Abstract
Synthetic pre-mRNAs containing the processing signals encoded by Drosophila melanogaster histone genes undergo efficient and faithful endonucleolytic cleavage in nuclear extracts prepared from Drosophila cultured cells and 0- to 13-h-old embryos. Biochemical requirements for the in vitro cleavage are similar to those previously described for the 3′ end processing of mammalian histone pre-mRNAs. Drosophila 3′ end processing does not require ATP and occurs in the presence of EDTA. However, in contrast to mammalian processing, Drosophila processing generates the final product ending four nucleotides after the stem-loop. Cleavage of the Drosophila substrates is abolished by depleting the extract of the Drosophila stem-loop binding protein (dSLBP), indicating that both dSLBP and the stem-loop structure in histone pre-mRNA are essential components of the processing machinery. Recombinant dSLBP expressed in insect cells by using the baculovirus system efficiently complements the depleted extract. Only the RNA-binding domain plus the 17 amino acids at the C terminus of dSLBP are required for processing. The full-length dSLBP expressed in insect cells is quantitatively phosphorylated on four residues in the C-terminal region. Dephosphorylation of the recombinant dSLBP reduces processing activity. Human and Drosophila SLBPs are not interchangeable and strongly inhibit processing in the heterologous extracts. The RNA-binding domain of the dSLBP does not substitute for the RNA-binding domain of the human SLBP in histone pre-mRNA processing in mammalian extracts. In addition to the stem-loop structure and dSLBP, 3′ processing in Drosophila nuclear extracts depends on the presence of a short stretch of purines located ca. 20 nucleotides downstream from the stem, and an Sm-reactive factor, most likely the Drosophila counterpart of vertebrate U7 snRNP.
Histone mRNAs, the only metazoan mRNAs that lack poly(A) tails, are generated from longer precursors (pre-mRNAs) by endonucleolytic cleavage after a conserved stem-loop, located 50 to 80 nucleotides downstream from the stop codon (10, 16, 20). The 3′ end processing of mammalian (34, 37) and sea urchin histone pre-mRNAs (55, 55, 59) requires two distinct cis-elements flanking the cleavage site: the stem-loop and a purine-rich element, also referred to as the histone downstream element (HDE). The stem-loop consists of a 6-bp stem and a four-nucleotide loop and is highly conserved in all metazoan organisms (10, 32, 47, 59). The absolutely invariant features of this structure include the second and sixth base pairs of the stem (G-C and U-A, respectively) and the first and third nucleotides of the loop, always uridines, with the only exception found in Caenorhabditis elegans histone mRNAs, which have cytidine in the first position of the loop (35, 41, 46). The first base pair of the stem in most organisms is G-C, although other base pairs are found in Drosophila melanogaster H1 and H2a genes (27).
The stem-loop and the five-nucleotide flanks, invariably AC-rich, are recognized by the stem-loop binding protein (SLBP) (64, 67), also known as the hairpin binding protein (31). In vertebrates, SLBP consists of a central RNA- binding domain (RBD), which mediates binding of the protein to the stem-loop, and two flanking domains (24, 64). The RBD of SLBP does not resemble any known motif involved in sequence-specific RNA recognition and binds the stem-loop with very high affinity (3). The gene encoding SLBP is essential for the viability of both Drosophila (57) and C. elegans (25, 41). In both organisms, deficiency in SLBP function results in reduced expression of histone mRNAs in the germ line and early embryonal lethality due to defects in chromosome condensation and segregation.
The HDE is highly conserved in sea urchins and is more variable in higher organisms, where it is usually limited to a short cluster of purines located ca. 15 nucleotides downstream from the cleavage site. The HDE interacts with U7 snRNP (6, 38, 55, 56), which contains an ca. 60-nucleotide U7 snRNA and associated proteins. These proteins include both some of the core Sm proteins found in the spliceosomal snRNPs, as well as U7 snRNP-specific proteins (49, 53). Among the U7-specific proteins are a zinc finger protein, hZFP100 (9), and a new Sm-like protein, Lsm10, which replaces the Sm D1 and D2 core proteins present in all other snRNPs (42). Binding of U7 snRNP to the pre-mRNA occurs by base pairing between the 5′ end of U7 snRNA and the HDE (5, 48) and is stabilized by interaction of SLBP bound to the histone pre-mRNA with a component of U7 snRNP, most likely hZFP100 (9). Formation of an active processing complex is followed by endonucleolytic cleavage four to five nucleotides downstream from the stem-loop structure. After cleavage, SLBP remains associated with the mature 3′ end (11) and is believed to participate in other steps in metabolism of histone mRNAs, including their nucleocytoplasmic transport (13, 66), translation (18, 58), and degradation (40, 66).
In mammalian cells the efficiency of 3′ end processing is maximal during S phase, resulting in rapid accumulation of mature histone mRNAs and thus allowing the synchronous synthesis of two major components of the chromatin: histones and DNA (22, 30, 52). Cell cycle regulation of histone mRNA formation in mammalian cells is at least in part achieved by cell cycle regulation of SLBP, which accumulates at the beginning of S phase and is destroyed by the proteasome pathway in early G2, after completion of DNA replication (65).
Study of the mechanism of histone pre-mRNA 3′ end processing has been greatly facilitated by the availability of efficient and accurate in vitro systems based on nuclear extracts from human and mouse cells (20, 38). Here we report the development and analysis of the histone 3′ end processing in vitro by using Drosophila components. Nuclear extracts from both Drosophila embryos and cultured cells specifically cleave histone pre-mRNAs containing the downstream sequence element from all five Drosophila histone genes. These nuclear extracts are very inefficient in processing the mouse H2a-614 pre-mRNA. Cleavage in Drosophila nuclear extracts occurs four nucleotides after the stem-loop. Although there is no obvious consensus sequence for an HDE in the Drosophila histone pre-mRNAs, small alterations in the purine-rich region downstream of the cleavage site abolish 3′ end processing in vitro. Neither the human nor the Drosophila SLBP (dSLBP) supports processing in the heterologous extract, suggesting that there has been coevolution of the components of the histone pre-mRNA processing machinery in the vertebrate and invertebrate lineages.
MATERIALS AND METHODS
Histone pre-mRNA processing substrates.
A 225-nucleotide fragment of Drosophila histone H3 gene was amplified by PCR from genomic DNA and subcloned into pBluescript II SK plasmid (Stratagene). This fragment contains 56 nucleotides of the H3 coding region, followed by the stop codon and 166 downstream nucleotides, including the stem-loop structure. Hybrid mouse-Drosophila histone genes were constructed by inserting an HDE from each of the five Drosophila histone genes into an EcoRI site engineered 8 to 13 nucleotides downstream from the stem-loop of mouse H2a-614 gene (Fig. 2). All mutations were generated either by ligating two complementary oligonucleotides carrying appropriate nucleotide substitutions into linearized vectors or by using QuikChange site-directed mutagenesis kit (Stratagene), as specified by the manufacturer.
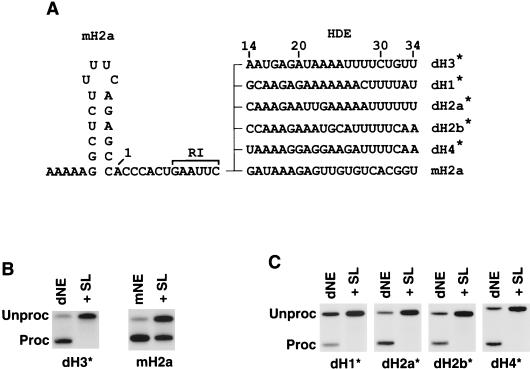
FIG. 2.
dSLBP is essential for 3′ end processing of pre-mRNAs encoded by all Drosophila histone genes. (A) The downstream sequence of the Drosophila H1, H2a, H2b, H3, and H4 histone pre-mRNAs was fused at EcoRI site to the stem-loop and the cleavage site of the mouse H2a pre-mRNA, giving rise to a set of hybrid substrates denoted by an asterisk. The nucleotides are numbered beginning from the first nucleotide after the stem-loop. (B) In vitro processing of dH3∗ (left panel) and mouse H2a (right panel) pre-mRNAs in Drosophila S-2 and mouse nuclear extracts, respectively. The reaction was carried out in the absence (dNE or mNE) or in the presence (+SL) of excess 26-nucleotide RNA containing the stem-loop. (C) In vitro processing of the hybrid pre-mRNAs in S-2 nuclear extract in the absence (dNE) or in the presence of the stem-loop RNA (+SL).
Recombinant dSLBP.
The wild type and deletion mutants of dSLBP were expressed in Sf9 insect cells by using the baculovirus expression system (Gibco-BRL) and purified by affinity chromatography on Ni-NTA agarose (Qiagen) as described previously (12).
Preparation and depletion of nuclear extracts.
Drosophila Schneider line 2 (S-2) cells were grown at room temperature in Shield's and Sung M3 medium (Sigma) containing 12.5% heat-inactivated fetal bovine serum and antibiotics (57). Nuclei were isolated as previously described for mouse myeloma cells and extracted for 1 h with a buffer containing 0.25 M NaCl (11, 33). The nuclear extract from 0 to 13 h embryos was prepared as previously described (45) and provided by L. Searles and K. Williamson (University of North Carolina at Chapel Hill). Depletion of dSLBP and anti-Sm-reactive particles was carried out as previously described for mammalian SLBP and U7 snRNP (9, 33).
RNA labeling and in vitro processing.
