Abstract
Bacterial speck disease in tomato is caused by Pseudomonas syringae pv. tomato. Resistance to this disease is conferred by the host Pto kinase, which recognizes P. s. pv. tomato strains that express the effector AvrPto. We report here that an AvrPto-dependent Pto-interacting protein 3 (Adi3) is a member of the AGC family of protein kinases. In mammals, AGC kinases are regulated by 3-phosphoinositide-dependent protein kinase-1 (Pdk1). We characterized tomato Pdk1 and showed that Pdk1 and Pto phosphorylate Adi3. Gene silencing of Adi3 in tomato causes MAPKKKα-dependent formation of necrotic lesions. Use of a chemical inhibitor of Pdk1, OSU-03012, also implicates Pdk1 and Adi3 in plant cell death regulation. Adi3 thus appears to function analogously to the mammalian AGC kinase protein kinase B/Akt by negatively regulating cell death via Pdk1 phosphorylation. We speculate that the negative regulatory function of Adi3 might be subverted by interaction with Pto/AvrPto, leading to host cell death that is associated with pathogen attack.
Keywords: Adi3, 3-phosphoinositide-dependent kinase-1, programmed cell death, protein kinase B
Introduction
Programmed cell death (PCD) is an essential process for development and immune responses in eukaryotic multicellular organisms. In mammalian systems, PCD is used for many processes from digit formation to removal of infected cells (Lam, 2004). In plants, leaf senescence, development of xylem tracheary elements, and host responses to pathogens all involve PCD (Greenberg and Yao, 2004; Lam, 2004). Some characteristics of PCD are shared between animals and plants, such as DNA fragmentation and organelle destruction, whereas others, such as apoptotic body formation and engulfment by neighboring cells, occur only in animals (Lam, 2004).
Localized PCD occurs in both resistant and susceptible plants during pathogen attack. In the former, PCD termed the hypersensitive response (HR) occurs rapidly (<12 h) upon attempted pathogen infection (Goodman and Novacky, 1994). The HR may inhibit further spread of the pathogen, but its biological significance has been difficult to verify due to the lack of plant mutants affected solely in cell death. In disease-susceptible plants, localized cell death referred to as ‘specks', ‘spots', or ‘blights' appears over the course of many days. Recent studies suggest that these disease symptoms also involve host-mediated PCD (Lincoln et al, 2002; Abramovitch and Martin, 2004; del Pozo et al, 2004). Thus, host-controlled PCD plays a critical role in determining both immunity and disease progression in plants. Despite this significance, relatively few plant genes have been identified that have a demonstrated role in PCD associated with pathogen attack, and upstream components that might regulate these cell death mediators are yet to be identified (Shah et al, 1999; del Pozo et al, 2004; Greenberg and Yao, 2004; Lam, 2004).
In mammals, PCD (apoptosis) has been studied extensively, and many proteins involved in this process are known (Nicholson and Thornberry, 2003). An important central regulator of mammalian apoptosis is protein kinase B (PKB; also known as Akt; Vivanco and Sawyers, 2002). PKB is a member of the AGC family of protein kinases, which often act as downstream effectors of second messengers such as cAMP, cGMP, phospholipids, or Ca2+. PKB negatively regulates apoptosis by phosphorylation and inactivation of proapoptotic factors such as BAD and activation of antiapoptotic factors such as CREB and IKK (Vivanco and Sawyers, 2002; Vara et al, 2004).
The activity of mammalian AGC kinases like PKB is regulated through phosphorylation by 3-phosphoinositide-dependent protein kinase-1 (Pdk1). Pdk1 phosphorylates some AGC kinases in a constitutive manner and others upon stimulation (Belham et al, 1999). The physical interaction of Pdk1 with its substrates is mediated through a hydrophobic amino-acid sequence termed the Pdk1-interacting fragment (PIF), found on the AGC kinase. A PIF-binding pocket found on Pdk1 binds the PIF, allowing Pdk1 to phosphorylate and activate the substrate (Biondi, 2004). Once activated, the AGC kinase and Pdk1 disassociate and the PIF is bound by a PIF-binding pocket on the AGC kinase, allowing for stabilization and phosphorylation of downstream substrates (Frödin et al, 2002; Biondi, 2004).
Little is known about the role of AGC kinases in plants. There are at least 39 AGC kinase genes in Arabidopsis (Bögre et al, 2003), but the functions of only five of them are known. PINOID (PID) is involved in regulating auxin signals (Christensen et al, 2000; Benjamins et al, 2001), phototropins 1 and 2 (PHOT1 and PHOT2) mediate blue-light signals (Briggs and Christie, 2002), and IRE and AGC2-1/OXI1 regulate root hair development (Oyama et al, 2002; Anthony et al, 2004; Rentel et al, 2004). AGC2-1/OXI1 also mediates oxidative burst signals (Rentel et al, 2004).
Bacterial speck disease in tomato is caused by Pseudomonas syringae pv. tomato (Pst). Resistance to Pst arises from a ‘gene-for-gene' interaction in which the product of the host Pto resistance gene, a Ser/Thr protein kinase, physically interacts with the Pseudomonas AvrPto protein upon its delivery into the plant cell via the bacterial type III secretion system (Pedley and Martin, 2003). Recognition of AvrPto by Pto elicits an HR-based resistance. This resistance requires another protein, Prf, although its role is unknown (Pedley and Martin, 2003). Several other components have been identified that appear to act in the Pto pathway (Ekengren et al, 2003; Pedley and Martin, 2003).
In order to investigate the role of the Pto/AvrPto interaction, we previously used a yeast three-hybrid (Y3H) screen to identify tomato proteins that interact only when both Pto and AvrPto are present (Bogdanove and Martin, 2000). From this screen, we identified AvrPto-dependent Pto-interacting protein 3 (Adi3). We report here the characterization of Adi3, which indicates that it plays a role as a negative regulator of plant cell death and therefore might act analogously to PKB in controlling plant cell death.
Results
Adi3 cDNA cloning and protein characterization
Adi3 was identified from a Y3H screen as requiring both Pto and AvrPto for its interaction (Bogdanove and Martin, 2000). The isolated cDNA, which was found to encode a full Ser/Thr protein kinase domain, lacked the 5′ end. The full-length Adi3 cDNA was isolated by using a tomato EST database at The Institute for Genomic Research (TIGR; EST TC102958) and 5′ RACE. The complete Adi3 cDNA has a 471 bp 5′ UTR, a 2103 bp coding region, and a 255 bp 3′ UTR.
Analysis of Adi3 revealed that it belongs to group VIIIa of plant AGC protein kinases described by Bögre et al (2003). Key features of this group include a large amino-acid insertion (74 aa for Adi3) between kinase subdomains VII and VIII (referred to as a T-loop extension), the conserved DFG motif of subdomain VII for Mg2+ coordination is changed to DFD, and a PIF consensus sequence of FD/ExF (Bögre et al, 2003). All other conserved protein kinase residues are present in Adi3. Although the Adi3 T-loop extension is similar to that of the Ndr family of AGC kinases (Tamaskovic et al, 2003), phylogenetic analysis indicates that Adi3 does not belong to the Ndr family (Bögre et al, 2003).
Mutations affecting Adi3 autophosphorylation
We examined the possible autophosphorylation activity of MBP-Adi3. MBP-Adi3 showed autophosphorylation activity, whereas MBP-Adi3K337Q, which has a substitution at the ATP-binding site, did not (Figure 1A). Adi3 activity was dependent on Mg2+ and was not observed with Mn2+ (data not shown). Substitution of S539 for Ala in the activation loop of Adi3 did not affect autophosphorylation (Figure 1A). However, substitution of S539 for Asp greatly increased the level of Adi3 autophosphorylation (Figure 1A), indicating that this protein is constitutively active and that phosphorylation of S539 likely contributes to full Adi3 activity.
Figure 1.
