Abstract
The Apn2 protein of Saccharomyces cerevisiae contains 3′→5′ exonuclease and 3′-phosphodiesterase activities, and these activities function in the repair of DNA strand breaks that have 3′-damaged termini and which are formed in DNA by the action of oxygen-free radicals. Apn2 also has an AP endonuclease activity and functions in the removal of abasic sites from DNA. Here, we provide evidence for the physical and functional interaction of Apn2 with proliferating cell nuclear antigen (PCNA). As indicated by gel filtration and two-hybrid studies, Apn2 interacts with PCNA both in vitro and in vivo and mutations in the consensus PCNA-binding motif of Apn2 abolish this interaction. Importantly, PCNA stimulates the 3′→5′ exonuclease and 3′-phosphodiesterase activities of Apn2. We have examined the involvement of the interdomain connector loop (IDCL) and of the carboxy-terminal domain of PCNA in Apn2 binding and found that Apn2 binds PCNA via distinct domains dependent upon whether the binding is in the absence or presence of DNA. In the absence of DNA, Apn2 binds PCNA through its IDCL domain, whereas in the presence of DNA, when PCNA has been loaded onto the template-primer junction by replication factor C, the C-terminal domain of PCNA mediates the binding.
Cellular DNA is continually being damaged by a variety of extrinsic and intrinsic factors. Oxygen-free radicals formed during the course of normal cellular metabolism produce modified miscoding bases, abasic (AP) sites, and DNA strand breaks. AP sites are some of the most common lesions that arise in cellular DNA. It has been estimated that a mammalian cell loses up to 10,000 purines per day spontaneously from its genome (18). AP sites are also formed from the action of DNA glycosylases on modified bases. Oxidative attack on DNA also results in DNA strand breaks that have a 3′-phosphate or 3′-phosphoglycolate (3′-PG) terminus (11, 13). The β-lyase activity of some DNA glycosylases also leads to the formation of such modified 3′ termini (28). Since these 3′-end groups are refractory to DNA synthesis, their removal is an essential early step in the repair of these lesions (3).
Class II AP endonucleases are multifunctional enzymes that function in the removal of AP sites as well as 3′-blocking termini. Two families of class II AP endonuclease and repair diesterase enzymes have been identified. The endonuclease IV family contains endonuclease IV of Escherichia coli and the Apn1 protein, the major AP endonuclease of Saccharomyces cerevisiae. In addition to AP endonuclease activity, these enzymes possess 3′-phosphatase and 3′-phosphodiesterase activities that can remove 3′-modified termini (22, 28). The exonuclease III (exoIII) family includes exoIII, the major AP endonuclease of E. coli, the Apn2 protein of S. cerevisiae, and Ape1, the dominant AP endonuclease in human cells. exoIII displays, in addition to the AP endonuclease activity, strong 3′→5′ exonuclease, 3′-phosphodiesterase, and 3′-phosphatase activities (28). Human Ape1, however, has a strong AP endonuclease activity but weak 3′→5′ exonuclease, 3′-phosphodiesterase, and 3′-phosphatase activities (23, 31). The Apn2 protein of S. cerevisiae exhibits a weak AP endonuclease activity and strong 3′→5′ exonuclease and 3′-phosphodiesterase activities that are 30- to 40-fold more active than the AP endonuclease activity (25, 26).
In S. cerevisiae, the apn1Δ strain exhibits a much higher sensitivity to the alkylating agent methyl methane sulfonate than the apn2Δ strain (14), and Apn1 accounts for more than 90% of the total AP endonuclease activity in yeast cells (28). In contrast, yeast cells show sensitivity to the oxidative agent H2O2 only in the absence of both the Apn1 and Apn2 proteins (26). These and other observations have suggested a more prominent role for Apn1 in the repair of AP sites, whereas for the repair of oxidative damage, Apn1 and Apn2 contribute more equally.
Proliferating cell nuclear antigen (PCNA) is a highly conserved eukaryotic homotrimeric protein that assembles around DNA to form a sliding clamp and acts as a processivity factor for the replicative polymerase δ (20, 24). PCNA interacts with a large variety of proteins involved in DNA replication, DNA repair, and cell cycle control, such as FEN1 and p21 (6, 17, 27). More recently, PCNA has been shown to interact with the various translesion synthesis DNA polymerases, yeast Polη, and human Polη, Polι, and Polκ (7-10). A conserved motif (Q1-x-x-[ILM]4-x-x-F7-[FY]8) in these proteins modulates their binding to PCNA (7, 9, 19, 21, 30).
Yeast Apn2 and its counterparts from other species constitute a distinct subfamily within the exoIII family (25). All Apn2-related proteins contain a C terminus not present in other members of the exoIII family (14, 25). The conserved motifs that confer AP endonuclease and/or exonuclease activities on the protein are found in the N-terminal region, while a putative PCNA-binding motif, NKSLDSFF (highly conserved residues are indicated in bold), is present at the C terminus of Apn2 (14, 25). In this study, we examine the interaction of Apn2 with PCNA and show that Apn2 interacts with PCNA in vitro and in vivo and that PCNA stimulates the 3′→5′ exonuclease and 3′-phosphodiesterase activities of Apn2. We also provide evidence for the involvement of different regions of PCNA in the binding to Apn2 depending upon whether Apn2 is on or off the DNA.
MATERIALS AND METHODS
Plasmids.
