Abstract
The array CGH technique (Array Comparative Genome Hybridization) has been developed to detect chromosomal copy number changes on a genome-wide and/or high-resolution scale. It is used in human genetics and oncology, with great promise for clinical application. Until recently primarily PCR amplified bacterial artificial chromosomes (BACs) or cDNAs have been spotted as elements on the array. The large-scale DNA isolations or PCR amplifications of the large-insert clones necessary for manufacturing the arrays are elaborate and time-consuming. Lack of a high-resolution highly sensitive (commercial) alternative has undoubtedly hindered the implementation of array CGH in research and diagnostics. Recently, synthetic oligonucleotides as arrayed elements have been introduced as an alternative substrate for array CGH, both by academic institutions as well as by commercial providers. Oligonucleotide libraries or ready-made arrays can be bought off-the-shelf saving considerable time and efforts. For RNA expression profiling, we have seen a gradual transition from in-house printed cDNA-based expression arrays to oligonucleotide arrays and we expect a similar transition for array CGH. This review compares the different platforms and will attempt to shine a light on the ‘BAC to the future’ of the array CGH technique.
ARRAY CGH IMPROVES SPATIAL RESOLUTION
Classical comparative genomic hybridization provided the possibility for detecting chromosomal copy number changes in cell and tissue samples, similar to karyotyping, without the need of culturing (1). Even formalin-fixed paraffin-embedded (FFPE) archival material could be analyzed, allowing for the exploration of large clinical tissue archives (2–4). Yet, resolution was limited and analysis required a high level of cytogenetic expertise. Array CGH was introduced in the late nineties (4) and overcame these two major drawbacks. Several excellent reviews have been written on the applications and current status of array CGH of which we highly recommend Pinkel and Alberston (5), among others (6–9).
The first array CGH platforms generally used large-insert clones, such as BAC (Bacterial Artificial Chromosomes), YAC (Yeast Artificial Chromosomes) or PAC (P1-derived Artificial Chromosomes) clones. Later, also the shorter cosmids and fosmids clones were introduced as spotted elements (5), as well as 130–600 bp single-stranded DNA molecules (10). Several laboratories used cDNA arrays, initially designed for expression profiling, as an alternative for measuring chromosomal copy number changes (2). Even though this approach certainly has yielded valuable information, it cannot compete with the current platforms in terms of its maximal achievable resolution. Advantages and disadvantages of the cDNA platform for array CGH are discussed in further detail by Davies et al. (8). Until recently, commercial alternatives have had very limited resolution, 3 Mb or less which is comparable with conventional CGH (1,11,12). As a result of the lack of widely accessible platforms, the possibilities of oligonucleotides as spotted elements on the arrays have been successfully explored. Oligonucleotide array CGH (oaCGH) now offers genome-covering resolution. To our knowledge, we are the only group that developed both oaCGH (13–15) as well as BAC-based array CGH (16–20), without a commercial interest. This allows us to discuss the advantages and disadvantages of the respective platforms, which we will do in this review. Reviews written so far are from laboratories that mainly perform BAC array CGH. Our goal is to evaluate the value of the different oaCGH platforms and compare it with the BAC array CGH platform. To our opinion oaCGH, and not BAC CGH arrays, is the platform that will prevail in the future.
DIFFERENT ARRAY CGH PLATFORMS
The vast majority of array CGH data available today has been generated using BAC CGH arrays. BACs vary in length from 150 to 200 kb and the DNA yield is generally low when isolated from Escherichia coli (5). Since high DNA concentration is mandatory for high quality results, most of the platforms use PCR amplification prior to spotting the arrays. Genome-wide BAC arrays vary in size from 2400 to ∼30 000 unique array elements. Difficulties when setting up BAC arrays parallel those of spotted cDNA arrays in terms of clone management and probe identity due to PCR contaminations. In addition, BAC array data suffer from mapping inaccuracies of the clones to the human genome. The venture for the production of a BAC array with a 1 Mb resolution, let alone a genome-covering array (21), is beyond the reach of most individual laboratories and as a consequence these arrays have not been widely available. Nevertheless, the BAC platform is outstandingly sensitive and precise.
