Abstract
Four 20 ns molecular dynamics simulations have been performed with two counterions, K+ or Na+, at two water contents, 15 or 20 H2O per nucleotide. A hexagonal simulation cell comprised of three identical DNA decamers [d(5′-ATGCAGTCAG) × d(5′-TGACTGCATC)] with periodic boundary condition along the DNA helix was used. The simulation setup mimics the DNA state in oriented DNA fibers or in crystals of DNA oligomers. Variation of counterion nature and water content do not alter averaged DNA structure. K+ and Na+ binding to DNA are different. K+ binds to the electronegative sites of DNA bases in the major and the minor grooves, while Na+ interacts preferentially with the phosphate groups. Increase of water causes a shift of both K+ and Na+ from the first hydration shell of O1P/O2P and of the DNA bases in the minor groove with lesser influence for the cation binding to the bases in the major groove. Mobility of both water and cations in the K–DNA systems is faster than in the Na–DNA systems: Na+ organizes and immobilizes water structure around itself and near DNA while for K+ water is less organized and more dynamic.
INTRODUCTION
Binding of different proteins through several layers of solvent to DNA is dependent on the DNA sequence and local structure, which in turn is affected by the presence and interactions with counterions and water (1–4). Therefore, efforts must be made in studying how counterions influence DNA structure and modify the electrostatic interactions with proteins in order to understand mechanisms and forces behind DNA–protein recognition and binding. Together with experimental methods (X-ray crystallography, NMR and fluorescent techniques), molecular dynamics (MD) computer simulations can address this question in much detail and many studies have been performed to this end with MD [see recent reviews (5–7)].
There have been many publications regarding sequence-specific and conformation-specific interactions of DNA with counterions (8–10) (and references cited therein). This was stimulated by the work by Young et al. (11), proposing that strongly electronegative pockets in the Dickerson–Drew dodecamer (DDD) (12) (where presence of solvent molecules was detected) harbour Na+ ions. By using MD simulation, relatively long residence times for ions were suggested in this (11) and following papers (13,14), in some cases matching or exceeding those of well-ordered water molecules. The reliability of MD simulation results depends on the accuracy of the model for the physical interactions and sufficiently long simulation times to obtain meaningful averages of equilibrium populations (14). Following recent advances in methodology and computational power, it is now possible to simulate nucleic acid fragments in explicit solvent over tenths of nanoseconds (5,7,15,16). Recent versions of the force fields demonstrate good agreement with experimental data on nucleic acid and solvent structure and dynamics (13,16,17) lending support to the general validity of the theoretical models.
In most MD simulation studies, DNA oligomers are placed in water solution with sodium as a counterion. However, potassium but not sodium dominates the cytoplasm of practically all living cells. Direct and detailed comparison of binding and dynamics of K+ and Na+ in interaction with DNA is important since in vitro and in silico studies are dominated by the less physiologically relevant sodium cation. Experimental data report small and contradictory selectivity of DNA for either Na+ or K+ under a variety of conditions. Methods based on NMR relaxation measurements report higher affinity of DNA for K+ relative to Na+ (18–20) whereas determination of the Na+/K+ amount absorbed on DNA in competitive experiments shows absence of K+/Na+ selectivity or small DNA selectivity for Na+ under a variety of conditions (21–23). Similar binding of the cations might be expected from their close similarity. However, experimentally determined equivalence in the binding averaged over all sites in DNA (or any other molecule) may actually hide actual different binding preferences of the K+ and Na+ in interaction with various electronegative sites of the complex DNA molecule. The minor differences in K+ and Na+ preferences of binding sites in DNA (or other biomolecules) can be crucially important when these differences are accumulated during cooperative structural transition or in reactions involving multiple residues (24). Examples include the selectivity of potassium channels of the cell membrane that separate between K+ and Na+ (25), the finding that a model anionic polypeptide, poly-l-glutamic acid (PGA), is unable to form α-helix in water–alcohol mixtures in the presence of K+ ions whereas Na-salts of PGA exhibit complete transformation from coil to α-helix above certain critical concentration of alcohol (26) and finally the strong dependence on the alkali cation nature of DNA compaction in crowded environment (27).
X-ray crystallography data obtained with DDD oligomers indicate higher probability for K+ than for Na+ to partially substitute water in the well-structured ‘spine of hydration’ (28,29) which might be explained not only by higher K+ affinity for the DNA sites but could be a consequence of higher sensitivity in detection of K+ by X-ray diffraction due to its higher electron density while Na+ is isoelectronic with oxygen and therefore may not be accurately distinguished from water.
So far there have been few simulation studies of nucleic acids with K+ as the counterion (30–33). The recent work by Varnai and Zakrzewska (33) points out variations in interaction of Na+ and K+ with DNA. However, that study does not address the important issue of the interplay between the dynamic cation–DNA interaction and DNA hydration. Another discrepancy between most MD simulation conditions and the situation in vivo or in crystals of DNA oligomers or in oriented DNA fibers, is that those simulations have mostly been carried out under dilute DNA solution condition and the lower water activity in the experimental systems as well as the explicit DNA–DNA interactions and the polymeric nature of this biopolymer are not captured.
In this work we compare the DNA interactions of the two counterions K+ and Na+ and how they influence DNA structure and water binding. Four systems have been studied in a periodic hexagonal cell containing three identical B-DNA decamers in explicit water environment, and containing either K+ or Na+ at two different water content, 15 and 20 H2O per nucleotide. These systems model an infinite array of parallel DNA in oriented DNA fibers or crystals. The presence of three DNA oligomers in the simulation cell automatically takes DNA–DNA interactions into consideration. Periodic conditions applied along the DNA helix axis mimics the conditions of the polymeric DNA or DNA crystals where the DNA oligomers are aligned parallel to each other. To our knowledge this work is the first systematic comparative MD simulation study of K–DNA and Na–DNA which includes analysis not only of K+/Na+–DNA interactions but also describe how these cations alter structure and dynamics of DNA hydration.
COMPUTATIONAL METHODS
We report the results of four MD simulations, using the CHARMM27 force field (34,35) with potassium and sodium as counterions. The MD simulations are abbreviated K/15, K/20, Na/15 and Na/20 where 15 or 20 denote the number of water molecules per nucleotide.
Real analogues of the Na/15 and Na/20 systems, oriented Na–DNA fibers prepared by the wet spinning technique (36) and with similar water content, have been studied previously by NMR diffusion and relaxation techniques (37) (and references cited therein). The Na/15 system corresponds to a Na–DNA fiber system in equilibrium with air of 85% relative humidity (RH), while the Na/20 system mimics Na–DNA fibers at 95% RH (37,38).
MD simulations have been carried out in a hexagonal cell with periodic conditions imposed in all directions. All the simulation cells contain three identical B-DNA decamers of the sequence d(5′-ATGCAGTCAG) × d(5′-TGACTGCATC) with ‘sticky’ ends which means that the 3′ end of the each DNA strand is connected to its periodic image along the Z-axis (periodic boundary condition). This setup mimics an infinite array of parallel ordered DNA in fibers. Water is represented by the flexible SPC model (39) with 900 and 1200 molecules (for the M/15 and M/20 system, respectively; M = Na or K) in the simulation cell. To neutralize the negative charge of DNA, 60 Na+ or K+ counterions were added in the M/15 systems and 64 Na+/K+ cations as well as 4 Cl− co-ions were included in the M/20 systems. Van der Waals parameters for the cations (Na+ and K+) were taken from the Refs (40) and (41), respectively; constants from Ref. (40) were used for Cl−. This choice of force constants in MD simulations has demonstrated good reproduction of transport properties as well as osmotic and ion activity parameters of water and salt solutions (41–43). Satisfactory agreement between MD calculated parameters and available experimental data have also been reported for oriented fibers of Na– and K–DNA (30,38,44).The starting configurations were taken from snapshots of earlier MD simulations carried in our research group on Na/15 (38) and Na/20 (45) systems.
