B-ZIP transcription factors (98) are exclusively eukaryotic proteins that bind to sequence-specific double-stranded DNA as homodimers or heterodimers to either activate or repress gene transcription (34). We have examined both of the recently published DNA sequences of the human genome (51, 95) and identified 56 genes that contain the B-ZIP motif. Three sequences were identical, giving a total of 53 unique B-ZIP domains with the potential to form 2,809 dimers. This creates the possibility for a tremendous range of transcriptional control (23, 50, 52). While significant effort has been directed at identifying dimerization partners of B-ZIP proteins, the full complement of dimerization partners remains to be elucidated. This review highlights two topics: (i) the known structural rules that regulate leucine zipper dimerization specificity and (ii) experimental data addressing mammalian B-ZIP dimerization partners.
We have annotated the leucine zippers of all human B-ZIP domains, highlighting amino acids in the a, d, e, and g positions that appear critical for leucine zipper dimerization specificity. These data were used to group B-ZIP proteins into 12 families with similar dimerization properties: (i) those that strongly favor homodimerization within the family (PAR, CREB, Oasis, and ATF6), (ii) those that have the ability to both homodimerize and heterodimerize with similar affinities (C/EBP, ATF4, ATF2, JUN, and the small MAFs), and (iii) those that favor heterodimerization with other families (FOS, CNC, and large MAFs).
BACKGROUND
In the late 1980s, several mammalian B-ZIP proteins were purified by double-stranded DNA affinity chromatography, and the genes encoding these proteins were cloned. Among the first cloned were the AP-1 (c-FOS and c-JUN) heterodimer, (4), the CREB homodimer (65), and the C/EBP homodimer (37, 53). These newly isolated genes were used as probes in low-stringency DNA hybridizations to identify new sequence-related B-ZIP proteins (5, 78, 101). In addition, new B-ZIP proteins (25, 36, 102) were isolated by screening lambda phage protein expression libraries with radiolabeled DNA binding elements (29, 85, 97). These functional DNA binding assays successfully isolated B-ZIP proteins because the B-ZIP motif is compact and refolds easily (87). The wealth of new sequences led to a confusing nomenclature, because multiple groups independently isolated and named the same B-ZIP proteins. Moreover, initial classification into families was often based on apparent DNA binding activity, resulting in grouping of proteins with different dimerization properties. Several reviews have helped clarify these issues, including a comprehensive review by Hurst in 1995 (34), one focusing on ATF proteins (24), and another focusing on FOS and JUN proteins (6). We hope this review will further contribute to a systematic B-ZIP classification.
B-ZIP STRUCTURE
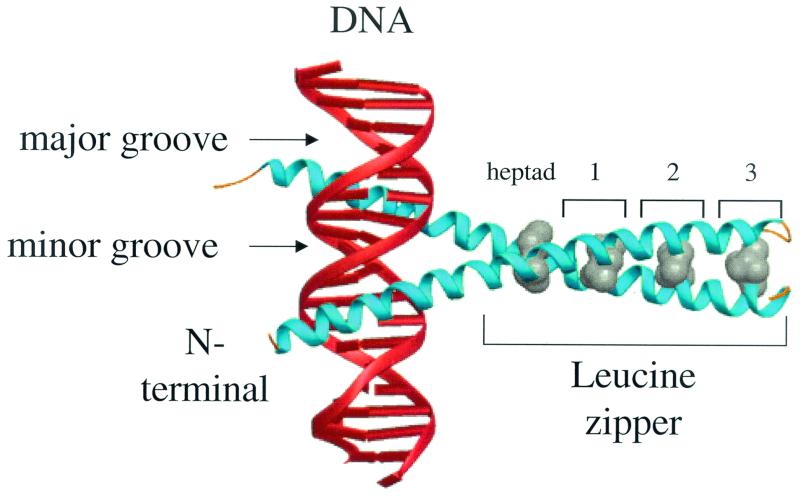
Amino acid alignment of B-ZIP proteins allowed the identification of the B-ZIP motif, a long bipartite α-helix that is 60 to 80 amino acids long (98). The N-terminal half contains two clusters of basic amino acids responsible for sequence-specific DNA binding, while the C-terminal half contains an amphipathic protein sequence of variable length with a leucine every seven amino acids. The shorter leucine zippers have less protein sequence flexibility, because amino acids must be optimized for dimerization stability. Longer leucine zippers allow better regulation of dimerization specificity, because they can contain amino acids that are suboptimal for stability but favor interaction with a particular partner. This amphipathic sequence, termed the “leucine zipper” (52), mediates homo- and heterodimerization of B-ZIP proteins (3, 20, 41, 44, 54, 77, 86, 91). Figure 1 shows the X-ray crystal structure of the B-ZIP domain from yeast GCN4 bound to DNA (15). B-ZIP DNA binding stabilizes the basic region inducing the random coil to form an α-helical extension of the leucine zipper (73, 84). Several B-ZIP proteins, including the small and large MAF proteins (13, 42), contain additional DNA binding elements N terminal to the basic region that increase the number of specific DNA bases that can be bound.
FIG. 1.
X-ray structure of the yeast B-ZIP homodimer, GCN4 (blue α-helices) bound to DNA (red helices). The N-terminal DNA recognition helix lies in the major groove of the DNA. An almost invariant leucine present every two turns of the C-terminal α-helix (at the d position) is shown in gray.
The leucine zipper dimerization domain forms a parallel coiled coil (75) that consists of four to five heptads, in which each heptad is composed of two α-helical turns or seven amino acids, labeled a, b, c, d, e, f, and g (61). Amino acids in the a, d, e, and g positions regulate leucine zipper oligomerization, dimerization stability, and dimerization specificity.
Amino acids in the a and d positions are on the same surface of the α-helix and are typically hydrophobic. The a and d amino acids from one monomer interact with the complementary a′ and d′ amino acid positions in the opposite monomer (′ refers to the second α-helix in the dimer). This interaction creates a hydrophobic core essential for dimer stability (87). The g and e positions typically contain charged amino acids (8, 96). X-ray crystallography reveals g↔e′ interhelical interactions between amino acids in the g position and oppositely charged amino acids in the e′, which is five amino acids C terminal (2, 15, 19, 21, 75, 80). Electrostatic interactions between the amino acids in the g↔e′ pair can be either attractive or repulsive and thus can regulate both homodimerization and heterodimerization. Furthermore, Van der Waal interactions between the g and e′ methylene groups and the underlying a and d amino acids contribute to stability (2).
LIST OF ALL HUMAN B-ZIP PROTEINS GROUPED BY DIMERIZATION PROPERTIES
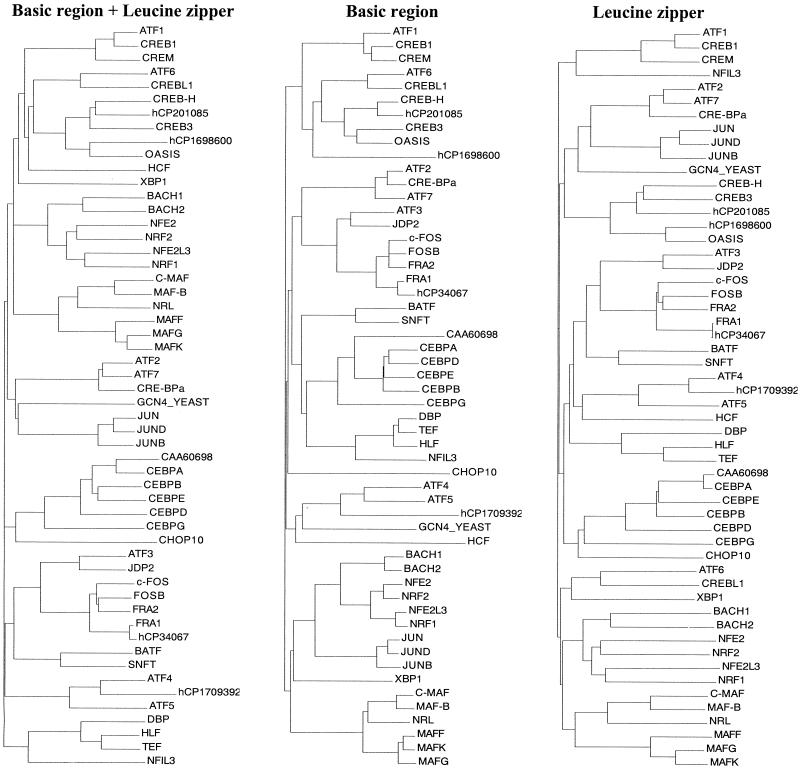
We have examined two versions of the DNA sequence of the human genome (51, 95) and identified 56 genes that contain the B-ZIP motif. Three of these motifs were identical, resulting in 53 unique B-ZIP domains. Four of the proteins were found only in the Celera database. Table 1 provides the chromosomal location, unique database search identifiers, and the number of amino acids found N terminal and C terminal of the B-ZIP domain. We have generated three dendrograms that examine the relatedness of the amino acid sequences of the 53 human B-ZIP domains. One dendrogram is based on the entire B-ZIP domain, one is based on the basic region that is critical for DNA binding, and the last is based on the leucine zipper region that regulates dimerization specificity (Fig. 2). The three dendrograms are similar. The differences are interesting, because they reveal whether similarities are based on DNA binding properties or dimerization specificities. For example, the basic region dendrogram places the CREB, ATF6, and Oasis families together, reflecting their binding to the CRE DNA sequence (5′-TGACGTCA-3′). In contrast, the leucine zipper dendrogram places the Oasis family separately from the CREB and ATF6 families, reflecting their different dimerization properties. It is possible that these families could compete for binding to the CRE DNA sequence to give a range of transcriptional control. Another example is the XBP protein, which is not related to any sequence when its basic region is examined, but clusters with the ATF6 proteins when its leucine zipper is examined.
TABLE 1.
