Abstract
Iron (Fe) is necessary for all living cells, but its bioavailability is often limited. Fe deficiency limits agriculture in many areas and affects over a billion human beings worldwide. In mammals, NRAMP2/DMT1/DCT1 was identified as a major pathway for Fe acquisition and recycling. In plants, AtNRAMP3 and AtNRAMP4 are induced under Fe deficiency. The similitude of AtNRAMP3 and AtNRAMP4 expression patterns and their common targeting to the vacuole, together with the lack of obvious phenotype in nramp3-1 and nramp4-1 single knockout mutants, suggested a functional redundancy. Indeed, the germination of nramp3 nramp4 double mutants is arrested under low Fe nutrition and fully rescued by high Fe supply. Mutant seeds have wild type Fe content, but fail to retrieve Fe from the vacuolar globoids. Our work thus identifies for the first time the vacuole as an essential compartment for Fe storage in seeds. Our data indicate that mobilization of vacuolar Fe stores by AtNRAMP3 and AtNRAMP4 is crucial to support Arabidopsis early development until efficient systems for Fe acquisition from the soil take over.
Keywords: Arabidopsis thaliana , germination, iron nutrition, NRAMP transporters, vacuole
Introduction
Iron (Fe) is essential for all living cells. This transition metal plays key roles in electron transfers in both photosynthetic and respiratory reactions in chloroplasts and mitochondria. In addition, a wealth of enzymatic activities require Fe as or within a cofactor. In mammals, most of the body Fe is associated with hemoglobin to transport oxygen through the blood system. Although, Fe is plentiful, representing 4.2% of the Earth crust, bioavailability of Fe is often very limited and Fe deficiency occurs frequently both in humans and in crop plants that feed them. World Health Organization reports indicate that over a billion human beings suffer from Fe deficiency, leading to lower resistance to infectious diseases and anemia (http://www.who.int/nut/publications.htm#ida). On 30% of the soils, low Fe availability limits plant productivity and may lead to chlorosis (Curie and Briat, 2003). Hence, the molecular mechanisms for Fe uptake and Fe homeostasis have been extensively studied in mammals and plants.
In plants, Fe mobilization and uptake from the soil occur through different mechanisms in grasses or dicots and nongraminaceous monocots (Curie and Briat, 2003). Graminaceous species secrete Fe chelators, called phytosiderophores, to mobilize Fe in the rhizosphere. Fe phytosiderophore complexes are then taken up by specific membrane transporters at the root surface. Mutations in Yellow Stripe 1 (YS1), the Fe–siderophore transporter, result in Fe deficiency symptoms in maize (Curie et al, 2001). In nongraminaceous species, a membrane-bound ferric chelate reductase mobilizes Fe from soil FeIII chelates by reducing FeIII to the more soluble FeII. Subsequently, FeII is taken up by metal transporters of the ZIP family (for Zinc-regulated transporter Fe-regulated transporter-like Proteins; Eide et al, 1996). In Arabidopsis thaliana, mutations in either FRO2, the root ferric chelate reductase, or IRT1, the main FeII uptake transporter, lead to Fe deficiency symptoms (Curie and Briat, 2003).
The search for Fe uptake systems in mammals led to the identification of DCT1/DMT1/NRAMP2. This transporter was identified by expression cloning in Xenopus oocytes on the basis of its ability to transport Fe (Gunshin et al, 1997) and by a genetic approach, because mutations in this transporter lead to severe anemia in both rat and mouse (Fleming et al, 1998). Interestingly, DCT1/DMT1/NRAMP2 plays multiple roles in mammalian Fe homeostasis: it is involved in Fe uptake into intestinal epithelial cells. It is also important for mobilization of Fe from endosomes after endocytosis of receptor-bound transferrin in erythrocyte precursors (Cellier and Gros, 2004). DCT1/DMT1/NRAMP2 belongs to the NRAMP family of metal transporters (Cellier and Gros, 2004). The first member of the family was named Natural Resistance Associated Macrophage Protein 1 (NRAMP1), because mutations in this macrophage-specific protein confer increased sensitivity to intracellular bacterial pathogens (Vidal et al, 1993). The metal transport function of NRAMP proteins was assigned later when Supek et al (1996), characterized SMF1, a yeast NRAMP homologue, as an Mn uptake system. NRAMP proteins are now recognized as a ubiquitous family of metal transporters with homologs in fungi, animals, plants and bacteria (Cellier and Gros, 2004).
We study the function of NRAMP homologs from plants. Previous work established that several NRAMP transporters from A. thaliana, rice, tomato and Glycine max encode broad-specificity metal transporters (Curie et al, 2000; Thomine et al, 2000; Bereczky et al, 2003; Kaiser et al, 2003). Expression studies have revealed that several plant NRAMP genes are upregulated upon Fe deficiency, suggesting a role in Fe homeostasis (Curie et al, 2000; Thomine et al, 2000; Bereczky et al, 2003). The subcellular localization of AtNRAMP3 on the vacuolar membrane suggested a function in intracellular metal homeostasis (Thomine et al, 2003). However, the analysis of AtNRAMP knockouts or AtNRAMP over expressing plants has so far not revealed strong phenotypes: AtNRAMP3 disruption as well as AtNRAMP3 and AtNRAMP4 over expression in A. thaliana mildly alter cadmium sensitivity without modifying the total cadmium content (Thomine et al, 2000; Lanquar et al, 2004). Overexpression of AtNRAMP1 increases resistance to toxic Fe concentrations (Curie et al, 2000). Disruption of AtNRAMP3 increases metal accumulation in response to Fe starvation, whereas overexpression of AtNRAMP3 downregulates Fe uptake mechanisms and decreases metal accumulation under Fe deficiency, without any growth phenotype under various Fe supply conditions (Thomine et al, 2003).
In the present work, the characterization of AtNRAMP4, a close homolog of AtNRAMP3, leads us to investigate a possible redundancy of AtNRAMP3 and AtNRAMP4 function. We show that subcellular localization and tissue-specific expression patterns of AtNRAMP3 and AtNRAMP4 exhibit strong similarities. Whereas neither AtNRAMP3 nor AtNRAMP4 knockouts display any obvious phenotypes, the nramp3 nramp4 double knockout mutant exhibits strong defects during seed germination under low Fe supply. We provide evidence that AtNRAMP3 and AtNRAMP4 participate specifically in Fe mobilization from vacuolar metal stores during seed germination.
Results
AtNRAMP4 is expressed in vascular tissues and is upregulated upon Fe starvation
To determine in which tissues AtNRAMP4 is expressed, we analyzed the activity of the AtNRAMP4 promoter in Arabidopsis using β-glucuronidase (GUS) as a reporter gene. Arabidopsis (Ecotype Columbia and Wassilewskija (WS)) was transformed with a vector-containing uidA gene downstream of 1928 base pairs of the genomic sequence upstream of AtNRAMP4 initiation codon (proNRAMP4). Figure 1A and B shows histochemical staining of GUS activity in plants transformed with proNRAMP4-GUS grown for 7 days either under Fe-replete conditions (100 μM FeHBED) or in an Fe-depleted medium (no Fe, 100 μM ferrozine). Fe-replete proAtNRAMP4-GUS plants do not show any staining (Figure 1A). In contrast, under Fe-depleted conditions, the AtNRAMP4 promoter drives the transcription of GUS in both roots and shoots (Figure 1B). The staining initiates in the early differentiation zone of the root and becomes stronger in older parts of the roots. The staining is stronger in the stele of roots (Figure 1C) and in the vascular bundles of leaves. Cross sections through the root revealed preferential expression in phloem tissues of the stele (Figure 1D). Similar expression patterns and regulations were observed in several transformed lines either in the Columbia (five lines) or WS ecotype (two lines). In comparison, plants transformed with proNRAMP4-GUS grown for 7 days either under manganese (Mn)-depleted conditions or Mn excess (1 mM) did not show any staining (Supplementary Figure S1A–C).
Figure 1.
