Abstract
ACF is a chromatin-remodeling complex that catalyzes the ATP-dependent assembly of periodic nucleosome arrays. This reaction utilizes the energy of ATP hydrolysis by ISWI, the smaller of the two subunits of ACF. Acf1, the large subunit of ACF, is essential for the full activity of the complex. We performed a systematic mutational analysis of Acf1 to elucidate the functions of specific subregions of the protein. These studies revealed DNA- and ISWI-binding regions that are important for the chromatin assembly and ATPase activities of ACF. The DNA-binding region of Acf1 includes a WAC motif, which is necessary for the efficient binding of ACF complex to DNA. The interaction of Acf1 with ISWI requires a DDT domain, which has been found in a variety of transcription and chromatin-remodeling factors. Chromatin assembly by ACF is also impaired upon mutation of an acidic region in Acf1, which may interact with histones during the deposition process. Lastly, we observed modest chromatin assembly defects on mutation of other conserved sequence motifs. Thus, Acf1 facilitates chromatin assembly via an N-terminal DNA-binding region with a WAC motif, a central ISWI-binding segment with a DDT domain, and a C-terminal region with an acidic stretch, a WAKZ motif, PHD fingers, and bromodomain.
The assembly of chromatin is a fundamental biological process (for reviews, see references 2, 6, 15-17, 29-32, 37, 46, and 48-50). In proliferating cells, chromatin is assembled onto newly synthesized DNA immediately after replication. In quiescent cells, chromatin assembly occurs during histone turnover and DNA repair. Thus, the process of chromatin assembly is critical for the proper growth and maintenance of cells.
The assembly of periodic nucleosome arrays is an ATP-dependent process that was originally observed in an extract derived from Xenopus oocytes (20). Subsequent studies of chromatin assembly in Drosophila embryo extracts led to the identification and purification of ACF as an ATP-utilizing factor that mediates chromatin assembly in conjunction with a core histone chaperone (24). In addition, ACF can catalyze the ATP-dependent mobilization of nucleosomes, and hence it is also a chromatin-remodeling factor.
ACF consists of the Acf1 and ISWI polypeptides (25). ISWI (for “imitation switch”) is an ATPase that was cloned as a Drosophila protein related to yeast Swi2/Snf2 (14). ISWI is also present in the NURF (43, 44) and CHRAC (47) chromatin-remodeling complexes. Acf1 was originally identified as the large subunit of ACF, and it has also been found in the CHRAC complex, which consists of two small (14- and 16-kDa) subunits termed CHRAC-14 and CHRAC-16, along with Acf1 and ISWI (9, 13). It is also notable that the largest subunit of NURF, termed NURF301, interacts with ISWI and is related to Acf1 (51).
Factors that are closely related to Drosophila ACF have been found in other species. For example, in humans, there are four complexes that are related to ACF, NURF, and/or CHRAC. First, hACF/WCRF complex consists of two subunits—WCRF/BAZ1A/hACF1, which is related to Acf1, and hSNF2H, which is related to ISWI (4, 34). Second, hCHRAC complex appears to be the human homologue of Drosophila CHRAC (39). Third, human RSF complex consists of a 325-kDa polypeptide and hSNF2H (33, 36). RSF was also found to mediate the ATP-dependent assembly of chromatin (36). Fourth, human NoRC complex contains TIP5 protein, which is related to WCRF/BAZ1A/hACF1, and hSNF2H (41). In Saccharomyces cerevisiae, there are two ISWI-containing complexes termed ISW1 and ISW2. The ISW2 complex is similar to Drosophila ACF, since it consists of two subunits, Itc1 and Isw2, which are related to Acf1 and ISWI (19, 45). In Xenopus laevis, four ISWI-containing complexes, including xACF, have been identified (21). In addition, a variety of eukaryotic genome databases contain predicted proteins that are related to Acf1 or ISWI. Thus, ACF and related factors appear to be present in a broad range of organisms.
The cloning of the factors that mediate chromatin assembly has allowed us to establish a purified chromatin assembly system that consists of purified recombinant Drosophila ACF, purified recombinant Drosophila NAP-1 (a core histone chaperone [23]), purified native Drosophila core histones, DNA, and ATP (25). We have also assembled chromatin with purified recombinant Drosophila core histones that are synthesized in bacteria and lack posttranslational modifications (35). Thus, it is possible to study the ability of ACF to assemble chromatin with a completely defined biochemical system.
Acf1 and ISWI function cooperatively in the assembly of chromatin (25), and it is likely that Acf1 imparts additional functionality to the general motor activity of the ISWI subunit. Acf1 is a 1,476-amino-acid polypeptide that contains several conserved motifs (WAC [25], WAKZ [25], DDT [12], bromodomain [22, 27], and PHD fingers [1]) and a 40-residue highly acidic region. In this study, we used the purified chromatin assembly system to investigate the function of Acf1. These experiments suggest that DNA binding by the N-terminal region of Acf1, which includes the WAC domain, is an important step in the assembly of nucleosomes by ACF.
MATERIALS AND METHODS
Baculovirus expression constructs.
cDNA fragments that contain the complete coding sequences of Drosophila Acf1 (25), Drosophila NAP-1 (23), and Drosophila ISWI (Drosophila EST GH08103 [Research Genetics, Inc.]) were each cloned into the pFastBac1 vector (Gibco-BRL). Appropriate restriction sites were introduced in the 5′ and 3′ ends of cDNAs by ligation with double-stranded oligonucleotides. In the Acf1 expression construct, a sequence that encodes the RGS-His FLAG tag (RGSHHHHHHDYKDDDDK) was fused in frame with the 3′ end of the Acf1 coding sequence. In the NAP-1 expression construct, a sequence that encodes the His6 tag was fused in frame with the 5′ end of the NAP-1 coding sequence. In the preparation of expression constructs for Acf1 deletion mutants, the pFastBac-Acf1 construct was digested with pairs of appropriate restriction enzymes, and double-stranded oligonucleotides with matching ends were ligated to replace the deleted DNA fragments. All of the constructs were sequenced across the newly created junctions. In the preparation of amino acid substitution mutants of Acf1, short fragments (from 500 to 1,500 bp) of Acf1 coding sequence were subcloned into the pGEM-7Zf(+) vector (Promega) and site-directed mutagenesis was performed with the U.S.E. kit (Pharmacia) as suggested by the manufacturer. (The second selection primer was GAGCTCCCAACGCGaTGGgTGCATAGCTTGAG, and the selection restriction enzyme was NsiI.) The mutagenized fragments were completely sequenced and subcloned into the pFastBac-Acf1 construct. Mutant constructs containing two or more different mutations or deletions (e.g., m[PHD1/2] and IBR) were prepared by subcloning restriction fragments from the original set of mutants.