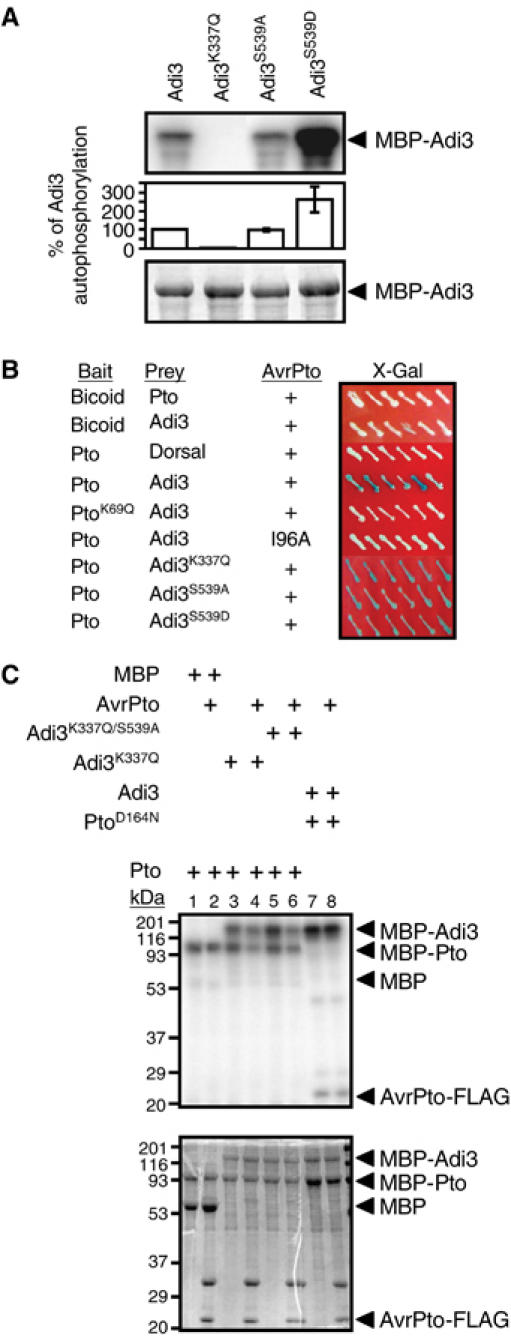
Adi3 autophosphorylation and protein interactions. In (A) and (C), the top panel shows the kinase assay (phosphorimage) and the bottom panel the assay input (Coomassie-stained gel). (A) Adi3 encodes a functional protein kinase. Analysis of kinase-active, -deficient, and constitutively active MBP-Adi3 fusion proteins by in vitro autophosphorylation assays is shown. (B) Adi3 interactions in the Y3H assay. The indicated bait and prey constructs were tested in the Y3H assay (Bogdanove and Martin, 2000) for expression of the lacZ gene on X-Gal plates (blue=interaction). The Drosophila proteins Bicoid and Dorsal were used as negative controls and indicate that the Pto/Adi3/AvrPto interaction is specific. + indicates the presence of AvrPto. (C) Analysis of cross-phosphorylation between Adi3 and Pto. Kinase-deficient MBP-Adi3 proteins were used as substrates for kinase-active MBP-Pto protein with or without AvrPto-FLAG protein. Kinase-deficient MBP-PtoD164N protein was used as a substrate for kinase-active MBP-Adi3 with or without AvrPto-FLAG protein. Molecular weight (kDa) markers are indicated.
Adi3 interactions in the yeast three-hybrid assay
To gain an insight into the specificity of the Pto/AvrPto/Adi3 interaction, the full-length Adi3 cDNA was tested in the Y3H assay with wild-type and mutant forms of Pto and AvrPto. Substitutions in Pto (K69Q) or AvrPto (I96A) are known to disrupt the Pto/AvrPto yeast two-hybrid (Y2H) interaction although they do not affect protein abundance (Loh and Martin, 1995; Tang et al, 1996; Frederick et al, 1998). The PtoK69Q mutation (subdomain II conserved Lys) also eliminates Pto kinase activity. We found that the Y3H interaction does not occur with PtoK69Q or AvrPtoI96A (Figure 1B). However, the kinase-deficient mutant Adi3K337Q as well as the constitutively active protein Adi3S539D interacted with Pto/AvrPto similar to wild-type Adi3 (Figure 1B). These data indicate that the kinase-active conformation of Pto, but not that of Adi3, is required for the Pto/AvrPto/Adi3 interaction.
Pto phosphorylates Adi3 independently of AvrPto
The requirement of the kinase-active conformation of Pto for the Y3H interaction suggested that Pto might phosphorylate Adi3. This was tested by using an in vitro kinase assay. When MBP-Pto was incubated with the kinase-deficient MBP-Adi3K337Q protein, phosphorylation of the kinase-inactive Adi3 protein was observed (Figure 1C, lane 3), which was independent of AvrPto (Figure 1C, lane 4). The ability of Adi3 to utilize Pto as a substrate was also tested. For these assays, PtoD164N was used because it is kinase-inactive but, unlike PtoK69Q, still interacts with AvrPto in the Y2H system (Rathjen et al, 1999). When MBP-Adi3 was incubated with MBP-PtoD164N, phosphorylation of PtoD164N was not seen regardless of the presence of AvrPto (Figure 1C, lanes 7 and 8). These data demonstrate that Adi3 is a substrate for Pto. The slight reduction in Pto phosphorylation of Adi3 in the presence of AvrPto (Figure 1C, lanes 4 and 6) appears to be nonspecific and due to the additional level of protein in the assay as Pto autophosphorylation level is reduced as well. Additionally, the same effect has been seen with Pdk1 phosphorylation of Adi3 (data not shown).
Isolation of the tomato Pdk1 gene and interactions with Adi3
AGC kinases are commonly regulated by Pdk1 phosphorylation. Thus, we identified a full-length Pdk1 cDNA in the tomato EST database using the Arabidopsis Pdk1 sequence (Supplementary Figure S1; Deak et al, 1999). Tomato Pdk1 encodes a protein of 494 aa, with a kinase domain and a pleckstrin homology domain. We found that it is an active protein kinase in autophosphorylation assays by using kinase-active MBP-Pdk1 and kinase-inactive MBP-Pdk1K77Q (subdomain II conserved Lys) proteins (Figure 2A). In the Y2H assay, Pdk1 did not interact with Pto or AvrPto, but did interact with Adi3 irrespective of the kinase activity of Adi3 or Pdk1 (Figure 2B).
Figure 2.
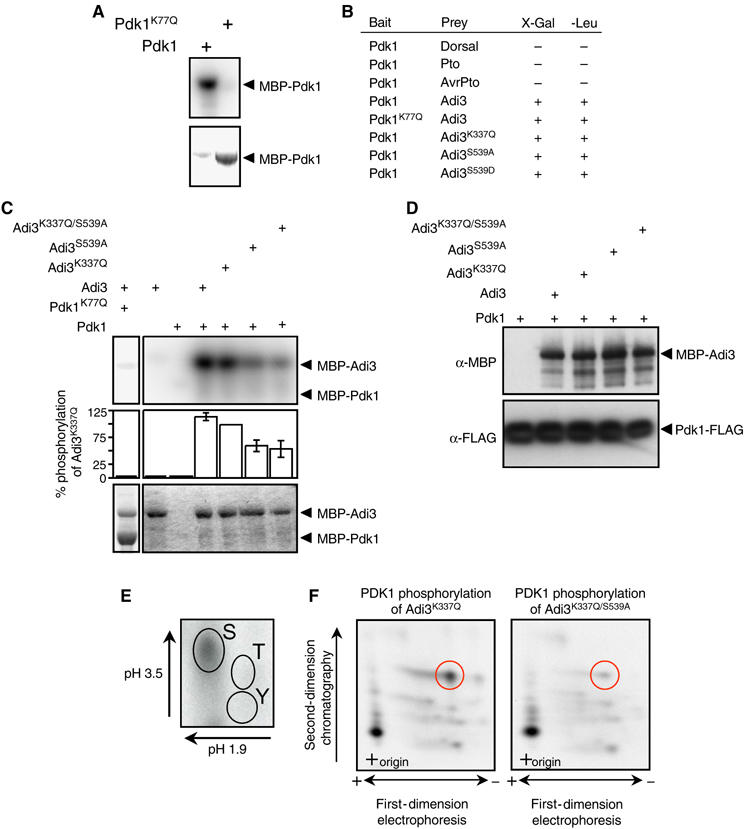
Pdk1 kinase activity and interaction with Adi3. In (A) and (C), the top panel represents the kinase assay (phosphorimage) and the bottom panel the assay input (Coomassie-stained gel). (A) Tomato Pdk1 is a functional protein kinase. Analysis of kinase-active and -deficient MBP-Pdk1 fusion proteins by in vitro autophosphorylation assays is shown. (B) Pdk1 interacts with Adi3 in the Y2H assay. The indicated bait and prey constructs were tested in the Y2H assay for expression of the lacZ gene on X-Gal plates or leucine prototrophy (+, interaction; −, no interaction). (C) Pdk1 phosphorylation of Adi3 in vitro. MBP-Adi3 proteins were used as substrates for kinase-active MBP-Pdk1 protein. (D) In vitro co-immunoprecipitation of Pdk1 and Adi3. MBP-Adi3 and Pdk1-FLAG proteins were co-precipitated using α-FLAG agarose and analyzed by Western blot. (E) Pdk1 phosphorylates Adi3 on Ser residues. Pdk1 phosphorylated Adi3 was analyzed by phosphoamino acid analysis; autoradiograph is shown. (F) Pdk1 phosphorylates Adi3 at S539 on one of two tryptic peptides. Pdk1 phosphorylated Adi3 was digested with trypsin and analyzed by phosphopeptide analysis; autoradiograph is shown.
