Abstract
Mitochondria are indispensable for cell viability; however, major mitochondrial functions including citric acid cycle and oxidative phosphorylation are dispensable. Most known essential mitochondrial proteins are involved in preprotein import and assembly, while the only known essential biosynthetic process performed by mitochondria is the biogenesis of iron–sulfur clusters (ISC). The components of the mitochondrial ISC-assembly machinery are derived from the prokaryotic ISC-assembly machinery. We have identified an essential mitochondrial matrix protein, Isd11 (YER048w-a), that is found in eukaryotes only. Isd11 is required for biogenesis of cellular Fe/S proteins and thus is a novel subunit of the mitochondrial ISC-assembly machinery. It forms a complex with the cysteine desulfurase Nfs1 and is required for formation of an Fe/S cluster on the Isu scaffold proteins. We conclude that Isd11 is an indispensable eukaryotic component of the mitochondrial machinery for biogenesis of Fe/S proteins.
Keywords: essential cellular functions, Fe/S proteins, mitochondria, Saccharomyces cerevisiae
Introduction
Mitochondria are essential for cell viability. Although mitochondria are best known for their function in oxidative phosphorylation and the citric acid cycle, many mitochondrial structures, including the respiratory chain complexes, ATP synthase, citric acid cycle enzymes and mitochondrial DNA, can be removed and yeast cells are still viable on fermentable medium (Contamine and Picard, 2000). Systematic gene deletion studies in yeast suggested that about 18% of all cellular proteins belong to the group of strictly essential proteins, that is, the deletion of each individual protein of this group is lethal for yeast under both fermentable and non-fermentable conditions (Giaever et al, 2002). In the case of mitochondria, however, less than 5% of the known proteins belong to this group of proteins that are strictly essential for cell viability as individual proteins (Rehling et al, 2003; Sickmann et al, 2003). Defining the molecular functions of these essential proteins is thus critical for understanding the cellular role of mitochondria.
To date, the only essential biosynthetic task performed by yeast mitochondria is the assembly of Fe/S proteins (Lill and Mühlenhoff, 2005). As a consequence, known essential mitochondrial proteins function either in this biochemical pathway or in the biogenesis of the organelle itself. Essential proteins involved in protein import have been found in the translocase of the outer mitochondrial membrane (TOM complex), the sorting and assembly machinery of the outer membrane (SAM complex), the import system of the intermembrane space, the translocases of the inner membrane (TIM), the presequence translocase-associated motor (PAM) and further molecular chaperones, as well as in the matrix processing peptidase (Neupert, 1997; Rehling et al, 2003; Koehler, 2004). Essential proteins of the iron–sulfur cluster (ISC)-assembly machinery in the mitochondrial matrix include the cysteine desulfurase Nfs1 as central component, as well as the ferredoxin reductase Arh1 and the ferredoxin Yah1 that probably function in an electron transfer chain for reduction of sulfur formed by Nfs1 (Kispal et al, 1999; Lange et al, 2000; Li et al, 2001; Lill and Mühlenhoff, 2005). Moreover, the homologous scaffold proteins Isu1 and Isu2, at which the de novo ISC formation takes place, can substitute for each other; yet, a double deletion of both proteins is lethal (Schilke et al, 1999; Mühlenhoff et al, 2003). The chaperone Jac1 is essential in most genetic backgrounds (Strain et al, 1998; Kim et al, 2001; Lutz et al, 2001; Voisine et al, 2001) and cooperates with the non-essential Hsp70, Ssq1, in ISC assembly (Schilke et al, 1996; Knight et al, 1998). The essential cochaperone Mge1 performs a dual role in protein import and Fe/S protein biogenesis (Dutkiewicz et al, 2003). These essential and further non-essential subunits of the ISC-assembly machinery contain homologs/correlates in the bacterial ISC, NIF and SUF systems for Fe/S cluster biogenesis, while the majority of essential components of the protein import machinery are specific for eukaryotic cells (Pfanner et al, 2004; Johnson et al, 2005; Lill and Mühlenhoff, 2005). The mitochondrial ISC-assembly machinery is also required for the biogenesis of Fe/S proteins in the eukaryotic cytosol and nucleus (Kispal et al, 1999; Lill and Mühlenhoff, 2005). For this process, components of the mitochondrial ISC-export machinery are needed, including the ABC transporter Atm1 (Kispal et al, 1999) and the essential intermembrane space protein Erv1, which is involved in both Fe/S protein biogenesis and protein import (Lange et al, 2001; Mesecke et al, 2005; Rissler et al, 2005).
Here, we report the identification of an essential mitochondrial matrix protein, designated Isd11. Isd11 is a novel subunit of the mitochondrial ISC-assembly machinery. It is conserved throughout eukaryotes but not found in prokaryotes, in contrast to the other members of the ISC-assembly machinery. Isd11 forms a stable complex with Nfs1 that we term the ISC-biogenesis desulfurase (ISD) complex. We define the function of Isd11 to be early in ISC biogenesis as it is required for de novo Fe/S cluster formation on the Isu scaffold proteins.
Results
Isd11 is an essential protein of the mitochondrial matrix
We performed a systematic comparison of high-throughput studies on gene deletion and subcellular protein localization in Saccharomyces cerevisiae. The open reading frame YER048w-a, now termed ISD11, was found to be essential for cell viability in a sporulation approach and code for an 11 kDa protein with mitochondrial localization (Winzeler et al, 1999; Huh et al, 2003). We confirmed the essential nature of ISD11 in a plasmid loss approach (see Materials and methods). We found homologs of Isd11 throughout the eukaryotic kingdom but not in prokaryotes (Figure 1A). A limited similarity of Isd11 is observed with the eukaryotic accessory subunits B22 and B14 of mitochondrial complex I of the respiratory chain. B22 and B14 have been grouped into the LYR family, named after the conserved tripeptide LYR (LYK). As the molecular functions of B22 and B14 are not known, the grouping of Isd11 into the LYR family did not provide information on its function.
Figure 1.
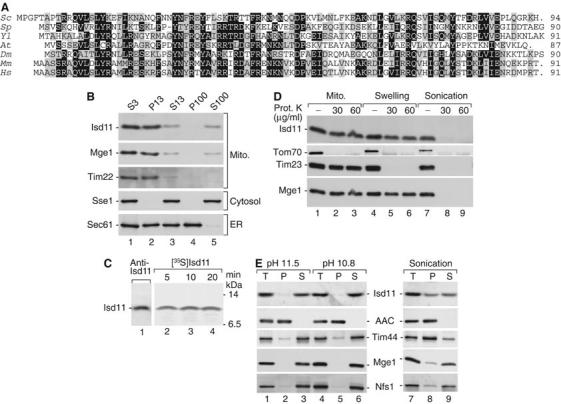
Isd11 is located in the mitochondrial matrix. (A) Deduced primary structures of Isd11 and homologs from S. cerevisiae (Sc), Schizosaccharomyces pombe (Sp), Yarrowia lipolytica (Yl), Arabidopsis thaliana (At), Drosophila melanogaster (Dm), Mus musculus (Mm) and Homo sapiens (Hs). Black, identical amino-acid residues in at least four proteins; gray, similar residues. (B) Yeast cells were fractionated by differential centrifugation and analyzed by SDS–PAGE and Western blotting. S, supernatant; P, pellet (multiples of 1000 g). (C) 35S-labeled Isd11 precursor was imported into isolated mitochondria. The mitochondria were swollen, treated with proteinase K and separated by SDS–PAGE. Analysis was performed by immunodecoration (lane 1) or digital autoradiography (lanes 2–4). (D) Mitochondria were either left untreated or subjected to swelling or sonication. Then, a treatment with proteinase K was performed as indicated. (E) Mitochondria were incubated at alkaline pH or subjected to sonication, followed by centrifugation at 100 000 g. T, total; AAC, ADP/ATP carrier.