OaCGH platforms are characterized by single-stranded 25 to 85mer oligonucleotide elements on the array. Different oligo arrays are combined with different labeling and hybridization techniques and all yield high-resolution copy number measurements.
Affymetrix is a commercial oaCGH platform, which contains short 25mer oligonucleotides photo-lithograhically synthesized on the arrays (http://www.affymetrix.com/) (22). These are single channel arrays, which means that only test DNA needs to be labeled and hybridized. The labeling of the test sample involves a restriction enzyme based complexity reduction procedure and requires 250 ng of DNA. Complexity reduction precludes the use of sub optimal DNA quality samples, as can be the case with DNA from archival FFPE specimens. The variation per element on the array is relatively high, which gets compensated by the large amount of elements on the array, currently 250 000 (250K) per array. A series of normal reference samples needs to be hybridized each time in parallel, which is used to calculate the chromosomal copy number changes across the entire genome, now implemented in the analysis program dChip (23). A big advantage of the Affymetrix system compared with any of the other systems is that SNPs are detected in parallel, allowing allelotyping (24).
Another commercial oaCGH platform was introduced by Agilent Technologies (25,26) (http://www.agilent.com/). They evaluated their original expression arrays for this purpose as well as arrays designed specifically for array CGH, which include oligonucleotides covering intergenic regions. Both Agilent array platforms constitute of 60mer oligonucleotides which are synthesized on the arrays and can be purchased with ∼200 K unique oligonucleotides on the array. The labeling protocol is basically the one used for the cDNA arrays (2) and requires 1 µg of input DNA which hampers the use of small clinical samples. To overcome this problem, a PCR amplification procedure was developed allowing as little as 10 ng of input DNA. Apart from the necessity to do amplifications of test and reference sample(s) in parallel, any PCR-based DNA amplification introduces some level of additional variation (8) and adds to the overall cost of arrays. The Agilent platform has already been proven highly valuable in a lung cancer study (27).
A third oligonucleotide platform offered commercially is by NimbleGen (http://www.nimblegen.com/). They provide arrays containing 385 K oligonucleotides photo-lithograhically synthesized on the array. The array production is extremely flexible such that each array produced can have a different set of oligonucleotides on it. The oaCGH oligonucleotides are designed to be isothermal and vary between 45 and 85 bp in length (28). For their labeling and hybridization procedures, Nimblegene adopted essentially the same conditions as Agilent (26) and the cDNA CGH platforms (2). Lucito et al. (29) applied an alternative method for labeling oaCGH arrays, which was applied for the NimbleGen arrays as well as for 70mer spotted oaCGH. The method called ROMA (representational oligonucleotide micro array analysis) implies a 98% complexity reduction of test and reference DNA. This is carried out by a digestion-amplification step, which allows starting with as little as 50 ng of input DNA. To warrant reproducibility, this labeling and amplification procedure requires test and reference samples to be amplified in parallel (8). ROMA combined with the NimbleGen array provides a high-resolution alternative that allows low amounts of sample DNA input and has already proven its value in the field of human genetics (30).
A fourth oaCGH platform is non-commercial and makes use of oligonucleotide libraries that are spotted as elements on the arrays (13,14). Currently, 60–70mer libraries of 30 K are used (14). A larger library of ∼50 K is currently being marketed, which was designed with oaCGH in mind (see http://alizadehlab.stanford.edu/). The labeling and hybridization procedures adapted for these ‘in-house printed’ oaCGH arrays is the one developed initially for the BAC arrays (18). This hybridization procedure uses random primer labeling which requires as little as 250 ng, which makes it very suitable for direct labeling of DNA from clinical samples and the procedure was proven to be functional for DNA of reduced quality, such as that isolated from archival FFPE specimens (14,31). The non-commercial in-house oaCGH platform has meanwhile proven its value in research (15). In our laboratories in Amsterdam we successfully tested random primer labeling combined with our hybridization procedures, on the 42 K oaCGH arrays from Agilent (data not shown). So far, this has worked for fresh DNA but not for DNA isolated from FFPE material.
