Abstract
T cells are produced in the thymus from progenitors of extrathymic origin. As no specific markers are available, the developmental pathway of progenitors preceding thymic colonization remains unclear. Here we show that progenitors in murine fetal liver and blood, which are capable of giving rise to T cells, NK cells and dendritic cells, but not B cells, can be isolated by their surface expression of paired immunoglobulin-like receptors (PIR). PIR expression is maintained until the earliest intrathymic stage, then downregulated before the onset of CD25 expression. Unlike intrathymic progenitors, generation of prethymic PIR+ progenitors does not require Hes1-mediated Notch signaling. These findings disclose a prethymic stage of T-cell development programmed for immigration of the thymus, which is genetically separable from intrathymic stages.
Keywords: clonal analysis, fetal liver, lineage commitment, T-cell development, thymus
Introduction
What kind of progenitors migrate to the thymus has been a longstanding question in developmental biology. Previous studies on early progenitors in adult thymus (AT) have shown that the cells in the earliest thymic population retain a potential to generate B cells in addition to T cells, NK cells and dendritic cells (DC) (Wu et al, 1991; Matsuzaki et al, 1993). Studies using mice with a compromised Notch-signaling pathway showed that, in the AT, B cells are generated at the cost of impaired T-cell development (Radtke et al, 1999; Koch et al, 2001; Han et al, 2002; Izon et al, 2002). These results were interpreted as follows: common lymphoid progenitors (CLP) in the bone marrow (BM) migrate to the AT and determine their cell fate to the T-cell lineage under Notch signaling. Recent studies on early progenitors in AT largely fall in line with this idea in that they retain B-cell potential (Benz and Bleul, 2005; Sambandam et al, 2005; Tan et al, 2005). However, conflicting results have been reported, in which the earliest AT progenitors are shown to be more specific to the T-cell lineage (Allman et al, 2003) or the frequency of progenitors having B-cell potential is extremely low (Porritt et al, 2004; Balciunaite et al, 2005), arguing that the B-cell potential of progenitors is lost prior to the thymic colonization. In the case of fetal thymus (FT), early studies on the earliest intrathymic cells with population level analyses indicated that they are multipotent for T, B and myeloid lineages (Peault et al, 1994; Hattori et al, 1996b). Using clonal analysis, however, we showed that the progenitors having T-cell potential in the earliest FT population are T-cell lineage restricted (Kawamoto et al, 1998). These studies on thymic progenitors faced the dilemma that the earliest intrathymic progenitors have already been exposed to the thymic microenvironment and thus do not necessarily represent the original thymic immigrants, and that it is difficult to determine which type of progenitors represent the majority of thymic immigrants (Petrie and Kincade, 2005).
For understanding the earliest stage of T-cell development, it is a prerequisite to clarify the prethymic differentiation pathway. Several studies indicated that T-cell lineage restricted progenitors expressing Thy-1 are present in the extrathymic organs such as BM (Dejbakhsh-Jones et al, 2001), spleen (Lancrin et al, 2002) or fetal blood (FB) (Rodewald et al, 1994; Carlyle and Zuniga-Pflucker, 1998). On the other hand, the lineage marker negative (Lin−) c-kit+ fraction of BM has been reported to contain B220+ progenitors having a potential to generate T and B cells (Martin et al, 2003). However, these cells might not represent major thymic immigrants, since no such population of cells sharing the same phenotype is found in the thymus (Matsuzaki et al, 1993; Hattori et al, 1996a). In the fetal liver (FL) of early embryonic stages, we have shown that the majority of progenitors in the Lin−c-kit+IL-7R+ (IL-7R+) population are T/NK/DC lineage restricted (Kawamoto et al, 2000). The corresponding population was found in FB and FT (Ikawa et al, 2004). Furthermore, we have recently found that progenitors that are just migrating into the thymus across the surrounding mesenchymal area are IL-7R+ and T/NK/DC lineage restricted (Masuda et al, 2005). These findings have led us to propose that the progenitors prethymically committed to the T/NK/DC lineage migrate into the thymus (Katsura, 2002).
To dissect the prethymic pathway of T/NK/DC lineage restriction, it is important to identify a differentiation marker specific to the prethymic progenitors and the recent thymic immigrants. In a search for such markers, we identified the paired immunoglobulin-like receptors (PIR) as a candidate marker. Two isoforms of PIR, PIR-A and PIR-B, are known to be coexpressed on mature myeloid cells, DC, mast cells and B cells, but not on T cells (Kubagawa et al, 1997, 1999). PIR-A pairs with the transmembrane FcRγ molecule that bears ITAM motifs, while PIR-B has its own ITIM motifs (Takai and Ono, 2001). In the present study, we show that PIR are specifically expressed on prethymic T/NK/DC progenitors in early fetal ages. We will further show genetic evidence that the production of PIR+ progenitors is regulated differently from that of intrathymic progenitors.
Results
Isolation of T/NK/DC progenitors in FL based on PIR expression
FL cells at various gestational ages were stained in four colors with anti-Lin, anti-c-kit, anti-IL-7R and anti-PIR, and the expression profiles for IL-7R versus PIR on Lin−c-kit+ cells were examined. We found that, at 11 and 12 days post coitum (dpc), a large proportion of FL IL-7R+ cells are PIR low to high positive (PIR+) (Figure 1A). IL-7R+PIR− cells begin to form a distinct population at 13 dpc, and the proportion of these cells increases along with the fetal age.
Figure 1.