Upon differential centrifugation of a cell extract, Isd11 cofractionated with the mitochondrial matrix protein Mge1 (Figure 1B), confirming the mitochondrial localization observed with a green fluorescent protein (GFP)-tagged Isd11 (Huh et al, 2003). Isd11 imported into isolated mitochondria was not proteolytically processed and showed the same electrophoretic mobility as endogenous Isd11 detected by immunodecoration (Figure 1C). To determine the submitochondrial localization of Isd11, isolated yeast mitochondria were treated with proteinase K. Isd11 remained protected, whereas the outer membrane receptor Tom70 was degraded (Figure 1D). Upon swelling of mitochondria, Isd11 remained intact like Mge1, whereas the intermembrane space-exposed Tim23 was degraded. Only when the matrix was opened by sonication, Isd11 was accessible to the protease (Figure 1D), indicating that Isd11 is either a matrix protein or an inner membrane protein accessible from the matrix side only. We subjected mitochondria to a treatment at alkaline pH. Isd11 was fully extracted from the membranes both at pH 11.5 and the milder condition of pH 10.8 (Truscott et al, 2003), whereas the integral membrane protein ADP/ATP carrier remained in the membrane pellet (Figure 1E). The lack of a predicted hydrophobic transmembrane segment supports the conclusion that Isd11 is not an integral membrane protein. When membranes and soluble fraction were separated after sonication of mitochondria, about 60% of Isd11 was found in the supernatant (Figure 1E). Thus, Isd11 shows an intermediate behavior in comparison to the peripheral inner membrane protein Tim44, which was largely found in the membrane fraction, and the matrix proteins Mge1 and Nfs1, which were mainly present in the soluble fraction. We conclude that Isd11 is an essential protein of the mitochondrial matrix loosely associated with the inner membrane.
A conditional yeast mutant of Isd11 reveals a relation to protein maturation but not to protein import
We generated conditional yeast mutants of ISD11 by low-fidelity PCR and selected the strain isd11-1 that was strongly impaired in growth on non-fermentable medium at 37°C (Figure 2A). Previous studies with temperature-conditional yeast mutants of mitochondrial proteins revealed that mitochondria isolated from cells grown at low, permissive temperature often showed kinetic deficiencies in specific processes that pointed to the function of the mutated protein (Truscott et al, 2002; Gabriel et al, 2003). As the majority of essential mitochondrial proteins are required for protein import, we first analyzed the import of radiolabeled preproteins. The matrix-targeted precursors of F1-ATPase subunit β and Su9-DHFR were imported and processed to the mature forms with similar efficiency in isd11-1 and wild-type mitochondria (Figure 2B and C). The inner membrane protein cytochrome c1 was similarly imported and processed by isd11-1 mitochondria as in wild-type mitochondria (Figure 2D). Moreover, isd11-1 mitochondria imported the outer membrane protein porin, as analyzed by formation of the assembled protein on blue native polyacrylamide gel electrophoresis (BN-PAGE) (Figure 2E).
Figure 2.
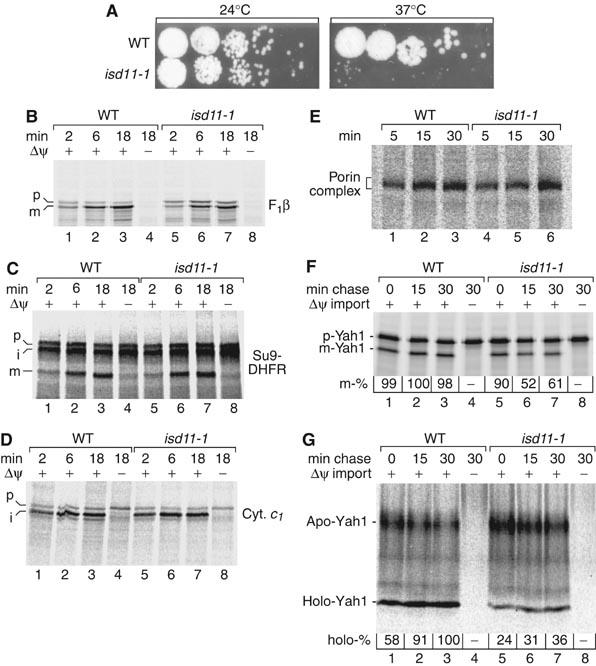
isd11-1 mutant mitochondria import preproteins but are impaired in maturation of Yah1. (A) A conditional yeast mutant of ISD11. Wild-type (WT) and isd11-1 yeast cells were spotted on YPG agar plates and grown for 3 days at 24 or 37°C. (B–D) Mitochondria were isolated from WT or isd11-1 cells grown at 19°C. 35S-labeled precursor proteins were imported and analyzed by SDS–PAGE and digital autoradiography. p, precursor; i, intermediate; m, mature; F1β, F1-ATPase subunit β. (E) [35S]porin was imported into isolated mitochondria and analyzed by BN-PAGE. (F) 35S-labeled precursor of Yah1 was imported into isolated mitochondria for 10 min, followed by dissipation of Δψ and a further incubation (chase). Imported mature-sized (m) Yah1 was analyzed by SDS–PAGE and digital autoradiography. The maximal amount formed was set to 100%. (G) The experiment was performed as described for panel F except that apo-Yah1 and the ISC-containing holo-Yah1 were analyzed by native PAGE (Lutz et al, 2001).
As no major effects on protein import were observed, we analyzed the maturation of Yah1 that contains an Fe/S cluster. The radiolabeled precursor of Yah1 was first imported into isolated mitochondria and then the membrane potential Δψ across the inner membrane was dissipated to prevent further import of new precursors. The formation of processed, mature-sized Yah1, as analyzed by SDS–PAGE, occurred with similar efficiency in wild-type and isd11-1 mitochondria (Figure 2F, lanes 1 and 5). To analyze the maturation of Yah1 to the holo-form containing the Fe/S cluster, we used the fact that holo-Yah1 migrates as a defined species on a native gel (Lutz et al, 2001). The formation of the holo-form was significantly diminished in isd11-1 mitochondria compared to wild-type mitochondria (Figure 2G, lanes 1 and 5). In a second incubation (chase), further maturation of Yah1 was allowed for up to 30 min; however, the holo-Yah1 levels in isd11-1 mitochondria remained far below the wild-type levels (Figure 2G). Rather a reduction of the mature-sized Yah1 was observed in isd11-1 mitochondria (Figure 2F, lanes 6 and 7), suggesting that the apo-form of imported mature-sized Yah1 is not stable without the Fe/S cluster. These results indicate a role of Isd11 in the formation of the holo-form of Yah1 but not in preprotein import.
Isd11 is required for the stability of the cysteine desulfurase Nfs1
To obtain evidence for the role of Isd11 in vivo, cells were grown under two conditions and the steady-state protein levels of isolated mitochondria were compared. One set of cells was grown at permissive temperature, whereas the other set was first grown at low temperature and then shifted to 37°C on non-fermentable medium. Upon growth of isd11-1 cells at low temperature, the mitochondrial levels of all marker proteins tested were in a similar range as those of wild-type mitochondria (Figure 3A and data not shown). The mutant protein Isd11-1 migrated faster on SDS–PAGE than the wild-type protein (the lower signal intensity of Isd11-1 can be due to two reasons, a reduction of the amount of Isd11 in the mutant or a lower reactivity of the polyclonal antiserum against the mutant protein) (Figure 3A, lower panel). After shift of isd11-1 cells to 37°C, the level of Jac1 of the ISC-assembly machinery was moderately affected (Figure 3B). A striking defect was observed for Nfs1 that was strongly diminished in isd11-1 mitochondria, similar to the strong reduction of the mutant protein Isd11-1 itself (Figure 3B).
Figure 3.