The specific details of the preparation of the Acf1 mutants are described below. Each mutant is represented as follows: common name; deletion boundaries or mutation positions relative to the initiating methionine residue of wild-type Acf1; restriction sites (or diagnostic restriction enzyme) used; sequence of the top-strand oligonucleotide; sequence of the bottom-strand oligonucleotide (for deletion mutants only); and the amino acid sequence across the deletion junction or mutated region. Altered amino acid residues and nucleotides introduced at substitution mutations are shown in lowercase letters. Deletion junctions are represented by a hyphen. Extra amino acid residues introduced at the deletion junction are underlined. Δ[WAC]; Δ[4-111]; NdeI-BalI; TATGCCCATTTTGG; CCAAAATGGGCA; MPI-L-AKF. Δ2;Δ[154-309]; PfIMI-MIuI; ATGGTATCGAA; CGCGTTCGATACCATTGA; NGI-ERV. Δ[DDT]; Δ[312-470]; MluI-BamHI; CGCGCTCTTG; GATCCAAGAG; IER-A-LGS. Δ4; Δ[476-626]; BamHI-AflII; GATCCGGAGCTATGC; TTAAGCATAGCTCCG; SGA-MLK. Δ5; Δ[631-694]; AflII-Bsu36I; TTAAGTTGAAGGACC; TTAGGTCCTTCAAC; LKL-KDL. Δ6; Δ[700-812]; Bsu36I-SphI; TAAGGGTAGAACAGCATG; CTGTTCTACCC; LRV-EQH. Δ7; Δ[818-1010]; SphI-PpuMI; CTAAGAAGTGG; GTCCCACTTCTTAGCATG; HAK-KWD. Δ[BRD]; Δ[1296-1464]; AvrII-StuI; CTAGGCGCCGGCACAGG; CCTGTGCCGGCGC; RRH-RPS. Δ[N-TRM]; Δ[7-309]; NdeI-MluI; TATGCCCATTTGCAAGCGGGAA; CGCGTTCCCGCTTGCAAATGGGCA; CKR-ERV. Δ[C-TRM]; Δ[1017-1461];PpuM-StuI; GACCCCAAGCAGCTACCGTTTAGG; CCTAAACGGTAGCTGCTTGGG; PKQ-LPF. m[WAKZ]; I1054A-W1056A-S1059A; ApaLI; CATCTTGCACGACTGTgcaCAGgcGAGGCGTgCCACCAATAAGTC; ILHDCaQaRRaTNKS. m[PHD1]; C1080S-C1083S-H1088A-C1091S; KpnI; GAGAAGATGCTGCTGTcCGATGAAaGCAACGCTGGtACcgcCATGTTCaGCCTGAAGCCTAAGC; EKMLLsDEsNAGTaMFsLKPK. m[PHD2]; C1257S-C1260S-H1265A-C1268S; XbaI; GGCGGTGAAATCAAATcTGTGCAATctAGaCTATTCTTTgcCCTGGAAaGTGTTCACCTCAAGC; GGEIKsVQsRLFFaLEsVHLKR.m[ACD]; D1145A-E1146A-E1147A-E1148A-E1149A-E1150A; SacII; GGATGATGAAGCTACAgcCgcggcAgcGgcAgcGAAAAAGGATGACGATATG;EDDEATaaaaaaKKDDDM.
Recombinant bacmids were generated from the expression constructs by recombination in Escherichia coli as recommended by the manufacturer (Bac-to-Bac system; Gibco-BRL). Bacmid DNA was isolated with a Qiagen Plasmid Mini Kit. The bacmid DNA preps were transfected into Sf9 insect cells with Lipofectin (Gibco-BRL), and baculoviruses were amplified three times to obtain high titer baculovirus stocks.
Recombinant protein synthesis and purification.
Recombinant wild-type ACF and mutant ACF complexes were produced by cosynthesis of untagged ISWI and FLAG-tagged Acf1 variants in Sf9 cells. Recombinant ISWI was synthesized and purified as a FLAG-tagged polypeptide in Sf9 cells (25). The proteins were purified by FLAG immunoaffinity chromatography, as described previously (25). The composition of the complexes was analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and silver staining. The protein concentrations were estimated by SDS-polyacrylamide gel electrophoresis and Coomassie brilliant blue R-250 staining along with a bovine serum albumin (BSA) standard (Pierce). Yields of recombinant ACF complexes varied from 500 ng (such as with IBR and Δ[C-TRM]) to 20 μg (such as with wild-type ACF) of total ACF protein per 150-mm tissue culture plate. The experiments whose results are shown in the figures were performed with baculovirus-synthesized ISWI. In addition, we purified bacterially synthesized ISWI (8) and found that its activity is indistinguishable from that of baculovirus-synthesized ISWI (data not shown).
For recombinant NAP-1 purification, 1 liter of Sf9 cell culture (106 cells/ml) was infected in a spinner flask at multiplicity of infection of 5 to 10 (which corresponded to about 20 to 25 ml of amplified virus). At 72 h postinfection, the cells were pelleted and washed with cold phosphate-buffered saline. Except where noted, the following operations were performed at 4°C. The washed pellet was suspended in 30 ml of lysis buffer (50 mM sodium phosphate [pH 7.0], 500 mM NaCl, 20 mM imidazole, 10 mM β-glycerophosphate, 15% glycerol, 0.01% NP-40, 0.2 mM phenylmethylsulfonyl fluoride (PMSF), 0.5 mM benzamidine-HCl) and homogenized thoroughly in a Dounce homogenizer (A pestle; 30 to 40 strokes over a period of 30 min on ice). After insoluble material was pelleted by centrifugation for 10 min at 14,500 × g (11,000 rpm; SS-34 rotor), the supernatant was combined with 2 ml of Ni-nitrilotriacetic acid resin (Qiagen) that had been equilibrated in the lysis buffer. The slurry was mixed on a rocking platform overnight. The resin was washed twice (20 ml each) with lysis buffer and then twice (12 ml each) with wash buffer (50 mM sodium phosphate [pH 7.0], 100 mM NaCl, 20 mM imidazole, 10 mM β-glycerophosphate, 15% glycerol, 0.01% NP-40, 0.2 mM PMSF, 0.5 mM benzamidine-HCl). The protein was eluted by four successive cycles of addition and removal of 2 ml of elution buffer (wash buffer + 480 mM imidazole). The elution time for each step was 5 min. The pooled protein sample (8 ml) was dialyzed for 4 h against 4 liters of buffer R (without MgCl2) plus 100 mM NaCl and then for 2 h against 4 liters of buffer R (10 mM potassium HEPES, [pH 7.6], 10 mM KCl, 1.5 mM MgCl2, 0.5 mM EGTA, 10% [vol/vol] glycerol, 10 mM β-glycerophosphate, 1 mM dithiothreitol, 0.2 mM PMSF) plus 100 mM NaCl. Significant precipitate formed at this step. The insoluble material was removed by centrifugation for 10 min at 14,500 × g (11,000 rpm; SS-34 rotor), and the supernatant was loaded onto a 1-ml Source 15Q (Pharmacia) column that was equilibrated with buffer R plus 100 mM NaCl. The column was washed with 5 column volumes of buffer R plus 100 mM NaCl followed by 10 column volumes of buffer R plus 200 mM NaCl. The NAP-1 protein was eluted with a 30-column-volume gradient from 200 to 500 mM NaCl in buffer R. Fractions (0.5 ml) were collected between NaCl concentrations of 250 to 400 mM. Recombinant NAP-1 eluted in two peaks—a minor peak at ≈250 mM NaCl, and a major peak at ≈300 mM NaCl. Fractions from the major peak (1.5 to 2 ml) were pooled and dialyzed against 4 liters of buffer R plus 100 mM NaCl for 4 h. The protein concentration was estimated by SDS-polyacrylamide gel electrophoresis and Coomassie brilliant blue R-250 staining along with a BSA mass standard (Pierce). Typical yields of recombinant NAP-1 were approximately 1 to 4 μg per ml of suspension culture. The protein was frozen in liquid nitrogen in small aliquots (100 to 200 μl) and stored at −80°C.
Chromatin assembly.
Chromatin was assembled onto a 3.2-kbp plasmid (pGIE-0) as described elsewhere (18, 25). A typical chromatin assembly reaction mixture contained 530 ng of plasmid DNA (0.25 pmol), 530 ng of core histones, 2.1 μg of NAP-1, and 44 fmol of ACF complex (wild type or mutant) or ISWI polypeptide. The reactions proceeded for 30 min at room temperature, and the products were analyzed by micrococcal nuclease digestion and DNA supercoiling assays (18). In the DNA supercoiling assays, the fluorescence of ethidium-stained DNA was quantitated with the Alpha Imager 2200 digital imaging system (Alpha Innotech Corp.). For each lane, the extent of chromatin assembly was assessed by dividing the amount of fluorescence in the supercoiled band by the combined amount of fluorescence in the relaxed plus supercoiled bands. (The amount of background fluorescence in the supercoiled band in the absence of ACF was also subtracted.) Then the chromatin assembly activity of each ACF mutant variant relative to that of wild-type ACF (defined to be 100%) was indicated.
