Abstract
In yeast, glucose depletion elicits a quick response in the transcription of stress-related genes. The main transcriptional activator that orchestrates this response is Msn2, whose nuclear localization and DNA binding are negatively controlled by the cAMP-dependent protein kinase (PKA). Msn2 activation by sudden glucose depletion correlates with a fast but transient decrease in phosphorylation of several sites in its nuclear localization signal (NLS). Here we show that protein phosphatase 1 (PP1) is the direct antagonist of PKA-dependent phosphorylation at the Msn2 nuclear import domain and therefore a potential mediator of glucose starvation signals that target this transcription factor. Apart from PKA, the protein kinase Snf1 can also directly modify one of the Msn2 phosphorylation sites (S582) and thereby repress Msn2 function. Consequently, in snf1 mutants, rephosphorylation of the NLS happens to be much slower during prolonged starvation. Thus, a second, Reg1-dependent form of PP1 indirectly influences Msn2 functionality by modulating Snf1 kinase activation and repression. Different activities of PP1 are therefore involved in shaping induction and adaptation of the transcriptional stress response during acute glucose starvation.
Keywords: glucose sensing, Msn2, PP1, Snf1, transcriptional regulation
Introduction
Within their natural habitat, yeast cells have to make many metabolic adjustments in response to changes in extracellular nutrient conditions. Such alterations are generally associated with broad effects on gene expression. In a detrimental growth environment genes involved in glucose metabolism are downregulated, whereas genes such as HSPs and CTT1, associated with stress protection mechanisms, become upregulated (Rep et al, 1999; Gasch et al, 2000). Still, yeasts must always be ready to quickly return to optimal metabolic means when the milieu becomes favorable, emphasizing the importance of adaptive mechanisms (Kaniak et al, 2004). Under standard culture conditions, when supplied with sufficient nutrients, including a fermentable carbon source such as glucose, yeast cells first grow exponentially until glucose levels become depleted. Ultimately, cells will reach stationary phase and enter into a quiescent state. All these relatively slow growth transitions are associated with major changes in transcription. A sudden lack of glucose in exponentially growing cells will also cause a major shift in gene expression. Its pattern mimics to a considerable extent the pattern observed under environmental stress situations (Gasch et al, 2000), suggesting that glucose deprivation and stress might target a common regulatory node.
Among the protein kinases implicated as key elements in the nutritional transitions mentioned above, PKA plays a central role in growth-related phenomena during fermentation. By targeting certain transcription factors, PKA also controls the induction of growth-related genes and the repression of stress-related genes. For instance, a defect in the PKA inhibitory subunit Bcy1 leads to high constitutive kinase activity and causes sensitivity to starvation and stress, whereas cells with weak cAMP production and therefore low PKA activity (e.g. ras2 mutants) display hyperaccumulation of glycogen and higher stress resistance (Rolland et al, 2002).
In contrast, Snf1, the ortholog of the mammalian AMP-activated protein kinase in Saccharomyces cerevisiae (S.c.), has no well-defined function for glucose-growing cells, although it assumes a central role during glucose derepression (Jiang and Carlson, 1997). High glucose levels normally inhibit the expression of multiple genes including those involved in the utilization of alternative carbon sources, gluconeogenesis, and respiration. Glucose repression is established and maintained by the transcription factor Mig1 and its corepressors (Carlson, 1999). While Mig1 is inactivated by the Snf1 kinase complex, Snf1 is in turn activated upon glucose depletion (Wilson et al, 1996). Additionally, the nutrient-regulated protein phosphatase 1 (PP1) complex plays an important role by negatively regulating Snf1 upon readdition of glucose (Sanz et al, 2000). Besides its catalytic subunit Glc7, the PP1-targeting subunit Reg1 plays a key role in this process, since reg1 cells are unable to establish glucose repression while exhibiting constitutive levels of active Snf1. The protein phosphatase acting on Mig1 upon addition of glucose has not been identified so far. Indirect evidence suggests PP1, as Glc7–Reg1 complex, to be a candidate because Mig1 is constitutively phosphorylated and cytosolic both in reg1 and glc7 cells (Treitel et al, 1998; McCartney and Schmidt, 2001). Alternatively, constitutive Snf1 activity could explain the constant phosphorylation of Mig1 in reg1 cells. Glucose signals, therefore, seem to have opposite effects towards activation of Snf1 and PKA. Indeed, many phenotypes associated with the loss of Snf1 function are similar to those allied with PKA hyperactivity (Hubbard et al, 1992).
During exponential growth and throughout the diauxic growth transition, the transcriptional adjustment of stress-protecting genes is coordinated by the two closely related transcription factors Msn2 and Msn4. Indeed, variations in Msn2 localization and activity have been used as a sensitive indicator of nutrient fluctuations and environmental stress conditions (Schmitt and McEntee, 1996). According to several DNA microarray analyses, the Msn2-regulon may consist of more than 150 genes (Gasch et al, 2000; Causton et al, 2001). High PKA activity normally holds this STress Responsive Element (STRE)-driven gene expression down and appears to set a threshold response to environmental stress signals (Marchler et al, 1993; Görner et al, 1998). Immune fluorescence studies with Msn2-GFP chimeras showed that most of Msn2 resides in the cytosol when PKA activity is high. However, either stress or acute glucose depletion induces rapid nuclear accumulation of Msn2 (Görner et al, 1998), an effect found to be reversible by simple removal of the stress factor or by increasing PKA activity. A nuclear export sequence (NES) was identified in the Msn2 protein, and its functionality was demonstrated to be dependent on the nuclear exportin Msn5 (Görner et al, 2002). Increased nuclear import can apparently also be achieved by dephosphorylation of sites in the Msn2 nuclear localization signal (NLS). This event occurs upon downregulation of PKA or under acute glucose deprivation. In fact, the identity of the phosphorylation sites has been determined and the direct involvement of PKA is well documented (Görner et al, 2002). Nuclear import of Msn2 is dependent on the karyopherin Kap123 that may preferentially interact with the nonphosphorylated form of the Msn2-NLS (W Görner, unpublished data). Finally, there is evidence that STRE binding of nuclear Msn2 is also induced by stress and antagonized by PKA, but the underlying mechanisms are not well defined (Görner et al, 1998).