The ability of Pdk1 to phosphorylate Adi3 was tested using MBP-Adi3 proteins as substrates for MBP-Pdk1. The Pdk1 phosphorylation site on AGC kinases is highly conserved (Bögre et al, 2003) and corresponds to S539 of Adi3. Mutation of S539 to Ala did not affect Adi3 autophosphorylation (Figure 1A), Adi3 Y3H interaction with Pto/AvrPto (Figure 1B), the ability of Pto to phosphorylate Adi3 (Adi3K337Q/S539A; Figure 1C, lane 5), or the Y2H interaction with Pdk1 (Figure 2B). Mutation of S539 to Asp, simulating a phosphorylation event, greatly increases the autophosphorylation activity of Adi3 (Figure 1A), suggesting that phosphorylation of S539 by Pdk1 is required for activation of Adi3. MBP-Pdk1 phosphorylated MBP-Adi3 but it phosphorylated MBP-Adi3S539A at a much reduced level (Figure 2C). Because MBP-Adi3S539A retains autophosphorylation (Figure 1A), MBP-Adi3K337Q/S539A kinase-deficient protein was used as a substrate for Pdk1 and was found to be phosphorylated by Pdk1 ∼45% less than MBP-Adi3, suggesting the presence of additional Pdk1 phosphorylation sites on Adi3 (Figure 2C). The reduced phosphorylation of the MBP-Adi3 mutant proteins by MBP-Pdk1 is not due to reduced protein–protein interactions, as MBP-Adi3 mutant proteins co-immunoprecipitated in vitro with Pdk1-FLAG in equal amounts (Figure 2D). This is also supported by the Pdk1/Adi3 Y2H assay where there was no difference in the interaction of Pdk1 with Adi3S539A (Figure 2B).
The likelihood that S539 of Adi3 is a Pdk1 phosphorylation site was further supported by phosphoamino acid and phosphopeptide analysis. MBP-Pdk1 phosphorylation of MBP-Adi3K337Q followed by phosphoamino acid analysis indicates that only Ser residues are phosphorylated on Adi3 by Pdk1 (Figure 2E). Phosphorylation of MBP-Adi3K337Q by MBP-Pdk1 followed by tryptic digestion and phosphopeptide mapping showed the presence of two major tryptic peptides (Figure 2F, left panel). The same analysis on the MBP-Adi3K337Q/S539A protein showed almost complete loss of one tryptic peptide (Figure 2F, right panel, compare red circles between panels). The small amount of radiolabel remaining in this tryptic peptide may be attributable to S537, which is contained in the same tryptic fragment. Taken together, these data indicate that Pdk1 phosphorylates Adi3 at S539 (Figure 2C, E, and F) and this phosphorylation event is required for Adi3 activity (Figure 1A). At least one additional Ser residue on Adi3 is phosphorylated by Pdk1 and remains to be determined.
Adi3 Pdk1-interacting fragment regulates interaction with Pdk1 and Adi3 autophosphorylation
In mammalian systems, the AGC kinase PIF motif regulates the interaction between Pdk1 and AGC kinases (Biondi and Nebreda, 2003; Biondi, 2004). The PIF is a short, hydrophobic amino-acid sequence motif, FxxFS/TF/Y, located close to or at the immediate carboxy-terminus of the protein. A PIF-binding pocket on Pdk1 (Figure 3A) binds the Phe residues and the phosphorylated Ser/Thr residue (Frödin et al, 2002). The minimal PIF sequence is defined as FxxF as found in PKA (Biondi, 2004). The Adi3 PIF (FExF) constitutes the last four residues of the protein (698–700; Figure 3A) and appears to be distinct from the PIF of PKA, as another group of plant AGC kinases (VIIIb) have a PIF consensus sequence identical to that of PKA (FxxF; Bögre et al, 2003).
Figure 3.
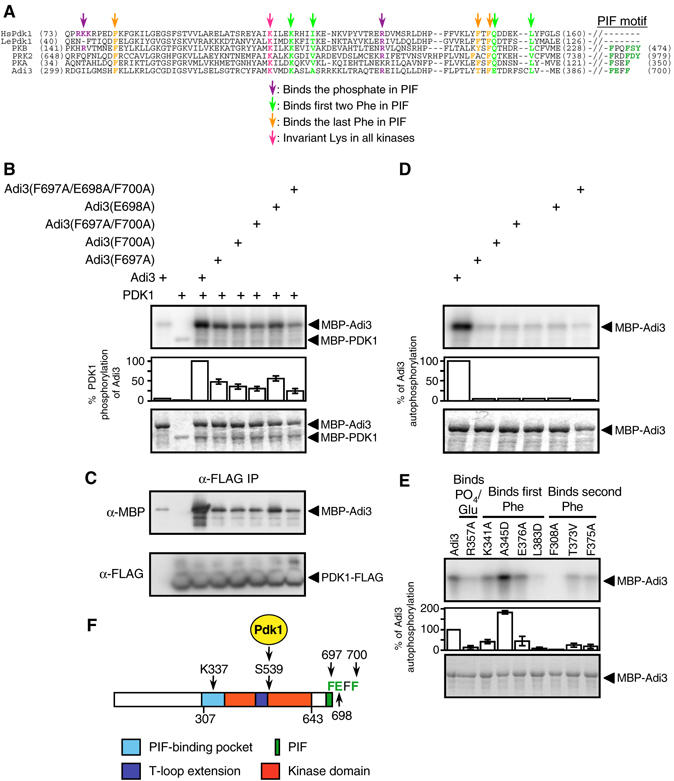
Adi3 PIF domain regulates interactions with Pdk1 and autophosphorylation. (A) Alignment of the PIF-binding pockets of human (Hs) and tomato (Le) Pdk1, PKBα, PKAα, and PRK2 with the corresponding region of Adi3 (adapted from Frödin et al, 2002). Binding of PIF residues is based on the mammalian PIF consensus of FxxFS/TF/Y. In (B), (D), and (E), the top panel shows kinase assay (phosphorimage) and the bottom panel the assay input (Coomassie-stained gel). (B) Adi3 PIF mutations reduce phosphorylation by Pdk1. Kinase assays were performed using MBP-Adi3 PIF mutant proteins as substrates for the MBP-Pdk1 protein. Adi3 PIF sequence is shown in (A) and (F). (C) Adi3 PIF mutations reduce interaction with Pdk1. MBP-Adi3 PIF mutant proteins and Pdk1-FLAG protein were co-immunoprecipitated as in Figure 2D. (D) Adi3 PIF mutations eliminate Adi3 autophosphorylation. MBP-Adi3 PIF mutant proteins were tested in autophosphorylation assays. Proteins used in the assay are the same as those in (B). (E) Mutations in the Adi3 PIF-binding pocket affect Adi3 autophosphorylation. MBP-Adi3 PIF-binding pocket mutant proteins were tested in autophosphorylation assays. (F) Diagram of the key features of the Adi3 protein. Total protein length, 700 aa; K337, invariant lysine in protein kinases; S539, Pdk1 phosphorylation site; F697, E698, F700, key PIF residues.
The role of the Adi3 PIF motif in regulating interaction with and phosphorylation by Pdk1 was tested by examining MBP-Adi3 PIF mutant proteins as possible substrates for Pdk1. Substitution of Adi3 F697 or F700 to Ala, individually or in combination, significantly reduced Pdk1 phosphorylation of Adi3 (Figure 3B). The E698A substitution also reduced Pdk1 phosphorylation of Adi3, but not to the extent of the F697A and F700A substitutions (Figure 3B). Combining all three substitutions gave the greatest loss of Pdk1 phosphorylation of Adi3 (Figure 3B). To confirm that the Adi3 PIF mutations affect the interaction with Pdk1, Adi3 and Pdk1 were co-immunoprecipitated in vitro. The binding of MBP-Adi3 PIF mutant proteins with Pdk1-FLAG correlated with the level of phosphorylation of MBP-Aid3 PIF mutant proteins by Pdk1 (Figure 3C), indicating that the Adi3 PIF regulates the interaction with Pdk1.