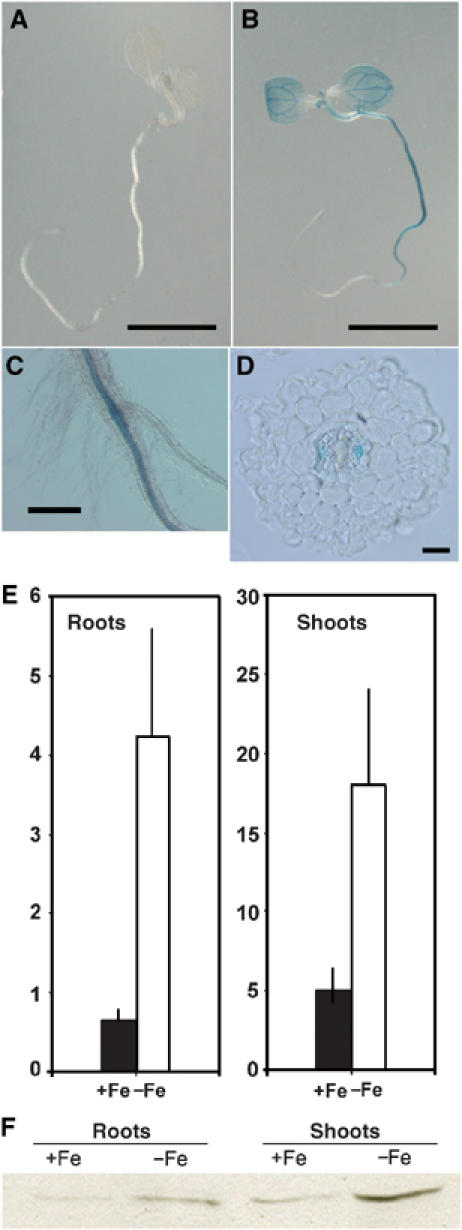
Tissue-specific expression of AtNRAMP4 is induced under Fe starvation. (A–D) Histochemical staining of GUS reporter activity in proAtNRAMP4-GUS seedlings grown for 7 days either on Fe replete medium (ABIS+100 μM FeHBED, A) or under Fe starvation (ABIS without Fe+100 μM ferrozine, B). Scale bar is 1 cm. (C) Detail of the junction between root and hypocotyl in a seedling grown under Fe starvation showing expression in vascular tissues. Scale bar is 0.6 mm. (D) Cross section through the root of a seedling grown under Fe starvation showing preferential expression in phloem tissues. Scale bar is 20 μm. (E) Quantitative analysis of GUS activity in protein extracts from roots or shoots of proAtNRAMP4-GUS seedlings. Seedlings were grown for a week on Fe-replete medium and then transferred either to +Fe (black bar) or −Fe (white bar) medium. Mean activities±s.e. (n=5 independent lines). (F) AtNRAMP4 protein abundance is upregulated under Fe starvation. Western blot experiment showing relative abundance of AtNRAMP4 protein in roots and shoots of Fe-starved (−Fe) or Fe-replete (+Fe) A. thaliana ecotype WS seedlings. 20 μg of proteins was loaded in each lane. AtNRAMP4 migrated at 48 kDa. Growth conditions as in (E).
To quantify the effect of Fe nutrition on AtNRAMP4 promoter activity, we measured GUS enzymatic activity in protein extracts from the roots or shoots of proAtNRAMP4-GUS Arabidopsis grown either in Fe-deficient or Fe-sufficient conditions. Figure 1E shows that GUS activity increased 6.5- and 3.4-fold upon Fe starvation in roots and shoots, respectively.
To determine whether the increase in AtNRAMP4 promoter activity is reflected at the protein level, we performed Western blot experiments using a polyclonal AtNRAMP4 antibody (Lanquar et al, 2004). Figure 1F shows that AtNRAMP4 was hardly detectable in root or shoot extracts from Arabidopsis WS plants grown under high Fe supply (100 μM Fe, n=6 independent biological replica). In contrast, AtNRAMP4 protein accumulated in shoots (in six out of six experiments) and in roots (in two out of six experiments) of plants grown under Fe deficiency (no Fe, 100 μM ferrozine). In roots, AtNRAMP4 levels were close to the detection limit, preventing clear detection of changes in protein accumulation. In addition, AtNRAMP4 protein levels also increased under Mn depletion, but were unaffected by Mn excess (Supplementary Figure S1D).
Taken together, our data show that AtNRAMP4 expression is upregulated under Fe deficiency. Both the tissue-specific expression pattern of AtNRAMP4 and its upregulation under Fe starvation match the regulation of AtNRAMP3 expression (Thomine et al, 2000, 2003). Furthermore, in A. thaliana ecotype WS, an increase in AtNRAMP4 protein correlates with the induction of AtNRAMP4 gene expression under low Fe supply.
AtNRAMP4 knockout displays no obvious phenotype under Fe-sufficient or -deficient conditions
To investigate the function of AtNRAMP4 in Arabidopsis, we screened collections of insertion mutants for mutants carrying a T-DNA in the coding region of AtNRAMP4 using PCR. An allele named nramp4-1 was identified with a T-DNA insertion 922 base pairs downstream of the initiation codon (Figure 2A). Western blots performed on the protein extracts of leaves and roots from either homozygous nramp4-1 mutant or wild type plants showed that AtNRAMP4 could not be detected in the nramp4-1 mutant protein extract (Figure 2B, n=2 independent biological replicates). This result indicates that nramp4-1 is a null allele of AtNRAMP4. Homozygous nramp4-1 mutants did not show any obvious phenotype either when grown on soil in the greenhouse or when grown in vitro on complete medium. Owing to the strong regulation of AtNRAMP4 expression by the plant Fe nutrition status, we tested whether nramp4-1 mutants exhibited any phenotype under various Fe nutrition conditions. Figure 2C shows that no major difference between the root length of nramp4-1 and wild type plants could be measured after germination and development for 16 days on synthetic media containing a range of Fe concentrations. No obvious difference in the level of chlorosis under Fe starvation between wild type and nramp4-1 could be detected. Similar results were obtained when wild type and nramp3-1 mutant, or plants overexpressing either AtNRAMP3 or AtNRAMP4 (Thomine et al, 2000; Lanquar et al, 2004), were compared in the same growth conditions (Figure 2C and Supplementary Figure S2). AtNRAMP4 protein levels and regulation upon Fe starvation were similar in nramp3-1 mutants and wild type Arabidopsis (n=2, data not shown).
Figure 2.
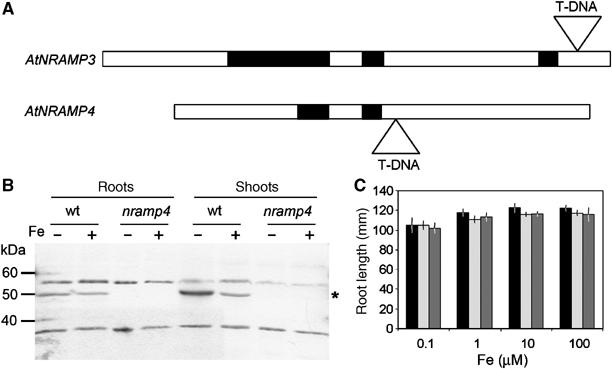
Identification of a knockout mutant carrying a T-DNA insertion in AtNRAMP4 gene. (A) Localization of the T-DNA insertions in nramp3-1 and nramp4-1 mutant alleles. (B) AtNRAMP4 protein is not detectable in nramp4-1 homozygous mutant line. Western blot showing abundance of AtNRAMP4 protein in roots and shoots of Fe-starved or -replete A. thaliana wild type or nramp4-1 homozygous seedlings. 20 μg of proteins was loaded in each lane. Growth conditions as in Figure 1E and F. Note that anti-AtNRAMP4 antibodies recognize two additional proteins that are not affected in nramp4-1 mutants. (C) Seedling development is not affected differentially in wild type, nramp3-1 and nramp4-1 grown under a range of Fe supply. Mean root length±s.e. (n=15–25) of wild type (black bars), nramp3-1 (light gray bars) or nramp4-1 mutant (dark gray bars) measured after 16 days of growth on ABIS media containing 0.1–100 μM FeHBED (as indicated) as the sole source of Fe.
