Abstract
Evidence has shown that the insulin and insulin receptor (IR) play a role in cognitive function. However, the detailed mechanisms underlying insulin's action on learning and memory are not yet understood. Here we investigated changes in long-term memory-associated expression of the IR and downstream molecules in the rat hippocampus. After long-term memory consolidation following a water maze learning experience, gene expression of IR showed an up-regulation in the CA1, but a down-regulation in the CA3 region. These were correlated with a significant reduction in hippocampal IR protein levels. Learning-specific increases in levels of downstream molecules such as IRS-1 and Akt were detected in the synaptic membrane accompanied by decreases in Akt phosphorylation. Translocation of Shc protein to the synaptic membrane and activation of Erk1/2 were also observed after long-term memory formation. Despite the clear memory-correlated alterations in IR signaling pathways, insulin deficits in experimental diabetes mellitus (DM) rats induced by intraperitoneal injections of streptozotocin resulted in only minor memory impairments. This may be due to higher glucose levels in the DM brain, and to compensatory mechanisms from other signaling pathways such as the insulin-like growth factor-1 receptor (IGF-1R) system. Our results suggest that insulin/IR signaling plays a modulatory role in learning and memory processing, which may be compensated for by alternative pathways in the brain when an insulin deficit occurs.
Insulin/insulin receptor plays diverse roles in brain functions including learning and memory. Cognition-enhancing effects of insulin have been reported in both human and animal experiments (Kern et al. 1999, 2001; Park et al. 2000), whereas impairments in insulin and the brain insulin receptor (IR) have been linked to development of aging-associated brain degenerative disorders such as Alzheimer's disease (Craft et al. 1996; Frolich et al. 1998; Gasparini and Xu 2003; Watson and Craft 2003). Unlike its peripheral counterpart, insulin/insulin receptor in the brain does not appear to exert a direct effect on neuronal glucose metabolism, but most likely plays a modulatory role, via activating specific signal transduction cascades, in synaptic activities involved in memory formation. Evidence has shown that insulin/insulin receptor is involved in regulation of neurotransmitter release (Bhattacharya and Saraswati 1991; Figlewicz and Szot 1991), receptor conductance and trafficking (Wan et al. 1997; Beattie et al. 2000; Lin et al. 2000; Man et al. 2000; Skeberdis et al. 2001; Zhou et al. 2001), and intracellular Ca2+ release and neuropeptide secretion (Jonas et al. 1997). As a receptor tyrosine kinase, the insulin receptor in the hippocampus was shown to respond to learning experiences by alterations in its gene expression and activation of downstream molecules such as Shc/Erk1/2 in an early stage of memory formation (Zhao et al. 1999).
Despite the accumulated evidence, contradictory results appear to exist regarding roles of insulin/insulin receptor in learning. On the one hand, specific deletion of the brain insulin receptor did not seem to affect animals' ability to form a spatial memory following water maze learning (Schubert et al. 2004). On the other hand, pharmacological disruption of the blood insulin supply by acute administrations of streptozotocin (STZ) that induced experimental diabetes mellitus (DM) resulted in spatial learning deficits and impaired long-term potentiation (LTP) (Biessels et al. 1996, 1998). Thus, the precise role of insulin/insulin receptor in learning and memory remains to be clarified. To gain better understanding of insulin/insulin receptor signaling in learning and memory formation, in this study we investigated long-term memory-associated changes in the brain insulin receptor and its downstream molecules. We also compared learning and the learning-induced biochemical changes in healthy animals to those in STZ-induced DM rats to evaluate the effects of circulating insulin supply on learning and insulin receptor signaling.
Results
Behavioral results
Rats that received the intraperitoneal injection of STZ developed DM ∼1 wk after the injection, showing typical symptoms including polydipsia, polyphagia, and polyuria, along with reduced body weight and elevated blood glucose levels (Table 1).
Table 1.
Changes in blood glucose levels and body weight in DM rats
| Group | Blood glucose (mg/DL) (mean ± SE) | P | Body weight (g) (mean ± SE) | P |
|---|---|---|---|---|
| Normal (n = 18) | 66.25 ± 2.455 | <0.001 | 481.9 ± 22.76 | <0.001 |
| DM (n = 23) | 372.8 ± 20.75 | 332.4 ± 25.72 |
Body weights and blood glucose from control and diabetic rats were measured on a weekly basis. The final measured results at the end of each experiment are shown.
DM rats showed normal acquisition
In the water maze training, DM rats showed no difference in acquisition compared to control animals. Like healthy rats, they were able to locate the hidden platform with progressively reduced search times after initial training (Fig. 1A). A two-way ANOVA analysis revealed significant time effects (F3,88 = 34, P < 0.001) but not significant group effects or interaction. These results suggest an apparently intact acquisition at 2 mo after the onset of DM.
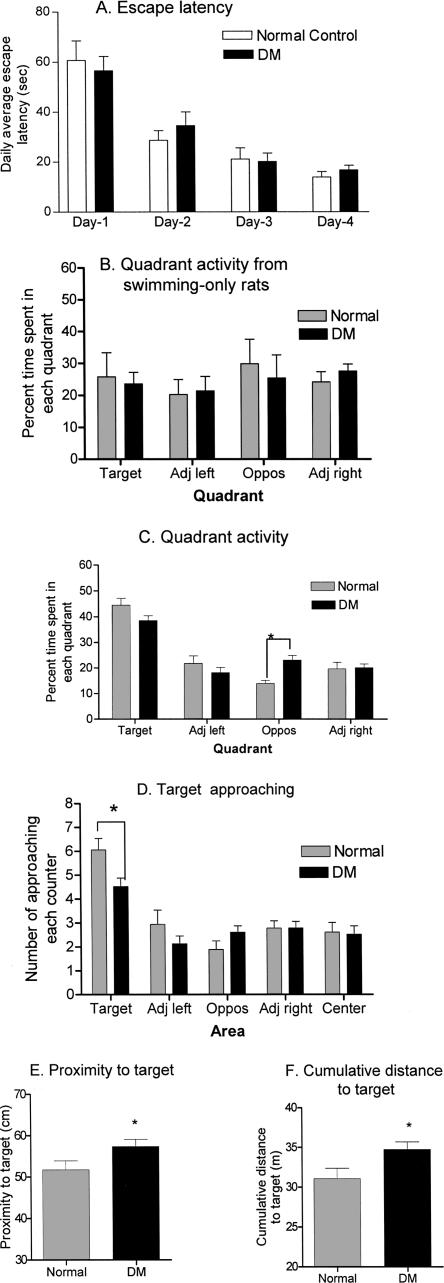
Figure 1.
Behavioral results. (A) Daily escape latency. Rats were trained to find a hidden platform in a water pool. The searching time from four training trials each day was averaged and analyzed with a two-way ANOVA. Escape latencies on days 2, 3, and 4 were significantly different from those on day 1 in both groups. No differences existed between the healthy and DM animals. (B) Quadrant activity from swimming-only rats in a probe test. In the probe test, healthy and DM rats that were subjected to swimming-only activities swam randomly in the pool spending a similar amount of time in each quadrant, as they had no knowledge of the platform for escape. (C) Quadrant activity of healthy and DM rats after training. Both healthy and DM rats spent the majority of their time searching in the target quadrant during a probe test, which was significantly different from time spent in the other three quadrants. There was a significant difference between healthy and DM rats in the time spent in the quadrant opposite the target quadrant. (D) Frequency of approaching the target region during a probe test. The numbers of times rats passed each defined area were compared between healthy and DM groups with a two-way ANOVA. Although rats from both groups showed a markedly higher rate approaching the target area compared to other areas, there was a significant difference between healthy and DM rats in the frequency of approaching the target area. (E) DM rats displayed higher values in proximity to target than healthy rats. (F) DM rats displayed higher values in cumulative distance to the target compared to healthy rats. (Adj left) adjacent quadrant to the left of the target quadrant; (Adj right) adjacent quadrant to the right of the target quadrant; (Oppos) quadrant opposite to the target quadrant. *, P < 0.05.
