Abstract
The NDT80 gene of Saccharomyces cerevisiae, which encodes a global activator of transcription of middle sporulation-specific genes, is first expressed after the activation of early meiotic genes but prior to activation of middle sporulation-specific genes. Both upstream repression sequence 1 (URS1) and mid-sporulation element (MSE) sites are present in the promoter region of the NDT80 gene; these elements have been shown previously to contribute to the regulation of expression of early and middle sporulation-specific genes, respectively, by mediating repression in growing cells and activation at specific times during sporulation. In this study, we have shown that the overlapping windows of URS1- and MSE-mediated repression and activation are responsible for the distinctive premiddle expression pattern of the NDT80 gene. Our data suggest that a Sum1-associated repression complex bound at the NDT80 MSE sites prevents Ime1 tethered at the NDT80 URS1 sites from activating transcription of the NDT80 gene at the time that Ime1-dependent activation of early URS1-regulated meiotic genes is occurring. We propose that a decrease in the efficiency of Sum1-mediated repression as cells progress through the early events of the sporulation program allows the previously inactive Ime1 tethered at the URS1NDT80 sites to promote a low level of expression of the NDT80 gene. This initial phase of URS1-dependent NDT80 expression is followed by Ndt80-dependent upregulation of its own expression, which requires the MSENDT80 sites and occurs concomitantly with Ndt80-dependent activation of a set of middle MSE-regulated sporulation-specific genes. Mutation of IME2 prevents expression of NDT80 in sporulating cells. We show in this study that NDT80 is expressed and that middle genes are activated in cells of an Δime2/Δime2 Δsum1/Δsum1 strain in sporulation medium. This suggests that Ime2 activates expression of NDT80 by eliminating Sum1-mediated repression.
The sporulation program of the yeast Saccharomyces cerevisiae provides a simple model system to study the temporal control of gene expression during development. On entry into the sporulation program, which is triggered by starvation, a diploid a/α cell completes one round of premeiotic DNA replication and then progresses through a lengthy prophase during which a high level of recombination occurs and homologous chromosomes pair. Cells then undergo the reductional and equational meiotic divisions, which result in a single four-lobed nucleus that contains the four haploid complements of chromosomes. The nuclear lobes are engulfed and ultimately pinched off by the prospore membranes that extend from the spindle pole bodies. The deposition of spore wall material within the prospore membrane completes the sporulation process, generating four mature spores arranged tetrahedrally within the ascus.
Studies aimed at identifying genes that are differentially expressed as cells progress through sporulation or that serve sporulation-specific roles have defined four temporally distinct classes of sporulation-specific genes: early, middle, mid-late, and late (reviewed in references 25 and 34). An additional three temporal classes have been identified by analysis of expression profiles obtained by a DNA microarray approach (6, 38). Progression through sporulation depends, at least in part, on the sequential completion of key genetic and morphological events that serve to regulate this transcriptional cascade. Various studies have identified transcriptional regulators that coordinate the expression of subsets of sporulation-specific genes (reviewed in reference 49) and have implicated other gene products as regulators of the transition from expression of one class to expression of the subsequent class (e.g., see references 10 and 46).
An understanding of some of the mechanisms that control the coordinate expression of early and middle sporulation-specific genes has been achieved through the characterization of promoter elements and identification of transcriptional regulatory factors (reviewed in reference 49). Expression of the early set of meiotic genes requires Ume6 and Ime1. The latter protein serves as a global regulator of entry into the sporulation program (43) and its expression is regulated both transcriptionally and posttranscriptionally by nutritional and mating-type signals (reviewed in references 25, 34, and 49). Ume6, which binds to the upstream repression sequence 1 (URS1) site that is present in the regulatory region of most early meiotic genes, prevents mitotic expression of these genes by recruitment of the Sin3-Rpd3 histone deacetylase complex (20, 21) and the Isw2 chromatin remodeling complex (13). Mutation of UME6 leads to an intermediate level of expression of early meiotic genes in cells during vegetative growth. At the onset of the sporulation program, the transcriptional activator Ime1 accumulates, interacts with URS1-bound Ume6 in a Rim11-, Mck1-dependent manner, and activates expression of early meiotic genes (2, 33, 40, 44, 50). Other general regulators, such as the RSC chromatin remodeling complex (54) and Abf1 (11), whose DNA-binding site is present in the promoter regions of several early meiotic genes, contribute to maximal expression. IME2, which encodes a protein kinase and is itself an early meiotic gene, is required for the normal kinetics and full expression of early meiotic genes and is essential for expression of middle sporulation-specific genes (reviewed in reference 34).
Ndt80 is a global activator of middle sporulation-specific gene expression that also upregulates its own expression (5, 6, 17). This activator binds to a short regulatory element (5), the mid-sporulation element (MSE) site (16, 36), which is found in the promoter region of 70% of the 158 genes that belong to the middle class of sporulation-specific genes as well as in the promoter region of the premiddle genes, SMK1 and NDT80 (5, 6, 37). NDT80, which was initially characterized as a gene required for exit from the pachytene stage of meiotic prophase (52), is regulated by the meiotic recombination checkpoint. Activation of this checkpoint by defects in recombination or synaptonemal complex formation reduces transcription of the NDT80 gene and leads to inactivation of Ndt80 as an activator of MSE-dependent gene expression (5, 17, 47). Thus, there is a low level of checkpoint-insensitive expression of NDT80 in checkpoint-arrested cells, but there is no Ndt80-dependent auto-upregulation of NDT80 expression or activation of middle sporulation-specific gene expression (5, 17).
A subset of MSEs confers repression during vegetative growth (16, 37, 51). SUM1 and HST1 were identified recently in a screen for genes that are required to maintain mitotic repression of a reporter gene containing the MSE site from the promoter of the SMK1 gene (51). SUM1 was also identified (J. Pak, unpublished data) in further analysis of mutant strains that had been isolated on the basis of allowing mitotic expression of a normally repressed reporter gene containing four copies of the MSE from the SPS4 gene (17). Originally identified as a dominant allele, SUM1-1, which bypasses the requirement for the SIR genes in silencing the HM mating-type loci (4, 22, 26, 28, 30), SUM1 has been shown more recently to encode an MSE-binding protein (51). HST1 is one of four genes that encode Sir2-related proteins (3, 7). Recently, Hst1 has been shown to have NAD+-dependent histone deacetylase activity and to interact with Sum1 (41, 45). Thus, Sum1 may compete with Ndt80 for binding to MSE sites (51) and, when bound, may lead to Hst1-mediated deacetylation of adjacent histones (41, 45), generating an inaccessible chromatin structure and effectively preventing transcription.
In the present study we have examined the regulation of expression of the NDT80 gene, the founding member of the premiddle class of sporulation-specific genes. Expression of the NDT80 gene begins after transcripts of the early meiotic genes are first detected but before transcripts of middle genes begin to accumulate. Our data suggest that early meiotic expression of NDT80 is prevented by Sum1 bound at the MSE sites that are present in the promoter region of the NDT80 gene. Ime2, the product of an early meiotic gene, however, leads to inactivation of Sum1, allowing a checkpoint-insensitive phase of NDT80 expression to proceed. This expression is promoted by the URS1 sites that are also present in the promoter region of the gene. Once this URS1-dependent phase of NDT80 expression has been initiated, a checkpoint-sensitive phase of expression occurs in which Ndt80 upregulates its own expression in an MSE-dependent manner.
MATERIALS AND METHODS
Yeast strains and genetic procedures.
Table 1 lists the S. cerevisiae strains used in this study. W303-derived strains were used for the experiment shown below in Fig. 3. SK1-derived strains were used for all other experiments. Strain JPY141 was derived from DKB407 by integrative transformation with a PCR product that replaced SUM1 with the kanMX6 cassette (31). Strain JPY187 (MATa Δsum1::kanMX6) was derived as a haploid spore segregant of the diploid strain obtained by mating JPY141 and DKB408. JPY215 was obtained by first mating a haploid MATa version of NKY2296 with JPY141. The resultant diploid strain was sporulated, and haploid MATa Δsum1::kanMX6 Δndt80::LEU2 and MATα Δsum1::kanMX6 Δndt80::LEU2 segregants were identified and mated to give the diploid JPY215. JPY211 was derived in a similar manner; a haploid MATa segregant of L213 was mated with JPY141 and the resultant diploid strain was sporulated giving rise to haploid MATa Δsum1::kanMX6 ime2::LEU2 and MATα Δsum1::kanMX6 ime2::LEU2 segregants, which were mated to generate the diploid strain JPY211.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| SK1 derivatives | ||
| DKB98 | MATa/MATα lys2/lys2 ho::LYS2/ho::LYS2 ura3/ura3 leu2::hisG/leu2::hisG his4X::LEU2/his4B::LEU2 arg4-NspI/arg4-BglII | D. Bishop (32) |
| DKB407 | MATα ho::LYS2 lys2 ura3 leu2 his4X-ADE2-his4B ade2 | D. Bishop (32) |
| DKB408 | MATaho::LYS2 lys2 ura3 leu2 his4X-ADE2-his4B ade2 | D. Bishop (32) |
| NKY2296 | MATa/MATα ho::LYS2/ho::LYS2 lys2/lys2 ura3/ura3 leu2/leu2 Δndt80::LEU2/Δndt80::LEU2 | N. Kleckner (52) |
| JPY141 | MATα ho::LYS2 lys2 ura3 leu2 his4X-ADE2-his4B ade2 Δsum1::kanMX6 | This study |
| JPY187 | MATaho::LYS2 lys2 ura3 leu2 his4X-ADE2-his4B ade2 Δsum1::kanMX6 | This study |
| JPY214 | MATa/MATα ho::LYS2/ho::LYS2 lys2/lys2 ura3/ura3 leu2/leu2 his4X-ADE2-his4B/his4X-ADE2-his4B ade2/ade2 Δsum1::kanMX6/Δsum1::kanMX6 | This study |
| JPY215 | MATa/MATα ho::LYS2/ho::LYS2 lys2/lys2 ura3/ura3 leu2/leu2 HIS4/aADE2/b Δndt80::LEU2/Δndt80::LEU2 Δsum1::kanMX6/Δsum1::kanMX6 | This study |
| L213 | MATa/MATα ho::LYS2/ho::LYS2 lys2/lys2 ura3/ura3 leu2/leu2 trp1/trp1 ime2::2::LEU2/ime2::2::LEU2 | B. Byers (8) |
| JPY211 | MATa/MATα ho::LYS2/ho::LYS2 lys2/lys2 ura3/ura3 leu2/leu2 TRP1/c HIS4/a ADE2/b ime2::2::LEU2/ime2::2::LEU2 Δsum1::kanMX6/Δsum1::kanMX6 | This study |
| W303 derivatives | ||
| LNY315 | MATaade2-1 trp1-1 his3-11,15 can1-100 ura3-1 leu2-3,112 | E. Winter (51) |
| JXY34 | MATα ade2-1 trp1-1 his3-11,15 can1-100 ura3-1 leu2-3,112 Δume6::HIS3 | E. Winter (51) |
| JXY3 | MATaade2-1 trp1-1 his3-11,15 can1-100 ura3-1 leu2-3,112 Δsum1::kanMX4 | E. Winter (51) |
| JXY15 | MATaade2 ade6 trp1-1 his3-11,15 can1-100 ura3-1 leu2-3,112 Δume6::HIS3 Δsum1::kanMX4 | E. Winter (51) |
This strain may carry his4X-ADE2-his4B.
