Abstract
Brd4 belongs to the BET family of nuclear proteins that carry two bromodomains implicated in the interaction with chromatin. Expression of Brd4 correlates with cell growth and is induced during early G1 upon mitogenic stimuli. In the present study, we investigated the role of Brd4 in cell growth regulation. We found that ectopic expression of Brd4 in NIH 3T3 and HeLa cells inhibits cell cycle progression from G1 to S. Coimmunoprecipitation experiments showed that endogenous and transfected Brd4 interacts with replication factor C (RFC), the conserved five-subunit complex essential for DNA replication. In vitro analysis showed that Brd4 binds directly to the largest subunit, RFC-140, thereby interacting with the entire RFC. In line with the inhibitory activity seen in vivo, recombinant Brd4 inhibited RFC-dependent DNA elongation reactions in vitro. Analysis of Brd4 deletion mutants indicated that both the interaction with RFC-140 and the inhibition of entry into S phase are dependent on the second bromodomain of Brd4. Lastly, supporting the functional importance of this interaction, it was found that cotransfection with RFC-140 reduced the growth-inhibitory effect of Brd4. Taken as a whole, the present study suggests that Brd4 regulates cell cycle progression in part by interacting with RFC.
The bromodomain is a conserved motif that is present in many nuclear regulatory factors and forms four α helices (13, 46; F. Jeanmougin, J. M. Wurtz, B. Le Douarin, P. Chambon, and R. Losson, Letter, Trends Biochem. Sci. 22:151-153, 1997). Structural studies of the bromodomains in PCAF, GCN5 (32), and TAF250 indicate that they have a binding affinity for acetylated lysines and acetylated histone H4 (13, 23), leading to the proposal that the domain acts as a chromatin-targeting module (46). Proteins that carry a bromodomain(s) generally possess additional unique motifs that are associated with distinct functions. Based on these and other features, bromodomain-carrying proteins are classified into several families (Jeanmougin et al., letter). They include the SWI/SNF family, the transcriptional coactivator family, and the histone acetylase family.
The BET family constitutes another distinct group of bromodomain proteins. Although their functions are poorly understood, proteins of this family share a characteristic structural feature: they have two bromodomains and an additional ET domain (37; Jeanmougin et al., letter). Drosophila melanogaster fsh is the oldest known member of the BET family (14, 21). In Saccharomyces cerevisiae, there are two BET family proteins, bdf1 and bdf2, which are similar in structure (7, 29). While each gene alone is not essential, disruption of both bdf1 and bdf2 is lethal (29). Bdf1 is part of a general transcription factor complex, playing a role in transcription (28). In addition, previous genetic analysis indicated that it plays a role in cell growth regulation (7). In mammals, there are five genes that encode members of the BET family (2, 12, 25, 35, 38). Under the guidance of the human and mouse genome nomenclature committees, these genes have recently been redesignated brd1 through brd5. Brd2, previously known as Ring3/fsrg1, is a component of the transcription mediator (24). It is expressed in proliferating cells in mouse tissues and leukemia cells in humans (35) and may have a kinase activity (9, 10). Less is known about Brd1, Brd3, and Brd5 (25, 38; Jeanmougin et al., letter). Brd4 is a novel member of the family, which we reported previously under the tentative designation of MCAP (mitotic chromosome-associated protein) (12). Both mouse and human Brd4/mcap encode a 200-kDa nuclear protein which has a signature BET motif in the N-terminal region and a large C-terminal region (see Fig. 6A). We have previously shown that Brd4 is expressed in dividing cells and tissues and is induced in response to growth stimuli. The Brd4 protein associates with condensed chromosomes during mitosis, a feature similarly found in the yeast bdf1 (7, 12).
FIG. 6.
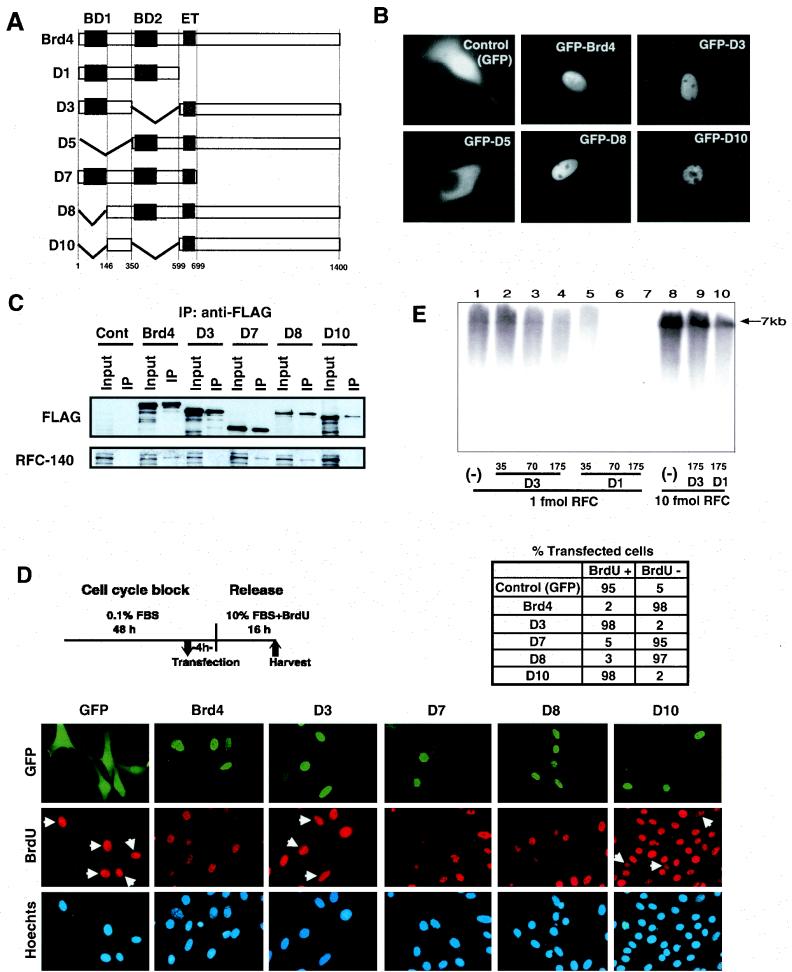
Brd4 domains required for the interaction with RFC and inhibition of S-phase progression. (A) Diagram of Brd4 deletion constructs. BD, bromodomain; ET, ET domain. Solid boxes represent the presence of indicated domains, and diagonal lines represent the absence of indicated domains. (B) Subcellular localization of GFP-Brd4. NIH 3T3 cells (105) were transfected with 2 μg of the indicated constructs, and GFP localization was visualized by microscopy 24 h after transfection. (C) HeLa cells synchronized at G1 were transfected with empty pCMV-Flag vector (Cont, control), pCMV-Flag full-length Brd4, or the indicated deletion constructs and harvested at 16 h. Extracts were precipitated with anti-Flag antibody and analyzed for RFC-140 and Flag-Brd4 by immunoblot analysis. Input represents Flag-Brd4 in 8% of the extracts. (D) Inhibition ofBrdU uptake by ectopically expressed Brd4 deletion constructs. Serum-starved NIH 3T3 cells were transfected with 4 μg of EGFP-Brd4 by Lipofectamine-Plus 4 h prior to release. Cells were allowed to proceed to S phase in the presence of BrdU (10 μM) for 16 h and were fixed and stained for BrdU (diagram at top left). FBS, fetal bovine serum. The number of cells with GFP-Brd4 (green) with or without BrdU incorporation (red) was counted from five independent fields. All cells were detected by Hoechst DNA stain (blue). A total of ∼150 cells was counted in each field. The table at top right indicates the percentage of BrdU-positive cells in the total number of GFP-positive cells. (E) Influence of recombinant Brd4 deletion constructs on the elongation of a singly primed DNA template by the DNA polymerase δ holoenzyme. DNA elongation reactions were carried out with 1 fmol (lanes 1 to 7) and 10 fmol (lanes 8 to 10) of RFC. Thirty-five, 70, 175, and 175 ng of D3 was added to lanes 2, 3, 4 and 9, respectively, and 35, 70, 175, and 175 ng of D1 was added to lanes 5, 6, 7 and 10, respectively. The DNA synthesized in each reaction (in picomoles) was as follows: 9.81 in lane 1, 13.7 in lane 2, 9.82 in lane 3, 9.51 in lane 4, 8.01 in lane 5, 5.01 in lane 6, 2.61 in lane 7, 17.0 in lane 8, 16.4 in lane 9, and 12.2 in lane 10. In the absence of RFC, no DNA synthesis was observed (data not shown).