COMPETING FOR SPATIAL RESOLUTION
Array CGH has thus been developed to increase the spatial resolution for the detection of chromosomal copy number changes. Different platforms therefore compete based on this spatial resolution. Spatial resolution is determined by the sensitivity of the hybridization of the elements on the array, the number of elements on the arrays, the chromosomal distribution of printed elements, the length of the elements as well as the amplitude of a chromosomal copy number change. Thus, the platform with the most and shortest array elements with least variance that are most evenly distributed across the genome and the most optimal sequence design has the best resolution.
None of the current oaCGH platforms can make a definite call for loss or gain using a single oligonucleotide, but rather 3–5 adjacent oligonucleotides are necessary for a reliable call (Table 1). For the BAC arrays, it is proclaimed one can make a call on a single arrayed element, such that after dye intensity normalization a log2ratio of plus or minus 0.5 represents a gain for the chromosomal region of the respective BAC. Thus, although BAC arrays have relatively few printed elements on the arrays, their spatial resolution is relatively high. The maximal resolution that can be obtained with BACs arrays, however, is finite because of their large size. We would like to illustrate this with the following calculation: a 150 kb BAC divided by the length of a 60 bp oligonucleotide results in 2500 adjacent oligonucleotides within one BAC. This would imply a 2500 times improved resolution. Practically, however, only unique oligonucleotide sequences will perform well such that cross-hybridizing and repetitive sequence elements need to be avoided in the design. Taking both the repetitive sequences as well as oligonucleotide sensitivity into account, we estimate that with oaCGH still a resolution of roughly 500 times or higher compared with BAC arrays can be obtained. Overlapping BACs can generate sub-BAC resolution, but still cannot match the resolution obtained by oligonucleotides.
Table 1.
Total variation expressed in standard deviation of raw log2-ratios for chromosomes without copy number aberrations
| Platform | Cell line | Chr. | Log2ratio Dev. |
|---|---|---|---|
| Agilent CGH | MB453 | 18q | 0.6600 |
| Agilent CGH | MB453 | 2 | 0.6196 |
| Agilent CGH | MB453 | 2 | 0.3085* |
| In-house printed | BT474a | 2 | 0.2442 |
| In-house printed | BT474b | 2 | 0.2514 |
| In-house printed | BT474c | 2 | 0.2498 |
| In-house printed | BT474c | 2 | 0.1480* |
| UCSF (BAC) | MB453 | 2 | 0.1828 |
| UCSF (BAC) | BT474a | 2 | 0.1708 |
| UCSF (BAC) | BT474b | 2 | 0.1403 |
| UCSF (BAC) | BT474c | 2 | 0.1371 |
Therefore the raw data provided as Supplementary Data to the Agilent (25), in-house printed (14) and UCSF BAC (18) arrays were used to calculate the standard deviation of the ratio of at least 70 consecutive elements in a chromosomal region without known copy number changes. Columns give, respectively, platform [Agilent, In-house printed and UCSF BAC (18); a, b and c represent the results from three independent arrays from that publication], cell line, chromosome (Chr.) and the data extracted from standard deviation (Log2ratio Dev.) for the arrayed elements (BAC or oligonucleotides). Total variations measured are rather consistent for the different hybridizations within one platform; the BAC platform with the large insert clones displays a considerable lower variation compared with the oligonucleotide platforms. Asterisk illustrates the effect of pooling; pooling was performed by moving the average of three arrayed elements before calculating standard deviations. The Affymetrix, ROMA or NimbleGen platforms could not be included, since raw log2 ratios were not available or comparable.
PCR amplification procedures reduce BACs to a mixture of genomic sequences and can as such be regarded as a ‘not-moving’ averaging window of 150–200 kb. Thus, a moving average on oligonucleotides could effectively be seen as the in silico synthesis of BACs, but providing repeated measurements on independent array elements resulting in reliable confidence intervals. As an alternative to the computational combining of oligonucleotides to mimic a BAC, oligonucleotides can be physically combined, but obviously confidence intervals are then lost.