The wild-type full-length APN2 gene from pBJ622 was cloned into pPM1088, which contains the glutathione S-transferase (GST) gene under the control of a galactose-inducible phosphoglycerate promoter. The GST tag can be proteolytically cleaved from the GST fusion protein produced from pPM1088. The plasmid containing the wild-type APN2 gene in pPM1088 was designated pPM1105. The APN2 gene was also cloned into pGBT9, a TRP1 2μm plasmid containing the GAL4 binding domain (BD) (residues 1 to 147) for yeast two-hybrid analyses, generating plasmid pPM1107. Three point mutations, L430A, F433A, and F434A, were generated in the wild-type APN2 gene in pBJ622 by PCR with the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) and the following oligonucleotides: LP-168 (GCAACATCAA AAACAAATCG GCCGACTCAG CTGCCCAGAA GGTAAATGG) and LP-169 (CCATTTACCT TCTGGGCAGC TGAGTCGGCC GATTTGTTTT TGATGTTGC). The resulting mutant apn2 A430A433A434 gene was cloned into the pPM1088 and pGBT9 vectors, generating plasmids pPM1106 and pPM1108, respectively.
Proteins.
The GST-Apn2 proteins were expressed and purified as described previously (26). For the in vitro interaction assays, the GST tags of the Apn2 proteins were removed by PreScission protease (Amersham Pharmacia Biotech). Wild-type PCNA and mutant pcna-79 and pcna-90 proteins were purified from E. coli strains as described previously (1, 4). A truncated form of replication factor C (RFC), from which residues 3 to 273 from Rfc1p were deleted, was purified as described previously (5).
Two-hybrid analysis.
The HF7c yeast strain harboring both the Gal4 BD and the Gal4 activation domain (AD) fusion constructs was grown on synthetic complete medium lacking leucine and tryptophan. The β-galactosidase activity of the different constructs was quantitated by liquid culture assay using o-nitrophenyl-β-d-galactopyranoside as substrate, as described in the Clontech yeast protocols handbook (PT3024-1), chapter VI. Experiments were performed at least three times in triplicate samples.
Gel filtration analysis of Apn2 with PCNA.
Gel filtration of wild-type or mutant Apn2 and PCNA proteins and their complexes was performed using a Superdex 200 PC 3.2/30 column (Amersham Pharmacia Biotech). To constitute complexes, wild-type or mutant Apn2 (3.5 μg) and PCNA (5 μg) or the mixture of these proteins was incubated in buffer containing 50 mM Tris (pH 7.5), 250 mM NaCl, 1 mM EDTA, 1 mM TCEP-HCl [Tris(2-carboxyethyl)-phosphine hydrochloride], 0.01% Nonidet P-40, and 10% glycerol for 30 min at 4°C and then for 10 min at 25°C. Samples were then gel filtrated through the column using a 40-μl/min flow rate. After 1.28 ml was eluted, fractions with a 40-μl volume were collected and analyzed on sodium dodecyl sulfate-10% polyacrylamide gels stained with Coomassie blue R-250.
DNA substrates.
DNA substrates used for the 3′→5′ exonuclease assays were generated by annealing a 75-nucleotide (nt) oligomer template (5′-AGC AAG TCA CCA ATG TCT AAG AGT TCG TAT TAT GCC TAC ACT GGA GTA CCG GAG CAT CGT CGT GAC TGG GAA AAC-3′) to the 44-nt (see Fig. 3), 31-nt (see Fig. 4 and 5), or 27-nt (see Fig. 6) 5′ 32P-labeled oligomer primers N4309 (5′-GTT TTC CCA GTC ACG ACG ATG CTC CGG TAC TCC AGT GTA GGC AT-3′), LP159 (5′-CGA CGA TGC TCC GGT ACT CCA GTG TAG GCA T-3′), or LP-158 (5′-CGA CGA TGC TCC GGT ACT CCA GTG TAG-3′), respectively. For the 3′-phosphodiesterase assay (Fig. 3), a 35-nt, 5′ 32P-labeled oligomer primer (5′-CAG TCA CGA TGC TCG GGA CTC TCT CGA GGA ATG CG-PG-3′) containing a 3′-PG terminus was hybridized to the 70-nt oligomer template (5′-AGC TGG ATT CGT GTC AGC CGC TAG CCA TTA GCT GGC GCA TTC CTC GAG AGA GTC CCG AGC ATC GTG ACT G-3′). DNA substrate for the AP endonuclease assay (Fig. 3) was generated by annealing a 75-nt 5′ 32P-labeled oligomer (5′-AGC TAC CAT GCC TGC CTC AAG AGT TCG TAA 0AT GCC TAC ACT GGA GTA CCG GAG CAT CGT CGT GAC TGG GAA AAC-3′) containing a single tetrahydrofuran, an abasic site analog (Midland Co., Midland, Tex.), at position 31 (0) to its complementary 75-nt oligomer containing a cytosine opposite the abasic site.
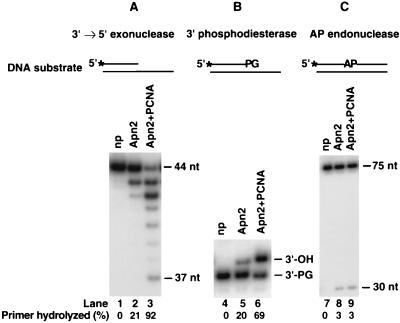
FIG. 3.