The rapid dephosphorylation of Msn2 under glucose depletion suggests that protein phosphatases must play an important role in the activation of this transcription factor. However, the relatively transient nature of dephosphorylation under prolonged glucose depletion hinted at a more complex regulatory network that could include additional players at the protein kinase and/or the protein phosphatase level. Indeed, previous genetic and cell biological observations have implicated PP1 as well as Snf1 in the regulatory networks around Msn2 function, as high copy numbers of the transcription factor could suppress certain Snf1 defects (Estruch and Carlson, 1993). More recently, Sanz and colleagues reported genetic evidence for the participation of Snf1 in the inhibition of Msn2 nuclear accumulation in response to glucose depletion (Mayordomo et al, 2002). However, neither the element responsive to the inactivation by Snf1 was identified, nor was a direct biochemical relationship to the kinase or its phosphatase regulator established. PP1 plays an important role in regulating various processes in eukaryotic cells. In budding yeast, Glc7 was demonstrated to be required for the regulation of, for instance, translation, cell cycle progression, chromosome segregation, glycogen accumulation, and glucose repression (Ceulemans and Bollen, 2004). In order to keep these regulation mechanisms independent, specialized targeting subunits are thought to restrict the function of the phosphatase to certain substrates (Bollen, 2001). For example, Reg1 was primarily identified as a cytosolic targeting factor that specifies inactivation of the Snf1 kinase by PP1 in glucose-growing cells. Glucose depletion has therefore been connected with the inactivation of the Reg1-dependent PP1 complex (Ludin et al, 1998; Sanz et al, 2000). More recently, the Glc7–Bud14 complex has been positively implicated in the regulation of Msn2-driven gene transcription during the diauxic shift (Lenssen et al, 2005); however, the target of this phosphatase complex remains unidentified. Here we focus on clarifying the function of Snf1 and PP1 towards one of the transport domains of Msn2, namely, the Msn2-NLS.
Results
The NLS of Msn2 contains a putative Snf1 kinase motif
The transcription factor Msn2 contains 704 amino acids with a NES, a NLS enclosing at least four phosphorylation sites, a zinc-finger region and several, but not well-defined, activation domains (Görner et al, 2002). In silico search for potential phosphorylation sites in Msn2 revealed that one PKA modification site overlapped with a Snf1 consensus motif (Dale et al, 1995) at amino acid 582 within the NLS. Since the NLS phosphorylation has been shown to correlate with inhibition of Msn2 import function, phosphorylation by Snf1 was also expected to attenuate Msn2 function. To further analyze the system, we raised antibodies specifically directed against phosphoserines of the Msn2 protein, namely, S582 and S620. Using these specific reagents on extracts from wild-type cells, we could demonstrate that both sites of Msn2-NLS, S582 and S620, are phosphorylated during growth on glucose, while rapidly becoming dephosphorylated upon acute glucose depletion. Both sites were rephosphorylated when glucose was added to the cell cultures, or after prolonged starvation (Figure 1A). According to previous data, it was thought that PKA was mainly responsible for the phosphorylation and inactivation of Msn2 (Görner et al, 2002). In agreement with this model, we found that in cells devoid of PKA activity (tpk1wtpk2tpk3 and tpk1tpk2tpk3yak1) Msn2 is still present but not phosphorylated on S620 (Figure 1A). Interestingly, a certain level of phosphorylation was still noticeable on S582 in these cells, suggesting that other protein kinases might also be able to recognize this site in vivo.
Figure 1.
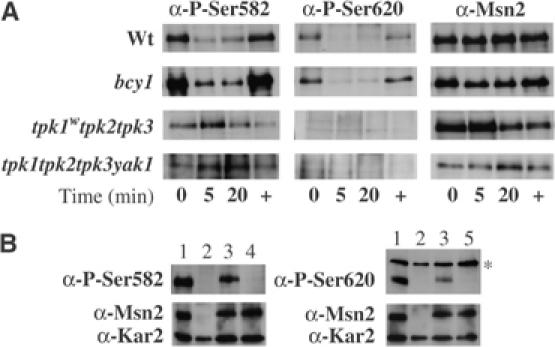
PKA activity is not the sole regulator for the phosphorylation pattern of the Msn2-NLS upon acute glucose depletion. (A) Western analyses were performed with crude protein extracts obtained from wild type (wt), bcy1-, tpk1wtpk2tpk3-, or tpk1tpk2tpk3yak1-mutant cells before (0) and during acute glucose depletion (5, 20), and 20 min after glucose re-addition (+). Endogenous levels of Msn2 were detected with polyclonal α-Msn2 antibodies. Msn2 phosphorylation was monitored with the specific antibodies α-P-S582 and α-P-S620. (B) Western analyses with protein extracts from wild type (1), msn2msn4-mutants (2), and mutants expressing Msn2 (3), Msn2-S582A (4), or Msn2-S620A (5) with α-P-S582, α-P-S620, and α-Msn2 antibodies. Loading was monitored with α-Kar2 antibodies. The α-P-S620 antibodies recognize an additional protein unspecifically at 150 kDa (*).
If PKA is solely responsible for the inactivation of Msn2, then, in a strain lacking the Bcy1 inhibitory subunit of PKA, Msn2 is expected to remain phosphorylated and inactive. Yet, we show that both S582 and S620 are still rapidly dephosphorylated in bcy1 cells (Figure 1A). Our observations suggest that either a phosphatase must activate the Msn2-NLS or that glucose starvation signals might also target the catalytic subunits of PKA. The loss of Western signal with protein extracts from msn2msn4 cells or from these cells expressing Msn2 with single alanine substitutions for S582 or S620 indicated that the antibodies used were specific for the phosphorylated forms of Msn2 at the respective sites (Figure 1B).
PP1 is involved in the regulation of Msn2
To identify Msn2-targeted protein phosphatases, we searched for mutant strains unable to import Msn2-GFP into the nucleus upon glucose depletion. An ordered collection of deletion strains served as source for assaying candidate mutants (Supplementary Figure S2). We found that most mutations tested had no noticeable effect on Msn2 localization upon glucose depletion. Many regulatory subunit mutants of PP1, such as glc8 or gac1, had no defect with regard to the regulation of Msn2 nuclear trafficking (data not shown). However, reg1 cells were severely affected in their response to acute glucose starvation, even though the cells could partially import Msn2-GFP upon osmotic shock (0.5 M NaCl). Interestingly, Reg1 was depicted as an important regulator of glucose signals towards Snf1 (Ludin et al, 1998) and was also identified as an effector of Msn2-driven transcription independently of our studies (Mayordomo et al, 2002). Overall, our observations were in good agreement with those reported by this lab.
We therefore tested whether an impediment of the dephosphorylation process at PKA-dependent modification sites could explain the cytoplasmic retention of Msn2. Figure 2A shows that in reg1 cells the Msn2-NLS remains predominantly phosphorylated even upon glucose depletion. Interestingly, this effect was noticeable for both of the sites tested. The role played by Reg1 in the regulation of the Msn2-NLS could be due to an indirect role of PP1 (Glc7-Reg1) through its effect on the Snf1 kinase, but also due to a direct role, possibly shared with other PP1 complexes. We therefore investigated the effects of a mutated version of Glc7, glc7-T152K, that is ineffective in binding Reg1 and supposedly affects the role of PP1 in Snf1 repression (Tu and Carlson, 1994, 1995). Once activated by glucose depletion, Snf1 was proposed to be maintained in its active state in this strain. As shown in Figure 2A, Msn2-S582 remained phosphorylated even when glucose was depleted from glc7-T152K cultures, whereas S620 was still dephosphorylated under the same conditions. Using a double-mutant strain glc7-T152Ksnf1, we could confirm that the effect on S582 was solely due to the activity of Snf1 (Figure 2A) and that the mutated form of the PP1 catalytic subunit did not affect Msn2-NLS dephosphorylation. Moreover, we could show that Bud14 had no role in the dephosphorylation of the Msn2-NLS under the condition tested (Figure 2A).
Figure 2.