This strain may carry ade2.
This strain may carry trp1.
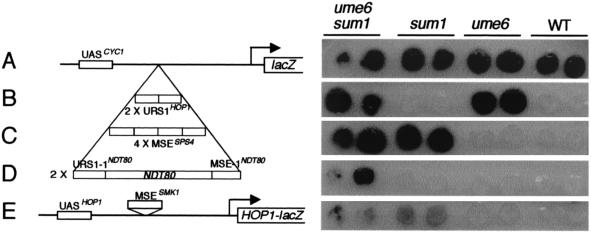
FIG. 3.
Derepression of a CYC1-(URS1-1-MSE-1)NDT80-lacZ reporter gene in mitotic cells requires mutation of both UME6 and SUM1. Duplicate patches of cells of a ume6 sum1 strain (first column), a sum1 strain (second column), a ume6 strain (third column), and a wild-type strain (fourth column) contained the following plasmids: (A) pLGΔ312(Bgl), which contains the CYC1-lacZ reporter gene described by Hepworth et al. (16); (B) pAV138-2, which contains two tandem copies of the URS1 site of the HOP1 gene inserted between the CYC1 UAS sites and TATA box in pLGΔ312(Bgl) (48); (C) pCYC1-SPS4-lacZ, which contains four tandem copies of a 29-bp fragment that spans the MSE site of the SPS4 gene inserted between the CYC1 UAS sites and TATA box in pLGΔ312(Bgl) (16, 17); (D) pCYC1-(URS1-1-MSE-1)NDT80-lacZ, which contains a 91-bp fragment that extends from the URS1-1 site to the MSE-1 site in the promoter region of the NDT80 gene (see Fig. 1), inserted between the CYC1 UAS sites and TATA box in pLGΔ312(Bgl) (see Materials and Methods); (E) pJX43, which contains a fragment spanning the MSE of the SMK1 promoter inserted into the URS1 site of a HOP1-lacZ reporter gene (37). The patches of cells were overlaid with X-Gal-containing agar (see Materials and Methods).
Yeast transformations were performed by the lithium acetate method (12). Standard genetic methods were used for mating, sporulation, and random spore disruption.
Media and growth conditions.
SD medium is a minimal medium (2% glucose, 0.7% yeast nitrogen base without amino acids) supplemented with 40 μg of adenine sulfate per ml, 20 μg of l-arginine per ml, 20 μg of l-histidine per ml, 60 μg of l-leucine per ml, 30 μg of l-lysine (mono-HCl) per ml, 20 μg of l-methionine per ml, 50 μg of l-phenylalanine per ml, 200 μg of l-threonine per ml, 40 μg of l-tryptophan per ml, 30 μg of l-tyrosine per ml, and 20 μg of uracil per ml. SD-X medium refers to SD medium that lacks supplement X. Rich medium (yeast-extract-peptone-dextrose [YEPD]), presporulation medium (yeast extract-peptone-acetate [YEPA]), and sporulation medium (SPO) were as described previously for SK1-derived strains (17). All yeast cultures were grown at 30°C.
Sporulation of SK1-derived strains was performed as follows. Cells were taken from a YEPA plate and were grown to late log phase in SD-uracil, and this culture was used to inoculate YEPA medium (1:100 dilution). When the YEPA culture reached a density of 1.0 × 107 to 2.0 × 107 cells per ml, the cells were harvested by centrifugation, washed once in 1% potassium acetate, and resuspended in SPO medium at a density of 2.0 × 107 cells per ml. The time of transfer of cells to sporulation medium is referred to as 0 h. The efficiency of ascus formation was assessed by examination of 200 or more cells by light microscopy after 24 h or more in SPO medium.
Plasmids.
A low-copy-number plasmid containing a truncated version of the NDT80 gene was constructed as follows. First, cSC131-1 was constructed by cloning the NotI-ClaI fragment spanning the GAL1-NDT80 fusion gene of cSC131 (5) into pRS316. Then, a PCR-amplified fragment that contained 505 bp of the sequence upstream of the initiator ATG of the NDT80 gene was generated with primers N80-505T (5′GGGGCGGCCGCCCATCAAGCGCTCCAAGC3′) and N80-1B (5′GGGGCATATGTTTAAGCGCTTTTTATAATATTGT3′) and pNKY1212 (52) as template. This PCR product was cloned into the SmaI site of pRS425 and then recovered from this plasmid as a NdeI-NotI fragment. This NdeI-NotI fragment was then used to replace the NdeI-NotI fragment of cSC131-1, which contains the GAL1 promoter region. The resultant plasmid, p1-1, contained 505 bp of sequence upstream of the NDT80 initiator ATG codon and the entire NDT80 coding region and downstream sequence. The initiator ATG codon of the NDT80 gene was within an engineered NdeI site. This plasmid was digested with EcoRI, and the resultant 304-bp and 6,262-bp fragments were recovered and religated to generate a plasmid, pRS316(−505)ndt80, that contained a truncated ndt80 gene which lacked the last 1,040 bp of the open reading frame (ORF).
Versions of pRS316(−505)ndt80 in which URS1 or MSE sites in the upstream region of the ndt80 gene were deleted were constructed as follows. The NdeI-NotI fragment containing the promoter region of the NDT80 gene was replaced with equivalent fragments, with the exception that a URS1 element or an MSE-1 element was deleted. These fragments were generated by the PCR-based overlap extension method (18). The outside primers were N80-505T and N80-1B (see above). The internal primers for deletion of 9 bp from the URS1-1 element to generate the (−505ΔU1)ndt80 minigene were N80-311T (5′CTTTACATTGTTACTATTTGACG3′) and N80-280B (5′CGTCAAATAGTAACAATGTAAAG3′); the internal primers for deletion of the 9 bp from the MSE-1 element to generate the (−505ΔM1)ndt80 minigene were N80-230T (5′AGGCCGTATGAGTAGAAAAC3′) and N80-202B (5′GTTTTCTACTCATACGGCCT3′); the internal primers for deletion of 8 bp from the URS1-2 element to generate the (−505ΔU2)ndt80 minigene were N80-176T (5′TCCTCTATACAGCTCTCTGA3′) and N80-149B (5′TCAGAGAGCTGTATAGAGGA3′); and the internal primers for deletion of 9 bp from the MSE-2 element to generate the (−505ΔM2)ndt80 minigene were N80-94T (5′GCCCTCCAACCTATTTAAGC3′) and N80-66B (5′GCTTAAATAGGTTGGAGGGC3′). See Fig. 1 and the text for a description of these elements and minigenes.
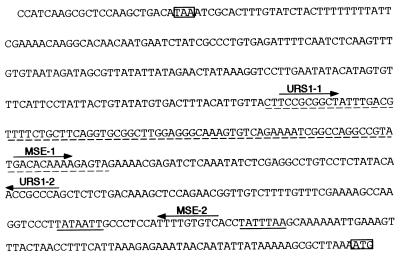
FIG. 1.
Sequence of the 5′-flanking region of the NDT80 gene. The sequence upstream of the initiator ATG codon of the NDT80 gene is given. All numbering in the text is with respect to the A of this ATG codon being nucleotide +1. This ATG codon and the stop codon of the upstream gene, EPT1, are boxed. Putative TATA boxes of the NDT80 gene are underlined. Arrows are over sequences that correspond to the consensus for a URS1 or an MSE and indicate the orientation of the element. See the text for the coding of the sites as URS1-1, URS1-2, MSE-1, and MSE-2. The NDT80 sequence inserted into pLGΔ312(Bgl) to create the CYC1-(URS1-1-MSE-1)NDT80-lacZ reporter gene (see Fig. 3) is underlined with a dashed line.
The (−505ΔU1ΔU2)ndt80 minigene that lacked both the URS1-1 and URS1-2 elements was constructed by ligating a 638-bp NaeI-BglII fragment from the plasmid containing the (−505ΔU1)ndt80 minigene with a 5,910-bp NaeI-BglII fragment from the plasmid containing the (−505ΔU2)ndt80 minigene. A plasmid containing the (−505ΔM1ΔM2)ndt80 minigene was constructed in the same manner from the plasmids containing the (−505ΔM1)ndt80 minigene and the (−505ΔM2)ndt80 minigene. The plasmids containing the (−505ΔU1ΔM1ΔM2)ndt80 minigene and the (−505ΔM1ΔU2ΔM2)ndt80 minigene were constructed by using the overlap extension method and the same primers that were used to delete the URS1-1 element and the URS1-2 element (see above), respectively, and the plasmid containing the (−505ΔM1ΔM2)ndt80 minigene as template. The resultant NdeI-NotI fragments were used to replace the equivalent fragment in pRS316(−505)ndt80.
