Abstract
Objective:
RNA interference (RNAi), mediated by small interfering RNA (siRNA), silences genes with a high degree of specificity and potentially represents a general approach for molecularly targeted anticancer therapy. The aim of this study was to evaluate the ability of systemically administered siRNA to silence gene expression in vivo and to assess the effect of this approach on tumor growth using a murine pancreatic adenocarcinoma xenograft model.
Summary Background Data:
Carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6) is widely overexpressed in human gastrointestinal cancer. Overexpression of CEACAM6 promotes cell survival under anchorage independent conditions, a characteristic associated with tumorigenesis and metastasis.
Methods:
CEACAM6 expression was quantified by real-time polymerase chain reaction (PCR) and Western blot. Mice (n = 10/group) were subcutaneously xenografted with 2 × 106 BxPC3 cells (which inherently overexpress CEACAM6). Tumor growth, CEACAM6 expression, cellular proliferation (Ki-67 immunohistochemistry), apoptosis, angiogenesis (CD34 immunohistochemistry), and survival were compared for mice administered either systemic CEACAM6-specific or control single-base mismatch siRNA over 6 weeks, following orthotopic tumor implantation.
Results:
Treatment with CEACAM6-specific siRNA suppressed primary tumor growth by 68% versus control siRNA (P < 0.05) and was associated with a decreased proliferating cell index, impaired angiogenesis and increased apoptosis in the xenografted tumors. CEACAM6-specific siRNA completely inhibited metastasis (0% of mice versus 60%, P < 0.05) and significantly improved survival, without apparent toxicity.
Conclusions:
Our data demonstrate the efficacy of systemically administered siRNA as a therapeutic modality in experimental pancreatic cancer. This novel therapeutic strategy may be applicable to a broad range of cancers and warrants investigation in patients with refractory disease.
RNA interference, mediated by small interfering RNA, induces potent and specific gene silencing. This in vivo study illustrates the application of this new technology as a novel approach for targeted cancer therapy, using a murine pancreatic adenocarcinoma xenograft model.
Pancreatic adenocarcinoma remains among the most refractory of malignancies. The majority of patients present with surgically incurable disease, and current chemotherapeutic agents offer little in terms of survival benefit or improved quality of life.1 There is clearly a need for new approaches to the treatment of this cancer. Recently, it has been recognized that double-stranded ribonucleic acid (RNA) 21–22 nucleotides in length, referred to as small interfering RNA (siRNA), when introduced into the cell induces potent gene silencing through a mechanism termed RNA interference (RNAi).2,3 The siRNA oligonucleotides associate with a multicomponent nuclease RNA-induced silencing complex, which targets and degrades mRNA complementary to the siRNA base sequence. RNAi appears to play important roles in many aspects of cellular biology. We hypothesized that this approach could be applied as a target-specific treatment of cancer.
Carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6) belongs to the immunoglobulin superfamily.4 This cell surface molecule is anchored to the outer leaflet of the cell membrane by a glycosylphosphatidylinositol anchor and is thought to function as an intercellular adhesion molecule.5 CEACAM6 is overexpressed in a variety of gastrointestinal malignancies.6,7 Despite lacking an intracellular domain, CEACAM6 is able to influence intracellular signaling events, and overexpression of this molecule appears to promote gastrointestinal cancer progression.7–10 Previously, we reported that pancreatic adenocarcinoma cells differentially express CEACAM6 and that overexpression of CEACAM6 is associated with a higher tolerance of anchorage-independent culture and greater in vivo metastatic ability.11 Furthermore, we have shown that posttranscriptional inhibition of CEACAM6 expression inhibits the ability of pancreatic adenocarcinoma cells to form liver metastases in vivo.11 Given that modulating levels of CEACAM6 expression can have marked effects on the phenotype of pancreatic adenocarcinoma cells, we selected CEACAM6 as a target for siRNA-mediated therapy.
Here, we evaluate the ability of systemically administered siRNA to silence tumor gene expression in vivo, using CEACAM6 as a target in a pancreatic adenocarcinoma nude mouse xenograft model. Our observations indicate that systemically administered siRNA can induce tumor gene silencing and alter tumor behavior in vivo.
MATERIALS AND METHODS
Cells and Cell Culture
BxPC3 human pancreatic ductal adenocarcinoma cells were obtained from American Type Culture Collection (ATCC) (Rockville, MD). Cells were maintained in Dulbecco Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS; Gibco BRL, Gaithersburg, MD) and were incubated in a humidified (37°C, 5% CO2) incubator, grown in 75-cm2 culture flasks and passaged upon reaching 80% confluence.
Proliferation and Colony Formation Assays
Cellular proliferation was quantified by 3-(4,5,-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Trevigen, Gaithersburg, MD) in accordance with the manufacturer's instructions. Results of the MTT assay have been shown to correlate well with [3H]-thymidine incorporation in pancreatic adenocarcinoma cell lines.12 Forty-eight hours following siRNA treatment, cells were seeded into 96-well plates at 104 cells per well and allowed to adhere overnight. Cells were cultured in medium containing 10% FBS. Cellular proliferation was determined after 48 hours. Plates were read using a Vmax microplate spectrophotometer (Molecular Devices, Sunnyvale, CA) at a wavelength of 570 nm, referenced to 650 nm. Ten samples were used for each experimental condition, and experiments were performed in triplicate.
To determine substrate-independent cell growth, soft agar assays were performed in 6-well plates with 1.5-mL bottom layer and 0.5-mL top layer. BxPC3 cells (1 × 104/well) were distributed as a single cell suspension in 0.34% (wt/vol) agar (Difco Laboratories, Detroit, MI) in assay medium and overlaid onto a 0.51% (wt/vol) bottom agar likewise dissolved in assay medium (DMEM + 10% [vol/vol] FBS). Cells were incubated for 2 to 3 hours at room temperature and then transferred to 37°C, 5% CO2. The cell layer was covered with 400 μL of fresh assay medium every second day. Colony formation was quantified after 10 days.
siRNA Preparation
siRNA was synthesized and purified by Qiagen-Xeragon (Germantown, MD). CEACAM6 sense (5′-CCGGACAGUUCCAUGUAUA-dTT-3′) and antisense (5′-UAUACAUGGAACUGUCCGG-dTT-3′) siRNAs and control siRNAs (sense 5′-CCCGACAGUUCCAUGUAUA-dTT-3′, antisense 5′-UAUACAUGGAACUGUCGGG-dTT-3′), which bear no homology with relevant human genes, were dissolved in buffer (100 mM potassium acetate, 30 mM HEPES-potassium hydroxide, 2 mM magnesium acetate, pH 7.4) to a final concentration of 20 μM, heated to 90°C for 60 seconds, and incubated at 37°C for 60 minutes prior to use to disrupt any higher aggregates formed during synthesis.
