Abstract
Clip-domain serine proteases (SPs) are the essential components of extracellular signaling cascades in various biological processes, especially in embryonic development and the innate immune responses of invertebrates. They consist of a chymotrypsin-like SP domain and one or two clip domains at the N-terminus. Prophenoloxidase-activating factor (PPAF)-II, which belongs to the noncatalytic clip-domain SP family, is indispensable for the generation of the active phenoloxidase leading to melanization, a major defense mechanism of insects. Here, the crystal structure of PPAF-II reveals that the clip domain adopts a novel fold containing a central cleft, which is distinct from the structures of defensins with a similar arrangement of cysteine residues. Ensuing studies demonstrated that PPAF-II forms a homo-oligomer upon cleavage by the upstream protease and that the clip domain of PPAF-II functions as a module for binding phenoloxidase through the central cleft, while the clip domain of a catalytically active easter-type SP plays an essential role in the rapid activation of its protease domain.
Keywords: clip domain, easter, innate immunity, melanin, three-dimensional structure
Introduction
Serine protease (SP) cascades amplify signals from physiological or pathological responses in the extracellular region of vertebrates and invertebrates. In mammals, the blood clotting and the complement system employ SP cascades in response to tissue damage and microbial infection, respectively (O'Brien and McVey, 1993). In invertebrates, SP cascades drive diverse biological processes, including embryonic development and immune responses (Krem and Cera, 2002). Many of these signal-amplifying reactions are accomplished by the clip domain-containing SP family, the number of which is expanding rapidly in public genome databases. However, the cellular function and the structure of any clip domain have been unknown. Clip-domain SPs were first identified as enzymes involved in the Toll signaling pathway required for the development of dorsal–ventral structures in Drosophila (Morisato and Anderson, 1995; Anderson, 1998). So far, these proteases have been found only in invertebrates. They consist of a chymotrypsin-like SP domain and one or two clip domain(s) at the N-terminus. The SP domain is responsible for signal amplification by cleaving and activating the downstream SP. The clip domain, usually composed of 37–55 amino-acid residues, is interlinked by three strictly conserved disulfide bonds (Jiang and Kanost, 2000) and connected to the SP domain by a linker of 23–92 residues. Like chymotrypsinogen, the SP domain of the clip-domain proteases begins with the Cys–Gly sequence, and this first cysteine residue forms a disulfide bond with a cysteine residue within the SP domain. The zymogen form of these proteases is converted into the active enzyme by cleavage at the arginine or lysine residue corresponding to Arg15 of chymotrypsinogen, which is located between the two cysteine residues forming the disulfide bond. Owing to this disulfide linkage, the N-terminal fragment including the clip domain remains covalently attached to the SP domain when the zymogens are activated.
Prophenoloxidase (ProPO) activation is one of the major immune responses in arthropods (Gillespie et al, 1997; Cerenius and Söderhäll, 2004). Upon injury or infection, proPO in the blood plasma is activated by clip-domain SPs, the so-called proPO-activating factors or enzymes (PPAFs or PPAEs), also known as proPO-activating proteins (PAPs) (Ashida and Brey, 1998; Lee et al, 1998a; Jiang and Kanost, 2000). Recognition of aberrant surfaces or foreign invaders triggers a cascade of PPAFs (Cerenius and Söderhäll, 2004), leading to the activation of proPO(s) (Ashida and Brey, 1998). Phenoloxidase (PO), the active form of proPO, catalyzes the production of quinones, which can crosslink neighboring molecules to form melanin at the injury site or around invading microorganisms. Quinones may also be involved in the production of cytotoxic molecules such as superoxides and hydroxyl radicals that could help to kill the invading microorganisms (Gillespie et al, 1997). The activation of proPO(s), however, needs to be tightly regulated as a local event, because excessive melanization could also cause fatal damages to hosts (Yu et al, 2003; Zhao et al, 2005). Consistently, melanization takes place locally at the site of injury or invading organisms (Söderhäll and Cerenius, 1998).
PPAFs have been identified in several insects and a crustacean, including the beetle Holotrichia diomphalia (Kwon et al, 2000), the mealworm Tenebrio molitor (Lee et al, 2002), the silkworm Bombyx mori (Satoh et al, 1999), the tobacco hornworm Manduca sexta (Yu et al, 2003), and the crayfish Pacifastacus leniusculus (Wang et al, 2001). PPAFs can be classified into catalytic and noncatalytic groups, based on the presence or absence of proteolytic activity. The catalytic group (referred to as the easter-type SPs) containing one or two clip domain(s) is structurally related to Drosophila easter, and includes H. diomphalia PPAF-I, -III, B. mori PPAE, and M. sexta PAP-I. The noncatalytic group (referred to as PPAF-II family) contains one clip domain and shares a similar sequence with the catalytic group, but lacks proteolytic activity due to the replacement of the active site serine residue by glycine. It includes PPAF-II of H. diomphalia, serine proteinase homolog (SPH)-I, -II of M. sexta (Jiang et al, 2003), and Drosophila CG5390.
Three PPAFs, PPAF-I, -II, and -III of H. diomphalia, were cloned and characterized (Lee et al, 1998a, 1998b; Kwon et al, 2000; Kim et al, 2002). A two-step cleavage mechanism for the activation of proPOs (two highly homologous isozymes proPO-I and -II) was put forth, in which full-length proPOs (79 kDa, ProPO79s) are cleaved into smaller forms (76 kDa, PO76s) by PPAF-I, and one of the smaller forms is converted into the active form of PO (66 kDa, PO66) by PPAF-I or unknown proteases (Kim et al, 2002). In this process, PPAF-III is the upstream protease that cleaves PPAF-II specifically at an arginine residue within the clip domain, and thereby generates the functional form of the protein. PPAF-II is an essential factor for the activation of PO despite the absence of an enzymatic activity (Lee et al, 1998b; Kim et al, 2002). The PPAF-II orthologs are found in a variety of insects, including Drosophila, H. diomphalia, T. molitor, and M. sexta (Lee et al, 1998b, 2002; Yu et al, 2003). However, the detailed mechanism of action of the PPAF-II family in the proPO activation pathway remains poorly understood.
To begin understanding the functions of the clip domains, we determined the crystal structure of PPAF-II from H. diomphalia. Structure-based biochemical analyses provided insights into the roles of the clip domains of the noncatalytic and the catalytic SPs in the activation of proPOs and PPAFs.
Results
Structure determination and overall structure of PPAF-II
Initially, we tried to produce easter (Morisato and Anderson, 1995; Anderson, 1998) and persephone (Ligoxygakis et al, 2002) of Drosophila and PPAF-I, -II, and -III of H. diomphalia in insect cells (Piao et al, 2005). Easter and persephone could not be expressed, but all PPAFs were successfully obtained in soluble forms with yields of 1–10 mg per 1 l of High-Five cell culture, which enabled us to attempt the initial screening of crystallization conditions. Only PPAF-II could be crystallized (Piao et al, 2005), and its crystal structure was determined by the molecular replacement method. The crystal-packing interactions between PPAF-II molecules were not extensive, indicating that the protein is monomeric in solution. The PPAF-II gene encodes a protein with 415 amino acids. The N-terminal 24-amino-acid segment of the protein functions as a signal peptide for secretion. The mature protein (392 amino acids) is made up of two parts: the clip domain including the flanking sequence composed of 114 amino acids, and the SP-like (SPL) domain composed of 278 amino acids (Figure 1). The final 2.0 Å resolution model contains residues 22–42 with an N-glycosylated residue, 47–116, 129–173, and 180–415, which include most of the clip and the SPL domains (Figure 2A and B). The missing residues constitute the loop segments flanking the clip domain and a flexible loop in the SPL domain. The small clip domain is bound to the larger SPL domain via hydrogen bonds, hydrophobic interactions, and a salt bridge (Figures 2A and 4B). While the structure of the SPL domain of PPAF-II is similar to chymotrypsin-like proteases (Figure 2C), that of the clip domain is a new fold according to a database search using the program DALI (Holm and Sander, 1993).
