Subjects) by using negative selection magnetic bead isolation kits (Miltenyi Biotec). Frozen primary human hepatocytes and HUVECs were obtained from Cambrex. Hepatocytes were thawed and placed in hepatocyte growth medium for 4 h before nuclei were prepared. HUVEC cells were grown in culture to generate the required number of cells. Mouse splenocytes were isolated from six C57Bl6/J mice, and CD4+ T cells were isolated by using a mouse magnetic bead isolation kit (Miltenyi Biotec). All lymphocyte populations were >90% pure and >99% viable, as detected by flow analysis.
Real-time PCR and identification of outlier DNase HS sites
Real-time PCR was used to verify that sequences from the DNase library represented valid DNase HS sites (McArthur et al. 2001). PCR primers were designed to flank DNase singlets, DNase clusters, or random regions of the genome. The 180 primer sets used for comparing different cell types were chosen in a nonbiased strategy around DNase singlets as well as DNase clusters of all cluster sizes. By using human/mouse orthology data available from the UCSC genome browser, sequences of orthologous mouse positions representing human DNase clusters of three or more were identified. Each primer set was designed to generate a 200-300 bp amplicon by using Primer3 (Rozen and Skaletsky 2000). For the DNase clusters, primers were designed around the center (mean) of the coordinates that comprise each cluster. DNase-treated and nondigested DNA was quantitated in triplicate by using pico-green and a fluorometer (Spectramax GeminiXS, Molecular Devices). Nine nanograms of DNase-treated and nondigested DNA was stamped onto 384 plates, and primer/SYBR green PCR mix (Qiagen) was added (Quadra 384, Tomtec). All PCR reactions were performed on a 7900 real-time PCR machine (Perkin Elmer).
To identify CD4+ T-cell HS sites that were not HS in other cell types, a statistical outlier method was used (Barnett and Lewis 1998). Briefly, Δ Ct values from non-CD4+ T cells were subtracted from Δ Ct values from CD4+ T cells. The absolute value differences were then sorted highest to lowest. Starting with the highest value, this number was subtracted from the average of the lower values. This number was divided by the standard deviation of the lower values to determine the number of standard deviations from the mean. If the highest number was greater than three standard deviations from the mean, this value was considered an outlier. This method was repeated for the next highest value, and so on.
Enhancer screen
Luciferase assays were used to determine the number of DNase HS sites that display enhancer activity. Seventy-seven PCR primer pairs, flanking sequences of ∼900-1100 bp, were designed to amplify and clone DNase clusters of three or more. Approximately one-third of these regions map within 1 kb of transcription start sites, while the remaining map to all other regions of the genome. Ten of these DNase clusters were taken from the Table 1 list, while the remaining were randomly chosen. To determine background enhancer activity, an additional 41 PCR primers were designed around random regions of the genome. PCR products were cloned into the pGL3 promoter luciferase vector (Promega) by using the InFusion cloning system (BD Biosciences). DNA was prepared by using mini-column purification kits (Qiagen) and cotransfected with a Renilla luciferase control vector in triplicate into Jurkat and HeLa cells. After 24 h, cells were lysed and screened for enhancer activity by detecting firefly luciferase-to-Renilla luciferase ratios using the Dual-Luciferase Reporter Assay System (Promega) and Centro LB 960 Luminometer (Berthold).
Acknowledgments
We thank Stacie Anderson and the NHGRI Microarray core for excellent technical assistance. We also thank Peter Scacheri, Mike Erdos, Larry Brody, and David Bodine for helpful suggestions and The Encode Consortium for interesting discussions and making data publicly available. This research was supported by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health (F.C.), and Na-tional Institutes of Health grant HL39639 (D.G.). D.G. is a Howard Hughes Medical Institute investigator.
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4074106.
References
- Barnett, V. and Lewis, T. 1998. Outliers in statistical data. John Wiley and Sons, West Sussex, UK.