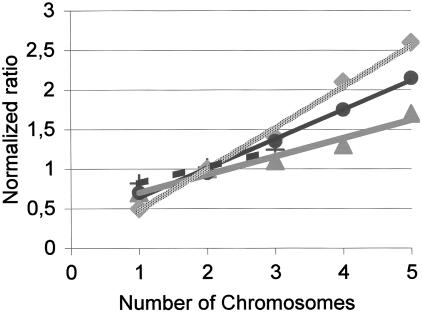
In Table 1, the standard deviations of arrayed elements on consecutive chromosomal positions are given for three different platforms for which datasets were publicly available. A standard deviation is used to illustrate how one could compare the variation of the different oaCGH platforms and (moving) averaging is used to give an impression how many array elements are required to make a call on a single copy number change. Non-overlapping standard deviations indicate a single copy number change call can be made (14). The amplitude of the single copy number change is platform dependent and is given in Figure 1. Figure 1 and Table 1 are meant to illustrate how current platforms can be compared with available raw data.
Figure 1.
Relation between theoretical and measured chromosomal copy number changes for several different platforms. Number of chromosomes on the horizontal axis and the dye normalized experimental ratios on the vertical axis. Agilent custom oaCGH (25), speckled gray line with diamonds, slope 0.53; BAC arrays (3), thin line with dots, slope 0.37; in-house printed oaCGH (14), short dashes line with plusses, slope 0.28; Agilent oaCGH on expression arrays (25), dark gray line with triangles slope 0.21. A single chromosomal copy gain in a diploid background theoretically shifts the ratio from 1 to 1.5 (two chromosomes/two chromosomes versus three chromosomes/two chromosomes). To detect a single copy gain in a diploid background, the total variance should therefore be smaller than 0.25 so that there is no overlap between normal and gain. The copy number values and slope for the Agilent CGH platform (25) is nearly identical to the theoretical values and slope. However, the variance for one or even three array elements combined is too high to unequivocally call a gain (Table 1). The relation between the theoretical and measured chromosomal copy number change for a given BAC array is slightly compressed, but the variance is maximally 0.18 (Table 1). This makes it possible to make a call on a single arrayed BAC element. For the in-house printed oaCGH platform the relation between theoretical and measured chromosomal copy number is also not ideal; however, three oligonucleotides are still sufficient to call a single copy number change, since the elements have a limited variance (Table 1). The ROMA or NimbleGen platforms could not be included, since raw log2 ratios were not available. The Affymetrix (22) platform was not included since it is a single channel array and this figure presents normalized ratios.
With regard to the CGH arrays one needs to be careful with the use of the word ‘variation’ as opposed to ‘noise’, since noise implies just technical noise. The total variation detected is the technical noise combined with biological variation consisting of segmental duplications and sequence variations between individuals (32–34). The consequence for array CGH of these sequence variations between individuals is discussed in detail by Pinkel and Albertson (5). Repeated hybridizations can separate the effects that sequence variation and technical noise have on the total variation (Table 1), which was carried out for both the BAC CGH and in-house printed oaCGH arrays using the BT474 cell line (14,18). BACs are less sensitive to large sequence variations compared with oligonucleotides, due to their length. The oligonucleotides, however, have the advantage that the flexibility in the design makes it possible to either include or exclude regions that vary between individuals. To this end, information on sequence of a high number of individuals needs to be obtained, which is now within our reach with the introduction of massive parallel sequencing (35). By excluding the biologically variable regions, the variation of oaCGH arrays can be reduced to technical noise, which would reduce the total variation by a factor of 2 for the in-house printed oligonucleotide platform (14). On the other hand, by using oligos that cover variable loci, the array CGH technique can be employed to measure these copy number variations (22,33,36,37).
