Abstract
Human papillomavirus (HPV) DNA replication requires the viral origin recognition protein E2 and the presumptive viral replicative helicase E1. We now report for the first time efficient DNA unwinding by a purified HPV E1 protein. Unwinding depends on a supercoiled DNA substrate, topoisomerase I, single-stranded-DNA-binding protein, and ATP, but not an origin. Electron microscopy revealed completely unwound molecules. Intermediates contained two single-stranded loops emanating from a single protein complex, suggesting a bidirectional E1 helicase which translocated the flanking DNA in an inward direction. We showed that E2 protein partially inhibited DNA unwinding and that Hsp70 or Hsp40, which we reported previously to stimulate HPV-11 E1 binding to the origin and promote dihexameric E1 formation, apparently displaced E2 and abolished inhibition. Neither E2 nor chaperone proteins were detected in unwinding complexes. These results suggest that chaperones play important roles in the assembly and activation of a replicative helicase in higher eukaryotes. An E1 mutation in the ATP binding site caused deficient binding and unwinding of origin DNA, indicating the importance of ATP binding in efficient helicase assembly on the origin.
Human and animal papillomaviruses are prevalent pathogens. Efficient origin (ori)-dependent replication of viral DNA requires the virus-encoded E1 and E2 proteins as well as cellular replication proteins (11, 27, 46, 59, 68). As such, these DNA viruses may serve as a model for higher eukaryotic DNA replication, as do simian virus 40 (SV40) and polyomavirus. The papillomavirus ori consists of several E2 binding sites (BS) flanking one E1 BS. E1 recruits the DNA polymerase α/primase (6, 12, 41) and the single-stranded-DNA-binding protein RPA (25). The human papillomavirus (HPV) E1 protein is required during initiation and elongation and is thought to be the replicative helicase (33). However, HPV E1 proteins are poor helicases in strand displacement assays (26, 65), and there has been no report of DNA-unwinding activity. In contrast, the bovine papillomavirus 1 (BPV-1) E1 exhibits helicase activity in both assays (51, 69).
We previously reported that purified HPV-11 E1 protein expressed in insect Sf9 cells binds to ori with low affinity and specificity and also binds to DNA nonspecifically (33). Electron microscopy (EM) shows that E1 binds ori primarily as a hexamer and, at a low frequency, as a dihexamer (34). The human heat shock proteins Hsp70, Hdj2, and Hdj1 greatly stimulate E1 binding to ori. Hdj1 and Hdj2 encode members of the Hsp40 family of proteins that normally function as cochaperones of the Hsp70 proteins and greatly stimulate the ATPase activity of Hsp70 (for a review, see reference 21). However, in the case of the HPV-11 E1-ori association, their effects are independent and additive. Most strikingly, EM has revealed that Hsp40 but not Hsp70 promotes E1 dihexamer formation on ori (34). The BPV-1 E1 has been reported to be a hexameric helicase (48). However, EM revealed a bilobed complex, which was presumed to be a dihexamer as well, but the size and the frequency of this complex were not reported (20). Two helicases functioning at divergent orientations are necessary for bidirectional DNA replication, but it has not been established whether E1 proteins function as a dihexameric, bidirectional helicase or whether the dihexamer separates into two independent hexameric helicases as the DNA flanking the ori unwinds.
From bacteria to higher eukaryotes, chaperone or heat shock proteins play crucial roles in protein folding, trafficking, and protein complex assembly and disassembly (21). The Escherichia coli chaperones DnaJ and DnaK serve important roles in the assembly of replicative helicases for bacteriophages λ , P1, and P7 (2, 3, 31, 66, 72). Human Hdj2 and Hsp70 are homologs of DnaJ and DnaK, respectively. In particular, the domain of Hsp40 which interacts with HPV-11 E1 has been mapped to a span of 20 amino acids within the highly conserved J domain (34). Moreover, incubation of HPV-11 E1, ori, and chaperones decreases the lag time in the onset of DNA replication and increases cell-free replication despite the presence of abundant chaperone proteins in the cell extracts (34). Along with SV40 T antigen, this provided the first indication that cellular chaperones have a role in DNA replication in higher eukaryotes.
The SV40/polymavirus T antigen functions as a dihexameric helicase (15, 17, 42, 52, 61, 64). Interestingly, its amino terminus is homologous to the J domain of DnaJ and interacts with Hsc70 (for a review, see reference 54). This domain is required for efficient SV40 DNA replication in vivo (7), and its truncation leads to incorrect oligomerization (62). Recently, Hsp40 and Hsp70 have also been reported to enhance the binding of UL9, the origin-binding protein of herpes simplex virus type 1, to oriS and the resultant ori opening (57). hTid-1, a human homolog of E. coli DnaJ, also binds to UL9 and promotes multimer formation from dimers (19). In addition, Hsp70 also interacts with Orc4p of Saccharomyces cerevisiae, dissociating the oligomerized Orc4p amino-terminal domain (23), a function which we previously proposed for Hsp70 in stimulating HPV-11 E1 binding to ori (34). Collectively, these observations strongly suggest that chaperone proteins play a role in the assembly and activation of replicative machinery in both prokaryotes and eukaryotes.
The papillomavirus E2 protein is also a multifunctional protein. Among its many functions, it is the primary ori recognition protein. It binds to the E2 BS with high specificity and high affinity and is crucial for assembly of the preinitiation complex by recruiting and targeting E1 to ori (27, 36, 44, 50, 68). In vivo, E2 prevents nucleosome formation around ori (30). Therefore, only plasmids containing the E2 BS function in transient replication (10, 35, 46). In addition, E2 also associates with the nuclear matrix and PML (a component of nuclear domain 10), where viral DNA replication takes place (56, 71). On the other hand, a stable association of E2 to E2 BSs that flank the E1 BS may act as a DNA clamp, preventing DNA unwinding. This latter possibility may explain why E2 is absent from the replication complex postinitiation (33). However, because HPV E1 DNA-unwinding activity has not been demonstrated, neither the E2 inhibition of DNA unwinding by E1 nor the mechanism of E2 release has been investigated.
In this study, we report that bacterially purified HPV-11 E1 protein exhibited a robust supercoiled DNA-unwinding activity. Using biochemical assays and EM examination, we characterized the requirements of the unwinding reaction, the nature of unwinding intermediates and products, and the effects of E2 protein and chaperone proteins. The implications of these findings are presented.
MATERIALS AND METHODS
Plasmids and proteins.
p7874-99 and p7874-20 are HPV-11 ori plasmids based on the pUC19 vector, containing HPV-11 nucleotides 7874 to 7933/1 to 99 and 7874 to 7933/1 to 20, respectively. p7730-99 (234M) contains mutations in E2 BS copies 2, 3, and 4 in the ori (spanning nucleotides 7730 to 7933/1 to 99) (10). Epitope-tagged HPV-11 E1 and E2 proteins were expressed and purified from E. coli. To express the EE-E1 protein (27), E. coli BL21(DE3) (Stratagene, La Jolla, Calif.) harboring pRSET-EE-E1 (32) was induced at mid-log phase with 0.3 mM isopropylthio-β-galactopyranoside (IPTG) for 24 h at 18°C. Cells were disrupted by sonication in lysis buffer (20 mM Tris-HCl [pH 7.0], 250 mM NaCl, 1 mM dithiothreitol). The soluble fraction was first passed through a Q-Sepharose column (Bio-Rad, Hercules, Calif.) and eluted with 20 mM Tris-HCl (pH 7.5)-800 mM NaCl. The eluant was then applied to an anti-EE immunoaffinity column, washed with 1 M NaCl-20 mM Tris-HCl (pH 7.5), and eluted with 100 mM triethylamine, as described previously (27, 34). The eluted protein was dialyzed overnight against Tris-HCl (pH 7.0)-50 mM NaCl-10% glycerol buffer and kept at −80°C. A P479S mutation of HPV-11 E1 was generated by site-directed mutagenesis with PCR amplification. This mutated protein was similarly tagged with the EE epitope and expressed and purified from E. coli as described for the wild-type E1 protein.
The induction of the HPV-11 E2 protein from pRSET-11E2, which was tagged with an epitope from a cytomegalovirus-encoded protein (32), in E. coli BL21(DE3)pLysS (Stratagene), was conducted as described previously for the E1 proteins except for the addition of 0.2 mM IPTG. To purify the E2 protein, the soluble fraction was diluted with buffer Q (20 mM Tris-HCl [pH 7.0], 10 mM 2-mercaptoethanol) to a final concentration of 50 mM NaCl and applied to a 10-ml Q-Sepharose column. The flowthrough was then applied to a 1-ml Macro-Prep High S column (Bio-Rad) and eluted with a 100 to 500 mM NaCl gradient in buffer Q. The fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot, and those containing E2 protein were pooled. The human Hsp70 (1) and Hdj2 (9, 34) proteins were expressed in and purified from E. coli as described previously (34). A recombinant baculovirus expressing human topoisomerase I was a gift from Jim Champoux, and protein was purified as described previously (53). An E. coli expression vector for His-tagged human RPA was a gift from Mike O'Donnell (70), and the protein complex was purified by nickel affinity chromatography (Qiagen, Valencia, Calif.). All proteins were purified to near homogeneity, as determined by Coomassie blue staining and Western blotting with antibodies (Fig. 3A and data not shown). For gel assays, the E. coli single-stranded-DNA-binding protein (SSB) was obtained from Amersham Pharmacia Biotech. For EM, SSB was purified as described previously (8).
