Abstract
Rho family G proteins, including Rac and Cdc42, regulate a variety of cellular functions such as morphology, motility, and gene expression. We developed fluorescent resonance energy transfer-based probes which monitored the local balance between the activities of guanine nucleotide exchange factors and GTPase-activating proteins for Rac1 and Cdc42 at the membrane. These probes, named Raichu-Rac and Raichu-Cdc42, consisted of a Cdc42- and Rac-binding domain of Pak, Rac1 or Cdc42, a pair of green fluorescent protein mutants, and a CAAX box of Ki-Ras. With these probes, we video imaged the Rac and Cdc42 activities. In motile HT1080 cells, activities of both Rac and Cdc42 gradually increased toward the leading edge and decreased rapidly when cells changed direction. Under a higher magnification, we observed that Rac activity was highest immediately behind the leading edge, whereas Cdc42 activity was most prominent at the tip of the leading edge. Raichu-Rac and Raichu-Cdc42 were also applied to a rapid and simple assay for the analysis of putative guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) in living cells. Among six putative GEFs and GAPs, we identified KIAA0362/DBS as a GEF for Rac and Cdc42, KIAA1256 as a GEF for Cdc42, KIAA0053 as a GAP for Rac and Cdc42, and KIAA1204 as a GAP for Cdc42. In conclusion, use of these single-molecule probes to determine Rac and Cdc42 activity will accelerate the analysis of the spatiotemporal regulation of Rac and Cdc42 in a living cell.
Ras superfamily G proteins function as molecular switches in a variety of signaling cascades (51). Among them, Rho family G proteins, including Rho, Rac, and Cdc42, are involved in the regulation of a variety of cellular processes, probably through actin cytoskeleton reorganization (1, 9, 13, 48). In a pioneering work by Nobes and Hall, it was shown that Rho regulates the assembly of the actin stress fiber, that Rac induces lamellipodia and membrane ruffles, and that Cdc42 triggers filopodium formation (41).
Rho family G proteins are regulated by three classes of protein, guanine nucleotide exchange factor (GEF), GTPase-activating protein (GAP), and guanine nucleotide dissociation inhibitor (GDI) (51). GEF promotes the exchange of GDP with GTP, which results in the binding of the G proteins to their effector proteins. A typical GEF protein of the Rho family of G proteins consists of a Dbl homology (DH) domain, which exhibits GEF activity, and additional domains that mediate interactions with peptides or lipids. DOCK180, originally isolated as a protein bound to adapter protein Crk (14), also promotes guanine nucleotide exchange of Rac, although it does not contain the DH domain (20). The GTP on the activated Rho family G protein is hydrolyzed in the presence of GAP to resume the GDP-bound inactive state. GDI not only competes with GEF but also holds the Rho family G proteins in the cytoplasm (43). Therefore, the dissociation of GDI is a prerequisite for the membrane association and activation of the Rho family G proteins. ERM family proteins, which are involved in cell-to-cell interaction, release Rho from RhoGDI, translocating Rho from the cytoplasm to membrane (50). Recently, del Pozo and colleagues have shown that integrin stimulation triggers the release of Rac from RhoGDI (5), showing for the first time the involvement of RhoGDI in Rac signaling.
Fluorescent resonance energy transfer (FRET) is a nonradiative transfer of energy between two fluorophores that are placed in close vicinity and in a proper relative angular orientation. Variants of green fluorescent protein (GFP) provide genetically encoded fluorophores that serve as the donor and the acceptor in FRET (15, 33, 35). With the GFP variants and FRET technology, several intracellular events have been visualized in a living cell. Activations of factor Xa and caspases have been monitored by the use of probes in which protease-sensitive peptides are placed between the pair of GFP variants (27, 33, 56). In these probes, activated proteases cleave the probes to dissociate the donor from the acceptor, terminating FRET. In other examples, two GFP variants are fused to a pair of proteins which form a dimer upon stimulation. The binding of Bcl-2 to Bax and the binding of transcription factor Pit-1 to Ets-1 have been demonstrated by detecting FRET between the two GFP variants (4, 28). To overcome the difficulty in controlling the relative expression levels of the two GFP variants in a cell, several research groups have developed single-molecule probes, in which the monitor peptides are sandwiched with the two GFP variants. In these probes, FRET efficiency between the two GFP variants varies depending on either the phosphorylation of (25, 37) or calcium binding to (34, 47) the monitor peptide.
Recently, activation of Rac has been successfully monitored by FRET technology. Kraynov and colleagues used Cdc42- and Rac-interactive binding motif (CRIB) of PAK labeled with Alexa 546 as the acceptor and GFP-tagged Rac as the donor of FRET (24). In a motile Swiss 3T3 cell microinjected with the pair of probes, Kraynov et al. observed an increasing gradient of Rac activity toward the leading edge. Graham and his colleagues expressed CRIB of PAK between two GFP variants (12). Upon binding to the active Rac in vitro, the probe decreases the efficiency of FRET between the two GFP variants.
We have previously reported a probe, called Ras and interacting chimeric unit (Raichu), for monitoring the Ras family G proteins (36). This probe consists of Ras, the Ras-binding domain of Raf, and yellow-emitting (YFP) and cyan-emitting (CFP) GFP mutants. Upon activation of Ras, the binding to Raf increases the efficiency of FRET between CFP and YFP. With Raichu, growth factor-induced activation of Ras and another Ras family G protein, Rap1, in living cells was video imaged. Here, we report on probes named Raichu-Rac and Raichu-Cdc42, which are derived from Raichu and which are applicable to the video imaging of active Rac and Cdc42, respectively. Using these probes, we show that the distributions of active Rac and Cdc42 mostly overlap each other but that high activity at the tip of the leading edge is specific to Cdc42.
MATERIALS AND METHODS
Plasmids.
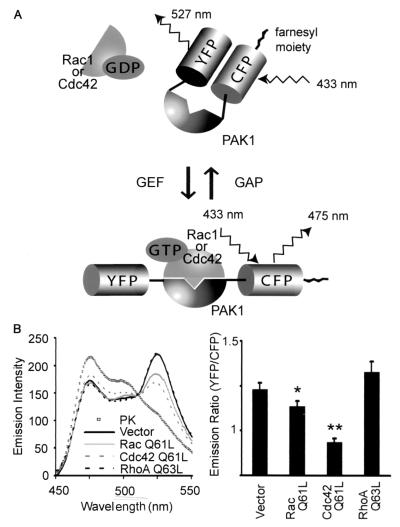
pRaichu-Ras and pRacihu-Rap1 have been described previously (36). pRaichu-Rac and pRaichu-Cdc42 were constructed using essentially the same procedure as was used to construct pRaichu-Ras (36). From the amino terminus, Raichu-Rac consists of YFP (amino acids [aa] 1 to 239), a spacer (Leu-Asp), CRIB of PAK1 (aa 68 to 150), a spacer (Ser-Gly-Gly-Thr-Gly-Gly-Gly-Gly-Thr), Rac1 (aa 1 to 176), a spacer (Gly-Gly-Arg), CFP (aa 1 to 237), a spacer (Gly-Arg-Ser-Arg), and the CAAX box of Ki-Ras (aa 169 to 188) (Fig. 1A). In this study, we used modified YFP (Thr66Gly, Val69Leu, Ser73Ala, Aln70Lys, and Thr204Tyr) and CFP (Lys27Arg, Tyr67Trp, Asp130Gly, Asn147Iso, Met154Thr, Val164Ala, Asn165His, and Ser176Gly) as the acceptor and the donor, respectively. In Raichu-Cdc42, Rac1 was replaced with Cdc42 (aa 2 to 176). In proteins denoted by suffixes V12, N17, and Y40C, Gly12, Thr17, Tyr40 of Rac1 or Cdc42 were replaced with Val, Asn, and Cys, respectively. pRaichu-CRIB was generated by removing the coding sequence of Rac1 from pRaichu-Rac. cDNAs of KIAA0053, KIAA0362, KIAA0793, KIAA1204, KIAA1256, and KIAA1391 were obtained from the Kazusa DNA Institute, Kisarazu, Japan. Synthesized cDNA of dsFP593 (8) was obtained from A. Miyawaki at the Brain Science Institute, RIKEN, Wako-shi, Japan. pIRM21 is an expression vector derived from pCAGGS (40) and contained the internal ribosomal entry site and the coding region of dsFP593 at the 3′ side of the multiple cloning site. Coding sequences of the KIAA proteins were subcloned into the cloning site of pIRM21. pCAGGS-mSos (11), pCAGGS-RasGRF1 (11), and pCAGGS-myc-DOCK180 (14) have been described previously. pSRα-Vav (18) and pcDNA3-HA-Tiam1(C1199) (30) were obtained from H. Mano at Jichi Medical School and J. Collard at The Netherlands Cancer Institute, respectively. In pCXN2-Flag-Rac and pCXN2-Flag-Cdc42, coding regions of human Rac1 and human Cdc42, respectively, were subcloned into eukaryotic expression vector pCXN2-Flag (40). Similarly, the coding regions of Rac1 and Cdc42 were subcloned into pCAGGS-EGFP (42) to generate pCAGGS-EGFP-Rac1 and pCAGGS-EGFP-Cdc42. In proteins denoted by suffixes Q61L and Q63L, Gln61 of Rac1 and Cdc42 and Gln63 of RhoA were replaced with Leu.