THE OPTIMAL LENGTH OF AN ARRAY CGH ELEMENT
It is important to evaluate what the minimal length of an array element is, which gives maximal performance. The ROMA group has evaluated influence of the length of oligonucleotides on variance from 30 to 70mers, and selected the 70mer as their standard (29). Of all the oaCGH platforms currently available, the short 25mer oligonucleotides on the Affymetrix arrays produce the highest noise level per element (22). No synthetic oligonucleotides longer than 85 bp (28) have been evaluated since oligonucleotides become increasingly impure with increasing length. A clue toward the sensitivity of arrays containing oligonucleotides longer than 100 bp can be obtained from the exon-arrays (10). Exon-arrays contain single-stranded PCR products ranging in size from 139 to 571 bp, essentially representing very long oligonucleotides, but with the same high sensitivity per arrayed element as BACs. Based on the fact that the 60–70mer ssDNA elements from the oligonucleotide arrays display considerably more variation on the CGH arrays compared with the 139mer ssDNA elements from the exon arrays, we conclude that oligonucleotides of ∼140 bp would be preferable. Regretfully, the exon-array cannot be produced on a genome-wide scale since they will by definition be lacking all ‘agenic’ portions of the genome and would be extremely labor intensive to produce (9). New developments for the synthesis of longer oligonucleotides are underway and could offer an excellent solution (38).
ADDITIONAL CONSIDERATIONS
An important consideration when selecting an array CGH platform is the price, especially when studying larger series of samples. Cost of the arrays, as with expression arrays, can be addressed as follows: in-house printed arrays are generally cheaper than commercial arrays and increasing amounts of elements on the array increase the price, whereby the resolution required is dependent on the research question. Furthermore, the amount of Cye dyes used for labeling reaction seriously influences the price as well as the amount of Cot-1 DNA.
Especially for diagnostic applications, standardization and reproducibility are important issues. Synthetic oligonucleotides obviously have the advantage that the exact sequence and length is known for each element on the arrays. For PCR-amplified BACs, this is not the case since the amplification procedure is not linear and variable for each amplification round (18,39). A second variable in array CGH is the Cot-1 DNA used to block repetitive DNA sequences in the arrayed elements (5). Each time Cot-1 DNA is isolated it is extracted from different human placentas and yields variable DNA fragments of 50–300 bp in length. Batch to batch variation makes Cot-1 DNA a highly variable element in the array procedure. For oaCGH, Cot-1 DNA would theoretically not be necessary, since the oligonucleotides are designed to be repeat-free. Cot-1 DNA has been included in all array CGH platforms, except for the NimbleGen arrays in which it was completely omitted (28). Also in our experience (14) omitting Cot-1 DNA is possible, but we observed a slight reduction in the quality of the measurements using the in-house printed oaCGH platform.
EXPECTATIONS
Based on the considerations discussed, odds are that the array CGH field is evolving towards oligonucleotide array CGH. Today oaCGH offers the highest resolution and is therefore slowly overtaking BAC arrays for the measurement of chromosomal copy number changes in human genetics and cancer. It is to be expected that the transition from BAC CGH arrays to oaCGH will gradually take place, analogous to the way cDNA arrays for expression profiling have been replaced by oligonucleotide arrays. For specific applications there will still be a place for BAC arrays, like in the case of methylation studies (40).
Each platform has its own advantages and disadvantages and arrays as well as protocols rapidly improve. Some oaCGH platforms can handle degraded DNA samples, some offer ultra-high density or are highly cost effective, others also give SNP information or are flexible in their design.