nramp3 nramp4 double knockout mutants display growth defects during early seedling development
In a previous study (Thomine et al, 2003), we have shown that AtNRAMP3 displays Fe regulation, tissue-specific expression pattern similar to AtNRAMP4. In addition, expression of a functional NRAMP4-GFP fusion protein in Arabidopsis shows that AtNRAMP4 is targeted to vacuolar membranes (Supplementary Figure S3), as is AtNRAMP3 (Thomine et al, 2003). Furthermore, AtNRAMP3 and AtNRAMP4 loci are located on duplicated regions of A. thaliana genome: At2g23150 and At5g67330 are included in the duplicated block 0204146800380 (http://wolfe.gen.tcd.ie/athal/dup). To test the hypothesis that AtNRAMP3 and AtNRAMP4 might encode transporters with redundant functions, we generated nramp3 nramp4 double mutants. The nramp3 nramp4 double mutants did not display any phenotype when grown on rich soil in the greenhouse and completed a normal life cycle. However, when mutant seed germination was observed more closely on complete synthetic medium containing 50 μM FeEDTA, they displayed an obvious retardation in cotyledon greening and in root growth during the first 4 days after sowing (Figure 3A). This phenotype was transient: the nramp3 nramp4 double mutant seedlings greened and completely recovered after another 6 days (Figure 3B). The mutant phenotype segregated with a 1:15 ratio in the F2 progenies consistent with the independent segregation of mutant alleles.
Figure 3.
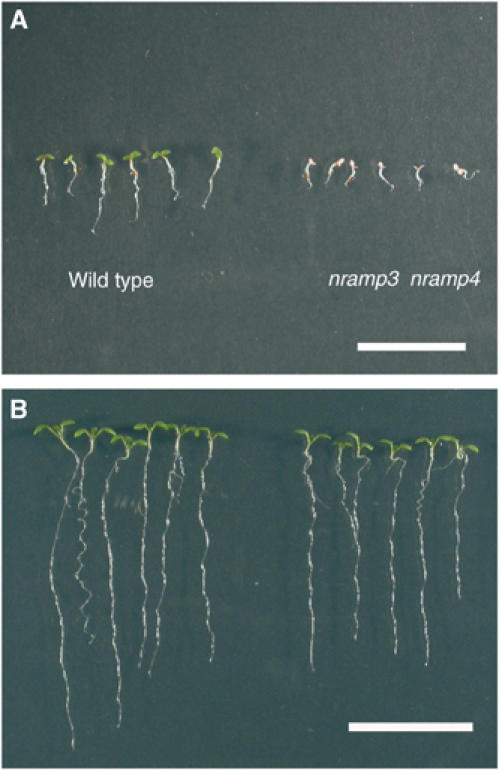
nramp3 nramp4 double knockout mutant exhibits a transient phenotype during germination on synthetic medium. (A) Four days after sowing on ABIS medium containing 50 μM FeEDTA as sole source of Fe, nramp3 nramp4 mutant exhibits pale cotyledon and reduced root elongation in comparison to wild type. (B) Ten days after sowing, the mutant phenotype is no longer obvious: the cotyledons have greened and the root growth almost caught up with that of the wild type seedlings. Scale bar is 20 mm.
To confirm that the observed phenotype was due to the disruption of both redundant transporters AtNRAMP3 and AtNRAMP4, we tested whether AtNRAMP3 and AtNRAMP4 overexpression could complement the phenotype of nramp3 nramp4 double mutants. Using GFP expression in seeds as a selection marker for transformation (Bensmihen et al, 2004), we selected 12–20 independent T1 fluorescent seeds from wild type transformed with the empty vector or from nramp3 nramp4 transformed either with empty vector or with AtNRAMP3 or AtNRAMP4 cDNA driven by the 35S promoter. The phenotype of the fluorescent seeds was scored during germination. Expression of either AtNRAMP3 or AtNRAMP4 rescued the mutant phenotype in 78 or 69% of the primary transformants, respectively. In contrast, all mutant seeds transformed with the empty vector displayed a mutant phenotype. The double mutant phenotype could also be rescued by transformation with either AtNRAMP3 or AtNRAMP4 genes (from ∼2 kb upstream of the initiation codon to the stop codon; Figure 4A) and with AtNRAMP4–GFP fusion cDNA driven by the 35S promoter.
Figure 4.
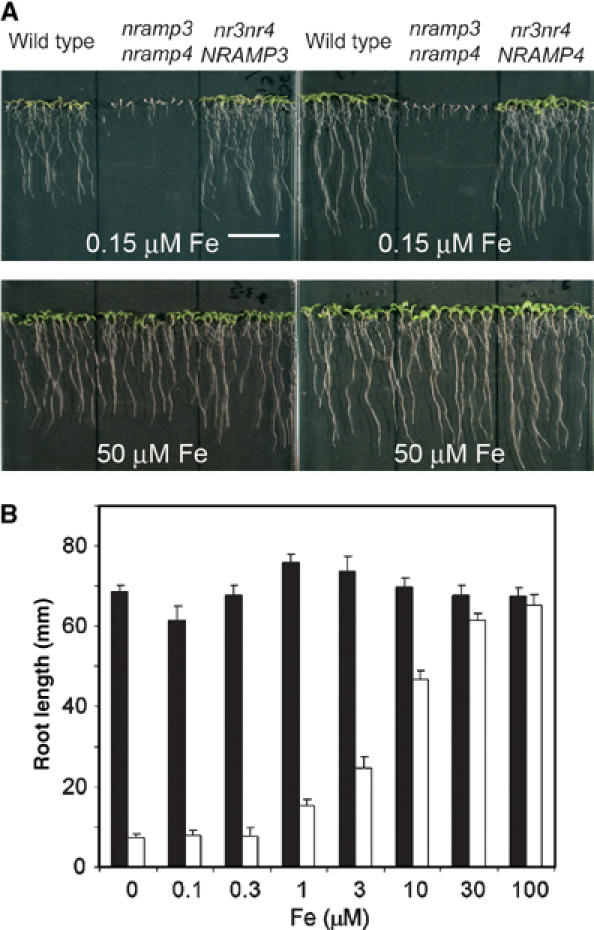
nramp3 nramp4 double mutant seedlings are hypersensitive to low Fe nutrition conditions. (A) Seedlings of wild type, nramp3 nramp4 and nramp3 nramp4 complemented by AtNRAMP3 gene (left panels) or AtNRAMP4 gene (right panels) grown for 10 days either with low Fe supply (upper panels, 0.15 μM FeHBED) or with high Fe supply (lower panels, 50 μM FeHBED). Under low Fe supply, nramp3 nramp4 seedling development is arrested. Fe supplementation or expression of either AtNRAMP3 or AtNRAMP4 gene rescues nramp3 nramp4 mutant phenotype. Scale bar: 2 cm. (B) Dose-dependent effect of Fe supply on nramp3 nramp4 mutant phenotype. Mean root length±s.e. (n=15–25) of wild type (black bars) or nramp3 nramp4 mutant (white bars) measured after 14 days of growth on ABIS media containing 0.1–100 μM FeHBED (as indicated) as the sole source of Fe.
nramp3 nramp4 double knockout mutants are hypersensitive to Fe deficiency during early seedling development
As both AtNRAMP3 and AtNRAMP4 are regulated by Fe and because chlorosis is often a symptom for Fe deficiency, we germinated nramp3 nramp4 double mutants under a range of Fe nutrition conditions. Figure 4A shows that under low Fe supply (0.15 μM Fe) nramp3 nramp4 double mutant development is arrested, their roots do not elongate and their cotyledons do not green, whereas wild type WS plants grown in the same conditions do not show developmental alterations during the first 10 days after germination. When nramp3 nramp4 mutants were germinated under high Fe supply, their growth and appearance was indistinguishable from that of wild type plants (Figure 4A). Figure 4B illustrates the progressive increase in severity of nramp3 nramp4 mutant root growth phenotype with progressive decrease of Fe availability in the medium. We tested whether increasing the level of other essential metals could also rescue nramp3 nramp4 phenotype. Neither an increase in Zn nor an increase in Mn supply rescued nramp3 nramp4 phenotype. Zn and Mn rather have inhibitory effects on early development, and nramp3 nramp4 mutants were even more sensitive to this inhibition than wild type seedlings. Conversely, Mn and Zn depletion of the growth medium (usually containing 30 μM Zn and 100 μM Mn) altered neither the wild type nor the nramp3 nramp4 mutant development during the first 10 days after sowing. Together, these results indicate that AtNRAMP3 and AtNRAMP4 act redundantly during seed germination and that the loss of AtNRAMP3 and AtNRAMP4 function results in dramatic and specific hypersensitivity to Fe starvation that can be fully rescued under high Fe supply.