Pre-mRNA substrates for in vitro 3′ end processing were synthesized in the presence of [α-32P]CTP (25 μCi) with T7 RNA polymerase (NEB) and linearized DNA templates. To achieve high specific activity labeling, the concentration of unlabeled CTP was reduced to 10 nM while the remaining ribonucleotides were at 100 nM. In some experiments, pre-mRNA substrates were prepared by 5′ end labeling of dephosphorylated RNA with [γ-32P]ATP and T4 polynucleotide kinase (NEB), as previously described (12). The labeled pre-mRNA substrates were separated from the unincorporated isotopes on G-50 spin columns and used for processing without further purification. All pre-mRNA substrates were synthesized in the absence of cap analogues and varied in length between 86 and 403 nucleotides depending on the clone used for transcription and the restriction enzyme used to cleave the template. Each processing reaction contained the following in a final volume of 10 μl: 5 × 103 to 10 × 103 cpm of 32P-labeled histone pre-mRNA, 5 μl of nuclear extract, 20 mM EDTA, and 10 μg of yeast tRNA. The reaction sample was assembled on ice and incubated at 22°C for 2 h. The RNA was purified by phenol extraction and analyzed on 6 to 8% denaturing (7 M urea) polyacrylamide gels. Some preparations of the S-2 nuclear extract contained a high level of nucleases and led to degradation of histone pre-mRNA even in the presence of EDTA. Incorporation of the cap analogue into the pre-mRNAs did not increase their stability in these nuclear extract preparations (not shown).
Dephosphorylation of dSLBP.
Recombinant dSLBP (5 μg) or the nuclear extract (25 μl) was treated for 30 min at 32°C with either 25 U of calf intestinal phosphatase (NEB) or 1,000 U of phage λ protein phosphatase (PPase; NEB) in buffers recommended by manufacturers. To confirm the extent of dephosphorylation, a portion of each sample before and after treatment was analyzed by a band shift assay and Western blotting, as previously described (12).
Protein molecular weight determination.
Prior to mass spectrometric analysis the proteins were purified on microcolumns (14) packed with POROS R1 (C4) resin (Applied Biosystems, Framingham, Mass.). A 5-μl bed volume of the POROS resin was packed in an Eppendorf Gel-loader tip, and the resin was incubated with a solution of 50:50 (vol/vol) acetonitrile-water and then equilibrated with 0.1% trifluoroacetic acid (TFA). A 5-μl aliquot of the protein (0.5 mg/ml) was diluted 1:1 (vol/vol) with 0.1% TFA and passed through the bed five times. The resin was washed with 0.1% TFA, and the protein was eluted with 5 μl of a solution which contained 75:24.8:0.2 acetonitrile-water-formic acid (vol/vol/vol). A 2-μl aliquot of the purified solution was used for measuring the molecular weight of the proteins in an Applied Biosystems/Perkin-Elmer Corp (Foster City, Calif.) API QSTAR-Pulsar (QSTAR) apparatus equipped with a Protana (Odense, Denmark) nanoelectrospray source.
RESULTS
Drosophila histone pre-mRNAs are efficiently processed in nuclear extracts from Drosophila cells.
Histone mRNAs in Drosophila, as in other metazoans, are not polyadenylated and instead contain a highly conserved stem-loop structure at the 3′ end (10, 27, 39). Although substantial progress has been made in studying histone mRNA 3′ end formation in vertebrates and sea urchins, there is little information about this process in Drosophila and other lower eukaryotes. It has been recently shown that dSLBP is required for correct 3′ end processing of all five histone pre-mRNAs in vivo (27, 57). Loss-of-function mutations in the dSLBP gene abolish correct 3′ end processing of histone pre-mRNAs in vivo and result in usage of cryptic polyadenylation signals and formation of polyadenylated histone mRNAs (27, 57). Unlike mammals, which have more than 50 different genes encoding the replication-dependent histone mRNAs (1, 2, 62, 63), in Drosophila there are only five different replication-dependent histone genes, one for each histone protein. These genes are in a cluster that is tandemly repeated ca. 100 times. The sequence of the stem-loop structure encoded by all five Drosophila histone genes differs from the previously established consensus with a major difference found in H1 and H2a sequences, both of which lack the GC base pair at the bottom of the stem, that is conserved in other metazoans. However, in spite of these differences, all five Drosophila stem-loop sequences are tightly bound by both dSLBP and the two Xenopus SLBPs (27). In order to characterize the components necessary for histone pre-mRNA processing in Drosophila, we developed an in vitro system from Drosophila nuclear extracts capable of processing radiolabeled synthetic pre-mRNAs containing Drosophila-specific processing signals.
Alignment of a 55-nucleotide region encompassing the stem-loop structure, the cleavage site, and the putative HDE of all five Drosophila histone pre-mRNAs and mouse H2a-614 pre-mRNA is shown in Fig. 1A. We generated a Drosophila histone H3 pre-mRNA substrate (dH3) containing a portion of the Drosophila H3 coding region and stop codon, followed by an additional 122 nucleotides, that contain the stem-loop and 101 downstream nucleotides, including 34 nucleotides shown in Fig. 1A. The dH3 pre-mRNA was efficiently and accurately processed in the nuclear extract from Drosophila S-2 cells (Fig. 1B, lane 1). Depending on the preparation of nuclear extract and amount of pre-mRNA used in the reaction, between 40 and 95% of the pre-mRNA was cleaved in a 2-h incubation at 22°C. Similar results were obtained with an extract prepared from 0- to 13-h-old embryos (Fig. 1B, lane 2) and Drosophila Kc cells (not shown). In addition to the mature mRNA, the downstream (designated “cutoff”) product of the cleavage reaction was detected when the pre-mRNA was uniformly labeled (Fig. 1B). This product was heterogeneous, most likely due to its subsequent partial degradation by a 5′ exonuclease associated with the processing machinery (60). The reaction in S-2 nuclear extract proceeded without a significant time lag (Fig. 1C), resembling 3′ end processing in mammalian nuclear extract (11, 20, 38). The mature product was readily detected after 15 min, and its amount increased steadily during the 2-h incubation. This suggests that a relatively small number of factors rapidly assemble on histone pre-mRNA to form an active processing complex.
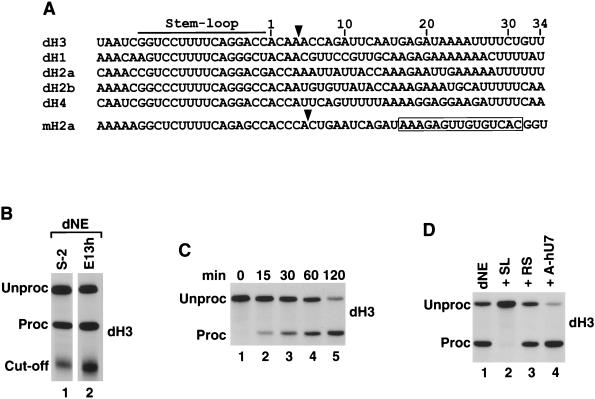
FIG. 1.
dH3 pre-mRNA is efficiently processed in nuclear extracts from Drosophila cells. (A) Sequence alignment of the 3′ end region of all five Drosophila histone pre-mRNAs and the mouse H2a-614 pre-mRNA. The stem-loop structure is indicated with the line, and the U7 binding site in the mouse H2a pre-mRNA is boxed. The known cleavage site used in mammalian processing and the deduced cleavage site used in Drosophila processing (see below) are indicated by the arrow. The nucleotides are numbered beginning from the first nucleotide after the stem-loop. (B) In vitro processing of radiolabeled dH3 pre-mRNA in dNE from S-2 cells (lane 1) and 0- to 13-h-old embryos (lane 2). The reaction was carried out for 2 h at 22°C, and RNA was resolved by electrophoresis in a 6% denaturing gel. “Unproc” and “Proc” correspond to the input pre-mRNA and the upstream cleavage product (mRNA), respectively. The band indicated as the “Cut-off” corresponds to the 3′ cleavage product. (C) Time course of Drosophila 3′ end processing. The dH3 pre-mRNA was incubated for the indicated times in nuclear extract from S-2 cells. (D) The dH3 pre-mRNA was incubated in the S-2 nuclear extract in the presence of excess of 26-nucleotide stem-loop RNA (SL, lane 2), reverse stem RNA (RS, lane 3), or a 2′ O-methyl oligonucleotide complementary to the 5′ end of human U7 snRNA (A-hU7, lane 4). The control processing reaction is shown in lane 1.
The addition of a vast molar excess of the stem-loop RNA, which sequesters dSLBP, to the nuclear extract abolished processing, whereas the addition of the reverse stem RNA, which is unable to bind dSLBP, had no effect (Fig. 1D, lanes 2 and 3, respectively). No inhibitory affect was exerted by a 2′ O-methyl oligonucleotide complementary to first 17 nucleotides of the human U7 snRNA (Fig. 1D, lane 4), a potent inhibitor of processing in human and mouse nuclear extracts. Similar to processing in mammalian nuclear extracts (20), processing in Drosophila nuclear extract does not require divalent ions, and all reactions were carried out in the presence of 20 mM EDTA.
dSLBP is a critical factor in processing of all five Drosophila histone pre-mRNAs.
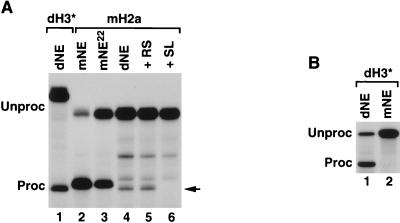
We determined whether dSLBP is also indispensable for in vitro 3′ end processing of the four remaining Drosophila histone pre-mRNAs. In mammalian nuclear extracts the importance of SLBP in 3′ end processing depends on the sequence of the downstream element and its potential to base pair with U7 snRNA (12, 51, 54). Since dSLBP binds efficiently to the stem-loop of the mouse histone H2a-614 pre-mRNA (27), we constructed a series of hybrid pre-mRNAs that contained a common sequence of stem-loop and the cleavage site from the mouse H2a-614 gene fused at an engineered EcoRI site to a 45-nucleotide variable downstream region (nucleotides 14 to 58, counting from the base of the stem) from Drosophila H1, H2a, H2b, and H4 genes (Fig. 2A; nucleotides 14 to 34 are shown). To ensure that the hybrid substrates are processed with efficiency similar to that of the original Drosophila pre-mRNAs, we also constructed a hybrid H3 pre-mRNA. The five hybrid pre-mRNAs (indicated with asterisks) were tested for processing in S-2 Drosophila nuclear extract (dNE) both in the absence and in the presence of excess stem-loop RNA (Fig. 2B and C). Processing of the hybrid dH3∗ pre-mRNA was as efficient as processing of the original dH3 substrate and was completely dependent on dSLBP (Fig. 2B, left panel). In contrast, processing of the mouse H2a-614 pre-mRNA in mouse nuclear extract (mNE) was only partially dependent on SLBP (Fig. 2B, right panel) as reported previously (11, 12). The efficiency of the processing of the hybrid pre-mRNAs varied between 20% (dH1∗) and 80% (dH3∗). The addition of excess stem-loop RNA abolished processing of all of the substrates (Fig. 2C), thus confirming earlier in vivo results that dSLBP is an essential component of the histone pre-mRNA processing machinery in Drosophila (57).
