Abstract
Certain DNA-binding repressors inhibit transcription by recruiting Rpd3 histone deacetylase complexes to promoters and generating domains of histone deacetylation that extend over a limited number of nucleosomes. Here, we show that the degree of Rpd3-dependent repression depends on the activator and the level of activation, not the extent of histone deacetylation. In all cases tested, activator binding is unaffected by histone deacetylation. In contrast, Rpd3-dependent repression is associated with decreased occupancy by TATA binding protein (TBP), the Swi/Snf nucleosome-remodeling complex, and the SAGA histone acetylase complex. Transcriptional repression is bypassed by direct recruitment of TBP and several TBP-associated factors, but not by natural activation domains or direct recruitment of polymerase II holoenzyme components. These results suggest that the domain of localized histone deacetylation generated by recruitment of Rpd3 mediates repression by inhibiting recruitment of chromatin-modifying activities and TBP.
A fundamental aspect of eukaryotic gene regulation is the ability of DNA-binding activators and repressors to recruit chromatin-modifying activities to specific promoters (60). Once recruited, such modifying activities generate local domains of altered chromatin structure that influence the level of gene activity. For example, certain activators recruit the SAGA histone acetylase complex to generate a domain of increased histone acetylation (1, 8, 31, 32, 33, 49), and histone acetylase activity is required for transcriptional activation in vivo (33, 68). Similarly, recruitment of the Esa1 histone acetylase complex is associated with coordinated induction of ribosomal protein genes in response to growth stimuli (51). Conversely, DNA-binding repressors often function by recruiting Rpd3 histone deacetylase complexes to promoters (50, 59). The recruitment of histone acetylases by activators and histone deacetylases by repressors is consistent with the long-standing correlation between histone acetylation and gene activity.
The yeast repressor Ume6 specifically binds DNA sequences (upstream repression sequence 1 [URS1]) in a variety of promoters, and it inhibits transcription by recruiting the Rpd3 histone deacetylase complex (26). Recruitment occurs through an interaction between the Ume6 repression domain and Sin3, a component of the Rpd3 complex (26). Histone deacetylase activity is important for repression (27), and targeted recruitment of Rpd3 leads to localized deacetylation of the N-terminal tails of histones H3 and H4 over a range of one to two nucleosomes (28, 53). In addition to creating local domains of histone deacetylation upon recruitment, Rpd3 deacetylates histones on a genomewide basis (66), and it appears to counteract heterochromatic silencing (10, 52, 64).
While the outline of repression by targeted recruitment of Rpd3 histone deacetylase is established, there is virtually no information on the mechanism by which localized histone deacetylation reduces transcription in vivo. In vitro, deacetylated nucleosomes are less accessible to certain activators (46, 65) and general transcription factors (38, 56), and they can inhibit transcription without altering nucleosome mobility (63). Deacetylated histones interact more strongly with DNA than acetylated histones (21), and the crystal structure of the nucleosome suggests that acetylation status affects nucleosome-nucleosome interactions (41). Deacetylated histones are less efficiently bound by the Swi/Snf nucleosome-remodeling complex (20) and perhaps other chromatin-modifying activities containing bromodomains, structural motifs that bind acetylated lysines (12, 70). However, the mechanistic relationship between histone acetylation status and transcriptional activity in vivo is poorly understood.
An initial hint of the repression mechanism has come from the analysis of TATA binding protein (TBP) mutants that increase expression from a promoter repressed by targeted recruitment of Rpd3 (17). These TBP mutants also increase transcription from a promoter lacking an enhancer, but not an equivalently weak promoter containing a mutated TATA element, suggesting that localized histone deacetylation represses transcription by inhibiting activator function. However, this suggestion is based on an indirect genetic argument, and it does not distinguish among various repression mechanisms.
Here, we examine the mechanism of repression mediated by targeted recruitment of the Rpd3 histone deacetylase complex. Our results suggest that (i) the degree of Rpd3-dependent repression depends on the activator and the level of activation, (ii) Rpd3 affects TBP occupancy but not activator binding, (iii) Rpd3 inhibits activator-dependent association of the Swi/Snf nucleosome-remodeling complex and the SAGA histone acetylase complex, and (iv) repression can be abolished by direct recruitment of TBP, but not polymerase II (Pol II) holoenzyme, to the promoter. We suggest that localized histone deacetylation by recruitment of Rpd3 represses transcription by inhibiting the association of chromatin-modifying activities and by creating a situation in which TBP association with the promoter becomes a more limiting step.
MATERIALS AND METHODS
DNAs and yeast strains.
All promoters were derived from YIp-his3A5, an allele in which all HIS3 sequences upstream of the core promoter region (positions −85 to −447) were replaced by various activator binding sites (23). Two copies of a URS (URS1) containing a Ume6 binding site from the IME2 promoter were inserted upstream of the activator binding sites by using complementary 34-bp oligonucleotides. To create the HIS3/LYS2 derivatives, 1.8 kb of the same HIS3 derivative lacking URS1 was fused at the ATG to the LYS2 open reading frame (ORF). The modified HIS3 and HIS3/LYS2 genes were inserted into FT5 (α ura3-53 trp1-Δ63 his3-Δ200 leu2::PET56) at their respective loci by two-step gene replacement. Strain JDY51 carries a Gcn4 site, JDY111 carries a poly(dA-dT) element, JDY121 carries a Rap1 site, JDY131 carries an Abf1 site, JDY141 carries a Leu3 site, JDY161 carries an Hsf1 site, JDY171 carries a Gal4 site, JDY151 carries a Put3 site, and JDY181 carries two Ace1 sites as the sole upstream activation sequence (UAS) in both reporter genes. The rpd3, sin3, ume6, and ace1 deletion strains were derived from the above-mentioned strains as described previously (9, 37). For the distance experiment, the Ume6 binding sites were inserted 100 (strain JDY70), 200 (JDY71), 400 (JDY72), or 800 (JDY73) bp upstream of the activator binding site. To measure TBP occupancy, a triple-hemagglutinin (HA)-tagged version of TBP was introduced into the above-mentioned strains by stable integration (35). Plasmid pAC-1, carrying the Ace1 DNA-binding domain (residues 1 to 124), and derivatives expressing Ace1 protein fusions are described elsewhere (37) and were obtained from John Lis. The plasmids containing the Ace1-TAF19 and Ace1-TAF23 fusion proteins were constructed by PCR amplifying the respective ORFs and ligating them in frame into pAC-1. Transcriptional activation was measured in ace1 and ace1 rpd3 strains isogenic to JDY181. For the SAGA and Swi/Snf occupancy experiments, Ada2-myc18 and Swi2-myc18 (8) were introduced into strains JDY51 and JDY171 and isogenic rpd3 deletion strains. The gcn5 and swi2 deletion strains were derived from these strains by using standard one-step gene replacement.
Yeast strains were grown in yeast-peptone-dextrose (YPD) medium unless otherwise indicated. To induce Gcn4-activated genes, strain JDY51 was grown in glucose minimal medium (SD) with essential amino acids to mid-log phase and then shifted to medium lacking histidine and containing 10 mM 3-aminotriazole for 4 h. Leu3-regulated genes were induced by growing strain JDY141 in synthetic complete medium containing 300 μg of leucine/ml and then shifting it to medium containing 30 μg of leucine/ml for 45 min. To activate the heat shock response, strain JDY161 was grown at 25°C and shifted to 39°C for 20 min. For galactose induction, strain JDY171 was grown in YPD medium and shifted to yeast-peptone (YP) medium containing 2% galactose for 8 h. To induce Put3-responsive genes, strain JDY151 was grown in SD and shifted to SD lacking ammonium sulfate and containing 0.1% proline. Copper-responsive genes were activated by growing strain JDY181 in SD and inducing it with 0.5 mM CuSO4 for 20 min. For the SAGA and Swi/Snf occupancy experiments and transcriptional analysis, derivatives of strain JDY171 were grown in YP medium containing 2% raffinose, and 2% galactose was added for 20 min. Likewise, derivatives of strain JDY51 were grown in SD and shifted to medium lacking histidine and containing 5 mM 3-aminotriazole for 4 h.
Transcriptional analysis.