Figure 1.
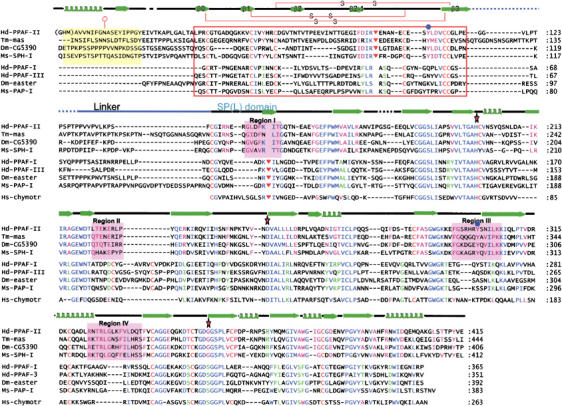
Sequence alignment and secondary structure assignment. The sequences of the mature forms of four PPAF-II family members, four easter-type SPs, and human chymotrypsin from top to bottom: Hd, Holotrichia diomphalia; Tm-mas, Tenebrio molitor masquerade; Dm, Drosophila; Ms, Manduca sexta; Hs, Homo sapiens are shown. The alignment was performed using CLUSTALW (Thompson et al, 1997), and then adjusted based on the conserved cysteine residues. The secondary structure of PPAF-II is shown above the sequences. The linker between the clip and SPL domains is shown as a blue line on the secondary structure. The red box indicates the clip domain whose disulfide bond linkages are drawn on the secondary structure. The PPAF-II family-specific N-terminal residues are shaded in yellow. Four signature sequences of the PPAF-II family are shaded in magenta and labeled. Conserved amino acids are color coded: blue letters, conserved in the both PPAF-II family and easter-type SPs; magenta, conserved only in the PPAF-II family; green, conserved only in the easter-type SPs. N-glycosylated asparagine is indicated by a hexagon with a tail, the catalytic triad residues (His, Asp, and Ser) by red stars, the cleavage sites by inverted triangles, and the strictly conserved tyrosine residues (Tyr108 and Tyr301) only in the PPAF-II family by blue circles.
Figure 2.
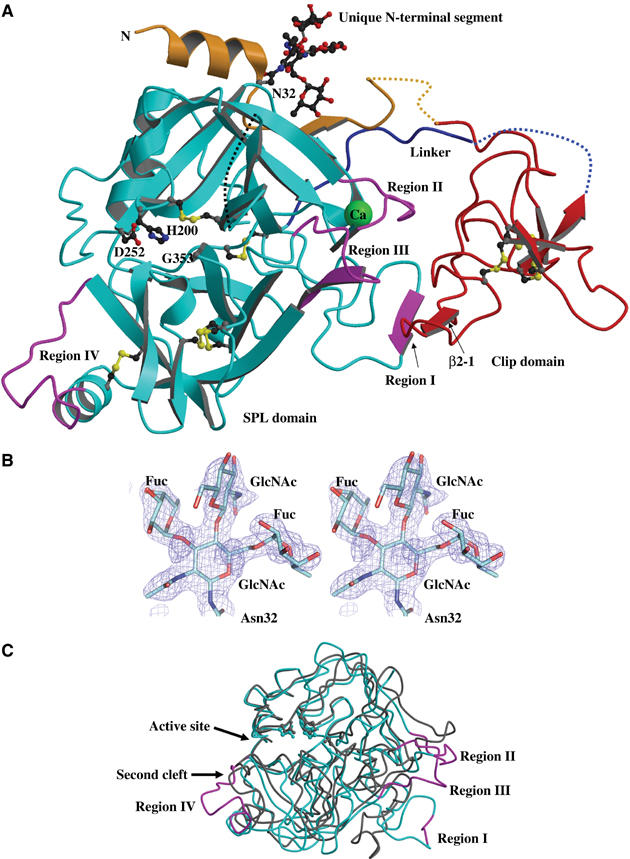
Overall structure of PPAF-II. (A) A ribbon representation of PPAF-II structure. The clip domain is shown in red, and the PPAF-II family-specific N-terminal segment is in yellow. The SP domain is shown in cyan except for the four signature sequences that are in magenta. Bound calcium ion is in green, and all the three disulfide bonds are drawn with sulfur atoms in yellow. The disordered regions are shown in dashed lines. The nonfunctional catalytic triad residues (Gly–His–Asp) are shown in the ball-and-stick representation. Asn32 is N-glycosylated in the structure. (B) Stereo view of N-glycosylated Asn32 with 2Fo−Fc electron density map contoured at 1.2σ (blue). The N-acetylglucosamine (GlcNAc) molecule covalently bonded to Asn32 is linked to two molecules of fucose (FUC) and one molecule of GlcNAc, which is a paucimannose (a major end product of glycosylation in insect cells) lacking three mannose molecules. (C) Superposition of the Cα traces of the SPL domain of PPAF-II and chymotrypsinogen (gray; PDB code 1CGI). The coloring scheme for PPAF-II and orientation of the proteins are the same as in (A). The signature sequences exhibit quite different Cα positions compared with chymotrypsinogen.
Structural feature of the clip domain
The clip domain is composed of a high portion of loops and a central, four-stranded, irregular β-sheet (Figure 3A left). The conserved three disulfide bonds knotting the loops and the β-strands together appear critical for the structural integrity of the central β-sheet that serves as the main framework of the clip domain structure. It has been suggested that clip domains may be structurally similar to antimicrobial proteins, β-defensins, based on the identical arrangement of the cysteine residues (Iwanaga et al, 1998). We show here that the clip domain structure is distinctively different from those of β-defensins. First, β-defensins contain three, instead of four, β-strands forming the central antiparallel β-sheet (Schneider et al, 2005) (Figure 3B right). Second, while β2 and β3 are antiparallel with each other in the structure of β-defensins, they are parallel in the structure of the clip domain (Figure 3B).
Figure 3.