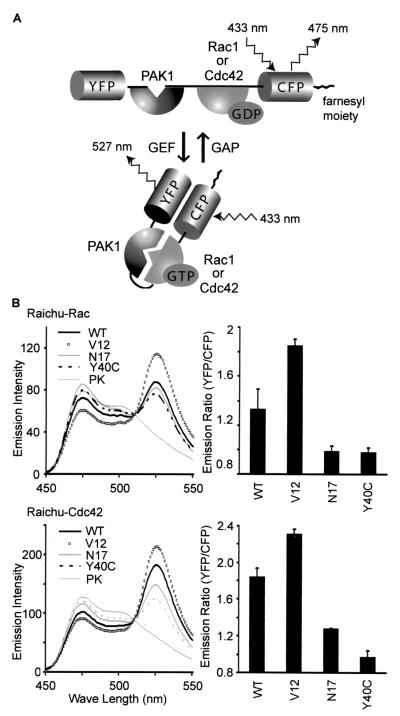
FIG. 1.
Fluorescence profiles of Raichu-Rac and Raichu-Cdc42. (A) Schematic representations of Raichu-Rac and Raichu-Cdc42 bound to GDP or GTP. When Rac1 or Cdc42 is bound to GDP, fluorescence of 475 nm emanates from CFP with excitation at 433 nm. When Rac1 or Cdc42 is bound to GTP, intramolecular binding to PAK1 brings YFP into close proximity to CFP, which causes FRET and fluorescence of 527 nm from YFP. (B) 293T cells expressing Raichu-Rac and Raichu-Cdc42 were lysed and analyzed with a fluorescence spectrometer at an excitation wavelength of 433 nm. WT, V12, N17, and Y40C, wild type, constitutively active mutant, dominant-negative mutant, and effector mutant, respectively. The lysates of the wild type were further treated with proteinase K (PK), which cleaved the probes between YFP and CFP. We used emission intensities of CFP at 475 nm and of YFP at 530 nm to calculate the emission ratio, YFP/CFP (right). Experiments were performed in duplicate, and data are shown with standard deviations. Representative results from three independent experiments are shown.
Recombinant adenoviruses.
Recombinant adenoviruses encoding Raichu-Ras, Raichu-Rap1, Raichu-Rac, and Raichu-Cdc42 were generated by the use of the Adeno-X expression system (Clontech), according to the manufacturer's instructions. First, coding regions of the probes were subcloned into pShuttle by restriction enzyme cleavage and ligation. Then, the expression units including the coding regions of the probes were cleaved out from these pShuttle-derived vectors by PI-SceI and I-CeuI and ligated with pAdeno-X cleaved with the same restriction enzymes. From these pAdeno-X-derived vectors, the genomes of the recombinant adenoviruses were cleaved out by PacI (New England Biolabs) and transfected into HEK293 cells. After 10 to 14 days, cells were harvested to propagate recombinant adenoviruses.
Cells and antibodies.
COS-1, HEK293, and HT1080 cells were purchased from the American Type Culture Collection or the Japan Cell Resource Bank. 293T cells were a gift from B. J. Mayer (University of Connecticut). Cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum. Anti-GFP rabbit serum was developed in our laboratory (36). The anti-Flag M2 monoclonal antibody was purchased from Sigma.
Protein expression, in vitro spectroscopy, and immunoblotting.
Plasmids were transfected into 293T cells by the calcium phosphate coprecipitation method. Thirty-six hours later, cells were harvested in lysis buffer (20 mM Tris-HCl [pH 7.5], 100 mM NaCl, 0.5% Triton X-100, 5 mM MgCl2) and clarified by centrifugation. A fluorescence spectrum was obtained by use of an excitation wavelength of 433 nm with an FP-750 spectrofluorometer (JASCO, Tokyo, Japan). For the demonstration of FRET, the cell lysates were incubated with 100 μg of proteinase K/ml at 37°C for 10 min and analyzed with a spectrometer.
Analysis of guanine nucleotides bound to G proteins.
Guanine nucleotides bound to Raichu-Rac, Raichu-Cdc42, Flag-Rac, and Flag-Cdc42 were analyzed essentially as described previously (29). Briefly, 293T cells that had been transfected with plasmids for 36 h were labeled with 32Pi in phosphate-free minimal essential medium (Life Technologies, Inc.) for 4 h. Cells were lysed in lysis buffer and clarified by centrifugation. Raichu-Rac and Raichu-Cdc42 were immunoprecipitated with an anti-GFP antibody, and Flag-tagged proteins were immunoprecipitated with an anti-Flag M2 monoclonal antibody. The immunoprecipitates were boiled and analyzed by thin-layer chromatography (TLC). The amount of 32Pi-labeled guanine nucleotides was quantitated with a BAS-1000 image analyzer (Fuji Film, Tokyo, Japan).
Imaging of HT1080 cells.
HT1080 cells were transfected with plasmids with FuGene6 (Roche). Forty-eight hours after transfection, cells were imaged on an Olympus IX70 inverted microscope equipped with a cooled charge-coupled device camera, CoolSNAP HQ (Roper Scientific, Trenton, N.J.), controlled by MetaMorph software (Universal Imaging, West Chester, Pa.) (34, 36). For dual-emission ratio imaging of Raichu-Rac and Raichu-Cdc42, we used a 440AF21 excitation filter, a 455DRLP dichroic mirror, and two emission filters, 480AF30 for CFP and 535AF25 for YFP (Omega Optical Inc., Brattleboro, Vt.). Cells were illuminated with a 75-W xenon lamp through a 10% neutral density (ND) filter (Omega Optical Inc.) and a 60× oil immersion objective lens. The exposure time was 0.5 s when binning was set to 3 by 3. After background subtraction, the ratio image of YFP/CFP was created with the MetaMorph software and used to represent FRET efficiency. For the photobleaching experiment, cells were illuminated without an ND filter for 10 min. In this experiment, we used an MX510 excitation filter (Asahi Spectra Co., Tokyo, Japan).
Imaging of moving HT1080 cells.
HT1080 cells were infected with recombinant adenoviruses as described previously (17) or transfected with plasmids by using FuGene6 (Roche). Forty-eight hours after infection or transfection, cells were replated onto a 35-mm-diameter collagen-coated glass base dish (Asahi Techno Glass Co., Tokyo, Japan). Beginning 1 h after replating, cells were imaged every 2 min for 2 h as already described.
Analysis of GEF and GAP activity in 96-well plates.
