Abstract
During macronuclear development the Tetrahymena thermophila ribosomal RNA gene is excised from micronuclear chromosome 1 by site-specific cleavage at chromosome breakage sequence (Cbs) elements, rearranged into a ‘palindromic’ 21 kb minichromosome and extensively amplified. Gene amplification initiates from origins in the 5′ non-transcribed spacer, and forks moving toward the center of the palindrome arrest at a developmentally regulated replication fork barrier (RFB). The RFB is inactive during vegetative cell divisions, suggesting a role in the formation or amplification of macronuclear rDNA. Using micronuclear (germline) transformation, we show that the RFB region facilitates Cbs-mediated excision. Deletion of the RFB inhibits chromosome breakage in a sub-population of developing macronuclei and promotes alternative processing by a Cbs-independent mechanism. Remarkably, the RFB region prevents spontaneous breakage of chromosome 1 in the diploid micronucleus. Strains heterozygous for ΔRFB and wild-type rDNA lose the ΔRFB allele and distal left arm of chromosome 1 during vegetative propagation. The wild-type chromosome is subsequently fragmented near the rDNA locus, and both homologs are progressively eroded, suggesting that broken micronuclear chromosomes are not ‘healed’ by telomerase. Deletion of this 363 bp segment effectively creates a fragile site in the micronuclear genome, providing the first evidence for a non-telomere cis-acting determinant that functions to maintain the structural integrity of a mitotic eukaryotic chromosome.
INTRODUCTION
The initiation of DNA replication is precisely regulated to assure that chromosomes are duplicated once and only once per cell division. Eukaryotic genomes contain hundreds to thousands of initiation sites that are relatively evenly spaced throughout chromosomes (1). The convergence of neighboring replication forks allows for the complete replication of chromosomes. Elongating replication forks can stall when they encounter DNA damage at random sites in the genome. Stalled forks are recognized by the ATR (Ataxia-Telangectasia and Rad3-related) kinase, which activates a checkpoint pathway that prevents the fork from collapsing and initiates the DNA repair response [reviewed in (2)]. Replication forks can also arrest at specified sites in the genome. Transient pausing of replication forks frequently occurs near coding regions and may help coordinate replication and transcription. Similar to sites of DNA damage, the paused replication machinery resumes DNA synthesis once the impediment has been removed. More pronounced replication fork barriers (RFBs) irreversibly block the movement of the replication fork, and require the activity of a converging fork to replicate sequences downstream of the RFB [reviewed in (3)].
Natural RFBs occur in both prokaryotes and eukaryotes, and have been shown to coordinate certain events with DNA replication. For example, unidirectional RFBs allow converging forks to enter, but not exit the replication terminus of circular bacterial chromosomes (3,4). RFBs in the intergenic regions of tandemly arrayed ribosomal RNA genes prevent head on collisions between the replication and transcription machinery (5–7). The contributions of RFBs to chromosome function are offset by the fact that they create hot spots for programmed or spontaneous genome rearrangement. For example, mating type switching in the fission yeast Schizosaccharomyces pombe requires that the mat locus be replicated from forks progressing in one direction (2,8). The Saccharomyces cerevisiae ribosomal DNA RFB promotes the excision of monomeric rDNA circles from head-to-tail tandem gene arrays, as well as spontaneous chromosome fragmentation (2,9–11). In addition to generating circular episomes, RFBs in S.cerevisiae rDNA arrays are susceptible to double-strand breaks (DSBs) in several mutant backgrounds, and specialized proteins help minimize the deleterious effects of RFBs (2,9–11).
Similar to yeast and metazoa, non-coding regions in Tetrahymena thermophila ribosomal RNA genes contain sequences that impede replication fork movement. Replication forks transiently pause at three conserved sequences in the 1.9 kb 5′ non-transcribed spacer (5′ NTS) during vegetative cell divisions (Figure 1A; PSE1, PSE2 and PSE3), and at a strong RFB during development (12,13). The role of fork arrest determinants in Tetrahymena rDNA must differ from those in yeast and metazoan rDNAs for two reasons. First, instead of being arranged in large head-to-tail arrays, the Tetrahymena rRNA genes reside in natural minichromosomes that contain just two gene copies in an inverted, repeated head-to-head configuration (Figure 1A). Since replication and transcription proceed in the same direction, head on collisions between the respective DNA and RNA polymerases cannot occur. Replication forks moving toward the Tetrahymena rDNA telomere transiently arrest at conserved pause site elements (PSEs), possibly coordinating replication and transcription during vegetative cell divisions. Second, the Tetrahymena RFB is developmentally regulated, and is only active during the formation of a new macronucleus, when palindromic rDNA minichromosomes are first generated and amplified. rDNA processing and amplification initiate before the onset of transcription of the new macronucleus, and the RFB is silent during vegetative cell divisions. These observations suggest a role for the RFB in the biogenesis and/or amplification of extrachromosomal rDNA.
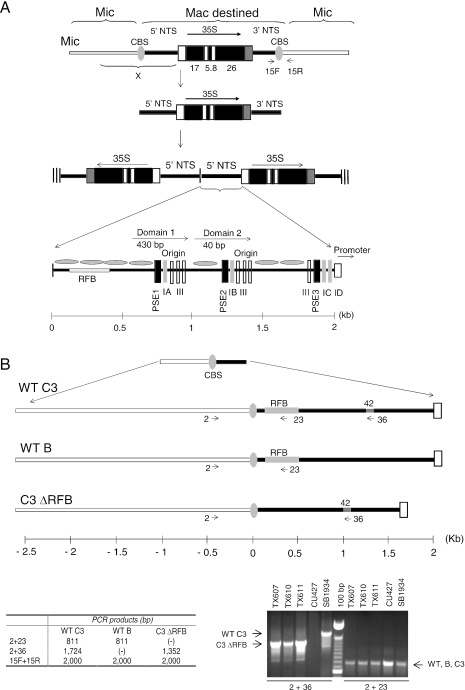
Figure 1.
rDNA processing pathway. (A) Organization of the rDNA in the micro- and macronucleus. The gene encoding the 35S precursor RNA for 17S, 5.8S and 26S ribosomal RNAs, exists as a single ‘integrated’ copy in micronuclear chromosome 1. Macronuclear-destined rDNA sequences are excised from this chromosome in the newly developing macronucleus by cleavage at flanking Cbs elements. The released rDNA monomer is subsequently rearranged into a giant head-to-head ‘palindrome’ with two inverted copies of the 5′ NTS at the center and telomeres at each terminus (hatched vertical lines). The 1.9 kb 5′ NTS (bottom panel) contains positioned nucleosomes that bracket the rRNA promoter and replication initiation sites, the later of which reside within the tandemly reiterated Domain 1 and 2 (D1, D2) regions. These nucleosome-free regions contain conserved cis-acting determinants that regulate replication initiation and fork progression, designated PSEs (PSE1, 2 and 3; vertical black boxes) and type 1 elements (1A–1D; vertical grey boxes). The approximate position of the developmentally regulated RFB is shown. (B) Polymorphisms for micronuclear rDNA allele analysis. Top and lower left panels: enlargement of the micronuclear chromosome 1 region designated as ‘X’ in panel A (top line), showing the RFB region and a 42 bp sequence that is present in wild-type C3 and C3ΔRFB alleles, but is absent in the naturally occurring B rDNA allele. The positions of PCR primers that anneal to the respective rDNA species are shown (arrows), along with the predicted sizes of PCR products (table). Primers 15F and 15R span the 3′ Cbs element and amplify the micronuclear rDNA species for all three alleles. Lower right panel: PCR analysis of micronuclear rDNA in wild-type C3 (SB1934) and wild-type B (CU427) rDNA strains, and heterozygous micronuclear transformants harboring the C3ΔRFB and B rDNA alleles (TX607, TX610 and TX611).
Tetrahymena thermophila, like all ciliated protozoa, separates its germline and somatic functions into two genetically related, but distinct compartments—the micronucleus and macronucleus [reviewed in (14)]. The diploid micronucleus is not transcribed throughout most of the life cycle and serves as the source of genetic information that is transferred during conjugation. In contrast, the polyploid macronucleus is actively transcribed, but is not sexually transmitted. The micronucleus contains five chromosomes that segregate by conventional mitosis and meiosis, and produces haploid pronuclei for genetic exchange. Following pronuclear exchange, a new macronucleus forms through the differentiation of a duplicated progeny micronucleus. The genome is subjected to extensive rearrangement at this time, including fragmentation at ∼300 specific sites, generating acentric chromosomes that endo-replicate to ∼45 copies.