The aim of this study was to delineate the role of Brd4 in cell growth regulation and to begin to address the possible mechanism of the regulation. As an initial approach to this goal, we have used ectopic-expression strategies. Transient, stable, and tetracycline (TET)-inducible Brd4 expressions all led to growth inhibition in NIH 3T3 cells. Analysis of synchronized cells showed that ectopic expression of Brd4 inhibits progression from G1 to S phase. Similar cell cycle inhibitory effects were seen in HeLa cells following Brd4 transfection. We show in this paper that Brd4 interacts with replication factor C (RFC) in vivo and in vitro. RFC is a component of the conserved replication machinery (6, 17, 30, 41, 44) that is essential for DNA replication (11, 15, 36). In vitro DNA elongation reactions that depend on RFC were inhibited by recombinant Brd4, providing a possible explanation for the effect of Brd4. Cotransfection with RFC-140 counteracted the growth-inhibitory effect of ectopic Brd4, further suggesting the functional significance of the Brd4-RFC interaction. As additional evidence supporting the significance of the interaction, it was found that deletion of a bromodomain that abolished the interaction with RFC also abolished inhibition of S-phase entry by ectopic Brd4. Together, the present results indicate that Brd4, by interacting with RFC, participates in the regulation of cell cycle progression from G1 to S.
MATERIALS AND METHODS
Plasmids and antibodies.
Full-length murine Brd4 cDNA (12) and Brd4 deletion mutants were subcloned into the following vectors: pCMV2-Flag (Kodak), pcxn2 (3), pTRE (Clontech), pEGFP-C1, pEGFP-N3 (Clontech), and pAcHLTC (PharMingen). Internal deletions of Brd4 were constructed from full-length MCAP cloned in pBluescript II SK+ (Stratagene) with EcoRI sites at both termini of the cDNA. Deletions were introduced by digestion through appropriate restriction sites with partial fill-in by T4 DNA polymerase, mung bean nuclease, and religation. Deletions and in-frame sequences without undesired mutations were confirmed by DNA sequencing. Murine RFC-140 cDNA was cloned into pcDNA3.1 (Invitrogen). Rabbit antibodies to Brd4 were as described previously (12). Rabbit antibody to RFC-140 was raised against the bacterial recombinant RFC-140 protein fused to the maltose-binding protein and affinity purified. Antibodies to small subunits of RFC were as described previously (41). Antibodies to PCNA, YY1 and green fluorescent protein (GFP) were obtained from Santa Cruz, Flag M2 was obtained from Sigma, bromodeoxyuridine (BrdU) was obtained from PharMingen, and second antibodies were obtained from Amersham.
Cell culture, transfection, and monitoring of cell growth.
NIH 3T3 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% calf serum and were synchronized by serum starvation. Cells were cultured in the presence of 0.1% serum for 48 to 72 h and then released to the complete medium for designated periods. Transfection was performed with Lipofectamine or Lipofectamine-Plus (Life Technologies). In some experiments, transfected cells were separated by cotransfecting cells with pCMX-IL-2R and then sorting transfected cells with antibody to IL-2R (3, 31). TET-inducible Brd4 expression was performed as follows. NIH 3T3 clones were generated following transfection of pTk-hygro and pTRE (Clontech) containing full-length Brd4 into cells that had been transfected with pTet-On vector (Clontech). Brd4 expression was induced by doxycyline (DOX) at specified doses for specified periods. Cell yields, BrdU incorporation, and cell cycle profiles were monitored by using previously described procedures (12, 31) and are detailed in the legends to the figures. HeLa cells were cultured in the same medium as described above supplemented with 10% fetal bovine serum and synchronized by double-thymidine block (12).
BrdU staining.
Synchronized NIH 3T3 cells transfected with GFP-Brd4 deletion mutants were incubated with 10 μM BrdU for 16 h and fixed with 4% paraformaldehyde. Cells were blocked in a blocking buffer (phosphate-buffered saline containing 0.1% Tween and 3% bovine serum albumin) for 1 h, incubated in 2 N HCl for 10 min, washed and permeabilized in 0.25% Triton X for 5 min, and incubated with anti-BrdU antibody at 37°C for 1 h and then with rhodamine-conjugated anti-mouse immunoglobulin G for 1 h. Cells were counterstained with Hoechst 33342 (1 μg/ml) for 3 min. Stained cells were viewed by using a Zeiss Axiophot microscope with a 63× planachromat objective.
Coimmunoprecipitation assays.
Preparations of nuclear extracts from NIH 3T3 cells and coimmunoprecipitation analysis were performed according to a previously described method (19), with minor modifications. Briefly, 60 μl (∼300 to 500 μg) of protein G precleared extracts were incubated with anti-Brd4 or anti-RFC-140 antibody overnight at 4°C and mixed with 50 μl of protein G- Sepharose beads for 1 h. HeLa cells transfected with Flag-Brd4 were synchronized at G1 and harvested at G1/S, and extracts were prepared following lysis in buffer containing 20 mM HEPES (pH 7.9), 0.2 M NaCl, 1 mM EGTA (pH 7), 1 mM EDTA (pH 8), 0.6% NP-40, 10% glycerol, 1 mM dithiothreitol, 1 mM Na3VO4, 50 mM NaF, 0.5 mM phenylmethylsulfonyl fluoride, and 1 mM sodium molybdate and dialyzed in buffer containing 20 mM HEPES (pH 7.9), 20% glycerol, 0.1 M KCl, and 0.2 mM EDTA (pH 8). About 150 μg of extracts was incubated with 30 μl of anti-Flag affinity gel (Sigma) at 4°C for 4 h. Bound materials were eluted with 2× sodium dodecyl sulfate (SDS) sample buffer, analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) on a 4 to 20% gradient gel, and immunoblotted with the designated antibodies.
Brd4-RFC interaction in vitro.
The in vitro coupled transcription-translation reaction of the cDNAs encoding the five subunits of human RFC were carried out with the TNT Quick Coupled Transcription/Translation System (Promega) according to the manufacturer's protocols. The cDNAs used in the reactions have been previously described and have been shown to produce biologically active RFC (41). Hexahistidine-tagged, recombinant Brd4 was produced by cloning corresponding cDNA fragments into pAcHLTC baculovirus vector (PharMingen) and was purified on Ni2+ resin beads (Qiagen) (3). The in vitro transcription-translation reaction was conducted by mixing specified amounts of purified recombinant Brd4, 0.35 μg of each RFC subunit plasmid, 60 μl of TNT Quick reticulocyte lysate, and 45 μCi of l-[35S]methionine (1,000 Ci/mmol; NEN) and incubating the mixture at 30°C for 90 min. DNase I (750 U; Boehringer) was then added, and the mixture was incubated for 10 min at 30°C. Antibodies (1 μl) specific for either the N-terminal sequence of Brd4 or the p37 subunit of RFC were added to 20-μl aliquots of the in vitro transcribed-translated RFC reaction mixture, which was then incubated on ice for 1 h. The immunocomplexes were diluted fivefold, bound to 10 μl of protein A-Sepharose beads (Upstate Biotechnology) equilibrated with wash buffer (25 mM Tris-Cl [pH 7.5], 10% glycerol, 5 mM EDTA, 1 mM dithiothreitol, 0.15 M NaCl, 0.5% NP-40, 1% bovine serum albumin), incubated for 30 min at 4°C, and then washed five times. Bound materials were separated by SDS-12% PAGE, and bands were visualized by autoradiography.
In vitro replication assay.
The replication assay using singly primed M13 DNA as the template was carried out as previously described (41). Six femtomoles of a singly primed DNA template, 0.5 pmol of PCNA, 250 ng of RPA, and 0.5 U of DNA polymerase δ were mixed with specified amounts of RFC and a recombinant Brd4 deletion mutant, D1 or D3, in a total reaction mixture of 20 μl for 30 min at 37°C. An aliquot was used to measure labeled nucleotide incorporation, and an aliquot was subjected to alkaline-agarose gel electrophoresis. After being dried, the gel was subjected to autoradiography.
RESULTS
Ectopic Brd4 expression inhibits cell growth.
