Abstract
In a screen to identify genes required for mRNA export in Saccharomyces cerevisiae, we isolated an allele of poly(A) polymerase (PAP1) and novel alleles encoding several other 3′ processing factors. Many newly isolated and some previously described mutants (rna14-48, rna14-49, rna14-64, rna15-58, and pcf11-1 strains) are defective in polymerase II (Pol II) termination but, interestingly, retain the ability to polyadenylate these improperly processed transcripts at the nonpermissive temperature. Deletion of the cis-acting sequences required to couple 3′ processing and termination also produces transcripts that fail to exit the nucleus, suggesting that all of these processes (cleavage, termination, and export) are coupled. We also find that several but not all mRNA export mutants produce improperly 3′ processed transcripts at the nonpermissive temperature. 3′ maturation defects in mRNA export mutants include improper Pol II termination and/or the previously characterized hyperpolyadenylation of transcripts. Importantly, not all mRNA export mutants have defects in 3′ processing. The similarity of the phenotypes of some mRNA export mutants and 3′ processing mutants indicates that some factors from each process may mechanistically interact to couple mRNA processing and export. Consistent with this assumption, we present evidence that Xpo1p interacts in vivo with several 3′ processing factors and that the addition of recombinant Xpo1p to in vitro processing reaction mixtures stimulates 3′ maturation. Of the core 3′ processing factors tested (Rna14p, Rna15p, Pcf11p, Hrp1p, Fip1p, and Cft1p), only Hrp1p shuttles. Overexpression of Rat8p/Dbp5p suppresses both 3′ processing and mRNA export defects found in xpo1-1 cells.
One of the defining features of eukaryotic cells is the physical separation of the nucleus, where mRNAs are synthesized, from the cytoplasm, where protein synthesis occurs. Gene expression and cell function require the efficient transport of macromolecules between these two compartments. All exchange between the nucleus and cytoplasm takes place through nuclear pore complexes (NPCs) that perforate the nuclear envelope and permit selective passage in both directions of molecules and complexes containing transport signals. Interactions between the import substrate or complex and NPC are mediated by import or export receptors that bind directly to specific transport signals on the substrate and NPCs. Directionality of this process is imparted through the activity of a small GTPase (Gsp1p in Saccharomyces cerevisiae and Ran in metazoans) that modulates the receptor's affinity for the substrate on opposite sides of the nuclear envelope.
Export of mRNA to the cytoplasm appears to be more complex than transport of proteins. mRNAs are exported as messenger ribonucleoprotein complexes (mRNPs) whose assembly begins during transcription (27, 55). mRNA biogenesis requires multiple processing steps including 5′ capping, splicing, and 3′ cleavage or polyadenylation. Most of these reactions are accomplished during transcription by several protein complexes, components of which associate physically with the transcribing polymerase (for a recent review, see reference 68). In addition, several general mRNA-binding proteins are deposited on the transcript during its synthesis. These proteins are thought to package the mRNA for efficient and accurate processing, and many are components of the RNA-protein complex physically transported to the cytoplasm. Prior to mRNA export, all pre-mRNA processing steps must be completed, and the complexes that process the pre-mRNA must be removed.
Characterization of splicing both in vivo and in vitro indicates that pre-mRNA processing has a complex linkage to mRNA transport. Inhibition of splicing (temperature-sensitive [ts] mutants of splicing factors or deletion of splicing signals) prevents transcripts from being exported unless the mutation affects the initial association of the pre-mRNA with the splicing machinery (11, 14, 36, 51). Both in vitro and in vivo, splicing results in the deposition of specific proteins onto the transcript which enhance the rate of mRNA export (45, 47, 52-54, 56, 93). These factors include the Aly-Ref complex, and efficient association of the complex with mRNAs requires the activity of UAP56, a spliceosome-associated RNA helicase (57, 82). Once Aly-Ref are deposited onto the mRNA, Aly-Ref interact with the essential export factor (TAP in metazoans; Mex67p in yeast) which mediates interactions with the nucleoporins required to translocate through the NPC (38, 77, 81-83).
Although splicing is an essential step in the formation of almost all metazoan mRNAs, about 95% of S. cerevisiae genes lack introns. Nevertheless, Yra1p, the yeast homologue of Aly, associates with transcribing RNA polymerase II (Pol II) in yeast, is transferred to both intron-containing and intron-lacking transcripts during transcription, and is required for mRNA export (55, 83). Yra1p interacts directly with Sub2p, the yeast homologue of UAP56, which is also essential for efficient mRNA export (55, 82). It has been suggested that Yra1p and Sub2p are loaded onto all yeast mRNAs via the THO complex (complex of Tho2p, Hpr1p, Mft1p, and Thp2p) previously implicated in transcriptional elongation (15, 16, 25, 66). This THO-Sub2p-Yra1p complex, collectively called the TREX (transcription and/or export) complex, is specifically recruited to activated genes during transcription and travels with the polymerase (84).
All eukaryotic mRNA transcripts undergo 3′ maturation before export. This maturation involves endonucleolytic cleavage within the 3′ untranslated region (3′UTR) followed by addition of a poly(A) tract which forms the 3′ end of the mRNA. cis-acting signals in the 3′UTR of the nascent transcript direct an evolutionarily conserved complex of proteins to the proper cleavage site (59, 90). More than a dozen polypeptides organized into three subcomplexes participate in 3′ processing. One of these subcomplexes, cleavage factor I (CFI) of S. cerevisiae, specifically recognizes these cis-acting elements and couples the cleavage reaction to polyadenylation and to the termination of Pol II transcription (8, 33). The finding that mRNAs accumulate in the nuclei of yeast cells carrying some temperature-sensitive (ts) alleles affecting components of CFI or poly(A) polymerase indicates that the addition of a poly(A) tail is required for efficient export (11). Defects in the β-globin polyadenylation signals cause these transcripts to be retained at the site of transcription (21). Furthermore, mRNAs whose 3′ ends are generated by mechanisms other than normal 3′ processing cannot be exported (24, 43).
Several factors specifically required for mRNA export have been identified by this and other laboratories (for recent reviews, see references 20 and 89). Whether a karyopherin and the Ran/Gsp1p system are directly involved in mRNA export is not known. In strains carrying ts mutations of Gsp1p's effector molecules (Prp20p and Rna1p), as well as the nuclear export signal (NES) export receptor, Xpo1p, mRNAs rapidly accumulate in nuclei following a shift to the nonpermissive temperature. However, this could be an indirect consequence of a block in protein transport. Consistent with the view that Xpo1p plays an indirect role in mRNA export, mRNAs microinjected into Xenopus oocytes can be exported under conditions where Ran (Gsp1) function and Ran-dependent protein export are severely compromised (19).
In this paper, we used a flow cytometry-based screen to identify mutants defective for mRNA export after stress, and we isolated novel alleles of genes encoding 3′ processing factors. We analyzed 3′ processing and export in these and other 3′ factor mutant strains and in a strain carrying an allele of CYC1 (cyc1-512) which lacks 3′ processing signals. We find that cleavage and polyadenylation at the normal 3′ processing site and mediated by the normal processing machinery are necessary but not sufficient for mRNA export. Some of the new 3′ processing mutations differ from previously characterized alleles in retaining some 3′ processing activity in vitro and in producing polyadenylated mRNAs in vivo whose accumulation in the nucleus can be readily detected at the nonpermissive temperature. This suggests that 3′ processing and mRNA export are coupled at a step in mRNA biogenesis dependent upon proper cleavage and polyadenylation.
We also analyzed 3′ pre-mRNA processing in cells carrying mutations affecting several proteins important for mRNA export. We observed strong defects in 3′ processing and Pol II termination in strains carrying ts mutations affecting Xpo1p, Mex67p, and Kap104p but not in strains carrying ts mutations affecting Rat7p/Nup159p, Yra1p, or Sub2p. Among mRNA export mutants defective for 3′ processing and termination, the defects of xpo1-1 cells were completely suppressed by overexpression of Rat8p/Dbp5p, as was the mRNA export defect of xpo1-1 cells (41). Rat8p/Dbp5p overexpression did not lead to suppression of these defects in any other mutant strains. Although most 3′ processing factors do not shuttle, we describe two-hybrid interactions between Xpo1p and several components of the 3′ processing machinery and report that addition of recombinant Xpo1p stimulates in vitro 3′ processing. These studies provide evidence that 3′ processing and mRNA export are not sequential but are interdependent.