ATPase assays.
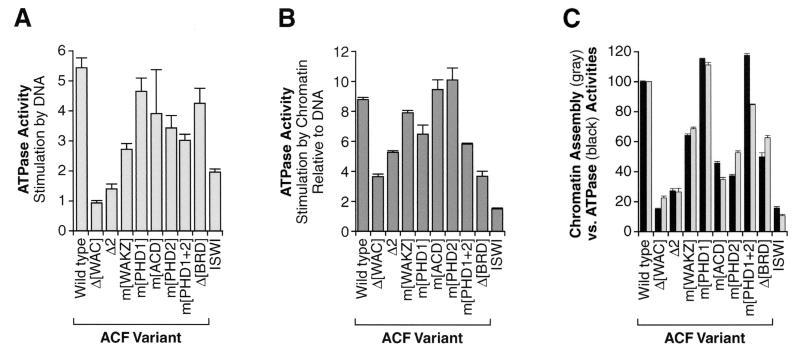
The ATPase activity of ACF, ACF mutants, and ISWI was measured by thin-layer chromatography of [γ-32P]ATP hydrolysis products. A typical ATPase reaction mixture contained 0.44 pmol of ACF complex or ISWI, 200 ng of BSA, and 0.1 μCi of [γ-32P]ATP in 10 μl of buffer containing 10 mM potassium HEPES (pH 7.6), 50 mM KCl, 5 mM MgCl2, and 0.3 mM (unlabeled) ATP. Where indicated, reaction mixtures additionally contained 350 ng of purified Drosophila core histones, 350 ng (0.17 pmol) of supercoiled plasmid pGIE-0, or 0.17 pmol of minichromosomes reconstituted from pGIE-0 by salt dialysis (28). The reactions were carried out for 10, 20, and 40 min at 27°C and then terminated by the addition of 1 μl of 0.5 M EDTA. For each reaction, an aliquot (0.5 μl) was applied to a polyethylenimine (PEI)-cellulose thin-layer chromatography plate (SelectoScientific) that had been prerun in water and air dried at room temperature for 60 min. ATP hydrolysis products were resolved by chromatography in thin-layer chromatography buffer (0.8 M LiCl, 0.8 M acetic acid) and quantitated by using a phosphorimager. In our ATPase experiments, such as those in Fig. 3, we observed less than twofold stimulation of the ATPase activity of ISWI polypeptide by chromatin (as circular minichromosomes) relative to covalently closed circular DNA (see Fig. 3B). In other studies (see, for example, reference 8), it was found that mononucleosomes stimulate the ATPase activity of ISWI polypeptide to a much greater extent (about 10-fold) than that seen with the corresponding 146-bp linear DNA fragment. Thus, the structure of the chromatin and DNA affects the magnitude of stimulation of ISWI ATPase by chromatin relative to naked DNA.
FIG. 3.
Analysis of the ATPase activity of mutant ACF complexes. (A) Stimulation of the ATPase activity of mutant ACF complexes by plasmid DNA. ATPase assays were performed with wild-type ACF, mutant ACF proteins, or ISWI alone in the presence or absence of plasmid DNA. The graph indicates the magnitude of stimulation of the ATPase activity by plasmid DNA. Error bars represent standard deviations (n = 4, except for m[ACD], where n = 3). ISWI-binding-defective Acf1 variants did not stimulate the basal level of ATPase activity of ISWI alone (data not shown). (B) Stimulation of the ATPase activity of mutant ACF complexes by chromatin relative to naked DNA. ATPase assays were performed with wild-type ACF, mutant ACF proteins, or ISWI alone in the presence of either salt-dialyzed chromatin or an equimolar amount of plasmid DNA. The graph indicates the amount of ATPase activity with chromatin relative to that with plasmid DNA. Error bars represent standard deviations (n = 4, except for m[ACD], where n = 3). (C) Comparison of the chromatin assembly and ATPase activities of mutant ACF complexes. The ATPase activity of wild-type ACF, mutant versions of ACF, and ISWI in the presence of chromatin was measured. The chromatin used in the ATPase experiments was prepared by salt dialysis reconstitution with supercoiled plasmid DNA and purified Drosophila core histones (24, 28). The relative ATPase activities of the mutant proteins were normalized to the activity of wild-type ACF (100%) and plotted (black bars) alongside their relative chromatin assembly activities (gray bars), as determined in the experiment in Fig. 2B. Error bars represent standard deviations (n ≥ 3 for ATPase data, and n = 2 for chromatin assembly data).
Sucrose gradient sedimentation analysis of ACF binding to DNA.
ACF (wild type or mutant) or ISWI polypeptide (2.5 pmol) was combined with 10 μg of recombinant human insulin (Roche; insulin was used as a nonspecific stabilizer of ACF activity) and 12 μg (5.7 pmol) of supercoiled pGIE-0 plasmid DNA in 100 μl of buffer containing 7.5 mM Tris-HCl (pH 7.9), 100 mM NaCl, 7.5% glycerol, 1.5 mM MgCl2, 0.15 mM EDTA, 0.75 mM DTT, and 0.01% NP-40. The binding-reaction mixtures were incubated for 30 min at 0°C and then applied to the top of a 5.5-ml tube containing a sucrose gradient (10 to 50% [wt/vol] sucrose in 25 mM potassium HEPES [pH 7.6], 100 mM KCl, 0.2 mM EDTA, 2 mM MgCl2, 1 mM DTT, 0.2 mM PMSF, 0.5 mM benzamidine-HCl, 0.01% NP-40, and 0.02% sodium azide). The sucrose gradients were subjected to centrifugation for 16 h at 225,000 × g (43,000 rpm; Beckman SW55 Ti rotor) at 4°C, and nine fractions (of 600 μl) were collected from each gradient. Precipitated material was not collected. An aliquot (35 μl) of each fraction was deproteinized by proteinase K digestion and phenol-chloroform extraction, and the resulting nucleic acids were precipitated with ethanol and subjected to agarose gel electrophoresis (1% agarose). The remainder of each fraction was precipitated with trichloroacetic acid and resuspended in 40 μl of SDS gel-loading buffer. The samples (10-μl aliquots) were subjected to SDS-polyacrylamide gel electrophoresis (7.5% polyacrylamide), transferred to nitrocellulose, and probed for the presence of ACF subunits with anti-Acf1 and anti-ISWI antibodies (25).
RESULTS AND DISCUSSION
The central region of Acf1 is important for its interaction with ISWI.