Mutational analysis of the downstream sequence element.
The mammalian and sea urchin U7 snRNPs bind to histone pre-mRNA primarily by base pairing between the 5′ end of U7 snRNA and the HDE. In sea urchin pre-mRNAs, the HDE is limited to the invariant GAAAGA sequence, beginning approximately 10 nucleotides downstream of the cleavage site, which base pair with nucleotides 2 to 7 of the sea urchin U7 snRNA (4, 56). In vertebrate histone pre-mRNAs, the HDE is more variable and the region complementary to the 5′ end of U7 snRNA extends beyond the purine-rich core, including a few additional downstream nucleotides (50, 51).
Comparison of the sequences downstream from the cleavage site in the five Drosophila histone pre-mRNAs did not reveal any highly conserved element, although they all contain a purine-rich region, similar to that in mammalian histone pre-mRNAs (Fig. 1A). In addition, in all Drosophila histone pre-mRNAs there is an interrupted track of uridines located immediately 3′ of the purine-rich element. In order to identify which sequences downstream from the cleavage site are important for Drosophila histone pre-mRNA processing, we introduced a number of nucleotide substitutions in dH3∗ pre-mRNA and analyzed the processing efficiency of the resultant mutants in the Drosophila S-2 nuclear extract. In the first round of mutagenesis, we made two six-nucleotide mutations (Fig. 3A). The first mutation (M1) was introduced into a purine-rich sequence, located in the region occupied in vertebrate pre-mRNAs by the HDE core (nucleotides 17 to 22 downstream from the stem-loop), whereas the second mutation (M2) changed six nucleotides just after the conserved patch of uridines. Replacing the GAGAUA sequence with CUCUAU (M1) abolished processing of dH3∗ histone pre-mRNA, while replacing GCCGAC with CGGCUG (M2) had no effect (Fig. 3B, lanes 2 and 3, respectively). We then made four three-nucleotide mutations spanning the region located between nucleotides 15 to 29 downstream from the stem-loop. One of these mutations, M4, was confined within the larger M1 mutation and changed the AGA sequence to UCU. This mutation reduced processing by more than 95% (Fig. 3B, lane 5) thus confirming the importance of the GAGAUA sequence in 3′ end processing. Two surrounding mutations, M3 and M5, that changed nucleotides immediately flanking the GAGAUA sequence did not significantly affect processing (Fig. 3B, lanes 4 and 6, respectively). The M6 mutation, which disrupted the tract of uridines conserved in all Drosophila histone pre-mRNAs, also had no effect on processing (Fig. 3B, lane 7). In conclusion, we identified in Drosophila histone pre-mRNA a second sequence element, in addition to the stem-loop, that is required for 3′ end processing and contains an essential purine-rich core located between nucleotides 17 and 22 after the stem-loop of the dH3∗ pre-mRNA.
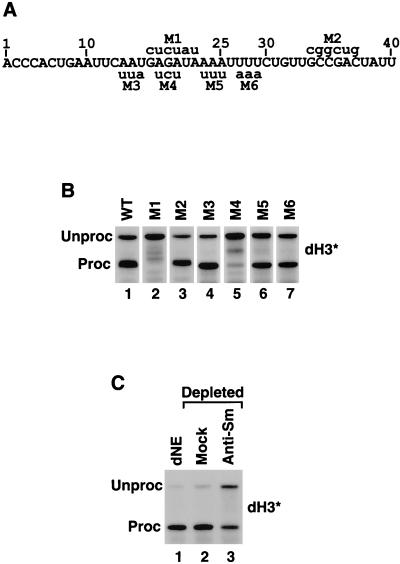
FIG. 3.
A sequence element downstream of the stem-loop is required for Drosophila histone pre-mRNA processing. (A) Sequence of the dH3∗ pre-mRNA starting first nucleotide after the stem-loop. The six mutations M1 to M6 are shown with the nucleotide substitutions indicated above and below the wild-type sequence. (B) In vitro processing of the wild-type (WT) and mutant dH3∗ pre-mRNAs (indicated above each lane) in the nuclear extract from S-2 cells. The processing of M1 and M2 and of M3 to M6 mutant pre-mRNAs was analyzed on two gels (lanes 1 to 3 and 4 to 7), accounting for the differences in separation of unprocessed and processed RNAs across the panel. (C) dNE from S-2 cells was incubated with a control monoclonal antibody (mock, lane 2) or anti-Sm antibody (lane 3), and the antibody complexes were removed from the extract with protein G beads. The processing activity of the depleted nuclear extract was assayed with dH3∗ pre-mRNA. Lane 1 represents untreated nuclear extract.
Both the location and the purine-rich sequence suggest that the downstream element in Drosophila histone pre-mRNAs interacts with the Drosophila equivalent of U7 snRNP. However, in spite of the availability of the complete sequence of the Drosophila genome, U7 snRNA from this organism has yet to be identified. To provide evidence that 3′ end processing in Drosophila requires an snRNP, the nuclear extract from S-2 cells (i.e., dNE) was incubated with the Y12 anti-Sm monoclonal antibody (28). This human antibody recognizes highly conserved epitopes on Sm proteins from such evolutionary distant organisms as mammals and yeast (15, 23). The complexes containing snRNPs were absorbed on protein G-agarose beads (9), and the supernatant was tested for in vitro processing activity with dH3∗ pre-mRNA (Fig. 3C). Silver staining of the RNA recovered from the beads confirmed that the Y12 antibody selectively precipitated a variety of RNA species, apparently including all major classes of the spliceosomal snRNAs from dNE (not shown). The control processing reaction proceeded with high efficiency, generating >95% of the mature product, and mock depletion carried out in the presence of a nonspecific monoclonal antibody had no detectable effect (Fig. 3C, lanes 1 and 2, respectively). However, depletion of the extract with anti-Sm antibody resulted in a reproducible reduction in processing efficiency, indicating the involvement of an snRNP (Fig. 3C, lane 3). A similar level of reduction was previously observed after treatment of the HeLa nuclear extract with the same preparation of anti-Sm monoclonal antibody (9). The failure to completely inhibit processing in both cases most likely results from limited accessibility of the antibody to the Sm epitopes and thus only partial removal of U7 snRNP from the nuclear extract (9).
In addition to depleting the anti-Sm reactive particles from the nuclear extract, we attempted to degrade the 5′ end of a putative Drosophila U7 snRNA by adding micrococcal nuclease in the presence of calcium ions. This treatment has been shown to completely eliminate the processing activity of mammalian nuclear extracts by preventing binding of U7 snRNP to the pre-mRNA (21, 38, 50). However, the same experiments in the Drosophila extract were inconclusive.
Mammalian and Drosophila nuclear extracts cleave histone pre-mRNAs at different sites.
In order to map the Drosophila cleavage site, dH3∗ pre-mRNA processing products generated in the nuclear extract from S-2 cells (dNE) were resolved on denaturing gels next to processing products of the mouse H2a-614 pre-mRNA generated in the nuclear extract from either mouse myeloma (mNE) or human HeLa cells (hNE). These two pre-mRNAs contain identical sequences upstream of the EcoRI site and differ only in the species-specific downstream sequences cleaved off during the processing reaction. Thus, the length of the final processing products generated from the two pre-mRNA substrates can be directly analyzed by comparing their relative electrophoretic mobilities. Surprisingly, the dH3∗ processing product generated in dNE (Fig. 4A, lane 1) migrated slightly faster than the H2a pre-mRNA cleaved in mouse and human nuclear extracts (Fig. 4A, lanes 2 and 3, respectively).
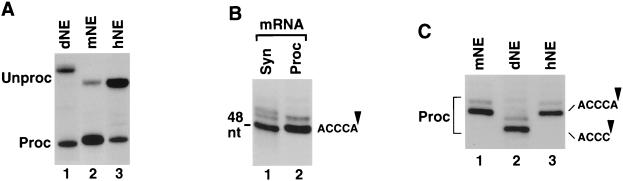
FIG. 4.
Drosophila and mammalian nuclear extracts cleave pre-mRNA at different sites. (A) dH3∗ pre-mRNA was processed in S-2 dNE (lane 1) and mouse H2a pre-mRNA was processed in either mouse myeloma (mNE, lane 2) or HeLa (hNE, lane 3) nuclear extracts. The length of the processing products (Proc) was compared after electrophoresis in an 8% low-resolution denaturing gel. The dH3∗ pre-mRNA (Unproc) substrate is longer from mouse H2a pre-mRNA at the 3′ end by 19 nucleotides, but the sequence of the 5′ end is identical. (B) The processing product of mouse H2a pre-mRNA generated in mNE (lane 2) was analyzed in high-resolution, 40-cm 12% polyacrylamide gels, next to a 48-nucleotide synthetic RNA ending precisely at the ACCCA sequence (lane 1). (C) Mammalian (lanes 1 and 3) and Drosophila (lane 2) processing products, generated as described for panel A, were analyzed side by side in a high-resolution gel, as described for panel B.
