Abstract
The polypyrimidine tract binding protein (PTB) is a 58 kDa protein involved in many aspects of RNA metabolism. In this study, we focused our attention on the structure of the two C-terminal RNA recognition motifs (RRM3 and RRM4) of PTB. In a previous study, it was found that the two RRMs are independent in the free state. We recently determined the structure of the same fragment in complex with RNA and found that the two RRMs interact extensively. This difference made us re-evaluate in detail the free protein structure and in particular the interdomain interface. We used a combination of NMR spectroscopy and segmental isotopic labeling to unambiguously study and characterize the interdomain interactions. An improved segmental isotopic labeling protocol was used, enabling us to unambiguously identify 130 interdomain NOEs between the two RRMs and to calculate a very precise structure. The structure reveals a large interdomain interface, resulting in a very unusual positioning of the two RRM domains relative to one another.
Keywords: isotopic labeling, NMR, protein ligation, RRM/RBD/RNP, segmental labeling
Introduction
The polypyrimidine tract binding protein (PTB) is a 58 kDa protein containing four RNA recognition motifs of the RNP/RRM/RBD type (Varani and Nagai, 1998; Maris et al, 2005). PTB is involved in many aspects of RNA metabolism, including splicing regulation (Wagner and Garcia-Blanco, 2001). PTB acts as a negative regulator of splicing for several alternative exons. In addition, PTB is involved in other processes: internal ribosome entry site (IRES)-mediated translation initiation (Hellen and Sarnow, 2001), 3′-end processing (Castelo-Branco et al, 2004) and mRNA stability (Knoch et al, 2004). How PTB acts in all these diverse cellular processes is poorly understood. PTB binds to single-stranded RNA polypyrimidine tracts, with an apparent preference for the sequences UCUUC (Perez et al, 1997), CUCUCU and UUCUCU that are usually present in multiple copies within introns (Zhang et al, 1996; Gooding et al, 1998; Southby et al, 1998; Modaferri and Black, 1999; Carstens et al, 2000; Yuan et al, 2002) or IRES sequences (Kolupaeva et al, 1996; Pilipenko et al, 2001).
RRMs are the most abundant RNA binding domains and are composed of a four-stranded antiparallel β-sheet packed against two α-helices (Nagai et al, 1995; Varani and Nagai, 1998). The structures of all four RRMs of PTB were determined by NMR spectroscopy. RRM1 and RRM4 adopt the canonical RRM structure, whereas RRM2 and RRM3 are extended by an additional fifth β-strand (Conte et al, 2000; Simpson et al, 2004). These structural studies suggested that all four RRMs of PTB are independent from each other and separated by flexible linkers. We recently determined the structure of PTB in complex with RNA and observed that RRM3 and RRM4 of PTB interact extensively, thereby positioning the bound RNAs in an antiparallel fashion (Oberstrass et al, 2005). Here, we aimed at elucidating whether this interaction is induced by RNA binding, as is the case for Nucleolin (Allain et al, 2000a, 2000b) and Sex-lethal (Handa et al, 1999), or whether it is present in the free protein as well like for hnRNPA1 (Deo et al, 1999).
To shed light on this structural question, we used NMR spectroscopy to study a protein construct containing RRM3 and RRM4 (RRM34). To make our study unambiguous and to precisely characterize the interdomain interactions, we prepared several segmentally labeled RRM34 by expressed protein ligation (EPL) (Muir et al, 1998; Severinov and Muir, 1998). Segmental isotopic labeling is potentially a very powerful method for the structure determination of large proteins by NMR, because the problem of overlapping signals can be partially overcome. Labeling a single domain within a multidomain protein requires the separate expression of two protein fragments that are subsequently ligated in order to generate the full-length protein. Protein ligation requires a C-terminal thioester group and an N-terminal α-cysteine at the ends of the protein fragments. Such protein termini can be generated by expressing the protein fragments in fusion with a full-length or truncated intein and subsequently inducing intein cleavage (Xu et al, 2000; Muir, 2003). The EPL approach was applied in a few cases to prepare segmentally labeled NMR samples (Yamazaki et al, 1998; Otomo et al, 1999a, 1999b; Xu et al, 1999; Camarero et al, 2002; Zuger and Iwai, 2005) but was never used to determine a protein structure.
Here, we adapted an intein-based protein ligation protocol originally developed to generate cyclic protein (Evans et al, 1999) to produce several PTB RRM34 constructs, in which only one of the two RRMs was 13C–15N-labeled. Our EPL approach is more efficient in terms of reaction time and overall yield than previously published EPL procedures used for NMR applications. Our segmentally labeled proteins enabled us to unambiguously identify 130 interdomain NOEs between RRM3 and RRM4 of PTB in its free state and to calculate a highly precise structure. The structure reveals a large interdomain interface resulting in a very unusual positioning of the two domains relative to each other.
Results
PTB RRM1 and RRM2 are independent whereas RRM3 and RRM4 are interdependent
The free form of PTB has been investigated by NMR spectroscopy (Conte et al, 2000; Simpson et al, 2004) and more recently by our group in complex with RNA (Oberstrass et al, 2005). We further studied the free protein in solution in order to compare the structures of free PTB and that bound to RNA. The full-length PTB (58 kDa) can be observed by NMR spectroscopy at low salt concentration, 30°C and acidic pH (5.8) (Figures 1 and 2A). In order to identify protein resonances of the full-length PTB, we expressed each RRM individually (Figure 1) and collected 15N-TROSY and/or 15N-HSQC spectra (Figure 2). Interestingly, when the 15N-TROSY spectra of RRM1, RRM2 and RRM34 are superimposed, the resulting 15N-TROSY spectrum (Figure 2B) is almost identical to the spectrum of the full-length protein (Figure 2A). This indicates that RRM1 and RRM2 are independent from the rest of PTB. When RRM3 and RRM4 are expressed separately (Figure 1), comparison of the single-domain 15N-HSQC spectra (Figure 2D) with the 15N-HSQC of RRM34 (Figure 2C) shows significant differences, especially for the resonances of RRM3. This suggests that RRM3 and RRM4 are not independent. The backbone resonances of RRM4 in isolation were assigned, and when compared with the assignments of RRM34 (Conte et al, 1999), we observed significant chemical shift differences in the resonances of the α-helix 2 of RRM4 (data not shown). Based on these results, we decided to re-evaluate the structure of RRM34 that was previously determined but did not reveal any interdomain interaction between the RRMs (Conte et al, 2000). In order to determine our structure with the highest precision, we prepared segmentally labeled RRM34 using the EPL approach (Severinov and Muir, 1998).
Figure 1.
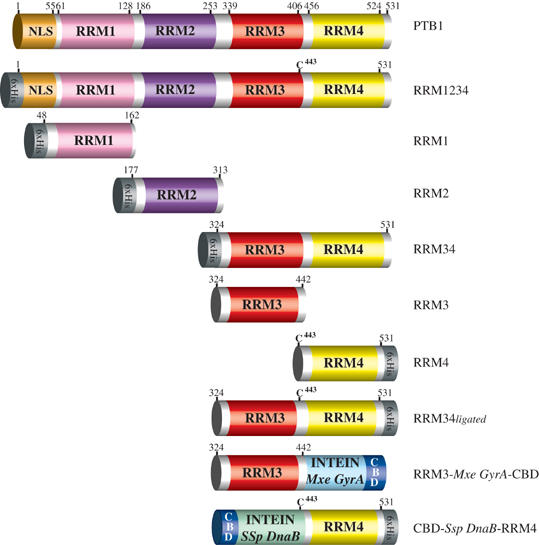
Protein constructs used in this study. Protein domains are represented by colored boxes: orange for the NLS sequence; white for linker regions; pink for PTB RRM1; violet for PTB RRM2; red for PTB RRM3 domain; yellow for PTB RRM4; gray for 6 × His tag; blue for CBD; light blue for the Mxe GyrA intein; and green for the Ssp DnaB intein.
Figure 2.