The multicopy plasmid pLGΔ312(Bgl) containing a CYC1-lacZ reporter gene has been described previously (16). pCYC1-(URS1-1-MSE-1)NDT80-lacZ contains a 91-bp fragment extending from just upstream of the URS1-1NDT80 site to just downstream of the MSE-1NDT80 site (Fig. 1) inserted into the BglII site of pLGΔ312(Bgl). The insert fragment was obtained by PCR amplification of the NDT80 sequence with the primers N80-298T-BglII (5′GGGAGATCTCTTCCGCGGCTATTTGACG3′) and N80-207B-BglII (5′GGGAGATCTCTACTCTTTTGTGTCATACGG3′), which had additional sequence at their 5′ ends introducing BglII sites in order to facilitate cloning.
pPHOP1-NDT80 contains the HOP1 promoter fused to the NDT80 ORF and was constructed as follows. pNH59-2 (19) was cut with EcoRI and the ends were filled in with Klenow. This linearized DNA was then cut with NdeI, releasing a fragment containing 990 bp of sequence upstream of the HOP1 ORF. This fragment was ligated to 1,688-bp and 4,764-bp fragments generated by cutting cSC131-1 with NotI, filling in with Klenow, and cutting with NdeI.
The integrity of all PCR-generated DNAs was confirmed by sequencing.
Assay for β-galactosidase activity.
β-Galactosidase expression from lacZ reporter genes was monitored by use of a colony overlay assay (1). Ten milliliters of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)-containing agar (0.5% agar, 0.5 M potassium phosphate [pH 7.0], 6% dimethyl formamide, 0.1% sodium dodecyl sulfate, 0.2 mg of X-Gal per ml) was poured over colonies on plates, and the plates were incubated at 30°C until blue color development was evident.
RNA isolation and Northern analysis.
RNA preparation from yeast and Northern blot analysis were performed as described previously (17). Gene-specific probes were prepared with the following templates: NDT80, a 1.2-kb Eco47III-BamHI fragment from pNKY1212 (52); CLB1, a 500-bp EcoRV-EcoRV fragment from pMT417 (provided by M. Tyers); SMK1, an 800-bp StyI-StyI fragment from pLAKK40 (Krisak et al. [24]); SPS1, a 550-bp ClaI-EcoRV fragment from pSPS1-URA3 (10); SPS4, a 521-bp MluI-ClaI fragment from p4LE159 (16); and pC4, which contains an uncharacterized gene whose expression is similar in growing cells and through sporulation (27).
RESULTS
A model for the temporal regulation of expression of the NDT80 gene.
Expression of NDT80, a premiddle sporulation-specific gene, is tightly regulated and requires its own gene product for upregulation of its expression (5, 17). Transcripts are first detected after the activation of early meiotic genes, such as HOP1 and IME2, but prior to the activation of middle sporulation-specific genes, such as SPS1 and SPS4 (5, 17). Transcript accumulation is maximal at 6 h after transfer of cells to sporulation medium and then declines such that transcripts are barely detectable at 15 h of sporulation. In setting out to study the temporal regulation of expression of the NDT80 gene, we focused on four candidate regulatory sites present in the ≈500-bp interval between the initiator methionine codon of the NDT80 gene and the stop codon of the upstream ORF. Sequence inspection revealed two URS1 sites, which we refer to as URS1-1NDT80 and URS1-2NDT80, present at nucleotide (nt) −295 and nt −165, respectively, and two MSE sites, which we refer to as MSE-1NDT80 and MSE-2NDT80, present at nt −221 and nt −86, respectively (Fig. 1) (5). The latter MSE is located between two potential TATA box sequences.
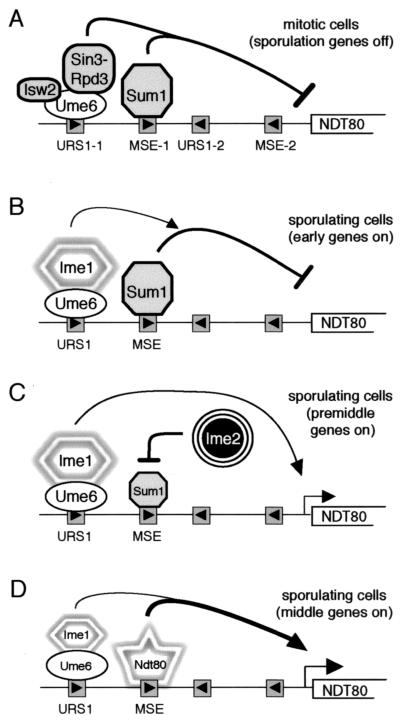
Based on the presence of both URS1 and MSEs in the upstream region of the NDT80 gene and the known roles of these elements in regulating early and middle sporulation-specific gene expression (see above), we put forth the following model for the temporal regulation of expression of this gene (Fig. 2). We suggest that in vegetatively growing cells, both Ume6 bound to URS1NDT80 and Sum1 bound to MSENDT80 assemble repression complexes that prevent expression of the NDT80 gene (13, 20, 21, 51) (Fig. 2A). We infer that early during sporulation, at the time that Ime1 is activating the expression of early meiotic genes, including IME2, Ime1 is also recruited to the putative Ume6-URS1NDT80 complex. We propose that activation of the NDT80 gene by the Ime1-Ume6-URS1NDT80 complex is initially prevented, however, by the continued presence of Sum1 at the MSENDT80 sites (Fig. 2B). Data presented in this study suggest that Ime2 promotes the first phase of NDT80 expression by inhibiting the repression activity of Sum1 and thereby allowing Ime1 to direct a low level of expression of the NDT80 gene (Fig. 2C). This is the premiddle phase of NDT80 expression. The newly synthesized Ndt80 would then compete with Sum1 for binding to the MSENDT80 sites and upregulate its own expression, leading to the middle phase of NDT80 expression (Fig. 2D). This would result in the peak of NDT80 transcript accumulation, coinciding with that of transcripts from MSE-regulated middle sporulation-specific genes. The transient decrease in the level of Sum1 that occurs during sporulation (29) may contribute to the regulated expression of the NDT80 gene. Finally, as the ability of Ime1 and Ndt80 to promote transcription is sequentially downregulated in a manner that is dependent on progression of the cells through the sporulation program (17, 35, 42), expression of NDT80 is shut off. As described below, we tested this model by assessing the contributions of the URS1NDT80 sites and of the MSENDT80 sites to the expression of the NDT80 gene.
FIG. 2.
Model for the temporal regulation of expression of the NDT80 gene. See text for details. The horizontal line represents the NDT80 promoter, with the URS1 and MSE sites depicted as shaded boxes with arrowheads (see Fig. 1). For simplicity, regulatory molecules are shown only at the URS1-1 and MSE-1 sites. Curved lines ending in an arrowhead indicate activation of transcription; curved lines ending with a bar indicate repression of transcription. URS1- or MSE-bound proteins, or complexes, that mediate repression are represented in gray; proteins, or complexes, that mediate activation are represented by shapes outlined with shadows. The reductions in size of the Sum1 symbol (C) and the Ime1-Ume6 symbols (D) denote a reduction in activity. (A) The NDT80 promoter in mitotic cells. (B to D) The NDT80 promoter in sporulating cells at the time that early meiotic genes, including IME2, are being expressed (B), after early genes, but before middle sporulation-specific genes have been activated (C), or at the time of middle sporulation-specific gene expression (D).
Role of Ume6 and Sum1 in mitotic repression of NDT80.
The observation that the chromosomal NDT80 gene is derepressed in vegetatively growing sum1 ume6 cells (51) implicates the URS1 sites and the MSE sites in the promoter region of the NDT80 gene as operator sites. We therefore tested the NDT80 sequence extending from nt −298, just upstream of the URS1-1NDT80 site, to nt −207, just downstream of the MSE-1NDT80 site (Fig. 1), for its ability to prevent expression of a heterologous reporter gene in vegetative cells. We cloned two copies of this fragment between the upstream activation sequence (UAS) and TATA box of a plasmid-borne CYC1-lacZ reporter gene and then monitored expression of the resultant CYC1-(URS1-1-MSE-1)NDT80-lacZ gene in wild-type, ume6, sum1, and ume6 sum1 cells (Fig. 3D). As controls, we also examined expression of a HOP1-lacZ reporter gene in which the URS1HOP1 site had been replaced with the MSE of the SMK1 gene (Fig. 3E) (37), a CYC1-lacZ reporter gene containing four copies of the MSE from the SPS4 gene (Fig. 3C) (16), and a CYC1-lacZ reporter gene containing two copies of the URS1 element from the HOP1 gene (Fig. 3B) (48). Each insert prevented expression of the plasmid-borne reporter gene in wild-type cells growing vegetatively as tested in a colony overlay assay for β-galactosidase expression (Fig. 3, WT column). As expected, mutation of UME6, but not mutation of SUM1, allowed expression of the CYC1-URS1HOP1-lacZ reporter gene (Fig. 3B) (48, 51); similarly, mutation of SUM1, but not mutation of UME6, allowed expression of the CYC1-4xMSESPS4-lacZ reporter gene (Fig. 3C) (17; J. Pak, unpublished observation) and the HOP1-MSESMK1-lacZ reporter gene (Fig. 3E) (51). However, mutation of both UME6 and SUM1 was required for expression of the CYC1-(URS1-1-MSE-1)NDT80-lacZ reporter gene in mitotic cells (Fig. 3D). This experiment showed that Ume6 and Sum1 serve redundant functions, presumably through URS1-1 and MSE-1, respectively, in supporting the ability of the NDT80-derived region extending from −298 to −207 to repress UASCYC1-driven expression in mitotic cells.
Expression of a plasmid-borne version of NDT80 parallels expression of the chromosomal NDT80 gene.