The 35S ribosomal RNA gene, encoding the 17S, 5.8S and 26S rRNAs, exists as a single integrated copy in micronuclear chromosome 1. During macronuclear development, this 10.3 kb segment is excised from the parental chromosome by site-specific cleavage at conserved 15 bp chromosome breakage sequence (Cbs) elements that flank both ends of the macronuclear-destined rDNA segment (15,16). The excised rDNA monomer is then rearranged into a 21 kb inverted repeat, termed the rDNA palindrome, through homologous recombination within a small inverted repeat at the beginning of the 5′ NTS (17,18). Telomeres are added de novo and the rDNA is amplified to ∼9000 copies in a single S phase (19,20). Once development is complete, cell cycle control is re-established and the rDNA is replicated once (on average) per cell division (19,20). rDNA monomers of 11 kb are also generated during development, but typically do not persist during prolonged vegetative propagation.
Here we examine the role of the developmentally programmed RFB region in macronuclear rDNA biogenesis and amplification by introducing an RFB deletion derivative into the germline micronucleus of Tetrahymena and following the fate of rDNA minichromosomes in progeny cells. We describe a role for the RFB in the formation of macronuclear rDNA minichromosomes and an unanticipated requirement for these sequences in the germline micronucleus.
MATERIALS AND METHODS
rDNA vectors and germline transformation
The rDNA plasmid AN101 carries a wild-type copy of the micronuclear C3 rDNA locus, and undergoes Cbs-mediated excision and palindrome formation in the developing macronucleus (21). The C3 rDNA origin confers a replication advantage over endogenous B rDNA during vegetative cell divisions (22). Plasmid pC3ΔRFB contains a C3 rDNA origin in which a 363 bp fragment corresponding to the RFB region was deleted (positions 131–494; C3ΔRFB). Plasmid pTTMN1 is a neomycin phosphotransferase co-transformation vector that confers resistance to paromomycin (pmr) upon induction of the upstream metallothionein promoter (MTT1) with cadmium (23). For germline transformation of Tetrahymena, 5 µg of each plasmid was digested with restriction enzymes to release the insert fragment for targeted homologous recombination to the rDNA or MTT1 locus, respectively (AN101:SalI, pC3ΔRFB:KpnI + SalI, pTTMN1:KpnI + SstI).
Vegetative cultures of the wild-type T.thermophila strains were propagated at 30°C in 2% protease peptone supplemented with 10 mM FeCl3 (PPYS) and PSF (penicillin 250 µg/ml, streptomycin 250 µg/ml, amphotericin B 25 µg/ml) (24). Tetrahymena strains used in this study are listed in Table 1. For conjugant germline transformation, log-phase cultures of strains CU427 and CU428 were harvested and starved in Tris-buffer (10 mM Tris–HCl, pH 7.4 + PSF) for 18 h at 30°C, at a density of 2.5 × 105 cells/ml. To induce conjugation, equal numbers of each strain were mixed and incubated at 30°C without shaking. The germline micronucleus was then co-transformed by biolistic bombardment at 4 h 50 min, using the DuPont Biolistic PDS-1000/He particle delivery system (Bio-Rad). PPYS (final concentration 1.8%) and cadmium chloride (1.2 µg/ml) were added to the culture 20–22 h after bombardment (25,26). Paromomycin (pm, 60 µg/ml) was added 5–6 h later and cells were plated in 96-well microtiter plates. Pmr progeny were typically observed within 3 days. To assure that transformants were clonal, single cells were isolated from each transformant and re-screened for micronuclear C3 rDNA by PCR (see below).
Table 1.
Tetrahymena strains used in this study
| Strain | Mic | Mac | pm-phenotype |
|---|---|---|---|
| CU427 | B | B | S |
| CU428 | B | B | S |
| SB1934 | C3 | B | S |
| SB210 | B | B | S |
| SF137 | C3 | C3 | S |
| A* III | – | B | S |
| A* V | – | B | S |
| TX607 | B/C3ΔRFB | ? | R |
| TX610 | B/C3ΔRFB | ? | R |
| TX611 | B/C3ΔRFB | ? | R |
| TX614 | B/C3 | C3 | R |
S, sensitive; R, resistant; pm, paromomycin.
Standard genetic and A* crosses
Mating of heterozygous germline C3ΔRFB/B rDNA transformant strains to one another or to wild-type tester strains were performed as described above. Clonal transformant lines were also mated to functionally amicronucleate A* strains (mating types III or V) (27). Progeny from the first round of conjugation contain DNA from just the transformant parental strain in their new micronucleus and are homozygous at all loci. Since a new macronucleus is not generated at this time, these cells can immediately pair with one another (round 2 genomic exclusion). These progeny will generate a new micronucleus and macronucleus, and consequently will be sexually immature (i.e. unable to form mating pairs for at least 70 fissions). Single pairs from Round 2 matings were isolated between 23 and 25 h post-mating, transferred to 100 µl fresh 2% PPYS and propagated for 3–5 days. Cells were then screened for pm-sensitivity, sexual immaturity and the presence of the C3ΔRFB allele in progeny micronucleus, using C3-specific PCR primers. This protocol resulted in the generation of new rDNA minichromosomes during macronuclear development and also ensured that new progeny strains were of clonal origin.
PCR amplification and Southern blotting
The complete macronuclear rDNA sequence (28,29) and flanking micronuclear DNA sequence upstream of the rDNA locus (30,31) were used to generate restriction maps and design PCR primers for distinguishing the various rDNA alleles (wild-type C3, C3ΔRFB and wild-type B). Additional flanking sequences were obtained from the Tetrahymena thermophila genome database (TIGR, http://www.tigr.org/). PCR primer sets that were used to monitor the fate of different arms of the five micronuclear chromosomes were kindly provided by Drs Eileen Hamilton and Eduardo Orias (32,33). These primer sets encompass sequences that are joined in the micronuclear genome, but are absent or separated in the macronucleus by Cbs-mediated chromosome fragmentation. Genomic DNA isolation and Southern blotting were performed as previously described (34). Probes A and B (Figure 1) were generated by PCR using primers 7 + 8 and 5 + 6, respectively, and were radiolabeled with [α-32P]dATP, using the Megaprime™ DNA labeling system (Amersham Life Sciences) according to manufacturer instructions, except that specific primers were used in place of random primers. For pulsed field gel electrophoresis (PFGE), undigested total genomic was electrophoresed on a BioRad CHEFII apparatus on 1% agarose gels in 0.5× TBE buffer (14°C) at 6 V/cm for 20–24 h (initial pulse time = 0.5 s, final pulse time = 3 s).
Fluorescence microscopy
For apofluor staining, a 0.1 ml sample of the conjugating cells was concurrently stained with 0.001% acridine orange and 5 mg/ml Hoechst 33342 (Sigma). After mixing briefly, the stained cells were fixed with 1% formaldehyde (final concentration) and observed immediately with fluorescence microscopy using filters for blue fluorescence as previously described. This combination of dyes allows for the detection of DNA and can be used to evaluate the acidification of non-exchanged pronuclei and the old macronucleus (an indicator of programmed nuclear death during development) (35,36).
RESULTS
Isolation of micronuclear rDNA fork barrier deletion mutants
Since the Tetrahymena rDNA RFB is only active in the macronucleus during macronuclear development, we set out to determine whether this region facilitates site-specific rDNA excision, rearrangement (palindrome formation) or amplification of rDNA minichromosomes. To assess the role of the RFB, we introduced a C3 rDNA derivative that was deleted for this region into the germline micronucleus of mating B rDNA strains and examined the fate of the mutant C3 allele in the newly formed macronucleus. All known determinants for rDNA excision and palindrome formation were present in the transforming rDNA fragment (Cbs elements and the M element inverted repeat) (16,17). The transgenic C3 rDNA derivative contained a 42 bp sequence proximal to the Domain 2 replication origin that confer a replication advantage over endogenous macronuclear B rDNA during vegetative cell divisions (22). The C3 allele also harbored a point mutation within the 17S rRNA coding region that confers resistance to paromomycin. Homologous recombination should result the replacement of one B rDNA allele in the diploid micronucleus of transformed progeny.