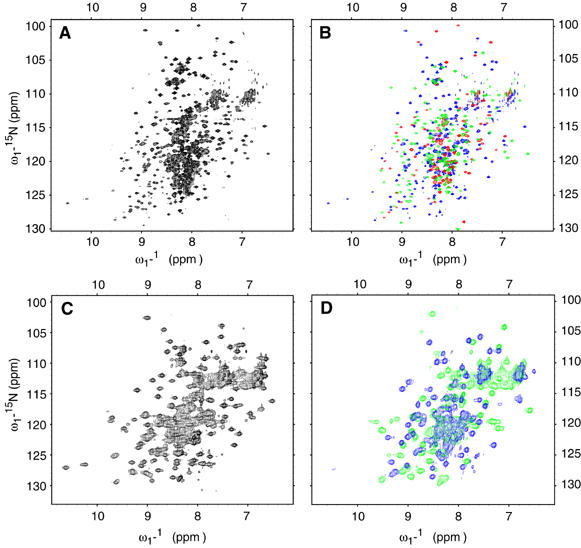
NMR spectra of full-length PTB and several PTB subfragments. (A) 15N-TROSY of full–length PTB recorded at 900 MHz, 303 K, pH 5.8. (B) Overlay of 15N-TROSY of PTB RRM1 (red), RRM2 (green) and RRM34 (blue) recorded under the same condition as in panel A. (C) 15N-HSQC of PTB RRM34 recorded at 500 MHz, 303 K, pH 6.5. (D) Overlay of 15N-HSQC recorded at 500 MHz, 303 K, pH 6.5 of PTB RRM3 (blue) and RRM4 (green).
Efficient segmental labeling of PTB34 by expressed protein ligation
Application of EPL requires a cysteine at the N-terminus of RRM4 in order to react with the thioester present at the C-terminus of RRM3 (Figure 3A). As there is no naturally occurring cysteine in the sequence of PTB RRM34, it was necessary to mutate one amino acid to a cysteine. Such an approach has already been applied in a number of proteins that were segmentally labeled (Severinov and Muir, 1998; Hondal et al, 2001; Wang and Cole, 2001). The interdomain linker in RRM34 offers four serines that could be mutated to cysteines: S409, S433, S443 and S453. The selection among the four possibilities was based on the identity of the preceding amino acid taking care not to have an Asp and Pro before the Ser (Xu et al, 2000). Based on these considerations, position 443, which is preceded by a glycine residue, was chosen as the ligation point and mutated to a cysteine.
Figure 3.
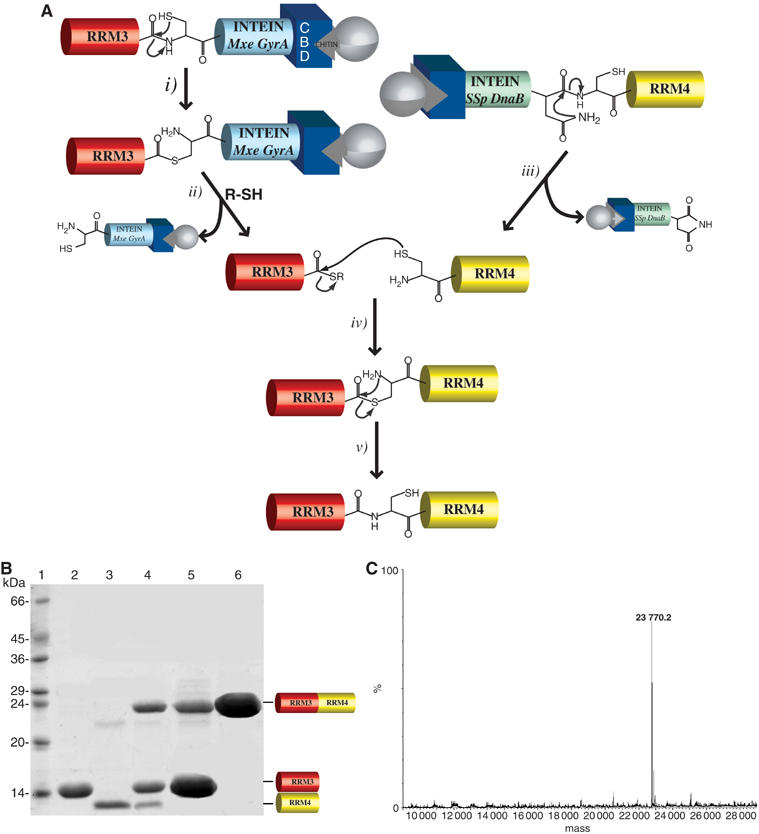
Segmental labeling of PTB RRM34. (A) Intein-mediated EPL. (i) N → S acyl shift of Cys at the intein N-terminus resulting in the formation of a reactive thioester. (ii) Thiol-mediated cleavage: a nucleophilic attack on the thioester by a small thiol compound (MESNA) cleaves the precursor protein and generates a new thioester at the C-terminus of the target protein. (iii) Cyclization of the Ssp DnaB intein C-terminal Asn releases the α-Cys domain from the succinamide derivative of the intein. (iv) The ligation reaction utilizes identical coupling chemistry as the native chemical ligation, and the activated thioester is attacked by the α-Cys residue. (v) An S → N acyl shift, a spontaneous rearrangement of the thioester, results in the formation of a peptide bond between the two domains. (B) SDS–PAGE of PTB RRM34 and subdomains. Lane 1: protein marker; lane 2: PTB RRM3 after chitin affinity column purification; lane 3: PTB RRM4 after chitin affinity column purification; lane 4: mixture of a PTB RRM34 ligation reaction following a standard EPL protocol (Severinov and Muir, 1998; Xu et al, 1999); lane 5: mixture of a PTB RRM34 ligation reaction performed on column; lane 6: PTB RRM34 ligated on column after the Ni-NTA purification. (C) Electrospray mass spectra of purified ligated PTB RRM34.
PTB RRM3 was fused to the N-terminus of the Mxe GyrA intein sequence (Southworth et al, 1999) and PTB RRM4 was fused to the C-terminus of the Ssp DnaB intein (Mathys et al, 1999) (Figure 1). A chitin binding domain (CBD) present at the C-terminus of the Mxe GyrA intein and at the N-terminus of the Ssp DnaB intein (Figure 1) is exploited to bind both RRM-intein constructs to the same affinity column (on column ligation). This approach is derived from a protocol used to generate a cyclic protein (Evans et al, 1999) that we adapted to ligate two protein fragments (Figure 3A). Cleavage of PTB-RRM3 from Mxe GyrA intein was carried out by adding 2-mercaptoethanesulfonic acid (MESNA) yielding the desired RRM3 thioester derivative (Figure 3B, lane 2). The cleavage reaction between RRM4 and Ssp DnaB intein was induced by a change of temperature and provided an α-Cys at the N-terminus of RRM4, as confirmed by SDS–PAGE (Figure 3B, lane 3) and ESI-MS analysis (not shown). As an initial trial, RRM3 and RRM4 were individually purified, concentrated and mixed in the presence of MESNA. After 16 h at room temperature, the reaction was analyzed by SDS–PAGE showing the formation of the ligation product (Figure 3B, lane 4).
Taking advantage of the presence of the same CBD at the C-terminus of Mxe GyrA intein and at the N-terminus of Ssp DnaB intein, we performed the ligation reaction directly on the chitin affinity column (‘on column' ligation). After loading the two PTB fragments fused to an intein, the column was flushed with MESNA to cleave the N-terminal intein and leave a thioester at the C-terminus of RRM3 (Figure 3A). Simultaneously, cleavage of the C-terminal intein is induced by elevating the temperature to 37°C, leaving an α-cysteine at the N-terminus of RRM4 (Figure 3A). Now, the chemical ligation between the two reactants occurs on the column, producing RRM34 efficiently (Figures 3A and 4A).
Figure 4.
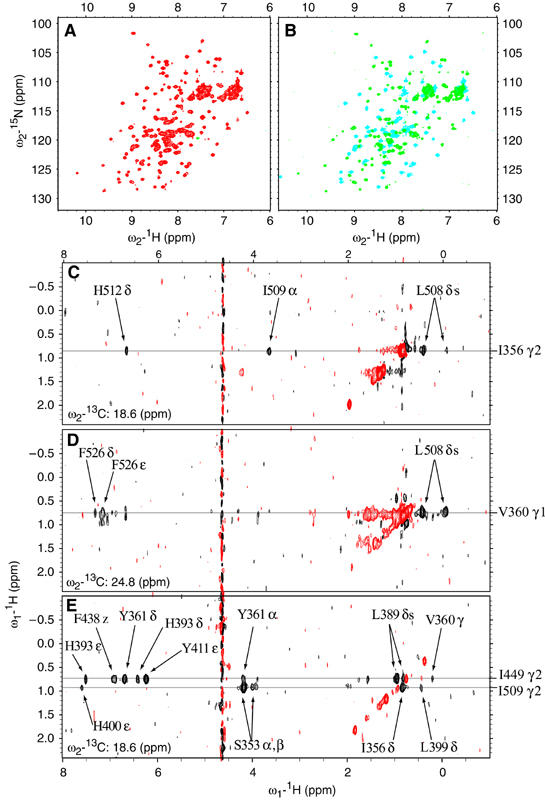
NMR spectra of the segmentally labeled PTB RRM34. (A) 15N-HSQC of RRM34 (S443C) with 15N,13C-labeled RRM3 ligated to 15N-only-labeled RRM4, recorded at 900 MHz, 303 K, pH 6.5. (B) Overlay of two 15N-HSQC of PTB RRM34 (S443C) segmentally labeled, under the same condition as panel A. 15N,13C–labeled RRM3 is indicated by blue peaks and 15N,13C–labeled RRM4 by green peaks. (C, D) Cross-sections of a 3D 13C-edited half-filter NOESY of the segmentally labeled RRM34 where RRM3 is 13C-labeled. (C) Several interdomain NOEs to I356 γ2 can be observed in the 13C plane at 18.6 ppm. (D) Several interdomain NOEs to V360 γ1 can be observed in the 13C plane at 24.8 ppm. (E) Cross-section of a 3D 13C-edited half-filter NOESY of the segmentally labeled RRM34 where RRM4 is 13C-labeled. Several interdomain NOEs to I449 and I509 γ2 can be observed in the 13C plane at 18.6 ppm.
