Abstract
Formation of iron/sulfur (Fe/S) clusters, protein translocation and protein folding are essential processes in the mitochondria of Saccharomyces cerevisiae. In a systematic approach to characterize essential proteins involved in these processes, we identified a novel essential protein of the mitochondrial matrix, which is highly conserved from yeast to human and which we termed Isd11. Depletion of Isd11 caused a strong reduction in the levels of the Fe/S proteins aconitase and the Rieske protein, and a massive decrease in the enzymatic activities of aconitase and succinate dehydrogenase. Incorporation of iron into the Fe/S protein Leu1 and formation of the Fe/S cluster containing holoform of the mitochondrial ferredoxin Yah1 were inhibited in the absence of Isd11. This strongly suggests that Isd11 is required for the assembly of Fe/S proteins. We show that Isd11 forms a stable complex with Nfs1, the cysteine desulfurase of the mitochondrial machinery for Fe/S cluster assembly. In the absence of Isd11, Nfs1 is prone to aggregation. We propose that Isd11 acts together with Nfs1 in an early step in the biogenesis of Fe/S proteins.
Keywords: cysteine desulfurase, Fe/S cluster formation, Fe/S protein, Isd11, mitochondria
Introduction
Large-scale deletion studies and single-gene analyses of the ca. 6600 open reading frames (ORF) of the Saccharomyces cerevisiae genome have identified about 1020 of them (17%) to be essential for cell viability. The mitochondria of this organism are composed of ca. 700–900 different proteins in yeast; yet, only a relatively small number of essential mitochondrial proteins have been described (Rehling et al, 2003; Sickmann et al, 2003; Prokisch et al, 2004; Reichert and Neupert, 2004). These latter proteins are mainly involved in three processes in mitochondria: protein translocation, protein folding/unfolding and biogenesis of iron/sulfur (Fe/S) proteins.
Essential proteins of the translocation machineries act in sorting of proteins into and across the mitochondrial membranes. Most of these proteins are constituents of the translocases of the outer membrane (TOM and TOB/SAM complex) and of the inner membrane, the TIM23 and the TIM22 translocases (Neupert and Brunner, 2002; Endo et al, 2003; Paschen et al, 2003; Koehler, 2004; Rehling et al, 2004). The second class of essential mitochondrial proteins is made up of the components of the Hsp70 and Hsp60 chaperone systems, which mediate the folding and unfolding of proteins in the mitochondrial matrix (Cheng et al, 1989; Hartl, 1996; Voos and Rottgers, 2002). The third group of essential mitochondrial proteins are required for the biogenesis of Fe/S proteins (Craig et al, 1999; Lill and Kispal, 2000). Fe/S clusters are present as cofactors in a variety of mitochondrial, cytosolic and nuclear proteins, which are crucial for numerous electron transfer processes and enzymatic reactions (Craig et al, 1999; Lill and Muhlenhoff, 2005). Recent studies suggest that maturation of all cellular Fe/S proteins in yeast depends on the function of a group of highly conserved mitochondrial proteins, the iron/sulfur cluster (ISC) assembly machinery (Kispal et al, 1999; Li et al, 2001; Gerber et al, 2004). Central players of the mitochondrial ISC assembly machinery are the cysteine desulfurase Nfs1 and the proteins Isu1 and Isu2 (Strain et al, 1998; Garland et al, 1999; Kispal et al, 1999; Li et al, 1999; Schilke et al, 1999; Gerber et al, 2004). Deletion of NFS1 as well as the double deletion of ISU1 and ISU2 are lethal for yeast cells (Li et al, 1999; Schilke et al, 1999). Nfs1, like its related bacterial cysteine desulfurases NifS, IscS and SufS, acts as a sulfur donor for the biogenesis of Fe/S proteins (Zheng et al, 1993; Schwartz et al, 2000; Loiseau et al, 2003; Muhlenhoff et al, 2004). It delivers the sulfur to the Isu proteins, which act as scaffolds for the formation of Fe/S clusters (Muhlenhoff et al, 2003). Further components required for this process include an electron transfer system consisting of the ferredoxin reductase Arh1 and the ferredoxin Yah1, as well as the yeast frataxin homolog (Yfh1), which facilitates the delivery of ferrous iron to the scaffold proteins (Babcock et al, 1997; Lange et al, 2000; Li et al, 2001; Gerber et al, 2003; Yoon and Cowan, 2003). Other components, like the chaperone Ssq1, its essential cochaperone Jac1 and the glutaredoxin Grx5, appear to function in a later step of the biogenesis of the Fe/S cluster-containing proteins (Strain et al, 1998; Kim et al, 2001; Lutz et al, 2001; Voisine et al, 2001; Craig and Marszalek, 2002; Rodriguez-Manzaneque et al, 2002; Dutkiewicz et al, 2003). In addition to the proteins of the ISC assembly machinery, two additional mitochondrial proteins, the ATP-binding cassette transporter Atm1 and the sufhydryl oxidase Erv1, are required for the biogenesis of cytoplasmic Fe/S clusters (Lill and Muhlenhoff, 2005). Despite the considerable increase of our knowledge of the ISC assembly machinery, its molecular mechanism and the interplay between the components are still poorly understood.
Here we describe the identification of Isd11 as a novel essential component of the ISC assembly machinery required for the biogenesis of Fe/S proteins. Isd11 forms a stable complex with Nfs1. We propose that Isd11 is essential for Nfs1 to fulfil its function of sulfur transfer for Fe/S cluster formation.
Results
The Isd11 protein is present in organisms throughout the eukaryotic kingdom
The ORF YER048w-a of S. cerevisiae was reported to be essential for viability and encodes a mitochondrial protein with a molecular mass of 11 kDa, further onwards referred to as Isd11 (ISC biogenesis desulfurase interacting protein). Homologs of ISD11 were found in the genomes in all eukaryotic species analyzed, such as fungi (Schizosaccharomyces pombe, Neurospora crassa) and higher organisms (e.g. Drosophila melanogaster and Homo sapiens) (Figure 1A). Sequence conservation is not restricted to certain segments of the proteins. Isd11 can be grouped among the LYR family (PF05347), which is characterized by the presence of the three amino-acid residues segment LYR/K (Bateman et al, 2004). Members of this family also include subunits of complex I of the respiratory chain of various organisms. A cladogram, including the Isd11 proteins and the related subunits of complex I, is presented as Supplementary data (Supplementary Figure S1). A possible evolutionary relationship between the Isd11 and complex I subunit proteins remains to be further investigated.
Figure 1.