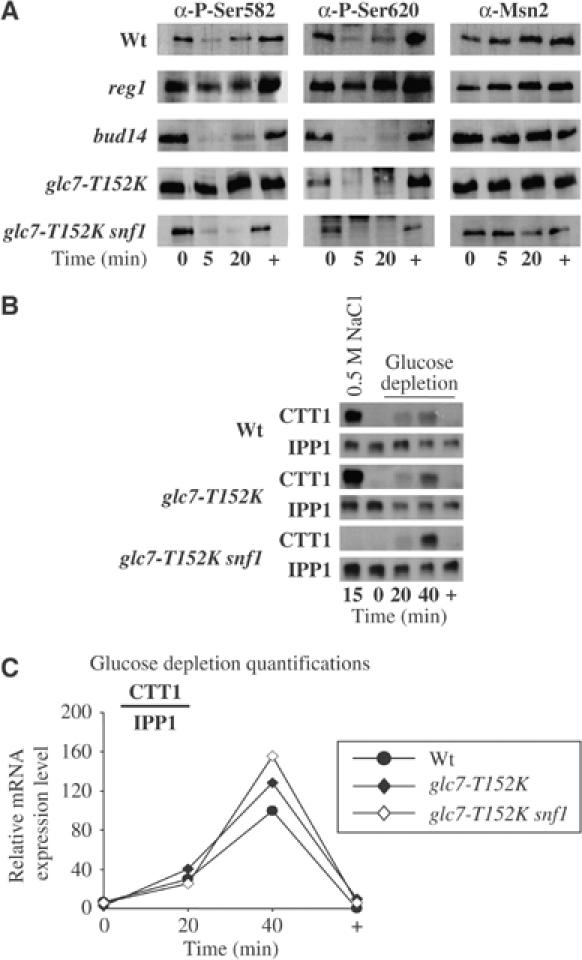
Glc7 and Snf1 participate in the regulation of Msn2. (A) Unregulated Snf1 activity (glc7-T152K and reg1) leads to aberrant phosphorylation of the Msn2-NLS. Wild type (wt), reg1-, bud14-, glc7-T152K- and glc7-T152Ksnf1-mutant cells were grown and treated as described in Figure 1, as were the subsequent Western analyses. (B) Constitutive Snf1 activity does not prevent Msn2 activation upon acute glucose depletion. Wt ( ), glc7-T152K (
), glc7-T152K ( ), and glc7-T152Ksnf1(
), and glc7-T152Ksnf1( )-mutant cells were grown to mid-exponential phase (0) and either treated with 0.5 M NaCl for 15 min (15) or subjected to acute glucose depletion for 20 and 40 min (20, 40), followed by immediate re-addition of glucose; a final sample was taken after 20 min (+). Total RNA was analyzed simultaneously for the expression of the STRE-driven gene CTT1 and the house-keeping gene IPP1. (C) The expression of CTT1 before, during, and after glucose depletion was normalized for each time point against the corresponding IPP1 expression level. Quantifications were plotted as a function of the highest expression level in wt cells, which was set to 100.
)-mutant cells were grown to mid-exponential phase (0) and either treated with 0.5 M NaCl for 15 min (15) or subjected to acute glucose depletion for 20 and 40 min (20, 40), followed by immediate re-addition of glucose; a final sample was taken after 20 min (+). Total RNA was analyzed simultaneously for the expression of the STRE-driven gene CTT1 and the house-keeping gene IPP1. (C) The expression of CTT1 before, during, and after glucose depletion was normalized for each time point against the corresponding IPP1 expression level. Quantifications were plotted as a function of the highest expression level in wt cells, which was set to 100.
To test whether the aberrant phosphorylation of Msn2 in glc7-T152K mutants had consequences for its physiological function, we followed the expression of CTT1 upon glucose depletion (Figure 2B and C). Our results show that Msn2 is still active in glc7-T152K mutants as compared to wild-type cells. This suggests that the inability of glc7-T152K to interact with Reg1 does not hinder Msn2 function. The additional deletion of SNF1 has no effect on CTT1 transcriptional induction upon glucose depletion, suggesting that Snf1 is specifically required for inactivation of Msn2 after prolonged glucose depletion. It is interesting to note that glc7-T152Ksnf1 double mutants are unable to respond to osmotic stress, supporting the idea that Snf1 plays a decisive role in stress-induced signal transduction (Sanz, 2003).
Snf1 directly phosphorylates serine 582 of Msn2-NLS
Considering that S582 was embedded in an Snf1 consensus motif, we attempted to show that Msn2 is a substrate of this kinase. We transformed snf1 cells with plasmids expressing either HA-tagged Snf1 or its catalytically inactive Snf1K84R counterpart. Figure 3A shows that only cells containing the active version of Snf1 were able to grow on ethanol or galactose as the sole carbon source. We isolated Snf1 complexes from these strains (Figure 3B) and tested their activity towards the Msn2-NLS in vitro. GST-extended Msn2 truncations were purified and used as substrate in a kinase assay with [γ-32P]ATP. Figure 3C clearly shows that Snf1 can specifically phosphorylate the Msn2-S582. The control with purified inactive Snf1K84R kinase demonstrates that the phosphorylation of the S582 was solely due to the activity of Snf1. Experiments with purified PKA illustrated the fact that PKA could phosphorylate all four serines of the NLS, namely, S582, S620, S625, and S633. Only those constructs with the serines substituted to alanines lacked the appropriate phosphorylation signals.
Figure 3.
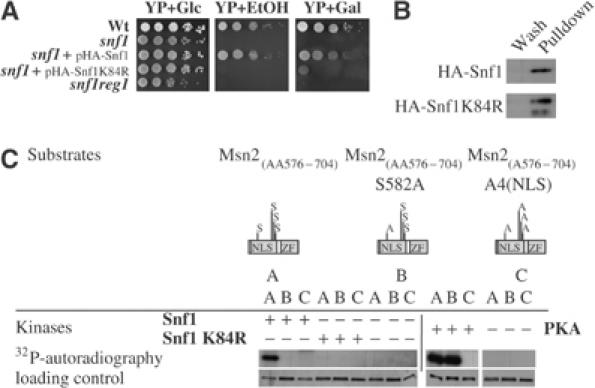
Msn2 is substrate for Snf1 in vitro. (A) Snf1 activity is required for growth on nonfermentable carbon sources. Wild type (wt), snf1-, snf1-expressing Snf1-HA, or Snf1-K84R-HA and reg1snf1-mutant cells were grown to OD600 nm=1.0. Then, 10-fold serial dilutions were spotted onto a rich medium (YP) with either a fermentable (glucose—Glc) or nonfermentable carbon source (ethanol—EtOH or galactose—Gal). Plates were incubated at 30°C for 3 days. (B) Snf1 and Snf1-K84R were purified from crude protein extracts obtained from exponentially grown snf1-mutant cells expressing either Snf1-HA or Snf1-K84R-HA and analyzed by Western blot of equal aliquots of the last washes and the eluates, with monoclonal α-HA antibodies. (C) Equal amounts of the substrates GST–Msn2(AA576–704) (A), GST–Msn2(AA576–704)S582A (B) and GST–Msn2(AA576–704)A4(NLS) (C) were incubated with [γ-32P]ATP and Snf1, Snf1-K84R, or PKA. Control assays (lanes 7–9 and 13–15) lacked the appropriate purified kinase. Samples were separated by SDS–PAGE and analyzed by autoradiography and silver staining.