Real-Time PCR
RNA was extracted from human cells using Trizol Reagent (Gibco BRL) according to the manufacturer's instructions. Two micrograms of RNA per sample was reverse transcribed into cDNA using reverse transcription (MMLV-RT; Gibco-BRL) in a standard protocol with random hexamer primers. For the negative control, MMLV was omitted. LUX primers for CEACAM6 were designed using the Invitrogen (Carlsbad, CA) online primer design software (https://pf.invitrogen.com/primerf/pages/Default.cfm?mL=1). 6-Carboxy-fluorescein (FAM) was chosen as reporter for CEACAM6. CEACAM6 primers were forward, 5′-GACGTTTGTGT-GGATTGCTGGAACG[FAM]C-3′; and reverse, 5′-TGCCACGCAGCCTCTAACC-3′. 6-Carboxy-4′,5′-dichloro-2′,7′-dimethoxy-fluorescein-labeled β-actin LUX primers were obtained from Invitrogen for use as an internal control. The PCR reaction was performed using the Platinum Quatitative RT-PCR Thermoscript One-Step System (Invitrogen) according to the manufacturer's instructions. The cycle consisted of 30 minutes at 50°C for cDNA synthesis followed by amplification for 45 cycles of 95°C, 15s; 55°C, 30s; 72°C, 30s. Reactions were performed and data analyzed using the GeneAmp Sequence Detection System (Applied Biosystems, Foster City, CA). Results are expressed as the ratio of CEACAM6 to β-actin. Measured threshold cycles were converted to relative copy numbers using primer-specific standard curves. Specificity was confirmed by electrophoretic analysis of the reaction products.
Western Blot Analysis
Cells were harvested and rinsed twice with PBS. Cell extracts were prepared with lysis buffer (20 mM Tris, pH 7.5, 0.1% Triton X-100, 0.5% deoxycholate, 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 μg/mL aprotinin, 10 μg/mL leupeptin) and cleared by centrifugation at 12,000g, 4°C. Total protein concentration was measured using the BCA assay kit (Sigma, St. Louis, MO) with bovine serum albumin as a standard, according to the manufacturer's instructions. Cell extracts containing 30 μg total protein were subjected to 10% sodium dodecyl sulfate polycrylamide gel electrophoresis (SDS/PAGE), and the resolved proteins transferred electrophoretically to polyvinylidene fluoride (PVDF) membranes (Invitrogen). Equal protein loading was confirmed by Coomassie (BioRad, Hercules, CA) staining of the gel. After blocking with phosphate buffered saline (PBS) containing 0.2% casein for 1 hour at room temperature, membranes were incubated with 3–5 μg/mL antibody in PBS containing 0.1% Tween 20 overnight at 4°C. Anti-CEACAM6 monoclonal antibody was obtained from InnoGenex (San Ramon, CA). Antiactin monoclonal antibody was obtained from LabVision (Freemont, CA). Chemiluminescent detection (Upstate, Lake Placid, NY) was performed in accordance with the manufacturer's instructions. The CEACAM6 signal was quantified using ImagePro Plus software version 4.0 and normalized to that of actin. Blots were performed in triplicate. Mean densitometric values (± SD) are shown.
Nude Mouse Xenograft Model
Male athymic nu/nu mice 5 weeks of age, weighing 20–22 g and specific pathogen-free were obtained from Charles River Laboratories (Wilmington, MA). Mice were housed in microisolator cages with autoclaved bedding in a specific pathogen-free facility with 12-hour light-dark cycles. Water and food were supplied ad libitum. Animals were observed for signs of tumor growth, activity, feeding, and pain in accordance with the guidelines of the Harvard Medical Area Standing Committee on Animals.
To determine the effect systemic siRNA on tumor growth and metastasis, mice were anesthetized with intraperitoneal ketamine (200 mg/kg) and xylazine (10 mg/kg) and subcutaneously implanted with 2 × 106 BxPC3 cells. Once tumors reached approximately 50 mm3 in volume, mice were allocated to receive either CEACAM6 or control siRNA (150 μg/kg by twice weekly tail vein injection). Tumor dimensions were measured weekly and the tumor volumes calculated using the formula: [1/2] × a × b2, where a and b represent the larger and smaller tumor diameters, respectively. After 6 weeks’ treatment, mice were killed by overdose of ketamine (400 mg/kg) and xylazine (50 mg/kg) and necropsy was performed. Tumors were weighed and liver metastases were counted13 and confirmed histologically. Tumor growth inhibition (TGI) was calculated using the formula TGI (%) = (1 − MT/MC) × 100, where MT and MC are the mean tumor masses in the treatment group and control group, respectively.
In a separate experiment, mice received 2 × 106 BxPC3 cells by orthotopic implantation to the body of the pancreas and 2 weeks later were allocated to treatment with either CEACAM6-specific or control siRNA for 6 weeks, as described previously. Following the treatment period, siRNA administration was discontinued. Mouse survival time was determined. Where necessary, mice were killed due to the presence of massive ascites or debilitating tumor growth in accordance with Harvard Medical Area Standing Committee on Animals protocols.
Immunohistochemistry
Tumor sections (5 μm) were deparaffinized, rehydrated through graded alcohol, washed with Tris-buffered saline, and processed using a streptavidin-biotin-peroxidase complex method. Antigen retrieval was performed by microwave heating sections in 10 mM sodium citrate buffer (pH 6) for 10 minutes. Following quenching of endogenous peroxidase activity and blocking of nonspecific binding, sections were incubated with mouse anti-CEACAM6 (By114; BioGenex, San Ramon, CA), anti-Ki-67 or anti-CD34 (DAKO, Carpinteria, CA) at 4°C overnight at a 1:200 dilution. The secondary antibody was biotinylated rabbit antimouse antibody (DAKO) used at a dilution of 1:200 for 30 minutes at 37°C. After further washing with Tris-buffered saline, sections were incubated with StrepABComplex/horseradish peroxidase (1:100; DAKO) for 30 minutes at 37°C. Immunolocalization was performed by exposure to 0.05% 3,3′-diaminobenzidine tetrahydrochloride as the chromogen. Normal serum was used in the place of primary antibody as a negative control. Slides were counterstained with hematoxylin before dehydration and mounting. The fraction of Ki-67-positive cells was determined by counting 100 cells in 5 random fields from each section. Proliferation indices (%) were derived and are presented as means (± SD). Angiogenesis was quantified by counting the number of CD34-positive structures in 5 randomly selected sections from each tumor section. Mean values (± SD) are presented.