Total RNAs from cells grown under the conditions indicated above were hybridized to completion with a mixture of oligonucleotide probes corresponding to the HIS3, LYS2, and DED1 coding regions, and the resulting RNA-DNA hybrids were treated with S1 nuclease (25). The degree of repression was determined by comparing the ratio of HIS3/LYS2 RNAs in an rpd3 mutant strain to the HIS3/LYS2 RNA ratio in a wild-type strain. When the degree of repression was calculated using sin3 or ume6 strains instead of rpd3 strains, the results were indistinguishable. In the absence of Rpd3 repression, there is approximately twofold more LYS2 RNA than HIS3 RNA, presumably because LYS2 RNA is more stable; this effect is unrelated to the Rpd3-dependent repression mechanism, and it is normalized out in our measurements of repression. To determine the level of activation mediated by the various activators, we measured the level of HIS3 RNA in an rpd3 mutant strain by using DED1 RNA as an internal control. The level of DED1 RNA was determined to be 25 molecules/cell when the cells are grown in YPD medium and some other media (25), and this value was used to calculate the number of HIS3 molecules/cell. We did not measure absolute DED1 RNA levels in these experiments, and it is likely that absolute DED1 RNA levels will differ to some extent in the various media used here. The error for the repression values is ±15%.
Chromatin immunoprecipitation.
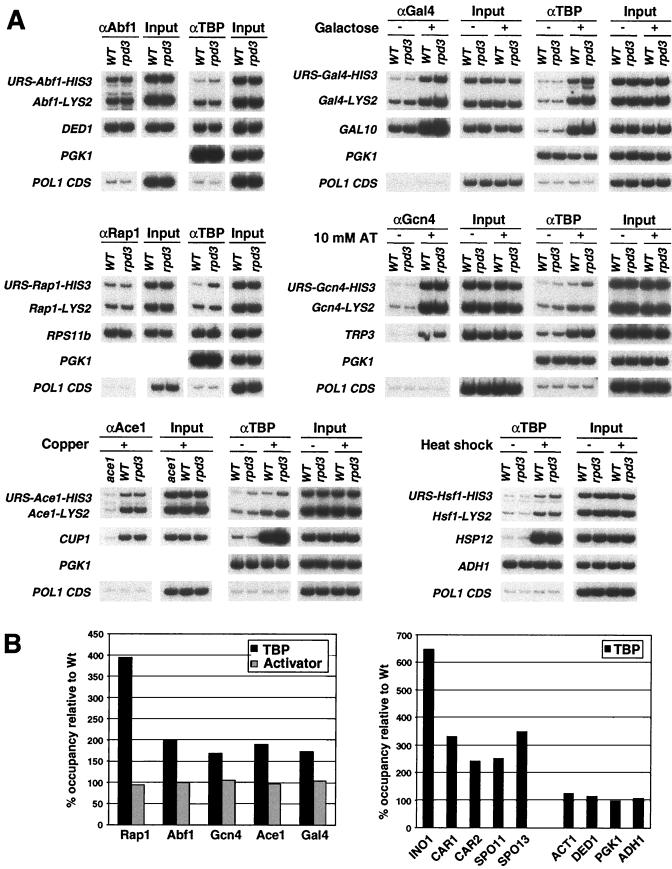
To measure histone acetylation levels, formaldehyde-cross-linked chromatin (35) was immunoprecipitated with antibodies against diacetylated (lysines 9 and 14) H3 and tetra-acetylated (lysines 5, 8, 12, and 16) H4 histone tails as described previously (9). To measure activator occupancy, chromatin was precipitated with the following antibodies: 20 μl of α-Abf1 (yC-20; Santa Cruz), 20 μl of α-Rap1 (yC-19; Santa Cruz), 20 μl of α-Gal4 (sc-577; Santa Cruz), 8 μl of α-Ace1 (a gift from Dennis Thiele), and 30 μl of monoclonal α-Gcn4. We used 20 μl of α-HA (F7; Santa Cruz) to measure (HA)3-TBP binding and 5 μl of polyclonal α-myc (Upstate Biotechnology) to measure Ada2-myc18 and Swi2-myc18 occupancy. For the TBP occupancy experiments, immunoprecipitated material was eluted with 1 mg of HA peptide (Roche)/ml for 30 min at room temperature, a procedure developed by Gauri Dhavan and Kevin Struhl. For almost all experiments (see Fig. 6B and 7 for exceptions), 1/100 of the precipitated chromatin and 1/10,000 to 1/100,000 of the input DNA was used as a template in a 26-cycle PCR. The PCR products were separated on 8% polyacrylamide gels and quantified using a Fujix BAS1000 phosphorimager. For the exceptional experiments (see Fig. 6B and 7), quantitative PCR analyses were performed in real time using an Applied Biosystems 7700 sequence detector. The degree of deacetylation was calculated as the ratio of HIS3-LYS2 PCR products in a mutant strain to the HIS3-LYS2 PCR product in a wild-type strain. As the degrees of deacetylation in rpd3, sin3, and ume6 strains were almost identical, an average of these values is presented. To calculate the levels of TBP occupancy at an individual promoter, we first determined the apparent cross-linking efficiency by dividing the amount of PCR product from the immunoprecipitated sample by the amount of PCR product in the input sample prior to immunoprecipitation and subtracting the apparent cross-linking efficiency of control DNA segments (an internal fragment of the POL1 structural gene and an ORF-free region), which was considered background. The ratios of TBP occupancies of the Rpd3-repressed (HIS3) and control (LYS2) promoters in a wild-type strain were set to 100%, and the relative values in an rpd3 strain were determined. The error in the promoter occupancy experiments is approximately ±10% except in cases where TBP occupancy is reduced to levels near those of control DNA segments.
FIG. 6.
TBP occupancy, but not activator occupancy, is reduced by recruitment of Rpd3. (A) Cross-linked chromatin from wild-type (WT) and rpd3 strains grown under the indicated conditions was immunoprecipitated with antibodies against various activators or the HA epitope (for TBP determining TBP occupancy) and analyzed by quantitative PCR with primers corresponding to the indicated promoter regions and the POL1 coding sequence (CDS). (B) Relative URS-HIS3 occupancy of TBP and transcriptional activators at the Rpd3-repressed derivatives of the HIS3 promoters (left-hand graph) and at natural promoters (right-hand graph). Occupancies in a wild-type strain are set to 100%, and the value in an rpd3 strain is presented relative to it.
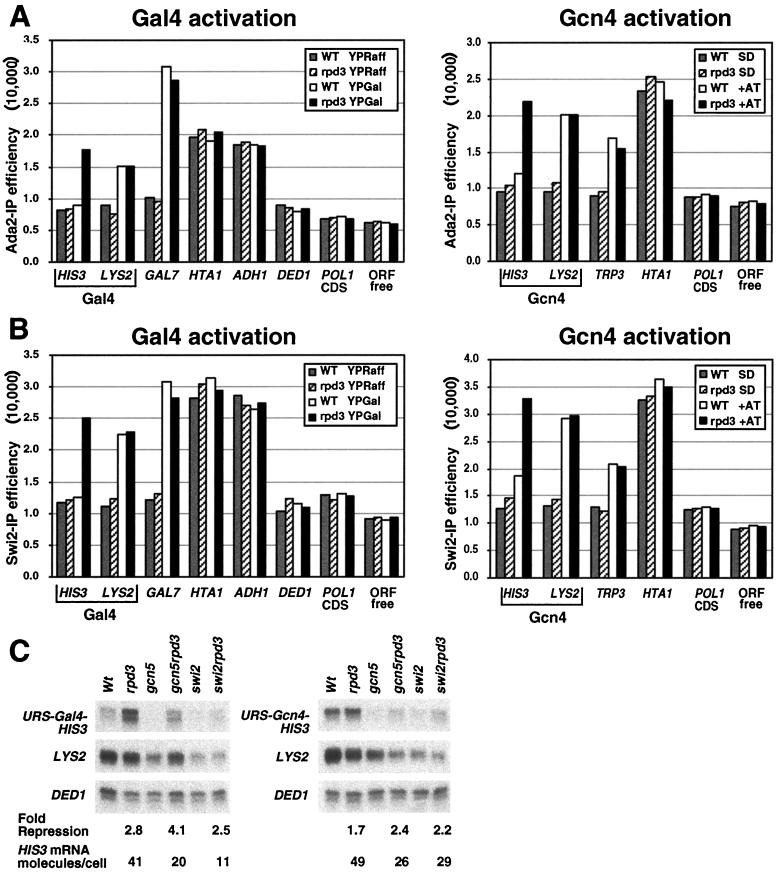
FIG. 7.