The Nose–Hoover thermostats and barostats (46) were applied in all the simulations to control the temperature at 300 K and the pressure at 1 atm. The Ewald summation method (47) was used to treat long-range electrostatic interactions. A double time step algorithm was implemented (48) with short time step of 0.2 fs for fast intramolecular vibrations and the short-range part (within 5 Å) of intermolecular interactions and a long time step of 2 fs for longer-range interactions. The simulation software was the M.DynaMix package developed for simulations of arbitrary mixtures of molecules and macromolecules in solutions using parallel computer architectures by Lyubartsev and Laaksonen (49). Each of the four systems was simulated for 20 ns after 0.5 ns of equilibration. Additional details of the MD simulation and analysis can be found in the previous work (38,45,50). Parameters of the DNA structure were calculated by the program Curves 5.2 (51) and trajectory analysis was performed by software included in the M.DynaMix package (49).
RESULTS AND DISCUSSION
General properties of the simulated systems
DNA structure
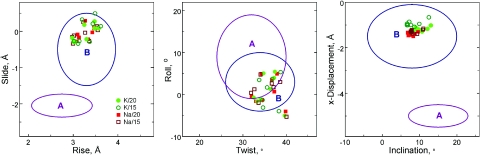
The six inter base pairs parameters (rise, twist, shift, roll, tilt and slide) of the DNA structure averaged over simulation time and over all three oligomers in the hexagonal cell have been compared with the parameters of standard A- and B-form DNA (Figure 1; additional data showing variation of these six parameters for the individual base pair steps are available in Supplementary Figure 1S). The results show that the simulations with the CHARMM27 force field, preserves the B-form of DNA with some sequence-specific flexibility. In the averaged structures (over 20 ns and three decamers), most features such as sugar pucker, torsion, groove width and depth are within values found for the B-DNA family (data not shown). Comparison of the averaged DNA structures of each of the four simulated systems shows that they are very similar with root mean square (r.m.s.) displacement below 1 Å between each other (determined for all non-hydrogen atoms). Superimposed structures coincide very well in the base positions with minor variations in the sugar-phosphate backbone. In our simulation setup, the B-form was stabilized due to topological constraints (10 bp per turn) arising from the periodic boundary conditions since the systems correspond to an infinite array of parallel DNA in oriented DNA fibers or crystals instead of individual DNA molecules in solution (38). Still, this setup leaves considerable range for variation of most of structural parameters. For example, fluctuations of the minor groove width (which may serve as indication of changes in backbone variables) are significant (typically, ±3–4 Å around averaged values displayed in Supplementary Figure 2S).
Figure 1.
Structural parameters calculated from averaged DNA coordinates by Curves 5.2 (51). Regions of the points are marked as follows: K/20, light green circles; K/15, green circles; Na/20, light red squares; Na/15, red squares; M/20 and M/15 systems are shown as closed and open symbols, respectively. Parameters were paired according to Ref. (74); the blue and purple ovals show regions typical for B- and A-DNA families, respectively, found from analysis of X-ray crystallography data. (More information is available in Supplementary Data where Supplementary Figure 2S displays the values shown in this figure in dependence of the DNA sequence).
It is of interest to study how ion and water positions affect DNA conformation. To explain the origin of DNA conformational heterogeneity two models have been proposed. The base-clash model (52,53) assumes that the conformation of the DNA duplex with a given sequence is primarily determined by the intrinsic geometry of the nucleotides and not influenced by the positions and movements of ions (at least for the case of alkali metal ions). The electrostatic model on the other hand (54) (and references cited therein), proposes that DNA conformation and dynamics are affected by the positions and fluctuations of ions through their association with electronegative sites in DNA. In the latter model ions would affect the time-dependent conformation of DNA. These two models have resulted in much debate in recent literature [e.g. (10,55)] but are somewhat difficult to test by experiments since both of them give similar average structures in agreement with experimental results. Careful analysis of correlations between cation presence and fluctuation of the local DNA structure reveal that such ions as Na+ or K+ are not capable of significant modulation of the DNA structure and observed changes of the local parameters are well within the range of thermal fluctuations (33,55–59) (and references cited therein). The data of the present work are in a full agreement with this conclusion. For example, in all systems the minor groove width shows great dynamic variation during the MD simulation. However, the dynamics of the minor groove width does not correlate with Na+ or K+ presence in the vicinity of the groove (data not shown). Furthermore, the minor groove width averaged over 20 ns and for three DNA decamers show little dependence on the nature of cation and water content (Supplementary Figure 2S). This result is in contrast to the case of polyamines as counterions, where the synthetic diaminopropane2+ and putrescine2+ and spermidine3+ were studied under the same conditions (57). In the cited work, significant alteration of the minor groove width was observed as a consequence of interaction with the polyamines. However this modulation of the DNA structure was caused by formation of multiple and relatively stable (nanoseconds) direct contacts between the polyamine and DNA.
Diffusion of water and ions
Table 1 lists some general properties of the simulated systems such as box size, density and species concentration. In addition, calculated r.m.s. displacement, 〈R〉, and diffusion coefficients, D, obtained from the simulations are presented. The data on transport properties determined in the present work using the CHARMM27 force field is in good agreement with our earlier data obtained for the Na/15 and Na/20 systems using the CHARMM22 force field and shorter (6 ns) simulation time (38) (Supplementary Table 1S). For the Na/15 and Na/20 systems reported in the work (38) we extended the simulations to 10 ns and also carried out 10 ns runs for the K/15 and K/20 systems using the CHARMM22 force field (60). With respect to ion mobility and diffusion, the results obtained by the CHARMM22 and CHARMM27 force fields in the same systems were nearly identical (Supplementary Table 1S). Water, K+, Na+ and Cl− are mobile and during the 20 ns MD run in all systems studied, the mobile species on the average sample the space exceeding the cell dimensions (see data on mean displacement in Table 1). The mobility of both water and cation in the systems with K+ is noticeably faster than that with Na+. This reflects the fundamental (although small) distinction between these alkali metal ions. Na+ is a weak ‘kosmotrope’ that organizes and immobilizes water structure around itself. K+ on the other hand is a weak ‘chaotrope’ in the sense that the hydration shell around the latter ion is less organized and more dynamic compared with the structure and mobility of pure water (61). This small but fundamental difference between K+ and Na+ is projected to their interaction with polar groups of biological systems and can result in important variations of the solvent structure and dynamics around the DNA. Comparing our results on water and cation mobility with available experimental and MD simulation results we conclude that the MD setups are in a good agreement with the data of NMR relaxation experiments with oriented DNA fibers and earlier MD simulations (37,38). The expected reduced mobility of water and ions in the present dense DNA systems relative to DNA and simple salt water solutions is well borne in the simulations. Diffusion coefficients for Na+ and K+ reported by Varnai and Zakrzewska (33) calculated from MD simulations for DNA solution (227 H2O per K+/Na+ ion) were found to exceed the experimental (62) and MD simulation (43) values obtained for dilute water solutions of KCl/NaCl (numerical data are presented in footnote to Supplementary Table 1S). It is known that the SPC/E and SPC (used here) models give a good description of transport properties in water (63), simple salt (43,64) and DNA systems (32) whereas, for example, the TIP3P model gives mobility values faster than the experimental ones, however, being correct in description of interaction potentials (65).