Mammalian histone pre-mRNAs, including the mouse H2a-614 pre-mRNA, are cleaved predominantly after an adenosine residue five nucleotides downstream from the stem-loop, generating mature histone mRNAs with the ACCCA sequence and a hydroxyl group at the 3′ end (16, 47). We first determined whether the 86-nucleotide derivative of the mouse H2a-614 pre-mRNA utilized in the present study is cleaved at the same site and gives rise to the predicted 48 nucleotides long final product. Using a PCR-generated DNA template containing the T7 RNA polymerase promoter and ending precisely at the ACCCA sequence, we synthesized a radiolabeled RNA standard corresponding in length and sequence to the correctly processed mRNA. High-resolution gel electrophoresis carried out in 40-cm 12% polyacrylamide gels capable of resolving RNA species differing by one nucleotide revealed that this synthetic mRNA comigrates with the processing product generated in mouse nuclear extract (Fig. 4B, lanes 1 and 2, respectively), confirming that the 86-nucleotide H2a pre-mRNA substrate is cleaved immediately 3′ of the ACCCA sequence. The same high-resolution gel electrophoresis revealed that the difference in mobility between Drosophila and mammalian processing products corresponds to approximately 1.5 nucleotides (Fig. 4C). We conclude that the final product of Drosophila processing (Fig. 4C, lane 2) is one nucleotide shorter than the product of mammalian processing (Fig. 4C, lanes 1 and 3) and most likely contains a 3′ phosphate group accounting for the additional half nucleotide higher mobility (7, 16). The cleavage of the hybrid pre-mRNAs in dNE must therefore occur after the last C in the ACCCA sequence. Interestingly, there is an A four nucleotides downstream from the stem-loop in all five Drosophila histone pre-mRNAs (Fig. 1A), suggesting that cleavage after an adenosine is also favored by the Drosophila histone processing system.
Processing of histone pre-mRNA in heterologous nuclear extracts.
We next tested whether the mouse H2a pre-mRNA could be processed in dNE and whether the cleavage site would be of Drosophila or mammalian specificity. The purine-rich region in the mouse H2a pre-mRNA does not differ significantly from the corresponding Drosophila region, which is relatively variable itself among five different Drosophila histone pre-mRNAs (Fig. 1A). Incubation of the mouse H2a pre-mRNA in Drosophila S-2 nuclear extract for 2 h at 22°C yielded a very small amount of RNA that comigrated with the dH3∗ processing product, as well as two additional larger RNA species (Fig. 5A, lane 4). The addition of excess stem-loop RNA (SL), but not reverse stem RNA (RS), to the processing reaction completely inhibited generation of the RNA comigrating with the dH3∗ processed mRNA, indicating that it represents a genuine processing product (Fig. 5A, lanes 6 and 5, respectively). Since the two remaining RNA species were still detected in the presence of the stem-loop competitor RNA, they must represent products of nonspecific degradation. The mouse H2a pre-mRNA incubated in mouse myeloma nuclear extract at both 22°C (Fig. 5A, lane 3) and the regular 32°C (Fig. 5A, lane 2) generated a product that was one nucleotide longer than the product of processing in dNE. The fact that the two substrates containing the downstream element from either mouse or Drosophila pre-mRNAs yield a final product of the same size in S-2 nuclear extract indicates that the Drosophila processing machinery directs cleavage four nucleotides after the stem-loop. Although mouse histone pre-mRNA was weakly recognized by the Drosophila processing machinery, the dH3∗ pre-mRNA was not processed in the mouse nuclear extract (Fig. 5B, lane 2).
FIG. 5.
Processing of histone pre-mRNA in the heterologous nuclear extracts. (A) The mouse H2a pre-mRNA is processed with low efficiency in S-2 dNE (lane 4). Generation of the processing product, indicated with the arrow, is inhibited by excess RNA containing the wild-type stem-loop (SL, lane 6) but not the reverse stem mutation (RS, lane 5). Processing of dH3∗ pre-mRNA in a limited amount of S-2 dNE (lane 1) and mouse H2a pre-mRNA in mNE at 32 and 22°C (lanes 2 and 3) are shown for comparison. Processing of dH3∗ pre-mRNA was carried out under suboptimal conditions to reduce amount of the final product and better assess its migration mobility. (B) mNE does not process dH3∗ pre-mRNA. Processing of dH3∗ pre-mRNA was tested in nuclear extract from mouse myeloma cells (mNE, lane 2). Processing of the same substrate in Drosophila S-2 nuclear extract (dNE, lane 1) is shown for comparison.
Minimal region of dSLBP proficient in processing.
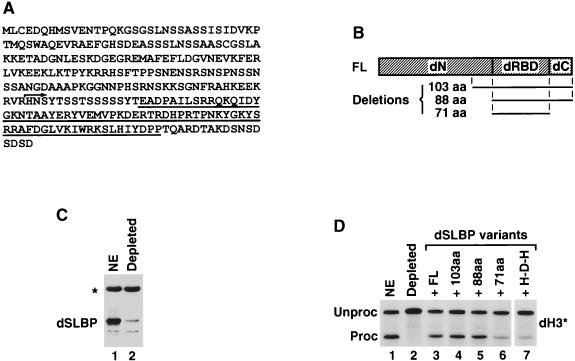
The mammalian SLBPs (64) and the Xenopus SLBPs (24, 61) are composed of three domains. Interaction of SLBPs with the histone stem-loop is mediated by the centrally located 73-amino-acid RNA-binding domain (RBD) (64). We have previously shown that the RBD and the first 35 amino acids of the C-terminal domain of human SLBP are sufficient for full processing activity in vitro (9, 12). The remainder of the protein, including the entire N-terminal domain, is dispensable for the processing and likely plays a role in other activities of SLBP, e.g., transport and translation of histone mRNAs and regulation of SLBP stability during the cell cycle. dSLBP has obvious similarity to the vertebrate SLBPs only in the RBD and differs from vertebrate SLBPs since it lacks the C-terminal domain and instead contains only 17 amino acids downstream from the RBD (Fig. 6A and B) (57). SLBPs from both Drosophila and vertebrates are ∼30 kDa since the N-terminal domain of dSLBP is much larger than the corresponding domain in vertebrate SLBPs.
FIG. 6.
Recombinant dSLBP restores processing activity to dSLBP-depleted nuclear extract. (A) Amino acid sequence of dSLBP. The RBD is underlined, and the arrows indicate the starting point of a 103-amino-acid deletion variant. (B) Diagram of the full-length dSLBP (FL) consisting of the N-terminal domain (dN), the Drosophila RBD (dRBD), and the 17-amino-acid end (dC). Regions included in three deletion mutants are shown below. The 103-amino-acid mutant starts 15 amino acids upstream from the Drosophila RBD, as shown in panel A, and contains the remainder of the protein, whereas the 88-amino-acid protein begins with the Drosophila RBD. The 71-amino-acid protein consists only of Drosophila RBD. (C) The S-2 nuclear extract was analyzed by Western blotting before (lane 1) and after depletion with dSLBP antibody (lane 2). The asterisk indicates a cross-reacting protein not removed from the extract during depletion. (D) The dH3∗ histone pre-mRNA was incubated in S-2 nuclear extract (lane 1), in dSLBP-depleted nuclear extract alone (lane 2), or in the presence of the baculovirus-expressed variants, as indicated (lanes 2 to 7). The H-D-H is a hybrid protein consisting of Drosophila RBD and two flanking domains from human SLBP (see Fig. 9A for details).
To define which regions of dSLBP (Fig. 6A) were required for 3′ end processing, we used an in vitro complementation assay. The nuclear extract was depleted of dSLBP with an antibody to dSLBP, and the SLBP-depleted nuclear extract was supplemented with various deletion mutants of dSLBP expressed in the baculovirus system (Fig. 6B). Incubation of the S-2 nuclear extract with anti-dSLBP antibody, followed by adsorption of immunocomplexes on protein A-agarose, resulted in the removal of >95% of the dSLBP, as analyzed by Western blotting (Fig. 6C, lane 2). The immunodepleted nuclear extract was virtually inactive in processing of dH3∗ pre-mRNA (Fig. 6D, lane 2), a finding consistent with those of previous competition experiments in which dSLBP was sequestered by excess stem-loop RNA. The addition of the full-length dSLBP expressed from a baculovirus vector restored the processing activity of the depleted extract (Fig. 6D, lane 3). Two deletion mutants, containing the last 103 or 88 amino acids, also restored processing (Fig. 6D, lanes 4 and 5, respectively). The 88-amino-acid mutant protein contains only the RBD and the C terminus. A 71-amino-acid protein consisting only of the RBD had very low activity (Fig. 6D, lane 6). To determine whether the inability of the RBD to efficiently function in processing reflected a requirement for a specific sequence in the C-terminal region of dSLBP or simply a requirement for additional amino acids, we constructed the hybrid protein H-D-H, which contains the RBD from dSLBP and both flanking domains from human SLBP (see Fig. 9A for graphic representation). The H-D-H protein had the same low activity in processing as the Drosophila RBD (Fig. 6D, lane 7). Thus, the RBD and at least some of the last 17 amino acids are necessary and sufficient for 3′ end processing.
FIG. 9.
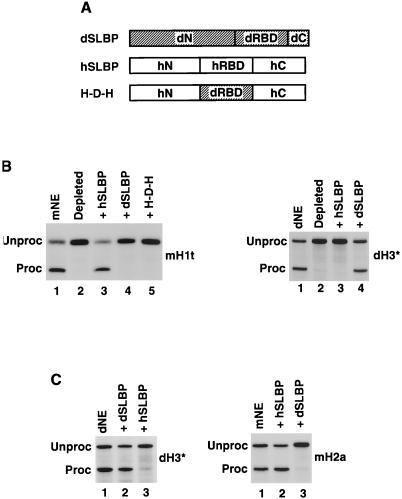
Drosophila and human SLBPs are not interchangeable. (A) Diagram of dSLBP and human SLBP (hSLBP) and a hybrid protein H-D-H composed of the RBD from dSLBP (dRBD) and both flanking domains from human SLBP (hN and hC). (B) The mouse H1t (left panel) and dH3∗ (right panel) pre-mRNAs were processed in the species-specific SLBP-depleted nuclear extracts alone (lane 2 in each panel) or in the presence of the baculovirus-expressed proteins, as indicated. Lane 1 in each panel shows processing in the control, undepleted nuclear extract. (C) Drosophila and human SLBPs inhibit processing in the heterologous nuclear extract. The Drosophila dH3∗ (left panel) and mouse H2a (right panel) pre-mRNAs were incubated in the Drosophila and mouse nuclear extracts, respectively (lane 1 in each panel) and in the same extracts in the presence of the indicated baculovirus-expressed SLBPs (lanes 2 and 3 in each panel).