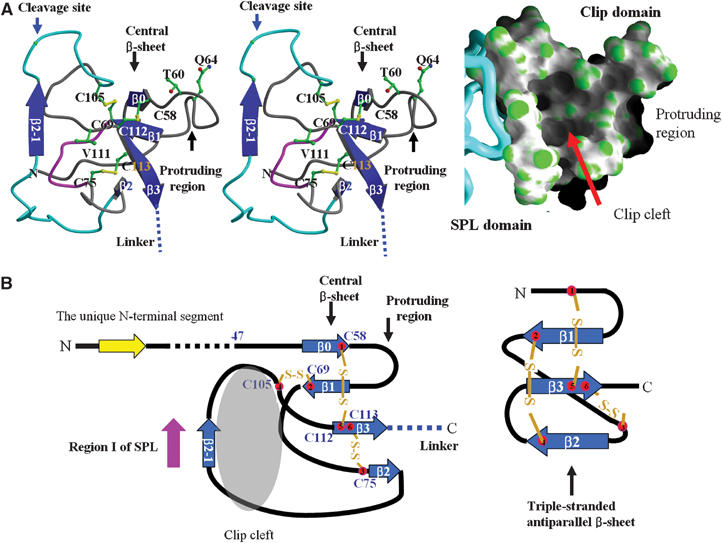
Structure of the clip domain. (A) Central cleft in the clip domain. Left, The secondary structures are shown in blue, and the disulfide bonds are in yellow. The cleavage site is indicated by a blue arrow. The central β-sheet is composed of β0, β1, β2, and β3. Residues substituted in the mutagenesis study and cysteine residues are shown in the ball-and-stick representation. The outer rim of the cleft lined by β2-1-containing loop is in cyan, while the bottom of the cleft composed of a loop between Cys69 and Cys75 is in magenta. Right, Surface representation of the clip domain with the SPL domain as a Cα worm. (B) Comparison of the clip domain with human β-defensin 1. Left, a schematic drawing of the clip domain. The secondary structural elements are numbered in accordance with β-defensin 1. Right, a schematic drawing of β-defensin 1. Note the identical arrangement of the disulfide bonds, but a different topology compared with the clip domain.
The clip domain has a noticeable protruding region and a prominent central cleft (Figure 3A). The protruding region consists of nonconserved amino acids between Cys58 (first Cys) and Cys69 (second Cys). The prominent cleft is found in the center of the clip domain (Figures 2C and 3A). The outer rim of the cleft is lined by the β2-1-containing loop (Asp80–Glu104), Cys112 (fifth Cys), and Cys113 (sixth Cys). The bottom of the cleft is composed of the loop flanked by Cys69 (second Cys) and Cys75 (third Cys), whose length in the number of amino acids is invariant in all the clip domains (Figures 1 and 3A). The loops forming the outer rim and the bottom are structurally fixed to the central β-sheet by the disulfide bonds between the four cysteine residues conserved among all the clip domains, suggesting that a similar cleft is present in all the other clip domains. The outer rim of the cleft predominantly consists of hydrophilic residues such as Asp80, Glu93, Glu104, Asp97, Thr91, and Arg99, while the interior surface of the cleft consists of hydrophobic residues including Tyr72, Val78, Phe96, and Val111 (Figure 3A). The shape and the electrostatic nature of the cleft appear to be well suited for binding a hydrophobic moiety.
Structural feature of SPL domain
The SPL domain of PPAF-II shares a relatively high sequence similarity with chymotrypsin, but it is devoid of a catalytic activity because of Gly353 in place of the invariant serine nucleophile in the catalytic SPs (Figures 1 and 2C). The domain contains two clefts, one of which corresponds to the defective active site cleft containing the Gly–His–Asp triad. It is elongated and shallow, similar to the active site cleft of chymotrypsin (Figure 4C). The cleft may serve as a docking site for binding a peptide segment of a protein. The other cleft is partly composed of a unique signature sequence (designated as Region IV; see below), and accommodates the N-terminal α-helix of a neighboring molecule in the crystal. This crystal packing interaction suggests that the cleft may also serve as a docking site for binding a protein.
Figure 4.
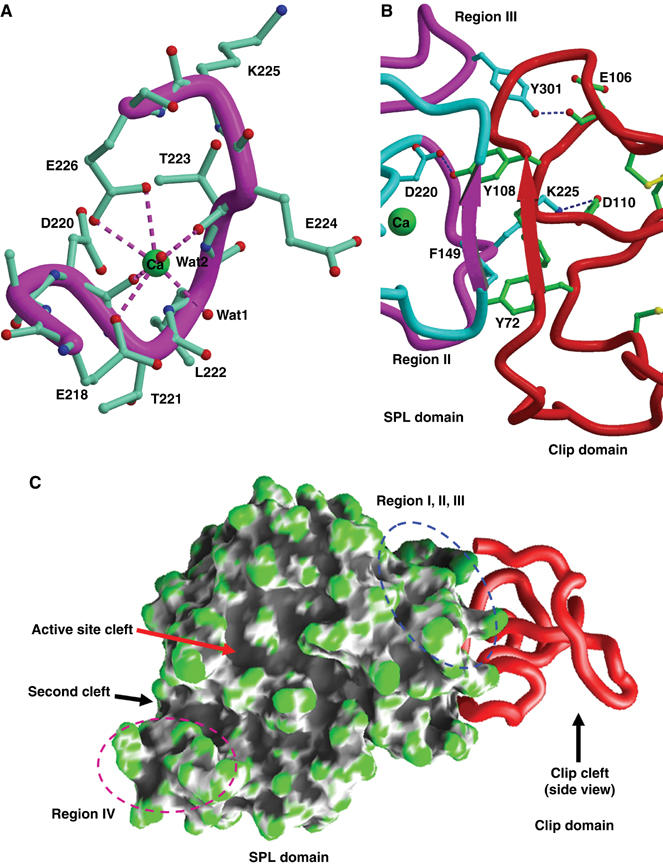
Structure of the SPL domain. (A) Calcium-binding site. A calcium ion bound to the SPL domain is in green, and the hepta-coordinations are indicated by dotted lines. Region II is represented as a Cα worm. (B) Representative interactions between the clip and SPL domains. The color scheme is the same as in Figure 2A. Hydrogen bonds and a salt bridge are shown in dashed lines. β2-1 of the clip domain and Region I of the SPL domain form a pair of the β-strands, which is stabilized by four hydrogen bonds (not shown). Phe96 and Tyr72 of the clip domain are involved in the hydrophobic interaction with Phe149 of the SPL domain. The conserved Tyr301 and Tyr108 form interdomain hydrogen bonds. (C) Clefts in the SPL domain. Surface representation of the SPL domain is shown with the clip domain as a Cα worm.
The structure of PPAF-II reveals two other unique features of the SPL domain compared with the structure of chymotrypsin: the presence of a calcium-binding site and four distinct loops composed of signature sequences. A calcium ion is bound in the loop composed of residues 218–228, and hence referred to as the calcium-binding loop. A similar calcium-binding site was predicted to be present in Drosophila easter based on a three-dimensional modeling study (Rose et al, 2003). The calcium ion is hepta-coordinated with a pseudo-octahedral geometry involving the carboxylates of Glu218 and Glu226, the carbonyl oxygens of Asp220 and Thr223, plus two water molecules (Figure 4A). The residues involved in the calcium coordination are strictly conserved among the PPAF-II family, suggesting the presence of a calcium cage in all the members of this family. The four distinct segments, designated as Regions I–IV, were identified by the superposition of the PPAF-II and chymotrypsinogen structures, showing obvious deviations of these segments from the structure of chymotrypsinogen (Figure 2C). The amino-acid sequences of these segments are signature sequences, since they are highly conserved in the PPAF-II family but distinguished from the corresponding sequences of the easter-type proteases (Figure 1).