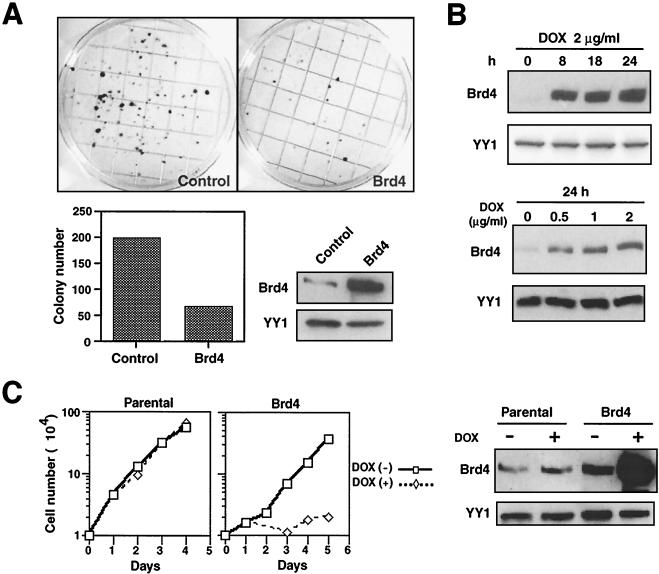
To explore the role of Brd4 in cell growth, NIH 3T3 cells were stably transfected with an expression vector for Brd4 and selected for G418 and colony formation was examined two weeks later. As shown in Fig. 1A, the number of colonies that arose from Brd4-transfected cells was much fewer than that from cells transfected with a control vector (<30%). Immunoblot analysis of pooled Brd4 transfectants (Fig. 1A) verified an increase in Brd4 expression compared to that in control transfectants. Similar growth-inhibitory effects were seen in other cell types including HeLa cells transfected with Brd4 (data not shown), providing the first indication that ectopic Brd4 expression is growth inhibitory.
FIG. 1.
Ectopic Brd4 expression inhibits cell growth. (A) Reduced colony formation by NIH 3T3 cells expressing ectopic Brd4. Cells (105) were transfected with an empty vector, pcxn (Control) or pcxn2-Brd4 and selected in G418 for 2 weeks (upper panels). Colonies were stained with methylene blue. The graph shows the number of colonies per plate in cells transfected with the control or Brd4. Also shown are the results of immunoblot analysis of Brd4 protein expression in control (pcxn2) or pcxn2-Brd4 transfectants (lower right panels). YY1 was used as a control for protein loading throughout this work. (B) TET-inducible Brd4 expression. TET-inducible cells were treated with DOX at the indicated doses for the indicated hours, and Brd4 expression was detected by immunoblot analysis. (C) TET-induced inhibition of cell growth. Parental or TET-inducible cells were treated with DOX (2 μg/ml) or left untreated, and cell numbers were counted on the indicated days (left panels). Inducible Brd4 expression was monitored by immunoblot analysis (right panels).
NIH 3T3 cells stably expressing ectopic Brd4 grew slowly and could not be maintained for extended periods of time, making a detailed study difficult. To further analyze the growth inhibition observed with Brd4, we established a TET-inducible Brd4 expression system (1) in NIH 3T3 cells. The results with a representative TET-inducible clone (Fig. 1B) showed that Brd4 expression was induced within 8 h after the addition of DOX in a dose-dependent manner. The low level of Brd4 expression seen before the addition of DOX represents the expression of the endogenous protein. Figure 1C shows a comparison of the cell yields from the parental clone with those from the inducible Brd4 clone over 4 days of DOX treatment. Cell growth was markedly attenuated in the inducible Brd4 clone following the addition of DOX, although the same clone grew rapidly in the absence of DOX. At the end of 4 days, cell yields from DOX-treated cultures were 20- to 50-fold less than those from untreated cultures. In contrast, parental cells grew exponentially over the same period, regardless of whether cells were treated with DOX or left untreated. The results of immunoblot analysis (Fig. 1C, right) confirmed that Brd4 was induced in a DOX-dependent manner in the inducible cells, while the endogenous Brd4 expression in parental cells was unaffected by DOX treatment during the same period. Three additional TET-inducible Brd4 clones also showed DOX-inducible inhibition of cell growth (data not shown). These results show that ectopic Brd4 expression negatively affects cell growth.
Ectopic Brd4 inhibits cell cycle progression from G1 to S phase.
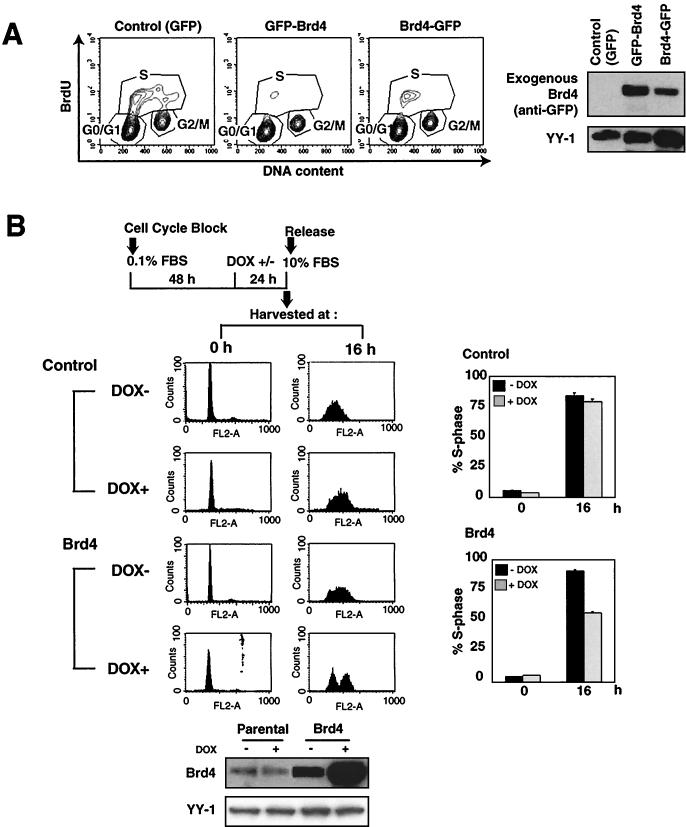
To study the role of Brd4 in cell cycle regulation, NIH 3T3 cells were transiently transfected with the Brd4 fusion constructs containing GFP either at the N or the C terminus (Fig. 2A) along with the IL-2R vector. Transfected cells were separated from untransfected cells by sorting IL-2R-positive cells (3, 31) and were then allowed to grow for an additional 10 h. They were pulse-labeled with BrdU, and cell cycle profiles were analyzed by flow cytometry for DNA content and BrdU incorporation (Fig. 2A). In the control transfection with GFP alone, 35% of the cells were in S phase and incorporated BrdU (Fig. 2A). In contrast, among cells transfected with Brd4 fusion constructs containing GFP (both GFP-Brd4 and Brd4-GFP), only about 8% to 13% of cells were BrdU positive, indicating an inhibition of S-phase entry. Immunoblot analysis of sorted cells confirmed ectopic Brd4 expression in transfected cells (Fig. 2A, right). This effect was reproducible and was caused by Brd4 itself and not by GFP, since Brd4 without GFP gave the same outcome (see Fig. 5). These data suggest that ectopic Brd4 expression negatively affects cell cycle progression from G1 to S.
FIG. 2.
Cell cycle analysis. (A) Reduced S-phase cells after transient Brd4 transfection. NIH 3T3 cells (2 × 106) were transfected with 30 μg of GFP-Brd4, Brd4-GFP, or control pEGFP-Cl (GFP only) along with 3 μg of pCMX-IL-2R (left panels). Twenty-four hours after transfection, cells were labeled with BrdU (10 μM) for 30 min and then sorted by anti-IL-2R antibody (GFP+ cells, ∼95%). Nuclei were double stained with fluorescein isothiocyanate-conjugated anti-BrdU antibody and PI and analyzed by flow cytometry. Also shown are the results of immunoblot detection of GFP-Brd4 with anti-GFP antibody (right panels). (B) The parental and TET-inducible cells were synchronized by serum starvation, and Brd4 was induced by DOX 24 h prior to release. Cells were harvested at 0 and 16 h after release, stained with PI, and analyzed by flow cytometry. The panels at the right show the percentage of the S-phase population. Values represent the averages from three independent experiments plus standard deviations. Panels at the bottom show the results of immunoblot detection of Brd4 expression.