Mammalian AGC kinases also contain a PIF-binding pocket that binds their own PIF for protein stabilization and autophosphorylation regulation (Frödin et al, 2002; Biondi and Nebreda, 2003). Most plant AGC kinases appear to lack a PIF-binding pocket (Bögre et al, 2003). Alignment of human and plant Pdk1 and other mammalian AGC kinase PIF-binding pockets with the corresponding region in Adi3 shows low overall sequence identity; however, most residues predicted to bind the PIF appear to be conserved (Figure 3A). If Adi3 does contain a PIF-binding pocket for interaction with its own PIF, mutation of PIF residues should affect autophosphorylation. This was analyzed using MBP-Adi3 PIF mutant proteins in kinase assays. Mutation of F697, E698, and F700 to Ala, singly or in combination, completely eliminated Adi3 autophosphorylation (Figure 3D). The role of the key residues in the potential Adi3 PIF-binding pocket toward autophosphorylation was investigated using MBP-Adi3 PIF-binding pocket mutants in kinase assays. While most of these mutations had reduced autophosphorylation, R357A, L383D, and F308A contained nearly complete loss of autophosphorylation (Figure 3E). These data suggest the presence of a PIF-binding pocket on Adi3 for interaction with its own PIF. Figure 3F summarizes the key features of the Adi3 protein as revealed from the studies above.
Gene silencing of Adi3 indicates that it negatively regulates plant cell death
To investigate the role of Adi3 in plants, we used virus-induced gene silencing (VIGS) to knock down expression of Adi3 (Figure 4). VIGS has been used previously to effectively silence signaling-related genes in tomato (Ekengren et al, 2003; del Pozo et al, 2004). A 937-bp fragment corresponding to the 3′ 687 bp of the Adi3 ORF (residues 472–700) plus an additional 250 bp of the 3′ UTR was cloned into a tobacco rattle virus (TRV) silencing vector. VIGS utilizing this fragment resulted in essentially complete silencing of the Adi3 gene, as shown by RT–PCR analysis of transcript levels (Figure 4C). Plants silenced for Adi3 developed spontaneous (pathogen-independent), localized regions of cell death on leaves and stems, and became stunted (Figure 4A and B). This phenotype was independent of Pto or Prf (data not shown), indicating that these proteins act upstream or in concert with Adi3. We inoculated TRV:Adi3-silenced susceptible tomato plants with Pst and observed increased resistance as compared to TRV-only-infected controls (data not shown). However, we do not ascribe much significance to this enhanced resistance because the cell death phenotype that occurred 3 weeks after TRV:Adi3 infection likely induces general defense responses. In fact, expression of the defense-related genes PR1 and PR3 was induced in Adi3-silenced plants (data not shown).
Figure 4.
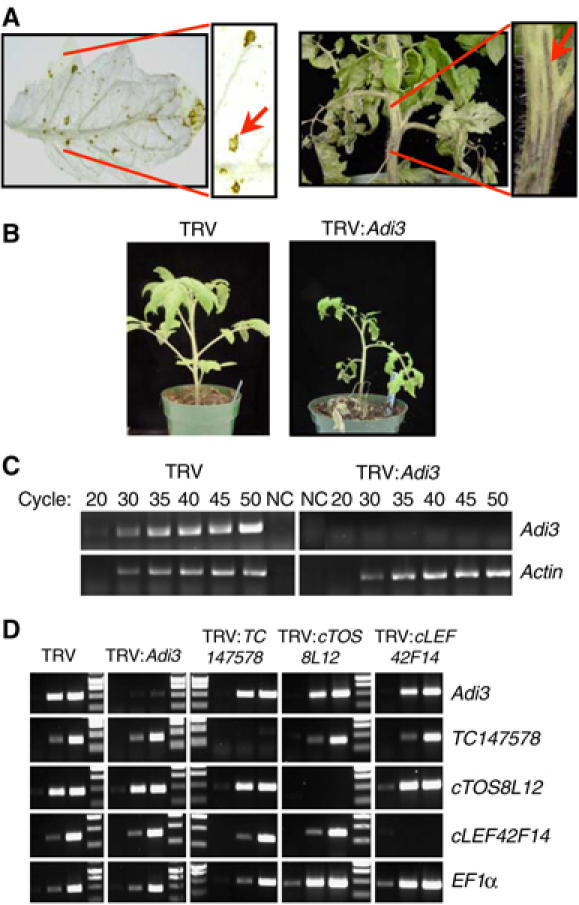
Gene silencing of Adi3 causes localized cell death. (A) Adi3 silencing causes cell death in leaves and stems. Cell death lesions were photographed 3 weeks after TRV:Adi3 infection. Arrows point to localized regions of cell death. In the left panel, leaves were cleared in 10% acetic acid, 30% chloroform, 60% EtOH. (B) Adi3 silencing causes stunting. (C) Adi3 transcripts are reduced by silencing. Control (TRV) and Adi3-silenced (TRV:Adi3) tissues were analyzed by RT–PCR for Adi3 transcripts. Aliquots were removed at the indicated cycles. Actin was the control. NC, negative control. (D) Adi3 silencing does not affect transcript levels of closely related AGC kinase genes. Tomato plants were silenced for the indicated genes (across top) and analyzed for transcript levels by RT–PCR (right side). EF1α was the control. Lanes show products from cycles 28, 34, and 40 and MW markers.
To check if unintended silencing of Adi3-related genes was responsible for the observed localized cell death phenotype, 17 genes in the tomato EST database that encode group VIIIa AGC kinases were identified for analysis of expression levels in Adi3-silenced plants (note that there are also 17 Arabidopsis genes in this group, although we were not able to establish possible orthologs). The DNA sequences were aligned with the Adi3 sequence and the three genes most similar to Adi3 were chosen for phenotypic and transcript analysis in silenced plants (i.e., TC147578 (64%; fragment amplified from gDNA), TC144536 (70%; EST cTOS8L12), TC138973 (69%; EST cLEF42F14)). RT–PCR using gene-specific primers showed that there was remarkable specificity in these silencing experiments—for each gene, only its corresponding transcript was knocked down by VIGS (Figure 4D). Importantly, we found that only Adi3, when silenced, caused localized cell death and stunting (Supplementary Figure S2). These experiments suggest that Adi3 acts as a negative regulator of host cell death and that silencing of Adi3 releases this inhibition.
Both Pdk1 and Adi3 regulate cell death in plants
In mammalian systems, Pdk1 phosphorylates PKB to promote its function in suppressing PCD (Vivanco and Sawyers, 2002; Vara et al, 2004; Zhu et al, 2004). Inhibition of Pdk1 does not allow for PKB activation and leads to PCD (Zhu et al, 2004). We therefore predicted that inhibition of Pdk1 might lead to PCD in tomato, and in fact, when a Pdk1 construct designed to silence both tomato Pdk1 genes (87.3 and 84.6% identical at the cDNA and protein level, respectively; data not shown) was used for VIGS, the plants died (data not shown).
The role of Pdk1 in plant cell death was further investigated using a specific inhibitor of mammalian Pdk1, OSU-03012 (Zhu et al, 2004). Tomato Pdk1 autophosphorylation was inhibited by OSU-03012 in a dose-dependent manner (Figure 5A). However, approximately 10 times more inhibitor was needed to achieve similar results seen with mammalian Pdk1 (Zhu et al, 2004), suggesting that OSU-03012 may be a less effective inhibitor of plant Pdk1.
Figure 5.
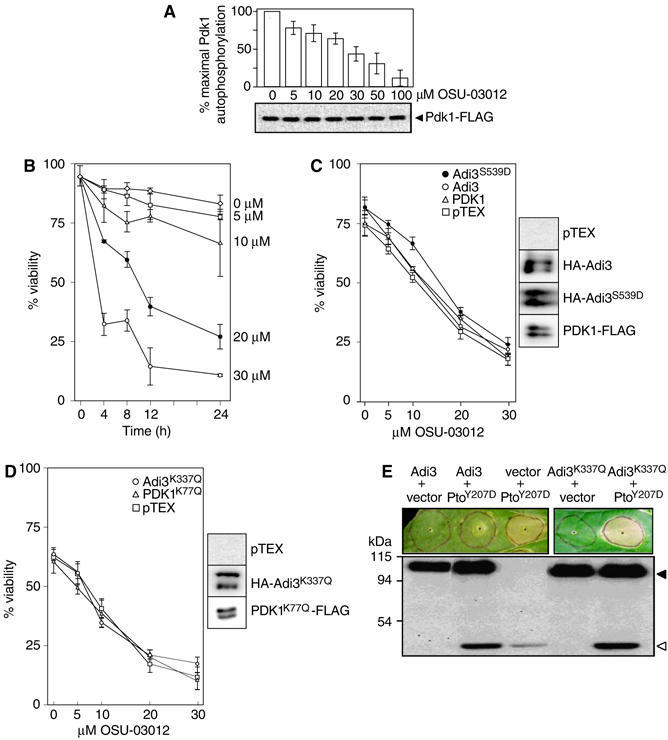
Pdk1 and Adi3 negatively regulate plant cell death. (A) OSU-03012 inhibition of tomato Pdk1. Top, dose-dependent inhibition of recombinant Pdk1-FLAG autophosphorylation. Bottom, α-FLAG Western blot showing assay input. (B) Pdk1 inhibitor OSU-03012 induces cell death in plants. Tomato protoplasts were treated with the Pdk1 inhibitor OSU-03012 and cell viability was monitored over 24 h. (C) Attenuation of OSU-03012-induced cell death by overexpression of Pdk1, Adi3, or Adi3S539D. Tomato protoplasts overexpressing proteins for 9 h were treated with OSU-03012 for an additional 12 h and cell viability was determined. The right panel shows Western blot detection of overexpressed proteins. (D) Lack of attenuation of OSU-03012-induced cell death by overexpression of kinase-deficient Pdk1K77Q or Adi3K337Q. The right panel shows Western blot detection of overexpressed proteins. (E) Agrobacterium-mediated overexpression of Adi3 can suppress cell death induced by PtoY207D in leaf tissue. The lower panel shows Western blot detection of overexpressed proteins; HA-Adi3, filled triangle; Pto-HA, open triangle.