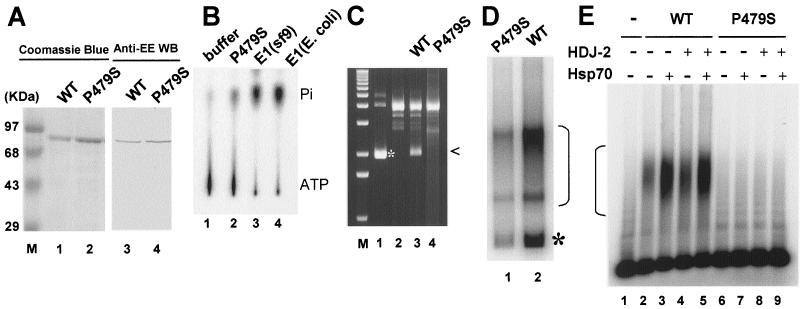
FIG. 3.
HPV-11 E1 P479S mutant protein is defective in multiple activities. Wild-type HPV-11 E1 proteins purified from Sf9 cells (27) and from E. coli were tested for comparison. (A) Wild-type (WT) E1 and mutated E1 P479S protein were analyzed by SDS-PAGE and stained with Coomassie blue (left) and also by Western blot with monoclonal antibody against the epitope tag (right). (B) Autoradiogram of ATPase assays. The positions of [γ-32P]ATP and free 32Pi are marked. (C) Unwinding of ori plasmid p7874-99 as revealed in an ethidium bromide-stained chloroquine-agarose gel. Lane 1, supercoiled input DNA (*) in reaction buffer. Lane 2, reaction without E1. Lane 3, reaction with wild-type E1 purified from E. coli. U-form DNA is marked (<). Lane 4, reaction with E1 P479S. (D) Autoradiogram of cell-free replication of ori DNA in the presence of purified HPV-11 E2 protein, wild-type HPV-11 E1, or E1 P479S and [α-32P]dCTP. Form I product (*) and replication intermediates (bracket) are marked. (E) Autoradiogram of EMSA of wild-type HPV-11 E1 or E1 P479S and 32P-labeled ori-containing fragment (nucleotides 7874 to 99) in the presence and absence of Hdj2 or Hsp70. DNA-protein complexes are marked (bracket).
DNA unwinding and chloroquine-agarose gel electrophoresis.
The conditions for unwinding were modified from those described previously (16). A standard reaction mixture of 60 μl contained 500 ng of supercoiled DNA, 300 ng to 1.0 μg of E1 protein, 25 ng of topoisomerase I, 300 ng of human RPA or E. coli SSB, 4 mM ATP, 40 mM creatine phosphate, 23.3 μg of creatine phosphate kinase (creatine phosphate kinase) per ml, 16.7 μg of bovine serum albumin per ml, 7 mM MgCl2, and 5 mM dithiothreitol. Variations in conditions were as described in Results. After incubation at 37°C for different lengths of time as indicated in each figure legend, reactions were terminated by adding 6.8 μl of stop mix, containing 13.5 mM EDTA, 30 μg of tRNA per ml, 0.3% N-lauroylsarcosine, and 0.45 mg of proteinase K per ml, and incubation was continued for 30 min at 37°C. After extraction with phenol-chloroform, DNA was separated by electrophoresis in 1.2% agarose gels containing 0.25 μg of chloroquine per ml (16). The gels were stained with ethidium bromide and documented with a Bio-Rad Gel Doc 2000 system.
Cell-free DNA replication and EMSA.
Cell-free replication of p7874-99 was conducted with human 293 cell extracts (27). Replication products were labeled with [α-32P]dCTP (Amersham Pharmacia Biotech), purified, and separated by agarose gel electrophoresis. Electrophoretic mobility shift assays (EMSAs) were performed as described previously (33, 34). The EcoRI-HindIII restriction fragment spanning HPV-11 ori (nucleotides 7874 to 7933/1 to 99) was labeled with [γ-32P]ATP (Amersham Pharmacia Biotech) and T4 polynucleotide kinase (Life Technology, Inc.) and used as a substrate. Data were acquired with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
ATPase assay.
ATPase activity was detected by release of 32Pi from [γ-32P]ATP as described previously (49). A 20-μl amount of reaction mixture contained 100 ng of HPV-11 E1 protein, 25 mM Tris-HCl (pH 8.0), 1 mM dithiothreitol, 10 mM MgCl2, 250 μg of bovine serum albumin per ml, 50 μM [γ-32P]ATP (1.5 × 104 cpm/pmol) (Amersham Pharmacia Biotech), and 50 ng of single-stranded M13 DNA. After incubation for 60 min at 37°C, 1 μl was spotted onto a polyethyleneimine-cellulose thin-layer chromatography plate (Sigma), which was developed with 1.0 M formic acid-0.5 M LiCl for 45 min. Data were captured with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
Electron microscopy.
Unwinding assays were conducted with 400 ng of ori plasmid p7874-99, 500 ng of E1, 25 ng of human topoisomerase I, and 180 ng of E. coli SSB in 60 μl of buffer containing 20 mM HEPES (pH 7.8), 80 mM NaCl, 4 mM ATP, and 2 mM MgCl2; 300 ng of E2, Hsp70, or Hdj2 was added as specified. For binding assays, ori plasmid and proteins were incubated in buffer containing 2 mM ATP or ATP-γ-S (33) for 20 min at 37°C. Reactions were terminated by adding EDTA to 10 mM on ice and then glutaraldehyde to 0.6% for 5 min at 20°C. The samples were chromatographed over 2 ml of BioGel A5m (Bio-Rad) columns equilibrated with TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). Fractions containing the DNA-protein complexes were collected, and aliquots were mounted on glow discharge-treated carbon supports as described previously (24). U-form DNA purified from a chloroquine-agarose gel was concentrated by ethanol precipitation, redissolved in TE, incubated with SSB (3 μg/ml), and prepared for EM as described above.
For immunogold EM, the DNA-protein complex-containing fractions from the A5m BioGel columns were pooled. After the NaCl concentration was adjusted to 80 mM, aliquots were incubated for 1 h at 37°C with primary rabbit polyclonal antibodies to E2 (11) or monoclonal antibodies to Hsp70 (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.) or Hdj2 (Neomarker, Fremont, Calif.) (1:1,000 to 1:5,000 dilution) and then for another 1 h with secondary goat anti-rabbit or anti-mouse immunoglobulin antibodies conjugated to 5- or 10-nm gold particles (Amersham Pharmacia Biotech), respectively. The reactions were stopped, and the mixtures were chromatographed and prepared for EM as described above. All samples were analyzed with a Philips CM12 electron microscope. The images were scanned with a Nikon LS-4500AF film scanner, and contrast was adjusted with Adobe Photoshop software.
RESULTS
Requirements for DNA unwinding by HPV-11 E1.
The supercoiled HPV-11 ori plasmid p7874-99, which contains three copies of the E2 BS flanking the single E1 BS, was used in unwinding reactions (Fig. 1A, lanes 1 to 8) based on the assay for the SV40 T antigen (16). The reactions were optimized by reiterative experiments with set amounts of DNA substrate, various amounts of purified E1, and lengths of reaction time at several concentrations of topoisomerase I and RPA (data not shown). DNA from the reactions was then purified, electrophoretically separated in a chloroquine-agarose gel, and revealed by ethidium bromide staining.
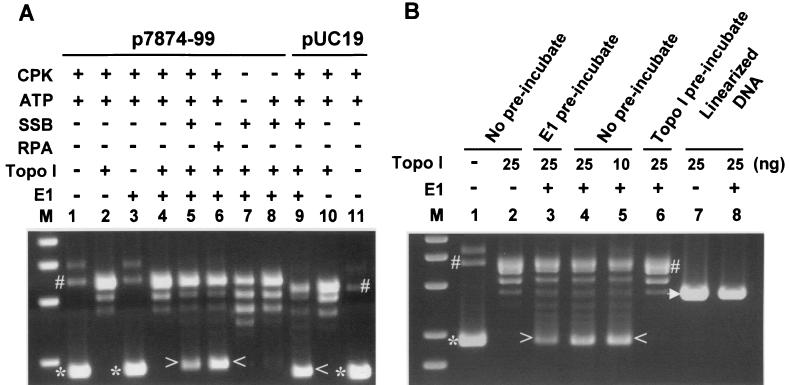
FIG. 1.
Requirements for DNA unwinding by HPV-11 E1 protein. Agarose gels were stained with ethidium bromide. The presence or absence of specific reagents (see Materials and Methods) is indicated by + or − above each lane. (A) Substrates were supercoiled plasmid DNA, p7874-99, which contains the HPV-11 ori cloned in pUC19 (lanes 1 to 8), or the vector (lanes 9 to 11). Incubation was for 15 min at 37°C. Similar results were obtained when incubation was for 10 min (data not shown). (B) Substrates were circular (lanes 1 to 6) or ScaI-linearized p7874-99 (lanes 7 and 8). All reagents were as described for panel A except in lane 5, in which topoisomerase I (Topo I) was reduced from 25 ng to 10 ng. Lane 1, input supercoiled DNA in reaction buffer. Preincubation of plasmid DNA with E1 (lane 3) or topoisomerase I (lane 6) was for 10 min at 37°C. The missing enzyme was then added, and incubation was continued for another 5 min at 37°C. Increasing the second incubation to 10 min yielded the same result (data not shown). Relaxed circular DNA (#), form I supercoiled DNA (*), U-form DNA (> or <), and linear DNA (arrowhead) are marked. Lane M, double-stranded size markers.
In the absence of E1, the addition of topoisomerase I converted the input, fast-migrating form I DNA (∗) to slow-migrating relaxed circular DNA (#) and a ladder of closed circular DNA with reduced superhelicity (collectively termed covalently closed [cc] DNA topoisomers) (compare lanes 1 to 2 and 10 to 11). Unwinding took place efficiently in the presence of E1, human topoisomerase I, human single-stranded-DNA-binding protein RPA, ATP, and creatine phosphate kinase, a component of an ATP-regenerating system. A fraction of the fast migrating form I DNA was converted to a slightly slower migrating unwound form (U-form DNA) (lane 6) (marked with > or < in this and other figures), as described previously (16, 43), the balance being the cc DNA topoisomers. Reactions reached a plateau by 10 to 15 min at 37°C (data not shown). An ori was, however, not necessary (Fig. 1A, lanes 9 to 11).