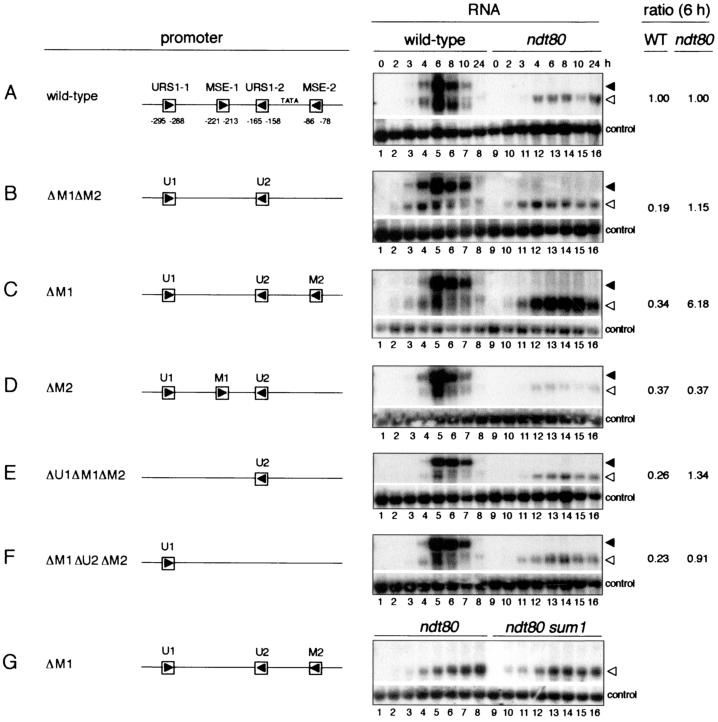
We used a truncated ndt80 gene present on a low-copy plasmid to investigate the contributions that the URS1NDT80 and MSENDT80 sites make to the expression of the NDT80 gene during sporulation. This reporter minigene, termed (−505WT)ndt80, which contained the NDT80 sequence from nt −505 to nt +880, with +1 being the start of the ORF, did not complement the sporulation deficiency of Δndt80/Δndt80 cells (data not shown). However, the presence of a full-length, plasmid-borne NDT80 gene with 505 bp of upstream region allowed Δndt80/Δndt80 cells to form spores (data not shown), indicating that 505 bp of the upstream region was sufficient to activate expression of the gene. We next monitored expression of the plasmid-borne (−505WT)ndt80 minigene by Northern blot analysis of RNA from diploid cells during vegetative growth and at various times after transfer to sporulation medium. Stable, minigene-encoded transcripts accumulated during sporulation, and these transcripts could be readily distinguished on the basis of size from transcripts of the wild-type NDT80 gene (Fig. 4A). This allowed us to compare expression of the plasmid-borne (−505WT)ndt80 minigene with expression of the chromosomal NDT80 gene in the same cells.
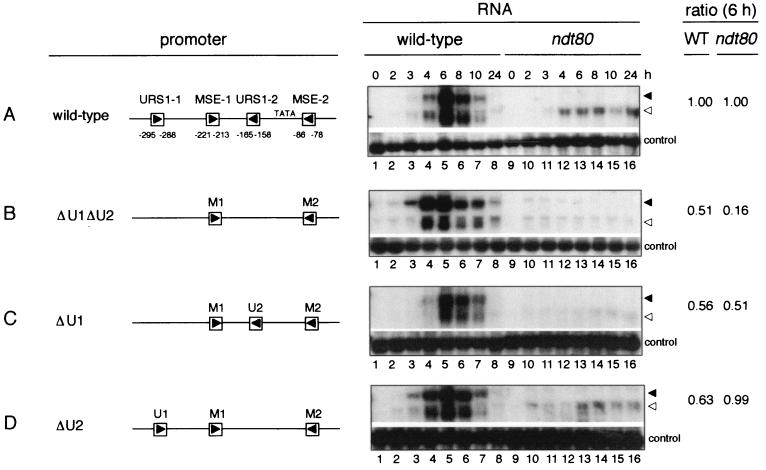
FIG. 4.
The URS1 sites are responsible for initial expression of the NDT80 gene. The Northern filters represented under the RNA heading contained RNA extracted from wild-type cells (lanes 1 to 8) and Δndt80/Δndt80 cells (lanes 9 to 16) harvested during vegetative growth (0 h) or at the indicated times, as noted above the top panel (in hours), after transfer of cells to sporulation medium. Cells used for the experiment of each panel harbored the following plasmid-borne ndt80 minigenes: (A) (−505WT)ndt80; (B) (−505ΔU1ΔU2)ndt80; (C) (−505ΔU1)ndt80; and (D) (−505ΔU2)ndt80. A schematic diagram of the promoter region of each of these minigenes is given under the promoter heading. Abbreviations: U1, URS1-1; M1, MSE-1; U2, URS1-2; M2, MSE-2. Δ denotes that the specified element has been deleted. The Northern filters were hybridized with a radioactively labeled NDT80-specific probe (top portion of each panel) and a control probe (bottom portion of each panel) prepared with pC4 (see Materials and Methods). The closed and open arrowheads denote the full-length chromosome-derived NDT80 transcripts and the truncated ndt80 minigene-derived transcripts, respectively. The ratio column presents a normalized ratio for the expression of each plasmid-borne (−505mutant)ndt80 minigene relative to the chromosomal NDT80 gene in the same cells, determined as follows. The WT column gives the intensity of the hybridization signal for transcripts derived from each plasmid-borne (−505mutant)ndt80 minigene at 6 h of sporulation (lane 5) relative to the intensity of the hybridization signal for transcripts derived from the chromosomal NDT80 gene in the same cells at 6 h of sporulation, normalized to the same ratio obtained for transcripts derived from cells containing the (−505WT)ndt80 plasmid-borne minigene (in panel A). The ndt80 ratio column gives the intensity of the hybridization signal for transcripts derived from each (−505mutant)ndt80 minigene in Δndt80/Δndt80 cells at 6 h after transfer to sporulation medium (lane 13), normalized to the loading control, relative to the amount of chromosome-derived NDT80 transcripts in wild-type cells at 6 h of sporulation (lane 5), normalized to the control probe, and then normalized to the same ratio obtained for transcripts derived from the (−505WT)ndt80 minigene (in panel A). Relative intensities of hybridization signals were determined by quantitation of phosphorimages obtained with a Molecular Dynamics STORM 860 PhosphorImager. The images were analyzed with ImageQuant (Molecular Dynamics, Inc.) and IPLab Gel (Signal Analytics Corporation) software. The images of autoradiograms presented in this figure are scans obtained with Adobe Photoshop 5.0 LE software and assembled with the use of Adobe Illustrator 10 and PowerPoint 98.
We found that the temporal pattern of expression of the plasmid-borne (−505WT)ndt80 minigene paralleled that of the chromosomal NDT80 gene (Fig. 4A, lanes 1 to 8). Transcripts were not detected in vegetative cells (Fig. 4A, lane 1). A low level of transcripts could first be detected between 3 and 4 h of sporulation (Fig. 4A, lanes 3 and 4), with maximal transcript accumulation occurring at 6 h (Fig. 4A, lane 5). This is the time at which Ndt80 is activating expression of middle sporulation-specific genes (6, 17). By 10 h of sporulation, transcript levels had declined to low levels (Fig. 4A, lane 7). At all times the amount of plasmid-borne minigene-derived transcripts that was present in the cells was similar to the amount of chromosome-derived NDT80 transcripts. Quantitation of the transcript levels at 6 h of sporulation indicated that transcript accumulation from the plasmid-borne minigene was 91% that of chromosome-derived NDT80 transcripts. The close correlation that we observed between the temporal patterns of expression of the plasmid-borne (−505WT)ndt80 minigene and the chromosomal NDT80 gene allowed us to use expression of the chromosomal gene as an internal control in monitoring the timing of expression of ndt80 minigenes containing promoter mutations. This internal comparison allowed us to ignore any small temporal differences in the overall pattern of gene expression that occurred between experiments. Such differences could result from variations in the timing of key regulatory events or in the synchrony of cells as the sporulation program proceeded.
We next monitored expression of the (−505WT)ndt80 minigene in Δndt80/Δndt80 cells transferred to sporulation medium (Fig. 4A, lanes 9 to 16). These cells, which lack functional Ndt80, arrest at pachytene (52) and do not express middle sporulation-specific genes (5, 6, 17). The onset of minigene expression in Δndt80/Δndt80 cells was the same as in wild-type cells, between 3 and 4 h after transfer of cells to sporulation medium, but there was no subsequent upregulation of expression (Fig. 4A, lanes 9 to 16). This pattern of expression was consistent with the notion that the initial phase of NDT80 expression is Ndt80 independent and that the subsequent upregulation of expression is Ndt80 dependent (5, 17). The continued presence of a low level of minigene transcripts in Δndt80/Δndt80 cells, even after 24 h in sporulation medium, was consistent with previous observations that early and premiddle genes continue to be expressed in cells that are unable to activate middle genes (e.g., references 17, 35, and 42).
The URS1 sites are required for the early phase of NDT80 gene expression.
Our model for the regulation of expression of the NDT80 gene proposed that the URS1 sites were responsible for the initial expression of the gene on transfer of cells to sporulation medium (Fig. 2C). Consistent with this model, expression of the plasmid-borne (−505ΔU1ΔU2)ndt80 minigene, which lacked both the URS1-1NDT80 and URS1-2NDT80 elements, appeared to be delayed relative to expression of the chromosomal wild-type gene (Fig. 4B, lanes 3 and 4). Transcripts from the wild-type chromosomal gene could be detected at 3 h after transfer of cells to sporulation medium, whereas minigene-derived transcripts were not readily detected in these cells until 4 h of sporulation (Fig. 4B, lane 4). We inferred that in the absence of URS1-driven expression, activation of the (−505ΔU1ΔU2)ndt80 minigene was delayed until Ndt80, expressed from the chromosomal NDT80 gene, promoted its expression. Consistent with this notion, expression of the plasmid-borne (−505ΔU1ΔU2)ndt80 minigene was barely detectable in Δndt80/Δndt80 cells (Fig. 4B, lanes 9 to 16), whereas the (−505WT)ndt80 minigene was expressed at a low level in these cells (Fig. 4A, lanes 9 to 16). In addition to regulating the time of onset of expression of the ndt80 minigene, the URS1NDT80 elements also contributed to its maximal expression. Transcript accumulation from the (−505ΔU1ΔU2)ndt80 minigene was 50% of that from the (−505WT)ndt80 minigene at 6 h of sporulation (Fig. 4A and B, lane 5). Both URS1-1NDT80 and URS1-2NDT80 contributed to maximal transcript accumulation in wild-type cells (Fig. 4C and D, lanes 1 to 8).