Recruitment of Rpd3 inhibits activator-dependent association of Swi/Snf and SAGA. (A) Cross-linked chromatin from wild-type (WT) and rpd3 strains grown under the indicated conditions was immunoprecipitated with α-myc antibodies to measure Ada2-myc occupancy. Immunoprecipitated and input material was analyzed by real-time quantitative PCR analysis with primers corresponding to the indicated promoters (HIS3 and LYS2 indicated the Rpd3-repressed and control promoters, respectively, that respond to the Gal4 and Gcn4 activators), the POL1 coding sequence (CDS), and an ORF-free region. (B) Cross-linked chromatin from wild-type and rpd3 strains grown under the indicated conditions was immunoprecipitated with α-myc antibodies to measure Swi2-myc occupancy as described for panel A. (C) Isogenic wild-type (Wt) and the indicated mutant strains carrying URS-HIS3 and LYS2 alleles activated by either Gal4 (left) or Gcn4 (right) were grown under inducing conditions, and the URS-HIS3, LYS2, and DED1 expression levels were determined by S1 nuclease assays. Repression of the URS-HIS3 alleles in wild-type strains and the relative expression (in HIS3 molecules/cell) of the nonrepressed URS-HIS3 allele are indicated.
RESULTS
Experimental design.
Natural Rpd3-repressed genes show localized deacetylation of histones H3 and H4 in their promoter regions (9, 53). However, it is difficult to address the mechanism by which localized histone deacetylation represses transcription at these natural promoters because the relevant activators and core promoter elements are poorly defined. In addition, natural promoters differ considerably with respect to the activators utilized; the relative locations of the binding sites for the activators, repressors, and core promoter elements; and the locations of these critical DNA sequences with respect to nucleosomes.
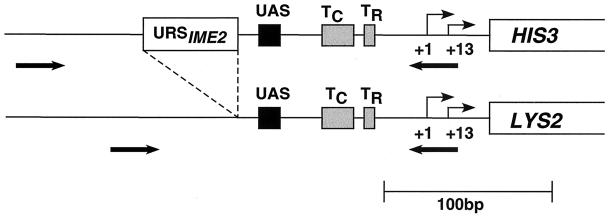
To systematically address the mechanism of repression by recruitment of Rpd3 and localized histone deacetylation, we generated a set of yeast strains, each of which contains an Rpd3-repressed and a control promoter responding to the same activator (Fig. 1). The control promoters contain a defined activator binding site upstream of the HIS3 TATA and initiator elements (23), and they drive expression of LYS2. The corresponding Rpd3-repressed promoters, which drive expression of HIS3, contain two Ume6 binding sites from the IME2 regulatory region (26) that are positioned upstream of the activator binding site. This arrangement permits an assay of Rpd3-dependent repression without the complication of steric inhibition that occurs when the repressor is bound between the activator binding site and core promoter elements (4, 29). Simultaneous monitoring of HIS3 and LYS2 expression permits a direct measurement of the degree of repression under defined experimental conditions, and it accounts for potential indirect effects on activation caused by the deletion of rpd3, sin3, or ume6. In addition, the experimental design permits internally controlled analysis of the Rpd3-repressed and control promoters for histone acetylation status, activator binding, and TBP, Swi/Snf, and SAGA association in vivo (Fig. 1). The use of a true internal control eliminates many sources of experimental error and hence permits much more accurate measurements of small quantitative differences.
FIG. 1.
Diagram of Rpd3-repressed and control his3 promoter derivatives. Both derivatives have a given activator binding site (UAS) upstream of the HIS3 TATA elements (TC and TR) and initiation sites (+1 and +13), with the Rpd3-repressed promoter containing two Ume6 binding sites from the IME2 regulatory region (URSIME2) upstream of the activator binding site. The activators tested were Abf1, Rap1, Leu3, Ace1, Put3, Gal4, Gcn4, Hsf1, and a poly(dA-dT) element. The Rpd3-repressed and control promoter derivatives are fused to the HIS3 and LYS2 protein-coding regions at the ATG translational initiation codons, respectively, such that simultaneous monitoring of HIS3 and LYS2 expression represents a direct measurement of repression. The thick arrows indicate the positions of PCR primers used to measure histone acetylation status, activator binding, and TBP, Swi/Snf, and SAGA association at the Rpd3-repressed and control promoters in an internally controlled manner. The diagram is drawn to scale.
The his3 promoter region used in these experiments lacks positioned nucleosomes and is preferentially accessible to enzymatic probes in vitro and in vivo (24, 40, 42, 47, 58, 61). The preferential accessibility of the his3 promoter region depends on a general property of the DNA sequence and is unaffected by the activator, TATA elements, Swi/Snf, Gcn5 histone acetylase, or Rad6 (42). Such preferential accessibility is typical of yeast promoter regions, so our experimental results are likely to pertain to many, but probably not all, natural situations.
Activator specificity of Rpd3-dependent repression.
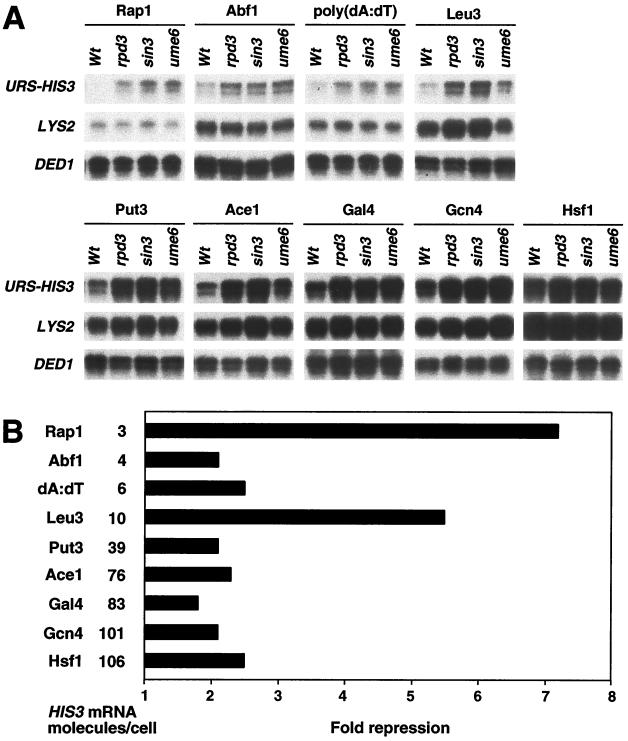
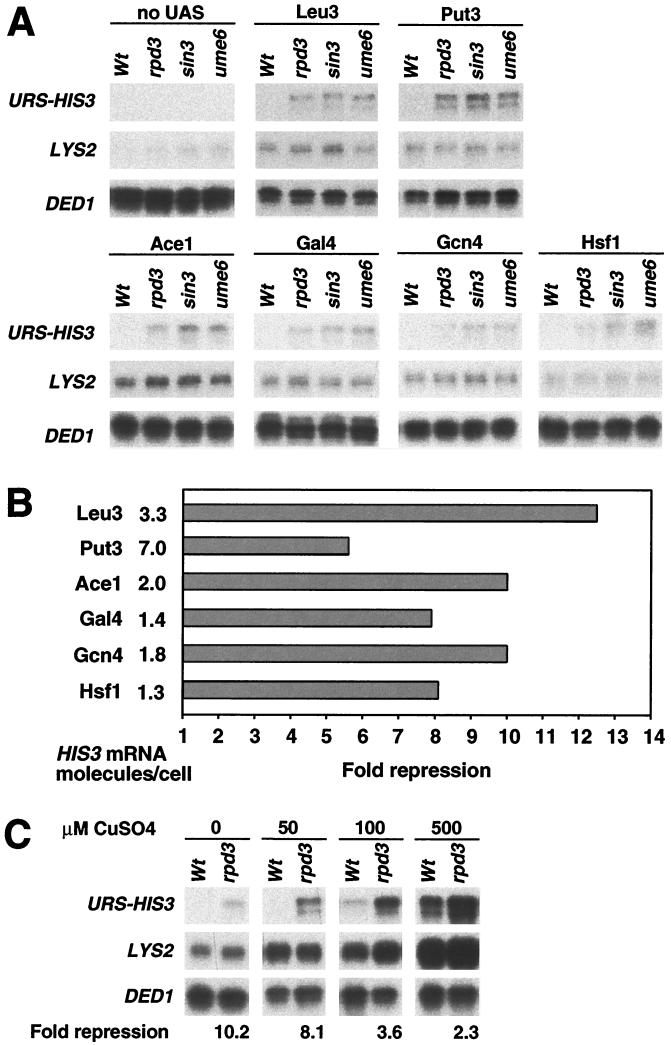
To address whether Rpd3-dependent repression is activator specific, we examined eight diverse activators: Abf1, Rap1, Leu3, Ace1, Put3, Gal4, Gcn4, and Hsf1. In addition, we examined a poly(dA-dT) sequence which stimulates transcription via its intrinsic structure and effect on chromatin, not by binding an activator protein (24). Transcriptional analysis was performed under inducing (Fig. 2A) and noninducing (Fig. 3A) conditions appropriate for each activator. The losses of repression in rpd3, sin3, and ume6 strains were indistinguishable, indicating that each of these proteins is required for repression.