The ori-independent DNA unwinding is consistent with the nonspecific association of E1 with DNA and with its ability to support ori-independent replication in the cell-free system when E1 or DNA is present at high concentrations (27, 33). Both conditions were also used in this study. Our result also agrees with reports that BPV-1 E1 unwinds DNA without an ori in the absence of a nonspecific inhibitor DNA (51, 69).
The E. coli single-stranded-DNA-binding protein SSB was less effective than the human RPA (Fig. 1A, compare lanes 5 and 6). Omission of one or more of the components abolished unwinding at the sensitivity limit of this detection method (lanes 1 to 4, 7, 8, 10, and 11) with one exception. Upon prolonged exposure, a faint U-form band was detected in the reaction from which creatine phosphate kinase was omitted (data not shown). This was confirmed by EM (see below). A reduction of E1 protein to below 300 ng abolished unwinding (data not shown). Neither preincubation of E1 with the supercoiled DNA nor reduction of topoisomerase I by 2.5-fold significantly affected the amount of U-form DNA produced (Fig. 1B, lanes 3 to 5, and data not shown). Thus, under our reaction conditions, binding of E1 to DNA was not rate limiting, nor was topoisomerase I in excess relative to E1 and DNA.
Because single-stranded DNA binds ethidium bromide poorly relative to double-stranded DNA, the percentage of DNA converted to the U form can best be approximated by comparing the remaining cc DNA topoisomers to those formed in the absence of E1 (for instance, Fig. 1A, compare lanes 2 and 4 to 6 and lane 10 to 9). Although the efficiency of generating U-form DNA varied with different E1 protein preparations, in some reactions, there was clearly more U-form DNA than remaining cc DNA topoisomers, indicating a conversion efficiency of over 50% (Fig. 1A, lanes 6 and 9; Fig. 1B, lane 5). These properties are similar to those reported for BPV-1 E1 (51, 68) except that unwinding with HPV-11 E1 protein was fast and highly efficient and did not require radioactive probes to reveal the U-form DNA.
Because of the poor helicase activity of HPV E1 proteins in strand displacement assays, we suspected that the nature of the substrate might be important. We tested this supposition. Indeed, linear ori DNA and ori DNA prerelaxed with topoisomerase I were poor substrates, as we detected no U-form DNA (Fig. 1B, lanes 8 and 6), which we showed to contain single-stranded circular and linear DNA (see below). However, a low level of partial unwinding of linear DNA probably did occur, as suggested by a reproducibly observed reduction in the substrate (compare lanes 7 and 8, and data not shown). We believe that unwinding of linear DNA was incomplete and that the products had heterogeneous migration rates and hence escaped detection.
U-form DNA contains single-stranded DNA.
U-form DNA was described previously as unwinding intermediates containing single-stranded regions of various lengths (16, 17, 51, 60, 67). We purified U-form DNA from an agarose gel, incubated it with E. coli SSB to extend the single-stranded regions, and then examined it by EM. We observed no partially unwound molecules or interlocked single strands. Rather, 60 to 70% of the molecules were circular or linear, SSB-coated single strands, with the remainder being double-stranded open circles (Fig. 2A, and data not shown). Since the slower-migrating open circles (and the cc DNA topoisomers) are not likely to contaminate the fast-migrating U-form DNA to a significant extent (Fig. 1, compare bands marked with #, >, and <), we suggest that the double-stranded open circles were generated by reannealing of single strands during purification of the U-form DNA from the agarose gel. We also suggest that single-strand breaks are introduced into the products by topoisomerase I or by physical strand breakage during DNA purification prior to agarose electrophoresis. Thus, any interlocked single-stranded circles would have been converted to separate single-stranded circles and linear molecules. Some of the single strands subsequently renature during purification from the agarose gel. Since we only examined DNA recovered from the U-form DNA band, unwinding intermediates that migrated more slowly in the agarose gels were not recovered and hence escaped detection.
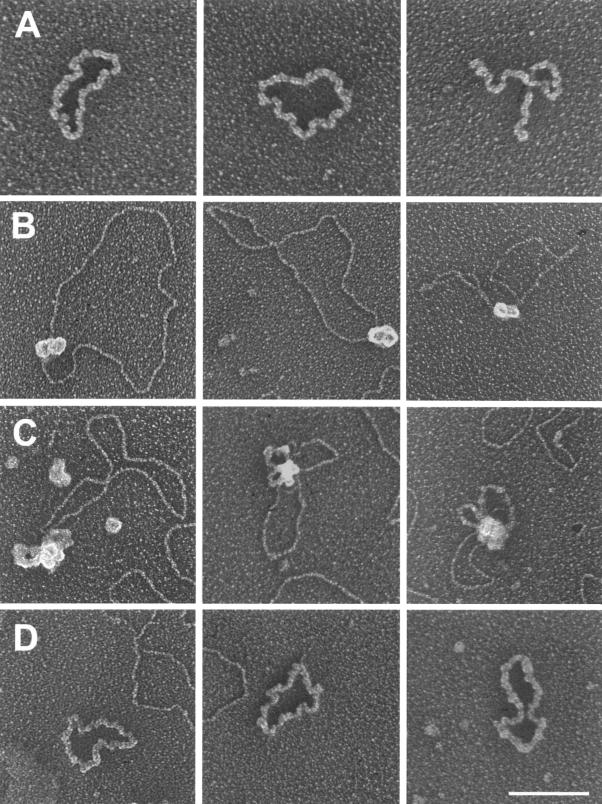
FIG. 2.
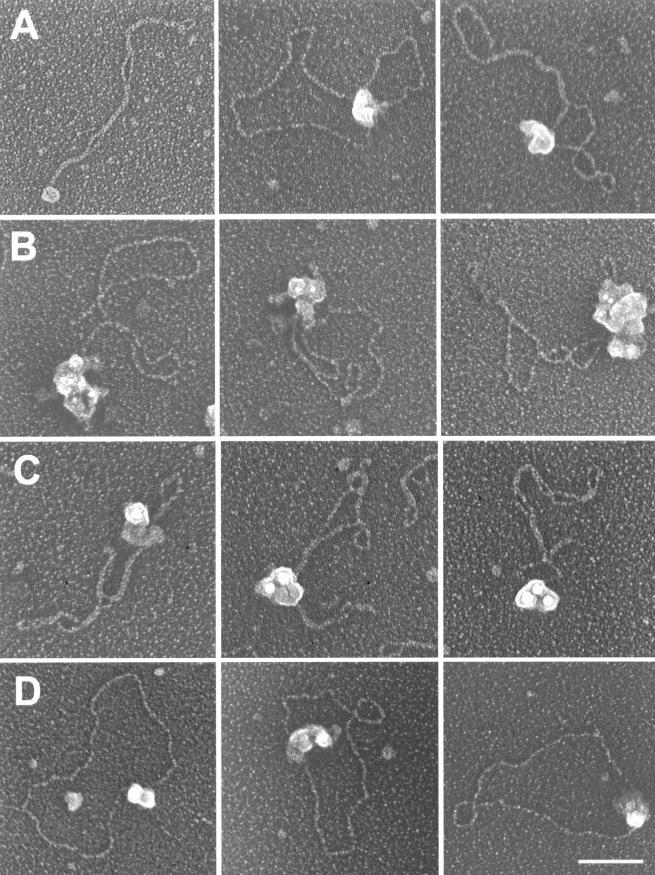
Electron micrographs of U-form DNA and protein-DNA complexes in unwinding reactions. Row A, circular and linear single strands recovered from the U-form DNA band from an unwinding reaction of p7874-99 under complete reaction conditions. The purified U-form DNA was coated with E. coli SSB and cross-linked with glutaraldehyde. Rows B to D, protein-DNA complexes present in the unwinding reactions with conditions modified for EM. Row B, DNA bound by an E1 dihexamer without discernible DNA unwinding. DNA bound by an E1 hexamer was more frequently observed (not shown). Row C, unwinding intermediates containing two rabbit ear-like, SSB-coated single-stranded DNA loops. Row D, completely unwound circular single-stranded DNA coated with SSB. Linear single strands were also observed (not shown). Images are shown in reverse contrast. Identical complexes were also visualized when E2, Hsp70, and Hsp40 were also present, but the relative frequencies of observation were affected (Table 1). Bar, 100 nm.
E1 mutation in the ATP binding site causes defects in ori binding and DNA unwinding.
To substantiate our interpretation that E1 was responsible for generating the single-stranded U-form DNA, we expressed and purified an epitope-tagged HPV-11 E1 mutation, P479S, from E. coli. Figure 3A shows a Coomassie blue-stained SDS-PAGE gel of the purified wild-type and mutated E1 proteins, along with a Western blot made with an antibody to the epitope tag. It can be seen that both proteins were purified to near homogeneity. This mutated HPV-11 E1 protein exhibited reduced ATPase activity (Fig. 3B), produced no U-form DNA in ethidium bromide gels (Fig. 3C), and had a very low activity in cell-free replication (Fig. 3D). These observations support the conclusion that the wild-type HPV E1 unwinds DNA.
To understand the molecular basis for these properties, we examined the ability of E1 P479S to bind ori DNA by EMSA (Fig. 3E). The wild-type HPV-11 E1 protein formed a faint smear of slowly migrating complexes with a radiolabeled ori fragment, and the presence of Hsp40 and Hsp70 greatly stimulated complex formation (Fig. 3E, lanes 1 to 5), as described previously (34). E1 P479S generated few or no detectable DNA-protein complexes in the absence or presence of either chaperone (Fig. 3E, lanes 6 to 9). EM confirmed that this mutated E1 protein did not form a discernible protein complex on ori DNA (data not shown).