FIG. 5.
RFC-140 overexpression alleviates Brd4 inhibition of G1/S transition. (A) Partial restoration of cell growth by RFC-140. TET-inducible Brd4 cells (2 × 106) were transfected with 30 μg of pcDNA-RFC-140 or empty vector along with 3 μg of pCMX-IL-2R. Transfected cells were separated by sorting and induced by treatment with DOX for 2 days. The graph indicates relative cell recovery, with the cell yield from untreated cells taken to be 100%. Values are the means from three independent experiments. The mean cell recoveries shown in lanes 1 to 4 were 6.3 × 106, 3.2 × 106, 7.2 × 106, and 6.1 × 106, respectively. The panels below the graph show the results of immunoblot detection of RFC-140 and Brd4. (B) Counterregulation of S-phase entry by RFC-140 and Brd4. Cells (2 × 106) were transfected with 15 μg of pcxn2-Brd4 and/or 15 μg of pcDNA-RFC-140 along with 3 μg of pcMX-IL-2R. Cells were allowed to proceed to S phase for 24 h andwere labeled with 10 μM BrdU for 30 min and then sorted as described in the legend to Fig. 2A. Cells were stained with fluorescein isothiocyanate-conjugated anti-BrdU antibody and PI and were analyzed by flow cytometry. The panels at the bottom show the results of immunoblot detection of Brd4 and RFC-140 in transfected cells.
To further assess a cell cycle stage at which Brd4 exerts its inhibitory activity, experiments were performed with synchronized, TET-inducible cells. As diagrammed in Fig. 2B, parental and TET-inducible Brd4 cells were serum starved for 72 h and treated with DOX beginning at 48 h of starvation. Cells were then released to a serum-rich medium and allowed to proceed to G1 and S for the subsequent periods. Figure 2B shows cell cycle profiles at 0 and 16 h after release. The great majority of parental cells that had been arrested at G0 at the end of starvation proceeded to S at 16 h after the addition of serum, irrespective of DOX treatment. In contrast, about half of the DOX-treated Brd4-inducible cells remained at G1 at 16 h, indicating an inhibition of S-phase entry (Fig. 2B, right graphs). The other half of the DOX-treated cells entered S at this time. The variability of the levels of Brd4 induction may account for the incomplete inhibition. Brd4-inducible cells not treated with DOX, however, proceeded to S phase, verifying that it is the ectopic Brd4 expression that inhibited S-phase entry. DOX induction of Brd4 expression was confirmed by immunoblot analysis (Fig. 2B, bottom). These results show that ectopic Brd4 expression during G0 and G1 inhibits cell cycle progression from G1 to S. We have also observed a notable inhibition of S-phase entry with synchronized HeLa cells transfected with Brd4 (data not shown).
Brd4 interacts with RFC in vivo.
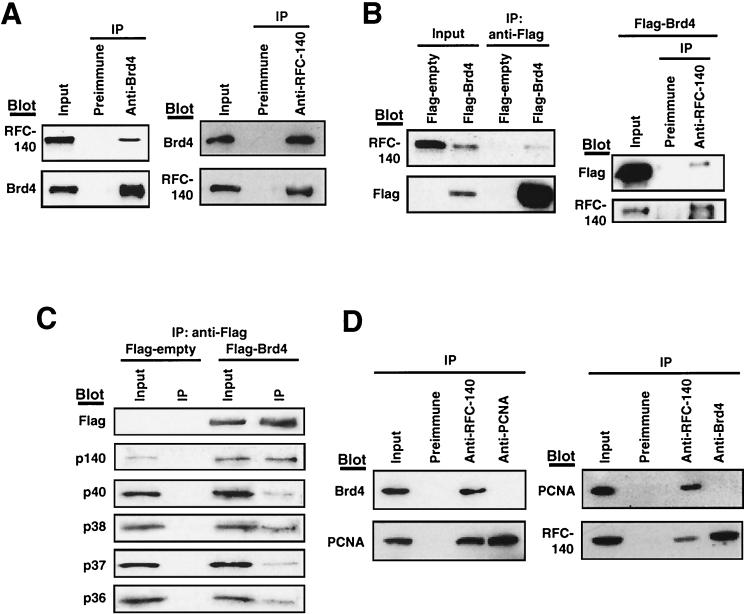
To examine the possibility that ectopic Brd4 inhibits entry into S phase by interacting with a protein involved in DNA synthesis, coimmunoprecipitation analyses were performed. Among several proteins tested, we found that RFC-140, the large subunit of RFC (4, 5, 44), interacts with Brd4. Extracts from NIH 3T3 cells were first precipitated with anti-Brd4 antibody, and the precipitated materials were tested for RFC-140 by immunoblot analysis. As shown in Fig. 3A, RFC-140 was found in the precipitates along with Brd4. Neither RFC-140 nor Brd4 was found in the precipitates from preimmune sera. Conversely, antibody to RFC-140, but not preimmune sera, precipitated Brd4 along with RFC-140, indicating that Brd4 and RFC-140 interacted with each other in vivo.
FIG. 3.
Brd4-RFC interaction in vivo. (A) Coimmunoprecipitation of endogenous Brd4 and RFC-140. Extracts (500 μg of protein) from NIH 3T3 cells were immunoprecipitated (IP) with the antibodies indicated above the panels. Precipitates were tested for RFC-140 and Brd4 by immunoblot analysis (Blot) using the indicated antibodies. Input represents 10% of the extracts used for precipitation. (B) Extracts from cells transfected with a Flag-Brd4 or empty vector were immunoprecipitated with anti-Flag antibody, and precipitates were blotted for RFC-140 or Flag-Brd4 (left panels). Extracts from cells transfected with Flag-Brd4 were immunoprecipitated with anti-RFC-140 antibody and blotted with anti-Flag antibody or anti-RFC-140 antibody (right panels). Input represents 10% of extracts. (C) Coimmunoprecipitation of RFC subunits. Extracts from cells transfected with Flag-Brd4 or the empty Flag vector were precipitated with anti-Flag antibody and blotted with antibodies to the indicated subunits of RFC. Input represents 10% of extracts. (D) Lack of Brd4-PCNA interaction. Extracts were immunoprecipitated with the antibodies indicated above the panels and blotted for Brd4, RFC-140, or PCNA.
To further substantiate the Brd4-RFC-140 interaction in vivo, coimmunoprecipitation analysis was performed with cells transfected with the Flag-tagged Brd4 (Fig. 3B). Extracts were precipitated with anti-Flag antibody, and precipitated materials were tested for RFC-140 by immunoblot analysis. Figure 3B shows that anti-Flag antibody precipitated the endogenous RFC-140 along with Flag-tagged Brd4. Supporting a specific interaction between Brd4 and RFC-140, it was found that anti-Flag antibody did not precipitate RFC-140 from samples transfected with the empty Flag vector. In the reciprocal coimmunoprecipitation assay, anti-RFC-140 antibody precipitated Flag-tagged Brd4 along with the endogenous RFC-140. The interaction between RFC-140 and the endogenous or Flag-tagged Brd4 was also observed with extracts from HeLa cells (see Fig. 6).
In addition to RFC-140, the complete RFC contains four smaller subunits, p40, p38, p37, and p36 (6, 8, 16, 18, 34, 43, 47). To assess whether Brd4 interacts with the complete RFC or only with RFC-140, materials precipitated by anti-Flag antibody were blotted with antibodies against all five subunits (41). As shown in Fig. 3C, all five RFC subunits were coprecipitated along with Flag-Brd4, indicating that Brd4 can interact with the RFC as a complete complex. Confirming this specificity, it was found that none of the subunits were precipitated from cells transfected with the empty vector. These results indicate that Brd4 associates with the complete RFC in vivo.
One of the important activities of RFC is binding to PCNA and loading it onto replicating DNA (26, 30, 39, 40). It was of interest to assess whether Brd4 also interacts with PCNA. To address this question, coimmunoprecipitation analysis was performed with antibodies to PCNA, RFC-140, and Brd4 (Fig. 3D). Anti-RFC-140 antibody coprecipitated Brd4 as well as PCNA, as expected. However, anti-PCNA antibody did not coprecipitate Brd4 (Fig. 3D, top left panel). Similarly, anti-Brd4 antibody (Fig. 3D, right panels) precipitated RFC-140 but not PCNA. Neither PCNA nor Brd4 was precipitated by preimmune antibody. These results indicate that Brd4 does not interact with PCNA.