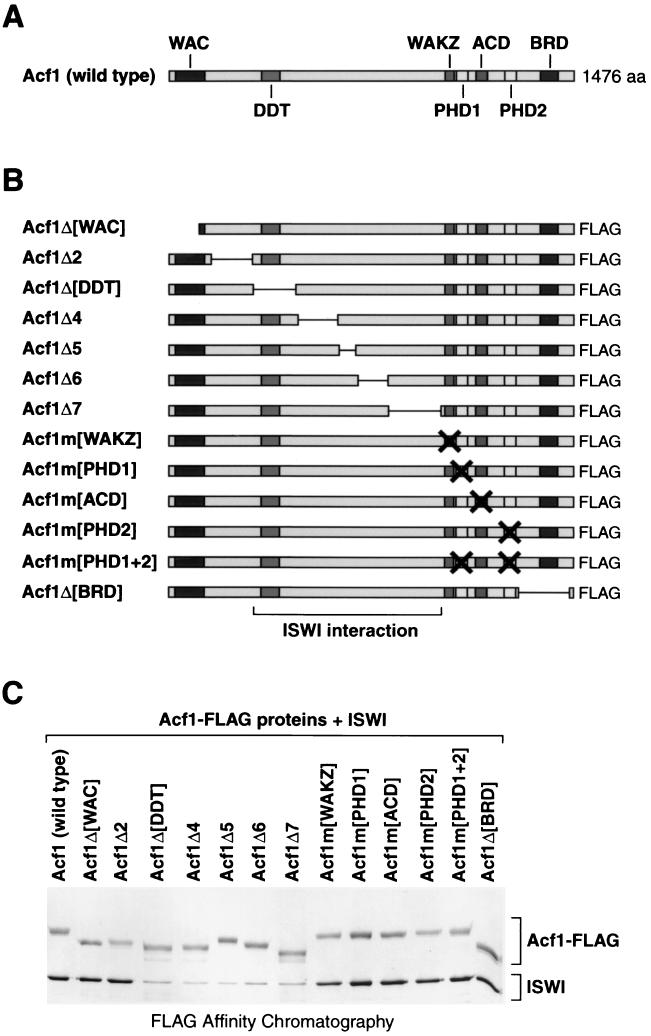
Acf1 contains a number of conserved sequence regions, which include WAC, WAKZ, DDT, bromodomain, and two PHD finger motifs, in addition to a highly acidic segment (denoted as ACD) (Fig. 1A). To investigate the functions of subregions of Acf1, we performed a systematic mutational analysis of the protein. To this end, we constructed a panel of mutant versions of Acf1, which are shown in Fig. 1B. In the design of these mutations, we sought to create a systematic set of deletions in Acf1 as well as to alter specific sequence motifs. The Acf1 mutant proteins (with C-terminal FLAG tags) were cosynthesized with wild-type (untagged) ISWI by using a baculovirus expression system. (Note that there was an excess of untagged ISWI relative to FLAG-tagged Acf1 in the baculovirus-infected cells.) The proteins were then purified to near homogeneity by FLAG affinity chromatography via the FLAG tag on the Acf1 variants. With wild-type Acf1 as well as with mutant Acf1 proteins in which the central region was not altered, we obtained copurification of the Acf1 and ISWI subunits of ACF (Fig. 1C). In contrast, however, deletion of any one of five adjoining fragments from amino acid residues 312 to 1010 of Acf1 resulted in a significant reduction in the association of ISWI with Acf1 (Fig. 1C; see Acf1Δ[DDT], Acf1Δ4, Acf1Δ5, Acf1Δ6, and Acf1Δ7). Thus, the deleted protein fragments may contain residues that are involved in the interaction of Acf1 with ISWI. Alternatively, the deletion of some of these fragments may alter the higher-order structure of the ISWI-interacting region of Acf1. In this segment of Acf1, only the DDT domain (amino acid residues 346 to 411) appears to be conserved between Acf1 and another ISWI-binding protein, NURF301 (51). Thus, the central region of Acf1 (residues 312 to 1010), which includes the DDT motif, is important for its binding to ISWI.
FIG. 1.
Identification of regions of Acf1 that are required for interaction with ISWI. (A) Sequence motifs in Acf1. The WAC domain (25), DDT domain (12), WAKZ domain (25), PHD fingers 1 and 2 (1), ACD (region containing acidic amino acid [aa] residues), and bromodomain (BRD) (22, 27) are indicated. (B) Diagram of deletion and substitution mutant variants of Acf1. The specific mutations in each of the Acf1 variants are given in Materials and Methods. These polypeptides contain a C-terminal FLAG tag. (The “wild-type” recombinant Acf1 polypeptide also contains a C-terminal FLAG tag.) The bracket at the bottom designates the putative ISWI interaction region. (C) Interaction of mutant Acf1 proteins with ISWI. Wild-type and mutant Acf1-FLAG polypeptides were cosynthesized with wild-type (untagged) ISWI and then subjected to FLAG affinity chromatography. The resulting samples were analyzed by SDS-polyacrylamide gel electrophoresis (6% polyacrylamide) and silver staining, which yields light yellow bands for Acf1 polypeptides and dark brown bands for ISWI polypeptide. Both Acf1 and ISWI polypeptides stain with approximately equivalent intensities with Coomassie brilliant blue R-250 (data not shown).
Different regions of Acf1 contribute to its chromatin assembly activity.
With the panel of mutant Acf1 proteins, we sought to determine which regions of Acf1 are important for chromatin assembly. To this end, we performed chromatin assembly reactions with the purified wild-type or mutant ACF proteins along with purified NAP-1, purified core histones, plasmid DNA, ATP (and the ATP-regenerating system), and the purified recombinant Drosophila topoisomerase I (in reactions subjected to DNA supercoiling analysis) (18, 25). To detect changes in the chromatin assembly activity of the mutant ACF proteins, we used limiting concentrations of ACF and terminated the reactions at early reaction times. Therefore, in these experiments, we used a 1:6 molar ratio of ACF protomers to plasmid DNA templates carried out the reactions for 30 min. Under these conditions, the templates were assembled to about 25% of full capacity with wild-type ACF.
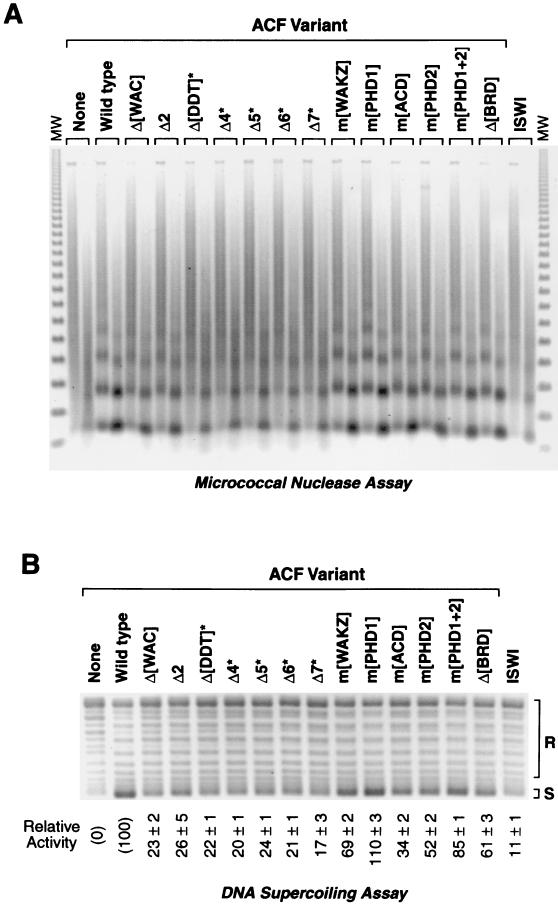
Chromatin assembly reactions were performed with wild-type or mutant ACF proteins, and the reaction products were subjected to micrococcal nuclease digestion and DNA supercoiling analyses (Fig. 2). The micrococcal nuclease digestion assay reveals the periodicity of the nucleosome arrays (Fig. 2A), whereas the DNA supercoiling assay reflects the extent of nucleosome assembly (Fig. 2B). The relative activities of the ACF proteins were quantitated from the DNA supercoiling data (Fig. 2B). There was also a good qualitative correlation between the extent of DNA supercoiling and the periodicity of the micrococcal nuclease digestion products. As controls and references, reactions were performed in the absence of ACF or ISWI (Fig. 2, left lanes) as well as with ISWI alone (in the absence of Acf1) (Fig. 2, right lanes).
FIG. 2.
Chromatin assembly activity of mutant ACF complexes. (A) Micrococcal nuclease digestion analysis of chromatin assembled with mutant ACF complexes. Chromatin was assembled for 30 min at a 6:1 molar ratio of plasmid DNA template to ACF (wild-type or mutant ACF, or ISWI alone) and then subjected to partial digestion by micrococcal nuclease (each reaction product was treated with two different concentrations of micrococcal nuclease). The samples were deproteinized, and the resulting DNA fragments were visualized by agarose gel electrophoresis (1.2% agarose) and staining with ethidium bromide. Mutant Acf1 proteins that do not interact with ISWI (indicated by asterisks) were supplemented with equimolar amounts of purified ISWI polypeptide. The size markers (MW) are the 123-bp DNA ladder (GIBCO-BRL). (B) DNA supercoiling analysis of chromatin assembled with mutant ACF complexes. Chromatin was assembled as in panel A, and the samples were deproteinized and visualized by agarose gel electrophoresis (0.8% agarose) and staining with ethidium bromide. The percent activity of each mutant ACF complex is relative to that of wild-type ACF (100%). The amount of signal obtained in the absence of ACF of ISWI (left lane) was subtracted as the background (0%). The relative activity is reported as the mean and standard deviation of two to four separate experiments. Mutant Acf1 proteins that do not interact with ISWI (indicated by asterisks) were supplemented with equimolar amounts of purified ISWI polypeptide.