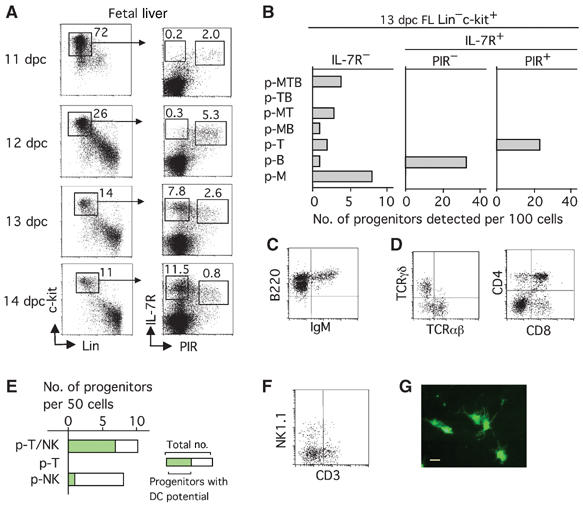
PIR+ cells in the IL-7R+ population of FL are T/NK/DC lineage restricted. (A) Emergence of PIR+ cells in the Lin−c-kit+IL-7R+ population during FL ontogeny. FL cells of various gestational days from Balb/c mice were four-color stained with anti-Lin, anti-c-kit, anti-PIR, and anti-IL-7R. Numbers in the left and right panels represent the percentage of cells gated in each box in whole FL cells, and that in the Lin−c-kit+ fraction, respectively. (B) Single cells of various populations from 13 dpc FL were individually examined by the MLP assay that covers T, B and myeloid lineages. The bars represent the numbers of progenitors detected among the 100 cells analyzed. (C) A profile of cells generated from a single 13 dpc PIR− FL progenitor on day 12 in simple FTOC conditions. (D) Profiles of cells generated on day 12 from a single 13 dpc PIR+ FL progenitor in FTOC conditions. (E) A total of 50 PIR+ FL cells from 13 dpc fetuses of EGFP Tg mice were individually analyzed for T, NK and DC potential by the MLP assay that covers T, NK and DC lineages. p-T/NK represent progenitors generating T and NK cells. The green portion of the columns indicates the number of progenitors that showed DC generation. (F) A profile of cells generated from a single PIR+ FL progenitor in the MLP assay conditions for T, NK and DC lineages. (G) A photomicrograph of EGFP+ DC generated in FTOC from a single PIR+ progenitor. Bar indicates 50 μm.
We examined the commitment status of cells in subpopulations of 13 dpc FL using the multilineage progenitor (MLP) assay, which can determine the developmental potential of individual cells toward the T, B and myeloid (M) lineages (Kawamoto et al, 1997). The progenitors are classified into seven types according to the cell types generated in each culture, which are the progenitors generating all three lineage cells (p-MTB), those generating two lineage cells (p-MT, p-MB and p-TB) and those generating one lineage cell (p-T, p-B and p-M). Conforming to our previous findings, all types of progenitors, except for p-TB, were detected in the IL-7R− population (Figure 1B). Strikingly, the PIR− and PIR+ subpopulations of IL-7R+ cells exclusively contained p-B and p-T, respectively. The isolation of T-cell progenitors by PIR expression was confirmed by other culture systems, that is, T-cell potential by conventional FT organ culture (FTOC), and B-cell potential and myeloid cell potential by coculture with stromal cells (Supplementary Figure 1). PIR− progenitors gave rise to B cells even in FTOC without adding any cytokines (Figure 1C). Single PIR+ FL cells almost always generate as many as 105 T cells in FTOC, containing both αβ T cells and γδ T cells, and can form CD4/CD8 thymic populations (Figure 1D).
We next examined the potential of individual PIR+ FL cells for the generation of NK cells and DC in addition to T cells in a modified MLP assay system (Ikawa et al, 1999; Shen et al, 2003). All PIR+ progenitors generating T cells were found to give rise to NK cells, and most of them exhibited DC potential (Figure 1E). A flow-cytometric profile of T and NK cells (Figure 1F) and a photomicrograph of DC (Figure 1G) generated from a single PIR+ progenitor are shown. These data indicate that the PIR+ T-cell progenitors in FL can be defined as common progenitors for T cells, NK cells and DC (T/NK/DC progenitors).
PIR expression is specific to the T/NK/DC progenitors
Multipotent progenitors, myeloid progenitors and erythroid progenitors have been shown to be enriched in the Sca-1 high positive (Sca-1hi), FcγRII/III (FcR)hi and CD45− subpopulations in the Lin−c-kit+ fraction, respectively (Kawamoto et al, 1999; Lu et al, 2002). It was found that all the FcRhi cells and CD45− cells are PIR− (Figure 2A). Thus, the expression of PIR at the progenitor stage in fetuses is specific to T/NK/DC common progenitors. RT–PCR analysis indicated that, whereas the expression levels of ikaros, PU.1, Gata2 and Gata3 are comparable between PIR− and PIR+ cells, transcription factors specific to the T-cell lineage (Tcf-1) and B-cell lineage (EBF, Pax5, mb-1 and λ5) are exclusively detected in PIR+ cells and PIR− cells, respectively (Figure 2B). Although Notch1 is expressed in both PIR− and PIR+ cells, molecules downstream of the Notch signal, such as Hes1 and Deltex1, were undetectable in these cells, indicating that the Notch signal is not so highly activated (see Figure 6A). Expression of Rag2 is seen in PIR− cells, but barely detectable in PIR+ cells. This is consistent with the previous finding that B-cell progenitors initiate D–J rearrangement of their Ig heavy (IgH) chain gene early in FL ontogeny (Chang et al, 1992), while D–J loci of the TCRβ chain gene are rearranged at a later intrathymic stage (Kawamoto et al, 2003). This is also in agreement with the finding made using the RAG1/GFP knockin mouse system (Yokota et al, 2003), since IL-7R+ FL cells are only partially RAG1/GFPhi at 13 dpc. Although the RAG protein begins to be expressed in PIR− cells, D–J loci of the IgH chain gene were not rearranged in them (Figure 2C). These results revealed that the genetic programs specific to T- and B-cell lineages independently progress in distinct cell populations of FL.
Figure 2.
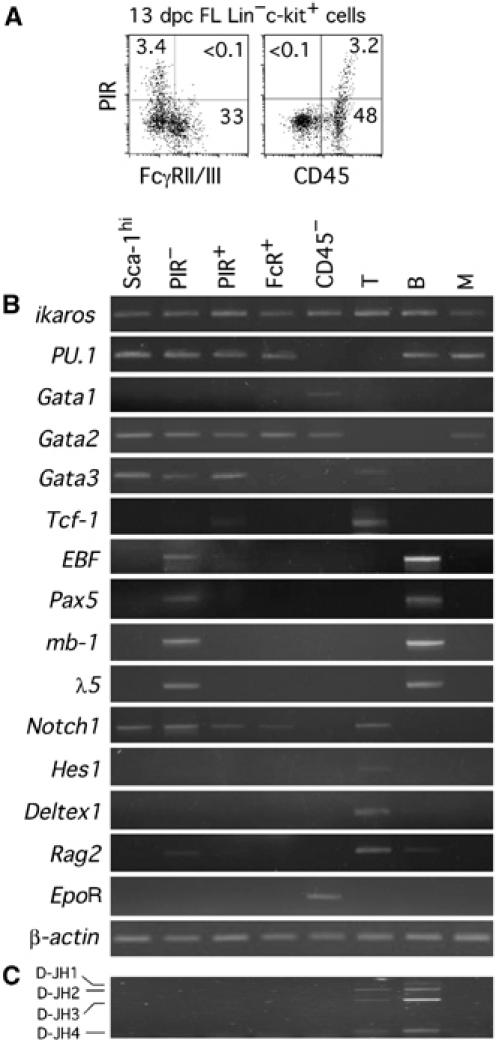
Expression of PIR at progenitors stages is specific to the T/NK/DC lineage. (A) FL cells of B6 mice were four-color stained with anti-Lin, anti-c-kit, anti-PIR, and anti-FcR, or with anti-Lin, anti-c-kit, anti-PIR, and anti-CD45. Profiles of cells gated on Lin−c-kit+ fraction are shown. The numbers in panels represent the percentage of cells in each quadrant. (B) RT–PCR analysis of various lineage-associated genes in the cells of different FL subpopulations from B6 mice. Thy-1+ cells from AT or 15 dpc FT (for Hes1 and Deltex1) (T), B220+ cells (B) and Mac-1+ cells (M) from BM were examined as controls. (C) Genomic DNA from the cells in the same populations as in (B) was analyzed for D–J rearrangement status of the IgH gene by PCR.