E1 complexes function as a bidirectional helicase.
We next used EM to examine DNA-protein complexes present in unwinding reactions conducted under modified conditions. E. coli SSB was used because SSB-coated single strands were more extended than RPA-coated molecules (unpublished observations). Creatine phosphate kinase and bovine serum albumin were omitted to reduce the amount of proteins, facilitating the visualization of protein-DNA complexes, especially unwinding intermediates, as the efficiency of unwinding was greatly reduced (Fig. 1). The majority of the DNA was present as relatively relaxed circles generated by topoisomerase I (Fig. 1). Some were bound by a single hexamer and, less frequently, by a dihexamer without visible unwinding (Fig. 2B). No ori DNA was associated with more than one E1 hexamer or dihexamer. Completely unwound SSB-coated single strands were also observed (Fig. 2D).
Importantly, we observed complexes at the early stages of unwinding (Fig. 2C) that contained two rabbit ear-like, single-stranded DNA loops emanating from a large protein complex. The E1 dihexamer was no longer discernible due to the presence of additional proteins and the overlay of SSB-coated single-stranded DNA. Nevertheless, the presence of double loops is entirely consistent with an interpretation that the functional helicase is a dihexamer and that DNA was translocated inwardly while being unwound. Other partially unwound complexes were uninformative due to a nonoptimal orientation on the EM grids or coalesence of SSB-coated single strands. It is notable that in no case did we observe unwinding intermediates in which a bubble of two SSB-coated single-stranded arms was flanked by one or two protein complexes. These structures would have suggested one or two unidirectional helicases that operate at one or both forks of two expanding single-stranded arms on an otherwise double-stranded circular DNA.
E2 inhibits DNA unwinding by E1.
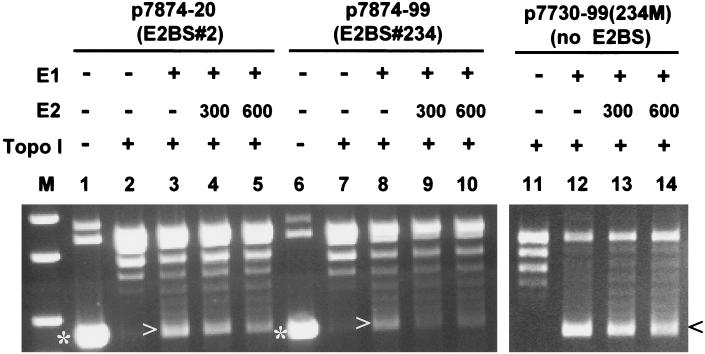
As we expected, addition of purified E2 protein, which was active in supporting HPV ori replication (Fig. 3D) and in binding to ori in EMSA (data not shown), reduced the amount of U-form DNA in a dose-dependent manner (Fig. 4). Inhibition was observed with an ori plasmid containing either one copy (p7874-20) or three copies (p7874-99) of the E2 BS (Fig. 4, left panel), in agreement with observations made with the BPV-1 system (36). Inhibition was also observed with p7730-99 (234M), in which all three E2 BSs are site mutated while maintaining a wild-type E1 BS (Fig. 4, right panel). This mutated ori no longer binds E2 in vitro, nor does it replicate in a transient or cell-free system (11, 27). Thus, protein-protein interactions may also contribute to the inhibition. EM confirmed that, in the presence of 300 ng of E2 protein, partially and completely unwound DNA molecules were reduced from 80% to 20% among the DNA-protein complexes scored (Table 1).
FIG. 4.
HPV-11 E2 inhibition of DNA unwinding by E1 is dose dependent but E2 BS independent. DNA substrates used were p7874-20, p7874-99, and p7730-99 (234M), which contain 1, 3, and 0 copies of the E2 BS, respectively. The presence (+) or absence (−) of HPV E1 and E2 and human topoisomerase I and the amounts of E2 protein (in nanograms) added are indicated. All other components were as described in Materials and Methods. Form I supercoiled DNA (*) and U-form DNA (> or <) are marked. Lane M, double-stranded size markers.
TABLE 1.
Modulation of E1 helicase by E2 and chaperones as visualized by EMa
| Protein(s) | No. of DNA-protein complexes
|
% Unwound (B + C / A + B + C) | ||
|---|---|---|---|---|
| No unwinding (A) | Fully unwound (B) | Partially unwound (C) | ||
| E1 | 34 | 15 | 120 | 80 |
| E1 + Hsp70 | 44 | 12 | 123 | 75 |
| E1 + Hdj2 | 49 | 13 | 109 | 71 |
| E1 + Hsp70 + Hdj2 | 46 | 10 | 113 | 73 |
| E1 + E2 | 120 | 3 | 28 | 21 |
| E1 + E2 + Hsp70 | 35 | 13 | 97 | 76 |
| E1 + E2 + Hdj2 | 44 | 11 | 112 | 74 |
| E1 + E2 + Hsp70 + Hdj2 | 49 | 22 | 98 | 71 |
All reactions contained 400 ng of p7874-99 ori DNA, 500 ng of HPV-11 E1, 180 ng of E. coli SSB, 25 ng of topoisomerase I, and 4 mM ATP but not creative phosphate kinase, which is a component of the ATP-regenerating system; 300 ng of E2, Hsp70, or Hdj2 was added, as specified. The reaction mixtures were fixed with 0.6% glutaraldehyde and chromatographed through 2-ml BioGel A5m (Bio-Rad) columns. Fractions containing protein-DNA complexes were examined by EM (see Materials and Methods). The majority of the DNA visualized was free double-stranded circles. Only protein-DNA complexes were scored as no unwinding, partially unwound, and completely unwound.
E2 protein does not affect E1 assembly on ori but remains associated with the E1-ori complex.
To investigate the mechanism by which the E1 helicase is inhibited by E2, protein-DNA complexes in binding reactions were examined by EM. In reactions containing both E1 and E2 proteins, the percentage of E1 dihexamer varied from 5 to 15% of the protein-ori complexes. The remaining complexes contained a single E1 hexamer (Fig. 5A, left panel). This distribution was similar to that in our previous report on E1-ori complexes formed in the absence of E2 (34). Furthermore, the frequency of observing protein-DNA complexes among all DNA molecules visualized was similar or only slightly reduced compared to that in the absence of E2 protein. However, because of the small size of the E2 protein (43 kDa), which binds to the E2 BS as a dimer, we were not able to ascertain the presence of bound E2.
FIG. 5.
Immunogold labeling of proteins associated with E1-ori. The binding reactions were conducted with E1 protein and ori DNA in the presence of E2, Hsp70, or Hdj2. Row A, left panel, E1 hexamer-ori complex assembled in the presence of E2 protein; middle and right panels, complexes assembled in the presence of E1 and E2 and then probed with primary antibodies (Ab) against E2 and secondary immunogold conjugates. The large complexes shown were coated with antibodies but not with gold particles. Row B, complexes assembled in the presence of E1 and E2 and then probed with primary antibodies against E2 and secondary immunogold conjugates. The complexes carried 5-nm gold-secondary antibody conjugates. Rows C and D, E1-ori complexes assembled in the presence of Hdj2 and Hsp70, respectively, were treated with primary antibodies against the respective chaperone and 10-nm gold-secondary antibody conjugates. Examples shown carried gold particles. Images are shown in reverse contrast. Bar, 100 nm.
To determine whether E2 remains associated with the E1-ori complex to account for its inhibitory effect on the E1 helicase, we performed immunogold EM with a rabbit polyclonal antibody against E2 and a secondary antibody reactive with the rabbit antibody. This secondary antibody had been electrostatically conjugated to 5-nm gold particles. In the absence of E2 protein or the primary antibody, immunogold particles were attached to about 2% or less of the E1-ori complexes. In contrast, among 126 protein-ori DNA complexes scored in a binding reaction containing E1, E2, the ori plasmid, the primary antibody, and the secondary immunogold conjugates, 19% of the protein-ori complexes were associated with immunogold particles (Table 2, Fig. 5B). Most of the complexes without attached immunogold particles were larger than those observed in the absence of any antibody, suggesting that these complexes were bound by antibodies lacking gold particles (compare middle and right panels of Fig. 5A to left panel). Thus, the percentage of immunogold labeling likely represents an underestimation as a result of dissociation of some of the gold conjugates during preparation for EM. Furthermore, the gold particles significantly retard the rate of association between the conjugates and the target complexes relative to secondary antibodies that had lost the gold particles, further reducing the efficiency of immunogold decoration.
TABLE 2.
Hsp70 and Hdj2 displace E2 from E1-orl complexes as visualized by immunogold EMa
| Protein(s) | % of E1-ori complexes carrying immunogold with antibody to:
|
||
|---|---|---|---|
| E2 | Hsp70 | Hdj2 | |
| E1 + E2 | 19 (126) | — | — |
| E1 + E2 + Hsp70 | 5 (240) | 17 (105) | — |
| E1 + E2 + Hdj2 | 7 (102) | — | 34 (107) |
| E1 + Hsp70 | — | 18 (124) | — |
| E1 + Hdj2 | — | — | 36 (135) |
The proteins were incubated with ori DNA in several parallel experiments. Primary antibody to E2, Hsp70, or Hdj2 and appropriate secondary immunogold conjugates (as indicated above the columns) were then incubated and processed for EM as described in Materials and Methods and in Results. From 100 to 200 protein-DNA complexes (numbers shown in parentheses) were scored in each reaction, and free DNA, which constituted the majority of the DNA, was not counted. In the absence of E2, chaperone proteins, or the primary antibodies, about 1 to 4% of the protein-DNA complexes carried gold particles. In the absence of any antibody, only in the reaction containing E1 and Hsp40 was the E1 dihexamer-ori complex observed at high frequency, in agreement with our previous observation (34), whereas primarily E1 hexamer-DNA was observed in all other reactions. —, not done.