MATERIALS AND METHODS
S. cerevisiae strains, cell culture, and isolation of mutants.
The S. cerevisiae strains used in this study are listed in Table 1. Strains were cultured by standard methods (78), using rich (yeast extract-peptone-dextrose [YPD]) or defined (complete synthetic [CS]) medium lacking the appropriate amino acids. Temperature-sensitive mutants were grown at 23°C (permissive temperature) or shifted to 37°C (nonpermissive temperature). Genetic techniques were performed by standard methods (70, 78). A mutant yeast library was created by subjecting yeast strain CHY167 [containing at the SSA4 locus an allele encoding a fully functional Ssa4p-GFP (pCH19) (green fluorescent protein) fusion] to UV light. To induce mutations, CHY167 cells were plated as single cells on YPD agar plates and inverted on an UV trans-illuminator (Fotodyne) (300-nm-wavelength light) for 9 or 10 s. This duration of UV irradiation resulted in the death of 50% of the cells. The remaining cells were allowed to grow at 23°C to form colonies. These colonies were then transferred from plates to fresh 15% glycerol, snap-frozen, and stored at −80°C.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Origin or reference |
|---|---|---|
| FY86 | MATatrp1Δ63 ura3-52 leu2Δ | F. Winston |
| FY23 | MATα ura3-53 leu2Δ1 his3Δ200 | F. Winston |
| LGy101 | MATα rat7-1/nup159-1 his3Δ200 ura3-52 leu2Δ1 | 30w |
| CSy550 | MATaura3-52 trp1Δ63 leu2Δ1 rat8-2 | 79 |
| CHY167 | MATα ura3-52 leu2Δ1 his3Δ200 + pCH19 (Ssa4p-GFP in YIPlac211) | This study |
| CHY119 | MATα ura3-53 leu2Δ1 his3Δ200 Δrip1::HIS3 | 76 |
| CMHy37.3B | MATarna14-37 ura3-52 leu2Δ1 his3Δ200 | This study |
| CMHy48.1B | MATarna14-48 ura3-52 leu2Δ1-63 | This study |
| CMHy49.1D | MATarna14-49 ura3-52 leu2Δ1 | This study |
| CMHy64.2A | MATarna14-64 ura3-52 leu2Δ1 his3Δ200 | This study |
| CMHy23.1B | MATapap1-23 ura3-52 leu2Δ1 | This study |
| CMHy58.1A | MATaura3-52 leu2Δ1 his3DΔ200 rna15-58 | This study |
| DAts389 | MATα ura3-52 leu2Δ1 hisΔ200 rna14-389 | This study |
| DAts244 | MATα ura3-52 leu2Δ1 hisΔ200 rna15-244 | This study |
| DAts433 | MATα ura3-52 leu2Δ1 hisΔ200 fip1-433 | This study |
| rna15-2 W | MATaura3-1 trp1-1 ade2-1 leu2-3,112 his3-11,15 rna15-2 | 7 |
| rna14-1 W | MATaura3-1 trp1-1 ade2-1 leu2-3,112 his3-11,15 rna14-1 | 7 |
| NA57 | MATaura3-1 trp1Δ ade2-1 leu2-3,112 his3-11,15 pcf11::TRP1 + pFL36(pcf11-1) | 7 |
| NA64 | MATaura3-1 trp1Δ ade2-1 leu2-3,112 his3-11,15 pcf11::TRP1 + pFL36(pcf11-2) | 7 |
| rna1-1 mutant | MATarna1-1 ura3− leu2− gal+ trp1− | 42 |
| prp20-1 mutant | MATatrp11Δ63 leu2Δ1 ura33-52 prp20-1 | 28 |
| xpo1-1 mutant | MATaade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 xpo1::leu2 + pRS313xpo1-1 | 80 |
| mex67-5 mutant | MATaade2 leu2 ura3 his3 trp1 mex67::HIS3 + pmex67-5 (TRP1 CEN) | 77 |
| yra1-1 mutant | MATaade2 leu2 ura3 his3 trp1 yra17::HIS3 + pyra1-1 (TRP1 CEN) | 83 |
| sub2-85 mutant | MATaura3 his3m leu2 trp1 sub2::kanMX4 + pTRP-sub2-85 | 82 |
| kap104-16 mutant | MATa, kap104::HIS3 trp1-1 ura3-52 his3Δ200 leu2-3 lys2-1 + pRS314kap104-16 | 2 |
| EGY48 | MATα trp1 ura3 LEU2::plexop6-LEU2 | E. Golemis |
| RFY206 | MATahis3 ura3 trp1 lys2 | E. Golemis |
Approximately 4 × 106 cells were sorted at 4°C using a FACSTAR cell sorter (Becton Dickinson). The sorting window was set to allow isolation of the darkest 1% cells, which would have included live cells which could not produce Ssa4p-GFP as well as dead cells and large cell fragments. Approximately 24,000 colonies grew on YPD plates. These colonies were then screened to identify the temperature-sensitive ones, and approximately 90 temperature-sensitive colonies were identified. These colonies were transformed with a high-copy-number plasmid encoding SSA4 and analyzed individually to identify those which accumulated SSA4 mRNA in nuclei after a shift to 42°C.
Approximately 400 colonies from our original collection of 1,200 temperature-sensitive strains were also screened for SSA4 mRNA accumulation (5). Each clone from the library was transformed with a high-copy-number SSA4 plasmid. To identify mutants defective for export of SSA4 mRNA after heat shock, we combined mutant strains (400 total transformants in the first pass) into 100 pools, each containing four separate strains, and used in situ hybridization to identify those which accumulated SSA4 mRNA in their nuclei after 1 h of heat shock treatment at 42°C. Strains from pools which contained positive cells were rescreened individually. Ten strains with the desired phenotypes were selected, and the defective genes were cloned by complementation.
In situ hybridization.
To localize poly(A) mRNA, in situ hybridization using an oligo(dT)50 probe was performed essentially as described previously (5). For CYC1 mRNA and SSA4 mRNA localization, PCR products containing digoxigenin-dUTP and complementary to the open reading frame (ORF) of CYC1 or the 3′UTR of SSA4 were used as previously described (75). To localize CYC1 and cyc1-512 mRNAs, FY23 cells were transformed with plasmids containing either the wild-type (pGCYC1) or mutant (pGcyc1-512) and grown overnight in SC medium with 2% dextrose or 2% galactose but lacking tryptophan.
Northern hybridization analysis.
RNA extraction was performed by the method of Pikielny and Rosbash (65). Northern hybridization analysis was performed using 10 μg of total RNA, in the same manner as outlined previously (5).
Poly(A) tail length measurement.
The method of Forrester et al. (28) was used. Ten micrograms of total RNA was digested with 2 μg of bovine pancreatic RNase per ml and 1,000 U of RNase T1 per ml to produce a RNA population consisting only of poly(A) tails. Each RNA sample was then end labeled with 100 μCi of [5′-32P]cytidine 3′,5′ bis(phosphate) (NEN Life Science Products, Boston, Mass.) in a solution containing 50 mM Tris-HCl (pH 7.9), 15 mM MgCl2, 3.3 mM dithiothreitol (DTT), 2% (vol/vol) dimethyl sulfoxide, 10 mg of bovine serum albumin per ml, 25 μM ATP, and 10 U of T4 RNA ligase for 21 h at 4°C. RNA samples were then extracted with phenol and precipitated with ethanol. Pellets were then resuspended in a solution containing 96% formamide, 0.1% bromophenol blue, and 0.1% xylene cyanol, denatured by boiling, and resolved on a 10% polyacrylamide-1 M urea-TBE (Tris-borate-EDTA) gel. Gels were dried and exposed to X-Omat Blue film (Kodak, Rochester, N.Y.).
Expression and purification of recombinant Xpo1p and Xpo1pT539C from Escherichia coli.
The ORF of XPO1 was amplified from genomic DNA and cloned into pGEX-3X to create a C-terminal fusion to glutathione S-transferase (GST) (pCMH036). A fragment containing the leptomycin-sensitive region of XPO1T539C was subcloned from pDCcrm1T539C and used to replace the wild-type portions of pCMH036, creating pCMH037. For protein expression, a one-liter culture of E. coli strain BL21(DE3) transformed with the appropriate plasmid was grown to an optical density at 600 nm of 0.7 at 37°C. Production of the fusion protein was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 1 h at 28°C. Cells were washed once in phosphate-buffered saline plus 1 mM phenylmethanesulfate fluoride (PMSF) and then resuspended in 7.5 ml of lysis buffer (250 mM KCl, 50 mM Tris [pH 8.0], 1 mM EDTA, 0.5 mM DTT, 10% glycerol, 0.5% Nonidet P-40) plus protease inhibitors (1 mM PMSF, 2 μM pepstatin A, 0.6 μM leupeptin). Cells were lysed by a freeze-thaw cycle, and the lysate was clarified by 30 min of centrifugation at 90,000 × g and then diluted 1:1 in lysis buffer without KCl. For affinity purification, 0.5 ml of glutathione-agarose beads (Pharmacia) was equilibrated with three 15-ml washes in bind/wash buffer (150 mM KCl, 20 mM Tris-Cl [pH 8.0], 0.2 mM EDTA, 0.5 mM DTT, 0.5% [vol/vol] Triton X-100, 10% [vol/vol] glycerol, and protease inhibitors). The lysate was brought to a volume of 15 ml with bind/wash buffer and incubated with beads at 4°C for 1 h. After extensive washing with bind/wash buffer, bound proteins were removed by incubation in 1 ml of bind/wash buffer plus 50 mM reduced glutathione for 1 h. Proteins were dialyzed twice for 90 min with buffer D (50 mM KCl, 20 mM Tris-Cl [pH 8.0], 0.2 mM EDTA, 0.5 mM DTT, 10% [vol/vol] glycerol, and protease inhibitors), frozen in liquid nitrogen, and stored at −80°C.
RNA processing assays.
Capped, 32P-labeled mRNAs were prepared by runoff transcription from plasmid pJCGAL7-1 by the method of Chen and Moore (17). RNA containing approximately 250,000 to 280,000 cpm was used per reaction mixture, equivalent to a final substrate concentration of 10 nM. Each reaction was performed in a volume of 20 μl, using 1 μl of whole-cell extract prepared as described previously (17). Each reaction mixture contained 1 mM ATP, 10 mM creatine phosphate, 1 mM magnesium acetate, 75 mM potassium acetate, 2% polyethylene glycol 8000, 1 mM DTT, 0.1 mg of bovine serum albumin (New England Biolabs) per ml, 0.4 U of RNasin (Promega), and 10 nM radioactive RNA precursor. For assays using leptomycin B (LMB), extracts were preincubated for 1 h with 10 nM LMB before addition to the reaction mixture. Reaction mixtures were assembled on ice and incubated at 4°C for 10 min and then at 30°C for 20 min. Reactions were stopped by the addition of proteinase K and sodium dodecyl sulfate as described previously (17), the reaction mixtures were brought to a volume of 30 μl with TE (10 mM Tris-Cl [pH 7.5], 1 mM EDTA), and RNA was extracted once with phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol/vol). One-tenth of the reaction mixture was resolved on a 5% acrylamide-8.3 M urea gel and visualized using a Storm 960 PhosphorImager (Molecular Dynamics).
Two-hybrid constructs.
ORFs were amplified using PCR with oligonucleotides which introduced restriction sites. The pEG202-CRM1 bait construct was obtained by cloning the coding region of CRM1 as an EcoRI-XhoI PCR fragment into the pEG202+PL vector (HIS3, 2μm) cut with EcoRI and XhoI. The LexA-Crm1p fusion protein encoded by this construct lacks the C-terminal region of Crm1p (about 100 amino acids). This pEG202-CRM1 bait construct fully rescues a CRM1 disruption, indicating that the fusion protein is functional. The pEG202-CRM1-T539C mutant construct (LMB sensitive) was obtained by replacing the ApaI-DraIII restriction fragment of pEG202-CRM1 by the corresponding restriction fragment from pDC-CRM1-T539C (63). pEG202-CSE1 was obtained by cloning the CSE1 coding region as a BamHI-SalI PCR fragment into pEG202 cut with the same enzymes.
Prey constructs were obtained by inserting the coding regions of PAB1 and SRP1 (as an XhoI PCR fragment), RNA15 (as an EcoRI-XhoI PCR fragment), and CLP1 (as an EcoRI PCR fragment) into the pJG4-5 (TRP1, 2μm) prey vector cut with the same enzymes, in frame with the B42 trans-activation domain and under the control of the galactose-inducible GAL1 promoter (71). The ORF of RNA14 was inserted as a XhoI PCR fragment into the prey vector pACTII (LEU2, 2μm) cut with XhoI, in frame with the Gal4 activating domain. pACTII-PCF11 encodes the C-terminal portion of Pcf11p (codons 271 to 452 [7]).
Two-hybrid strains and assay.