A role for PP1 in the regulation of Msn2 activity
Our in vivo observations in reg1 and glc7T152K cells suggest a dual role for PP1 in the regulation of Msn2: (1) indirect via regulation of the Snf1 kinase complex and (2) direct via dephosphorylation of Msn2. As in S.c. the gene GLC7 is essential, we chose to work with a temperature-sensitive mutant strain (Andrews and Stark, 2000). Our results demonstrate that in glc7-10ts cells the Msn2-NLS did not become dephosphorylated upon glucose depletion at restrictive temperature (Figure 4A). This effect seems to be independent of the presence of Snf1, since similar results were observed in glc7-10tssnf1 cells. In wild-type cells at restrictive temperature, Snf1 activity may mask the dephosphorylation of S582. We also tested the effect of Glc7 inactivation on the expression of CTT1, demonstrating that upon glucose depletion Msn2 transcriptional activity is greatly affected by the inactivation of Glc7 (Figure 4B).
Figure 4.
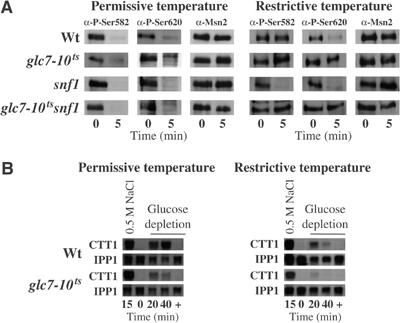
Glc7 is necessary for the dephosphorylation of the Msn2-NLS upon acute glucose depletion. (A) Inactivation of Glc7 prevents dephosphorylation of the Msn2-NLS upon acute glucose depletion. Wild-type (wt), glc7-10ts-, snf1-, and glc7-10tssnf1-mutant cells were grown to OD600 nm=0.6. Then, cultures were split and each half incubated at permissive (26°C) or restrictive (37°C) temperature for 2.5 divisions. Samples were taken before (0) and 5 min after (5) acute glucose depletion. Western analyses of crude protein extracts were performed as described in Figure 1. (B) Total RNA was analyzed simultaneously for the expression of CTT1 and IPP1 as in Figure 2.
In an attempt to determine whether the Msn2 dephosphorylation defects in glc7-10ts cells were due to a direct interaction or due to a more indirect effect caused by a general change in cell physiology, we established an in vitro phosphorylation/dephosphorylation assay. We obtained the Msn2 substrates as bacterially expressed GST fusions. These proteins were radioactively labeled with affinity-purified yeast PKA. For GST–Msn2(AA576–704), most of the phosphate groups are targeted to the four PKA sites within the NLS because a mutant product collectively affecting these sites remained virtually unphosphorylated (Görner et al, 2002). Enzyme assays were performed with PP1 phosphatase pulled down from glc7 cells expressing a PrA-tagged version of Glc7 or wild-type cells expressing the PrA-tag only. The phosphorylated substrates GST–Msn2(AA576–704), GST–Msn2, and calf histone were monitored for loss of radioactivity (Figure 5A). Control experiments showed that PKA-phosphorylated calf histone was not a substrate for PP1, while λ-phosphatase removed the modifications in all substrates. As shown in Figure 5A, the purified Glc7 complex can dephosphorylate both Msn2 substrates in vitro. However, a large amount of residual radioactivity remained in phospho-labeled full-length Msn2. The discrepancy observed between both Msn2 substrates in dephosphorylation efficiency reveals that Msn2 may contain sites that are nonspecifically phosphorylated by PKA, that Glc7 action on Msn2 is specifically targeted towards the NLS, or that the full-length protein is generally less recognized as substrate. Definitely, when we used the full-length GST–Msn2 as a substrate, we observed that PKA phosphorylated more sites than the strict consensus motifs proposed by Görner et al (2002). Incubation of phospho-Msn2 with a protein complex pulled down via the Reg1 subunit (Reg1-HA) did not lead to variations in the intensity of Msn2 phosphorylation even though the complex contained Glc7 (Figure 5B) and was positive in a phosphatase activity assay on myelin basic protein (data not shown). Together with our in vivo analysis (Figure 4A and B), these data fully support our contention that Reg1-independent PP1 activity is specifically directed against the Msn2-NLS.
Figure 5.
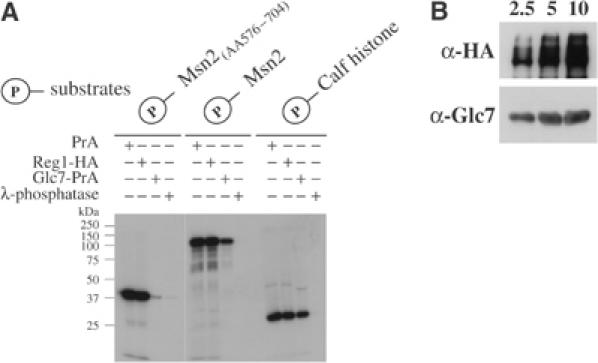
(A) Phosphorylated Msn2-NLS is substrate for Glc7 in vitro. Purified substrates GST–Msn2(AA576–704), GST–Msn2, and calf histone were first phosphorylated in vitro with yeast PKA and subsequently incubated with excess of affinity-purified PrA, Reg1-HA, Glc7-PrA, or with λ-phosphatase. Samples were separated by SDS–PAGE and remaining phosphorylation was determined by autoradiography. (B) Reg1-HA complex contains Glc7. Western analysis of increasing amounts of Reg1-HA-attached beads, 2.5, 5, and 10 μl, for the presence of Reg1-HA and Glc7.
A direct role for Snf1 in stress adaptation
The involvement of Snf1 in Msn2 regulation would explain that in reg1 cells Msn2 is unable to activate STRE-gene transcription upon glucose depletion (Mayordomo et al, 2002). We therefore studied the phosphorylation pattern of Msn2 both in snf1 and in reg1snf1 cells. In wild-type cells, the phosphorylated serines of the NES (S288) and NLS (S582 and S620) are rapidly dephosphorylated and become rephosphorylated by an unknown kinase about 15 min after glucose depletion. Figure 6A shows that phosphorylation of S288 is still regulated in the mutants. In contrast, rephosphorylation of the NLS is greatly delayed after longer glucose depletion in snf1-mutant cells. This observation supports a crucial role for Snf1 in the inactivation of Msn2-NLS on S582, which would be followed by the action of PKA on S620 and the consequent adaptation of the cells to the absence of glucose. When glucose was added to wild-type cells, Msn2 was rapidly exported from the nucleus and rephosphorylated probably by the reactivated PKA, which seems to be partially able to phosphorylate S582. This hypothesis is confirmed by the analysis of the double mutant reg1snf1, in which the Msn2-NLS remained dephosphorylated until glucose was added to the cells. In order to confirm our assumption on Msn2 activity, we analysed the expression of CTT1. In wild-type cells, CTT1 transcripts were detectable 30 min after glucose depletion for about 2 h, whereas in reg1 cells no CTT1 transcript could be detected. In snf1 cells, expression of the CTT1 reporter gene did not cease even 3 h later (Figure 6B). However, when glucose was added, all responding cells stopped to transcribe CTT1. This demonstrates the involvement of two independent regulation mechanisms to inactivate Msn2: (1) an Snf1-dependent mechanism in the absence of glucose and (2) an Snf1-independent process in the presence of glucose (Figure 6B). The latter is most likely attributed to a constant PKA activity in glucose-growing cells.