Stimulation of the 3′→5′ exonuclease and 3′-phosphodiesterase activities of Apn2 by PCNA. Apn2 (5 nM) was incubated with DNA substrate (20 nM) in a standard reaction mixture containing 0 mM NaCl for 10 min at 30°C. Reactions were carried out in the absence (lanes 2, 5, and 8) or presence (lanes 3, 6, and 9) of PCNA (50 nM). No protein (np) was added to the reactions in lanes 1, 4, and 7. Reaction products were analyzed on an 8 M urea-10% polyacrylamide gel. The 3′→5′ exonuclease activity was assayed on a 44- and 75-nt partial DNA duplex containing a 5′ 32P-labeled strand with a 3′ recessed terminus (A). Phosphodiesterase activity was tested on a 35- and 70-nt partial DNA duplex containing a 5′ 32P-labeled strand with a recessed 3′-PG terminus (B). The AP endonuclease assay was examined on a 75-nt DNA duplex containing a single abasic site at position 31 on the 5′ 32P-labeled strand (C). The asterisks indicate the radioactively labeled termini.
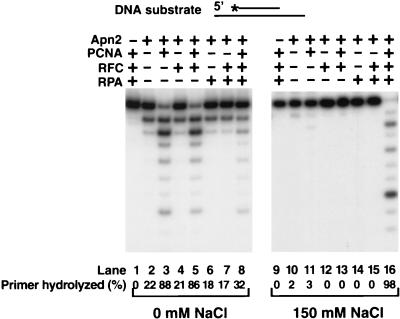
FIG. 4.
PCNA, RFC, and RPA cooperatively stimulate the 3′→5′ exonuclease activity of Apn2. Apn2 was incubated with DNA substrate in standard reaction buffer containing 0 mM NaCl (lanes 1 to 8) or 150 mM NaCl (lanes 9 to 16) in the presence or absence of PCNA, RFC, and/or RPA for 10 min at 30°C. The complete reaction mixture contained Apn2 (5 nM), 31- and 75-nt partial duplex DNA substrate (20 nM) containing a 5′ 32P-labeled primer recessed at both ends, and PCNA (50 nM), RFC (5 nM), and RPA (50 nM). No Apn2 was added in lanes 1 and 9. PCNA, RFC, RPA, and combinations of these proteins were omitted from the reaction mix as indicated at the top.
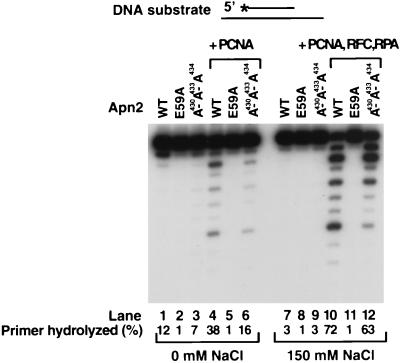
FIG. 5.
Effect of mutations in Apn2 on the stimulation of its 3′→5′ exonuclease activity by PCNA. Standard Apn2 exonuclease assays were carried out on a 31- and 75-nt primer-template partial duplex DNA substrate (20 nM) in the absence (lanes 1 to 6) or presence (lanes 7 to 12) of 150 mM NaCl for 10 min at 30°C. The primer was recessed on both ends and labeled at its 5′ end. The reactions were performed in the presence of wild-type Apn2 (lanes 1 and 7), Apn2 E59A (lanes 2 and 8), or Apn2 A430A433A434 (lanes 3 and 9) alone (2 nM each), or the wild-type and mutant Apn2 proteins were assayed for activity in the presence of PCNA (50 nM) (lanes 4 to 6) or PCNA (50 nM) together with RFC (5 nM) and RPA (50 nM) (lanes 10 to 12).
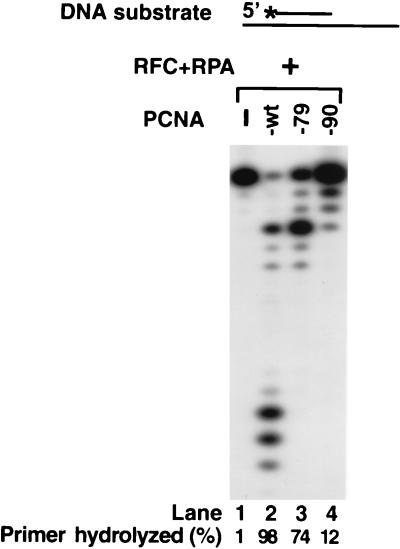
FIG. 6.
Effect of pcna-79 and pcna-90 on the 3′→5′ exonuclease activity of Apn2. Apn2 (2 nM) was assayed on a 27- and 75-nt primer-template partial duplex DNA (20 nM), wherein the 5′ 32P-labeled primer was recessed on both ends, for 10 min at 30°C. Reactions were carried out at a 150 mM NaCl concentration and in the presence of RFC (5 nM) and RPA (50 nM) plus wild-type PCNA (lane 2), pcna-79 (lane 3), or pcna-90 (lane 4) (50 nM each), as indicated. No PCNA was added in lane 1. The percentage of primer hydrolyzed by Apn2 was quantitated by a phosphorimager.
3′→5′ exonuclease, 3′-phosphodiesterase, and AP endonuclease assays.
A standard Apn2 reaction mixture (10 μl) contained 40 mM Tris-HCl (pH 7.5), 8 mM MgCl2, 1 mM dithiothreitol, 100 μg of bovine serum albumin per ml, 500 μM ATP, 20 nM DNA substrate, and 2 to 5 nM Apn2. For the AP endonuclease activity, Mg2+ was omitted. Where indicated, the reaction was supplemented with 150 mM NaCl. Reactions were performed in the absence or presence of different combinations of PCNA, pcna-79, or pcna-90 (50 nM each), RFC (5 nM), and/or replication protein A (RPA) (50 nM). Assays were assembled on ice, incubated at 30°C for 10 min, and stopped by the addition of loading buffer (40 μl) containing EDTA (20 mM), 95% formamide, and 0.3% cyanol blue. The reaction products were resolved on 10% polyacrylamide gels containing 8 M urea. Quantitation of the results was done using Molecular Dynamics STORM PhosphorImager and ImageQuant software.