nramp3 nramp4 hypersensitivity to Fe starvation leads to lethality on soils with low Fe availability
To study the consequence of nramp3 nramp4 double mutant hypersensitivity to Fe starvation revealed by in vitro experiments on plant survival in soil, we germinated wild-type A. thaliana and nramp3 nramp4 mutants either on humic soil or on calcareous soil. Humic soils contain Fe chelated to humic acids facilitating Fe acquisition (Mackowiak et al, 2001). In contrast, calcareous soils often limit Fe acquisition because of their alkaline pH (Marschner and Romheld, 1994). When wild-type and nramp3 nramp4 mutant Arabidopsis were sown on humic soil, both genotypes germinated and developed green cotyledons after 6 days (Figure 5A). After 12 days, seedlings of both genotypes grew to healthy plants (Figure 5C). In contrast, when wild type and nramp3 nramp4 were sown on calcareous soil, the wild type seedlings developed green cotyledons, whereas the mutant seedlings did not green and appeared bleached after 6 days (Figure 5B). Most nramp3 nramp4 mutants did not develop further than 3 mm bleached seedlings and, after 12 days, extremely few nramp3 nramp4 mutant plants had survived. In contrast, most of the wild type seedlings had developed to a level comparable to the wild type plants grown on humic soil (Figure 5D). Disruption of both AtNRAMP3 and AtNRAMP4 genes results in early lethality for over 90% of Arabidopsis plants when they are grown on Fe-limiting soil.
Figure 5.

AtNRAMP3 and AtNRAMP4 are critical for plant survival on Fe-limiting soils. Wild type (front) or nramp3 nramp4 mutant (back) seedlings 6 days after sowing either on humic soil (A) or on calcareous soil (B). Note the bleached appearance of nramp3 nramp4 seedlings on calcareous soil (B, back). Wild type (front) or nramp3 nramp4 mutant (back) plants 12 days after sowing either on humic soil (C) or on calcareous soil (D). nramp3 nramp4 seedlings did not develop into adult plants on calcareous soil (D, back).
AtNRAMP3, AtNRAMP4 and IRT1 are expressed sequentially during seed germination
As the phenotype of nramp3 nramp4 mutants is particularly severe during germination under Fe-limiting conditions, we studied the expression of AtNRAMP3 and AtNRAMP4 in these conditions. In addition, we monitored the expression of IRT1, a plasma membrane transport protein essential for Fe acquisition under Fe starvation (Vert et al, 2002). Seeds containing either AtNRAMP3 or AtNRAMP4 or IRT1 promoter-GUS constructs were germinated under Fe-limiting conditions. The seedlings were stained for GUS activity 1, 2 and 3 days after sowing. Figure 6 shows that AtNRAMP3 and AtNRAMP4 were expressed during the early stages of germination (days 1 and 2). In contrast, IRT1 was not expressed during the first 2 days but became strongly induced on the third day. Similar results were obtained under high Fe supply, except that expression of IRT1 and AtNRAMP4 was lower on day 3 and AtNRAMP4 staining disappeared on day 7 (Figure 1A). The timing of AtNRAMP3 and AtNRAMP4 expression early during germination is thus consistent with the severe phenotype of the nramp3 nramp4 double mutant at this developmental stage.
Figure 6.

AtNRAMP3, AtNRAMP4 and IRT1 are expressed sequentially during germination. Histochemical staining of proAtNRAMP3-GUS (A, D, G), proAtNRAMP4-GUS (B, E, H) and proIRT1-GUS (C, F, I) seedlings germinated under Fe starvation (ABIS without Fe+100 μM ferrozine) one (A–C), two (D–F) or three (G–I) days after sawing. Scale bars are 0.5 mm (A–C), 1 mm (D–F) and 3 mm (G–I). At days 1 and 2, the embryos have been extracted from the seed teguments. Similar results were obtained with independent lines of proAtNRAMP3-GUS and proAtNRAMP4-GUS.
AtNRAMP4 localizes to vacuolar membranes in cotyledon cells during seed germination
The expression of AtNRAMP3 and AtNRAMP4 in cotyledons coincides with an early developmental arrest during germination in nramp3 nramp4 double mutants. To understand the cellular basis of this phenotype, we investigated the subcellular localization of AtNRAMP4 protein in cotyledon cells by immunolocalization using AtNRAMP4 antibodies. Figure 7 illustrates the localization of AtNRAMP4 in 2-day-old germinating seeds. Two different intracellular localization patterns were observed, sometimes within the same cotyledon: a punctuate localization within the vacuoles (Figure 7B) and a tonoplast localization surrounding the vacuoles corresponding to protein bodies in dry seeds (Figure 7A). No specific labelling was observed in cotyledons from nramp4-1 mutants (Figure 7C). At this developmental stage, γTIP, a marker for lytic vacuoles, also localized around the vacuoles corresponding to protein bodies in dry seeds (Figure 7D) as well as on punctuate structures within the vacuoles (Figure 7F). To confirm that AtNRAMP4 and γTIP reside on the same membranes, coimmunolocalization experiments were performed. Figure 7E–G shows that AtNRAMP4 colocalized with γTIP in the tonoplast (E) and in the punctuate structures within the vacuoles (F) of cotyledon cells.
Figure 7.
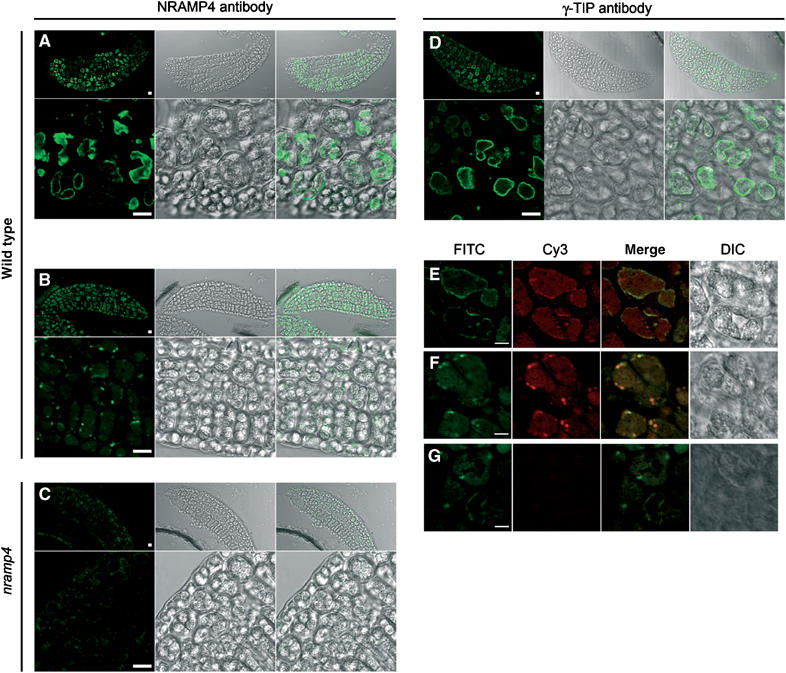
AtNRAMP4 is localized on vacuolar and punctuate structures in cotyledon cells. (A–G) Immunolocalization in PEG sections of 2-day-old Arabidopsis seedling cotyledons. (A–D) Fluorescence detection with anti-rabbit antibody coupled to FITC (green images). Upper panels: overviews of the cotyledons; lower panels: staining at the cellular level. Scale bars are 10 μm. (A, B) AtNRAMP4 staining of internal membranes of wild type seeds. (C) AtNRAMP4 staining of nramp4-1 mutants: nramp4-1 seeds are devoid of staining. (D) γ-TIP staining of wild type seeds. (E–G) Colocalization of AtNRAMP4 and γTIP. AtNRAMP4 was detected with FITC (green fluorescence). γ-TIP was detected using Cy3 (red fluorescence), colocalization (yellow) is shown in the merged picture. (E) Vacuolar and (F) punctuate patterns of AtNRAMP4/γ-TIP staining. (G) AtNRAMP4 staining of cells in the absence of γ-TIP antibody. No Cy3 staining was detectable. Differential interference contrast pictures are shown at the right side of each panel.