Human B-ZIP proteinsaa
| Family and protein | Alternate name | Chromosomal no.b | Accession no.c | Tail length (no. of amino acids)d
|
||
|---|---|---|---|---|---|---|
| N-terminal | C-terminal | |||||
| Homodimer | ||||||
| PAR | ||||||
| TEF | 22 | NP_003207.1 | 231 | 5 | ||
| TEF paralog | 22 | hCP1717043 | 77 | 5 | ||
| DBP | DABP | 19 | NP_001343 | 253 | 5 | |
| HLF | 17 | NP_002117.1 | 223 | 5 | ||
| NFIL3 | NFIL3A, E4BP4 | 9 | NP_005375 | 71 | 324 | |
| CREB | ||||||
| CREB | CREB1 | 2 | NP_004370.1 | 267 | 0 | |
| ATF1 | 12 | NP_005162.1 | 211 | 0 | ||
| CREM | HCREM-1, ICER1 | 10 | AAC60617.2 | 272 | 0 | |
| Oasis | ||||||
| Oasis | BBF-2 | 11 | XP_054856.1 | 178 | 161 | |
| CREB-H | 19 | NP_115996 | 241 | 155 | ||
| CREB3 | LZIP, Luman | 9 | NP_006359 | 146 | 156 | |
| hCP201085 | 1 | hCP201085 | 248 | 118 | ||
| hCP1698600 | 7 | hCP1698600 | 135 | 161 | ||
| ATF6 | ||||||
| ATF6 | 1 | BAA34722 | 304 | 299 | ||
| CREBL1 | 6 | XP_004202.3 | 320 | 313 | ||
| XBP1 | TREB5 | 22 | NP_005071 | 68 | 126 | |
| Homo- and heterodimer | ||||||
| C/EBP | ||||||
| CEBPA | CEBP | 19 | XP_009180.2 | 280 | 11 | |
| CEBPB | NFIL6, IL6DBP, LAP, TCF5 | 20 | XP_009510.1 | 269 | 9 | |
| CEBPD | CRP3 | 8 | NP_005186.1 | 189 | 13 | |
| CEBPE | CRP1 | 14 | NP_001796.1 | 202 | 12 | |
| CAA60698 | HP8 peptide | 10 | CAA60698 | 268 | 11 | |
| CEBPG | GCSF | 19 | S26300 | 145 | 23 | |
| CHOP10 | DDIT3, GADD153, GA15 | 12 | NP_004074 | 96 | 5 | |
| ATF4 | ||||||
| ATF4 | CREB2, TAXREB67 | 22 | NP_001666 | 276 | 8 | |
| ATF4 paralog | 17 | hCP1640238M | 361 | 8 | ||
| hCP1709392 | X | hCP1709392 | 354 | 8 | ||
| ATF5 | ATFX | 19 | NP_036200 | 206 | 9 | |
| ATF2 | ||||||
| ATF2 | CREBP1, (CRE82) | 2 | XP_002767.2 | 350 | 88 | |
| ATF7 | ATFA | 12 | NP_006847.1 | 330 | 86 | |
| CRE-BPa | 7 | XP_004938.3 | 366 | 68 | ||
| JUN | ||||||
| JUND | 19 | NP_005345.2 | 266 | 14 | ||
| JUN | AP1 | 1 | NP_002219.1 | 250 | 14 | |
| JUNB | 19 | NP_002220.1 | 266 | 14 | ||
| S-MAF | ||||||
| MAFK | NFE2U, p18 | 7 | NP_002351.1 | 49 | 40 | |
| MAFG | 17 | NP_002350.1 | 49 | 46 | ||
| MAFF | 22 | NP_036455.1 | 49 | 48 | ||
| Heterodimer | ||||||
| FOS | ||||||
| c-FOS | FOS | 14 | NP_005243 | 136 | 178 | |
| FOSB | 19 | XP_009143.2 | 153 | 118 | ||
| FRA1 | FOSL1 | 11 | NP_005429.1 | 103 | 101 | |
| FRA2 | FOSL2 | 2 | NP_005244.1 | 122 | 137 | |
| hCP34067 | 4 | hCP34067 | 149 | 0 | ||
| ATF3 | LRF-1, LRG-21, CRG-5 | 1 | NP_001665 | 84 | 30 | |
| JDP2 | 14 | AAF21148.1 | 3 | 26 | ||
| JDP1 | p21SNFT, SNFT | 1 | NP_061134 | 33 | 27 | |
| BATF | SFA-2 | 14 | NP_006390.1 | 24 | 34 | |
| CNC | ||||||
| BACH1 | 21 | NP_001177.1 | 555 | 114 | ||
| NRF1 | NFE2L1, TCF11, LCR-F1 | 17 | NP_003195.1 | 652 | 53 | |
| NFE2 | p45 | 12 | XP_028656 | 264 | 42 | |
| BACH2 | 6 | NP_068585.1 | 644 | 130 | ||
| NRF2 | NFE2L2 | 2 | XP_002548.1 | 479 | 43 | |
| Family and protein | Alternate name | Chromosomal no.b | Accession no.c | Tail length (no. of amino acids)d
|
||
| N-terminal | C-terminal | |||||
| NFE2L3 | NRF3 | 7 | AAC09039.1 | 576 | 51 | |
| NFE2L3 paralog | 18 | hCP46847 | 281 | 42 | ||
| L-MAF | ||||||
| NRL | 14 | NP_006168.1 | 157 | 13 | ||
| MAF-B | MAFB, KRML | 20 | XP_009665.2 | 236 | 20 | |
| C-MAF | MAF | 16 | AAC27037 | 286 | 20 | |
| HCF | ||||||
| ZF, C1, VCAF, CFF | 11 | NP_067035 | 127 | 77 | ||
NCBI LocusLink gene names are given in boldface (http://www.ncbi.nlm.nih.gov/Locuslink).
Chromosomal number at which the B-ZIP protein is located.
Accession number to uniquely identify the B-Zip domain in the NCBI or Celera database.
Number of amino acids N terminal and C terminal of the B-ZIP domain.
FIG. 2.
Dendrogram of 53 human B-ZIP proteins. Phylogenetic trees were generated by Align X module of Vector NTI, 7.0 with default parameters. (A) Alignment based on the entire B-ZIP domain. (B) Alignment based on the basic region. (C) Alignment based on the leucine zipper defined from the first leucine shown in the consensus on Fig. 3.
To achieve a functional classification of the B-ZIP proteins, we have considered the dimerization properties of the B-ZIP domains. We have examined the amino acids in the g, a, d, and e positions of the leucine zipper region of the B-ZIP domains to rationalize the known and predict the unknown dimerization properties of the 53 human B-ZIP domains whose amino acid sequences are presented in Fig. 3. Using this analysis, we have grouped B-ZIP proteins with similar dimerization properties into 12 families. Our grouping is consistent with the dendrograms and agrees particularly with the dendrogram based on the leucine zipper sequence. An examination of the human genome by Green and colleagues (90) grouped B-ZIP proteins into eight previously identified families, (PAR, CREB, C/EBP, FOS, JUN, Maf, CNC, and ATF6) based on amino acid similarity throughout the B-ZIP domain. Our analysis divides their FOS family into FOS, ATF2, and ATF4; the MAF family into the small MAFs and large MAFs; and the CREB family into the CREB and Oasis families. We also reassigned two sequences, XBP1 was moved from the FOS family to the ATF6 family, and NFLI3 was moved from the C/EBP to the PAR family.
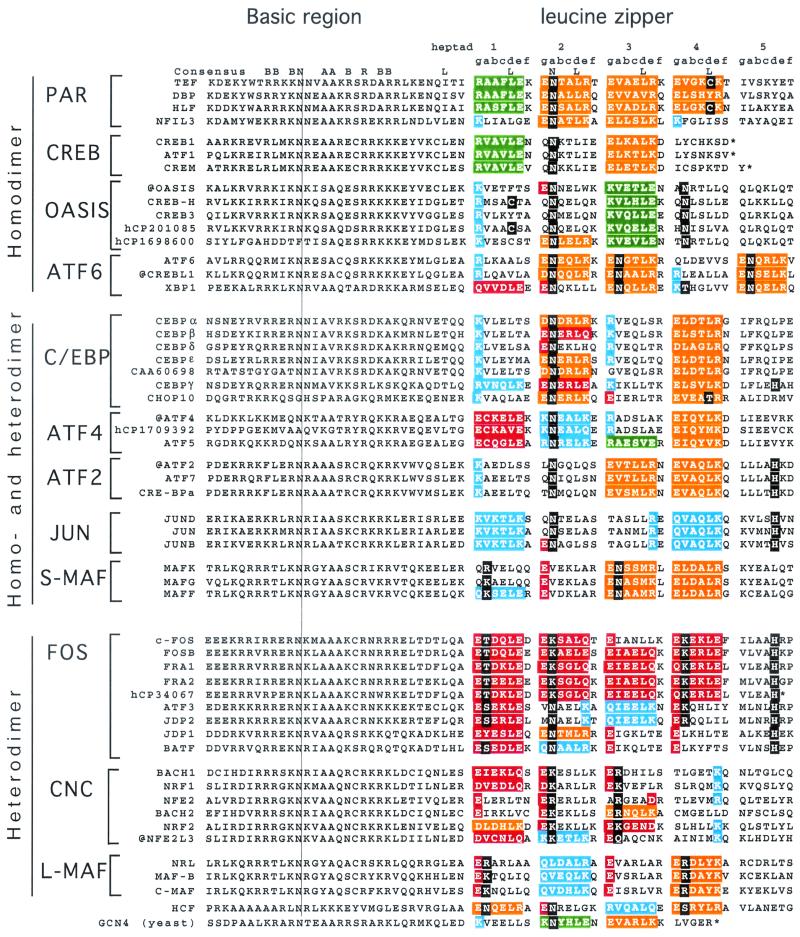
FIG. 3.
Amino acid sequence of all 53 human B-ZIP domains. Proteins are placed in to 12 groups based on predicted dimerization properties. We have placed the well-studied yeast B-ZIP protein GCN4 at the bottom of the figure for reference. The natural C terminus is denoted with an asterisk. Leucine zipper heptads are grouped (gabcdef) to help visualize potential g↔e′ pairs. If both the g and e positions contain charged amino acids, we have colored the gabcde amino acids. Our description of “attractive” and “repulsive” refers to pairs that would be present in the homodimer. We use the four colors to describe the g↔e′ pairs. Green is for the attractive basic-acidic pairs (R↔E and K↔E), orange is for the attractive acidic-basic pairs (E↔R, E↔K, D↔R, and D↔K), red is for the repulsive acidic pairs (E↔E, E↔D, E↔Q, and Q↔E), and blue is for the repulsive basic pairs (K↔K, R↔K, Q↔K, R↔Q, and K↔Q (Table 3). If only one of the two amino acids in the g↔e′ pair is charged, we color only that amino acid: red if it is acidic and blue if it is basic. If the a or d positions contain polar or charged amino acids, they are colored black.
We divide these 12 families into three general groups: (i) those that strongly favor homodimerization within the family (PAR, CREB, Oasis, and ATF6), (ii) those that homodimerize and heterodimerize (C/EBP, ATF4, ATF2, JUN, and the small MAFs), and (iii) those that strongly favor heterodimerization with other families (FOS, CNC, and large MAFs). Amino acids in the leucine zipper region of each human B-ZIP domain that regulate attractive and repulsive interactions are color-coded in Fig. 3 and reveal similar interaction patterns within each family. Figure 4 presents a helical wheel representation of homo- and heterodimers to help explain the color code used in Fig. 3. A similar annotation for Drosophila B-ZIP proteins indicates that the 12 families we have identified are conserved in the insects (17).