Sum1 prevents premature expression of the NDT80 gene.
As discussed above, we inferred that in vegetatively growing cells URS1NDT80-bound Ume6 and MSENDT80-bound Sum1 assembled repression complexes that prevented expression of the NDT80 gene (Fig. 2A). Early during sporulation, Ime1 would presumably be recruited to the Ume6-URS1NDT80 complexes in the promoter region of the NDT80 gene at the same time that it was recruited to similar complexes in the promoter region of early meiotic genes. Whereas early meiotic genes would be expressed immediately, we proposed that activation of the NDT80 gene by the Ime1-Ume6-URS1NDT80 complex was initially prevented by the continued presence of the Sum1 repression complexes at the MSENDT80 sites (Fig. 2B). We tested this idea by assessing the effect of the deletion of SUM1 on the expression profile of NDT80. We note that SUM1 has been shown previously to have no essential role in spore formation (29).
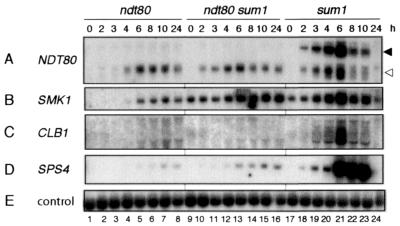
Examination of the temporal profile of gene expression during sporulation of a Δsum1/Δsum1 strain showed that both the chromosomal NDT80 gene and the (−505WT)ndt80 minigene were expressed prematurely in the absence of Sum1 (Fig. 5A, lanes 17 to 24). Whereas NDT80 transcripts were generally first detected at 3 to 4 h in wild-type cells (Fig. 4, lanes 3 to 6; chromosomal transcript), transcripts from the chromosomal NDT80 gene and the (−505WT)ndt80 minigene could be readily detected at 2 h after transfer of Δsum1/Δsum1 cells to sporulation medium (Fig. 5A, lane 18). Similarly, transcripts of the (−505WT)ndt80 minigene accumulated earlier in cells of the Δndt80/Δndt80 Δsum1/Δsum1 strain than in cells of the Δndt80/Δndt80 strain on transfer to sporulation medium (Fig. 5A, compare lanes 9 to 12 with lanes 1 to 4). We also monitored expression of the premiddle gene, SMK1, and the middle sporulation-specific genes, CLB1 and SPS4, whose expression is maximal around 6 to 8 h of sporulation in wild-type cells (5, 17). As reported previously, we found that SMK1 was expressed in mitotic Δsum1/Δsum1 cells (Fig. 5B, lanes 9 and 17) (29, 51) and in Δndt80/Δndt80 cells in sporulation medium, albeit at a reduced level (Fig. 5B, lanes 1 to 8) (5, 17). The middle sporulation-specific genes, CLB1 and SPS4, were expressed in Δsum1/Δsum1 cells but not in Δndt80/Δndt80 cells (Fig. 5C and D, lanes 1 to 8 and 17 to 24) (5, 17, 51) or in Δndt80/Δndt80 Δsum1/Δsum1 cells in sporulation medium (Fig. 5C and D, lanes 9 to 16). This analysis supported our suggestion that Sum1 prevents premature expression of the NDT80 gene in sporulating cells.
FIG. 5.
SUM1 prevents premature expression of the NDT80 gene. Northern filters were prepared with RNA from Δndt80/Δndt80 cells (lanes 1 to 8), Δndt80/Δndt80 Δsum1/Δsum1 cells (lanes 9 to 16), and Δsum1/Δsum1 cells (lanes 17 to 24) that contained the plasmid-borne (−505WT)ndt80 minigene. Cells were harvested during vegetative growth (0 h) or at the indicated time (in hours), as noted above panel A, after transfer of cells to sporulation medium. The filter was hybridized sequentially with the gene-specific probes denoted on the left. The closed and open arrowheads on the right of panel A denote the full-length chromosome-derived NDT80 transcripts and the truncated ndt80 minigene-derived transcripts, respectively.
The MSE sites prevent premature expression of the NDT80 gene.
To test the notion that Sum1 prevents early expression of the (−505WT)ndt80 minigene by maintaining repression through the MSENDT80 sites, we monitored expression of a minigene in which both MSEs had been deleted. As noted above, expression of the plasmid-borne (−505WT)ndt80 minigene was coincident with expression of the chromosomal NDT80 gene (Fig. 6A). Deletion of both MSEs advanced transcript accumulation to a modest extent; transcript accumulation from the (−505ΔM1ΔM2)ndt80 minigene was slightly higher than that from the chromosomal NDT80 gene at 3 h of sporulation (Fig. 6B, lane 3, and data not shown). A longer autoradiographic exposure indicated that this was also the case at 2 h of sporulation (data not shown). The earlier onset of expression of the (−505ΔM1ΔM2)ndt80 minigene relative to the (−505WT)ndt80 minigene was particularly evident in Δndt80/Δndt80 cells (Fig. 6, compare panels A and B, lanes 9 to 16). We note that the temporal expression profiles of the MSE-less (−505ΔM1ΔM2)ndt80 minigene in wild-type cells (Fig. 6B, lanes 1 to 8) and in Δndt80/Δndt80 cells (Fig. 6B, lanes 9 to 16) were similar to those of the (−505WT)ndt80 minigene in Δsum1/Δsum1 cells (Fig. 5A, lanes 17 to 24) and in Δndt80/Δndt80 Δsum1/Δsum1 cells (Fig. 5A, lanes 9 to 16), respectively. These observations support our suggestion (Fig. 2B) that Sum1 acts through the MSEs to prevent premature expression of the NDT80 gene in sporulating cells. In addition, the MSE sites are required for Ndt80 to upregulate expression of the NDT80 gene. The maximal level of expression of the plasmid-borne (−505ΔM1ΔM2)ndt80 minigene was only one-fifth that of the (−505WT)ndt80 minigene (Fig. 6A and B, lane 5).
FIG. 6.
The MSE sites prevent premature expression of the NDT80 gene and mediate upregulation of its expression midway through sporulation. (A to F) The Northern filters represented under the RNA column heading contained RNA extracted from wild-type cells (lanes 1 to 8) and Δndt80/Δndt80 cells (lanes 9 to 16) harvested during vegetative growth (0 h) or at the indicated times, as noted above the top panel (in hours), after transfer of cells to sporulation medium. (G) The Northern filter contained RNA extracted from Δndt80/Δndt80 cells (lanes 1 to 8) and Δndt80/Δndt80 Δsum1/Δsum1 cells (lanes 9 to 16). Cells used for the experiment of each panel harbored the following plasmid-borne ndt80 minigenes: (A) (−505WT)ndt80; (B) (−505ΔM1ΔM2)ndt80; (C and G) (−505ΔM1)ndt80; (D) (−505ΔM2)ndt80; (E) (−505ΔU1ΔM1ΔM2)ndt80; (F) (−505ΔM1ΔU2ΔM2)ndt80. The closed and open arrowheads on the right of panel A denote the full-length chromosome-derived NDT80 transcripts and the truncated ndt80 minigene-derived transcripts, respectively. For experimental details and explanations of nomenclature, see the legend to Fig. 4. The filter represented in panel A is the same as that shown in Fig. 4A.
Comparison of the expression patterns of ndt80 minigenes lacking either MSE-1 or MSE-2 suggested that only MSE-1NDT80 had a role in preventing premature expression of the NDT80 gene in wild-type cells, whereas both MSEs contributed to Ndt80-dependent upregulation of minigene expression (Fig. 6, compare panels A to D, lanes 1 to 8). Transcript accumulation from the (−505ΔM1)ndt80 minigene (Fig. 6C, lane 5) and from the (−505ΔM2)ndt80 minigene (Fig. 6D, lane 5) was approximately one-third that from the (−505WT)ndt80 minigene in wild-type cells at 6 h of sporulation (Fig. 6A, lane 5). We also monitored the effect of deletion of each URS1 element on expression of the (−505ΔM1ΔM2)ndt80 minigene. The levels of expression of the (−505ΔU1ΔM1ΔM2)ndt80 minigene (Fig. 6E, lanes 1 to 8) and the (−505ΔM1ΔU2ΔM2)ndt80 minigene (Fig. 6F, lanes 1 to 8) in wild-type cells were similar to each other and to that of the (−505ΔM1ΔM2)ndt80 minigene (Fig. 6B, lanes 1 to 8). This was also the case for expression in Δndt80/Δndt80 cells (Fig. 6B, E, and F, lanes 9 to 16). This suggested, as noted above, that both URS1NDT80 sites were able to promote expression of the NDT80 gene; we could not, however, distinguish the individual contributions made by each URS1NDT80 site. It is possible that we could not reliably quantify the low levels of expression observed in Δndt80/Δndt80 cells.
Unexpectedly, we found that not only was the (−505ΔM1)ndt80 minigene expressed prematurely in Δndt80/Δndt80 cells but also that it was expressed at a sixfold-higher level than the (−505WT)ndt80 minigene in Δndt80/Δndt80 cells (Fig. 6, compare panels A and C, lanes 9 to 16). This high level of expression of the (−505ΔM1)ndt80 minigene required that MSE-2NDT80 be present (Fig. 6B and C, lanes 9 to 16). Thus, MSE-1NDT80 appeared to act as an operator that prevented Ndt80-independent activation from MSE-2NDT80. To test the possibility that Sum1 might form a novel activation complex at MSE-2NDT80 in the absence of Ndt80, we compared expression of the (−505ΔM1)ndt80 minigene in Δndt80/Δndt80 cells and in Δndt80/Δndt80 Δsum1/Δsum1 cells in sporulation medium. We found that the minigene was expressed at similar levels in both Δndt80/Δndt80 cells and in Δndt80/Δndt80 Δsum1/Δsum1 cells (Fig. 6G). Thus, Sum1 was not required for the Ndt80-independent, MSE-2NDT80-mediated activation that was revealed on removal of the MSE-1NDT80 site. We have not explored this unusual expression pattern further.
