Abstract
The eukaryotic Y-box-binding protein YB-1 functions in various biological processes, including DNA repair, cell proliferation, and transcriptional and translational controls. To gain further insight into how human YB-1 plays its role in pleiotropic functions, we here used two-hybrid screenings to identify partners of this protein; the results showed that YB-1 itself, iron-regulatory protein 2 (IRP2), and five ribosomal proteins each served as partners to YB-1. We then examined the biological effect of the interaction of YB-1 and IRP2 on translational regulation. Both in vitro binding and coimmunoprecipitation assays showed the direct interaction of YB-1 and IRP2 in the presence of a high concentration of iron. RNA gel shift assays showed that YB-1 reduced the formation of the IRP2-mRNA complex when the iron-responsive element of the ferritin mRNA 5′ untranslated region (UTR) was used as a probe. By using an in vitro translation assay using luciferase mRNA ligated to the ferritin mRNA 5′UTR as a reporter construct, we showed that both YB-1 and IRP2 inhibited the translation of the mRNA. However, coadministration of YB-1 and IRP2 proteins abrogated the inhibition of protein synthesis by each protein. An In vivo coimmunoprecipitation assay showed that IRP2 bound to YB-1 in the presence of iron and a proteasome inhibitor. The direct interaction of YB-1 and IRP2 provides the first evidence of the involvement of YB-1 in the translational regulation of an iron-related protein.
The Y-box protein family, which is widely distributed from bacteria to mammals, contains a cold shock domain, which is highly conserved from prokaryotic cold shock proteins. The human Y-box-binding protein, YB-1, which is located on chromosome 1p34, (23, 41) was initially identified as a transcriptional factor which associates with the Y-box sequence appearing in the major histocompatibility complex class II genes (5). It has been hypothesized that YB-1 might play a role in promoting cell proliferation through transcriptional regulation of various relevant genes, including epidermal growth factor receptor, thymidine kinase, DNA topoisomerase II, and DNA polymerase (21, 43). The recent demonstration of the biological roles of YB-1 showed modification of chromatin, translational masking of mRNA, participation in the redox signaling pathway, RNA chaperoning, and stress response regulation (38).
It has also been demonstrated that eukaryotic Y-box proteins regulate gene expression at translational levels by recognizing RNA (25, 36). The rabbit Y-box protein, p50, which is 97% identical to human YB-1, is found in cytoplasmic messenger ribonucleoprotein particles (mRNP) in somatic cells and regulates translation by interacting with mRNA (7). The murine MSY1 protein and chicken Y-box protein both regulate transcription and translation (18, 37). Furthermore, the Y-box family proteins,Xenopus mRNP3/mRNP4 and mouse MSY2, are also found as mRNA-masking proteins in germinal cells (10, 12, 24). Chen et al. (2) have reported that YB-1 is involved in mRNA stability of the cdk4 gene through binding to a specific sequence of the mRNA. YB-1 also stabilizes cap-dependent mRNA, since depletion of YB-1 results in accelerated mRNA decay (6). Y-box binding proteins thus appear to play critical roles in both mRNA turnover and translational control.
Cellular levels of iron are regulated posttranslationally by mRNA-protein interactions. Iron-regulatory protein 2 (IRP2) is an RNA-binding protein that regulates the homeostatic binding of intracellular iron to its specific cognate mRNA hairpin structures, known as iron-responsive elements (IREs). IREs are present in the 5′ untranslated region (UTR) of ferritin heavy and light chains (39), erythroid 5-aminolevulinate synthase (4), mammalian mitochondrial aconitase (9), and Drosophia melanogaster succinate dehydrogenase subunit b (20), and they mediate the translational efficiency of these mRNAs. Five individual IRE motifs present in the 3′ UTR of the transferrin receptor mediate mRNA stability (19). Under low-iron conditions, IRP binding keeps the ferritin mRNA in mRNPs by preventing the interaction between the cap-binding complex eIF4F and the small ribosomal subunit, resulting in the formation of the efficient translation complex. In iron-replete cells, IRP2 affects an iron-dependent oxidative modification that increases degradation by means of a ubiquitin-mediated proteasome pathway (11, 14). After IRP2 is released from IRE, the translation of ferritin protein is stimulated.
Y-box-binding proteins are expected to exert their pleiotropic roles through association with other partner proteins in mammalian cells. YB-1 associates with various factors, such as RelA, YY-1, AP2, Purα, proliferating-cell nuclear antigen, and p53, to modulate gene expression (13, 22, 26, 29, 31, 34). Rabbit p50 interacts with actin to modulate mRNA transport, anchoring, and localization (33). To gain more insight into how YB-1 proteins exert their multiple functions, two-hybrid screening using full-length YB-1 as bait should be useful. In our present study, we used two-hybrid screening to first identify several proteins as partners of YB-1; the identified partners included YB-1 itself, several ribosomal proteins, IRP2, and several cellular proteins, which include the transcription factor. YB-1 is a potent cap-dependent mRNA stabilizer and involves mRNA stabilization and translational repression in a non-sequence-specific manner. IRP2 inhibits the translation of ferritin mRNA through the IRE, which is located near the cap structure. In this experiment, we then focused on IRP2 in an attempt to clarify how YB-1 plays its role in translation through interaction with this protein by ferritin mRNA, whose 5′ UTR contains IRE. The role of YB-1 in protein synthesis is therefore discussed in its plausible association with IRP2-IRE complex formation on the ferritin mRNA 5′ UTR.
MATERIALS AND METHODS
Plasmids for two-hybrid screening.
cDNA encoding the full-length YB-1 protein was inserted into multiple cloning sites in frame with the GAL4 sequence encoding the DNA-binding domain in the vector pDBLeu (Life Technologies, Rockville, Md.). The HeLa cell cDNA library inserted in frame with the GAL4 sequence encoding the activation domain in the vector pPC86 for the two-hybrid screen was obtained from Life Technologies.
Plasmids.
Because the IRP2 cDNA construct detected by the two-hybrid screening was partial, we amplified full-length human IRP2 from human liver cDNA by PCR using the primer pair 5′-ATGGACGCCCCAAAAGCA-3′ and 5′-GCAAAGCCAAACTCGAA-3′. The full-length IRP2 cDNA amplified was cloned into pThioHis-C (Invitrogen, Carlsbad, Calif.) and into pGEX-4T-2 (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom) for expression in bacteria. ThioHis-IRP2 deletion mutants were constructed as follows. To construct Thio-IRP2 ΔN, the cDNA construct detected by the two-hybrid screening was digested with NotI-SalI and cloned into pThioHis-C digested with NotI-XhoI. To construct Thio-IRP2 Δ2, full-length IRP2 was digested with BamHI-StuI and ligated into pThioHis-C (BamHI-StuI). To construct Thio-IRP2Δ3, full-length IRP2 was digested with BamHI-MunI and ligated into pThioHis-C, which was digested with BamHI-EcoRI. To construct Thio-IRP2 Δ4 and Δ5, full-length IRP2 was digested with BamHI-PvuII or BamHI-HpaI, respectively, and ligated into pThioHis-C (BamHI-StuI). For pCMV/Flag-IRP2, the full-length IRP2 cDNA was ligated into pCMV-Flag (XhoI site) (Stratagene). The plasmids containing full-length glutathione S-transferase (GST)-YB-1 cDNA, GST-YB-1 deletion mutants (GST-YB-1Δ1, Δ2, Δ3, Δ4, Δ5, Δ11), and ThioHis-YB-1 have been described previously (13, 29).