Table 1.
Properties of the simulated systems
| System | Density (g/cm3) | Box volume (nm3) | Species concentration (M) | Diffusion parameters: D·1010 (m2/s) [〈R〉 (Å/ns)]a |
|---|---|---|---|---|
| K/15 | 1.375 | 44.7 | 33.21 (H2O) | 4.35 (16.2) H2O |
| 2.214 (K+) | 1.76 (10.3) K+ | |||
| K/20 | 1.318 | 53.85 | 37.52 (H2O) | 7.0 (20.6) H2O |
| 2.001 (K+) | 3.52 (14.6) K+ | |||
| 0.125 (Cl−) | 3.0 (13.5) Cl− | |||
| Na/15 | 1.382 | 43.9 | 34.82 (H2O) | 2.82 (13.0) H2O |
| 2.321 (Na+) | 0.58 (5.93) Na+ | |||
| Na/20 | 1.326 | 52.2 | 38.39 (H2O) | 5.5 (18.2) H2O |
| 2.048 (Na+) | 1.33 (8.9) Na+ | |||
| 0.128 (Cl−) | 3.22 (10.3) Cl− |
a〈R〉, r.m.s. displacement.
Averaged binding and dynamics of ions and water
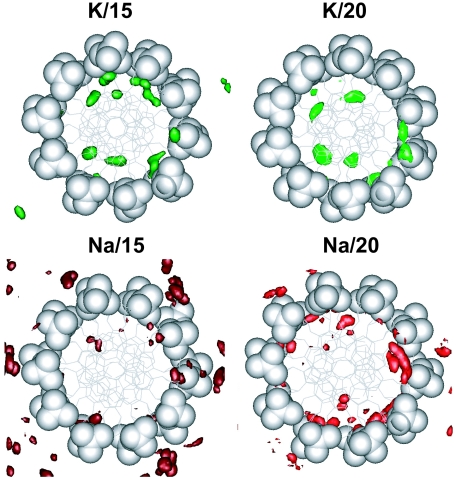
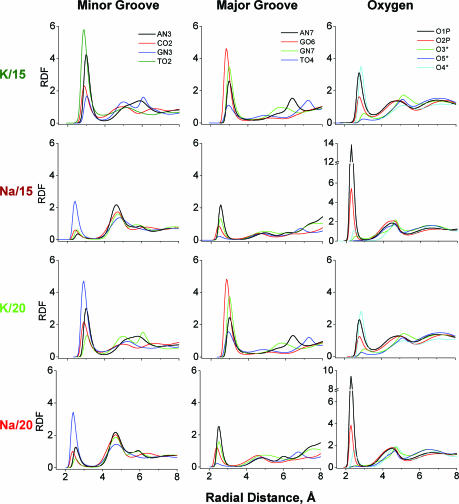
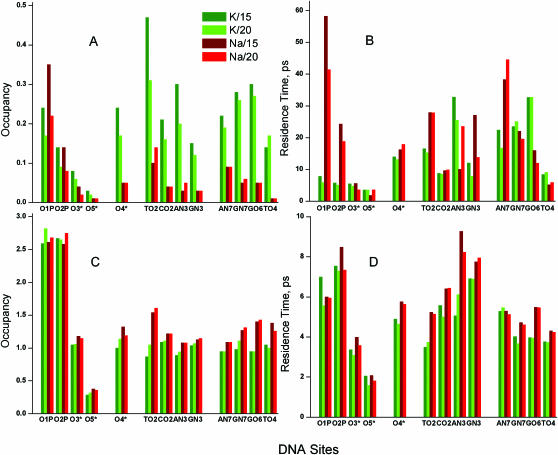
In this section we analyse K+/Na+ binding and hydration averaged over electronegative sites of DNA. Some sequence-specific binding is discussed in the following section. We consider three groups of DNA binding sites: (i) oxygen atoms of the phosphate group and the pentose ring (O1P, O2P, O3*, O5*, O4*), (ii) minor (TO2, CO2, AN3, GN3) and (iii) major (AN7, GN7, GO6, TO4) groove sites. Occupancy of ions and oxygen atom of water (Ow) of a binding site was determined by integration of the first maximum of the corresponding radial distribution function (RDF). Average residence time was calculated ignoring fast (<1 ps) fluctuations of particle across the boundary. (Detailed results are available in Supplementary Table 2S with data on occupancies, average and maximal residence times). Figure 2 displays spatial distribution functions (SDFs) of the cations around averaged DNA structures seen along the DNA helix axis. Figure 3 presents cation RDFs around the electronegative sites of DNA. In Figure 4, occupancies of ions (Figure 4A) and water (Figure 4C) as well as their averaged residence times (Figure 4B and D, respectively) are summarized.
Figure 2.
SDFs of K+ (red and dark red), Na+ (green and bright green) around averaged DNA structure seen along the DNA helix axis. To highlight the preferential presence of Na+ ions, phosphate groups have been displayed as CPK spheres with the rest of DNA shown in stick model. For the best display, the SDFs of ions in the M/15 and M/20 systems are drawn for densities >18 and >27 particles/nm3, respectively.
Figure 3.
RDFs of cations (K+ or Na+) relative to the electronegative atoms of DNA. The first and second groups of columns give sites in the minor and major grooves, respectively, with the third group showing the oxygen atoms of phosphate groups and pentose. The magnitude of RDF is units of relative density compared with uniform distribution of the given type of particles in the simulation cell.
Figure 4.
Averaged occupancy (A and C) and averaged residence time (B and D) of cations (A and B) and oxygen atoms of water (C and D) around electronegative atoms of DNA. K/15 and K/20 systems are shown in dark green and light green columns, respectively; Na/15 and Na/20 are represented by dark red and light red columns. (Numerical data on occupancies and residence times are given in Supplementary Table 2S).
Distribution of cations between phosphate group, minor and major grooves
Figure 2 clearly demonstrates that K+ ions prefer binding to the grooves of DNA while Na+ ions reside near the phosphate groups outside the DNA or on the edges of the DNA grooves separated from the sites on the DNA bases by water. Increase of water content from 15 to 20 water molecules/nucleotide results in larger hydration of the phosphate groups of DNA and Na+ is pushed from these sites and shifted closer to the grooves of DNA. The same clear difference between K+ and Na+ interaction with DNA is also seen from the RDF curves (Figure 3) and occupancy values (Figure 4). Potassium ions prefer direct binding to the electronegative sites of the DNA bases and to the O4* atom of pentose; whereas for sodium ion the major site for direct binding is the O1P atom of the phosphate group. The high occupancy of K+ near O4* is caused by coordination of the cation both to base sites and to the O4* when K+ resides in the minor groove (see below). Interaction of Na+ with DNA via water bridges is common with a second maximum appearing in the majority of the Na+ RDFs shown in Figure 3. In the Na–DNA systems, Na+ is involved in organization of solvent structure around phosphate groups and in the minor groove. In the K–DNA systems water-mediated interaction of K+ with electronegative sites of DNA is observed only around the phosphate group near the O1P atom (Figure 3).