DM rats showed subtle impairment of spatial memory retention
When tested on a probe trial after four consecutive days of training, rats subjected to swimming-only activity showed no significant difference in the amount of time swimming in the four quadrants (Fig. 1B). In contrast, both healthy and DM rats demonstrated similarly high retention levels after training measured by various parameters. In quadrant activities (Fig. 1C), both healthy and DM rats spent significantly higher percent times during the probe trial swimming within the quadrant where the platform was previously placed (target quadrant). A two-way ANOVA revealed significant quadrant effects (F3,156 = 56, P < 0.001) and a significant interaction (F3,156 = 5.04, P < 0.01), but not significant group effects. A post hoc test showed no significant differences between healthy and DM rats in time spent in the target and adjacent quadrants. However, it showed that DM rats spent significantly longer times in the quadrant opposite the target quadrant (P < 0.01), suggesting a possible higher error rate among DM rats. When analyzed independently with one-way ANOVA, both groups showed that the searching time spent in the target quadrant was significantly higher than that spent in other quadrants. To raise the measurement sensitivity, we dissected the first 15 and 30 sec of the probe test duration (1 min) and compared differences in rats' searching behavior between the two groups. The results showed no significant difference in the time spent in the target quadrant between healthy and DM rats during either the first 15 or 30 sec (data not shown). A second parameter used to assess the rats' retention levels is the frequency of target approaching (i.e., the number of times a rat passes through the target area during the probe test). Among the five defined areas (target, adjacent left, adjacent right, opposite, and the pool center) in the pool, rats from both groups approached the target area (40 cm in diameter surrounding the center of the platform) with significantly higher frequency (Fig. 1D). A two-way ANOVA revealed significant area effects (F4,195 = 22.3, P < 0.0001) and a significant interaction (F4,195 = 2.65, P < 0.05), but no significant group effects. However, a post hoc test comparing values between healthy and DM rats in each individual area showed that the target approaching rate of the healthy rats was significantly higher (t = 2.91, P < 0.05) than that of the DM rats (Fig. 1D). Furthermore, rats from both groups showed a lower cumulative animal-to-target distance and target approaching latency. Proximity measurements and cumulative animal-to-target distance during the probe trial are parameters known as Gallagher measures, with the former measuring the average rat-to-target distance and the latter the sum of rat-to-target distance. Low values of Gallagher measures indicate high retention levels for place learning, because if the rat has knowledge of platform location, it will spend time searching close to the platform (Gallagher et al. 1993). As shown in Figure 1E,F, healthy rats showed significantly lower values in Gallagher proximity measures (t = 2.04, P = 0.048) and cumulative rat-to-target distance (t = 2.34, P = 0.025) than DM rats, indicating a better knowledge of the platform location. On the other hand, there were no significant differences between healthy and DM rats in the latency to reach the target, and other general behaviors such as swimming speed and thigmotaxis (Table 2). These results suggest that although DM rats were able to learn and retain spatial information, their memory consolidation is compromised. Compared with healthy controls, the impairment was behaviorally subtle at 2 mo after DM onset.
Table 2.
Comparison of probe test results from healthy and DM rats
| Parameters | Control (n = 18) (mean ± SE) | DM (n = 23) (mean ± SE) | P |
|---|---|---|---|
| Percent time spent in the target quadrant | 44.53 ± 2.7 | 38.57 ± 1.86 | 0.068 |
| Latency to reach target (Sec) | 9.056 ± 2.2 | 9.56 ± 2.01 | 0.85 |
| Thigmotaxis | 18.25 ± 2.46 | 17.50 ± 1.85 | 0.28 |
| Swimming speed (cm/sec) | 23.53 ± 1.1 | 22.07 ± 0.86 | 0.30 |
Several parameters from healthy and DM rats for target searching during the probe test were compared by t-test.
Changes in gene expression of IR in the brains of DM rats
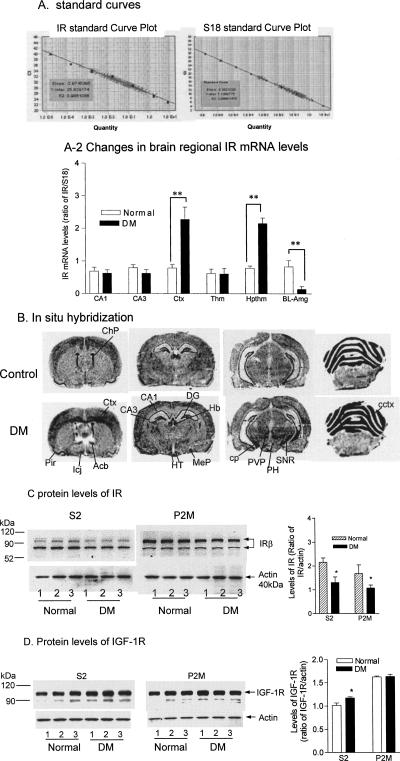
In this experiment we investigated DM-induced changes in IR gene expression and protein levels in the rat brain. Total RNA from multiple brain regions of the DM rats including the hippocampus, somatosensory cortex, thalamic nuclei, hypothalamic nuclei, and basal lateral amygdala (BLA) area were subjected to real-time RT-PCR. In these PCR runs, a series of dilutions of cDNA produced linear standard curves for both IR and S18 (Fig. 2A), which covered concentrations of the entire sample population (crosses spread on the standard curve). As shown in Figure 2A-2, changes in IR gene expression in the DM brain were not observed universally but only in specific regions. For example, slightly lower but not significant levels of IR mRNA were shown in hippocampal regions of DM rats. In contrast, markedly increased IR mRNA levels were detected in the somatosensory cortex and hypothalamus of the DM brain. Certain regions such as the BLA showed a decrease in IR gene expression. A two-way ANOVA revealed highly significant regional effects (F5,24 = 16.9, P < 0.0001), group effects (F1,24 = 10, P < 0.01), and interaction (F5,24 = 15.9, P < 0.0001). Post hoc tests indicated that there were significant differences in the IR mRNA levels from cortex (t = 6.43, P < 0.001), hypothalamus (t = 5.93, P < 0.001), and BLA (t = 3.54, P < 0.05) between healthy and DM rats. In addition, in situ hybridization was performed to verify regional distributions of brain IR gene expression changes, and the results were generally consistent with those obtained with real-time PCR (Fig. 2B); elevated IR mRNA signals were seen in the cerebral cortex and hypothalamic nuclei. In the cerebral cortex, up-regulation of IR gene expression was mainly seen in the deep layer (layers 5–6) of the cerebral cortical neurons, which is particularly evident in the frontal region of the cortex. Other areas that showed increased IR mRNA levels in DM included the paraventricular nuclei, substantia nigra, and cerebellar cortex. These results indicate that distinct brain regions respond to an insulin deficiency differently.
Figure 2.