To construct pT7-Luc, luciferase cDNA of pGL3 basic vector (Promega) were digested with EcoRI, blunted with Klenow enzyme, and ligated into pT7Blue3 (Novagen). To construct pT7-ferritin-5′ UTR-Luc, cDNA of the ferritin heavy-chain 5′ UTR region amplified by PCR from human liver cDNA was ligated into pT7-Luc (EcoRV site). To construct the plasmid of pT7Blue-IRE, double-strand oligonucleotides for IRE (5′-TCTTGCTTCAACAGTGTTTGAACGGAAC-3′) were ligated into pT7Blue3. All constructs were confirmed by sequencing.
Two-hybrid screening.
The two-hybrid screening was carried out based on modifications by Chevray and Nathans (3) and Vidal et al. (42). The pDBLeu-YB-1 plasmid was introduced into yeast strain MaV203 by transformation. After MaV203 cells containing pDBLeu-YB-1 were tested for self-activation, transformed MaV203 cells containing pDBLeu-YB-1 with the pPC86-HeLa cDNA library were selected for cells that contain both plasmids and that induce the HIS3 reporter gene. Transformants were selected by determining which of three reporter genes (HIS3, URA3, and lacZ) was induced. Positive clones were then retested with fresh pDBLeu-YB-1 by either a retransformation assay into MaV203 or a version of plasmid shuffling.
Antibodies.
Antibodies to Flag-tag, GST, and thioredoxin were purchased from Sigma, Santa Cruz Biotechnology, and Invitrogen, respectively. Antibody to YB-1 was generated as described previously (28).
In vitro binding assays.
For the GST pull-down assay against the ThioHis fusion protein, GST and ThioHis fusion protein expression in bacteria was induced by incubation with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), at an optical density of 0.8 to 1.0, for 2 to 3 h at 30°C. Cells were collected by centrifugation and sonicated in 300 μl of binding buffer (50 mM Tris-HCl ([pH 8.0], 1 mM EDTA, 10% glycerol, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride). The cell lysates were cleared by centrifugation, and the supernatants were screened by Western blot analysis using antibodies directed against GST or thioredoxin. ThioHis fusion proteins were loaded onto 15 μl of affinity resin containing GST fusion proteins and incubated for 1 h at 4°C. After the resin was washed three times with 1 ml of binding buffer, the column resin was boiled in Western blot sample buffer. Column eluates and starting materials were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins contained in the binding complex were detected by immunoblot analysis with anti-thioredoxin antibody.
Cells.
RAW264.7 murine macrophages were obtained from the American Type Culture Collection (Rockville, Md.) and grown in a humidified atmosphere of 95% air and 5% CO2 at 37°C. The cells were maintained in Dulbecco modified essential medium containing 10% fetal bovine serum and antibiotics.
Coimmunoprecipitation from cell extracts.
RAW264.7 murine macrophages were grown at a concentration of 106 cells/ml. For iron treatments, 600 μM ferric ammonium citrate (FAC) was added to cells and incubated in the presence or absence of the proteasome inhibitor MG132 (1 μM) for 6 h. Cell extract was incubated for 1 h at 4°C with 5 μl of anti-IRP2 antibody in TNE buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, 1 mM phenylmethyl sulfonyl fluoride, 10 μg of leupeptin per ml, 10 μg of aprotinin per ml) and then with protein A-Sepharose (Pharmacia) for 2 h at 4°C. The beads were washed three times with TNE buffer. After centrifugation, the precipitate and starting material were boiled in Western sample buffer for SDS-PAGE and Western blot analysis.
In vitro coimmunoprecipitation assay.
Flag-IRP2 protein was expressed in the TNT-T7 Quick coupled in vitro transcription-translation system as specified by the manufacturer (Promega), using pCMV/Flag-IRP2. A 30-μl volume of the reticulocyte lysate expressing Flag-IRP2 fusion protein was incubated for 4 h at 4°C in the presence or absence of 100 μM deferrioxamine (DFO) or FAC (50 μM) in 500 μl of phosphate-buffered saline (PBS) (pH 7.3) with 20 μl of beads that had been precoated with anti-Flag antibody (Sigma). The beads were then washed with PBS three times. After centrifugation, the precipitate and starting material were subjected to SDS-PAGE and the interaction of Flag-IRP2 and endogenous YB-1 was analyzed by Western blotting.
RNA band shift assays.
The RNA band shift assays (REMSA) were performed using established techniques (27). Briefly, a 32P-labeled IRE probe was transcribed in vitro by T7 RNA polymerase in the presence of [α-32P]UTP from the plasmid pT7blue3-IRE, which contains the sequence corresponding to the IRE of human ferritin heavy chain. The DNA template was removed by digestion with DNase I, and the IRE probe was then extracted by column chromatography. To form RNA-protein complexes, 5 μg of cytoplasmic protein or 0.5 to 20 pmol of purified GST-YB-1 or GST-IRP2 was incubated for 10 min at room temperature with 32P-labeled IRE probe (50,000 cpm). Heparin (5 mg/ml) was added for another 10 min to prevent nonspecific binding. RNA-protein complexes were analyzed in 6% nondenaturing polyacrylamide gels.
In vitro transcription and translation.
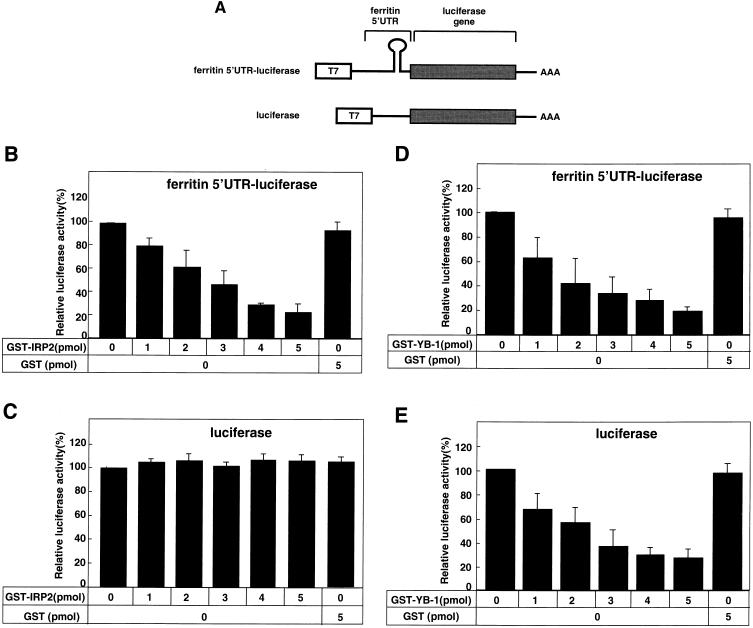
Plasmid pT7Blue3 (2 μg), which encodes luciferase cDNA ligated to the ferritin heavy-chain mRNA 5′ UTR region (ferritin 5′ UTR-luciferase) or luciferase cDNA that is not ligated (luciferase) (see Fig. 6A) were transcribed in vitro using an in vitro transcription system (Promega). The DNA template was then removed by digestion with DNase I, and the RNA was purified with phenol-chloroform. Next, 2 μg of each RNA was translated in the rabbit reticulocyte lysate system (Promega). The indicated amount of purified GST-IRP2, GST-YB-1, or GST was added to the system in the presence or absence of 0.5 mM DFO with 2 μg of ferritin 5′ UTR RNA. The luciferase assay was performed after incubation for 1.5 h at 30°C as described previously (29). All data are shown as the mean and standard deviation for three independent experiments.