These experiments revealed that several regions throughout Acf1 are important for its chromatin assembly activity. First, mutations that impair the interaction of Acf1 with ISWI (indicated by asterisks in Fig. 2) cause a defect in chromatin assembly activity. (Note that reaction mixtures containing these mutant Acf1 proteins, which did not copurify with ISWI [see Fig. 1C], were supplemented with equimolar amounts of purified ISWI.) The reduced activity of these ISWI-binding-defective Acf1 variants is consistent with the low level of activity by ISWI alone in the absence of Acf1 (≈11% of ACF [25] [Fig. 2, right lanes]). Second, we observed a significant reduction in the chromatin assembly activity upon mutation of the acidic region of Acf1 (m[ACD]). It is possible, for instance, that the acidic region may be important for interactions with histones. There were also moderate but distinct defects in chromatin assembly upon mutation of the second PHD finger (m[PHD2]) or the bromodomain (m[BRD]). It is interesting that mutation of the second PHD finger causes a reduction in chromatin assembly activity whereas mutation of the first PHD finger leads to a slight increase in activity. Lastly, aside from the mutations in the ISWI interaction region, the strongest defects in chromatin assembly were seen in the two mutant proteins containing deletions in the N-terminal region of Acf1 (Δ[WAC] and Δ2). These results indicate that multiple regions of Acf1, rather than a single protein motif, contribute to its chromatin assembly activity.
The ATPase activity of ACF correlates with its chromatin assembly activity.
ACF has an ATPase activity that is stimulated by DNA or chromatin but not by free core histones (reference 24 and unpublished data). To investigate the effect of Acf1 on the basal ATPase activity of the ISWI subunit of ACF, we analyzed the extent to which DNA stimulates the ATPase activity of the various mutant ACF complexes (Fig. 3A). We observed that deletions in the N-terminal region of Acf1 (Δ[WAC] and Δ2) resulted in a near complete loss of DNA-mediated stimulation of the ACF ATPase activity. These findings suggest that the N-terminus of Acf1 is involved in the interaction of ACF with DNA. This hypothesis is further explored below.
We also examined the stimulation of the ATPase activity by chromatin. The ATPase activity of wild-type ACF is about nine times higher in the presence of chromatin than in the presence plasmid DNA. We therefore determined the ratio of chromatin-mediated to DNA-mediated stimulation of the ATPase activity of the ACF variants and ISWI polypeptide (Fig. 3B). The ATPase activity of ISWI alone was not enhanced by chromatin (relative to DNA), and thus the Acf1 subunit is required for the chromatin-specific enhancement of the ATPase activity of ACF. Regions of Acf1 that appear to be particularly important for this chromatin-specific ATPase enhancement include the N-terminal WAC domain as well as the C-terminal bromodomain. The bromodomain binds to nucleosomes with specifically acetylated histones (11, 26). In addition, the WAC domain is present in the N-terminal fragment of cbp146, a mouse protein related to Acf1, which associates with pericentric heterochromatin (42). Thus, the bromodomain and WAC domain may be involved in the interaction of ACF with chromatin.
To see whether there is a relation between the ATPase and chromatin assembly activities of ACF, we plotted the ATPase activities of the various mutant ACF proteins (in the presence of chromatin) alongside their chromatin assembly activities. As shown in Fig. 3C, there is a strong correlation between the chromatin assembly and chromatin-stimulated ATPase activities of the various mutant ACF complexes. Thus, the ATPase data suggest that Acf1 may facilitate chromatin assembly, at least in part by enhancement of the ATP-driven motor activity of ISWI. These results additionally implicate the WAC domain and the bromodomain of Acf1 in the interaction of ACF with DNA and/or chromatin.
The N-terminal region of Acf1 is important for the binding of ACF to DNA.
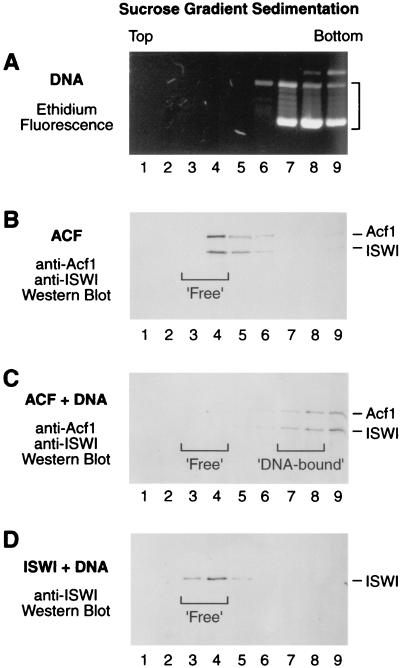
The ATPase experiments suggested that the N-terminal region of Acf1 is important in the interaction of ACF with DNA. It is possible, for instance, that this region of Acf1 mediates the recruitment of ACF to DNA during chromatin assembly. We therefore investigated the binding of ACF to DNA. In these assays, ACF or ACF-DNA mixture was subjected to sucrose gradient sedimentation and the Acf1 and ISWI subunits were detected by Western blot analysis. These experiments indicated that ACF forms a stable ACF-DNA complex that sediments significantly faster than free ACF (Fig. 4). In contrast, we did not observe a stable interaction of ISWI polypeptide with DNA (Fig. 4D). Under the same conditions, Acf1 polypeptide alone (in the absence of ISWI) formed an insoluble precipitate and was therefore not used in these studies.
FIG. 4.
Sucrose gradient sedimentation analysis of ACF binding to DNA. (A) Sedimentation of plasmid DNA in a 10 to 50% sucrose gradient. The fractions were subjected to agarose gel electrophoresis (0.8% agarose), and the DNA was visualized by staining with ethidium bromide. (B) Sedimentation of wild-type ACF in a 10 to 50% sucrose gradient. The fractions were subjected to SDS-polyacrylamide gel electrophoresis (7.5% polyacrylamide) followed by Western blot analysis with antibodies against Acf1 and ISWI. The designation of fractions 3 and 4 as ‘Free’ ACF is indicated by a bracket. (C) Sedimentation of ACF plus plasmid DNA in a 10 to 50% sucrose gradient. The molar ratio of ACF protomers to plasmid DNA molecules was 1:2.3. The fractions were subjected to SDS-polyacrylamide gel electrophoresis (7.5% polyacrylamide) followed by Western blot analysis with antibodies against Acf1 and ISWI. The designation of fractions 3 and 4 as ‘Free’ ACF and fractions 7 to 8 as ‘DNA-bound’ ACF is indicated by brackets. (D) Sedimentation of ISWI plus plasmid DNA in a 10 to 50% sucrose gradient. The molar ratio of ISWI polypeptides to plasmid DNA molecules was 1:2.3. The fractions were subjected to SDS-polyacrylamide gel electrophoresis (7.5% polyacrylamide) followed by Western blot analysis with antibodies against ISWI. The migration of ‘Free’ ISWI is indicated by a bracket. ISWI polypeptide exhibited the same rate of sedimentation in the presence and absence of plasmid DNA (data not shown).