COS-1 cells were plated to collagen-coated, glass bottom 96-well plates (Asahi Techno Glass Co.). Cells in each well were transfected with 50 ng of pRaichu-Rac or pRaichu-Cdc42 in combination with 100 ng of expression vectors encoding GEFs or GAPs with Polyfect (Qiagen). Eight wells were used for each sample. After 24 h, cells were fed with serum-free minimal essential medium and incubated for an additional 6 h, followed by imaging as already described, except that a 10× objective lens was used. From a single viewing field, FRET images of more than 30 cells were routinely obtained by using the autothreshold function of Metamorph. Values of area and fluorescence intensity for each cell were saved after background reduction. Further analysis was performed with Excel software (Microsoft). The following five steps were automated by a custom-made macro program that ran on the Excel program. First, data from cellular debris or aggregated cells were automatically removed by gating with area values. Second, the ratio of fluorescent intensity of YFP to that of CFP was calculated for each cell. Third, the average of the YFP/CFP ratio was calculated for each well. Fourth, the average and standard deviation were calculated from the YFP/CFP ratios for the eight wells that were transfected with the same set of plasmids. Finally, these data were used to draw a bar graph.
Nucleotide sequence accession numbers.
The nucleotide sequences of the coding regions of pRaichu-Rac and pRaichu-Cdc42 were deposited in the DDBJ/EMBL/GenBank data base with accession no. AB074145 and AB074146, respectively.
RESULTS
In vivo probes for Rac and Cdc42 activities.
We designed a protein named Raichu-Rac that consisted of Rac1, CRIB of PAK, and the pair of YFP and CFP (Fig. 1A). At the carboxy terminus, the CAAX box of Ki-Ras was fused to locate the probe to the membrane. In this probe, we expected that the intramolecular binding of GTP-Rac1 to CRIB brought CFP close to YFP, increasing FRET from CFP to YFP. By replacing Rac1 with Cdc42, we also made a probe named Raichu-Cdc42. To examine whether GTP loading increased FRET efficiency, we prepared several mutants. In the mutants denoted with the suffix V12, Gly12 was replaced with Val to inactivate the GTPase activity, resulting in the increased GTP loading in the mutants. In N17 mutants, Thr17 was replaced with Asn; this mutation is known to reduce the affinity of G proteins for guanine nucleotides (32). In Y40C mutants, Cys was substituted for Tyr40 in the effector domain of Rac or Cdc42, which is essential for the binding to Pak1 (26).
The probes were expressed in 293T cells, and their fluorescence emission profiles were obtained by the use of an excitation wavelength of 433 nm (Fig. 1B). Because FRET was most typically observed as an increase in an emission peak of 530 nm and a decrease in an emission peak of 475 nm, the emission ratio of 530 nm/475 nm is used to represent the FRET efficiency. As shown in Fig. 1B, right, the emission ratios for wild-type Raichu-Rac and Raichu-Cdc42 were lower than those for V12 mutants and higher than those for the N17 and Y40C mutants. The results demonstrated that increases in the emission ratios of Raichu-Rac and Raichu-Cdc42 reflected the binding of GTP-Rac and GTP-Cdc42 to CRIB. The cell lysates were further incubated with proteinase K, which cleaved the probes between YFP and CFP. The disappearance of the 530-nm peak confirmed FRET from CFP to YPF in the absence of proteinase K (Fig. 1B, left).
GTP loading of the probes and the authentic G proteins.
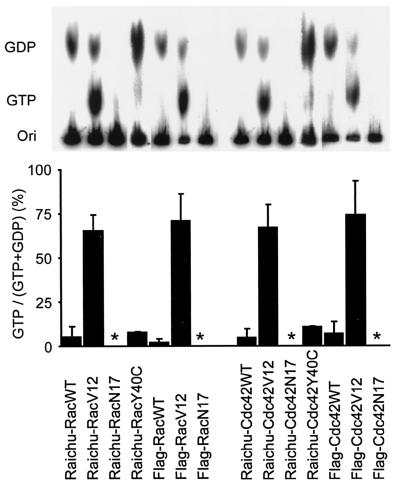
To examine whether the GTP loading on Raichu probes correlated with those of the authentic proteins, 293T cells expressing Raichu-Rac, Flag-Rac, Raichu-Cdc42, or Flag-Cdc42 were labeled with 32Pi. Then, the labeled recombinant G proteins were immunoprecipitated, and the guanine nucleotides bound to them were analyzed by TLC (Fig. 2). The amount of GTP on Raichu-Rac-WT, Flag-Rac-WT, and Raichu-Rac-Y40C was less than 10%, whereas the amount of GTP on Raichu-Rac-V12 and Flag-Rac-V12 was about 80% (Fig. 2, left). In Rac N17 mutants, the binding of guanine nucleotides was not detectable, which reflected the reduced affinity for the guanine nucleotides due to the Asn17 mutation (7). Very similar results were obtained with Raichu-Cdc42 and Flag-Cdc42 (Fig. 2, right). These results validated the use of Raichu-Rac and Raichu-Cdc42 for monitoring Rac and Cdc42 activities in living cells.
FIG. 2.
GTP/GDP loading of Raichu-Rac and Raichu-Cdc42. 293T cells expressing Raichu-Rac, Raichu-Cdc42, Flag-tagged Rac, and Flag-tagged Cdc42 were labeled with 32Pi. The G proteins were precipitated with either anti-GFP rabbit serum or an anti-Flag monoclonal antibody, followed by TLC analysis. GTP and GDP were quantitated with a BAS-1000 image analyzer, and values of GTP/(GTP + GDP) with standard deviations were plotted. Asterisks indicate that no 32Pi-labeled guanine nucleotides were detected.
Imaging of Rac1 and Cdc42 activities in HT1080 cells.
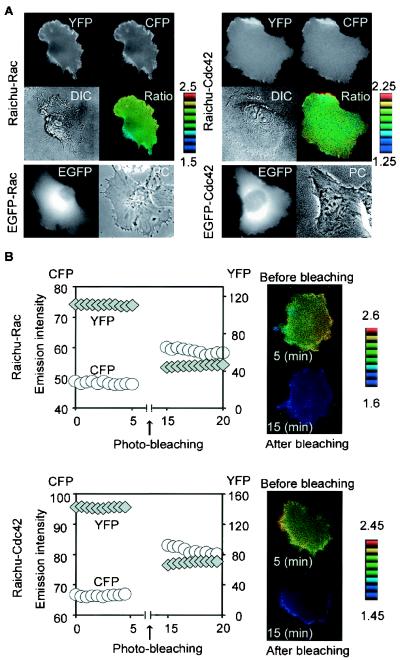
Next, we used Raichu-Rac and Raichu-Cdc42 for cell imaging. HT1080 cells expressing Raichu-Rac or Raichu-Cdc42 were imaged for YFP (535 ± 12 nm) and CFP (480 ± 15 nm) with an excitation wavelength of 440 ± 10 nm (Fig. 3A). In both the YFP and CFP images, Raichu-Rac and Raichu-Cdc42 were detected diffusely within the cells, suggesting that they were localized mostly to the plasma membrane via the farnesyl moiety at the carboxyl terminus (Fig. 3A). The YFP/CFP ratio image was used to show the FRET efficiency in the intensity-modulated display mode.
FIG. 3.
Imaging of Rac and Cdc42 activities in HT1080 cells. (A) HT1080 cells were transfected with expression vectors as indicated on the left. After 48 h, cells expressing EGFP-Rac or EGFP-Cdc42 were photographed for EGFP and phase contrast (PC) images. Cells expressing Raichu-Rac or Raichu-Cdc42 were imaged for YFP, CFP, and differential interference contrast (DIC). In the IMD mode images (Ratio), eight colors from red to blue are used to represent the YFP/CFP ratio, with the intensity of each color indicating the mean intensity of YFP and CFP. The upper and lower limits of the ratio range are shown on the right. (B) Increase in the CFP emission by YFP photobleaching. HT1080 cells expressing Raichu-Rac or Raichu-Cdc42 were photobleached at an excitation wavelength of 510 nm for 10 min. Emission intensities of CFP and YFP in this cell were monitored every 30 s and are shown at the left. Cell images of pre- and postbleaching are shown at the right.
In contrast to Raichu-Rac and Raichu-Cdc42, the authentic Rac1 and Cdc42 tagged with enhanced GFP (EGFP) were condensed around the nucleus (Fig. 3A). This distribution agreed with the cytoplasmic localization of GDP-Rac1 and GDP-Cdc42 coupled with RhoGDI (43). Thus, for the interpretation of the following image data, we emphasize that Raichu-Rac and Raichu-Cdc42 do not directly detect active Rac1 and Cdc42 but that they image the local balance of GEFs and GAPs for Rac1 and Cdc42 at the membrane compartments.