FIG. 2.
Rpd3 repression is activator specific. (A) Isogenic wild-type (Wt), rpd3, sin3, and ume6 strains carrying URS-HIS3 and LYS2 alleles controlled by the indicated activators were grown under appropriate inducing conditions, and the URS-HIS3, LYS2, and DED1 RNA levels were determined by quantitative S1 nuclease protection assays. (B) Repression of the URS-HIS3 alleles in wild-type strains and relative expression (in HIS3 molecules/cell) of the nonrepressed URS-HIS3 allele.
FIG. 3.
Rpd3 efficiently represses weakly activated transcription. (A) Isogenic wild-type (Wt), rpd3, sin3, and ume6 strains carrying URS-HIS3 and LYS2 alleles controlled by the indicated activators (“no UAS” indicates control promoters lacking an activator binding site) were grown under noninducing conditions, and URS-HIS3, LYS2, and DED1 expression levels were determined by S1 nuclease assays. (B) Repression of the URS-HIS3 alleles in wild-type strains and relative expression (in HIS3 molecules/cell) of the nonrepressed URS-HIS3 allele. Expression in the absence of an activator binding site is barely detected, and it corresponds to approximately 0.25 HIS3 molecules/cell, which is 5- to 10-fold below the level observed for the activators under nonoptimal conditions. (C) JDY181 and the isogenic rpd3 strain were grown in the presence of the indicated concentrations of CuSO4.
It is apparent that the level of repression due to Rpd3 recruitment depends on the activator. Under conditions optimal for the activator, the degree of repression varies from two- to sevenfold (Fig. 2B). In general, stronger activators, such as Gal4, Ace1, Gcn4, and Hsf1, are mildly affected by Rpd3 repression, while weaker activators are repressed more strongly. However, the level of repression is not related simply to the strength of the activator. For example, Abf1 and the poly(dA-dT) element are relatively weak activators, yet Rpd3-dependent repression of these promoters occurs at the same modest level as that observed for the strongest activators. Conversely, Rpd3 represses Rap1-dependent activation much more efficiently than Abf1- or poly(dA-dT)-dependent activation even though the levels of activation in these cases are similar. As the core promoters are identical in these experiments, the results suggest that the transcriptional consequence of recruiting Rpd3 histone deacetylase to a promoter depends on the activator.
Rpd3-dependent repression is more efficient at less active promoters.
Although repression by targeted recruitment of Rpd3 is often more pronounced at promoters responding to weak activators, this conclusion is based on a comparison of activators that are likely to have different functional properties. To directly address the relationship between activator strength and the degree of Rpd3-dependent repression, we analyzed repression under conditions where the activators are less functional (Fig. 3A). Under these suboptimal conditions, the activation mechanisms are compromised due to a low protein level (Gcn4 and Gal4), reduced DNA-binding function (Ace1), or reduced transcriptional activation function (Gal4, Hsf1, Put3, and Leu3), although the activators do enhance transcription to a small extent (11, 16, 24, 43, 57, 67). The activators function on the relevant promoters under these suboptimal conditions, because transcription levels are at least fivefold above the barely detectable level of the comparable promoter lacking activator binding sites.
In all cases tested, Rpd3-dependent repression under poor activation conditions ranged from 6- to 12-fold (Fig. 3B). This degree of repression is considerably more pronounced than that observed for the same activators under optimal conditions (Fig. 3B). Furthermore, Rpd3-dependent repression of Ace1 activation becomes progressively less efficient as a function of increased copper concentration (Fig. 3C). As Ace1 activity is regulated solely by the copper-dependent folding of the DNA-binding domain (16), the magnitude of Rpd3-dependent repression can be strongly affected by the amount of the activator bound at the promoter. Thus, Rpd3-dependent repression is more efficient at less active promoters, and strong activators can override Rpd3-dependent repression to some extent.
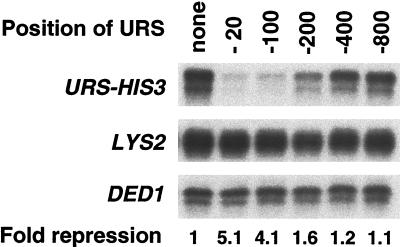
Rpd3-dependent repression operates over a limited distance.
Recruitment of Rpd3 creates a localized domain of deacetylated histones that extends one to two nucleosomes to either side of the recruitment site (28, 53). However, it is unclear where an Rpd3 recruitment site needs to be located on the promoter in order to exert its repressive effect on transcription. To examine this issue, we inserted the Rpd3 recruitment site at distances from 30 to 800 bp upstream of the Gcn4 binding site and HIS3 core promoter (Fig. 4). Recruitment of Rpd3 leads to strong repression at distances of 30 bp (5-fold) and 100 bp (4-fold), to weak repression at 200 bp (1.5-fold), and to no repression at larger distances. Thus, Rpd3-dependent repression is observed only when the recruitment site is located at a distance of <200 bp relative to the region containing the activator binding site and core promoter elements, which is in accord with the size of the domain of histone deacetylation. Mapping experiments with one of these promoters show that histone deacetylation peaks at the Rpd3 recruitment site and extends 200 to 300 bp in both directions.
FIG. 4.
Rpd3 repression acts at a limited distance. Strains carrying a Gcn4-LYS2 allele and a Gcn4-HIS3 allele with a URS element at the indicated distance from the Gcn4 site were grown in glucose minimal medium supplemented with all essential amino acids. The HIS3, LYS2, and DED1 RNA levels were determined by S1 nuclease assays; the degree of repression is indicated.
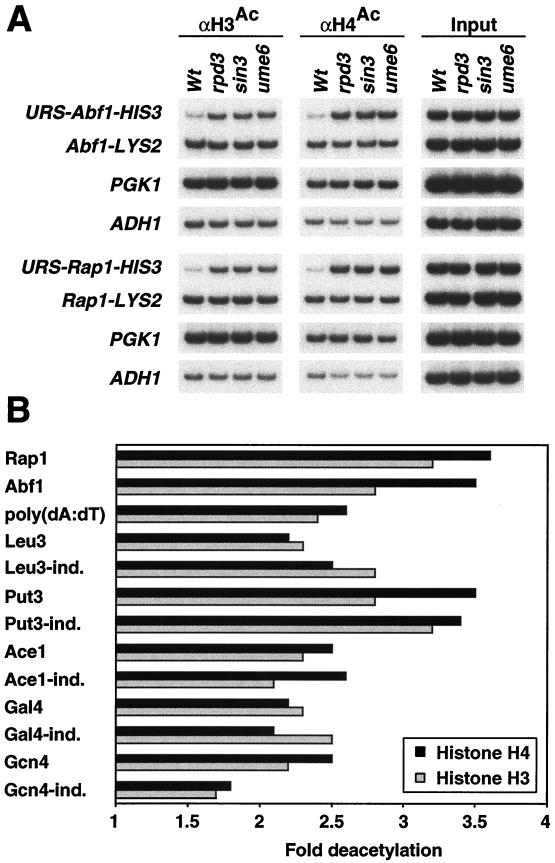
Histone deacetylation does not correlate with the degree of repression.
The different degrees of repression observed with various activators could be due to a differential response of the activator to the deacetylated chromatin or to a difference in the extent of deacetylation. We therefore analyzed histone H3 and H4 acetylation at all promoters in wild-type and mutant strains by using chromatin immunoprecipitation and a PCR primer pair that amplifies both the Rpd3-repressed and the control promoter (Fig. 5A and data not shown). For all Rpd3-repressed promoters under inducing and noninducing conditions, we observed a 2- to 3.5-fold decrease in H3 and H4 acetylation that required the presence of Rpd3, Sin3, and Ume6 (Fig. 5B). The acetylation levels at the control promoters and the unrelated PGK1 and ADH1 loci were comparable in wild-type and mutant strains. Thus, the reductions of H3 and H4 acetylation caused by targeted recruitment of Rpd3 histone deacetylase were similar at every promoter regardless of the degree of transcriptional repression. This indicates that activators respond differently to the same level of histone deacetylation.
FIG. 5.