DM-induced changes in IR gene expression and protein levels. Total RNA was prepared and reverse transcribed to cDNA, from which real-time quantitative PCR was performed with specific primers for IR and S18 rRNA, respectively. (A) Standard curves of IR and S18 rRNA. (A-2) DM-induced changes in IR mRNA levels in different brain regions. After being normalized with S18, IR PCR results from three pools of tissues of each brain region were compared with a two-way ANOVA followed by a post hoc test between normal and DM rats. CA1, CA1 region of the hippocampus; CA3, CA3 region of the hippocampus; Ctx, primary somatosensory cortex; Thm, thalamic nuclei; Hpthm, hypothalamic nuclei; BL-Amg, basal lateral amygdala area. **, P < 0.01; n = 3. (B) In situ hybridization results. Cryostat sections from three healthy and three DM rats were prepared. In situ hybridization using an IR riboprobe labeled with S35 was performed as described in Materials and Methods, followed by film autoradiography. The brain regions were defined according to the rat brain stereotaxic coordinates by Paxinos and Watson (1998). Acb, accumbens nucleus; CA1, CA1 region of the hippocampus; CA3, CA3 region of the hippocampus; cctx, cerebellar cortex; ChP, choroids plexus; cp, cerebral peduncle, basal part; Ctx, cerebral cortex; DG, dentate gyrus; Hb, habenular nucleus; HT, hypothalamic nuclei; Icj, islands of Calleja; MeP, medial amygdaloid nucleus; PH, posterior hypothalamic area; PVP, periventricular nuclei, posterior; SNR, substantia nigra. (C,D) DM-induced changes in levels of IR and IGF-1R. IR and IGF-IR from cytosolic and synaptic membrane fractions from individual hippocampi of healthy and DM rats was measured with Western blotting. Ratios of IR and IRG-1R relative to actin were compared, respectively, with a t-test between healthy and DM rats. S2, cytosolic fractions; P2M, synaptic membrane fractions. *, P < 0.05; **, P < 0.01; n = 3.
Because the hippocampus is critically involved in the spatial memory processing of rodents, we measured changes in IR levels in this region using immunoblotting. The anti-IRβ antibody recognized two IR species in the rat hippocampus, which showed apparent molecular weights of ∼95 and 83 kDa. Although the 83-kDa IR species was predominantly localized in the S2 fraction, the majority of the 95-kDa IR species was seen in the P2M fraction (Fig. 2C). Although no significant changes in IR gene expression were detected in the hippocampus, the amount of IR protein in this region was reduced in both the cytosolic (S2) and synaptic membrane (P2M) fractions of DM rats. The t-tests of the data indicated that the reductions were significant in both fractions (S2: t = 2.91, P < 0.05; P2M: t = 3.72, P < 0.05). To test whether DM also affected the insulin-like growth factor-1 receptor (IGF-1R), we also measured levels of the IGF-1R in hippocampal subfractions. In comparison, the hippocampus contained high concentrations of IGF-1R, and no changes were seen in the P2M fraction from DM rats. However, a small but significant increase was detected in the S2 fraction from DM rats (Fig. 2C).
Learning-induced changes in IR gene expression and protein levels
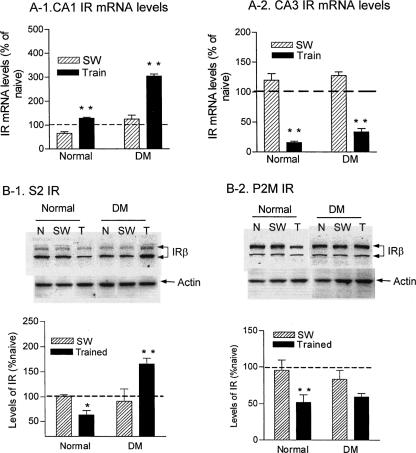
We previously reported a short-term memory-associated IR gene up-regulation following water maze training (Zhao et al. 1999). Here we tested changes in IR gene expression and protein levels in the hippocampus of healthy and DM rats associated with long-term memory. At 24 h after 4 consecutive days of training, IR gene expression from both healthy and DM rats showed a significant up-regulation in the CA1 region (Fig. 3A-1), with DM rats showing a markedly higher increase in IR mRNA levels under both swimming-only and training conditions. A two-way ANOVA indicated highly significant group (normal vs. DM) effects (F1,8 = 322, P < 0.0001) and training effects (F1,8 = 306, P < 0.0001), and interaction (F1,8 = 89, P < 0.001). On the other hand, a marked down-regulation in IR mRNA (P < 0.01) was detected in the CA3 region after training (Fig. 3A-2), with a two-way ANOVA revealing significant training effects (F1,8 = 352, P < 0.0001). These results suggest that IR gene expression was differentially regulated in subregions of the hippocampus in spatial memory processing.
Figure 3.
Learning-induced changes in IR gene expression and protein levels in healthy and DM rats. (A) Total RNA was prepared from CA1 and CA3 regions from naive, swimming control and trained rats. Ratios of IR mRNA levels over those of S18 controls from swimming control and trained animals were converted to percentages from those of naive animals and compared by two-way ANOVA. (A-1) IR gene expression in the CA1 was up-regulated in long-term memory formation. (A-2) IR gene expression in the CA3 was down-regulated in long-term memory formation. **, P < 0.01; n = 3. (B) Learning-induced changes in IR protein levels in the hippocampus. S2 and P2M fractions were prepared from three naive, three swimming controls, and six trained rats. The IR-β subunit levels were measured with Western blotting and normalized with levels of actin. The IR/actin ratios from swimming controls and trained rats were converted to percent ratios from naive animals and then analyzed with t-tests between healthy and DM groups. (B-1) Cytosolic IR levels after learning. (B-2) Synaptic membrane IR levels after learning. N, naive controls; SW, swimming controls; Train, trained animals. *, P < 0.05; **, P < 0.01.
We next examined learning-induced IR protein levels. As shown in Figure 3B-1 and B-2, the amount of IR protein was significantly down-regulated in both S2 (t = 2.8, P < 0.05) and P2M (t = 2.9, P < 0.05) fractions from healthy rats after training. In DM rats, IR showed a decrease that was not significant in the P2M fraction, but a significant increase in the S2 fraction (t = 4.0, P < 0.01).
Learning-induced changes in molecules involved in the IR-Shc-Erk pathway
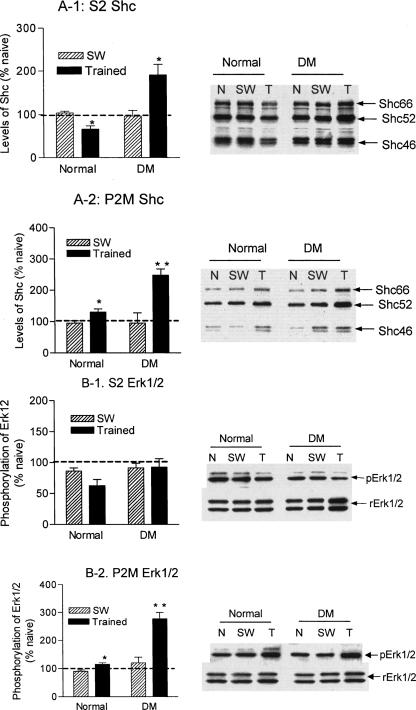
IR is known to activate Erk1/2 via the Shc/Grb-2/Ras pathway. As shown in Figure 4A, Shc species prepared from the rat hippocampus are predominantly cytosolic proteins. Learning induced a small but significant decrease (t = 3.05, P < 0.05) in S2 fractions from normal animals (Fig. 4A-1, normal). This was correlated with an increase in the P2M fraction (t = 2.5, P < 0.05; Fig. 4A-2, normal), indicating a translocation of Shc from the cytosol to the membrane. In the DM brains, on the other hand, Shc showed more substantial increases in both the S2 (t = 4.8, P < 0.01) and P2M fractions (t = 4.07, P < 0.01; Fig. 4A-1 DM and 4A-2 DM), indicating that a net increase in the amount of Shc species may be induced in the DM hippocampus in response to learning. Farther downstream, we measured changes in phosphorylation of Erk1/2 after learning. Whereas no significant changes in Erk1/2 were shown in the S2 fractions (Fig. 4B-1, normal), a significant increase in Erk1/2 phosphorylation (t = 3.47, P < 0.01) was detected in the P2M fraction (Fig. 4B-2, normal) from normal rats. This is consistent with our previous findings (Zhao et al. 1999). In DM rats, a substantial increase in Erk1/2 was seen in the S2 fraction (Fig. 4B-1, DM, rErk1/2), and a multifold increase in the phosphorylation of Erk1/2 was detected in the P2M fractions (t = 4.82, P = 0.002) (Fig. 4B-2, DM). Despite the marked elevation in pErk2 (42 kDa), pErk1 (44 kDa) was abolished from the P2M fractions of DM rats.