Clip domain is tightly associated with SPL domain
Notably, Regions I–III form the major interface for interaction with the clip domain (Figure 2A). Region I forms paired β-strands with β2-1 of the clip domain. Region II, which is the calcium-binding loop, interacts with the clip domain via Asp220 and Lys225 (Figure 4B). Especially, Asp220 forms a hydrogen bond with Tyr108 that is uniquely conserved in the PPAF-II family (Figure 4B). The calcium-binding loop is fairly rigid, as indicated by a low average temperature factor (24.1 Å2), and is likely to be built to interact with the clip domain through the calcium coordination. Tyr301 in Region III, which is strictly conserved in the PPAF-II family, forms a hydrogen bond with the backbone carbonyl atom of Glu106 in the clip domain. The interaction at the interface is also strengthened by hydrophobic interactions consisting of Phe149 in Region I, the aliphatic part of Lys225 in Region II, and Tyr72 and Phe96 in the clip domain (Figure 4B). Most residues at the interface are conserved in the PPAF-II family, but not in the easter-type SPs. Due to these interactions, the domain boundaries between the clip and SPL domains are barely distinguishable, strongly suggesting that the clip domain should be tightly associated with the SPL domain in solution. In support of this notion, we did not observe the cleavage at Region I by PPAF-III or a preferential cleavage by other proteases, including trypsin and lysyl endopeptidase (data not shown), although it corresponds to the activation loop of the easter-type SPs and contains the cleavage sequence 150Lys-Ile that is conserved in most of the SPs. Presumably, the interdomain interactions between Region I and β-2-1 (Figure 2A) prevent these proteases from accessing Region I.
The clip domain also interacts with the SPL domain via the N-terminal segment. Compared with the sequences of the easter-type SPs, the PPAF-II family has a unique insertion of 33 amino acids at the N-terminus (Figure 1). Of these, 20 amino acids interact with the SPL domain at a site remote from the interface formed by Regions I–III (Figure 2A). The presence of this unique N-terminal segment should further restrict movement of the clip domain from the SPL domain.
Oligomerization of cleaved, functional PPAF-II
PPAF-II is specifically cleaved by PPAF-III at Arg99 within the clip domain and is subsequently converted into the functional form (Kim et al, 2002). The residue is right next to β2-1, which interacts with Region I of the SPL domain (Figure 3A left). Given the availability of recombinant PPAF-II and -III, we inspected whether the cleavage may accompany a quaternary structural change by using a size-exclusion chromatographic column. The analysis revealed that cleaved and functional PPAF-II is a large oligomer with ∼600 kDa molecular mass, whereas uncleaved PPAF-II is an ∼45 kDa monomer (Figure 5A top). Therefore, cleaved PPAF-II becomes functional by forming the quaternary structure. A cleavage-induced conformational change would be small and confined to the region of the paired β2-1/Region I, considering the tight interaction of the clip and SPL domains in uncleaved PPAF-II and no loss of the clip domain following the cleavage. The change into the quaternary structure is likely to be a common property of most PPAF-II family members, as judged based on the high degree of sequence similarity and an observation of a similar oligomerization of a PPAF-II family protein masquerade from T. molitor (S Piao and N-C Ha, unpublished data). In contrast, the striking difference between the two forms was not observed for PPAF-I and -III, whose pro- and active forms were eluted as a monomer from the size-exclusion column (data not shown).
Figure 5.
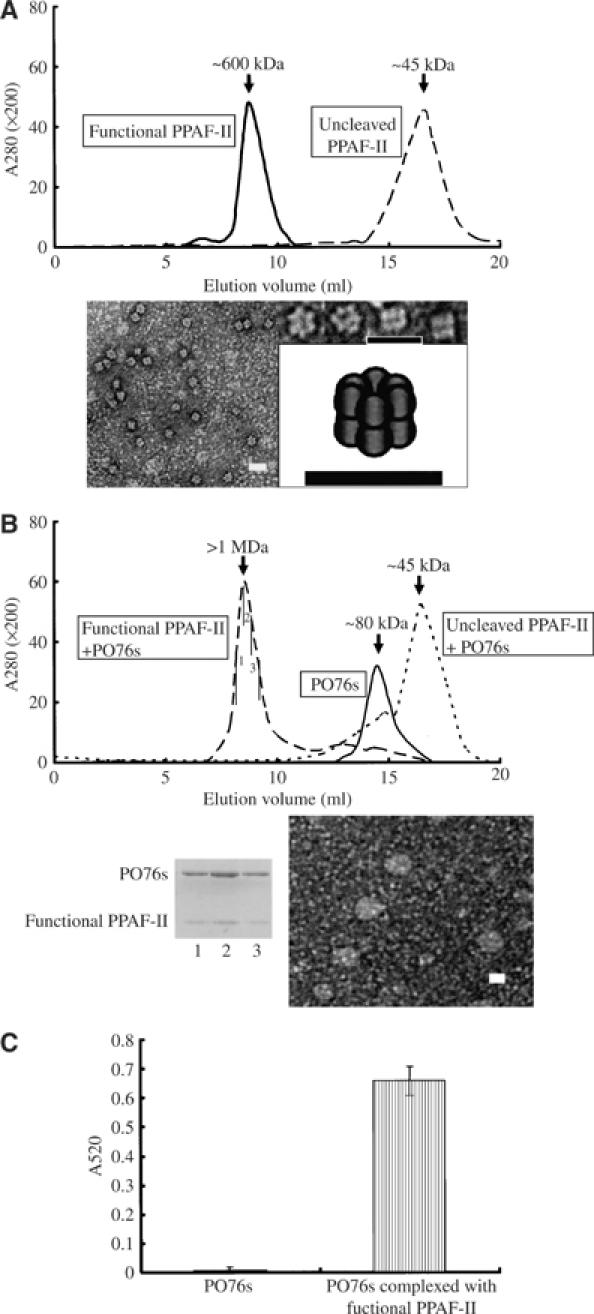
Biochemical and mutagenesis analyses of PPAF-II and proPO. (A) Oligomerization of PPAF-II. Top, size-exclusion chromatography of uncleaved PPAF-II and functional PPAF-II. The estimated molecular weight is shown above the peaks. Elution profile is shown and the identity of each peak was confirmed by SDS–PAGE. Bottom, electron microscopic picture of functional PPAF-II. Selected particles are displayed with a cartoon showing the arrangement of the protein molecules at the right side. Scale bars represent 20 nm. (B) Functional PPAF-II forms a complex with PO76s. Three different samples, analyzed using a Superdex S-200 size-exclusion column, are indicated in the rectangular boxes. The uncleaved or functional form of PPAF-II was preincubated with excess amount of PO76s for 30 min at 4°C in the presence of 5 mM CaCl2 before loading the sample on the column. PO76s were eluted as monomeric proteins. The bottom panels show the SDS–PAGE analysis of the fraction containing proteins with >1 MDa molecular weight (left) and an electron microscopic image (right; scale bar: 20 nm). The numbers (1–3) correspond to the subfractions indicated on the chromatogram. Similar results were obtained when 1 mM EDTA was added instead of 5 mM CaCl2 (data not shown). (C) PO activity of PO76s. Fractions of PO76s and the proteins in complex with functional PPAF-II were separated using Superdex S-200 as in (B), and the PO activity of each sample was measured. The amounts of PO76s in the two samples were adjusted to be equal based on the band intensities on an SDS–PAGE. The results represent the mean±s.d. of three separate experiments.