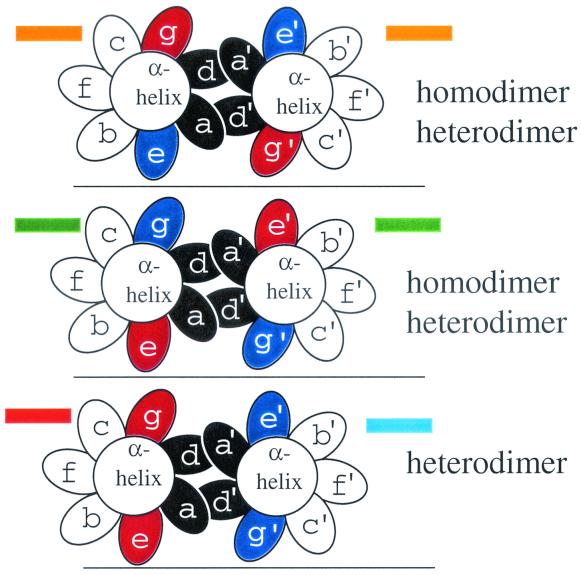
FIG. 4.
Schematic of leucine zipper dimerization according to the color code used in Fig. 3. End view of a coiled coil with the seven unique positions of the heptad presented as ellipses, looking from the N terminus to the C terminus. The a and d positions are colored black. Three possible combinations of acidic and basic amino acids in the g and e positions are presented and color-coded as in Fig. 3. (Top panel) A coiled coil with a g↔e′ pair containing an acidic amino acid in the g position and a basic amino acid in the following e position (orange in Fig. 3) can form a homo- or heterodimer with a similarly charged α-helix. (Middle panel) An α-helix with a g↔e′ pair containing a basic amino acid in the g position and an acidic amino acid in the following e position (green in Fig. 3) can form a homo- or heterodimer with a similarly charged α-helix. (Bottom panel) A heterodimer between an acidic g↔e′ pair (red in Fig. 3) and a basic g↔e′ pair (blue in Fig. 3).
An examination of the g↔e′ pairs in the first four heptads of human B-ZIP proteins results in several general observations. Thirty percent are attractive, with acidic-basic pairs (orange) predominating; 23% are repulsive with acidic-acidic (red) pairs predominating; and 30% contain a single charged amino acid that can stabilize either homodimerization or heterodimerization.
The d position of the hydrophobic interface typically contains leucine with very few polar and no charged amino acids. The 2nd heptad a position typically contains asparagine. The a position in other heptads typically contains hydrophobic amino acids, but asparagine and basic amino acids are occasionally observed, which are critical for regulating dimerization specificity. The surprisingly limited diversity of amino acids in the a, d, e, and g positions suggests that a limited set of rules might regulate leucine zipper dimerization specificity.
STRUCTURAL RULES REGULATING DIMERIZATION SPECIFICITY
In this section, we will review what is known about the contribution of individual amino acids to leucine zipper dimerization specificity and apply these rules to all human B-ZIP proteins. B-ZIP dimerization stability can be measured by circular dichroism spectroscopy (CD). Ellipticity at 222 nm indicates the presence of α-helical structure, and ellipticity decreases as the B-ZIP structure is unfolded by denaturants such as heat or urea. Thermal- or denaturant-induced unfolding of B-ZIP dimers occurs cooperatively and reversibly from an α-helical dimer to an unfolded monomer. The homodimerizing B-ZIP domains typically have a melting temperature (Tm [midpoint of thermal transition]) of approximately 50°C, which represents 10 kcal/mol/dimer or 100 nM affinity at 37°C (1, 48, 49). The thermal denaturation is well described by a two-state model so that mutational analysis of specific amino acids allows their contribution to dimerization stability to be measured (47, 87).
The g and e positions.
Seventy-six percent of the g and e positions in the leucine zipper of human B-ZIP proteins contain one of four long-side-chain amino acids: the acidic glutamic acid (E), the basic arginine (R) or lysine (K), or the polar glutamine (Q) (2, 8, 74, 96). Changing the g and e positions in two heptads of the PAR B-ZIP domain from charged amino acids to alanine resulted in the formation of tetramers instead of dimers (48)[]. Thus, charged amino acids in the g and e positions inhibit the higher-order oligomerization that would occur if the g and e positions formed a continuous hydrophobic surface with the a and d positions.
Charged amino acids in the g and e positions contribute to leucine zipper stability. The most common g↔e′ pair has glutamic acid (E) in the g position and an oppositely charged arginine (R) or lysine (K) in the following e′ position (E↔R and E↔K). These oppositely charged amino acids produce a g↔e′ pair that contributes −1.3 (E↔R) or −0.9 (E↔K) kcal/mol/pair more energy to dimer stability than a reference alanine pair (A↔A) (47). The g↔e′ pairs R↔R and K↔K are stabilizing, contributing −0.1 and −0.3 kcal/mol/pair, respectively, relative to the A↔A pair. Presumably the repulsive electrostatic energies between the like charges in arginine or lysine pairs are overcome by the favorable Van der Waals interactions between the methylenes of arginine or lysine side chains and hydrophobic amino acids in the a and d positions (2). E↔E is the only pair less stable than A↔A, contributing +0.4 kcal/mol/pair.
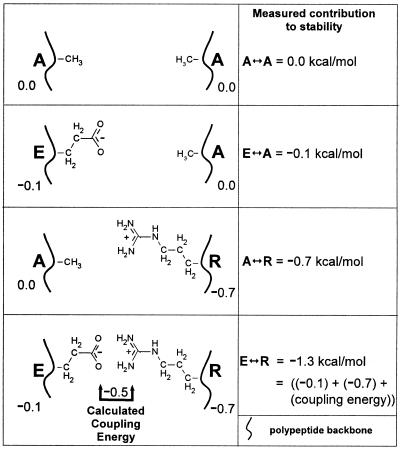
In addition to affecting stability, charged amino acids in the g and e positions also regulate dimerization specificity (2, 3, 48, 57, 67, 74, 96, 104). In the analysis of stability presented above, we considered the stability of a charged g↔e′ pair relative to an A↔A pair without addressing any potential energetic interaction between the amino acids in the g↔e′ pair; the contribution of the individual E or R side chain to stability can be determined by examining the E↔A and A↔R pairs. Any excess stability conferred by the E↔R pair is due to the energetic interaction between E and R, termed the “coupling energy.” This can be calculated by using a double-mutant thermodynamic cycle (31, 81) that involves the analysis of four proteins. This idea is presented graphically in Fig. 5. For example, the coupling energy of the E↔R pair is derived from a comparison of a protein containing the E↔R pairs with three additional proteins mutated to contain the pairs A↔R, E↔A, and A↔A (47). The levels of stability of the pairs relative to A↔A are as follows: E↔R, −1.3 kcal/mol; E↔A, −0.1 kcal/mol; and A↔R, −0.7 kcal/mol. The additional energy of the E↔R pair compared to the A↔R and E↔A pairs is −0.5 kcal/mol/salt-bridge [E↔R − (A↔R + E↔A) = coupling energy] [−1.3 − (−0.7 + −0.1) = −0.5] and represents the energy of interaction (coupling energy) between E and R. Table 2 lists the stability of each g↔e′ pair relative to A↔A, and Table 3 gives their coupling energy.
FIG. 5.
Schematic describing the four proteins used for a double-mutant thermodynamic cycle. The top panel depicts two alanines in the g and e′ positions of a g↔e′. The second panel shows that an E↔A pair is 0.1 kcal/mol more stable than an A↔A pair. The third panel shows that a A↔R pair is 0.7 kcal/mol more stable than an A↔A pair. The fourth panel shows a E↔R pair. Instead of being 0.8 kcal/mol more stable than A↔A, as would be expected if the two amino acids did not interact, E↔R is 1.3 kcal/mol more stable. The additional 0.5 kcal/mol is described as the coupling energy indicative of an physical interaction between the E and R side chains.
TABLE 2.
Thermodynamic differences for g↔e′ pairs relative to A↔A (ΔΔGAA in kcal mol−1/pair) a
| g |
e′
|
||||
|---|---|---|---|---|---|
| A | E | Q | R | K | |
| A | 0.0 | −0.2 | −0.8 | −0.7 | −0.5 |
| E | −0.1 | +0.4 | −0.7 | −1.3 | −0.9 |
| Q | −0.4 | −0.5 | −1.2 | −0.8 | −0.8 |
| R | −0.2 | −1.6 | −0.6 | −0.1 | −0.1 |
| K | −0.2 | −1.4 | −0.7 | −0.3 | −0.3 |
| D | >+1.6 | +0.4 | +0.6 | ||
Thermodynamic differences were determined with 150 mM KCl and 12.5 mM phosphate (pH 7.4) (47). Negative numbers indicate more stability than A↔A.
TABLE 3.
Coupling energy of interaction for g↔e′ pairs (ΔΔGint in kcal mol−1/pair) a
| g |
e′
|
|||
|---|---|---|---|---|
| E | Q | R | K | |
| E | +0.7 | +0.2 | −0.5 | −0.3 |
| Q | +0.2 | −0.0 | −0.3 | +0.3 |
| R | −1.1 | +0.4 | +0.8 | +0.8 |
| K | −0.9 | +0.3 | −0.6 | +0.6 |
Values were calculated from Table 2.
The larger coupling energy for the E↔R pair (−0.5 kcal/mol) than that of the E↔K pair (−0.3 kcal/mol) indicates that the E↔R pair contributes more to dimerization specificity than E↔K. The like-charged E↔E, K↔K, and R↔R pairs all have destabilizing coupling energies (+0.7, +0.6, and +0.8 kcal/mol, respectively) that are larger than the E↔R and E↔K attractive coupling energies (Table 3). This suggests that preventing a repulsive g↔e′ pair is more important for driving dimerization specificity than forming an attractive pair. The energetic basis of the coupling energy for g↔e′ pairs is a subject of ongoing debate in the literature (47, 56, 59), with some studies suggesting the measured coupling energy does not have a strong electrostatic component.
The suggestion that coupling energy may not be driven by charge interactions is highlighted by the polar glutamine that has a repulsive coupling energy in pairs with either acidic E or basic K (Table 3). Because of the positive calculated coupling energies, we have color-coded E↔Q and Q↔E pairs in red as depicting repulsive acidic pairs and K↔Q, R↔Q, and Q↔K in blue as depicting repulsive basic pairs (Fig. 3).
Many B-ZIP families, including JUN, CNC, and C/EBP, have only one charged amino acid in the g↔e′ pair. These charged amino acids contribute to the stability of the homodimer, as seen in the A↔R and E↔A pairs in the double-mutant thermodynamic analysis. Heterodimers, however, may be preferred if they form an attractive g↔e pair, because of the coupling energy. Thus, incomplete g↔e′ pairs can stabilize both homodimer and heterodimeric interactions.
The a and d positions.
Amino acids in the a and d positions of the leucine zipper are typically hydrophobic, with a variety of amino acids in the a positions and leucine in 84% of the d positions. An exception is the 2nd heptad a position, which contains asparagine in most homodimerizing B-ZIP proteins. The a and d amino acids are packed in the “holes and knobs” pattern predicted by Crick in 1953 (10), which is essential for leucine zipper dimer stability (87). When the GCN4 leucine zipper a and d positions were changed from valine and leucine to other aliphatic residues, trimers and tetramers formed (26). Similar to the role of amino acids in the e and g positions, amino acids in the a and d positions function to prevent higher-order oligomerization.