Isd11 is a protein of the mitochondrial matrix. (A) Sequence alignment of Isd11 proteins of various species. The alignment was generated using ClustalX (1.8). Amino-acid residues identical in at least four of the aligned proteins are shown in black and similar residues are shown in gray: S. cerevisiae (Sc), S. pombe (Sp), N. crassa (Nc), H. sapiens (Hs) and D. melanogaster (Dm). (B) Isd11 is located in the mitochondria. Yeast cells were subfractionated and equal amounts of protein were subjected to SDS–PAGE and immunodecoration with antibodies against Isd11, a cytosolic protein (Bmh2), a protein of the ER (Erp1) and a mitochondrial protein (Mge1). (C) Isd11 is located in the mitochondrial matrix. Isolated mitochondria, mitoplasts prepared by osmotic shock and Triton-solubilized mitochondria were treated with or without proteinase K (PK). Mitochondria were subjected to carbonate extraction and supernatant (S) and pellet (P) fractions were separated. Samples were analyzed by SDS–PAGE and immunodecoration with antibodies against the indicated proteins. (D) Isd11 is attached to the inner membrane. WT mitochondria were resuspended in buffer containing different concentrations of KCl and opened by sonication. Supernatant fractions (Sup) containing soluble proteins and the membrane fractions were separated by centrifugation. Proteins in the supernatant fraction were collected by TCA precipitation. Laemmli buffer was added to both fractions and subjected to SDS–PAGE and immunodecoration with antibodies against Isd11, the mitochondrial desulfurase Nfs1, a protein of the inner membrane (ADP/ATP carrier, Aac2) and a matrix protein (Mge1). (E) Isd11 is imported into isolated mitochondria in a membrane-potential-dependent manner. Reticulocyte lysate containing 35S-labelled Isd11 was incubated with mitochondria in the presence or absence of membrane potential ΔΨ. After import, mitochondria were splitted and treated with or without PK under isotonic or hypotonic swelling (Sw) conditions, as indicated. Samples were analyzed by SDS–PAGE and autoradiography. 50% Input, 50% of the amount of lysate used per lane.
Isd11 is a membrane-associated protein of the mitochondrial matrix space
Subcellular fractionation of yeast cells confirmed the mitochondrial location of Isd11, which was previously analyzed in a genome-wide screen using a GFP-tagged version and in an integrative study of the mitochondrial proteome (Huh et al, 2003; Prokisch et al, 2004) (Figure 1B). In order to analyze the submitochondrial location of Isd11, mitochondria were prepared. Mitoplasts were obtained by incubation of mitochondria in hypotonic medium to rupture the outer membrane. Mitochondria and mitoplasts were treated with proteinase K (PK) (Figure 1C). Neither in mitochondria nor in mitoplasts the protein was degraded. On the other hand, the protein was degraded by the added protease upon opening of the inner membrane by the addition of Triton X-100. When the mitochondria were treated with sodium carbonate at pH 11.5, Isd11 was largely recovered in the supernatant (Figure 1C). This indicates that Isd11 is not an integral membrane protein. However, Isd11 was present in the membrane fraction, when membranes and soluble material were separated after sonication of mitochondria in the presence of low concentrations of salt (Figure 1D). In conclusion, Isd11 is a protein of the mitochondrial matrix space associated with the inner membrane. This topology is in agreement with the hydrophilic nature of the protein, with no recognizable predicted transmembrane segments.
The N-terminal sequence of Isd11 does not display a characteristic cleavable N-terminal mitochondrial targeting sequence. Most of the Isd11 homologs have only two positively charged residues between the start methionine and the conserved LYR motif. We therefore studied the import of Isd11 preprotein synthesized in reticulocyte lysate into isolated mitochondria. Isd11 was imported into mitochondria in a ΔΨ-dependent manner (Figure 1E). After import it was present, like the endogenous protein, in a protease-protected location in mitoplasts. The size of the precursor form was indistinguishable from the size of the imported protein. Apparently, a cleavable presequence is not present in the precursor, but the N-terminal sequence is sufficient to direct the protein into the mitochondrial matrix.
Downregulation of Isd11 leads to a complete stop of cell growth
In order to study the function of Isd11, we constructed a yeast strain (GAL10-ISD11) in which the gene was under control of the GAL10 promoter, allowing the regulated expression of Isd11. Growth of this strain was tested on YP-agar plates containing either galactose or glucose. In the presence of glucose, cells stopped growing (Figure 2A). A similar observation was made upon cultivation of cells in liquid medium. GAL10-ISD11 cells and wild-type (WT) cells were grown in medium containing galactose and then shifted to medium containing glucose (Figure 2B). Cells depleted of Isd11 slowed down significantly in growth compared to WT cells 15 h after the shift to glucose-containing medium, and virtually stopped growing at 25 h after the shift (Figure 2B). These results confirm the essential nature of the ISD11 gene (Giaever et al, 2002).
Figure 2.
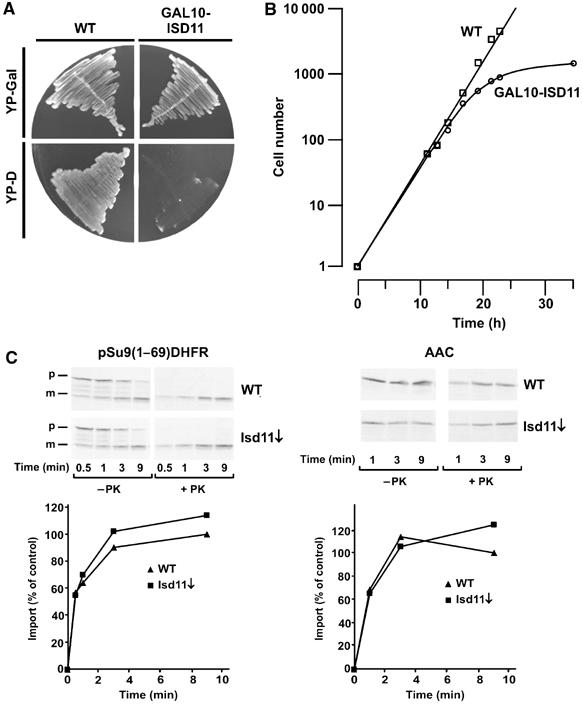
Downregulation of Isd11 in yeast cells affects cell growth, but not the capacity of the mitochondria to import preproteins. (A) Cells carrying the ISD11 gene under control of the GAL10 promoter and WT cells were grown for two rounds on plates containing rich medium with galactose (YP-Gal) or glucose (YP-D). (B) GAL10-ISD11 cells and WT cells were first grown on galactose and then incubated in glucose-containing medium for the indicated times. At time zero, the cell number was set to 1. (C) Isd11 is not required for the import of mitochondrial precursor proteins. The indicated radiolabelled precursor proteins of the TIM23 complex pathway (pSu9(1–69)DHFR) and of the TIM22 complex pathway (AAC) were imported into WT mitochondria and mitochondria depleted of Isd11 (Isd11↓) for different time periods. Mitochondria were reisolated, treated with or without PK and analyzed by SDS–PAGE and autoradiography. The mature forms of the proteins were quantified after PK treatment of mitochondria. Import into WT mitochondria at the longest time point was set to 100% (control). p, precursor form; m, mature form.