Two-hybrid interactions were examined by transforming the pEG202 bait constructs (HIS3, 2μm) and pJG4-5 prey constructs (TRP1, 2μm) into strains RFY206 and EGY48, respectively. Strains EGY48 and RFY206 each contain the LacZ reporter pSH18-34 (URA3, 2μm). The transformed EGY48 and RFY206 strains were mated, and diploids were selected on medium lacking uracil, tryptophan, and histidine. The diploids were subsequently tested for galactose-dependent activation of the reporter gene by replica plating to selective plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and either 3% galactose or 1% raffinose in the presence or absence of 100 ng of LMB per ml. Two-hybrid interactions between pEG202 bait and pACTII prey (LEU2, 2μm) constructs were determined by cotransforming the corresponding plasmids into a wild-type W303 strain containing the LacZ reporter plasmid pSH18-34. Triple transformants were selected on medium lacking uracil, histidine, and leucine and replica plated to selective plates containing X-Gal and 2% glucose in the presence or absence of 100 ng of LMB per ml to allow visualization of β-galactosidase reporter gene activity.
RESULTS
Identification of a novel class of mRNA export mutants.
Eukaryotic cells respond to stress in a variety of ways that enable them to survive rapid environmental changes and recover once these conditions no longer exist (26, 75). One of these responses involves the inhibition of bulk mRNA export while permitting transcriptional induction and efficient export of heat shock mRNAs (75). Over the years, we and others have identified several proteins important for mRNA export, including a subset of nucleoporins, specific mRNA export factors, mRNA-binding proteins, and components of the general nuclear transport machinery (89). Export of heat shock mRNAs requires almost all of the proteins required for export of mRNA under normal conditions. In addition, yeast cells carrying deletions of the nonessential nucleoporin RIP1/NUP42 grow normally and do not accumulate poly(A) RNA at temperatures below 37°C but are completely defective for export of all transcripts, including heat shock mRNAs, at 42°C or following a 10% ethanol shock (76, 85).
SSA4 encodes an S. cerevisiae Hsp70 protein whose transcription is not activated until yeast cells sense stress (10). To screen for additional mutants defective in mRNA export after stress, we constructed a reporter protein by fusing the SSA4 ORF to GFP (Fig. 1A). To obtain uniform expression in all cells, we inserted the construct encoding Ssa4p-GFP into the SSA4 locus along with URA3. We monitored the expression of Ssa4p-GFP using a fluorescence-activated cell sorter (FACS). Wild-type cells containing the integrated SSA4-GFP fusion were not fluorescent when grown to early log phase (Fig. 1B). Transfer of these cultures to 42°C rapidly induced expression of Ssa4p-GFP, and the fluorescent reporter could be detected within 2 to 5 min after the temperature shift (C. V. Heath and C. N. Cole, unpublished results). In contrast to wild-type cells (Fig. 1B), Ssa4p-GFP was not detectable when Δrip1/nup42 cells were heat shocked for 1 h (Fig. 1C). Wild-type cells were, on average, more than 100 times brighter than Δrip1/nup42 cells. To determine whether we could use flow cytometry to separate cells defective for heat shock mRNA export from other cells, we mixed equal proportions of wild-type and Δrip1/nup42 cells, induced Ssa4p-GFP expression by shifting to 42°C, and sorted the mixture. Two fluorescent peaks were observed (Fig. 1D). Since the wild-type cells had TRP1/his3 genes and the Δrip1/nup42cells had trp1/HIS3 genes, we could readily determine the proportion of each strain in the dark and bright pools by plating cells on selective medium. Of the cells in the dark pool, 83% were Δrip1/nup42 cells, indicating that flow cytometry could be used to enrich for mutants defective for mRNA export.
FIG. 1.
Flow cytometry of Ssa4-GFP reporter strains. (A) Schematic representation of the inducible fluorescent reporter used in this screen. Wild-type (B) or Δrip1 cells (C) containing an integrated SSA4-GFP reporter were sorted by FACS at 23°C or 1 h after the temperature shift to 42°C. In panels B to D, relative fluorescence is plotted on the x axis, and cell counts are plotted on the y axis. (D) An equal mixture of wild-type cells and Δrip1 cells were mixed, incubated at 42°C for 1 h, and sorted. The two peak fractions were plated on selective media to determine the relative proportion of each strain in each peak. Among those cells isolated in the dark fraction will be cells of either type which were no longer alive at the time of sorting.
To prepare new mutants, wild-type cells containing the integrated SSA4-GFP fusion were randomly mutagenized by UV radiation, incubated at 42°C for 1 h, and sorted at 4°C to prevent expression of the reporter gene while the samples were being sorted. We isolated the darkest ∼1% of the cells. From ∼3 × 106 cells, we obtained 24,000 colonies, which were then replica plated and incubated at 23 and 37°C to identify ts strains. Approximately 90 strains, which grew well at 23°C and did not grow at 37°C, were selected for further study.
From the ts population, we expected several classes of “dark” mutants. Some should be defective in transcription, some should be defective in pre-mRNA processing, others should be defective in translation, and a fourth group should be defective in mRNA export. To identify strains which accumulated mRNA in their nuclei under nonpermissive conditions, we transformed ts strains with a high-copy-number SSA4 plasmid and screened them by in situ hybridization to identify those defective in SSA4 mRNA export at 42°C. Some strains failed to produce detectable SSA4 mRNA following heat shock, presumably reflecting defects in transcription or, possibly, processing; some strains showed a strong cytoplasmic signal for SSA4 mRNA and were presumably defective in translation; and some strains accumulated SSA4 mRNA in their nuclei after heat shock. This last class, comprising 10 strains, was studied further.
Standard genetic analyses showed that these 10 mutations were recessive and fell into four complementation groups. These strains were transformed with a CEN-based library and then selected for complementation to restore growth at 37°C. Three of the four complementation groups encoded 3′ processing factors. These 3′ processing factors included the poly(A) polymerase Pap1p (one allele) and two components of CFIA, Rna14p (four alleles) and Rna15p (one allele). These strains are listed in Table 1. The four strains in the remaining complementation group had a high rate of reversion. The defective gene could not be identified, and these strains were not studied further.
At the same time, we also screened 400 ts strains, prepared for our initial mRNA export mutant screen (5), for defects in export of SSA4 mRNA. By using this screen, we identified additional alleles of RNA14 and RNA15 and an allele of FIP1, another 3′ processing factor (Table 1).
New ts alleles of CFI components accumulate polyadenylated RNA in their nuclei.
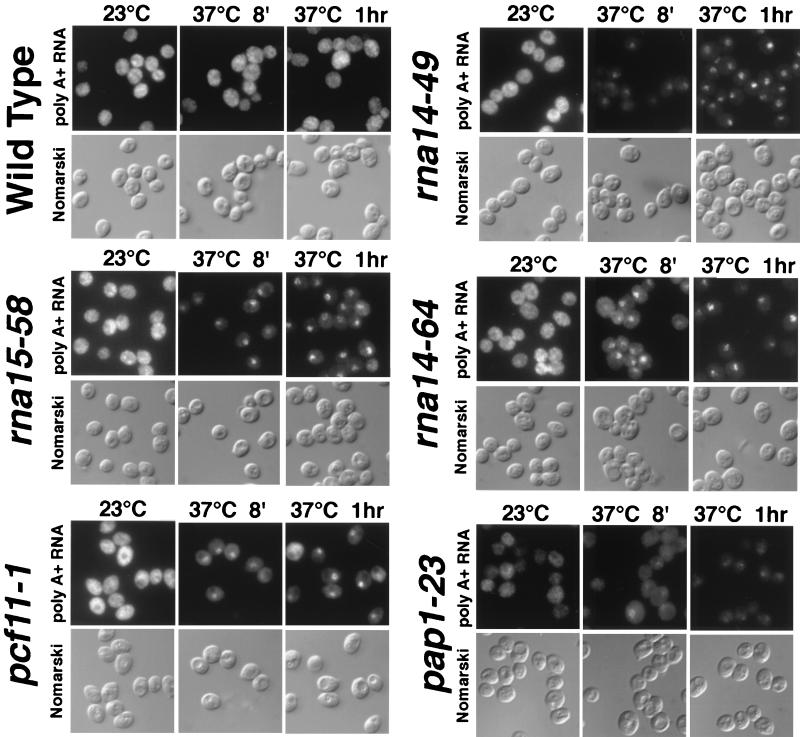
We analyzed mRNA export in these strains at 37°C using in situ hybridization with an oligo(dT)50 probe. In several of the strains (rna15-58, rna14-48, rna14-49, and rna14-64), accumulation of poly(A) RNA could be detected as soon as 8 min after a shift to 37°C (Fig. 2) and in at least 90% of cells. Other alleles (rna14-37, rna14-389, rna15-244, fip1-433, and pap1-23) did not show significant nuclear accumulation of poly(A) RNA (Fig. 2 and data not shown). This group, like previously characterized alleles affecting CFI proteins (rna14-1, rna15-1, rna15-2, pap1-1, pcf11-2, and all hrp1 mutant alleles) and two ts alleles of another 3′ processing factor (pta1-1 and pta1-2) rapidly lost both nuclear and cytoplasmic signals for poly(A) RNA after a shift to 37°C. This lack of in vivo poly(A) tail production is consistent with previous characterization of these alleles in other laboratories (7, 60, 67, 92). Interestingly, one allele of PCF11, pcf11-1, did accumulate nuclear poly(A) mRNA (Fig. 2). This allele has been shown to produce hyperadenylated transcripts at its nonpermissive temperature (7).
FIG. 2.
New alleles affecting the 3′ processing machinery accumulate poly(A) RNA at the nonpermissive temperature. Wild-type or mutant cells were grown at 23°C and shifted to 37°C for 8 min (8′) or 1 h. Samples were then fixed, permeabilized, and probed with an oligo(dT)50 probe to localize poly(A) RNAs. Both FITC [oligo(dT)] and Normarski images of the same fields are shown.
Nuclear accumulation of heat shock mRNA at 42°C was seen in all cases, including those strains which did not show nuclear accumulation of polyadenylated mRNA at 37 or 42°C. In the latter strains, the defect in 3′ processing factors most likely prevented formation of a poly(A) tail. Both Northern blotting (data not shown) and the detection of SSA4 mRNA at 42°C indicate that the mRNAs were not rapidly degraded. In summary, all of the strains we examined which were defective for 3′ processing were also defective for export, including those which produced polyadenylated mRNA. This indicates that the presence of a poly(A) tail is essential but not sufficient for mRNA export.
New ts alleles of CFI components are temperature sensitive for 3′ processing in vitro.