It was not clear whether the C3ΔRFB allele would generate a functional macronuclear chromosome. Consequently, we co-transformed the C3ΔRFB construct with a second linear DNA fragment encoding the neomycin phosphotransferase gene under the control of an inducible metallothionein (MTT1) promoter (neophosphotransferase inactivates pm). Pm-resistant transformants were selected and subjected to allele-specific PCR to identify co-transformants that were heterozygous for the C3ΔRFB and wild-type B rDNA alleles in the germline micronucleus (Figure 1B). Three co-transformants were identified (TX607, TX610, TX611), and single cells were isolated and expanded to assure that each strain was of clonal origin. Heterozygous C3ΔRFB/B rDNA strains were obtained from three independent transformations, and 18 clonal lines derived from strains TX607, TX610 and TX611 were propagated vegetatively and subjected to detailed molecular analysis. As a control, mating B rDNA strains were transformed with a wild-type C3 rDNA fragment and a germline transformant (TX614) was identified by RFLP analysis of the macronuclear DNA and micronuclear PCR analysis (data not shown). Table 1 summarizes the relevant micronuclear genotypes and macronuclear phenotypes of wild-type and mutant strains used in this study.
The RFB region is required for proper excision of the rDNA
Since conjugating germline transformants go on to generate a new progeny macronucleus, we examined the fate of macronuclear rDNA minichromosomes in heterozygous progeny. Total genomic DNA was isolated from C3ΔRFB/B and wild-type C3/B heterozygotes, and homozygous wild-type C3 or B rDNA strains, and digested with EcoRV to distinguish between palindromic (P) and non-palindromic (NP) forms of each macronuclear rDNA species (Figure 2A, schematic; WT: wild-type C3 or B rDNA, ΔRFB: C3ΔRFB rDNA). Southern blot analysis was performed with a PCR-generated probe that was common to each predicted macronucleus rDNA species (Figure 2A, probe B). Macronuclear B (CU428) and C3 rDNA (TX614: wild-type C3 transformant) strains produced a strong 4.4 kb band corresponding to the central fragment of palindromic rDNA minichromosomes (P), and a weaker ∼2.2 kb band corresponding to monomeric (non-palindromic, NP) rDNA minichromosomes that are transiently observed in newly formed progeny (Figure 2B, left panel, lanes 1 and 5) (37).
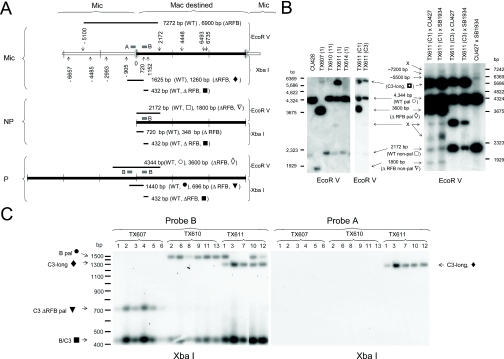
Figure 2.
Heterozygous C3ΔRFB/B strains undergo stochastic excision of the rDNA in the developing macronucleus. (A) Restriction map and fragment sizes for products following digestion with EcoRV (downward arrowheads) or XbaI (upward arrowheads) for micronuclear rDNA locus (Mic), excised non-palindromic macronuclear rDNA (NP) and palindromic (P) rDNA minichromosomes derived from wild-type B rDNA (WT) or C3ΔRFB deletion alleles. Hybridization probes A and B: thick black lines in micronuclear rDNA diagram. (B) Stochastic appearance and structure of macronuclear C3 rDNA in clonal C3ΔRFB transformants. Left panel: Southern blot analysis of EcoRV digested genomic DNA with probe B. Wild-type B rDNA strain; CU428, heterozygous C3ΔRFB/B germline transformant strains; TX607(1), TX610(11) and TX611(1), and wild-type C3 rDNA transformant strain; TX614(1). The position of wild-type B and wild-type C3 rDNA monomers (WT non-pal) and palindromes (WT pal), predicted C3ΔRFB species (ΔRFB pal, ΔRFB non-pal), and unexpected products (C3-long and X) are indicated. The corresponding restriction fragments for expected products are symbol coded (see panel A). TX611(C1) and TX611(C3) are clonal progeny derived from a cross between TX611(1) and amicronucleate strains A* III and A* V, respectively, and have generated a new macronucleus by round II genomic exclusion (see text). Note the presence of the C3ΔRFB rDNA palindrome in TX611(C1) and absence of this species in its parent, TX611(1). Right panel: Southern blot analysis of genomic DNA with probe B from mating progeny harvested 24 h after mixing strains of opposite mating type. Crosses: TX611(C1) or TX611(C3) were mated with tester strains, CU427 (wild B rDNA in micronucleus) or SB1934 (wild-type C3 rDNA in micronucleus). Control mating: CU427 × SB1934. X indicates rDNA species that were not predicted by conventional processing at the Cbs element upstream of the rDNA 5′ NTS. (C) The C3-long rDNA species is fragmented at a novel site upstream of the 5′ Cbs element. Southern blot of XbaI digested genomic DNA hybridized with probe B (left) or probe A (right). Numbers correspond to clonal lines established from each heterozygous C3ΔRFB/B rDNA germline transformant.
EcoRV digestion of the C3ΔRFB allele should generate a 3.6 kb palindromic fragment and 1.8 kb monomeric fragment if the mutant allele was correctly excised in the developing macronucleus (Figure 2A). Both species were detected in just one of the three C3ΔRFB transformant strains, for which no macronuclear B rDNA was observed [Figure 2B, left panel, lane 2: TX607(1)]. This result is consistent with the rapid loss of macronuclear B rDNA during vegetative propagation of C3/B rDNA heterozygotes (22), and suggests that properly processed C3ΔRFB minichromosomes are fully competent for vegetative DNA replication. In contrast, C3ΔRFB strain TX610 contained just wild-type B rDNA in its macronucleus, while TX611 contained B rDNA and a novel ∼5.5 kb species, hereafter designated ‘C3-long’ (Figure 2B, left panel, lanes 3 and 4; see below for more detailed analysis). The macronuclear composition of TX610 (B rDNA only) is indicative of failed excision of the C3ΔRFB allele, while the unexpected ∼5.5 kb EcoRV fragment in TX611 is consistent with alternative DNA processing. The collective results show that the RFB region promotes rDNA excision, but functions stochastically in the developing macronucleus.
To examine the fate of macronuclear rDNA during vegetative cell divisions, clonal lines were propagated for ∼70 fissions. DNA samples were digested with XbaI to monitor the fate of macronuclear C3 and B rDNA minichromosomes (Figure 2A micronuclear rDNA schematic), and to further examine the structure of unexpected rDNA species. Consistent with the results from earlier fissions, only C3ΔRFB palindromes were detected in TX607 (696 bp). No palindromic C3 rDNA minichromosomes were observed in TX610 at 70 fissions (Figure 2C). Instead, only B rDNA palindromes (1440 bp) were observed, along with a fragment common to both rDNA alleles (B/C3; 432 bp). No variation was detected between clonal TX607 and TX610 lines. In contrast, while all five TX611 clones retained the C3-long rDNA species, three clones contained palindromic B rDNA as well. Thus, the vegetative C3 rDNA replication advantage (22) is somewhat compromised in C3-long minichromosomes.
The novel XbaI fragment detected with probe B in TX611 was smaller than the B rDNA palindrome rather than larger (Figure 2C), while the relative size of EcoRV C3 and B rDNA products was reversed (Figure 2B). This observation suggested that the new rDNA species contained sequences upstream of the first XbaI site adjacent to the rDNA 5′ NTS (Figure 2A, micronuclear rDNA schematic, −905 bp). Southern blot hybridization with the micronuclear-limited rDNA probe A confirmed this prediction (Figure 2A, micronuclear rDNA schematic; Figure 2C, right panel). We conclude that this new rDNA species was produced by aberrant rDNA processing, and propose that the RFB facilitates the proper excision of the rDNA from its parental chromosome.
PFGE was used to examine the size and organization of the novel rDNA species in TX611. Southern blot analysis of undigested DNA revealed that the C3-long minichromosome is ∼2 kb larger than the 21 kb wild-type palindrome (Figure 3A). To determine if this molecule is palindromic (Figure 3B, schematic), genomic DNA was digested with NcoI or MluI. The rDNA contains a single site for each enzyme, and there are no sites in the 30 kb interval upstream of the rDNA locus (E. Orias, personal communication). Hybridization with a 5′ NTS probe detected a 7.3 kb NcoI fragment and 6.4 kb MluI fragment in TX611 DNA samples (Figure 3C, lanes 5 and 6), consistent with the expectations for palindromic C3ΔRFB minichromosomes (Figure 3C, see table for expected fragment lengths). The size of the C3ΔRFB fragments relative to wild-type controls indicated that mutant rDNA contains an additional 1.6 kb segment.