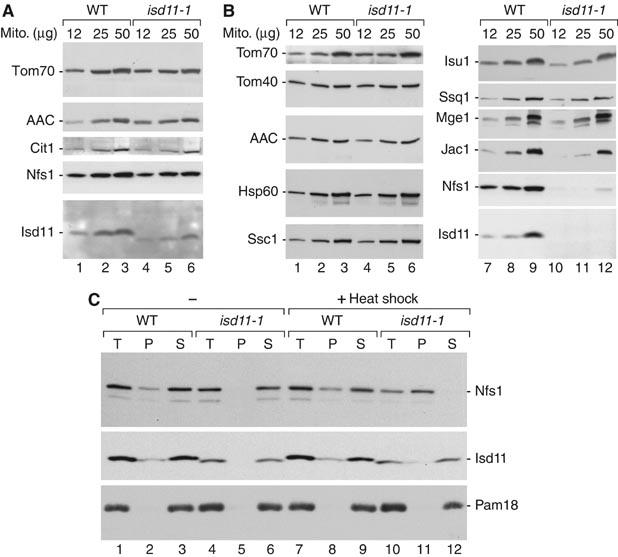
Reduced mitochondrial levels of Nfs1 in isd11-1 cells grown under non-permissive conditions. (A) Wild-type (WT) and isd11-1 cells were grown at 19°C. Isolated mitochondria (μg of protein) were analyzed by SDS–PAGE and Western blotting. Cit1, citrate synthase. (B) Cells were grown at 19°C and shifted to 37°C for 8 h on non-fermentable medium. (C) Mitochondria isolated from cells grown at 19°C were either kept on ice or heat shocked for 15 min at 37°C, lysed with digitonin and separated into pellet and supernatant by centrifugation.
To obtain further insight into the role of Isd11, mitochondria isolated from cells grown at low temperature were shifted to non-permissive conditions in vitro. Nfs1 was still detectable (Figure 3C, lane 10). Mitochondria were solubilized by digitonin and subjected to centrifugation. Isd11 from both wild-type and isd11-1 mitochondria was mainly found in the solubilized fraction like a control protein, the inner membrane protein Pam18 (Figure 3C). In wild-type mitochondria, the majority of Nfs1 was released to the supernatant, whereas a small fraction remained in the non-solubilized pellet (Figure 3C, lanes 2, 3, 8 and 9). In isd11-1 mitochondria, Nfs1 was released to the supernatant at low temperature but was fully found in the pellet after the heat shock (Figure 3C, lanes 6 and 11). We conclude that after a heat shock of isolated mitochondria, Nfs1 aggregates in isd11-1 mitochondria. After a prolonged shift of cells to non-permissive conditions, the aggregated Nfs1 is apparently degraded. Thus, Isd11 is involved in maintaining the stability of Nfs1.
Isd11 is required for the biogenesis of mitochondrial, cytosolic and nuclear Fe/S proteins
To analyze a possible function of Isd11 in Fe/S protein biogenesis, we measured the enzyme activities of mitochondrial and cytosolic Fe/S proteins. Mitochondria were isolated from wild-type and isd11-1 cells grown at 24–30°C. The activities of aconitase, succinate dehydrogenase (complex II) and cytochrome c reductase (complex III) were diminished by 75–95% in isd11-1 mitochondria (Figure 4A). In contrast, the activity of malate dehydrogenase as a non-Fe/S control enzyme was not impaired. The steady-state levels of the Fe/S proteins aconitase, subunit 2 of succinate dehydrogenase and Rieske Fe/S protein of complex III were moderately diminished in the isd11-1 mitochondria whereas those of Nfs1, Isu1 and control proteins like porin were not reduced at these growth conditions (Figure 4B). The strong reduction of the enzyme activities of Fe/S proteins supports the view that Isd11 functions in mitochondrial Fe/S protein assembly. In agreement with previous observations in various ISC-assembly mutants, Fe/S apoproteins are often sensitive to degradation (see, e.g., Balk et al, 2004).
Figure 4.

isd11-1 cells show decreased activities of mitochondrial and cytosolic Fe/S proteins. (A) Mitochondria were isolated from WT and isd11-1 cells grown for 24 h in rich medium supplemented with glucose at 24 and 30°C. The enzymatic activities of aconitase, respiratory complexes II and III and malate dehydrogenase (MDH) were measured. The activity of aconitase in isd11-1 mitochondria isolated from cells grown at 19°C (as used in Figure 2B–G) was similarly diminished by 75% compared to WT mitochondria. (B) Levels of mitochondrial proteins detected by Western blotting. Aco1, aconitase; Rip1, Rieske Fe/S protein; Sdh2, subunit 2 of succinate dehydrogenase. (C, D) WT and isd11-1 cells were grown in minimal medium supplemented with glucose at 24 or 30°C for 24 h. In addition, Gal-ISD11 cells were cultured for 18 h in minimal medium containing galactose (Gal) or glucose (Glc) to induce or repress, respectively, synthesis of Isd11. Extracts were prepared by disruption of cells with glass beads, and aliquots were analyzed (C) for the enzymatic activities of Leu1 and alcohol dehydrogenase (ADH), or (D) for the protein levels of Isd11, Leu1 and the cytosolic marker protein Pgk1 by Western blotting.
The activity of the cytosolic Fe/S protein isopropylmalate isomerase (Leu1) was two- to three-fold decreased in extracts of isd11-1 cells, whereas the activity of the control enzyme alcohol dehydrogenase did not change significantly (Figure 4C, left panels). The protein levels of Leu1 in isd11-1 cells were comparable to those in wild-type cells (Figure 4D, lanes 1–4), indicating a requirement of Isd11 for Leu1 function. We also constructed a regulatable mutant strain in which the ISD11 gene was under control of the galactose-inducible and glucose-repressible promoter of GAL1-10. After depletion of Isd11 upon growth of the resulting Gal-ISD11 cells in glucose-containing medium, Leu1 activity was diminished four-fold compared to Isd11-containing cells, whereas the alcohol dehydrogenase activity was unchanged (Figure 4C, right panels). The protein levels of Leu1 were not significantly altered upon depletion of Isd11 (Figure 4D, lane 6).
To analyze whether Isd11 was involved in the de novo maturation of cellular Fe/S proteins, we determined the incorporation of 55Fe into various Fe/S proteins (Kispal et al, 1999; Mühlenhoff et al, 2002). Wild-type and isd11-1 cells were grown in iron-poor minimal medium and incubated with 55Fe in the presence of ascorbate. A cell extract was prepared, Fe/S proteins were immunoprecipitated and the amounts of incorporated Fe/S clusters were estimated by scintillation counting. Fe/S cluster assembly was significantly impaired in cytosolic Leu1 (Figure 5A). An almost quantitative loss in Fe/S cluster incorporation into the cytosolic Rli1 (Kispal et al, 2005) and the nuclear DNA repair enzyme Ntg2 was found using Isd11-depleted Gal-ISD11 cells (Figure 5B and C). The levels of the Fe/S apoproteins were decreased, but not as strongly as the assembly of the Fe/S clusters (Figure 5B and C, insets). Thus, Isd11 is required for the formation of Fe/S proteins in mitochondria, cytosol and nucleus, identifying Isd11 as a novel component of the mitochondrial ISC-assembly machinery.
Figure 5.