The requirement for IME2 for expression of the NDT80 gene can be bypassed by mutation of SUM1.
IME2 is an early sporulation-specific gene that encodes a kinase (23, 53) that regulates several meiotic events, including the correct timing of premeiotic DNA synthesis (8, 9, 14), maximal expression of early meiotic genes (35, 42), and expression of middle sporulation-specific genes (35, 42) and of NDT80 (17). We have investigated further the role of IME2 in controlling the expression of NDT80.
Because it has been suggested that Ime2 promotes premeiotic DNA synthesis in early sporulating cells by causing the destruction of Sic1, a Cdk inhibitor, we first tested whether the absence of Sic1 might bypass the need for IME2 not only for DNA synthesis (8) but also for activation of expression of NDT80. Northern blot analysis showed that NDT80 is not expressed in Δime2/Δime2 Δsic1/Δsic1 cells after transfer to sporulation medium (data not shown). Thus, mutation of SIC1 did not allow Ime2-independent expression of NDT80. Control experiments showed, as expected, that NDT80 was expressed in Δsic1/Δsic1 cells and was not expressed in Δime2/Δime2 cells on transfer to sporulation medium (data not shown).
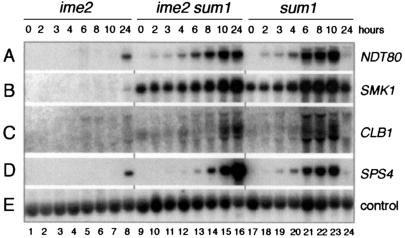
Taking into consideration our suggestion that the first phase of NDT80 expression depended on a reduction in Sum1-mediated repression, we next tested whether the requirement for IME2 in NDT80 expression might reflect a role for Ime2 as a regulator of the activity of Sum1. We therefore monitored gene expression in cells of an Δime2/Δime2 Δsum1/Δsum1 strain. Whereas NDT80 was expressed at a low level only after extended incubation of Δime2/Δime2 cells in sporulation medium (Fig. 7A, lanes 1 to 8), NDT80 transcripts could be detected at 2 h in Δime2/Δime2 Δsum1/Δsum1 cells (Fig. 7A, lanes 9 to 16), as was also the case in Δsum1/Δsum1 cells (Fig. 7A, lanes 17 to 24; Fig. 5A, lanes 17 to 24), and by 8 h significant transcript accumulation had occurred (Fig. 7A, lane 14). This suggested that Ime2 promoted the early phase of NDT80 expression by inhibiting Sum1-dependent repression. Additionally, the Ndt80 that was expressed in the Δime2/Δime2 Δsum1/Δsum1 cells was active, as both CLB1 and SPS4 were also expressed in these cells but not in Δime2/Δime2 cells in sporulation medium (Fig. 7C and D). Consistent with the observation that Ime2 plays multiple roles during sporulation (reviewed in reference 25), Δime2/Δime2 Δsum1/Δsum1 cells did not form spores. We concluded that IME2 activates middle gene expression, at least in part, by relieving Sum1-mediated repression of NDT80, as noted in Fig. 2C.
FIG. 7.
IME2 activates middle sporulation-specific gene expression by alleviating Sum1-mediated repression of the NDT80 gene. A Northern filter was prepared with RNA from ime2/ime2 cells (lanes 1 to 8), ime2/ime2 Δsum1/Δsum1 cells (lanes 9 to 16), and Δsum1/Δsum1 cells (lanes 17 to 24) harvested during vegetative growth (lanes 1, 9, and 17) or at the indicated time after transfer of cells to sporulation medium, as noted above panel A. The filter was hybridized sequentially with the gene-specific probes denoted on the right.
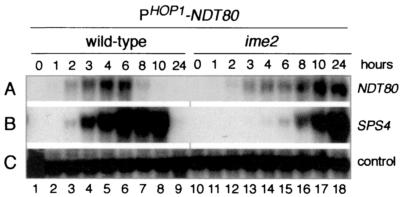
The activity of Ndt80 is posttranslationally inhibited on activation of the meiotic recombination checkpoint (5, 17, 47). Although Ime2 is a candidate kinase for posttranslational activation of Ndt80, the experiment described above (Fig. 7) suggested that regulation of NDT80 by IME2 occurs primarily at the transcriptional level. Moreover, ectopically produced Ndt80 is capable of activating MSE-dependent gene expression in vegetatively growing cells, which do not express IME2 (5). To test further the suggestion that Ime2 has no essential role in posttranscriptional regulation of Ndt80 in sporulating cells, we constructed a HOP1-NDT80 fusion gene in which the ORF of the NDT80 gene was under the control of the promoter of the early meiotic gene, HOP1 (see Materials and Methods). Neither Ime2 nor Ndt80 are required for expression of HOP1, although Ime2 is involved in upregulation of its expression (reviewed in reference 49). We found that expression of NDT80 in an Ime2-independent manner from the HOP1-NDT80 fusion gene (Fig. 8A, lanes 10 to 18) was sufficient to activate expression of the middle sporulation-specific gene, SPS4, in cells of a Δime2/Δime2 strain in sporulation medium (Fig. 8B, lanes 10 to 18). We note that although NDT80 transcripts derived from the HOP1-NDT80 gene could be detected at 2 to 3 h in the Δime2/Δime2 strain, maximal accumulation of these transcripts occurred only at 10 h. This is consistent with the known requirement for IME2 for the normal kinetics and full expression of early meiotic genes (35, 42). We infer that the delayed expression of Ndt80 accounted, at least in part, for the delayed expression of the Ndt80-dependent middle sporulation-specific gene SPS4 in this experiment. Although it is possible that Ime2-dependent phosphorylation of Ndt80 contributes to its full activity, our results indicated that Ime2 kinase is not essential for the transcription factor activity of Ndt80.
FIG. 8.
Ndt80 that is ectopically expressed in ime2/ime2 cells in sporulation medium is active. A Northern filter was prepared with RNA extracted from wild-type cells (lanes 1 to 9) and ime2/ime2 cells (lanes 10 to 18) during vegetative growth (lanes 1 and 9) or at the indicated time after transfer of cells to sporulation medium, as noted above panel A. Both the wild-type cells and the ime2/ime2 cells harbored a plasmid that contained a HOP1-NDT80 fusion gene, in which the NDT80 coding region was fused to the promoter region (see Materials and Methods). The filter was hybridized sequentially with the gene-specific probes denoted on the right.
DISCUSSION
Roles of the URS1NDT80 and MSENDT80 sites in generating the premiddle expression profile of the NDT80 gene.
In this study we have examined the role of the URS1 and MSE sites that are present in the promoter region of the NDT80 gene in regulation of its expression. We postulated that the overlapping windows of URS1- and MSE-mediated repression and activation were responsible for the distinctive premiddle expression pattern of the NDT80 gene (Fig. 2). Our experiments showed that a plasmid-borne ndt80 minigene lacking the URS1 elements was expressed in NDT80/NDT80 cells with the same kinetics as a middle gene; the premiddle phase of expression did not occur and expression of the URS1-less minigene was entirely Ndt80 dependent. In contrast, a plasmid-borne ndt80 minigene lacking the MSEs was expressed prematurely and was not significantly upregulated. Indeed, the levels of expression of the MSE-less minigene in wild-type cells and of the (−505WT)ndt80 minigene in Δndt80/Δndt80 cells were similar. We also found that the (−505WT)ndt80 minigene was expressed prematurely on mutation of SUM1 and that this early expression was independent of NDT80.
These observations support the idea that a Sum1-associated repression complex bound at MSENDT80 prevents URS1NDT80-tethered Ime1 from activating transcription of the NDT80 gene at the time that Ime1 is activating the expression of early meiotic genes. Our data are consistent with the idea that a decrease in the efficiency of Sum1-mediated repression as cells progress through the early events of the sporulation program allows the previously inactive Ime1 tethered at the URS1NDT80 sites to promote a low level of expression of the NDT80 gene. This initial expression would account for the Ime1 dependence of NDT80 expression (5). This proposal is also supported by the observations that a transient reduction in the level of Sum1 protein occurs from 6 to 8 h of sporulation (29) and that mutation of SUM1 leads to premature expression of NDT80 (this study). This initial phase of URS1-dependent NDT80 expression is followed by Ndt80-dependent upregulation of its own expression, which requires the MSENDT80 sites and occurs concomitantly with Ndt80-dependent activation of a set of middle MSE-regulated sporulation-specific genes. The presumed replacement of Sum1 by Ndt80, particularly at the MSE-1NDT80 site, as cells progress into the middle phase of sporulation could be a reflection of a higher affinity of MSE-1NDT80 for Ndt80 than for Sum1 as well as a reduction in the level of Sum1 (29, 51).
We note that although Xie et al. (51) reported that MSE-2NDT80 acts as a more efficient Sum1-dependent operator than does MSE-1NDT80, we found that MSE-1NDT80, but not MSE-2NDT80, was responsible for preventing expression of the (−505WT)ndt80 minigene at early times of sporulation. It is possible that differing experimental approaches account for these differing observations. Whereas Xie et al. (51) monitored repression by testing the ability of MSE elements to substitute for the URS1HOP1 operator element in preventing expression of a HOP1-lacZ reporter gene in mitotic cells, we monitored the effect of deletion of the MSE on the temporal expression pattern of the (−505)ndt80 minigene during sporulation. We note that examination of the role of MSE-2NDT80 in its natural context is complicated by its close juxtaposition to the putative TATA box of the NDT80 gene. We cannot exclude the possibility that deletion of MSE-2NDT80 may have indirectly affected the function of the promoter (see below). However, we were also unable to detect operator activity for MSE-2NDT80 in the context of a CYC1-lacZ reporter gene in mitotic cells (data not shown).