The occurrence of FRET in the living cells was confirmed by a photobleaching experiment as described previously (36). Intensities of both YFP and CFP were monitored before and after photobleaching. As expected, photobleaching of YFP resulted in a marked increase in CFP intensity (Fig. 3B).
Activities of Rac, Cdc42, Ras, and Rap1 in motile HT1080 cells.
To understand the role of Rac and Cdc42 in cell migration, we examined the activities of Rac, Cdc42, Ras, and Rap1 in motile HT1080 cells that were infected with the recombinant adenoviruses encoding the Raichu probes. All of the cell images presented below are extracted from video images that are presented on our website (http://www-tv/biken.osaka-u.ac.jp/rei/).
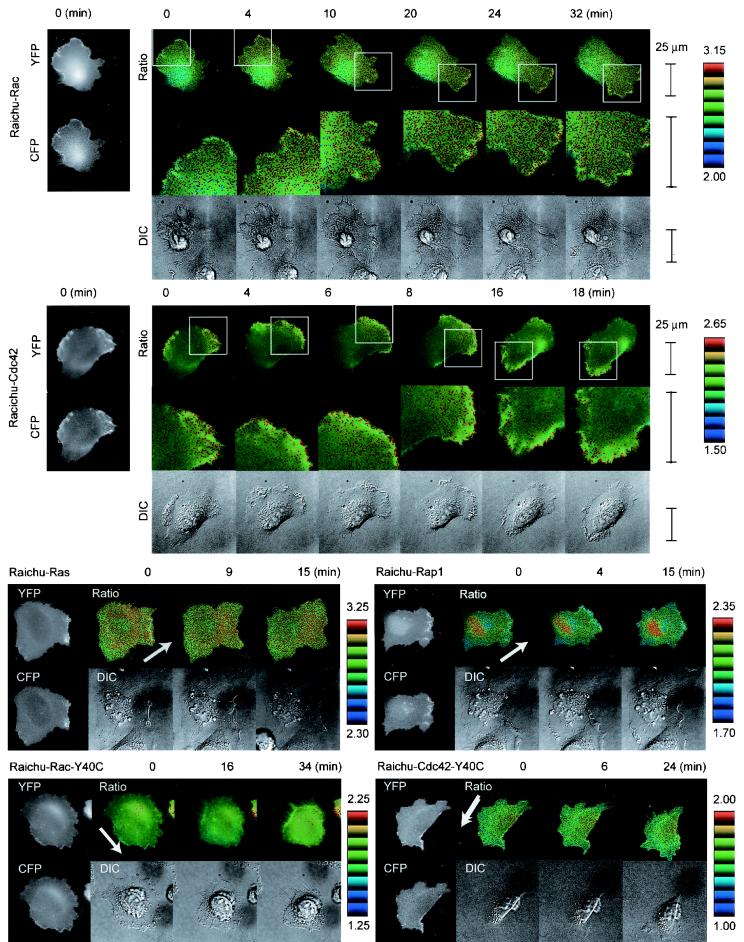
For the video imaging of HT1080 cells for up to several hours, we minimized the exposure time to prevent YFP from photobleaching. Consequently, we had to compromise with the resolution of the images and fluctuation of the ratio data of each pixel. Therefore, the mosaic of ratio images was made up of pixels from blue to red, which corresponded from the lowest to the highest FRET efficiency. We interpreted the increasing number of red pixels toward the leading edge as reflecting the gradual increase in Rac activity (Fig. 4). When the cell changed direction and the plasma membrane started to withdraw, the Rac activity decreased rapidly. In a higher magnification, we noticed that Rac activity was highest immediately behind the leading edge (Fig. 4).
FIG. 4.
Activity of Rac and Cdc42 in a motile HT1080 cell video imaged by using Raichu-Rac and Raichu-Cdc42. HT1080 cells infected with recombinant adenoviruses coding Raichu probes were replated onto a collagen-coated glass base dish. Beginning 1 h after replating, CFP, YFP, and differential interference contrast (DIC) images were obtained every 2 min with a time-lapse microscope. YFP/CFP ratio images were created to represent FRET efficiency, which correlated with the activities of the G proteins. Representative YFP, CFP, YFP/CFP (Ratio), and DIC images obtained at the indicated time points are shown. Arrows point in the direction of cell migration. The middle panels show magnified FRET images of the leading edges (boxes in the top panels). The upper and lower limits of the ratio range are shown on the right. The original video images are presented on our website (http://www-tv/biken.osaka-u.ac.jp/rei/). Experiments were performed at least five times for each probe, and similar results were obtained.
Cdc42 activity also increased toward the leading edge (Fig. 4). In the higher magnification, however, high Cdc42 activity is most concentrated at the tip of the leading edge (Fig. 4). In cells expressing Raichu-Rac-Y40C or Raichu-Cdc42-Y40C, we could not detect any gradient of the emission ratio, confirming that the intramolecular binding of CRIB to Rac or Cdc42 generated the gradient of FRET efficiency.
The activity of Ras was higher at the peripheral region, but there was no difference between the leading and the tailing edges. Rap1 activity was higher around the nucleus. Again, we could not observe a remarkable difference between the leading and the tailing edges, indicating that the gradual increase toward the leading edge was specific to the activities of Rac and Cdc42.
Development of Raichu-CRIB.
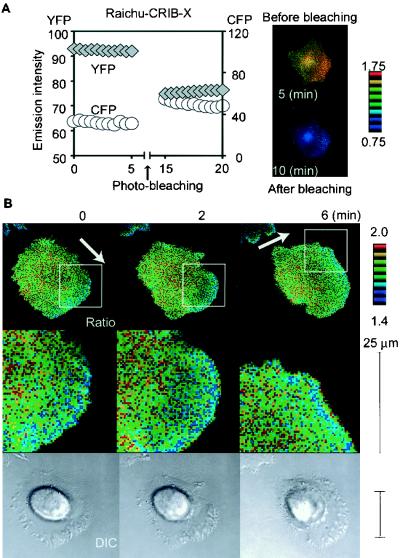
RhoGDI recognizes the carboxy termini of geranyl-geranylated Rho family G proteins (10, 16). Because the carboxy terminus of Raichu-Rac and Raichu-Cdc42 consisted of the CAAX box of Ki-Ras, it was predictable that RhoGDI did not bind to Raichu-Rac or Raichu-Cdc42. Indeed, we did not observe the effect of overexpressed RhoGDI on the FRET efficiency of Raichu-Rac or Raichu-Cdc42 (data not shown). To compensate for this defect, we prepared another probe consisting of CRIB, YFP, and CFP as reported previously (Fig. 5A) (12). This probe, named Raichu-CRIB, was coexpressed with constitutively active Rac1, Cdc42, or RhoA in 293T cells. As shown in Fig. 5B, the FRET efficiency of Raichu-CRIB was decreased only in the presence of the active Rac1 or active Cdc42, not in the presence of the active RhoA. Therefore, in this Raichu-CRIB probe, the decrease in FRET indicated the binding to the active Cdc42 or Rac1. Notably, Rac1 decreased the FRET efficiency of Raichu-CRIB less efficiently than did Cdc42. This probably reflected the previous observation that the affinity of Rac1 for Pak is weaker than that of Cdc42 (52). The occurrence of FRET was confirmed by the proteinase K treatment as already described.
FIG. 5.
Fluorescence profile of Raichu-CRIB. (A) Schematic representations of Raichu-CRIB and Raichu-CRIB-X with or without GTP-Rac or -Cdc42. The farnesyl moiety was fused only to Raichu-CRIB-X. When the probe is free, fluorescence at 527 nm by FRET is observed. When the probe is bound to GTP-Rac or GTP-Cdc42, YFP is displaced from CFP and FRET is inhibited. (B) 293T cells transfected with pRaichu-CRIB and pEB6-Rac1-Q61L, pEB6-Cdc42-Q61L, or pEB6-RhoA-Q63L were lysed and analyzed with a fluorescence spectrometer at an excitation wavelength of 433 nm (left). The lysates of the wild type were further treated with proteinase K (PK), which cleaved the probes between YFP and CFP. Mean YFP/CFP ratios are shown with standard error bars at the right. Single and double asterisks indicate that the differences between control and Rac1-Q61L and between control and Cdc42-Q61L, respectively, were statistically significant by t test (P < 0.05 and P < 0.01, respectively).