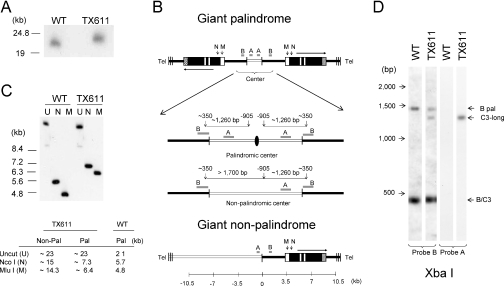
Figure 3.
The macronuclear rDNA species, C3-long, is a giant head-to-head inverted repeat with a palindromic central region. (A) Pulse field gel analysis of undigested total genomic DNA from wild-type (CU428) and ‘C3-long’ ΔRFB [TX611(1)] strains hybridized with probe B. (B) Restriction maps for three possible ‘C3-long’ macronuclear rDNA configurations: giant inverted repeat with a palindromic center (top), giant inverted repeat with a non-palindromic center (middle) and giant monomer (bottom). Black lines: macronuclear-destined 5′ NTS sequences in wild-type minichromosomes; open lines: upstream sequences that are not associated with wild-type macronuclear rDNA. The enlarged central region in the rDNA palindrome maps exhibit the position of XbaI sites (vertical arrows) and predicted restriction fragments that would be detected with probes A or B for molecules with palindromic or non-palindromic central cores. The black oval corresponds to the axis of symmetry for the species with a palindromic center. Note that probe B spans the XbaI site in the normal 5′ NTS. (C) Southern blot hybridization of uncut (U), MluI (M) or NcoI (N) digested DNA with probe B. Note: The apparent slower migration of uncut wild-type control DNA is a gel electrophoresis artifact (‘smiling’). The table shows the expected fragment sizes for monomeric (non-pal) and palindromic C3-long species. (D) Southern blot hybridization of XbaI- digested DNA with probes A and B (see panel B map for predicted product sizes for molecules with palindromic or non-palindromic centers).
Taking into account the size of the ΔRFB deletion (0.37 kb), the upstream rearrangement site would be ∼1.2 kb upstream of the normal Cbs fragmentation site if the additional C3-long sequences had rearranged into a palindrome (Figure 3B, giant palindrome/palindromic center). The size of the ∼5.5 kb band detected in EcoRV digested genomic DNA (Figure 2B) is also consistent with a palindromic organization. This band contains the ∼2 kb palindromic center (micronuclear sequences) and is flanked by ∼1.8 kb (2172–363 bp) (see also Figure 3). Alternatively, the breakpoint would be ∼2.0 kb upstream of the normal excision site if the additional segment was not rearranged (Figure 3B, giant palindrome/non-palindromic center). XbaI digestion was used to distinguish between these possibilities. A single 1.3 kb species was detected with probe A (Figure 2C). The additional 1.7 kb product that would be detected with probe B for a non-palindromic center was not observed. Thus, the additional sequences at the center of C3-long have a palindromic configuration.
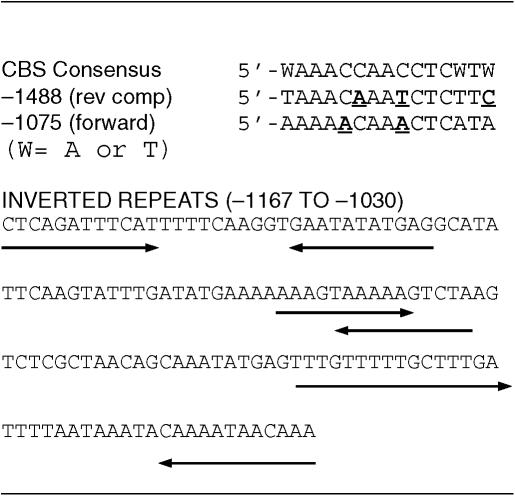
Examination of the DNA sequence around the predicted C3-long fragmentation site revealed two segments of partial homology with the Cbs consensus: 5′-WAAACCAACCTCWTW (where W stands for A or T) (Table 2) (33). The distal homology at position −1488 (5′-GAAGAGATTTGTTTA, reverse complement 5′-TAAACAAATCTCTTC) contains three positions that deviate from the consensus (underlined bases), two of which render the Cbs non-functional when placed in competition with an adjacent wild-type element (38). The proximal Cbs homology at position −1075 (forward: 5′-TATGAGTTGTTTTT, reverse complement: 5′-AAAAACAAACTCATA) includes two mismatches with the consensus, one of which is known to ablate function. Wild-type palindromes are formed by intramolecular recombination involving inverted, repeated 42 bp segments (M elements) that are separated by a 29 bp non-palindromic spacer (18). Therefore, we used the M-fold algorithm to look for inverted repeats. Several short repeats were found, 10–12 bp in length, in the proposed region for C3-long palindrome formation (spanning nucleotide positions −1167 to −1030) (Table 2). The tight clustering of three inverted repeats raises the possibility that these sequences act concertedly to promote homologous recombination. We propose that the RFB deletion derivative is fragmented in the vicinity of the clustered inverted repeats, and that these sequences direct the rearrangement of monomeric rDNA into a giant palindrome analogous to endogenous rDNA. Alternatively, chromosome 1 may be broken at one or more sites further upstream and eroded to expose sequences that promote palindrome formation.
Table 2.
DNA sequences proximal to the C3-long 5′ rDNA processing site
Alternative chromosome fragmentation in rDNA replication fork barrier deletion mutants
The stochastic rDNA excision defect associated with the RFB deletion allele precluded us from assessing whether this segment plays a direct role in rDNA gene amplification. For example, population analysis of mass matings involving germline ΔRFB transformant strains would not be informative if two sub-populations were present, one that failed to excise the ΔRFB rDNA allele and another that excised the rDNA at normal or alternative site(s). Additional genetic crosses allowed us to verify the role of the RFB region in rDNA excision and gain further insight into alternative processing pathways. We were particularly interested in determining whether normal C3ΔRFB palindromes could be produced from the C3-long mutant, and whether the aberrant rDNA excision site detected in TX611 was preferentially utilized in subsequent generations.
To this end, we first mated TX611 with A* strains (mating type III or V) to generate new progeny that could be mated to one another and to various tester strains. A* strains lack a functional micronucleus and induce an alternative developmental program, termed genomic exclusion, in which the parental macronucleus is retained and the progeny micronucleus becomes homozygous at all loci (27). Since no new macronucleus is created, exconjugants are sexually mature (round 1 genomic exclusion) and can re-mate with other cells in the culture. For reasons that are unclear, none of the clonal lines established after the first round of mating contained the genotype predicted for the products of round 1 genomic exclusion. One possible explanation is that a significant fraction of TX611 cells had lost one or more essential genes from their micronucleus during the vegetative cell divisions required to achieve sexual maturity (minimum of 70 fissions).
To overcome this problem, conjugating TX611 × A* cells were allowed to go through two rounds of mating, the later of which essentially functions as a selfing cross. Paired cells were isolated at 24 h (in all likelihood round 2 mating pairs) and several clonal lines were established. The sexual immaturity of these strains indicated that they had generated a new macronucleus. Southern blot analysis revealed that the C3-long rDNA species was regenerated in the two examined strains, [TX611(C1) and TX611(C3) (Figure 2B, left panel, lanes 6 and 7)]. Furthermore, C3ΔRFB palindromes were detected in the TX611(C1) macronucleus. The reappearance of C3-long rDNA in both strains indicates that the alternative processing site is preferentially utilized. The formation of C3ΔRFB palindromes in TX611(C1) demonstrates that the TX611 germline deletion allele can undergo normal excision and palindrome formation. It also implies that rDNA excision occurs after cells have replicated micronuclear chromosome 1 to at least a 4C content (two C3ΔRFB and two B rDNA alleles) (39,40).