Apoptosis Staining
Following preparation of 5-μm tumor sections, apoptosis was quantified using a commercially available fluorescent terminal deoxynucleotidyl transferase nick-end labeling (TUNEL) kit, in accordance with the manufacturer's protocol (Roche Diagnostics Corporation, Indianapolis, IN). The fractions of apoptotic cells in 5 random fields from each tumor section were counted, scoring 100 cells in each field, and expressed as an apoptotic fraction (%). Values presented are means (± SD).
Statistical Analysis
Differences between groups were analyzed using Student t test, multifactorial ANOVA of initial measurements and Mann-Whitney U test, for nonparametric data, as appropriate, using GraphPad InStat (GraphPad Software, Inc., San Diego, CA). In cases in which averages were normalized to controls, the standard deviations of each nominator and denominator were taken into account in calculating the final standard deviation. P < 0.05 was considered statistically significant.
RESULTS
Reduction of CEACAM6 Expression in BxPC3 Cells by RNAi
BxPC3 pancreatic adenocarcinoma cells inherently express high levels of CEACAM6. This cell line was therefore chosen to evaluate the effects of CEACAM6-specific siRNA. To confirm the ability of CEACAM6-specific siRNA to silence CEACAM6 expression, cells were transfected with either CEACAM6-specific or control siRNA. The ability of CEACAM6-specific siRNA to inhibit CEACAM6 expression was quantified by Western blot analysis 48 hours following siRNA exposure. CEACAM6 expression was suppressed by up to 80% at this time in cells transfected with CEACAM6-specific siRNA but not control siRNA. This level of CEACAM6 protein suppression persisted at 96 hours post-transfection (Fig. 1A). Suppression of CEACAM6 expression by CEACAM6-specific siRNA, but not control siRNA, was confirmed at the transcript level using real-time PCR at the same time point (Fig. 1B). Cellular proliferation in monolayer culture was unaffected by either siRNA (Fig. 2A). However, colony formation in soft agar was strongly inhibited by treatment with CEACAM6-specific, but not control, siRNA (Fig. 2B).
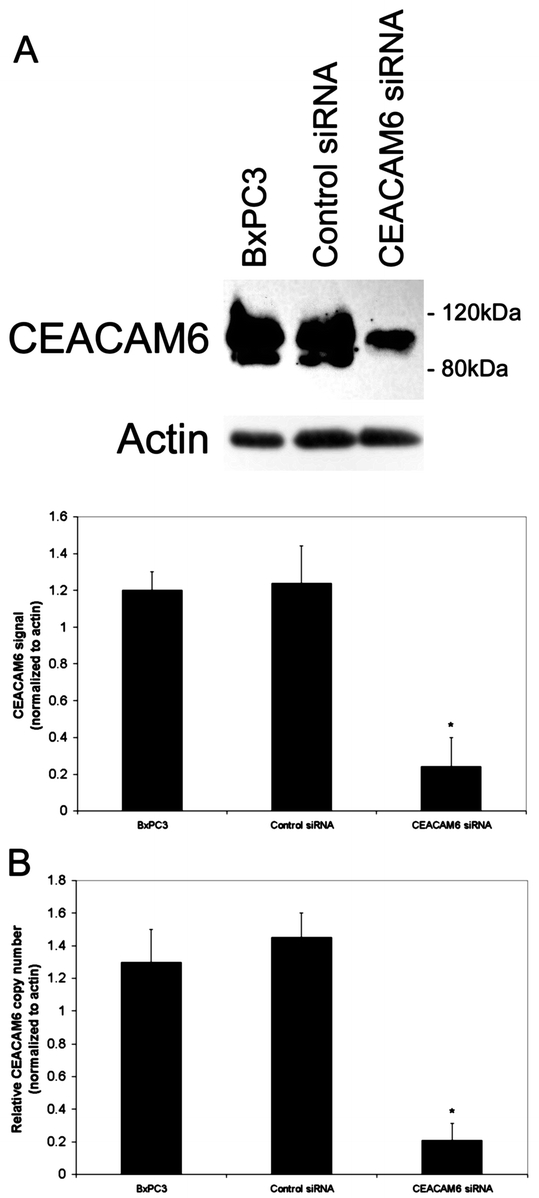
FIGURE 1. CEACAM6 gene silencing by siRNA. Suppression of CEACAM6 expression was confirmed at protein level by Western blot analysis (A) and at transcript level by real-time PCR (B). Analysis was performed 96 hours following siRNA transfection. CEACAM6 protein expression was suppressed by approximately 80% in CEACAM6-specific siRNA treated cells, relative to control siRNA and untransfected cells, which did not differ in their level of CEACAM6 expression. Representative blots are shown. Densitometric values are means (± SD) from triplicate blots, normalized to actin. * P < 0.05 versus control siRNA-treated cells.
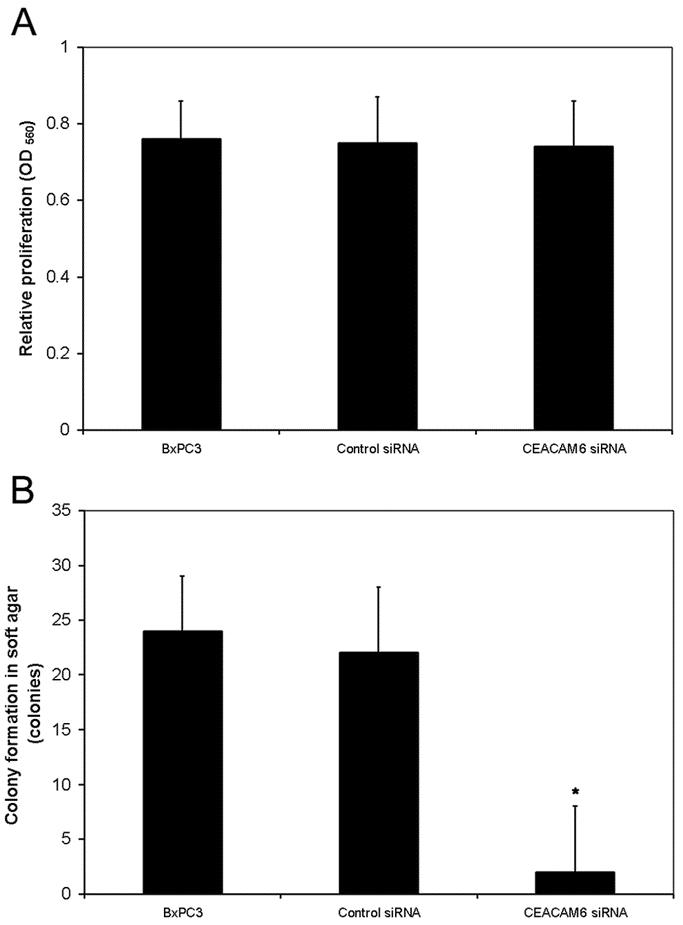
FIGURE 2. Effect of CEACAM6 gene silencing on cellular proliferation in monolayer culture and soft agar. A, Suppression of CEACAM6 expression had no significant effect on cellular proliferation in monolayer culture, as quantified by MTT assay. B, Colony formation in soft agar was significantly attenuated by CEACAM6 gene silencing. Values are means (± SD) from triplicate experiments. * P < 0.05 versus control siRNA.