Several groups have examined the contribution of amino acids in the a and d positions to leucine zipper stability (63, 88). Leucine in the 4th d position is 9.2 kcal/mol/dimer more stable than alanine and 5.9 kcal/mol/dimer more stable than isoleucine. The additional stability conferred by leucine relative to isoleucine, an amino acid of similar size, likely results from the unique packing interactions of the two leucines with each other and with neighboring amino acids (63). The preferential energetic contribution of leucine over other aliphatic amino acids is not observed in the a position (99, 105; Vinson laboratory, unpublished data).
In addition to g↔e′ pairs, amino acids in the a position can also regulate dimerization specificity. The asparagine side chains, which are common in the 2nd heptad a position (Fig. 3), interact interhelically to form a polar pocket in the hydrophobic interface that limits oligomerization and directs the orientation of the α-helices (69). Hu and coworkers (103) mutated various a position asparagines to isoleucine and found that the asparagine-containing molecules interact preferentially with each other; thus, asparagine a position interactions are important for homodimerization specificity. In contrast, the basic amino acids lysine and arginine in the a position result in repulsive interactions that destabilize homodimerization and thus favor heterodimerization.
Coupling energies for the g↔e′ pair can be measured in the context of a homodimerizing system because the g and e positions in the monomer (and thus the g and e' positions in the dimer) can be changed independently. In contrast, changing a and a′ or d and d′ independently requires a heterodimerizing system. The contribution of the d position to heterodimer formation remains to be examined. However, the prevalence of leucine in the d position in mammalian B-ZIPs suggests that this position contributes to stability rather than to specificity. The a position in contrast is more variable than the d position, suggesting this position may be important for regulating dimerization specificity.
We used a heterodimerizing leucine zipper system to determine the contribution of alanine, leucine, isoleucine, valine, asparagine, and lysine in the a position to dimerization specificity. Pairs comprising any combination of the three aliphatic amino acids leucine, isoleucine, and valine have similar coupling energies. Asparagine in contrast, prefers to interact with itself and not with the aliphatic amino acids. The N↔N and V↔V interactions are more stable than the N↔V interactions by 2.3 and 5.3 kcal/mol, respectively. Therefore, asparagine drives interactions with asparagine in the a position. Asparagines in the a position of the 4th heptads of the Oasis family and the 3rd and 5th heptads of the ATF6 families appears to be critical for their predicted homodimerization. In contrast, K↔K interactions in the a position are repulsive, so that lysine preferentially interacts with asparagine and the aliphatic amino acids (Vinson laboratory, unpublished data). The FOS, CNC, large MAF, and small MAF B-ZIP families use basic amino acids in the a position to create heterodimerizing B-ZIP families
REPULSION BETWEEN LEUCINE ZIPPERS: USING A-ZIP DOMINANT NEGATIVES
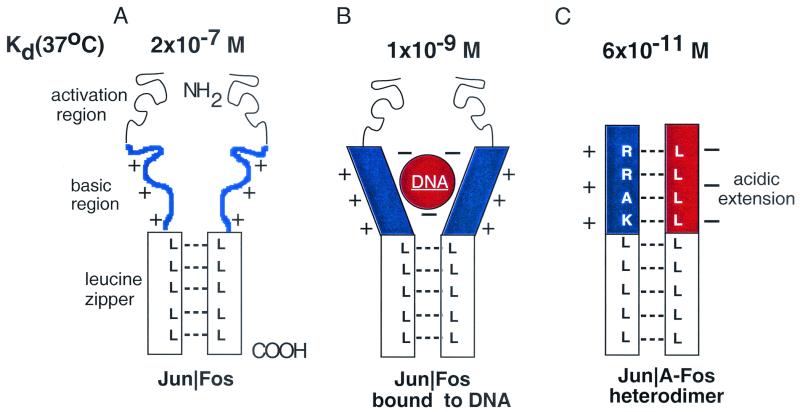
A problem with analyzing leucine zipper interactions by CD spectroscopy is that the stability of heterodimers can only be measured if the heterodimer is significantly more stable than either homodimer. To gain further insights into dimerization specificity, we have developed an A-ZIP protein consisting of a leucine zipper and a designed amphipathic acidic α-helical sequence that replaces the B-ZIP basic region (Fig. 6). This acidic extension forms a coiled-coil structure with the basic region in the B-ZIP|A-ZIP heterodimer and stabilizes the complex by up to 8 kcal/mol (1, 22, 49, 64, 71). The B-ZIP|A-ZIP heterodimer drives interactions between weakly attractive or even somewhat repulsive leucine zippers and gives us access to measuring a range of dimerization affinities. This is important because in vivo dimerization is often driven by DNA binding. The B-ZIP|A-ZIP heterodimer is more stable than the B-ZIP protein bound to DNA, so that the A-ZIPs specifically prevent B-ZIP DNA binding at equimolar concentrations. Because the acidic extension interacts with all basic regions, the specificity of interaction between A-ZIP and B-ZIP domains is primarily leucine zipper dependent (1, 22, 64, 70). The inhibition of DNA binding provides an assay for dimerization.
FIG. 6.
Schematic of the A-ZIP dominant negative. (A) A model of a B-ZIP dimer with the basic region (blue) unstructured. (B) A B-ZIP dimer bound to DNA with the basic region now α-helical. (C) A B-ZIP|A-ZIP heterodimer with the protein-protein interface extending N terminally into the basic region. The basic region (blue) and the designed acidic amphipathic region (red) interact as α-helices to extend the coiled-coil domain.
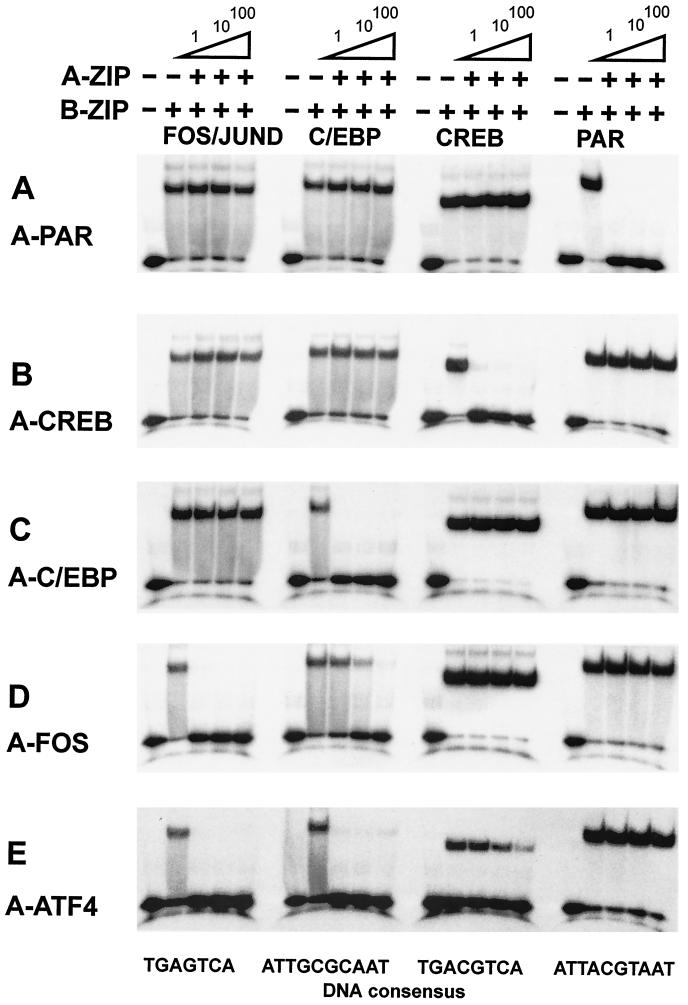
Competition between A-ZIP protein and DNA for interaction with a B-ZIP protein can be analyzed with the gel shift assay. Figure 6 shows the FOS|JUND heterodimer and the C/EBPα, CREB, and PAR homodimers binding to their canonical DNA binding sites. One molar equivalent of A-PAR inhibits the DNA binding of PAR; it does not inhibit the DNA binding of FOS|JUND, C/EBPα, or CREB, even at 100 molar equivalences (Fig. 7, top panel). Similarly, 1 molar equivalent of A-CREB, A-C/EBP, and A-FOS specifically inhibit the DNA binding of CREB, C/EBP, and FOS|JUND, respectively. In contrast, A-ATF4 has somewhat promiscuous dimerization properties, inhibiting the DNA binding of FOS|JUND and C/EBPα at 1 molar equivalent (Fig. 7E).
FIG. 7.
B-ZIP DNA binding is inhibited by specific A-ZIP dominant negatives. Four B-ZIP dimers, the PAR, CREB, and C/EBPα homodimers, and the FOS|JUND heterodimer are bound to a 28-bp DNA containing the optimal binding site for each B-ZIP dimer. One, 10, or 100 molar equivalents of the A-ZIP is added to the B-ZIP before addition of DNA, and the solution is electrophoresed with an electrophoretic mobility shift assay. The sequence of the 28-mer DNA probes is shown below with the consensus binding sites underlined: GTCAGTCAGAATGACTCATATCGGTCAG (AP-1), GTCAGTCAGATGACGTCATATCGGTCAG (CREB), GTCAGTCAGATTACGTAATATCGGTCAG (VBP), and GTCAGTCAGATTGCGCAATATCGGTCAG (C/EBP).
We can rationalize the specificity seen in Fig. 7 based on the leucine zipper sequence of each protein. The homodimerizing PAR, CREB, and C/EBP families have similar a and d positions with an asparagine in the a position of the 2nd heptad. The families differ in their attractive g↔e′ pairs, PAR has four attractive g↔e′ pairs, while CREB and C/EBP each have two attractive g↔e′ pairs that are subsets of the PAR pattern. Dimerization between PAR and CREB or PAR and C/EBP is prevented, because PAR preferentially homodimerizes due to the 4.0-kcal/mol/dimer of coupling energy from the eight attractive g↔e′ pairs in the four heptads. Therefore, CREB or C/EBP remain to homodimerize. C/EBP and CREB do not interact because their g↔e′ pairs are in different heptads. To test the validity of this idea, we mutated only three amino acids in the g and e positions of the C/EBP leucine zipper to confer the PAR pattern of g↔e′ pairs. The mutated C/EBP displays dimerization properties similar to those of PAR (64).
THE 13 B-ZIP FAMILIES
We can predict dimerization partners from Fig. 3 by using the following strategy. Heptads that interact to produce attractive g↔e′ pairs and drive dimerization are green-green, orange-orange, red-blue, and blue-red. Heptads that interact to give repulsive g↔e′ pairs and discourage dimerization are green-orange, orange-green, red-red, or blue-blue. As noted previously, biophysical measurements show that repulsive g↔e′ pairs are more important than attractive g↔e′ pairs in driving dimerization specificity. The a and d positions that affect dimerization are colored black. Asparagine in the a position will preferentially dimerize with another a position asparagine, while basic amino acids in the a position repel each other and thus drive heterodimerization. It should be appreciated that these rules are an oversimplification! We expect more detailed work will show more interactions that are critical for mediating dimerization specificity.