When the ligation reaction is performed on the same chitin column with Ssp DnaB intein-RRM4 and RRM3-Mxe GyrA intein (in about two-fold excess over Ssp DnaB intein-RRM4), RRM4 is found to be completely ligated to RRM3. The sample eluted from the chitin column after ligation shows the presence of only two protein bands on an SDS–PAGE, one corresponding to the ligated PTB RRM34 and the other to the unreacted RRM3 (Figure 3B, lane 5). With a His tag at the C-terminus of RRM4, the ligation product (RRM34) can be easily separated from RRM3 with a single Ni-NTA affinity purification step (Figure 3B, lane 6). The identity of the ligated product was confirmed by ESI-MS analysis (Figure 3C). Using this ‘on column' ligation procedure, proteins with different labeling schemes were prepared for our NMR study. Yields of 2–3 mg/l media of segmentally labeled RRM34 could be obtained.
NMR of the segmentally labeled RRM34
Three segmentally labeled RRM34 constructs were generated in order to study the interaction between the two RRMs. The first sample contains a 15N,13C-labeled RRM3 and an unlabeled RRM4, the second sample an unlabeled RRM3 and a 15N,13C-labeled RRM4 and the third sample a 15N,13C-labeled RRM3 and a 15N-only-labeled RRM4. With the third sample, we can show that the Ser443 to Cys mutation does not affect the structure of RRM34, as the 15N-HSQC spectra of the ligated RRM34 of the uniformly labeled RRM34 are basically identical (compare Figure 4A with Figure 2C). Moreover, spectral comparison between the three segmentally labeled samples shows that the resonance overlap can be considerably reduced if only one domain is isotopically labeled (compare Figure 4A with Figure 4B).
More importantly, we could record several 2D homonuclear and 3D 13C-edited NOESY half-filter experiments in order to identify NOE crosspeaks between the two RRMs. In cross-sections of the 3D 13C-edited half-filter NOESY, one can clearly see several interdomain NOEs to I356 (Figure 4C) and V360 (Figure 4D) of RRM3, and to I449 and I509 of RRM4 (Figure 4E). Using these spectra, 130 interdomain NOEs could be assigned resulting in long-range interproton distance constraints useful in the structure determination of PTB RRM34.
Structure determination of PTB RRM34
With unambiguous evidence that RRM3 and RRM4 of PTB interact in the free state, we decided to re-determine the structure of RRM34. A total of 3294 NOE-derived distance constraints including 130 NOEs between the two RRMs made it possible to obtain a precise structure of RRM34 with an RMSD of 0.52 Å for the backbone atoms of the two domains and of 0.65 Å for the 27 side chains involved in the interdomain interaction (Figure 5A and Table I). Each RRM on its own adopts the same structure as previously determined. In particular, the presence of the unusual fifth β-strand in RRM3 that lies antiparallel to β2 was confirmed (Conte et al, 2000). The novelty of our structure lies in the fixed relative orientation of RRM3 and RRM4. The RRMs interact through their helices, namely helix 1 and 2 of RRM3 and helix 2 of RRM4 (Figure 5B). This unusual topology of RRM3 and RRM4 is due to a large interdomain interface involving 27 protein side chains from the two RRMs and from the interdomain linker (Figure 5B).
Figure 5.
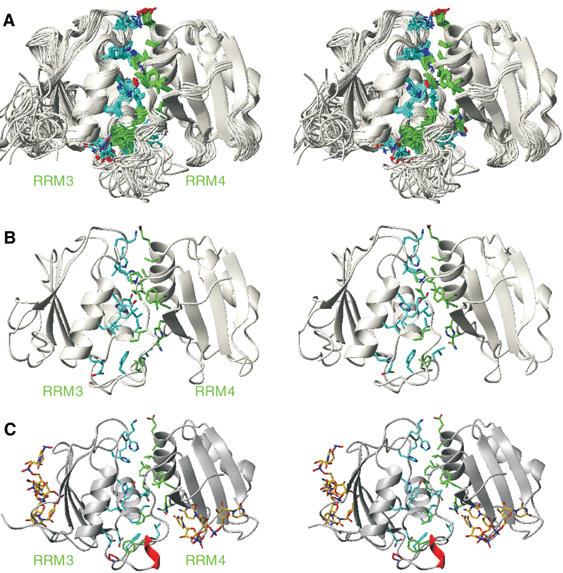
Structure of free and bound PTB RRM34. (A) Superimposition of the 20 lowest energy conformers of RRM34 in its free form. Protein side chains contributing to the interdomain interaction are displayed in blue (residues 324–442 that include RRM3) and in green (residues 443–531 that include RRM4). (B) Stereo view of the lowest energy conformer of PTB RRM34. (C) Stereo view of PTB RRM34 in complex with RNA (Oberstrass et al, 2005). The RNA is shown in yellow.
Table 1.
Structural statistics of PTB RRM34
| NMR restraints | ||
| Distance restraints | 3303 | |
| Protein | ||
| Intraresidual | 746 | |
| Sequential (∣i−j∣=1) | 814 | |
| Medium range (1<∣i−j∣⩽4) | 518 | |
| Long range (∣i−j∣>4) | 1035 | |
| Interdomain NOEs | 130 | |
| Hydrogen bondsa | 60 | |
| Energy statistics (20 structures) | ||
| NOE violations >0.4 (Å) | 4.5±1.4 | |
| Mean constraint violation energy (kcal/mol) | 200.0±7.4 | |
| Mean AMBER energy (kcal/mol) | −7426±30 | |
| Mean deviation from ideal covalent geometry | ||
| Bond length (Å) | 0.0103±1E−4 | |
| Bond angle (deg) | 2.73±0.02 | |
| Ramachandran plot analysis | ||
| Residues in most favored regions (%) | 71.6±2.4 | |
| Residues in additional allowed regions (%) | 22.3±2.2 | |
| Residues in generously allowed regions (%) | 3.7±1.3 | |
| Residues in disallowed regions (%) | 2.4±0.9 | |
| RMSD from the mean structure (Å) | ||
| Backbone heavy atoms (N, Cα, C′, O)b | 0.54±0.08 | |
| All heavy atomsb | 0.93±0.13 | |
| RRM3c | RRM4d | |
| Backbone heavy atoms (N, Cα, C′, O) |
0.51±0.08 |
0.34±0.08 |
| All heavy atoms |
0.98±0.18 |
0.70±0.12 |
| aBased on slow-exchanging amide protons in D2O. | ||
| bRMSD is based on protein residues N336–V414, L426–G431, S453–I531. | ||
| cRMSD is based on protein residues N336–V414, L426–G431. | ||
| dRMSD is based on protein residues S453–I531. | ||
An unusually large interdomain interface between RRM3 and RRM4
The interdomain interactions involve 27 residues forming a large hydrophobic core. This interaction is additionally stabilized by an ion pair (K398–E502) (Figure 6A). Two main sets of interactions can be observed. The first set is composed of direct inter-RRM contacts and involves α-helix 2 of RRM4 (Figure 6A, in green). The N-terminal part of helix 2 of RRM4 (V501, E502 and V505) interacts with the loop between helix 2 and β4 of RRM3 (H397, K398, L399 and H400) and its C-terminal part (L508, I509, H512 and N513) interacts with the N-terminal part of helix 1 of RRM3 (Q352, F355, I356 and V360) and L335 of the linker (Figure 6A). These interactions result in the positioning of helix 2 of RRM4 perpendicular to helix 1 of RRM3 (Figure 6A, in blue). The second set of interactions involves contacts mediated by the interdomain linker (Figure 6B, in red). Several side chains of the interdomain linker (F438, F446, I449, P451 and P452) contact helix 2 of RRM3 (Q386, L389, H393) and H397 of the following loop (Figure 6B, in blue). In this latter interaction, it is particularly interesting to note the hydrophobic interaction between P452 and H397 and between P451 and H393, the five-membered rings stacking over one another (Figure 6B). This region of the interdomain linker also interacts with helix 1 of RRM3 (V360, Y361 and D363; Figure 6B, in blue) and F526 of β4 of RRM4 (Figure 6B, in green). As most of the interdomain linker contributes to the interdomain interactions, it is well structured from G431 to F438 and from F446 to A454. The remaining part of the linker, from K439 to N445, shows no contact to the structured regions in RRM34 and has therefore a much higher degree of conformational variability (Figure 5A).