Because the protein-DNA complexes reacted with antibodies were rather large, we were not able to discern simultaneously E1 hexamers or dihexamers in the complexes. Even though we could not rule out the possibility that some of these complexes contained only E2, others must also contain E1 to account for the inhibitory effect of the E2 protein on the E1 helicase (Fig. 4 and Table 1). We suggest that, at the concentrations used, E2 remains associated with the E1-ori complexes without affecting E1 assembly on the ori.
Chaperone proteins abolish E2 inhibition of E1 helicase.
To determine whether chaperones might play a role in activating the E1 helicase inhibited by the E2 protein, we added 300 ng of Hsp70 or Hdj2 to the p7874-99 ori plasmid unwinding reactions in the presence of 300 ng of the E2 protein. The conditions used were those modified for EM, as described previously. EM examination showed that, among the DNA-protein complexes scored, the inhibitory effect of the E2 protein on DNA unwinding was largely alleviated by either or both chaperone proteins (Table 1). However, the overall unwinding remained very inefficient, as the majority of the DNA visualized was still free DNA relaxed by topoisomerase I. This is because the reaction was conducted in the absence of creatine phosphate kinase, where the ATP supply is limiting. Furthermore, the assembly of E1 on the ori DNA requires ATP (Fig. 3), which was also consumed by Hsp70.
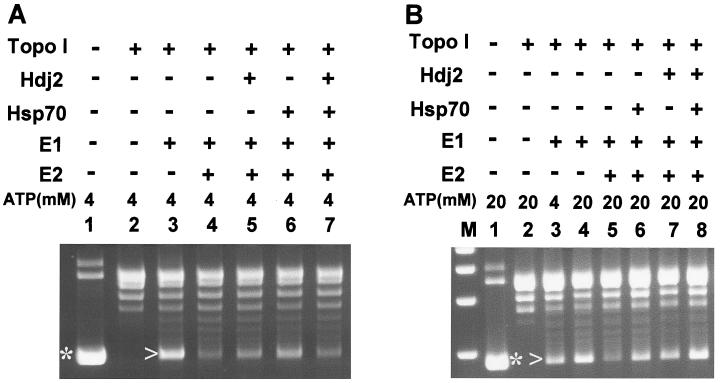
To obtain more quantitative results on the effects of chaperones on p7874-99 unwinding by E1 in the presence of E2, we conducted unwinding reactions in the presence of all the necessary components and analyzed the products by chloroquine-agarose gel electrophoresis. Addition of 200 ng of Hsp70 or Hdj2 to the unwinding reaction, which was partially inhibited by 300 ng of HPV-11 E2, indeed partially restored the unwinding activity, based on the amounts of U-form DNA observed. However, when both chaperones were present, the increase in U-form DNA was marginal relative to that achieved in the presence of either chaperone alone (Fig. 6A, compare lanes 3 through 7), as also observed by EM (Table 1). We attributed this result to a highly stimulated ATPase of Hsp70 when Hdj2 was also present (28). A depletion of ATP would then impede E1 assembly on the ori DNA (33) and the E1 helicase activity (Fig. 1).
FIG. 6.
Effects of chaperone proteins on HPV E1 helicase inhibited by HPV E2. (A) Unwinding reactions were conducted in the presence (+) or in the absence (−) of 200 ng of E2, Hsp70, or Hdj2. All reactions contained 4 mM ATP. (B) Unwinding reactions were conducted in the presence (+) or in the absence (−) of 200 ng of E2 and 50 ng of Hsp70, Hdj2, or both. The concentration of ATP was 4 or 20 mM, as indicated. Lane M, size markers.
To substantiate this interpretation, we modified the reaction conditions. As shown in Fig. 6B, increasing the ATP concentration to 20 mM in the otherwise standard reaction conditions significantly increased the amount of U-form DNA (Fig. 6B, compare lane 4 to 3). Although we did not test additional ATP concentrations, this experiment clearly showed that the 4 mM ATP in the standard reaction was limiting for the amount of E1 protein used. The inhibitory effect of 200 ng of E2 protein persisted under this condition (Fig. 6B, lane 5). In the presence of 20 mM ATP, the addition of only 50 ng of Hsp70 or Hdj2 was sufficient to abolish much of the E2 inhibition (Fig. 6B, lanes 6 and 7). When both chaperones were present, the amount of U-form DNA exceeded that observed in the absence of E2 protein (Fig. 6B, compare lane 8 to 4). Thus, chaperone proteins indeed abolish E2 inhibition as long as ATP is not limiting. In vivo, the supply of ATP will be continuously replenished, and ATP should not be a limiting factor as it is in vitro.
Chaperone proteins displace E2 from the E1-ori complex.
To investigate the mechanism by which chaperones reactivate the E1 helicase in the presence of the E2 protein, we proceeded to examine the E1-ori (p7874-99) complexes formed in a series of binding reactions in the presence of either chaperone alone or when E2 was also added with immunogold by EM (see Materials and Methods). The data are summarized in Table 2. In the presence of chaperones but in the absence of primary antibodies to either chaperone, only 2 to 4% of the protein-ori complexes carried immunogold conjugates targeted to the chaperones. In another control, in the absence of chaperones but in the presence of primary antibodies to chaperones, only 1% of the protein-ori complexes carried immunogold particles. These control experiments demonstrate a lack of nonspecific association of the primary and second antibodies with the protein-DNA complexes, in agreement with our previous report (34). In contrast, in binding reactions containing chaperones and primary antibodies, 18% and 36%, respectively, of the protein-ori complexes carried immunogold conjugates (Table 2), indicating that both chaperones can independently associate with the E1-protein complexes. Examples are presented in Fig. 5C and D.
Since the immunogold conjugates obscured the E1-ori complexes, we analyzed the nature of protein-ori complexes after incubation with either chaperone in the absence of any antibodies. Only Hdj2 induced efficient formation of dihexameric E1 complexes on over 90% of the E1-ori complexes observed, in agreement with our previous data (34). Because Hdj2 coimmunoprecipitates with E1 and also remains associated with E1-ori in EMSA (34), the high efficiency of dihexameric E1-ori complexes would suggest that most of these complexes also contain the Hdj2 protein. Thus, these data support our interpretation that immunogold EM indeed underestimated the presence of the target protein in the protein-ori complexes.
We then examined the effect of chaperones on the association of the E2 protein with the E1-ori complexes and, conversely, the effect of the E2 protein on the association of either chaperone with the E1-ori complexes. We incubated E1, the p7874-99 ori plasmid, E2, and either chaperone in parallel experiments, which we then probed with primary antibodies to E2, Hsp70, or Hdj2 along with the appropriate secondary immunogold conjugate. Interestingly, the frequencies of chaperone association with E1-ori complexes were not affected in the presence of the E2 protein (Table 2). In contrast, the frequency of E2 association with E1-ori complexes was significantly reduced to near background levels in the presence of either chaperone (Table 2). Collectively, these data suggest that chaperone proteins displace E2 protein from the E1-ori complex and that this E2 displacement is the most likely mechanism for E1 helicase reactivation. The alternative possibility, that chaperones shields E2 from primary antibody, cannot explain the reactivation of the DNA unwinding (Fig. 6, Table 1).
Finally, we examined the protein-ori complexes in an unwinding reaction containing both E2 and chaperones by immunogold EM. Three parallel experiments were conducted under the reaction conditions modified for EM. To each we added one of the primary antibodies, followed by the appropriate secondary immunogold conjugates. Only 2% (n = 265), 4% (n = 361), or 2% (n = 357) of the unwinding intermediates carried E2, Hsp70, and Hdj2 immunogold conjugates, respectively, within the range of background labeling, when the protein or the primary antibody was omitted. We conclude that neither E2 nor chaperone remains associated with E1-ori complexes in active unwinding complexes.
DISCUSSION
In this study, we demonstrate for the first time that HPV E1 possesses a highly efficient DNA-unwinding activity on supercoiled substrates, generating U-form DNA (Fig. 1), which contains completely unwound single-stranded molecules (Fig. 2). EM conducted under modified conditions with reduced efficiency of unwinding revealed unwinding intermediates with two single-stranded loops extending from a large protein complex on the DNA substrate (Fig. 2). This structure suggests that the HPV E1 dihexamer functions as an integral helicase. We propose that each E1 hexameric subunit is an active helicase and that DNA on both sides of ori is unwound while being translocated in an inward direction.
A bidirectional helicase is required for bidirectional HPV replication (4). Interestingly, unlike the SV40 T antigen, which has strong helicase activities in unwinding circular or linear DNA and in strand displacement assays, DNA unwinding by HPV E1 is dependent on supercoiled DNA, as no U-form DNA was generated from either a prerelaxed circular DNA or a linear DNA substrate detectable by ethidium bromide staining (Fig. 1 and 2). Thus, energy stored in supercoils helps the E1 dihexamer untwist duplex DNA to initiate unwinding. This observation would explain the poor helicase activities exhibited by HPV E1 proteins in strand displacement assays. We also showed that the E2 protein partially inhibited this E1 activity, but inhibition was abolished by Hsp70 and Hdj2 (Fig. 4 and 6 and Table 1). A systematic EM examination of the binding and unwinding reactions provided strong evidence that chaperone proteins displace E2 from the E1-ori complexes and are themselves released during unwinding (Fig. 5, Table 2).