Figure 6.
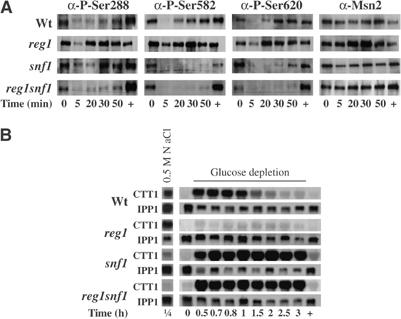
Snf1 is required for the inactivation of Msn2 during prolonged glucose depletion. (A) Snf1 is needed to rephosphorylate the Msn2-NLS during glucose depletion. Western blot analyses were performed with crude protein extracts obtained from wild type (wt), reg1-, snf1-, and reg1snf1-mutant cells before (0) and during acute glucose depletion (5, 20, 30, 50) and 20 min after glucose re-addition (+). Extracts were analyzed as described in Figure 1, with the additional use of α-P-S288 antibodies. (B) Snf1 is required to inactivate STRE-driven gene expression during prolonged glucose depletion. Wt, reg1-, snf1-, and reg1snf1-mutant cells were grown to mid-exponential phase (0) and either treated with 0.5 M NaCl for 15 min (1/4) or subjected to acute glucose depletion for the indicated time points, followed by immediate re-addition of glucose with the final sample taken after 20 min (+). Total RNA was analyzed simultaneously for the expression of CTT1 and IPP1.
To test whether Snf1 activity is responsible for the lack of Msn2 activity in reg1 cells, we also analyzed the CTT1 expression levels in reg1snf1 cells. In the absence of REG1, Glc7 cannot act on Snf1; therefore, the kinase is constitutively active. With the additional absence of SNF1, Msn2 should not be instantaneously inactivated; consequently, the effect of the REG1 deletion should be directly visualized on CTT1 expression pattern. Indeed, our Northern analyses corroborate this proposition, while no CTT1 expression can be detected in reg1 cells; reg1snf1 cells present a 2.5-fold higher expression than wild-type cells under the same conditions (Figure 6B). It is noteworthy that reg1snf1 cells are unresponsive to stress, as shown by the absence of CTT1 transcript upon osmotic shock.
Snf1 initiates adaptation to the lack of glucose at the Msn2-NLS level
To illustrate whether Snf1 is required for Msn2 cytoplasmic relocalization under prolonged glucose starvation, we followed the location of Msn2-GFP in vivo. To distinguish msn2msn4 from msn2msn4snf1 cells, we additionally expressed the DsRed fused mitochondrial protein ATPase subunit 9 only in msn2msn4 cells. We mixed both cell cultures just before observation and directly followed their fluorescence. Figure 7A shows that about 150 min after glucose depletion cells with DsRed labeling had relocalized their Msn2-GFP to the cytoplasm, whereas in snf1 mutants GFP remained fully nuclear. The observation demonstrates that Snf1 function is important for the decrease of Msn2 nuclear import rates. We therefore asked whether single amino-acid substitutions in Msn2 at position 582 could mimic the absence or presence of Snf1 activity. We expressed Msn2-S582A and Msn2-S582D in msn2msn4 cells and followed phosphorylations at S288 and S620, as well as the transcriptional activity of Msn2. As shown in Figure 7B and C, the status of S582 has a considerable influence on the overall phosphorylation pattern and function of Msn2. In the context of S582A, which mimics a dephosphorylated site, Msn2-S620 rephosphorylation was highly retarded, whereas, in the context of S582D, Msn2-S620 was prematurely rephosphorylated. To confirm that Msn2 activity was impaired by these mutations, we also followed the expression of CTT1 in the depicted strains. As predicted, cells expressing Msn2-S582A produced higher CTT1 transcript levels than cells expressing Msn2, whereas cells expressing Msn2-S582D produced much lower CTT1 transcript levels. Our data strongly suggest that the phosphorylation of S582 by Snf1 constitutes a priming event required for the full rephosphorylation of the Msn2-NLS and the decrease in Msn2 nuclear concentration.
Figure 7.
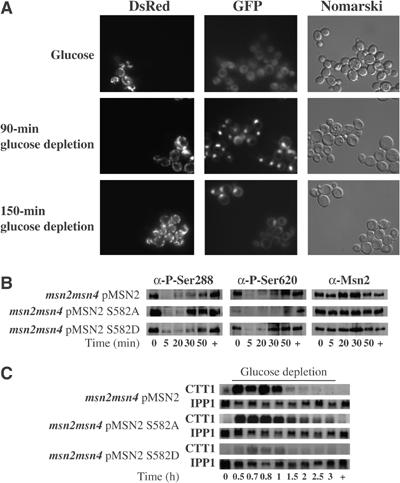
Snf1 is required for the export of Msn2 from the nucleus and its inactivation upon prolonged glucose depletion. (A) msn2msn4-mutant cells expressing ATPaseSU9-DsRed and Msn2-GFP and msn2msn4snf1-mutant cells expressing only Msn2-GFP were grown to midexponential phase and mixed while subjected to acute glucose depletion for the indicated time points. (B, C) msn2msn4-mutant cells expressing Msn2, Msn2-S582A, or Msn2-S582D were grown and treated as described in Figure 6, as were the subsequent Western (B) and Northern (C) analyses.
Discussion
Since the level and turnover of the transcription factor Msn2 remain stable through short periods of stress and even through complex alterations in nutrient supply, most changes in Msn2 activity must be attributed to the regulation of either its post-translational modifications or its interactions with regulatory partners. Msn2 was already described to be hyperphosphorylated upon heat shock (Garreau et al, 2000) and rapidly dephosphorylated after acute glucose removal (Görner et al, 2002), indicating that different independent mechanisms participate in its regulation. Similarly, the function of PP2A was proposed to be essential to regulate Msn2 activity under stress conditions, but not under glucose depletion (Santhanam et al, 2004). In this study, we further investigated the influence of glucose signaling on the activation state of Msn2. As documented previously, PKA controls at least partially the activity of Msn2 via direct phosphorylation (Görner et al, 2002). Here we develop this theme in several important directions. Our results show that PP1 plays a crucial role in the regulation of Msn2 by directly antagonizing phosphorylation at consensus motifs that regulate its NLS. We could clarify the role of Snf1 in the downregulation of Msn2 during prolonged glucose starvation, as it modifies the S582, a site recognized by PKA during exponential growth with glucose. Finally, the results of our study may serve as a paradigm to describe the complexity of resolving a network of signaling communications in which both kinases and phosphatases participate in multiple regulatory interactions.