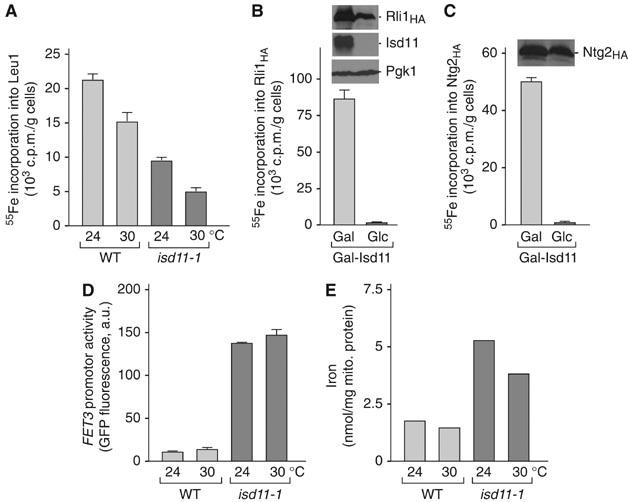
isd11-1 cells are impaired in the de novo formation of Fe/S proteins and iron homeostasis. (A) Wild-type (WT) and isd11-1 cells were grown for 24 h in iron-poor minimal medium supplemented with glucose at 24 and 30°C. Cells were labeled with 55Fe, washed and a cell extract was prepared using glass beads. Leu1 was immunoprecipitated. The radioactivity associated with the immuno-beads was quantified by liquid scintillation counting. (B, C) A similar analysis as described in panel A was performed using Gal-ISD11 cells after growth for 40 h in iron-poor minimal medium containing galactose or glucose. After radiolabeling, cell extracts were analyzed for the de novo assembly of the Fe/S clusters on cytosolic Rli1HA (Kispal et al, 2005) and nuclear Ntg2HA. (D) Cellular iron homeostasis is perturbed in isd11-1 cells. WT and isd11-1 cells carrying plasmid p415-FET3-GFP were grown in minimal medium supplemented with 200 μM ferric ammonium citrate at 24 or 30°C. Cells were harvested at an optical density of 0.5 and the transcriptional activity of the FET3 promoter was determined by recording the fluorescence emission of the cell suspension at 513 nm (excitation at 480 nm). The signal of cells lacking plasmid p415-FET3-GFP was subtracted. (E) Mitochondrial iron accumulation in isd11-1 cells. Mitochondria were isolated from cells grown in rich medium containing glucose and the iron content was determined.
An impairment of the mitochondrial ISC-assembly or ISC-export systems is generally accompanied by an increased uptake of iron into the yeast cells owing to the activation of the iron-dependent transcription factors Aft1/Aft2 (Rutherford et al, 2001, 2005; Chen et al, 2004). These proteins sense the cellular iron concentration, and, under iron-limiting conditions or upon Fe/S protein maturation defects, they induce the expression of genes of the ‘iron regulon' resulting in increased iron uptake. We analyzed the induction of the Aft1-dependent FET3 gene, encoding a copper-dependent ferroxidase, in isd11-1 cells using a fusion construct of the FET3 promoter and the GFP gene. A plasmid encoding the FET3-GFP fusion gene was transformed into wild-type and isd11-1 cells, and the cells were analyzed for GFP fluorescence. A strongly increased fluorescence intensity under iron-replete conditions was observed in isd11-1 cells as compared to wild-type cells (Figure 5D). We conclude that isd11-1 cells display a defect in cellular iron homeostasis.
We further asked whether cells defective in Isd11 accumulate iron within mitochondria, as this is seen for mutants in the ISC-assembly and ISC-export systems (Kispal et al, 1999; Schilke et al, 1999; Lange et al, 2000; Li et al, 2001). The content of non-heme and non-Fe/S iron was quantified in isolated mitochondria. isd11-1 mitochondria contained a three-fold higher amount of iron (Figure 5E). The dramatic effect of the isd11 mutation on cellular iron uptake and mitochondrial iron accumulation supports the notion that Isd11 is a novel component of the ISC-assembly machinery.
An active cysteine desulfurase complex containing Nfs1 and Isd11
We used a yeast strain expressing Isd11 with a tandem affinity purification (TAP) tag and performed an IgG affinity purification after lysis of mitochondria under native conditions. We detected a prominent protein band of about 50 kDa and several smaller bands in the eluate that were not found with control mitochondria lacking a TAP tag. A mass spectrometric analysis of these bands led to the identification of numerous peptides derived from Nfs1 and Isd11 (Figure 6A). The predicted size of 51 kDa of mature-sized Nfs1 (Kispal et al, 1999; Li et al, 1999) agrees with the size of the prominent band. To obtain evidence for an interaction of Isd11 and Nfs1 in intact mitochondria, radiolabeled Isd11 was imported into mitochondria containing Nfs1 with a protein A tag (Figure 6B). After crosslinking with m-maleimidobenzoyl-N-hydroxysuccinimide ester (MBS), Nfs1ProtA was purified by IgG affinity purification. Upon elution with SDS, a radiolabeled band of about 75 kDa was released (Figure 6B, lane 2). The size of 75 kDa corresponds to a crosslinking product between Nfs1ProtA (65 kDa) and Isd11. When the crosslinking product was released from the IgG affinity column by TEV protease that specifically removes the protein A tag of Nfs1, the size of the crosslinking product was about 62 kDa, corresponding to the removal of the 14 kDa protein A tag (Figure 6B, lane 3). Thus, Isd11 imported into mitochondria is in close proximity to Nfs1.
Figure 6.
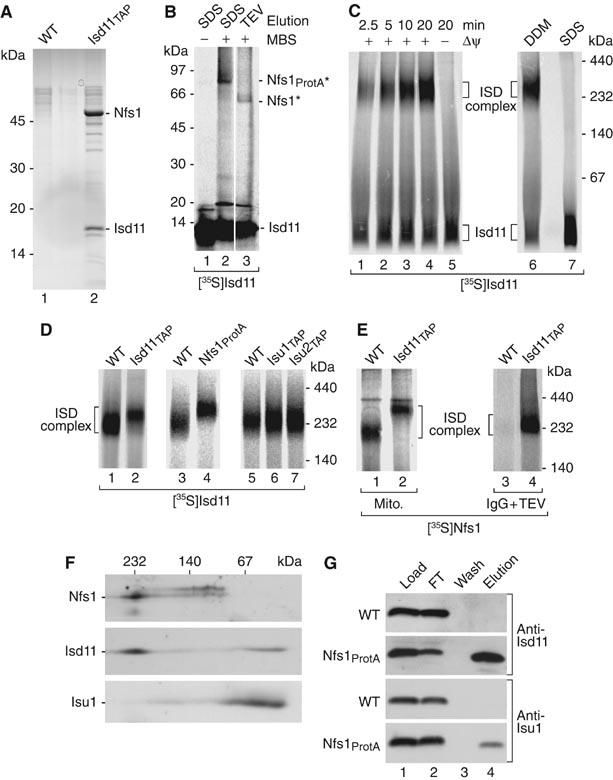
Nfs1 and Isd11 form a stable complex. (A) Mitochondria were lysed in 0.1% n-dodecyl-β-D-maltoside (DDM) and incubated with IgG-Sepharose. After washing of the beads and incubation with TEV protease, eluates were separated by SDS–PAGE, stained with colloidal Coomassie and analyzed by tandem mass spectrometry. (B) 35S-labeled Isd11 (non-tagged) was imported into Nfs1ProtA mitochondria. After incubation with MBS and quenching, mitochondria were lysed and subjected to IgG affinity chromatography. Elution was performed with SDS sample buffer or TEV protease, followed by SDS–PAGE and digital autoradiography. Asterisks, crosslinking products. (C) [35S]Isd11 was imported into WT mitochondria, followed by lysis with 0.1% DDM (samples 1–6) or 0.1% SDS (sample 7) and BN-PAGE analysis. (D) [35S]Isd11 was imported into energized mitochondria. Upon lysis with DDM, the samples were analyzed by BN-PAGE. (E) [35S]Nfs1 (non-tagged) was imported into energized mitochondria. After lysis with DDM, samples 1 and 2 were directly analyzed by BN-PAGE, whereas samples 3 and 4 were first subjected to IgG affinity chromatography, followed by elution with TEV protease and BN-PAGE. (F) WT mitochondria were lysed and proteins were separated by BN-PAGE, followed by a second-dimension SDS–PAGE and Western blotting. (G) WT and Nfs1ProtA mitochondria were subjected to IgG affinity chromatography. Isd11 and Isu1 were analyzed by SDS–PAGE and Western blotting. FT, flow-through.