In work by Fouts et al. (20), a bilobed BPV-1 E1 bound to ori was observed, but the bidirectional unwinding intermediates contained two single-stranded arms rather than two loops. Both types of unwinding intermediates have been reported for the SV40 T antigen (15, 17, 42, 52, 61, 64). We believe that single-stranded arms were converted from single-stranded loops when the dihexamer dissociated during subsequent manipulations in those studies. The origin-binding protein of herpes simplex virus type 1, UL9, also forms binary complexes consisting of two dimers and induces bidirectional DNA unwinding from oriS (39). This mode of DNA unwinding by a replicative helicase implies that bidirectional DNA replication would occur on two expanding central loops. DNA translocation through centrally located replication machinery is far more favorable in terms of energy consumption and speed relative to sliding the large replication machinery along the DNA strands. The commonly depicted Cairn's or θ form replication intermediates with divergently expanding arms on which bidirectional replication takes place would then be the result of deproteination of the replication complexes.
The mutated HPV-11 E1 P479S in the putative ATP binding site shows dramatically reduced ATPase activity (Fig. 3) (65). It exhibits no discernible DNA-unwinding activity and supports cell-free HPV ori replication very poorly, in agreement with an analogous BPV E1 mutation (38, 55). Our EMSA and EM assays further show that the defect stems from its poor ability to bind ori DNA (Fig. 3 and data not shown). Thus, binding of ATP may have induced a conformational change in E1, allowing it to bind and assemble as a hexamer or dihexamer on the ori. This interpretation is in agreement with our previous observation that HPV-11 E1 binds to ori only in the presence of ATP, although ATP hydrolysis is not necessary (33, 34), a property shared with SV40 T antigen (22, 42). In contrast, the BPV E1 protein binds to BPV-1 ori in the absence of Mg2+ and ATP, although ATP greatly stimulates binding (47, 50). The HPV-11 E1 P479S was previously reported to exhibit reduced ATPase activity (65) but nearly wild-type activity in transient replication (58). The discrepancy may have arisen from the use of PCR amplification of a DNA fragment to demonstrate replication in that study rather than Southern blot hybridization to reveal full-length, newly replicated DNA (11, 59) or cell-free replication, as in this study.
Replicative helicases are substrates of cyclin-dependent kinases or other kinases that regulate cell cycle progression and initiation of DNA replication. For instance, the MCM proteins essential for initiating cellular DNA replication are phosphorylated (29, 40). Thus, the issues of whether and how phosphorylation may affect helicase activity have been of considerable interest. SV40 T antigen purified from E. coli does not function properly for lack of phosphorylation (45). In particular, phosphorylation by cdk at T124 of T antigen purified from insect cells is critical, and a T124A mutation was shown to cripple the protein for dihexamer formation and ori unwinding (5, 43, 63). Both the BPV-1 and HPV-11 E1 proteins are also substrates of cyclin E/cdk2 and other cdk complexes in vitro and in vivo (14, 37). In particular, the cyclin E-cdk2 complex is critical for efficient HPV-11 ori replication in vitro (32, 37). Our results do not rule out the possibility that E1 phosphorylation by cdk may modulate its DNA-unwinding activity. However, they clearly demonstrate that phosphorylation is not essential for dihexamer formation or for efficient DNA unwinding. E1 phosphorylation must then affect its interactions with other proteins. This issue remains to be investigated.
The papillomavirus E2 protein targets E1 to the viral ori. Our EM data suggest that, at the concentrations used, HPV E2 does not promote E1 dihexamer formation, nor does it disrupt E1 hexamer or dihexamer formation on ori. Rather, E2 remains bound to the E1-ori complexes (Fig. 5) and inhibits the HPV E1 helicase (Fig. 4). Since inhibition was observed with plasmids containing 0, 1, or 3 copies of the E2 BS, we further suggest that unwinding of DNA on both sides of the ori might be a concerted reaction or that interactions between E2 and E1 or cellular proteins also adversely affect the E1 helicase. Indeed, E1 and E2 interactions have been previously demonstrated for HPV-11 and BPV-1 proteins in vitro (13, 35, 50, 68), consistent with their function in recruiting E1 to the ori. In contrast to our observations, inhibition of the BPV-1 E1 helicase by BPV-1 E2 has been attributed to interference with E1 oligomer assembly on the ori (36). Also, unlike our results, in the presence of ATP and Mg2+, BPV-1 E2 dissociates from the E1-ori complex (47). The reasons for the distinctions between the HPV and BPV proteins are not understood.
Hdj2 associates with purified HPV-11 E1 expressed in insect cells and promotes dihexamer formation on ori (34). These observations were reproduced in the present study with bacterially expressed E1 protein and were further substantiated by immunogold EM. We have now demonstrated by immunogold EM that Hsp70 also associates with the E1-ori complex. Our EM data further suggest that chaperones can displace E2 and reactivate an E2-inhibited E1 helicase and that chaperones are then released during DNA unwinding (Fig. 6, Tables 1 and 2). It is intriguing that, although chaperones stimulate E1 hexamer and dihexamer formation on the ori, they are not required for E1 helicase activity unless E2 is also present (Table 1, Fig. 6). Nevertheless, the structure of the unwinding intermediates clearly suggests that DNA unwinding is associated with E1 dihexamers (Fig. 2). Either the E1 helicase activity originates from the low percentage of dihexamers that formed in the absence of the chaperones, or there is another mechanism by which dihexameric E1 assembles on the ori. Possibilities include an interaction of E1 with topoisomerase or RPA and the recruitment of a second hexamer upon initial limited DNA opening by the first hexamer. We further propose that, when E1 protein concentration is low, as it is in vivo, chaperone proteins may indeed assist in the assembly of dihexamers on ori.
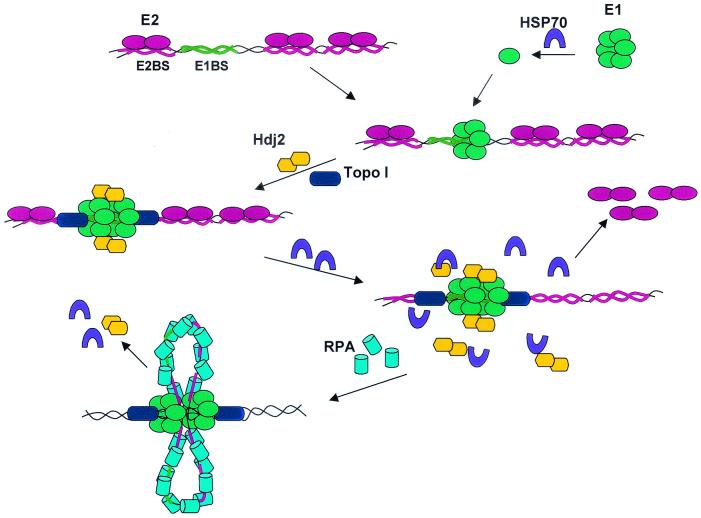
Based on previous and present reports, we propose a working model for the role of chaperone proteins in the assembly and activation of the E1 helicase on the HPV ori (Fig. 7). The E2 protein targets E1 to the ori by virtue of its highly specific association with E2 BSs and its interaction with E1. Hsp70 may facilitate E1 binding to the ori by dissociating previously formed oligomers into monomers, which can then reassemble on the ori as hexamers. Hsp40 promotes E1 dihexamer formation on the ori. The E1 dihexamer then unwinds DNA bidirectionally by translocating DNA on both sides of the ori in an inward direction, generating two centrally located, single-stranded loops. The loops are stabilized by RPA, while topoisomerase I removes the resultant positive supercoiling. However, the high-affinity association between E2 and E2 BSs may prevent the translocation and unwinding of DNA sequences beyond the E2 BSs that flank the E1 dihexamer. An interaction between E2 and E1 or possibly a cellular protein(s) could also inhibit E1 activity (not illustrated for simplicity).
FIG. 7.
Proposed model to illustrate the functions of E2 and chaperone proteins in the assembly and activation of the HPV E1 helicase on the origin. The E1 BS is flanked by E2 BSs. For simplicity, not illustrated is the E1/E2 interaction, which helps recruit E1 to the ori and may also contribute to inhibition of E1 helicase. The binding of E1 to ori, the activity of Hsp70, and DNA unwinding by E1 dihexamer all require ATP. The two single-stranded loops are shown to emerge from the center of the dihexamer. Not shown is the possibility that the two E1 hexamers each encircle the opposing single strands (as depicted in reference 20). Hsp70 and Hdj2 can independently displace E2 from the E2 BS. The stoichiometry of Hjd2 and E1 during the assembly of E1 dihexamer is not known. Upon completion of the unwinding in vitro or coupled DNA replication in vivo, chaperone proteins may promote E1 dihexamer disassembly from the substrate or template, and the process is then repeated on another ori DNA molecule.
In either case, DNA unwinding beyond the ori region occurs only when E2 is released from the complex. Our data show that the release can be accomplished by chaperone proteins, although other mechanisms cannot be excluded. Chaperones may also promote recycling of E1 onto new substrates by dissociating E1 dihexamers into monomers after each round of unwinding and replication (33). These activities of chaperones show similarities but also differences with the known functions of E. coli DnaJ and DnaK during replication of bacteriophages λ , P1, and P7 (2, 3, 18, 66, 72).
To our knowledge, our study is one of a few examples that demonstrate how heat shock proteins can play a role in chaperoning the assembly and activation of the unwinding activity of a replicative helicase in higher eukaryotes. We are not certain which members of the family of chaperones and cochaperones preferentially interact with E1 and E2 proteins in vivo. The mechanisms by which chaperones promote the assembly or disassembly of protein complexes are not understood at present. Finally, because E1 protein is the only enzyme encoded by papillomaviruses, an in-depth understanding of its function as well as a robust helicase assay are prerequisite to the development of efficient antiviral compounds for this large family of medically important human pathogens.
Acknowledgments
This research was supported by USPHS grants CA CA83679 to L.T.C. and T.R.B. and GM 31819 to J.D.G.
We thank Jim Champoux, Mike McDonnell, Richard Morimoto, T. Mohanakumar, and Douglas Cyr for sharing expression vectors for various cellular proteins.