Acknowledgments
This work was supported by VU Unversity Medical Center intramural funds, the EU-sixth framework programme and the Centre for Medical Systems Biology (CMSB), a centre of excellence approved by the Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research (NWO). The authors wish to thank Paul van Hummelen (UZ Gasthuisberg, Leuven, BE) for critically reading the manuscript prior to submission and Paul P. Eijk for evaluating random prime labeling combined with Agilent arrays. The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kallioniemi A., Kallioniemi O.P., Sudar D., Rutovitz D., Gray J.W., Waldman F., Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 2.Pollack J.R., Perou C.M., Alizadeh A.A., Eisen M.B., Pergamenschikov A., Williams C.F., Jeffrey S.S., Botstein D., Brown P.O. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nature Genet. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- 3.Pinkel D., Segraves R., Sudar D., Clark S., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nature Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 4.Solinas-Toldo S., Lampel S., Stilgenbauer S., Nickolenko J., Benner A., Dohner H., Cremer T., Lichter P. Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes Chromosomes Cancer. 1997;20:399–407. [PubMed] [Google Scholar]
- 5.Pinkel D., Albertson D.G. Array comparative genomic hybridization and its applications in cancer. Nature Genet. 2005;37(Suppl.):S11–S17. doi: 10.1038/ng1569. [DOI] [PubMed] [Google Scholar]
- 6.Oostlander A.E., Meijer G.A., Ylstra B. Microarray-based comparative genomic hybridization and its applications in human genetics. Clin. Genet. 2004;66:488–495. doi: 10.1111/j.1399-0004.2004.00322.x. [DOI] [PubMed] [Google Scholar]
- 7.Vissers L.E., Veltman J.A., van Kessel A.G., Brunner H.G. Identification of disease genes by whole genome CGH arrays. Hum. Mol. Genet. 2005;14:R215–R223. doi: 10.1093/hmg/ddi268. [DOI] [PubMed] [Google Scholar]
- 8.Davies J.J., Wilson I.M., Lam W.L. Array CGH technologies and their applications to cancer genomes. Chromosome Res. 2005;13:237–248. doi: 10.1007/s10577-005-2168-x. [DOI] [PubMed] [Google Scholar]
- 9.Mantripragada K.K., Buckley P.G., de Stahl T.D., Dumanski J.P. Genomic microarrays in the spotlight. Trends Genet. 2004;20:87–94. doi: 10.1016/j.tig.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 10.Dhami P., Coffey A.J., Abbs S., Vermeesch J.R., Dumanski J.P., Woodward K.J., Andrews R.M., Langford C., Vetrie D. Exon array CGH: detection of copy-number changes at the resolution of individual exons in the human genome. Am. J. Hum. Genet. 2005;76:750–762. doi: 10.1086/429588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ulger C., Toruner G.A., Alkan M., Mohammed M., Damani S., Kang J., Galante A., Aviv H., Soteropoulos P., Tolias P.P., et al. Comprehensive genome-wide comparison of DNA and RNA level scan using microarray technology for identification of candidate cancer-related genes in the HL-60 cell line. Cancer Genet. Cytogenet. 2003;147:28–35. doi: 10.1016/s0165-4608(03)00155-9. [DOI] [PubMed] [Google Scholar]
- 12.Hidalgo A., Baudis M., Petersen I., Arreola H., Pina P., Vazquez-Ortiz G., Hernandez D., Gonzalez J., Lazos M., Lopez R., et al. Microarray comparative genomic hybridization detection of chromosomal imbalances in uterine cervix carcinoma. BMC Cancer. 2005;5:77. doi: 10.1186/1471-2407-5-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carvalho B., Ouwerkerk E., Meijer G.A., Ylstra B. High resolution microarray comparative genomic hybridisation analysis using spotted oligonucleotides. J. Clin. Pathol. 2004;57:644–646. doi: 10.1136/jcp.2003.013029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van den IJssel P., Tijssen M., Chin S.