Import of mitochondrial preproteins is not affected in cells depleted of Isd11
Since most of the essential proteins of mitochondria have a function in protein translocation, we analyzed the import of mitochondrial preproteins into isolated mitochondria from cells depleted of Isd11 (Isd11↓) and from WT cells as control. In view of the location of Isd11, preproteins using the TIM23 translocation pathway, such as the fusion protein of the first 69 amino-acid residues of subunit 9 of the F1Fo-ATPase from N. crassa and mouse DHFR (pSu9(1–69)DHFR), as well as substrates of the TIM22 translocase, such as the ATP/ADP carrier (AAC), were tested. No significant import defect was observed in cells depleted of Isd11 (Figure 2C). Thus, Isd11 does not appear to play a role in the translocation of proteins into mitochondria and the stop of cell growth is not caused by a defect in the protein import machinery.
Isd11 is required for the biogenesis of Fe/S proteins
Isd11 was hardly detectable in the mitochondria of cells of the GAL10-ISD11 strain 12 h after shift from galactose to glucose (Figure 3A). At 18 and 24 h after the shift, Isd11 was not detectable anymore. The steady-state levels of various proteins were determined in mitochondria isolated from cells depleted of Isd11 (Isd11↓) for 12, 18 and 24 h. Aconitase and Rieske FeS, two proteins containing Fe/S clusters, showed strongly reduced levels upon 18 h depletion of Isd11 and were almost not detectable after 24 h of downregulation of Isd11. In contrast, the levels of other mitochondrial proteins, such as the outer membrane protein Tom40, the inner membrane translocation component Tim23 and the matrix protein Mge1 were not altered. Thus, depletion of Isd11 appears to affect specifically Fe/S proteins.
Figure 3.
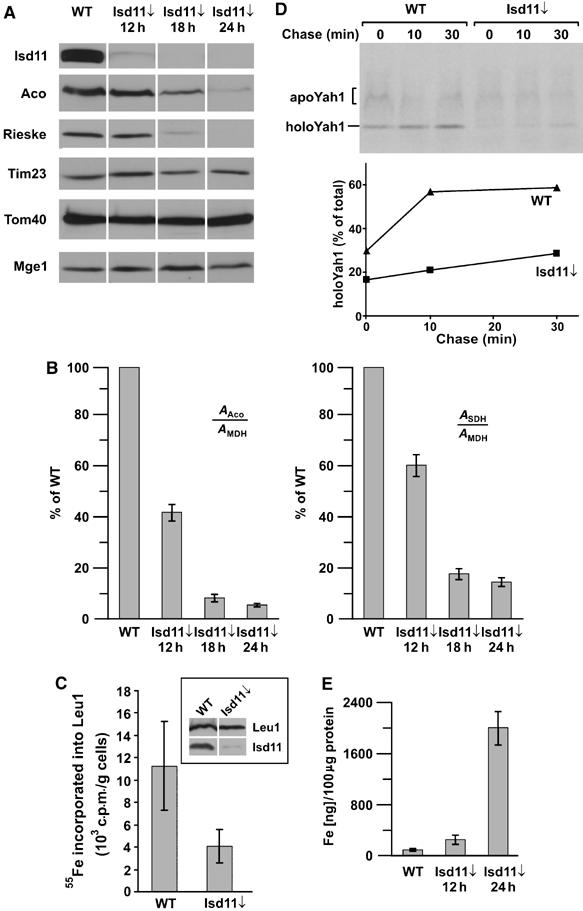
Downregulation of Isd11 affects the levels and activities of FeS cluster-containing proteins. (A) GAL10-ISD11 cells were grown at 30°C for 12, 18 and 24 h after shift to glucose-containing medium. Protein levels of mitochondria (100 μg) isolated from these cells and from WT cells were analyzed by SDS–PAGE and immunodecoration with antibodies against the indicated proteins. (B) The activities of SDH and aconitase (Aco) were measured in mitochondria isolated from WT cells and cells downregulated of Isd11 (Isd11↓) for the indicated time periods. As a standard, the non-FeS cluster-containing protein MDH was measured. The ratio of the activities of aconitase and MDH or SDH and MDH was calculated and expressed as percent of the ratio in WT cells. (C) The in vivo assembly of the Fe/S cluster of the cytoplasmic Leu1 protein was inhibited in the absence of Isd11. GAL-Isd11 cells were depleted of Isd11 by growth for 18 h in glucose-containing medium. These cells and WT cells were incubated with 55Fe for 2 h. Cell lysates were prepared and an immunoprecipitation with antibodies against Leu1 was performed. Incorporated 55Fe into Leu1 was determined by liquid scintillation counting. The inset shows the immunostaining of Leu1 and Isd11 present in these cell lysates. (D) The formation of holoYah1 was dependent on Isd11. The 35S-labelled precursor protein of Yah1 was imported into the mitochondria isolated of WT and Isd11↓ cells for 10 min at 25°C. The import reactions were stopped by the addition of valinomycin and incubated further for the indicated time periods (‘chase'). Aliquots were taken and the nonimported precursor proteins were removed by treatment with PK. The samples were analyzed by native gel electrophoresis and autoradiography. At the indicated time points, the amounts of apo- and holoYah1 were quantified and their ratios were expressed as percent holoYah1 of the sum of both forms (total). (E) The iron content of 100 μg of mitochondria isolated from WT and Isd11 downregulated cells was determined.
In addition, we measured the enzymatic activities of aconitase and succinate dehydrogenase (SDH) in cells depleted of Isd11. The activities were standardized in relation to the activity of the non-Fe/S protein, malate dehydrogenase (MDH) and compared to the relative activities in WT cells. A strong reduction in the activities of aconitase and SDH was observed in cells depleted of Isd11 (Figure 3B). Downregulation of Isd11 for only 12 h led to a significant decrease of the activities to about 40–60%. At this time point, endogenous protein levels of Fe/S proteins were not significantly altered, indicating that the depletion of Isd11 results in the reduction of the active form of the Fe/S enzymes. This shows that Isd11 is crucial for the formation or stability of functional Fe/S proteins.