nramp3 nramp4 double knockout mutant fails to mobilize vacuolar Fe during seed germination
To further understand the function of AtNRAMP3 and AtNRAMP4 during seed germination, we compared the metal content in wild type and nramp3 nramp4 plants. As nramp3 nramp4 mutants display a strong phenotype during germination, we first analyzed the metal content in seeds. Table I shows the metal content in seeds, roots and shoots of wild type, single nramp3 and nramp4 mutants and the nramp3 nramp4 double mutant determined by Inductively Coupled Plasma Optical Emission Spectrometry (ICP-OES). No significant difference in the seed contents in Fe, Mn and Zn could be detected between the wild type and any of the mutants (n=3, Table I). We then compared the metal content of wild type and nramp3 nramp4 seedlings grown for 14 days on complete synthetic medium. Table I shows that nramp3 nramp4 seedlings accumulated about 20% more Fe in their roots than wild type (n=3, Table I). Apart from this moderate increase in Fe, the accumulation of metals was not significantly altered in the nramp3 nramp4 double mutant.
Table 1.
Metal concentrations in seeds, roots and shoots of nramp mutants (ppm)
| Metal | Fe | Mn | Zn |
|---|---|---|---|
| Seed | |||
| Wild type | 73±4 | 131±11 | 56±3 |
| nramp3 | 85±9 | 183±57 | 52±5 |
| nramp4 | 86±18 | 176±23 | 50±9 |
| nramp3 nramp4 | 70±6 | 128±13 | 62±9 |
| Root | |||
| Wild type | 105±12 | 4954±989 | 1730±410 |
| nramp3 | 105±12 | 4563±720 | 1723±415 |
| nramp4 | 97±39# | 4796±1166# | 1849±706# |
| nramp3 nramp4 | 134±12* | 5409±656 | 2132±407 |
| Shoot | |||
| Wild type | 91±10 | 543±29 | 186±13 |
| nramp3 | 86±9 | 596±41 | 192±22 |
| nramp4 | 84±7 | 505±36 | 174±20 |
| nramp3 nramp4 |
103±13 |
562±18 |
220±21 |
| Dry seeds were harvested from Arabidopsis grown on rich soil in the greenhouse. Arabidopsis seedlings were grown vertically on plates containing complete ABIS synthetic medium for 14 days. Values are given as mean±s.e. All measurements were performed on three independent biological replicates, except values labelled #performed on two independent biological replicates. *Significant difference between wild type and nramp3 nramp4 mutants (P<0.005). All other tests yield P>0.05. | |||
As AtNRAMP3 and AtNRAMP4 are targeted to the vacuolar membrane (Supplementary Figure S3 and Figure 7; Thomine et al, 2003) and the nramp3 nramp4 mutant phenotype is rescued by Fe, we compared the fate of vacuolar Fe during germination of wild type and nramp3 nramp4 double mutants. Figure 8A and B shows the localization of Fe in cells of dry Arabidopsis seeds determined by electron microscopy coupled to Fe ESI (imaging of inelastically scattered electrons). In both wild type and nramp3 nramp4 cotyledon seeds, Fe could be localized to the electron dense globoids included in the protein bodies in agreement with earlier studies on different species (Buttrose, 1978). This was confirmed by X ray analysis of individual globoids (Figure 8E and F): it revealed the presence of phosphate together with Ca, Mg as well as Zn and Fe, but, in agreement with a previous report (Otegui et al, 2002), almost no Mn. Figure 8C and D shows the localization of Fe in the globoids of cotyledon cells in 2-day-old seedlings. Whereas Fe was almost undetectable in globoids of the wild type cotyledon cells at this stage, Fe still mapped to the globoids of nramp3 nramp4 mutant cotyledon vacuoles 2 days after germination. Figure 8G and H shows representative Energy Dispersive X ray (EDX) spectra confirming the presence of Fe in nramp3 nramp4 globoids 2 days after germination, while Fe is missing in wild type globoids. Altogether, these results show that Fe is retained in globoids of the nramp3 nramp4 mutant during germination, while globoid Fe is rapidly mobilized from the vacuole in wild type cotyledons.
Figure 8.
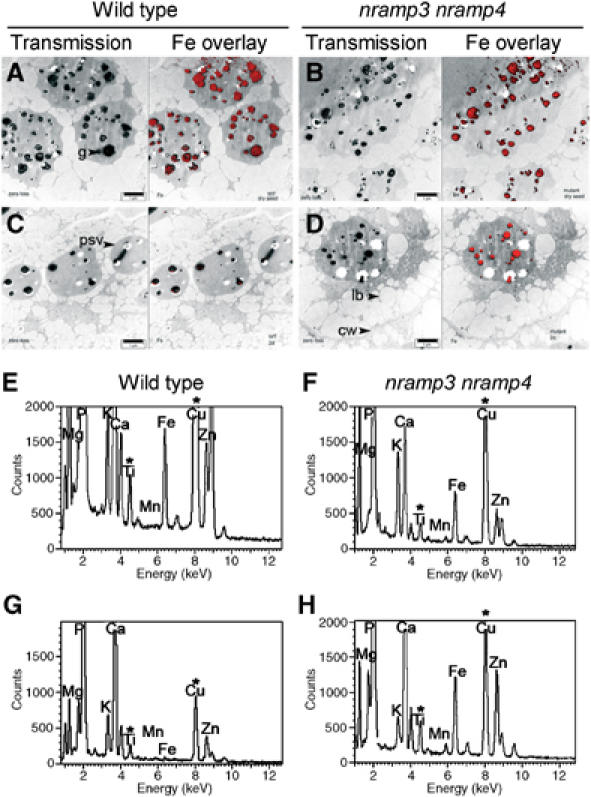
nramp3 nramp4 double mutants fail to retrieve Fe from seed vacuole globoids. (A–D) Transmission electron microscopy images of cotyledon parenchyma cells from wild type (A, C) or nramp3 nramp4 (B, D) either in dry seeds (A, B) or 2 days after sowing (C, D). Left panels: transmission image; right panels: Fe localization (red) obtained through imaging of inelastically scattered electrons (ESI) superimposed on the transmission image. Visible structures are labelled according to the Arabidopsis Atlas (Mansfield and Bowman, 1994): cw, cell wall; lb, lipid body; g, globoid; psv, protein storage vacuole. Scale bar is 1 μm. Empty globoid cavities correspond to globoid compartments from which the globoid crystal has been ejected during sectioning. (E–H) Representative EDX spectra of globoids from wild type (E, G) or nramp3 nramp4 (F, H) either in dry seeds (E, F) or 2 days after sowing (G, H). *Titanium and copper originate mostly from the grids holding the sections.
Discussion
The results reported in this study identify AtNRAMP4 and AtNRAMP3 as redundant metal transporters essential for early seedling development under low Fe availability. The analysis of the nramp3 nramp4 double mutant phenotype and the targeting of both AtNRAMP3 and AtNRAMP4 proteins to the vacuolar membrane lead us to propose that AtNRAMP3 and AtNRAMP4 mediate Fe mobilization from the vacuolar stores during seed germination.