Brd4 interacts with RFC in vitro.
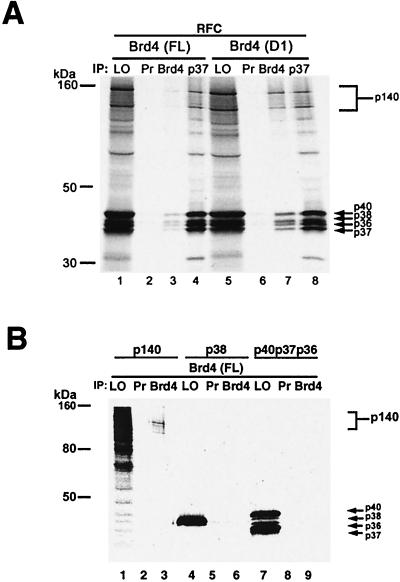
To determine whether Brd4 directly binds to RFC or interacts with RFC by binding to a third-party protein, immunoprecipitation assays were performed in vitro. The cDNAs encoding all five subunits of RFC (41) were translated in vitro and 35S labeled in the presence of recombinant full-length Brd4 or a Brd4 deletion mutant, D1, and reactions were precipitated with antibody against Brd4. Like D7, D1 lacks the C-terminal region but retains both bromodomains (see Fig. 6A). As shown in Fig. 4A, anti-Brd4 antibody precipitated all five subunits of RFC both when they were incubated with full-length Brd4 and when they were incubated with D1. Control preimmune serum did not precipitate any of the RFC subunits with either Brd4 or D1. D1 was much more efficient than full-length Brd4 in precipitating RFC under these conditions, although the basis for this is not clear at present. Thus, Brd4 binds to the entire RFC as a complex rather than to separate subunits. To identify the RFC subunit responsible for binding to Brd4, the large subunit (p140) and the small subunit p38 as well as the mixture of p40, p37, and p36 were separately translated in vitro, 35S labeled, incubated with full-length Brd4, and examined with an immunoprecipitation assay as described above. As shown in Fig. 4B, only p140, but not p38 or the combination of p40, p37, and p36, was precipitated by anti-Brd4 antibody. These results indicate that the large subunit of RFC, p140, but none of the small subunits, directly binds to Brd4. Combining this result with the data presented in Fig. 3C, it appears that binding of Brd4 to RFC-140 does not disrupt the formation of the five-subunit complex.
FIG. 4.
Brd4-RFC interaction in vitro. (A) Either full-length Brd4 (lanes 1 to 4) or D1 (lanes 5 to 8) (both 0.5 μg) was added to in vitro transcribed-translated RFC. 35S-labeled RFC was immunoprecipitated with preimmune sera (Pr) (lanes 2 and 6), Brd4 antisera (lanes 3 and 7), or p37 antisera (lanes 4 and 8). Precipitated materials were resolved by SDS-12%PAGE. LO, load on, represents 10% of the aliquots used in the immunoprecipitation. (B) Full-length Brd4 (0.4 μg) was added to in vitro transcribed-translated RFC-p140, p40, p38, p37, and p36. Material shown in lanes 1, 4, and 7 was immunoprecipitated with either preimmune sera (lanes 2, 5, and 8) or anti-Brd4 antisera (lanes 3, 6, and 9). Fifty percent of the immunoprecipitated products was subjected to SDS-12%PAGE.
Overexpression of RFC-140 counteracts the growth-inhibitory effect of Brd4.
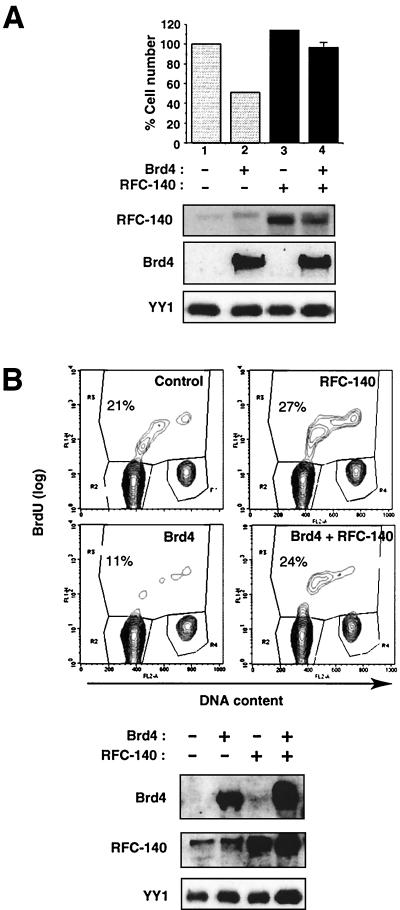
The above-mentioned data suggested that the interaction of Brd4 with RFC-140 is part of the mechanism by which ectopic Brd4 inhibits progression to S, as overexpression of Brd4 may have interfered with the activity of RFC. If this is the case, then ectopic expression of RFC-140 may offset the negative effect of Brd4. Previously, Haque et al. showed that overexpression of RFC-140 in NIH 3T3 cells stimulated proliferation, increasing the percentage of cells in S (20). With that report in mind, we first tested whether transfection with RFC-140 could ameliorate the growth-inhibitory effect seen with ectopic Brd4. TET-inducible Brd4 cells were transiently transfected with an RFC-140 expression vector along with IL-2R vector. Transfected cells were isolated by sorting, treated with DOX for induction of Brd4 expression, and allowed to grow for 48 h. Figure 5A shows the data for cell recovery. Transfecting cells with a control vector and treating them with DOX (hence overexpressing Brd4) led to a significant reduction in cell yields (∼50%) relative to what was seen without DOX induction, as was expected from the data presented in Fig. 1C. In contrast, transfecting cells with RFC-140 led to a lower reduction (<18%) in cell yields following DOX treatment. The results of immunoblot analysis presented in Fig. 5A showed that transfection with RFC-140 increased the expression of RFC-140 protein, irrespective of the addition of DOX. As expected, DOX treatment induced Brd4 expression at comparable levels in control and RFC-140-transfected cells. These results indicate that RFC-140 counteracts the growth-inhibitory effect of Brd4.
To ascertain whether ectopic RFC-140 relieves inhibition of S-phase entry caused by ectopic Brd4, the following experiments were performed. NIH 3T3 cells were transiently transfected with Brd4 and RFC-140 along with IL-2R for 24 h and pulse-labeled with BrdU. Transfected cells were separated by sorting and were analyzed by flow cytometry for BrdU uptake and propidium iodide (PI) staining. As seen in Fig. 5B, whereas transfection with Brd4 alone reduced the percentage of cells in S, transfection with RFC-140 alone led to an increase in S-phase cells, which is in agreement with the results of the report by Haque et al. (20). Importantly, compared with transfection with Brd4 alone, cotransfection with Brd4 and RFC-140 led to a significant increase in the percentage of S-phase cells. Immunoblot analysis showed that transfection with Brd4 and RFC-140 increased the expression of the respective proteins and that RFC-140 did not inhibit Brd4 expression and vice versa (Fig. 5B). Several independent transfection experiments gave similar outcomes (data not shown). These results are consistent with the data presented in Fig. 5A and indicate that RFC-140 relieved the inhibition of S-phase entry by Brd4.
A bromodomain is required for the interaction with RFC-140 and for the inhibition of progression from G1 to S.
To assess the domains of Brd4 involved in the interaction with RFC and inhibition of S-phase entry, a series of Brd4 deletion mutants was generated. As diagrammed in Fig. 6A, D3 lacked the second bromodomain but retained the first bromodomain, D8 lacked the first bromodomain but retained the second one, D10 lacked both bromodomains, and D7 and D1 lacked the large C-terminal region but retained both bromodomains. These deletion mutants all carried the spacer region between the two bromodomains. D5, on the other hand, lacked the spacer and the first bromodomain. These deletion mutants were expressed as GFP fusion constructs in NIH 3T3 cells, and their intracellular localization was examined (Fig. 6B). Full-length Brd4 fused to GFP localized to the nucleus with uniform distribution, except for the nucleoli, which is consistent with the pattern of the endogenous Brd4 (12). Removal of one or both bromodomains did not change the nuclear localization (Fig. 6B, panels showing D3, D8, and D10). D7 and D1 also localized to the nucleus (data not shown). However, D5, which lacked the spacer region, localized exclusively to the cytoplasm (Fig. 6B). These data show that the spacer region separating the two bromodomains is at least partly responsible for the nuclear localization of Brd4.