We studied in vivo and in organello whether the incorporation of Fe/S clusters into the apoproteins is affected in cells depleted of Isd11. First, we employed an assay developed by Lill and co-workers, which determines the uptake of radioactive 55Fe in the Fe/S cluster of Leu1, a cytoplasmic Fe/S protein of the biosynthesis pathway of leucine (Kispal et al, 1999). Following growth in iron-free medium, the cells were incubated in minimal medium containing radioactive 55Fe. Cell lysates were prepared and Leu1 was precipitated with specific antibodies against Leu1. The incorporated radioactivity in the Fe/S cluster of Leu1 was determined by liquid scintillation counting. Compared to WT cells, much less 55Fe incorporated into Leu1 was detected in cells depleted of Isd11, albeit similar levels of Leu 1 protein were present in these cells (Figure 3C). This indicates that cells depleted of Isd11 lack the Fe/S cluster of Leu1 and suggests that the absence of the enzymatic activities of the Fe/S proteins was due to the lack of their clusters. Next, we analyzed the incorporation of the Fe/S cluster into the apoform of the mitochondrial ferredoxin Yah1 in the mitochondria of WT cells and cells depleted of Isd11. The Yah1 protein was synthesized in vitro in the presence of 35S-methionine and incubated with isolated mitochondria. The import reaction was stopped by addition of valinomycin to deplete the membrane potential and the samples were further incubated (‘chase'). Aliquots were withdrawn at the indicated time points, nonimported material was digested by PK and mitochondria were reisolated. The conversion of the apoform to the Fe/S cluster containing holoform was analyzed by separation of these forms on a native gel and subsequent autoradiography (Figure 3D, upper panel). The holoform migrates as a distinct band and runs faster than the apoform, which migrates in a broader range in the native gel (Lutz et al, 2001). During the import reaction, much more holoYah1 was formed in the mitochondria of WT cells than in the mitochondria depleted of Isd11 (Figure 3D). The ratio of the two forms after import and the indicated times of chase were determined by quantification using the phosphor imaging system. In contrast to WT mitochondria, the conversion of the apoform into the holoform of Yah1 was strongly affected in mitochondria depleted of Isd11 (Figure 3D, lower panel). Thus, the incorporation of the Fe/S cluster into the apoform of Yah1 is inhibited in the absence of Isd11. In summary, the results obtained in vivo and in organello indicate that Fe/S proteins are defective in the absence of Isd11 due to the lack of assembly of their Fe/S clusters. The results suggest that Isd11 is required in an early step in the biogenesis of the Fe/S cluster containing holoforms of Fe/S proteins.
In view of the connection between Fe/S cluster formation and the iron metabolism, we determined the iron contents in mitochondria depleted of Isd11 (Figure 3E). Compared to the mitochondria of WT cells, the iron content was increased about 30-fold after 24 h of depletion. Such mitochondrial iron accumulation was previously observed for many mutant cells defective in the pathway of assembly of Fe/S proteins (Babcock et al, 1997; Foury and Cazzalini, 1997; Knight et al, 1998; Garland et al, 1999; Kispal et al, 1999; Li et al, 1999, 2001; Schilke et al, 1999; Lange et al, 2000; Kim et al, 2001). Mutants of this class show further defects in cellular iron homeostasis, such as decreased cytoplasmic levels of iron and an increased cellular iron uptake (Li et al, 1999, 2001). Our results suggest that Isd11 is required for the biogenesis of Fe/S clusters and that it might play a role for the cellular iron homeostasis.
Isd11 forms a complex with Nfs1
To address at which step in the biogenesis of Fe/S proteins Isd11 is required, we constructed a strain expressing Isd11 with a C-terminal heptahistidinyl tag and performed pulldown experiments. Mitochondria isolated from this strain and from a WT strain were solubilized with dodecylmaltoside (DDM) and the extracts obtained were incubated with NiNTA agarose beads. Bound material was eluted and analyzed by SDS–PAGE and Coomassie Blue staining. Significant amounts of two proteins with a molecular weight of about 45 and 14 kDa were coeluted in a specific manner from the beads incubated with the extract containing his-tagged Isd11 (Figure 4A). These proteins were absent in the eluate from the WT strain, in contrast to a few additional other proteins. Mass spectroscopy identified the 45 kDa protein as the mitochondrial desulfurase of the ISC machinery, Nfs1, and the 14 kDa protein as Isd11.
Figure 4.
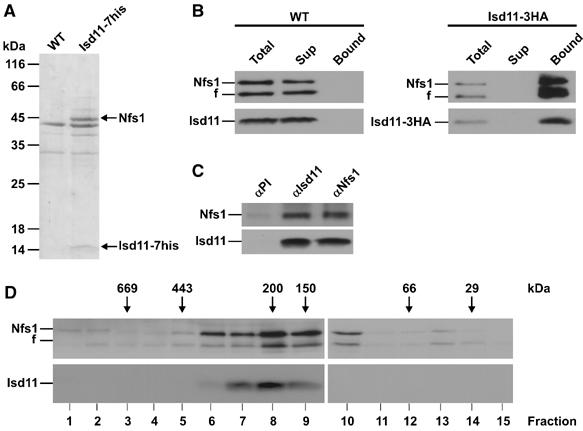
Isd11 is present in a complex with Nfs1. (A) Coisolation of Nfs1 with His-tagged Isd11. Isolated mitochondria of WT cells or cells expressing Isd11 with a C-terminal heptahistidinyl tag (Isd11-7his) were solubilized with DDM and incubated with NiNTA beads. Bound material was eluted and analyzed by SDS–PAGE and Coomassie staining. Nfs1 and Isd11 were identified by mass spectrometry. (B) Copurification of Nfs1 with Isd11. Mitochondria of a WT strain and a strain carrying a triple HA-tagged Isd11 were solubilized with digitonin. Following clarifying spin, the supernatants were incubated with antibodies against the HA tag coupled to protein A-Sepharose beads. The beads were harvested, washed and bound proteins were eluted with Laemmli buffer. Samples were analyzed by SDS–PAGE and immunodecoration with antibodies against Nfs1 and Isd11. In all, 10% of total and supernatant were loaded. f, Fragment of Nfs1. (C) Co-immunoprecipitation of Isd11 and Nfs1. WT mitochondria were solubilized with DDM. The co-immunoprecipitation was performed and analyzed as in (B), using antibodies against either Nfs1 or Isd11. (D) Isd11 and Nfs1 cofractionate on a gel-filtration column. Mitochondrial membranes were opened in the presence of 250 mM KCl by sonication. After a clarifying spin, the supernatant was subjected to a Superose-12 gel-filtration column. Fractions were analyzed by SDS–PAGE and immunodecoration with antibodies against Isd11 and Nfs1.
In order to check whether Isd11 forms a specific complex with Nfs1, we generated a strain expressing Isd11 with a C-terminal triple HA-tag from a chromosomal copy. Mitochondria were isolated from this strain, as well as from WT, and were solubilized with digitonin. The mitochondrial extracts were subjected to co-immunoprecipitation with antibodies against the HA peptide. These antibodies depleted the mitochondrial extract of the HA-tagged Isd11, but not of WT Isd11 (Figure 4B). Together with HA-tagged Isd11, Nfs1 was co-precipitated from the cell extract and was not detectable in the supernatant fraction of the precipitation. The interaction is obviously specific, as Nfs1 was not precipitated from the mitochondrial extract of WT cells using antibodies against the HA peptide. Parts of the Nfs1 in these experiments were present as a slightly smaller fragment. This fragment was generated by a proteolytic event occurring after lysis of mitochondria with detergents through protease activity present in the mitochondrial preparation. This degradation was pronounced when the mitochondrial lysates were processed for longer time periods and could be partly controlled by addition of a mixture of a variety of protease inhibitors. This phenomenon has been observed before (Gerber et al, 2003). When mitochondrial extracts of WT cells were subjected to co-immunoprecipitation with antibodies against Isd11 and Nfs1, Isd11 was co-precipitated with Nfs1 and vice versa (Figure 4C). Neither Nfs1 nor Isd11 were detected in the nonbound fraction of the co-precipitations (data not shown). Thus, the Nfs1 appears to be in a stable complex with Isd11.