Because several mutants produced polyadenylated transcripts at their nonpermissive temperature, we tested their abilities to cleave and polyadenylate transcripts in vitro. Extracts prepared from several strains (pap1-23, rna15-58, and rna14-64 cells) were able to carry out 3′ processing at 23°C (Fig. 3A and B), though their activity was lower than that seen in wild-type extracts. The processing activity of these mutant extracts was thermolabile: extracts heated to 37°C for 5 min lost all processing activity, while wild-type extracts retained activity after this treatment (Fig. 3C and D). However, when extracts from rna14-64 and rna15-58 mutant strains were combined and assayed at 30°C, these extracts could complement each other (Fig. 3C and D). This contrasts with results obtained using extracts of previously described rna14-1 and rna15-2 alleles where no complementation between mutant extracts was seen (60). It is known that Rna14p and Rna15p associate in vivo and that each requires the other for stability (9).
FIG. 3.
In vitro 3′ processing analysis of new 3′ processing mutants and in vivo poly(A) tail lengths of mRNA export mutants and 3′ processing mutants identified in this screen. (A) Extracts from wild-type (WT), pap1-23, rna15-58, or rna14-64 cells were incubated with an in vitro-transcribed GAL7 3′UTR and dATP at 23°C to measure the in vitro cleavage activity of these strains. Lane (−), negative control. (B) Extracts from the same strains were incubated with a precleaved, in vitro-transcribed GAL7 precursor at 23°C to measure polyadenylation efficiencies. (C and D) Coupled cleavage and polyadenylation reactions (C) of the GAL7 3′UTR or polyadenylation reactions (D) of the same extracts heated to 37°C for 5 min before addition to the reaction mixture at 30°C. In panels C and D, extracts from the various mutants were also mixed in equal proportions and incubated with the same GAL7 precursors to measure each extract's ability to complement other defective extracts. (E and F) The poly(A) tail length distribution of each mutant was determined by isolating total RNA from each strain that had been grown continuously at 23°C or shifted to 37°C for 30 min. The resulting RNAs were digested to completion with RNases T1 and A. The remaining oligo(A) and poly(A) fragments were then end labeled with [32P]pCp and RNA ligase and resolved on 9% polyacrylamide-7 M urea-TBE gels. Poly(A) lengths of mRNAs from several CFIA mutants isolated in this screen (E) or from several mRNA export mutants (F) are shown.
Although rna14-1 and rna15-2 extracts retain Pap1p activity, it is likely that each strain alone exhibits reduced Rna14p and Rna15p activity. In contrast, in rna14-64 cells, Rna15p appears to be relatively stable because an rna14-64 extract can complement an rna15-58 extract, while in rna15-58 cells, Rna14p appears to be stable. Most likely, the mutations in the new strains do not result in complete lability of the mutant protein or assembled complex, consistent with their retaining the ability to produce some poly(A) RNA in vivo at 37°C.
New ts alleles have 3′ processing and polyadenylation defects in vivo.
The signals for 3′-end formation in S. cerevisiae have been identified as several loosely conserved consensus sequence elements and have been well characterized (34, 35, 90). These signals are also important for transcription termination. Recognition of these signal elements by the CFIA complex (Rna14p, Rna15p, Pcf11p, and Clp1p) couples 3′ processing to Pol II termination (8, 72, 73). Mutations affecting CFIA components result in the production of mRNA transcripts with elongated 3′UTRs, consistent with the role of CFIA in transcriptional termination.
Improper 3′ processing usually produces unstable mRNAs, most likely due to rapid turnover of RNAs which lack a poly(A) tail (28, 60, 62). Defects in transcriptional termination also lead to unstable mRNAs whose extended 3′UTRs are actively targeted by the nonsense-mediated decay pathway (22). However, some yeast transcripts, including CUP1 mRNA, encoding copper metallothionein, are stable even when there are defects in 3′ cleavage and/or polyadenylation (28). Properly processed CUP1 mRNAs are ∼600 nucleotides long and are efficiently capped and polyadenylated in wild-type cells (Fig. 4). Defective 3′ processing of CUP1 mRNAs produces elongated transcripts of various sizes which can be visualized by Northern hybridization (28, 58).
FIG. 4.
Transcriptional termination phenotypes of 3′ processing and mRNA export mutants. Temperature-sensitive CFIA mutants (A), mRNA export mutants (B), or mutants of two karyopherin b family members (C) were grown at 23°C and then shifted to 37°C for 30 min (30′). Ten micrograms of total RNA was extracted, separated on a 1% formaldehyde gel, transferred to a membrane, and probed with an antisense CUP1 riboprobe. Panel C also compares the effects of Rat8p/Dbp5p overexpression on the termination defects of xpo1-1 and kap104-16 strains. The mature form of CUP1 mRNA transcripts is approximately 600 nucleotides. The asterisks indicate improperly processed CUP1 transcripts that are approximately 1.8 kb long. WT, wild type.
We examined the production of CUP1 mRNA in several yeast strains carrying ts mutations affecting CFIA components. All lanes of the gel were loaded to contain an identical amount of rRNA (data not shown). When wild-type cells and several of the mutant strains were shifted to 37°C, there was a dramatic increase in the overall level of CUP1 mRNA. In mutant cells, elongated and improperly terminated CUP1 transcripts were produced at the nonpermissive temperature, consistent with the essential role of 3′ processing factors in cleavage and termination (Fig. 4A). Some mutants also produced a substantial amount of normal-length CUP1 mRNA at 37°C, although the extended transcripts were generally more abundant. CUP1 transcripts produced at both temperatures were polyadenylated (data not shown). Although we detected nuclear accumulation of poly(A) RNA in these strains (Fig. 2), we were not able to localize CUP1 transcripts by in situ hybridization.
We also measured the lengths of the poly(A) tails produced in cells carrying these new alleles. Figure 3E shows that the tails produced in these mutants after a shift to 37°C for 30 min are not much longer than those produced in wild-type cells. In addition, the data confirm that mutant cells contain much less polyadenylated mRNA at 37°C than do wild-type cells, consistent with reduced mRNA stability.
Deletion of the cis-acting element for mRNA termination and polyadenylation results in production of extended, polyadenylated mRNAs which accumulate in nuclei.
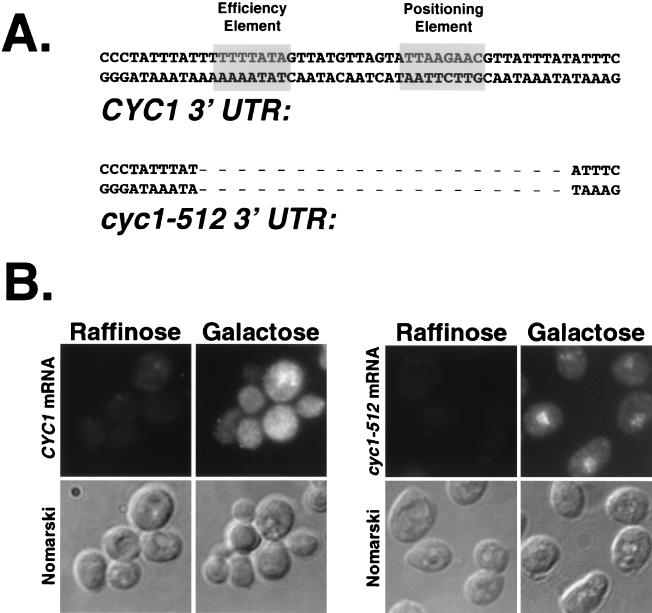
The CYC1 gene of S. cerevisiae encodes isocytochrome c. The cis-acting elements required for its expression have been studied extensively (72-74, 87, 88). The cyc1-512 mutation is a 38-bp deletion of 3′ processing and termination signals located in the CYC1 3′UTR (Fig. 5A). Transcription from this mutant locus produces unstable cyc1-512 mRNAs (less than 10% of wild-type transcript levels) (87, 88). Furthermore, cyc1-512 transcripts are improperly terminated and, as a result, contain elongated 3′UTRs (up to 3 to 4 kb longer that normal) and are polyadenylated (22, 87, 88). The almost complete absence of Cyc1-512p (<10@0025) has been attributed to the unstable nature of its mRNA and inefficient translation of these aberrantly long mRNAs (>22) correlates with the very low level of cyc1-512 mRNA.
FIG. 5.
Deletion of cis-acting processing and termination elements leads to a block to mRNA export. (A) Comparison of 3′UTR sequence of CYC1 and cyc1-512 genes. cyc1-512 locus lacks a 38-bp region (dashes) required for normal cleavage, polyadenylation, and Pol II termination. (B) Wild-type cells were transformed with galactose-inducible CYC1 or cyc1-512 constructs. Cells were then grown in noninducing (raffinose) or inducing (galactose) conditions overnight, fixed, and permeabilized. Samples were then probed with a digoxigenin-labeled PCR probe complementary to the CYC1 coding region (present in both transcripts). Anti-digoxigenin-FITC antibodies were then used to localize mRNAs which hybridized with the PCR-labeled CYC1 probe.
Because defects in CFIA activity and function yielded mRNAs which were exported inefficiently, we reasoned that defects in the cis-acting 3′ processing elements might have the same effect. We transformed wild-type cells with plasmids that place either wild-type CYC1 or cyc1-512 under control of a galactose-inducible promoter (8) and performed message-specific in situ hybridization (Fig. 5). No expression of either CYC1 or cyc1-512 mRNA was detected in the absence of galactose. Expression from the genomic CYC1 locus was not detectable by in situ hybridization under any conditions. After galactose induction, wild-type CYC1 mRNA was exported efficiently, but there was strong nuclear accumulation of cyc1-512 mRNA (Fig. 5B). This shows that efficient mRNA export requires that cleavage and polyadenylation be directed by the normal 3′ processing and termination signals.
Analysis of 3′ processing and production of 3′ extended mRNAs in strains defective for mRNA export.
Interestingly, earlier studies showed that mutations in Gsp1p's effector molecules, encoded by PRP20 and RNA1, also lead to severe defects in termination, within 90 s of a shift to 37°C. These strains produce hyperadenylated mRNAs (28) and are also defective in mRNA export (4, 28). To determine whether defects in polyadenylation and termination were common phenotypes of mRNA export mutants, we examined CUP1 mRNAs produced in strains carrying deletions or ts mutations of several proteins required for efficient mRNA export. Overall, defects in termination leading to production of 3′ extended mRNAs were seen in some but not all strains which accumulated mRNA in their nuclei (Fig. 4B and C). Some of these (e.g., Kap104p) affect mRNA biogenesis indirectly: export defects seen in some kap104-16 cells likely reflect defects in the import of shuttling RNA-binding proteins, including Hrp1p, required for efficient 3′ processing, and Nab2p, required for mRNA export and poly(A) tail length control (2, 37, 46). Consistent with this, nuclear mRNA accumulation occurs asynchronously and in only a fraction of kap104-16 cells (2). In many other strains examined, however, the defective protein is thought to participate directly in mRNA export. Little or no extended mRNA was detected in rat7-1/nup159-1, yra1-1, or sub2-85 cells shifted to 37°C (Fig. 4B). This indicates that improper transcriptional termination is not an automatic consequence of defective mRNA export or accumulation of poly(A) mRNAs in the nucleus. In contrast, a significant level of improperly terminated CUP1 transcripts was detected in mex67-5 and xpo1-1 cells at the nonpermissive temperature (Fig. 4B and C). Extended CUP1 transcripts were also produced in kap104-16 cells (Fig. 4C).