Figure 6.
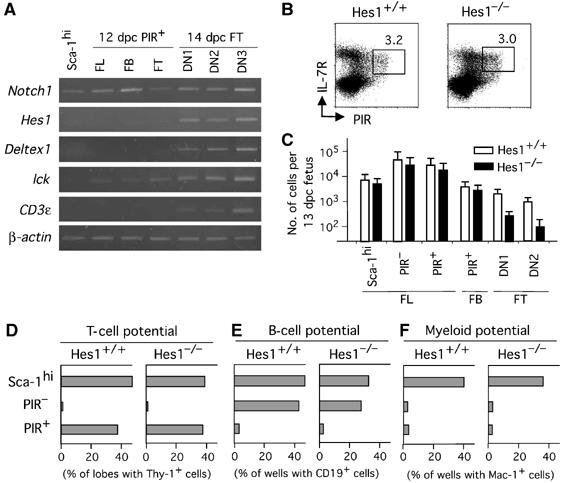
PIR+ T-cell progenitors in FL and FB emerge independently of Hes1. (A) RT–PCR analysis for expression of Notch-signal-related genes in cells of various stages of T-cell development. (B) PIR+ population in FL from 13 dpc fetuses of Hes1−/− mice. Cells were four-color stained in the same way as in Figure 1A, and profiles of Lin−c-kit+ cells are shown. (C) Numbers of Sca-1hi, PIR− and PIR+ cells of FL, PIR+ cells of FB, and DN1 and DN2 cells of FT from 13 dpc fetuses of Hes1−/− mice and Hes1+/+ littermate mice, are shown. Error bars indicate SD in each group. (D–F) PIR+ cells in FL from Hes1−/− fetuses are restricted to the T-cell lineage. Sca-1hi cells, PIR− cells and PIR+ cells of 13 dpc FL (a total of 100 cells for each group) from wild-type mice and Hes1−/− mice were examined for T, B and myeloid cell potential by coculture with a dGuo lobe for detection of T-cell potential (D), coculture with TSt-4 for B-cell potential (E), and coculture with PA6 for myeloid cell potential (F), respectively, except that Thy-1 was used as a marker for the T-lineage cells in (D).
Expression of PIR on FB and FT cells
We have previously shown that IL-7R+ T/NK/DC progenitors circulate in FB during 11–14 dpc (Ikawa et al, 2004). Most of these IL-7R+ cells in FB were found to express PIR (Figure 3A). In the FL of 13 dpc fetuses, the proportion of PIR− cells is even higher than PIR+ cells (Figure 1A), but they are rare in FB (Figure 3A), strongly suggesting that the PIR+ cells are selectively released from FL into the circulation. Emergence of the PIR+ cells in FL and FB is independent of the thymus, since the number as well as commitment status of PIR+ cells in FL and FB of nude mice were comparable to those of normal mice (Supplementary Figure 2).
Figure 3.

Expression of PIR visualizes the thymic immigration pathway. (A) FB cells at 11 and 13 dpc were four-color stained in the same manner as in Figure 1A. Profiles of cells gated on Lin−c-kit+ fraction (1.2 and 0.5% of whole FB cells at 11 and 13 dpc, respectively) are shown. The numbers in the panels represent the percentage of cells gated in each box in Lin−c-kit+ FB cells. (B) Tissue containing the 11 dpc thymic anlage and surrounding mesenchyme was taken, digested and stained in four colors with anti-CD45, anti-c-kit, anti-PIR and anti-IL-7R. (C) A frozen section of an 11 dpc fetus containing the thymic anlage region was stained in two colors with an anti-PIR antibody (green) and an anti-Keratin antibody (red), and a serial section was stained with the anti-PIR antibody (green) and an anti-IKAROS antibody (red).
Progenitors in the 11 dpc thymic anlage region may represent the genuine thymic immigrants, since they reside in the mesenchymal area surrounding the thymic epithelium and have not yet been influenced by the thymic epithelial components (Itoi et al, 2001). We have recently reported that these immigrating cells are IL-7R+ and T/NK/DC lineage restricted (Masuda et al, 2005). Flow-cytometric analysis of these cells revealed that virtually all IL-7R+ cells in the 11 dpc FT anlage region highly express PIR (Figure 3B). Using immunohistochemistry, we also analyzed sections of 11 dpc FT anlage region in two colors with anti-Keratin and anti-PIR, and with anti-IKAROS and anti-PIR (Figure 3C). IKAROS+ cells represent the progenitors of the hematopoietic lineage. It was confirmed that virtually all IKAROS+ cells in the mesenchyme adjacent to the Keratin+ epithelial primordium are PIR+.
It is known that all 12 dpc FT cells are CD44+c-kit+CD25− (double negative 1 (DN1)) cells, and CD44+c-kit+CD25+ (DN2) cells appear at 13 dpc. Intrathymic progenitors at 12 dpc still express PIR, but they downregulate PIR and begin to express CD25 at 13 dpc (Figure 4A). We recapitulated this pathway in vitro, by culturing sorted PIR+ cells from 12 dpc FL on a monolayer of the stromal cell line TSt-4 enforced to express the Notch ligand Delta-like-1 (Dll-1) (Figure 4B), which can support T-cell development from the early progenitor stage to the CD4+CD8+ stage (Miyazaki et al, 2005). The differentiation order was further confirmed by culturing PIR+ and PIR− cells of the 14 dpc FT DN1 population, in which PIR+ cells generated PIR− cells, but not vice versa (Figure 4C). These data indicate that the downregulation of PIR takes place at the DN1 stage.
Figure 4.