RESULTS
Mutant Apn2 and PCNA proteins.
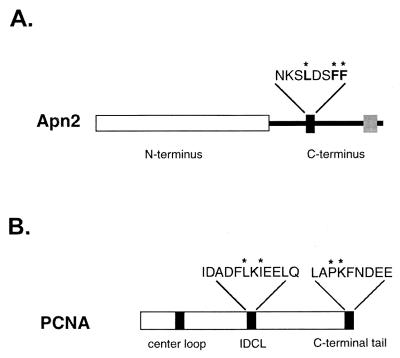
A consensus PCNA-binding motif, NKSLDSFF (highly conserved residues are indicated in bold), is present in the middle of the C-terminal portion of Apn2, spanning amino acids 427 to 434 in the 520-amino-acid protein. To test whether this motif mediates interaction of Apn2 with PCNA, we generated a mutant Apn2 protein, Apn2 A430A433A434, in which the highly conserved leucine residue and the two consecutive phenylalanine residues of the PCNA-binding motif have been changed to alanine residues (Fig. 1A). The mutant protein was purified to apparent homogeneity and during purification it behaved similarly to the wild-type protein and displayed an identical electrophoretic mobility to the wild-type protein on a sodium dodecyl sulfate-polyacrylamide gel (data not shown).
FIG. 1.
Schematic representation of mutations in the PCNA-binding motif of Apn2 and in PCNA. (A) Mutations in Apn2. The N terminus of Apn2, which contains the conserved sequence motifs characteristic of this family, is shown as a white box. In the C terminus, the sequence of the PCNA-binding motif is shown as a black box, and the conserved amino acids are shown above. The asterisks indicate the amino acid residues that were changed to alanines in the Apn2 A430A433A434 protein. The gray box represents the sequence that bears homology to the tandem repeat of topoisomerase III. (B) Mutations in PCNA. The sequences of the IDCL and the C-terminal tail of PCNA are shown. The amino acid residues that were changed to alanines in the IDCL region in pcna-79 and in the C-terminal region in pcna-90 are indicated by asterisks.
The conserved PCNA-binding motif, similar to that in Apn2, is present in a number of proteins with different functions, and it has been shown to interact with the interdomain connector loop (IDCL) of PCNA (15, 29). The crystal structure of Archae FEN1 shows that the conserved QSTLESWF PCNA-binding motif (highly conserved residues are indicated in bold) in this protein is at the end of an α-helix that extends away from the folded protein and that permits the docking of this region to the IDCL of PCNA via the insertion of the highly conserved hydrophobic residues into the hydrophobic pocket of IDCL (12). To identify the regions of PCNA involved in interaction with Apn2, we have examined the effect of two mutations in PCNA, pcna-79 and pcna-90 (4). In the pcna-79 protein, the hydrophobic residues L126 and I128 that are present in the IDCL have been changed to alanine residues, whereas in the pcna-90 mutant, the P252 and K253 residues that are present in the carboxyl-terminal region have been changed to alanine residues (Fig. 1B) (4).
Physical interaction of Apn2 with PCNA.
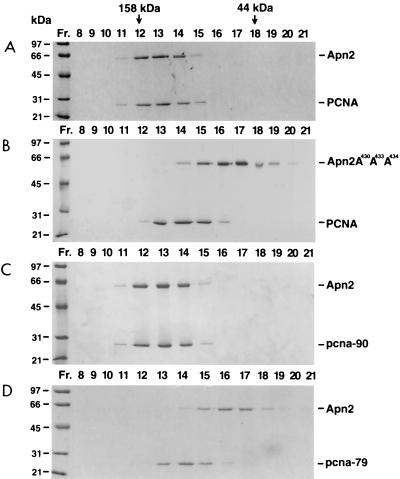
We examined the interaction of Apn2 with PCNA in vitro by gel filtration during which only strong protein-protein interactions survive under the solution conditions used. Purified wild-type Apn2 or the Apn2 A430A433A434 protein was incubated with PCNA and then passed through the gel filtration column. When alone, Apn2 eluted in fractions 15 to 17 and PCNA eluted in fractions 13 to 15 (data not shown), but when the two proteins were mixed together, Apn2 and PCNA coeluted in the higher molecular mass range, in peak fractions 12 to 14 (Fig. 2A). However, the Apn2 A430A433A434 protein failed to interact with PCNA, as the mutant Apn2 protein and PCNA eluted separately, in fractions 15 to 17 and 13 to 15, respectively (Fig. 2B), which is coincident with the positions of the wild-type Apn2 protein and PCNA when they were gel filtered alone. These observations indicate that Apn2 interacts with PCNA in vitro, that this interaction is mediated through the conserved PCNA-binding motif of Apn2, and that mutations in this site abolish the interaction. The Apn2-PCNA interaction is strong, as it is stable in 300 mM NaCl (data not shown).
FIG. 2.
Physical interaction of Apn2 with PCNA by gel filtration. In the reactions, 3.5 μg of the Apn2 protein and 5 μg of PCNA were incubated in different combinations and gel filtered. (A) Wild-type Apn2 with PCNA. (B) Mutant Apn2 A430A433A434 with PCNA. (C) Wild-type Apn2 with pcna-90. (D) Wild-type Apn2 with pcna-79. The fraction numbers and the elution positions of the molecular mass markers for the gel filtration column are shown on the top. The positions of molecular mass standards on the protein gels are indicated on the left.