We used BN-PAGE to further analyze the Nfs1–Isd11 complex. Radiolabeled Isd11 was imported into wild-type mitochondria that were lysed with non-ionic detergent. In addition to Isd11 running in the front of the gel, we observed a time-dependent formation of a complex migrating at about 230 kDa that we term the ISD complex (Figure 6C, lanes 1–4). When mitochondria were lysed by SDS, the ISD complex was dissociated (Figure 6C, lane 7). To determine if the ISD complex contained both Isd11 and Nsf1, radiolabeled Isd11 was imported into Nfs1ProtA mitochondria. The ISD complex was indeed shifted to a higher molecular mass compared to the corresponding wild-type complex (Figure 6D, lanes 3 and 4). Moreover, [35S]Nfs1 imported into wild-type mitochondria was found in a BN complex of the same mobility as the [35S]Isd11-containing ISD complex (Figure 6E, lane 1, compared to Figure 6D, lane 1). When mitochondria containing Isd11TAP were used, the [35S]Nfs1-containing complex was shifted to a higher molecular mass (Figure 6E, lane 2) and could be purified via IgG affinity chromatography (Figure 6E, lane 4). These results prove that Isd11 and Nfs1 are present in the same ISD complex on BN-PAGE.
As Nfs1 was shown to interact with Isu1/Isu2 (Gerber et al, 2003), we asked if the 230 kDa ISD complex contained the Isu proteins. The mobility of the ISD complex on BN-PAGE was not significantly changed in mitochondria containing TAP-tagged versions of the Isu proteins (Figure 6D, lanes 6 and 7), suggesting that Isu1/Isu2 are not stoichiometric subunits of the ISD complex. For a direct analysis, we performed a two-dimensional gel analysis by first separating mitochondrial proteins by BN-PAGE, followed by a second-dimension SDS–PAGE and Western blotting (Figure 6F). Nfs1 migrated mainly in the 230 kDa BN region and a smaller amount was present in a region below 140 kDa, corresponding to the size of an Nfs1 dimer. The majority of Isd11 was also found in the ISD complex region and a smaller fraction in a low molecular mass region below 100 kDa. In contrast, Isu1 was not observed in the 230 kDa ISD complex but mainly in a low molecular mass region.
How can the discrepancy with the previously reported interaction between Nfs1 and Isu proteins (Gerber et al, 2003) be resolved? Previous studies with mitochondrial translocases showed that BN-PAGE analysis typically reveals the stable and stoichiometric association of proteins in core complexes, whereas transient, non-stoichiometric or loose interactions can be detected by affinity purification approaches (Chacinska et al, 2003, 2005). We purified Nfs1ProtA from mitochondria lysed under non-denaturing conditions by IgG affinity purification and performed a Western blot with antibodies against Isd11 and Isu1. As expected from the stable association in BN-PAGE, Isd11 was efficiently copurified with Nfs1 (Figure 6G). Isu1 was also specifically found in the eluate, but the yield was about 10-fold lower than that of Isd11 (Figure 6G). As the Coomassie-stained protein bands, which were copurified in amounts roughly stoichiometric with those of Isd11 in Figure 6A, were found to contain Nfs1, it is likely that Nfs1 and Isd11 represent the major components of the ISD complex. The Isu scaffold proteins are not stoichiometric subunits of the ISD complex but can interact with the complex.
We asked if the ISD complex observed on BN-PAGE represents a functional desulfurase complex. Isolated mitochondria were treated with chloramphenicol to inhibit protein synthesis in the matrix. Mitochondrial lysates were generated with non-ionic detergent and incubated with [35S]cysteine or [35S]methionine. Only in the presence of [35S]cysteine, a radiolabeled complex of the size of the ISD complex was observed on BN-PAGE (Figure 7A, lanes 1 and 3). In mitochondria containing TAP-tagged Isd11, this complex was shifted, confirming that it represented the ISD complex (Figure 7A, lane 2). During the desulfurase action of Nfs1, the sulfur derived from cysteine is bound to Nfs1 in the form of a persulfide intermediate (Zheng et al, 1994). To obtain evidence that the 35S label observed in the ISD complex was present in a persulfide, we added dithiothreitol (DTT) before analyzing the samples by BN-PAGE. Thereby, the [35S]cysteine-labeled ISD complex was no longer observed by digital autoradiography (Figure 7B, lane 2). To exclude that the ISD protein complex itself was dissociated by DTT, we used mitochondria with imported [35S]methionine-labeled Isd11; DTT did not dissociate the ISD complex (Figure 7B, lane 4). We conclude that the ISD complex observed on BN-PAGE is functional in forming a persulfide intermediate. As total mitochondrial lysates were used in the labeling reaction with [35S]cysteine, the preferential labeling of the ISD complex points to its major role as a mitochondrial desulfurase.
Figure 7.
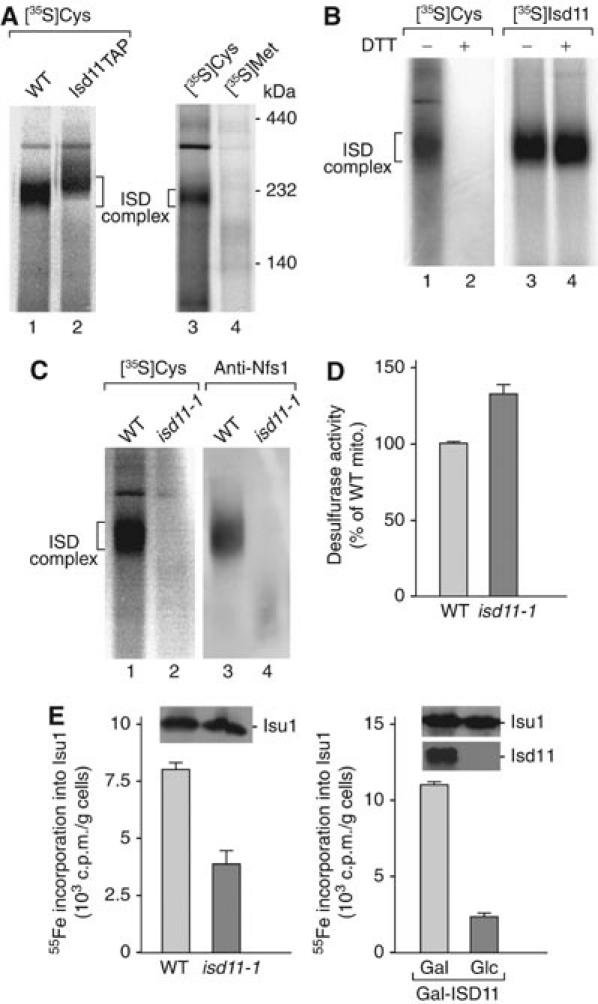
Isd11 is part of the active desulfurase complex and required for ISC formation on Isu1. (A) Wild-type (WT) and Isd11TAP mitochondria were treated with chloramphenicol. Mitochondria were lysed, incubated with [35S]cysteine or [35S]methionine and subjected to BN-PAGE and digital autoradiography. (B) Samples 1 and 2, WT mitochondrial lysate was incubated with [35S]cysteine as described for panel A. Samples 3 and 4, [35S]Isd11 was imported into WT mitochondria. Before BN-PAGE analysis, half of the samples were treated with 10 mM DTT. (C) WT and isd11-1 mitochondrial lysates were either directly subjected to BN-PAGE and Western blotting (samples 3 and 4) or incubated with [35S]cysteine and analyzed by BN-PAGE and digital autoradiography (samples 1 and 2). (D) isd11-1 cells are not impaired in cysteine desulfurase activities. Mitochondria isolated from cells grown at 30°C were lysed and incubated in the presence of 4 mM cysteine and 1 mM DTT for 20 min at 30°C. The amount of sulfide formed from cysteine by Nfs1 was determined (Mühlenhoff et al, 2004). The signal recorded in samples lacking cysteine was subtracted. (E) Isd11 is required for Fe/S cluster assembly on Isu1. WT and isd11-1 cells overexpressing ISU1 from vector p426-GPD were grown for 24 h at 30°C. Gal-ISD11 cells overexpressing ISU1 were grown in the presence of galactose or glucose. Fe/S cluster assembly on Isu1 was determined by 55Fe labeling and immunoprecipitation assay described in Figure 5A.