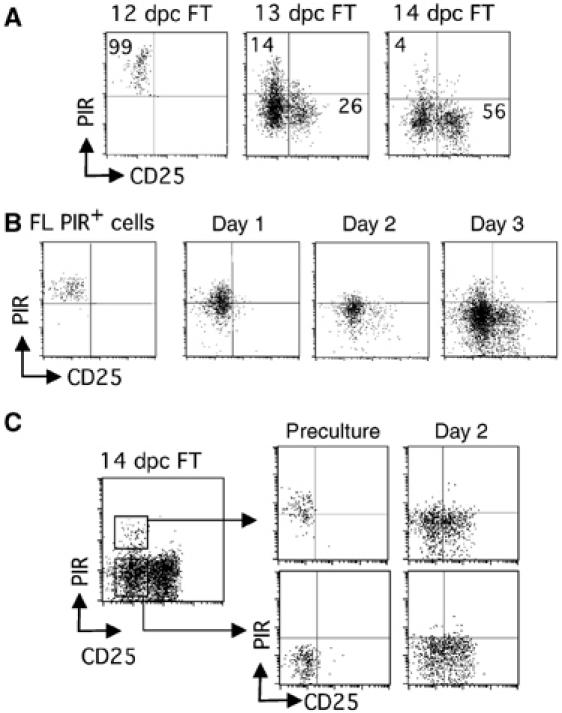
Expression of PIR is downregulated in thymocytes before they express CD25. (A) Profiles of cells from FT of various ages gated on the CD3−CD4−CD8−c-kit+ fraction are shown. The numbers in panels represent percentage of cells in each quadrant. (B, C) PIR+ cells from 12 dpc FL (B), and PIR+ and PIR− cells from 14 dpc FT (C), were cultured with monolayered stromal cells (TSt-4) enforced to express Dll-1. Profiles of precultured cells, and those of cells recovered from culture on indicated days of cultivation, all gated on the CD3−CD4−CD8−c-kit+ fraction, are shown.
αβ T cells and γδ T cells generated from individual PIR+ progenitors
Single PIR+ progenitors from FL, FB and FT were found to produce a comparable number of T-lineage cells in FTOC (data not shown). All clones from PIR+ cells of different sources exhibited rearranged gene sequences for almost all the Dβ–Jβ loci analyzed, indicating that each of these progenitors have a potential to produce a highly diversified TCRβ chain repertoire (Figure 5A). Assessment of the clonal expansion size of Rag2−/− progenitors in FTOC indicated that PIR+ progenitors of FL, FB and FT retain very high progenitor activities to proliferate prior to TCRβ chain gene rearrangement, which are almost equivalent to each other, although those in FT are somehow lower (Figure 5B). These results provided supporting evidence that the PIR+ progenitors in FL and FB represent the thymic immigrants. The usage of Vγ genes was not restricted to the fetal type (Vγ3) in all these clones (Figure 5C).
Figure 5.
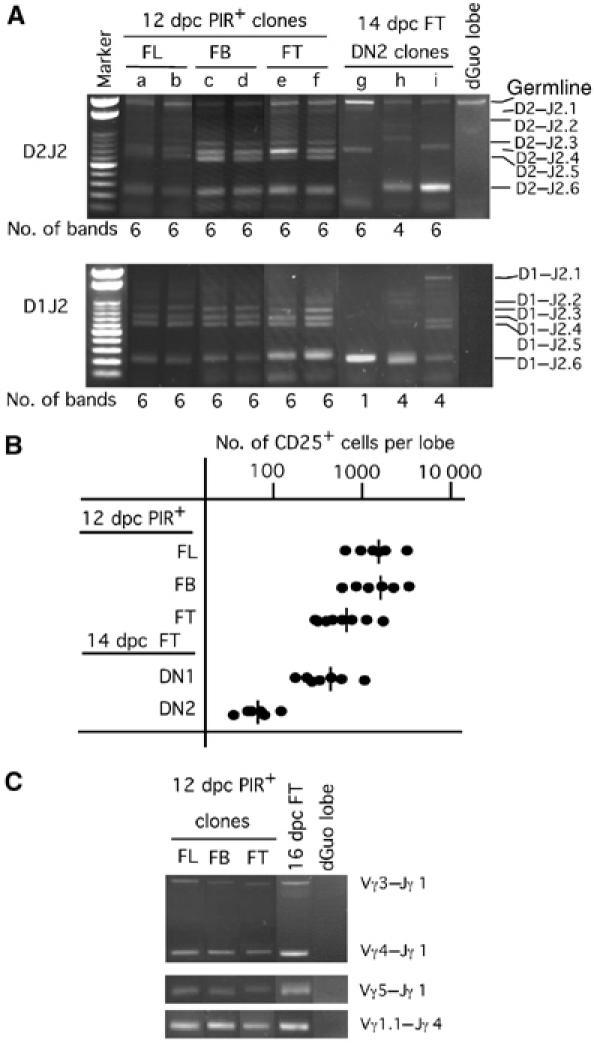
The potential of individual PIR+ progenitors for the production of αβ T cells and γδ T cells. (A) PIR+ cells from FL, FB and FT of 12 dpc fetuses and 14 dpc CD44+CD25+ FT cells were individually cultured with a dGuo lobe for 12 days. Generated cells were examined for their rearrangement status for D1–J2 and D2–J2 loci of the TCRβ chain gene. (B) PIR+ cells from FL, FB and FT of 12 dpc Rag2−/− fetuses, as well as DN1 and DN2 FT cells from 14 dpc Rag2−/− fetuses, were individually cultured with a dGuo-treated FT lobe for 10 days. Numbers of CD25+ cells generated in each clonal culture (6–8 clones per group) are shown. Bars represent the average in each group. (C) Cells of indicated populations were similarly cultured as in (A), and generated cells were examined for their rearrangement status of Vγ–Jγ loci. Unfractionated 16 dpc FT cells were used as a positive control.
Difference in requirement for Hes1-mediated Notch signaling between prethymic and intrathymic development
We and another group have reported that the disruption of Hes1 results in severe impairment in T-cell development (Tomita et al, 1999; Kaneta et al, 2000). Harman et al (2003) found that Hes1 is upregulated in progenitors only when they encounter thymic epithelial cells, and proposed that lineage restriction directed by the Notch signal takes place after thymic colonization. It is thus important to examine whether Notch/Hes1 signaling participates in prethymic T-cell lineage restriction. RT–PCR indicated that prethymic progenitors as well as PIR+ intrathymic progenitors do not highly express Hes1 nor Deltex1, while they express Notch1 (Figure 6A). These results are in line with the findings by Harman et al (2003). We then investigated whether PIR+ FL and FB cells from Hes1−/− fetuses exhibit any defects or not. Flow-cytometric profiles and numbers of PIR+ FL and FB cells from 13 dpc Hes1−/− fetuses were indistinguishable from those of wild-type fetuses (Figure 6B and C). A significant reduction in number was seen only in intrathymic populations.