Accumulation of AtNRAMP4 under Fe deficiency
The increased accumulation of AtNRAMP4 mRNA under Fe deficiency was reported earlier (Thomine et al, 2000). In this work, using AtNRAMP4 promoter fusions, we further confirm that AtNRAMP4 expression is upregulated under Fe starvation and provide arguments that this regulation occurs at the transcriptional level. Neither AtNRAMP3 nor AtNRAMP4 upregulation under Fe starvation has been observed in microarray experiments (Wintz et al, 2003; Colangelo and Guerinot, 2004). Differences in the experimental conditions, such as the age of the plants and the duration of Fe starvation, may explain this discrepancy. In addition, using AtNRAMP4 antibodies, we show that upregulation of AtNRAMP4 transcription leads to AtNRAMP4 protein accumulation under Fe deficiency. AtNRAMP4 protein accumulation was observed in the WS ecotype of A. thaliana and not in the Columbia ecotype (Lanquar et al, 2004) although accumulation of AtNRAMP4 mRNA occurred in both ecotypes under Fe deficiency (Thomine et al, 2000). This suggests an additional post-transcriptional level of regulation of AtNRAMP4 protein level and requires further investigations. Strong post-transcriptional regulation of metal transport proteins has already been reported for IRT1 in Arabidopsis (Connolly et al, 2002) and for SMF1 in yeast (Liu and Culotta, 1999).
AtNRAMP4 is targeted to the vacuolar membrane
We provide two lines of evidence that the AtNRAMP4 resides on vacuolar membranes. An AtNRAMP4-GFP fusion protein is targeted to the membrane of the vacuole after transient expression in Arabidopsis protoplasts and in stably transformed Arabidopsis plants. Immunolocalization of AtNRAMP4 in cotyledon cells of young seedlings also indicates targeting to the vacuole (Supplementary Figure S3). In addition, AtNRAMP4 was identified by a third approach during the proteomic analysis of vacuoles from Arabidopsis leaves (Carter et al, 2004). The localization of AtNRAMP4 determined from transient expression in Arabidopsis protoplasts is undistinguishable from that of AtNRAMP3 (Thomine et al, 2003).
The analysis of nramp3 nramp4 double mutant suggests that AtNRAMP3 and AtNRAMP4 function to mobilize Fe from cotyledon parenchyma storage vacuoles. However, AtNRAMP3 and AtNRAMP4 GFP fusion proteins are targeted to lytic vacuoles. Seed storage vacuoles have within them a membrane-bound compartment that surrounds globoids and harbors γTIP, a marker of the lytic vacuole (Jiang et al, 2001). The localization of AtNRAMP4 to the globoid compartment of protein storage vacuoles would reconciliate the phenotype of nramp3 nramp4 mutants with the targeting of AtNRAMP3 and AtNRAMP4 GFP fusion proteins. Consistent with this localization, the punctuate structures labelled by AtNRAMP4 and γTIP antibodies in cotyledon cells (Figure 7B) have the size and distribution of seed globoids observed by electron microscopy (Figure 8; Otegui et al, 2002). However, a membrane-bound globoid compartment has not been clearly identified in Arabidopsis seeds so far (Gillespie et al, 2005). In addition, AtNRAMP4 and γTIP, a marker of the lytic vacuole, colocalize to the tonoplast of cotyledon of storage vacuoles. We propose that during germination the membrane surrounding storage vacuoles rapidly acquires the properties of lytic vacuoles. This localization is also consistent with the function of AtNRAMP3 and AtNRAMP4 in vacuolar Fe mobilization.
Function of AtNRAMP3 and AtNRAMP4 in seed germination in environments with low Fe availability
AtNRAMP3 and AtNRAMP4 act redundantly to provide sufficient Fe for Arabidopsis development during germination. Indeed, the germination arrest of nramp3 nramp4 double mutant under low Fe supply can be rescued either by individual expression of AtNRAMP3 or AtNRAMP4, or by supplying high Fe nutrition to the seedlings. The germination phenotype of the nramp3 nramp4 double mutant is observed not only under extreme Fe deficiency generated in vitro but also when the mutant is grown on calcareous soils, which are Fe limiting (Marschner and Romheld, 1994). Thus, the critical function of AtNRAMP3 and AtNRAMP4 during germination is relevant to widely represented Fe-limiting soils. It is likely that even under Fe sufficiency, the transient delay in root growth and greening of nramp3 nramp4 seedlings make them more sensitive to environmental stresses. The nramp3 nramp4 double mutants are arrested early during germination. In comparison, other Arabidopsis mutants that are hypersensitive to low Fe nutrition, such as irt1 and fit1 mutants, are able to develop into plants although they are strongly chlorotic (Vert et al, 2002; Colangelo and Guerinot, 2004). The development of nramp3 nramp4 double mutant is arrested before IRT1 expression initiates. Thus, the first 2 days of seedling development when AtNRAMP3 and AtNRAMP4 but not IRT1 are expressed determine a critical period when AtNRAMP3 or AtNRAMP4 function is crucial. It will be interesting to test whether constitutive expression of IRT1 can rescue the nramp3 nramp4 phenotype under low Fe nutrition.
Fe storage in seeds: globoids versus ferritins
The detailed analysis of nramp3 nramp4 double mutant supports the hypothesis that AtNRAMP3 and AtNRAMP4 function to mobilize vacuolar Fe during seed germination. Indeed, nramp3 nramp4 and wild type seeds contain the same amount of total Fe and the phenotype is linked to the embryo genotype rather than to the maternal genotype. Taken together, these results indicate that metal storage is not affected in nramp3 nramp4 mutant seeds. In contrast, microanalysis of Fe in seed storage vacuoles shows that Fe remains associated to the vacuole globoids in nramp3 nramp4 double mutant, while it is efficiently removed from the globoids in wild type germinating seeds. This result suggests that AtNRAMP3 and AtNRAMP4 are required to retrieve globoid-associated Fe during germination and to make Fe bioavailable for seedling development. Accordingly, the slight increase in Fe in the roots of 15-day-old plants can be interpreted as increased retention of Fe in vacuoles.
On the basis of early studies on legume seeds, it has been widely accepted that plastid ferritin is the main site for Fe storage in seeds (Lobreaux and Briat, 1991). Our results indicate that vacuole globoids represent an important site for Fe storage in Arabidopsis seeds. It is possible that the intracellular site for Fe storage in seeds differs according to species. Pea seeds contain a lot of ferritin-associated Fe and only few globoids (Lott et al, 1984; Lobreaux and Briat, 1991). The globoids contain phytate (inositol hexakis phosphate) associated with cations such as K, Mg and Ca, and divalent metals such as Zn and Fe in lower amounts (Buttrose, 1978). Fe associated with phytate is poorly available for animal and human nutrition (Hallberg, 2001). In contrast, ferritin-associated Fe is bioavailable (Hallberg, 2001). Increasing ferritin content has been used as a biotechnological strategy to improve the Fe content in rice grains (Goto et al, 1999). Our results suggest that combining overexpression of NRAMP and ferritin in seed might further improve the balance between bioavailable Fe associated with ferritin and nonbioavailable Fe associated with phytate in crops.
In this work, we identify for the first time the vacuole as a major compartment for Fe storage in plant seeds and show that retrieval of the Fe stored in vacuoles is an essential step for successful germination in a wide range of environments.
Materials and methods
T-DNA insertion mutants
The isolation of the insertion mutant in AtNRAMP3 gene has been described previously (Thomine et al, 2000): nramp3-1 was identified from T-DNA insertion lines of A. thaliana ecotype WS. The T-DNA insertion in AtNRAMP3 gene is located in the last exon between GLU467 and VAL468. The insertion mutant in AtNRAMP4 gene was identified by screening the T-DNA insertion mutant collection of the Center for Biotechnology at the University of Wisconsin (http://www.biotech.wisc.edu/Arabidopsis/Index2.asp) by PCR using Ex-Taq polymerase (Takara). Two pairs of primers were used, each including an AtNRAMP4 gene-specific primer, NR4-KOS or NR4-KOR, and the T-DNA primer, JL202 (Supplementary Table S1) specific for the left border of the T-DNA. The PCR product from one pool (NR4-KOS/JL202), which gave a positive signal after southern hybridization with an AtNRAMP4 cDNA probe, was re-amplified by nested PCR using NR2-4S and JL270 (Supplementary Table S1). Sequencing of the nested PCR product revealed that the T-DNA insertion in AtNRAMP4 is localized 922 bp from the initiation codon on the genomic sequence. The T-DNA insertion in AtNRAMP4 is predicted to truncate the protein after glutamine 234.