Figure 6.
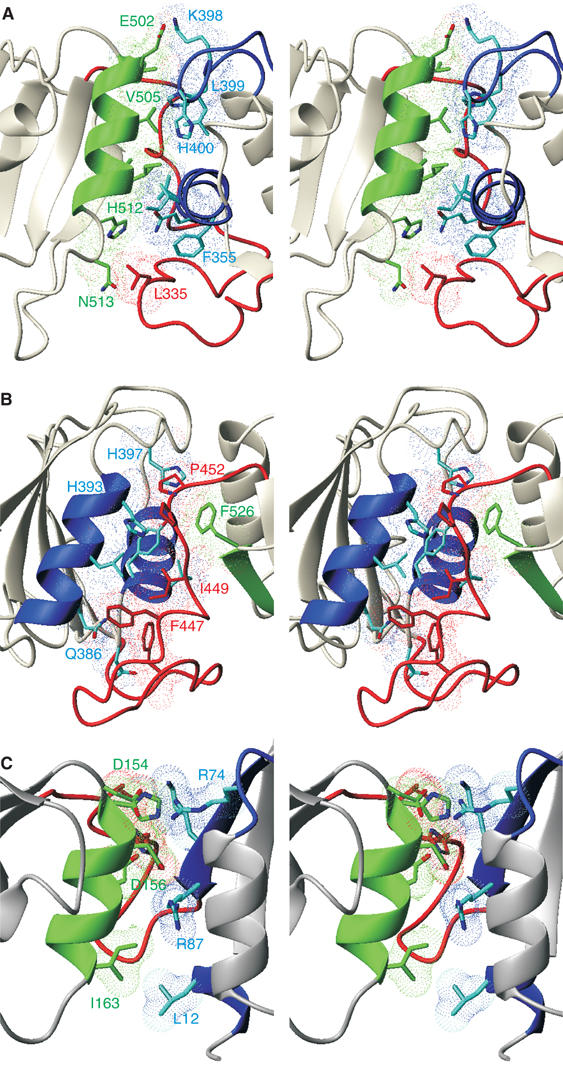
Close-up view of the interdomain interface in PTB RRM34 and hnRNPA1 RRM12. (A) Stereo view of the interaction between helix 2 of RRM4 and helix 1 of RRM3. Side chains for RRM3 and RRM4 and the interdomain linker are represented by sticks and dotted surfaces colored in blue, green and red, respectively. (B) Stereo view of the interaction between helix 2 of RRM3 with the interdomain linker and F526 from RRM4. (C) Stereo view of the interdomain interface found in the structure of hnRNPA1 RRM12 (Shamoo et al, 1997).
Dynamics and mutagenesis of PTB RRM34
In order to further confirm this unusual interdomain interaction, we performed a dynamic study for both the wild-type PTB RRM34 and for one protein embedding side-chain mutations at the interdomain interface. 1H–15N NOE, T1 and T2 were measured for RRM34 (Figure 7). Owing to resonance overlap or line broadening, only 115 of the 208 amide resonances were sufficiently separated to be analyzed. The amide resonances of the β2–β3 loop of RBD4 (F487–R491) and of part of the interdomain linker (L435, R437, K439, G441, K443–N448) are very broad, making it difficult to determine the relaxation parameters accurately. Except for the β2–β3 and β4–β5 loops of RBD3 and for the N- and C-termini that both show low 1H–15N NOE values indicative of mobility, all amides of the two RBD domains showed a 1H–15N NOE between 0.75 and 0.85 indicative of rigidity (Figure 7A). In the interdomain linker (G431–A454), the amide resonances that could be accurately measured have a 1H–15N NOE between 0.68 and 0.94 for an average of 0.80±0.09. This indicates that part of the interdomain linker is rigid, in agreement with the structure where part of the linker interacts with the two RRMs (Figure 6). For the most rigid residues (with a 1H–15N NOE equal or above 0.75), we obtained a T1/T2 ratio of 25.0±4.0 (Table II). From this value, an overall correlation time (τc) of 10.4 ns could be estimated assuming isotropic symmetry (Fushman et al, 1994). This value is within the range of values that one would expect for a 24 kDa protein fragment (Wagner, 1997).
Figure 7.
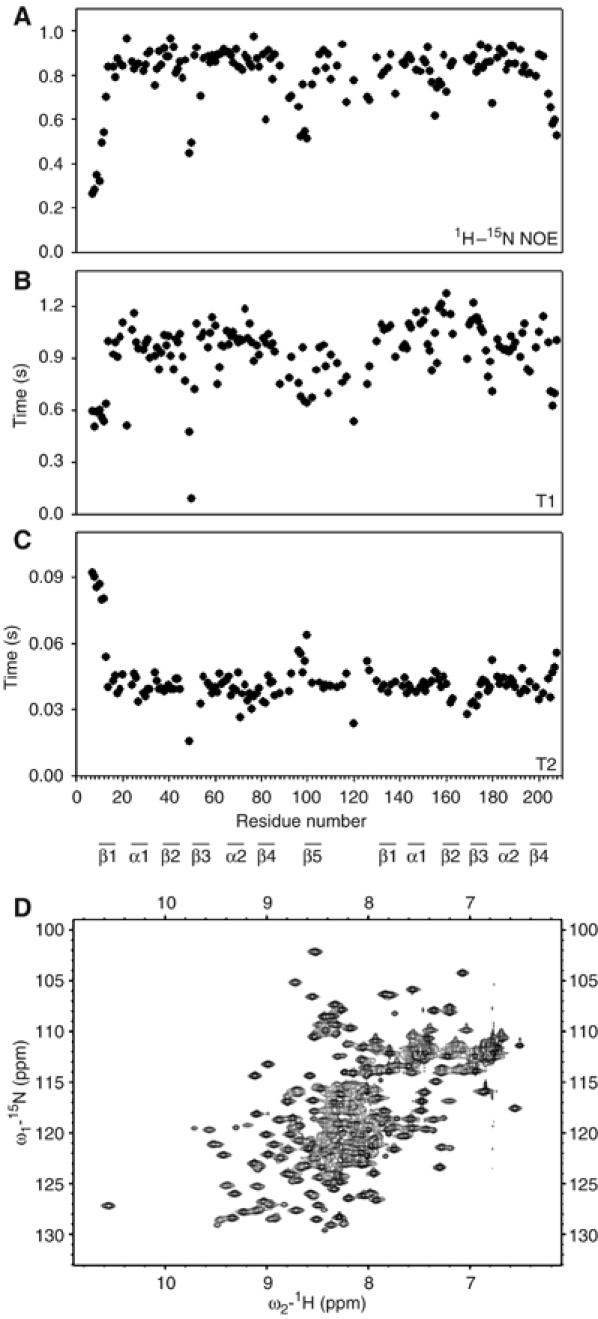
Relaxation measurement and mutagenesis of PTB RRM34. (A) 15N–1H NOE values of the backbone amide resonances of PTB RRM34 plotted against the residue number. (B) T1 values of the backbone amides. (C) T2 values of the backbone amides. (D) 15N-HSQC spectra of PTB RRM34 containing six side-chain mutations (I356K, F446E, I449K, E502K, V505E and I509K) at the interdomain interface.
Table 2.