The increase of water content from 15 to 20 H2O/nucleotide reduces the direct binding of both Na+ and K+ to the phosphate group (Figure 4A). A drop in K+ occupancy with increase of water content is also observed in the minor groove. In previous works (31–33) no K+ binding was observed in more dilute systems. Since we see a reduced potassium binding in the minor groove with reduced DNA concentration (an effect which continues in a similar system with 75 water/nucleotide; N. Korolev and L. Nordenskiöld, unpublished data) this is in part likely to be due to a concentration effect. However, the difference compared with other work may be a result of sequence difference and variation in force field between this study (CHARMM27) and that of the cited work (AMBER) (31–33).
For Na–DNA, the picture of ion binding in the minor groove is a bit more complicated. Substantial amount of Na+ is released from the phosphate group (especially from the O1P atom) and this leads to redistribution of Na+ with a slight increase of Na+ occupancy near the TO2 and AN3 sites. In the major groove, the cation–DNA interaction displays little sensitivity to the change in water content. It is likely that the high affinity of the GO6/N7 and AN7 sites for the cations is the origin of this effect. Only a small reduction of the K+ occupancy in the major groove is observed in a system with 75 H2O/nucleotide (N. Korolev and L. Nordenskiöld, unpublished data) as compared with the present K/20 system. Furthermore, data of the MD simulations reported in the literature and carried out at much higher water content (>100 H2O/nucleotide) show significant binding of K+ (31–33) and Na+ (33,55,66) in the major groove.
Additionally, using the method described in earlier work (38), we investigated whether cation binding in the minor and in the major grooves of the same base are correlated and found no correlation (data not shown). That means an ion coordinated to the site in the minor groove ‘does not feel’ whether another ion interacts with the electronegative site of the same base facing to the major groove.
Among the oxygen atoms of the phosphate group, the O1P atom is the major binding site both for K+ and Na+. The O5* atom is hidden both for ion interaction and for hydration. In the minor groove K+ and Na+ preferably interact with the TO2 site; the other sites of the minor groove do not attract Na+ while the presence of K+ is above 10% for the AN3, CO2 and GN3 sites in the K/15 and K/20 systems. In the major groove coordination of K+ to the GN7/O6 atoms is significant; the AN7 and TO4 sites have lower (than GN7/O6) but still substantial binding of K+. The TO4 atom is the site least occupied by Na+.
Interplay between cation binding and hydration
Despite a relatively low amount of water available in the system, Na+ and K+ remain substantially hydrated with average hydration numbers 5.30 and 5.47 for Na+ and 7.28 and 7.56 for K+ in the systems with 15 and 20 H2O/nucleotide, respectively. With a few exceptions, hydration of the DNA sites is relatively uniform and does not show large dependence on number of water molecules in the system and on the nature of the cation (Figure 4C). The O1P and O2P atoms are the most hydrated sites with 2.5 water molecules or above in the first solvation shell which (with addition of occupancies of the cations) gives a coordination number of about three. Except for the TO2 site, the electronegative atoms of the DNA bases are uniformly hydrated by about 1–1.5 H2O molecules with larger hydration number for the Na–DNA systems which may be explained by higher occupancy of K+ in the K–DNA systems. The TO2 site is more exposed to the solvent than the other atoms of the DNA bases and on overall these sites interact with up to two particles of the solvent. In general, cations and water molecules compete with each other for the presence in the first solvation shell of the DNA atoms carrying partial charge with roughly constant total number of particles within this layer. The major difference between Na– and K–DNA is observed in a more pronounced organization of solvent in the second and third solvation shells for the Na/15 and Na/20 systems.
Residence times of the cations
Figure 4B displays values of average residence times RTav for Na+/K+ in the first solvation shell of the selected sites. Average and maximal residence times are also tabulated in Supplementary Table 2S. The electrostatic forces play a more important role for the mobility of the cations than do their mass and size since the lighter and smaller Na+ exchanges at a slower rate than the heavier and larger K+ (Figure 4B). These data for Na+ and water in the Na–DNA systems reflect the stronger Na+–water interaction and the more pronounced structuring of water in this system.
Residence times of Na+ near the O2P and O1P atoms of the phosphate group are longer than those of K+. The increase of water content from 15 to 20 H2O/nucleotide accelerates the exchange of the cations near the exposed O1P and O2P atoms where the most pronounced competition between cations and water is observed. Addition of water pushes Na+ and to a lesser extent K+ away from O1P (Figure 4A) and accelerates the ion–water exchange in the first shell of this site (Figure 4B). O1P is the site where the longest residence time was registered in Na–DNA with RTmax = 11.88 and 4.75 ns in the Na/15 and Na/20 systems, respectively. In the K/15 and K/20 systems the corresponding values are RTmax = 1.57 and 0.51 ns. K+ exchange is fast near all oxygen atoms of the phosphate group (Figure 4B) and similar to the RTav values of water (Figure 4D).
The O4* atom of the sugar ring is an important site for coordination with the cations in the minor groove. More details on O4* coordination with K+ and Na+ are given below in discussion of site-specific cation binding. In the minor groove, the CO2 site is characterized by relatively fast dynamics of both Na+ and K+ exchange (RTav < 10 ps).In the major groove, exchange near the TO4 site is fast in all systems (RTav is below 10 ps). Sodium ions form the most long-lived pairs with the AN7 site while K+ has the longest residence time near the GO6.
Comparing our data on residence times for Na+ and K+ with the same values reported by Varnai and Zakrzewska (33) we note that both RTav and RTmax values determined in the present work are considerably longer than those observed in Ref. (33). To some extent this difference may be explained by the lower water content of the systems studied in this work. However, the discrepancy between our results and this data may also arise from the differences in the DNA force fields as well as arising from the different models of water [SPC in the present and TIP3P in the cited work (33)]. Good agreement between experimental and MD simulated values of water and ion diffusion coefficients obtained in this and in the earlier (38,50) works is an indication of the reliability of the residence times reported in this paper. We also observe several long-lived ion–DNA and water–DNA pairs in agreement with experimentally obtained estimations of nanosecond time-scale exchange between the small fraction of ‘bound’ and the large population of loose and mobile bulk ions (20) and water (67) (see below).
Residence times of water
The exchange of water from the ion hydration site is fast with K+ exchange rate about three times faster than the corresponding data for Na+ (with RTav close to 20 ps for Na+) although some long-lived water–ion pairs exist for both of the ions. For the Na+···Ow pair, RTmax = 5.8 ns in the Na/15 system, while for the K+···Ow pair, RTmax = 3.1 ns in the K/15 system (Supplementary Table 2S). In general, Na+ attracts and organizes water molecules around itself and near the DNA. The presence of Na+ slows down the exchange of water near the DNA sites as compared with the K+ system. This effect manifests itself clearly by comparing maximum residence times. For example, in the case of the GN3 site RTmax = 0.86 ns in the K/15 system while RTmax = 3.0 ns in the Na/15 system (Supplementary Table 2S). Average residence times of water near the binding sites of the DNA are of the order 5–10 ps (Figure 4D). Recent data from femtosecond-resolved fluorescence measurements give an estimation of RTav ≈ 10–20 ps (68,69). Given the differences in systems and in experimental and theoretical criteria of measuring the fraction of bound water and in determination of RTav, this correspondence between MD and experiment is reasonable.