As cells progress beyond the middle portion of the sporulation program, NDT80 expression is downregulated in an NDT80-dependent manner. This downregulation does not occur in cells that arrest at pachytene (17). The turn-down of NDT80 expression could occur in several ways. For example, it is possible that the transcriptional activation function or DNA-binding function of Ndt80 is inactivated by the product of a middle sporulation-specific gene. Alternatively, MSE-bound Ndt80 could once again be replaced by Sum1 or by a yet-to-be identified MSE-binding or MSE-tethered repressor that is encoded by a middle gene.
Additional regulators of expression of the NDT80 gene?
Although the key aspects of our model are supported by our data, some aspects of our data remain unexplained. For example, the observation that the (−505ΔM1)ndt80 minigene was expressed to a relatively high level in Δndt80/Δndt80 cells in sporulation medium, a situation which has little biological relevance, was unexpected. This unusual expression pattern, which was observed only with this minigene, depended on the MSE-2 site and did not occur in NDT80/NDT80 cells. Because Δndt80/Δndt80 cells, which arrest at pachytene, fail to turn down Ime1-mediated expression of early meiotic genes, it is possible that continued URS1-mediated expression of this (−505ΔM1)ndt80 minigene allowed continued transcript accumulation. The dependence on MSE-2 might be a fortuitous reflection of the close juxtaposition of this site to the putative TATA element of the NDT80 gene. As mentioned above, it is possible that deletion of MSE-2 might nonspecifically reduce the strength of the promoter. Although none of our experiments directly addressed this possibility, we note that several versions of the ndt80 minigenes that lacked the MSE-2 site were expressed and therefore contained a functional promoter, but none was expressed at a high level. It is also possible that MSE-2 functions as an Ndt80-independent UAS. We have ruled out the possibility that Sum1 served a novel role as an activator when bound at MSE-2; expression of the (−505ΔM1)ndt80 minigene was high in both Δndt80/Δndt80 cells and Δndt80/Δndt80 Δsum1/Δsum1 cells.
In the course of this study we were surprised to find that multiple copies of a 21-bp fragment spanning URS1-1NDT80 or URS1-2NDT80 or a 22-bp fragment spanning MSE-1NDT80 or MSE-2NDT80 failed to act as operator elements when inserted into the CYC1-lacZ reporter gene (data not shown). The observation that mutation of both UME6 and SUM1 was required to allow expression of the CYC1-(URS1-1-MSE-1)NDT80-lacZ reporter gene in mitotic cells (Fig. 3) suggested that both URS1-1NDT80 and MSE-1NDT80 promoted the assembly of a repression complex. It is possible that repression complexes were efficiently assembled at these sites only in the context of the NDT80-derived sequence present in the 91-bp fragment. For example, it is possible that an expanded site provided increased affinity for binding of the protein or an accessory factor relative to the 21-bp or 22-bp sites. It is also possible that an additional site within the 91-bp NDT80-derived sequence acted in conjunction with either the URS1-1NDT80 site or the MSE-1NDT80 site to direct efficient repression.
It is clear that there are additional complexities in the NDT80 promoter that remain to be elucidated. As suggested by Xie et al. (51), the relative affinities of Ndt80 and Sum1 for MSE sites could be sequence and context dependent. The DNA-binding and regulatory activities of Ndt80 and Sum1 could be affected by posttranslational modifications as well as by cofactors. Other DNA elements, including cryptic UAS sites in the vector sequence and sequences within the first 880 bp of the coding region of NDT80, could influence expression of the minigene. Also, unidentified regulators may exist that bind to the URS1, MSE, or other elements in the promoter region of the NDT80 gene and contribute to its regulated expression.
Coordinate regulation of expression of the NDT80 and SMK1 genes.
The sporulation-specific gene SMK1, which encodes a mitogen-activated protein kinase that is required for spore wall development, was initially considered to be expressed as a middle gene (24, 37). Other studies, however, indicate that SMK1 is a member of the premiddle class of sporulation genes. SMK1 transcripts are first detected after the onset of expression of early meiotic genes and prior to activation of middle sporulation-specific genes and the expression of SMK1 is subsequently upregulated in an Ndt80-dependent manner (5, 17). Consistent with their pattern of coregulation, both NDT80 and SMK1, but not middle sporulation-specific genes, are expressed at a low level in cells that arrest at pachytene in response to defects in meiotic recombination (5, 17, 29).
Three regulatory elements have been identified in the promoter region of the SMK1 gene: an Abf1-binding site, a URS1 element, and an MSE (37). The Abf1-binding site acts nonspecifically to upregulate expression of the SMK1 gene (37). The MSESMK1 site prevents SMK1 expression in mitotic and early meiotic cells and upregulates its expression midway into the sporulation program (37). On mutation of the MSESMK1 site, the URS1SMK1 site acts early during sporulation to direct a low level of expression of the SMK1 gene (37). Thus, for both the NDT80 gene and the SMK1 gene, a combination of URS1 and MSE sites appears to be responsible for setting the premiddle pattern of gene expression. It will be interesting to analyze in greater detail the expression pattern of other sporulation-specific genes that contain both URS1 and MSE sites in their promoter region (6, 38)
Roles of IME2 and SUM1 in NDT80 expression.
IME2 encodes a protein kinase that is required for multiple processes during sporulation, including full expression and subsequent downregulation of expression of early meiotic genes (35), the degradation of Sic1 (8), the correct timing of premeiotic DNA replication (9, 14), expression of NDT80 (17), and activation of middle sporulation-specific gene expression (35, 42). We have shown that the requirement for Ime2 in the activation of middle sporulation-specific genes is a consequence of its key role in expression of NDT80. The middle sporulation-specific gene SPS4 was expressed in Δime2/Δime2 cells that ectopically expressed NDT80 from the HOP1 promoter. Based on our observation that NDT80 is expressed in Δime2/Δime2 Δsum1/Δsum1 cells, we have speculated that Ime2 inactivates Sum1. Moreover, our observation that mutation of SUM1 bypasses the requirement for Ime2 for both the expression and transcriptional activity of NDT80 indicates that Ime2 has no essential role as a direct activator of Ndt80. It remains possible, however, that Ime2-dependent phosphorylation of Ndt80 contributes to its full activity.
How does Ime2 inactivate Sum1? Lindgren et al. (29) demonstrated that although the level of SUM1 mRNA remains constant through sporulation, the level of Sum1 protein fluctuates, reaching its lowest level around the time that middle sporulation-specific genes are induced. This suggests that Ime2 might be a regulator of degradation of Sum1, with Sum1 being stabilized in Δime2/Δime2 cells. It is tantalizing to suggest that Ime2 might serve this function directly by phosphorylating Sum1 in a manner that targets it for degradation. This is analogous to the recent observation that the availability of Ime1 is regulated by Ime2-dependent phosphorylation targeting it for degradation (15).
Regulation of NDT80 expression and activity by the meiotic recombination checkpoint.
Defects in certain aspects of meiotic recombination and chromosome synapsis during sporulation generate a checkpoint signal that is transmitted to downstream targets and leads to arrest at pachytene, prior to entry into the meiotic divisions (reviewed in reference 39). NDT80 is one of the targets of this checkpoint. The expression of NDT80 is reduced and the ability of Ndt80 to activate expression of middle sporulation-specific genes is inhibited when the meiotic recombination checkpoint is triggered (5, 17, 52). On the basis of the present study, we suggest that the low level of NDT80 expression that is observed in checkpoint-arrested cells reflects URS1NDT80-dependent expression.
Our data suggest that Sum1 is regulated by Ime2 (see above). Interestingly, Sum1 is also a target of the meiotic recombination checkpoint; on activation of this checkpoint, Sum1 is stabilized (29). An intriguing possibility is that Ime2 transmits the checkpoint signal to Sum1; in this case, the checkpoint would prevent Ime2 from acting to destabilize Sum1. If this were the case, then putative checkpoint-mediated inactivation of Ime2 would occur only after Ime2 had served its role in promoting the initial phase of NDT80 expression. Subsequent checkpoint-mediated stabilization of Sum1 would allow Sum1-dependent repression of NDT80 gene expression to be reestablished. Additionally, any Ndt80 that had been synthesized would have its transcription factor activity inhibited. Further study will lead to a more complete understanding of the regulatory events that coordinate the sporulation-specific transcriptional cascade with progression through the sporulation program.
Acknowledgments
We thank Ed Winter and his coworkers for communicating results prior to publication and for helpful discussions. We thank Helena Friesen and Ghadeer Shubassi for their helpful comments on the manuscript.
This work was supported by a Canadian Institute of Health Research grant (MOP-6826) to J.S. J.P. was supported in part by an Ontario Government Scholarship for Science and Technology and by a University of Toronto Open Fellowship.