By using various culture systems, we examined the commitment status of PIR+ FL cells from Hes1−/− fetuses. PIR− and PIR+ cells, from 13 dpc FL of Hes1−/− and wild-type littermate mice, were individually analyzed for T, B and myeloid cell potential. For comparison, cells in the Sca-1hi fraction of 13 dpc FL were examined. The frequencies of cells in Sca-1hi, PIR− and PIR+ populations that gave rise to Thy-1+ cells were almost the same between Hes1+/+ and Hes1−/− fetuses (Figure 6D). No significant difference was seen between Hes1+/+ and Hes1−/− fetuses with regard to the frequencies of progenitors having B cell or myeloid potential in all Sca1hi, PIR− and PIR+ populations (Figure 6E and F). These data indicate that Hes1−/− PIR+ FL progenitors are T-cell lineage restricted. We thus concluded that the commitment to the T/NK/DC lineage takes place in extrathymic organs independently of the Hes1-mediated Notch-signaling pathway. These results disclosed the existence of a prethymic stage that is genetically distinct from the intrathymic stages.
Discussion
Previous studies have suggested that there are two phases of immigration of progenitors into the thymus during the embryonic development of mice (Jotereau et al, 1987; Dunon et al, 1999). The emergence of PIR+ T/NK/DC progenitors in FL and FB at 11–13 dpc may represent the first phase of thymic immigration. Progenitor populations restricted to the T/NK cell lineage, such as B220+ cells in FL (Douagi et al, 2002) and Thy-1+c-kitlowNK1.1+ cells in FB (Rodewald et al, 1994; Carlyle and Zuniga-Pflucker, 1998), were reported to be present during 13–15 dpc. Using RAG1/GFP knockin mice, Yokota et al (2003) recently showed that the 14 dpc FL contains progenitors that preferentially give rise to T cells, but not B or myeloid cells. These cells may also participate in the second or later phase of thymic colonization, although the majority of cells in the earliest thymic population are Thy-1−B220−c-kithiRag1− cells even after 13 dpc (data not shown).
While we found that the initial thymic progenitors are almost exclusively PIR+, this does not necessarily mean that all PIR+ cells in prethymic tissues are determined to immigrate into the thymus. It is probable that some of the prethymic PIR+ T/NK/DC progenitors may serve as progenitors of extrathymic T cells, NK cells and DC. Similarly, our present findings do not preclude the possibility of migration of other types of progenitors to the thymus. Indeed, we have detected small but substantial numbers of B and myeloid cell progenitors in FT (Kawamoto et al, 1998). It has previously been shown that a small number of multipotent progenitors are present in the FB of early (10–12 dpc) fetuses (Delassus and Cumano, 1996), and it was thus speculated that the circulating multipotent progenitors also migrate into the thymus. The possibility thus formally remains that a very small number of multipotent progenitors also migrate into the thymus in addition to the T/NK/DC progenitors, although our present findings do not support this speculation.
Whereas previous studies on early progenitors in the AT have suggested that it is colonized by common progenitors for T, B, NK and DC lineages (Wu et al, 1991; Matsuzaki et al, 1993), pieces of evidence are accumulating that the earliest progenitors in AT have already lost B-cell potential (Porritt et al, 2004; Balciunaite et al, 2005). On the other hand, very recent studies argue that the earliest thymic progenitors retain B-cell potential (Benz and Bleul, 2005; Sambandam et al, 2005; Tan et al, 2005). It was further suggested that multipotent progenitors in Lin−c-kit+Sca-1+IL-7R− cells in adult blood represent thymic immigrants (Schwarz and Bhandoola, 2004). One possible explanation is that various types of progenitors migrate into the AT (Petrie and Kincade, 2005). We have recently found that T-cell progenitors in BM can be isolated based on PIR expression, and AT contains a small number of PIR+ progenitors (unpublished data). Further studies on the prethymic pathway in adult animals are ongoing.
It is of importance that PIR expression is downregulated within the DN1 stage. This finding makes it clear that PIR+ cells are distinct from the conventional DN1 cells, which have been considered to represent the cells at the first stage of T-cell development. It is thus supposed that a differentiation program specified to the thymic immigrant stage exists. We further showed that prethymic T/NK/DC progenitors do not require Hes1 for their generation, but intrathymic ones do. Such difference in requirement for Hes1 also supports the idea that PIR+ stages are genetically distinct from intrathymic stages. Our finding as to the Hes1 independence of prethymic progenitors, however, does not necessarily exclude the possibility that cascades of Notch signaling other than those mediated by the Hes1 molecule are involved in the prethymic production of T/NK/DC progenitors.
Our results also indicate that very early B-cell lineage committed progenitors can be isolated in the IL-7R+PIR− subpopulation. It is of note that these PIR− cells can give rise to B cells even in coculture with fetal thymic lobes, without adding any exogenous cytokines (Figure 1C). Although some studies reported that a small number of B cells are produced in the thymus (Akashi et al, 2000), the thymic environment is thought to suppress B-cell generation by some soluble factor(s) (Hashimoto et al, 2002) or Notch ligands expressed on thymic epithelial cells (Radtke et al, 1999; Izon et al, 2002). Therefore, the experimental results that PIR− progenitors produce B cells in the fetal thymic organ culture environment may indicate that their commitment status is stable enough to sustain B-cell potential in a condition biased to T-cell production. The PIR− cells are negative for the known B-cell markers such as B220 and CD19, and thus can be regarded as the earliest B-cell progenitors in ontogeny. However, the environment of the thymic organ culture is not necessarily inductive exclusively for cells towards the T-cell lineage, because the thymic environment is heterogeneous, allowing differentiation of various lineage cells. We are now studying how B-cell lineage specification proceeds along with surface phenotypic progression, by culturing progenitors in more homogenously specified environments.
The function of PIR at the progenitor stage remains to be clarified. PIR-A and PIR-B are usually coexpressed, and PIR-A can act as a activation signal through a paired ITAM-bearing FcRγ element, while PIR-B conducts inhibitory signals through ITIM motifs that locate in the intracellular portion of the PIR-B molecule. PIR-B KO mice were reported to exhibit constitutive B-cell activation, impairment in DC maturation and severe GVHD (Ujike et al, 2002; Nakamura et al, 2004). While physiological ligands for PIR have not yet been identified, it is reported that ITIM motifs of PIR-B on macrophages and B cells are constitutively phosphorylated (Ho et al, 1999). Since the phosphorylation status of ITIM motifs of PIR-B is significantly reduced in β2m−/− mice, some MHC class I-like molecules that are ubiquitously expressed are assumed to serve as a ligand (Ho et al, 1999). If this is the case also at progenitor stages, it may be that PIR plays a role in conducting mainly inhibitory signals to T-cell progenitors.