OSU-03012 induces cell death in mammalian systems and this response can be reduced by overexpression of constitutively active Pdk1 or PKB (Zhu et al, 2004). We found that OSU-03012 reduced viability of tomato protoplasts in a dose-dependent manner over a 24-h period (Figure 5B). The ability of Pdk1, Adi3, and constitutively active Adi3S539D overexpression to reduce OSU-03012-induced cell death was examined in tomato protoplasts. Overexpression of the proteins was confirmed by Western blot (Figure 5C, right panels). Adi3S539D offered partial yet significant reduction in cell death, whereas Pdk1 and Adi3 gave marginal cell death reduction in response to OSU-03012 (Figure 5C). Other studies using a constitutively active Pdk1 showed more reduction in cell death (Kulp et al, 2004; Zhu et al, 2004). Our Pdk1 was not constitutively active and this may explain the low level of protection against OSU-03012. We have yet to identify the residues required to make such a tomato Pdk1 protein. One study using a Pdk1 inhibitor and overexpression of wild-type Pdk1 did show a similar level of protection against the inhibitor as we have seen (Arico et al, 2002). Induction of cell death at high levels of OSU-03012 prevented overexpression of any protein from protecting against the inhibitor. Overexpression of kinase-inactive Pdk1K77Q and Adi3K337Q did not protect protoplasts from OSU-03012-induced cell death (Figure 5D). These results are consistent with PDK1 inhibitor studies on mammalian PKB and Pdk1 and suggest that OSU-03012-induced plant cell death is related to the inhibition of Pdk1 activity and that Adi3 function lies downstream of Pdk1 (Arico et al, 2002; Kulp et al, 2004; Zhu et al, 2004).
The ability of Adi3 to suppress cell death in leaves was investigated by using Agrobacterium-mediated transient overexpression of Adi3 in the presence of PtoY207D, a constitutively active Pto that produces HR-like cell death in the absence of AvrPto (Rathjen et al, 1999). Overexpression of Adi3 suppressed PtoY207D-induced cell death in 28 of 34 trials (Figure 5E). Adi3 kinase activity was required for this cell death suppression because Adi3K337Q was not able to suppress PtoY207D-induced cell death in 22 of 25 trials (Figure 5E).
Adi3 silencing-induced cell death is abolished by silencing of a MAPKKK gene known to play a role in pathogen-associated cell death
To investigate whether cell death lesions formed upon Adi3 silencing are related to PCD associated with pathogen attack, Adi3 was co-silenced with MAPKKKα. The latter gene has recently been shown to regulate PCD associated with both disease susceptibility and Pto/AvrPto-mediated disease resistance (del Pozo et al, 2004). Adi3 and MAPKKKα were silenced individually and together in separate tomato plants and the formation of cell death lesions was monitored as silencing progressed. As expected, silencing of Adi3 alone caused cell death, whereas silencing of MAPKKKα alone did not. Significantly, co-silencing of Adi3/MAPKKKα completely eliminated the spontaneous formation of cell death lesions (Figure 6A) and the usual stunting caused by Adi3 silencing was not observed (Supplementary Figure S3). Adi3 and MAPKKKα transcript levels were not affected when analyzed from MAPKKKα- and Adi3-silenced plants, respectively, and in co-silenced plants both genes were effectively silenced (Figure 6B). Overexpression of Adi3 in leaf tissue did not affect the Pto/AvrPto induction of a MAPK known to be activated by MAPKKKα (Supplementary Figure S4; del Pozo et al, 2004). Also, Adi3 and MAPKKKα do not interact in a Y2H assay (data not shown). These data suggest that Adi3 and MAPKKKα act in parallel, interdependent pathways.
Figure 6.
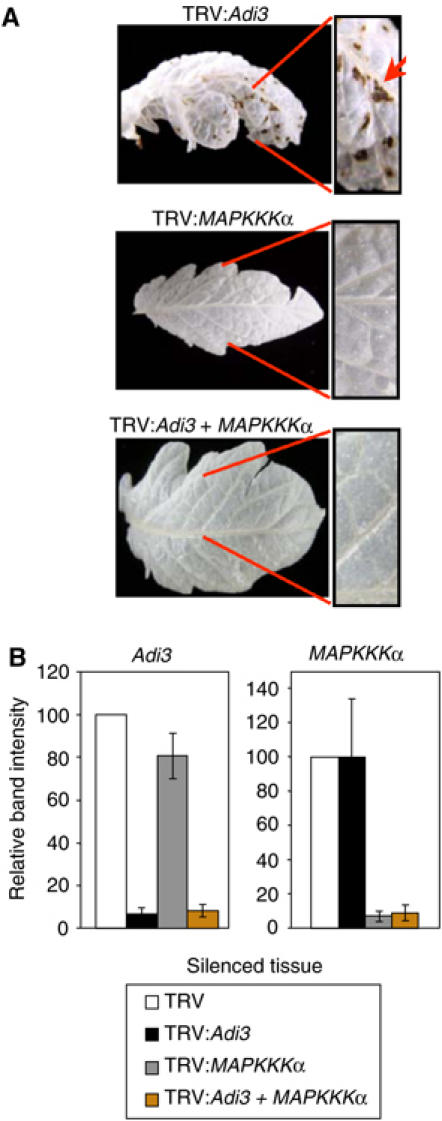
Co-silencing of Adi3 and MAPKKKα abolishes Adi3 silencing-induced cell death. (A) Adi3/MAPKKKα co-silencing eliminates cell death caused by Adi3 silencing. Adi3 and MAPKKKα were silenced in tomato plants. Pictures of Adi3/MAPKKKα co-silencing represent two independent silencing experiments. Leaves were cleared in 10% acetic acid, 30% chloroform, 60% EtOH. (B) RT–PCR analysis of Adi3/MAPKKKα co-silencing. Tomato plants were silenced for the indicated gene and analyzed for transcript levels by RT–PCR. Abundance of PCR products from PCR cycle 32 was quantitated by spot densitometry and graphed.
Discussion
Adi3 and Pdk1 regulate plant cell death
We present several lines of evidence that indicate that Pdk1/Adi3 are part of a general pathway regulating cell death in plants. Loss-of-function analysis of Adi3 and Pdk1 using VIGS results in localized cell death and whole plant death, respectively. The Pdk1 inhibitor OSU-03012 induces cell death in tomato protoplasts that can be attenuated by overexpression of wild-type Pdk1, Adi3, or a constitutively active Adi3. These findings bear striking similarities to observations with mammalian Pdk1 and PKB proteins. PKB knockout mice display spontaneous apoptosis in several different tissues, whereas Pdk1 knockout mice do not survive past embryonic day 9.5 (Chen et al, 2001; Lawlor et al, 2002). Expression of a kinase-deficient form of PKB produces cell death in cerebellar neurons (Dudek et al, 1997; Luo et al, 2003). In addition, the Pdk1 inhibitors celecoxib and OSU-03012 induce mammalian cell death, which can be attenuated by the expression of wild-type Pdk1 and constitutively active forms of Pdk1 and PKB (Arico et al, 2002; Kulp et al, 2004; Zhu et al, 2004). Overexpression of wild-type PKB can also reduce cell death in the presence of other apoptosis-inducing conditions (low KCl, minus serum, or plus NMDA; Dudek et al, 1997; Luo et al, 2003). These convergent observations support a role for Pdk1 and Adi3 as negative regulators of plant cell death, analogous to the role of Pdk1 and PKB in mammalian systems. In spite of these functional similarities, Adi3 and PKB have little sequence similarity (21.4% amino-acid identity over the full protein, 34.6% over the kinase domains, data not shown), suggesting that there are important regulatory and functional differences between plant and animal systems.