Figure 4.
Learning-induced changes in Shc and Erk1/2 in healthy and DM rats. (A) Levels of Shc protein species from different groups were measured on Western blots that were normalized with those of actin. The Shc/actin ratios from swimming controls and trained animals were converted to percent ratios from naive animals and compared with t-tests between healthy and DM animals. (A-1) Learning-induced changes in Shc in cytosolic fractions. (A-2) Learning-induced changes in Shc in synaptic membrane fractions. (B) Changes in levels and phosphorylation of Erk1/2 after learning. The Erk protein levels were measured with immunoblotting and normalized with levels of actin. Activation of Erk1/2 was detected with an anti-pErk1/3 antibody, and normalized with the total amount of Erk1/2 to determine the extent of its phosphorylation. The normalized ratios from swimming controls and trained rats were converted to percent ratios from naive animals and compared with t-tests between healthy and DM animals. (B-1) Changes in the total and phosphorylated Erk1/2 in the cytosolic fraction. (B-2) Changes in the total and phosphorylated Erk1/2 in the synaptic membrane fraction. Normal, healthy rats; DM, diabetic rats; N, naive rats; SW, swimming control rats; T, trained rats; S2, cytosolic fractions; P2M, synaptic membrane fractions. *, P < 0.05; **, P < 0.01.
Learning-induced changes in molecules involved in the IR-IRS-1 pathway
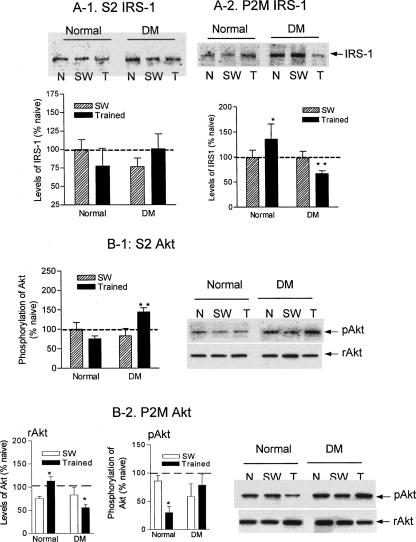
Additionally, we measured learning-induced changes in molecules involved in the IRS-1/PI3 kinase/Akt chain, which constitutes another major IR downstream pathway. Learning induced different changes in IRS-1 between the healthy and DM hippocampus. Similar to Shc, IRS-1 showed a possible learning-induced membrane translocation manifested by a reduction in the S2 fraction (Fig. 5A-1, normal) and a significant increase (t = 2.69, P < 0.05) in the P2M fraction (Fig. 5A-2, normal). In contrast, IRS-1 from DM rats showed an opposite change: an increase in the S2 fraction (Fig. 5A-1, DM) and a significant decrease (t = 3.56, P < 0.01) in the P2M fraction (Fig. 5A-2, DM). We next measured changes in the amount and phosphorylation of Akt, a downstream molecule of IRS-1. In healthy rats no changes in Akt were detected in S2 fractions after learning. However, Akt underwent significant dephosphorylation in the P2M fractions after training (P < 0.01) that was accompanied by an increase (P < 0.05) in the total amount of Akt (rAkt). In contrast, DM induced an overall increase in Akt phosphorylation in both S2 and P2M fractions, under basal conditions and after training (Fig. 5B-1 and B-2, DM).
Figure 5.
Learning-induced changes in IRS-1 and Akt in healthy and DM rats. Levels of IRS-1 from cytosolic and synaptic membrane from different groups were measured on Western blots that were normalized by those of actin. The normalized ratios from swimming controls and trained animals were converted to percent ratios from naive animals and compared with t-tests. (A-1) Changes in the cytosolic IRS-1 after learning. (A-2) Changes in the synaptic membrane IRS-1 after learning. (B) Levels and phosphorylation of Akt from cytosolic and synaptic membrane fractions were measured and analyzed similar to those of Erk1/2 in Figure 4. (B-1) Changes in the total and phosphorylated Akt in the cytosolic fractions; (B-2) Changes in the total and phosphorylated Akt in the synaptic membrane fractions. Normal, healthy rats; DM, diabetic rats; N, naive rats; SW, swimming control rats; T, trained rats; S2, cytosolic fractions; P2M, synaptic membrane fractions. *, P < 0.05; **, P < 0.01.
Discussion
Learning-induced changes in IR gene expression and downstream cascades under normal physiology
We previously reported that spatial learning induced biochemical changes in IR gene expression and specific downstream signaling molecules such as Shc and Erk1/2 during short-term memory processing (Zhao et al. 1999). In the present study, we measured these changes during long-term memory formation. Our results consistently showed that IR displayed learning-specific increases in gene expression not only in short-term but also in long-term memory formation. Interestingly, the learning-responding IR gene expression showed a regional specificity with a persistent up-regulation only in the CA1 region, similar to our previous findings (Zhao et al. 1999), and a profound down-regulation in the CA3 region. Although the precise roles of the CA1 and the CA3 region in memory processing are not yet fully understood, the CA1 neurons have been shown to be the most reactive cells in response to both physiological input such as learning (Frank et al. 2004; Knafo et al. 2004; Nolan et al. 2004) and pathological stimuli such as ischemia, hypoxia (Lukyanetz et al. 2003; Englund et al. 2004; Olson and McKeon 2004), and hyperglycemia (the present study). It is not surprising, therefore, that CA1 neurons are equipped with active signal transduction systems including those regulated by tyrosine receptor kinases such as the insulin receptor. In contrast, a reduction in IR protein levels was detected after long-term memory formation, possibly due to a down-regulation of the receptor after initial increases of receptor activities. Two species of IR were detected in the hippocampus, with IR95 predominantly in the synaptic membrane and IR83 in the cytosol. IR in the brain has been known to have molecular weights different from those of the periphery due to alternative splicing and differences in glycosylation (Heidenreich and Brandenburg 1986; Simon and Leroith 1986; Kenner et al. 1995). However, it remains to be determined whether the IR83 is a differently glycosylated isoform, or a proteolysis product of IR95 after IR internalization.
Among downstream molecules, a possible translocation of Shc to the synaptic membrane was shown in the hippocampus specifically associated with learning, suggesting an increase in Shc-IR binding. Subsequently, Erk1/2 showed a persistent activation after learning. Both Shc and Erk1/2 have been shown to be involved in spatial learning (Selcher et al. 1999, 2003; Zhao et al. 1999; Morozov et al. 2003; Tomizawa et al. 2003). Sustained Erk1/2 activation may play a role in consolidation of long-term memory through activation of gene transcription. Learning also induced changes in the IRS-1/PI3 kinase/Akt pathway, including possible membrane translocation of IRS-1 and Akt. Depletion of brain IR was shown to abolish the majority of Akt phosphorylation in the brain (Schubert et al. 2004), suggesting that IR is a major upstream kinase in the brain regulating Akt's phosphorylation. Increases in the amount of synaptic membrane Akt are consistent with the up-regulation of Akt gene expression in spatial learning reported by Robles et al. (2003). As a downstream molecule of PI-3 kinase, Akt has been reported to be phosphorylated in the amygdala shortly after fear conditioning, and inactivated 60 min after training (Lin et al. 2001). This suggests that activation of Akt is a rapid but temporary event in response to learning. Not surprisingly, in the present study we also observed Akt dephosphorylation associated with long-term memory formation. Although the precise roles of the IRS-1/PI3 kinase/Akt pathway in long-term memory storage remain to be understood, they may be involved in regulation of memory-associated neuronal metabolism, activation of nitric oxide synthesis (Montagnani et al. 2002), and interactions with the Ras/Erk1/2 system (Lin et al. 2001; Sweatt 2001) leading to regulation of gene transcription.