To identify the domain of PCNA which mediates the interaction with Apn2, we gel filtered Apn2 with the IDCL mutant pcna-79 protein or with the C-terminal mutant pcna-90 protein (Fig. 1B). The interaction of Apn2 with pcna-90 was not noticeably affected as, analogous to wild-type PCNA, both the Apn2 and pcna-90 proteins coeluted in fractions 12 to 14 (Fig. 2, compare panels A and C). However, the interaction of Apn2 with pcna-79 was eliminated, as the two proteins eluted separately from the column (Fig. 2, compare panels B and D). These results indicate that in the absence of DNA, the IDCL of PCNA and the PCNA-binding motif of Apn2 are responsible for sustaining the interaction between these two proteins.
Interaction of Apn2 with PCNA in the two-hybrid system.
To examine whether Apn2 interacts with PCNA in vivo, we performed a two-hybrid analysis. The wild-type APN2 and the apn2 A430A433A434 open reading frames were cloned in fusion with the GAL4 BD in the pGBT9 two-hybrid vector. The HF7c yeast reporter strain was transformed with a plasmid containing the GAL4 BD fusion construct expressing either the wild-type or mutant Apn2 protein and the plasmid harboring the GAL4 AD in fusion with PCNA. The interaction between the GAL4 BD-Apn2 protein and GAL4 AD-PCNA was measured in liquid β-galactosidase assays, and the results of these experiments are summarized in Table 1. The wild-type Apn2 protein showed strong interaction with PCNA, resulting in a 70-fold higher β-galactosidase activity than the background level. The Apn2 A430A433A434 mutant protein yielded only a sevenfold increase in the β-galactosidase activity, indicating that mutations in the PCNA-binding motif strongly impair, but do not abolish, the interaction of Apn2 with PCNA. These results verify that Apn2 and PCNA interact in vivo and that this interaction is predominantly mediated through the conserved PCNA-binding motif of Apn2.
TABLE 1.
Interaction of Apn2 with PCNA in the yeast two-hybrid system
| DNA BD fusion | AD fusion | Mean β-galactosidase activity (Miller units) ± SD | Fold activation |
|---|---|---|---|
| GAL4 BD-Apn2 (wt) | GAL4 AD | 0.02 ± 0.003 | 1 |
| GAL4 BD-Apn2 (wt) | GAL4 AD-PCNA | 1.4 ± 0.1 | 70 |
| GAL4 BD-Apn2 A430A433A434 | GAL4 AD-PCNA | 0.14 ± 0.03 | 7 |
Stimulation of the 3′→5′ exonuclease and 3′-phosphodiesterase activities of Apn2 by PCNA.
Yeast Apn2 has a strong 3′→5′ exonuclease activity, specific for double-stranded DNA, and a 3′-phosphodiesterase activity that can remove a 3′-terminal group such as a 3′-PG from the DNA (26). In addition, Apn2 has a weak AP endonuclease activity (25). To assess the functional significance of Apn2 interaction with PCNA for each of these activities, we examined whether the presence of PCNA would affect the activity of Apn2 on the various DNA substrates shown in Fig. 3. On linear DNA templates, PCNA can slide onto the DNA, albeit less efficiently than when loaded by RFC (2).
Apn2 shows a 3′→5′ exonuclease activity on the partial DNA duplex in which it digests the 5′-labeled strand, with a recessed 3′ terminus, from the 3′ end (Fig. 3A). PCNA strongly stimulated the 3′→5′ exonuclease activity of Apn2, as there was greatly enhanced hydrolysis of the labeled strand as well as the generation of shorter digestion products (Fig. 3A). A double-stranded DNA substrate containing a 3′-PG residue in the 5′-labeled strand was used to monitor the removal of the 3′-PG group by Apn2 (Fig. 3B). Enzymatic removal of the 3′-PG group results in a 3′-hydroxyl terminus, which confers a slower electrophoretic mobility to DNA on a denaturing polyacrylamide gel. The addition of PCNA strongly stimulated the removal of the 3′-PG by Apn2 (Fig. 3B). The AP endonuclease activity of Apn2 was assayed using a 75-nt duplex DNA substrate in which the 5′-labeled strand contains a single abasic site (Fig. 3C). Apn2 catalyzes the hydrolysis of the 5′-phosphodiester bond at the abasic site, generating a 30-nt labeled oligomer. Surprisingly, the addition of PCNA did not stimulate the AP endonuclease activity of Apn2 (Fig. 3C). However, because the endonuclease assay was carried out in the absence of Mg2+ and we do not know whether PCNA is able to slide efficiently onto linear DNA under those conditions, we cannot draw definite conclusions regarding the stimulation of the endonuclease activity of Apn2 by PCNA. Therefore, we have focused our further studies on the 3′→5′ exonuclease activity of Apn2, which is stimulated by PCNA.
PCNA cooperates with RFC and RPA to stimulate the 3′→5′ exonuclease activity of Apn2.