Activities of Rac and Cdc42 monitored by Raichu-CRIB-X.
In HT1080 cells, Raichu-CRIB localized diffusely in the cytoplasm and there was no difference in its FRET efficiency throughout the cells (data not shown). We speculated that the excess of the probe and/or the limited binding to Rac and/or Cdc42 resulted in the failure to detect the Rac and Cdc42 activities. To increase the percentage of the probe that participated in the binding to Rac and Cdc42, the CAAX box of Ki-Ras was fused to the carboxy terminus of CFP, generating Raichu-CRIB-X (Fig. 5A). First, the occurrence of FRET in Raichu-CRIB-X-expressing cells was confirmed by a photobleaching experiment as already described. As expected, photobleaching of YFP resulted in a marked increase in the CFP intensity of the cells expressing Raichu-CRIB-X (Fig. 6A). Then, using Raichu-CRIB-X, we imaged the activities of Rac and Cdc42 in a motile HT1080 cell (Fig. 6B). We found that the FRET efficiency gradually decreased toward the leading edge, indicating that the binding of CRIB to the endogenous Rac and/or Cdc42 gradually increased toward the leading edge. This observation was in complete agreement with findings obtained by using Raichu-Rac and Raichu-Cdc42 and also implied that RhoGDI did not contribute significantly to the spatial gradient of the Rac and Cdc42 activity in a motile HT1080 cell. Alternately, RhoGDI activity may have been cooperatively regulated with GEFs and GAPs to produce a gradient of Rac and Cdc42 activity.
FIG. 6.
Activity of Rac and Cdc42 video imaged with Raichu-CRIB-X. (A) Increase in the CFP emission by YFP photobleaching. HT1080 cells expressing Raichu-CRIB-X were photobleached at an excitation wavelength of 510 nm for 10 min. Emission intensities of CFP and YFP in these cells (left) were monitored every 30 s. Cell images of pre- and postbleaching are also shown (right). (B) HT1080 cells transfected with pRaichu-CRIB-X were replated onto a collagen-coated glass base dish. Beginning 1 h after replating, CFP, YFP, and differential interference contrast (DIC) images were obtained every 2 min with a time-lapse microscope as described in the legend to Fig. 4. Arrows point in the direction of cell migration. The upper and lower limits of the ratio range are shown on the right. The original video images are presented on our website (:http://www-tv/biken.osaka-u.ac.jp/rei/).
Analysis of GEF and GAP in 96-well plates with Raichu-Rac and Raichu-Cdc42.
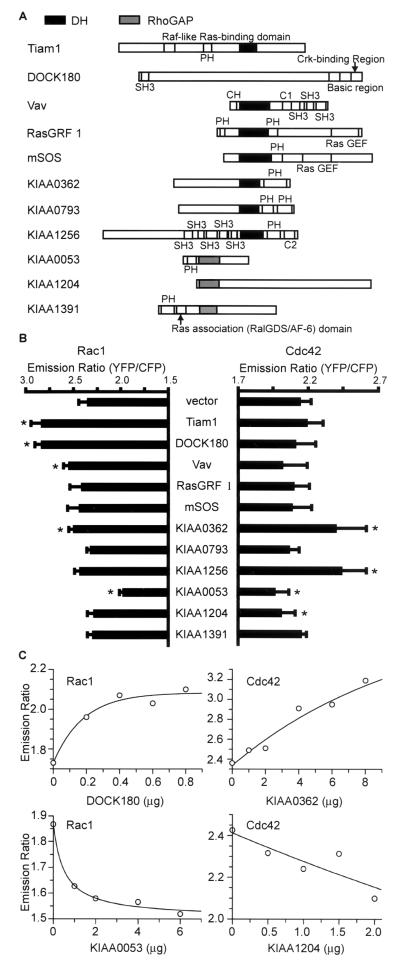
We set up a rapid and simple cell-based assay for the measurement of GEF and GAP activities by the use of Raichu-Rac and Raichu-Cdc42. For this purpose, we removed the CAAX box from Raichu-Rac and Raichu-Cdc42, because CAAX-negative probes were expressed more abundantly in 293T cells than were CAAX-positive probes. To test the validity of our assay system, we obtained six putative GEFs and GAPs from the HUGE cDNA library of the Kazusa DNA Institute. As controls, we used Tiam-1, DOCK180, Vav, Ras-GRF1, and mSos-1, all of which contain the DH domain. The deduced domain structures of these GEFs and GAPs are shown schematically in Fig. 7A. The cDNAs were cotransfected with CAAX-negative pRaichu-Rac or pRaichu-Cdc42 into COS-1 cells, which were grown in glass bottom 96-well plates (Fig. 7B). The emission ratio of Raichu-Rac increased remarkably in the presence of Tiam-1 and DOCK180 and slightly in the presence of Vav and KIAA0362, demonstrating the GEF activity of these proteins toward Rac. GAP activity in response to Rac was detected only in KIAA0053. GEF activity in response to Cdc42 was detected in KIAA0362 and KIAA1256, and GAP activity in response to Cdc42 was detected in KIAA0053 and KIAA1204. Thus, we could examine the specificity of putative GEFs and GAPs by this simple method with Raichu probes.
FIG. 7.
Analysis of GEF and GAP activities. (A) Schematic representation of GEFs and GAPs used in this study. Abbreviations: PH, pleckstrin homology domain; DH, DH domain; SH2 and SH3, Src homology domains 2 and 3. (B) COS-1 cells plated on glass bottom 96-well plates were transfected with CAAX-negative pRaichu-Rac or pRaichu-Cdc42 and expression vectors for the proteins listed at the center. Cells were imaged, and the YFP/CFP ratio was determined as described in the text. Error bars, standard deviations. Asterisks, samples that show a difference from the control vector with statistical significance by t test (P < 0.01). (C) CAAX-negative pRaichu-Rac or pRaichu-Cdc42 was coexpressed in 293T cells with various quantities of pCAGGS-DOCK180, pIRM21-KIAA0362/DBS, pIRM21-KIAA0053, and pIRM21-KIAA1204. Cells were lysed and examined for emission ratio as for Fig. 1B.
Dose response to GEFs and GAPs.
Finally, we confirmed the results obtained by the 96-well-plate-based assay with a fluorescence spectrometer. We expressed in 293T cells various quantities of DOCK180, KIAA0362/DBS, KIAA0053, and KIAA1204 with Raichu probes and examined the dose response. As shown in Fig. 7C, the FRET efficiency of Raichu-Rac increased with an increasing amount of DOCK180. Similarly, the FRET efficiency of Raichu-Cdc42 correlated with the amount of KIAA0362/DBS. In contrast, expression of KIAA0053 and KIAA1204 effectively decreased the emission ratio of Raichu-Rac and Raichu-Cdc42, respectively, in a dose-dependent manner.
DISCUSSION
Probes based on the GFP and FRET technology have enormous potential for monitoring the intracellular signal transduction cascades in living cells (53). Nevertheless, the number of probes developed has not been increasing as might be expected. There are two problems to be overcome. When the assay system consists of two probes which are bound covalently to their respective donor and acceptor fluorophores, the relative amounts of the two probes cannot be controlled in most systems. Thus, bleed-through fluorescence, which emanates from the donor and which is detected in the acceptor channel, must be accurately compensated for in the estimation of FRET efficiency (2). A single-gene-encoded probe, in contrast, contains both the donor and acceptor; therefore, simply monitoring the ratio of donor fluorescence to acceptor fluorescence is sufficient. The problem with this type of probe resides in its construction. Typically, the probe is designed so that the signal-induced conformational change of the probe brings CFP into close proximity with YFP. The increases in FRET efficiency for most prototype probes, however, are less than 10% and do not apply to cell imaging. As for the first in vivo probe for Ras family G protein Raichu-Ras (36), where Ras is placed at the amino terminus of the Ras-binding domain of Raf, we originally placed Rac at the amino terminus of CRIB. In this construct, the difference in FRET efficiency between the wild-type and the G12V mutants was less than 10%. Only when we placed Rac or Cdc42 at the carboxy terminus of CRIB did we detect an increase in FRET in the G12V mutants. Thus, currently we cannot tell whether G proteins should be placed at the amino terminus or carboxy terminus of the binding partner in probe designs.