Sequence-specific cation binding and hydration
In ‘typical’ MD simulations (i.e. for a single short DNA oligomer in a large simulation cell at high water content), site- and sequence-specific cation binding may be influenced by the end effects (70,71) and these simulations do also not mimic the conditions of condensed DNA fibres and crystals where most experimental data on DNA structure, hydration and ion binding have been obtained. That means that site- and structure-specific affinities of the cations in interaction with DNA might be biased by the relative position of a particular site in the middle or at the end of the DNA oligomer. For example, in Ref. (33) most of the cation site-specific interaction is observed in the middle of the DNA oligomer with several attractive sites (e.g. two consecutive GN6/O6 sites in the major groove at one end of the DNA oligomer) being rarely visited.
Site-specific cation binding and hydration of DNA is dependent on the DNA sequence and on local structural specificity of DNA. At the same time, the small (of order 1 nm) size of the first solvation shell of the alkali metal ion or water molecule creates quite limited number of combinations for the ‘mosaic’ of hydrogen bond donor/acceptor sites on the DNA surface. In particular, by analysis of databases of X-ray structures of DNA oligomers, it has been shown that hydration of the DNA bases (72) and phosphate group (73) is local, i.e. it is possible to predict with good accuracy the structure of the hydration shell of any DNA sequence from building blocks, using the patterns of hydration of DNA base pairs and nucleotides (72,73). The above conclusion may be reasonably correct provided that the first solvation shell of DNA consists of only water molecules. It may be not true in the case of DNA crystals and fibers with low water content where substantial inclusion of counterions in the first shell may take place. Na+ and K+ have different structures of the solvation shell and therefore the structure of the DNA solvation may vary in dependence of the nature of counterion. Indeed, superposition of ‘solvent peaks’ around the DNA bases and nucleotides from various crystals of the DNA oligomers (crystallized from solutions with sometimes very different concentration and composition of salts) shows substantial scatter in the data (72,73) (http://beta-ndb.rutgers.edu/h2o/). This scatter may be caused not only by intrinsic features of the X-ray diffraction method (limits of resolution, B-factor values and differences in crystal packaging) but may reflect the influence from the presence of different type of counterions.
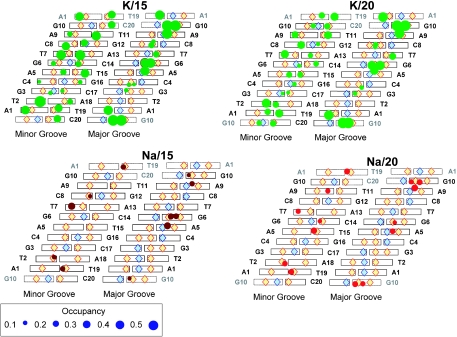
Figure 5 shows the distribution of cation occupancies near the DNA bases in the minor and in the major grooves in the four simulated systems. Much higher occupancy of the major and especially of the minor groove is observed in the systems of K–DNA compared with Na–DNA. With the increase of water content, K+ occupancies in the minor groove decrease but those in the major groove are less affected. The Na+ occupancies increase in the two grooves but only a slight increment can be seen in the major groove (Figure 4A). In both the K/15 and K/20 systems a quite uniform presence of the cation is seen in the minor groove whereas in the major groove, two similar ‘clusters’ of high K+ presence are seen. One is near the A5N7-G6O6-G6N7 atoms and the second near the A9N7-G10O6-G10N7 atoms. The latter location can be extended by one more site, T19O4 from the periodic image of the decamer. Although occupancies are lower, sequence-specific preferences of Na+ are similar to those of K+ both in the minor and in the major grooves. Below we compare in more detail cation binding and DNA hydration of the four studied systems for two regions, in the minor groove, A1T2/A18T19; and in the major groove, A5G6.
Figure 5.
Sequence-specific binding of counterions (K+ and Na+) to the sites in the minor and major grooves of DNA. Hydrogen-bond donors and acceptors of the base pairs are indicated as blue and orange rhombi, respectively. The area of the circle is proportional to the occupancy of the counterion; (sites with occupancies <0.08 are omitted). Top and bottom periodic images of the base pairs are shaded.
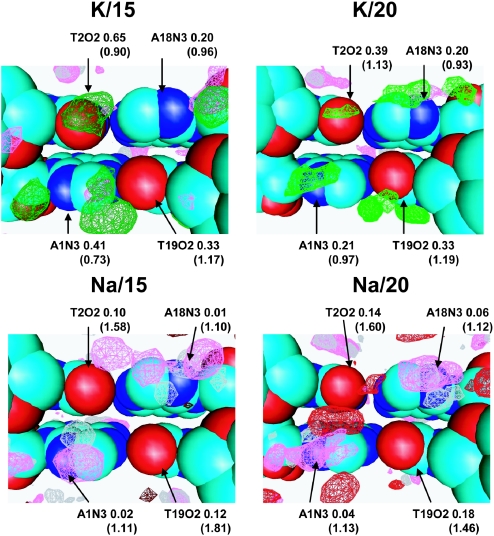
Cation binding and hydration in the minor groove of the (AT)2 fragment
Figure 6 displays SDF of the cations, water hydrogen and oxygen atoms inside the minor groove at the A1T2/A18T19 region. Comparing hydration and K+ coordination of K/15 and K/20, it can be observed that the structure of the solvation shell is similar in both systems (two top cartoons of Figure 6). There are peaks near each of the electronegative sites of the bases A1N3, T2O2, A18N3 and T19O2. Coordination of K+ at each site is similar; the cation sits in the pocket formed by the site of the base and is coordinated to the O4* of the next nucleotide. Water structure is less organized compared with systems of Na–DNA. Peaks of water SDF overlap with those of K+. Such structural arrangement of the hydration shell, AN3/TO2/GN3···Ow···O4* (with possible partial substitution of water for the counterions) is a typical motif found in the minor groove of crystals of B-DNA oligomers (72) and observed also in other MD simulations (31,32,56).
Figure 6.
SDFs of K+ (light red and dark red), Na+ (light green and dark green), oxygen (magenta) and hydrogen (grey) atoms of water in the minor groove of the A1T2/A18T19 region. Occupancies of cation and oxygen of water (in parentheses) are indicated near the corresponding AN3 and TO2 sites.
The major difference between SDFs of K– and Na–DNA is that the presence of cation in the Na/15 (and to a lesser extent also the Na/20) system is substantially lower than the corresponding potassium system. However, the positions of the SDF peaks are similar. Increase of Na+ occupancy in the Na/20 system compared with that of the Na/15 system is caused by the shift of Na+ from the phosphate group to the sites of the DNA bases. The central T2O2-T19O2 hydration site in Na/20 shows a significant presence of Na+ (bottom-right cartoon of Figure 6).
It is worth mentioning that the average picture shown in the figures is a superposition of all possible structures observed for three equivalent fragments during 20 ns of the simulations. Unlike the situation with cation coordination to the opposite sides of the DNA double helix, very rarely can two cations simultaneously reside in the vicinity of the A1T2/A18T19 fragment. For this kind of local interaction with DNA (in the same groove or near the phosphate groups), cations repel each other. We carried out an analysis of the cation–cation correlation using the method described in earlier work (38) and it was found that situations when the ions are located closer than 7–8 Å within each other are very rare and short-lived (data not shown).