T1, T2 and overall correlation time of wild-type (wt) and mutant (mut) PTB RRM34
| Protein fragment | T1 (s) | T2 (s) | T1/T2 | τc (ns) |
|---|---|---|---|---|
| RRM34 wt (all) | 0.99±0.11 | 0.040±0.0042 | 25.0±4.0 | 10.4±0.85 |
| RRM34 wt (RRM3) | 0.96±0.11 | 0.040±0.0040 | 24.4±3.7 | 10.3±0.77 |
| RRM34 wt (RRM4) | 1.02±0.11 | 0.040±0.0040 | 25.8±4.4 | 10.6±0.93 |
| RRM34 mut (RRM3) | 0.76±0.07 | 0.052±0.0073 | 15.1±2.8 | 7.97±0.76 |
| RRM34 mut (RRM4) | 0.67±0.06 | 0.058±0.0082 | 11.7±2.1 | 6.90±0.67 |
In order to disrupt the interface, charged residues were introduced instead of several hydrophobic side chains and the K398–E502 salt bridge was disrupted by mutating E502 into a lysine. Two mutant proteins were recombinantly expressed, one containing three mutations in RRM4 α-helix 2 (E502K, V505E and I509K) and the other construct comprising six mutations (the same three in RRM4 plus I356K in RRM3 and F446E and I449K in the linker). The spectra of both RRM34 mutants are very similar (not shown) and closely resemble the spectra of the individually expressed RRM3 and RRM4 (compare Figure 7D with Figure 2D). This suggests that the interdomain interaction is abolished with such mutations, with the result that the two RBDs are now independent. To further prove the independence of the two RRMs, we measured the amide 15N–1H NOE, T1 and T2 for the mutant protein comprising six mutations. From these data, we could estimate the overall correlation time of RRM3 and RRM4 to be 8.0 and 6.9 ns, respectively (Table II). These results unambiguously confirm that the two domains are independent in the mutant protein, as the two correlation times are smaller than the wild type (τc=10.6 ns) and the correlation time of RRM3 is higher than that of RRM4, which reflects on the slightly higher molecular weight of RRM3 (13 kDa) compared to RRM4 (10.7 kDa).
Discussion
An efficient protocol for segmental isotopic labeling of interacting proteins domains
Although segmental isotopically labeled proteins have been generated previously (Yamazaki et al, 1998; Otomo et al, 1999a, 1999b; Xu et al, 1999; Camarero et al, 2002; Zuger and Iwai, 2005), EPL was never used to determine the structure of a protein using NMR spectroscopy. The difficulty in obtaining high amounts of the ligated protein product might explain why the method has not been used widely for this application. For PTB RRM34, we have obtained 2–3 mg of the ligated protein per liter of culture, allowing us to perform a structural study of the protein. The efficient ligation yield that we obtained can be explained by two reasons: an improved protocol and conformational assistance. By taking advantage of the CBD presence in the two expressed constructs, we performed our ligation reaction on a single column. Consequently, the number of steps from the protein overexpression to the synthesis of the ligated product is considerably reduced compared to previous EPL protocol (Severinov and Muir, 1998; Xu et al, 1999). Moreover, carrying out the ligation on column allowed us to work at a high local concentration of the reactants, which is a prerequisite for an efficient ligation reaction (Muir, 2003). Furthermore, we exploited the fact that RRM3 and RRM4 can interact in trans (F Vitali and FHT Allain, unpublished data). As a consequence, the thioester at the C-terminus of RRM3 and the α-cysteine at the N-terminus of RRM4 are likely to be brought in close proximity by this interaction. Consequently, the ligation rate is likely to be increased. In support of this hypothesis, ligation experiments of peptides interacting in trans showed that the rate of reaction was dramatically increased if assisted conformationally (Beligere and Dawson, 1999). It is likely that this simple protocol could be generalized to the segmental isotopic labeling of most multidomain proteins.
Exploiting segmental isotopic labeling for the structure determination of PTB RRM34
We used our segmentally labeled proteins to re-determine the structure of PTB RRM34 that was previously investigated by others (Conte et al, 2000) with uniform labeling. With uniform labeling of RRM34, even at 900 MHz, the spectral overlap is so significant that we could only unambiguously assign about 30 interdomain NOEs. The previous structure determination (Conte et al, 2000) was performed at 500 MHz; therefore, it is very likely that even less interdomain NOEs could be observed, explaining that no interdomain contacts were found. Exploiting segmental labeling of RRM34, 130 interdomain NOEs were identified resulting in a very precise structure (Table I and Figures 4C–E and 5A). This is the first time that segmental isotopic labeling is used for the structure determination of a protein, promising a wider use of EPL for protein structure determination in the future.
An unusual and conserved interdomain interaction between RRM3 and RRM4 of PTB
The structure of PTB RRM34 reveals a very unusual interaction between the two RRMs and also a new role for the interdomain linker. Sequence alignment of human PTB with its homologs in pig, rat, mouse, cow, fly and worm and with its paralogs human and rat nPTB (Markovtsov et al, 2000), rat smPTB (Gooding et al, 2003) and human and rat ROD1 (Yamamoto et al, 1999) shows that 24 out of 27 residues contributing to the interdomain contact in PTB RRM34 are either conserved (14 aa) or exchanged by similar amino acids (10 aa). Only three amino acids are not well conserved, namely Q352, H397 and N513, but examination of their position in the structure indicates that none of the mutations in homologs or paralogs would abolish the interdomain interaction. The salt bridge formed by K398 and E502 is conserved, E502 being absolutely conserved and K398 being conserved except in ROD1 and fly where it is an arginine. Consequently, the interdomain contact and the resulting structure of RRM34 are very likely to be conserved in all homologs and paralogs of PTB.
Five other structures of tandem RRM proteins have been determined: Sex-Lethal free (Lee et al, 1994a; Inoue et al, 1997) and bound to RNA (Handa et al, 1999), Nucleolin free (Allain et al, 2000b) and bound to RNA (Allain et al, 2000a), HuD bound to RNA (Wang and Tanaka Hall, 2001), PABP bound to RNA (Deo et al, 1999) and hnRNP A1 free (Shamoo et al, 1997) and in complex with DNA (Ding et al, 1999). The two RRMs of nucleolin and sex-lethal are not interacting in the free proteins and their interdomain linker is mostly unstructured. While in complex, the two RRMs interact and the interdomain linker is significantly involved in RNA binding. Only hnRNP A1 presents an interaction between its two N-terminal RRMs, both in its free and bound form. In the free hnRNPA1 (Figure 6C), the interdomain contact is mediated by 10 side chains, with four forming two salt bridges (Shamoo et al, 1997). Interestingly, like for PTB RRM34, the interdomain contact is mediated by helix 2 of the most C-terminal RRM (i.e. RRM2; Figure 6C, in green); however, in hnRNPA1, the helix 2 interacts with other protein elements compared to PTB, such as the β4-strand of RRM1 (Figure 6C, in blue). Therefore, the positioning of the two domains relative to one another in PTB is very different from hnRNPA1, and unique among RRM-containing proteins (Maris et al, 2005). This comparison demonstrates the general versatility of the RRM (Maris et al, 2005) in order to fulfill a specific biological function (see below).
Biological implication of the structure of PTB
With this new structure determination of PTB RRM34 in its free state, we can now compare the structure with RRM34 in its bound form (compare Figure 5B with Figure 5C). This comparison reveals that the structure of RRM34 is mostly unchanged upon binding to RNA. It should be noted that RRM34 in complex was determined without segmental labeling (Oberstrass et al, 2005), which resulted in an interface between the two domains being less precise (the RMSD for the side chains at the interdomain interface is equal to 1.1 Å in the complex compared to 0.65 Å in the free protein). The single difference between the free and bound RRM34 structures lies in a small region of the interdomain linker (P441–F446). This region is unstructured in the free RRM34 and becomes structured upon RNA binding, forming a short α-helix (Figure 5C). The presence of a strong inter-RRM interaction for PTB RRM34 in its free form differs from what was observed in most tandem RRM protein structures where interdomain contacts are formed only upon RNA binding (Maris et al, 2005).
Overall, PTB has RRM1 and RRM2 being totally independent (Figure 2), and RRM3 and RRM4 being very tightly associated (Figure 5). As all four RRMs bind short pyrimidine tracts sequence specifically (Oberstrass et al, 2005), one might wonder what this structural arrangement implies for PTB functions. We speculate that the independence of RRM1 and RRM2 will allow PTB to bind a large variety of substrates, as the spacing between the bound pyrimidine tract can vary. Indeed, the location of PTB functional binding sites, found either in intronic or IRES sequences, varies widely. On the contrary, the restricted positioning of RRM3 and RRM4 relative to each other may play a very different role. As suggested from the structure of RRM34 in complex with RNA (Oberstrass et al, 2005) and further confirmed here with the free protein structure, it appears that the unusual orientation and tight interaction of PTB RRM3 and RRM4 would induce the formation of RNA loops. Thus, PTB could repress splicing by sequestering either a short alternative-exon or a branch point within these RNA loops. This model is supported by the structure of PTB RRM34 shown here, and would explain the function of PTB as a general, alternative-splicing repressor.