Rho family G proteins are regulated by three classes of proteins, GEF, GAP, and GDI. GDI controls the partitioning of the Rho family G proteins between the cytoplasm and cell membrane (43). By placing the probes constitutively at the membrane, the effect of GDI could not be monitored by Raichu-Rac and Raichu-Cdc42. Therefore, the activities of Rac and Cdc42 monitored by these probes are correctly expressed as the balance between GEF and GAP activities at the cell membrane. Although the regulatory role of GDI has been recently emerging (5, 50), it is widely accepted that many signaling cascades regulate Rac and Cdc42 by changing the balance of GEF and GAP activities at the cell membrane (51). Thus, Raichu-Rac and Raichu-Cdc42 provide useful tools to unravel the signaling cascades connected to Rac and Cdc42 pathways.
Irrespective of the loss of sensitivity to GDI, we fused the probes to the CAAX box of Ki-Ras for the following four reasons. First, active Rac and Cdc42 are detected only at the membrane compartments (43), providing a rationale for placing the probe only at the membrane. Second, we have found that an excess of cytosolic probes profoundly decreases the signal-to-noise ratio during the development of a probe for tyrosine phosphorylation (25). The same happened to Raichu-CRIB. To improve the signal-to-noise ratio, we needed to place Raichu-CRIB at the membrane. Third, CAAX-less probes diffuse throughout the cytoplasm within 10 s. Considering that the rate constants of most GEFs are between 10−3 and 10−2/s, we needed to restrict the movement of probes to obtain the spatial information (25). Last, we found that probes fused with the CAAX box of H-Ras or the myristylation signal of Src often accumulated at the Golgi apparatus, deteriorating the ratio image in the central region. In our hands, the CAAX box of Ki-Ras delivered the probes most uniformly to the membrane compartments.
We observed that the FRET efficiency of wild-type Raichu-Rac or Raichu-Cdc42 was remarkably higher than those of N17 and Y40C mutants (Fig. 1B) or than those in the presence of GAPs (Fig. 7B). This observation appears inconsistent with biochemical data. Because the GTP/GDP ratio of the wild-type Rac or Cdc42 was less than 10% in TLC analysis (Fig. 2), a further decrease in the GTP/GDP ratio of the wild-type Rac or Cdc42 should not significantly decrease their FRET efficiencies. This discrepancy probably reflects high GAP activity in the cell lysates, which results in the hydrolysis of GTP bound to Rac and Cdc42 during the immunoprecipitation before TLC analysis (3). Indeed, we observed that the FRET efficiency of the wild-type Raichu-Rac or Raichu-Cdc42 in the cell lysates decreased with time (data not shown).
It has been richly documented that Rac and Cdc42 induce lamellipodia and filopodia, respectively (51). Nevertheless, the mechanism underlying these unique morphological outputs remains elusive (51). Notably, both Rac and Cdc42 activate the two well-characterized signaling cascades that directly regulate actin polymerization, the PAK-LIM kinase-cofilin (49, 57) and N-WASP/WAVE-Arp2/3 (31, 46) pathways. Thus, it is likely that a difference in the distributions of active Rac and Cdc42 leads to distinct morphological changes. Indeed, such a possibility was supported by our observation that the activity of Rac was highest immediately behind the leading edge, whereas the Cdc42 activity was highest at the tip of the leading edge (Fig. 4). Moreover, this observation suggests that the leading-edge formation may be initiated by Cdc42 activation, followed by Rac activation. On the other hand, the gradual increase toward the leading edge is a feature common to Rac and Cdc42. This finding agrees with the presence of many GEFs and GAPs that act on both Rac and Cdc42 (51). In conclusion, our findings imply that both dual- and monospecific GEFs and GAPs contribute to the spatial regulation of Rac and Cdc42 in a motile HT1080 cell.
Raichu-Rac and Raichu-Cdc42 enabled us to rapidly identify the substrate specificity of putative GEFs and GAPs, the numbers of members of which have been expanding enormously with the progress of genome sequencing projects (51). For the demonstration of GEF or GAP activity in vitro, purified proteins are required; however, GEFs and GAPs are often insoluble in Escherichia coli. Alternately, to avoid their expression in E. coli, the putative GEFs or GAPs are expressed in mammalian cells and partially purified by immunoprecipitation for the in vitro analysis (3). In this case, preparation of a specific antibody or epitope tagging is required. We subcloned cDNAs of putative GEFs and GAPs into a eukaryotic expression vector using the same restriction enzymes that were employed for the construction of the KIAA cDNA libraries (38), minimizing the task of subcloning. The substrate specificities of the putative GEFs and GAPs were readily determined by coexpressing them with Raichu-Rac or Raichu-Cdc42 and examining FRET efficiency in COS-1 cells; no further manipulation was necessary. We are currently developing Raichu-Rho, a monitor for RhoA (H. Yoshizaki and M. Matsuda, unpublished data); therefore, the use of Raichu-Rho, Rac, and Cdc42 will cover the majority of Rho family G proteins and accelerate the characterization of GEFs and GAPs for the Rho family G proteins.
Among the three putative GEF proteins tested, KIAA0362 and KIAA1256 are found to be identical to DBS (54) and intersectin-2 (44), respectively. DBS/KIAA0362 has been shown to promote the guanine nucleotide exchange of RhoA and Cdc42 (55). Although GEF activity of intersectin-2/KIAA1256 has not been reported, a related protein, intersectin-1, has been demonstrated to be a GEF for Cdc42. Our observation is in agreement with these previous reports, except that KIAA0362/DBS slightly stimulated Rac in our assay. This difference may be because we expressed the entire protein, whereas only the Dbl domain was used in the previous study (55). Alternately, this observation may simply reflect the fact that the substrate specificities of some GEFs and GAPs under in vitro conditions differ from those under and in vivo conditions. For instance, p50 RhoGAP stimulates GTPase activities of Rho, Rac, and Cdc42 in vitro but it stimulates only Rho in vivo (45).
Among the known activators of Rac or Cdc42 tested, we failed to detect the GEF activity of Ras-GRF1 and Sos-1. This observation probably reflects tight regulation of these GEFs in the cells. Ras-GRF1 is activated only in the presence of the βγ subunit or by phosphorylation (21, 22). Similarly, Rac GEF activity of Sos-1 requires phosphatidylinositol 3-kinase activation (39). Therefore, the assay system with Raichu-Rac or Raichu-Cdc42 appears to reflect the in vivo enzyme activity of GEFs and GAPs. This property will enable us to study the regulatory mechanism of GEFs and GAPs in living cells by a simple imaging technique with Raichu-Rac and Raichu-Cdc42.
Finally, it should be remembered that every monitoring system for Rho family G proteins has some defects. For example, due to the high GAP activity in the cell lysates, the GTP/GDP ratios of the authentic Rac and Cdc42 are barely measurable by the classical TLC analysis (3, 20). As a simple and safer alternative, Bos's pull-down method has been applied to detect the activation of Rac and Cdc42 in cells (6, 23); however, none has ever shown that the activity detected by this method correlates with the increases in the GTP/GDP ratios of Rac and Cdc42 in the cells. Considering the inefficient recovery rate of the GTP-bound form (19), the results obtained by the pull-down method have to be interpreted with careful attention. Raichu-Rac and Raichu-Cdc42 also suffer from the defect that they cannot monitor RhoGDI activity. Raichu-CRIB complements this defect, but possesses the flaw that it may competitively interfere the endogenous Rac and Cdc42 signaling cascades. Indeed, we found that overexpression of Raichu-CRIB was apparently toxic to the cells. Another demerit of Raichu-CRIB is its reactivity with both Rac and Cdc42, rendering the interpretation of the data complicated. Thus, Raichu-CRIB may be used as an auxiliary method for Raichu-Rac and Raichu-Cdc42 to evaluate the effect of RhoGDI.