Dephosphorylation of dSLBP reduces its activity in processing.
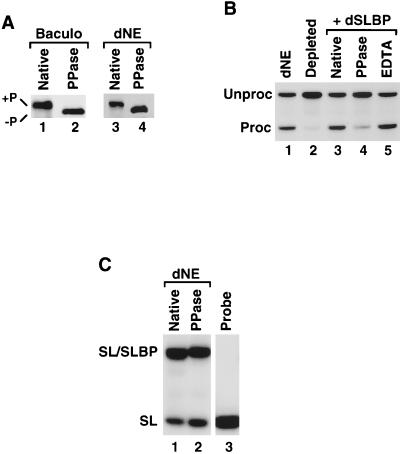
Endogenous dSLBP present in S-2 cells and in Drosophila embryos is quantitatively phosphorylated (27), as is recombinant dSLBP expressed both in the baculovirus system and in the rabbit reticulocyte lysate. Treatment of either the recombinant dSLBP or the nuclear extract with PPase (Fig. 7A, lanes 2 and 4, respectively) or calf intestinal phosphatase (not shown) resulted in an increased mobility of dSLBP on sodium dodecyl sulfate (SDS)-gels.
FIG. 7.
Phosphorylation of dSLBP is required for processing. (A) Western blotting of baculovirus-expressed dSLBP (left panel) and the dSLBP in the S-2 nuclear extract (right panel) before (lanes 1 and 3) and after (lanes 2 and 4) treatment with PPase. (B) Processing of the dH3∗ pre-mRNA was analyzed in dNE (lane 1), in the dSLBP-depleted dNE (lane 2), after the addition of the untreated baculovirus-expressed dSLBP (lane 3), in dSLBP treated with PPase in the presence of Mn2+ (lane 4), or in dSLBP treated with PPase in the presence of Mn2+ and 20 mM EDTA (lane 5). (C) Equal amounts of the dNE (lane 1) or the dNE treated with PPase (lane 2) were analyzed by a mobility shift assay for the ability to bind to a 26-nucleotide stem-loop RNA labeled with [32P]ATP at the 5′ end. Lane 3 contains only the free probe.
To determine whether phosphorylation is required for activity of dSLBP in processing, the baculovirus expressed protein was treated with PPase. The extent of dephosphorylation was monitored by comparison of the electrophoretic mobility of dSLBP before and after the treatment (Fig. 7A). Dephosphorylation of the baculovirus-expressed protein reduced its activity in the complementation assay to 20 to 25% of the original activity (Fig. 7B, lane 4). Treatment of the dSLBP with PPase in the presence of EDTA, which chelates essential manganese ions, thus preventing PPase function, did not result in detectable removal of phosphate groups from dSLBP (not shown) and did not affect processing activity (Fig. 7B, lane 5). As shown in Fig. 7C, native (lane 1) and dephosphorylated (lane 2) dSLBP from dNE bound to the stem-loop probe with comparable efficiency. Thus, the reduced processing activity of dephosphorylated dSLBP was not a result of its inability to bind the stem-loop. Note that, whereas dephosphorylation of dSLBP increased its electrophoretic mobility in SDS-polyacrylamide gels (Fig. 7A), it reduced the migration of the dSLBP-RNA complex in the native gel (Fig. 7C).
SLBP is phosphorylated on the C-terminal 17 amino acids.
As judged by the increase in mobility on an SDS-gel after treatment with PPase, both the 103- and 88-amino-acid fragments of dSLBP (Fig. 6B) were quantitatively phosphorylated (Fig. 8A, 1lanes 2 and 4). The mobility of the 71-amino-acid RBD was not altered by treatment with PPase (Fig. 8A, lane 6), suggesting that the phosphates were present on the C-terminal 17 amino acids.
FIG. 8.
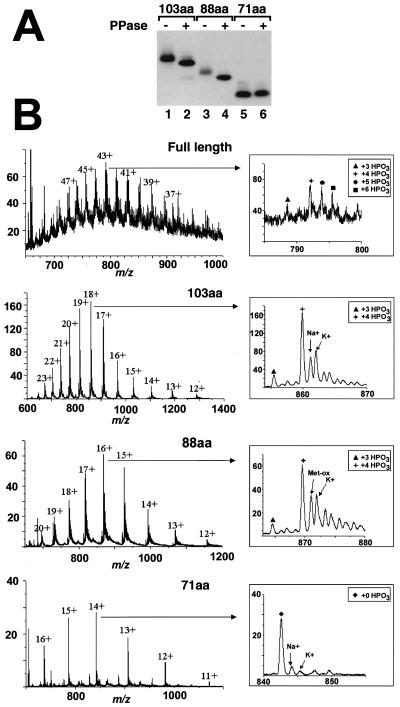
dSLBP contains four phosphates on the C-terminal region. (A) The deletion mutants of dSLBP shown in Fig. 5B were expressed by using the baculovirus system and purified. Aliquots of each variant were treated with PPase, and the untreated (lanes 1, 3, and 5) and dephosphorylated (lanes 2, 4, and 6) proteins were separated by gel electrophoresis and then detected by Western blotting. The mobility of the 103 (lanes 1 and 2)- and 88 (lanes 3 and 4)-amino-acid dSLBPs was increased after PPase treatment, whereas the mobility of the 71-amino-acid protein containing only the RBD was unchanged (lanes 5 and 6). (B) The full-length dSLBP and the 103-, 88-, and 71-amino-acid dSLBPs expressed in insects cells were analyzed by mass spectrometry as described in Materials and Methods. The charge/mass (m/z) spectrum is shown, and the charges on the individual clusters of peaks are indicated. The insets show a blowup of the most abundant ion. The various phosphorylated forms and proteins associated with sodium or potassium ions are indicated. The major peaks in the full-length, 103- and 88-amino-acid dSLBPs correspond to a molecular mass of 320 Da higher than that predicted from the amino acid sequence, corresponding to the four phosphate groups. The two satellite peaks indicated by the circle and square in the full-length dSLBP correspond to additional partially phosphorylated molecules containing five and six phosphates, respectively, whereas the peak indicated by the triangle corresponds to a protein containing three phosphates. The major peak in the 71-amino-acid SLBP corresponds to the molecular weight predicted from the amino acid sequence, indicating that the RBD is not phosphorylated.
In order to localize the sites of phosphorylation in dSLBP, we determined the molecular weights of the full-length protein and the deletion mutants by nanoelectrospray ionization-mass spectrometry (nanoESI-MS) (Fig. 8B). All of the proteins were expressed in the baculovirus system and thus contained on the N terminus additional 25 amino acids from the His tag and vector sequences. The molecular weight determination of the full-length protein (Fig. 8B, top panel) showed that the most abundant form of the protein had four phosphates, whereas a small amount of the protein contained three phosphates. In addition, the molecular weight determination of the full-length SLBP revealed two additional series of peaks, which correspond to the partial incorporation of a fifth and sixth phosphoryl group on some molecules. The molecular weight determination of the 103- and 88-amino-acid dSLBP mutants revealed one abundant series of ions for each species, demonstrating that predominantly one protein isoform was present. The most abundant ion series in each case corresponded to additional mass increments of 319.12 and 320.11 Da compared to the molecular mass expected for the unmodified protein. These mass differences correspond very closely to the addition of four phosphoryl groups (319.87 Da) and demonstrate that each of the analyzed proteins is almost completely phosphorylated on four sites. The spectrum of the RBD also showed one abundant series of multiply charged ions, with a molecular mass of 11,782 Da (standard deviation, ±0.3 Da), a finding that is in good agreement with the calculated molecular mass of nonphosphorylated RBD (monoisotopic molecular mass, 11,785 Da; average molecular mass, 11,792 Da; Fig. 8B, bottom panel). These results are consistent with the C-terminal region of dSLBP being phosphorylated on four sites (amino acids 285 to 301 in the baculovirus-expressed full-length protein). There are six potential phosphorylation sites within the sequence of the C terminus. Thr285 can be excluded because the nonphosphorylated tryptic peptide (amino acids 277 to 288) has been identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis in a separate experiment, and the phosphorylated peptide was not detected (data not shown). These results demonstrate that there are four quantitatively phosphorylated sites on the C-terminal 13 amino acids (i.e., amino acids 289 to 301) and two additional partially phosphorylated sites in the full-length dSLBP.
Components of mammalian and Drosophila processing machinery are not interchangeable.
We have previously shown by using both in vitro and in vivo assays that Xenopus SLBP1 (xSLBP1) and human SLBP can substitute for each other in 3′ end processing (24, 61). This interchangeability was not surprising given the strong amino acid conservation of SLBP in vertebrates throughout the entire length of the proteins. We determined whether the Drosophila and human SLBPs, which are both involved in processing, were also interchangeable. While the RBDs from Drosophila and mammalian SLBPs are highly conserved, retaining 56% identity and 80% similarity over the 71-amino-acid total length (57), the remaining regions of the proteins share no obvious homology. As shown previously (8, 12), the mouse nuclear extract (mNE) efficiently cleaved mouse H1t pre-mRNA and depletion of the endogenous SLBP abolished processing of this substrate (Fig. 9B, left panel, lanes 1 and 2, respectively). High efficiency of processing was restored by the addition of the baculovirus-expressed human SLBP (Fig. 9B, left panel, lane 3) but not dSLBP (Fig. 9B, left panel, lane 4), indicating that the proteins from these two evolutionary distant organisms are not interchangeable.
The inability of dSLBP to function in mammalian 3′ end processing could be explained by the absence of the important C-terminal residues in dSLBP, shown to be essential for processing activity of human SLBP (12). To test this possibility, we constructed a hybrid protein H-D-H in which the RBD of human SLBP was replaced by the RBD from dSLBP (Fig. 9A). The H-D-H protein contains the entire C-terminal domain of human SLBP, including the 35-amino-acid region important for processing in mammalian nuclear extracts. However, when tested in the complementation assay, the H-D-H protein failed to restore processing activity to the mouse SLBP-depleted nuclear extract (Fig. 9B, left panel, lane 5). Thus, the Drosophila RBD cannot substitute for the human RBD in processing.