In the following section, we describe the known dimerization properties of each B-ZIP family and rationalize these properties based on the leucine zipper amino acid sequences.
The homodimerizing B-ZIP families.
The homodimerizing B-ZIP families are PAR, CREB, Oasis, and ATF6. Homodimerizing leucine zippers have two defining properties. First, each B-ZIP family has a distinct pattern of attractive g↔e′ pairs. Second, all have asparagine in the a position of the 2nd heptad. The Oasis family has an additional a position asparagine in the 4th heptad, while the ATF6 family has two additional a position asparagines in the 3rd and 5th heptads that help prevent heterodimerization with other B-ZIP proteins.
(i) PAR family.
The PAR family, named for a conserved proline- and acid-rich domain N terminal of the basic region (14), consists of four family members, TEF/VBP, HLF, DBP, and the more distant NFIL3. PAR family members dimerize within the family (14, 33), but not with C/EBP family members (36). The dimers bind the palindromic DNA consensus sequence (5′-ATTACGTAAT-3′), which differs from the CREB binding site by 1 bp per half-site. We view PAR with its four attractive g↔e′ pairs as the canonical homodimerizing leucine zipper. The 1st heptad contains an R↔E pair (green), and the 2nd, 3rd, and 4th heptads contain E↔R or E↔K pairs (orange). The a and d positions contain aliphatic amino acids, except for an asparagine in the 2nd a position. The attractive g↔e′ pairs and the absence of destabilizing a and d amino acids suggest that these proteins will homodimerize.
(ii) CREB family.
The CREB family, which has one of the shortest leucine zippers, contains three members ATF1, CREM, and CREB. ICRE is an alternative splice product of the CREB gene. These proteins form dimers within the family (11, 12, 60). CREB homodimers bind the palindromic DNA consensus sequence 5′-TGACGTCA-3′, termed the cyclic AMP responsive element (CRE).
This family has conserved attractive R↔E pairs (green) in the 1st heptad and E↔K pairs (orange) in the 3rd heptad. The crystal structure of CREB bound to a consensus CRE identified the R↔E and E↔K interhelical interactions and an additional Y↔E interaction one heptad N-terminal of the leucine zipper, as being important in dimer stability (80). A single asparagine in the 2nd heptad a position and the unique pattern of g↔e′ pairs results in homodimerization among CREB members. Although the g↔e′ pairs are a subset of those found in the PAR family, heterodimerization between PAR and CREB is not predominant, because the PAR|PAR homodimer has greater coupling energy than a PAR|CREB heterodimer.
(iii) Oasis family.
The Oasis family has five proteins, Oasis (30), CREB-H (72), CREB3 (18, 55), hcp201085, and hcp1698600. These proteins also bind the CRE DNA sequence. The 2nd and 3rd heptads contain attractive g↔e′ pairs, and the a positions of the 2nd and 4th heptads contain asparagine. Two characteristics are consistent with this being a homodimerizing family. First, there is a unique pattern of attractive g↔e′ pairs. Second, the asparagine in the 4th a position will encourage dimerization within the family, but not with the other homodimerizing families without an asparagine in the 4th a position.
(iv) ATF6 family.
ATF6 is a group of three proteins, ATF6, CREBL1, and Xbp-1 (7). ATF6 binds to three DNA sequences, termed ERSE-, SRE-, and CRE-like sites (24, 100).
ATF6 proteins have a unique pattern of attractive g↔e′ pairs (orange) in the 2nd, 3rd, and 5th heptads. These three heptads also contain asparagine in the a position. This novel combination of attractive g↔e′ pairs and asparagine placements is likely to promote dimerization within the family and discourage interactions with other B-ZIP proteins. Xbp-1 has a 4th heptad a position threonine that may result in exclusive homodimerization of this protein. However, the interactions in the 2nd, 3rd, and 5th heptads remain the same, so that interaction of Xbp-1 with other ATF6 proteins is a possibility.
The homodimerizing and heterodimerizing B-ZIP families.
The C/EBP, ATF4, ATF2, JUN, and the small MAF families homodimerize and heterodimerize with other B-ZIP families. These proteins have properties found in both the homodimerizing and heterodimerizing proteins.
(i) C/EBP family.
The C/EBP family consists of seven members: C/EBPα, C/EBPβ, C/EBPγ, C/EBPδ, C/EBPɛ, CAA60698, and CHOP10. C/EBP proteins form dimers within the family (5, 101) that bind to the palindromic DNA consensus sequence 5′-ATTGCGCAAT-3′ and the related CRE and PAR sites (16). The CHOP10 basic region is unique among the listed B-ZIP proteins with a proline in the basic region that likely distorts the α-helical basic region and alters DNA binding. The CHOP10|C/EBP dimer binds a unique DNA sequence (92). Dimerization within the family is driven by attractive g↔e′ (orange) pairs in the 2nd and 4th heptads and a single asparagine at the 2nd heptad a position.
Heterodimeric interactions have been reported between C/EBP and other B-ZIP families, such as the FOS, JUN (32), ATF4 (93, 96), and ATF2 (83) families. C/EBPβ has also been reported to interact with CREB (89), however, A-C/EBP and A-CREB fail to inhibit the DNA binding of CREB and C/EBP, respectively (Fig. 6), suggesting repulsion between the leucine zippers of C/EBP and CREB. Heterodimerization between C/EBP and other B-ZIP proteins may be promoted by incomplete g↔e′ pairs found in the 1st and 3rd heptad g positions of C/EBP.
One of the C/EBP members, C/EBPγ, may have dimerization properties similar to those found in the FOS and JUN families described later. These properties include a repulsive R↔K (blue) pair in the 1st heptad found in JUN, a repulsive E↔E (red) pair in the 2nd heptad found in FOS, and a histidine in the 5th d position, as found in JUN, FOS, and ATF2.
(ii) ATF4.
The ATF4 family has three members, ATF4, ATF5, and hcp1709392. Besides homodimerizing, ATF4 heterodimerizes with C/EBP (93, 96), FOS (23), and NRF2 (27). This family is noteworthy in having acidic and basic repulsive g↔e′ pairs as well as attractive g↔e′ pairs, which may explain their promiscuous dimerization properties. The interface contains the 2nd heptad asparagine in the a position found in homodimerizing B-ZIP proteins.
(iii) ATF2 family.
The ATF2 family contains three proteins: ATF2, ATF7, and CRE-BPa (68). These proteins contain an asparagine in the 2nd heptad a position and attractive g↔e′ pairs (orange) in the 3rd and 4th heptads, structural features that favor homodimerization. The 5th heptad d position contains a histidine that is also found in the FOS and JUN families and may be important for interaction with both of these families.
ATF2 has been reported to heterodimerize with JUN (23, 58), FOS (28), and C/EBP (83) family members. Heterodimerization with FOS could be driven by an incomplete g↔e′ pair in the 1st heptad of C/EBP interacting with the E↔E pair in the 1st heptad of FOS to form an attractive K↔E pair.
Heterodimerization between C/EBP and ATF2 has been observed on a chimeric DNA sequence composed of a C/EBP half site and an ATF2 half site (83). CEBPα and ATF2 homodimers have four attractive interhelical salt bridges and no repulsive g↔e′ pairs, while a ATF2|C/EBP heterodimer has two attractive and one repulsive g↔e′ pair, suggesting that these two proteins would prefer to homodimerize. The formation of heterodimerization on a chimeric site demonstrates the importance of DNA sequence in modulating dimerization specificity.
(iv) JUN family.
The JUN family is comprised of three proteins, c-JUN, JUND, and JUNB. The best-studied JUN partner is FOS. JUN and FOS heterodimerize to form the AP-1 transcription factor, originally isolated as a biochemical activity, that binds the 5′-TGAGTCA-3′ DNA sequence, termed the “TRE” (12-O-tetradecanoylphorbol-13-acetate response element) (79, 82). Many other proteins have been reported to heterodimerize with JUN family members, among them CNC, ATF2 (58, 68), ATF3, and c-Maf. JUN family members can also homodimerize, but these complexes bind DNA poorly, bringing into question their biological function (70).
The amino acid sequence of the JUN leucine zipper is consistent with the experimentally observed promiscuous dimerization. Properties that drive heterodimerization are repulsive K↔K (blue) and Q↔K pairs (blue) in the 1st and 4th heptads, respectively. An incomplete g↔e′ pair in the 3rd heptad also promotes promiscuous heterodimerization. In contrast, the asparagine in the 2nd heptad a position is commonly found in homodimerizing B-ZIP proteins. Heterodimerization with ATF2 creates a canonical interface and attractive g↔e′ pairs. An elegant study (94) showed that by changing amino acids in the e and g positions, the promiscuous dimerization of JUN could be restricted to either FOS or ATF2 with a corresponding change in the biological activity of the JUN mutants. The large number of basic amino acids in the g and e positions encourages heterodimerization with acidic proteins such as FOS. A histidine (H) in the 5th heptad d position is conserved among the JUN, FOS, and ATF2 proteins and contributes to dimer stability (9), but its contribution to dimerization specificity has yet to be elucidated.
(v) Small MAF family.
There are three small MAF (S-MAF) musculoaponeurotic fibrosarcome proteins, MafF, MafG, and MafK. The S-MAFs homodimerize, but do not contain, a transactivation domain and thus repress transcription. However, they also heterodimerize with CNC and FOS family members to activate gene expression (35, 40, 43). The S-MAF leucine zipper contains attractive E↔R (orange) in the 3rd and 4th heptads that favor homodimerization (also observed in the ATF2 family) and an asparagine in the 3rd a heptad. Features that promote heterodimerization include a lysine in the 1st heptad a position (also found in the L-MAFs) and an incomplete glutamic acid g↔e′ pair (red) in the 2nd heptad.
The heterodimerizing B-ZIP families.
The heterodimerizing B-ZIP families are FOS, CNC, and the large Maf. Three general properties are apparent for these proteins: (i) repulsive g↔e′ pairs that inhibit homodimerization; (ii) amino acids in the a positions include lysine or arginine, which discourages homodimerization; (iii) incomplete g↔e′ pairs that promote promiscuous heterodimerization.
(i) FOS family.
There are nine FOS family proteins: c-Fos, FosB, Fra1, Fra2, hcp34067, ATF3, JDP2, SNFT, and BATF. FOS family members heterodimerize with the JUN, CNC, and small Maf families (reviewed in reference 6). The FOS family has acidic amino acids in the g and e positions and heterodimerize with basic JUN zippers (74). Specifically, FOS dimerization properties can be divided into three sets of characteristics that are slightly variable. Five of the proteins (c-Fos, FosB, Fra1, Fra2, and hcp34067) contain conserved repulsive E↔E or Q↔E pairs (red) in the 1st and 4th heptads and repulsive E↔Q or E↔E pairs (red) in the 2nd and 3rd heptads. The hydrophobic interface is composed of a threonine in the 1st a position, lysines in the 2nd and 4th a positions, and a histidine in the 5th heptad. The lysines in the a position inhibit homodimerization and drive heterodimerization.