TX611(C1) and TX611(C3) were grown to sexual maturity and mated with tester strains CU427 (wild-type B rDNA) and SB1934 (wild-type C3 rDNA). DNA was harvested from mass matings late in macronuclear development to assess macronuclear rDNA composition prior to the selection for rDNA fitness during vegetative cell divisions. Since the pair efficiency was 70–80%, most of the rDNA detected by Southern blotting was derived from progeny cells. The C3ΔRFB long rDNA species was abundantly represented in all four matings (Figure 2B, right panel, lanes 1–4). Two of the four matings also generated palindromic ΔRFB molecules (lanes 1 and 2), as well as fainter bands (designated X) that appear to result from fragmentation at new sites. Palindromic C3ΔRFB molecules were noticeably absent in the other two mating cell populations. Instead, the larger of the two new rDNA species was much more abundant (Figure 2B, right panel, lanes 3 and 4). We conclude that the C3ΔRFB allele is fragmented at a limited number of new sites, and that these events take place during macronuclear development.
The C3ΔRFB allele destabilizes chromosome 1 in the germline micronucleus
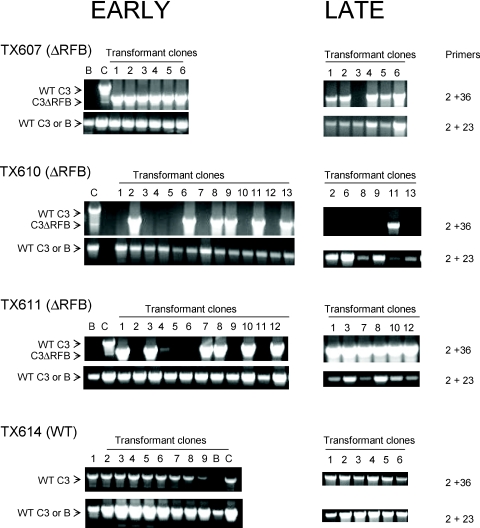
Our initial PCR screen of clonal transformant lines identified strains that were heterozygous for the C3ΔRFB and B rDNA alleles in the germline micronucleus (Figure 1B). Unexpectedly, micronuclear genotyping at ∼10 passages (∼70 fissions) revealed that the C3ΔRFB allele was lost from the micronucleus in a substantial fraction of these clones [Figure 4, early passages (C3 rDNA: primers 2 + 36); TX610 7/13 clones; TX611 6/12 clones; see Figure 1B for primer positions]. This observation suggested that the original clonal population became heterogeneous over time, consisting of cells that had either lost or retained the C3 rDNA allele in the germline micronucleus. Continued cultivation for 10 more passages (>150 fissions) revealed the loss of the C3ΔRFB allele in additional clonal lines (Figure 4, late passages, TX607 1/6 additional clones; TX610: 5/6 additional clones). While not quantitative, PCR analysis indicated that the product derived from the B rDNA allele was diminished and more variable in intensity at the later passages (Figure 4, B rDNA: primers 2 + 23). In contrast to C3ΔRFB clones, the wild-type C3 rDNA transformant showed no apparent loss of C3 or B rDNA alleles in the germline micronucleus (Figure 4, TX614).
Figure 4.
Loss of the micronuclear C3ΔRFB rDNA allele during vegetative propagation. Heterozygous germline transformants (C3ΔRFB/B or wild-type C3/B) were continually propagated following the establishment of clonal lines (transformant clones 1–13). Expanded clones were serially passaged by transferring ∼1000 cells into fresh media (1:100 dilution). DNA was isolated at ∼70 (early) and ∼150 (late) fissions and subjected to PCR amplification with primers that selectively amplify micronuclear C3 rDNA (wild-type C3 or C3ΔRFB alleles, primers 2 + 36), or wild-type C3 and B rDNA (primers 2 + 23). Control PCR templates—B: wild-type B rDNA strain (CU428), C: wild-type C3 rDNA strain (SB1934). The micronuclear genotype of clonal TX611 lines could not be unambiguously established due to the potential presence of macronuclear rDNA minichromosomes derived from aberrant rDNA excision (Figures 2 and 3, C3-long).
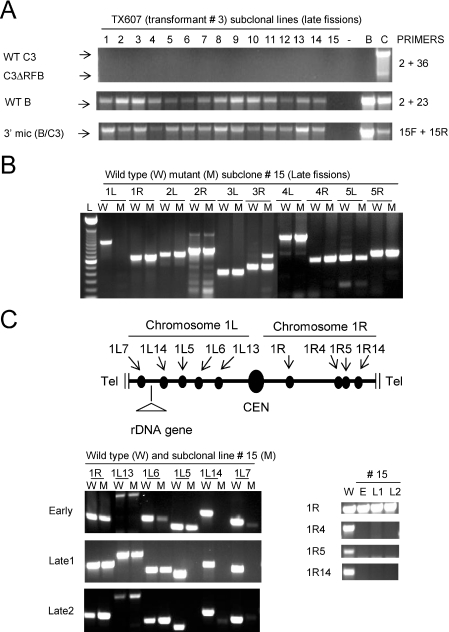
To explore the possibility that the micronuclear B rDNA allele was being deleted in heterozygous B/C3ΔRFB mutants, 15 subclones of TX607(3) were established and screened for micronuclear C3ΔRFB and B rDNA. All of these lines had lost the germline C3ΔRFB rDNA copy, while the subclone 15 was missing the B rDNA allele as well [Figure 5A, C3ΔRFB (primers 2 + 36) and B rDNA (primers 2 + 23)]. PCR analysis with primers that span the Cbs element at the 3′ end of the rDNA (Figure 1A, primers 15F + 15R) revealed that the entire rDNA locus was absent from the micronucleus of line 15 (Figure 5A). Additional primer sets derived from the left and right arms of each micronuclear chromosome (1–5L, 1–5R) yielded products in wild-type and mutants strains, with the exception of primer set 1L, which spans a Cbs fragmentation site distal to the rDNA locus on chromosome 1L (32,33). A robust product was obtained with wild-type DNA, but no signal was detected for TX607(3) subclone 15 (Figure 5B). These results are inconsistent with a local breakage and re-joining event. Instead, they suggest that chromosome 1 was fragmented such that the entire 1L arm distal to the rDNA was lost in this subclone.
Figure 5.
Progressive loss of the chromosome 1L arm and loss of the distal 1R region. (A) Germline PCR analysis of newly established clonal lines derived from TX607(3) at ∼150 fissions (see Figures 1A and 1B for PCR primer positions). The germline C3ΔRFB allele was absent in all the 15 lines and the B rDNA homolog was also missing in the subclone 15. PCR primers 15F and 15R span the Cbs element at the 3′ end of the micronuclear rDNA locus in intact micronuclear chromosomes. B: wild-type B rDNA strain, CU428; C: wild-type C3 rDNA strain, SB1934. (B) PCR analysis of micronuclear-limited DNA fragments from the left and right arms of the five micronuclear chromosomes. While most of the analyzed Cbs junctions have not been precisely positioned on their respective micronuclear chromosome, the primer sets used to examine chromosomes 4 and 5 map close to micronuclear telomeres (E. Hamilton and E. Orias, personal communication). W: wild-type B rDNA strain CU428, M: TX607(3) subclone 15. Lane 1 (L): 100 bp DNA ladder. (C) Loss of 1L and 1R markers during prolonged propagation of TX607(3) subclone 15. Top panel: map depicting the relative order of chromosome 1 Cbs junctions that were subjected to PCR analysis in wild-type (CU428) and mutant (TX607(3) subclone 15) strains (CEN: centromere, Tel: telomere). TX607(3) subclone 15 (derived from TX607(3) late passage) was serially propagated and subjected to PCR analysis at early (E: ∼70 fissions) and late [late 1 (L1): ∼150, late 2 (L2): ∼250] fissions (bottom panels).
The chromosome 1L and 1R arms are unstable during vegetative propagation of the C3ΔRFB mutant strain TX607(3) subclone 15
The loss of B and C3 rDNA alleles from micronuclear chromosome 1 allowed us to assess whether these chromosome 1 derivatives were stable or subjected to further erosion or rearrangement during vegetative cell divisions. To examine these possibilities, TX607(3) subclone 15 was continually propagated and DNA was isolated at defined intervals: early (E, ∼70 fissions), late 1 (L1, ∼150 fissions) and late 2 (L2, ∼250 fissions). PCR analysis with primer sets that span different Cbs elements in the micronuclear chromosome 1L arm (Figure 5C, schematic), revealed the progressive loss of 1L DNA within the entire cell population. For example, primer sets 1L7 and 1L14 (distal and proximal to the rDNA, respectively) failed to generate PCR products at both the early and late 1 time points (Figure 5C, lower left panel). The next marker, 1L-5, was present at the early time point and absent in the late 1 population. Only centromere-proximal Cbs markers were stably detected (primer sets 1L6, 1L13 and 1R).