We also examined poly(A) tail lengths in some of these mutant strains. The longest poly(A) tails were seen in rat7-1/nup159-1 and mex67-5 cells (Fig. 3F) and in prp20-1 and rna1-1 cells (data not shown), as has been shown previously (28, 40, 44). In contrast, poly(A) tails in yra1-1, sub2-85, kap104-16, and mtr10Δ cells were the same length as seen in wild-type cells. These results show that hyperpolyadenylation is not always linked with defects in transcriptional termination or mRNA export and suggest that the various mRNA export mutants exert their effect on 3′-end formation at different levels.
CFI proteins interact in vivo with Xpo1p.
Because inefficient 3′ cleavage and termination were observed in cells carrying mutations affecting CFI components and in xpo1-1 cells, we wondered whether 3′ processing factors might interact with Xpo1p. To examine this, we employed the two-hybrid system and prepared LexA-Xpo1p fusions as baits. One of these encodes the LMB-sensitive T539C substitution allele of Xpo1p that prevents Xpo1p-substrate interactions in vivo and in vitro in the presence of LMB (48, 63, 86). The prey constructs encode 3′ processing factors fused at their N termini to the B42 or Gal4 trans-activating domains (Gal4AD). For controls for Xpo1p interactions, we used previously characterized Gal4AD fusions with Rev, a bona fide NES-containing substrate for Xpo1p, and the NES-deficient RevM10 mutant (64). We also constructed a LexA fusion with Cse1p, another protein export receptor of the karyopherin β family (49).
As expected, Cse1p interacted strongly with its known cargo, Srp1p (importin α), but not with any other constructs tested (Table 2). In contrast, several positive two-hybrid interactions between Xpo1p and 3′ processing factors were observed. The results from multiple experiments are summarized in Table 2. A strong interaction, approximately the same strength as with a wild-type Rev prey, was seen between Xpo1p and Pab1p. The interactions with Rev and Pab1p were equally strong using either wild-type Xpo1p or the Xpo1pT539C LMB-sensitive form. Addition of LMB disrupted Xpo1pT539C's interactions with both Pab1p and Rev to equal extents. The interactions between Xpo1p and Rna15p, Pcf11p, and Hrp1p were weaker than with Pab1p, while the interaction with Rna14p was even weaker but significantly above the level seen using vector alone or the RevM10 negative control. The effect of LMB on these in vivo interactions varied. The interactions between Xpo1pT539C and Pcf11p or Hrp1p was sensitive to but not completely eliminated by LMB, while the interaction between Xpo1p and Rna15p was not affected. Since all of the CFI components function as a complex, it is likely that Xpo1p does not interact directly with all of these factors, and there could be other proteins mediating the Xpo1p-CFIA interactions (32). Known two-hybrid interactions among different CFI components also scored positive in this assay (data not shown). However, the fact that LMB affected the interactions to different degrees suggests that interactions between 3′ processing factors and Xpo1p could involve other regions of Xpo1p in addition to the normal NES-binding domain.
TABLE 2.
Two-hybrid interactions between Xpo1p and 3′ processing factorsa
| Construct | β-Galactosidase activityb
|
|||||
|---|---|---|---|---|---|---|
| Xpo1p
|
Xpo1pT539C
|
Cse1p
|
||||
| −LMB | +LMB | −LMB | +LMB | −LMB | +LMB | |
| pJG4 bait | ||||||
| Vector (control) | − | − | − | − | − | − |
| Rev | +++ | +++ | +++ | − | − | − |
| RevM10 | − | − | − | − | − | − |
| Pab1p | +++ | +++ | +++ | − | − | − |
| Rna15p | ++ | ++ | ++ | ++ | − | − |
| Cse1p | − | − | − | − | ++++ | ++++ |
| pACTII bait | ||||||
| Vector (control) | − | − | − | − | − | − |
| Pcf11p | ++ | ++ | ++ | + | − | − |
| Rna14pc | + | + | − | − | − | − |
| Hrp1pc | ++ | ++ | ++ | + | − | − |
Diploids containing LexA-export receptor fusions, 3′ processing factor-Gal4AD fusions, and a LacZ reporter were grown on selective medium containing 3% galactose or 1% raffinose. LMB was added (+) to the solid growth medium to a final concentration of 100 ng/ml or not added (−) to the growth medium.
The β-galactosidase activities are expressed relative to that of the vector control in each experiment. The activity measurements are based on the color plate assay, where ++++ is very dark blue and − is white.
Strains containing Rna14p-Gal4AD and Hrp1-Gal4AD fusions grow very slowly.
Xpo1p stimulates 3′-end processing in vitro.
Since temperature-sensitive mutations in XPO1 lead to the production of improperly terminated Pol II transcripts in vivo (Fig. 4C) and because Xpo1p interacted with CFI proteins known to play a critical role in 3′ cleavage and termination, we wondered how the presence of Xpo1p would affect 3′ processing in vitro. First, we prepared extracts from cells carrying mutations in known export factors (xpo1-1, prp20-1, and rna1-1 cells) and examined 3′ processing in a coupled cleavage and polyadenylation reaction. We found that these extracts had activities comparable to that of a wild-type extract prepared under the same conditions (S. Gross, C. M. Hammell, and C. Moore, unpublished results), indicating that Xpo1p, Prp20p, and Rna1p are not essential for in vitro 3′ processing. We next tested whether addition of recombinant GST-Xpo1p would stimulate 3′ processing. The efficiency of processing was measured by determining the fraction of total radioactivity in each lane present in the cleaved and polyadenylated products. Preincubating the processing extract with increasing amounts of GST before addition of the precursor did not change the efficiency of processing (Fig. 6A, lanes 2 to 6). However, incubation with GST-Xpo1p reproducibly stimulated processing two- to threefold (Fig. 6A, lanes 7 to 11). The lane-to-lane variation in total counts was less than 10%, indicating that the exogenously added GST-Xpo1p also did not contain RNases nor did it activate any that might be present in the extract. Increasing amounts of GST-Kap95p, another transport receptor, had no effect on 3′ processing activity (data not shown). Furthermore, no degradation or specific cleavage of the precursor was observed upon incubation of the precursor RNA with GST-Xpo1p or GST-Kap95p alone (data not shown).
FIG. 6.
Xpo1p stimulates 3′ processing efficiency in vitro. (A) Extracts prepared from wild-type cells were preincubated with 8, 24, 74, 220, or 660 ng of recombinant GST (lanes 2 to 6) or GST-Xpo1p (lanes 7 to 11) prior to addition of a labeled in vitro-transcribed RNA precursor containing sequences flanking the GAL7 poly(A) site. Coupled cleavage and polyadenylation reactions were then carried out at 30°C for 30 min, and the products were resolved by electrophoresis on a 5% acrylamide-urea gel. Lane 1 contains unreacted precursor. (B) Effect of LMB addition on the 3′ processing activity of extracts made from strains resistant (R; XPO1) or sensitive (S; xpo1T539C) to LMB (lanes 1 to 4). Recombinant GST-Xpo1 (R) or GST-Xpo1T539C (S) was added (lanes 5 to 12) or not added (lanes 1 to 4) to the extracts, with (lanes 3, 4, and 9 to 12) or without (lanes 1, 2, and 5 to 8) LMB, as indicated above the gel.
We also prepared extracts from isogenic strains differing solely at the XPO1 locus; one has the wild-type XPO1 gene, and the other has the LMB-sensitive XPO1T539C gene (63). The two extracts carried out 3′ processing with approximately the same efficiency (Fig. 6B, compare lanes 1 and 2). Preincubation of the extracts with LMB prior to adding the labeled 3′ processing substrate reduced 3′ processing in the Xpo1T539Cp extract by about 40% but had no effect on extracts prepared from wild-type cells (Fig. 6B, lanes 2 and 4).
Processing was stimulated to approximately the same extent by the addition of either wild-type or LMB-sensitive GST-Xpo1p (Fig. 6B, lanes 5 to 8). Added wild-type GST-Xpo1p stimulated 3′ processing in wild-type extracts equally well both in the presence or absence of LMB (Fig. 6B, lanes 5 and 9). The final level of processing was lower when both wild-type GST-Xpo1p and LMB were added to an extract prepared from LMB-sensitive cells, reflecting the effect of LMB on extracts prepared from LMB-sensitive cells (Fig. 6B, compare lanes 4, 6, and 10). LMB prevented the LMB-sensitive Xpo1T539Cp from stimulating 3′ processing (Fig. 6B, lanes 3, 4, 7, 8, 11, and 12). The data do not indicate whether the effect of Xpo1p is direct or indirect (see discussion below).
Unlike Hrp1p (CFIB), the CFIA subunits Rna14p, Rna15p, and Pcf11p do not shuttle.
The experiments described above indicate that similar defects in 3′ processing occur in strains with mutations affecting 3′ processing factors, the Ran/Gsp1p system, some importin β family members, and several mRNA export factors. Because of these similarities and the two-hybrid interactions seen between CFIA proteins and Xpo1p, we reasoned that 3′ processing factors might also function directly during mRNA export and remain associated with the mRNA until export was completed. To determine whether any CFIA proteins shuttle, we performed the standard shuttling assay developed by Lee et al. (50). This assay uses the nup49-313 allele which allows efficient export of shuttling proteins but is defective at the nonpermissive temperature in protein import of all classes of proteins examined. The nup49-313 mutation does not affect mRNA export. We made GFP fusions to several 3′ processing factors (Rna14p, Rna15p, Pcf11p, Cft1p, and Fip1p). Each was functional, as judged by its ability to complement a deletion or ts mutation in the corresponding gene (data not shown). Each GFP fusion was produced from 2μm plasmids. Because excess Rna14p or Rna15p can accumulate only when both are overexpressed (9), cells producing Rna15p-GFP were engineered to overproduce Rna14p, and cells producing Rna14p-GFP were engineered to overproduce Rna15p. Each 3′ processing factor (Rna14p, Rna15p, Pcf11p, Cft1p, or Fip1p) is a nuclear protein, and we detected the GFP fusions solely in nuclei in wild-type cells (data not shown). In nup49-313 cells, these proteins remained in the nuclei at both the permissive and nonpermissive temperatures (Fig. 7). This suggests that these 3′ processing factors are resident nuclear proteins which do not shuttle. In contrast, Hrp1p, an RNA-binding protein required for both cleavage and polyadenylation, has been shown to be exported in a transcription-dependent manner, consistent with Hrp1p's export as a passenger with mRNA (46). As expected, Hrp1p-GFP accumulated in the cytoplasm of nup49-313 cells shifted to 36°C (Fig. 7).