In conclusion, we have substantiated the prethymic stage of the T/NK/DC common pathway in the fetal period as a genetically distinct stage from the intrathymic stages by defining it on the basis of PIR expression. The definition of a novel stage of T-cell development will not only be a guidepost for further studies in this field but also provide useful tools to isolate target cells for gene therapy or for regeneration medicine.
Materials and methods
Mice
Balb/c mice and C57BL/6 (B6) mice were purchased from SLC (Shizuoka, Japan). B6Rag2−/− mice, enhanced GFP transgenic (EGFP Tg) mice of B6 background (Ikawa et al, 1998) and Hes1−/− mice of ICR background (Tomita et al, 1999) were maintained in our animal facility. Balb/c mice were used in most experiments as progenitor sources unless described otherwise in the legend to the figures. The day of finding the vaginal plug was designated as 0 dpc.
Antibodies
The following antibodies were used: anti-Ly5.1 (A20), anti-Ly5.2 (104), anti-c-kit (2B8), anti-erythroid lineage cells (TER119), anti-Mac-1 (M1/70), anti-Gr-1 (RB6-8C5), anti-B220 (RA3-6B2), anti-Thy1.2 (53-2.1), anti-CD8 (53-6.7), anti-CD4 (H129.19), anti-NK1.1 (PK136), anti-TCRγδ (GL-3), anti-TCRαβ (H57-597), anti-CD3ɛ (145-2C11), anti-CD19 (1D3), anti-CD25 (PC61), anti-CD44 (IM7), anti-CD45 (30-F11) and anti-FcR (2.4G2) were purchased from BD PharMingen (San Jose, CA). Anti-IL-7R (A7R34) and anti-IgM (1B4B1) were purchased from eBioscience (San Diego, CA). Anti-PIR (6C1) was produced by us (Kubagawa et al, 1997). TER119, anti-Gr-1, anti-B220, anti-CD19, anti-NK1.1 and anti-Thy-1.2 were used as Lin markers. For immunohistological detection of the thymic anlage and progenitors, the following antibodies were used: rabbit anti-Keratin (DAKO, Glostrup, Denmark) and rabbit anti-IKROS (Hattori et al, 1996a). Alexa 594-donkey anti-rabbit IgG (Molecular Probes, Eugene, OR) and Alexa 488-goat anti-rat IgG (Molecular Probes) were used as secondary reagents.
Growth factors
Recombinant murine (rm) SCF, rm IL-2, rm IL-3, rm Flt-3 ligand and rm IL-7 were purchased from Genzyme-Techne (Cambridge, MA).
Preparation of fetal cells
Embryos were separated from the placenta by pinching and cutting the umbilical cord using fine forceps. The placenta was not removed from the uterus in order to reduce contamination with maternal blood. The embryo was washed twice to remove any contamination of maternal blood, and then placed in medium to allow bleeding until it became completely pale. The embryo was then removed, washed once and placed in another dish containing medium, where it was dissected to obtain FL and FT. Single-cell suspension of FL was prepared by pipetting the FL lobes. FT lobes were minced between glass slides using the frosted portion. All fetal cells were then passed through 40 μm nylon mesh, washed, and resuspended in medium. Viable cells were counted using trypan blue exclusion. Tissues containing the 11 dpc FT anlage and surrounding region were taken together, and were digested for 30 min at 37°C in the presence of collagenase (1 mg/ml) (Wako, Osaka, Japan).
FTOC and flow-cytometric analysis
The basic procedures for the coculture with dGuo-treated FT lobe under high oxygen submersion (HOS) condition, and analysis for generated cells has been described previously (Kawamoto et al, 1997). In the MLP assay for T, B and myeloid lineages, single cells were individually cultured for 10 days with a dGuo lobe, in the presence of SCF (5 ng/ml), IL-3 (3 ng/ml) and IL-7 (5 ng/ml). In the MLP assay modified for detection of T, NK and DC potentials, the cells for examination were obtained from EGFP Tg mice, and IL-2 (1 ng/ml) and Flt-3 ligand (10 ng/ml) were added in addition to the above-described cytokines (Shen et al, 2003).
Coculture with stromal cells
TSt-4 cells were retrovirally transduced with the murine Dll-1 gene (TSt-4/Dll-1) (Miyazaki et al, 2005). To assess the potential of single cells for their ability to give rise to B cells, cells were individually cultured in 96-well plates monolayered with the stromal cell line TSt-4 (Ohmura et al, 1999) for 10 days, and B-cell generation was determined by examining the CD19 expression. Myeloid potential was assessed similarly, but the stromal cell line used was PA6 (Nishikawa et al, 1988), and Mac-1 was used as the myeloid cell marker.
RT–PCR
RT–PCR was performed as described previously (Kawamoto et al, 2000). Primers and PCR conditions are available online. PCR product was electrophoresed through 1.2% agarose and stained with ethidium bromide.
PCR analysis of TCRβ, TCRγ and IgH chain gene rearrangement
The basic procedures for PCR and primers have been described previously (Kawamoto et al, 2000, 2003). Primers and PCR conditions are available online. Amplified DNA products were applied to a 1.2% agarose gel, electrophoresed and stained with ethidium bromide.
Immunohistochemistry
Fetuses (11 dpc) were embedded in an OCT compound and snap-frozen using Leica Histomolds (Leica Microsystems, Wetzlar, Germany). Frozen blocks were cut into serial 5 μm sections, using a Leica M3050S cryostat, and mounted onto MAS-coated microscope slides (Matsunami, Osaka, Japan). After acetone fixation for 1 min, sections were incubated with primary antibodies, washed with PBS/0.01% Tween, followed by incubation with the proper secondary reagent. For fluorescence microscopic analysis, antibody binding was visualized by the appropriate fluorescent reagents, while nuclei were counterstained with DAPI (Molecular Probes).