- Brenner, S., Johnson, M., Bridgham, J., Golda, G., Lloyd, D.H., Johnson, D., Luo, S., McCurdy, S., Foy, M., Ewan, M., et al. 2000. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 18: 630-634. [DOI] [PubMed] [Google Scholar]
- Collins, F.S., Green, E.D., Guttmacher, A.E., and Guyer, M.S. 2003. A vision for the future of genomics research. Nature 422: 835-847. [DOI] [PubMed] [Google Scholar]
- Crawford, G.E., Holt, I.E., Mullikin, J.C., Tai, D., National Institutes of Health Intramural Sequencing, Blakesley, R., Bouffard, G., Young, A., Masiello, C., Green, E.D., et al. 2004. Identifying gene regulatory elements by genome-wide recovery of DNase hypersensitive sites. Proc. Natl. Acad. Sci. 101: 992-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Encode Consortium. 2004. The ENCODE (Encyclopedia of DNA Elements) Project. Science 306: 636-640. [DOI] [PubMed] [Google Scholar]
- Gross, D.S. and Garrard, W.T. 1988. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 57: 159-197. [DOI] [PubMed] [Google Scholar]
- Hebbes, T.R., Thorne, A.W., Clayton, A.L., and Crane-Robinson, C. 1992. Histone acetylation and globin gene switching. Nucleic Acids Res. 20: 1017-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewish, D. 1977. Features of the structure of replicating and non-replicating chromatin in chicken erythroblasts. Nucleic Acids Res. 4: 1881-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry, R.A., Bolstad, B.M., Collin, F., Cope, L.M., Hobbs, B., and Speed, T.P. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik, D., Baertsch, R., Diekhans, M., Furey, T.S., Hinrichs, A., Lu, Y.T., Roskin, K.M., Schwartz, M., Sugnet, C.W., Thomas, D.J., et al. 2003. The UCSC Genome Browser Database. Nucleic Acids Res. 31: 51-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T.H., Barrera, L.O., Qu, C., Van Calcar, S., Trinklein, N.D., Cooper, S.J., Luna, R.M., Glass, C.K., Rosenfeld, M.G., Myers, R.M., et al. 2005. Direct isolation and identification of promoters in the human genome. Genome Res. 15: 830-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies, E.H., Blanchette, M., Haussler, D., and Green, E.D. 2003. Identification and characterization of multi-species conserved sequences. Genome Res. 13: 2507-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur, M., Gerum, S., and Stamatoyannopoulos, G. 2001. Quantification of DNaseI-sensitivity by real-time PCR: Quantitative analysis of DNaseI-hypersensitivity of the mouse β-globin LCR. J. Mol. Biol. 313: 27-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, S. and Skaletsky, H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132: 365-386. [DOI] [PubMed] [Google Scholar]
- Sabo, P.J., Humbert, R., Hawrylycz, M., Wallace, J.C., Dorschner, M.O., McArthur, M., and Stamatoyannopoulos, J.A. 2004. Genome-wide identification of DNaseI hypersensitive sites using active chromatin sequence libraries. Proc. Natl. Acad. Sci. 101: 4537-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, R., Bannister, A.J., Myers, F.A., Thorne, A.W., Crane-Robinson, C., and Kouzarides, T. 2004. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 6: 73-77. [DOI] [PubMed] [Google Scholar]
- Su, A.I., Cooke, M.P., Ching, K.A., Hakak, Y., Walker, J.R., Wiltshire, T., Orth, A.P., Vega, R.G., Sapinoso, L.M., Moqrich, A., et al. 2002. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. 99: 4465-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein, N.D., Aldred, S.J., Saldanha, A.J., and Myers, R.M. 2003. Identification and functional analysis of human transcriptional promoters. Genome Res. 13: 308-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C., Wong, Y.C., and Elgin, S.C. 1979. The chromatin structure of specific genes, II: Disruption of chromatin structure during gene activity. Cell 16: 807-814. [DOI] [PubMed] [Google Scholar]
Web site references
- http://genome.ucsc.edu/ENCODE/; UCSC genome browser for ENCODE regions
- http://genome.ucsc.edu/; UCSC genome browser
- http://www.ncbi.nlm.nih.gov/geo; Gene Expression Omnibus
- http://www.bioconductor.org; Bioconductor
- http://research.nhgri.nih.gov/DNaseHS/; List of DNase HS sites described in this paper