The previous contention that the inhibition of PKA was the main cause for Msn2 activation during glucose starvation was based on experiments with bcy1 cells. These mutants seemed to have much less pronounced effects on phospho-Msn2 after glucose withdrawal than the corresponding wild-type cells (Görner et al, 2002). While repeating the analysis of bcy1 cells, we observed a much less drastic effect on Msn2 regulation. The results presented here would suggest that glucose deprivation is either affecting the access and function of the PKA catalytic subunits towards their substrates, or that a sizeable part of Msn2-NLS dephosphorylation could ensue through an increase in specific phosphatase activity. The predominant regulation of a C2H2 zinc-finger transcription factor by a phosphatase is not without precedent in yeast. Crz1, a factor quite related to Msn2 with regard to its overall structure, is also modified and inhibited by PKA in its nuclear import domain. In this case, the activation of the Ca2+/calmodulin-dependent phosphatase calcineurin seems to exert the main control on the behavior of the transcription factor (Kafadar and Cyert, 2004).
The differences in results and conclusions between the study from Görner et al (2002) and ours might have a trivial cause. We suspect that their quantifications might have been distorted because their measurements were within or close to the nonlinear range of Western analysis. For instance, in exponentially growing cells, the authors did not observe a distinct increase of Msn2-NLS phosphorylation in bcy1 cells. In contrast, we have consistently measured a four- to five-fold increase in phosphorylation in bcy1 cells compared to wild-type cells.
The assignment of a direct role for a protein phosphatase has been notoriously difficult. Assessments based on genetic data can be misleading due to the rather pleiotropic effects on the available mutants, while biochemical approaches can be hampered by either the loss of necessary adaptors for the substrate or a highly unstable behavior of the enzyme in vitro. These problems have been particularly notable for PP1 function, owing to the large number and variety of regulatory subunits. A role for PP1 in the regulation of Msn2 has already been projected by previous work (Mayordomo et al, 2002). However, in this study, the genetic evidence was obscured by the fact that Snf1 appeared as the main regulatory component affected by the loss of the PP1 subunit Reg1. Although reg1 mutants presented a severe defect in Msn2 localization, snf1reg1 double mutants behaved again like wild-type cells. Still, the authors never investigated whether the absence of Glc7 activity would mimic the reg1-mutant effects and if so whether these were suppressed by the loss of SNF1. Our studies show that, in contrast to reg1snf1 mutants, in glc7-10tssnf1 mutants the Msn2-NLS still exhibits a strong de-phosphorylation defect in response to glucose starvation. Therefore, the reasonable assumption is that PP1 is indeed the phosphatase that antagonizes PKA activity at Msn2-NLS. It should be noted, however, that the Msn2 dephosphorylation defect in glc7-10ts cells was difficult to document, first of all because it takes some time to inactivate the catalytic subunit and second because prolonged heat stress itself appears to eventually cause an increase in PKA and Snf1 kinase activities. Nevertheless, our data illustrate that the phosphatase must have a role that is independent of Reg1 and its substrate Snf1, a conclusion that was corroborated by our biochemical analysis. Immune purification of the protein complex using tagged Reg1 did not yield any material with phosphatase activity towards Msn2, whereas the complex purified via Glc7 could dephosphorylate its target (Figure 5). Thus, there should be little doubt that the phosphatase directly acts on these sites.
Bud14 has recently been proposed as putative Msn2-regulating factor during or after diauxic shift (Lenssen et al, 2005). However, our study shows that bud14 cells had no defect during acute glucose depletion and we still do not know the regulatory subunits required for the recognition of Msn2. Since a large set of additional candidate mutants did not yield the expected phenotype (Supplementary Figures S1 and S2), it could be that lethality or redundancy prevented us from identifying such factors in a genetic approach. One can even envisage the existence of multiple docking sites with different recognition features for different PP1 complexes, possibly distributed throughout the whole Msn2 protein. Nevertheless, structures recognized by the phosphatase must be embedded within a 120 amino-acid domain containing the NLS- and DNA-binding domain, since PP1-dependent dephosphorylation is specific for such a fragment both in vitro and in vivo. In fact, the Msn2-NLS might be one of the first genuine targets that can be used as a specific in vitro substrate of PP1, perhaps opening a biochemical approach towards the identification of the regulatory subunits.
The effects of glucose starvation on the phosphorylation level of Msn2 are only transient (Figure 6). One can notice significant rephosphorylation of Msn2 about 20 min after glucose withdrawal, an effect that will eventually result in a decrease of stress-related gene expression. According to our results, Snf1 must not only play a crucial role in this adaptive response but also a direct one. First, we demonstrate a correlation between the transcriptional response and Msn2-S582 dephosphorylation in snf1 cells. Second, we show that Snf1 could directly phosphorylate Msn2-S582 in vitro. Yet, the consequences of S582 phosphorylation by Snf1 are quite dramatic as cells with high Snf1 activity lack any Msn2 response to glucose starvation (Mayordomo et al, 2002). We suspect that this result is due to a rather complete phosphorylation of the pool of Msn2 owing to a high affinity of Snf1 kinase for this specific site. At normal levels, PKA seems to lead to only a smaller amount of molecules carrying the S582 modification (about 20%) if we assume that bcy1 mutants are close to saturation. Such a view is also supported by the fact that S582 phosphorylation levels are elevated to the same extent in reg1 and bcy1 mutants. Moreover, we observed that modification at S582 partly functions as a priming event for the phosphorylation of other PKA-dependent sites in Msn2-NLS. Subtle regulation of a transcription factor via post-translational modifications has already been described for Pho4 (Springer et al, 2003). Mutating specific sites within its localization domains, Springer et al (2003) could point out a negative correlation between the phosphorylation state of Pho4 and its binding capacity to promoters of the Pho-regulon. Conceptually similar to Pho4 regulation, differential phosphorylation of Msn2 could lead to activation of different subsets of STRE-containing genes, especially since the Msn2-dependent expression of a given gene differs according to environmental conditions (Amoros and Estruch, 2001).
Several recent observations seemed to connect the function of Snf1 kinase with PKA activity and stress signals particularly in cells under nutrient limitation, salt exposure and heat shock (Sanz, 2003; Hahn and Thiele, 2004; Hedbacker et al, 2004). A mutant analysis demonstrated that the absence of PKA activity resulted in the relocalization of some Snf1 kinase complexes, thereby raising the possibility that the PKA pathway participated in the regulation of Snf1 (Hedbacker et al, 2004). DNA microarray studies documented that about 30% of the heat shock factor (HSF) target genes are also induced by the diauxic shift. The authors showed that Snf1 directly phosphorylated HSF, and using chromatin immunoprecipitation they demonstrated that this phosphorylation event enhanced chromosomal HSF DNA binding to low-affinity target promoters, for example, SSA3 and HSP30 (Hahn and Thiele, 2004). Although we found some unexpected evidence for a positive role of Snf1 in Msn2-driven transcription upon osmotic shock, the mechanism and physiological relevance for this effect await further investigations. More importantly, in our study, we could clearly document an adaptive function for Snf1 kinase with regard to the general stress response. From classical genetic studies of the PKA pathway as well as the use of Msn2 hyperactive alleles, it has become apparent that a prolonged stress response has a rather detrimental effect on cellular growth and proliferation. To liberate the cells from growth arrest, several mechanisms have been proposed for the downregulation of Msn2. One study favored the view that the export rate of Msn2 might be increased by Srb10-induced phosphorylation (Chi et al, 2001). Other groups rather contented that higher degradation rates of Msn2 in the nucleus were responsible for adaptation (Durchschlag et al, 2004; Lallet et al, 2004; Bose et al, 2005). Here we add yet another adaptation mechanism that is specifically geared towards the situation after glucose deprivation. It relies on the direct phosphorylation of Msn2 by Snf1, a modification that may attenuate its nuclear import. Our current model (Figure 8) summarizes the complicated interplay between PP1 and the different protein kinases for the regulation of Msn2 function during acute glucose fluctuations.