To assess the domains required for the interaction with RFC-140, Flag-tagged Brd4 deletion mutants were transiently transfected in synchronized HeLa cells, harvested at G1/S, precipitated with anti-Flag antibody, and tested for endogenous RFC-140 by immunoblot analysis. As shown in the upper panel in Fig. 6C, Brd4 deletion mutants of expected sizes were expressed in transfected cells and precipitated by the antibody. The lower panel in Fig. 6C shows that RFC-140 was coprecipitated with the wild-type Brd4, D7, and D8 but not with D3 and D10, indicating that the second bromodomain, but not the first one, is critical for the interaction with RFC-140.
We next examined whether the domain required for the interaction with RFC-140 is also required for the inhibition of S-phase entry by ectopic Brd4. NIH 3T3 cells were first synchronized by serum starvation for 48 h and transfected with GFP-Brd4 deletion mutants 4 h prior to release. Then they were allowed to proceed through G1 and to enter S in the subsequent 16 h in the presence of BrdU. As shown in Fig. 6D, cells that entered S were detected by anti-BrdU antibody. Cells expressing Brd4 deletion mutants were monitored with GFP, and all cells were detected by Hoechst DNA stain. The use of Lipofectamine-Plus, a high-efficiency transfection reagent, enabled microscopic inspection without the sorting of transfected cells. On average, the transfection efficiency was ∼50% for all constructs tested here, as about half of the cells on the fields were GFP positive. Untransfected, GFP-negative cells incorporating BrdU on the same field served as internal controls. The majority of control cells expressing only GFP incorporated BrdU, indicating that they entered S. However, the majority of cells expressing full-length Brd4, D7, or D8 failed to incorporate BrdU, indicating that they failed to enter S. In contrast, cells expressing D3 and D10 incorporated BrdU, indicating that these deletion mutants no longer inhibited S-phase entry. The table presented in Fig. 6D shows the percentage of BrdU-positive cells within transfected, GFP-positive cells, indicating that deletion of the second bromodomain abolishes inhibition of S-phase entry. In experiments performed with HeLa cells, it was also found that only the wild-type Brd4, D7, and D8, but not D3 or D10, inhibited BrdU uptake (data not shown). These results indicate that both the interaction with RFC-140 and the inhibition of S-phase entry are dependent on the second bromodomain of Brd4.
Brd4 inhibits DNA elongation reaction in vitro.
To begin to address the mechanism by which Brd4 inhibits BrdU incorporation at S phase, we investigated the activity of recombinant Brd4 and its deletion mutants (D1 and D3) in the previously established in vitro DNA replication system (41). While D1 and full-length Brd4 bind to RFC, D3 does not (Fig. 6C). The influence of D1 and D3 on the elongation of a singly primed M213 DNA substrate catalyzed by DNA polymerase δ holoenzyme (including RFC and PCNA) was examined (Fig. 6E). Increasing levels of D1 (Fig. 6E, lanes 5 to 7) markedly inhibited DNA replication, whereas the addition of D3 hardly affected the reaction (lanes 2 to 4). The inhibitory effects of D1 were reversed substantially by the addition of higher levels of RFC (Fig. 6E, compare lanes 7 and 10). Inhibition by D1, however, was not reversed by increased levels of PCNA and/or DNA polymerase δ (data not shown). These findings suggest that D1 selectively affected the action of RFC. Full-length Brd4 was also found to inhibit the elongation reaction, and this effect was also reversed by increased levels of RFC (data not shown). Thus, in keeping with the in vivo findings, the second bromodomain of Brd4 appears to play a key role in affecting the action of RFC.
Coexpression of Brd4 and RFC-140 during cell growth.
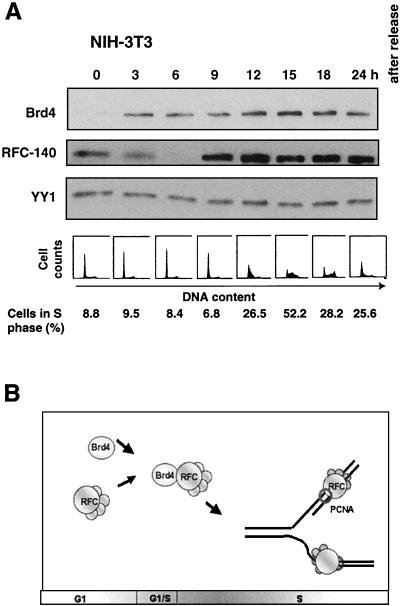
Because the interaction between Brd4 and RFC-140 appeared to exert a function when cells progress from G1 to S, we were interested in assessing whether the expression of the two proteins is regulated during cell growth. NIH 3T3 cells were synchronized by serum starvation, and the expression of the proteins was detected by immunoblot analysis (Fig. 7A). Brd4 expression was not detected at G0 but was induced during early G1 and was observed throughout the remainder of the cell cycle. RFC-140 expression was detected in quiescent cells (22). The levels were reduced in early G1, followed by an increase in late G1 just prior to entry into S, and RFC-140 remained expressed in G2 and M. These results indicate that Brd4 and RFC-140 are coexpressed through late G1 to M.
FIG. 7.
Cell cycle-regulated interaction between Brd4 and RFC-140. (A) NIH 3T3 cells synchronized by serum starvation were released and harvested at the indicated times and analyzed by immunoblot analysis. Cell synchronization was confirmed by flow cytometry analysis (bottom). (B) A model for Brd4 action. Levels of Brd4 and RFC-140 increase during G1, and Brd4 interacts efficiently with RFC during G1 to S. This interaction may influence the activity of RFC in supporting DNA replication.
DISCUSSION
Role of Brd4 in cell cycle progression.
The present study established that Brd4 has a regulatory role in cell cycle progression from G1 to S. Our initial observation was that ectopic Brd4 expression inhibits cell growth, as verified in three independent systems, the stable and transient transfections as well as the TET-inducible Brd4 expression (Fig. 1, 2, and 5). In these experiments, the proportion of cells in S phase was reduced while that in G1 increased. Consistent with this result, analysis with synchronized cells showed that ectopic Brd4 inhibited entry into S phase, leaving cells in G1 that did not incorporate BrdU (Fig. 2 and 6). A similar inhibition of progression to S phase was observed with ectopic Brd4 expression in synchronized HeLa cells (data not shown). Although the present study relied on an ectopic-expression strategy, it is important to mention that the endogenous Brd4 plays a role in cell growth and the results presented here are not due to an artifact of Brd4 overexpression. This has been ascertained by our ongoing study employing other strategies. For example, preliminary analysis of cells expressing morpholino-modified antisense Brd4 oligomers suggests that a reduction of Brd4 protein expression during G1 phase promotes S-phase entry (A. Farina, A. Dey, and K. Ozato, unpublished data). In view of the previously presented finding indicating that Brd4 regulates G2-to-M transition (12), it seems that Brd4 has a complex growth-regulatory activity that affects multiple steps of the cell cycle. An additional piece of evidence supporting the role of Brd4 in cell growth is the observation that disruption of Brd4 alleles in embryonic stem cells is growth inhibitory (A. Nishiyama and K. Ozato, unpublished data). Together, our findings add to and extend previous reports pointing to a role for the BET family in cell growth; it has been shown that disruption of bdf1, a yeast BET member, causes a slow-growth phenotype and a defect in sporulation (7). Also suggesting a role in the cell cycle, Ring3/Brd2 has been reported to be capable of regulating cell cycle-dependent transcription (10).
Interaction with RFC.