In Vivo Tumor Growth, Metastasis and Survival
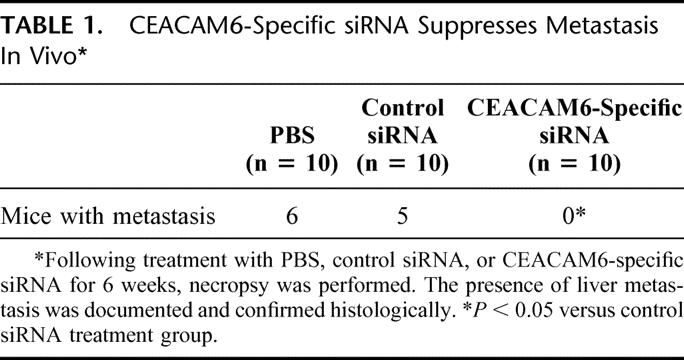
Once tumor xenografts reached 50 mm3 in size, treatment was commenced with either CEACAM6-specific or control siRNA or PBS, administered twice weekly by tail vein injection. Tumor volumes were calculated weekly. Mice receiving CEACAM6-specific siRNA exhibited a 68% reduction in final tumor size, relative to control siRNA treated mice (Fig. 3A). Neither group exhibited signs of systemic toxicity. While 60% of PBS-treated mice and 50% of control siRNA-treated mice developed liver metastases, no metastasis occurred in the CEACAM6-specific siRNA-treated group (Table 1).
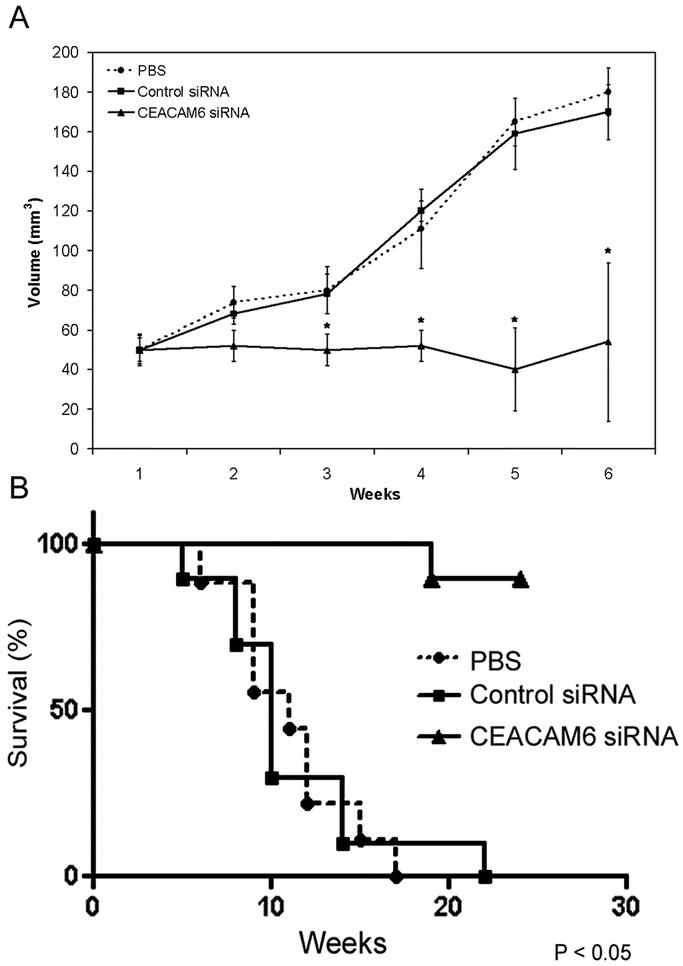
FIGURE 3. Effect of systemic siRNA on xenograft tumor growth and mouse survival. A, Subcutaneous tumor xenograft-bearing nude mice were treated systemically with either CEACAM6-specific siRNA, control siRNA, or PBS for 6 weeks (n = 10 per group), during which time tumor volume was calculated weekly using the formula: [1/2] × a × b2, where a and b represent the larger and smaller tumor diameters, respectively. CEACAM6-specific siRNA suppressed mean tumor growth by 68% at 6 weeks. The volume of tumors from mice treated with control siRNA did not differ significantly from those from mice receiving PBS. Values are means (± SEM). B, In a separate experiment, mice received 2 × 106 BxPC3 cells by surgical orthotopic implantation. Two weeks following implantation, mice were treated systemically with either CEACAM6-specific siRNA, control siRNA, or PBS for 6 weeks (n = 10 per group). Mouse survival was analyzed using the Kaplan-Meier method. The survival rate of mice treated with CEACAM6-specific siRNA was significantly greater than that of mice treated with either control siRNA or PBS (P < 0.05 by log-rank test).
TABLE 1. CEACAM6-Specific siRNA Suppresses Metastasis In Vivo
In a separate experiment, mice were implanted with 2 × 106 BxPC3 cells orthotopically into the body of the pancreas. Two weeks later, treatment was commenced twice-weekly with either CEACAM6-specific siRNA, control siRNA or PBS by tail vein injection in the same manner as before. Treatment was discontinued after 6 weeks, following which survival was analyzed. Treatment with CEACAM6 siRNA for 6 weeks resulted in a significant improvement in survival, relative to mice receiving control siRNA or PBS (Fig. 3B).
In Vivo CEACAM6 Gene Silencing
We confirmed suppression of CEACAM6 expression in tumors from mice receiving CEACAM6-specific siRNA by Western blot and CEACAM6 immunohistochemisty (Fig. 4A). Control siRNA treatment had no effect on CECAM6 expression, relative to PBS-treated mice.