Deacetylation of histones H3 and H4 by Rpd3. (A) Cross-linked chromatin from wild-type (Wt), rpd3, sin3, and ume6 strains carrying URS-HIS3 and LYS2 alleles controlled by the indicated activators was immunoprecipitated with antibodies against acetylated H3 and H4 histone tails (αH3Ac and αH4Ac, respectively), and the amounts of immunoprecipitated and input material for the indicated promoter regions were determined by quantitative PCR. (B) Deacetylation of the URS-HIS3 allele under inducing and noninducing conditions.
Targeted recruitment of Rpd3 does not affect activator binding.
Using the same chromatin preparations utilized to measure histone acetylation, we analyzed the occupancies of five different activators (Abf1, Rap1, Ace1, Gal4, and Gcn4) at the relevant Rpd3-repressed and control promoters (Fig. 6A). In each case, binding of the activator was unaffected by targeted recruitment of Rpd3; the difference in activator occupancy between the repressed and control promoters was <10% (Fig. 6B). Thus, for the five different activators tested, hypoacetylation does not alter accessibility of the activator binding sites and Rpd3-dependent repression functions at a step that occurs after activator binding.
Recruitment of Rpd3 reduces TBP occupancy in accordance with transcription.
To examine whether TBP occupancy correlates with Rpd3-mediated repression, we simultaneously monitored the binding of TBP to the Rpd3-repressed and control promoters in vivo. To maximize the signal-to-noise ratios, we introduced an HA-tagged version of TBP (35) into the relevant strains and eluted the immunoprecipitated material with an HA peptide. This additional purification step for TBP-bound DNA fragments is important, because TBP occupancy at weakly transcribed promoters is very low and hence near the background level of binding to any genomic region.
In accord with the strong relationship between TBP occupancy and transcriptional activity (35, 39), and in contrast to the lack of effect on activator binding, recruitment of Rpd3 caused a decrease in TBP occupancy at promoters activated by six different activators, Abf1, Rap1, Ace1, Gal4, Gcn4, and Hsf1 (Fig. 6). The reduction in TBP occupancy is clearly beyond the experimental error (±10%), and it correlates well with the degree of Rpd3-dependent repression. The Rap1-activated promoter, in which Rpd3-dependent repression is most pronounced, showed the largest difference in TBP occupancy (about fourfold, although a precise measurement is difficult because TBP occupancy at the repressed promoter is near the background level). The degree of repression at the remaining promoters tested was about 2-fold, and the corresponding change in TBP occupancy ranged from 1.7- to 2-fold.
We next extended these results to natural promoters in which transcription is repressed by recruitment of Rpd3. To obtain a meaningful TBP occupancy ratio, our analysis was limited to promoters in which TBP binding in a wild-type strain is above the background level observed at inactive promoters and at protein-coding regions. At all Rpd3 target genes tested, TBP binding was low in a wild-type strain and increased in an rpd3 strain (Fig. 6B). The largest effect on TBP binding (six- to sevenfold) at the INO1 promoter is in agreement with the tight repression of this gene, and the two- to threefold effect at the CAR1, CAR2, SPO11, and SPO13 promoters corresponds well with the reported degree of repression (2). Thus, decreased TBP occupancy upon recruitment of Rpd3 was observed for all artificial and natural promoters tested. Moreover, the excellent correlation between the decrease in TBP occupancy and the degree of transcriptional repression argues that inhibition occurs primarily (and perhaps exclusively) at or prior to the step of preinitiation complex formation.
Recruitment of Rpd3 decreases activator-dependent association of Swi/Snf and SAGA.
To identify steps between activator binding and TFIID recruitment that might be affected by Rpd3-mediated repression, we analyzed the occupancy of the Swi/Snf nucleosome-remodeling and SAGA histone acetylase complexes by using strains containing myc-tagged versions of Swi2 and Ada2, respectively (8). Gal4 and Gcn4 require both chromatin-modifying activities for full transcriptional activation (6, 14, 15, 44, 54), and they recruit SAGA to promoters independently of transcription (3, 32, 36). We therefore analyzed wild-type and rpd3 strains with Rpd3-repressed and control promoters containing an upstream Gal4 or Gcn4 site.
Following galactose induction, Ada2 (Fig. 7A) and Swi2 (Fig. 7B) are recruited to the GAL7 and control (Gal4-LYS2) promoters (signals upon induction are two- to threefold above those prior to induction), and the levels of recruitment are comparable in wild-type and rpd3 strains. However, at the Rpd3-repressed (Gal4-HIS3) promoter, the levels of Ada2 and Swi2 occupancy are indistinguishable from the levels observed under noninducing conditions and are roughly comparable to the levels at genomic regions presumed not to recruit Swi/Snf or SAGA (e.g., the POL1 coding region, the DED1 promoter, and an ORF-free region). Similarly, during amino acid starvation, Ada2 and Swi2 are recruited to the TRP3 and the Gcn4-LYS2 control promoters in both wild-type and rpd3 strains, whereas Ada2 and Swi2 occupancy at the Rpd3-repressed promoter (Gcn4-HIS3) is reduced nearly to the level under noninducing conditions. Importantly, the Rpd3-dependent effect on Ada2 and Swi2 occupancy is only observed for the URS1-containing promoters that target Rpd3 (i.e., those fused to the HIS3 structural gene [Fig. 7A and B]). In all other cases tested (including the HTA1 and ADH1 promoters, which appear to be bound by Swi2 and Ada2), the signals at a given promoter varied by <10% in the wild-type and rpd3 strains. Thus, recruitment of Rpd3 significantly reduces activator-dependent association of Swi/Snf and SAGA at the target promoters.
Given these results, we analyzed the degree of Rpd3-dependent repression of Gcn4- and Gal4-activated transcription in gcn5 and swi2 mutant strains. The LYS control genes allow us to observe and account for the defects in transcriptional activation in the various strains, thereby permitting a direct measurement of Rpd3-dependent repression. In all strains, Rpd3 represses Gcn4-activated transcription twofold and represses Gal4-activated transcription two- to threefold (Fig. 7B). Thus, repression is not influenced by deletion of either Gcn5 or Swi2, suggesting that inhibition of Swi/Snf or SAGA recruitment is not the sole mechanism of Rpd3 repression.
Repressive effect of Rpd3 can be bypassed by direct recruitment of TFIID.
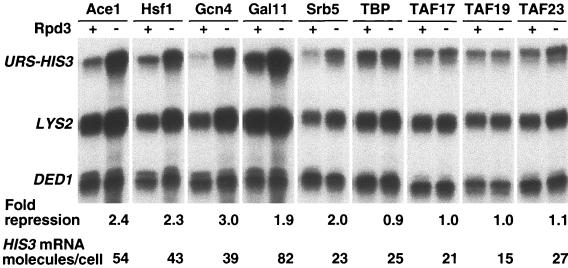
If Rpd3 represses transcription by inhibiting the association of a particular factor(s) with the promoter, it should be possible to override Rpd3-dependent repression by directly recruiting this component(s) to a promoter in an alternate fashion. We therefore analyzed the effect of Rpd3 on promoters activated by protein fusions between the Ace1 DNA-binding domain and natural activation domains or various components of the Pol II machinery. As folding of the Ace1 DNA-binding domain requires copper (16), the Ace1 fusions can be analyzed in the context of deacetylated chromatin that was established prior to copper induction.
As with natural activators, activation by Ace1 fusions to classical activation domains is repressed by Rpd3 (Fig. 8). Similarly, Rpd3 can repress activation by direct recruitment of Pol II holoenzyme via Ace1 fusions to the Gal11 or Srb5 components. In contrast, activation through direct recruitment of the TFIID component TBP, TBP-associated factor 17 (TAF17), TAF19, or TAF23 is not affected by Rpd3. These Ace1 fusions to TFIID components clearly have a special property with respect to Rpd3 repression, because they are less potent activators than other Ace1 fusions and natural activators that are repressible by Rpd3. The observation that direct recruitment of TBP or associated factors, but not Pol II holoenzyme, can bypass Rpd3 repression suggests that Rpd3 acts by limiting activator-dependent recruitment of TBP or TFIID.
FIG. 8.
Rpd3 repression can be bypassed by direct recruitment of TFIID components. JDY181 derivatives lacking Ace1 that contain (+) or lack (−) Rpd3 expressing a fusion of the indicated protein to the Ace1 DNA-binding domain and grown in the presence of 0.5 mM CuSO4 were used. The URS-HIS3, LYS2, and DED1 RNA levels were determined by quantitative S1 nuclease protection assays; repression of the URS-HIS3 alleles in wild-type strains and the relative expression (in HIS3 molecules/cell) of the nonrepressed URS-HIS3 allele are indicated.
DISCUSSION
Localized histone deacetylation inhibits the association of TBP/TFIID with promoters.