In isd11-1 mutant mitochondria, the [35S]cysteine-derived labeling of the ISD complex was blocked (Figure 7C, lane 2). However, the ISD protein complex itself was dissociated in isd11-1 mitochondria (Figure 7C, lane 4) and thus it could not be decided whether functional Isd11 was required for formation of the persulfide intermediate at Nfs1 (the released Nfs1 did not migrate as a defined band on BN-PAGE, a behavior previously observed with several Tom or Tim proteins upon release from translocase complexes; Chacinska et al, 2003, 2005). We therefore directly determined the enzyme activity of the cysteine desulfurase Nfs1 by following the sulfide production in extracts of isolated mitochondria (Mühlenhoff et al, 2004). Surprisingly, the Nfs1 enzyme activity was even moderately increased in isd11-1 cells (Figure 7D). Thus, functional Isd11 and the formation of the ISD complex are not required for the activity of Nfs1 in sulfide production in vitro, yet both Isd11 and the ISD complex are needed for the formation of the sulfur present in Fe/S cluster-containing proteins.
We therefore asked at which stage of mitochondrial Fe/S protein biogenesis Isd11 might be necessary. We previously developed an in vivo assay, discriminating whether ISC components are required early in the synthesis of the transient Fe/S cluster on Isu1 or whether they perform their function after cluster assembly on Isu1 (Mühlenhoff et al, 2003). Wild-type and isd11-1 cells were transformed with a plasmid overproducing Isu1. After radiolabeling the cells with 55Fe, Isu1 was immunoprecipitated from a cell extract and the associated radioactivity was quantified by scintillation counting. Fe/S cluster formation on Isu1 was significantly diminished in isd11-1 cells (Figure 7E). A five-fold reduction in Fe/S cluster assembly on Isu1 was seen after depletion of Isd11 in Gal-ISD11 cells (Figure 7E). The protein levels of Isu1 in these experiments were hardly altered (insets). These data suggest that Isd11, like Nfs1 (Mühlenhoff et al, 2003), is necessary for Fe/S cluster formation on Isu1, that is, Isd11 is required at an early stage of biogenesis.
Discussion
We report the identification of an essential protein of the mitochondrial matrix. Isd11 shows all characteristics of a component of the mitochondrial machinery for Fe/S cluster assembly. Yeast mutant mitochondria of Isd11 display strong defects in the activity of Fe/S enzymes under conditions that permit efficient import of precursor proteins. The de novo formation of Fe/S clusters on cytosolic and nuclear proteins is decreased in isd11 mutant cells, in agreement with the essential role of the mitochondrial ISC-assembly machinery in the formation of extra-mitochondrial Fe/S proteins (Lill and Mühlenhoff, 2005). Moreover, the iron homeostasis is disturbed in isd11 mutant cells, including activation of the iron-dependent transcription factor Aft1 and accumulation of iron in mitochondria, two typical observations in cells with a defective ISC-assembly machinery (Kispal et al, 1999; Li et al, 1999, 2001; Schilke et al, 1999; Lange et al, 2000; Kim et al, 2001; Mühlenhoff et al, 2002; Chen et al, 2004; Gerber et al, 2004).
Isd11 is well conserved in eukaryotes but not in prokaryotes. This is unprecedented for an ISC-assembly component of the mitochondrial matrix because so far all subunits are either derived from the bacterial isc operon (Nfs1, Isu1/2, Yah1, Nfu1, Isa1/2, Ssq1, Jac1) or possess homologs in bacteria (Arh1, Yfh1, Grx5) (Zheng et al, 1998; Johnson et al, 2005; Lill and Mühlenhoff, 2005). Isd11 belongs to the small group of mitochondrial proteins that are essential for cell viability, emphasizing the essential nature of mitochondrial Fe/S biogenesis. So far, only one essential mitochondrial protein, Erv1 of the intermembrane space, has been known that is involved in Fe/S protein biogenesis but is not conserved in prokaryotes. Erv1, however, is required for both biogenesis of cytosolic Fe/S proteins (mitochondrial ISC-export machinery) and import of precursor proteins (Lange et al, 2001; Mesecke et al, 2005; Rissler et al, 2005). Although it cannot be excluded that prokaryotes possess a distant homolog or analog of Isd11, the current results suggest that Isd11 is a eukaryotic addition to the prokaryote-derived ISC-assembly machinery of the mitochondrial matrix.
Isd11 forms a stable complex of about 230 kDa with the cysteine desulfurase Nfs1. Our results suggest that the resulting ISD complex rather than Nfs1 alone represents the active mitochondrial desulfurase that serves as sulfur donor to the Isu scaffold proteins. When the ISD complex is dissociated in isd11 mutant mitochondria, Nfs1 is still enzymatically active as a desulfurase (S2− release) in vitro, but this activity does not appear to represent the physiological sulfur donor in vivo, as Isd11 is additionally required for the transfer of the Nfs1-bound persulfide sulfur to the Isu proteins. In bacteria, SufS and SufE have also been shown to function as a two-component cysteine desulfurase (Johnson et al, 2005). SufE stimulates the SufS desulfurase activity and sulfur is directly transferred from SufS to a conserved cysteine of SufE (see, e.g., Ollagnier-de-Choudens et al, 2003). However, Isd11 does not show sequence similarity to SufE and apparently cannot function as an intermediate sulfur carrier as it does not contain any cysteine. Upon treatment of isd11 mutant mitochondria at high temperature, Nfs1 aggregates and, upon prolonged incubation of the mutant cells at non-permissive conditions, Nfs1 is degraded, indicating that Isd11 is also required for the stabilization of Nfs1. We suggest that Isd11 functions as adapter and stabilizer of Nfs1.
The 230 kDa ISD complex contains Nfs1 and Isd11 in stoichiometric amounts and is stable on BN-PAGE. A fraction of the Isu proteins is found in association with the ISD complex in an affinity purification, whereas they are released from the complex on BN-PAGE. This behavior is characteristic for proteins that are only loosely or transiently associated with a stable core complex (Chacinska et al, 2003, 2005). Although the bulk of Nfs1 and Isd11 are present in the ISD complex, minor fractions of non-assembled Isd11 and dimeric Nfs1 are also observed, possibly representing a dynamic pool for assembly reactions. The existence of distinct pools is also supported by the observation that a fraction of Isd11 but not Nfs1 is loosely associated with the inner membrane. It is tempting to speculate that the proposed sulfur transfer from an enzyme-bound persulfide intermediate at Nfs1 to the Isu proteins (Smith et al, 2001; Urbina et al, 2001) involves a dynamic assembly and disassembly of Isu proteins with Nfs1 and Isd11.
In summary, Isd11 is a novel subunit of the essential mitochondrial machinery for Fe/S protein biogenesis. Isd11 interacts with Nfs1 in the mitochondrial desulfurase complex and is required at an early stage of ISC biogenesis that leads to Fe/S cluster formation on the Isu scaffold proteins.
Materials and methods
Yeast strains
Yeast strains used in this study were BY4741, Isd11TAP (Yer048w-aTAP), Isu1TAP and Isu2TAP (Open Biosystems), Nfs1ProtA (BY4741, Nfs1 tagged by homologous recombination), YPH499 and W303-1A (MATa, ura3-1, ade2-1, trp1-1, his3-11,15, leu2-3,112). An isd11-deletion strain (derived from YPH499) carrying a URA3-plasmid with the ISD11 (Yer048w-a/FOMP6/EDI1) open reading frame was viable like yeast wild-type cells but was unable to grow on 5-fluoroorotic acid, that is, the plasmid loss approach did not lead to a viable isd11-deletion strain. Deletion of ISD11 caused inviability of cells on any medium and temperature tested. Mutant forms of ISD11 were generated by low-fidelity PCR and cloned by gap repair into the plasmid pFL39 (Truscott et al, 2002). In the isd11-deletion strain, the URA3-plasmid with wild-type ISD11 was exchanged against pFL39-fomp6Ts1(isd11-1) by plasmid shuffling, yielding the temperature-conditional yeast strain isd11-1 (YBG-0601a; MATa, ade2-101, his3-Δ200, leu2-Δ1, ura3-52, trp1-Δ63, lys2-801, isd11∷ADE2, pFL39-fomp6Ts1-CEN). Amino-acid substitutions in Isd11-1 are F4V, K45E, N57D, R71G, M78K and V86E, yielding a pI of 9.8 for the mutant protein compared to a pI of 10.3 of wild-type Isd11. The strain Gal-ISD11 was generated in the W303-1A background by exchanging the region −290 to −1 of ISD11 for the GAL1-10 promoter by PCR-mediated DNA replacement (Mühlenhoff et al, 2002). Yeast strains were cultivated in YP media (1% (w/v) yeast extract, 2% (w/v) bactopeptone) with 2% (w/v) dextrose (YPD), 2% (w/v) sucrose (YPS) or 3% (w/v) glycerol (YPG) or in rich (YP), minimal (SC) or ‘iron-poor' minimal medium lacking added iron chloride.