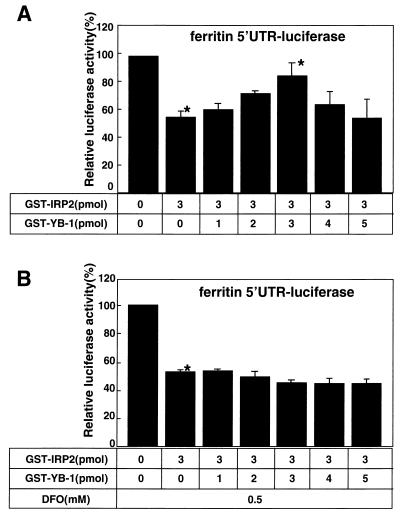
FIG. 6.
Functional interactions between YB-1 and IRP2 were revealed by their effects on the ferritin 5′ UTR-containing mRNA. (A) The indicated amounts of GST-IRP2 and GST-YB-1 were added to the rabbit reticulocyte lysate system using 2 μg of ferritin 5′ UTR-luciferase RNA. The in vitro translation was performed for 1.5 h at 30°C, and the luciferase assays were performed after incubation. Data are shown as the mean and standard deviation of three independent experiments. ∗, Analyzed by Student's t test, P < 0.01. (B) Coadministration of GST-YB-1 and GST-IRP2 proteins in the presence of DFO.
RESULTS
Identification of new proteins interacting with YB-1 using a yeast two-hybrid system.
To identify new protein partners of YB-1, we first performed two-hybrid screening using full-length YB-1 protein as bait and the HeLa cell cDNA library as prey. The Gal4BD-YB-1 fusion protein by itself did not demonstrate transactivation activity when expressed in yeast. The yeast strain MaV203 expressing the Gal4BD-YB-1 protein was transformed using the pGAD424-HeLa cell cDNA library. Transformants were then selected as described in Materials and Methods. Putative positive clones were retransformed into MaV203 yeast strains and tested. After sequencing positive clones, we identified several new protein partners of YB-1. These were YB-1 itself, IRP2, and the ribosomal proteins S3A, L18A, L5, L23A, and S5. We also observed several cellular proteins, which include transcription factor (data not shown). We previously confirmed the interaction of YB-1 with YB-1 itself by means of an in vitro pull-down assay (16), which suggested that YB-1 works as a homomultimer. The interaction of the YB-1 protein with each of five positive clones encoding ribosomal proteins and with IRP2 suggests that YB-1 may participate in translational regulation. Of these translation-related proteins, we further examined how YB-1 could modulate IRP2-dependent translational regulation.
Interaction of YB-1 protein with IRP2.
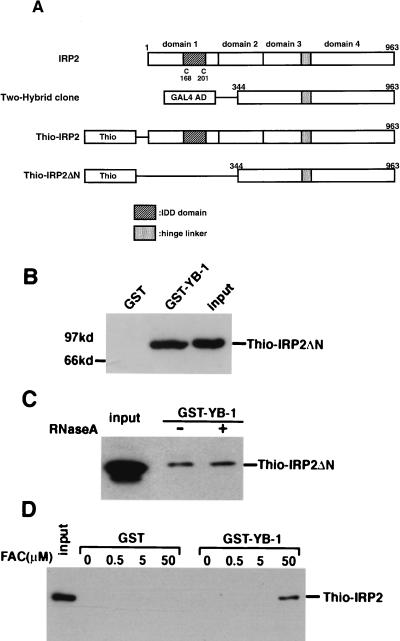
To confirm the results of the two-hybrid yeast screening, we examined whether IRP2 could directly interact with YB-1 by an in vitro binding assay. The IRP2 cDNA identified from the two-hybrid screening included the partial sequence encoding amino acids 344 to 963, which deleted the N-terminal domain (Fig. 1A). We constructed Thio-IRP2 using the partial IRP2 cDNA from the two-hybrid system (Thio-IRP2ΔN) and full-length IRP2 cDNA (Fig. 1A). The GST-YB-1 fusion protein immobilized on glutathione-agarose was used in an affinity column for Thio-IRP2 protein. In agreement with the result of the two-hybrid screening, interaction between YB-1 and IRP2ΔN was observed by the in vitro pull-down assay (Fig. 1B). In contrast, GST protein itself did not interact with IRP2ΔN (Fig. 1B). Since RNA often mediates the protein-protein interactions of RNA-binding proteins, we tested whether RNA molecules were involved in the interaction between YB-1 and IRP2. The binding of YB-1 to IRP2ΔN did not appear to change after addition of RNase A (Fig. 1C).
FIG. 1.
Interaction of YB-1 with IRP2 in vitro. (A) Schematic illustration of modular domains of IRP2. The positions of cysteines that are thought to be iron sensors are indicated by the letter C and by the numbers under the schema. Abbreviations: IDD domain, iron-dependent degradation domain responsible for the iron-dependent IRP2 degradation; Two-Hybrid clone, plasmids containing partial cDNA of human IRP2 fused to the GAL4 activation domain were rescued from true positive clones from the yeast strain MaV203; Thio-IRP2 and Thio/IRP2ΔN, full or partial IRP2 were cloned into the ThioHis plasmid as described in Materials and Methods. (B to D) GST-pull down assay. (B) 1 μg of GST-YB-1 and GST which were bound to 30 μl of glutathione-Sepharose 4B beads were incubated with a 100-fold excess of Thio-IRP2ΔN. The beads were washed three times with washing buffer containing 1% NP-40. Proteins bound to the beads were resolved by SDS-PAGE and analyzed by Western blotting using an anti-Thio antibody. (C) 1 μg of GST-YB-1 protein which binds to glutathione-Sepharose 4B was incubated with Thio-IRP2ΔN in the absence or presence of 100 U of RNase A. Beads were washed, and then bound proteins were eluted and analyzed as described above. (D) 1 μg of GST-YB-1 protein which binds to glutathione-Sepharose 4B was incubated with full-length Thio-IRP2 in the presence of various concentrations of FAC as indicated. The beads were washed, and then bound proteins were eluted and analyzed as described above.
We next examined whether the full-length IRP2 protein could bind to YB-1 and found that it could not (Fig. 1D). We also observed that the full-length IRP2 did not bind to full-length YB-1 in the yeast two-hybrid system (data not shown). Since IRP2 is a unique protein whose interaction with IRE is abolished under high-iron conditions, we hypothesized that iron affects the interaction of YB-1 and IRP2. We therefore performed an in vitro binding assay using both full-length proteins under various concentrations of FAC (Fig. 1D). GST-YB-1 was found to interact with IRP2 in the presence of 50 μM FAC, but GST alone did not interact with IRP2 in the presence of FAC (Fig. 1D). YB-1 could interact with IRP2ΔN in the absence of FAC, whereas YB-1 could interact with full-length IRP2 only in the presence of FAC.
Mapping the regions of YB-1 or IRP2 involved in the mutual interaction with IRP2 or YB-1.