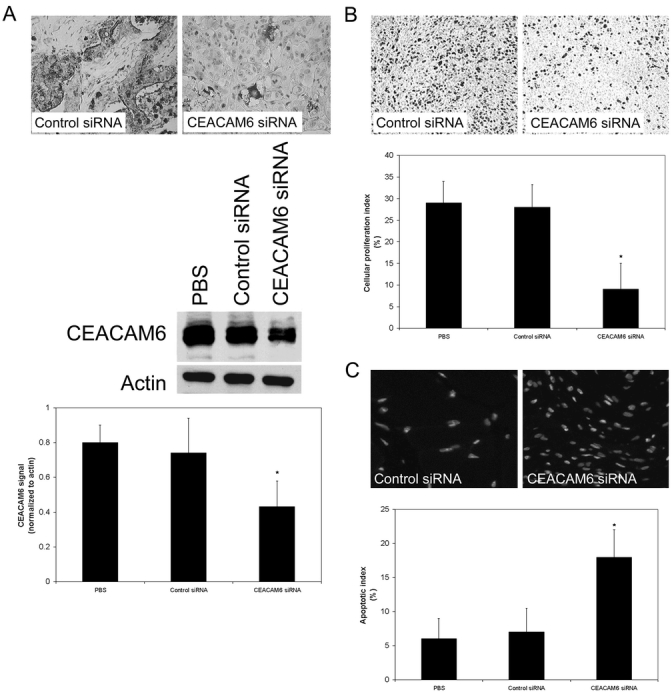
FIGURE 4. A, Suppression of CEACAM6 expression by systemically administered CEACAM6-specific siRNA, but not control siRNA, was confirmed by performing CEACAM6 immunohistochemistry on 5-μm tumor sections and by Western blot analysis of tumor homogenates. CEACAM6 expression was suppressed by 42%, relative to control siRNA and PBS treated tumors. Representative blot shown with mean densitometric values (± SD) from triplicate blots. * P < 0.05 versus control siRNA-treated tumors. B, Tumor xenograft cellular proliferation was quantified by Ki-67 immunohistochemistry. The fraction of Ki-67-positive cells (proliferation index) was determined by counting 100 cells in 5 random fields from each tumor section. Tumors from mice treated with CEACAM6-specific siRNA exhibited significantly lower cellular proliferation indices than those from mice treated with control siRNA. Values are means (± SD). * P < 0.05 versus control siRNA-treated tumors. C, Tumor xenograft cellular apoptosis was quantified by TUNEL staining 5-μm tumor sections. The apoptotic fraction was derived by counting the TUNEL-positive fraction of 100 cells from 5 random fields in each tumor section. Tumors from mice treated with CEACAM6-specific siRNA exhibited significantly higher levels of apoptosis than those from mice treated with control siRNA. Values are means (± SD). * P < 0.05 versus control siRNA-treated tumors.
Tumor Cellular Proliferation and Apoptosis
Cellular proliferation, quantified by Ki-67 immunohistochemistry, was reduced by 67% in the tumors from CEACAM6-specific siRNA-treated mice, relative to their control siRNA-treated counterparts (Fig. 4B). Cellular proliferation indices of tumors derived from control siRNA-treated mice did not differ significantly from those of tumors derived from PBS-treated mice. Tumor apoptosis, determined by TUNEL staining, was increased 2.6-fold in mice treated with CEACAM6-specific siRNA, relative to control siRNA-treated mice (Fig. 4C). Apoptotic indices of tumors derived from control siRNA-treated mice did not differ significantly from those of tumors derived from PBS-treated mice.
Tumor Angiogenesis
Tumor angiogenesis was quantified by counting CD34-positive structures detected immunohistochemically in 5-μm tumor sections. Tumors from mice treated with CEACAM6-specific siRNA exhibited a 6.6-fold reduction in angiogenesis, relative to those from control siRNA-treated mice (Fig. 5). Angiogenesis in tumors from control siRNA-treated mice did not differ significantly from than observed in tumors from PBS-treated mice.
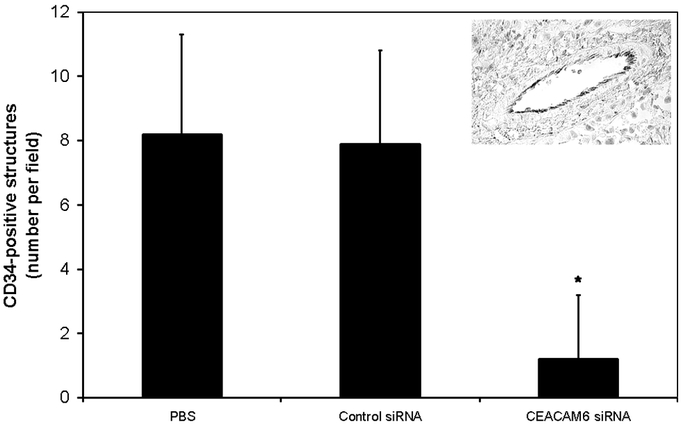
FIGURE 5. Tumor angiogenesis was quantified by immunohistochemical staining for CD34. The number of CD34-positive structures was quantified in 5 random fields in each tumor section. CEACAM6-specific siRNA substantially reduced the number of CD34-positive structures, suggesting reduced levels of tumor angiogenesis, relative to control siRNA-treated cells. Values are means (± SD). * P < 0.05 versus control siRNA-treated tumors. A typical CD34-positive structure containing erythrocytes is shown in the inset.
DISCUSSION
The purpose of this study was to evaluate the applicability of systemically administered siRNA as a target-specific cancer treatment, using pancreatic adenocarcinoma as a model. We have shown that posttranscriptional suppression of CEACAM6 expression inhibits BxPC3 pancreatic adenocarcinoma cell colony formation in soft agar, while leaving cellular proliferation in monolayer culture unaffected. In the nude mouse xenograft model, systemically administered CEACAM6-specific siRNA suppresses tumor CEACAM6 expression and cellular proliferation, as well as promoting tumor cellular apoptosis and inhibiting angiogenesis. These effects are associated with a significant inhibition of tumor growth and improved survival of xenograft-bearing mice, without signs of systemic toxicity.
The kinetics of 3-dimensional colony formation in vitro resemble those of in vivo tumor growth more closely than those of cells in monolayer culture.14 The normal cellular response to culture under conditions of inadequate or inappropriate contact with substratum is to undergo apoptosis which, in this context, is termed anoikis.15 Transformed cells are generally less susceptible to anoikis and proliferate more readily in soft agar.16 Resistance to anoikis has been reported to contribute to tumorigenesis and metastasis in a range of cancers,17–19 and CEACAM6 plays a role in this process in gastrointestinal malignancy.9,11 While the proliferation rate of cells transfected with CEACAM6-specific siRNA equaled that of cells treated with control siRNA or PBS in monolayer culture, posttranscriptional suppression of CEACAM6 expression resulted in a significant inhibition of colony formation in soft agar, further supporting the hypothesis that high levels of CEACAM6 expression represent a mechanism by which cells survive in conditions that would normally induce anoikis. The fact that growth in monolayer culture is not affected by CEACAM6 gene silencing indicates that overexpression of CEACAM6 does not simply act as a mitogenic signal.