We examined whether RFC, a multisubunit clamp loader which couples the hydrolysis of ATP to the loading of PCNA onto DNA, and RPA, a single-stranded DNA-binding protein, affect the PCNA-dependent stimulation of the 3′→5′ exonuclease activity of Apn2. In these studies we used a short linear 31- and 75-nt primer-template DNA substrate, designed such that both ends of the template DNA had a single-stranded region. On this DNA substrate, the PCNA ring remains more stably bound than on a blunt-ended DNA substrate. We examined the effect of PCNA in the presence of RFC and RPA on Apn2 activity at two different salt concentrations, 0 and 150 mM NaCl. The Apn2 exonuclease activity is very sensitive to salt, exhibiting the highest activity in the absence of any salt, whereas at a high salt concentration (150 mM NaCl), which more closely reflects the physiological salt concentration (16), Apn2 exhibits little exonuclease activity (Fig. 4, compare lanes 2 and 10). At a low salt concentration (0 mM NaCl), PCNA alone highly stimulated the exonuclease activity of Apn2, and the addition of RFC and RPA did not stimulate this activity further (Fig. 4, lanes 1 to 8). This probably reflects that at a low salt concentration, PCNA alone passively slides onto the DNA substrate from the end of the template. However, at a high salt concentration, no significant enhancement of the exonuclease activity of Apn2 occurred, unless PCNA was added together with RFC and RPA (Fig. 4, lanes 9 to 16). This suggests that at a high salt concentration, PCNA is loaded onto DNA by RFC, and the coating of the single-stranded DNA with RPA may either prevent PCNA from sliding off the DNA or stabilize interactions with the other proteins.
Disruption of the PCNA-binding motif does not inactivate the PCNA-dependent stimulation of 3′→5′ exonuclease activity of Apn2.
Next, we examined the contribution of the PCNA-binding motif in Apn2 to PCNA-dependent stimulation of its 3′→5′ exonuclease activity. The exonuclease activity of the wild-type Apn2 protein and the Apn2 A430A433A434 protein with mutations in the PCNA-binding motif was assayed with PCNA at a low salt concentration and with the combination of PCNA, RFC, and RPA at a high salt concentration (Fig. 5). Surprisingly, PCNA stimulated the exonuclease activity of the Apn2 A430A433A434 protein, in spite of the fact that this mutant protein displays a severe defect in the binding of PCNA in the absence of DNA in gel filtration experiments. However, the Apn2 A430A433A434 protein is stimulated by PCNA to a somewhat lower level than the wild-type protein (Fig. 5, compare lanes 4 and 6 and lanes 10 and 12). As a control, we also tested the E59A mutant protein, in which the change of the active site Glu59 residue to Ala inactivates the Apn2 exonuclease activity (26). This mutant protein showed no exonuclease activity in the presence of PCNA, indicating that the exonuclease activity being stimulated by PCNA was in fact due to Apn2 (Fig. 5, lanes 2, 5, 8, and 11).
Contribution of the IDCL and C-terminal regions of PCNA to stimulation of 3′→5′ exonuclease activity of Apn2.
Next, we examined whether the IDCL mutant pcna-79 and the C-terminal mutant pcna-90 are able to stimulate the exonuclease activity of Apn2 (Fig. 6). At the physiological salt concentration, where the stimulation of Apn2 exonuclease activity by PCNA requires the cooperative action of RFC and RPA, the pcna-79 protein was reduced in its proficiency, while the pcna-90 protein was highly defective (Fig. 6). The deficiency of pcna-90 does not arise from a defect in the loading of this mutant protein onto DNA, since previous experiments have shown that pcna-90 is efficiently loaded onto singly primed DNA by RFC and stimulates the processive DNA synthesis activity of DNA polymerase δ (4).
DISCUSSION
Physical and functional interaction of Apn2 with PCNA.
Here we show that purified Apn2 cofractionates with PCNA in gel filtration studies, and two-hybrid analyses indicate that the two proteins interact in vivo. Importantly, PCNA stimulates the 3′→5′ exonuclease and 3′-phosphodiesterase activities of Apn2. Also, while PCNA alone can stimulate the Apn2 exonuclease at a low salt concentration (0 mM NaCl), at a higher salt concentration (150 mM NaCl) which approximates physiological levels, RFC and RPA are required, in addition to PCNA, to effect the stimulation. The lack of requirement of RFC and RPA for the PCNA-dependent stimulation of Apn2 exonuclease activity at low salt concentrations is consistent with the view that under these conditions, PCNA can slide onto the ends of linear DNA without the assistance of RFC, but that at high salt concentrations, PCNA is actively loaded onto DNA by RFC (2).
Contribution of the conserved motif in Apn2 to PCNA binding.
The conserved motif NKSLDSFF present in the C-terminal region of Apn2 mediates the interaction of Apn2 with PCNA, since interaction with PCNA is greatly impaired both in vitro and in vivo with the mutant Apn2 protein in which the highly conserved L and FF residues have all been changed to alanine residues. Surprisingly, however, PCNA was able to stimulate the exonuclease activity of the Apn2 A430A433A434 mutant protein, although the efficiency of stimulation was somewhat reduced. These observations suggest that in the absence of DNA, Apn2 associates with PCNA via its conserved PCNA-binding motif, whereas when the proteins are assembled onto the substrate DNA, the mode of Apn2 binding to PCNA changes and is not that dependent on the conserved PCNA-binding motif in Apn2.
Distinct domains in PCNA modulate its binding to Apn2 in the absence or presence of DNA.
Two PCNA mutant proteins, pcna-79 and pcna-90, have enabled us to identify the respective roles of the IDCL and the C-terminal regions of PCNA in Apn2 binding. Our gel filtration studies indicate that mutations in the IDCL of PCNA inactivate the interaction with Apn2, whereas mutations in the C-terminal region of PCNA have no effect on this interaction. Because mutations in the PCNA-binding motif of Apn2, as well as in the IDCL region of PCNA, impair the interaction of Apn2 with PCNA in gel filtration studies, these results support the premise that in the absence of DNA, the conserved PCNA-binding motif of Apn2 binds the IDCL region of PCNA. The mode of binding changes, however, in the presence of DNA. At the physiological salt concentration, the pcna-90 protein displays a much greater defect in its ability to stimulate the Apn2 activity than pcna-79. From these observations, we infer that when PCNA is actively recruited to the primer-template junction by RFC, the IDCL region of PCNA is not as necessary for interaction with Apn2 but the C-terminal region of PCNA is essential for this interaction.