Effects of DM on learning and the brain IR signaling
Disruption of IR signaling can be caused by diabetes, in which the production of insulin becomes insufficient (type 1 DM) or the activation of IR is impaired, leading to insulin resistance (type 2 DM). To further assess the role of insulin signaling in learning and memory, we investigated the effects of experimental type 1 DM on spatial learning and the correlative changes in the brain IR signaling. Our results showed that disruption in the insulin supply did not seem to affect rats' acquisition in a spatial learning task, as DM rats displayed significantly reduced escape latencies correlated to training progress that were not different from those of healthy rats. Furthermore, the probe test indicated that DM rats formed a clear spatial memory following the training experience indicated by measures of various parameters such as quadrant activities and latency to reach the target that were comparable to those of healthy rats. However, in-depth comparison of certain measures revealed significantly poorer performance by DM rats during testing. For example, although the DM rats spent the majority of the time searching in the target quadrant, their frequency of traveling across the target spot was significantly lower than that of the healthy rats, and the DM rats spent a markedly longer time than healthy rats searching in the opposite quadrant. Moreover, compared to the healthy rats, the DM rats showed significantly greater values in the Gallagher proximity measures and a cumulative rat-to-target distance, indicating that their knowledge of the searching target location was less precise than that of the healthy controls. These results indicate that memory impairment, although behaviorally subtle, was associated with DM. Other investigators have also reported effects of DM on learning and memory (Biessels et al. 1996; Li et al. 2002). Although several factors such as an animal's age, STZ delivery route (intravenous vs. intraperitoneal), and disease duration may be responsible for the severity of DM-associated memory impairment, our results are basically in agreement with those previously reported (Biessels et al. 1996; Li et al. 2002).
Despite subtle changes in behavior, evident DM-associated biochemical alterations were detected in the brain. Elevated IR gene expression was shown in several brain regions including the hypothalamus, substantia nigra, and cerebral and cerebellar cortices, whereas no significant changes in IR gene expression were detected in the hippocampus. This suggests that the circulating insulin dependency is different among brain regions, and that mechanisms regulating IR gene expression in the hippocampus are different from those in the hypothalamus and cortex. IR protein levels were significantly reduced in the hippocampus of DM animals. This reduction could be due to a persistent deficiency of insulin leading to a disuse receptor down-regulation. Alternatively, an increased IR turnover and/or more frequent receptor internalization due possibly to hyperglycemia and changes in its binding affinity may have contributed to a lower level of IR protein. The DM hippocampus also showed striking increases in the CA1 IR mRNA and the cytosolic IR proteins' levels responding to learning. Similarly, rigorous learning-induced responses were also observed in Shc and Erk1/2 from DM hippocampus, which displayed more than twofold learning-induced increases in the amount of Shc species and Erk1/2 phosphorylation compared to those from normal rats. On the other hand, the learning-induced increase in the amount of IRS-1 and dephosphorylation of Akt were interrupted in the DM hippocampus.
Although the striking biochemical changes in the brain did not appear to reconcile with the minor memory impairment in the DM animals, the following reasons may explain such a dissociation: First, a major consequence of insulin deficits is systemic hyperglycemia that inevitably causes a high glucose concentration in the brain. Because glucose is the major energy fuel for neuronal activity including memory formation (Korol and Gold 1998; Benton and Nabb 2003), it is not surprising that higher metabolic activities (including active signaling transduction such as protein phosphorylation) may exist in the hyperglycemic brain. Indeed, when we applied high concentrations of glucose similar to those from the blood of DM rats to primary hippocampal cultures for 24–72 h, significant increases in Akt phosphorylation and levels of other proteins such as IGF-1R, C-Src, and NMDA receptors were detected, whereas IR levels were increased in a short-term (24-h) exposure but decreased after a longer term (72-h) high glucose exposure (data not shown). Thus, it is conceivable that in a relatively early stage of DM when the brain is able to compensate, high concentrations of glucose and increased cellular activities in the brain may help memory formation following an acute learning experience, although long-term and untreated hyperglycemia will ultimately lead to neuronal damage (Russell et al. 1999; Vincent et al. 2002).
Second, to overcome the disrupted IR signaling, the brain may utilize alternative mechanisms to accomplish its functions. For example, it may adjust IR expression and its ligand-binding affinity in the neuronal membrane to compensate for an insulin deficiency. Moreover, other insulin-like neuronal hormones such as IGF-1 may help activate analog downstream signaling cascades mediated by the IGF-1 receptor (IGF-1R) and/or the IR/IGF-1R hybrids that exhibit higher binding affinity for IGF-1 (Bailyes et al. 1997). Our finding of an increased cytosolic IGF-1R level in the DM hippocampus may suggest compensation from IGF-1R for deficient IR signaling. Therefore, the higher learning-induced responses in the DM brain such as IR gene up-regulation and Erk1/2 phosphorylation might just reflect the efforts of the brain to cope with an abnormal molecular environment that enabled the animals to achieve apparent learning in a water maze task. Additionally, the hyperglycemia-induced high neuronal activities might augment learning-induced signals following activation of neurotransmitter receptors such as NMDA receptors that could in turn result in a stronger activation of Erk1/2.
Third, although persistent insulin deficits and hyperglycemia are likely to lead to degenerative alterations in the brain, impairment of memory may progress at a slow pace, analogous to that in aging. In fact, the higher values of Gallagher proximity measures and cumulative distance in the DM rats of the present study were also shown to be highly sensitive measurements in age-related impairment (Gallagher et al. 1993). Furthermore, results from Li et al. (2002) showed that rats suffering from spontaneous DM did not develop detectable memory impairment until 4 mo after onset of the disease.
Finally, the finding that learning and memory formation can still be achieved under disrupted IR signaling suggests that IR activation may not be a necessary mechanism required for memory processing. This appears to be particularly true for the IRS-1 and Akt cascade, which was shown to be specifically regulated by IR in the brain (Schubert et al. 2004). Although DM rats showed opposite changes in IRS-1 and Akt phosphorylation following learning compared to normal animals, they still achieved relatively high levels of memory formation. Similarly, Schubert et al. (2004) showed that mice with a specific deletion of the brain IR were able to learn in a water maze task although they showed significant reductions in Akt and GSK-3β phosphorylation in the brain. Thus, the IRS-1/PI3K/Akt pathway may not be directly involved in the regulation of memory processing, but rather may play a role in neuronal protection and survival important for neuronal recovery after activation. On the other hand, the Shc/Ras/MAP kinase pathway is known to be critical for memory formation, and this pathway can also be activated by other upstream kinases and neurotransmitter receptors.
In conclusion, insulin receptors in the hippocampus showed clear changes in both short-term and long-term memory formation. However, unlike those neurotransmitter receptors such as NMDA, AMPA, and cholinergic receptors, the functions of which are essentially required for memory formation-associated synaptic activities, IR signaling may play a modulating role to aid functions of the mainstream neurotransmitter receptors during memory processing. The brain appears to express redundant signaling systems with similar modulatory functions.
Thus, when IR signaling is disrupted, an existing alternative pathway(s) may be used for compensation in order for the brain to achieve its functions important for survival. Because using the type 1 DM model to study roles of IR in learning is limited by the resulting hyperglycemia that may complicate the interpretation, future development of an animal model with local IR activity disruption in the brain may offer a clearer picture in such functional studies.
Materials and Methods
Animals
Male adult Wistar rats were purchased from Charles River Laboratories.
Production of DM rats
Male Wistar rats (220–250 g, 2.5- to 3-mo-old) were purchased from Charles River Laboratories. DM rats were produced by two intraperitoneal (i.p.) injections of streptozotocin (STZ, 40 mg/kg body weight) with an interval of 3 d, or by i.p. injection of a single dose (60 mg/kg) of STZ. Control rats receiving i.p. injections of physiological saline were housed under the same conditions. The blood glucose levels and body weights for control and DM groups were monitored on a weekly basis, with the former performed using a OneTouch blood glucose monitor (LifeScan) with a drop of blood from a cut tail tip.