CEACAM6 appears to play a significant role in the cellular behavior and clinical outcome of a range of human cancers.6–8,10 While CEACAM6 is known to act as an intercellular adhesion molecule,5 it is also being increasingly recognized to have additional functions in malignant disease.9,11 We have recently reported that overexpression of CEACAM6 is associated with an increased ability of cells to survive under anchorage-independent conditions and that posttranscriptional silencing of CEACAM6 expression suppresses this ability. Furthermore, pancreatic adenocarcinoma cells in which CEACAM6 expression is suppressed exhibit lower metastatic ability in the nude mouse.11 In view of considerable evidence supporting CEACAM6 as an important determinant of pancreatic adenocarcinoma tumor progression, this molecule was selected as a target for siRNA-mediated therapy.
The ability of RNAi mediated by siRNA to silence individual gene expression with a high degree of specificity presents a unique opportunity to study gene function, but RNAi is also showing promise as a strategy for target-directed therapies in a range of diseases.20–25 Targeted suppression of individual gene expression would allow therapies to be tailored to the expression profile of an individual patient's tumor. A theoretical benefit of such target-specific therapies is a lower risk of systemic side effects. siRNA oligonucleotides have advantages over DNA oligonucleotides in terms of their resistance to nucleases,26 and RNAi appears to have greater potency than antisense-based approaches.27 Tumor gene silencing by systemically administered siRNA has been reported by other groups.23 However, the ability of systemically administered siRNA to induce tumor gene silencing remains poorly understood. The inherently high permeability of tumor neovasculature28 may be contributory to the activity of systemically administered siRNA; however, the potency of siRNA remains limited by intratumoral bioavailability and the transient nature of gene silencing. While repeated administration of siRNA overcomes this latter limitation to some extent, plasmid and viral vectors producing siRNA using the polymerase III promoter29,30 may offer more efficient siRNA delivery and can theoretically induce stable gene silencing. Such strategies are showing promise both in vitro and in vivo.29
Although CEACAM6 expression is not completely inhibited by siRNA treatment, the effects on malignant cellular behavior and tumor progression were marked, emphasizing the importance of CEACAM6 expression as a determinant of pancreatic adenocarcinoma tumorgenicity in the nude mouse xenograft model. The effects of CEACAM6 gene silencing on anchorage-independent cell survival are clear and may contribute to impaired tumorigenesis and metastasis. The effect of CEACAM6-specific siRNA on tumor angiogenesis is intriguing and may underlie an as yet unrecognized function of CEACAM6. There are clearly limitations to the nude mouse as a model for human cancer, but our results support further investigation of siRNA as a therapeutic strategy in human patients. We observed no adverse effects in the mice treated with siRNA, and CEACAM6-specific therapy would be expected to have little toxicity in normal adult human tissues where CEACAM6 expression is generally low.
In summary, this study indicates that CEACAM6 plays a significant role in the colony-forming ability of pancreatic adenocarcinoma cells. Targeting this oncoprotein using systemically administered siRNA induces tumor gene silencing. Suppression of CEACAM6 expression impairs pancreatic adenocarcinoma xenograft growth in vivo and improves the survival of tumor-bearing nude mice. These effects are associated with impaired cellular proliferation, increased apoptosis, and lower levels of tumor angiogenesis. While this study used CEACAM6 and pancreatic adenocarcinoma as model target and system, respectively, this approach has broader applicability to a range of tumors. RNAi warrants further evaluation as a target-directed therapeutic strategy in human cancer.
ACKNOWLEDGMENTS
The authors acknowledge the technical assistance of Jan Rounds.
Discussions
Dr. Howard A. Reber (Los Angeles, California): Dr. Whang, what a beautiful presentation, very nice work. Thank you for asking me to discuss it.
Dr. Whang didn't mention in the very beginning, probably because I guess most of us know, that pancreatic cancer is an extraordinarily lethal disease, and although we as surgeons have made considerable progress in its management over the years, with Whipple mortality rates down now to around 1%, I think most of us as well, as surgeons, realize we are not likely to make much more of an impact on survival by subtle variations in surgical technique. So, improvements in management are likely to come from these kinds of novel studies, of which you just showed a very nice example.
I have a number of questions. Most of them relate to the animal work that you just presented and maybe a couple relate to the clinical use of these siRNAs.
First of all, I am wondering whether you studied or have information about any other pancreatic cancer cell lines. You used the BxPC3 for obvious reasons. Do you know whether the other cell lines that are generally available also express CEACAM3? I am interested, for example, if there is one that doesn't express it, or that expresses it much more weakly. Wouldn't that make also a good control to use in some of these studies to see whether those that don't express it respond or do not respond to the siRNAs that you used?
What tissues in the mice normally do express CEACAM6? You indicated that there were really no deleterious effects of the treatment, but I wonder whether if there are other tissues that expressed it you might have taken a look at them histologically to see whether there were any alterations that didn't show up in any other obvious way.
I am wondering as well whether the siRNA effect that you described on angiogenesis is a specific effect. Do you know whether siRNAs have any effect on endothelial cells? Did you look at that? Or do you think the effect on angiogenesis that you observed might be related in a more general way to the effect on apoptosis that you also saw?
You indicated in the second series of animal experiments that those animals that had orthotopic tumor implantation lived longer. And I noticed that you didn't say anything at all about whether or not they had liver metastases, which you were able to essentially eliminate in the subcutaneous model. Did they? Did you look at that? If they did have liver metastases, do you have any explanation for why they lived longer? What was the effect that resulted in greater survival?
Do you know anything about how long these siRNA molecules are effective once they are given? Can they be stably transfected into the cells? How did you come around to choosing this twice-weekly dose schedule?
Then finally just a couple of, I guess, conjectural questions about clinical use. And you touched briefly on this. How would you think that these things might be delivered in patients? What would be the best way to do that? And I guess most importantly, before one could hope to do even a clinical study, how would you test for safety? How would you look at other tissues and get information about whether the target of the siRNA might very well hit other tissues in ways that would be potentially very dangerous?
Dr. Whang, this is a beautiful piece of work. Thank you again very much for the privilege of discussing it.
Dr. Edward E. Whang (Boston, Massachusetts): Thank you very much, Dr. Reber, for discussing this paper.