Materials and methods
Cloning and expression of PTB RRM1, RRM2, RRM34 and mutant and full-length PTB
All four DNAs coding for the protein constructs (full-length PTB, RRM1, RRM2, RRM34; Figure 1) were inserted into a pET-28a(+) vector and transformed into an Escherichia coli BL21(DE3) codonplus strain (Stratagene). The proteins were uniformly labeled by overexpression in M9 minimal medium, containing 15N-NH4Cl and 13C-glucose as the only source of nitrogen and carbon, respectively. After two affinity purification steps, the proteins were dialyzed against 20 mM NaCl and 10 mM NaH2PO4 at pH 5.8 and concentrated by centricon to 1–2 mM. Mutations at the interdomain interface in RRM34 were introduced by site-directed mutagenesis (Quickchange, Stratagene).
Cloning and expression of PTB RRM3
The DNA encoding RRM3 of PTB (residues G324–G442) was isolated by PCR from human PTB-1 (Swissprot P26599) gene with the oligonucleotide primers PTB324-442Nde (5′-GCT GGT CAT ATG GGT CGG ATC GCC ATC C-3′) and PTB324-442Sap (5′-CGT CCG CTC TTC CGC AGC CCG GCT TCT TG-3′). The PCR product was cloned into the vector pTWIN1 (New England Biolabs) between the NdeI and SapI sites.
The resulting plasmid, pTWIN1-PTB RRM3, expresses PTB RRM3 fused to the Mxe GyrA intein and the CBD from an isopropyl-1-thio-β-D-galactopyranoside-inducible T7 promoter (Figure 1). The pTWIN1-PTB RRM3 plasmid was shown to be free of mutations in the RRM3 coding region by DNA sequencing. E. coli BL21(DE3) cells transformed with pTWIN1-PTB RRM3 were grown to mid-log phase in Luria–Bertani (LB) medium and induced with 1 mM IPTG at 30°C for 4 h. After centrifugation, cells were resuspended in 20 ml of buffer A (25 mM Hepes/pH 8.0/0.1 mM EDTA/500 mM NaCl) and disrupted by two passages through a French pressure cell at 12 000 psi. The lysate was centrifuged and the clarified lysate was loaded onto a 10-ml chitin column that was pre-equilibrated with buffer A. The chitin column was used with gravity flow, and the flow was such that it allowed 20–30 min for the binding of the CBD fusion protein to chitin beads. After extensive washing of the column, RRM3 is released from the column by inducing the intein to undergo self-splicing in the presence of MESNA. The column was quickly flushed with 3 column volumes of buffer B (25 mM Hepes, pH 8.0/0.1 mM EDTA/500 mM NaCl/100 mM MESNA/10 mM DTT) and its flow was stopped. The column was left overnight at 37°C to allow the cleavage reaction to proceed. To examine the cleavage efficiency, 20 μl of resin slurry was removed from the column and mixed with 20 μl of 4 × SDS–PAGE buffer (187.5 mM Tris–HCl/pH 6.8/6% SDS/30% glycerol/0.03% bromophenol blue) and boiled for 10 min. The resin was pelleted by centrifugation and a sample of supernatant was used for SDS–PAGE analysis. The eluted protein showed a single band by SDS–PAGE, and gave an electrospray mass spectrum corresponding to the loss of N-terminal Met ([M+H]+=13 043.1 m/z). All the electrospray mass spectrometry analyses were performed on a Waters Q-TOF Ultima API mass spectrometer. Expression of PTB RRM3 in 1 l of 15N,13C-labeled M9 medium yields 12 mg of pure protein.
Cloning and expression of PTB RRM4
The DNA encoding the PTB RRM4 domain (residues K444–I531) was isolated by PCR from human PTB-1 (Swissprot P26599) gene with the oligonucleotide primers PTB443-531SapF (5′-GCT GGT TGC TCT TCC AAC TGT AAG AAC TTC CAG AAC ATA TTC C-3′) and PTB443-531TWNrev (5′-GGT GGT CTG CAG TTA GTG GTG GTG GTG GTG GTG GAT GGT GGA CTT-3′). This oligonucleotide creates a Ser443 to Cys mutation point and an insertion of a C-terminal His6 tag in the coding sequence. The PCR product was purified and digested simultaneously with SapI and PstI and then cloned into plasmid pTWIN1. The sequence was confirmed by DNA sequencing. The resulting plasmid, pTWIN1-PTB RRM4, expresses the PTB RRM4 domain fused to the Ssp DnaB intein and the CBD from an IPTG-inducible T7 promoter (Figure 1). The cell growth and expression procedures were essentially the same as in the protocols described for the generation of a PTB RRM3, except that all purification steps were performed at 4°C and buffers kept at 4°C to minimize the premature cleavage of the target protein. To induce C-terminal cleavage, chitin beads loaded with PTB RRM4-Ssp DnaB-CBD were flushed at 4°C with 3 column volumes of buffer C (25 mM Hepes/pH 7.0/0.1 mM EDTA/500 mM NaCl/10 mM DTT) and left at 37°C overnight. The eluted protein showed a single band by SDS–PAGE, and gave an electrospray mass spectrum corresponding to the α-Cys N-terminus protein ([M+H]+=10 744.2 m/z). Expression of PTB RRM4 in 1 l of 15N,13C-labeled M9 medium yields 3 mg of pure protein.
Standard ligation reaction
Freshly isolated PTB RRM3 thioester and RRM4 α-Cys were concentrated at 4°C with a Centricon (Vivaspin) to a final concentration of 1.5 and 0.7 mM, respectively. The same volumes of each protein solution were then mixed (i.e. with a 2.1:1.0 ratio between RRM3 and RRM4), and in order to start the ligation reaction, MESNA and DTT were added to the solution with a final concentration of 100 and 10 mM, respectively. The reaction was left at 37°C overnight. The formation of the expected product was checked by SDS–PAGE (Figure 3B, lane 4).
Ligation reaction on column
The cell pellets of the thioester protein and of the α-Cys protein from 1 l culture each were resuspended together in 30 ml of ice-cold buffer A and lysated by French press. Chitin purification step was performed at 4°C, as already mentioned, to minimize premature protein cleavage from the column. The mixed cell extract was clarified by centrifugation for 30 min and the supernatant was applied to a pre-equilibrated chitin beads column. After extensive washing of the column, it was quickly flushed with 3 bed volumes of buffer B and the flow was stopped. The column was left at 37°C to allow the cleavage reactions of the two proteins and their subsequent ligation to proceed at the same time. After ∼16 h of reaction, the desired ligation product was eluted from the chitin beads with buffer D (50 mM NaPi/pH 8.0/300 mM NaCl) and directly applied to a pre-equilibrated Ni-NTA column. After loading, the column was washed with buffer E (50 mM NaPi/pH 8.0/1 M NaCl) and successively with buffer F (50 mM NaPi/pH 8.0/300 mM NaCl/10 mM imidazole), and finally the His6-tagged ligated protein was eluted with buffer G (50 mM NaPi/pH 8.0/300 mM NaCl/500 mM imidazole). The protein showed a single band by SDS–PAGE and gave an electrospray mass spectrum consistent with the calculated mass without the N-terminal Met ([M+H]+=23 770 m/z).
Yields of all the three segmentally labeled PTB RRM34 NMR are as follows: 9 mg of pure segmentally labeled PTB RRM34 (with RRM4 15N,13C-labeled) was obtained from 0.4 l of LB medium culture of RRM3-Mxe GyrA and 2 l of 15N,13C-labeled M9 medium culture of SSp DnaB-RRM4; 7.5 mg of pure segmentally labeled PTB RRM34 (with RRM3 15N,13C-labeled) was obtained from 1 l of 15N,13C-labeled M9 medium culture of RRM3-Mxe GyrA and 1 l of LB medium culture of SSp DnaB-RRM4; and 8 mg of pure segmentally labeled PTB RRM34 (with RRM3 15N-labeled and RRM4 15N,13C-labeled) was obtained from 2 l of 15N-labeled M9 medium culture of RRM3-Mxe GyrA and 1 l of 15N,13C-labeled M9 medium culture of SSp DnaB-RRM4.
NMR spectroscopy
NMR spectra were acquired at 303 K on four-channel Bruker DRX-500, DRX-600 and Avance-900 spectrometers. NMR data were processed using XWINNMR (Bruker) and analyzed using Sparky (http://www.cgl.ucsf.edu/home/sparky/). Sequence-specific backbone assignment of PTB RRM34 and RRM4 was obtained using HNCA, HN(CO)CA, CBCA(CO)NH, HNCACB, 15N-HSQC, 13C-HSQC (Cavanagh et al, 1996) and 15N-TROSY (Pervushin et al, 1998) spectra. The aliphatic side chains of PTB RRM34 were assigned based on 3D 15N- and 13C-edited NOESYs and a 3D HCCH-TOCSY (Bax et al, 1990). The assignment of side chain amides was achieved by analyzing the 15N 3D NOESY and the assignment of the aromatic side chains with a 2D TOCSY and a 2D NOESY in 100% D2O. Distance restraints used in the structure calculations of RRM34 were extracted from the 15N- and 13C-edited NOESY and the 2D NOESY in D2O. Hydrogen-bonded NH groups were identified by the presence of amide resonances in 15N-HSQC spectra, which were recorded immediately after lyophilizing and dissolving the sample in D2O. Using the RRM34 segmentally labeled sample with only one RRM 13C-labeled, 2D half-filter (Peterson et al, 2004) and 3D 13C-edited half-filter NOESY (Lee et al, 1994b) spectra were recorded in order to identify interdomain NOEs.