Adi3 interaction with Pdk1 shows similarities and differences with mammalian systems
The activity of PKB is regulated by the induced phosphorylation of S473 in the PIF motif (FxxFSY; known as the PDK2 site), which allows for interaction with and activation by Pdk1 (Vara et al, 2004). Other AGC kinases such as S6K and SGK are regulated in a similar manner. However, the interaction of AGC kinases PRK2 and PKCζ with Pdk1 does not appear to be regulated by induced PIF phosphorylation, as they contain an Asp and Glu, respectively, in place of the Ser/Thr in the PIF. These acidic residues appear to mimic the phosphorylated Ser/Thr, as they are absolutely required for Pdk1 interaction and phosphorylation (Balendran et al, 2000).
Our results indicate that Pdk1 and Adi3 interact through the Adi3 PIF. This interaction with Pdk1 appears to be similar to PRK2/PKCζ and not to the induced PIF phosphorylation of PKB, as the Adi3 PIF (FExF) contains an acidic Glu (E698) instead of a phosphorylatable Ser/Thr. However, Adi3 residue E698 appears to be only partially required for Pdk1 interaction, whereas F697 and F700 appear to be more important. Mutation of all three residues to Ala did not completely abolish Pdk1 interaction, suggesting the involvement of additional amino acids. The only other plant PIF motifs to be analyzed are those of AGC1-1 (FDxF) and AGC2-1 (FxxF) (Anthony et al, 2004). The Phe residues in these PIFs were shown to be absolutely required for interaction with Pdk1, confirming that the need of acidic or phosphorylated residues in this motif is not necessarily required for plant AGC kinases.
The mammalian model for Pdk1/AGC kinase interaction predicts that, after PIF binding, the AGC kinase is phosphorylated by Pdk1, which stabilizes and activates the AGC kinase. This stabilization makes the PIF-binding pocket of the AGC kinase available for binding its own PIF, allowing for release from Pdk1 (Biondi, 2004). In this manner, Pdk1 can only bind an inactive kinase with a free PIF. Our Y2H data do not support this model for Adi3, as the constitutively active Adi3S539D protein retains interaction with Pdk1. This suggests that even when activated, the Adi3 PIF is still available for Pdk1 interaction or that phosphorylation of other Adi3 residues contributes to regulation of Pdk1 interaction. We have shown that at least one additional Ser residue in Adi3, in addition to S539, is phosphorylated by Pdk1. Identification and analysis of this residue may show that it is involved in the interaction with Pdk1. Alternatively, other factors distinct from the PIF might regulate the interaction with Pdk1. In support of this idea, some plant AGC kinases such as PHOT1 and PHOT2 contain the conserved Pdk1 phosphorylation site but not a PIF (Bögre et al, 2003), suggesting alternate mechanisms for interaction. This represents a potentially significant difference in the underlying mechanism for the interaction of Pdk1 and plant AGC kinases as compared with mammalian systems.
The Adi3 PIF also regulates autophosphorylation (Figure 3C), suggesting the presence of an Adi3 PIF-binding pocket. A recent review suggested that most plant AGC kinases lack a PIF-binding pocket (Bögre et al, 2003). Owing to low sequence identity, a PIF-binding pocket is not obvious in the Adi3 sequence (Figure 3A). However, specific residues in mammalian PIF-binding pockets known to interact with PIF residues are conserved in Adi3 (Figure 3A). Of these residues, R357, L383, and F308 appear to be the most important for Adi3 autophosphorylation, suggesting that they are important for self-binding of the PIF. This is in agreement with previous studies, which indicate that L155 and F82 of human Pdk1 (L383 and F308 in Adi3) interact with the first and last Phe, respectively, in the PIF (Biondi et al, 2000; Frödin et al, 2002). Human Pdk1 R131 (R357 in Adi3) is one of four residues that interact with the phosphorylated Ser/Thr in the PIF (Frödin et al, 2002) and is the only residue of the four conserved in Adi3. This difference may be due to the substitution of phosphorylatable Ser/Thr with a Glu (E698) in the Adi3 PIF, as discussed above. Mutation of the other potentially important residues in the Adi3 PIF-binding pocket also affects autophosphorylation (Figure 3E). However, as conformational changes due to these mutations cannot be discounted, a more detailed study is required to determine the true binding nature of these residues to the Adi3 PIF.
In contrast to most mammalian AGC kinases, mutation of the Pdk1 phosphorylation site in Adi3 (S539) to Ala does not abolish autophosphorylation activity (Figure 1A; Parekh et al, 2000). This result raises the question as to whether Adi3 is a true substrate of Pdk1. Two observations suggest that it is. First, mutation of Adi3 S539 to Asp greatly enhances its autophosphorylation activity (Figure 1A), indicating that phosphorylation of this site is required for full activity of Adi3. Second, we have shown that S539 is one of at least two Pdk1 phosphorylation sites on Adi3 (Figure 2C and F) and that Adi3 and Pdk1 interact in a PIF-dependent manner (Figure 3A and B). Interestingly, mutation of the Pdk1 phosphorylation site of PKCδ (T505) to Ala also does not affect catalytic activity (Stempka et al, 1997; Parekh et al, 2000) although PKCδ is known to be a Pdk1 substrate (Le Good et al, 1998; Parekh et al, 2000).
Our Y2H data suggest that the Adi3/Pdk1 interaction is constitutive (Figure 2B), making it possible that Pdk1 phosphorylation of Adi3 is constitutive. Other AGC kinases, such as conventional PKC family members, are constitutively phosphorylated by Pdk1 but remain in an inactive conformation until interaction with second messengers (Newton, 2003). Adi3 may not be regulated in this manner, as the Adi3S539D protein, which mimics Pdk1 phosphorylation, is constitutively active (Figure 1A). Thus, the activity of Pdk1-phosphorylated Adi3 may be regulated through the action of phosphatases.
Adi3 function may be subverted for induction of cell death in response to pathogen attack
Four observations suggest that loss of Adi3 function contributes to cell death associated with the Pto/AvrPto-mediated host response. First, Adi3 interacts specifically in the Y3H system only when functional forms of both AvrPto and Pto are present. Second, Pto phosphorylates Adi3. Third, suppression of Adi3 gene expression causes formation of spontaneous necrotic lesions on plant leaves; similar localized cell death develops during the defense response (the HR) and disease progression. Fourth, co-silencing of MAPKKKα with Adi3 suppresses Adi3-related cell death, indicating that this MAPKKK plays a role downstream of Adi3; MAPKKKα is known to be a positive regulator of cell death associated with Pst-related immunity and disease (del Pozo et al, 2004).
AGC kinases are central regulators of many eukaryotic cellular processes and it is perhaps not surprising that these proteins are targeted for manipulation by pathogens. In mammals, type III effector proteins YopH from Yersinia enterocolitica and ExoS and OdDHL from Pseudomonas aeruginosa inhibit activation of PKB, presumably for induction of apoptosis (Henriksson et al, 2000; Sauvonnet et al, 2002; Li et al, 2004). In addition, the Y. enterocolitica effector protein YopM forms a complex with AGC kinases PRK2 and RSK, although the function of this interaction is not yet known (McDonald et al, 2003). Our discovery that the Pseudomonas type III effector AvrPto interacts with Adi3/Pto suggests that pathogens may also target plant AGC kinases to manipulate PCD in a manner similar to the targeting of PKB by mammalian bacterial pathogens. Considering that PKB regulates cell death in response to many different stimuli (Luo et al, 2003), it would not be surprising for Adi3 to be involved in cell death control during both disease and resistance, given its apparent upstream regulatory position.
A model for Adi3 control of plant cell death
Based on our results and by analogy with Pdk1/PKB, we propose a model that suggests that in unchallenged plant cells, Adi3 is phosphorylated and activated by Pdk1 (Figure 7). This phosphorylation would activate the negative regulatory function of Adi3 and inhibit PCD through the activation of PCD inhibitors or the inactivation of PCD initiators. PKB acts similarly by phosphorylating substrates ranging from the apoptotic inhibitors NF-κB, CREB, and IKK to proapoptotic proteins such as BAD, caspase-9, and the Forkhead family transcription factor FKHR (Vivanco and Sawyers, 2002). However, the Adi3 recognition site in potential substrates may be different from that of PKB, as Adi3 is not capable of phosphorylating an artificial PKB substrate (data not shown). The mechanisms regulating the Pdk1/Adi3 pathway, and the identity and role of Adi3 substrates, are unknown and will be important goals for future research.