Previously, a strong PO activity was observed only in the presence of functional PPAF-II, when proPO79s were incubated with active PPAF-I (Lee et al, 2002). To test whether PPAF-II binds PO76s, which are the major cleaved products generated from proPO79-I and -II by PPAF-I, we purified the PO76 proteins from H. diomphalia plasma and carried out size-exclusion chromatography after incubation of the sample with functional PPAF-II. Indeed, functional PPAF-II formed a tight complex with PO76s (Figure 5B). In a control experiment, PPAF-I or -III did not form a complex with PO76s (data not shown). The intensities of the protein bands on a denaturating gel indicated 1:1 stoichiometry of binding between functional PPAF-II and PO76s (Figure 5B bottom). Therefore, the homo-oligomerization of functional PPAF-II and its interaction with PO76s induce clustering of many molecules of PO76s in close proximity to each other. Consistently, an electron microscopic analysis revealed that 12 molecules of functional PPAF-II form two stacked hexameric rings (Figure 5A bottom), and the quaternary structure of the protein turns into a much bigger ball-like supramolecular assembly in the presence of PO76s (Figure 5B bottom).
Clip-domain-dependent activation of PO76s
We suspected that the clip domain may play an important role in the interaction of functional PPAF-II with PO76s. If it is true, we thought that the protruding region and/or the prominent central cleft of the clip domain may be the interaction surface for PO76s, since the unusual surface feature of a protein is often involved in the molecular interactions. We substituted Thr60 and Gln64, surface-exposed and located in the protruding region, with alanine. Like the other amino acids in this region, the two residues are poorly conserved. The resulting T60A or Q64A mutants of PPAF-II did not affect the cleavage, oligomerization, and the PO activity of PO76s (Table I). However, Val111, which is located in the central cleft and strictly conserved among all the clip domains, plays a critical role in the interaction with PO76s. When Val111 was mutated into alanine, the binding of PPAF-II to PO76s was abolished, although the protein could oligomerize following the proteolytic cleavage by PPAF-III (Table I). The data demonstrate that the central cleft of the clip domain, but not the protruding region, mediates the binding of functional PPAF-II to PO76s.
Table 1.
Biochemical characterization of clip domain mutants of PPAF-II
| PPAF-II | Cleavage by PPAF-III | Oligomerization | Complex with PO76s | PO activity |
|---|---|---|---|---|
| Wild type | ++ | ++ | ++ | ++ |
| T60A | + | ++ | ++ | ++ |
| Q64A | ++ | ++ | ++ | ++ |
| V111A |
++ |
++ |
− |
− |
| Mutant PPAF-IIs were tested if they can be cleaved by PPAF-III, and the cleaved mutants oligomerize, form a complex with PO76s, and yield the PO activity. All experiments were carried out in the presence of 5 mM CaCl2 because calcium ion is required for the PO activity. | ||||
Previously, it has been suggested that proPO79s are converted into PO76s and then into PO67 by proteolytic cleavages, in a two-step cleavage mechanism where active PPAF-I plays an essential role (Lee et al, 2002). However, the PO activity of PO76s in the absence of active PPAF-I was not tested because of unavailability of purified PO76s. In order to test whether PO76s yield the PO activity without the further cleavage into PO67, PO76s were purified from the H. diomphalia plasma. The purified PO76s remained intact and showed a strong PO activity in the presence of functional PPAF-II and calcium ion (Figure 5C). The purified PO76s sample used for these experiments did not exhibit any PO activity in the absence of functional PPAF-II (Figure 5C), demonstrating a crucial role played by functional PPAF-II in yielding the PO activity, as well as excluding a possibility that a trace amount of contaminating proteins led to the PO activity. Based on these results, we conclude that PO76s in complex with functional PPAF-II are active, and PO67, which has been believed to be the active form of PO, could be simply a nonspecifically cleaved product from PO76s by a prolonged incubation with active PPAF-I.
Lack of tethering of clip domain with SP domain in easter-type SPs
The clip domains of the easter-type SPs and the PPAF-II family members are likely to share a similar structural framework, considering the strictly conserved cysteine residues forming the disulfide bonds, in addition to the high sequence similarity between the two groups. Likewise, the overall structure of the SP domains of the easter-type SPs is expected to be similar to that of chymotrypsin-like SPs. However, the easter-type SPs differ from PPAF-II in several aspects. Their clip domains lack the N-terminal 33 residues present in the PPAF-II clip domain, and their SP domains do not exhibit a sequence similarity in Regions I–III of PPAF-II. Secondly, while the easter-type proteases are cleaved at the activation loop within their SP domain, the PPAF-II family members are cleaved within the clip domain (Kim et al, 2002). In addition, Tyr72, Phe96, and Tyr108, conserved in the PPAF-II family and involved in the interaction with the SPL domain, are substituted in the easter-type proteases (Figure 1). These differences suggested that the interaction between the clip and SP domains of the easter-type proteases might be different from that observed in the PPAF-II structure. We investigated this possibility using proPPAF-I, a member of the easter-type proteases, as a model. We opportunely found that purified proPPAF-I was specifically cleaved into two fragments when it was stored at 4°C for about a week, probably owing to a residual activity of the zymogen or a trace amount of nonspecific proteases present in the protein sample. By N-terminal amino-acid sequencing and Western blotting, one peptide fragment was identified as a segment including the SP domain plus a few amino acids of the interdomain linker, and the other fragment as a segment including the clip domain (Figure 6A). The cleavage at the interdomain linker is different from the cleavage within the SP domain, resulting in the activation of the zymogen form. The serendipitous finding suggested that the clip domain is connected to the SP domain by a flexible linker that is sensitive to proteolysis. From a size-exclusion column, the clip and SP domains were eluted as two separate proteins with lower molecular weight compared with the full-length protein (Figure 6A). A similar observation was observed for H. diomphalia PPAF-III, which is also an easter-type SP (data not shown). We conclude that the two domains of the easter-type SPs are likely to move freely as two rigid bodies within the length of the flexible interdomain linker, in sharp contrast with the two domains of PPAF-II tethered together.
Figure 6.