Structure calculation
Initial structure calculation was performed using the CANDID/DYANA package (Guntert et al, 1997; Herrmann et al, 2002) based on distance constraints derived from NOE peak intensities found in the two 3D NOESY (15N- and 13C-edited) and the 2D homonuclear NOESY (all with a τm of 150 ms), all recorded at 900 MHz on the uniformly labeled 15N, 13C spectrum. Altogether seven iterations were performed. At each iteration step, 100 independent structures were calculated. At this stage, the first family of converging conformers had well-defined structures for RRM3 and RRM4 in isolation but with a poor definition of the interdomain interface, owing to the presence of only 30 interdomain distance constraints. Analysis of the 2D and 3D half-filter NOESYs recorded on the segmentally labeled samples allows the unambiguous identification of 100 more interdomain NOEs. The addition of these 100 additional interdomain distance constraints into the structure calculation resulted in a well-defined interdomain interface.
The structure was further refined with the inclusion of several hydrogen-bond constraints, derived from slow-exchanging amides. Altogether, 3294 distance constraints including 128 hydrogen-bond constraints were used to calculate the structure of PTB RRM34. A final refinement step was performed in AMBER (Wang et al, 2004) using the same protocol as for the calculation of PTB RRM34 in complex with RNA (Oberstrass et al, 2005).
NMR dynamics
For the NMR dynamics study, 15N–1H NOE, 15N T1 and T2 measurements were recorded at 899.37 MHz and 91.13 MHz, 1H and 15N frequency respectively (Kay et al, 1989; Skelton et al, 1993). For the 15N–1H NOE measurement, a relaxation delay of 2 s and a 1H presaturation of 3 s were used in the NOE experiment and a 5 s relaxation delay was used in the reference experiment. 15N T1 values were derived from seven 15N–1H spectra with different delays: 10.01, 105.25, 255.63, 506.26, 756.89, 1007.52 and 1508.78 ms and an interscan delay of 3 s. Similarly, 15N T2 values were derived from 15N–1H spectra with seven different delays: 12.42, 24.84, 37.25, 49.67, 62.09, 74.51 and 86.93 ms and an interscan delay of 3 s. T1 and T2 values were extracted by a curve-fitting subroutine included in the program Sparky (Goddard and Kneeler, 1999). Overall correlation times (τc) were derived from the average T1/T2 ratio of the rigid amide resonances (1H–15N NOE>0.75), assuming isotropic motion as described (Fushman et al, 1994).
Structure deposition
The coordinates of the ensemble of conformers of PTB RRM34 have been deposited in the PDB databank under the accession code 2EVZ.
Acknowledgments
We are grateful to Dr Fred Damberger, Dr Christian Hilty, Lars Dreier and Professor Gerhard Wider for their assistance in the NMR instrumentation and Dr Mario Schubert for helpful discussion about the dynamic study. We thank Professor Kurt Wüthrich (ETH, Zurich) for insightful discussion and Dr Stephen Curry and Dr Steven Matthews (Imperial College, London, UK) for helpful discussions. We also thank Dr P Hunziker and Dr S Chesnov (Protein Analysis Unit, University of Zurich) for the mass spectrometry measurements. This investigation was performed in part in the Structural Biology National Center of Competence in Research (NCCR) ISO-laboratory co-directed by Professor K Wüthrich. The investigation was supported by a predoctoral fellowship from the Roche Research Fund for Biology to FCO, grants from the Swiss National Science Foundation and the ETH Zurich through the NCCR Structural Biology and by the Roche Research Fund for Biology at the ETH Zurich to FHTA. FHTA is an EMBO Young Investigator.
References
- Allain FH, Bouvet P, Dieckmann T, Feigon J (2000a) Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin. EMBO J 19: 6870–6881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain FH, Gilbert DE, Bouvet P, Feigon J (2000b) Solution structure of the two N-terminal RNA-binding domains of nucleolin and NMR study of the interaction with its RNA target. J Mol Biol 303: 227–241 [DOI] [PubMed] [Google Scholar]
- Bax A, Clore GM, Gronenborn AM (1990) 1H–1H correlation via isotropic mixing of 13C magnetization, a new three-dimensional approach for assigning 1H and 13C spectra of 13C-enriched proteins. J Magn Reson 88: 425–431 [Google Scholar]
- Beligere GS, Dawson PE (1999) Conformationally assisted protein ligation using C-terminal thioester peptides. J Am Chem Soc 121: 6332–6333 [Google Scholar]
- Camarero JA, Shekhtman A, Campbell EA, Chlenov M, Gruber TM, Bryant DA, Darst SA, Cowburn D, Muir TW (2002) Autoregulation of a bacterial σ factor explored by using segmental isotopic labeling and NMR. Proc Natl Acad Sci USA 99: 8536–8541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstens RP, Wagner EJ, Garcia-Blanco MA (2000) An intronic splicing silencer causes skipping of the IIIb exon of fibroblast growth factor receptor 2 through involvement of polypyrimidine tract binding protein. Mol Cell Biol 20: 7388–7400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelo-Branco P, Furger A, Wollerton M, Smith C, Moreira A, Proudfoot N (2004) Polypyrimidine tract binding protein modulates efficiency of polyadenylation. Mol Cell Biol 24: 4174–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh J, Fairbrother WJ, Palmer AG, Skelton NJ (1996) Protein NMR Spectroscopy, Principle and Practice. San Diego: Academic Press [Google Scholar]
- Conte MR, Gruene T, Curry S, Matthews S (1999) Resonance assignment and topology of a 22 kDa C-terminal fragment of the polypyrimidine tract binding protein (PTB) containing two RNA binding domains. J Biomol NMR 14: 383–384 [Google Scholar]
- Conte MR, Gruene T, Ghuman J, Kelly G, Ladas A, Matthews S, Curry S (2000) Structure of tandem RNA recognition motifs from polypyrimidine tract binding protein reveals novel feature of the RRM fold. EMBO J 19: 3132–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo RC, Bonanno JB, Sonenberg N, Burley SK (1999) Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell 98: 835–845 [DOI] [PubMed] [Google Scholar]
- Ding J, Hayashi MK, Zhang Y, Manche L, Krainer AR, Xu RM (1999) Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev 13: 1102–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans TC Jr, Benner J, Xu MQ (1999) The cyclization and polymerization of bacterially expressed proteins using modified self-splicing inteins. J Biol Chem 274: 18359–18363 [DOI] [PubMed] [Google Scholar]
- Fushman D, Weisemann R, Thuring H, Ruterjans H (1994) Backbone dynamics of ribonuclease T1 and its complex with 2′GMP studied by two-dimensional heteronuclear NMR spectroscopy. J Biomol NMR 4: 61–78 [DOI] [PubMed] [Google Scholar]
- Goddard TD, Kneeler DG (1999) Sparky 3, 3rd edn. San Francisco: University of California [Google Scholar]
- Gooding C, Kemp P, Smith CWJ (2003) A novel polypyrimidine tract-binding protein paralog expressed in smooth muscle cells. J Biol Chem 278: 15201–15207 [DOI] [PubMed] [Google Scholar]
- Gooding C, Roberts GC, Smith CWJ (1998) Role of an inhibitory pyrimidine element and polypyrimidine tract binding protein in repression of a regulated a-tropomyosin exon. RNA 4: 85–100 [PMC free article] [PubMed] [Google Scholar]
- Guntert P, Mumenthaler C, Wuthrich K (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol 273: 283–298 [DOI] [PubMed] [Google Scholar]
- Handa N, Nureki O, Kurimoto K, Kim I, Sakamoto H, Shimura Y, Muto Y, Yokoyama S (1999) Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature 398: 579–585 [DOI] [PubMed] [Google Scholar]
- Hellen CU, Sarnow P (2001) Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev 15: 1593–1612 [DOI] [PubMed] [Google Scholar]
- Herrmann T, Guntert P, Wuthrich K (2002) Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J Biomol NMR 24: 171–189 [DOI] [PubMed] [Google Scholar]
- Hondal RJ, Nilsson BL, Raines RT (2001) Selenocysteine in native chemical ligation and expressed protein ligation. J Am Chem Soc 123: 5140–5141 [DOI] [PubMed] [Google Scholar]