The dNE depleted of the endogenous dSLBP can be efficiently complemented by dSLBP expressed in the baculovirus system (Fig. 9B, right panel, lane 4) but not by human SLBP (Fig. 9B, right panel, lane 3), further confirming that these two proteins are not interchangeable. Human SLBP and dSLBPs strongly inhibit efficiency of processing when added in excess to the heterologous nuclear extract (Fig. 9C). This dominant-negative effect most likely results from the exogenous SLBP competing with the endogenous SLBP for binding to histone pre-mRNA but being unable to form a productive processing complex with other components of processing machinery from the evolutionary distant organism.
DISCUSSION
Development of an in vitro system based on nuclear extracts from human and mouse cells was a major advance that allowed a molecular analysis of 3′ end processing of mammalian histone pre-mRNA (12, 20, 38, 50). We describe here the development of an in vitro system based on nuclear extract from Drosophila S-2 cells and Drosophila histone pre-mRNAs. This system was utilized for mapping the structural features in Drosophila histone pre-mRNA and dSLBP essential for 3′ end processing.
In vitro 3′ end processing in dNE.
Drosophila cultured cells are a convenient and relatively inexpensive source of nuclear extracts proficient in transcription (44) or splicing (45) and were recently used for large-scale purification of spliceosomal snRNPs (26). The nuclear extract from Drosophila S-2 cells and 0- to 20-h-old embryos are also very efficient in 3′ end processing of all five Drosophila histone pre-mRNAs. Since Drosophila contains only one gene (present in multiple copies) for each of the five different histone proteins, it is essential that all five Drosophila histone pre-mRNAs be efficiently processed. In mammalian cells there are multiple nonallelic copies of each histone gene (2, 62), and the processing efficiency of different pre-mRNAs encoded by these copies significantly varies in vivo and in vitro (29).
Drosophila histone pre-mRNA processing in vitro has biochemical properties similar to processing in mammalian cells. The reaction does not require divalent ions or ATP and generates the final product without a significant lag time, suggesting that a relatively small number of factors assemble to form a functional processing complex. The presence of the cleaved 3′ fragment indicates that generation of the mature 3′ end in Drosophila histone mRNAs occurs through endonucleolytic cleavage and not by the activity of a 3′ exonuclease. In dNE, histone pre-mRNAs are processed four nucleotides after the stem-loop, whereas in mammalian nuclear extracts cleavage occurs one nucleotide farther downstream. There is an adenosine residue four nucleotides after the stem-loop in all five Drosophila histone pre-mRNAs, whereas most mammalian histone mRNAs end in ACCCA, suggesting that cleavage after an A has been conserved in evolution.
Development of the in vitro 3′ end processing system of histone pre-mRNAs based on Drosophila components has been previously reported by Price and Parker (43). In contrast to the present study, processing in their system required magnesium ions, which could be partially substituted by calcium and manganese. Since sequence and biochemical requirements have not been studied in the previous work, its relevance to the processing system described here is unknown. It is possible that the correct RNA product detected in the presence of magnesium was in fact generated by a powerful 3′ to 5′ exonuclease stalled near the stem-loop structure tightly bound by dSLBP rather than by an endonucleolytic cleavage. Such nucleases are also present in our preparations of Drosophila and mammalian nuclear extracts, but their activity is completely inhibited by the presence of 20 mM EDTA (20), which was routinely added to all of the processing reactions described here.
A second cis-acting sequence required for Drosophila H3 pre-mRNA processing.
Nuclear extract from Drosophila S-2 cells was very active in cleaving histone pre-mRNAs containing the downstream element encoded by all five Drosophila histone genes. In sea urchins an invariant sequence (CAAGAAAGA) was readily identified in the downstream element of all histone pre-mRNAs (19), and this sequence was later shown to base pair with the 5′ end of sea urchin U7 snRNA (48, 56). The Drosophila histone genes do not share any highly conserved sequences downstream of the stem-loop, although they are all generally purine-rich. Mutagenesis studies of the downstream sequence from Drosophila H3 pre-mRNA revealed that a GAGAUA element plays a critical role in processing; substitution of this sequence with the complementary nucleotides (mutant M1) abolished in vitro processing, whereas mutation of adjacent nucleotides had no effect. In addition to identifying the downstream processing element in Drosophila H3 pre-mRNA, the M1 mutant provides further evidence that our in vitro system reproduces a genuine processing event and is not a result of 3′ to 5′ exonucleases activity stalled by the stem-loop associated with dSLBP. The sequence requirements of the downstream element must be more complex than simply the presence of the purine-rich element at a proper distance from the stem-loop since the mouse H2a pre-mRNA contains a similar purine-rich sequence in the same location and is processed in the Drosophila extract very inefficiently, in contrast to all five Drosophila histone pre-mRNAs. Perhaps there are other sequence elements in Drosophila histone pre-mRNAs that are not conserved in pre-mRNAs of higher organisms and which contribute to high efficiency of processing in dNE.
While the low efficiency of processing of mouse H2a pre-mRNA in Drosophila extract is puzzling, it is easier to explain the inability of the mNE to process Drosophila H3 pre-mRNA. The downstream element from Drosophila H3 pre-mRNA has a very limited complementarity to the 5′ end of mouse U7 snRNA and in the optimal configuration the two RNAs can form only 10 bp over a 19-nucleotide region, with the longest uninterrupted stretch of duplex RNA consisting of only 4 bp. For comparison, the mouse H1t pre-mRNA, previously shown to be a poor and completely SLBP-dependent substrate, can form either 11 or 13 bp in two alternative alignments with the U7 snRNA, and the longest uninterrupted duplex consists of 7 and 6 bp, respectively. In contrast, the mouse H2a-614 pre-mRNA, a good mammalian processing substrate, forms 14 base pairs with mouse U7 snRNA interrupted by only one mismatch. Thus, given the requirement for an extensive duplex between U7 snRNA and the downstream element for mammalian 3′ end processing, the inability of the mNE to process Drosophila H3 pre-mRNA is not surprising.
The downstream purine-rich sequence identified in these studies as essential for processing in the dNE is most likely recognized by the Drosophila equivalent of U7 snRNP. This interpretation is supported by the finding that Sm antibodies, but not control monoclonal antibodies, reproducibly reduce the efficiency of histone pre-mRNA processing. However, despite sequencing the entire Drosophila genome, U7 snRNA has not yet been identified in this organism. Both the small size and the limited evolutionary conservation precludes a search for this RNA based on sequence similarity to known vertebrate and sea urchin U7 snRNAs.
In mammalian extracts the importance of SLBP in 3′ end processing in vitro varies considerably between multiple histone pre-mRNAs and depends on the strength with which U7 snRNA base pairs with the downstream element (12, 51, 54). When there is limited complementarity between the downstream element and 5′ end of U7 snRNA, as in the case of mouse H1t pre-mRNA, there is a complete dependence of processing on SLBP (12). Processing of some mammalian histone mRNAs that are capable of extensive base pairing with the U7 snRNA can occur in the absence of SLBP (12, 51, 54). In contrast, in vitro processing of all five Drosophila histone pre-mRNAs is virtually completely dependent on the presence of SLBP. This dependence most likely results from the relatively short downstream element in Drosophila histone pre-mRNA, which in dH3 pre-mRNA appears to be ca. six nucleotides long, and the 5′ end of a putative Drosophila U7 snRNA. In sea urchins the region of complementarity between pre-mRNA and U7 snRNA is limited to only 6 bp and the formation of rather short duplexes between the two RNAs and thus a requirement for additional interaction involving U7 snRNP and SLBP may be a general feature of 3′ end processing in lower metazoans.
We have previously studied a series of mutations in the dSLBP gene that result in a large reduction in dSLBP concentration in vivo. In the mildest of the mutants, which are viable and female sterile and still express some dSLBP, there is a great reduction in the amount of histone mRNA synthesized during oogenesis (27, 57), resulting in embryonic lethality due to a lack of histone proteins and the inability to complete the syncytial cell cycles. More severe mutants are zygotically lethal, with death occurring in the larval stages. Interestingly, zygotic mutants express a significant amount of polyadenylated histone mRNA during embryogenesis as a result of transcription past the stem-loop and usage of cryptic polyadenylation sites present 3′ of each of the Drosophila histone genes (27). However, even the most severe dSLBP mutants generate during embryogenesis a substantial amount of histone mRNA that ends at or near the stem-loop (27, 57). Whether these histone mRNAs are formed by a small amount of dSLBP remaining in these embryos (which in at least one mutant cannot be detected by biochemical assays) (27, 57) or whether there is an alternative mechanism for forming and stabilizing mRNAs ending at the stem-loop is not known. If the latter situation is true, then this mechanism does not function in nuclear extracts from Drosophila cultured cells.
Differences between mammalian and Drosophila 3′ end processing.
A striking feature of dSLBP not shared by vertebrate SLBPs is its hyperphosphorylation. dSLBP overexpressed in insect cells is quantitatively phosphorylated on four sites within the C-terminal region. Based on electrophoretic mobility, a similar level of phosphorylation is present both in embryonic dSLBP and dSLBP expressed in Drosophila cultured cells. Phosphorylation of dSLBP is essential for complete processing activity of the protein in vitro. Since the dephosphorylated and the phosphorylated dSLBP bind to the stem-loop with similar affinity, the phosphorylation must be required for interaction of dSLBP with other factors involved in histone pre-mRNA processing. In the last 13 amino acids of dSLBP there are four serines, which alternate with aspartic acid residues, and it is likely that dSLBP is phosphorylated on these four serines, creating a highly acidic C terminus. Full-length dSLBP also contains at least two partially phosphorylated sites. Detailed mutational analysis will be required to determine which of these sites are critical for histone pre-mRNA processing.