RFC is a component of conserved DNA replication machinery (18, 41, 44). Composed of five subunits (6, 8), RFC loads PCNA onto DNA (40, 47) and plays an essential role in DNA replication (16, 34, 42). We found that Brd4 interacts with RFC in vivo, as demonstrated by reciprocal coimmunoprecipitation of the endogenous proteins. RFC-140 and small subunits of RFC were also coimmunoprecipitated along with transfected, Flag-tagged Brd4 (Fig. 3). In vitro studies performed with recombinant Brd4 and individual subunits of RFC showed that Brd4 interacts with the complete RFC by binding to the largest subunit, RFC-140 (Fig. 4). Several lines of evidence support the view that this interaction accounts, at least in part, for the inhibition of progression to S phase caused by ectopic Brd4. First, indicative of a functional interaction, inhibition of S-phase entry by ectopic Brd4 was alleviated by cotransfection with RFC-140 (Fig. 5). In these experiments, transfection with RFC-140 alone promoted S-phase entry while that with Brd4 alone caused an opposite effect. Second, Brd4 deletion mutants that lost the ability to interact with RFC-140 also lost the ability to inhibit S-phase entry, which shows a good concordance between the RFC-Brd4 interaction and inhibition of entry into S phase. Conversely, deletion mutants that retained the ability to interact with RFC retained the ability to inhibit S-phase entry (Fig. 6). Further supporting the functionality of the interaction, RFC-dependent DNA elongation reactions in vitro were inhibited by recombinant Brd4, indicating that Brd4 is capable of interfering with the function of RFC in supporting DNA replication (Fig. 6).
Despite the convincing evidence of the Brd4-RFC interaction and the functional implications obtained in this study, the overall biological significance of the interaction is not completely apparent at this time. At present, however, it may be surmised that Brd4 plays a role in fine-tuning the function of RFC in DNA replication (Fig. 7B). Brd4 might adjust the timing and intranuclear location of RFC action and might help prevent premature onset of DNA replication.
It seems plausible that Brd4-RFC interaction is influenced by various factors in vivo. First, the interaction may be stoichiometrically controlled, and abnormal levels of either protein may distort the balance of interaction as well as the activity of the two proteins. Brd4 overexpression may be deleterious to the function of RFC in supporting DNA replication. Or, overexpression of Brd4 may alter its overall interaction with chromatin (see below), thereby negatively influencing the activity of RFC. The fact that RFC-140 overexpression alleviated the Brd4-imposed G1 arrest appears consistent with this view. Second, Brd4 and RFC may not always be complexed with each other. Brd4 may interact with RFC transiently, perhaps depending on the cell cycle stages and other factors. The fact that PCNA was not coprecipitated with Brd4 may support this notion. In addition, our preliminary coimmunoprecipitation data with synchronized cells also appear to support this possibility (Farina et al., unpublished). Moreover, RFC has been shown to interact with other factors, including the BRCA-1 complexes (45) and C/EBPα (22). It is possible that interactions with these factors may influence the ability of RFC to interact with Brd4. Furthermore, Brd4-RFC interaction may be influenced by posttranslational modification of either protein. Since both RFC-140 and Brd4 are phosphoproteins and the status of phosphorylation may be under cell cycle control (12), such a posttranslational modification may contribute to transient interaction of the two proteins.
Role for bromodomain.
Bromodomains have been shown to interact with acetylated histones and have been proposed to act as chromatin-targeting modules (13, 46). Consistent with a link with chromatin, the yeast bdf1 has been reported to interact with histones in vitro (33). It has been shown that two tandem bromodomains present in TAF250 act synergistically to interact with acetylated histones (23). Given that all BET family proteins have two bromodomains, Brd4 and related proteins may have preferential affinity for acetylated chromatin. Based on this idea, it seems attractive to envisage that Brd4 plays a role in recruiting RFC to specific regions of chromatin during DNA replication, thereby contributing to cell cycle progression (27).
Brd4 deletion analysis revealed that only one of the two bromodomains, namely the second bromodomain, is essential for inhibiting progression to S phase. In our analysis, the first bromodomain did not appear to be critical for the interaction with RFC and the inhibition of S-phase entry. From these results, it is evident that (i) a bromodomain has a role in interacting not only with histones but also with nonhistone proteins and (ii) the first and second bromodomains in Brd4 possess distinct functions. The latter result may not be surprising, since the first and second bromodomains of Brd4 are less than 40% identical to each other in their primary amino acid sequences (12). Likewise, the two bromodomains present in other BET members are relatively dissimilar. In contrast to this relative dissimilarity, the first bromodomains of different BET members exhibit a marked similarity across species. The second bromodomains of different members also show a notable similarity (Jeanmougin et al., letter). For example, the first bromodomain of Brd4 is 80% identical to that of Brd2 and 75% identical to that of Drosophila fsh. In light of this and the fact that bromodomains within the BET family are more similar to each other than to those of other families, it is possible that other proteins of the BET family are also involved in cell growth control, employing a mechanism similar to that of Brd4.
In summary, we investigated the role of Brd4 in cell growth, particularly during progression from G1 to S, and described the interaction between Brd4 and RFC as a possible mechanistic basis for this role. This work represents an initial delineation of the complex growth-regulatory activities of Brd4 and the BET family. Further studies utilizing other approaches are required for full elucidation of the function of Brd4 and related proteins.
Acknowledgments
We thank M. DePamphilis and B. Howard for valuable comments on this work and critical reading of the paper. We also thank S. Sekiyama and A. Abbasi for technical assistance and T. Howard, V. Ogryzko, and H. Sabe for advice on the experiments.
T.M. was supported by the Japan Society for Promotion of Science and Yasuda Medical Foundation.
T.M., A.F., and A.D. contributed equally to this work.
REFERENCES
- 1.Baron, U., and H. Bujard. 2000. Tet repressor-based system for regulated gene expression in eukaryotic cells: principles and advances. Methods Enzymol. 327:401-421. [DOI] [PubMed] [Google Scholar]
- 2.Beck, S., I. Hanson, A. Kelly, D. J. Pappin, and J. Trowsdale. 1992. A homologue of the Drosophila female sterile homeotic (fsh) gene in the class II region of the human MHC. DNA Sequence 2:203-210. [DOI] [PubMed] [Google Scholar]
- 3.Blanco, J. C., S. Minucci, J. Lu, X. J. Yang, K. K. Walker, H. Chen, R. M. Evans, Y. Nakatani, and K. Ozato. 1998. The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev. 12:1638-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bunz, F., R. Kobayashi, and B. Stillman. 1993. cDNAs encoding the large subunit of human replication factor C. Proc. Natl. Acad. Sci. USA 90:11014-11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burbelo, P. D., A. Utani, Z. Q. Pan, and Y. Yamada. 1993. Cloning of the large subunit of activator 1 (replication factor C) reveals homology with bacterial DNA ligases. Proc. Natl. Acad. Sci. USA 90:11543-11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai, J., E. Gibbs, F. Uhlmann, B. Phillips, N. Yao, M. O'Donnell, and J. Hurwitz. 1997. A complex consisting of human replication factor C p40, p37, and p36 subunits is a DNA-dependent ATPase and an intermediate in the assembly of the holoenzyme. J. Biol. Chem. 272:18974-18981. [DOI] [PubMed] [Google Scholar]
- 7.Chua, P., and G. S. Roeder. 1995. Bdf1, a yeast chromosomal protein required for sporulation. Mol. Cell. Biol. 15:3685-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cullmann, G., K. Fien, R. Kobayashi, and B. Stillman. 1995. Characterization of the five replication factor C genes of Saccharomyces cerevisiae. Mol. Cell. Biol. 15:4661-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denis, G. V., and M. R. Green. 1996. A novel, mitogen-activated nuclear kinase is related to a Drosophila developmental regulator. Genes Dev. 10:261-271. [DOI] [PubMed] [Google Scholar]