We further analyzed the apparent native molecular mass of Isd11 and Nfs1 by gel filtration. Mitochondria from WT cells were resuspended in buffer containing 250 mM KCl and the mitochondrial membranes were opened by sonication. Following a clarifying spin, the supernatant was subjected to gel filtration over a Superose-12 column. Nfs1 and Isd11 eluted from the column in the same fraction corresponding to a molecular mass of about 180–200 kDa (Figure 4D). A minor amount of Nfs1 was present as a monomer, which might indicate instability of the Nfs1•Isd11 complex or the presence of a minor pool of Nfs1, which is available for interactions with other proteins. In summary, the results indicate that the majority of Isd11 and Nfs1 forms a high-molecular-weight complex. We were able to show that 35S of radiolabelled cysteine was incorporated in intact mitochondria into the Nfs1, which was in complex with Isd11 (K Hell, unpublished data). This suggests that the complex of Nfs1 and Isd11 is active in persulfide formation as a step in the sulfur transfer reaction in vivo.
In the absence of Isd11, Nfs1 has desulfurase activity, but is prone to aggregation
We analyzed the properties of Nfs1 in the presence and absence of its complex partner Isd11. In the absence of Isd11, the endogenous levels of Nfs1 as determined by immunodecoration were slightly reduced compared to the levels in mitochondria of WT cells (Figure 5A). In order to test the stability of Nfs1, mitochondria from WT cells and from cells lacking Isd11 were incubated on ice and at 37°C. They were solubilized in Triton X-100 and insoluble material was separated from soluble material by centrifugation. Fractions were analyzed by SDS–PAGE and immunodecoration. With mitochondria depleted of Isd11, but not with WT mitochondria, Nfs1 was largely found in the fraction containing insoluble material following treatment at 37°C. In contrast, treatment on ice did not affect the solubility of Nfs1. Other proteins such as Tim16 stay soluble, even after treatment at 37°C (Figure 5B). Thus, Nfs1 is prone to form insoluble higher oligomeric aggregates in the absence of Isd11. In addition, following lysis of mitochondria, Nfs1 was more sensitive to proteolytic degradation by endogenous proteases in the absence of Isd11 (data not shown). Notably, in WT mitochondria, Nfs1 and Isd11 were present in the membrane fraction after sonication of mitochondria in the presence of low salt up to about 100 mM (see Figure 1D). In contrast, when mitochondria of cells depleted of Isd11 were opened by sonication, less Nfs1 was found in the membrane fraction (data not shown). Thus, in the absence of Isd11, the membrane association of Nfs1 seems to be destabilized, which might increase the tendency of Nfs1 to aggregate.
Figure 5.
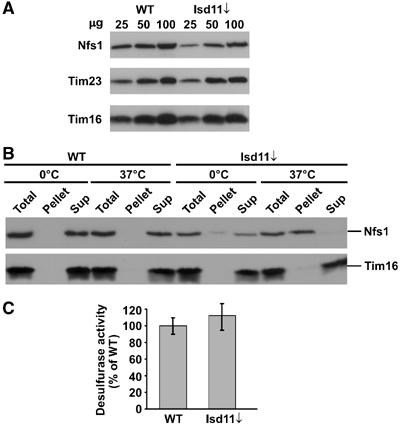
Nfs1 in Isd11-depleted mitochondria has desulfurase activity, but is prone to aggregation. (A) Protein levels of Nfs1 in mitochondria (25, 50, 100 μg) isolated from cells depleted of Isd11 for 18 h (Isd11↓) and from WT cells were analyzed by SDS–PAGE and immunodecoration. (B) Aggregation of Nfs1 in cells lacking Isd11. Mitochondria isolated from WT and Isd11-depleted cells were incubated for 20 min at 0 or 37°C. Mitochondria were lysed with 0.5% Triton X-100 and aggregated material was pelleted by centrifugation. Total and the supernatant fractions (Sup) were precipitated with TCA. Samples were analyzed by SDS–PAGE and immunodecoration. (C) Nfs1 has desulfurase activity in the absence of Isd11. Mitochondria (100 μg) from WT and Isd11↓ cells were lysed with 0.2% DDM and then incubated for 30 min at 30°C in buffer containing 5 mM DTT and 4 mM cysteine. Sulfide produced was converted to methylene blue, whose absorption was measured at 667 nm. Background activity was determined by incubation of the mitochondrial extracts in the absence of cysteine and subtracted. The desulfurase activity was expressed as percent of activity present in WT mitochondria.
Nfs1 is a desulfurase that acts in the abstraction of sulfur from cysteine for the formation of Fe/S clusters in the mitochondria (Lill and Muhlenhoff, 2005). We tested the desulfurase activity in mitochondrial extracts from cells depleted of Isd11 and from WT cells. To this end, we employed an assay with mitochondrial extracts, which measures desulfurase activity by determination of sulfide production from cysteine in vitro (Muhlenhoff et al, 2004). In the absence of Isd11, sulfide production was similar to that of WT extracts (Figure 5C). Thus, Nfs1 seems to have normal or even slightly enhanced desulfurase activity in the absence of Isd11.
In summary, the results suggest that Isd11 stabilizes Nfs1, but is not required for the desulfurase activity of Nfs1. The data presented argue for a function of Isd11 at the level of Nfs1 at a step following the desulfurase reaction of Nfs1.
Discussion
We describe here the identification and characterization of a novel protein, Isd11, in the matrix of mitochondria. Isd11 has a molecular mass of about 11 kDa and is highly conserved among virtually all eukaryotic organisms. We show that Isd11 fulfills an essential role in the formation of iron sulfur clusters and therefore is essential for viability of yeast cells. Reduced protein levels and activities of Fe/S proteins and accumulation of iron in mitochondria were observed in cells depleted of Isd11. The loss of the enzymatic activity of the Fe/S proteins was due to the lack of their Fe/S clusters. We demonstrate that the incorporation of the Fe/S clusters into the apoform of the mitochondrial ferredoxin Yah1 was strongly decreased in mitochondria depleted of Isd11. Depletion of Isd11 affects not only mitochondrial but also cytoplasmic Fe/S proteins, as shown for the Leu1 protein. This is a feature well known for other mitochondrial proteins involved in the assembly of Fe/S proteins, such as Nfs1, Yah1, Isu1 and Isu2 (Kispal et al, 1999; Lange et al, 2000; Gerber et al, 2004).