In conclusion, because none of the other methods can currently obtain spatiotemporal information on Rac and Cdc42 activity in living cells, the FRET-based probes reported in this study will accelerate research to delineate the spatiotemporal regulation of Rac and Cdc42 in living cells.
Acknowledgments
We thank A. Miyawaki, Y. Takai, H. Mano, J. M. Adams, J. Collard, S. Hattori, L. Feig, J. Miyazaki, and B. J. Mayer for plasmids and N. Fujimoto, N. Yoshida, and Y. Matsuura for technical assistance.
This work was supported in part by grants from the Ministry of Education, Science, Sports and Culture, the Ministry of Health and Welfare, and the Health Science Foundation of Japan.
REFERENCES
- 1.Bar-Sagi, D., and A. Hall. 2000. Ras and Rho GTPases: a family reunion. Cell 103:227-238. [DOI] [PubMed] [Google Scholar]
- 2.Chamberlain, C. E., V. S. Kraynov, and K. M. Hahn. 2000. Imaging spatiotemporal dynamics of Rac activation in vivo with FLAIR. Methods Enzymol. 325:389-400. [DOI] [PubMed] [Google Scholar]
- 3.Crespo, P., K. E. Schuebel, A. A. Ostrom, J. S. Gutkind, and X. R. Bustelo. 1997. Phosphotyrosine-dependent activation of Rac-1 GDP/GTP exchange by the vav proto-oncogene product. Nature 385:169-172. [DOI] [PubMed] [Google Scholar]
- 4.Day, R. N. 1998. Visualization of Pit-1 transcription factor interactions in the living cell nucleus by fluorescence resonance energy transfer microscopy. Mol. Endocrinol. 12:1410-1419. [DOI] [PubMed] [Google Scholar]
- 5.del Pozo, M. A., W. B. Kiosses, N. B. Alderson, N. Meller, K. M. Hahn, and M. A. Schwartz. 2002. Integrins regulate GTP-Rac localized effector interactions through dissociation of Rho-GDI. Nat. Cell Biol. 4:232-239. [DOI] [PubMed] [Google Scholar]
- 6.del Pozo, M. A., L. S. Price, N. B. Alderson, X. D. Ren, and M. A. Schwartz. 2000. Adhesion to the extracellular matrix regulates the coupling of the small GTPase Rac to its effector PAK. EMBO J. 19:2008-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feig, L. A., and G. M. Cooper. 1988. Inhibition of NIH3T3 cell proliferation by a mutant ras protein with preferential affinity for GDP. Mol. Cell. Biol. 8:3235-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fradkov, A. F., Y. Chen, L. Ding, E. V. Barsova, M. V. Matz, and S. A. Lukyanov. 2000. Novel fluorescent protein from Discosoma coral and its mutants possesses a unique far-red fluorescence. FEBS Lett. 479:127-130. [DOI] [PubMed] [Google Scholar]
- 9.Geiger, B., and A. Bershadsky. 2001. Assembly and mechanosensory function of focal contacts. Curr. Opin. Cell Biol. 13:584-592. [DOI] [PubMed] [Google Scholar]
- 10.Gosser, Y. Q., T. K. Nomanbhoy, B. Aghazadeh, D. Manor, C. Combs, R. A. Cerione, and M. K. Rosen. 1997. C-terminal binding domain of Rho GDP-dissociation inhibitor directs N-terminal inhibitory peptide to GTPases. Nature 387:814-819. [DOI] [PubMed] [Google Scholar]
- 11.Gotoh, T., Y. Niino, M. Tokuda, O. Hatase, S. Nakamura, M. Matsuda, and S. Hattori. 1997. Activation of R-Ras by ras-guanine nucleotide-releasing factor. J. Biol. Chem. 272:18602-18607. [DOI] [PubMed] [Google Scholar]
- 12.Graham, D. L., P. N. Lowe, and P. A. Chalk. 2001. A method to measure the interaction of Rac/Cdc42 with their binding partners using fluorescence resonance energy transfer between mutants of green fluorescent protein. Anal. Biochem. 296:208-217. [DOI] [PubMed] [Google Scholar]
- 13.Hall, A., and C. D. Nobes. 2000. Rho GTPases: molecular switches that control the organization and dynamics of the actin cytoskeleton. Phil. Trans. R. Soc. Lond. B Biol. Sci. 355:965-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasegawa, H., E. Kiyokawa, S. Tanaka, K. Nagashima, N. Gotoh, M. Shibuya, T. Kurata, and M. Matsuda. 1996. DOCK180, a major CRK-binding protein, alters cell morphology upon translocation to the membrane. Mol. Cell. Biol. 16:1770-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heim, R., and R. Y. Tsien. 1996. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr. Biol. 6:178-182. [DOI] [PubMed] [Google Scholar]
- 16.Hoffman, G. R., N. Nassar, and R. A. Cerione. 2000. Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell 100:345-356. [DOI] [PubMed] [Google Scholar]
- 17.Kanegae, Y., G. Lee, Y. Sato, M. Tanaka, M. Nakai, T. Sakaki, S. Sugano, and I. Saito. 1995. Efficient gene activation in mammalian cells by using recombinant adenovirus expressing site-specific Cre recombinase. Nucleic Acids Res. 23:3816-3821. [DOI] [PMC free article] [PubMed]
- 18.Katzav, S., D. Martin-Zanca, and M. Barbacid. 1989. vav, a novel human oncogene derived from a locus ubiquitously expressed in hematopoietic cells. EMBO J. 8:2283-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura, K., T. Tsuji, Y. Takada, T. Miki, and S. Narumiya. 2000. Accumulation of GTP-bound RhoA during cytokinesis and a critical role of ECT2 in this accumulation. J. Biol. Chem. 275:17233-17236. [DOI] [PubMed] [Google Scholar]
- 20.Kiyokawa, E., Y. Hashimoto, S. Kobayashi, H. Sugimura, T. Kurata, and M. Matsuda. 1998. Activation of Rac1 by a Crk SH3-binding protein, DOCK180. Genes Dev. 12:3331-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiyono, M., Y. Kaziro, and T. Satoh. 2000. Induction of rac-guanine nucleotide exchange activity of Ras-GRF1/CDC25(Mm) following phosphorylation by the nonreceptor tyrosine kinase Src. J. Biol. Chem. 275:5441-5446. [DOI] [PubMed] [Google Scholar]
- 22.Kiyono, M., T. Satoh, and Y. Kaziro. 1999. G protein betagamma subunit-dependent rac-guanine nucleotide exchange activity of ras-GRF1/CDC25(Mm). Proc. Natl. Acad. Sci. USA 96:4826-4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi, S., T. Shirai, E. Kiyokawa, N. Mochizuki, M. Matsuda, and Y. Fukui. 2001. Membrane recruitment of DOCK180 by binding to PtdIns(3,4,5,)P3. Biochem. J. 354:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kraynov, V. S., C. Chamberlain, G. M. Bokoch, M. A. Schwartz, S. Slabaugh, and K. M. Hahn. 2000. Localized rac activation dynamics visualized in living cells. Science 290:333-337. [DOI] [PubMed] [Google Scholar]
- 25.Kurokawa, K., N. Mochizuki, Y. Ohba, H. Mizuno, A. Miyawaki, and M. Matsuda. 2001. A pair of FRET-based probes for tyrosine phosphorylation of the CrkII adapter protein in vivo. J. Biol. Chem. 276:31305-31310. [DOI] [PubMed] [Google Scholar]
- 26.Lamarche, N., N. Tapon, L. Stowers, P. D. Burbelo, P. Aspenstrom, T. Bridges, J. Chant, and A. Hall. 1996. Rac and Cdc42 induce actin polymerization and G1 cell cycle progression independently of p65(PAK) and the JNK/SAPK MAP kinase cascade. Cell 87:519-529. [DOI] [PubMed] [Google Scholar]
- 27.Mahajan, N. P., D. C. Harrison-Shostak, J. Michaux, and B. Herman. 1999. Novel mutant green fluorescent protein protease substrates reveal the activation of specific caspases during apoptosis. Chem. Biol. 6:401-409. [DOI] [PubMed] [Google Scholar]