FIG. 7.
Most components of the core 3′ processing machinery do not shuttle. nup49-313 cells individually expressing GFP fusions of Hrp1p, Rna14p, Rna15p, Pcf11p, Fip1p, or Cft1p were incubated at 23°C or shifted to 36°C for 6 h to assay the nuclear export of these proteins. Living cells were either photographed with Nomarski optics or examined for GFP fluorescence.
Suppression of production of extended transcripts in xpo1-1 cells by overexpression of Rat8p/Dbp5p.
We reported previously that the strong block to mRNA export seen when xpo1-1 cells were shifted to 37°C could be prevented if cells overexpressed Rat8p/Dbp5p (41). xpo1-1 mutants mislocalize Rat8/Dbp5p under these conditions. This finding suggests that mRNA export required Rat8p/Dbp5p on the cytoplasmic side of the nuclear envelope and that this depends on Xpo1p activity. It is also possible that overexpression of Rat8p/Dbp5p in the nucleus is required to suppress nuclear defects in xpo1-1 cells. To examine how overexpression of Rat8p/Dbp5p affected the termination and 3′ processing defects of xpo1-1 cells, we analyzed CUP1 mRNAs produced in xpo1-1 cells containing a RAT8/DBP5 high-copy-number plasmid. Overexpression of Rat8p/Dbp5p completely prevented the production of 3′ extended, CUP1 mRNAs (Fig. 4C). Overexpression of Rat8p/Dbp5p had no effect on improper 3′ processing in kap104-16 cells (Fig. 4C) or in any other strains with 3′ processing defects (data not shown). Thus, Rat8p/Dbp5p shows specificity in suppressing these processing defects solely in xpo1-1 mutant cells.
DISCUSSION
A link between 3′ processing and mRNA export.
The experiments presented here provide important information about the connection between efficient mRNA processing and mRNA export. Previous studies in both S. cerevisiae and metazoans have shown that posttranscriptional addition of a poly(A) tail is essential for efficient mRNA export (11, 24, 29, 43). In two reports, mRNA produced in vivo from reporter constructs accumulated in nuclei if the normal 3′ processing signals were replaced with a cis-acting ribozyme (24, 43). Inclusion of an encoded poly(A) tract (90 A's) immediately upstream of the ribozyme cleavage site did not enhance export, indicating that the context of these posttranscriptional processing events affect downstream functions in mRNA export (43). Together, these studies suggest that passage “through” the proper 3′ processing machinery greatly facilitates access to the mRNA export pathway but do not indicate how mRNA export depends on 3′ processing. This view is supported by recent observations that mRNA export is inefficient in some mutant yeast strains affecting 3′ processing factors (11).
In a novel screen for additional mutants affecting mRNA export, we identified new alleles affecting 3′ processing factors. Our analysis of pap1-23 confirms previous reports that addition of a poly(A) tail is required for efficient mRNA export. What is noteworthy about our screen is that we failed to isolate mutations affecting previously characterized mRNA export factors. This probably reflects the fact that the FACS method can distinguish cells which produce low levels of Ssa4p-GFP from those which fail to produce any fluorescent reporter mRNA. The Δrip1 strain which served as our control is unique in its apparent complete export block: it fails to produce detectable cytoplasmic heat shock mRNAs after heat shock, and inducible heat shock proteins cannot be detected by gel analysis. Other previously characterized export mutants accumulate mRNAs in the nucleus, but after a short lag (15 to 20 min), reporter mRNAs are exported and translated at low levels (40). Mutations affecting 3′ processing may prevent mRNAs from entering the export pathway much more effectively than mutations affecting nucleoporins or other mRNA export factors. However, the specific degradation of mRNAs triggered by defective transcription termination may also contribute to the lack of reporter protein production in these mutants (22).
Another surprising finding is that a majority of mutations identified in this screen affected components of a single subcomplex of 3′ processing factors (CFIA), which is required to couple 3′ processing and Pol II termination (8). mRNA transport defects in these strains occur sufficiently rapidly that little or no induced heat shock mRNA is exported, indicating that the cell becomes defective for the function of the mutant protein with little delay. Although these mutations cause noticeable defects in polyadenylation in vivo and in vitro, the mRNAs which are polyadenylated are not exported (Fig. 2). This indicates that polyadenylation is necessary but not sufficient for efficient mRNA transport and further implies a functional interaction between the 3′ processing machinery and mRNA export factors. The notion that mRNA export depends on interactions between CFIA proteins and the mRNA is supported by our finding that deletion of cis-acting signals recognized by CFIA (and required for proper 3′ processing) yields elongated, polyadenylated cyc1-512 mRNAs which accumulate in nuclei (Fig. 5B).
Analysis of 3′-end formation in mutant strains revealed two distinct defects in 3′ processing. One was the production of CUP1 mRNAs polyadenylated at sites downstream from the normal 3′ processing site. The other was production of mRNAs with poly(A) tails much longer than normal. The extended CUP1 transcripts were found in all of the strains with mutations affecting CFIA proteins, consistent with CFIA involvement in accurate cleavage, termination, and poly(A) addition. However, these phenotypes were also seen in some mRNA export mutant strains.
The production of hyperpolyadenylated mRNAs with extended 3′UTRs was reported previously for prp20-1 and rna1-1 mutants (28). The production of hyperpolyadenylated transcripts was also seen in some but not all mRNA export mutants (Fig. 3F). In wild-type cells, tails become slightly longer following a shift to 37°C, and a similar slight increase was observed for yra1-1, sub2-85, kap104-16, and Δmtr10 cells. As reported previously by Jensen et al. (44) and Hilleren and Parker (40), hyperpolyadenylated transcripts were abundant in rat8-2/dbp5-2, rat7-1/nup159-1, and mex67-5 cells (Fig. 3F). It was suggested that hyperadenylation might be a general consequence of defects in mRNA export (40). Our studies indicate that this is not the case. Strong nuclear accumulation of mRNA occurs in sub2-85 and yra1-1 cells at the nonpermissive temperature, yet in both cases poly(A) tails were the same length as in the wild-type cells (Fig. 3F) (82, 83). Tails of normal length were also seen in two strains with mutations affecting protein import receptors, Δmtr10 and kap104-16 cells (Fig. 3F). These strains show modest mRNA export defects and are thought to affect mRNA transport indirectly as a consequence of their inability to reimport shuttling transport factors. These results suggest that there is a common mechanistic defect in rat7-1/nup159-1, rat8/dbp5-2, and mex67-5 cells which does not occur in other mRNA export-defective strains.
Interestingly, a defect in polyadenylation leading to long poly(A) tails was not necessarily linked to defects in cleavage and termination that result in production of mRNAs with extended 3′UTRs. Improperly terminated mRNAs were produced in some but not all mRNA export mutants examined (Fig. 4B). Among mutants producing long poly(A) tails, defective termination was strongest in mex67-5 cells and not seen at all in rat7-1/nup159-1 or rat8-2/dbp5-2 cells. A very strong termination defect was also seen in xpo1-1 cells. Among mutants which did not produce long poly(A) tails, kap104-16 cells showed a strong cleavage or termination defect, but no such defect was seen in yra1-1 or sub2-85 cells. We conclude that two different (and perhaps overlapping) deficiencies must underlie the production of long poly(A) tails and the defects in cleavage and termination (Fig. 3F and Fig. 4B and C).
One possibility is that the availability or activity of CFIA is affected in some but not all mRNA export mutant strains. Another possibility, which is not mutually exclusive, is that some defects in cleavage or termination and polyadenylation may reflect direct involvement of mRNA export factors in 3′ processing. Defects in Kap104p activity can easily account for termination defects, as kap104-16 mutants mislocalize Hrp1p, a critical component required for both cleavage and polyadenylation, and Nab2p, a general mRNA-binding protein recently implicated in mRNA export and poly(A) tail length control (31, 37, 46). Both Nab2p and Hrp1p are shuttling proteins and are predominantly found in nuclei under steady-state conditions (2). They are passively exported and move to the cytoplasm only when associated with mRNA (46). However, we found that the location of Hrp1p and Nab2p was not altered in rat8-2dbp5-2, rat7-1/nup159-1, xpo1-1, or mex67-5 cells (C. M. Hammell and C. N. Cole, unpublished observations). This indicates that the 3′ processing defects seen in these strains reflect nuclear events.
Jensen et al. have reported that defects in mRNA export result in the production of hyperpolyadenylated transcripts that accumulate at or near the sites of transcription (44). Even though cleaved, unadenylated transcripts can be found in actively translating polyribosomes, hypopolyadenylated transcripts are also retained at the site of transcription (23, 39, 69). Improperly processed transcripts (both hypo- and hyperpolyadenylated) are actively retained by a novel function of the nuclear exosome (39). It appears that significant distinctions between exportable and nonexportable transcripts are made at the level of interactions on the poly(A) tail. Presumably, mutations affecting any one of a subset of mRNA export factors prevent efficient coupling of 3′ processing to mRNA export. One potential coupling point is the loading of poly(A)-binding protein Pab1p onto the poly(A) tail.
Do Pab1p and Xpo1p function in both 3′ processing and mRNA export?
We have shown that a mutation of XPO1 causes mRNA 3′-end processing defects in vivo. Furthermore, a strong two-hybrid interaction was found between Xpo1p and Pab1p. These results suggest a model in which Pab1p and Xpo1p are important for both 3′ processing and mRNA export and interact in the nucleus during these events. The strength and LMB sensitivity of the two-hybrid interaction between Xpo1p and Pab1p was equal to those of the interaction between Xpo1p and Rev. Our recent experiments indicate that Pab1p shuttles in an Gsp1p/Xpo1p-dependent manner (Hammell and Cole, unpublished). Physical interactions between Pab1p and Xpo1p can also be detected using recombinant proteins (K. Weis, personal communication). Xpo1p also exhibits a weaker two-hybrid interaction with CFIA components, in agreement with an equimolar amount of Pab1p purifying with CFIA from yeast whole-cell extract (6, 61).