Figure 7.
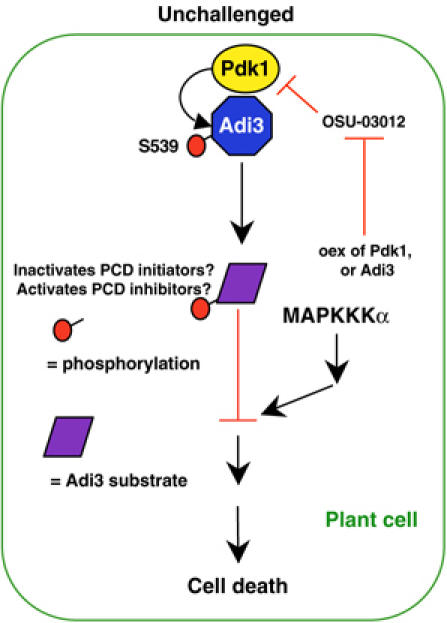
Model for Pdk1/Adi3 cell death regulation. Negative regulation of plant cell death is initiated by Pdk1 phosphorylation of Adi3 at S539. Adi3 may then activate/inactivate PCD inhibitors/initiators by phosphorylation, preventing cell death through a parallel MAPKKKα pathway. OSU-03012-induced cell death can be overcome by overexpression of Pdk1 or the constitutively active Adi3S539D.
Our co-silencing and MAPK assay results suggest that Adi3 acts in parallel to MAPKKKα to inhibit a MAPK cascade. This role is consistent with evidence from both mammalian and plant systems that AGC kinases signal through MAPK cascades. In mammalian cells, EGF activation of the Raf-1/MEK1,2 cascade has been shown to signal through PKCζ in a Pdk1-dependent manner, with PKCζ acting upstream of Raf-1 (Corbit et al, 2000, 2003). Other studies contradict PKCζ involvement, showing that Pdk1 directly phosphorylates MEK1 and MEK2 (Sato et al, 2004). Arabidopsis AGC kinase OXI1/AGC2-1 signaling utilizes MPK3 and MPK6 to mediate oxidative bust signals generated during pathogen defense and cellular development (Rentel et al, 2004). Further study is needed to determine the extent to which regulation of the Adi3 pathway might be a master switch controlling various MAPK-controlled host cell death pathways.
Materials and methods
Adi3 and Pdk1 cDNA cloning
The full-length Adi3 cDNA was cloned using a combination of screening the tomato EST database with the 776-bp Adi3 Y3H cDNA fragment (Bogdanove and Martin, 2000) and 5′ RACE. More specifics on Adi3 cDNA cloning can be found in Supplementary data.
Tomato Pdk1 was identified by searching the tomato EST database (www.tigr.org) using the Arabidopsis Pdk1 sequence (Deak et al, 1999). More specifics on Pdk1 cDNA cloning can be found in Supplementary data.
Yeast two- and three-hybrid assays
Y2H and Y3H assays were carried out as previously reported (Bogdanove and Martin, 2000) and detailed in Supplementary data using the pEG202 vector for bait constructs and the pJG4-5 vector for prey constructs.
Protein expression, kinase assays, and in vitro co-immunoprecipitation
MBP translational fusion proteins were produced using the pMAL-c2 vector (New England Biolabs) and FLAG translational fusion proteins were produced using the pFLAG-CTC vector (Sigma). In vitro kinase assays were carried out on MBP translational fusion proteins. MBP-Adi3 and Pdk1-FLAG proteins were co-immunoprecipitated using α-FLAG M2 agarose. Specifics can be found in Supplementary data.
Phosphoamino acid analysis and phosphopeptide mapping
Phosphoamino acid analysis and phosphopeptide mapping were carried out as described in Supplementary data.
Virus-induced gene silencing
Silencing was carried out using the pTRV1 and pTRV2 vectors with cloning by the Gateway system (Invitrogen) as described (Ekengren et al, 2003). Gene fragments used for VIGS are described in Supplementary data.
OSU-03012 inhibition of Pdk1 and treatment of protoplast
Tomato protoplast isolation was performed as described in Supplementary data using 3-week-old Rio Grande-PtoR leaves. OSU-03012 was obtained from Dr Ching-Shih Chen at The Ohio State University and dissolved in DMSO. Protoplast treatment and transformation are described in Supplementary data.
Agrobacterium-mediated transient expression
Agrobacterium tumefaciens strain GV2260 was used for Agrobacterium-mediated transient expression in Nicotiana benthamiana leaves as reported (del Pozo et al, 2004). HA-Adi3 (0.5 OD600) and PtoY207D-HA (0.05 OD600) were infiltrated at a final density of 0.55 OD600 for expression from the constitutive 35S CaMV promoter in the binary vector pCAMBIA (HA-Adi3) or pBTEX (PtoY207D-HA).
Supplementary Material
Supplementary Data
Acknowledgments
We thank Dr Patrick Giavalisco, Rob Abramovitch, Jeff Anderson, and Tracy Rosebrock for critical reading of the manuscript. This work was supported, in part, by USDA-NRI grant 2002-3501 (TPD and GBM) and NSF grant DBI-0116076 (GBM).
References
- Abramovitch RB, Martin GB (2004) Strategies used by bacterial pathogens to suppress plant defenses. Curr Opin Plant Biol 7: 356–364 [DOI] [PubMed] [Google Scholar]
- Anthony RG, Henriques R, Helfer A, Meszaros T, Rios G, Testerink C, Munnik T, Deak M, Koncz C, Bögre L (2004) A protein kinase target of a PDK1 signalling pathway is involved in root hair growth in Arabidopsis. EMBO J 23: 572–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arico S, Pattingre S, Bauvy C, Gane P, Barbat A, Codogno P, Ogier-Denis E (2002) Celecoxib induces apoptosis by inhibiting 3-phosphoinositide-dependent protein kinase-1 activity in the human colon cancer HT-29 cell line. J Biol Chem 277: 27613–27621 [DOI] [PubMed] [Google Scholar]
- Balendran A, Biondi RM, Cheung PC, Casamayor A, Deak M, Alessi DR (2000) A 3-phosphoinositide-dependent protein kinase-1 (PDK1) docking site is required for the phosphorylation of protein kinase Cζ (PKCζ) and PKC-related kinase 2 by PDK1. J Biol Chem 275: 20806–20813 [DOI] [PubMed] [Google Scholar]
- Belham C, Wu S, Avruch J (1999) Intracellular signalling: PDK1—a kinase at the hub of things. Curr Biol 9: R93–R96 [DOI] [PubMed] [Google Scholar]
- Benjamins R, Quint A, Weijers D, Hooykaas P, Offringa R (2001) The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128: 4057–4067 [DOI] [PubMed] [Google Scholar]
- Biondi RM (2004) Phosphoinositide-dependent protein kinase 1, a sensor of protein conformation. Trends Biochem Sci 29: 136–142 [DOI] [PubMed] [Google Scholar]
- Biondi RM, Cheung PC, Casamayor A, Deak M, Currie RA, Alessi DR (2000) Identification of a pocket in the PDK1 kinase domain that interacts with PIF and the C-terminal residues of PKA. EMBO J 19: 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biondi RM, Nebreda AR (2003) Signalling specificity of Ser/Thr protein kinases through docking-site-mediated interactions. Biochem J 372: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanove AJ, Martin GB (2000) AvrPto-dependent Pto-interacting proteins and AvrPto-interacting proteins in tomato. Proc Natl Acad Sci USA 97: 8836–8840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bögre L, Okresz L, Henriques R, Anthony RG (2003) Growth signalling pathways in Arabidopsis and the AGC protein kinases. Trends Plant Sci 8: 424–431 [DOI] [PubMed] [Google Scholar]
- Briggs WR, Christie JM (2002) Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci 7: 204–210 [DOI] [PubMed] [Google Scholar]
- Chen WS, Xu PZ, Gottlob K, Chen ML, Sokol K, Shiyanova T, Roninson I, Weng W, Suzuki R, Tobe K, Kadowaki T, Hay N (2001) Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt1 gene. Genes Dev 15: 2203–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SK, Dagenais N, Chory J, Weigel D (2000) Regulation of auxin response by the protein kinase PINOID. Cell 100: 469–478 [DOI] [PubMed] [Google Scholar]
- Corbit KC, Soh J-W, Yoshida K, Eves EM, Weinstein IB, Rosner MR (2000) Different protein kinase C isoforms determine growth factor specificity in neuronal cells. Mol Cell Biol 20: 5392–5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbit KC, Trakul N, Eves EM, Diaz B, Marshall M, Rosner MR (2003) Activation of Raf-1 signaling by protein kinase C through a mechanism involving Raf kinase inhibitory protein. J Biol Chem 278: 13061–13068 [DOI] [PubMed] [Google Scholar]