These data suggest that the left arm is progressively eroded. While the relative order of the retained markers is known, their physical distances from 1L-5 have not been established. Additional PCR analysis with distal chromosome 1R markers (1R4, 1R5 and 1R14) failed to generate products at early and late passages, indicating that instability was not restricted to the 1L (rDNA-containing) chromosome arm. The early loss of several 1R markers in this line is consistent with stochastic fragmentation at a centromere-proximal site prior to subcloning.
C3ΔRFB strains exhibit an abnormal transition from conjugal development to the first vegetative cell division
The C3ΔRFB allele induces stochastic, progressive deterioration of micronuclear chromosome 1 during vegetative cell divisions, but has no obvious effect on macronuclear rDNA copy number or rRNA expression. To further explore the effect of this mutation on micronuclear chromosome fitness, we propagated heterozygous mutant strains vegetatively and then examined their ability to generate viable progeny by microscopic examination of developmental landmarks. Mating proficiency and macronuclear biogenesis were examined in crosses involving heterozygous mutants [TX607(1), TX610(11) and TX611(1)]. Whereas these parental strains tested positive for micronuclear C3ΔRFB and B rDNA alleles immediately prior to use, we anticipated that they contained sub-populations that had lost one or both micronuclear rDNA alleles.
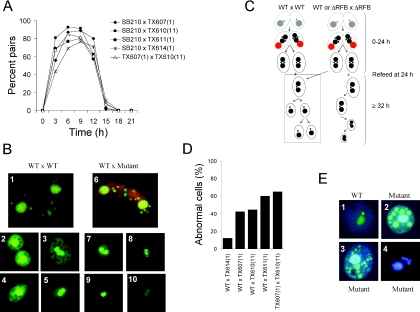
The kinetics of pair formation and separation in matings involving C3ΔRFB/B mutants and tester strains, SB210 or CU428, were comparable with the wild C3 rDNA transformant [TX614(1)] and with matings between two wild-type tester strains (CU427 × SB1934) (Figure 6A and data not shown). Cytological examination of wild-type [TX614(1)] or mutant-containing mating pairs with apofluor revealed the expected progression through developmental landmarks (Figure 6C; wild-type schematic T = 3–24 h), including the generation of mating partners with four pronuclei (Figure 6B, micrograph 1), acidification and degradation of the old parental macronucleus (Figure 6C, step 2), pair separation (Figure 6B, micrograph 2) and formation of the typical exconjugants, which contain two new macronuclei (macronuclear anlagen) and two micronuclei (Figure 6B, micrograph 5) [reviewed in (14)]. Since these events are controlled by the parental macronucleus, we expected that the C3ΔRFB mutation would not disrupt these processes. Occasionally, mating cells with an elevated number of pronuclei were observed in the mutant [Figure 6B, compare micrographs 1 (4 pronuclei) and 6 (8 or 9 pronuclei)].
Figure 6.
Abnormal transition from conjugal development to the first vegetative cell division in heterozygous C3ΔRFB/B progeny. (A) Normal kinetics for pair formation and separation in C3ΔRFB mutant progeny. Microscopic analysis of pair formation and separation in wild-type crosses [SB210 × TX614(1)], crosses between wild-type and mutant strains [SB210 × TX607(1), SB210 × TX610(11), SB210 × TX611(1)] and cross between two mutant strains [TX607(1) × TX610(11)]. (B) Cytological examination of crosses between wild-type strain SB210 and wild-type C3 rDNA transformant, TX614(1) (WT × WT),or SB210 and C3ΔRFB transformant, TX607(1) (WT × Mutant) with the DNA staining dye apofluor. The vast majority of cells displayed normal progression through development (data not shown), with a small percentage of mutant mating partners containing extra micronuclei/pronuclei prior to genetic exchange (compare micrographs 1 and 6). Panels 2–5 (WT × WT) and 7 ×10 (WT × Mutant) depict representative cells in exconjugant mating populations (24 h mating followed by 7–9 h re-feeding). (C) Cartoon depicting the progression of mating cells during development (0–24 h) and the fate of exconjugants after re-feeding at 24 h for (WT × WT), and (WT × mutant) or (mutant × mutant) crosses. See text for details. (D) Quantification of the number of ‘abnormal cells’ for different mating 7–9 h after re-feeding (see text for details). (E) Progeny of crosses between TX611(1) and A* mating type III obtained by single pair isolation after 23–25 h and subsequent propagation for 5–10 passages. Note the appearance of extra nuclei in the mutant (micrographs 2–4) compared with wild-type (micrograph 1). Staining: apofluor (micrographs 1–3), DAPI (micrograph 4).
Although conjugal events associated with parental micro- and macronuclei were largely unaffected, the ability of exconjugants to propagate was significantly compromised in C3ΔRFB mutants. This was evident when exconjugants were refed and allowed to complete macronuclear development, at which time they were solely dependent on gene expression from the new progeny macronucleus. When wild-type mating cultures were examined 8 h after re-feeding (32 h post-mating), the vast majority of cells contained two macronuclei and one micronucleus, and had a large cytoplasmic diameter expected for exconjugant progeny (Figure 6B, micrograph 3, fourth diagram in Figure 6C). Actively dividing and non-dividing cells with a single large macronucleus and large cytoplasm were also observed (Figure 6B, micrographs 2 and 4); the latter of which could be new progeny or parental cells that failed to mate. Approximately 10% of the cells from wild-type matings had not yet reabsorbed one of the progeny micronuclei (Figure 6B, micrograph 5). These cells were classified as abnormal due to their temporal developmental delay or arrest [Figure 6D, WT × TX614(1) graph]. The frequency of abnormal cells in wild-type × C3ΔRFB/B mutant mating was significantly elevated at the exconjugant stage (Figure 6D, 40–65%). Aberrant cells consisted primarily of arrested exconjugants (2 macronuclei and 1 micronucleus) with an extremely small cytoplasm (Figure 6B, micrographs 7–10). These small cells eventually disappeared from the culture, indicating that the ΔRFB parental strains are less fertile.
Vegetative cultures derived from crosses involving TX607(1) and CU427 contained a small, but significant percentage of viable cells that had an elevated number of nuclear structures that stained with apofluor or DAPI. These cultures appeared normal at early cell divisions, but progressively worsened over time. Whereas extra nuclei were not observed in primary C3ΔRFB/B transformants, this cytological phenotype occurred at a high frequency in subsequent generations (Figure 6E). These results suggest that essential genes are not transmitted to progeny. Thus, in a sub-population of cells, the deletion of the RFB region initiates a cascade of events that destabilize the wild-type and mutant homologs of chromosome 1 and render the micronucleus insufficient to support normal development in the next generation.
Prolonged vegetative propagation of ΔRFB parental strains (TX610 and TX611) resulted in even more severe defects in development and progeny formation. While virtually all of the TX610 × TX611 mating pairs examined in Figure 6 produced exconjugants, only 10–20% of the mating pairs generated exconjugants in a subsequent mating with older parental strains (data not shown). Instead, cells typically arrested randomly at earlier developmental stages. The reduced viability of exconjugants derived from ‘young’ RFB deletions strains and diminished fertility of ‘older’ RFB deletion strains (i.e. fewer exconjugants) is consistent with the progressive deterioration of the micronuclear genome during vegetative cell divisions.