The stimulation of in vitro 3′ processing by Xpo1p and the sensitivity of this stimulation to LMB are also consistent with an interaction (direct or indirect) between Xpo1p and 3′ processing factors (Table 2 and Fig. 6A and B). We cannot rule out the possibility that Xpo1p affects 3′ processing by titrating an inhibitor present in the extract. However, loss of function of Xpo1p activity results in a termination phenotype similar to those found in CFIA mutants (Fig. 4A and C). One possibility is that interactions between Xpo1p and 3′ processing factors enhance processing in vitro by stabilizing the processing complex. Overexpression of Pab1p in vivo suppresses specific temperature-sensitive defects of RNA15 mutants, even though pab1 mutants do not appear to directly affect cleavage activity in vitro (6).
Several lines of evidence suggest that Pab1p may play an important role in mRNA export. Overexpression of Pab1p can suppress both the mRNA export and polyadenylation defects seen in nab2 mutants (37). Yeast Pab1p and both metazoan poly(A)-binding proteins (PABI and PABII) shuttle (1, 12) in an Xpo1p-dependent manner (Hammell and Cole, unpublished). Yeast Pab1p interacts with specific nucleoporins (directly or indirectly) in a manner that is insensitive to RNase and stimulated by the addition of Gsp1-GTP (3). In metazoan cells, the influenza virus protein NS1A prevents host mRNA export and has recently been shown to interact with the nuclear poly(A)-binding protein (PABII) and CPSF30, a component of the metazoan polyadenylation machinery (18, 29). NS1A can bind CPSF30 and PABII via nonoverlapping domains, suggesting that NS1A may exert its dominant effect on host export by preventing the two proteins from interacting functionally with one another in vivo (18).
In addition to the formation of the mRNA 3′ ends, another role for the cleavage and polyadenylation machinery may be to facilitate the interaction of Pab1p with the newly synthesized poly(A) tail. We suggest that proper recruitment of Pab1p onto the poly(A) tail is an important contribution of 3′ processing to mRNA export and that this loading is essential for efficient mRNA export. Once loaded, Pab1p is positioned to participate functionally in the export process. Interestingly, mutations of pab1 also result in a short temporal lag in the association of mRNA with the cytoplasmic degradation and translation machinery, as is the case in strains with mutations affecting certain mRNA export factors (13, 40).
Overexpression of Rat8/Dbp5p completely suppresses the 3′ processing defect seen in xpo1-1 cells (Fig. 4C). Previously, we showed that this also suppresses the mRNA export defect of xpo1-1 cells (41). This may reflect the need for Rat8/Dbp5p to localize to the cytoplasmic face of NPCs where it could function in the removal of hnRNP proteins during export of mRNPs. However, Rat8p/Dbp5p is a shuttling protein and may also have essential functions in the nucleus (41, 91). If Hrp1p is the only component of CFI which is exported, CFIA (Rna14p-Rna15p-Clp1p-Pcf11p) must be dissociated from Hrp1p and the processed mRNA in order for both efficient export of the transcript and intranuclear recycling of CFIA to proceed. Defects in dissociation of these factors would prevent efficient mRNA export by sequestering CFIA, preventing future rounds of processing by these factors. Perhaps the RNA-dependent ATPase activity of Rat8p/Dbp5p is required for this disassociation and, in fact, couples correct 3′-end formation and export.
Evidence for mechanistic coupling among the multiple stages of mRNA biogenesis and function has increased dramatically in the past few years. Determining how 3′ processing and mRNA export are linked functionally will require a much better understanding of when and where mRNA export factors act, what they do, and how their functions are affected, spatially and mechanistically. This endeavor is complicated by the very large number of other proteins important for the production and metabolism of mRNA. An important next step will be the isolation and characterization of mRNP complexes associated with these and other improperly processed mRNAs. Understanding the differences between the RNA-protein complexes formed under normal and processing-defective conditions should greatly increase our understanding of how improperly processed transcripts are distinguished from those that are processed correctly and efficiently transported to the cytoplasm.
Acknowledgments
We thank members of the Cole, Moore, and Stutz laboratories for discussions and for critical review of this manuscript. We thank Ewan Dunn for consistent discussion and input into this project. We also thank Francoise Wyers, N. Proudfoot, Pamela Silver, Ed Hurt, Marvin Wickens, Steve Elledge, and Erica Golemis for strains and plasmids.
This research was supported in part by grants to C. N. Cole from the National Science Foundation (9983378) and National Institute of General Medical Sciences, National Institutes of Health (GM33998), by a grant to C. A. Moore from the National Institutes of Health (GM41752), and by a grant to F. Stutz from the Swiss National Science Foundation (3100-61378). C. M. Hammell was supported by a training grant (CA09658) from the National Cancer Institute, National Institutes of Health.
REFERENCES
- 1.Afonina, E., R. Stauber, and G. N. Pavlakis. 1998. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 273:13015-13021. [DOI] [PubMed] [Google Scholar]
- 2.Aitchison, J. D., G. Blobel, and M. P. Rout. 1996. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science 274:624-627. [DOI] [PubMed] [Google Scholar]
- 3.Allen, N. P., L. Huang, A. Burlingame, and M. Rexach. 2001. Proteomic analysis of nucleoporin interacting proteins. J. Biol. Chem. 276:29268-29274. [DOI] [PubMed] [Google Scholar]
- 4.Amberg, D. C., M. Fleischmann, I. Stagljar, C. N. Cole, and M. Aebi. 1993. Nuclear PRP20 protein is required for mRNA export. EMBO J. 12:233-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amberg, D. C., A. L. Goldstein, and C. N. Cole. 1992. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 6:1173-1189. [DOI] [PubMed] [Google Scholar]
- 6.Amrani, N., M. Minet, M. Le Gouar, F. Lacroute, and F. Wyers. 1997. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol. Cell. Biol. 17:3694-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amrani, N., M. Minet, F. Wyers, M. E. Dufour, L. P. Aggerbeck, and F. Lacroute. 1997. PCF11 encodes a third protein component of yeast cleavage and polyadenylation factor I. Mol. Cell. Biol. 17:1102-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birse, C. E., L. Minvielle-Sebastia, B. A. Lee, W. Keller, and N. J. Proudfoot. 1998. Coupling termination of transcription to messenger RNA maturation in yeast. Science 280:298-301. [DOI] [PubMed] [Google Scholar]
- 9.Bonneaud, N., L. Minvielle-Sebastia, C. Cullin, and F. Lacroute. 1994. Cellular localization of RNA14p and RNA15p, two yeast proteins involved in mRNA stability. J. Cell Sci. 107:913-921. [DOI] [PubMed] [Google Scholar]
- 10.Boorstein, W. R., and E. A. Craig. 1990. Structure and regulation of the SSA4 HSP70 gene of Saccharomyces cerevisiae. J. Biol. Chem. 265:18912-18921. [PubMed] [Google Scholar]
- 11.Brodsky, A. S., and P. A. Silver. 2000. Pre-mRNA processing factors are required for nuclear export. RNA 6:1737-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calado, A., U. Kutay, U. Kuhn, E. Wahle, and M. Carmo-Fonseca. 2000. Deciphering the cellular pathway for transport of poly(A)-binding protein II. RNA 6:245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caponigro, G., and R. Parker. 1995. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev. 9:2421-2432. [DOI] [PubMed] [Google Scholar]
- 14.Chang, D. D., and P. A. Sharp. 1989. Regulation by HIV Rev depends upon recognition of splice sites. Cell 59:789-795. [DOI] [PubMed] [Google Scholar]
- 15.Chang, M., D. French-Cornay, H. Y. Fan, H. Klein, C. L. Denis, and J. A. Jaehning. 1999. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol. Cell. Biol. 19:1056-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chavez, S., T. Beilharz, A. G. Rondon, H. Erdjument-Bromage, P. Tempst, J. Q. Svejstrup, T. Lithgow, and A. Aguilera. 2000. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J. 19:5824-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen, J., and C. Moore. 1992. Separation of factors required for cleavage and polyadenylation of yeast pre-mRNA. Mol. Cell. Biol. 12:3470-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen, Z., Y. Li, and R. M. Krug. 1999. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J. 18:2273-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clouse, K. N., M. J. Luo, Z. Zhou, and R. Reed. 2001. A Ran-independent pathway for export of spliced mRNA. Nat. Cell Biol. 3:97-99. [DOI] [PubMed] [Google Scholar]
- 20.Cole, C. N. 2001. Choreographing mRNA biogenesis. Nat Genet. 29:6-7. [DOI] [PubMed] [Google Scholar]
- 21.Custodio, N., M. Carmo-Fonseca, F. Geraghty, H. S. Pereira, F. Grosveld, and M. Antoniou. 1999. Inefficient processing impairs release of RNA from the site of transcription. EMBO J. 18:2855-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das, B., Z. Guo, P. Russo, P. Chartrand, and F. Sherman. 2000. The role of nuclear cap binding protein Cbc1p of yeast in mRNA termination and degradation. Mol. Cell. Biol. 20:2827-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duvel, K., O. Valerius, D. A. Mangus, A. Jacobson, and G. H. Braus. 2002. Replacement of the yeast TRP4 3′ untranslated region by a hammerhead ribozyme results in a stable and efficiently exported mRNA that lacks a poly(A) tail. RNA 8:336-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eckner, R., W. Ellmeier, and M. L. Birnstiel. 1991. Mature mRNA 3′ end formation stimulates RNA export from the nucleus. EMBO J. 10:3513-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan, H. Y., R. J. Merker, and H. L. Klein. 2001. High-copy-number expression of Sub2p, a member of the RNA helicase superfamily, suppresses hpr1-mediated genomic instability. Mol. Cell. Biol. 21:5459-5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feder, M. E., and G. E. Hofmann. 1999. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu. Rev. Physiol. 61:243-282. [DOI] [PubMed] [Google Scholar]
- 27.Fong, N., and D. L. Bentley. 2001. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev. 15:1783-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forrester, W., F. Stutz, M. Rosbash, and M. Wickens. 1992. Defects in mRNA 3′-end formation, transcription initiation, and mRNA transport associated with the yeast mutation prp20: possible coupling of mRNA processing and chromatin structure. Genes Dev. 6:1914-1926. [DOI] [PubMed] [Google Scholar]
- 29.Fortes, P., A. Beloso, and J. Ortin. 1994. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks mRNA nucleocytoplasmic transport. EMBO J. 13:704-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorsch, L. C., T. C. Dockendorff, and C. N. Cole. 1995. A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J. Cell Biol. 129:939-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Green, D. M., K. A. Marfatia, E. B. Crafton, X. Zhang, X. Cheng, and A. H. Corbett. 2002. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J. Biol. Chem. 4:4. [DOI] [PubMed] [Google Scholar]