- 10.Denis, G. V., C. Vaziri, N. Guo, and D. V. Faller. 2000. RING3 kinase transactivates promoters of cell cycle regulatory genes through E2F. Cell Growth Differ. 11:417-424. [PMC free article] [PubMed] [Google Scholar]
- 11.DePamphilis, M. L. 2000. Review: nuclear structure and DNA replication. J. Struct. Biol. 129:186-197. [DOI] [PubMed] [Google Scholar]
- 12.Dey, A., J. Ellenberg, A. Farina, A. E. Coleman, T. Maruyama, S. Sciortino, J. Lippincott-Schwartz, and K. Ozato. 2000. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G2-to-M transition. Mol. Cell. Biol. 20:6537-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dhalluin, C., J. E. Carlson, L. Zeng, C. He, A. K. Aggarwal, and M. M. Zhou. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491-496. [DOI] [PubMed] [Google Scholar]
- 14.Digan, M. E., S. R. Haynes, B. A. Mozer, I. B. Dawid, F. Forquignon, and M. Gans. 1986. Genetic and molecular analysis of fs(1)h, a maternal effect homeotic gene in Drosophila. Dev. Biol. 114:161-169. [DOI] [PubMed] [Google Scholar]
- 15.Donaldson, A. D., and J. J. Blow. 1999. The regulation of replication origin activation. Curr. Opin. Genet. Dev. 9:62-68. [DOI] [PubMed] [Google Scholar]
- 16.Ellison, V., and B. Stillman. 1998. Reconstitution of recombinant human replication factor C (RFC) and identification of an RFC subcomplex possessing DNA-dependent ATPase activity. J. Biol. Chem. 273:5979-5987. [DOI] [PubMed] [Google Scholar]
- 17.Fotedar, R., and A. Fotedar. 1995. Cell cycle control of DNA replication. Prog. Cell Cycle Res. 1:73-89. [DOI] [PubMed] [Google Scholar]
- 18.Fotedar, R., R. Mossi, P. Fitzgerald, T. Rousselle, G. Maga, H. Brickner, H. Messier, S. Kasibhatla, U. Hubscher, and A. Fotedar. 1996. A conserved domain of the large subunit of replication factor C binds PCNA and acts like a dominant negative inhibitor of DNA replication in mammalian cells. EMBO J. 15:4423-4433. [PMC free article] [PubMed] [Google Scholar]
- 19.Grueneberg, D. A., R. W. Henry, A. Brauer, C. D. Novina, V. Cheriyath, A. L. Roy, and M. Gilman. 1997. A multifunctional DNA-binding protein that promotes the formation of serum response factor/homeodomain complexes: identity to TFII-I. Genes Dev. 11:2482-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haque, J., H. van der Kuip, A. Kumar, W. Aulizky, M. Rutherford, C. Huber, T. Fisher, and B. Williams. 1996. Overexpression of mouse p140 subunit of replication factor C accelerates cellular proliferation. Cell Growth Differ. 7:319-326. [PubMed] [Google Scholar]
- 21.Haynes, S. R., B. A. Mozer, N. Bhatia-Dey, and I. B. Dawid. 1989. The Drosophila fsh locus, a maternal effect homeotic gene, encodes apparent membrane proteins. Dev. Biol. 134:246-257. [DOI] [PubMed] [Google Scholar]
- 22.Hong, S., S. J. Park, H. J. Kong, J. D. Shuman, and J. Cheong. 2001. Functional interaction of bZIP proteins and the large subunit of replication factor C in liver and adipose cells. J. Biol. Chem. 276:28098-28105. [DOI] [PubMed] [Google Scholar]
- 23.Jacobson, R. H., A. G. Ladurner, D. S. King, and R. Tjian. 2000. Structure and function of a human TAFII250 double bromodomain module. Science 288:1422-1425. [DOI] [PubMed] [Google Scholar]
- 24.Jiang, Y. W., P. Veschambre, H. Erdjument-Bromage, P. Tempst, J. W. Conaway, R. C. Conaway, and R. D. Kornberg. 1998. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc. Natl. Acad. Sci. USA 95:8538-8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, M. H., M. Numata, and M. Shimane. 1997. Identification and characterization of BRDT: a testis-specific gene related to the bromodomain genes RING3 and Drosophila fsh. Genomics 45:529-534. [DOI] [PubMed] [Google Scholar]
- 26.Kelman, Z., and J. Hurwitz. 1998. Protein-PCNA interactions: a DNA-scanning mechanism? Trends Biochem. Sci. 23:236-238. [DOI] [PubMed] [Google Scholar]
- 27.Krude, T. 1999. Chromatin replication: finding the right connection. Curr. Biol. 9:R394-R396. [DOI] [PubMed] [Google Scholar]
- 28.Lygerou, Z., C. Conesa, P. Lesage, R. N. Swanson, A. Ruet, M. Carlson, A. Sentenac, and B. Seraphin. 1994. The yeast BDF1 gene encodes a transcription factor involved in the expression of a broad class of genes including snRNAs. Nucleic Acids Res. 22:5332-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matangkasombut, O., R. M. Buratowski, N. W. Swilling, and S. Buratowski. 2000. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 14:951-962. [PMC free article] [PubMed] [Google Scholar]
- 30.Mossi, R., Z. O. Jonsson, B. L. Allen, S. H. Hardin, and U. Hubscher. 1997. Replication factor C interacts with the C-terminal side of proliferating cell nuclear antigen. J. Biol. Chem. 272:1769-1776. [DOI] [PubMed] [Google Scholar]
- 31.Ogryzko, V. V., P. Wong, and B. H. Howard. 1997. WAF1 retards S-phase progression primarily by inhibition of cyclin-dependent kinases. Mol. Cell. Biol. 17:4877-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Owen, D. J., P. Ornaghi, J. C. Yang, N. Lowe, P. R. Evans, P. Ballario, D. Neuhaus, P. Filetici, and A. A. Travers. 2000. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J. 19:6141-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pamblanco, M., A. Poveda, R. Sendra, S. Rodriguez-Navarro, J. E. Perez-Ortin, and V. Tordera. 2001. Bromodomain factor 1 (Bdf1) protein interacts with histones. FEBS Lett. 496:31-35. [DOI] [PubMed] [Google Scholar]
- 34.Podust, V. N., N. Tiwari, R. Ott, and E. Fanning. 1998. Functional interactions among the subunits of replication factor C potentiate and modulate its ATPase activity. J. Biol. Chem. 273:12935-12942. [DOI] [PubMed] [Google Scholar]
- 35.Rhee, K., M. Brunori, V. Besset, R. Trousdale, and D. J. Wolgemuth. 1998. Expression and potential role of Fsrg1, a murine bromodomain-containing homologue of the Drosophila gene female sterile homeotic. J. Cell Sci. 111:3541-3550. [DOI] [PubMed] [Google Scholar]
- 36.Stillman, B. 2001. DNA replication. Genomic views of genome duplication. Science 294:2301-2304. [DOI] [PubMed] [Google Scholar]
- 37.Thorpe, K. L., S. Abdulla, J. Kaufman, J. Trowsdale, and S. Beck. 1996. Phylogeny and structure of the RING3 gene. Immunogenetics 44:391-396. [DOI] [PubMed] [Google Scholar]
- 38.Thorpe, K. L., P. Gorman, C. Thomas, D. Sheer, J. Trowsdale, and S. Beck. 1997. Chromosomal localization, gene structure and transcription pattern of the ORFX gene, a homologue of the MHC-linked RING3 gene. Gene 200:177-183. [DOI] [PubMed] [Google Scholar]
- 39.Tsurimoto, T. 1999. PCNA binding proteins. Front. Biosci. 4:D849-D858. [DOI] [PubMed] [Google Scholar]
- 40.Tsurimoto, T., and B. Stillman. 1991. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J. Biol. Chem. 266:1950-1960. [PubMed] [Google Scholar]
- 41.Uhlmann, F., J. Cai, H. Flores-Rozas, F. B. Dean, J. Finkelstein, M. O'Donnell, and J. Hurwitz. 1996. In vitro reconstitution of human replication factor C from its five subunits. Proc. Natl. Acad. Sci. USA 93:6521-6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uhlmann, F., J. Cai, E. Gibbs, M. O'Donnell, and J. Hurwitz. 1997. Deletion analysis of the large subunit p140 in human replication factor C reveals regions required for complex formation and replication activities. J. Biol. Chem. 272:10058-10064. [DOI] [PubMed] [Google Scholar]
- 43.Uhlmann, F., E. Gibbs, J. Cai, M. O'Donnell, and J. Hurwitz. 1997. Identification of regions within the four small subunits of human replication factor C required for complex formation and DNA replication. J. Biol. Chem. 272:10065-10071. [DOI] [PubMed] [Google Scholar]
- 44.Waga, S., and B. Stillman. 1998. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67:721-751. [DOI] [PubMed] [Google Scholar]
- 45.Wang, Y., D. Cortez, P. Yazdi, N. Neff, S. J. Elledge, and J. Qin. 2000. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 14:927-939. [PMC free article] [PubMed] [Google Scholar]
- 46.Winston, F., and C. D. Allis. 1999. The bromodomain: a chromatin-targeting module? Nat. Struct. Biol. 6:601-604. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, G., E. Gibbs, Z. Kelman, M. O'Donnell, and J. Hurwitz. 1999. Studies on the interactions between human replication factor C and human proliferating cell nuclear antigen. Proc. Natl. Acad. Sci. USA 96:1869-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]