DISCUSSION
The rDNA minichromosome of Tetrahymena thermophila is generated de novo by developmentally programmed excision of the single integrated rDNA gene copy. Origins in the 5′ NTS repeatedly fire to amplify macronuclear rDNA minichromosomes, and forks moving toward the center of the rDNA palindrome arrest at a strong replication fork barrier. Once development is complete, replication initiates once per cell cycle and forks no longer arrest at the RFB. Our analysis of germline transformants deleted for the RFB revealed that this segment not only acts in cis to promote programmed excision of the rDNA in the developing macronucleus, but is also needed to prevent spontaneous chromosome breakage in the mitotic germline micronucleus. Our ability to uncover and study the fragmentation and progressive degeneration of micronuclear chromosome 1 was possible because the diploid micronucleus of ciliated protozoa is not transcribed during vegetative cell divisions, and consequently is not required for cell viability. The loss of both chromosome 1L arms, for example, would be lethal in conventional eukaryotes that contain a single transcriptionally-active diploid nucleus.
rDNA excision in the developing for macronucleus
The sites for programmed cleavage of chromosomes in the developing Tetrahymena macronucleus share a common 15 bp motif in intervening micronuclear DNA, the Cbs (15). Cbs elements are remarkably conserved and sequence variation is poorly tolerated (15,32,41). Functional studies of clustered Cbs' immediately upstream of the rDNA revealed that these elements dictate the position of site-specific chromosome breakage and telomere addition and that these processes are mechanistically coupled (16). Our analysis of an rDNA segment encompassing the developmentally regulated RFB indicates that this region promotes excision at the proximal Cbs elements upstream of the rRNA gene. Correct excision is observed in a sub-population of cells, while other cells fail to produce C3 rDNA minichromosomes altogether or utilize an alternative processing pathway. Molecules that are properly excised subsequently rearrange into palindromic minichromosomes that replace endogenous B rDNA during vegetative propagation.
To our surprise cells that failed to properly excise the mutant rDNA allele could still generate macronuclear rDNA minichromosomes. The novel rDNA species that were observed bypassed the proximal Cbs elements entirely and were ‘processed’ at a limited number of positions, including a preferred site ∼1.1 kb upstream of the rDNA. Cbs-like sequences were present near the new ‘fragmentation sites’; however, they contain mismatches that have been shown to ablate Cbs function when placed in competition with wild-type Cbs elements (38). Since these alternative processing sites would be in competition with two fully functional Cbs elements at the 5′ end of the rDNA, we predict that excision occurs by a Cbs-independent mechanism.
The 5′ end of excised rDNA in the most prominent new macronuclear rDNA species, C3-long, is proximal to three short inverted repeats. These rDNA molecules had rearranged into a palindrome, similar to wild-type minichromosomes. The proximal inverted repeat sequences that were detected in the rearrangement region are not homologous to the M sequence repeat at the center of wild-type rDNA. Previous studies revealed that primary sequence is not important for palindrome formation; only a minimal repeat length of 20 bp is required (18). We propose that the three 10–12 bp clustered repeats near the C3-long breakpoint are sufficient to promote homology-induced recombination (17). Finally, in contrast to wild-type C3 minichromosomes (22) or properly processed C3ΔRFB palindromes, C3-long palindromes failed to out-replicate endogenous B rDNA. The added segment might contain sequences that repress origin activation or could diminish replication efficiency by increasing the distance between origins on opposite sides of the palindrome. In support of the later model, a previous study revealed that mutations that ablate origin function on monomeric circular episomes are fully functional in palindromic minichromosomes (42). In summary, these studies identify a new genetic determinant for chromosome breakage and uncover alternative sites and possibly mechanisms for chromosome fragmentation and rearrangement.
Deletion of the RFB region induces micronuclear chromosome instability
The most unexpected finding of this work is the discovery that the rDNA RFB promotes chromosome stability in the micronucleus. The Tetrahymena rDNA RFB region is the first example of a cis-acting DNA segment that actively protects a chromosome from spontaneous fragmentation. How this short sequence serves as a safeguard is unknown. One possible mechanism is that the RFB maintains the proper chromatin organization in the micronucleus. Micro- and macronuclear chromatin exhibit many distinguishing features, including the size of the nucleosome repeat length (200 versus 170 bp) (43), post-translational modification of histone subunits [reviewed in (44)] and biochemical makeup of the linker histone (45–47). Since the macronucleus is derived from a micronucleus, the entire genome must be remodeled during macronuclear development. While the RFB region is dispensable for macronuclear rDNA replication, it may be required to utilize the rDNA origin in the micronucleus. Deletion of the RFB could alter origin utilization, replication timing and/or the direction of replication fork movement1. We propose that the RFB region acts as a replication fork barrier in both the micronucleus and developing macronucleus, and represents the ‘ground state’ for chromatin that is remodeled later in development. Previous studies showed that the RFB is active prior to onset of rRNA transcription during development, while transient fork arrest at other sites (Figure 1A, PSE1–3) does not occur until the chromatin is competent for transcription (13).
The RFB deletion creates a hot spot for chromosome breakage, analogous to fragile sites in mammalian genomes. In the Tetrahymena case, removal of <400 bp renders chromosome 1 sensitive to breakage. By comparison, 0.6–5.5 kb CGG/CCG repeat expansions pre-dispose the human FRAXA locus to spontaneous breakage [reviewed in (48)]. Similarly, 10–70 kb expansions of a 33 bp microsatellite sequence promote breakage at another rare fragile site, FRA16B. In contrast to rare fragile sites, common fragile sites in the human genome are devoid of highly repetitive DNA sequence, analogous to the Tetrahymena rDNA locus.
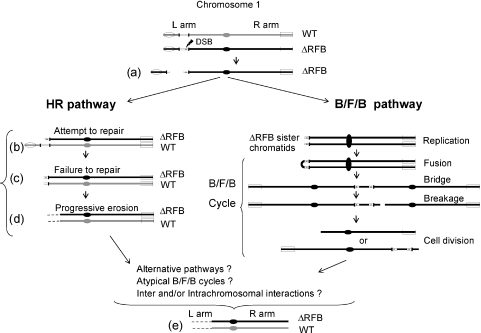
A recent S.pombe study demonstrated that fork arrest at a non-repetitive RFB induces genome instability, including homologous recombination and global chromosome rearrangement (2). Remarkably, removal of the Tetrahymena rDNA RFB appears to have the same effect. In both instances, activation of the DNA repair response is observed without the apparent involvement of the S phase checkpoint pathway (2) (J. S. Yakisich and G. M. Kapler, unpublished data). In the Tetrahymena case, fragmentation of micronuclear chromosome 1 initiates a cascade of events that not only affect the mutant ΔRFB chromosome (cis-acting effects), but also compromise the wild-type homolog as well (trans-acting effects). Two different mechanisms can account for most of the observed micronuclear genome instability. In both pathways the deleted RFB region triggers the formation of a DSB in the mutant (ΔRFB) chromosome [Figure 7, (a)]. In the first model, breakage of the mutant chromosome activates a homologous recombination (HR) repair pathway (Figure 7, HR). A second model proposes that the exposed DSB triggers breakage/fusion/bridge (B/F/B) cycles (Figure 7, B/F/B pathway).
Figure 7.
Models for the loss of the rDNA gene, progressive loss of the 1L chromosome arm and subsequent loss of 1R DNA sequences in heterozygous C3ΔRFB/B rDNA strains. Each micronucleus contains one copy of wild-type (WT) and mutant (C3 ΔRFB) rDNA in the respective chromosome 1 homologs. The jagged arrow in the mutant chromosome indicates a fragile site that induces a DSB (a). In one model (left), the attempted repair the DSB in the mutant chromosome by homologous recombination (HR) involves alignment of both homologs (b). Failure to repair the damaged chromosome induces DSB in the WT chromosome (c). The lack of telomere addition leads to progressive loss of 1L sequences on both homologs (d). The alternative model involves of B/F/B cycles on the fragmented C3ΔRFB chromosome. The DNA complexity generated by B/F/B cycles can be the same as that generated by homologous recombination for the C3ΔRFB allele. Loss of 1R DNA sequences (e) (see text for details).
Fragmentation of wild-type chromosome 1 must be mediated in trans, since this rDNA allele contains an intact RFB region. Consequently, the wild-type and ΔRFB homologs must interact during mitotic cell cycles. In the HR pathway, a process that requires pairing between homologs, the wild-type allele attempts to repair the fragmented mutant chromosome [Figure 7, (b)]. This futile effort generates a DSB in the wild-type chromosome [Figure 7 (c)] [reviewed in (49)]. Initial loss of the micronuclear C3ΔRFB locus and subsequent loss of the B rDNA allele (Figure 4) are consistent with the notion that the fragmented mutant chromosome promotes the loss of the wild-type homolog. In the next step, broken chromosomes are progressively eroded, possibly due to the inability of telomerase to add telomeres de novo onto exposed chromosome ends [Figure 7, (d)]. Accordingly, chromosome 1 markers proximal to the rDNA-proximal are lost from the population prior to centromere-proximal markers, a prediction supported by our analysis of numerous clonal lines (Figure 5C). The quantitative loss of a given DNA segment within the cell population indicates that fragmentation and subsequent erosion are common events.