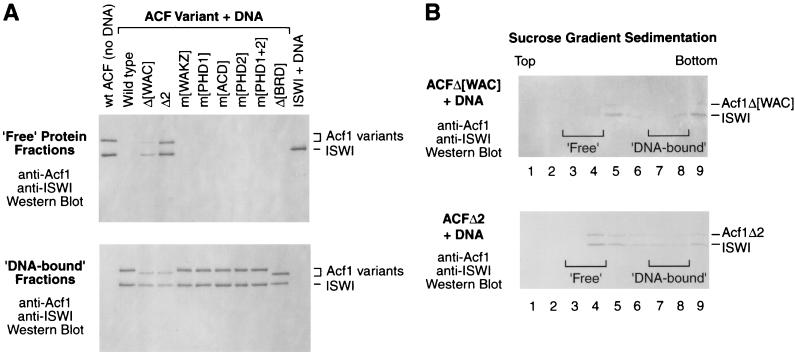
We then tested the binding of the mutant ACF complexes to DNA. As depicted in Fig. 4 and 5A, we analyzed gradient fractions that corresponded to free ACF (‘Free’ Protein Fractions) and to ACF-DNA complexes (‘DNA-bound’ Fractions). We observed that the binding of ACF to DNA was not affected by Acf1 mutations in the WAKZ domain, either or both PHD fingers, the acidic region, or the bromodomain (Fig. 5A). In contrast, deletion of the WAC domain or the adjacent N-terminal region (in the Acf1Δ2 mutant) caused a significant reduction in the interaction of ACF with DNA, as indicated by the presence of ‘Free’ ACF (Fig. 5A, upper panel) as well as the reduced amount of ‘DNA-bound’ ACF (lower panel). We also analyzed each of the gradient fractions with the ACFΔ[WAC] and ACFΔ2 complexes and observed that the majority of these mutant complexes did not remain stably associated with the DNA during sucrose gradient sedimentation (Fig. 5B). Thus, these data indicate that the N-terminal region of Acf1, which includes the WAC domain, is important for the binding of ACF to DNA. In addition, the N terminus of Acf1 with DNA was required for DNA-mediated stimulation of the ATPase activity of ACF (Fig. 3A). Together, these results suggest that the N terminus of Acf1 has a DNA-binding function that is required for the stimulation of the ATPase activity of ACF by DNA.
FIG. 5.
The N terminus of Acf1 is important for binding of ACF to DNA. (A) Interaction of wild-type and mutant versions of ACF with DNA. Wild-type ACF, mutant ACF proteins, and ISWI alone were each subjected to 10 to 50% sucrose gradient sedimentation in the presence of plasmid DNA (except in the no-DNA control [left lane]). For each sample, fractions 3 and 4, which correspond to the sedimentation of free ACF, were pooled and analyzed by Western blotting with antibodies against Acf1 and ISWI (upper panel). In parallel, fractions 7 and 8, which correspond to the sedimentation of DNA-bound ACF, were pooled and analyzed by Western blotting (lower panel). (B) Interaction of ACFΔ[WAC] and ACFΔ2 with DNA. ACFΔ[WAC] and ACFΔ2 were each subjected to 10 to 50% sucrose gradient sedimentation and Western blot analysis. The brackets correspond to the sedimentation of free and DNA-bound wild-type ACF (as indicated) under identical conditions.
Multiple domains of Acf1 function in chromatin assembly.
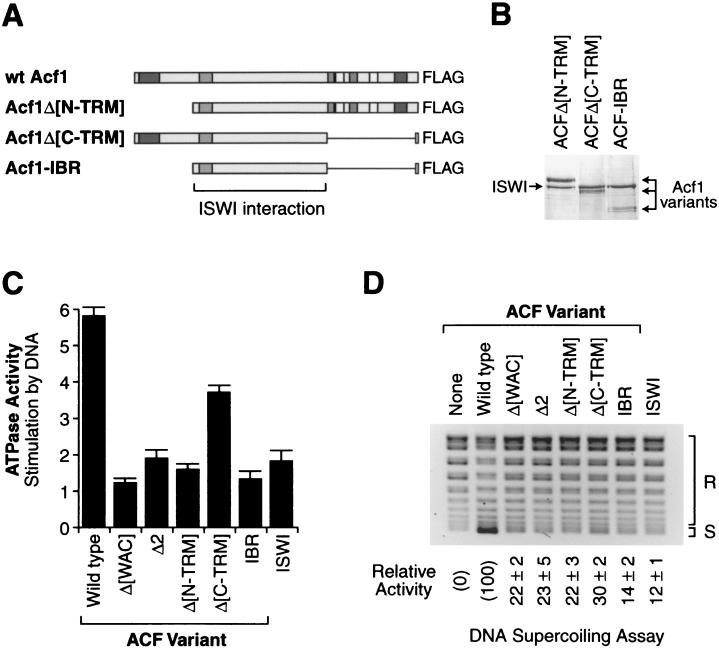
To analyze the N- and C-terminal segments of Acf1 in relation to the central region that is required for binding to ISWI, we constructed and analyzed three additional mutant versions of Acf1, which are depicted in Fig. 6A). Acf1-IBR consists of the region that spans the five adjoining fragments of Acf1 that were each found to be important for binding to ISWI (Fig. 1C; see Acf1Δ[DDT], Acf1Δ4, Acf1Δ5, Acf1Δ6, and Acf1Δ7). Acf1Δ[N-TRM] is an N-terminal deletion of Acf1 that is lacking both the WAC domain and the adjacent region 2 (i.e., the region of Acf1 that is deleted in the Acf1Δ2 mutant). Lastly, Acf1Δ[C-TRM] lacks residues that are C terminal to the essential ISWI binding region.
The Acf1 proteins were cosynthesized with wild-type ISWI by using baculovirus vectors, and the mutant ACF complexes were purified by affinity chromatography via the FLAG tag on the Acf1 polypeptides. As shown in Fig. 6B, each of the new mutant proteins copurified with ISWI. The ability of Acf1-IBR to associate with ISWI indicates that this region of Acf1 not only is required for binding of Acf1 to ISWI but also is sufficient for binding to ISWI. We then tested the DNA-stimulated ATPase activity (Fig. 6C) and chromatin assembly activity (Fig. 6D) of the mutant proteins. ACF-IBR has ATPase and chromatin assembly activities that are similar to those of ISWI alone. Thus, the binding of Acf1-IBR to ISWI does not appear to affect the enzymatic activity of ISWI. ACFΔ[N-TRM], which lacks both the WAC domain and region 2 of Acf1, exhibits defects in DNA-stimulated ATPase and chromatin assembly activities that are essentially identical to those of either ACFΔ[WAC] or ACFΔ2. These results, combined with the DNA- binding data (Fig. 4 and 5), suggest that the WAC domain and region 2 are each constituents of a broader N-terminal DNA-binding domain. Lastly, ACFΔ[C-TRM] has intermediate levels of ATPase and chromatin assembly activities, and hence the removal of all of the C-terminal protein motifs in Acf1 does not result in a complete loss of its activity.
FIG. 6.
Analysis of mutant ACF complexes lacking the N- and/or C-terminal region of Acf1. (A) Diagram of the Acf1Δ[N-TRM], Acf1Δ[C-TRM], and Acf1-IBR mutants of Acf1. The sequence motifs in Acf1 correspond to those depicted in Fig. 1A. The specific residues that are deleted in each of the Acf1 variants are indicated in Materials and Methods. These polypeptides also contain a C-terminal FLAG tag. (B) Purification of mutant ACF complexes containing Acf1Δ[N-TRM], Acf1Δ[C-TRM], or Acf1-IBR polypeptides. The proteins were subjected to SDS-polyacrylamide gel electrophoresis and staining with Coomassie brilliant blue R-250. (C) Stimulation of the ATPase activity of mutant ACF complexes by plasmid DNA. ATPase assays were performed with wild-type ACF, mutant ACF proteins, or ISWI alone in the presence or absence of plasmid DNA. The graph indicates the magnitude of stimulation of the ATPase activity by plasmid DNA. Error bars represent standard deviations (n = 3). (D) DNA supercoiling analysis of chromatin assembled with mutant ACF complexes. Chromatin was assembled and analyzed as in the experiment in Fig. 2B. The percent activity of the mutant ACF complexes is relative to that of wild-type ACF (100%). The amount of signal obtained in the absence of ACF or ISWI (left lane) was subtracted as the background (0%). The relative activity is reported as the mean ± standard deviation of two separate experiments.