Our experiments provide information about the function of Isd11 in the biogenesis of Fe/S cluster-containing proteins. Isd11 is required for an early step in this process. It is present in a tight complex with the cysteine desulfurase Nfs1. In the absence of Isd11, Nfs1 showed a tendency for aggregation that was not observed in WT mitochondria. Thus, Isd11 seems to stabilize Nfs1 in mitochondria. Nfs1 extracts sulfur from cysteine and transfers it to the Isu1/Isu2 complex for the formation of Fe/S cluster. We observed that Nfs1 is able to produce sulfide from cysteine in vitro and thus has still desulfurase activity in the absence of Isd11. This is consistent with results of a previous report demonstrating such activity for recombinant Nfs1 protein (Muhlenhoff et al, 2004). However, in vivo, a role of Isd11 in the processes of sulfur transfer mediated by Nfs1 for the formation of the Fe/S clusters is supported by the inhibition of Fe/S cluster formation in the absence of Isd11 and the strongly reduced activities of Fe/S proteins at early time points of downregulation of Isd11. We indeed observed in intact mitochondria that the Nfs1•Isd11 complex forms the persulfide intermediate as a step in the sulfur transfer reaction in vivo (K Hell, unpublished data). Isd11 might mediate the assembly/interaction of Nfs1 with other proteins. Candidates might be the Isu proteins, since Nfs1 was partly found in interaction with Isu1, as expected from the function of Nfs1 as sulfur donor of the Isu1/2 complex (Gerber et al, 2003). Another potential interaction partner of the Nfs1•Isd11 complex is Yah1, which is thought to transfer electrons to Nfs1 to reduce the sulfur and thus might transiently interact with Nfs1.
Certain steps in the formation of Fe/S clusters in mitochondria might occur at the matrix side of the inner membrane, since Isd11 and Nfs1 are attached to the inner membrane in significant amounts. It has been reported that the ferredoxin reductase Arh1 is associated with the inner membrane (Manzella et al, 1998). Thus, at least part of the pathway of the formation of Fe/S clusters might be located at the matrix side of the inner membrane. Such membrane localization might also play a role in the regulation of the iron homeostasis of mitochondria and the cell.
Most of the mitochondrial proteins of the ISC assembly machinery have homologs in bacteria. These are encoded by operons specifying the nitrogen-fixation (NIF) machinery, the iron–sulfur cluster (ISC) assembly machinery, the sulfur mobilization (SUF) machinery and the CSD system (Zheng et al, 1998; Takahashi and Tokumoto, 2002; Frazzon and Dean, 2003; Outten et al, 2004; Loiseau et al, 2005). An apparent homolog of Isd11 was not found in these operons and we could not identify a bacterial homolog of Isd11 in various NCBI BLAST searches against all available bacterial sequences (McGinnis and Madden, 2004). Proteins other than the Isu homologs interacting with the cysteine desulfurase have been reported in bacteria. In the SUF and the CSD system, the activity of the cysteine desulfurase is stimulated by the proteins SufE and CsdE, respectively, which are suggested to act as sulfur acceptors (Loiseau et al, 2003, 2005; Outten et al, 2003). Since Isd11 lacks cysteine residues, the mitochondrial system is unlikely to function in a similar manner. It appears that the bacterial homologs of the mitochondrial Nfs1 act without a homolog of Isd11, although the presence of a functional homolog cannot be completely excluded. In Arabidopsis thaliana, part of the chloroplastic Nfs1 was found in a large 600 kDa complex, suggesting an interaction with other proteins (Ye et al, 2005). Some of these appear to stimulate the activity of the chloroplast NifS-like cysteine desulfurase (Ye et al, 2005).
The presented results suggest that Isd11 functions in the process of Nfs1-mediated steps in the biogenesis of Fe/S proteins. Future studies will have to elucidate the structure of the Nfs1•Isd11 oligomer to clarify why Isd11 is absolutely required in this process.
Materials and methods
Yeast strains and cell growth
The WT strain YPH499 was used (Sikorski and Hieter, 1989). The GAL10-ISD11 strain was generated by replacement of the sequence comprising of 100 bp upstream of the Isd11 coding region with a GAL10 promoter-containing cassette according to published procedures (Lafontaine and Tollervey, 1996). In order to generate the strains expressing Isd11 with either a C-terminal heptahistidinyl tag or a triple HA tag, the nucleotides encoding the tags were inserted on the chromosome of the WT strain YPH499 after the coding sequence of Isd11 by homologous recombination according to published methods (Knop et al, 1999). For depletion of Isd11, GAL10-ISD11 cultures grown at 30°C were shifted from lactate medium containing 0.5% galactose to lactate medium containing 0.5% glucose for the indicated time periods. Control WT cells were treated in the same way. Mitochondria were isolated as described previously (Daum et al, 1982).
Purification of the Isd11–Nfs1 complex
Mitochondria (10 mg) from WT yeast and yeast cells containing His-tagged Isd11 were solubilized in buffer A containing 20 mM Tris/HCl, pH 7.4, 80 mM KCl, 0.5% DDM, 20 mM imidazole and 1 mM phenylmethylsulfonylfluoride (PMSF) for 30 min at 4°C. The clarifying spin was carried out at 100 000 g for 20 min. The supernatant was incubated with 50 μl Ni-NTA agarose beads for 60 min at 4°C. Then, the beads were washed three times with 1 ml buffer A prior to elution with Laemmli buffer containing 300 mM imidazole. Samples were analyzed by SDS–PAGE and staining with Coomassie Blue. Protein bands were excised from the gel and analyzed by mass spectrometry.
Gel filtration
Mitochondria (1 mg) from WT were resuspended in buffer containing 20 mM Tris/HCl, pH 7.4, 250 mM KCl, 1 mM PMSF and opened by sonication. After clarifying spin (100 000 g, 20 min), the supernatant was loaded on a Superose-12 column (25 ml column volume, Pharmacia) equilibrated with running buffer (20 mM Tris/HCl, pH 7.4, 250 mM KCl). Fractions of 500 μl volume were collected and precipitated with trichloroacetic acid (TCA). Samples were then analyzed by SDS–PAGE and immunodecoration. The following marker proteins were used for calibration: bovine thyroglobulin (669 kDa), horse spleen apoferritin (443 kDa), sweet potato β-amylase (200 kDa), S. cerevisiae alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa) and bovine erythrocyte carbonic anhydrase (29 kDa).
Miscellaneous methods
The following procedures were applied essentially as published: subcellular and submitochondrial fractionation (Rowley et al, 1994), preparation of mitochondria (Daum et al, 1982), import of preproteins into isolated mitochondria (Mokranjac et al, 2003), affinity purification of antibodies (Harlow and Lane, 1988), measurements of activities of MDH (Englard and Siegel, 1969), SDH (Robinson et al, 1991), aconitase (Fansler and Lowenstein, 1969), desulfurase activity (Muhlenhoff et al, 2004), in vivo labelling of Leu1 with 55Fe (Kispal et al, 1999), in vitro assay of Fe/S cluster incorporation into apoYah1 (Lutz et al, 2001) and determination of mitochondrial iron levels (Smith et al, 1984). Co-immunoprecipitations were performed following solubilization of mitochondria in buffer (20 mM Tris/HCl, pH 7.4, 80 mM KCl, 1 mM PMSF) containing either 1% DDM or 1% digitonin. In the aggregation assay, mitochondria were solubilized with 0.5% Triton X-100 in 50 mM HEPES, pH 7.4, 150 mM KCl, and 1 mM PMSF containing protease inhibitor cocktail (Roche). Pellet and supernatant fractions were separated by centrifugation for 10 min at 30 000 g.