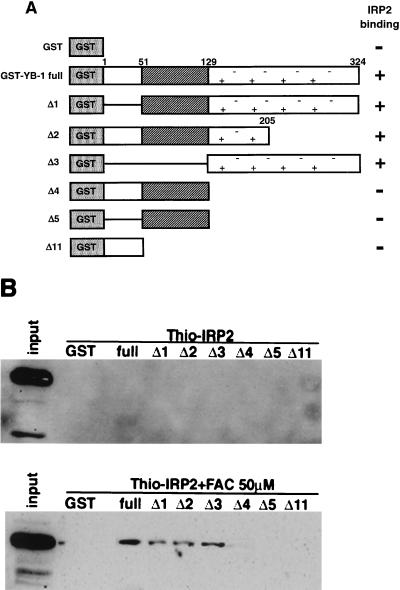
To identify the IRP2-binding region in YB-1, we performed a pull-down assay using a recombinant full-length Thio-IRP2 fusion protein and GST fusion proteins containing deletion mutants of YB-1 (Fig. 2A) in the presence or absence of 50 μM FAC. Thio-IRP2 bound to YB-1Δ1, YB-1Δ2, and YB-1Δ3 in the presence of FAC (Fig. 2B). In contrast, Thio-IRP2 did not bind to YB-1 in the absence of FAC (Fig. 2B). These findings suggest that the YB-1 protein domain from amino acids 129 to 205 is involved in complex formation with IRP2 in the presence of FAC.
FIG. 2.
Identification of the IRP2-binding domain in YB-1. (A) Schematic illustration of the full-length and deleted GST-YB-1 proteins used in this study. The striped box indicates the cold shock domain. (B) GST-pull down assay. 1 μg of each GST-YB-1 deletion mutant and GST which binds to 30 μl of glutathione-Sepharose 4B beads were incubated with a 100-fold excess of full-length Thio-IRP2 in the presence or absence of 50 μM FAC. Proteins bound to the beads were resolved by SDS-PAGE and analyzed by Western blotting using an anti-Thio antibody.
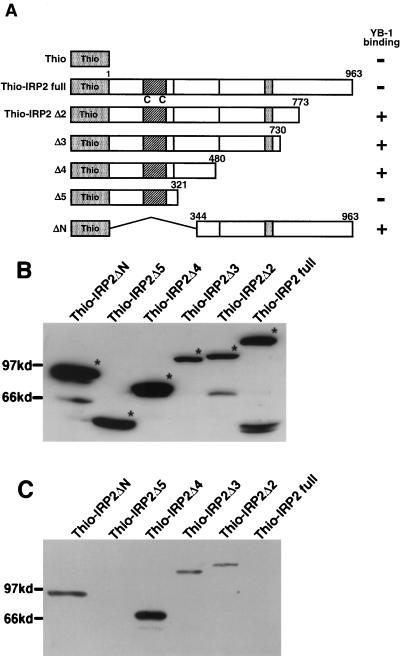
To identify the specific region in IRP2 that binds to the YB-1 protein, we created several IRP2 deletion mutants (Fig. 3A). We performed a GST pull-down assay using the GST-YB-1 fusion protein and Thio fusion proteins containing deletion mutants of IRP2 (Fig. 3B). GST-YB-1 interacted with Thio-IRP2Δ2, IRP2Δ3, IRP2Δ4, and IRP2ΔN but not with IRP2Δ5 or full IRP2 in the absence of FAC (Fig. 3C). These pull-down assays demonstrated that the GST-YB-1 fusion protein bound to residues 344 to 480 of IRP2, which collectively were designated domain 2.
FIG. 3.
Identification of the YB-1-binding domain in IRP2. (A) Schematic illustration of the ThioHis-IRP2 deletion mutants used in this study. (B) Expression of recombinant proteins. Deletion mutants were expressed in bacteria and analyzed by Coomassie brilliant blue staining of SDS-PAGE gels. (C) GST pull-down assay. 1 μg of GST-YB-1 or GST which binds to 30 μl of glutathione-Sepharose 4B beads was incubated with a 100-fold excess of ThioHis-IRP2 deletion mutants. Proteins bound to the beads were resolved by SDS-PAGE and analyzed by Western blotting using an anti-Thio antibody.
Effect of YB-1-IRP2 complex on the binding to IRE of ferritin mRNA 5′ UTR.
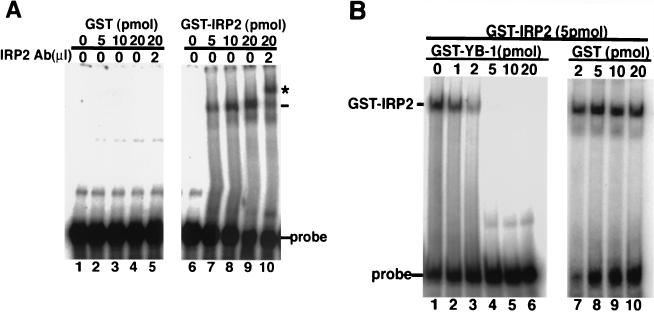
Previous studies have shown that IRP2 specifically binds to the IRE of ferritin mRNA 5′ UTR and regulates the efficiency of the protein synthesis of ferritin mRNA. In the presence of a high dose of iron, IRP2 was rapidly targeted to proteasome-mediated degradation and released from the IRE, resulting in the stimulation of ferritin protein synthesis. To identify whether GST-IRP2 could interact with IRE in the presence or absence of YB-1, we performed an RNA gel shift assay using purified GST fusion proteins and an mRNA probe of IRE from the ferritin heavy chain (Fig. 4). GST-IRP2 bound to IRE of ferritin 5′ UTR in a dose-dependent manner (Fig. 4A, lanes 6 to 9). The presence of IRP2 was further demonstrated by the ability of IRP2-specific antibody to shift the mobility of the GST-IRP2 complex (lane 10).
FIG. 4.
YB-1 inhibits IRP2 binding to RNA. (A) The indicated amounts of GST and GST fusion IRP2 were incubated with 5 × 104 cpm of 32P-labeled ferritin IRE for 15 min. IRP2-specific antibody (Ab) was added to lanes 5 and 10. IRP2-IRE complexes were separated by REMSA using a 6% native polyacrylamide gel. (B) 5 pmol of GST-IRP2 was incubated with the indicated concentration of GST or GST-YB-1 for 10 min and with 5 × 104 counts of 32P-labeled ferritin IRE for another 15 min. Samples were assessed by REMSA.
To identify whether YB-1 affects the binding capacity of IRP2 to IRE, we performed an RNA gel shift assay in the presence of various doses of GST-YB-1. A shifted band of IRE was observed in the presence of 5 pmol of IRP2. The retarded band that formed a complex with IRP2 was decreased by the addition of GST-YB-1, while the retarded band was not affected by GST (Fig. 4B). We did not observe that GST-YB-1 alone could bind to IRE in 5 pmol of GST-YB-1 (data not shown). These results suggested that IRP2 could bind to IRE but additional YB-1 interfered with the interaction of IRP2 with IRE.
Effect of YB-1-IRP2 on ferritin mRNA translation.