- Inoue M, Muto Y, Sakamoto H, Kigawa T, Takio K, Shimura Y, Yokoyama S (1997) A characteristic arrangement of aromatic amino acid residues in the solution structure of the amino-terminal RNA-binding domain of Drosophila sex-lethal. J Mol Biol 272: 82–94 [DOI] [PubMed] [Google Scholar]
- Kay LE, Torchia DA, Bax A (1989) Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry 28: 8972–8979 [DOI] [PubMed] [Google Scholar]
- Knoch KP, Bergert H, Borgonovo B, Saeger HD, Altkruger A, Verkade P, Solimena M (2004) Polypyrimidine tract-binding protein promotes insulin secretory granule biogenesis. Nat Cell Biol 6: 207–214 [DOI] [PubMed] [Google Scholar]
- Kolupaeva VG, Hellen CUT, Shatsky IN (1996) Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA 2: 1199–1212 [PMC free article] [PubMed] [Google Scholar]
- Lee AL, Kanaar R, Rio DC, Wemmer DE (1994a) Resonance assignments and solution structure of the second RNA-binding domain of sex-lethal determined by multidimensional heteronuclear magnetic resonance. Biochemistry 33: 13775–13786 [DOI] [PubMed] [Google Scholar]
- Lee W, Revington MJ, Arrowsmith C, Kay LE (1994b) A pulsed field gradient isotope-filtered 3D 13C HMQC-NOESY experiment for extracting intermolecular NOE contacts in molecular complexes. FEBS Lett 350: 87–90 [DOI] [PubMed] [Google Scholar]
- Maris C, Dominguez C, Allain FHT (2005) The RNA recognition motif, a plastic RNA binding platform to regulate post-transcriptional gene expression. FEBS J 272: 2118–2131 [DOI] [PubMed] [Google Scholar]
- Markovtsov V, Nikolic JM, Goldman JA, Turck CW, Chou MY, Black DL (2000) Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol Cell Biol 20: 7463–7479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathys S, Evans TC, Chute IC, Wu H, Chong S, Benner J, Liu XQ, Xu MQ (1999) Characterization of a self-splicing mini-intein and its conversion into autocatalytic N- and C-terminal cleavage elements: facile production of protein building blocks for protein ligation. Gene 231: 1–13 [DOI] [PubMed] [Google Scholar]
- Modaferri EF, Black DL (1999) Combinatorial control of a neuron-specific exon. RNA 5: 687–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir TW (2003) Semisynthesis of proteins by expressed protein ligation. Annu Rev Biochem 72: 249–289 [DOI] [PubMed] [Google Scholar]
- Muir TW, Sondhi D, Cole PA (1998) Expressed protein ligation: a general method for protein engineering. Proc Natl Acad Sci USA 95: 6705–6710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai K, Oubridge C, Ito N, Avis J, Evans P (1995) The RNP domain: a sequence-specific RNA-binding domain involved in processing and transport of RNA. Trends Biochem Sci 20: 235–240 [DOI] [PubMed] [Google Scholar]
- Oberstrass FC, Auwerter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, Allain FHT (2005) Structure of PTB bound to RNA: specific binding and implications for splicing repression. Science 309: 2054–2057 [DOI] [PubMed] [Google Scholar]
- Otomo T, Ito N, Kyogoku Y, Yamazaki T (1999a) NMR observation of selected segments in a larger protein: central-segment isotope labeling through intein-mediated ligation. Biochemistry 38: 16040–16044 [DOI] [PubMed] [Google Scholar]
- Otomo T, Teruya K, Uegaki K, Yamazaki T, Kyogoku Y (1999b) Improved segmental isotope labeling of proteins and application to a larger protein. J Biomol NMR 14: 105–114 [DOI] [PubMed] [Google Scholar]
- Perez I, Lin CH, McAfee JG, Patton JG (1997) Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA 3: 764–778 [PMC free article] [PubMed] [Google Scholar]
- Pervushin K, Riek R, Wider G, Wuthrich K (1998) Transverse relaxation-optimized spectroscopy (TROSY) for NMR studies of aromatic spin systems in C-13-labeled proteins. J Am Chem Soc 120: 6394–6400 [Google Scholar]
- Peterson RD, Theimer CA, Wu H, Feigon J (2004) New applications of 2D filtered/edited NOESY for assignment and structure elucidation of RNA and RNA–protein complexes. J Biomol NMR 28: 59–67 [DOI] [PubMed] [Google Scholar]
- Pilipenko EV, Viktorova EG, Guest ST, Agol VI, Roos RP (2001) Cell-specific proteins regulate viral RNA translation and virus-induced disease. EMBO J 20: 6899–6908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severinov K, Muir TW (1998) Expressed protein ligation, a novel method for studying protein–protein interactions in transcription. J Biol Chem 273: 16205–16209 [DOI] [PubMed] [Google Scholar]
- Shamoo Y, Krueger U, Rice LM, Williams KR, Steitz TA (1997) Crystal structure of the two RNA binding domains of human hnRNP A1 at 1.75 Å resolution. Nat Struct Biol 4: 215–222 [DOI] [PubMed] [Google Scholar]
- Simpson PJ, Monie TP, Szendroi A, Davydova N, Tyzack JK, Conte MR, Read CM, Cary PD, Svergun DI, Konarev PV, Curry S, Matthews S (2004) Structure and RNA interactions of the N-terminal RRM domains of PTB. Structure 12: 1631–1643 [DOI] [PubMed] [Google Scholar]
- Skelton NJ, Palmer AG III, Akke M, Kordel J M R, Chazin WJ (1993) Practical aspects of two-dimensional proton-detected 15N spin relaxation measurements. J Magn Reson B 102: 253–264 [Google Scholar]
- Southby J, Gooding C, Smith Ch (1998) Polypyrimidine tract binding protein functions as a repressor to regulate alternative splicing of -actinin mutually exclusive exons. Mol Cell Biol 19: 2699–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southworth MW, Amaya K, Evans TC, Xu M-Q, Perler FB (1999) Purification of proteins fused to either the amino or carboxy terminus of the Mycobacterium xenopi gyrase A intein. BioTechniques 27: 110–120 [DOI] [PubMed] [Google Scholar]
- Varani G, Nagai K (1998) RNA recognition by RNP proteins during RNA processing. Annu Rev Biophys Biomol Struct 27: 407–445 [DOI] [PubMed] [Google Scholar]
- Wagner EJ, Garcia-Blanco MA (2001) Polypyrimidine tract binding protein antagonizes exon definition. Mol Cell Biol 21: 3281–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G (1997) An account of NMR in structural biology. Nat Struct Biol 4 (Suppl): 841–844 [PubMed] [Google Scholar]
- Wang D, Cole PA (2001) Protein tyrosine kinase Csk-catalyzed phosphorylation of Src containing unnatural tyrosine analogues. J Am Chem Soc 123: 8883–8886 [DOI] [PubMed] [Google Scholar]
- Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general amber force field. J Comput Chem 25: 1157–1174 [DOI] [PubMed] [Google Scholar]
- Wang X, Tanaka Hall TM (2001) Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat Struct Biol 8: 141–145 [DOI] [PubMed] [Google Scholar]
- Xu MQ, Paulus H, Chong S (2000) Fusions to self-splicing inteins for protein purification. Methods Enzymol 326: 376–418 [DOI] [PubMed] [Google Scholar]
- Xu R, Ayers B, Cowburn D, Muir TW (1999) Chemical ligation of folded recombinant proteins: segmental isotopic labeling of domains for NMR studies. Proc Natl Acad Sci USA 96: 388–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Tsukahara K, Kanaoka Y, Jinno S, Okayama H (1999) Isolation of a mammalian homologue of a fission yeast differentiation regulator. Mol Cell Biol 19: 3829–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Otomo T, Oda N, Kyogoku Y, Uegaki K, Ito N, Ishino Y, Nakamura H (1998) Segmental isotope labeling for protein NMR using peptide splicing. J Am Chem Soc 120: 5591–5592 [Google Scholar]
- Yuan X, Davydova N, Conte MR, Curry S, Matthews S (2002) Chemical shift mapping of RNA interactions with the polypyrimidine tract binding protein. Nucleic Acids Res 30: 456–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ashiya M, Sherman TG, Grabowski PJ (1996) Essential nucleotides direct neuron-specific splicing of 2 pre-mRNA. RNA 2: 682–698 [PMC free article] [PubMed] [Google Scholar]
- Zuger S, Iwai H (2005) Intein-based biosynthetic incorporation of unlabeled protein tags into isotopically labeled proteins for NMR studies. Nat Biotechnol 23: 736–740 [DOI] [PubMed] [Google Scholar]