Supplementary Material
Supplementary Information
Acknowledgments
We are very grateful to Alexandra Stiegler, Ulrike Gärtner and Marica Malesic for excellent technical assistance. We thank Drs Roland Lill and Ulrich Mühlenhoff and Eugen Urzica for advice, Dr Andreas Reichert for critically reading the manuscript, and Drs Johannes Herrmann and Thomas Lutz for discussions and the antibody against Nfs1. This work was supported by grants from the Deutsche Forschungsgemeinschaft, SFB 594 (B3, B13), the German–Israeli Foundation, the German–Israeli Project Cooperation DIP F.5.1, the Fonds der Chemischen Industrie and the German National Genome Network (grant 01GR0411).
References
- Babcock M, de Silva D, Oaks R, Davis-Kaplan S, Jiralerspong S, Montermini L, Pandolfo M, Kaplan J (1997) Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science 276: 1709–1712 [DOI] [PubMed] [Google Scholar]
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR (2004) The Pfam protein families database. Nucleic Acids Res 32 (database issue): D138–D141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MY, Hartl FU, Martin J, Pollock RA, Kalousek F, Neupert W, Hallberg EM, Hallberg RL, Horwich AL (1989) Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature 337: 620–625 [DOI] [PubMed] [Google Scholar]
- Craig EA, Marszalek J (2002) A specialized mitochondrial molecular chaperone system: a role in formation of Fe/S centers. Cell Mol Life Sci 59: 1658–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig EA, Voisine C, Schilke B (1999) Mitochondrial iron metabolism in the yeast Saccharomyces cerevisiae. Biol Chem 380: 1167–1173 [DOI] [PubMed] [Google Scholar]
- Daum G, Bohni PC, Schatz G (1982) Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem 257: 13028–13033 [PubMed] [Google Scholar]
- Dutkiewicz R, Schilke B, Knieszner H, Walter W, Craig EA, Marszalek J (2003) Ssq1, a mitochondrial Hsp70 involved in iron–sulfur (Fe/S) center biogenesis. Similarities to and differences from its bacterial counterpart. J Biol Chem 278: 29719–29727 [DOI] [PubMed] [Google Scholar]
- Endo T, Yamamoto H, Esaki M (2003) Functional cooperation and separation of translocators in protein import into mitochondria, the double-membrane bounded organelles. J Cell Sci 116: 3259–3267 [DOI] [PubMed] [Google Scholar]
- Englard S, Siegel L (1969) Mitochondrial L-malate dehydrogenase of beef heart. Methods Enzymol 12: 99–106 [Google Scholar]
- Fansler B, Lowenstein JM (1969) Aconitase from pig heart. Methods Enzymol 12: 26–30 [Google Scholar]
- Foury F, Cazzalini O (1997) Deletion of the yeast homologue of the human gene associated with Friedreich's ataxia elicits iron accumulation in mitochondria. FEBS Lett 411: 373–377 [DOI] [PubMed] [Google Scholar]
- Frazzon J, Dean DR (2003) Formation of iron–sulfur clusters in bacteria: an emerging field in bioinorganic chemistry. Curr Opin Chem Biol 7: 166–173 [DOI] [PubMed] [Google Scholar]
- Garland SA, Hoff K, Vickery LE, Culotta VC (1999) Saccharomyces cerevisiae ISU1 and ISU2: members of a well-conserved gene family for iron–sulfur cluster assembly. J Mol Biol 294: 897–907 [DOI] [PubMed] [Google Scholar]
- Gerber J, Muhlenhoff U, Lill R (2003) An interaction between frataxin and Isu1/Nfs1 that is crucial for Fe/S cluster synthesis on Isu1. EMBO Rep 4: 906–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber J, Neumann K, Prohl C, Muhlenhoff U, Lill R (2004) The yeast scaffold proteins Isu1p and Isu2p are required inside mitochondria for maturation of cytosolic Fe/S proteins. Mol Cell Biol 24: 4848–4857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Hartl FU (1996) Molecular chaperones in cellular protein folding. Nature 381: 571–579 [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Kim R, Saxena S, Gordon DM, Pain D, Dancis A (2001) J-domain protein, Jac1p, of yeast mitochondria required for iron homeostasis and activity of Fe–S cluster proteins. J Biol Chem 276: 17524–17532 [DOI] [PubMed] [Google Scholar]
- Kispal G, Csere P, Prohl C, Lill R (1999) The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J 18: 3981–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SA, Sepuri NB, Pain D, Dancis A (1998) Mt-Hsp70 homolog, Ssc2p, required for maturation of yeast frataxin and mitochondrial iron homeostasis. J Biol Chem 273: 18389–18393 [DOI] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15: 963–972 [DOI] [PubMed] [Google Scholar]
- Koehler CM (2004) New developments in mitochondrial assembly. Annu Rev Cell Dev Biol 20: 309–335 [DOI] [PubMed] [Google Scholar]
- Lafontaine D, Tollervey D (1996) One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res 24: 3469–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H, Kaut A, Kispal G, Lill R (2000) A mitochondrial ferredoxin is essential for biogenesis of cellular iron–sulfur proteins. Proc Natl Acad Sci USA 97: 1050–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Kogan M, Knight SA, Pain D, Dancis A (1999) Yeast mitochondrial protein, Nfs1p, coordinately regulates iron–sulfur cluster proteins, cellular iron uptake, and iron distribution. J Biol Chem 274: 33025–33034 [DOI] [PubMed] [Google Scholar]
- Li J, Saxena S, Pain D, Dancis A (2001) Adrenodoxin reductase homolog (Arh1p) of yeast mitochondria required for iron homeostasis. J Biol Chem 276: 1503–1509 [DOI] [PubMed] [Google Scholar]
- Lill R, Kispal G (2000) Maturation of cellular Fe–S proteins: an essential function of mitochondria. Trends Biochem Sci 25: 352–356 [DOI] [PubMed] [Google Scholar]
- Lill R, Muhlenhoff U (2005) Iron–sulfur–protein biogenesis in eukaryotes. Trends Biochem Sci 30: 133–141 [DOI] [PubMed] [Google Scholar]
- Loiseau L, Ollagnier-de Choudens S, Lascoux D, Forest E, Fontecave M, Barras F (2005) Analysis of the heteromeric CsdA–CsdE cysteine desulfurase, assisting Fe–S cluster biogenesis in Escherichia coli. J Biol Chem 280: 26760–26769 [DOI] [PubMed] [Google Scholar]
- Loiseau L, Ollagnier-de-Choudens S, Nachin L, Fontecave M, Barras F (2003) Biogenesis of Fe–S cluster by the bacterial Suf system: SufS and SufE form a new type of cysteine desulfurase. J Biol Chem 278: 38352–38359 [DOI] [PubMed] [Google Scholar]