Reversible changes in phosphorylation status of dSLBP would provide an attractive mechanism for regulating function of dSLBP during embryogenesis and/or the cell cycle. Dephosphorylation would convert the active protein into an inactive form that would effectively inhibit processing. In mammals SLBP accumulates to the highest level in S phase and is degraded by the proteasome pathway immediately after completion of DNA replication (65). It is not known whether dSLBP displays the same pattern of accumulation and disappearance during the cell cycle in Drosophila cells, but reversible changes in phosphorylation status could provide an equally efficient mechanism of adjusting dSLBP activity. While regulation of dSLBP phosphorylation is a possible attractive mechanism that could contribute to regulation of histone pre-mRNA processing, we have observed only hyperphosphorylated SLBP in Drosophila cultured cells (Fig. 7) and embryos (27). However, we would not necessarily have detected changes in dSLBP phosphorylation that affected only one or two sites, particularly if the modification did not alter the electrophoretic mobility.
Rapid evolution of the histone pre-mRNA processing machinery.
The initial characterization of histone pre-mRNA processing in sea urchins was made possible because one of the sea urchin histone pre-mRNAs was not processed in frog oocytes as a result of differences in the HDE (17, 56). The Drosophila histone pre-mRNAs are not processed in mammalian extracts, and the mammalian mRNAs are processed very inefficiently in Drosophila extracts. Moreover, Drosophila and human SLBPs are not interchangeable: dSLBP does not function in 3′ end processing in mammalian nuclear extracts, and human SLBP fails to complement dSLBP-depleted nuclear extract from Drosophila S-2 cells. Instead, each protein has a strong inhibitory effect on processing in the heterologous nuclear extract by competing with the endogenous SLBP for binding to the stem-loop in histone pre-mRNA. The processing activity of both dSLBP and human SLBP requires the RBD and the adjacent amino acids of the C-terminal region. Amino acid conservation between Drosophila and mammalian SLBPs is limited only to the RBD and does not extend into the C-terminal region, thus explaining the inability of each protein to substitute for each other. Interestingly, the RBDs are also not interchangeable. The hybrid H-D-H SLBP containing both flanking domains from human SLBP and the RBD from dSLBP does not support processing in mammalian nuclear extracts. We have recently shown that the human RBD contains a nine-amino-acid region dispensable for RNA binding but necessary for processing (8). This region, together with the C-terminal residues, is involved in interaction of the SLBP-pre-mRNA complex with a novel 100-kDa zinc finger protein (hZFP100) associated with the U7 snRNP (9). The nine-amino-acid region includes the critical DR dipeptide, which is changed in dSLBP to ER. It is possible that this single amino acid substitution, while allowing dSLBP to function efficiently in dNE, is largely responsible for inability of the hybrid H-D-H protein to function in mammalian nuclear extract. These data are consistent with coevolution of the machinery independently in the vertebrate and invertebrate lineages.
Conclusions.
The data presented in the present study suggest that there are many similarities between the processing machineries in Drosophila and higher organisms, including the requirement for SLBP and the purine-rich sequence downstream from the cleavage site. However, the components have diverged significantly during evolution and are no longer recognized in the heterologous systems. Formally, it is still possible that there is no U7 snRNA in Drosophila, since there is no U12 snRNA, although there are ATAC introns and a U11 snRNA (36). This would imply that the mechanism of histone pre-mRNA processing in Drosophila is substantially different from that in sea urchins and vertebrates. It might be significant that three other proteins required for mammalian histone pre-mRNA processing—hZFP100 (9) and two U7 specific proteins, Lsm10 (42) and Lsm11 (R. S. Pillai and D. Schumperli, unpublished data)—have no obvious homologues in Drosophila genomes. Undoubtedly, future studies with dNE will be very helpful in providing more information about the mechanism of histone pre-mRNA processing in this organism.
Acknowledgments
This work was supported by NIH grant GM58921 to W.F.M. and an NSF Career Award to R.J.D.
We thank L. Searles and K. Williamson (University of North Carolina at Chapel Hill) for the nuclear extract from Drosophila embryos.
REFERENCES
- 1.Albig, W., and D. Doenecke. 1997. The human histone gene cluster at the D6S105 locus. Hum. Genet. 101:284-294. [DOI] [PubMed] [Google Scholar]
- 2.Albig, W., P. Kioschis, A. Poustka, K. Meergans, and D. Doenecke. 1997. Human histone gene organization: nonregular arrangement within a large cluster. Genomics 40:314-322. [DOI] [PubMed] [Google Scholar]
- 3.Battle, D. J., and J. A. Doudna. 2001. The stem-loop binding protein forms a highly stable and specific complex with the 3′ stem-loop of histone mRNAs. RNA 7:123-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnstiel, M. L., and F. J. Schaufele. 1988. Structure and function of minor snRNPs, p. 155-182. In M. L. Birnstiel (ed.), Structure and function of major and minor small ribonucleoprotein particles. Springer-Verlag, Berlin, Germany.
- 5.Bond, U. M., T. A. Yario, and J. A. Steitz. 1991. Multiple processing-defective mutations in a mammalian histone premessenger RNA are suppressed by compensatory changes in U7 RNA both in vivo and in vitro. Genes Dev. 5:1709-1722. [DOI] [PubMed] [Google Scholar]
- 6.Cotten, M., O. Gick, A. Vasserot, G. Schaffner, and M. L. Birnstiel. 1988. Specific contacts between mammalian U7 snRNA and histone precursor RNA are indispensable for the in vitro RNA processing reaction. EMBO J. 7:801-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cruz-Reyes, J., K. J. Piller, L. N. Rusche, M. Mukherjee, and B. Sollner-Webb. 1998. Unexpected electrophoretic migration of RNA with different 3′ termini causes a RNA sizing ambiguity that can be resolved using nuclease P1-generated sequencing ladders. Biochemistry 37:6059-6064. [DOI] [PubMed] [Google Scholar]
- 8.Dominski, Z., J. A. Erkmann, J. A. Greenland, and W. F. Marzluff. 2001. Mutations in the RNA binding domain of stem-loop binding protein define separable requirements for RNA binding and histone pre-mRNA processing. Mol. Cell. Biol. 21:2008-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dominski, Z., J. A. Erkmann, X. Yang, R. Sanchez, and W. F. Marzluff. 2002. A novel zinc finger protein is associated with U7 snRNP and interacts with the stem-loop binding protein in the histone pre-mRNP to stimulate 3′-end processing. Genes Dev. 16:58-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dominski, Z., and W. F. Marzluff. 1999. Formation of the 3′ end of histone mRNA. Gene 239:1-14. [DOI] [PubMed] [Google Scholar]
- 11.Dominski, Z., J. Sumerel, R. J. Hanson, and W. F. Marzluff. 1995. The polyribosomal protein bound to the 3′ end of histone mRNA can function in histone pre-mRNA processing. RNA 1:915-923. [PMC free article] [PubMed] [Google Scholar]
- 12.Dominski, Z., L.-X. Zheng, R. Sanchez, and W. F. Marzluff. 1999. The stem-loop binding protein facilitates 3′ end formation by stabilizing U7 snRNP binding to the histone pre-mRNA. Mol. Cell. Biol. 19:3561-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckner, R., W. Ellmeier, and M. L. Birnstiel. 1991. Mature mRNA 3′ end formation stimulates RNA export from the nucleus. EMBO J. 10:3513-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erdjument-Bromage, H., M. Lui, L. Lacomis, A. Grewal, R. S. Annan, D. E. McNulty, S. A. Carr, and P. Tempst. 1998. Examination of micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J. Chromatogr. A 826:167-181. [DOI] [PubMed] [Google Scholar]
- 15.Fabrizio, P., S. Esser, B. Kastner, and R. Lührmann. 1994. Isolation of S. cerevisiae snRNPs: comparison of U1 and U4/U6.U5 to their human counterparts. Science 264:261-265. [DOI] [PubMed] [Google Scholar]
- 16.Furger, A., A. Schaller, and D. Schümperli. 1998. Functional importance of conserved nucleotides at the histone RNA 3′ processing site. RNA 4:246-256. [PMC free article] [PubMed] [Google Scholar]
- 17.Galli, G., H. Hofstetter, H. G. Stunnenberg, and M. L. Birnstiel. 1983. Biochemical complementation with RNA in the Xenopus oocyte: a small RNA is required for the generation of 3′ histone mRNA termini. Cell 34:823-828. [DOI] [PubMed] [Google Scholar]
- 18.Gallie, D. R., N. J. Lewis, and W. F. Marzluff. 1996. The histone 3′-terminal stem-loop is necessary for translation in Chinese hamster ovary cells. Nucleic Acids Res. 24:1954-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Georgiev, O., and M. L. Birnstiel. 1985. The conserved CAAGAAAGA spacer sequence is an essential element for the formation of 3′ termini of the sea urchin H3 histone mRNA by RNA processing. EMBO J. 4:481-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gick, O., A. Krämer, W. Keller, and M. L. Birnstiel. 1986. Generation of histone mRNA 3′ ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J. 5:1319-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilmartin, G. M., F. Schaufele, G. Schaffner, and M. L. Birnstiel. 1988. Functional analysis of the sea urchin U7 small nuclear RNA. Mol. Cell. Biol. 8:1076-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris, M. E., R. Böhni, M. H. Schneiderman, L. Ramamurthy, D. Schümperli, and W. F. Marzluff. 1991. Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Mol. Cell. Biol. 11:2416-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hermann, H., P. Fabrizio, V. A. Raker, K. Foulaki, H. Hornig, H. Brahms, and R. Lührmann. 1995. snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein-protein interactions. EMBO J. 14:2076-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ingledue, T. C., Z. Dominski, R. Sanchez, J. A. Erkmann, and W. F. Marzluff. 2000. Dual role for the RNA binding domain of Xenopus laevis SLBP1 in histone pre-mRNA processing. RNA 6:1635-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kodama, Y., J. H. Rothman, A. Sugimoto, and M. Yamamoto. 2002. The stem-loop binding protein CDL-1 is required for chromosome condensation, progression of cell death and morphogenesis in Caenorhabditis elegans. Development 129:187-196. [DOI] [PubMed] [Google Scholar]
- 26.Labourier, E., and D. C. Rio. 2001. Purification of Drosophila snRNPs and characterization of two populations of functional U1 particles. RNA. 7:457-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lanzotti, D. J., H. Kaygun, X. Yang, R. J. Duronio, and W. F. Marzluff. 2002. Developmental control of histone mRNA and dSLBP synthesis during Drosophila embryogenesis and the role of dSLBP in histone mRNA 3′ processing in vivo. Mol. Cell. Biol. 22:2267-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lerner, E. A., M. R. Lerner, C. A. Janeway, Jr., and J. A. Steitz. 1981. Monoclonal antibodies to nucleic acid containing cellular constituents: probes for molecular biology and autoimmune disease. Proc. Natl. Acad. Sci. USA 78:2737-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, T.-J., B. J. Levine, A. I. Skoultchi, and W. F. Marzluff. 1989. The efficiency of 3′-end formation contributes to the relative levels of different histone mRNAs. Mol. Cell. Biol. 9:3499-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lüscher, B., and D. Schümperli. 1987. RNA 3′ processing regulates histone mRNA levels in a mammalian cell mutant: a processing factor becomes limiting in G1-arrested cells. EMBO J. 6:1721-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin, F., A. Schaller, S. Eglite, D. Schümperli, and B. Müller. 1997. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 16:769-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marzluff, W. F. 1992. Histone 3′ ends: essential and regulatory functions. Gene Expr. 2:93-97. [PMC free article] [PubMed] [Google Scholar]
- 33.Marzluff, W. F., M. L. Whitfield, Z. Dominski, and Z.-F. Wang. 1997. Identification of the protein that interacts with the 3′ end of histone mRNA, p. 163-193. In J. D. Richter (ed.), mRNA formation and function. Academic Press, Inc., New York, N.Y.