Thus, the findings of this study suggest a view of Acf1 as a protein with an N-terminal DNA-binding region containing a WAC motif, a central ISWI-binding segment with a DDT domain, and a C-terminal region with multiple sequence motifs. It is interesting to compare the structures of Drosophila Acf1 and its Saccharomyces cerevisiae ortholog, Itc1 (19). Itc1 and Isw2 (which is related to Drosophila ISWI) form a chromatin-remodeling complex termed ISW2. Itc1 contains an N-terminal WAC domain, a region with a DDT domain that is similar to the ISWI-binding region of Acf1, and stretches of acidic amino acid residues that may be related to the ACD segment in Acf1. On the other hand, Itc1 appears to lack WAKZ, PHD finger, and bromodomain motifs. Notably, the regions that are conserved between Acf1 and Itc1 were found to be most important for the chromatin assembly activity of Acf1.
Binding of chromatin-remodeling complexes to DNA via the WAC domain or other motifs.
The WAC domain is present at the N-termini of Acf1-related proteins in a variety of organisms, and it seems likely that it plays a general role in the binding of ACF-related factors to DNA. In addition, components of other chromatin-remodeling complexes interact with DNA. For example, the p270 subunit (BAF250) of the human SWI-SNF complex contains a DNA-binding ARID motif (10, 38); the Snf6 subunit of yeast SWI-SNF complex binds to DNA (40); and the Rsc1 and Rsc2 subunits of yeast RSC (7), as well as the hBrm and BRG1 subunits of human SWI-SNF, contain DNA-binding AT-hook motifs (5). The Rsc3 and Rsc30 subunits of RSC also contain several leucine zippers and zinc clusters (3). Finally, the NF-terminus of NURF301, the largest subunit of NURF, contains a composite DNA-binding domain that is closely related to HMGA proteins (51).
DNA binding could play several distinct roles in chromatin assembly and remodeling. Some possible functions that would involve DNA binding include the bending or twisting of DNA, the formation of DNA loops, translocation along DNA via an inchworm mechanism, the establishment of nucleosome spacing and linker DNA (e.g., by transient occupation of internucleosomal DNA during assembly), and recruitment of the protein complex to the DNA substrate. In addition, DNA-binding motifs, such as the WAC domain, could act in conjunction with chromatin-binding motifs, such as the bromodomain.
In summary, the structure-function analysis of Acf1 has revealed DNA-binding as well as ISWI-binding regions that are important for the chromatin assembly activity of ACF. The DNA-binding region of Acf1 includes a WAC motif, which is important for the DNA-binding activity. It seems probable that the WAC motif will be involved in DNA binding in other related factors. In addition, the binding of Acf1 to ISWI requires the DDT domain, which has been found in a variety of transcription and chromatin-remodeling factors. It has been suggested that the DDT domain is involved in binding to DNA (12), and our results suggest an involvement of the DDT domain in protein-protein interactions. Chromatin assembly by ACF is also impaired upon mutation of an acidic region in Acf1. This segment of Acf1 may interact with histones during the deposition process. Lastly, we observed modest chromatin assembly defects on mutation of other conserved sequence motifs, such as the WAKZ domain, PHD fingers, and bromodomain. These motifs are not apparently conserved in Acf1-related proteins from Drosophila to S. cerevisiae. It is possible, for instance, that these sequence motifs are involved in functions that are distinct from the intrinsic assembly process, such as the interaction of ACF with other factors, the localization of ACF within the nucleus, the coupling of assembly with DNA synthesis, or the regulation of ACF activity. Thus, in conclusion, these studies provide a useful and important perspective on ACF function that could be extended to the further analysis of ACF and other related chromatin assembly and chromatin-remodeling factors.
Acknowledgments
We thank Buyung Santoso, Arthur Hsu, Vassili Alexiadis, Karl Haushalter, Tammy Juven-Gershon, Alexandra Lusser, Tom Boulay, and June Parsons for critical reading of the manuscript. We thank Davide Corona and Peter Becker for providing an ISWI bacterial expression construct and Buyung Santoso for purification of the bacterially expressed ISWI. DNA sequencing was performed by the Molecular Pathology Shared Resource, UCSD Cancer Center, which is funded in part by NCI Cancer Center Support Grant 5P0CA23100-16.
This work was supported by a grant from the National Institutes of Health (GM58272) to J.T.K. D.V.F. is an American Cancer Society Postdoctoral Fellow (fellowship PF-99-329-01-CCG).
REFERENCES
- 1.Aasland, R., T. J. Gibson, and A. F. Stewart. 1995. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20:56-59. [DOI] [PubMed] [Google Scholar]
- 2.Adams, C. R., and R. T. Kamakaka. 1999. Chromatin assembly: biochemical identities and genetic redundancy. Curr. Opin. Genet. Dev. 9:185-190. [DOI] [PubMed] [Google Scholar]
- 3.Angus-Hill, M. L., A. Schlichter, D. Roberts, H. Erdjument-Bromage, P. Tempst, and B. R. Cairns. 2001. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol. Cell 7:741-751. [DOI] [PubMed] [Google Scholar]
- 4.Bochar, D. A., J. Savard, W. Wang, D. W. Lafleur, P. Moore, J. Côté, and R. Shiekhattar. 2000. A family of chromatin remodeling factors related to Williams syndrome transcription factor. Proc. Natl. Acad. Sci. USA 97:1038-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourachot, B., M. Yaniv, and C. M. Muchardt. 1999. The activity of brm/Snf2a is dependent on a high-mobility group protein I/Y-like DNA-binding domain. Mol. Cell. Biol. 19:3931-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cairns, B. R. 1998. Chromatin remodeling machines: similar motors, ulterior motives. Trends Biochem. Sci. 23:20-25. [DOI] [PubMed] [Google Scholar]
- 7.Cairns, B. R., A. Schlichter, H. Erdjument-Bromage, P. Tempst, R. D. Kornberg, and F. Winston. 1999. Two functionally distinct forms of RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol. Cell 4:715-723. [DOI] [PubMed] [Google Scholar]
- 8.Corona, D. F. V., G. Längst, C. R. Clapier, E. J. Bonte, S. Ferrari, J. W. Tamkun, and P. B. Becker. 1999. ISWI is an ATP-dependent nucleosome remodeling factor. Mol. Cell 3:239-245. [DOI] [PubMed] [Google Scholar]
- 9.Corona, D. V. F., A. Eberharter, A. Budde, R. Deuring, S. Ferrari, P. Varga-Weisz, M. Wilm, J. Tamkun, and P. B. Becker. 2000. Two histone fold proteins, CHRAC-14 and CHRAC-16, are developmentally regulated subunits of chromatin accessibility complex (CHRAC). EMBO J. 19:3049-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dallas, P. B., S. Pacchione, D. Wilsker, V. Bowrin, R. Kobayashi, and E. Moran. 2000. The human SWI-SNF complex protein p270 is an ARID family member with non-sequence-specific DNA binding activity. Mol. Cell. Biol. 9:3137-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dhalluin, C., J. E. Carlson, L. Zeng, C. He, A. K. Aggarwal, and M.-M. Zhou. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491-496. [DOI] [PubMed] [Google Scholar]
- 12.Doerks, T., R. Copley, and P. Bork. 2001. DDT—a novel domain in different transcription and chromosome remodeling factors. Trends Biochem. Sci. 26:145-146. [DOI] [PubMed] [Google Scholar]