- Lutz T, Westermann B, Neupert W, Herrmann JM (2001) The mitochondrial proteins Ssq1 and Jac1 are required for the assembly of iron sulfur clusters in mitochondria. J Mol Biol 307: 815–825 [DOI] [PubMed] [Google Scholar]
- Manzella L, Barros MH, Nobrega FG (1998) ARH1 of Saccharomyces cerevisiae: a new essential gene that codes for a protein homologous to the human adrenodoxin reductase. Yeast 14: 839–846 [DOI] [PubMed] [Google Scholar]
- McGinnis S, Madden TL (2004) BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res 32: W20–W25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokranjac D, Paschen SA, Kozany C, Prokisch H, Hoppins SC, Nargang FE, Neupert W, Hell K (2003) Tim50, a novel component of the TIM23 preprotein translocase of mitochondria. EMBO J 22: 816–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlenhoff U, Balk J, Richhardt N, Kaiser JT, Sipos K, Kispal G, Lill R (2004) Functional characterization of the eukaryotic cysteine desulfurase Nfs1p from Saccharomyces cerevisiae. J Biol Chem 279: 36906–36915 [DOI] [PubMed] [Google Scholar]
- Muhlenhoff U, Gerber J, Richhardt N, Lill R (2003) Components involved in assembly and dislocation of iron–sulfur clusters on the scaffold protein Isu1p. EMBO J 22: 4815–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W, Brunner M (2002) The protein import motor of mitochondria. Nat Rev Mol Cell Biol 3: 555–565 [DOI] [PubMed] [Google Scholar]
- Outten FW, Djaman O, Storz G (2004) A suf operon requirement for Fe–S cluster assembly during iron starvation in Escherichia coli. Mol Microbiol 52: 861–872 [DOI] [PubMed] [Google Scholar]
- Outten FW, Wood MJ, Munoz FM, Storz G (2003) The SufE protein and the SufBCD complex enhance SufS cysteine desulfurase activity as part of a sulfur transfer pathway for Fe–S cluster assembly in Escherichia coli. J Biol Chem 278: 45713–45719 [DOI] [PubMed] [Google Scholar]
- Paschen SA, Waizenegger T, Stan T, Preuss M, Cyrklaff M, Hell K, Rapaport D, Neupert W (2003) Evolutionary conservation of biogenesis of beta-barrel membrane proteins. Nature 426: 862–866 [DOI] [PubMed] [Google Scholar]
- Prokisch H, Scharfe C, Camp II DG, Xiao W, David L, Andreoli C, Monroe ME, Moore RJ, Gritsenko MA, Kozany C, Hixson KK, Mottaz HM, Zischka H, Ueffing M, Herman ZS, Davis RW, Meitinger T, Oefner PJ, Smith RD, Steinmetz LM (2004) Integrative analysis of the mitochondrial proteome in yeast. PLoS Biol 2: e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehling P, Brandner K, Pfanner N (2004) Mitochondrial import and the twin-pore translocase. Nat Rev Mol Cell Biol 5: 519–530 [DOI] [PubMed] [Google Scholar]
- Rehling P, Pfanner N, Meisinger C (2003) Insertion of hydrophobic membrane proteins into the inner mitochondrial membrane—a guided tour. J Mol Biol 326: 639–657 [DOI] [PubMed] [Google Scholar]
- Reichert AS, Neupert W (2004) Mitochondriomics or what makes us breathe. Trends Genet 20: 555–562 [DOI] [PubMed] [Google Scholar]
- Robinson KM, von Kieckebusch-Guck A, Lemire BD (1991) Isolation and characterization of a Saccharomyces cerevisiae mutant disrupted for the succinate dehydrogenase flavoprotein subunit. J Biol Chem 266: 21347–21350 [PubMed] [Google Scholar]
- Rodriguez-Manzaneque MT, Tamarit J, Belli G, Ros J, Herrero E (2002) Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol Biol Cell 13: 1109–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley N, Prip-Buus C, Westermann B, Brown C, Schwarz E, Barrell B, Neupert W (1994) Mdj1p, a novel chaperone of the DnaJ family, is involved in mitochondrial biogenesis and protein folding. Cell 77: 249–259 [DOI] [PubMed] [Google Scholar]
- Schilke B, Voisine C, Beinert H, Craig E (1999) Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 96: 10206–10211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz CJ, Djaman O, Imlay JA, Kiley PJ (2000) The cysteine desulfurase, IscS, has a major role in in vivo Fe–S cluster formation in Escherichia coli. Proc Natl Acad Sci USA 97: 9009–9014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schonfisch B, Perschil I, Chacinska A, Guiard B, Rehling P, Pfanner N, Meisinger C (2003) The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci USA 100: 13207–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FE, Herbert J, Gaudin J, Hennessey DJ, Reid GR (1984) Serum iron determination using ferene triazine. Clin Biochem 17: 306–310 [DOI] [PubMed] [Google Scholar]
- Strain J, Lorenz CR, Bode J, Garland S, Smolen GA, Ta DT, Vickery LE, Culotta VC (1998) Suppressors of superoxide dismutase (SOD1) deficiency in Saccharomyces cerevisiae. Identification of proteins predicted to mediate iron–sulfur cluster assembly. J Biol Chem 273: 31138–31144 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Tokumoto U (2002) A third bacterial system for the assembly of iron–sulfur clusters with homologs in archaea and plastids. J Biol Chem 277: 28380–28383 [DOI] [PubMed] [Google Scholar]
- Voisine C, Cheng YC, Ohlson M, Schilke B, Hoff K, Beinert H, Marszalek J, Craig EA (2001) Jac1, a mitochondrial J-type chaperone, is involved in the biogenesis of Fe/S clusters in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 98: 1483–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voos W, Rottgers K (2002) Molecular chaperones as essential mediators of mitochondrial biogenesis. Biochim Biophys Acta 1592: 51–62 [DOI] [PubMed] [Google Scholar]
- Ye H, Garifullina GF, Abdel-Ghany SE, Zhang L, Pilon-Smits EA, Pilon M (2005) The chloroplast NifS-like protein of Arabidopsis thaliana is required for iron–sulfur cluster formation in ferredoxin. Planta 220: 602–608 [DOI] [PubMed] [Google Scholar]
- Yoon T, Cowan JA (2003) Iron–sulfur cluster biosynthesis. Characterization of frataxin as an iron donor for assembly of [2Fe–2S] clusters in ISU-type proteins. J Am Chem Soc 125: 6078–6084 [DOI] [PubMed] [Google Scholar]
- Zheng L, Cash VL, Flint DH, Dean DR, Outten FW, Djaman O, Storz G (1998) Assembly of iron–sulfur clusters. Identification of an iscSUA–hscBA–fdx gene cluster from Azotobacter vinelandii. J Biol Chem 273: 13264–13272 [DOI] [PubMed] [Google Scholar]
- Zheng L, White RH, Cash VL, Jack RF, Dean DR (1993) Cysteine desulfurase activity indicates a role for NIFS in metallocluster biosynthesis. Proc Natl Acad Sci USA 90: 2754–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