- 34.Melin, L., D. Soldati, R. Mital, A. Streit, and D. Schümperli. 1992. Biochemical demonstration of complex formation of histone pre-mRNA with U7 small nuclear ribonucleoprotein and hairpin binding factors. EMBO J. 11:691-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michel, F., D. Schumperli, and B. Muller. 2000. Specificities of Caenorhabditis elegans and human hairpin binding proteins for the first nucleotide in the histone mRNA hairpin loop. RNA 6:1539-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mount, S. M., and H. K. Salz. 2000. Pre-messenger RNA processing factors in the Drosophila genome. J. Cell Biol. 150:F37-F43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mowry, K. L., R. Oh, and J. A. Steitz. 1989. Each of the conserved sequence elements flanking the cleavage site of mammalian histone pre-mRNAs has a distinct role in the 3′-end processing reaction. Mol. Cell. Biol. 9:3105-3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mowry, K. L., and J. A. Steitz. 1987. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA's. Science 238:1682-1687. [DOI] [PubMed] [Google Scholar]
- 39.Muller, B., and D. Schumperli. 1997. The U7 snRNP and the hairpin binding protein: key players in histone mRNA metabolism. Semin. Cell Dev. Biol. 8:567-576. [DOI] [PubMed] [Google Scholar]
- 40.Pandey, N. B., and W. F. Marzluff. 1987. The stem-loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol. Cell. Biol. 7:4557-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pettitt, J., C. Crombie, D. Schümperli, and B. Müller. 2002. The Caenorhabditis elegans histone hairpin-binding protein is required for core histone expression and is essential for embryonic and postembryonic cell division. J. Cell Science 115:857-866. [DOI] [PubMed] [Google Scholar]
- 42.Pillai, R. S., C. L. Will, R. Lührmann, D. Schümperli, and B. Müller. 2001. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14-kDa Sm D1-like protein. EMBO J. 20:5470-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price, D. H., and C. S. Parker. 1984. The 3′ end of Drosophila histone mRNA is produced by a 3′ processing activity in vitro. Cell 38:423-429. [DOI] [PubMed] [Google Scholar]
- 44.Price, D. H., A. E. Sluder, and A. L. Greenleaf. 1987. Fractionation of transcription factors for RNA polymerase II from Drosophila Kc cell nuclear extracts. J. Biol. Chem. 262:3244-3255. [PubMed] [Google Scholar]
- 45.Rio, D. C. 1988. Accurate and efficient pre-mRNA splicing in Drosophila cell-free extracts. Proc. Natl. Acad. Sci. USA 85:2904-2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts, S. B., S. W. Emmons, and G. Childs. 1989. Nucleotide sequences of Caenorhabditis elegans core histone genes: genes for different histone classes share common flanking sequence elements. J. Mol. Biol. 206:567-577. [DOI] [PubMed] [Google Scholar]
- 47.Scharl, E. C., and J. A. Steitz. 1994. The site of 3′ end formation of histone messenger RNA is a fixed distance from the downstream element recognized by the U7 snRNP. EMBO J. 13:2432-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schaufele, F., G. M. Gilmartin, W. Bannwarth, and M. L. Birnstiel. 1986. Compensatory mutations suggest that base-pairing with a small nuclear RNA is required to form the 3′ end of H3 messenger RNA. Nature 323:777-781. [DOI] [PubMed] [Google Scholar]
- 49.Smith, H. O., K. Tabiti, G. Schaffner, D. Soldati, U. Albrecht, and M. L. Birnstiel. 1991. Two-step affinity purification of U7 small nuclear ribonucleoprotein particles using complementary biotinylated 2′-O-methyl oligoribonucleotides. Proc. Natl. Acad. Sci. USA 88:9784-9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soldati, D., and D. Schümperli. 1988. Structural and functional characterization of mouse U7 small nuclear RNA active in 3′ processing of histone pre-mRNA. Mol. Cell. Biol. 8:1518-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spycher, C., A. Streit, B. Stefanovic, D. Albrecht, T. H. W. Koning, and D. Schümperli. 1994. 3′ end processing of mouse histone pre-mRNA: evidence for additional base-pairing between U7 snRNA and pre-mRNA. Nucleic Acids Res. 22:4023-4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stauber, C., and D. Schümperli. 1988. 3′ processing of pre-mRNA plays a major role in proliferation-dependent regulation of histone gene expression. Nucleic Acids Res. 16:9399-9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stefanovic, B., W. Hackl, R. Lührmann, and D. Schümperli. 1995. Assembly, nuclear import and function of U7 snRNPs studied by microinjection of synthetic U7 RNA into Xenopus oocytes. Nucleic Acids Res. 23:3141-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Streit, A., T. W. Koning, D. Soldati, L. Melin, and D. Schümperli. 1993. Variable effects of the conserved RNA hairpin element upon 3′ end processing of histone pre-mRNA in vitro. Nucleic Acids Res. 21:1569-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strub, K., and M. L. Birnstiel. 1986. Genetic complementation in the Xenopus oocyte: co-expression of sea urchin histone and U7 RNAs restores 3′ processing of H3 pre-mRNA in the oocyte. EMBO J. 5:1675-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strub, K., G. Galli, M. Busslinger, and M. L. Birnstiel. 1984. The cDNA sequences of the sea urchin U7 small nuclear RNA suggest specific contacts between histone mRNA precursor and U7 RNA during RNA processing. EMBO J. 3:2801-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sullivan, E., C. Santiago, E. D. Parker, Z. Dominski, X. Yang, D. J. Lanzotti, T. C. Ingledue, W. F. Marzluff, and R. J. Duronio. 2001. Drosophila stem-loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes Dev. 15:173-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun, J.-H., D. R. Pilch, and W. F. Marzluff. 1992. The histone mRNA 3′ end is required for localization of histone mRNA to polyribosomes. Nucleic Acids Res. 20:6057-6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vasserot, A. P., F. J. Schaufele, and M. L. Birnstiel. 1989. Conserved terminal hairpin sequences of histone mRNA precursors are not involved in duplex formation with the U7 RNA but act as a target site for a distinct processing factor. Proc. Natl. Acad. Sci. USA 86:4345-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walther, T. N., K. T. Wittop, D. Schümperli, and B. Muller. 1998. A 5′-3′ exonuclease activity involved in forming the 3′ products of histone pre-mRNA processing in vitro. RNA 4:1034-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, Z.-F., T. C. Ingledue, Z. Dominski, R. Sanchez, and W. F. Marzluff. 1999. Two Xenopus proteins that bind the 3′ end of histone mRNA: implications for translational control of histone synthesis during oogenesis. Mol. Cell. Biol. 19:835-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, Z.-F., T. Krasikov, M. R. Frey, J. Wang, A. G. Matera, and W. F. Marzluff. 1996. Characterization of the mouse histone gene cluster on chromosome 13: 45 histone genes in three patches spread over one megabase. Genome Res. 6:688-701. [DOI] [PubMed] [Google Scholar]
- 63.Wang, Z.-F., R. Tisovec, R. W. Debry, M. R. Frey, A. G. Matera, and W. F. Marzluff. 1996. Characterization of the 55 kilobase mouse histone gene cluster on chromosome 3. Genome Res. 6:702-714. [DOI] [PubMed] [Google Scholar]
- 64.Wang, Z.-F., M. L. Whitfield, T. I. Ingledue, Z. Dominski, and W. F. Marzluff. 1996. The protein which binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 10:3028-3040. [DOI] [PubMed] [Google Scholar]
- 65.Whitfield, M. L., L.-X. Zheng, A. Baldwin, T. Ohta, M. M. Hurt, and W. F. Marzluff. 2000. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 20:4188-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williams, A. S., T. C. Ingledue, B. K. Kay, and W. F. Marzluff. 1994. Changes in the stem-loop at the 3′ terminus of histone mRNA affects its nucleocytoplasmic transport and cytoplasmic regulation. Nucleic Acids Res. 22:4660-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Williams, A. S., and W. F. Marzluff. 1995. The sequence of the stem and flanking sequences at the 3′ end of histone mRNA are critical determinants for the binding of the stem-loop binding protein. Nucleic Acids Res. 23:654-662. [DOI] [PMC free article] [PubMed] [Google Scholar]