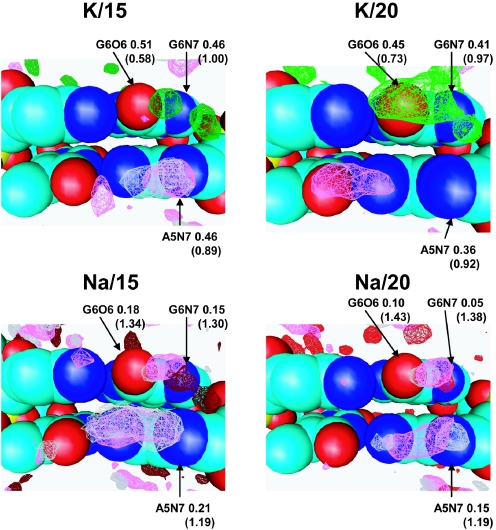
Cation binding and hydration of the A5N7-G6O6-G6N7 site
Figure 7 presents SDFs of water and cations near the ‘triangle’ of electronegative sites, A5N7-G6O6-G6N7. In all systems except Na/15, significant presence of the cations is observed at a position of coordination of a cation to all three electronegative atoms of the A5G6 bases (Figure 5). In the K/15, Na/15 and to some extent in the K/20 systems additional peaks are resolved, which correspond to coordination separately either to the G6O6 or to the G6N7 site. For the Na–DNA systems, peaks of water SDF co-localize with those of Na+. In general, as for the case of the minor groove of the A1T2/A18T19 fragment, the structure of the solvent near K– and Na–DNA is similar to water peak intensities in the K–DNA systems lower than the corresponding ones in the Na–DNA systems which display more ordered and less dynamic solvation of the DNA.
Figure 7.
SDFs of K+ (light red and dark red), Na+ (light green and dark green, oxygen (magenta) and hydrogen (grey) atoms of water in the major groove of the A5G6 region. Occupancies of cation and oxygen of water (in parentheses) are indicated near the corresponding AN7, GN7 and GO6 sites.
CONCLUSIONS
We have carried out a comparative MD simulation study of K+ and Na+ binding and dynamics in a system of parallel oriented double helical DNA molecules with low water content, mimicking conditions of oriented DNA fibers or crystals of DNA oligomers. The major results are the following:
The nature of the monovalent cation (Na+ or K+) and variation of water content (15 or 20 H2O/nucleotide) does not significantly influence the average DNA structure.
Binding preferences of K+ and Na+ are different: the major binding site of Na+ is the oxygen atoms of the phosphate group (mostly O1P) whereas K+ interacts with the electronegative sites of the DNA bases in the major and in the minor groove. Increase of water content leads to the displacement of ions (both Na+ and K+) from the first hydration shell of the phosphate group and of the DNA bases in the minor groove. Binding of the cation to the DNA bases in the major groove is less affected by competition with water.
Good agreement with experimental data on water diffusion is observed. Potassium mobility and diffusion is faster than that of sodium. Na+ ions make water structure around DNA more organized and less mobile compared with potassium.
K+ ions interact with most sites of the DNA bases in a water-sensitive way; coordination of K+ to the base is usually accompanied by direct interaction with the O4* of the pentose ring. In the major groove, both Na+ and K+ (K+ with higher occupancies) prefer to interact with clusters of electronegative sites such as combination of 5′-AN7-GN7/O6 sites on one of the DNA strands.
Analysis of the solvent structure shows that in general there is much similarity in location of water and/or cations around the DNA. However this similarity does not mean similar extent of water-to-cation substitution for Na+ and K+. Mobility of the water/counterions is noticeably faster in the K–DNA systems.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The NTU Bioinformatics Research Centre (BIRC) is gratefully acknowledged for computer resources. This work has been supported by a Singapore Ministry of Education University Research Committee (URC) grant and by a grant from the Singapore Agency for Science Technology and Research (A*STAR) through the Biomedical Research Council (BMRC). Funding to pay the Open Access publication charges for this article was provided by BMRC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kalodimos C.G., Biris N., Bonvin A.M.J.J., Levandoski M.M., Guennuegues M., Boelens R., Kaptein R. Structure and flexibility adaptation in nonspecific and specific protein–DNA complexes. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 2.Sun J., Viadiu H., Aggarwal A.K., Weinstein H. Energetic and structural considerations for the mechanism of protein sliding along DNA in the nonspecific BamHI–DNA complex. Biophys. J. 2003;84:3317–3325. doi: 10.1016/S0006-3495(03)70056-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang F., Ouporov I.V., Fernandes C., Motriuk D., Thomasson K.A. Brownian dynamics simulating the ionic-strength dependence of the nonspecific association of 434 cro repressor binding B-DNA. J. Phys. Chem. B. 2001;105:12601–12608. [Google Scholar]
- 4.Jen-Jacobson L. Protein–DNA recognition complexes: conservation of structure and binding energy in the transition state. Biopolymers. 1997;44:153–180. doi: 10.1002/(SICI)1097-0282(1997)44:2<153::AID-BIP4>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 5.Cheathan T.E., III Simulation and modeling of nucleic structure, dynamics and interactions. Curr. Opin. Struct. Biol. 2004;14:360–367. doi: 10.1016/j.sbi.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Makarov V., Pettitt B.M., Feig M. Solvation and hydration of proteins and nucleic acids: a theoretical view of simulation and experiment. Acc. Chem. Res. 2002;35:376–384. doi: 10.1021/ar0100273. [DOI] [PubMed] [Google Scholar]
- 7.Cheathan T.E., III, Young M.A. Molecular dynamics simulations of nucleic acids: successes, limitations, and promise. Biopolymers. 2001;56:232–256. doi: 10.1002/1097-0282(2000)56:4<232::AID-BIP10037>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 8.Hud N.V., Polak M. DNA–cation interactions: the major and minor grooves are flexible ionophores. Curr. Opin. Struct. Biol. 2001;11:293–301. doi: 10.1016/s0959-440x(00)00205-0. [DOI] [PubMed] [Google Scholar]
- 9.Subirana J.A., Soler-Lopez M. Cations as hydrogen bond donors: a view of electrostatic interactions in DNA. Annu. Rev. Biophys. Biomol. Struct. 2003;32:27–45. doi: 10.1146/annurev.biophys.32.110601.141726. [DOI] [PubMed] [Google Scholar]
- 10.Egli M. DNA–cation interactions: Quo vadis? Chem. Biol. 2002;9:277–286. doi: 10.1016/s1074-5521(02)00116-3. [DOI] [PubMed] [Google Scholar]
- 11.Young M.A., Jayaram B., Beveridge D.L. Intrusion of counterions into the spine of hydration in the minor groove of B-DNA: fractional occupancy of electronegative pockets. J. Am. Chem. Soc. 1997;119:59–69. [Google Scholar]
- 12.Drew H.R., Dickerson R.E. Structure of a B-DNA dodecamer III. Geometry of hydration. J. Mol. Biol. 1981;151:535–556. doi: 10.1016/0022-2836(81)90009-7. [DOI] [PubMed] [Google Scholar]
- 13.Young M.A., Jayaram B., Beveridge D.L. Local dielectric environment of B-DNA in solution: results from a 14 ns molecular dynamics trajectory. J. Phys. Chem. B. 1998;102:7666–7669. [Google Scholar]