Figure 8.

A switch diagram for the regulation of Msn2 upon acute glucose depletion. (1) Msn2 is phosphorylated and inactivated by PKA during growth on glucose, while the Snf1 kinase is kept in an inactive state by PP1 (Glc7-Reg1). (2) Glucose depletion results in Msn2 dephosphorylation by PP1-Glc7 associated with unknown regulatory subunit(s), followed by Msn2 nuclear accumulation and activation of STRE-driven genes. It also causes the rapid activation of Snf1 kinase. (3) Cells adapt to the loss of glucose as the rising level of Snf1 kinase throttles nuclear entry of Msn2.
Materials and methods
Strains, plasmids, growth conditions, immuno- and Northern blotting
Supplementary data contain detailed information on the strains (Supplementary Figures S1 and S2), plasmids, and growth conditions used in this study, and a description of the applied Northern and Western blot analysis procedures.
GST purification
Escherichia coli (DH5α) cells transformed with plasmids coding for GST-extended proteins were grown to mid-exponential phase in 50 ml 2 × YT and induced with 1 mM IPTG for 4 h. GST substrates were isolated using MicroSpin GST columns (Amersham) according to the manufacturer's instructions. Proteins were separated by SDS–PAGE and visualized by silver staining.
Yeast PKA purification
The purification of yeast PKA using a bcy1 mutant expressing a TAP-tagged version of Bcy1 was described before (Görner et al, 2002).
Snf1 purification
snf1-mutant cells expressing either pHA-Snf1 or pHA-Snf1K84R were grown in 500 ml YPD (1.2 × 107 cells/ml), harvested at 4000 g for 5 min, washed twice with water and resuspended in 5 ml ice-cold lysis buffer (IPP150, complete protease inhibitor cocktail EDTA-free (Roche), 2 mM PMSF). Cells were lysed with glass beads and centrifuged at 5000 g for 10 min (4°C). Lysates were clarified further by centrifugation at 10 000 g for 10 min (4°C) and incubation on a rotator for 12–14 h (4°C) with PanMouse IgG Dynabeads (DYNAL Biotech), chemically crosslinked (DMP-Sigma) with culture medium. Cleared lysates were incubated for 6 h on a rotator (4°C) with PanMouse IgG Dynabeads crosslinked with C12A5 hybridoma supernatant (α-HA). Subsequently, beads were collected by centrifugation (4°C), washed twice in 2 vol. IPP150, twice in 2 vol. washing buffer (IPP150, 0.5 mM EDTA, and 1 mM DTT) and resuspended in 400 μl Snf1-kinase buffer (20 mM HEPES, pH 7.0; 0.5 mM EDTA, pH8.0; 0.5 mM DTT; 5 mM Mg-acetate) containing 10% glycerol ready for storage.
Kinase assays
For each reaction, 10 μg of purified GST–Msn2 substrate was incubated with [γ-32P]ATP (5 μCi, 3000 Ci/mmol) and purified PKA, Snf1, or Snf1K84R for 30 min (30°C) in either PKA-elution buffer supplemented with 100 mM Tris, pH 7.4 and 2 mM MgCl2, or in Snf1-kinase buffer, respectively. Simultaneously, control assays were performed by omitting the respective purified kinase from the reaction. Reactions were terminated by the addition of an equal volume of preheated SDS-loading buffer and boiled for 10 min. Proteins were separated by SDS–PAGE, loadings were analyzed by silver staining, and incorporation of radioactive phospho-groups visualized by autoradiography.
PP1 assay
GST–Msn2 fusions and Calf Histone (Calbiotech) were phosphorylated in vitro with PKA. Phosphorylated substrates were separated by SDS–PAGE, eluted from the gel, and the amount of incorporated [γ-32P]ATP was quantified (Cerenkov). Each phosphatase assay was initiated with 1500 c.p.m. of phosphorylated substrate. Purification of Reg1-HA was performed as described for Snf1-HA. TAP purifications were carried out simultaneously from the yeast strains PWY3 and LKY150 using the same amounts of total proteins and IgG-Sepharose beads, according to Walsh et al (2002). Briefly, cells were grown in 500 ml YPD (1.2 × 107 cells/ml), harvested at 4000 g for 5 min and shock-frozen. Pellets were resuspended in 5 ml of lysis buffer (100 mM KCl, 0.1% Triton X-100, 0.1 mM EDTA, pH 8.0, 1 mM MgCl2, 50 mM HEPES–KOH, pH 7.5), supplemented with complete protease inhibitors (Roche). Cell suspensions were lysed with glass beads, centrifuged at 5000 g for 20 min, and cleared further by centrifugation for 30 min at 15 000 g. Cleared lysates were used immediately for overnight incubation (4°C), each with 500 μl IgG-Sepharose beads, washed three times in lysis buffer. The phosphatase assays were performed with 25 μl PrA-TEV-bound beads, PrA-TEV-Glc7-bound beads, Reg1-HA-bound beads, or 1 μl λ-phosphatase on phosphorylated GST–Msn2(AA576–704), GST–Msn2, and calf histone. Aliquots were analyzed for the presence of Glc7 by Western blot.
Fluorescence microscopy
Glucose depletion was performed as described in Supplementary data, and cells were visualized live unfixed. Cells were visualized with an × 63 objective from a Zeiss Axioplan 2 fluorescence microscope and images were collected with a Quantix CCD camera using the Lightview software and processed in Adobe Photoshop 8.0.
Supplementary Material
Fig. S1
Fig. S2
Acknowledgments
This publication is dedicated to the memory of H Ruis, who died during the progress of this work. We thank MJR Stark for supplying the strains PW3 and LKY150, M Schmidt for providing the Snf1-HA encoding plasmid, M Carlson for providing the Snf1K84R-HA and yeast strains, F Estruch for the α-Msn2 antibodies, and E Ogris for help with phosphatase assays. We are grateful to K Stanslicki for excellent technical assistance and J Norbeck, C Schüller for fruitful discussions. We thank K Mechtler for help with the antibody production. This work was supported by the Austrian Fonds zur Förderung der wissenschaftlichen Forschung P14653. WR was a STREP project ‘QUASI' (EU) LSHG-CT-2003-503230 fellow.