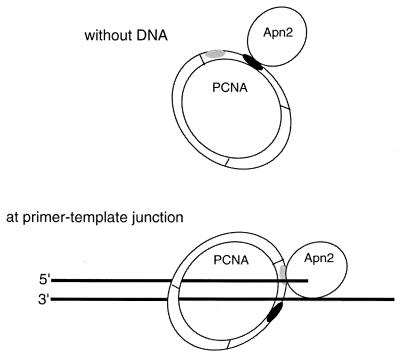
These observations suggest a model in which Apn2 interacts with different regions of PCNA, dependent upon whether Apn2 binds PCNA in the absence of DNA or whether Apn2 is bound to the DNA at the primer-template junction (Fig. 7). In the absence of DNA, association of Apn2 with PCNA is mediated by the interaction of the conserved PCNA-binding motif of Apn2 with the IDCL of PCNA (Fig. 2). The preformed Apn2-PCNA complex can then load onto the ends of linear DNA and slide on such DNA. Upon reaching the primer-template junction, however, the mode of Apn2 binding to PCNA switches from the IDCL region to the C-terminal region (Fig. 7), and the consensus PCNA-binding domain of Apn2 is no longer required for productive interaction.
FIG. 7.
Schematic model for the interaction of Apn2 with PCNA. In the absence of DNA (top), the conserved PCNA-binding motif of Apn2 and the IDCL of PCNA mediate the interaction between the two proteins. Upon encountering a 3′ primer-template junction (bottom), the C-terminal tail of PCNA becomes engaged in the interaction. The IDCL part of PCNA is shown in black, and the C-terminal portion is shown in gray.
Possible significance of Apn2 binding to different domains of PCNA.
Previously, FEN1, a 5′→3′ flap endonuclease which functions in the removal of RNA-DNA primers during Okazaki fragment maturation, has been shown to interact with PCNA in two distinct modes (5) in a manner similar to that shown here for Apn2. For both these proteins, the initial binding to PCNA in the absence of DNA is effected via the interaction of the consensus PCNA-binding motif with the IDCL of PCNA, whereas the stimulation of their respective nuclease activities requires their interaction with the C-terminal domain of PCNA. This suggests that both these proteins complex with PCNA in the absence of DNA via interaction with the IDCL portion, which raises the possibility that such preformed complexes are then loaded onto the DNA by RFC. Subsequently, both proteins might undergo a conformational change which enables them to bind the C-terminal portion of PCNA. However, in striking contrast to both these nucleases, S. cerevisiae Polδ interacts with the DNA-bound PCNA via its IDCL domain. Mutations in the IDCL eliminate the interaction of PCNA with Polδ, and they also cause a dramatic reduction in processivity, particularly on a DNA substrate with secondary structure, whereas mutations in the C terminus of PCNA have no effect on the binding of Polδ or on its activity (4). These observations indicating that nucleases such as FEN1 and Apn2 bind the DNA-bound PCNA at its C terminus, whereas Polδ binds PCNA in the IDCL region, suggest the possibility that a nuclease and Polδ could simultaneously bind PCNA, which might enable the two proteins to function in a highly coordinated fashion. Thus, Apn2 bound to PCNA together with Polδ may be able to remove only the nucleotide with the damaged 3′ terminus because of the subsequent efficient filling in of the 1-nt gap by the associated Polδ. In such a scenario, PCNA would play a key role in promoting efficient short patch repair of 3′-damaged termini by coordinating the degradative and polymerizing activities of two different proteins.
Acknowledgments
This work was supported by NIH grants CA41261 and GM32431.
I.U. and L.H. contributed equally to this work.
REFERENCES
- 1.Ayyagari, R., K. J. Impellizzeri, B. L. Yoder, S. L. Gary, and P. M. J. Burgers. 1995. A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and repair. Mol. Cell. Biol. 15:4420-4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burgers, P. M. J., and B. L. Yoder. 1993. ATP-independent loading of the proliferating cell nuclear antigen requires DNA ends. J. Biol. Chem. 268:19923-19926. [PubMed] [Google Scholar]
- 3.Demple, B., A. Johnson, and D. Fung. 1986. Exonuclease III and endonuclease IV remove 3′ blocks from DNA synthesis primers in H2O2-damaged Escherichia coli. Proc. Nat. Acad. Sci. USA 83:7731-7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eissenberg, J. C., R. Ayyagari, X. V. Gomes, and P. M. J. Burgers. 1997. Mutations in yeast proliferating cell nuclear antigen define distinct sites for interaction with DNA polymerase δ and DNA polymerase ɛ. Mol. Cell. Biol. 17:6367-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gomes, X. V., and P. M. J. Burgers. 2000. Two modes of FEN1 binding to PCNA regulated by DNA. EMBO J. 19:3811-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gulbis, J. M., Z. Kelman, J. Hurwitz, M. O. O'Donnell, and J. Kuriyan. 1996. Structure of the C-terminal region of p21WAF1CIP1 complexed with human PCNA. Cell 87:297-306. [DOI] [PubMed] [Google Scholar]
- 7.Haracska, L., R. E. Johnson, I. Unk, B. Phillips, J. Hurwitz, L. Prakash, and S. Prakash. 2001. Physical and functional interactions of human DNA polymerase η with PCNA. Mol. Cell. Biol. 21:7199-7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haracska, L., R. E. Johnson, I. Unk, B. B. Phillips, J. Hurwitz, L. Prakash, and S. Prakash. 2001. Targeting of human DNA polymerase ι to the replication machinery via interaction with PCNA. Proc. Natl. Acad. Sci. USA 98:14256-14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haracska, L., C. M. Kondratick, I. Unk, S. Prakash, and L. Prakash. 2001. Interaction with PCNA is essential for yeast DNA polymerase η function. Mol. Cell 8:407-415. [DOI] [PubMed] [Google Scholar]