F2 populations from reciprocal crosses between nramp3-1 and nramp4-1 single mutants were generated. Several double knockout mutants were isolated after genotyping of 48 F2 plants using the primers designed for single knockout allele identification. Note that the wild type and single nramp3 and nramp4 mutants used in this study originate from the same F2 progeny.
Mutant complementation
To complement nramp3 nramp4 double mutants, AtNRAMP3 and AtNRAMP4 coding sequences were amplified using Advantage cDNA Taq polymerase (Clontech) with NR3BamH1S and NR3-SalR, or NR4BamH1S and NR4-SalR (Supplementary Table S1). The products were cloned between the BamH1 and Sal1 restriction sites in pFP101. pFP101, which gave rise to the pALLIGATOR plasmid family, includes GFP driven by the At2S3 seed-specific promoter as a selectable marker for transformed seeds, but does not contain a triple HA tag and Gateway cassette (Bensmihen et al, 2004). Empty pFP101, pFP101-AtNRAMP3 and pFP101-AtNRAMP4 were introduced into nramp3 nramp4 and WS plants through Agrobacterium-mediated transformation. Primary transformant seeds were selected on the basis of GFP fluorescence (Bensmihen et al, 2004).
Alternately, AtNRAMP3 and AtNRAMP4 genes were amplified from BAC T20D16 and K8K14 using Advantage cDNA Taq polymerase (Clontech) with the primers proNR1S and cmycNR1-R for AtNRAMP3 or proNR2S and cmycNR2-R for AtNRAMP4 (Supplementary Table S1). The PCR products were cloned in the pRD400 binary vector. Empty pRD400 and pRD400 containing AtNRAMP genes were introduced in nramp3 nramp4 mutants through Agrobacterium-mediated transformation. Primary transformants were selected on the basis of the complementation of the mutant phenotype (growth on ABIS medium containing 0.3 μM Fe). No complemented seedling was recovered after transformation with the empty pRD400.
Growth conditions
Arabidopsis seedlings were grown on plates containing 2.5 mM H3PO4, 5 mM KNO3, 2 mM MgSO4, 1 mM Ca(NO3)2, Murashige and Skoog microelements, 1% sucrose, 1% Phytagel, 1 mM MES adjusted with KOH to pH 6.1 and 50 μM FeEDTA (ABIS medium). To control Fe nutrition, Fe EDTA was substituted with a range of concentrations of FeHBED as indicated in the figures. FeHBED was prepared as a 10 mM stock solution from FeCl3 (Sigma, Saint Louis, MO) and HBED (N,N′-di(2-hydroxybenzyl)ethylene diamine-N,N′-diacetic acid monochloride hydrate (Strem Chemicals, Newburyport, MA). HBED was added with a 10% excess to ensure that all Fe is chelated. To deplete the medium with Fe, Fe was omitted and 100 μM ferrozine was added to the ABIS medium (‘−Fe').
Promoter β-glucuronidase fusions, β-glucuronidase activity measurements and histological analysis
The genomic sequence upstream of AtNRAMP4 initiation codon (1928 base pairs) was amplified by PCR from the BAC K8K14 using Advantage cDNA Taq polymerase (Clontech) and the primers ProNR2S and ProNR2R (Supplementary Table S1). The PCR fragment was cloned in pCAMBIA 1381Z. Constructs in which the AtNRAMP4 promoter was driving the expression of the uidA gene were used in parallel with empty pCAMBIA 1381Z for Agrobacterium tumefaciens-mediated Arabidopsis transformation. For histological analysis, Arabidopsis seedlings were grown on +Fe or −Fe plates (see growth conditions) and stained as described (Thomine et al, 2003). To generate sections, seedlings were embedded according to Technovit 7100 instructions (EBSciences) and as described (Beeckman and Viane, 1999). Cross sections of 15 μm through the roots were realized and observed.
For GUS activity assays, plants were grown for 7 days on plates containing Fe-replete medium and then either on the same medium (+Fe) or on −Fe medium and grown for another for 7 days. GUS activity was quantified independently in roots and shoots as described (Thomine et al, 2003).
Plant protein isolation and immunoblot analysis
A. thaliana, ecotype WS, wild type or nramp4-1 mutant were grown vertically for 7 days on ABIS medium contaning 100 μM FeHBED and transferred for another 7 days on a medium containing either 100 μM FeHBED (+Fe) or no Fe and 100 μM ferrozine (−Fe). Total proteins from shoot or root tissues were extracted and separated as described (Lanquar et al, 2004). The proteins were detected by anti-NRAMP4 antibodies and then revealed by goat anti-rabbit, conjugated to horseradish peroxidase (Amersham Biosciences) and visualized using the enhanced chemioluminescence ECL+kit (Amersham Biosciences).
Immunofluorescence
Immunostaining procedures were performed on sections of 2-day-old A. thaliana seedlings embedded in PEG. Simple labelling was performed as described previously (Couchy et al, 2003; Bolte et al, 2004). Double labelling of cells with two rabbit primary antibodies was performed using Fab fragments in the first step and a fixation before the second hybridization (Bolte et al, 2004). After incubation with the first primary antibody, cells were incubated with an excess of the secondary antibody anti-rabbit Fab fragment coupled to FITC diluted to 1/50 (Interchim). Sections were then washed and post-fixed for 1 h with 3.7% para-formaldehyde in PBS and rinsed overnight with buffer alone. Nonspecific binding sites were blocked with buffer containing 1% (w/v) BSA and the cells were then treated with the second primary antibody, followed by the second secondary antibody (anti-rabbit IgG conjugated with Cy3, 1:800, Interchim). Primary antibodies were rabbit polyclonal antibody AtNRAMP4 1:200 (Lanquar et al, 2004) and the lytic vacuole marker rabbit polyclonal γTIP 1:200 (Jauh et al, 1999). Images were collected with a Leica laser-scanning confocal microscope TCS SP2 (Leica Microsystems, Heidelberg, Germany). Fluorochromes were detected sequentially using laser lines 488 and 543 nm.
Metal content
Seedlings were grown vertically for 12 days on plates containing ABIS medium with 1% agar. Roots and shoots were harvested separately and washed. The dry weight (DW) of the samples was measured after drying at 60°C for 3 days. The dried samples were digested in a mixture of 1 ml of 65% nitric acid and 0.5 ml ultrapure H2O in a MARS5 microwave (CEM GmbH, Kamp-Lintford, Germany) at 200°C at 15 bar for 10 min. After dilution in trace metal-free water, the metal content of the samples was determined by inductively coupled plasma optical emission spectroscopy using an IRIS Advantage Duo ER/S (Thermo Jarrell Ash, Franklin, MA, USA).
Dry seeds were treated as dry samples. Fe content in seeds was determined as described previously (Lobreaux and Briat, 1991). Copper, manganese and zinc contents were determined by inductively coupled plasma optical emission spectroscopy using a PQ II Turbo+ quadrupole inductively coupled plasma mass spectrometer (VG Elemental, Cambridge, UK).
Electron microscopy and microanalysis
Electron microscopy experiments were performed as described (Neumann et al, 1995): dry seeds or seeds 2 days after hydration were chilled in liquid propane (JFD 030, Balzers, Lichtenstein) and freeze substituted in methanol for 4 days (CS auto, Leica, Bensheim, Germany). The temperature was then increased slowly (1°C/h) to −20°C and the material was embedded in Lowicryl 4KM. UV polymerization was performed at −20°C for 2 days. Sections (100 nm) were observed with a transmission electron microscope equipped with an energy filter (EM912 Omega, Zeiss, Oberkochen, Germany) and an EDX analyser (Link exIII, Oxford Instruments, High Wycombe, Bucks, UK). For ESI, images were recorded with a SIT-TV camera with an energy resolution of about 5 eV and processed with an image-processing system (Analysis 3.0, SIS software GmbH, Münster, Germany). For colocalization, the ESI images of Fe were background-subtracted, colored and overlaid with a zero loss image.