Supplementary Material
Supplementary Materials and Methods
Supplementary Figure 1
Supplementary Figure 2
Acknowledgments
We thank WTV Germeraad (University Hospital Maastricht, Maastricht, the Netherlands) for critical reading of the manuscript. This work was supported by the Special Coordination Funds for Promoting Science and Technology from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Akashi K, Richie LI, Miyamoto T, Carr WH, Weissman IL (2000) B lymphopoiesis in the thymus. J Immunol 164: 5221–5226 [DOI] [PubMed] [Google Scholar]
- Allman D, Sambandam A, Kim S, Miller JP, Pagan A, Well D, Meraz A, Bhandoola A (2003) Thymopoiesis independent of common lymphoid progenitors. Nat Immunol 4: 168–174 [DOI] [PubMed] [Google Scholar]
- Balciunaite G, Ceredig R, Rolink AG (2005) The earliest subpopulation of mouse thymocytes contains potent T, significant macrophage, and natural killer cell but no B-lymphocyte potential. Blood 105: 1930–1936 [DOI] [PubMed] [Google Scholar]
- Benz C, Bleul CC (2005) A multipotent precursor in the thymus maps to the branching point of the T versus B lineage decision. J Exp Med 202: 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlyle JR, Zuniga-Pflucker JC (1998) Requirement for the thymus in αβ T lymphocyte lineage commitment. Immunity 9: 187–197 [DOI] [PubMed] [Google Scholar]
- Chang Y, Paige CJ, Wu GE (1992) Enumeration and characterization of DJH structures in mouse fetal liver. EMBO J 11: 1891–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejbakhsh-Jones S, Garcia-Ojeda ME, Chatterjea-Matthes D, Zeng D, Strober S (2001) Clonable progenitors committed to the T lymphocyte lineage in the mouse bone marrow; use of an extrathymic pathway. Proc Natl Acad Sci USA 98: 7455–7460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delassus S, Cumano A (1996) Circulation of hematopoietic progenitors in the mouse embryo. Immunity 4: 97–106 [DOI] [PubMed] [Google Scholar]
- Douagi I, Colucci F, Di Santo JP, Cumano A (2002) Identification of the earliest prethymic bipotent T/NK progenitor in murine fetal liver. Blood 99: 463–471 [DOI] [PubMed] [Google Scholar]
- Dunon D, Allioli N, Vainio O, Ody C, Imhof BA (1999) Quantification of T-cell progenitors during ontogeny: thymus colonization depends on blood delivery of progenitors. Blood 93: 2234–2243 [PubMed] [Google Scholar]
- Han H, Tanigaki K, Yamamoto N, Kuroda K, Yoshimoto M, Nakahata T, Ikuta K, Honjo T (2002) Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int Immunol 14: 637–645 [DOI] [PubMed] [Google Scholar]
- Harman BC, Jenkinson EJ, Anderson G (2003) Entry into the thymic microenvironment triggers Notch activation in the earliest migrant T cell progenitors. J Immunol 170: 1299–1303 [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Montecino-Rodriguez E, Leathers H, Stephan RP, Dorshkind K (2002) B-cell development in the thymus is limited by inhibitory signals from the thymic microenvironment. Blood 100: 3504–3511 [DOI] [PubMed] [Google Scholar]
- Hattori N, Kawamoto H, Fujimoto S, Kuno K, Katsura Y (1996a) Involvement of transcription factors TCF-1 and GATA-3 in the initiation of the earliest step of T cell development in the thymus. J Exp Med 184: 1137–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori N, Kawamoto H, Katsura Y (1996b) Isolation of the most immature population of murine fetal thymocytes that includes progenitors capable of generating T, B, and myeloid cells. J Exp Med 184: 1901–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho LH, Uehara T, Chen CC, Kubagawa H, Cooper MD (1999) Constitutive tyrosine phosphorylation of the inhibitory paired Ig-like receptor PIR-B. Proc Natl Acad Sci USA 96: 15086–15090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawa M, Yamada S, Nakanishi T, Okabe M (1998) ‘Green mice' and their potential usage in biological research. Febs Lett 430: 83–87 [DOI] [PubMed] [Google Scholar]
- Ikawa T, Kawamoto H, Fujimoto S, Katsura Y (1999) Commitment of common T/Natural killer (NK) progenitors to unipotent T and NK progenitors in the murine fetal thymus revealed by a single progenitor assay. J Exp Med 190: 1617–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawa T, Masuda K, Lu M, Minato N, Katsura Y, Kawamoto H (2004) Identification of the earliest prethymic T-cell progenitors in murine fetal blood. Blood 103: 530–537 [DOI] [PubMed] [Google Scholar]
- Itoi M, Kawamoto H, Katsura Y, Amagai T (2001) Two distinct steps of immigration of hematopoietic progenitors into the early thymus anlage. Int Immunol 13: 1203–1211 [DOI] [PubMed] [Google Scholar]
- Izon DJ, Aster JC, He Y, Weng A, Karnell FG, Patriub V, Xu L, Bakkour S, Rodriguez C, Allman D, Pear WS (2002) Deltex1 redirects lymphoid progenitors to the B cell lineage by antagonizing Notch1. Immunity 16: 231–243 [DOI] [PubMed] [Google Scholar]
- Jotereau F, Heuze F, Salomon VV, Gascan H (1987) Cell kinetics in the fetal mouse thymus: precursor cell input, proliferation, and emigration. J Immunol 138: 1026–1030 [PubMed] [Google Scholar]
- Kaneta M, Osawa M, Sudo K, Nakauchi H, Farr AG, Takahama Y (2000) A role for Pref-1 and HES-1 in thymocyte development. J Immunol 164: 256–264 [DOI] [PubMed] [Google Scholar]
- Katsura Y (2002) Redefinition of lymphoid progenitors. Nat Rev Immunol 2: 127–132 [DOI] [PubMed] [Google Scholar]
- Kawamoto H, Ikawa T, Ohmura K, Fujimoto S, Katsura Y (2000) T cell progenitors emerge earlier than B cell progenitors in the murine fetal liver. Immunity 12: 441–450 [DOI] [PubMed] [Google Scholar]
- Kawamoto H, Ohmura K, Fujimoto S, Katsura Y (1999) Emergence of T cell progenitors without B cell or myeloid differentiation potential at the earliest stage of hematopoiesis in the murine fetal liver. J Immunol 162: 2725–2731 [PubMed] [Google Scholar]
- Kawamoto H, Ohmura K, Fujimoto S, Lu M, Ikawa T, Katsura Y (2003) Extensive proliferation of T cell lineage-restricted progenitors in the thymus: an essential process for clonal expression of diverse T cell receptor β chains. Eur J Immunol 33: 606–615 [DOI] [PubMed] [Google Scholar]