Rpd3 recruitment to a promoter leads to various degrees of repression depending on the activator (Fig. 2). Although weaker activators tend to be more strongly affected by Rpd3-dependent repression (see below), the degree of repression is not simply related to the strength of the activator. For example, the promoters controlled by Abf1 or the poly(dA-dT) element are poorly repressed by Rpd3 in comparison to all the other promoters involving comparably weak activators. Similarly, Ace1 fusions to TBP and TAFs are immune to Rpd3-dependent repression, even though they are less efficient activators than Ace1 fusions to natural activation domains or to Gal11, which are subject to Rpd3 repression. Importantly, the variation in repression occurs even though the levels of histone deacetylation are comparable at essentially all promoters tested (Fig. 5). Therefore, the activator-specific response to targeted recruitment of Rpd3 reflects the individual response of the activator to the same chromatin context.
Localized histone deacetylation does not affect the binding of all five activators tested (Fig. 6), indicating that Rpd3-mediated repression affects a later step in the activation process. Efficient binding of activators to deacetylated chromatin is likely to be a common phenomenon in vivo, because the DNA-binding domains of the five activators tested interact with DNA in structurally different manners. In this regard, Hsf1 binds in the context of hypoacetylated nucleosomes at the mating-type silencer (55), and several activators bind their target sites prior to recruitment of SAGA and increased histone acetylation at the promoter (3, 8, 19, 32, 36). As targeted histone deacetylation at the artificial promoters analyzed here is comparable in magnitude and extent to that observed at natural Rpd3-repressed promoters (9, 53), our results suggest that Rpd3-dependent repression of natural promoters will generally occur at a step after activator binding. However, association of the SBF activator with the HO promoter appears to require increased histone acetylation (8, 31), suggesting that Rpd3-dependent repression could occur at the activator binding step in certain situations. In addition, localized histone deacetylation could affect activator binding depending on the location of the binding site within the nucleosome. As is the case for many (but not all) yeast promoter regions, the activator binding sites in our promoter derivatives lie in a region of chromatin that is preferentially accessible and is not covered by a positioned nucleosome (24, 40, 42, 47, 58, 61).
For all natural and artificial promoters tested, recruitment of Rpd3 reduces TBP occupancy in accord with the degree of repression (Fig. 6). This reduction in TBP occupancy is notable, because the TATA element in our promoter derivatives also lies in an accessible region of chromatin lacking positioned nucleosomes (24, 40, 42, 47, 58, 61). As TBP occupancy is strictly correlated with association of TFIIA and TFIIB (34), this result indicates that Rpd3 inhibits transcription prior to or during the step of preinitiation complex formation. In this regard, Rpd3 behaves similarly to the Cyc8-Tup1 corepressor and the vast majority of activators in that transcriptional activity is strongly correlated with TBP occupancy in yeast cells (35, 39). Thus, targeted recruitment of Rpd3 represses transcription primarily by restricting the access of the Pol II machinery to the promoter. While our results do not exclude inhibitory effects after assembly of the preinitiation complex, such effects are unlikely to significantly contribute to the overall level of repression.
A key observation pertaining to how localized histone deacetylation inhibits transcription is that Rpd3-dependent repression is eliminated by direct recruitment of TFIID components (Fig. 8). In contrast, Rpd3 represses transcription under all other circumstances tested; these include eight natural activators, three Ace1 fusions to classical activation domains, and direct recruitment of the Gal11 and Srb5 components of Pol II holoenzyme. These results are strikingly similar to the observation that transcription from TATA-defective promoters can be activated by direct recruitment of TFIID components but not by direct recruitment of Pol II holoenzyme or natural activation domains (7, 18, 30). In the case of TATA-defective promoters, the limiting step for transcription is association of TBP with the TATA element, and this limitation can be overcome only by direct recruitment of TFIID components. By analogy, the observations here strongly argue that localized histone deacetylation at the promoter creates a situation in which TBP association with the TATA element becomes a more limiting step. Direct recruitment of TBP or its associated factors specifically overrides this limiting step, indicating that Rpd3 represses transcription by inhibiting recruitment of TBP or TFIID to the promoter.
The properties of the Ace1 fusions with respect to Rpd3-mediated repression are also analogous to the observation that direct recruitment of TFIID, but not Pol II holoenzyme, activates transcription in mammalian cells (13). This previous observation prompted the suggestion that association of TFIID with promoters is more limiting in mammalian cells than in yeast cells (13). Perhaps this distinction between yeast and mammalian cells is related to the fact that chromatin in yeast cells is generally more acetylated than chromatin in mammalian cells (69).
Recruitment of Rpd3 inhibits activator-dependent association of Swi/Snf and SAGA.
Under inducing conditions, Gal4 and Gcn4 recruit the Swi/Snf and SAGA complexes to target promoters independently of TBP association and transcriptional activity (3, 32, 36) (Fig. 7). Such activator-dependent association of Swi/Snf and SAGA is significantly inhibited by the localized domain of histone deacetylation caused by recruitment of Rpd3 (Fig. 7). Indeed, Rpd3 reduces Swi/Snf and SAGA association to (for Gal4 activation) or near (for Gcn4 activation) the level observed under noninducing conditions. As activator binding is not affected by recruitment of Rpd3, and as activator-dependent association of Swi/Snf and SAGA occurs independently of and prior to preinitiation complex formation (3, 8, 31, 32, 36), this result defines the first step in the activation process that is sensitive to histone deacetylation. Further, it supports the notion that Swi/Snf and SAGA occupancies are influenced by the chromatin environment at the promoter.
Gal4 and Gcn4 interact directly with Swi/Snf and SAGA in vitro (5, 14, 44, 45, 71), and such protein-protein interactions undoubtedly are critical for recruitment to target promoters in vivo. However, we do not favor the idea that targeted recruitment of the Rpd3 deacetylase complex sterically blocks the interactions of activators with Swi/Snf and SAGA. Rpd3 is not required for Ume6-dependent recruitment of the complex in vivo (26), and mutations in the active site of Rpd3 histone deacetylase have phenotypes indistinguishable from those of gene deletion (27). In addition, recruitment of the Rpd3 complex does not inhibit activator binding, and we have never observed Rpd3 at promoters by chromatin immunoprecipitation assays, suggesting that the physical association of Rpd3 with promoters is transient.
For these reasons, we favor the idea that localized deacetylation weakens the interactions of Swi/Snf and SAGA with nucleosomes in vivo. In support of this idea, histone acetylation per se stabilizes the association of Swi/Snf with nucleosomes in vitro (20). Moreover, Swi/Snf, SAGA, and other chromatin-modifying complexes contain subunits with bromodomains that can interact with acetylated lysine residues in vitro (12, 22, 48, 70) and coordinate nucleosome remodeling in vivo (62). Thus, an attractive hypothesis is that the domain of histone deacetylation caused by targeting of Rpd3 locally inhibits the association of Swi/Snf, SAGA, and perhaps other chromatin-modifying activities by blocking the bromodomain-dependent interaction with acetylated lysine residues.
Rpd3-dependent repression is most effective at weakly activated promoters.
For all six activators tested (Fig. 3), repression by Rpd3 is more efficient under conditions of weak activation (6- to 12-fold) than under conditions of strong activation (2-fold). The experiments involving Ace1 and Gcn4 provide the best evidence that the degree of Rpd3-dependent repression can be affected solely by the level of activation. For both activators, changes in the experimental conditions do not affect the activation mechanism per se but rather the amount of the activator that can bind the promoter. These observations are consistent with, and help explain, the tendency of weaker activators to be more strongly repressed by targeted recruitment of Rpd3. Thus, the degree of repression due to localized histone deacetylation depends both on the specific activator (and hence the activation mechanism) and on the level of activation.
These considerations indicate that a strong activator can partially override the negative effect of histone deacetylation. A strong activator is likely to stabilize the association of the Pol II machinery with the promoter through multiple protein-protein interactions, and it is also likely to cause longer-lasting changes in chromatin structure by efficient recruitment of chromatin-modifying activities. By either or both of these properties, strong activators might partially overcome the limitation on TBP association imposed by histone deacetylation. It should be noted that all the experiments described in this paper involve Rpd3-dependent repression of promoters responding to activators; hence, we do not know the extent to which Rpd3 represses “basal” transcription that occurs in the absence of any activator. In the experiments here, such basal transcription is barely detectable, and even this very low level might arise from cryptic activator binding sites located a few hundred base pairs upstream from the core promoter.