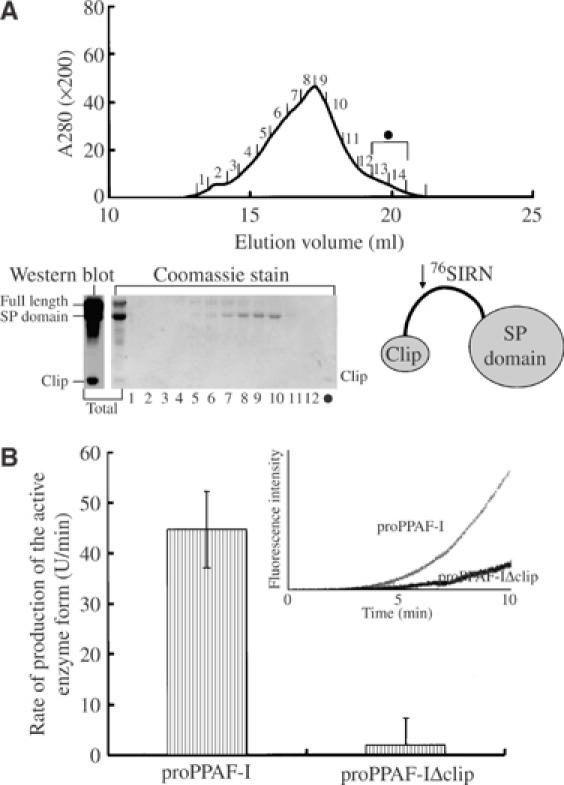
Separable domains of PPAF-I and the role of its clip domain. (A) Separation of the clip and SP domains by a proteolytic cleavage. Purified PPAF-I incubated for a week at 4°C was loaded on a Superdex S-200 column, and subfractions indicated by numbers on the chromatogram (top) were analyzed by SDS–PAGE and Western blotting (bottom left). N-terminal sequencing of the band labeled as SP domain identified the internal 76Ser–Ile–Arg–Asn sequence of PPAF-I. Fractions 13 and 14 loaded on the last lane (•) were concentrated before the analysis. Schematic drawing of the PPAF-I structure based on the two experiments is shown (bottom right). Note that the clip and SP domains are linked by a flexible linker and lacks interdomain interactions. (B) Clip domain is essential for the activation of PPAF-I. The production of the active enzyme in the proPPAF-I and proPPAF-IΔclip samples was monitored by recording the intensity of fluorescence from the cleaved artificial substrate (inset). The increase of the fluorescence intensity as a function of time, which is the rate of production of the active enzyme form, is presented with an error bar for each sample. The error bars represent standard deviations of three separate experiments.
Clip domain is essential for rapid activation of an easter-type SP
To explore the role of the clip domain of the catalytically active easter-type SPs, we produced a deletion mutant of proPPAF-I lacking the clip domain (proPPAF-IΔclip). The upstream factor of PPAF-I has not been characterized yet, mainly due to the presence of the protein at an extremely low concentration in the hemolymph. We therefore used the plasma of H. diomphalia to activate proPPAF-I, since the plasma is known to contain the uncharacterized upstream factor(s) whose activity depends on calcium ion (Lee et al, 2002). We first confirmed that both the proPPAF-I and proPPAF-IΔclip samples did not exhibit a noticeable proteolytic activity (data not shown). Next, after adding the plasma solution and calcium ion, we monitored how fast the active PPAF-I was produced from proPPAF-I and proPPAF-IΔclip by measuring the apparent amidase activities of the two samples in the presence of a peptide substrate. Remarkably, the proPPAF-I sample exhibited a highly increasing amidase activity compared with the proPPAF-IΔclip sample (Figure 6B inset). Accordingly, the calculated rates of the PPAF-I production showed a sharp difference between the two protein samples (Figure 6B), demonstrating that the clip domain of the easter-type SPs plays an essential role in the activation of the zymogen form of its SP domain, presumably through a direct or indirect interaction with the upstream factor(s).
Discussion
The clip domain is ubiquitously found in SPs involved in the extracellular signaling cascades, especially in those that are locally amplified. The function of the clip domain has been unknown. We demonstrate that the clip domain of PPAF-II is a protein-interaction module that plays an essential role in the binding and activation of PO76s via its central cleft. The interaction between the two is likely to induce a conformational change of PO76, because the conversion of PO76 into PO67 was observed only in the presence of functional PPAF-II (Kim et al, 2002). Probably, the conformational change of PO76s leads to the exposure of an otherwise hidden segment for cleavage by PPAF-I, and, at the same time, it may result in a rearrangement of the active site residues into the functional state.
Remarkably, the clip domain of PPAF-II remains tightly associated with the SPL domain after the cleavage of the protein by PPAF-III. The ‘nicked' PPAF-II oligomerizes into the two-hexameric-toroidal structure, which serves as a hub for the docking of many molecules of PO76s. In the homo-oligomerization, the defective active site and/or the second cleft in the SPL domain may mediate the intermolecular interactions. Other than the binding-induced activation of PO76s, the oligomerization of PO76s by the PPAF-II hub should facilitate the enzyme reactions, which are the oxygenation of monophenols to o-diphenols (the intermediates) and the oxidation of the intermediates into o-quinones (the final products) (Gillespie et al, 1997). The intermediate molecules released from one PO76 molecule can easily diffuse into the active site of another PO76 molecule, both on the large assembly of functional PPAF-II and PO76s.
We extended our scope of the study into the characterization of the clip domain in the easter-type SPs that are involved in not only innate immune defense but also developmental process (Krem and Cera, 2002). A striking difference between the easter-type protease PPAF-I and PPAF-II is that the clip domain of the former is separable from the SP domain by a single proteolytic cleavage at the interdomain linker. Neither of the two domains formed an oligomer alone or together. Despite lacking tight physical interaction with the SP domain, the clip domain of PPAF-I was required for a rapid activation of the enzyme in the plasma solution of H. diomphalia. The high sequence similarity among the easter-type proteases suggests that the clip domain of the other members is not tethered to its SP domain either, and it may play a common role of the rapid activation of the proteolytic activity of the SP domain. What would be the role of the separable clip domain in the activation of the easter-type proteases in the proPO cascade? Based on the localized production of melanin and the two rigid bodies-like domain organization, we propose that the clip domain of these proteases binds unknown stereotypical molecular structure on a pathogen surface. Through this interaction, many molecules of the easter-type proteases would be brought in close contact with each other. Once the initial activator activates a molecule of these proteases bound to the pathogen surface, it subsequently activates its downstream proteases within a distance covered by the flexible linker between the clip and SP domains. Likewise, the activated proteases sequentially activate their neighboring downstream proteases, allowing signal amplification localized at the pathogen surface. This model is analogous to dominoes in that the SP cascade will be activated rapidly only when the density of the upstream and downstream protease is high enough to allow physical interactions with each other on a pathogen surface. In this model, we expect that the cleavage at the activation loop results in the juxtaposition of the catalytic triad residues in the same manner as the activation of the chymotrypsin zymogen (Wang et al, 1985), and that the freely moving clip domain has no direct effect in this process.
In conclusion, we presented the first three-dimensional structure of a clip-domain SP, that of PPAF-II, and demonstrated a tight association of the clip domain of the protein with the SPL domain and its role as a protein-interacting module essential for the proPO activation. The domain organization of the easter-type SPs, which is reminiscent of two rigid bodies linked by a string, was further demonstrated. Based on the biochemical studies, we proposed the domino model that provides a plausible explanation to how the proPO activation cascade, whose components are diffusible soluble proteins, can be localized on the pathogen surface.