We next examined the role of YB-1-IRP2 complex in the protein synthesis of ferritin, which is known to be regulated by IRP2 at the translational level. We prepared two plasmids: one that carried the ferritin 5′ UTR (300 bp) ligated to the luciferase coding sequence, and one that carried only the luciferase reporter gene in pT7-Blue (Fig. 5A). We examined how YB-1 or IRP2 could affect the translational activity of ferritin 5′ UTR-luciferase by using a rabbit reticulocyte lysate translation assay system. In this assay, the translation activity of luciferase mRNA was determined in the presence of GST-IRP2 or GST. As shown in Fig. 5B, translation activity by the ferritin 5′ UTR-luciferase construct was inhibited to less than 20% of the initial baseline activity in the presence of 5 pmol of IRP2 or higher. By contrast, there appeared to be no translational inhibition by IRP2 when only luciferase RNA was used as a template (Fig. 5C). The ferritin 5′ UTR-luciferase RNA-driven translation activity was blocked in a dose-dependent manner by exogenous addition of YB-1. Luciferase activity was inhibited to less than 40% in the presence of 5 pmol of YB-1 (Fig. 5D). The luciferase activity driven by luciferase RNA was also blocked by YB-1 in a dose-dependent manner, and the inhibition rates were similar to those when the ferritin 5′ UTR-luciferase construct was used (Fig. 5E; Table 1). IRP2 could thus inhibit the translation of luciferase RNA only when the ferritin 5′ UTR element was present. However, YB-1 could inhibit the translation of luciferase RNA independently of the ferritin 5′ UTR element.
FIG. 5.
IRP2 or YB-1 represses translation. (A) Schematic illustration of the reporter plasmid containing luciferase cDNA ligated to the ferritin heavy-chain mRNA 5′ UTR region 300 bp (ferritin 5′UTR-luciferase) or containing unligated luciferase cDNA (luciferase). (B and C) Each reporter plasmid was transcribed in vitro, which was driven by T7 RNA polymerase and then translated in the rabbit reticulocyte lysate system. The indicated amount of GST-IRP2 was added to the system with 2 μg of ferritin 5′ UTR-luciferase RNA (B) or luciferase RNA (C). The luciferase assay was performed after incubation for 1.5 h at 30°C. All data are shown as the mean ± standard deviation from three independent experiments. (D and E) The indicated amount of GST-YB-1 was added to the reticulocyte lysate system with 2 μg of ferritin 5′UTR-luciferase RNA (D) or luciferase RNA (E).
TABLE 1.
Percent inhibition of luciferase activity by YB-1 and IRP2 on in vitro translation.
| RNA | % Inhibition of luciferase activitya
|
|||||||
|---|---|---|---|---|---|---|---|---|
| GST (5 pmol) | YB-1 (5 pmol) | IRP2 (5 pmol) | IRP2 (3 pmol) plus YB-1 at:
|
|||||
| 0 pmol | 3 pmol | 5 pmol | 3 pmol + DFOb | 5 pmol + DFOb | ||||
| Ferritin 5′ UTR-luciferase | 5.8 ± 4.2 | 79.5 ± 8.5 | 78.9 ± 7.3 | 49.2 ± 4.8 | 18.9 ± 13.5 | 46.1 ± 15.3 | 47.5 ± 2.6 | 46.9 ± 3.8 |
| Luciferase | 4.6 ± 3.5 | 71.5 ± 6.8 | −1.2 ± 5.8 | NDc | ND | ND | ND | ND |
Expressed as a percentage decrease with respect to the control.
DFO (0.5 mM) was coadministrated.
ND., not done.
We next examined how the translation activity driven by ferritin 5′ UTR was affected by the combined presence of YB-1 and IRP2. Consistent with Fig. 5D, the luciferase activity in the presence of 3 pmol of IRP2 was reduced to about 50% of the initial activity (Fig. 6A; Table 1). However, the inhibitory activity by IRP2 was restored by coadministration of YB-1 in a dose-dependent manner up to 3 pmol. The addition of YB-1 at 4 to 5 pmol could not restore IRP2-induced inhibition of the translational activity. These results suggested that an equimolar amount of YB-1 influenced the IRP2-mediated translational activity through ferritin 5′ UTR.
We next hypothesized that IRP2 protein could be modified by free iron in rabbit reticulocyte lysate, resulting in decreased inhibition by YB-1 through modified IRP2. We added DFO, an iron chelator, to the rabbit reticulocyte system and incubated it with the ferritin 5′ UTR-luciferase RNA in the presence of YB-1. The luciferase activity in the presence of 3 pmol of IRP2 with 0.5 mM DFO was reduced to about 50% of its initial activity (Fig. 6B). However, exogenous addition of YB-1 did not induce any change in the translational activity in the presence of DFO. The presence of iron appeared to require the abrogatory effect by exogenous addition of YB-1 on the IRP2-dependent inhibition of protein synthesis. However, we did not observe an additional effect of YB-1 at 4 to 5 pmol on translational activity in the presence of DFO. We also observed that DFO alone did not affect the translational inhibition of YB-1 alone in this assay system (data not shown).
Coimmunoprecipitation of YB-1 and IRP2 in the rabbit reticulocyte system.
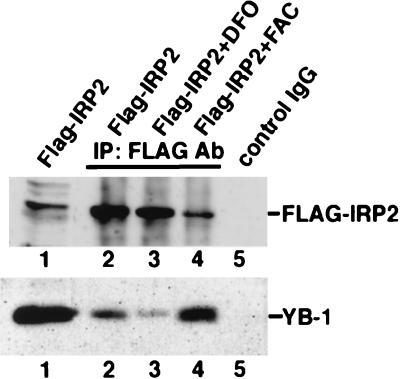
To determine whether YB-1 can interact with IRP2 in a rabbit reticulocyte lysate, we performed an in vitro coimmunoprecipitation assay as shown in Materials and Methods. A 30-μl volume of the reticulocyte lysate expressing the Flag-IRP2 fusion protein was incubated with beads that had been precoated with anti-Flag antibody in the presence or absence of 100 μM DFO or 50 μM FAC in 500 μl of PBS (pH 7.3). Because the sequence of rabbit YB-1 has a high homology (more than 90%) to human YB-1, the protein complexes were examined by sequential immunoblotting with human YB-1 antibody. The endogenous YB-1 in rabbit reticulocyte lysate was coprecipitated with Flag-IRP2 (Fig. 7, lane 2). The interaction of YB-1 and IRP2 appeared to be stronger in the presence of FAC (lane 4) than in its absence (lane 2), but the amount of YB-1 interacting with IRP2 was reduced in the presence of an iron chelator, DFO (lane 3). These results suggest that the inhibitory effect of IRP2-dependent protein synthesis in reticulocyte lysate is due to the increased interaction of these two YB-1 and IRP2 proteins.
FIG. 7.
Flag-IRP2 binds YB-1 in a rabbit reticulocyte lysate. Rabbit reticulocyte lysate incubated with pCMV/Flag-IRP2 plasmid or control vector was immunoprecipitated (IP) with either control IgG (lane 5) or anti-Flag M2-agarose (lanes 2 to 4) in the presence of DFO (lane 3) or FAC (lane 4). The immunocomplexes were separated by SDS-PAGE and transferred to an Immobilon-P membrane. Blots were probed with anti-YB-1 antibody (Ab).
Coimmunoprecipitation of YB-1 and IRP2 from cell extracts.