Of biological significance, Rpd3-dependent repression extends the magnitude of transcriptional activation, because transcription under “nonactivating” conditions is repressed to a larger degree than fully activated transcription. For the promoters examined here, the degrees of activation by Gal4, Hsf1, Gcn4, and Ace1 are about 250-fold in a wild-type strain but only about 50-fold in the absence of Rpd3. Thus, targeted recruitment of Rpd3 and localized histone deacetylation allows transcription to be effectively blocked in the absence of external induction yet still permits relatively high levels of transcription upon activation. This property reflects the need for precise control of natural Rpd3 target genes, such as those involved in sporulation and meiosis that must be tightly repressed during vegetative growth to prevent premature meiosis.
Multiple mechanisms for transcriptional repression by localized histone deacetylation.
Our results strongly suggest that targeted recruitment of Rpd3 represses transcription by at least two distinct mechanisms. One repression mechanism involves inhibition of activator-dependent recruitment of Swi/Snf and SAGA to promoters. In our experiments, Rpd3 reduces Swi/Snf and SAGA association to or near the level that occurs under noninducing conditions. As activator-mediated recruitment of Swi/Snf and SAGA can be important to achieve the fully activated level of transcription, it follows that inhibition of Swi/Snf and SAGA recruitment in such cases will result in repression. Repression by this mechanism should also result in decreased TBP occupancy, because Swi/Snf and SAGA recruitment is often required for and precedes TBP association (1, 3, 8, 36).
Several observations indicate that inhibition of SAGA and Swi/Snf association is not the only mechanism of Rpd3-mediated repression in vivo. First, Rpd3-mediated repression is observed when activation is achieved by direct recruitment of Pol II holoenzyme (i.e., the Ace1-Gal11 and Ace1-Srb5 fusions), a situation that bypasses the recruitment of chromatin-modifying activities. Second, Rpd3-mediated repression is more effective at weakly activated promoters, a condition in which Swi/Snf and SAGA are poorly associated with the promoter (3, 36) (Fig. 7). Third, Rpd3-mediated repression is not compromised by the absence of Gcn5 or Swi2, the catalytic subunits of SAGA and Swi/Snf, respectively. Under all these circumstances where Swi/Snf and Gcn5 histone acetylase play a minimal role in activation, transcription is still repressible by Rpd3.
These considerations suggest that targeted recruitment of Rpd3 can inhibit TBP association with promoters at a step that occurs after activator binding and recruitment of Swi/Snf and SAGA. The most likely mechanism is that localized histone deacetylation directly inhibits TBP/TFIID binding to the TATA element. This mechanism is strongly suggested by our results showing that Rpd3-dependent repression is alleviated when TBP/TFIID, but not Pol II holoenzyme, is directly recruited to the promoter and that repression is most effective under conditions when activators are least effective. Further, it is supported by the observation in vitro that histone acetylation can facilitate TBP binding on a chromatin template containing a positioned nucleosome over the TATA element (56).
Direct blocking of TBP association and inhibition of activator-dependent recruitment of Swi/Snf and SAGA are both likely to contribute to the activator specificity of Rpd3-mediated repression. The transcriptional requirement for Swi/Snf and SAGA depends on the activator as well as the individual promoter. By analogy with the artificial recruitment experiments (Fig. 8), differences among the various activators in the ability to recruit TBP or TFIID should contribute to the activator specificity of Rpd3 repression. The poor Rpd3-dependent repression of the poly(dA-dT)-dependent promoter might reflect the ability of poly(dA-dT) to generally increase the accessibility of proteins in the context of chromatin (24, 72) and perhaps to partially counteract the effect of histone deacetylation. Although direct inhibition of TBP does not directly involve activators, it should contribute to activator specificity and activator strength effects, because activators directly or indirectly increase TBP recruitment to promoters in vivo and might vary in the ability to override the inherent inhibitory effects of the TBP-TATA association. Thus, the relative importance of the two mechanisms by which localized histone deacetylation represses transcription will depend on the individual promoter.
Acknowledgments
We thank Gauri Dhavan for the peptide elution protocol necessary to improve the measurements of TBP occupancy, Dong-Ki Lee and John Lis for Ace1 plasmids, and Dennis Thiele for Ace1 antibodies.
This work was supported by grant GM 53720 to K.S. from the National Institutes of Health.
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-b promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein, B. E., J. K. Tong, and S. L. Schreiber. 2000. Genomewide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. USA 97:13708-13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhaumik, S. R., and M. R. Green. 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 15:1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brent, R., and M. Ptashne. 1985. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell 43:729-736. [DOI] [PubMed] [Google Scholar]
- 5.Brown, C. E., L. Howe, K. Sousa, S. C. Alley, M. J. Carrozza, S. Tan, and J. L. Workman. 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292:2333-2337. [DOI] [PubMed] [Google Scholar]
- 6.Burns, L. G., and C. L. Peterson. 1997. The yeast SWI-SNF complex facilitates binding of a transcriptional activator to nucleosomal sites in vivo. Mol. Cell. Biol. 17:4811-4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatterjee, S., and K. Struhl. 1995. Connecting a promoter-bound protein to the TATA-binding protein overrides the need for a transcriptional activation region. Nature 374:820-822. [DOI] [PubMed] [Google Scholar]
- 8.Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97:299-311. [DOI] [PubMed] [Google Scholar]
- 9.Deckert, J., and K. Struhl. 2001. Histone acetylation at promoters is differentially affected by activators and repressors. Mol. Cell. Biol. 21:2726-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Rubertis, F., D. Kadosh, S. Henchoz, D. Pauli, G. Reuter, K. Struhl, and P. Spierer. 1996. The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature 384:589-591. [DOI] [PubMed] [Google Scholar]
- 11.des Etages, S. A., D. A. Falvey, R. J. Reece, and M. C. Brandriss. 1996. Functional analysis of the PUT3 transcriptional activator of the proline utilization pathway in Saccharomyces cerevisiae. Genetics 142:1069-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhalluin, C., J. E. Carlson, L. Zeng, C. He, A. K. Aggarwal, and M. M. Zhou. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491-496. [DOI] [PubMed] [Google Scholar]
- 13.Dorris, D. R., and K. Struhl. 2000. Artificial recruitment of TFIID, but not RNA polymerase II holoenzyme, activates transcription in mammalian cells. Mol. Cell. Biol. 20:4350-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drysdale, C. M., B. M. Jackson, E. R. Klebanow, Y. Bai, T. Kokubo, M. Swanson, Y. Nakatani, A. Weil, and A. G. Hinnebusch. 1998. The Gcn4p activation domain interacts specifically in vitro with RNA polymerase II holoenzyme, TFIID, and the Adap-Gcn5p coactivator complex. Mol. Cell. Biol. 18:1711-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dudley, A. M., C. Rougeulle, and F. Winston. 1999. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 13:2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furst, P., S. Hu, R. Hackett, and D. Hamer. 1988. Copper activates metallothionein gene transcription by altering the conformation of a specific DNA binding protein. Cell 55:705-717. [DOI] [PubMed] [Google Scholar]
- 17.Geisberg, J. V., and K. Struhl. 2000. TATA-binding protein mutants that increase transcription from enhancerless and repressed promoters in vivo. Mol. Cell. Biol. 20:1478-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez-Couto, E., N. Klages, and M. Strubin. 1997. Synergistic and promoter-selective activation of transcription by recruitment of TFIID and TFIIB. Proc. Natl. Acad. Sci. USA 94:8036-8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gregory, P. D., A. Schmid, M. Zavari, M. Munsterkotter, and W. Horz. 1999. Chromatin remodeling at the PHO8 promoter requires SWI-SNF and SAGA at a step subsequent to activator binding. EMBO J. 18:6407-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hassan, A. H., K. E. Neely, and J. L. Workman. 2001. Histone acetyltransferase complexes stabilize Swi/Snf binding to promoter nucleosomes. Cell 104:817-827. [DOI] [PubMed] [Google Scholar]
- 21.Hong, L., G. P. Schroth, H. R. Matthews, P. Yau, and E. M. Bradbury. 1993. Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 “tail” to DNA. J. Biol. Chem. 268:305-314. [PubMed] [Google Scholar]
- 22.Hudson, B. P., M. A. Martinez-Yamout, H. J. Dyson, and P. E. Wright. 2000. Solution structure and acetyl-lysine binding activity of the GCN5 bromodomain. J. Mol. Biol. 304:355-370. [DOI] [PubMed] [Google Scholar]