We have tested a panel of the commonly studied human pancreatic cancer cell lines and have found all of them to express CEACAM6, although at varying levels, under baseline conditions. We have also found that reductions in anoikis-resistance (that is, apoptosis induced by anchorage deprivation) and metastatic potential induced by siRNA-mediated suppression of CEACAM6 expression are generalizable features of all of these cell lines. The magnitude of the effects of CEACAM6 gene silencing appear to be roughly proportional to the baseline expression level of CEACAM6.
Regarding CEACAM6 expression in other tissues, available data suggests that CEACAM6 expression is limited to higher primates, including humans. In these species CEACAM6 expression is known to be expressed in normal colonic epithelium and in neutrophils. The physiological role of CEACAM6 in normal tissues remains to be defined.
I believe the inhibition in angiogenesis we observed to occur with suppression of CEACAM6 expression is related to the effects of suppressing CEACAM6 signaling. We have recently shown that activating CEACAM6 signaling in pancreatic cancer cells through antibody-mediated crosslinking of CEACAM6 results in the induction in alpha v beta 3 integrin-mediated cell adhesion to extracellular matrix components fibronectin and vitronectin. In animal models, inhibition of this integrin results in the inhibition of angiogenesis. These observations suggest a plausible mechanism by which suppressing CEACAM6 expression might inhibit angiogenesis. Obviously, further work is required to test this hypothesis.
Regarding the fate of the mice entered into our second protocol, mice that were treated with PBS or control siRNA died early and were found to have liver metastases and ascites. Mice treated with CEACAM6-specific siRNA exhibited prolonged survival; most were sacrificed by us rather than dying spontaneously from the effects of their cancers. None of these mice had ascites. Some, but not all, had liver metastases.
Regarding the duration of action of siRNAs, in our in vitro studies, siRNA-mediated suppression of CEACAM6 expression persisted for up to 96 hours after transfection of cells with siRNA. Our in vivo siRNA dosing schedule was based on these findings. In other studies, we have achieved stable, or long-term, siRNA-mediated gene silencing using approaches based on either plasmid expression vectors containing selectable markers or retroviral vectors.
Finally, what is the next step and how do we get this therapy to patients with pancreatic cancer? Clearly, we will need to test potential toxicities induced by this approach. We will need to choose our targets carefully. For CEACAM6, given that it is expressed only in higher primates, we will need to carefully test potential toxicities associated with targeting CEACAM6 in primates before considering clinical trials.
Dr. Richard H. Bell, Jr. (Chicago, Illinois): The importance of this work obviously depends on how critical the CEACAM molecule is for the behavior of pancreatic cancer cells. The fact your CEACAM negative patients live longer doesn't necessarily mean that their enhanced survival is because of their CEACAM status. It doesn't satisfy all of Koch's postulates. Therefore, I want to ask you some more questions about your cell line experiments. When you orthotopically transplant pancreatic cancer cell lines there is tremendous variability. Some cell lines don't metastasize at all, some cell lines metastasize within a few days. Can you correlate the level of CEACAM expression with biological behavior in the 10 or 11 pancreatic cell lines that are available? Secondly, could you potentially overexpress CEACAM in a non-expressing or low-expressing cell line and demonstrate that biological behavior is changed when those cells are implanted in an orthotopic model?
Dr. Edward E. Whang (Boston, Massachusetts): Thank you, Dr. Bell, for those questions. In the panel of human pancreatic cancer cell lines we examined, we have found that degree of CEACAM6 overexpression is indeed correlated with expression of the malignant phenotype, for example degree of anoikis-resistance and metastatic potential.
We have forcibly overexpressed CEACAM6 in Capan2 cells (pancreatic cancer cells that inherently express low levels of CEACAM6 under baseline conditions). The consequence of CEACAM6 overexpression in these cells is an increase in the features of the malignant phenotype, as you correctly predicted.
Dr. Henry A. Pitt (Milwaukee, Wisconsin): Dr. Whang, congratulations on a very nice study, as the others have suggested.
My first question has to do with the fact that your tumors stayed constant, they didn't reduce in size. You showed very nice data on prevention, progression of growth, and improvement in survival. But what about the fact that you really didn't eliminate the tumors?
My second question relates to your relatively blanket statement about toxicity. Would you tell us a little bit more about how hard you looked for toxicity to not find any?
Finally, if we get to the point where clinical trails are warranted, do you think that this treatment is going to make more sense as an adjuvant when the gross tumor is gone or more in the patients with extensive disease?
Dr. Edward E. Whang (Boston, Massachusetts): Regarding your question on toxicities, I have to be honest. We didn't assess toxicities in a comprehensive manner, as the primary endpoints of our study related to treatment-related efficacy. As I mentioned before, toxicities will be important to assess before embarking on clinical testing.
As you noted, we observed that CEACAM6-specific siRNA inhibited the progression of the xenografted cancers but did not seem to consistently induce shrinkage of the primary tumors. I would envision that targeted therapy directed against CEACAM6 would be used in the adjuvant setting following surgical resection of grossly apparent disease. One of our current interests is developing polymeric systems for delivering siRNA in very high concentration to tumor resection beds. This approach may be particularly applicable to pancreatic cancers. I don't believe siRNA-based approaches will make surgical resection for pancreatic cancer obsolete.
Dr. Graeme L. Hammond (New Haven, Connecticut): This is an extremely interesting paper in a new and very rapidly moving field. These siRNAs were discovered in plants and invertebrate species. To my knowledge this is the first time they have been shown to work in an intact mammalian species. This could have wide therapeutic application.
In the research reported in sub-mammalian species and plants the siRNAs, processed from larger non coding RNAs, works either by methylating the CpG islands in DNA or by complementary binding to messenger RNA preventing transcription. We have reported, in mammalian cells, this may be a possible mechanism for MHC suppression. I wonder if you have any indication that 1 of these 2 mechanisms is working in your model.
Dr. Edward E. Whang (Boston, Massachusetts): We've known about RNA interference in mammalian cells for several years. In mammalian cells, the primary mechanism of gene silencing induced by siRNAs appears to be siRNA-mediated cleavage of their cognate mRNAs. Recent reports, including this one, suggest that in vivo systemic delivery of siRNAs to effect therapeutic gene silencing may be feasible.
Footnotes
Grant support: National Pancreas Foundation, American Cancer Society (RSG-04-221-01-CCE) and Departmental funding from the Department of Surgery, Brigham and Women's Hospital.
Reprints: Edward E. Whang, MD, Department of Surgery, Brigham and Women's Hospital, Harvard Medical School, 75 Francis Street, Boston, MA 02115. E-mail: ewhang1@partners.org.