Materials and methods
Construction, expression, and purification of proteins
Overexpression and purification of recombinant PPAF-II based on the baculovirus vector expression system (Invitrogen) were reported previously (Piao et al, 2005). The protein contained three additional amino acids (Gly–His–Met) at the N-terminus of the mature protein as a cloning artifact (Piao et al, 2005). Recombinant proPPAF-I, -III, and proPPAF-IΔclip were overexpressed and purified according to the method used for the purification of PPAF-II (Piao et al, 2005), except that the N-terminal (His)6-tag was not removed. Mutant PPAF-II proteins were constructed using the QuikChange Mutagenesis Kit (Stratagene, USA) and purified according to the same protocol used for the wild-type PPAF-II. To generate functional PPAF-II, 10 mg of PPAF-II was incubated at 30°C for 1 h with less than 10 μg of recombinant PPAF-III, which was activated during the preincubation of the protein with 5 μg of plasma proteins from H. diomphalia in the presence of calcium ion. The plasma of H. diomphalia was prepared as reported (Kim et al, 2002). The functional form of PPAF-II was further purified with a Mono-Q column (Amersham, USA) using a 0–1 M NaCl linear gradient in 20 mM Tris buffer (pH 8.0). PO76s were purified from the H. diomphalia plasma. Freshly thawed plasma solution was treated with final 0.2 mM diisopropyl fluorophosphates, a protease inhibitor, and then dialyzed against 20 mM Tris buffer (pH 8.0). The proteins were purified from the solution by a combination of Sephacryl S-300, Hitrap-Q, and Superdex S-200 columns in the presence of 5 mM EDTA, primarily based on a previous report (Kwon et al, 1997).
Structure determination and analysis
Crystallization of PPAF-II was reported (Piao et al, 2005). In brief, the crystals were obtained by the hanging-drop vapor-diffusion method at 14°C in a precipitation solution containing 0.15 M ammonium sulfate, 1.25 M lithium sulfate, and 0.1 M sodium citrate (pH 5.5). Before data collection, the crystals were immersed briefly in a cryoprotectant solution, which was the precipitant solution supplemented with 10% glycerol. An X-ray diffraction data set was collected using the synchrotron radiation from the beamline NW12A at Photon Factory (Japan), and processed using the program HKL2000 (Otwinosky and Minor, 1997). Initial phases were determined by the molecular replacement package MOLREP (CCP4, 1994) using the coordinates of the catalytic domain of plasminogen activator (PDB code 1A5I) as a search model. The initial electron density map allowed model-building of the SPL domain only. After many rounds of structure refinement of the SPL domain, the electron density corresponding to the clip domain started to emerge. The crystals contained one molecule of PPAF-II in the asymmetric unit. Model building and structure refinement were carried out using the programs O (Jones et al, 1991) and CNS (Brünger et al, 1998). Crystallographic data statistics are summarized in Table II. Structures were superposed and analyzed using the program SUPERPOSE (CCP4, 1994), and structure presentations were generated using MOLSCRIPT (Kraulis, 1991), RASTER3D (Merritt and Murphy, 1994), and GRASP (Nicholls et al, 1991).
Table 2.
X-ray data collection and refinement statistics
| Data set | |
| Source | NW12A at photon factory |
| Wavelength (Å) | 0.9496 |
| Resolution limit (Å) | 50–2.0 (2.07–2.00)a |
| Space group | C2 |
| Unit cell (Å) | a=106.78, b=76.587, c=69.833, and β=113.447° |
| Reflections | |
| Measured | 206 704 |
| Unique | 31 743 |
| Rsym (%) | 3.0 (8.2)a |
| Completeness (%) | 90.8 (76.2)a |
| Refinement | |
| Resolution range | 100–2.0 |
| R-factor (%) | 20.6 |
| Rfree (%)b | 23.4 |
| Average B-value (Å2) | 31.9 |
| R.m.s.d. for bonds (Å) | 0.005 |
| R.m.s.d. for angles (deg) | 1.3 |
| Total number of atoms | 3215 |
| No. of protein atoms | 2911 |
| No. of water atoms | 268 |
| No. of sulfate molecules | 5 |
| No. of metal atoms | 1 |
| Ramachandran plot | |
| Most favored regions | 87.7% |
| Additional allowed regions | 11.4% |
| Generously disallowed region | 0.6% |
| Disallowed region |
0.3% |
| aThe numbers in parentheses are statistics for the highest resolution shell. | |
| bRfree was calculated with 10% of the data. | |
Size-exclusion column chromatography
To determine the molecular weight of proteins or to learn whether the two proteins form a complex in solution, size-exclusion chromatography was performed at a flow rate of 0.2 ml/min on Superdex S-200 HR 10/30 (Amersham, USA) equilibrated with 20 mM Tris buffer (pH 8.0) containing 100 mM NaCl and 5 mM CaCl2 (or 1 mM EDTA).
Electron microscopy
Each sample diluted to a concentration of 100 μg/ml in 20 mM Tris buffer (pH 8.0) containing 100 mM NaCl and 5 mM CaCl2 was adsorbed to a thin carbon foil, and negatively stained with 2% uranyl acetate for 1 min. Transmission electron microscopy was carried out using an EF-TEM (EM912Ω, Zeiss) at a magnification of × 100 000.
PPAF-I activity assay
The rate of production of active PPAF-I was determined by measuring the apparent amidase activity of the protein. Before the measurement, 3 μM of proPPAF-I or proPPAF-IΔclip was preincubated with 50 μg of H. diomphalia plasma proteins in 50 μl of 20 mM Tris buffer (pH 8.0) at 30°C for 10 min. The mixture was added to the final 1 ml of 20 mM Tris buffer containing 2 mM artificial substrate, Boc–Phe–Ser–Arg–MCA. Immediately after adding CaCl2 into the reaction mixture to 5 mM concentration, the florescence intensity from the sample was measured for 10 min with excitation and emission wavelengths of 380 and 460 nm, respectively.
PO assay
PO activity was measured using 4-methylcatechol and 4-hydroxyproline ethylester as substrates according to a method reported previously (Lee et al, 1998a). Briefly, 50 μl of each sample was added into 450 μl of 20 mM Tris buffer (pH 8.0) containing 2 mM 4-methylcatechol, 2 mM 4-hydroxylproline, and 5 mM CaCl2. The production of the prolyl adduct of 4-methylquinone, the final product of this reaction, was monitored by recording the increase of the absorbance at 520 nm.
Activation of wild-type and mutant PPAF-IIs by PPAF-III
In all, 30 μg of a wild-type or a mutant PPAF-II protein was incubated with 0.1 μg of activated PPAF-III in 60 μl of reaction buffer containing 20 mM Tris (pH 8.0), 5 mM CaCl2. An aliquot of 20 μl of the reaction mixture was taken at a given time (0, 30, and 60 min) and boiled immediately for a denaturating gel electrophoretic analysis.
Coordinates
The coordinates and structure factors have been deposited into the Protein Data Bank (PDB code 2B9L).
Acknowledgments
We thank the staff members at the Photon Factory beamline NW12A (Japan) for the data collection, Beamline 6B at PLS (Korea) for preliminary analysis, and the EM team at Korea Basic Science Institute (Daejeon, Korea) for electron microscopy. We are grateful to Deukmi Kim and Jung Won Han for assisting biochemical experiments, and to Dr Jaewon Lee for helpful comments on the manuscript. This work was supported by the National Research Laboratory (M10400000028-04J0000-02) grant from the Korea Ministry of Science and Technology to BLL and N-CH, and the Creative Research Initiative of Korea Ministry of Science and Technology to B-HO.
Competing interests statement The authors declare that they have no competing financial interests.