- 14.Ponomarev S.Y., Thayer K.M., Beveridge D.L. Ion motion in molecular dynamics simulations on DNA. Proc. Natl Acad. Sci. USA. 2004;101:14771–14775. doi: 10.1073/pnas.0406435101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noy A., Perez A., Lankas F., Luque F.J., Orozco M. Relative flexibility of DNA and RNA: a molecular dynamics study. J. Mol. Biol. 2004;343:627–638. doi: 10.1016/j.jmb.2004.07.048. [DOI] [PubMed] [Google Scholar]
- 16.Norberg J., Nilsson L. Molecular dynamics applied to nucleic acids. Acc. Chem. Res. 2002;35:465–472. doi: 10.1021/ar010026a. [DOI] [PubMed] [Google Scholar]
- 17.Beveridge D.L., Dixit S.B., Barriero G., Thayer K.M. Molecular dynamics simulations of DNA curvature and flexibility: helix phasing and premelting. Biopolymers. 2004;73:380–403. doi: 10.1002/bip.20019. [DOI] [PubMed] [Google Scholar]
- 18.Bleam M.L., Anderson C.F., Record M.T. Relative binding affinities of monovalent cations for double-stranded DNA. Proc. Natl Acad. Sci. USA. 1980;77:3085–3089. doi: 10.1073/pnas.77.6.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halle B., Denisov V.P. Water and monovalent ions in the minor groove of B-DNA oligonucleotides as seen by NMR. Biopolymers. 1998;48:210–233. doi: 10.1002/(sici)1097-0282(1998)48:4<210::aid-bip3>3.3.co;2-p. [DOI] [PubMed] [Google Scholar]
- 20.Denisov V.P., Halle B. Sequence-specific binding of counterions to B-DNA. Proc. Natl Acad. Sci. USA. 2000;97:629–633. doi: 10.1073/pnas.97.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strauss U.P., Helfgott C., Pink H. Interactions of polyelectrolyte with simple electrolytes. II. Donnan equilibria obtained with DNA in solutions of 1-1 electrolytes. J. Phys. Chem. 1967;71:2550–2556. doi: 10.1021/j100867a024. [DOI] [PubMed] [Google Scholar]
- 22.Korolev N., Lyubartsev A.P., Rupprecht A., Nordenskiöld L. Competitive binding of Mg2+, Ca2+, Na+, and K+ to DNA in oriented DNA fibers: experimental and Monte Carlo simulation results. Biophys. J. 1999;77:2736–2749. doi: 10.1016/s0006-3495(99)77107-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuznetsov I.A., Gorshkov V.I., Ivanov V.A., Kargov S.I., Korolev N.I., Filippov S.M., Khamisov R.K. Ion exchange properties of immobilized DNA. React. Polym. 1984;3:37–49. [Google Scholar]
- 24.Korolev N., Nordenskiöld L. Influence of alkali cation nature on structural transitions and reactions of biopolyelectrolytes. Biomacromolecules. 2000;1:648–655. doi: 10.1021/bm000042f. [DOI] [PubMed] [Google Scholar]
- 25.Doyle D.A., Cabral J.M., Pfuetzner R.A., Kuo A., Gulbis J.M., Cohen S.L., Chait B.T., MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 26.Satoh M., Fujii Y., Kato F., Komiyama J. Solvent- and salt-induced coil-helix transition of alkali metal salts of poly(l-glutamic acid) in aqueous organic solvents. Biopolymers. 1991;31:1–10. [Google Scholar]
- 27.Zinchenko A.A., Yoshikawa K. Na+ shows a markedly higher potential than K+ in DNA compaction in a crowded environment. Biophys. J. 2005;88:4118–4123. doi: 10.1529/biophysj.104.057323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tereshko V., Minasov G., Egli M. A ‘hydrate-ion’ spine in a B-DNA minor groove. J. Am. Chem. Soc. 1999;121:3590–3595. [Google Scholar]
- 29.Shui X., Sines C.C., McFail-Isom L., VanDerveer L., Williams L.D. Structure of the potassium form of CGCGAATTCGCG: DNA deformation by electrostatic collapse around inorganic cations. Biochemistry. 1998;37:16877–16887. doi: 10.1021/bi982063o. [DOI] [PubMed] [Google Scholar]
- 30.Lyubartsev A.P., Laaksonen A. Effective potentials for ion–DNA interactions. J. Chem. Phys. 1999;111:11207–11215. [Google Scholar]
- 31.Auffinger P., Westhof E. Water and ion binding around RNA and DNA (C,G) oligomers. J. Mol. Biol. 2000;300:1113–1131. doi: 10.1006/jmbi.2000.3894. [DOI] [PubMed] [Google Scholar]
- 32.Auffinger P., Westhof E. Water and ion binding around r(UpA)12 and d(TpA)12 oligomers—comparison with RNA and DNA (CpG)12 duplexes. J. Mol. Biol. 2001;305:1057–1072. doi: 10.1006/jmbi.2000.4360. [DOI] [PubMed] [Google Scholar]
- 33.Varnai P., Zakrzewska K. DNA and its counterions: a molecular dynamics study. Nucleic Acids Res. 2004;32:4269–4280. doi: 10.1093/nar/gkh765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foloppe N., MacKerell A.D., Jr All-atom empirical force field for nucleic acids: I. Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Comput. Chem. 2000;21:86–104. [Google Scholar]
- 35.MacKerell A.D., Jr, Banavali N. All-atom empirical force field for nucleic acids: II. Application to molecular dynamics simulations of DNA and RNA in solution. J. Comput. Chem. 2000;21:105–120. [Google Scholar]
- 36.Lee S.A., Lindsay S.M., Powel J.W., Weidlich T., Tao N.J., Lewen G.D., Rupprecht A. A Brillouin scattering study of the hydration of Li– and Na–DNA films. Biopolymers. 1987;26:1637–1665. doi: 10.1002/bip.360261002. [DOI] [PubMed] [Google Scholar]
- 37.van Dam L., Korolev N., Nordenskiöld L. Polyamine mobility and effect on DNA structure in oriented DNA fibers. Nucleic Acids Res. 2002;30:419–428. doi: 10.1093/nar/30.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korolev N., Lyubartsev A.P., Laaksonen A., Nordenskiöld L. A molecular dynamics simulation study of oriented DNA with polyamine and sodium counterions:diffusion and averaged binding of water and cations. Nucleic Acids Res. 2003;31:5971–5981. doi: 10.1093/nar/gkg802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toukan K., Rahman A. Molecular-dynamics study of atomic motions in water. Phys. Rev. B Condens Matter. 1985;31:2643–2648. doi: 10.1103/physrevb.31.2643. [DOI] [PubMed] [Google Scholar]
- 40.Smith D.E., Dang L.X. Computer-simulations of NaCl association in polarizable water. J. Chem. Phys. 1994;100:3757–3766. [Google Scholar]
- 41.Heinzinger K. Computer simulations of aqueous electrolyte solutions. Physica B. 1985;131B:196–216. [Google Scholar]
- 42.Lyubartsev A.P., Laaksonen A. Osmotic and activity coefficient from effective potentials for hydrated ions. Phys. Rev. E. 1997;55:5689–5696. [Google Scholar]
- 43.Koneshan S., Rasaiah J.C., Lynden-Bell R.M., Lee S.H. Solvent structure, dynamics, and ion mobility an aqueous solutions at 25°C. J. Phys. Chem. B. 1998;102:4193–4204. [Google Scholar]
- 44.Lyubartsev A.P., Nordenskiöld L. Computer simulations of polyelectrolytes. In: Tripathy S.K., Kumar J., Nalwa H.S., editors. Handbook of Polyelectrolytes and Their Applications v. 3. LA: American Scientific Publishers; 2002. pp. 309–326. [Google Scholar]