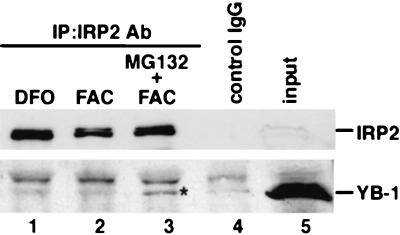
To determine whether YB-1 can interact with IRP2 in cultured cells, we used macrophages that expressed the IRP2 protein. Total cellular extracts were prepared to examine for the presence of YB-1-IRP2 complex by coimmunoprecipitation with anti-IRP2 antibody. The protein complexes were examined by sequential immunoblotting with YB-1 antibody. The binding of IRP2 with YB-1 was very weak in the presence of DFO (Fig. 8, lane 1). After treatment with FAC in macrophages for 6 h, IRP2 was degraded by the proteasome pathway (lane 2). Under these conditions, we did not observe any binding activity of IRP2 to YB-1, presumably due to the low level of IRP2 protein (lane 2). Treatment with a proteasome inhibitor, MG132, in the presence of FAC for 6 h resulted in a level of IRP2 protein similar to that seen in the presence of the iron chelator DFO (lane 3). YB-1 was coprecipitated with IRP2 only when treated with both FAC and MG132 (lane 3). There appeared to be no coimmunoprecipitation of YB-1 when treated with control mouse immunoglobulin G (IgG) (lane 4). YB-1 thus appeared to interact with IRP2 more strongly in the presence of FAC and MG132 in vivo, suggesting that YB-1 can bind to IRP2 before degradation by the ubiquitin-proteasome pathway in the presence of a high concentration of iron.
FIG. 8.
Coimmunoprecipitation of IRP2 and YB-1 from cell extracts. RAW264.7 cells were incubated in 100 μM DFO (lane 1), 600 μM FAC (lane 2), or FAC and MG132 (1 μM) (lane 3) for 6 h. Extracts from each treated cell were immunoprecipitated (IP) with immune anti-IRP2 (lanes 1-3) or control mouse IgG (lane 4). The immunocomplex was subjected to SDS-PAGE and transferred to an Immobilon-P membrane. Blots were probed with anti-YB-1 antibody (Ab). YB-1 is indicated by an asterisk.
DISCUSSION
We isolated five ribosomal proteins and IRP2 as well as YB-1 itself as partner proteins of YB-1 by two-hybrid analysis using full-length YB-1 as bait. To our surprise, most of these partner proteins were found to be translational machinery proteins. We previously identified the homomultimer interaction of YB-1 by means of an in vitro pull-down assay (16), and in the present study we found that YB-1 exhibited at least heterodimeric interaction in vivo, suggesting that this method of two-hybrid analysis works well. However, we could not observe the binding to previously reported proteins such as YY-1, AP2, and proliferating-cell nuclear antigen in this screening, suggesting that we need to perform further screening using a different cDNA library or a different yeast condition in two-hybrid screening. Of these translational proteins, we focused further on the interaction between IRP2 and YB-1 and its possible role in the modulation of protein synthesis. In this study, we found that IRP2 bound to IRE and inhibited the translational activity of ferritin protein under low-iron conditions. However, at high iron levels, YB-1 bound directly to oxidized modified IRP2 and inhibited the IRP2 binding to the IRE of ferritin mRNA before the degradation of IRP2 through a ubiquitin-proteasome pathway, resulting in enhanced translational activity of ferritin mRNA. We first presented a novel and unique function of YB-1 involving the regulation of ferritin gene expression through its modulation of the interaction of IRP2 and ferritin IRE.
YB-1 could interact with IRP-2 in vitro as well as in vivo, and this interaction was stimulated in the presence of iron. A GST pull-down assay showed binding of amino acid residues 321 to 480 in the IRP2 protein to YB-1 (Fig. 3A); this domain of YB-1 was involved in complex formation with IRP2. In our previous study, we reported that the central region was also responsible for interaction with dimerization (16). We also observed that YB-1 could bind to IRP2ΔN, but not to full-length IRP2, in both the two-hybrid assay and the in vitro pull-down assay (Fig. 1 and 3). Although YB-1 could interact with IRP2ΔN in the absence of FAC, YB-1 could interact with full-length IRP2 only in the presence of FAC, suggesting that the domain of IRP2 interacting with YB-1 is masked in the absence of FAC. When oxidized modification of IRP2 is induced by FAC (14), the modified IRP2 is susceptible to binding to YB-1 due to its conformational change. We observed that the full-length YB-1 can interact with IRP2Δ2, IRP2Δ3, IRP2Δ4, and IRP2ΔN in the absence of FAC (Fig. 3), suggesting that a domain of IRP2 which is highly interactive with YB-1 is conformationally changed by such deletion mutations. These results indicated that conformational changes of IRP2 are needed for its binding to YB-1.
In our present study, IRP2 alone bound to ferritin mRNA, but the binding of IRP2 to IRE on the ferritin mRNA was almost completely blocked by the presence of YB-1 (Fig. 5). Moreover, ferritin mRNA translation activity was blocked in a dose-dependent manner by the presence of either YB-1 or IRP2 alone when ferritin 5′ UTR-luciferase RNA was used (Fig. 5B and D). However, there appeared to be no inhibition of the translation by IRP2 when only luciferase RNA was used (Fig. 5C), suggesting that this inhibitory effect of IRP2 is mediated through a specific sequence, such as an IRE sequence. Coadministration of YB-1 was shown to abrogate the IRP2-induced inhibition of the translation activity by ferritin 5′ UTR-luciferase RNA (Fig. 6A). This abrogatory effect of YB-1 could be due to its blocking of IRP2 binding to IRE on ferritin mRNA. Since IRP1/IRP2 has been shown to block the binding of ribosome initiation complex when bound to the 5′ UTR IRE of ferritin mRNA (40), the block by IRP2 could be annihilated by the presence of YB-1, possibly through the interaction of IRP2 and YB-1.
The ferritin 5′ UTR including IRE contains stem-loop structures, and interaction of the 5′ UTR with IRP2 alters both the stem-loop structures and translation (40). YB-1 and its related proteins also interact with RNA (30). It remains unclear if YB-1 preferentially binds to the stem-loop structures to compete binding to such an altered RNA structure with IRP2. Several studies have reported that YB-1 and related proteins can change the secondary structures of RNA and play critical roles in translation and RNA splicing. Bacterial cold shock protein A (CspA) binds cooperatively to mRNA longer than 74 bp, destabilizing secondary structures (17). This RNA chaperone activity might stimulate the translation of certain mRNAs under a cold shock condition. Like the prokaryotic homologue CspA, the Y-box protein might destabilize the RNA secondary structures. Interaction of YB-1 with modified IRP2 might destabilize the secondary structure of IRE and facilitate attachment of the ribosomal large subunit or scanning of the 5′ UTRs of mRNA by the 43S preinitiation complex after releasing IRP2 from IRE.
Shnyreva et al. (35) showed that heterogeneous nuclear RNP (hnRNP) K, a component of the hnRNP particles, could also interact with YB-1 through the C-terminal domain. This hnRNP K promotes the formation of the pre-mRNA secondary structure and influences splice site selection. Chansky et al. (1) also reported that YB-1 influences the splice site selection of E1A pre-mRNA transcripts and that this splicing function of YB-1 is inhibited by oncogenic translocation liposarcoma protein and Ewing's sarcoma protein fusion proteins. These results suggested that the Y-box protein influences either stabilization or destabilization of the RNA secondary structure in splicing and translation machinery.