-F., Eijk P.P., Carvalho B., Hopmans E., Holstege H., Bangarusamy D.K., Jonkers J., Meijer G.A., et al. Human and mouse oligonucleotide-based array CGH. Nucleic Acids Res. 2005;33:e192. doi: 10.1093/nar/gni191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van de Wiel M.A., Costa J.L., Smid K., Oudejans C.B., Bergman A.M., Meijer G.A., Peters G.J., Ylstra B. Expression microarray analysis and oligo array comparative genomic hybridization of acquired gemcitabine resistance in mouse colon reveals selection for chromosomal aberrations. Cancer Res. 2005;65:10208–10213. doi: 10.1158/0008-5472.CAN-05-0760. [DOI] [PubMed] [Google Scholar]
- 16.Weiss M.M., Snijders A.M., Kuipers E.J., Ylstra B., Pinkel D., Meuwissen S.G., van Diest P.J., Albertson D.G., Meijer G.A. Determination of amplicon boundaries at 20q13.2 in tissue samples of human gastric adenocarcinomas by high-resolution microarray comparative genomic hybridization. J. Pathol. 2003;200:320–326. doi: 10.1002/path.1359. [DOI] [PubMed] [Google Scholar]
- 17.Weiss M.M., Kuipers E.J., Postma C., Snijders A.M., Siccama I., Pinkel D., Westerga J., Meuwissen S.G., Albertson D.G., Meijer G.A. Genomic profiling of gastric cancer predicts lymph node status and survival. Oncogene. 2003;22:1872–1879. doi: 10.1038/sj.onc.1206350. [DOI] [PubMed] [Google Scholar]
- 18.Snijders A.M., Nowak N., Segraves R., Blackwood S., Brown N., Conroy J., Hamilton G., Hindle A.K., Huey B., Kimura K., et al. Assembly of microarrays for genome-wide measurement of DNA copy number. Nature Genet. 2001;29:263–264. doi: 10.1038/ng754. [DOI] [PubMed] [Google Scholar]
- 19.Schreurs M.W., Hermsen M.A., Geltink R.I., Scholten K.B., Brink A.A., Kueter E.W., Tijssen M., Meijer C.J., Ylstra B., Meijer G.A., et al. Genomic stability and functional activity may be lost in telomerase-transduced human CD8+ T lymphocytes. Blood. 2005;106:2663–2670. doi: 10.1182/blood-2004-09-3742. [DOI] [PubMed] [Google Scholar]
- 20.Smeets S.J., Braakhuis B.J., Abbas S., Snijders P.J., Ylstra B., van de Wiel M.A., Meijer G.A., Leemans C.R., Brakenhoff R.H. Genome-wide DNA copy number alterations in head and neck squamous cell carcinomas with or without oncogene-expressing human papillomavirus. Oncogene. 2006 doi: 10.1038/sj.onc.1209275. in press, doi:10.1038/sj.onc.1209275. [DOI] [PubMed] [Google Scholar]
- 21.Ishkanian A.S., Malloff C.A., Watson S.K., DeLeeuw R.J., Chi B., Coe B.P., Snijders A., Albertson D.G., Pinkel D., Marra M.A., et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nature Genet. 2004;36:299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- 22.Zhao X., Li C., Paez J.G., Chin K., Janne P.A., Chen T.H., Girard L., Minna J., Christiani D., Leo C., et al. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res. 2004;64:3060–3071. doi: 10.1158/0008-5472.can-03-3308. [DOI] [PubMed] [Google Scholar]
- 23.Zhao X., Weir B.A., LaFramboise T., Lin M., Beroukhim R., Garraway L., Beheshti J., Lee J.C., Naoki K., Richards W.G., et al. Homozygous deletions and chromosome amplifications in human lung carcinomas revealed by single nucleotide polymorphism array analysis. Cancer Res. 2005;65:5561–5570. doi: 10.1158/0008-5472.CAN-04-4603. [DOI] [PubMed] [Google Scholar]
- 24.Raghavan M., Lillington D.M., Skoulakis S., Debernardi S., Chaplin T., Foot N.J., Lister T.A., Young B.D. Genome-wide single nucleotide polymorphism analysis reveals frequent partial uniparental disomy due to somatic recombination in acute myeloid leukemias. Cancer Res. 2005;65:375–378. [PubMed] [Google Scholar]