- 32.Gross, S., and C. Moore. 2001. Five subunits are required for reconstitution of the cleavage and polyadenylation activities of Saccharomyces cerevisiae cleavage factor I. Proc. Natl. Acad. Sci. USA 98:6080-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gross, S., and C. L. Moore. 2001. Rna15 interaction with the a-rich yeast polyadenylation signal is an essential step in mRNA 3′-end formation. Mol. Cell. Biol. 21:8045-8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo, Z., and F. Sherman. 1996. 3′-end-forming signals of yeast mRNA. Trends Biochem. Sci. 21:477-481. [DOI] [PubMed] [Google Scholar]
- 35.Guo, Z., and F. Sherman. 1995. 3′-end-forming signals of yeast mRNA. Mol. Cell. Biol. 15:5983-5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamm, J., and I. W. Mattaj. 1990. Monomethylated cap structures facilitate RNA export from the nucleus. Cell 63:109-118. [DOI] [PubMed] [Google Scholar]
- 37.Hector, R. E., K. R. Nykamp, S. Dheur, J. T. Anderson, P. J. Non, C. R. Urbinati, S. M. Wilson, L. Minvielle-Sebastia, and M. S. Swanson. 2002. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J. 21:1800-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herold, A., M. Suyama, J. P. Rodrigues, I. C. Braun, U. Kutay, M. Carmo-Fonseca, P. Bork, and E. Izaurralde. 2000. TAP (NXF1) belongs to a multigene family of putative RNA export factors with a conserved modular architecture. Mol. Cell. Biol. 20:8996-9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hilleren, P., T. McCarthy, M. Rosbash, R. Parker, and T. H. Jensen. 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413:538-542. [DOI] [PubMed] [Google Scholar]
- 40.Hilleren, P., and R. Parker. 2001. Defects in the mRNA export factors Rat7p, Gle1p, Mex67p, and Rat8p cause hyperadenylation during 3′-end formation of nascent transcripts. RNA 7:753-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hodge, C. A., H. V. Colot, P. Stafford, and C. N. Cole. 1999. Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1-1 cells. EMBO J. 18:5778-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hopper, A. K., H. M. Traglia, and R. W. Dunst. 1990. The yeast RNA1 gene product necessary for RNA processing is located in the cytosol and apparently excluded from the nucleus. J. Cell Biol. 111:309-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang, Y., and G. C. Carmichael. 1996. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell. Biol. 16:1534-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jensen, T. H., K. Patricio, T. McCarthy, and M. Rosbash. 2001. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell 7:887-898. [DOI] [PubMed] [Google Scholar]
- 45.Kataoka, N., J. Yong, V. N. Kim, F. Velazquez, R. A. Perkinson, F. Wang, and G. Dreyfuss. 2000. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell 6:673-682. [DOI] [PubMed] [Google Scholar]
- 46.Kessler, M. M., M. F. Henry, E. Shen, J. Zhao, S. Gross, P. A. Silver, and C. L. Moore. 1997. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 11:2545-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim, V. N., J. Yong, N. Kataoka, L. Abel, M. D. Diem, and G. Dreyfuss. 2001. The Y14 protein communicates to the cytoplasm the position of exon-exon junctions. EMBO J. 20:2062-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kudo, N., N. Matsumori, H. Taoka, D. Fujiwara, E. P. Schreiner, B. Wolff, M. Yoshida, and S. Horinouchi. 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 96:9112-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kunzler, M., and E. C. Hurt. 1998. Cse1p functions as the nuclear export receptor for importin alpha in yeast. FEBS Lett. 433:185-190. [DOI] [PubMed] [Google Scholar]
- 50.Lee, M. S., M. Henry, and P. A. Silver. 1996. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 10:1233-1246. [DOI] [PubMed] [Google Scholar]
- 51.Legrain, P., and M. Rosbash. 1989. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell 57:573-583. [DOI] [PubMed] [Google Scholar]
- 52.Le Hir, H., D. Gatfield, E. Izaurralde, and M. J. Moore. 2001. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 20:4987-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Le Hir, H., E. Izaurralde, L. E. Maquat, and M. J. Moore. 2000. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19:6860-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Le Hir, H., M. J. Moore, and L. E. Maquat. 2000. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev. 14:1098-1108. [PMC free article] [PubMed] [Google Scholar]
- 55.Lei, E. P., H. Krebber, and P. A. Silver. 2001. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev. 15:1771-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luo, M. J., and R. Reed. 1999. Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl. Acad. Sci. USA 96:14937-14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luo, M. L., Z. Zhou, K. Magni, C. Christoforides, J. Rappsilber, M. Mann, and R. Reed. 2001. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413:644-647. [DOI] [PubMed] [Google Scholar]
- 58.McNeil, J. B., H. Agah, and D. Bentley. 1998. Activated transcription independent of the RNA polymerase II holoenzyme in budding yeast. Genes Dev. 12:2510-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Minvielle-Sebastia, L., and W. Keller. 1999. mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr. Opin. Cell Biol. 11:352-357. [DOI] [PubMed] [Google Scholar]
- 60.Minvielle-Sebastia, L., P. J. Preker, and W. Keller. 1994. RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3′-end processing factor. Science 266:1702-1705. [DOI] [PubMed] [Google Scholar]
- 61.Minvielle-Sebastia, L., P. J. Preker, T. Wiederkehr, Y. Strahm, and W. Keller. 1997. The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in premessenger RNA 3′-end formation. Proc. Natl. Acad. Sci. USA 94:7897-7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Minvielle-Sebastia, L., B. Winsor, N. Bonneaud, and F. Lacroute. 1991. Mutations in the yeast RNA14 and RNA15 genes result in an abnormal mRNA decay rate: sequence analysis reveals an RNA-binding domain in the RNA15 protein. Mol. Cell. Biol. 11:3075-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neville, M., and M. Rosbash. 1999. The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 18:3746-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Neville, M., F. Stutz, L. Lee, L. I. Davis, and M. Rosbash. 1997. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr. Biol. 7:767-775. [DOI] [PubMed] [Google Scholar]
- 65.Pikielny, C. W., and M. Rosbash. 1985. mRNA splicing efficiency in yeast and the contribution of non-conserved sequences. Cell 41:119-126. [DOI] [PubMed] [Google Scholar]
- 66.Piruat, J. I., and A. Aguilera. 1998. A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J. 17:4859-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Preker, P. J., M. Ohnacker, L. Minvielle-Sebastia, and W. Keller. 1997. A multisubunit 3′ end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J. 16:4727-4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 69.Proweller, A., and S. Butler. 1994. Efficient translation of poly(A)-deficient mRNAs in Saccharomyces cerevisiae. Genes Dev. 8:2629-2640. [DOI] [PubMed] [Google Scholar]
- 70.Rose, M. D. 1987. Isolation of genes by complementation in yeast. Methods Enzymol. 152:481-504. [DOI] [PubMed] [Google Scholar]
- 71.Ruden, D. M., J. Ma, Y. Li, K. Wood, and M. Ptashne. 1991. Generating yeast transcriptional activators containing no yeast protein sequences. Nature 350:250-252. [DOI] [PubMed] [Google Scholar]
- 72.Russo, P. 1995. Saccharomyces cerevisiae mRNA 3′ end forming signals are also involved in transcription termination. Yeast 11:447-453. [DOI] [PubMed] [Google Scholar]
- 73.Russo, P., W. Z. Li, Z. Guo, and F. Sherman. 1993. Signals that produce 3′ termini in CYC1 mRNA of the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 13:7836-7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Russo, P., W. Z. Li, D. M. Hampsey, K. S. Zaret, and F. Sherman. 1991. Distinct cis-acting signals enhance 3′ endpoint formation of CYC1 mRNA in the yeast Saccharomyces cerevisiae. EMBO J. 10:563-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saavedra, C., K. S. Tung, D. C. Amberg, A. K. Hopper, and C. N. Cole. 1996. Regulation of mRNA export in response to stress in Saccharomyces cerevisiae. Genes Dev. 10:1608-1620. [DOI] [PubMed] [Google Scholar]
- 76.Saavedra, C. A., C. M. Hammell, C. V. Heath, and C. N. Cole. 1997. Yeast heat shock mRNAs are exported through a distinct pathway defined by Rip1p. Genes Dev. 11:2845-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Segref, A., K. Sharma, V. Doye, A. Hellwig, J. Huber, R. Luhrmann, and E. Hurt. 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 16:3256-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 79.Snay-Hodge, C. A., H. V. Colot, A. L. Goldstein, and C. N. Cole. 1998. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 17:2663-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stade, K., C. S. Ford, C. Guthrie, and K. Weis. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90:1041-1050. [DOI] [PubMed] [Google Scholar]
- 81.Strasser, K., J. Bassler, and E. Hurt. 2000. Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J. Cell Biol. 150:695-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Strasser, K., and E. Hurt. 2001. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature 413:648-652. [DOI] [PubMed] [Google Scholar]
- 83.Strasser, K., and E. Hurt. 2000. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 19:410-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Strasser, K., S. Masuda, P. Mason, J. Pfannstiel, M. Oppizzi, S. Rodriguez-Navarro, A. G. Rondon, A. Aguilera, K. Struhl, R. Reed, and E. Hurt. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 28:28. [DOI] [PubMed] [Google Scholar]
- 85.Stutz, F., J. Kantor, D. Zhang, T. McCarthy, M. Neville, and M. Rosbash. 1997. The yeast nucleoporin rip1p contributes to multiple export pathways with no essential role for its FG-repeat region. Genes Dev. 11:2857-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wolff, B., J. J. Sanglier, and Y. Wang. 1997. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 4:139-147. [DOI] [PubMed] [Google Scholar]
- 87.Zaret, K. S., and F. Sherman. 1982. DNA sequence required for efficient transcription termination in yeast. Cell 28:563-573. [DOI] [PubMed] [Google Scholar]
- 88.Zaret, K. S., and F. Sherman. 1984. Mutationally altered 3′ ends of yeast CYC1 mRNA affect transcript stability and translational efficiency. J. Mol. Biol. 177:107-135. [DOI] [PubMed] [Google Scholar]
- 89.Zenklusen, D., and F. Stutz. 2001. Nuclear export of mRNA. FEBS Lett. 498:150-156. [DOI] [PubMed] [Google Scholar]
- 90.Zhao, J., L. Hyman, and C. Moore. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63:405-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhao, J., S. B. Jin, B. Bjorkroth, L. Wieslander, and B. Daneholt. 2002. The mRNA export factor Dbp5 is associated with Balbiani ring mRNP from gene to cytoplasm. EMBO J. 21:1177-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao, J., M. Kessler, S. Helmling, J. P. O'Connor, and C. Moore. 1999. Pta1, a component of yeast CF II, is required for both cleavage and poly(A) addition of mRNA precursor. Mol. Cell. Biol. 19:7733-7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou, Z., M. J. Luo, K. Straesser, J. Katahira, E. Hurt, and R. Reed. 2000. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407:401-405. [DOI] [PubMed] [Google Scholar]