IRP1 and IRP2 proteins, members of the aconitase superfamily, are each conserved at ∼90% sequence identity within the organisms in which they share 60 to 70% identity (32). The IRP-binding activity of the IRPs may be activated by stimuli other than iron depletion, including exposure to nitric oxide or hydrogen peroxide. In addition, hypoxic conditions appear to increase IRP2-binding activity while simultaneously decreasing IRP1-binding activity. Thus different cell types may be differentially affected by various physiologic stimuli depending on which of the two IRPs predominates within them. IRP2 differs from IRP1 with respect to the mechanism by which iron levels are sensed (11, 15). Analysis of the IRP2 degradation process reveals that this protein undergoes iron-dependent oxidation, and the oxidized form of the protein is a target for ubiquitination and proteasomal degradation (14). In the present study, the cellular levels of IRP2 were found to be much lower in the absence than in the presence of MG132, suggesting that IRP2 is involved in the proteasomal degradation pathway. An immunoprecipitation assay with anti-IRP2 antibody showed complex formation of IRP2 and YB-1 in growing cells in the presence of iron and MG132 (Fig. 8). However, it remains unclear whether YB-1 in complex with IRP2 could be degraded by ubiquitination-proteasome after being released from the mRNAs of both proteins in the presence of iron.
YB-1 is a potent cap-dependent mRNA stabilizer in vitro as well as in vivo in various mammalian cell lines, and mRNA stabilization and translational repression facilitated by YB-1 are general and sequence nonspecific (6). In the present study, IRP2 also inhibited the translation of ferritin mRNA through IRE, which is located near the cap structure (8). We hypothesized that in the presence of a high concentration of FAC, YB-1 can interact with oxidized IRP2 and release IRP2 from the IRE of ferritin mRNA. Subsequently, IRP2 was degraded by the ubiquitine-proteosome pathway, and translation of ferritin mRNA was activated. In conclusion, we observed a direct interaction between YB-1 and IRP2 and showed that this interaction was involved in the translational regulation of ferritin mRNA and other iron-related proteins.
Acknowledgments
We thank L.C. Kühn for kind provision of the IRP2 antibody.
This work was supported by a Grant-in-Aid for Scientific Research on the Priority Area of ABC Proteins, by CREST (Core Research for Evolutional Science and Technology) of the Japan Science and Technology Corporation (JST), by the Second-Term Comprehensive Ten-Year Strategy for Cancer Control from the Ministry of Health and Welfare of Japan, and by the Cancer Research Fund from the Ministry of Education, Culture, Sports, Science, and Technology.
REFERENCES
- 1.Chansky, H. A., M. Hu, D. D. Hickstein, and L. Yang. 2001. Oncogenic TLS/ERG and EWS/Fli-1 fusion proteins inhibit RNA splicing mediated by YB-1 protein. Cancer Res. 61:3586-3590. [PubMed] [Google Scholar]
- 2.Chen, C. Y., R. Gherzi, J. S. Andersen, G. Gaietta, K. Jurchott, H. D. Royer, M. Mann, and M. Karin. 2000. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 14:1236-1248. [PMC free article] [PubMed] [Google Scholar]
- 3.Chevray, P. M., and D. Nathans. 1992. Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc. Natl. Acad. Sci. USA 89:5789-5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox, T. C., M. J. Bawden, A. Martin, and B. K. May. 1991. Human erythroid 5-aminolevulinate synthase: promoter analysis and identification of an iron-responsive element in the mRNA. EMBO J. 10:1891-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Didier, D. K., J. Schiffenbauer, S. L. Woulfe, M. Zacheis, and B. D. Schwartz. 1988. Characterization of the cDNA encoding a protein binding to the major histocompatibility complex class II Y box. Proc. Natl. Acad. Sci. USA 85:7322-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evdokimova, V., P. Ruzanov, H. Imataka, B. Raught, Y. Svitkin, L. P. Ovchinnikov, and N. Sonenberg. 2001. The major mRNA-associated protein YB-1 is a potent 5′ cap-dependent mRNA stabilizer. EMBO J. 20:5491-5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evdokimova, V. M., E. A. Kovrigina, D. V. Nashchekin, E. K. Davydova, J. W. Hershey, and L. P. Ovchinnikov. 1998. The major core protein of messenger ribonucleoprotein particles (p50) promotes initiation of protein biosynthesis in vitro. J. Biol. Chem. 273:3574-3581. [DOI] [PubMed] [Google Scholar]
- 8.Goossen, B., and M. W. Hentze. 1992. Position is the critical determinant for function of iron-responsive elements as translational regulators. Mol. Cell. Biol. 12:1959-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gray, N. K., K. Pantopoulous, T. Dandekar, B. A. Ackrell, and M. W. Hentze. 1996. Translational regulation of mammalian and Drosophila citric acid cycle enzymes via iron-responsive elements. Proc. Natl. Acad. Sci. USA 93:4925-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gu, W., S. Tekur, R. Reinbold, J. J. Eppig, Y. C. Choi, J. Z. Zheng, M. T. Murray, and N. B. Hecht. 1998. Mammalian male and female germ cells express a germ cell-specific Y-Box protein, MSY2. Biol. Reprod. 59:1266-1274. [DOI] [PubMed] [Google Scholar]
- 11.Guo, B., J. D. Phillips, Y. Yu, and E. A. Leibold. 1995. Iron regulates the intracellular degradation of iron regulatory protein 2 by the proteasome. J. Biol. Chem. 270:21645-21651. [DOI] [PubMed] [Google Scholar]
- 12.Herbert, T. P., and N. B. Hecht. 1999. The mouse Y-box protein, MSY2, is associated with a kinase on non-polysomal mouse testicular mRNAs. Nucleic Acids Res. 27:1747-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ise, T., G. Nagatani, T. Imamura, K. Kato, H. Takano, M. Nomoto, H. Izumi, H. Ohmori, T. Okamoto, T. Ohga, T. Uchiumi, M. Kuwano, and K. Kohno. 1999. Transcription factor Y-box binding protein 1 binds preferentially to cisplatin-modified DNA and interacts with proliferating cell nuclear antigen. Cancer Res. 59:342-346. [PubMed] [Google Scholar]
- 14.Iwai, K., S. K. Drake, N. B. Wehr, A. M. Weissman, T. LaVaute, N. Minato, R. D. Klausner, R. L. Levine, and T. A. Rouault. 1998. Iron-dependent oxidation, ubiquitination, and degradation of iron regulatory protein 2: implications for degradation of oxidized proteins. Proc. Natl. Acad. Sci. USA 95:4924-4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwai, K., R. D. Klausner, and T. A. Rouault. 1995. Requirements for iron-regulated degradation of the RNA binding protein, iron regulatory protein 2. EMBO J. 14:5350-5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izumi, H., T. Imamura, G. Nagatani, T. Ise, T. Murakami, H. Uramoto, T. Torigoe, H. Ishiguchi, Y. Yoshida, M. Nomoto, T. Okamoto, T. Uchiumi, M. Kuwano, K. Funa, and K. Kohno. 2001. Y box-binding protein-1 binds preferentially to single-stranded nucleic acids and exhibits 3′→5′ exonuclease activity. Nucleic Acids Res. 29:1200-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang, W., Y. Hou, and M. Inouye. 1997. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 272:196-202. [DOI] [PubMed] [Google Scholar]