The first two authors contributed equally to this work.
REFERENCES
- 1.Abravaya, K., M. P. Myers, S. P. Murphy, and R. I. Morimoto. 1992. The human heat shock protein hsp70 interacts with HSF, the transcription factor that regulates heat shock gene expression. Genes Dev. 6:1153-1164. [DOI] [PubMed] [Google Scholar]
- 2.Alfano, C., and R. McMacken. 1989. Heat shock protein-mediated disassembly of nucleoprotein structures is required for the initiation of bacteriophage lambda DNA replication. J. Biol. Chem. 264:10709-10718. [PubMed] [Google Scholar]
- 3.Alfano, C., and R. McMacken. 1989. Ordered assembly of nucleoprotein structures at the bacteriophage lambda replication origin during the initiation of DNA replication. J. Biol. Chem. 264:10699-10708. [PubMed] [Google Scholar]
- 4.Auborn, K. J., R. D. Little, T. H. Platt, M. A. Vaccariello, and C. L. Schildkraut. 1994. Replicative intermediates of human papillomavirus type 11 in laryngeal papillomas: site of replication initiation and direction of replication. Proc. Natl. Acad. Sci. USA 91:7340-7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbaro, B. A., K. R. Sreekumar, D. R. Winters, A. E. Prack, and P. A. Bullock. 2000. Phosphorylation of simian virus 40 T antigen on Thr 124 selectively promotes double-hexamer formation on subfragments of the viral core origin. J. Virol. 74:8601-8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonne-Andrea, C., S. Santucci, P. Clertant, and F. Tillier. 1995. Bovine papillomavirus E1 protein binds specifically DNA polymerase alpha but not replication protein A. J. Virol. 69:2341-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, K. S., K. P. Mullane, I. A. Aksoy, H. Stubdal, J. Zalvide, J. M. Pipas, P. A. Silver, T. M. Roberts, B. S. Schaffhausen, and J. A. DeCaprio. 1997. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes Dev. 11:1098-1110. [DOI] [PubMed] [Google Scholar]
- 8.Chase, J. W., R. F. Whittier, J. Auerbach, A. Sancar, and W. D. Rupp. 1980. Amplification of single-strand DNA binding protein in Escherichia coli. Nucleic Acids Res. 8:3215-3227. [DOI] [PMC free article] [PubMed]
- 9.Chellaiah, A., A. Davis, and T. Mohanakumar. 1993. Cloning of a unique human homologue of the Escherichia coli DnaJ heat shock protein. Biochim. Biophys. Acta 1174:111-113. [DOI] [PubMed] [Google Scholar]
- 10.Chiang, C. M., G. Dong, T. R. Broker, and L. T. Chow. 1992. Control of human papillomavirus type 11 origin of replication by the E2 family of transcription regulatory proteins. J. Virol. 66:5224-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiang, C. M., M. Ustav, A. Stenlund, T. F. Ho, T. R. Broker, and L. T. Chow. 1992. Viral E1 and E2 proteins support replication of homologous and heterologous papillomaviral origins. Proc. Natl. Acad. Sci. USA 89:5799-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conger, K. L., J. S. Liu, S. R. Kuo, L. T. Chow, and T. S. Wang. 1999. Human papillomavirus DNA replication. Interactions between the viral E1 protein and two subunits of human DNA polymerase alpha/primase. J. Biol. Chem. 274:2696-2705. [DOI] [PubMed] [Google Scholar]
- 13.Cooper, C. S., S. N. Upmeyer, and P. L. Winokur. 1998. Identification of single amino acids in the human papillomavirus 11 E2 protein critical for the transactivation or replication functions. Virology 241:312-322. [DOI] [PubMed] [Google Scholar]
- 14.Cueille, N., R. Nougarede, F. Mechali, M. Philippe, and C. Bonne-Andrea. 1998. Functional interaction between the bovine papillomavirus virus type 1 replicative helicase E1 and cyclin E-Cdk2. J. Virol. 72:7255-7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dean, F. B., J. A. Borowiec, T. Eki, and J. Hurwitz. 1992. The simian virus 40 T antigen double hexamer assembles around the DNA at the replication origin. J. Biol. Chem. 267:14129-14137. [PubMed] [Google Scholar]
- 16.Dean, F. B., P. Bullock, Y. Murakami, C. R. Wobbe, L. Weissbach, and J. Hurwitz. 1987. Simian virus 40 (SV40) DNA replication: SV40 large T antigen unwinds DNA containing the SV40 origin of replication. Proc. Natl. Acad. Sci. USA 84:16-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dodson, M., F. B. Dean, P. Bullock, H. Echols, and J. Hurwitz. 1987. Unwinding of duplex DNA from the SV40 origin of replication by T antigen. Science 238:964-967. [DOI] [PubMed] [Google Scholar]
- 18.Dodson, M., H. Echols, S. Wickner, C. Alfano, K. Mensa-Wilmot, B. Gomes, J. LeBowitz, J. D. Roberts, and R. McMacken. 1986. Specialized nucleoprotein structures at the origin of replication of bacteriophage lambda: localized unwinding of duplex DNA by a six-protein reaction. Proc. Natl. Acad. Sci. USA 83:7638-7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eom, C.-Y., and I. R. Lehman. 2002. The human DnaJ protein, hTid-1, enhances binding of a multimer of the herpes simplex virus type 1 UL9 protein to oris, an origin of viral DNA replication. Proc. Natl. Acad. Sci. USA 99:1894-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fouts, E. T., X. Yu, E. H. Egelman, and M. R. Botchan. 1999. Biochemical and electron microscopic image analysis of the hexameric E1 helicase. J. Biol. Chem. 274:4447-4458. [DOI] [PubMed] [Google Scholar]
- 21.Frydman, J. 2001. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem. 70:603-647. [DOI] [PubMed] [Google Scholar]
- 22.Gai, D., R. Roy, C. Wu, and D. T. Simmons. 2000. Topoisomerase I associates specifically with simian virus 40 large-T-antigen double hexamer-origin complexes. J. Virol. 74:5224-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giraldo, R., and R. Diaz-Orejas. 2001. Similarities between the DNA replication initiators of Gram-negative bacteria plasmids (RepA) and eukaryotes (Orc4p)/archaea (Cdc6p). Proc. Natl. Acad. Sci. USA 98:4938-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffith, J. D., and G. Christiansen. 1978. Electron microscope visualization of chromatin and other DNA-protein complexes. Annu. Rev. Biophys. Bioeng. 7:19-35. [DOI] [PubMed] [Google Scholar]
- 25.Han, Y., Y. M. Loo, K. T. Militello, and T. Melendy. 1999. Interactions of the papovavirus DNA replication initiator proteins, bovine papillomavirus type 1 E1 and simian virus 40 large T antigen, with human replication protein A. J. Virol. 73:4899-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenkins, O., D. Earnshaw, G. Sarginson, A. Del Vecchio, J. Tsai, H. Kallender, B. Amegadzie, and M. Browne. 1996. Characterization of the helicase and ATPase activity of human papillomavirus type 6b E1 protein. J. Gen. Virol. 77:1805-1809. [DOI] [PubMed] [Google Scholar]
- 27.Kuo, S. R., J. S. Liu, T. R. Broker, and L. T. Chow. 1994. Cell-free replication of the human papillomavirus DNA with homologous viral E1 and E2 proteins and human cell extracts. J. Biol. Chem. 269:24058-24065. [PubMed] [Google Scholar]
- 28.Laufen, T., M. P. Mayer, C. Beisel, D. Klostermeier, A. Mogk, J. Reinstein, and B. Bukau. 1999. Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc. Natl. Acad. Sci. USA 96:5452-5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lei, M., and B. K. Tye. 2001. Initiating DNA synthesis: from recruiting to activating the MCM complex. J. Cell Sci. 114:1447-1454. [DOI] [PubMed] [Google Scholar]
- 30.Li, R., and M. Botchan. 1994. Acidic transcription factors alleviate nucleosome-mediated repression of DNA replication of bovine papillomavirus type 1. Proc. Natl. Acad. Sci. USA 91:7051-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liberek, K., J. Osipiuk, M. Zylicz, D. Ang, J. Skorko, and C. Georgopoulos. 1990. Physical interactions between bacteriophage and Escherichia coli proteins required for initiation of lambda DNA replication. J. Biol. Chem. 265:3022-3029. [PubMed] [Google Scholar]
- 32.Lin, B. Y., T. Ma, J. S. Liu, S. R. Kuo, G. Jin, T. R. Broker, J. W. Harper, and L. T. Chow. 2000. HeLa cells are phenotypically limiting in cyclin E/CDK2 for efficient human papillomavirus DNA replication. J. Biol. Chem. 275:6167-6174. [DOI] [PubMed] [Google Scholar]
- 33.Liu, J. S., S. R. Kuo, T. R. Broker, and L. T. Chow. 1995. The functions of human papillomavirus type 11 E1, E2, and E2C proteins in cell-free DNA replication. J. Biol. Chem. 270:27283-27291. [DOI] [PubMed] [Google Scholar]
- 34.Liu, J. S., S. R. Kuo, A. M. Makhov, D. M. Cyr, J. D. Griffith, T. R. Broker, and L. T. Chow. 1998. Human Hsp70 and Hsp40 chaperone proteins facilitate human papillomavirus-11 E1 protein binding to the origin and stimulate cell-free DNA replication. J. Biol. Chem. 273:30704-30712. [DOI] [PubMed] [Google Scholar]