Water maze training
Rats were subjected to a water maze task according to a procedure described by Dou et al. (2003). In brief, rats were trained in a 5 × 1.5-ft water pool to find a platform hidden below the water surface at a fixed location. On each day, four training trials were given with an interval of 10 min. Rats were given a 2-min maximum to find the platform, and their performance was recorded by an HVS video camera (HVS Image) installed above the center of the swimming pool. The time rats spent searching for the platform (i.e., their escape latency) in the four trials each day was averaged and used to assess their acquisition ability. The training was applied for 4 consecutive days. On the fifth day, ∼24 h after the last day's training, the rats underwent a 60-sec probe trial to test their retention levels, during which the platform was removed from the pool and the rats' searching behavior was recorded with the HVS video camera. To investigate the learning-induced changes in IR gene expression in the brain, rats from different groups were sacrificed after the test, and their brains were rapidly removed and frozen on dry ice. To prepare hippocampal synaptic fractions, hippocampi from a subpopulation of rats from each group were rapidly removed after decapitation and frozen on dry ice. Tissues were stored at -80°C until fractionation. To control for nonspecific factors such as stress- and locomotor activity-induced biochemical changes, separate groups of control and DM rats were subjected to swimming activities without the platform present in the pool. To assure physical activity equivalency, the swimming time of each rat in the swimming-only group was yoked with those in training groups. In a particular trial, for example, if the #1 rat spent 20 sec to find the platform, the same 20-sec swimming time would be given to the #1 rat in the swimming-only group. On the fifth day, swimming-only rats were also subjected to a probe test.
Dissections of brain regions
Hippocampal tissues from those rats that clearly showed high retention levels after 4 consecutive days of training were used for biochemical measurements. To quantify brain IR gene expression in different brain regions, a frozen brain was mounted on a cryostat set at -20°C. The brain was sectioned to an appropriate depth, and tissues (∼1 mm3 in volume) from different regions were dissected using a fine-pointed surgical scalpel. The dissection process was guided with the rat brain atlas by Paxinos and Watson (1998). The dissecting area was well lighted and the experimenter wore magnifying glasses to aid vision of brain regions. Nine rats from each group (naive, swimming control, and trained) of both healthy and DM rats were used for this experiment. Tissues from each brain region of three animals were pooled to generate three pools for each group. These tissues were stored at -80°C until preparation of total RNA.
Real-time quantitative PCR
Total RNA from dissected brain tissues was prepared using Tri-Agent (Sigma-Aldrich). After treatment with DNase-I (Ambion) to remove the genomic DNA, RNA was reverse transcribed to single-strand cDNA using a single-strand cDNA synthesis kit (Novagen) with random hexamer primers. Real-time quantitative PCR for IR was performed on the ABI Taqman 9700 platform (Applied Biosystems) with primer pairs (forward: GCCTGGGCAACTGTTCAGA; reverse: GTTTCGACAGGCCACACACTT). The amount of PCR product was detected with a FAM-labeled oligo probe (ACCCGATGACCCTACC) specific to the amplified IR PCR segment. For internal normalization, a segment of S18 rRNA was amplified and measured by a FAM-labeled probe. The amplifying conditions were: stage 1, 50°C for 2 min; stage 2, 95°C for 10 min; stage 3, 40 cycles of denature at 95°C for 15 sec, followed by anneal/extend at 60°C for 1 min. The basal line for fluorescence signal measurement was defaulted at cycles 3–15, with a threshold placed in a region of exponential amplification. Amounts of IR and S18 rRNA PCR product were automatically calculated according to their respective standard curves generated from a series dilution of cDNA templates. Final IR mRNA levels were determined after normalization with the amount of S18 rRNA PCR product from the same sample. Samples were applied to PCR with duplications, and results from three PCR runs for each brain region were compared between control and DM rats.
Protein chemistry experiments
To ensure the reliability of results, these experiments were performed under a double-blind condition. The hippocampi from each rat and from each group were individually coded. Tissue fractionation and subsequent immunoblotting experiments were performed by an experimenter who was unaware of the coding identity of samples. Decoding was made after densitometric scanning results were obtained.
Subcellular fractionation of the hippocampal tissues
Subcellular fractions including the homogenate, cytosolic, and crude synaptic membrane fractions were prepared from hippocampi of individual rats using sucrose centrifugation according to a previously described procedure (Zhao et al. 1999). In brief, the frozen hippocampal tissues from each rat were homogenized in pre-cooled Buffer made of 0.32 M sucrose, 2 mM HEPES, pH 7.4, and 1% protease inhibitor cocktail (Sigma-Aldrich). The homogenate was diluted to 10% (w/v) with the same buffer, and centrifuged at 1000g for 5 min. After collecting the supernatant (S1), the pellet (P1) was resuspended in the same buffer and centrifuged at 1000g again for 5 min. The supernatant was combined with S1 and centrifuged at 10,000g for 20 min. The supernatant (S2) from this step was used as the cytosolic fraction. The pellet (P2) was osmotically lysed (10 mL/g original tissue) on ice for 30 min in a buffer containing 2 mM HEPES, pH 7.4, 50 μm Ca2+, and 1% protease inhibitor cocktail. The lysed P2 fraction was then centrifuged at 20,000g for 30 min. The pellet from this step was collected as a crude synaptic membrane fraction (P2M), and resuspended in a small volume of 2 mM HEPES, pH 7.4 containing 1% protease inhibitor cocktail. Protein concentrations were measured using a BCA protein assay reagent (Pierce Biotechnology). Tissue fractions were stored in small aliquots at -80°C until use.
Western blotting
Western blotting was used to detect changes in protein levels and phosphorylation. Protein concentrations of subcellular fractions across individual samples were equalized. S2 and P2M fractions from each rat were individually loaded for SDS-PAGE for separation, followed by transfer onto a nitrocellulose membrane. IR protein levels were assessed with an anti-IR β-subunit antibody (Santa Cruz Biotechnology). Other antibodies used in this experiment included anti-IR substrate-1 (IRS-1), anti-Shc, anti-Erk1/2 (all from Upstate Biotechnology); anti-Akt, anti-phospho-Akt, and anti-phospho-Erk1/2 (all from Cell Signaling Technology). Nitrocellulose membranes were incubated with individual primary antibodies diluted (1:300 to 1:2000) with 1X PBS, pH 7.4 containing 0.05% Tween-20 (PBS-T) and 0.1% dry milk at 4°C overnight. After washes with PBS-T, membranes were then incubated with a secondary antibody conjugated with horseradish peroxidase (HRP) at room temperature for 1 h. Following washes with PBS-T, membranes were reacted with chemiluminescent reagents (Pierce Biotechnology) and exposed to an x-ray film. To control for sample loading variations, levels of β-actin were measured from each blot by an anti β-actin antibody (Sigma-Aldrich) and used as a reference protein for normalization.