The remaining FOS family members, ATF3, JDP2, SNFT, and BATF, are not as acidic and do not contain as many repulsive lysines in the a position as the prototypical FOS leucine zipper. They can be further divided into two groups: SNFT and BATF, which do not contain the repulsive 4th heptad a position basic amino acid found in ATF3 and JDP2.
(ii) CNC family.
The second acidic leucine zipper family is named after the founding member, cap'n'collar (CNC), a Drosophila protein (62). There are six members: BACH1, CNC1/NRF1, NF-E2, BACH2, CNC2/NRF2, and NF-E2L3. These proteins heterodimerize with the S-MAF family (35, 66). These proteins have either a repulsive acidic g↔e′ pair or an acidic incomplete g↔e′ pair in the 1st heptad. The presence of multiple incomplete g↔e′ pairs is expected to confer promiscuous dimerization properties. Like the FOS family, the CNC family has lysines in the a positions that drive heterodimerization. However, in contrast to the FOS family, where the lysines are in the 2nd and 4th heptad a positions, CNC proteins contain a basic amino acid in the 2nd and 3rd heptad a positions. This arrangement should alter the heterodimerization partners for the CNC proteins compared to FOS proteins. Interestingly, the CNC|S-MAF heterodimer forms a 3rd heptad a position interaction between N and K, similar to the 2nd heptad interaction found between FOS and JUN in the 2nd heptad.
(iii) Large MAF family.
The large MAF (L-MAF) family contains three members: NRL, c-MAF, and MAF-B. These proteins can homodimerize and heterodimerize with FOS and JUN proteins (39, 43).
L-MAF family members form heterodimers driven by the repulsive Q↔K (blue) or Q↔R pairs (blue) in the 2nd heptad, while homodimerization is mediated through attractive E↔K pairs (orange) in the 4th heptad. Along with the small MAF proteins, these are the only B-ZIP proteins with aliphatic amino acids in the 2nd a position. The 1st and 4th heptad a position lysine or arginine discourages homodimerization. Features that promote promiscuous heterodimerization include incomplete g↔e′ pairs composed of glutamic acid (red) in the 1st and 3rd heptads.
(iv) HCF family.
The HCF (host cell factor) protein, also known as C1 (45), VCAF (76), or CFF (38), exists in human cells as a family of proteolytic cleavage products of the primary HCF protein (46).
HCF has a unique pattern of interactions that suggest it will homodimerize. This protein contains attractive g↔e′ pairs (orange) in the 1st and 4th heptads. The a positions contain asparagine in the 1st and 2nd heptad a positions and serine (a small polar amino acid similar to asparagine) in the 4th heptad. HCF was found to interact with Luman (18, 55).
SUMMARY
We have reviewed the literature on the dimerization properties of mammalian B-ZIP proteins and made predictions about the amino acids in the a, d, e, and g positions of the leucine zipper that mediate their known dimerization specificities. We have extended these predictions to all the identified B-ZIP proteins in the human genome. These predictions appear more robust for the homodimerizing proteins than for the heterodimerizing proteins. This type of analysis will be valuable in predicting the dimerization properties of B-ZIP proteins from newly sequenced genomes for which much less experimental data exists. Additional experimental data is needed to quantify the attraction and repulsion between different B-ZIP leucine zippers to gain insight into which dimers may form in vivo. In addition, the contribution of DNA binding to B-ZIP stability needs to be quantified to gain insight into how DNA sequences can regulate B-ZIP dimer partner choice.
Acknowledgments
We thank Tsonwin Hai, Jon Shuman, and an anonymous reviewer for comments on the manuscript.
REFERENCES
- 1.Ahn, S., M. Olive, S. Aggarwal, D. Krylov, D. D. Ginty, and C. Vinson. 1998. A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol. Cell. Biol. 18:967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alber, T. 1992. Structure of the leucine zipper. Curr. Opin. Genet. Dev. 2:205-210. [DOI] [PubMed] [Google Scholar]
- 3.Baxevanis, A. D., and C. R. Vinson. 1993. Interactions of coiled coils in transcription factors: where is the specificity? Curr. Opin. Genet. Dev. 3:278-285. [DOI] [PubMed] [Google Scholar]
- 4.Bohmann, D., T. J. Bos, A. Admon, T. Nishimura, P. K. Vogt, and R. Tjian. 1987. Human proto-oncogene c-jun encodes a DNA binding protein with structural and functional properties of transcription factor AP-1. Science 238:1386-1392. [DOI] [PubMed] [Google Scholar]
- 5.Cao, Z., R. M. Umek, and S. L. McKnight. 1991. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 5:1538-1552. [DOI] [PubMed] [Google Scholar]
- 6.Chinenov, Y., and T. K. Kerppola. 2001. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene 20:2438-2452. [DOI] [PubMed] [Google Scholar]
- 7.Clauss, I. M., M. Chu, J. L. Zhao, and L. H. Glimcher. 1996. The basic domain/leucine zipper protein hXBP-1 preferentially binds to and transactivates CRE-like sequences containing an ACGT core. Nucleic Acids Res. 24:1855-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen, C., and D. Parry. 1990. A-helical coiled coils and bundles: how to design an α-helical protein. Protein 7:1-14. [DOI] [PubMed] [Google Scholar]
- 9.Cohen, D. R., and T. Curran. 1990. Analysis of dimerization and DNA binding functions in Fos and Jun by domain-swapping: involvement of residues outside the leucine zipper/basic region. Oncogene 5:929-939. [PubMed] [Google Scholar]
- 10.Crick, F. 1953. The packing of α-helices: simple coiled-coils. Acta Crystallogr. 6:689-697. [Google Scholar]
- 11.Daniel, P. B., W. H. Walker, and J. F. Habener. 1998. Cyclic AMP signaling and gene regulation. Annu. Rev. Nutr. 18:353-383. [DOI] [PubMed] [Google Scholar]
- 12.De Cesare, D., G. M. Fimia, and P. Sassone-Corsi. 1999. Signaling routes to CREM and CREB: plasticity in transcriptional activation. Trends Biochem. Sci. 24:281-285. [DOI] [PubMed] [Google Scholar]
- 13.Dlakic, M., A. V. Grinberg, D. A. Leonard, and T. K. Kerppola. 2001. DNA sequence-dependent folding determines the divergence in binding specificities between Maf and other bZIP proteins. EMBO J. 20:828-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drolet, D. W., K. M. Scully, D. M. Simmons, M. Wegner, K. T. Chu, L. W. Swanson, and M. G. Rosenfeld. 1991. TEF, a transcription factor expressed specifically in the anterior pituitary during embryogenesis, defines a new class of leucine zipper proteins. Genes Dev. 5:1739-1753. [DOI] [PubMed] [Google Scholar]
- 15.Ellenberger, T., C. Brandl, K. Struhl, and S. Harrison. 1992. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted α helices: crystal structure of the protein-DNA complex. Cell 71:1223-1237. [DOI] [PubMed] [Google Scholar]
- 16.Falvey, E., L. Marcacci, and U. Schibler. 1996. DNA-binding specificity of PAR and C/EBP leucine zipper proteins: a single amino acid substitution in the C/EBP DNA-binding domain confers PAR-like specificity to C/EBP. Biol. Chem. 377:797-809. [PubMed] [Google Scholar]
- 17.Fassler, J., D. Landsman, A. Acharya, J. Moll, M. Bonovich, et al. B-ZIP proteins encoded by the Drosophila genome: evaluation of potential dimerization partners. Genome Res., in press. [DOI] [PMC free article] [PubMed]
- 18.Freiman, R. N., and W. Herr. 1997. Viral mimicry: common mode of association with HCF by VP16 and the cellular protein LZIP. Genes Dev. 11:3122-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujii, Y., T. Shimizu, T. Toda, M. Yanagida, and T. Hakoshima. 2000. Structural basis for the diversity of DNA recognition by bZIP transcription factors. Nat. Struct. Biol. 7:889-893. [DOI] [PubMed] [Google Scholar]
- 20.Gentz, R., F. J. Rauscher III, C. Abate, and T. Curran. 1989. Parallel association of Fos and Jun leucine zippers juxtaposes DNA binding domains. Science 243:1695-1699. [DOI] [PubMed] [Google Scholar]
- 21.Glover, J. N., and S. C. Harrison. 1995. Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature 373:257-261. [DOI] [PubMed] [Google Scholar]
- 22.Greenwel, P., S. Tanaka, D. Penkov, W. Zhang, M. Olive, J. Moll, C. Vinson, M. Di Liberto, and F. Ramirez. 2000. Tumor necrosis factor alpha inhibits type I collagen synthesis through repressive CCAAT/enhancer-binding proteins. Mol. Cell. Biol. 20:912-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hai, T., and T. Curran. 1991. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc. Natl. Acad. Sci. USA 88:3720-3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hai, T., and M. G. Hartman. 2001. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene 273:1-11. [DOI] [PubMed] [Google Scholar]
- 25.Hai, T., F. Liu, W. Coukos, and M. Green. 1989. Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 3:2083-2090. [DOI] [PubMed] [Google Scholar]
- 26.Harbury, P. B., T. Zhang, P. S. Kim, and T. Alber. 1993. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262:1401-1417. [DOI] [PubMed] [Google Scholar]
- 27.He, C. H., P. Gong, B. Hu, D. Stewart, M. E. Choi, A. M. Choi, and J. Alam. 2001. Identification of activating transcription factor 4 (ATF4) as an Nrf2-interacting protein. Implication for heme oxygenase-1 gene regulation. J. Biol. Chem. 276:20858-20865. [DOI] [PubMed] [Google Scholar]
- 28.Hoeffler, J. P., J. W. Lustbader, and C. Y. Chen. 1991. Identification of multiple nuclear factors that interact with cyclic adenosine 3′,5′-monophosphate response element-binding protein and activating transcription factor-2 by protein-protein interactions. Mol. Endocrinol. 5:256-266. [DOI] [PubMed] [Google Scholar]