- 28.Mahajan, N. P., K. Linder, G. Berry, G. W. Gordon, R. Heim, and B. Herman. 1998. Bcl-2 and Bax interactions in mitochondria probed with green fluorescent protein and fluorescence resonance energy transfer. Nat. Biotechnol. 16:547-552. [DOI] [PubMed] [Google Scholar]
- 29.Matsuda, M., Y. Hashimoto, K. Muroya, H. Hasegawa, T. Kurata, S. Tanaka, S. Nakamura, and S. Hattori. 1994. CRK binds to two guanine nucleotide-releasing proteins for the Ras family and modulates nerve growth factor-induced activation of Ras in PC12 cells. Mol. Cell. Biol. 14:5495-5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michiels, F., G. G. Habets, J. C. Stam, R. A. van der Kammen, and J. G. Collard. 1995. A role for Rac in Tiam1-induced membrane ruffling and invasion. Nature 375:338-340. [DOI] [PubMed] [Google Scholar]
- 31.Miki, H., T. Sasaki, Y. Takai, and T. Takenawa. 1998. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature 391:93-96. [DOI] [PubMed] [Google Scholar]
- 32.Miki, T. 1995. Interaction of ect2 and Dbl with Rho-related GTPases. Methods Enzymol. 256:90-98. [DOI] [PubMed] [Google Scholar]
- 33.Mitra, R. D., C. M. Silva, and D. C. Youvan. 1996. Fluorescence resonance energy transfer between blue-emitting and red-shifted excitation derivatives of the green fluorescent protein. Gene 173:13-17. [DOI] [PubMed] [Google Scholar]
- 34.Miyawaki, A., J. Llopis, R. Heim, J. M. McCaffery, J. A. Adams, M. Ikura, and R. Y. Tsien. 1997. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388:882-887. [DOI] [PubMed] [Google Scholar]
- 35.Mizuno, H., A. Sawano, P. Eli, H. Hama, and A. Miyawaki. 2001. Red fluorescent protein from Discosoma as a fusion tag and a partner for fluorescence resonance energy transfer. Biochemistry 40:2502-2510. [DOI] [PubMed] [Google Scholar]
- 36.Mochizuki, N., S. Yamashita, K. Kurokawa, Y. Ohba, T. Nagai, A. Miyawaki, and M. Matsuda. 2001. Spacio-temporal images of growth factor-induced activation of Ras and Rap1. Nature 411:1065-1068. [DOI] [PubMed] [Google Scholar]
- 37.Nagai, Y., M. Miyazaki, R. Aoki, T. Zama, S. Inouye, K. Hirose, M. Iino, and M. Hagiwara. 2000. A fluorescent indicator for visualizing cAMP-induced phosphorylation in vivo. Nat. Biotechnol. 18:313-316. [DOI] [PubMed] [Google Scholar]
- 38.Nagase, T., N. Seki, K. Ishikawa, M. Ohira, Y. Kawarabayasi, O. Ohara, A. Tanaka, H. Kotani, N. Miyajima, and N. Nomura. 1996. Prediction of the coding sequences of unidentified human genes. VI. The coding sequences of 80 new genes (KIAA0201-KIAA0280) deduced by analysis of cDNA clones from cell line KG-1 and brain. DNA Res. 3:321-354. [DOI] [PubMed] [Google Scholar]
- 39.Nimnual, A. S., B. A. Yatsula, and D. Bar-Sagi. 1998. Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science 279:560-563. [DOI] [PubMed] [Google Scholar]
- 40.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-200. [DOI] [PubMed] [Google Scholar]
- 41.Nobes, C. D., and A. Hall. 1995. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81:53-62. [DOI] [PubMed] [Google Scholar]
- 42.Ohba, Y., N. Mochizuki, K. Matsuo, S. Yamashita, M. Nakaya, Y. Hashimoto, M. Hamaguchi, T. Kurata, K. Nagashima, and M. Matsuda. 2000. Rap2 as a slowly responding molecular switch in the Rap1 signaling cascade. Mol. Cell. Biol. 20:6074-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olofsson, B. 1999. Rho guanine dissociation inhibitors: pivotal molecules in cellular signalling. Cell Signal. 11:545-554. [DOI] [PubMed] [Google Scholar]
- 44.Pucharcos, C., C. Casas, M. Nadal, X. Estivill, and S. de la Luna. 2001. The human intersectin genes and their spliced variants are differentially expressed. Biochim. Biophys. Acta 1521:1-11. [DOI] [PubMed] [Google Scholar]
- 45.Ridley, A. J., A. J. Self, F. Kasmi, H. F. Paterson, A. Hall, C. J. Marshall, and C. Ellis. 1993. rho family GTPase activating proteins p190, bcr and rhoGAP show distinct specificities in vitro and in vivo. EMBO J. 12:5151-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rohatgi, R., L. Ma, H. Miki, M. Lopez, T. Kirchhausen, T. Takenawa, and M. W. Kirschner. 1999. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell 97:221-231. [DOI] [PubMed] [Google Scholar]
- 47.Romoser, V. A., P. M. Hinkle, and A. Persechini. 1997. Detection in living cells of Ca2+-dependent changes in the fluorescence emission of an indicator composed of two green fluorescent protein variants linked by a calmodulin-binding sequence. A new class of fluorescent indicators. J. Biol. Chem. 272:13270-13274. [DOI] [PubMed] [Google Scholar]
- 48.Sastry, S. K., and K. Burridge. 2000. Focal adhesions: a nexus for intracellular signaling and cytoskeletal dynamics. Exp. Cell Res. 261:25-36. [DOI] [PubMed] [Google Scholar]
- 49.Sumi, T., K. Matsumoto, Y. Takai, and T. Nakamura. 1999. Cofilin phosphorylation and actin cytoskeletal dynamics regulated by Rho- and Cdc42-activated LIM-kinase 2. J. Cell Biol. 147:1519-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takahashi, K., T. Sasaki, A. Mammoto, K. Takaishi, T. Kameyama, S. Tsukita, and Y. Takai. 1997. Direct interaction of the Rho GDP dissociation inhibitor with ezrin/radixin/moesin initiates the activation of the Rho small G protein. J. Biol. Chem. 272:23371-23375. [DOI] [PubMed] [Google Scholar]
- 51.Takai, Y., T. Sasaki, and T. Matozaki. 2001. Small GTP-binding proteins. Physiol Rev. 81:153-208. [DOI] [PubMed] [Google Scholar]
- 52.Thompson, G., D. Owen, P. A. Chalk, and P. N. Lowe. 1998. Delineation of the Cdc42/Rac-binding domain of p21-activated kinase. Biochemistry 37:7885-7891. [DOI] [PubMed] [Google Scholar]
- 53.Tsien, R. Y., and A. Miyawaki. 1998. Seeing the machinery of live cells. Science 280:1954-1955. [DOI] [PubMed] [Google Scholar]
- 54.Whitehead, I., H. Kirk, and R. Kay. 1995. Retroviral transduction and oncogenic selection of a cDNA encoding Dbs, a homolog of the Dbl guanine nucleotide exchange factor. Oncogene 10:713-721. [PubMed] [Google Scholar]
- 55.Whitehead, I. P., Q. T. Lambert, J. A. Glaven, K. Abe, K. L. Rossman, G. M. Mahon, J. M. Trzaskos, R. Kay, S. L. Campbell, and C. J. Der. 1999. Dependence of Dbl and Dbs transformation on MEK and NF-κB activation. Mol. Cell. Biol. 19:7759-7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu, X., A. L. V. Gerard, B. C. Huang, D. C. Anderson, D. G. Payan, and Y. Luo. 1998. Detection of programmed cell death using fluorescence energy transfer. Nucleic Acids Res. 26:2034-2035. [DOI] [PMC free article] [PubMed]
- 57.Yang, N., O. Higuchi, K. Ohashi, K. Nagata, A. Wada, K. Kangawa, E. Nishida, and K. Mizuno. 1998. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature 393:809-812. [DOI] [PubMed] [Google Scholar]