In situ hybridization experiments
In situ hybridization was performed to show regional changes of IR mRNA in the brain. For this purpose, a specific riboprobe corresponding to an IR mRNA sequence of 760–1140 (GenBank accession number M29014) was synthesized in an in vitro transcriptional reaction containing S35 UTP, and hybridized with brain cryosections as described previously (Zhao et al. 1999) with minor modifications. Briefly, brain cryosections (12 μM) were fixed with 4% formaldehyde/PBS, pH 7.4 at room temperature for 10 min. After three washes with PBS, pH 7.4, slices were penetrated with 0.1% Triton X-100/PBS, pH 7.4 for 10 min, followed by washes with 1X PBS, pH 7.4. Slices were then acetylated and dehydrated with graded ethanol. Three brain sections on each slide were hybridized with 1 × 106 cpm IR riboprobe in a 50-μL mixture containing 20 mM Tris-HCl, pH 7.4, 1 mM EDTA, 300 mM NaCl, 50% formamide, 10% dextran sulfate, 1× Denhardt's solution, 4μg/mL salmon sperm DNA, 10 μg/mL yeast total RNA, 10 μg/mL yeast tRNA, 100 mM DTT, 0.1% SDS, and 0.1% sodium thiosulfate. For negative controls, slices were hybridized with the sense riboprobe or pretreated with RNase-A before hybridization with the antisense probe. The hybridization was carried out for 48 h at 55°C. Following a post-hybridization treatment including RNase-A digestion and a series of high-stringency washes, slices were dehydrated with grade ethanol, air-dried, and exposed to x-ray films.
Data analyses
Data analysis for behavioral experiments
Daily escape latency averaged from four training trials was used to assess rats' acquisition and analyzed with one-way ANOVA followed by the Newman-Keuls post hoc test. For retention levels, several parameters were measured during the probe trial. These included percent time spent in the target area, frequency of target approach, and proximity measures. Measured values from each parameter were analyzed with one-way, two-way ANOVAs and t-test.
Analysis of gene expression data from real-time PCR and in situ hybridization
For real-time PCR, values of insulin receptor mRNA levels were normalized with those of S18. The normalized values from each brain region were compared with a t-test between healthy and DM rats. For in situ hybridization, autoradiograms for the brain IR gene expression were scanned with a densitometry scanner. The gray levels for IR mRNA signals were calibrated with a C14 standard and measured with the Scion image program (Scion).
Analyses of biochemistry data
Protein chemistry data from subcellular fractions of individual animals were collected from 2–3 repeated experiments. Chemiluminescent signals on Western blots were acquired with densitometric scanning, and the gray levels for each signal were measured with UN-SCAN-IT software (Silk Scientific). After normalization with values of β-actin, the basal protein levels for each target protein were expressed as target/β-actin ratios for subsequent statistical analyses. For learning-induced changes, quantitated values for IR gene expression and each protein from swimming control and trained rats were converted into percent values of naive animals, which were subsequently compared with ANOVA or t-test. All statistical analyses were performed with GraphPad Prism software (GraphPad Prism Software).
Article published online ahead of print. Article and publication date are at http://www.learnmem.org/cgi/doi/10.1101/lm.88005.
References
- Bailyes, E.M., Navé, B.T., Soos, M.A., Orr, S.R., Hayward, A.C., and Siddle, K. 1997. Insulin receptor/IGF-I receptor hybrids are widely distributed in mammalian tissues: Quantification of individual receptor species by selective immunoprecipitation and immunoblotting. J. Biochem. 327: 209-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie, E.C., Carroll, R.C., Yu, X., Morishita, W., Yasuda, H., von Zastrow, M., and Malenka, R.C. 2000. Regulation of AMPA receptor endocytosis by a signaling mechanism shared with LTD. Nat. Neurosci. 3: 1291-1300. [DOI] [PubMed] [Google Scholar]
- Benton, D. and Nabb, S. 2003. Carbohydrate, memory, and mood. Nutr. Rev. 61: S61-S67. [DOI] [PubMed] [Google Scholar]
- Bhattacharya, S.K. and Saraswati, M. 1991. Effect of intracerebroventricularly administered insulin on brain monoamines and acetylcholine in euglycaemic and alloxan-induced hyperglycaemic rats. Indian J. Exp. Biol. 29: 1095-1100. [PubMed] [Google Scholar]
- Biessels, G.J., Kamal, A., Ramakers, G.M., Urban, I.J., Spruijt, B.M., Erkelens, D.W., and Gispen, W.H. 1996. Place learning and hippocampal synaptic plasticity in streptozotocin-induced diabetic rats. Diabetes 45: 1259-1266. [DOI] [PubMed] [Google Scholar]
- Biessels, G.J., Kamal, A., Urban, I.J.A., Spruijt, B.M., Erkelens, D.W., and Gispen, W.H. 1998. Water maze learning and hippocampal synaptic plasticity in streptozotocin-diabetic rats: Effects of insulin treatment. Brain Res. 800: 125-135. [DOI] [PubMed] [Google Scholar]
- Craft, S., Newcomer, J., Kanne, S., Dagogo-Jack, S., Cryer, P., Sheline, Y., Luby, J., Dagogo-Jack, A., and Alderson, A. 1996. Memory improvement following induced hyperinsulinemia in Alzheimer's disease. Neurobiol. Aging. 17: 123-130. [DOI] [PubMed] [Google Scholar]
- Dou, J., Cui, C., Dufour, F., Alkon, D.L., and Zhao, W.Q. 2003. Gene expression of α-endosulfine in the rat brain: Correlative changes with aging, learning and stress. J. Neurochem. 87: 1086-1100. [DOI] [PubMed] [Google Scholar]
- Englund, M., Bjurling, M., Edin, F., Hyllienmark, L., and Brismar, T. 2004. Hypoxic excitability changes and sodium currents in hippocampal CA1 neurons. Cell Mol. Neurobiol. 24: 685-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figlewicz, D.P. and Szot, P. 1991. Insulin stimulates membrane phospholipid metabolism by enhancing endogenous α 1-adrenergic activity in the rat hippocampus. Brain Res. 550: 101-107. [DOI] [PubMed] [Google Scholar]
- Frank, L.M., Stanley, G.B., and Brown, E.N. 2004. Hippocampal plasticity across multiple days of exposure to novel environment. J. Neurosci. 24: 7681-7689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolich, L., Blum-Degen, D., Bernstein, H.G., Engelsberger, S., Humrich, J., Laufer, S., Muschner, D., Thalheimer, A., Turk, A., Hoyer, S., et al. 1998. Brain insulin and insulin receptors in aging and sporadic Alzheimer's disease. J. Neural Transm. 105: 423-438. [DOI] [PubMed] [Google Scholar]
- Gallagher, M., Burwell, R., and Burchinal, M. 1993. Severity of spatial learning impairment in aging: Development of a learning index for performance in the Morris water maze. Behav. Neurosci. 107: 618-626. [DOI] [PubMed] [Google Scholar]
- Gasparini, L. and Xu, H. 2003. Potential roles of insulin and IGF-1 in Alzheimer's disease. Trends Neurosci. 26: 404-406. [DOI] [PubMed] [Google Scholar]
- Heidenreich, K.A. and Brandenburg, D. 1986. Oligosaccharide heterogeneity of insulin receptors. Comparison of N-linked glycosylation of insulin receptors in adipocytes and brain. Endocrinology 118: 1835-1842. [DOI] [PubMed] [Google Scholar]