- 10.Haracska, L., I. Unk, R. E. Johnson, B. B. Phillips, J. Hurwitz, L. Prakash, and S. Prakash. 2002. Stimulation of DNA synthesis activity of human DNA polymerase κ by PCNA. Mol. Cell. Biol. 22:784-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henle, E. S., and S. Linn. 1997. Formation, prevention, and repair of DNA damage by iron/hydrogen peroxide. J. Biol. Chem. 272:19095-19098. [DOI] [PubMed] [Google Scholar]
- 12.Hosfield, D. J., C. D. Mol, and J. A. Tainer. 1998. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell 95:135-146. [DOI] [PubMed] [Google Scholar]
- 13.Imlay, J. A., and S. Linn. 1988. DNA damage and oxygen radical toxicity. Science 240:1302-1309. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, R. E., C. A. Torres-Ramos, T. Izumi, S. Mitra, S. Prakash, and L. Prakash. 1998. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev. 12:3137-3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jonsson, Z. O., R. Hindges, and U. Hubscher. 1998. Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen. EMBO J. 17:2412-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kao-Huang, Y., A. Revzin, A. P. Butler, P. O'Conner, D. W. Noble, and P. H. von Hippel. 1977. Nonspecific DNA binding of genome-regulating proteins as a biological control mechanism: measurement of DNA-bound Escherichia coli lac repressor in vivo. Proc. Natl. Acad. Sci. USA 74:4228-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, X., J. Li, J. Harrington, M. R. Lieber, and P. M. Burgers. 1995. Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by PCNA. J. Biol. Chem. 270:22109-22112. [DOI] [PubMed] [Google Scholar]
- 18.Lindahl, T., and B. Nyberg. 1972. Rate of depurination of native deoxyribonucleic acid. Biochemistry 11:3610-3617. [DOI] [PubMed] [Google Scholar]
- 19.Nakanishi, M., R. S. Robetorye, O. M. Pereira-Smith, and J. R. Smith. 1995. The C-terminal region of p21SD11/WAF1/CIP1 is involved in proliferating cell nuclear antigen binding but does not appear to be required for growth inhibition. J. Biol. Chem. 270:17060-17063. [DOI] [PubMed] [Google Scholar]
- 20.Prelich, G., C. K. Tan, M. Kostura, M. B. Mathews, A. G. So, K. M. Downey, and B. Stillman. 1987. Functional identity of proliferating cell nuclear antigen and a DNA polymerase-delta auxiliary protein. Nature 326:517-520. [DOI] [PubMed] [Google Scholar]
- 21.Reynolds, N., E. Warbrick, P. A. Fantes, and S. A. MacNeill. 2000. Essential interaction between the fission yeast DNA polymerase δ subunit Cdc27 and Pcn1 (PCNA) mediated through a C-terminal p21Cip1-like PCNA binding motif. EMBO J. 19:1108-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seeberg, E., L. Eide, and M. Bjorås. 1995. The base excision repair pathway. Trends Biochem. Sci. 20:391-396. [DOI] [PubMed] [Google Scholar]
- 23.Suh, D., D. M. Wilson III, and L. F. Povirk. 1997. 3′-phosphodiesterase activity of human apurinic/apyrimidinic endonuclease at DNA double-strand break ends. Nucleic Acids Res. 25:2495-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan, C. K., C. Castillo, A. G. So, and K. M. Downey. 1986. An auxiliary protein for DNA polymerase δ from fetal calf thymus. J. Biol. Chem. 261:12310-12316. [PubMed] [Google Scholar]
- 25.Unk, I., L. Haracska, R. E. Johnson, S. Prakash, and L. Prakash. 2000. Apurinic endonuclease activity of yeast Apn2 protein. J. Biol. Chem. 275:22427-22434. [DOI] [PubMed] [Google Scholar]
- 26.Unk, I., L. Haracska, S. Prakash, and L. Prakash. 2001. 3′-phosphodiesterase and 3′→5′ exonuclease activities of yeast Apn2 protein and requirement of these activities for repair of oxidative DNA damage. Mol. Cell. Biol. 21:1656-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waga, S., G. J. Hannon, D. Beach, and B. Stillman. 1994. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature 369:574-578. [DOI] [PubMed] [Google Scholar]
- 28.Wallace, S. 1997. Oxidative damage to DNA and its repair, p. 49-90, Oxidative stress defenses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Warbrick, E., D. P. Lane, D. M. Glover, and L. S. Cox. 1997. Homologous regions of Fen1 and p21Cip1 compete for binding to the same site on PCNA: a potential mechanism to coordinate DNA replication and repair. Oncogene 14:2313-2321. [DOI] [PubMed] [Google Scholar]
- 30.Warbrick, E., D. P. Lane, D. M. Glover, and L. S. Cox. 1995. A small peptide inhibitor of DNA replication defines the site of interaction between the cyclin-dependent kinase inhibitor p21WAF1 and proliferating cell nuclear antigen. Curr. Biol. 5:275-282. [DOI] [PubMed] [Google Scholar]
- 31.Wilson, D. M., III, M. Takeshita, A. P. Grollman, and B. Demple. 1995. Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. J. Biol. Chem. 270:16002-16007. [DOI] [PubMed] [Google Scholar]