- Deak M, Casamayor A, Currie RA, Downes CP, Alessi DR (1999) Characterization of a plant 3-phosphoinositide-dependent protein kinase-1 homologue which contains a pleckstrin homology domain. FEBS Lett 451: 220–226 [DOI] [PubMed] [Google Scholar]
- del Pozo O, Pedley KF, Martin GB (2004) MAPKKKα is a positive regulator of cell death associated with both plant immunity and disease. EMBO J 23: 3072–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudek H, Datta SR, Franke TF, Birnbaum MJ, Yao R, Cooper GM, Segal RA, Kaplan DR, Greenberg ME (1997) Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science 275: 661–665 [DOI] [PubMed] [Google Scholar]
- Ekengren SK, Liu Y, Schiff M, Dinesh-Kumar SP, Martin GB (2003) Two MAPK cascades, NPR1, and TGA transcription factors play a role in Pto-mediated disease resistance in tomato. Plant J 36: 905–917 [DOI] [PubMed] [Google Scholar]
- Frederick RD, Thilmony RL, Sessa G, Martin GB (1998) Recognition specificity for the bacterial avirulence protein AvrPto is determined by Thr-204 in the activation loop of the tomato Pto kinase. Mol Cell 2: 241–245 [DOI] [PubMed] [Google Scholar]
- Frödin M, Antal TL, Dummler BA, Jensen CJ, Deak M, Gammeltoft S, Biondi RM (2002) A phosphoserine/threonine-binding pocket in AGC kinases and PDK1 mediates activation by hydrophobic motif phosphorylation. EMBO J 21: 5396–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman RN, Novacky AJ (1994) The Hypersensitive Reaction in Plants to Pathogens: A Resistance Phenomenon. St Paul: The American Phytopathological Society Press [Google Scholar]
- Greenberg JT, Yao N (2004) The role and regulation of programmed cell death in plant–pathogen interactions. Cell Microbiol 6: 201–211 [DOI] [PubMed] [Google Scholar]
- Henriksson ML, Rosqvist R, Telepnev M, Wolf-Watz H, Hallberg B (2000) Ras effector pathway activation by epidermal growth factor is inhibited in vivo by exoenzyme S ADP-ribosylation of Ras. Biochem J 347 (Part 1): 217–222 [PMC free article] [PubMed] [Google Scholar]
- Kulp SK, Yang Y-T, Hung C-C, Chen K-F, Lai J-P, Tseng P-H, Fowble JW, Ward PJ, Chen C-S (2004) 3-Phosphoinositide-dependent protein kinase-1/Akt signaling represents a major cyclooxygenase-2-independent target for celecoxib in prostate cancer cells. Cancer Res 64: 1444–1451 [DOI] [PubMed] [Google Scholar]
- Lam E (2004) Controlled cell death, plant survival and development. Nat Rev Mol Cell Biol 5: 305–315 [DOI] [PubMed] [Google Scholar]
- Lawlor MA, Mora A, Ashby PR, Williams MR, Murray-Tait V, Malone L, Prescott AR, Lucocq JM, Alessi DR (2002) Essential role of PDK1 in regulating cell size and development in mice. EMBO J 21: 3728–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Good JA, Ziegler WH, Parekh DB, Alessi DR, Cohen P, Parker PJ (1998) Protein kinase C isotypes controlled by phosphoinositide 3-kinase through the protein kinase PDK1. Science 281: 2042–2045 [DOI] [PubMed] [Google Scholar]
- Li L, Hooi D, Chhabra SR, Pritchard D, Shaw PE (2004) Bacterial N-acylhomoserine lactone-induced apoptosis in breast carcinoma cells correlated with down-modulation of STAT3. Oncogene 23: 4894–4902 [DOI] [PubMed] [Google Scholar]
- Lincoln JE, Richael C, Overduin B, Smith K, Bostock R, Gilchrist DG (2002) Expression of the antiapoptotic baculovirus p35 gene in tomato blocks programmed cell death and provides broad-spectrum resistance to disease. Proc Natl Acad Sci USA 99: 15217–15221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh Y-T, Martin GB (1995) The Pto bacterial resistance gene and the Fen insecticide sensitivity gene encode functional protein kinases with serine/threonine specificity. Plant Physiol 108: 1735–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo HR, Hattori H, Hossain MA, Hester L, Huang Y, Lee-Kwon W, Donowitz M, Nagata E, Snyder SH (2003) Akt as a mediator of cell death. Proc Natl Acad Sci USA 100: 11712–11717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald C, Vacratsis PO, Bliska JB, Dixon JE (2003) The Yersinia virulence factor YopM forms a novel protein complex with two cellular kinases. J Biol Chem 278: 18514–18523 [DOI] [PubMed] [Google Scholar]
- Newton AC (2003) Regulation of the ABC kinases by phosphorylation: protein kinase C as a paradigm. Biochem J 370: 361–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson DW, Thornberry NA (2003) Apoptosis. Life and death decisions. Science 299: 214–215 [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K (2002) The IRE gene encodes a protein kinase homologue and modulates root hair growth in Arabidopsis. Plant J 30: 289–299 [DOI] [PubMed] [Google Scholar]
- Parekh DB, Ziegler W, Parker PJ (2000) Multiple pathways control protein kinase C phosphorylation. EMBO J 19: 496–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedley KF, Martin GB (2003) Molecular basis of Pto-mediated resistance to bacterial speck disease. Annu Rev Phytopathol 41: 215–243 [DOI] [PubMed] [Google Scholar]
- Rathjen JP, Chang JH, Staskawicz BJ, Michelmore RW (1999) Constitutively active Pto induces a Prf-dependent hypersensitive response in the absence of AvrPto. EMBO J 18: 3232–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentel MC, Lecourieux D, Ouaked F, Usher SL, Peterson L, Okamoto H, Knight H, Peck SC, Grierson CS, Hirt H, Knight MR (2004) OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis. Nature 427: 858–861 [DOI] [PubMed] [Google Scholar]
- Sato S, Fujita N, Tsuruo T (2004) Involvement of 3-phosphoinositide-dependent protein kinase-1 in the MEK/MAPK signal transduction pathway. J Biol Chem 279: 33759–33767 [DOI] [PubMed] [Google Scholar]
- Sauvonnet N, Lambermont I, van der Bruggen P, Cornelis GR (2002) YopH prevents monocyte chemoattractant protein 1 expression in macrophages and T-cell proliferation through inactivation of the phosphatidylinositol 3-kinase pathway. Mol Microbiol 45: 805–815 [DOI] [PubMed] [Google Scholar]
- Shah J, Kachroo P, Klessig DF (1999) The Arabidopsis ssi1 mutation restores pathogenesis-related gene expression in npr1 plants and renders defensin gene expression salicylic acid dependent. Plant Cell 11: 191–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stempka L, Girod A, Muller HJ, Rincke G, Marks F, Gschwendt M, Bossemeyer D (1997) Phosphorylation of protein kinase Cδ (PKCδ) at threonine 505 is not a prerequisite for enzymatic activity. Expression of rat PKCδ and an alanine 505 mutant in bacteria in a functional form. J Biol Chem 272: 6805–6811 [DOI] [PubMed] [Google Scholar]
- Tamaskovic R, Bichsel SJ, Hemmings BA (2003) NDR family of AGC kinases—essential regulators of the cell cycle and morphogenesis. FEBS Lett 546: 73–80 [DOI] [PubMed] [Google Scholar]
- Tang X, Frederick RD, Zhou J, Halterman DA, Jia Y, Martin GB (1996) Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science 274: 2060–2063 [DOI] [PubMed] [Google Scholar]
- Vara JAF, Casado E, de Castro J, Cejas P, Belda-Iniesta C, Gonzales-Baron M (2004) PI3K/Akt signalling pathway and cancer. Cancer Treat Rev 30: 193–204 [DOI] [PubMed] [Google Scholar]
- Vivanco I, Sawyers CL (2002) The phosphatidylinositol 3-kinase–AKT pathway in human cancer. Nat Rev Cancer 2: 489–501 [DOI] [PubMed] [Google Scholar]
- Zhu J, Huang J-W, Tseng P-H, Yang Y-T, Fowble J, Shiau C-W, Shaw Y-J, Kulp SK, Chen C-S (2004) From the cyclooxygenase-2 inhibitor celecoxib to a novel class of 3-phosphoinositide-dependent protein kinase-1 inhibitors. Cancer Res 64: 4309–4318 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data