- 35.Lu, J., Y. Sun, R. Rose, W. Bonnez, and D. McCance. 1993. Two E2 binding sites (the E2 BS) alone or one E2 BS plus an A/T-rich region are minimal requirements for the replication of the human papillomavirus type 11 origin. J. Virol. 67:7131-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lusky, M., J. Hurwitz, and Y. S. Seo. 1994. The bovine papillomavirus E2 protein modulates the assembly of but is not stably maintained in a replication-competent multimeric E1-replication origin complex. Proc. Natl. Acad. Sci. USA 91:8895-8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma, T., N. Zou, B. Y. Lin, L. T. Chow, and J. W. Harper. 1999. Interaction between cyclin-dependent kinases and human papillomavirus replication-initiation protein E1 is required for efficient viral replication. Proc. Natl. Acad. Sci. USA 96:382-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacPherson, P., L. Thorner, L. M. Parker, and M. Botchan. 1994. The bovine papilloma virus E1 protein has ATPase activity essential to viral DNA replication and efficient transformation in cells. Virology 204:403-408. [DOI] [PubMed] [Google Scholar]
- 39.Makhov, A. M., P. E. Boehmer, I. R. Lehman, and J. D. Griffith. 1996. The herpes simplex virus type 1 origin-binding protein carries out origin specific DNA unwinding and forms stem-loop structures. EMBO J. 15:1742-1750. [PMC free article] [PubMed] [Google Scholar]
- 40.Masai, H., E. Matsui, Z. You, Y. Ishimi, K. Tamai, and K. Arai. 2000. Human Cdc7-related kinase complex. In vitro phosphorylation of MCM by concerted actions of Cdks and Cdc7 and that of a criticial threonine residue of Cdc7 by Cdks. J. Biol. Chem. 275:29042-29052. [DOI] [PubMed] [Google Scholar]
- 41.Masterson, P. J., M. A. Stanley, A. P. Lewis, and M. A. Romanos. 1998. A C-terminal helicase domain of the human papillomavirus E1 protein binds E2 and the DNA polymerase alpha-primase p68 subunit. J. Virol. 72:7407-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mastrangelo, I. A., P. V. Hough, J. S. Wall, M. Dodson, F. B. Dean, and J. Hurwitz. 1989. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature 338:658-662. [DOI] [PubMed] [Google Scholar]
- 43.Moarefi, I. F., D. Small, I. Gilbert, M. Hopfner, S. K. Randall, C. Schneider, A. A. Russo, U. Ramsperger, A. K. Arthur, and H. Stahl. 1993. Mutation of the cyclin-dependent kinase phosphorylation site in simian virus 40 (SV40) large T antigen specifically blocks SV40 origin DNA unwinding. J. Virol. 67:4992-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mohr, I. J., R. Clark, S. Sun, E. J. Androphy, P. MacPherson, and M. R. Botchan. 1990. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science 250:1694-1699. [DOI] [PubMed] [Google Scholar]
- 45.Mohr, I. J., Y. Gluzman, M. P. Fairman, M. Strauss, D. McVey, B. Stillman, and R. D. Gerard. 1989. Production of simian virus 40 large tumor antigen in bacteria: altered DNA-binding specificity and DNA-replication activity of underphosphorylated large tumor antigen. Proc. Natl. Acad. Sci. USA 86:6479-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Remm, M., R. Brain, and J. R. Jenkins. 1992. The E2 binding sites determine the efficiency of replication for the origin of human papillomavirus type 18. Nucleic Acids Res. 20:6015-6021. [DOI] [PMC free article] [PubMed]
- 47.Sanders, C. M., and A. Stenlund. 1998. Recruitment and loading of the E1 initiator protein: an ATP-dependent process catalysed by a transcription factor. EMBO J. 17:7044-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sedman, J., and A. Stenlund. 1998. The papillomavirus E1 protein forms a DNA-dependent hexameric complex with ATPase and DNA helicase activities. J. Virol. 72:6893-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seo, Y. S., S. H. Lee, and J. Hurwitz. 1991. Isolation of a DNA helicase from HeLa cells requiring the multisubunit human single-stranded DNA-binding protein for activity. J. Biol. Chem. 266:13161-13170. [PubMed] [Google Scholar]
- 50.Seo, Y. S., F. Müller, M. Lusky, E. Gibbs, H. Y. Kim, B. Phillips, and J. Hurwitz. 1993. Bovine papilloma virus (BPV)-encoded E2 protein enhances binding of E1 protein to the BPV replication origin. Proc. Natl. Acad. Sci. USA 90:2865-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seo, Y. S., F. Müller, M. Lusky, and J. Hurwitz. 1993. Bovine papilloma virus (BPV)-encoded E1 protein contains multiple activities required for BPV DNA replication. Proc. Natl. Acad. Sci. USA 90:702-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smelkova, N. V., and J. A. Borowiec. 1997. Dimerization of simian virus 40 T-antigen hexamers activates T-antigen DNA helicase activity. J. Virol. 71:8766-8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stewart, L., and J. J. Champoux. 2001. Assaying DNA topoisomerase I relaxation activity. Methods Mol. Biol. 95:1-11. [DOI] [PubMed] [Google Scholar]
- 54.Sullivan, C. S., and J. M. Pipas. 2001. The virus-chaperone connection. Virology 287:1-8. [DOI] [PubMed] [Google Scholar]
- 55.Sun, S., L. Thorner, M. Lentz, P. MacPherson, and M. Botchan. 1990. Identification of a 68-kilodalton nuclear ATP-binding phosphoprotein encoded by bovine papillomavirus type 1. J. Virol. 64:5093-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swindle, C. S., N. Zou, B. A. Van Tine, G. M. Shaw, J. A. Engler, and L. T. Chow. 1999. Human papillomavirus DNA replication compartments in a transient DNA replication system. J. Virol. 73:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanguy Le Gac, N., and P. E. Boehmer. 2002. Activation of the herpes simplex virus type-1 origin-binding protein (UL9) by heat shock proteins. J. Biol. Chem. 277:5660-5666. [DOI] [PubMed] [Google Scholar]
- 58.Titolo, S., A. Pelletier, F. Sauve, K. Brault, E. Wardrop, P. W. White, A. Amin, M. G. Cordingley, and J. Archambault. 1999. Role of the ATP-binding domain of the human papillomavirus type 11 E1 helicase in E2-dependent binding to the origin. J. Virol. 73:5282-5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ustav, M., and A. Stenlund. 1991. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 10:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walter, J., and J. Newport. 2000. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol. Cell 5:617-627. [DOI] [PubMed] [Google Scholar]
- 61.Wang, E. H., and C. Prives. 1991. ATP induces the assembly of polyoma large tumor antigen into hexamers. Virology 184:399-403. [DOI] [PubMed] [Google Scholar]
- 62.Weisshart, K., M. K. Bradley, B. M. Weiner, C. Schneider, I. Moarefi, E. Fanning, and A. K. Arthur. 1996. An N-terminal deletion mutant of simian virus 40 (SV40) large T antigen oligomerizes incorrectly on SV40 DNA but retains the ability to bind to DNA polymerase alpha and replicate SV40 DNA in vitro. J. Virol. 70:3509-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weisshart, K., P. Taneja, A. Jenne, U. Herbig, D. T. Simmons, and E. Fanning. 1999. Two regions of simian virus 40 T antigen determine cooperativity of double-hexamer assembly on the viral origin of DNA replication and promote hexamer interactions during bidirectional origin DNA unwinding. J. Virol. 73:2201-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wessel, R., J. Schweizer, and H. Stahl. 1992. Simian virus 40 T-antigen DNA helicase is a hexamer which forms a binary complex during bidirectional unwinding from the viral origin of DNA replication. J. Virol. 66:804-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.White, P. W., A. Pelletier, K. Brault, S. Titolo, E. Welchner, L. Thauvette, M. Fazekas, M. G. Cordingley, and J. Archambault. 2001. Characterization of recombinant HPV6 and 11 E1 helicases: effect of ATP on the interaction of E1 with E2 and mapping of a minimal helicase domain. J. Biol. Chem. 276:22426-22438. [DOI] [PubMed] [Google Scholar]
- 66.Wickner, S., J. Hoskins, and K. McKenney. 1991. Function of DnaJ and DnaK as chaperones in origin-specific DNA binding by RepA. Nature 350:165-167. [DOI] [PubMed] [Google Scholar]
- 67.Wold, M. S., J. J. Li, and T. J. Kelly. 1987. Initiation of simian virus 40 DNA replication in vitro: large-tumor-antigen- and origin-dependent unwinding of the template. Proc. Natl. Acad. Sci. USA 84:3643-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang, L., R. Li, I. J. Mohr, R. Clark, and M. R. Botchan. 1991. Activation of BPV-1 replication in vitro by the transcription factor E2. Nature 353:628-632. [DOI] [PubMed] [Google Scholar]
- 69.Yang, L., I. Mohr, E. Fouts, D. A. Lim, M. Nohaile, and M. Botchan. 1993. The E1 protein of bovine papilloma virus 1 is an ATP-dependent DNA helicase. Proc. Natl. Acad. Sci. USA 90:5086-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang, D., L. Frappier, E. Gibbs, J. Hurwitz, and M. O'Donnell. 1998. Human RPA (hSSB) interacts with EBNA1, the latent origin binding protein of Epstein-Barr virus. Nucleic Acids Res. 26:631-637. [DOI] [PMC free article] [PubMed]
- 71.Zou, N., B. Y. Lin, F. Duan, K. Y. Lee, G. Jin, R. Guan, G. Yao, E. J. Lefkowitz, T. R. Broker, and L. T. Chow. 2000. The hinge of the human papillomavirus type 11 E2 protein contains major determinants for nuclear localization and nuclear matrix association. J. Virol. 74:3761-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zylicz, M., D. Ang, K. Liberek, and C. Georgopoulos. 1989. Initiation of lambda DNA replication with purified host- and bacteriophage-encoded proteins: the role of the DnaK, DnaJ and GrpE heat shock proteins. EMBO J. 8:1601-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]