References
- Anderson KV (1998) Pinning down positional information: dorsal–ventral polarity in the Drosophila embryo. Cell 95: 439–442 [DOI] [PubMed] [Google Scholar]
- Ashida M, Brey PT (1998) Recent advances on the research of the insect prophenoloxidase cascade. In: Brey PT, Hultmart D (eds), Molecular Mechanism of Immune Responses in Insects, pp 135–172. London: Bios Scientific Press [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54 (Part 5): 905–921 [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Cerenius L, Söderhäll K (2004) The prophenoloxidase-activating system in invertebrates. Immunol Rev 198: 116–126 [DOI] [PubMed] [Google Scholar]
- Gillespie JP, Kanost MR, Trenczek T (1997) Biological mediators of insect immunity. Annu Rev Entomol 42: 611–643 [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C (1993) Protein structure comparison by alignment of distance matrices. J Mol Biol 233: 123–138 [DOI] [PubMed] [Google Scholar]
- Iwanaga S, Kawabata S, Muta T (1998) New types of clotting factors and defense molecules found in horseshoe crab hemolymph: their structures and functions. J Biochem (Tokyo) 123: 1–15 [DOI] [PubMed] [Google Scholar]
- Jiang H, Kanost MR (2000) The clip-domain family of serine proteinases in arthropods. Insect Biochem Mol Biol 30: 95–105 [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Yu XQ, Zhu Y, Kanost M (2003) Prophenoloxidase-activating proteinase-3 (PAP-3) from Manduca sexta hemolymph: a clip-domain serine proteinase regulated by serpin-1J and serine proteinase homologs. Insect Biochem Mol Biol 33: 1049–1060 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard L (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47 (Part 2): 110–119 [DOI] [PubMed] [Google Scholar]
- Kim MS, Baek MJ, Lee MH, Park JW, Lee SY, Söderhäll K, Lee BL (2002) A new easter-type serine protease cleaves a masquerade-like protein during prophenoloxidase activation in Holotrichia diomphalia larvae. J Biol Chem 277: 39999–40004 [DOI] [PubMed] [Google Scholar]
- Kraulis PJ (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr 24: 946–950 [Google Scholar]
- Krem MM, Cera ED (2002) Evolution of enzyme cascades from embryonic development to blood coagulation. Trends Biochem Sci 27: 67–74 [DOI] [PubMed] [Google Scholar]
- Kwon TH, Kim MS, Choi HW, Joo CH, Cho MY, Lee BL (2000) A masquerade-like serine proteinase homologue is necessary for phenoloxidase activity in the coleopteran insect, Holotrichia diomphalia larvae. Eur J Biochem 267: 6188–6196 [DOI] [PubMed] [Google Scholar]
- Kwon TH, Lee SY, Lee JH, Choi JS, Kawabata S, Iwanaga S, Lee BL (1997) Purification and characterization of prophenoloxidase from the hemolymph of coleopteran insect, Holotrichia diomphalia larvae. Mol Cells 7: 90–97 [PubMed] [Google Scholar]
- Lee KY, Zhang R, Kim MS, Park JW, Park HY, Kawabata S, Lee BL (2002) A zymogen form of masquerade-like serine proteinase homologue is cleaved during pro-phenoloxidase activation by Ca2+ in coleopteran and Tenebrio molitor larvae. Eur J Biochem 269: 4375–4383 [DOI] [PubMed] [Google Scholar]
- Lee SY, Cho MY, Hyun JH, Lee KM, Homma KI, Natori S, Kawabata SI, Iwanaga S, Lee BL (1998a) Molecular cloning of cDNA for pro-phenol-oxidase-activating factor I, a serine protease is induced by lipopolysaccharide or 1,3-β-glucan in coleopteran insect, Holotrichia diomphalia larvae. Eur J Biochem 257: 615–621 [DOI] [PubMed] [Google Scholar]
- Lee SY, Kwon TH, Hyun JH, Choi JS, Kawabata SI, Iwanaga S, Lee BL (1998b) In vitro activation of pro-phenol-oxidase by two kinds of pro-phenol-oxidase-activating factors isolated from hemolymph of coleopteran, Holotrichia diomphalia larvae. Eur J Biochem 254: 50–57 [DOI] [PubMed] [Google Scholar]
- Ligoxygakis P, Pelte N, Hoffmann JA, Reichhart JM (2002) Activation of Drosophila Toll during fungal infection by a blood serine protease. Science 297: 114–116 [DOI] [PubMed] [Google Scholar]
- Merritt EA, Murphy ME (1994) Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr D 50: 869–873 [DOI] [PubMed] [Google Scholar]
- Morisato D, Anderson KV (1995) Signaling pathways that establish the dorsal–ventral pattern of the Drosophila embryo. Annu Rev Genet 29: 371–399 [DOI] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11: 281–296 [DOI] [PubMed] [Google Scholar]
- O'Brien D, McVey J (1993) Blood coagulation, inflammation, and defense. In: Sim E (ed), The Natural Immune Systems, Humoral Factors, pp 257–280. New York: IRL Press [Google Scholar]
- Otwinosky Z, Minor W (1997) Processing of X-ray crystallization data in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Piao S, Kim D, Won Park J, Leul Lee B, Ha NC (2005) Overexpression and preliminary X-ray crystallographic analysis of prophenoloxidase activating factor II, a clip domain family of serine proteases. Biochim Biophys Acta 1752: 103–106 [DOI] [PubMed] [Google Scholar]
- Rose T, LeMosy EK, Cantwell AM, Banerjee-Roy D, Skeath JB, Di Cera E (2003) Three-dimensional models of proteases involved in patterning of the Drosophila embryo. Crucial role of predicted cation binding sites. J Biol Chem 278: 11320–11330 [DOI] [PubMed] [Google Scholar]
- Satoh D, Horii A, Ochiai M, Ashida M (1999) Prophenoloxidase-activating enzyme of the silkworm, Bombyx mori. Purification, characterization, and cDNA cloning. J Biol Chem 274: 7441–7453 [DOI] [PubMed] [Google Scholar]
- Schneider JJ, Unholzer A, Schaller M, Schafer-Korting M, Korting HC (2005) Human defensins. J Mol Med 83: 587–595 [DOI] [PubMed] [Google Scholar]
- Söderhäll K, Cerenius L (1998) Role of the prophenoloxidase-activating system in invertebrate immunity. Curr Opin Immunol 10: 23–28 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) CLUSTALX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Bode W, Huber R (1985) Bovine chymotrypsinogen A X-ray crystal structure analysis and refinement of a new crystal form at 1.8 Å resolution. J Mol Biol 185: 595–624 [DOI] [PubMed] [Google Scholar]
- Wang R, Lee SY, Cerenius L, Söderhäll K (2001) Properties of the prophenoloxidase activating enzyme of the freshwater crayfish, Pacifastacus leniusculus. Eur J Biochem 268: 895–902 [DOI] [PubMed] [Google Scholar]
- Yu XQ, Jiang H, Wang Y, Kanost MR (2003) Nonproteolytic serine proteinase homologs are involved in prophenoloxidase activation in the tobacco hornworm, Manduca sexta. Insect Biochem Mol Biol 33: 197–208 [DOI] [PubMed] [Google Scholar]
- Zhao M, Soderhall I, Park JW, Ma YG, Osaki T, Ha NC, Wu CF, Soderhall K, Lee BL (2005) A novel 43-kDa protein as a negative regulatory component of phenoloxidase-induced melanin synthesis. J Biol Chem 280: 24744–24751 [DOI] [PubMed] [Google Scholar]