- 23.Iyer, V., and K. Struhl. 1995. Mechanism of differential utilization of the his3 TR and TC TATA elements. Mol. Cell. Biol. 15:7059-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iyer, V., and K. Struhl. 1995. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic structure. EMBO J. 14:2570-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iyer, V., and K. Struhl. 1996. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:5208-5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kadosh, D., and K. Struhl. 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89:365-371. [DOI] [PubMed] [Google Scholar]
- 27.Kadosh, D., and K. Struhl. 1998. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 12:797-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kadosh, D., and K. Struhl. 1998. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol. 18:5121-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keleher, C. A., M. J. Redd, J. Schultz, M. Carlson, and A. D. Johnson. 1992. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell 68:709-719. [DOI] [PubMed] [Google Scholar]
- 30.Klages, N., and M. Strubin. 1995. Stimulation of RNA polymerase II transcription initiation by recruitment of TBP in vivo. Nature 374:822-823. [DOI] [PubMed] [Google Scholar]
- 31.Krebs, J. E., M.-H. Kuo, C. D. Allis, and C. L. Peterson. 1999. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 13:1412-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuo, M.-H., E. vom Baur, K. Struhl, and C. D. Allis. 2000. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell 6:1309-1320. [DOI] [PubMed] [Google Scholar]
- 33.Kuo, M.-H., J. Zhou, P. Jambeck, M. E. A. Churchill, and C. D. Allis. 1998. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 12:627-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuras, L., P. Kosa, M. Mencia, and K. Struhl. 2000. TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science 288:1244-1248. [DOI] [PubMed] [Google Scholar]
- 35.Kuras, L., and K. Struhl. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399:609-612. [DOI] [PubMed] [Google Scholar]
- 36.Larschan, E., and F. Winston. 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15:1946-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, D. K., S. Kim, and J. T. Lis. 1999. Different upstream transcriptional activators have distinct coactivator requirements. Genes Dev. 13:2934-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, D. Y., J. J. Hayes, D. Pruss, and A. P. Wolffe. 1993. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell 72:73-84. [DOI] [PubMed] [Google Scholar]
- 39.Li, X.-Y., A. Virbasius, X. Zhu, and M. R. Green. 1999. Enhancement of TBP binding by activators and general transcription factors. Nature 389:605-609. [DOI] [PubMed] [Google Scholar]
- 40.Losa, R., S. Omari, and F. Thoma. 1990. Poly(dA)·poly(dT) rich sequences are not sufficient to exclude nucleosome formation in a constitutive yeast promoter. Nucleic Acids Res. 18:3495-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 angstrom resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 42.Mai, X., S. Chou, and K. Struhl. 2000. Preferential accessibility of the yeast his3 promoter is determined by a general property of the DNA sequence, not by specific elements. Mol. Cell. Biol. 20:6668-6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melcher, K., and H. E. Xu. 2001. Gal80-Gal80 interaction on adjacent Gal4 binding sites is required for complete GAL gene repression. EMBO J. 20:841-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Natarajan, K., B. M. Jackson, H. Zhou, F. Winston, and A. G. Hinnebusch. 1999. Transcriptional activation by Gcn4 involves independent interactions with the Swi/Snf complex and the Srb/mediator. Mol. Cell 4:657-664. [DOI] [PubMed] [Google Scholar]
- 45.Neely, K. E., A. H. Hassan, A. E. Wallberg, D. J. Steger, B. R. Cairns, A. P. Wright, and J. L. Workman. 1999. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol. Cell 4:649-655. [DOI] [PubMed] [Google Scholar]
- 46.Nightingale, K. P., R. E. Wellinger, J. M. Sogo, and P. B. Becker. 1998. Histone acetylation facilitates RNA polymerase II transcription of the Drosophila hsp26 gene in chromatin. EMBO J. 17:2865-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oettinger, M. A., and K. Struhl. 1985. Suppressors of Saccharomyces cerevisiae promoter mutations lacking the upstream element. Mol. Cell. Biol. 5:1901-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Owen, D. J., P. Ornaghi, J. C. Yang, N. Lowe, P. R. Evans, P. Ballario, D. Neuhaus, P. Filetici, and A. A. Travers. 2000. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase Gcn5. EMBO J. 19:6141-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parekh, B. S., and T. Maniatis. 1999. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-b promoter. Mol. Cell 3:125-129. [DOI] [PubMed] [Google Scholar]
- 50.Pazin, M. J., and J. T. Kadonaga. 1997. What's up and down with histone deacetylation and transcription? Cell 89:325-328. [DOI] [PubMed] [Google Scholar]
- 51.Reid, J. L., V. R. Iyer, P. O. Brown, and K. Struhl. 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6:1297-1307. [DOI] [PubMed] [Google Scholar]
- 52.Rundlett, S. E., A. A. Carmen, R. Kobayashi, S. Bavykin, B. M. Turner, and M. Grunstein. 1996. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes. Proc. Natl. Acad. Sci. USA 93:14503-14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rundlett, S. E., A. A. Carmen, N. Suka, B. M. Turner, and M. Grunstein. 1998. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392:831-835. [DOI] [PubMed] [Google Scholar]
- 54.Ryan, M. P., R. Jones, and R. H. Morse. 1998. SWI-SNF complex participation in transcriptional activation at a step subsequent to activator binding. Mol. Cell. Biol. 18:1774-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sekinger, E. A., and D. S. Gross. 2001. Silenced chromatin is permissive to activator binding and PIC recruitment. Cell 105:403-414. [DOI] [PubMed] [Google Scholar]
- 56.Sewack, G. F., T. W. Ellis, and U. Hansen. 2001. Binding of TATA-binding protein to a naturally positioned nucleosome is facilitated by histone acetylation. Mol. Cell. Biol. 21:1404-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sorger, P. K., and H. R. B. Pelham. 1988. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell 54:855-864. [DOI] [PubMed] [Google Scholar]
- 58.Struhl, K. 1982. Promoter elements, regulatory elements, and chromatin structure of the yeast his3 gene. Cold Spring Harbor Symp. Quant. Biol. 47:901-910. [DOI] [PubMed] [Google Scholar]
- 59.Struhl, K. 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12:599-606. [DOI] [PubMed] [Google Scholar]
- 60.Struhl, K. 1999. Fundamentally different logic of gene expression in eukaryotes and prokaryotes. Cell 98:1-4. [DOI] [PubMed] [Google Scholar]
- 61.Suter, B., M. Livingstone-Zatchej, and F. Thoma. 1997. Chromatin structure modulates DNA repair by photolyase in vivo. EMBO J. 16:2150-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Syntichaki, P., I. Topalidou, and G. Thireos. 2000. The Gcn5 bromodomain coordinates nucleosome remodeling. Nature 404:414-417. [DOI] [PubMed] [Google Scholar]
- 63.Ura, K., H. Kurumizaka, S. Dimitrov, G. Almouzni, and A. P. Wolffe. 1997. Histone acetylation: influence on transcription, nucleosome mobility and positioning, and linker histone-dependent transcriptional repression. EMBO J. 16:2096-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vannier, D., D. Balderes, and D. Shore. 1996. Evidence that the transcriptional regulators, SIN3 and RPD3, and a novel gene (SDS3) with similar functions, are involved in transcriptional silencing in S. cerevisiae. Genetics 144:1343-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vettese-Dadey, M., P. A. Grant, T. R. Hebbes, C. Crane-Robinson, C. D. Allis, and J. L. Workman. 1996. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 15:2508-2518. [PMC free article] [PubMed] [Google Scholar]
- 66.Vogelauer, M., J. Wu, N. Suka, and M. Grunstein. 2000. Global histone acetylation and deacetylation in yeast. Nature 408:495-498. [DOI] [PubMed] [Google Scholar]
- 67.Wang, D., Y. Hu, F. Zheng, K. Zhou, and G. B. Kohlhaw. 1997. Evidence that intramolecular interactions are involved in masking the activation domain of transcriptional activator Leu3. J. Biol. Chem. 272:19383-19392. [DOI] [PubMed] [Google Scholar]
- 68.Wang, L., L. Liu, and S. L. Berger. 1998. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev. 12:640-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Waterborg, J. H. 2000. Steady-state levels of histone acetylation in Saccharomyces cerevisiae. J. Biol. Chem. 275:13007-13011. [DOI] [PubMed] [Google Scholar]
- 70.Winston, F., and C. D. Allis. 1999. The bromodomain: a chromatin-targeting module? Nat. Struct. Biol. 6:601-604. [DOI] [PubMed] [Google Scholar]
- 71.Yudkovsky, N., C. Logie, S. Hahn, and C. L. Peterson. 1999. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 13:2369-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu, Z., and D. J. Thiele. 1996. A specialized nucleosome modulates transcription factor access to a C. glabrata metal responsive promoter. Cell 87:459-470. [DOI] [PubMed] [Google Scholar]