REFERENCES
- 1.Barral, Y., S. Jentsch, and C. Mann. 1995. G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev. 9:399-409. [DOI] [PubMed] [Google Scholar]
- 2.Bowdish, K. S., H. E. Yuan, and A. P. Mitchell. 1994. Analysis of RIM11, a yeast protein kinase that phosphorylates the meiotic activator IME1. Mol. Cell. Biol. 14:7909-7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brachmann, C. B., J. M. Sherman, S. E. Devine, E. E. Cameron, L. Pillus, and J. D. Boeke. 1995. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 9:2888-2902. [DOI] [PubMed] [Google Scholar]
- 4.Chi, M. H., and D. Shore. 1996. SUM1-1, a dominant suppressor of SIR mutations in Saccharomyces cerevisiae, increases transcriptional silencing at telomeres and HM mating-type loci and decreases chromosome stability. Mol. Cell. Biol. 16:4281-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu, S., and I. Herskowitz. 1998. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell 1:685-696. [DOI] [PubMed] [Google Scholar]
- 6.Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699-705. [DOI] [PubMed] [Google Scholar]
- 7.Derbyshire, M. K., K. G. Weinstock, and J. N. Strathern. 1996. HST1, a new member of the SIR2 family of genes. Yeast 12:631-640. [DOI] [PubMed] [Google Scholar]
- 8.Dirick, L., L. Goetsch, G. Ammerer, and B. Byers. 1998. Regulation of meiotic S phase by Ime2 and a Clb5.6-associated kinase in Saccharomyces cerevisiae. Science 281:1854-1857. [DOI] [PubMed] [Google Scholar]
- 9.Foiani, M., E. Nadjar-Boger, R. Capone, S. Sagee, R. Hashimshoni, and Y. Kassir. 1996. A meiosis-specific protein kinase, Ime2, is required for the correct timing of DNA replication and for spore formation in yeast meiosis. Mol. Gen. Genet. 253:278-288. [DOI] [PubMed] [Google Scholar]
- 10.Friesen, H., R. Lunz, S. Doyle, and J. Segall. 1994. Mutation of the SPS1-encoded protein kinase of Saccharomyces cerevisiae leads to defects in transcription and morphology during spore formation. Genes Dev. 8:2162-2175. [DOI] [PubMed] [Google Scholar]
- 11.Gailus-Durner, V., J. Xie, C. Chintamaneni, and A. K. Vershon. 1996. Participation of the yeast activator Abf1 in meiosis-specific expression of the HOP1 gene. Mol. Cell. Biol. 16:2777-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gietz, D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldmark, J. P., T. G. Fazzio, P. W. Estep, G. M. Church, and T. Tsukiyama. 2000. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103:423-433. [DOI] [PubMed] [Google Scholar]
- 14.Guttmann-Raviv, N., E. Boger-Nadjar, I. Edri, and Y. Kassir. 2001. Cdc28 and Ime2 possess redundant functions in promoting entry into premeiotic DNA replication in Saccharomyces cerevisiae. Genetics 159:1547-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guttmann-Raviv, N., S. Martin, and Y. Kassir. 2002. Ime2, a meiosis-specific kinase in yeast, is required for destabilization of its transcriptional activator, Ime1. Mol. Cell. Biol. 22:2047-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hepworth, S. R., L. K. Ebisuzaki, and J. Segall. 1995. A 15-base-pair element activates the SPS4 gene midway through sporulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:3934-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hepworth, S. R., H. Friesen, and J. Segall. 1998. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:5750-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 19.Hollingsworth, N. M., L. Goetsch, and B. Byers. 1990. The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell 61:73-84. [DOI] [PubMed] [Google Scholar]
- 20.Kadosh, D., and K. Struhl. 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89:365-371. [DOI] [PubMed] [Google Scholar]
- 21.Kadosh, D., and K. Struhl. 1998. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 12:797-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klar, A. J., S. N. Kakar, J. M. Ivy, J. B. Hicks, G. P. Livi, and L. M. Miglio. 1985. SUM1, an apparent positive regulator of the cryptic mating-type loci in Saccharomyces cerevisiae. Genetics 111:745-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kominami, K., Y. Sakata, M. Sakai, and I. Yamashita. 1993. Protein kinase activity associated with the IME2 gene product, a meiotic inducer in the yeast Saccharomyces cerevisiae. Biosci. Biotech. Biochem. 57:1731-1735. [DOI] [PubMed] [Google Scholar]
- 24.Krisak, L., R. Strich, R. W. Winters, J. P. Hall, M. J. Mallory, D. Kreitzer, R. S. Tuan, and E. Winter. 1994. SMK1, a developmentally regulated MAP kinase, is required for spore wall assembly in Saccharomyces cerevisiae. Genes Dev. 8:2151-2161. [DOI] [PubMed] [Google Scholar]
- 25.Kupiec, M., B. Byers, R. E. Esposito, and A. P. Mitchell. 1997. Meiosis and sporulation in Saccharomyces cerevisiae, p. 899-1036. In J. R. Pringle, J. R. Broach, and E. W. Jones (ed.), The molecular biology of the yeast Saccharomyces: cell cycle and cell biology. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 26.Laurenson, P., and J. Rine. 1991. SUM1-1: a suppressor of silencing defects in Saccharomyces cerevisiae. Genetics 129:685-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Law, D. T. S., and J. Segall. 1988. The SPS100 gene of Saccharomyces cerevisiae is activated late in the sporulation process and contributes to spore wall maturation. Mol. Cell. Biol. 8:912-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, C. I., G. P. Livi, J. M. Ivy, and A. J. Klar. 1990. Extragenic suppressors of mar2 (sir3) mutations in Saccharomyces cerevisiae. Genetics 125:321-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindgren, A., D. Bungard, M. Pierce, J. Xie, A. Vershon, and E. Winter. 2000. The pachytene checkpoint in Saccharomyces cerevisiae requires the Sum1 transcriptional repressor. EMBO J. 19:6489-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livi, G. P., J. B. Hicks, and A. J. Klar. 1990. The SUM1-1 mutation affects silent mating-type gene transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:409-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 32.Lydall, D., Y. Nikolsky, D. K. Bishop, and T. Weinert. 1996. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature 383:840-843. [DOI] [PubMed] [Google Scholar]
- 33.Malathi, K., Y. Xiao, and A. P. Mitchell. 1997. Interaction of yeast repressor-activator protein Ume6p with glycogen synthase kinase 3 homolog Rim11p. Mol. Cell. Biol. 17:7230-7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitchell, A. P. 1994. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol. Rev. 58:56-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitchell, A. P., S. E. Driscoll, and H. E. Smith. 1990. Positive control of sporulation-specific genes by the IME1 and IME2 products in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:2104-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozsarac, N., M. J. Straffon, H. E. Dalton, and I. W. Dawes. 1997. Regulation of gene expression during meiosis in Saccharomyces cerevisiae: SPR3 is controlled by both ABFI and a new sporulation control element. Mol. Cell. Biol. 17:1152-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pierce, M., M. Wagner, J. Xie, V. Gailus-Durner, J. Six, A. K. Vershon, and E. Winter. 1998. Transcriptional regulation of the SMK1 mitogen-activated protein kinase gene during meiotic development in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:5970-5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Primig, M., R. M. Williams, E. A. Winzeler, G. G. Tevzadze, A. R. Conway, S. Y. Hwang, R. W. Davis, and R. E. Esposito. 2000. The core meiotic transcriptome in budding yeast. Nat. Genet. 26:415-423. [DOI] [PubMed] [Google Scholar]
- 39.Roeder, G. S., and J. M. Bailis. 2000. The pachytene checkpoint. Trends Genet. 16:395-403. [DOI] [PubMed] [Google Scholar]
- 40.Rubin-Berjerano, I., S. Mandel, K. Robzyk, and Y. Kassir. 1996. Induction of meiosis in Saccharomyces cerevisiae depends on conversion of the transcriptional repressor Ume6 to a positive regulator by its regulated association with the transcriptional activator Ime1. Mol. Cell. Biol. 16:2518-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rusché, L. N., and J. Rine. 2001. Conversion of a gene-specific repressor to a regional silencer. Genes Dev. 15:955-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sia, R. A. L., and A. P. Mitchell. 1995. Stimulation of later functions of the yeast meiotic protein kinase Ime2p by the IDS2 gene product. Mol. Cell. Biol. 15:5279-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith, H. E., S. S. Y. Su, L. Neigeborn, S. E. Driscoll, and A. P. Mitchell. 1990. Role of IME1 expression in regulation of meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:6103-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steber, C. M., and R. E. Esposito. 1995. UME6 is a central component of a developmental regulatory switch controlling meiosis-specific gene expression. Proc. Natl. Acad. Sci. USA 92:12490-12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sutton, A., R. C. Heller, J. Landry, J. S. Choy, A. Sirko, and R. Sternglanz. 2001. A novel form of transcriptional silencing by Sum1-1 requires Hst1 and the origin recognition complex. Mol. Cell. Biol. 21:3514-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tachikawa, H., A. Bloecher, K. Tatchell, and A. M. Neiman. 2001. A Gip1p-Glc7p phosphatase complex regulates septin organization and spore wall formation. J. Cell Biol. 155:797-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tung, K.-S., E.-J. E. Hong, and G. S. Roeder. 2000. The pachytene checkpoint prevents accumulation and phosphorylation of the meiosis-specific transcription factor Ndt80. Proc. Natl. Acad. Sci. USA 97:12187-12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vershon, A. K., N. M. Hollingsworth, and A. D. Johnson. 1992. Meiotic induction of the yeast HOP1 gene is controlled by positive and negative regulatory sites. Mol. Cell. Biol. 12:3706-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vershon, A. K., and M. Pierce. 2000. Transcriptional regulation of meiosis in yeast. Curr. Opin. Cell Biol. 12:334-339. [DOI] [PubMed] [Google Scholar]
- 50.Xiao, Y., and A. P. Mitchell. 2000. Shared roles of yeast glycogen synthase kinase 3 family members in nitrogen-responsive phosphorylation of meiotic regulator Ume6p. Mol. Cell. Biol. 20:5447-5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie, J., M. Pierce, V. Gailus-Durner, M. Wagner, E. Winter, and A. K. Vershon. 1999. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 18:6448-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu, L., M. Ajimura, R. Padmore, C. Klein, and N. Kleckner. 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:6572-6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshida, M., H. Kawaguchi, Y. Sakata, K. Kominami, M. Hirano, H. Shima, R. Akada, and I. Yamashita. 1990. Initiation of meiosis and sporulation in Saccharomyces cerevisiae requires a novel protein kinase homologue. Mol. Gen. Genet. 221:176-186. [DOI] [PubMed] [Google Scholar]
- 54.Yukawa, M., S. Katoh, T. Miyakawa, and E. Tsuchiya. 1999. Nps1/Sth1p, a component of an essential chromatin-remodeling complex of Saccharomyces cerevisiae, is required for the maximal expression of early meiotic genes. Genes Cells 4:99-110. [DOI] [PubMed] [Google Scholar]