Mitochondrial fractionation, in vitro import and complex isolations
Isolated mitochondria were sonicated in high-salt buffer (0.5 M NaCl, 10 mM Tris, pH 7.2). For aggregation assays, mitochondria were either kept on ice or heat shocked for 15 min at 37°C. After centrifugation for 5 min at 20 000 g and 2°C, mitochondrial pellets were resuspended in digitonin buffer (1% digitonin, 20 mM Tris, pH 7.4, 0.1 mM EDTA, 10% glycerol, 0.1 M NaCl), incubated for 15 min on ice and clarified by centrifugation for 5 min at 20 000 g and 2°C. The samples were separated into pellet and supernatant by centrifugation for 30–60 min at 100 000 g.
Precursor proteins were synthesized in rabbit reticulocyte lysate in the presence of [35S]methionine/cysteine and incubated with mitochondria at 20–25°C as described (Ryan et al, 2001). Dissipation of Δψ, treatment with proteinase K and extraction with 0.1 M Na2CO3 (pH 11.5 or 10.8) were performed as published (Ryan et al, 2001; Truscott et al, 2003). Crosslinking was performed with 1 mM MBS for 30 min on ice, followed by quenching with 0.1 M Tris and 20 mM cysteine for 5 min. For IgG affinity chromatography, mitochondria were lysed in 0.1% DDM or 1% digitonin with 20 mM Tris, pH 7.4, 0.1 mM EDTA, 10% glycerol, 0.25 M NaCl, 1 mM PMSF and complete protease inhibitor (Roche) (Truscott et al, 2003).
55Fe incorporation into Fe/S cluster apoproteins in vivo
Yeast cells were labeled with radioactive iron (55FeCl3) in vivo and 55Fe incorporation into various Fe/S proteins was determined as described (Kispal et al, 1999, Mühlenhoff et al, 2002). isd11-1 cells were grown in iron-poor minimal medium supplemented with glucose (SD) for 16 h at various temperatures before analysis. Gal-ISD11 cells were cultivated for 24 h in SC medium and for 16 h in iron-poor SC medium supplemented with either glucose or galactose before analysis. Hemagglutinin (HA)-tagged Rli1 and Ntg2 (overproduced from the 2μ plasmid p426GPD using the TDH3 promoter) and Leu1 were used as Fe/S reporter proteins.
In vitro labeling of the ISD complex
Mitochondria (100 μg protein) were resuspended in 100 μl labeling buffer (0.6 M sorbitol, 20 mM Tris, pH 7.2, 0.15 M KCl, 15 mM KH2PO4, 12.5 mM MgCl2, 3 mg/ml fatty acid-free BSA) with 0.875 mg/ml chloramphenicol and incubated for 15 min at 20°C. Mitochondria were reisolated by centrifugation for 5 min at 20 000 g and 2°C, resuspended in 50 μl digitonin buffer (0.4% digitonin, 20 mM Tris/HCl, pH 7.4, 0.1 mM EDTA, 10% glycerol, 50 mM NaCl) and incubated for 5 min on ice. After addition of 1 mM NADH, 2 mM ATP and 200 kBq [35S]cysteine or [35S]methionine, the samples were incubated for 15 min at 20°C.
Miscellaneous
Homologs of Isd11 were identified by BLAST search against all publicly available DNA sequences (GenBank) and selected homologs were aligned with MultAlin. Published methods were used to determine induction of the iron regulon by following the fluorescence levels of GFP expressed from a FET3-GFP fusion gene (Rutherford et al, 2005) and to measure the activities of malate dehydrogenase, aconitase, succinate dehydrogenase, complex III, Leu1 and alcohol dehydrogenase (Kispal et al, 1999; Mühlenhoff et al, 2002). Swelling of mitochondria (Ryan et al, 2001), native PAGE (Lutz et al, 2001) and BN-PAGE (Chacinska et al, 2003) were performed as described. Mitochondrial iron contents were determined by the bathophenanthroline method (Li et al, 1999).
Acknowledgments
We thank Drs S Rospert and T Sommer for antisera. This work was supported by the Deutsche Forschungsgemeinschaft, Gottfried Wilhelm Leibniz Program, Max Planck Research Award, Alexander von Humboldt Foundation, Bundesministerium für Bildung und Forschung, Sonderforschungsbereiche 388 and 593, Nationales Genomforschungsnetz, Australian Research Council Linkage International and the Fonds der Chemischen Industrie.
References
- Balk J, Pierik AJ, Netz DJ, Mühlenhoff U, Lill R (2004) The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron–sulphur proteins. EMBO J 23: 2105–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Lind M, Frazier AE, Dudek J, Meisinger C, Geissler A, Sickmann A, Meyer HE, Truscott KN, Guiard B, Pfanner N, Rehling P (2005) Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell 120: 817–829 [DOI] [PubMed] [Google Scholar]
- Chacinska A, Rehling P, Guiard B, Frazier AE, Schulze-Specking A, Pfanner N, Voos W, Meisinger C (2003) Mitochondrial translocation contact sites: separation of dynamic and stabilizing elements in formation of a TOM–TIM–preprotein supercomplex. EMBO J 22: 5370–5381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen OS, Crisp RJ, Valachovic M, Bard M, Winge DR, Kaplan J (2004) Transcription of the yeast iron regulon does not respond directly to iron but rather to iron–sulfur cluster biosynthesis. J Biol Chem 279: 29513–29518 [DOI] [PubMed] [Google Scholar]
- Contamine V, Picard M (2000) Maintenance and integrity of the mitochondrial genome: a plethora of nuclear genes in the budding yeast. Microbiol Mol Biol Rev 64: 281–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutkiewicz R, Schilke B, Knieszner H, Walter W, Craig EA, Marszalek J (2003) Ssq1, a mitochondrial Hsp70 involved in iron–sulfur (Fe/S) center biogenesis: similarities to and differences from its bacterial counterpart. J Biol Chem 278: 29719–29727 [DOI] [PubMed] [Google Scholar]
- Gabriel K, Egan B, Lithgow T (2003) Tom40, the import channel of the mitochondrial outer membrane, plays an active role in sorting imported proteins. EMBO J 22: 2380–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber J, Mühlenhoff U, Lill R (2003) An interaction between frataxin and Isu1/Nfs1 that is crucial for Fe/S cluster synthesis on Isu1. EMBO Rep 4: 906–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber J, Neumann K, Prohl C, Mühlenhoff U, Lill R (2004) The yeast scaffold proteins Isu1p and Isu2p are required inside mitochondria for maturation of cytosolic Fe/S proteins. Mol Cell Biol 24: 4848–4857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Güldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kötter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Johnson DC, Dean DR, Smith AD, Johnson MK (2005) Structure, function, and formation of biological iron–sulfur clusters. Annu Rev Biochem 74: 247–281 [DOI] [PubMed] [Google Scholar]
- Kim R, Saxena S, Gordon DM, Pain D, Dancis A (2001) J-domain protein, Jac1p, of yeast mitochondria required for iron homeostasis and activity of Fe–S cluster proteins. J Biol Chem 276: 17524–17532 [DOI] [PubMed] [Google Scholar]
- Kispal G, Csere P, Prohl C, Lill R (1999) The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J 18: 3981–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispal G, Sipos K, Lange H, Fekete Z, Bedekovics T, Janaky T, Bassler J, Aguilar Netz DJ, Balk J, Rotte C, Lill R (2005) Biogenesis of cytosolic ribosomes requires the essential iron–sulphur protein Rli1p and mitochondria. EMBO J 24: 589–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SA, Sepuri NB, Pain D, Dancis A (1998) Mt-Hsp70 homolog, Ssc2p, required for maturation of yeast frataxin and mitochondrial iron homeostasis. J Biol Chem 273: 18389–18393 [DOI] [PubMed] [Google Scholar]