- 18.Kelm, R. J., Jr., J. G. Cogan, P. K. Elder, A. R. Strauch, and M. J. Getz. 1999. Molecular interactions between single-stranded DNA-binding proteins associated with an essential MCAT element in the mouse smooth muscle alpha-actin promoter J. Biol. Chem. 274:14238-14245. [DOI] [PubMed] [Google Scholar]
- 19.Klausner, R. D., T. A. Rouault, and J. B. Harford. 1993. Regulating the fate of mRNA: the control of cellular iron metabolism. Cell 72:19-28. [DOI] [PubMed] [Google Scholar]
- 20.Kohler, S. A., B. R. Henderson, and L. C. Kuhn. 1995. Succinate dehydrogenase b mRNA of Drosophila melanogaster has a functional iron-responsive element in its 5′-untranslated region. J. Biol. Chem. 270:30781-30786. [DOI] [PubMed] [Google Scholar]
- 21.Ladomery, M., and J. Sommerville. 1995. A role for Y-box proteins in cell proliferation. Bioessays 17:9-11. [DOI] [PubMed] [Google Scholar]
- 22.Li, W. W., Y. Hsiung, V. Wong, K. Galvin, Y. Zhou, Y. Shi, and A. S. Lee. 1997. Suppression of grp78 core promoter element-mediated stress induction by the dbpA and dbpB (YB-1) cold shock domain proteins. Mol. Cell. Biol. 17:61-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makino, Y., T. Ohga, S. Toh, K. Koike, K. Okumura, M. Wada, M. Kuwano, and K. Kohno. 1996. Structural and functional analysis of the human Y-box binding protein (YB-1) gene promoter. Nucleic Acids Res. 24:1873-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsumoto, K., F. Meric, and A. P. Wolffe. 1996. Translational repression dependent on the interaction of the Xenopus Y-box protein FRGY2 with mRNA. Role of the cold shock domain, tail domain, and selective RNA sequence recognition. J. Biol. Chem. 271:22706-22712. [DOI] [PubMed] [Google Scholar]
- 25.Matsumoto, K., and A. P. Wolffe. 1998. Gene regulation by Y-box proteins: coupling control of transcription and translation. Trends Cell Biol. 8:318-323. [DOI] [PubMed] [Google Scholar]
- 26.Mertens, P. R., M. A. Alfonso-Jaume, K. Steinmann, and D. H. Lovett. 1998. A synergistic interaction of transcription factors AP2 and YB-1 regulates gelatinase A enhancer-dependent transcription. J. Biol. Chem. 273:32957-32965. [DOI] [PubMed] [Google Scholar]
- 27.Mullner, E. W., B. Neupert, and L. C. Kuhn. 1989. A specific mRNA binding factor regulates the iron-dependent stability of cytoplasmic transferrin receptor mRNA. Cell 58:373-382. [DOI] [PubMed] [Google Scholar]
- 28.Ohga, T., K. Koike, M. Ono, Y. Makino, Y. Itagaki, M. Tanimoto, M. Kuwano, and K. Kohno. 1996. Role of the human Y box-binding protein YB-1 in cellular sensitivity to the DNA-damaging agents cisplatin, mitomycin C, and ultraviolet light. Cancer Res. 56:4224-4228. [PubMed] [Google Scholar]
- 29.Okamoto, T., H. Izumi, T. Imamura, H. Takano, T. Ise, T. Uchiumi, M. Kuwano, and K. Kohno. 2000. Direct interaction of p53 with the Y-box binding protein, YB-1: a mechanism for regulation of human gene expression. Oncogene 19:6194-6202. [DOI] [PubMed] [Google Scholar]
- 30.Pelletier, M., M. M. Miller, and L. K. Read. 2000. RNA-binding properties of the mitochondrial Y-box protein RBP16. Nucleic Acids Res. 28:1266-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raj, G. V., M. Safak, G. H. MacDonald, and K. Khalili. 1996. Transcriptional regulation of human polyomavirus JC: evidence for a functional interaction between RelA (p65) and the Y-box-binding protein, YB-1. J. Virol. 70:5944-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rouault, T. A., D. J. Haile, W. E. Downey, C. C. Philpott, C. Tang, F. Samaniego, J. Chin, I. Paul, D. Orloff, J. B. Harford, et al. 1992. An iron-sulfur cluster plays a novel regulatory role in the iron-responsive element binding protein. Biometals 5:131-140. [DOI] [PubMed] [Google Scholar]
- 33.Ruzanov, P. V., V. M. Evdokimova, N. L. Korneeva, J. W. Hershey, and L. P. Ovchinnikov. 1999. Interaction of the universal mRNA-binding protein, p50, with actin: a possible link between mRNA and microfilaments. J. Cell Sci. 112:3487-3496. [DOI] [PubMed] [Google Scholar]
- 34.Safak, M., G. L. Gallia, S. A. Ansari, and K. Khalili. 1999. Physical and functional interaction between the Y-box binding protein YB-1 and human polyomavirus JC virus large T antigen. J. Virol. 73:10146-10157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shnyreva, M., D. S. Schullery, H. Suzuki, Y. Higaki, and K. Bomsztyk. 2000. Interaction of two multifunctional proteins. Heterogeneous nuclear ribonucleoprotein K and Y-box-binding protein. J. Biol. Chem. 275:15498-15503. [DOI] [PubMed] [Google Scholar]
- 36.Sommerville, J. 1999. Activities of cold-shock domain proteins in translation control. Bioessays 21:319-325. [DOI] [PubMed] [Google Scholar]
- 37.Swamynathan, S. K., A. Nambiar, and R. V. Guntaka. 2000. Chicken Y-box proteins chk-YB-1b and chk-YB-2 repress translation by sequence-specific interaction with single-stranded RNA. Biochem. J. 348:297-305. [PMC free article] [PubMed] [Google Scholar]
- 38.Swamynathan, S. K., A. Nambiar, and R. V. Guntaka. 1998. Role of single-stranded DNA regions and Y-box proteins in transcriptional regulation of viral and cellular genes. FASEB J. 12:515-522. [DOI] [PubMed] [Google Scholar]
- 39.Theil, E. C. 1998. The iron responsive element (IRE) family of mRNA regulators. Regulation of iron transport and uptake compared in animals, plants, and microorganisms. Metal Ions Biol. Syst. 35:403-434. [PubMed] [Google Scholar]
- 40.Thomson, A. M., J. T. Rogers, and P. J. Leedman. 1999. Iron-regulatory proteins, iron-responsive elements and ferritin mRNA translation. Int. J. Biochem. Cell Biol. 31:1139-1152. [DOI] [PubMed] [Google Scholar]
- 41.Toh, S., T. Nakamura, T. Ohga, K. Koike, T. Uchiumi, M. Wada, M. Kuwano, and K. Kohno. 1998. Genomic organization of the human Y-box protein (YB-1) gene. Gene 206:93-97. [DOI] [PubMed] [Google Scholar]
- 42.Vidal, M., R. K. Brachmann, A. Fattaey, E. Harlow, and J. D. Boeke. 1996. Reverse two-hybrid and one-hybrid systems to detect dissociation of protein-protein and DNA-protein interactions. Proc. Natl. Acad. Sci. USA 93:10315-10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolffe, A. P. 1994. Structural and functional properties of the evolutionarily ancient Y-box family of nucleic acid binding proteins. Bioessays 16:245-251. [DOI] [PubMed] [Google Scholar]