References
- Amoros M, Estruch F (2001) Hsf1p and Msn2/4p cooperate in the expression of Saccharomyces cerevisiae genes HSP26 and HSP104 in a gene- and stress type-dependent manner. Mol Microbiol 39: 1523–1532 [DOI] [PubMed] [Google Scholar]
- Andrews PD, Stark MJ (2000) Type 1 protein phosphatase is required for maintenance of cell wall integrity, morphogenesis and cell cycle progression in Saccharomyces cerevisiae. J Cell Sci 113 (Part 3): 507–520 [DOI] [PubMed] [Google Scholar]
- Bollen M (2001) Combinatorial control of protein phosphatase-1. Trends Biochem Sci 26: 426–431 [DOI] [PubMed] [Google Scholar]
- Bose S, Dutko JA, Zitomer RS (2005) Genetic factors that regulate the attenuation of the general stress response of yeast. Genetics 169: 1215–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M (1999) Glucose repression in yeast. Curr Opin Microbiol 2: 202–207 [DOI] [PubMed] [Google Scholar]
- Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA (2001) Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell 12: 323–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceulemans H, Bollen M (2004) Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol Rev 84: 1–39 [DOI] [PubMed] [Google Scholar]
- Chi Y, Huddleston MJ, Zhang X, Young RA, Annan RS, Carr SA, Deshaies RJ (2001) Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev 15: 1078–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale S, Wilson WA, Edelman AM, Hardie DG (1995) Similar substrate recognition motifs for mammalian AMP-activated protein kinase, higher plant HMG-CoA reductase kinase-A, yeast SNF1, and mammalian calmodulin-dependent protein kinase I. FEBS Lett 361: 191–195 [DOI] [PubMed] [Google Scholar]
- Durchschlag E, Reiter W, Ammerer G, Schüller C (2004) Nuclear localization destabilizes the stress-regulated transcription factor Msn2. J Biol Chem 279: 55425–55432 [DOI] [PubMed] [Google Scholar]
- Estruch F, Carlson M (1993) Two homologous zinc finger genes identified by multicopy suppression in a SNF1 protein kinase mutant of Saccharomyces cerevisiae. Mol Cell Biol 13: 3872–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garreau H, Hasan RN, Renault G, Estruch F, Boy-Marcotte E, Jacquet M (2000) Hyperphosphorylation of Msn2p and Msn4p in response to heat shock and the diauxic shift is inhibited by cAMP in Saccharomyces cerevisiae. Microbiology 146 (Part 9): 2113–2120 [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görner W, Durchschlag E, Martinez-Pastor MT, Estruch F, Ammerer G, Hamilton B, Ruis H, Schüller C (1998) Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev 12: 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görner W, Durchschlag E, Wolf J, Brown EL, Ammerer G, Ruis H, Schüller C (2002) Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J 21: 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn JS, Thiele DJ (2004) Activation of the Saccharomyces cerevisiae heat shock transcription factor under glucose starvation conditions by Snf1 protein kinase. J Biol Chem 279: 5169–5176 [DOI] [PubMed] [Google Scholar]
- Hedbacker K, Townley R, Carlson M (2004) Cyclic AMP-dependent protein kinase regulates the subcellular localization of Snf1-Sip1 protein kinase. Mol Cell Biol 24: 1836–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard EJ, Yang XL, Carlson M (1992) Relationship of the cAMP-dependent protein kinase pathway to the SNF1 protein kinase and invertase expression in Saccharomyces cerevisiae. Genetics 130: 71–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Carlson M (1997) The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol Cell Biol 17: 2099–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafadar KA, Cyert MS (2004) Integration of stress responses: modulation of calcineurin signaling in Saccharomyces cerevisiae by protein kinase A. Eukaryot Cell 3: 1147–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniak A, Xue Z, Macool D, Kim JH, Johnston M (2004) Regulatory network connecting two glucose signal transduction pathways in Saccharomyces cerevisiae. Eukaryot Cell 3: 221–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallet S, Garreau H, Poisier C, Boy-Marcotte E, Jacquet M (2004) Heat shock-induced degradation of Msn2p, a Saccharomyces cerevisiae transcription factor, occurs in the nucleus. Mol Genet Genomics 272: 353–362 [DOI] [PubMed] [Google Scholar]
- Lenssen E, James N, Pedruzzi I, Dubouloz F, Cameroni E, Bisig R, Maillet L, Werner M, Roosen J, Petrovic K, Winderickx J, Collart MA, De Virgilio C (2005) The Ccr4–Not complex independently controls both Msn2-dependent transcriptional activation—via a newly identified Glc7/Bud14 type I protein phosphatase module—and TFIID promoter distribution. Mol Cell Biol 25: 488–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludin K, Jiang R, Carlson M (1998) Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 95: 6245–6250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler G, Schuller C, Adam G, Ruis H (1993) A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J 12: 1997–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayordomo I, Estruch F, Sanz P (2002) Convergence of the target of rapamycin and the Snf1 protein kinase pathways in the regulation of the subcellular localization of Msn2, a transcriptional activator of STRE (Stress Response Element)-regulated genes. J Biol Chem 277: 35650–35656 [DOI] [PubMed] [Google Scholar]
- McCartney RR, Schmidt MC (2001) Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J Biol Chem 276: 36460–36466 [DOI] [PubMed] [Google Scholar]
- Rep M, Reiser V, Gartner U, Thevelein JM, Hohmann S, Ammerer G, Ruis H (1999) Osmotic stress-induced gene expression in Saccharomyces cerevisiae requires Msn1p and the novel nuclear factor Hot1p. Mol Cell Biol 19: 5474–5485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F, Winderickx J, Thevelein JM (2002) Glucose-sensing and -signalling mechanisms in yeast. FEMS Yeast Res 2: 183–201 [DOI] [PubMed] [Google Scholar]
- Santhanam A, Hartley A, Duvel K, Broach JR, Garrett S (2004) PP2A phosphatase activity is required for stress and Tor kinase regulation of yeast stress response factor Msn2p. Eukaryot Cell 3: 1261–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz P (2003) Snf1 protein kinase: a key player in the response to cellular stress in yeast. Biochem Soc Trans 31: 178–181 [DOI] [PubMed] [Google Scholar]
- Sanz P, Alms GR, Haystead TA, Carlson M (2000) Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol Cell Biol 20: 1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AP, McEntee K (1996) Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 93: 5777–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M, Wykoff DD, Miller N, O'Shea EK (2003) Partially phosphorylated Pho4 activates transcription of a subset of phosphate-responsive genes. PLoS Biol 1: E28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treitel MA, Kuchin S, Carlson M (1998) Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol Cell Biol 18: 6273–6280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu J, Carlson M (1994) The GLC7 type 1 protein phosphatase is required for glucose repression in Saccharomyces cerevisiae. Mol Cell Biol 14: 6789–6796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu J, Carlson M (1995) REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J 14: 5939–5946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh EP, Lamont DJ, Beattie KA, Stark MJ (2002) Novel interactions of Saccharomyces cerevisiae type 1 protein phosphatase identified by single-step affinity purification and mass spectrometry. Biochemistry 41: 2409–2420 [DOI] [PubMed] [Google Scholar]
- Wilson WA, Hawley SA, Hardie DG (1996) Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr Biol 6: 1426–1434 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1
Fig. S2