- Koehler CM (2004) New developments in mitochondrial assembly. Annu Rev Cell Dev Biol 20: 309–335 [DOI] [PubMed] [Google Scholar]
- Lange H, Kaut A, Kispal G, Lill R (2000) A mitochondrial ferredoxin is essential for biogenesis of cellular iron–sulfur proteins. Proc Natl Acad Sci USA 97: 1050–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H, Lisowsky T, Gerber J, Mühlenhoff U, Kispal G, Lill R (2001) An essential function of the mitochondrial sulfhydryl oxidase Erv1p/ALR in the maturation of cytosolic Fe/S proteins. EMBO Rep 2: 715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Kogan M, Knight SA, Pain D, Dancis A (1999) Yeast mitochondrial protein, Nfs1p, coordinately regulates iron–sulfur cluster proteins, cellular iron uptake, and iron distribution. J Biol Chem 274: 33025–33034 [DOI] [PubMed] [Google Scholar]
- Li J, Saxena S, Pain D, Dancis A (2001) Adrenodoxin reductase homolog (Arh1p) of yeast mitochondria required for iron homeostasis. J Biol Chem 276: 1503–1509 [DOI] [PubMed] [Google Scholar]
- Lill R, Mühlenhoff U (2005) Iron–sulfur-protein biogenesis in eukaryotes. Trends Biochem Sci 30: 133–141 [DOI] [PubMed] [Google Scholar]
- Lutz T, Westermann B, Neupert W, Herrmann JM (2001) The mitochondrial proteins Ssq1 and Jac1 are required for the assembly of iron sulfur clusters in mitochondria. J Mol Biol 307: 815–825 [DOI] [PubMed] [Google Scholar]
- Mesecke N, Terziyska N, Kozany C, Baumann F, Neupert W, Hell K, Herrmann JM (2005) A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell 121: 1059–1069 [DOI] [PubMed] [Google Scholar]
- Mühlenhoff U, Balk J, Richhardt N, Kaiser JT, Sipos K, Kispal G, Lill R (2004) Functional characterization of the eukaryotic cysteine desulfurase Nfs1p from Saccharomyces cerevisiae. J Biol Chem 279: 36906–36915 [DOI] [PubMed] [Google Scholar]
- Mühlenhoff U, Gerber J, Richhardt N, Lill R (2003) Components involved in assembly and dislocation of iron–sulfur clusters on the scaffold protein Isu1p. EMBO J 22: 4815–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlenhoff U, Richhardt N, Ristow M, Kispal G, Lill R (2002) The yeast frataxin homolog Yfh1p plays a specific role in the maturation of cellular Fe/S proteins. Hum Mol Genet 11: 2025–2036 [DOI] [PubMed] [Google Scholar]
- Neupert W (1997) Protein import into mitochondria. Annu Rev Biochem 66: 863–917 [DOI] [PubMed] [Google Scholar]
- Ollagnier-de-Choudens S, Lascoux D, Loiseau L, Barras F, Forest E, Fontecave M (2003) Mechanistic studies of the SufS–SufE cysteine desulfurase: evidence for sulfur transfer from SufS to SufE. FEBS Lett 555: 263–267 [DOI] [PubMed] [Google Scholar]
- Pfanner N, Wiedemann N, Meisinger C, Lithgow T (2004) Assembling the mitochondrial outer membrane. Nat Struct Mol Biol 11: 1044–1048 [DOI] [PubMed] [Google Scholar]
- Rehling P, Pfanner N, Meisinger C (2003) Insertion of hydrophobic membrane proteins into the inner mitochondrial membrane—a guided tour. J Mol Biol 326: 639–657 [DOI] [PubMed] [Google Scholar]
- Rissler M, Wiedemann N, Pfannschmidt S, Gabriel K, Guiard B, Pfanner N, Chacinska A (2005) The essential mitochondrial protein Erv1 cooperates with Mia40 in biogenesis of intermembrane space proteins. J Mol Biol 353: 485–492 [DOI] [PubMed] [Google Scholar]
- Rutherford JC, Jaron S, Ray E, Brown PO, Winge DR (2001) A second iron-regulatory system in yeast independent of Aft1p. Proc Natl Acad Sci USA 98: 14322–14327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford JC, Ojeda L, Balk J, Mühlenhoff U, Lill R, Winge DR (2005) Activation of the iron regulon by the yeast Aft1/Aft2 transcription factors depends on mitochondrial but not cytosolic iron–sulfur protein biogenesis. J Biol Chem 280: 10135–10140 [DOI] [PubMed] [Google Scholar]
- Ryan MT, Voos W, Pfanner N (2001) Assaying protein import into mitochondria. Methods Cell Biol 65: 189–215 [DOI] [PubMed] [Google Scholar]
- Schilke B, Forster J, Davis J, James P, Walter W, Laloraya S, Johnson J, Miao B, Craig E (1996) The cold sensitivity of a mutant of Saccharomyces cerevisiae lacking a mitochondrial heat shock protein 70 is suppressed by loss of mitochondrial DNA. J Cell Biol 134: 603–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilke B, Voisine C, Beinert H, Craig E (1999) Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 96: 10206–10211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schönfisch B, Perschil I, Chacinska A, Guiard B, Rehling P, Pfanner N, Meisinger C (2003) The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci USA 100: 13207–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AD, Agar JN, Johnson KA, Frazzon J, Amster IJ, Dean DR, Johnson MK (2001) Sulfur transfer from IscS to IscU: the first step in iron–sulfur cluster biosynthesis. J Am Chem Soc 123: 11103–11104 [DOI] [PubMed] [Google Scholar]
- Strain J, Lorenz CR, Bode J, Garland S, Smolen GA, Ta DT, Vickery LE, Culotta VC (1998) Suppressors of superoxide dismutase (SOD1) deficiency in Saccharomyces cerevisiae: identification of proteins predicted to mediate iron–sulfur cluster assembly. J Biol Chem 273: 31138–31144 [DOI] [PubMed] [Google Scholar]
- Truscott KN, Voos W, Frazier AE, Lind M, Li Y, Geissler A, Dudek J, Müller H, Sickmann A, Meyer HE, Meisinger C, Guiard B, Rehling P, Pfanner N (2003) A J-protein is an essential subunit of the presequence translocase-associated protein import motor of mitochondria. J Cell Biol 163: 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truscott KN, Wiedemann N, Rehling P, Müller H, Meisinger C, Pfanner N, Guiard B (2002) Mitochondrial import of the ADP/ATP carrier: the essential TIM complex of the intermembrane space is required for precursor release from the TOM complex. Mol Cell Biol 22: 7780–7789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbina HD, Silberg JJ, Hoff KG, Vickery LE (2001) Transfer of sulfur from IscS to IscU during Fe/S cluster assembly. J Biol Chem 276: 44521–44526 [DOI] [PubMed] [Google Scholar]
- Voisine C, Cheng YC, Ohlson M, Schilke B, Hoff K, Beinert H, Marszalek J, Craig EA (2001) Jac1, a mitochondrial J-type chaperone, is involved in the biogenesis of Fe/S clusters in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 98: 1483–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M'Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Véronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]
- Zheng L, Cash VL, Flint DH, Dean DR (1998) Assembly of iron–sulfur clusters: identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J Biol Chem 273: 13264–13272 [DOI] [PubMed] [Google Scholar]
- Zheng L, White RH, Cash VL, Dean DR (1994) Mechanism for the desulfurization of L-cysteine catalyzed by the nifS gene product. Biochemistry 33: 4714–4720 [DOI] [PubMed] [Google Scholar]