REFERENCES
- 1.Burris HA III, Moore MJ, Andersen J, et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol. 1997;15:2403–2413. [DOI] [PubMed] [Google Scholar]
- 2.Fire A, Xu S, Montgomery MK, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. [DOI] [PubMed] [Google Scholar]
- 3.Hannon GJ. RNA interference. Nature. 2002;418:244–251. [DOI] [PubMed] [Google Scholar]
- 4.Thompson JA, Grunert F, Zimmermann W. Carcinoembryonic antigen gene family: molecular biology and clinical perspectives. J Clin Lab Anal. 1991;5:344–366. [DOI] [PubMed] [Google Scholar]
- 5.Kuroki M, Abe H, Imakiirei T, et al. Identification and comparison of residues critical for cell-adhesion activities of two neutrophil CD66 antigens, CEACAM6 and CEACAM8. J Leukoc Biol. 2001;70:543–550. [PubMed] [Google Scholar]
- 6.Jantscheff P, Terracciano L, Lowy A, et al. Expression of CEACAM6 in resectable colorectal cancer: a factor of independent prognostic significance. J Clin Oncol. 2003;21:3638–3646. [DOI] [PubMed] [Google Scholar]
- 7.Kodera Y, Isobe K, Yamauchi M, et al. Expression of carcinoembryonic antigen (CEA) and nonspecific crossreacting antigen (NCA) in gastrointestinal cancer; the correlation with degree of differentiation. Br J Cancer. 1993;68:130–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hasegawa T, Isobe K, Tsuchiya Y, et al. Nonspecific crossreacting antigen (NCA) is a major member of the carcinoembryonic antigen (CEA)-related gene family expressed in lung cancer. Br J Cancer. 1993;67:58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ilantzis C, DeMarte L, Screaton RA, et al. Deregulated expression of the human tumor marker CEA and CEA family member CEACAM6 disrupts tissue architecture and blocks colonocyte differentiation. Neoplasia. 2002;4:151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scholzel S, Zimmermann W, Schwarzkopf G, et al. Carcinoembryonic antigen family members CEACAM6 and CEACAM7 are differentially expressed in normal tissues and oppositely deregulated in hyperplastic colorectal polyps and early adenomas. Am J Pathol. 2000;156:595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duxbury MS, Ito H, Zinner MJ, et al. CEACAM6 gene silencing impairs anoikis resistance and in vivo metastatic ability of pancreatic adenocarcinoma cells. Oncogene. 2004;23:465–473. [DOI] [PubMed] [Google Scholar]
- 12.Raitano AB, Scuderi P, Korc M. Binding and biological effects of tumor necrosis factor and gamma interferon in human pancreatic carcinoma cells. Pancreas. 1990;5:267–277. [DOI] [PubMed] [Google Scholar]
- 13.Sun FX, Tohgo A, Bouvet M, et al. Efficacy of camptothecin analog DX-8951f (Exatecan Mesylate) on human pancreatic cancer in an orthotopic metastatic model. Cancer Res. 2003;63:80–85. [PubMed] [Google Scholar]
- 14.Demicheli R, Foroni R, Ingrosso A, et al. An exponential-Gompertzian description of LoVo cell tumor growth from in vivo and in vitro data. Cancer Res. 1989;49:6543–6546. [PubMed] [Google Scholar]
- 15.Frisch SM, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol. 1994;124:619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westermark B. The deficient density-dependent growth control of human malignant glioma cells and virus-transformed glia-like cells in culture. Int J Cancer. 1973;12:438–451. [DOI] [PubMed] [Google Scholar]
- 17.Shanmugathasan M, Jothy S. Apoptosis, anoikis and their relevance to the pathobiology of colon cancer. Pathol Int. 2000;50:273–279. [DOI] [PubMed] [Google Scholar]
- 18.Streuli CH, Gilmore AP. Adhesion-mediated signaling in the regulation of mammary epithelial cell survival. J Mammary Gland Biol Neoplasia. 1999;4:183–191. [DOI] [PubMed] [Google Scholar]
- 19.Yawata A, Adachi M, Okuda H, et al. Prolonged cell survival enhances peritoneal dissemination of gastric cancer cells. Oncogene. 1998;16:2681–2686. [DOI] [PubMed] [Google Scholar]
- 20.Duxbury MS, Ito H, Zinner MJ, et al. RNA interference targeting the M2 subunit of ribonucleotide reductase enhances pancreatic adenocarcinoma chemosensitivity to gemcitabine. Oncogene. 2003;311:786–792. [DOI] [PubMed] [Google Scholar]
- 21.Duxbury MS, Ito H, Benoit E, et al. RNA interference targeting focal adhesion kinase enhances pancreatic adenocarcinoma gemcitabine chemosensitivity. Biochem Biophys Res Commun. 2003;311:786–792. [DOI] [PubMed] [Google Scholar]
- 22.Duxbury MS, Ito H, Zinner MJ, et al. EphA2: a determinant of malignant cellular behavior and a potential therapeutic target in pancreatic adenocarcinoma. Oncogene. 2004;23:1448–1456. [DOI] [PubMed] [Google Scholar]
- 23.Filleur S, Courtin A, Ait-Si-Ali S, et al. SiRNA-mediated inhibition of vascular endothelial growth factor severely limits tumor resistance to antiangiogenic thrombospondin-1 and slows tumor vascularization and growth. Cancer Res. 2003;63:3919–3922. [PubMed] [Google Scholar]
- 24.Song E, Lee SK, Wang J, et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med. 2003;9:347–351. [DOI] [PubMed] [Google Scholar]
- 25.Zender L, Hutker S, Liedtke C, et al. Caspase 8 small interfering RNA prevents acute liver failure in mice. Proc Natl Acad Sci U S A. 2003;100:7797–7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bertrand JR, Pottier M, Vekris A, et al. Comparison of antisense oligonucleotides and siRNAs in cell culture and in vivo. Biochem Biophys Res Commun. 2002;296:1000–1004. [DOI] [PubMed] [Google Scholar]
- 27.Aoki Y, Cioca DP, Oidaira H, et al. RNA interference may be more potent than antisense RNA in human cancer cell lines. Clin Exp Pharmacol Physiol. 2003;30:96–102. [DOI] [PubMed] [Google Scholar]
- 28.Dvorak HF. Leaky tumor vessels: consequences for tumor stroma generation and for solid tumor therapy. Prog Clin Biol Res. 1990;354A:317–330. [PubMed] [Google Scholar]
- 29.Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243–247. [DOI] [PubMed] [Google Scholar]
- 30.Wilda M, Fuchs U, Wossmann W, et al. Killing of leukemic cells with a BCR/ABL fusion gene by RNA interference (RNAi). Oncogene. 2002;21:5716–5724. [DOI] [PubMed] [Google Scholar]