- 45.Korolev N., Lyubartsev A.P., Nordenskiöld L., Laaksonen A. Spermine: an ‘invisible’ component in the crystals of B-DNA. A grand canonical Monte Carlo and molecular dynamics simulation study. J. Mol. Biol. 2001;308:907–917. doi: 10.1006/jmbi.2001.4642. [DOI] [PubMed] [Google Scholar]
- 46.Martyna G.J., Tobias D.J., Klein M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- 47.Allen M.P., Tildesley D.J. Computer Simulations of Liquids. Oxford: Clarendon; 1987. [Google Scholar]
- 48.Tuckerman M., Berne B., Martyna G.J. Reversible multiple time scale molecular-dynamics. J. Chem. Phys. 1992;97:1990–2001. [Google Scholar]
- 49.Lyubartsev A.P., Laaksonen A. M.DynaMix—a scalable portable parallel MD simulation package for arbitrary molecular mixtures. Comput. Phys. Commun. 2000;128:565–589. [Google Scholar]
- 50.Korolev N., Lyubartsev A.P., Laaksonen A., Nordenskiöld L. On the competition between water, sodium ions, and spermine in binding to DNA. A molecular dynamics computer simulation study. Biophys. J. 2002;82:2860–2875. doi: 10.1016/S0006-3495(02)75628-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stofer E., Lavery R. Measuring the geometry of DNA grooves. Biopolymers. 1994;34:337–346. doi: 10.1002/bip.360340305. [DOI] [PubMed] [Google Scholar]
- 52.El Hassan M.A., Calladine C.R. Conformational characteristics of DNA: empirical classifications and hypothesis for the conformational behaviour of dinucleotide steps. Phil. Trans. R Soc. Lond. A. 1997;355:43–100. [Google Scholar]
- 53.Chiu T.K., Kaczor-Grzeskowiak M., Dickerson R.E. Absence of minor groove monovalent cations in the crosslinked dodecamer C-G-C-G-A-A-T-T-C-G-C-G. J. Mol. Biol. 1999;292:589–608. doi: 10.1006/jmbi.1999.3075. [DOI] [PubMed] [Google Scholar]
- 54.Williams L.D., Maher J.L., III Electrostatic mechanisms of DNA deformation. Annu. Rev. Biophys. Biomol. Struct. 2000;29:497–521. doi: 10.1146/annurev.biophys.29.1.497. [DOI] [PubMed] [Google Scholar]
- 55.McConnell K.J., Beveridge D.L. DNA structure: what's in charge? J. Mol. Biol. 2000;304:803–820. doi: 10.1006/jmbi.2000.4167. [DOI] [PubMed] [Google Scholar]
- 56.Madhumalar A., Bansal M. Structural insights into the effect of hydration and ions on A-tract DNA: a molecular dynamics study. Biophys. J. 2003;85:1805–1816. doi: 10.1016/S0006-3495(03)74609-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Korolev N., Lyubartsev A.P., Laaksonen A., Nordenskiöld L. A molecular dynamics simulation study of polyamine– and sodium–DNA. Interplay between polyamine binding and DNA structure. Eur. Biophys. J. 2004;33:671–682. doi: 10.1007/s00249-004-0410-7. [DOI] [PubMed] [Google Scholar]
- 58.Korolev N., Lyubartsev A.P., Laaksonen A., Nordenskiöld L. Molecular dynamics simulation study of oriented polyamine– and Na–DNA: sequence specific interactions and effects on DNA structure. Biopolymers. 2004;73:542–555. doi: 10.1002/bip.10583. [DOI] [PubMed] [Google Scholar]
- 59.Rueda M., Cubero E., Laughton C.A., Orozco M. Exploring the counterion atmosphere around DNA: what can be learned from molecular dynamics simulations? Biophys. J. 2004;87:800–811. doi: 10.1529/biophysj.104.040451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.MacKerell A.D., Jr, , Bashford D., Belloott M., Dunbrack R.L., Evanseck J.D., Field M.J., Fischer S., Gao J., Guo H., Ha S., et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 61.Collins K.D. Charge density-dependent strength of hydration and biological structure. Biophys. J. 1997;72:65–76. doi: 10.1016/S0006-3495(97)78647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harned H.S., Robinson R.A. The International Encyclopedia of Physical Chemistry and Chemical Physics. Oxford: Pergamon Press; 1968. [Google Scholar]
- 63.Mark P., Nilsson L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A. 2001;105:9954–9960. [Google Scholar]
- 64.Bastug T., Kuyucak S. Temperature dependence of the transport coefficients of ions from molecular dynamics simulations. Chem. Phys. Lett. 2005;408:84–88. [Google Scholar]
- 65.Mark P., Nilsson L. Structure and dynamics of liquid water with different long range interaction truncation and temperature control methods in molecular dynamics simulations. J. Comput. Chem. 2002;23:1211–1219. doi: 10.1002/jcc.10117. [DOI] [PubMed] [Google Scholar]
- 66.Hamelberg D., Williams L.D., Wilson W.D. Influence of the dynamic positions of cations on the structure of the DNA minor groove: sequence-dependent effects. J. Am. Chem. Soc. 2001;123:7745–7755. doi: 10.1021/ja010341s. [DOI] [PubMed] [Google Scholar]
- 67.Denisov V.P., Carlström G., Venu K., Halle B. Kinetics of DNA hydration. J. Mol. Biol. 1997;268:118–136. doi: 10.1006/jmbi.1996.0862. [DOI] [PubMed] [Google Scholar]
- 68.Pal S.K., Zhao L., Xia T., Zewail A.H. Site- and sequence-selective ultrafast hydration of DNA. Proc. Natl Acad. Sci. USA. 2003;100:13746–13751. doi: 10.1073/pnas.2336222100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pal S.K., Zhao L., Zewail A.H. Water at DNA surfaces: ultrafast dynamics in minor groove recognition. Proc. Natl Acad. Sci. USA. 2003;100:8113–8118. doi: 10.1073/pnas.1433066100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang W., Bond J.P., Anderson C.F., Lohman T.M., Record M.T. Large electrostatic differences in the binding thermodynamics of a cationic peptide to oligomeric and polymeric DNA. Proc. Natl Acad. Sci. USA. 1996;93:2511–2516. doi: 10.1073/pnas.93.6.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang W., Ni H., Capp M.W., Anderson C.F., Lohman T.M., Record M.T. The importance of coulombic end effects: experimental characterization of the effects of oligonucleotide flanking charges on the strength and salt dependence of oligocation (L8+) binding to single-stranded DNA oligomers. Biophys. J. 1999;76:1008–1017. doi: 10.1016/S0006-3495(99)77265-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schneider B., Berman H.M. Hydration of the DNA bases is local. Biophys. J. 1995;69:2661–2669. doi: 10.1016/S0006-3495(95)80136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schneider B., Patel K., Berman H.M. Hydration of the phosphate group in double-helical DNA. Biophys. J. 1998;75:2422–2434. doi: 10.1016/S0006-3495(98)77686-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vargason J.M., Eichman B.F., Ho P.S. The extended and eccentric E-DNA structure induced by cytosine methylation or bromination. Nature Struct. Biol. 2000;7:758–761. doi: 10.1038/78985. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.