- Kawamoto H, Ohmura K, Katsura Y (1997) Direct evidence for the commitment of hematopoietic stem cells to T, B and myeloid lineages in murine fetal liver. Int Immunol 9: 1011–1019 [DOI] [PubMed] [Google Scholar]
- Kawamoto H, Ohmura K, Katsura Y (1998) Presence of progenitors restricted to T, B, or myeloid lineage, but absence of multipotent stem cells, in the murine fetal thymus. J Immunol 161: 3799–3802 [PubMed] [Google Scholar]
- Koch U, Lacombe TA, Holland D, Bowman JL, Cohen BL, Egan SE, Guidos CJ (2001) Subversion of the T/B lineage decision in the thymus by lunatic fringe-mediated inhibition of Notch-1. Immunity 15: 225–236 [DOI] [PubMed] [Google Scholar]
- Kubagawa H, Burrows PD, Cooper MD (1997) A novel pair of immunoglobulin-like receptors expressed by B cells and myeloid cells. Proc Natl Acad Sci USA 94: 5261–5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubagawa H, Cooper MD, Chen CC, Ho LH, Alley TL, Hurez V, Tun T, Uehara T, Shimada T, Burrows PD (1999) Paired immunoglobulin-like receptors of activating and inhibitory types. Curr Top Microbiol Immunol 244: 137–149 [DOI] [PubMed] [Google Scholar]
- Lancrin C, Schneider E, Lambolez F, Arcangeli ML, Garcia-Cordier C, Rocha B, Ezine S (2002) Major T cell progenitor activity in bone marrow-derived spleen colonies. J Exp Med 195: 919–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Kawamoto H, Katsube Y, Ikawa T, Katsura Y (2002) The common myelolymphoid progenitor: a key intermediate stage in hemopoiesis generating T and B cells. J Immunol 169: 3519–3525 [DOI] [PubMed] [Google Scholar]
- Martin CH, Aifantis I, Scimone ML, von Andrian UH, Reizis B, von Boehmer H, Gounari F (2003) Efficient thymic immigration of B220+ lymphoid-restricted bone marrow cells with T precursor potential. Nat Immunol 4: 866–873 [DOI] [PubMed] [Google Scholar]
- Masuda K, Itoi M, Amagai T, Minato N, Katsura Y, Kawamoto H (2005) Thymic anlage is colonized by progenitors restricted to T, NK, and dendritic cell lineages. J Immunol 174: 2525–2532 [DOI] [PubMed] [Google Scholar]
- Matsuzaki Y, Gyotoku J, Ogawa M, Nishikawa S, Katsura Y, Gachelin G, Nakauchi H (1993) Characterization of c-kit positive intrathymic stem cells that are restricted to lymphoid differentiation. J Exp Med 178: 1283–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki M, Kawamoto H, Kato Y, Itoi M, Miyazaki K, Masuda K, Tashiro S, Ishihara H, Igarashi K, Amagai T, Kanno R, Kanno M (2005) Polycomb group gene mel-18 regulates early T progenitor expansion by maintaining the expression of Hes-1, a target of the Notch pathway. J Immunol 174: 2507–2516 [DOI] [PubMed] [Google Scholar]
- Nakamura A, Kobayashi E, Takai T (2004) Exacerbated graft-versus-host disease in Pirb−/− mice. Nat Immunol 5: 623–629 [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Ogawa M, Nishikawa S, Kunisada T, Kodama H (1988) B lymphopoiesis on stromal cell clone: stromal cell clones acting on different stages of B cell differentiation. Eur J Immunol 18: 1767–1771 [DOI] [PubMed] [Google Scholar]
- Ohmura K, Kawamoto H, Fujimoto S, Ozaki S, Nakao k, Katsura Y (1999) Emergence of T, B and myeloid lineage-committed as well as multipotent hematopoietic progenitors in the aoorta-gonad-mesonephros region of day 10 fetuses of the mouse. J Immunol 163: 4788–4795 [PubMed] [Google Scholar]
- Peault B, Khazaal I, Weissman IL (1994) In vitro development of B cells and macrophages from early mouse fetal thymocytes. Eur J Immunol 24: 781–784 [DOI] [PubMed] [Google Scholar]
- Petrie HT, Kincade PW (2005) Many roads, one destination for T cell progenitors. J Exp Med 202: 11–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porritt HE, Rumfelt LL, Tabrizifard S, Schmitt TM, Zuniga-Pflucker JC, Petrie HT (2004) Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity 20: 735–745 [DOI] [PubMed] [Google Scholar]
- Radtke F, Wilson A, Stark G, Bauer M, van MJ, MacDonald HR, Aguet M (1999) Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 10: 547–558 [DOI] [PubMed] [Google Scholar]
- Rodewald HR, Kretzschmar K, Takeda S, Hohl C, Dessing M (1994) Identification of pro-thymocytes in murine fetal blood: T lineage commitment can precede thymus colonization. EMBO J 13: 4229–4240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandam A, Maillard I, Zediak VP, Xu L, Gerstein RM, Aster JC, Pear WS, Bhandoola A (2005) Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat Immunol 6: 663–670 [DOI] [PubMed] [Google Scholar]
- Schwarz BA, Bhandoola A (2004) Circulating hematopoietic progenitors with T lineage potential. Nat Immunol 5: 953–960 [DOI] [PubMed] [Google Scholar]
- Shen HQ, Lu M, Ikawa T, Masuda K, Ohmura K, Minato N, Katsura Y, Kawamoto H (2003) T/NK bipotent progenitors in the thymus retain the potential to generate dendritic cells. J Immunol 171: 3401–3406 [DOI] [PubMed] [Google Scholar]
- Takai T, Ono M (2001) Activating and inhibitory nature of the murine paired immunoglobulin-like receptor family. Immunol Rev 181: 215–222 [DOI] [PubMed] [Google Scholar]
- Tan JB, Visan I, Yuan JS, Guidos CJ (2005) Requirement for Notch1 signals at sequential early stages of intrathymic T cell development. Nat Immunol 6: 671–679 [DOI] [PubMed] [Google Scholar]
- Tomita K, Hattori M, Nakamura E, Nakanishi S, Minato N, Kageyama R (1999) The bHLH gene Hes1 is essential for expansion of early T cell precursors. Genes Dev 13: 1203–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujike A, Takeda K, Nakamura A, Ebihara S, Akiyama K, Takai T (2002) Impaired dendritic cell maturation and increased TH2 responses in PIR-B−/− mice. Nat Immunol 3: 542–548 [DOI] [PubMed] [Google Scholar]
- Wu L, Antica M, Johnson GR, Scollay R, Shortman K (1991) Developmental potential of the earliest precursor cells from the adult mouse thymus. J Exp Med 174: 1617–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T, Kouro T, Hirose J, Igarashi H, Garrett KP, Gregory SC, Sakaguchi N, Owen JJ, Kincade PW (2003) Unique properties of fetal lymphoid progenitors identified according to RAG1 gene expression. Immunity 19: 365–375 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods
Supplementary Figure 1
Supplementary Figure 2