- 25.Barrett M.T., Scheffer A., Ben Dor A., Sampas N., Lipson D., Kincaid R., Tsang P., Curry B., Baird K., Meltzer P.S., et al. Comparative genomic hybridization using oligonucleotide microarrays and total genomic DNA. Proc. Natl Acad. Sci. USA. 2004;101:17765–17770. doi: 10.1073/pnas.0407979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brennan C., Zhang Y., Leo C., Feng B., Cauwels C., Aguirre A.J., Kim M., Protopopov A., Chin L. High-resolution global profiling of genomic alterations with long oligonucleotide microarray. Cancer Res. 2004;64:4744–4748. doi: 10.1158/0008-5472.CAN-04-1241. [DOI] [PubMed] [Google Scholar]
- 27.Tonon G., Wong K.K., Maulik G., Brennan C., Feng B., Zhang Y., Khatry D.B., Protopopov A., You M.J., Aguirre A.J., et al. High-resolution genomic profiles of human lung cancer. Proc. Natl Acad. Sci. USA. 2005;102:9625–9630. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selzer R.R., Richmond T.A., Pofahl N.J., Green R.D., Eis P.S., Nair P., Brothman A.R., Stallings R.L. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer. 2005;44:305–319. doi: 10.1002/gcc.20243. [DOI] [PubMed] [Google Scholar]
- 29.Lucito R., Healy J., Alexander J., Reiner A., Esposito D., Chi M., Rodgers L., Brady A., Sebat J., Troge J., et al. Representational oligonucleotide microarray analysis: a high-resolution method to detect genome copy number variation. Genome Res. 2003;13:2291–2305. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jobanputra V., Sebat J., Troge J., Chung W., Anyane-Yeboa K., Wigler M., Warburton D. Application of ROMA (representational oligonucleotide microarray analysis) to patients with cytogenetic rearrangements. Genet. Med. 2005;7:111–118. doi: 10.1097/01.gim.0000153661.11110.fb. [DOI] [PubMed] [Google Scholar]
- 31.Devries S., Nyante S., Korkola J., Segraves R., Nakao K., Moore D., Bae H., Wilhelm M., Hwang S., Waldman F. Array-based comparative genomic hybridization from formalin-fixed, paraffin-embedded breast tumors. J. Mol. Diagn. 2005;7:65–71. doi: 10.1016/S1525-1578(10)60010-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharp A.J., Locke D.P., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tuzun E., Sharp A.J., Bailey J.A., Kaul R., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., et al. Fine-scale structural variation of the human genome. Nature Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 34.van Ommen G.J. Frequency of new copy number variation in humans. Nature Genet. 2005;37:333–334. doi: 10.1038/ng0405-333. [DOI] [PubMed] [Google Scholar]
- 35.Rogers Y.H., Venter J.C. Genomics: massively parallel sequencing. Nature. 2005;437:326–327. doi: 10.1038/437326a. [DOI] [PubMed] [Google Scholar]
- 36.Snijders A.M., Nowak N.J., Huey B., Fridlyand J., Law S., Conroy J., Tokuyasu T., Demir K., Chiu R., Mao J.H., et al. Mapping segmental and sequence variations among laboratory mice using BAC array CGH. Genome Res. 2005;15:302–311. doi: 10.1101/gr.2902505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 38.Egeland R.D., Southern E.M. Electrochemically directed synthesis of oligonucleotides for DNA microarray fabrication. Nucleic Acids Res. 2005;33:e125. doi: 10.1093/nar/gni117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fiegler H., Carr P., Douglas E.J., Burford D.C., Hunt S., Scott C.E., Smith J., Vetrie D., Gorman P., Tomlinson I.P., et al. DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer. 2003;36:361–374. doi: 10.1002/gcc.10155. [DOI] [PubMed] [Google Scholar]
- 40.Ching T.T., Maunakea A.K., Jun P., Hong C., Zardo G., Pinkel D., Albertson D.G., Fridlyand J., Mao J.H., Shchors K., et al. Epigenome analyses using BAC microarrays identify evolutionary conservation of tissue-specific methylation of SHANK3. Nature Genet. 2005;37:645–651. doi: 10.1038/ng1563. [DOI] [PubMed] [Google Scholar]