- 29.Hoeffler, J. P., T. E. Meyer, Y. Yun, J. L. Jameson, and J. F. Habener. 1988. Cyclic AMP-responsive DNA-binding protein: structure based on a cloned placental cDNA. Science 242:1430-1433. [DOI] [PubMed] [Google Scholar]
- 30.Honma, Y., K. Kanazawa, T. Mori, Y. Tanno, M. Tojo, H. Kiyosawa, J. Takeda, T. Nikaido, T. Tsukamoto, S. Yokoya, and A. Wanaka. 1999. Identification of a novel gene, OASIS, which encodes for a putative CREB/ATF family transcription factor in the long-term cultured astrocytes and gliotic tissue. Mol. Brain Res. 69:93-103. [DOI] [PubMed] [Google Scholar]
- 31.Horovitz, A., L. Serrano, B. Avron, M. Bycroft, and A. R. Fersht. 1990. Strength and cooperativity of contributions of surface salt bridges to protein stability. J. Mol. Biol. 216:1031-1044. [DOI] [PubMed] [Google Scholar]
- 32.Hsu, W., T. K. Kerppola, P.-L. Chen, T. Curran, and S. Chen-Kiang. 1994. Fos and Jun repress transcription activation by NF-IL6 through association at the basic zipper region. Mol. Cell. Biol. 14:268-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hunger, S. P., K. Ohyashiki, K. Toyama, and M. L. Cleary. 1992. Hlf, a novel hepatic bZIP protein, shows altered DNA-binding properties following fusion to E2A in t(17;19) acute lymphoblastic leukemia. Genes Dev. 6:1608-1620. [DOI] [PubMed] [Google Scholar]
- 34.Hurst, H. C. 1995. Transcription factors 1: bZIP proteins. Protein Profile 2:101-168. [PubMed] [Google Scholar]
- 35.Igarashi, K., K. Kataoka, K. Itoh, N. Hayashi, M. Nishizawa, and M. Yamamoto. 1994. Regulation of transcription by dimerization of erythroid factor NF-E2 p45 with small Maf proteins. Nature 367:568-572. [DOI] [PubMed] [Google Scholar]
- 36.Iyer, S. V., D. L. Davis, S. N. Seal, and J. B. E. Burch. 1991. Chicken vitellogenin gene-binding protein, a leucine zipper transcription factor that binds to an important control element in the chicken vitellogenin II promoter, is related to rat DBP. Mol. Cell. Biol. 11:4863-4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson, P. F., W. H. Landschulz, B. J. Graves, and S. L. McKnight. 1987. Identification of a rat liver nuclear protein that binds to the enhancer core element of three animal viruses. Genes Dev. 1:133-146. [DOI] [PubMed] [Google Scholar]
- 38.Katan, M., A. Haigh, C. P. Verrijzer, P. C. van der Vliet, and P. O'Hare. 1990. Characterization of a cellular factor which interacts functionally with Oct-1 in the assembly of a multicomponent transcription complex. Nucleic Acids Res. 18:6871-6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kataoka, K., K. T. Fujiwara, M. Noda, and M. Nishizawa. 1994. MafB, a new Maf family transcription activator that can associate with Maf and Fos but not with Jun. Mol. Cell. Biol. 14:7581-7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kataoka, K., K. Igarashi, K. Itoh, K. T. Fujiwara, M. Noda, M. Yamamoto, and M. Nishizawa. 1995. Small Maf proteins heterodimerize with Fos and may act as competitive repressors of the NF-E2 transcription factor. Mol. Cell. Biol. 15:2180-2190. (Erratum, 15: 3461.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kerppola, T., and T. Curran. 1991. Transcription factor interactions: basics on zippers. Curr. Opin. Struct. Biol. 1:71-79. [Google Scholar]
- 42.Kerppola, T. K., and T. Curran. 1994. A conserved region adjacent to the basic domain is required for recognition of an extended DNA binding site by Maf/Nrl family proteins. Oncogene 9:3149-3158. [PubMed] [Google Scholar]
- 43.Kerppola, T. K., and T. Curran. 1994. Maf and Nrl can bind to AP-1 sites and form heterodimers with Fos and Jun. Oncogene 9:675-684. [PubMed] [Google Scholar]
- 44.Kouzarides, T., and E. Ziff. 1988. The role of the leucine zipper in the fos-jun interaction. Nature 336:646-651. [DOI] [PubMed] [Google Scholar]
- 45.Kristie, T. M., J. H. LeBowitz, and P. A. Sharp. 1989. The octamer-binding proteins form multi-protein-DNA complexes with the HSV alpha TIF regulatory protein. EMBO J. 8:4229-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kristie, T. M., J. L. Pomerantz, T. C. Twomey, S. A. Parent, and P. A. Sharp. 1995. The cellular C1 factor of the herpes simplex virus enhancer complex is a family of polypeptides. J. Biol. Chem. 270:4387-4394. [DOI] [PubMed] [Google Scholar]
- 47.Krylov, D., J. Barchi, and C. Vinson. 1998. Inter-helical interactions in the leucine zipper coiled coil dimer: pH and salt dependence of coupling energy between charged amino acids. J. Mol. Biol. 279:959-972. [DOI] [PubMed] [Google Scholar]
- 48.Krylov, D., I. Mikhailenko, and C. Vinson. 1994. A thermodynamic scale for leucine zipper stability and dimerization specificity: e and g interhelical interactions. EMBO J. 13:2849-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krylov, D., M. Olive, and C. Vinson. 1995. Extending dimerization interfaces: the bZIP basic region can form a coiled coil. EMBO J. 14:5329-5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lamb, P., and S. L. McKnight. 1991. Diversity and specificity in transcriptional regulation: the benefits of heterotypic dimerization. Trends Biochem. Sci. 16:417-422. [DOI] [PubMed] [Google Scholar]
- 51.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, R. Funke, D. Gage, K. Harris, A. Heaford, J. Howland, L. Kann, J. Lehoczky, R. LeVine, P. McEwan, K. McKernan, J. Meldrim, J. P. Mesirov, C. Miranda, W. Morris, J. Naylor, C. Raymond, M. Rosetti, R. Santos, A. Sheridan, C. Sougnez, N. Stange-Thomann, N. Stojanovic, A. Subramanian, D. Wyman, J. Rogers, J. Sulston, R. Ainscough, S. Beck, D. Bentley, J. Burton, C. Clee, N. Carter, A. Coulson, R. Deadman, P. Deloukas, A. Dunham, I. Dunham, R. Durbin, L. French, D. Grafham, S. Gregory, T. Hubbard, S. Humphray, A. Hunt, M. Jones, C. Lloyd, A. McMurray, L. Matthews, S. Mercer, S. Milne, J. C. Mullikin, A. Mungall, R. Plumb, M. Ross, R. Shownkeen, S. Sims, R. H. Waterston, R. K. Wilson, L. W. Hillier, J. D. McPherson, M. A. Marra, E. R. Mardis, L. A. Fulton, A. T. Chinwalla, K. H. Pepin, W. R. Gish, S. L. Chissoe, M. C. Wendl, K. D. Delehaunty, T. L. Miner, A. Delehaunty, J. B. Kramer, L. L. Cook, R. S. Fulton, D. L. Johnson, P. J. Minx, S. W. Clifton, T. Hawkins, E. Branscomb, P. Predki, P. Richardson, S. Wenning, T. Slezak, N. Doggett, J. F. Cheng, A. Olsen, S. Lucas, C. Elkin, E. Uberbacher, M. Frazier et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 52.Landschultz, W. H., P. F. Johnson, and S. L. McKnight. 1988. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240:1759-1764. [DOI] [PubMed] [Google Scholar]
- 53.Landschulz, W. H., P. F. Johnson, E. Y. Adashi, B. J. Graves, and S. L. McKnight. 1988. Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev. 2:786-800. [DOI] [PubMed] [Google Scholar]
- 54.Landschulz, W. H., P. F. Johnson, and S. L. McKnight. 1989. The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science 243:1681-1688. [DOI] [PubMed] [Google Scholar]
- 55.Lu, R., P. Yang, P. O'Hare, and V. Misra. 1997. Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol. Cell. Biol. 17:5117-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lumb, K. J., and P. S. Kim. 1995. Measurement of interhelical electrostatic interactions in the GCN4 leucine zipper. Science 268:436-439. [DOI] [PubMed] [Google Scholar]
- 57.Lupas, A. 1996. Coiled coils: new structures and new functions. Trends Biochem. 21:375-382. [PubMed] [Google Scholar]
- 58.Macgregor, P. F., C. Abate, and T. Curran. 1990. Direct cloning of leucine zipper proteins: Jun binds cooperatively to the CRE with CRE-BP1. Oncogene 5:451-458. [PubMed] [Google Scholar]
- 59.Marti, D. N., I. Jelesarov, and H. R. Bosshard. 2000. Interhelical ion pairing in coiled coils: solution structure of a heterodimeric leucine zipper and determination of pKa values of Glu side chains. Biochemistry 39:12804-12818. [DOI] [PubMed] [Google Scholar]
- 60.Mayr, B., and M. Montminy. 2001. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2:599-609. [DOI] [PubMed] [Google Scholar]
- 61.McLachlan, A., and M. Stewart. 1975. Tropomyosin coiled-coil interactions: evidence for an unstaggered structure. J. Mol. Biol. 98:293-304. [DOI] [PubMed] [Google Scholar]
- 62.Mohler, J., K. Vani, S. Leung, and A. Epstein. 1991. Segmentally restricted, cephalic expression of a leucine zipper gene during Drosophila embryogenesis. Mech. Dev. 34:3-9. [DOI] [PubMed] [Google Scholar]
- 63.Moitra, J., L. Szilak, D. Krylov, and C. Vinson. 1997. Leucine is the most stabilizing aliphatic amino acid in the d position of a dimeric leucine zipper coiled coil. Biochemistry 36:12567-12573. [DOI] [PubMed] [Google Scholar]
- 64.Moll, J. R., M. Olive, and C. Vinson. 2000. Attractive interhelical electrostatic interactions in the proline- and acidic-rich region (PAR) leucine zipper subfamily preclude heterodimerization with other basic leucine zipper subfamilies. J. Biol. Chem. 275:34826-34832. [DOI] [PubMed] [Google Scholar]
- 65.Montminy, M. R., and L. M. Bilezikjian. 1987. Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature 328:175-178. [DOI] [PubMed] [Google Scholar]
- 66.Motohashi, H., J. A. Shavit, K. Igarashi, M. Yamamoto, and J. D. Engel. 1997. The world according to Maf. Nucleic Acids Res. 25:2953-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nicklin, M., and G. Casari. 1991. A single site mutation in a truncated Fos protein allows it to interact with the TRE in vitro. Oncogene 6:173-179. [PubMed] [Google Scholar]
- 68.Nomura, N., Y. L. Zu, T. Maekawa, S. Tabata, T. Akiyama, and S. Ishii. 1993. Isolation and characterization of a novel member of the gene family encoding the cAMP response element-binding protein CRE-BP1. J. Biol. Chem. 268:4259-4266. [PubMed] [Google Scholar]