- Jonas, E.A., Knox, R.J., Smith, T.C., Wayne, N.L., Connor, J.A., and Kaczmarek, L.K. 1997. Regulation by insulin of a unique neuronal Ca2+ pool and of neuropeptide secretion. Nature 385: 343-346. [DOI] [PubMed] [Google Scholar]
- Kenner, K.A., Kusari, J., and Heidenreich, K.A. 1995. cDNA sequence analysis of the human brain insulin receptor. Biochem. Biophys. Res. Commun. 217: 304-312. [DOI] [PubMed] [Google Scholar]
- Kern, W., Born, J., Schreiber, H., and Fehm, H.L. 1999. Central nervous system effects of intranasally administered insulin during euglycemia in men. Diabetes 48: 557-563. [DOI] [PubMed] [Google Scholar]
- Kern, W., Peters, A., Fruehwald-Schultes, B., Deininger, E., Born, J., and Fehm, H.L. 2001. Improving influence of insulin on cognitive functions in humans. Neuroendocrinology 74: 270-280. [DOI] [PubMed] [Google Scholar]
- Knafo, S., Ariav, G., Barkai, E., and Libersat, F. 2004. Olfactory learning-induced increase in spine density along the apical dendrites of CA1 hippocampal neurons. Hippocampus 4: 819-825. [DOI] [PubMed] [Google Scholar]
- Korol, D.L. and Gold, P.E. 1998. Glucose, memory, and aging. Am. J. Clin. Nutr. 67: 764S-771S. [DOI] [PubMed] [Google Scholar]
- Li, Z.G., Zhang, W., Grunberger, G., and Sima, A.A. 2002. Hippocampal neuronal apoptosis in type 1 diabetes. Brain Res. 946: 221-231. [DOI] [PubMed] [Google Scholar]
- Lin, J.W., Ju, W., Foster, K., Lee, S.H., Ahmadian, G., Wyszynski, M., Wang, Y.T., and Shen, M. 2000. Distinct molecular mechanisms and divergent endocytotic pathways of AMPA receptor internalization. Nat. Neurosci. 3: 1282-1290. [DOI] [PubMed] [Google Scholar]
- Lin, C.H., Yeh, S.H., Lin, C.H., Lu, K.T., Leu, T.H., Chang, W.C., and Gean, P.W. 2001. Role for the PI-3 kinase signaling pathway in fear conditioning and synaptic plasticity in the amygdala. Neuron 31: 841-851. [DOI] [PubMed] [Google Scholar]
- Lukyanetz, E.A., Shkryl, V.M., Kravchuk, O.V., and Kostyuk, P.G. 2003. Action of hypoxia on different types of calcium channels in hippocampal neurons. Biochim. Biophys. Acta. 1618: 33-38. [DOI] [PubMed] [Google Scholar]
- Man, H.Y., Lin, J.W., Ju, W.H., Ahmadian, G., Liu, L., Becker, L.E., Sheng, M., and Wang, Y.T., 2000. Regulation of AMPA receptor-mediated synaptic transmission by clathrin-dependent receptor internalization. Neuron 25: 649-662. [DOI] [PubMed] [Google Scholar]
- Montagnani, M., Ravichandran, L.V., Chen, H., Esposito, D.L., and Quon, M.J. 2002. Insulin receptor substrate-1 and phosphoinositide-dependent kinase-1 are required for insulin-stimulated production of nitric oxide in endothelial cells. Mol. Endocrinol. 16: 1931-1942. [DOI] [PubMed] [Google Scholar]
- Morozov, A., Muzzio, I.A., Bourtchouladze, R., Van-Strien, N., Lapidus, K., Yin, D., Winder, D.G., Adams, J.P., Sweatt, J.D., Kandel, E.R. 2003. Rap1 couples cAMP signaling to a distinct pool of p42/44MAPK regulating excitability, synaptic plasticity, learning, and memory. Neuron 39: 309-325. [DOI] [PubMed] [Google Scholar]
- Nolan, M.F., Malleret, G., Dudman, J.T., Buhl, D.L., Santoro, B. Gibbs, E., Vronskaya, S., Buzsaki, G., Siegelbaum, S.A., Kandel, E.R., et al. 2004. A behavioral role for dendritic integration: NCN1 channels constrain spatial memory and plasticity at inputs to distal dendrites of CA1 pyramidal neurons. Cell 119: 719-732. [DOI] [PubMed] [Google Scholar]
- Olson, E.E. and McKeon, R.J. 2004. Characterization of cellular and neurological damage following unilateral hypoxia/ischemia. J. Neurol. Sci. 227: 7-19. [DOI] [PubMed] [Google Scholar]
- Park, C.R., Seeley, R.J., Craft, S., and Woods, S.C. 2000. Intracerebroventricular insulin enhances memory in a passive-avoidance task. Physiol. Behav. 68: 509-514. [DOI] [PubMed] [Google Scholar]
- Paxinos, G. and Watson, C. 1998. The rat brain: In stereotaxic coordinates, 4th ed. Academic Press, San Diego.
- Robles, Y., Vivas-Mejia, P.E., Ortiz-Zuazaga, H.G., Felix, J., Ramos, X., and Pena de Ortiz, S. 2003. Hippocampal gene expression profiling in spatial discrimination learning. Neurobiol. Learn. Mem. 80: 80-95. [DOI] [PubMed] [Google Scholar]
- Russell, J.W., Sullivan, K.A., Windebank, A.J., Herrmann, D.N., and Feldman, E.L. 1999. Neurons undergo apoptosis in animal and cell culture models of diabetes. Neurobiol. Dis. 6: 347-363. [DOI] [PubMed] [Google Scholar]
- Schubert, M., Gautam, D., Surjo, D., Ueki, K., Baudler, S., Schubert, D., Kondo, T., Alber, J., Galldiks, N., Kustermann, E., et al. 2004. Role for neuronal insulin resistance in neurodegenerative diseases. Proc. Natl. Acad. Sci. 101: 3100-3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selcher, J.C., Atkins, C.M., Trzaskos, J.M., Paylor, R., and Sweatt, J.D. 1999. A necessity for MAP kinase activation in mammalian spatial learning. Learn. Mem. 6: 478-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selcher, J.C., Weeber, E.J., Christian, J., Nekrasova, T., Landreth, G.E., and Sweatt, J.D. 2003. A role for ERK MAP kinase in physiologic temporal integration in hippocampal area CA1. Learn. Mem. 10: 26-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, J. and Leroith, D. 1986. Insulin receptors of chicken liver and brain: Characterization of α and β subunit properties. Eur. J. Biochem. 158: 125-132. [DOI] [PubMed] [Google Scholar]
- Skeberdis, V.A., Lan, J., Zheng, X., Zukin, R.S., and Bennett, M.V. 2001. Insulin promotes rapid delivery of N-methyl-d-aspartate receptors to the cell surface by exocytosis. Proc. Natl. Acad. Sci. 98: 3561-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt, J.D. 2001. The neuronal MAP kinase cascade: A biochemical signal integration system subserving synaptic plasticity and memory. J. Neurochem. 76: 1-10. [DOI] [PubMed] [Google Scholar]
- Tomizawa, K., Iga, N., Lu, Y.F., Moriwaki, A., Matsushita, M., Li, S.T., Miyamoto, O., Itano, T., and Matsui, H. 2003. Oxytocin improves long-lasting spatial memory during motherhood through MAP kinase cascade. Nat. Neurosci. 6: 384-390. [DOI] [PubMed] [Google Scholar]
- Vincent, A.M., Brownlee, M., and Russell, J.W. 2002. Oxidative stress and programmed cell death in diabetic neuropathy. Ann. NY Acad. Sci. 959: 368-383. [DOI] [PubMed] [Google Scholar]
- Wan, Q., Xiong, Z.G., Man, H.Y., Ackerley, C.A., Braunton, J., Lu, W.Y., Becker, L.E., MacDonald, J.F., and Wang, Y.T. 1997. Recruitment of functional GABA(A) receptors to postsynaptic domains by insulin. Nature 388: 686-690. [DOI] [PubMed] [Google Scholar]
- Watson, G.S. and Craft, S. 2003. The role of insulin resistance in the pathogenesis of Alzheimer's disease: Implications for treatment CNS. Drugs 17: 27-45. [DOI] [PubMed] [Google Scholar]
- Zhao, W., Chen, H., Xu, H., Moore, E., Meiri, N., Quon, M.J., and Alkon, D.L. 1999. Brain insulin receptors and spatial memory. Correlated changes in gene expression, tyrosine phosphorylation, and signaling molecules in the hippocampus of water maze trained rats. J. Biol. Chem. 274: 34893-34902. [DOI] [PubMed] [Google Scholar]
- Zhou, Q., Xiao, M.Y., and Nicoll, R.A. 2001. Contribution of cytoskeleton to the internalization of AMPA receptors. Proc. Natl. Acad. Sci. 98: 1261-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]