Supplementary Material
Figure S1
Figure S2
Figure S3
Table S1
Acknowledgments
We thank François Parcy for sharing pFP101 vector before publication. This work was funded by GENOPLANTE (Project AF 2001065), French ministry of research and the CNRS. Exchanges between the ISV and U Krämer's research group were supported by Procope project 04567VM and the EU Research Training Network HPRN CT2002 00243.
References
- Beeckman T, Viane R (1999) Embedding thin plant specimens for oriented sectioning. Biotechn Histochem 75: 23–26 [DOI] [PubMed] [Google Scholar]
- Bensmihen S, To A, Lambert G, Kroj T, Giraudat J, Parcy F (2004) Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsisseeds. FEBS Lett 561: 127–131 [DOI] [PubMed] [Google Scholar]
- Bereczky Z, Wang HY, Schubert V, Ganal M, Bauer P (2003) Differential regulation of nramp and irt metal transporter genes in wild type and iron uptake mutants of tomato. J Biol Chem 278: 24697–24704 [DOI] [PubMed] [Google Scholar]
- Bolte S, Brown S, Satiat-Jeunemaitre B (2004) The N-myristoylated Rab-GTPase m-Rabmc is involved in post-Golgi trafficking events to the lytic vacuole in plant cells. J Cell Sci 117: 943–954 [DOI] [PubMed] [Google Scholar]
- Buttrose MS (1978) Manganese and iron in globoid crystals of protein bodies from Avena and Casuarina. Aust J Plant Physiol 5: 631–639 [Google Scholar]
- Carter C, Pan S, Zouhar J, Avila EL, Girke T, Raikhel NV (2004) The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16: 3285–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cellier M, Gros P (eds) (2004) The Nramp Family. New York: Eurekah.com and Kluwer Academic/Plenum [Google Scholar]
- Colangelo EP, Guerinot ML (2004) The essential basic helix–loop–helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly EL, Fett JP, Guerinot M-L (2002) Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell 14: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couchy I, Bolte S, Crosnier MT, Brown S, Satiat-Jeunemaitre B (2003) Identification and localization of a beta-COP-like protein involved in the morphodynamics of the plant Golgi apparatus. J Exp Bot 54: 2053–2063 [DOI] [PubMed] [Google Scholar]
- Curie C, Alonso JM, Le Jean M, Ecker R, Briat J-F (2000) Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochem J 347: 749–755 [PMC free article] [PubMed] [Google Scholar]
- Curie C, Briat JF (2003) Iron transport and signaling in plants. Annu Rev Plant Physiol Plant Mol Biol 54: 183–206 [DOI] [PubMed] [Google Scholar]
- Curie C, Panaviene Z, Loulergue C, Dellaporta SL, Briat JF, Walker EL (2001) Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409: 346–349 [DOI] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93: 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming MD, Romano MA, Su MA, Garrick LM, Garrick MD, Andrews NC (1998) Nramp2 is mutated in the anemic Belgrade (b) rat: evidence of a role for Nramp2 in endosomal iron transport. Proc Natl Acad Sci USA 95: 1148–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie J, Rogers SW, Deery M, Dupree P, Rogers JC (2005) A unique family of proteins associated with internalized membranes in protein storage vacuoles of the Brassicaceae. Plant J 41: 429–441 [DOI] [PubMed] [Google Scholar]
- Goto F, Yoshihara T, Shigemoto N, Toki S, Takaiwa F (1999) Iron fortification of rice seed by the soybean ferritin gene. Nat Biotechnol 17: 282–286 [DOI] [PubMed] [Google Scholar]
- Gunshin H, Mackenzie B, Berger UV, Gunshin Y, Romero MF, Boron WF, Nussberger S, Gollan JL, Hediger MA (1997) Cloning and characterization of a mammalian proton-coupled metal–ion transporter. Nature 388: 482–488 [DOI] [PubMed] [Google Scholar]
- Hallberg L (2001) Perspective on nutritional iron deficiency. Annu Rev Nutr 21: 1–21 [DOI] [PubMed] [Google Scholar]
- Jauh G-Y, Phillips TE, Rogers JC (1999) Tonoplast intrinsic protein isoforms as markers for vacuolar functions. Plant Cell 11: 1867–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Phillips TE, Hamm CA, Drozdowicz YM, Rea PA, Maeshima M, Rogers SW, Rogers JC (2001) The protein storage vacuole: a unique compound organelle. J Cell Biol 155: 991–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser BN, Moreau S, Castelli J, Thomson R, Lambert A, Bogliolo S, Puppo A, Day DA (2003) The soybean NRAMP homologue, GmDMT1, is a symbiotic divalent metal transporter capable of ferrous iron transport. Plant J 35: 295–304 [DOI] [PubMed] [Google Scholar]
- Lanquar V, Lelièvre F, Barbier-Brygoo H, Thomine S (2004) Regulation and function of AtNRAMP4 metal transporter protein. Soil Sci Plant Nutr 50: 477–484 [Google Scholar]
- Liu XF, Culotta VC (1999) Post-translation control of Nramp metal transport in yeast. Role of metal ions and the BSD2 gene. J Biol Chem 274: 4863–4868 [DOI] [PubMed] [Google Scholar]
- Lobreaux S, Briat JF (1991) Ferritin accumulation and degradation in different organs of pea (Pisum-sativum) during development. Biochem J 274: 601–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott JNA, Goodchild DJ, Craig S (1984) Studies of mineral reserves in pea (Pisum sativum) cotyledons using low-water-content procedures. Aust J Plant Physiol 11: 459–469 [Google Scholar]
- Mackowiak CL, Grossl PR, Bugbee BG (2001) Beneficial effects of humic acid on micronutrient availability to wheat. Soil Sci Soc Am J 65: 1744–1750 [DOI] [PubMed] [Google Scholar]
- Mansfield SG, Bowman JL (1994) Embryogenesis. In: Arabidopsis: An Atlas of Morphology and Development, Bowman JL (ed) pp 350–401. New York: Springer Verlag [Google Scholar]
- Marschner H, Romheld V (1994) Strategies of plants for acquisition of iron. Plant Soil 165: 261–274 [Google Scholar]
- Neumann D, Zur Nieden U, Lichtenberger O, Leopold I (1995) How does Armeria maritima tolerate high heavy metal concentrations? J Plant Physiol 146: 704–717 [Google Scholar]
- Otegui MS, Capp R, Staehelin LA (2002) Developing seeds of Arabidopsis store different minerals in two types of vacuoles and in the endoplasmic reticulum. Plant Cell 14: 1311–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Supekova L, Nelson H, Nelson N (1996) A yeast manganese transporter related to the macrophage protein involved in conferring resistance to mycobacteria. Proc Natl Acad Sci USA 93: 5105–5110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomine S, Lelièvre F, Debarbieux E, Schroeder JI, Barbier-Brygoo H (2003) AtNRAMP3, a multispecific vacuolar metal transporter involved in plant responses to iron deficiency. Plant J 34: 685–695 [DOI] [PubMed] [Google Scholar]
- Thomine S, Wang R, Ward JM, Crawford NM, Schroeder JI (2000) Cadmium and iron transport by members of a plant transporter gene family in Arabidopsis with homology to NRAMP genes. Proc Nat Acad Sci USA 97: 4991–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dedaldechamp F, Gaymard F, Guerinot ML, Briat JF, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal SM, Malo D, Vogan K, Skamene E, Gros P (1993) Natural resistance to infection with intracellular parasites—isolation of a candidate for Bcg. Cell 73: 469–485 [DOI] [PubMed] [Google Scholar]
- Wintz H, Fox T, Wu YY, Feng V, Chen W, Chang HS, Zhu T, Vulpe C (2003) Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis. J Biol Chem 278: 47644–47653 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Figure S3
Table S1