- 69.Oakley, M. G., and P. S. Kim. 1998. A buried polar interaction can direct the relative orientation of helices in a coiled coil. Biochemistry 37:12603-12610. [DOI] [PubMed] [Google Scholar]
- 70.Olive, M., D. Krylov, D. R. Echlin, K. Gardner, E. Taparowsky, and C. Vinson. 1997. A dominant negative to activation protein-1 (AP1) that abolishes DNA binding and inhibits oncogenesis. J. Biol. Chem. 272:18586-18594. [DOI] [PubMed] [Google Scholar]
- 71.Olive, M., S. C. Williams, C. Dezan, P. F. Johnson, and C. Vinson. 1996. Design of a C/EBP-specific, dominant-negative bZIP protein with both inhibitory and gain-of-function properties. J. Biol. Chem. 271:2040-2047. [DOI] [PubMed] [Google Scholar]
- 72.Omori, Y., J. Imai, M. Watanabe, T. Komatsu, Y. Suzuki, K. Kataoka, S. Watanabe, A. Tanigami, and S. Sugano. 2001. CREB-H: a novel mammalian transcription factor belonging to the CREB/ATF family and functioning via the box-B element with a liver-specific expression. Nucleic Acids Res. 29:2154-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.O'Neil, K. T., R. H. Hoess, and W. F. DeGrado. 1990. Design of DNA binding peptides based on the leucine zipper motif. Science 243:774-778. [DOI] [PubMed] [Google Scholar]
- 74.O'Shea, E., R. Rutkowski, and P. Kim. 1992. Mechanism of specificity in the fos-jun oncoprotein heterodimer. Cell 68:699-708. [DOI] [PubMed] [Google Scholar]
- 75.O'Shea, E. K., J. D. Klemm, P. S. Kim, and T. Alber. 1991. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled-coil. Science 254:539-544. [DOI] [PubMed] [Google Scholar]
- 76.Popova, B., P. Bilan, P. Xiao, M. Faught, and J. P. Capone. 1995. Transcriptional activation by DNA-binding derivatives of HSV-1 VP16 that lack the carboxyl-terminal acidic activation domain. Virology 209:19-28. [DOI] [PubMed] [Google Scholar]
- 77.Ransone, L. J., J. Visvader, P. Sassone-Corsi, and I. M. Verma. 1989. Fos-Jun interaction: mutational analysis of the leucine zipper domain of both proteins. Genes Dev. 3:770-781. [DOI] [PubMed] [Google Scholar]
- 78.Ryder, K., A. Lanahan, E. Perez-Albuerne, and D. Nathans. 1989. jun-D: a third member of the jun gene family. Proc. Natl. Acad. Sci. USA 86:1500-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ryseck, R. P., and R. Bravo. 1991. c-JUN, JUN B, and JUN D differ in their binding affinities to AP-1 and CRE consensus sequences: effect of FOS proteins. Oncogene 6:533-542. [PubMed] [Google Scholar]
- 80.Schumacher, M. A., R. H. Goodman, and R. G. Brennan. 2000. The structure of a CREB bZIP.somatostatin CRE complex reveals the basis for selective dimerization and divalent cation-enhanced DNA binding. J. Biol. Chem. 275:35242-35247. [DOI] [PubMed] [Google Scholar]
- 81.Serrano, L., A. Horovitz, B. Avron, M. Bycroft, and A. R. Fersht. 1990. Estimating the contribution of engineered surface electrostatic interactions to protein stability by using double-mutant cycles. Biochemistry 29:9343-9352. [DOI] [PubMed] [Google Scholar]
- 82.Shaulian, E., and M. Karin. 2001. AP-1 in cell proliferation and survival. Oncogene 20:2390-2400. [DOI] [PubMed] [Google Scholar]
- 83.Shuman, J. D., J. Cheong, and J. E. Coligan. 1997. ATF-2 and C/EBPalpha can form a heterodimeric DNA binding complex in vitro. Functional implications for transcriptional regulation. J. Biol. Chem. 272:12793-12800. [DOI] [PubMed] [Google Scholar]
- 84.Shuman, J. D., C. R. Vinson, and S. L. McKnight. 1990. Evidence of changes in protease sensitivity and subunit exchange rate on DNA binding by C/EBP. Science 249:771-774. [DOI] [PubMed] [Google Scholar]
- 85.Singh, H., J. H. LeBowitz, A. S. Baldwin, Jr., and P. A. Sharp. 1988. Molecular cloning of an enhancer binding protein: isolation by screening of an expression library with a recognition site DNA. Cell 52:415-423. [DOI] [PubMed] [Google Scholar]
- 86.Smeal, T., P. Angel, J. Meek, and M. Karin. 1989. Different requirements for formation of Jun:Jun and Jun:Fos complexes. Genes Dev. 3:2091-2100. [DOI] [PubMed] [Google Scholar]
- 87.Thompson, K. S., C. R. Vinson, and E. Freire. 1993. Thermodynamic characterization of the structural stability of the coiled-coil region of the bZIP transcription factor GCN4. Biochemistry 32:5491-5496. [DOI] [PubMed] [Google Scholar]
- 88.Tripet, B., K. Wagschal, P. Lavigne, C. T. Mant, and R. S. Hodges. 2000. Effects of side-chain characteristics on stability and oligomerization state of a de novo-designed model coiled-coil: 20 amino acid substitutions in position “d.” J. Mol. Biol. 300:377-402. [DOI] [PubMed] [Google Scholar]
- 89.Tsukada, J., K. Saito, W. R. Waterman, A. C. Webb, and P. E. Auron. 1994. Transcription factors NF-IL6 and CREB recognize a common essential site in the human prointerleukin 1β gene. Mol. Cell. Biol. 14:7285-7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tupler, R., G. Perini, and M. R. Green. 2001. Expressing the human genome. Nature 409:832-833. [DOI] [PubMed] [Google Scholar]
- 91.Turner, R., and R. Tjian. 1989. Leucine repeats and an adjacent DNA binding domain mediate the formation of functional cFos-cJun heterodimers. Science 243:1689-1694. [DOI] [PubMed] [Google Scholar]
- 92.Ubeda, M., X.-Z. Wang, H. Zinszner, I. Wu, J. F. Habener, and D. Ron. 1996. Stress-induced binding of the transcriptional factor CHOP to a novel DNA control element. Mol. Cell. Biol. 16:1479-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vallejo, M., D. Ron, C. P. Miller, and J. F. Habener. 1993. C/ATF, a member of the activating transcription factor family of DNA-binding proteins, dimerizes with CAAT/enhancer-binding proteins and directs their binding to cAMP response elements. Proc. Natl. Acad. Sci. USA 90:4679-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.van Dam, H., S. Huguier, K. Kooistra, J. Baguet, E. Vial, A. J. van der Eb, P. Herrlich, P. Angel, and M. Castellazzi. 1998. Autocrine growth and anchorage independence: two complementing Jun-controlled genetic programs of cellular transformation. Genes Dev. 12:1227-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Venter, J. C., M. D. Adams, E. W. Myers, P. W. Li, R. J. Mural, G. G. Sutton, H. O. Smith, M. Yandell, C. A. Evans, R. A. Holt, J. D. Gocayne, P. Amanatides, R. M. Ballew, D. H. Huson, J. R. Wortman, Q. Zhang, C. D. Kodira, X. H. Zheng, L. Chen, M. Skupski, G. Subramanian, P. D. Thomas, J. Zhang, G. L. Gabor Miklos, C. Nelson, S. Broder, A. G. Clark, J. Nadeau, V. A. McKusick, N. Zinder, A. J. Levine, R. J. Roberts, M. Simon, C. Slayman, M. Hunkapiller, R. Bolanos, A. Delcher, I. Dew, D. Fasulo, M. Flanigan, L. Florea, A. Halpern, S. Hannenhalli, S. Kravitz, S. Levy, C. Mobarry, K. Reinert, K. Remington, J. Abu-Threideh, E. Beasley, K. Biddick, V. Bonazzi, R. Brandon, M. Cargill, I. Chandramouliswaran, R. Charlab, K. Chaturvedi, Z. Deng, V. Di Francesco, P. Dunn, K. Eilbeck, C. Evangelista, A. E. Gabrielian, W. Gan, W. Ge, F. Gong, Z. Gu, P. Guan, T. J. Heiman, M. E. Higgins, R. R. Ji, Z. Ke, K. A. Ketchum, Z. Lai, Y. Lei, Z. Li, J. Li, Y. Liang, X. Lin, F. Lu, G. V. Merkulov, N. Milshina, H. M. Moore, A. K. Naik, V. A. Narayan, B. Neelam, D. Nusskern, D. B. Rusch, S. Salzberg, W. Shao, B. Shue, J. Sun, Z. Wang, A. Wang, X. Wang, J. Wang, M. Wei, R. Wides, C. Xiao, C. Yan, et al. 2001. The sequence of the human genome. Science 291:1304-1351. [DOI] [PubMed] [Google Scholar]
- 96.Vinson, C. R., T. Hai, and S. M. Boyd. 1993. Dimerization specificity of the leucine zipper-containing bZIP motif on DNA binding: prediction and rational design. Genes Dev. 7:1047-1058. [DOI] [PubMed] [Google Scholar]
- 97.Vinson, C. R., K. L. LaMarco, P. F. Johnson, W. H. Landschulz, and S. L. McKnight. 1988. In situ detection of sequence-specific DNA-binding activity specified by a recombinant bacteriophage. Genes Dev. 2:801-806. [DOI] [PubMed] [Google Scholar]
- 98.Vinson, C. R., P. B. Sigler, and S. L. McKnight. 1989. A scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science 246:911-916. [DOI] [PubMed] [Google Scholar]
- 99.Wagschal, K., B. Tripet, P. Lavigne, C. Mant, and R. S. Hodges. 1999. The role of position a in determining the stability and oligomerization state of alpha-helical coiled coils: 20 amino acid stability coefficients in the hydrophobic core of proteins. Protein Sci. 8:2312-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang, Y., J. Shen, N. Arenzana, W. Tirasophon, R. J. Kaufman, and R. Prywes. 2000. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J. Biol. Chem. 275:27013-27020. [DOI] [PubMed] [Google Scholar]
- 101.Williams, S. C., C. A. Cantwell, and P. F. Johnson. 1991. A family of C/EBP-related proteins capable of forming covalently linked leucine zipper dimers in vitro. Genes Dev. 5:1553-1567. [DOI] [PubMed] [Google Scholar]
- 102.Yoshimura, T., J. Fujisawa, and M. Yoshida. 1990. Multiple cDNA clones encoding nuclear proteins that bind to the tax-dependent enhancer of HTLV-1: all contain a leucine zipper structure and basic amino acid domain. EMBO J. 9:2537-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zeng, X., A. M. Herndon, and J. C. Hu. 1997. Buried asparagines determine the dimerization specificities of leucine zipper mutants. Proc. Natl. Acad. Sci. USA 94:3673-3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhou, N., C. Kay, and R. Hodges. 1994. The net energetic contribution of interhelical electrostatic attractions to coiled-coil stability. Protein Eng. 7:1365-1372. [DOI] [PubMed] [Google Scholar]
- 105.Zhu, H., S. A. Celinski, J. M. Scholtz, and J. C. Hu. 2000. The contribution of buried polar groups to the conformational stability of the GCN4 coiled coil. J. Mol. Biol. 300:1377-1387. [DOI] [PubMed] [Google Scholar]