- 13.Eberharter, A., S. Ferrari, G. Längst, T. Straub, A. Imhof, P. Varga-Weisz, M. Wilm, and P. B. Becker. 2001. Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodeling. EMBO J. 20:3781-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elfring, L. K., R. Deuring, C. M. McCallum, C. L. Peterson, and J. W. Tamkun. 1994. Identification and characterization of Drosophila relatives of the yeast transcriptional activator SNF2/SWI2. Mol. Cell. Biol. 14:2225-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flaus, A., and T. Owen-Hughes. 2001. Mechanisms for ATP-dependent chromatin remodeling. Curr. Opin. Genet. Dev. 11:148-154. [DOI] [PubMed] [Google Scholar]
- 16.Fry, C. J., and C. L. Peterson. 2001. Chromatin remodeling enzymes: who's on first? Curr. Biol. 11:R185-R197. [DOI] [PubMed] [Google Scholar]
- 17.Fyodorov, D. V., and J. T. Kadonaga. 2001. The many faces of chromatin remodeling: SWItching beyond transcription. Cell 106:523-525. [DOI] [PubMed] [Google Scholar]
- 18.Fyodorov, D. V., and M. E. Levenstein. 2002. Chromatin assembly in Drosophila systems. Curr. Protocols Mol. Biol. 21.7.1-21.7.27. [DOI] [PubMed]
- 19.Gelbart, M. E., T. Rechsteiner, T. J. Richmond, and T. Tsukiyama. 2001. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol. Cell. Biol. 21:2098-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glikin, G. C., I. Ruberti, and A. Worcel. 1984. Chromatin assembly in Xenopus oocytes: in vitro studies. Cell 37:33-41. [DOI] [PubMed] [Google Scholar]
- 21.Guschin, D., T. M. Geiman, N. Kikyo, D. J. Tremethick, A. P. Wolffe, and P. A. Wade. 2000. Multiple ISWI ATPase complexes from Xenopus laevis. J. Biol. Chem. 275:35248-35255. [DOI] [PubMed] [Google Scholar]
- 22.Haynes, S. R., C. Dollard, F. Winston, S. Beck, J. Trowsdale, and I. B. Dawid. 1992. The bromodomain: a conserved sequence found in human, Drosophila, and yeast proteins. Nucleic Acids Res. 20:2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito, T., M. Bulger, R. Kobayashi, and J. T. Kadonaga. 1996. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly-spaced nucleosomal arrays. Mol. Cell. Biol. 16:3112-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito, T., M. Bulger, M. J. Pazin, R. Kobayashi, and J. T. Kadonaga. 1997. ACF, and ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90:145-155. [DOI] [PubMed] [Google Scholar]
- 25.Ito, T., M. E. Levenstein, D. V. Fyodorov, A. K. Kutach, R. Kobayashi, and J. T. Kadonaga. 1999. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 13:1529-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobson, R. H., A. G. Ladurner, D. S. King, and R. Tjian. 2000. Structure and function of a human TAFII250 double bromodomain module. Science 288:1422-1425. [DOI] [PubMed] [Google Scholar]
- 27.Jeanmougin, F., J.-M. Wurtz, B. Le Douarin, P. Chambon, and R. Losson. 1997. The bromodomain revisited. Trends Biochem. Sci. 22:151-153. [DOI] [PubMed] [Google Scholar]
- 28.Jeong, S. W., J. D. Lauderdale, and A. Stein. 1991. Chromatin assembly on plasmid DNA in vitro. Apparent spreading of nucleosome alignment from one region of pBR327 by histone H5. J. Mol. Biol. 222:1131-1147. [DOI] [PubMed] [Google Scholar]
- 29.Kaufman, P. D. 1996. Nucleosome assembly: the CAF and the HAT. Curr. Opin. Cell Biol. 8:369-373. [DOI] [PubMed] [Google Scholar]
- 30.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 31.Kornberg, R. D., and Y. Lorch. 1999. Chromatin-modifying and -remodeling complexes. Curr. Opin. Genet. Dev. 9:148-151. [DOI] [PubMed] [Google Scholar]
- 32.Längst, G., and P. B. Becker. 2001. Nucleosome mobilization and positioning by ISWI-containing chromatin-remodeling factors. J. Cell Sci. 114:2561-2568. [DOI] [PubMed] [Google Scholar]
- 33.LeRoy, G., G. Orphanides, W. S. Lane, and D. Reinberg. 1998. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science 282:1900-1904. [DOI] [PubMed] [Google Scholar]
- 34.LeRoy, G., A. Loyola, W. S. Lane, and D. Reinberg. 2000. Purification and characterization of a human factor that assembles and remodels chromatin. J. Biol. Chem. 275:14787-14790. [DOI] [PubMed] [Google Scholar]
- 35.Levenstein, M. E., and J. T. Kadonaga. 2002. Biochemical analysis of chromatin containing recombinant Drosophila core histones. J. Biol. Chem. 277:8749-8754. [DOI] [PubMed] [Google Scholar]
- 36.Loyola, A., G. LeRoy, Y. H. Wang, and D. Reinberg. 2001. Reconstitution of recombinant chromatin establishes a requirement for histone-tail modifications during chromatin assembly and transcription. Genes Dev. 15:2837-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mello, J. A., and G. Almounzi. 2001. The ins and outs of nucleosome assembly. Curr. Opin. Genet. Dev. 11:136-141. [DOI] [PubMed] [Google Scholar]
- 38.Nie, Z., Y. Xue, D. Yang, S. Zhou, B. J. Deroo, T. K. Archer, and W. Wang. 2000. A specificity and targeting subunit of human SWI/SNF family-related chromatin-remodeling complex. Mol. Cell. Biol. 20:8879-8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poot, R. A., G. Dellaire, B. B. Hülsmann, M. A. Grimaldi, D. F. V. Corona, P. B. Becker, W. A. Bickmore, and P. D. Varga-Weisz. 2000. HuCHRAC, a human ISWI chromatin remodeling complex contains hACF1 and two novel histone-fold proteins. EMBO J. 19:3377-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sengupta, S. M., M. VanKanegan, J. Persinger, C. Logie, B. R. Cairns, C. L. Peterson, B. Bartholomew. 2001. The interactions of yeast SWI/SNF and RSC with the nucleosome before and after chromatin remodeling. J. Biol. Chem. 275:12636-12644. [DOI] [PubMed] [Google Scholar]
- 41.Strohner, R., A. Nemeth, P. Jansa, U. Hofmann-Rohrer, R. Santoro, G. Längst, and I. Grummt. 2001. NoRC—a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 20:4892-4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tate, P., M. Lee, S. Rweedle, W. C. Skarnes, and W. A. Bickmore. 1998. Capturing novel mouse genes encoding chromosomal and other nuclear proteins. J. Cell Sci. 111:2575-2585. [DOI] [PubMed] [Google Scholar]
- 43.Tsukiyama, T., and C. Wu. 1995. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell 83:1011-1020. [DOI] [PubMed] [Google Scholar]
- 44.Tsukiyama, T., C. Daniel, J. Tamkun, and C. Wu. 1995. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell 83:1021-1026. [DOI] [PubMed] [Google Scholar]
- 45.Tsukiyama, T., J. Palmer, C. C. Landel, J. Shiloach, and C. Wu. 1999. Characterization of the Imitation Switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 13:686-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turner, B. M. 2001. Chromatin and gene regulation: mechanisms in epigenetics. Blackwell Science Ltd., Oxford, United Kingdom.
- 47.Varga-Weisz, P. D., M. Wilm, E. Bonte, K. Dumas, M. Mann, and P. B. Becker. 1997. Chromatin-remodeling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature 388:598-602. [DOI] [PubMed] [Google Scholar]
- 48.Verreault, A. 2000. De novo nucleosome assembly: new pieces in an old puzzle. Genes Dev. 14:1430-1438. [PubMed] [Google Scholar]
- 49.Vignali, M., A. H. Hassan, K. E. Neely, and J. L. Workman. 2000. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20:1899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolffe, A. P. 1998. Chromatin: structure and function, 3rd ed. Academic Press, Inc., San Diego, Calif.
- 51.Xiao, H., R. Sandaltzopoulos, H.-M. Wang, A. Hamiche, R. Ranallo, K.-M. Lee, D. Fu, and C. Wu. 2001. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol. Cell 8:531-543. [DOI] [PubMed] [Google Scholar]