In the second pathway broken C3ΔRFB sister chromatids fuse, generating a dicentric chromosome that initiates B/F/B cycles (Figure 7, B/F/B pathway), similar to those observed following the loss of a single telomere (50). Repeated cycles could lead to the loss of ordered markers on the C3ΔRFB chromosome; however, random breakage at each cell division would generate enormous heterogeneity within large cell populations. Furthermore, since fusion events are not driven by sequence homology, the wild-type homolog should not be selectively destabilized. Instead of deletions and translocations involving multiple chromosomes, we observed the fragmentation of the wild-type 1L arm, followed by the loss of markers between this breakpoint and the centromere. Finally, since the RFB mutation did not induce the massive global micronuclear genome instability like that associated with the absence of the origin binding protein, TIF1 (34) (T. L. Morrison, P. Sandoval, J. S. Yakisich and G. M. Kapler, unpublished data), the molecular data strongly support a homology-driven mechanism for the loss of the wild-type homolog and progressive erosion of exposed chromosome end.
Neither HR nor B/F/B cycles alone or in combination can account for the loss of the 1R arm [Figure 7, (e)]. While, circularization of macronuclear rDNA has been reported in a particular telomerase RNA mutant (51), we have no evidence for circularization of chromosome 1. Instead, the loss of 1R sequences suggests that the two chromosome 1 arms interact at some point. Recombination between repetitive sequences or non-homologous joining of a broken 1L arm with the 1R telomere could be involved.
Whatever the mechanism for chromosome degeneration, it is clear that the broken 1L chromosome is not stabilized. This deficiency may reflect intrinsic properties of Tetrahymena telomerase. Telomerase has been shown to elongate pre-existing macro- and micronuclear telomeric tracts in vivo (52,53). While de novo telomere addition is a hallmark of macronuclear development, it is temporally and mechanistically coupled to Cbs-mediated cleavage in the developing macronucleus (54). The efficiency of ‘chromosome healing’ in the mitotic micronucleus must be low at best, since chromosome 1 sequences were continually lost from entire mutant cell populations.
In conclusion, the Tetrahymena rDNA RFB region promotes programmed site-specific DNA fragmentation of the rDNA locus in the developing macronucleus and safeguards its micronuclear chromosomal counterpart from spontaneous breakage during vegetative cell divisions. The compartmentalization of gene expression and transmission functions into separate nuclei places different demands on the respective chromosomes. The long-term propagation of the species is dependent on the transmission of a full complement of genetic information during conjugation. Thus, the unexpected role of the RFB in the micronucleus must be a strong driving force for the acquisition and retention of these DNA sequences.
Acknowledgments
The authors thank Audrey Nicholson for the construction of the C3ΔRFB plasmid. They acknowledge the members of the Kapler laboratory for discussions, and thank Linda Guarino and Dorothy Shippen for advice and comments on the manuscript. This work was supported by NIH grant GM53572 to GMK. Funding to pay the Open Access publication charges for this article was provided by NIH grant GM53572.
Conflict of interest statement. None declared.
REFERENCES
- 1.Edenberg H.J., Huberman J.A. Eukaryotic chromosome replication. Annu. Rev. Genet. 1975;9:245–284. doi: 10.1146/annurev.ge.09.120175.001333. [DOI] [PubMed] [Google Scholar]
- 2.Lambert S., Watson A., Sheedy D.M., Martin B., Carr A.M. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell. 2005;121:689–702. doi: 10.1016/j.cell.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 3.Hyrien O. Mechanisms and consequences of replication fork arrest. Biochimie. 2000;82:5–17. doi: 10.1016/s0300-9084(00)00344-8. [DOI] [PubMed] [Google Scholar]
- 4.Bussiere D.E., Bastia D. Termination of DNA replication of bacterial and plasmid chromosomes. Mol. Microbiol. 1999;31:1611–1618. doi: 10.1046/j.1365-2958.1999.01287.x. [DOI] [PubMed] [Google Scholar]
- 5.Olavarrieta L., Hernandez P., Krimer D.B., Schvartzman J.B. DNA knotting caused by head-on collision of transcription and replication. J. Mol. Biol. 2002;322:1–6. doi: 10.1016/s0022-2836(02)00740-4. [DOI] [PubMed] [Google Scholar]
- 6.Sanchez-Gorostiaga A., Lopez-Estrano C., Krimer D.B., Schvartzman J.B., Hernandez P. Transcription termination factor reb1p causes two replication fork barriers at its cognate sites in fission yeast ribosomal DNA in vivo. Mol. Cell Biol. 2004;24:398–406. doi: 10.1128/MCB.24.1.398-406.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimizu N., Hashizume T., Shingaki K., Kawamoto J.K. Amplification of plasmids containing a mammalian replication initiation region is mediated by controllable conflict between replication and transcription. Cancer Res. 2003;63:5281–5290. [PubMed] [Google Scholar]
- 8.Dalgaard J.Z., Klar A.J. swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell. 2000;102:745–751. doi: 10.1016/s0092-8674(00)00063-5. [DOI] [PubMed] [Google Scholar]
- 9.Ivessa A.S., Zhou J.Q., Zakian V.A. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell. 2000;100:479–489. doi: 10.1016/s0092-8674(00)80683-2. [DOI] [PubMed] [Google Scholar]
- 10.Weitao T., Budd M., Campbell J.L. Evidence that yeast SGS1, DNA2, SRS2, and FOB1 interact to maintain rDNA stability. Mutat. Res-Fund. Mol. Mech. Mut. 2003;532:157–172. doi: 10.1016/j.mrfmmm.2003.08.015. [DOI] [PubMed] [Google Scholar]
- 11.Weitao T., Budd M., Hoopes L.L.M., Campbell J.L. DNA2 helicase/nuclease causes replicative fork stalling and double-strand breaks in the ribosomal DNA of Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:22513–22522. doi: 10.1074/jbc.M301610200. [DOI] [PubMed] [Google Scholar]
- 12.MacAlpine D.M., Zhang Z., Kapler G.M. Type I elements mediate replication fork pausing at conserved upstream sites in the Tetrahymena thermophila ribosomal DNA minichromosome. Mol. Cell Biol. 1997;17:4517–4525. doi: 10.1128/mcb.17.8.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z., MacAlpine D.M., Kapler G.M. Developmental regulation of DNA replication: Replication fork barriers and programmed gene amplification in Tetrahymena thermophila. Mol. Cell Biol. 1997;17:6147–6156. doi: 10.1128/mcb.17.10.6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karrer K.M. Tetrahymena genetics: two nuclei are better than one. Methods Cell Biol. 2000;Vol. 62:127–186. doi: 10.1016/s0091-679x(08)61529-0. [DOI] [PubMed] [Google Scholar]
- 15.Yao M.C., Zheng K.Q., Yao C.H. A conserved nucleotide-sequence at the sites of developmentally regulated chromosomal breakage in Tetrahymena. Cell. 1987;48:779–788. doi: 10.1016/0092-8674(87)90075-4. [DOI] [PubMed] [Google Scholar]
- 16.Yao M.C., Yao C.H., Monks B. The controlling sequence for site-specific chromosome breakage in Tetrahymena. Cell. 1990;63:763–772. doi: 10.1016/0092-8674(90)90142-2. [DOI] [PubMed] [Google Scholar]
- 17.Butler D.K., Yasuda L.E., Yao M.C. An intramolecular recombination mechanism for the formation of the ribosomal-RNA gene palindrome of Tetrahymena thermophila. Mol. Cell Biol. 1995;15:7117–7126. doi: 10.1128/mcb.15.12.7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yasuda L.F., Yao M.C. Short inverted repeats at a free end signal large palindromic DNA formation in Tetrahymena. Cell. 1991;67:505–516. doi: 10.1016/0092-8674(91)90525-4. [DOI] [PubMed] [Google Scholar]
- 19.King B.O., Yao M.C. Tandemly repeated hexanucleotide at Tetrahymena rDNA free end is generated from a single copy during development. Cell. 1982;31:177–182. doi: 10.1016/0092-8674(82)90417-2. [DOI] [PubMed] [Google Scholar]
- 20.Yao M.C., Blackburn E., Gall J.G. Amplification of the ribosomal-RNA genes in Tetrahymena. Cold Spring Harb. Symp. Quant. Biol. 1978;43:1293–1296. doi: 10.1101/sqb.1979.043.01.147. [DOI] [PubMed] [Google Scholar]
- 21.Saha S., Nicholson A., Kapler G.M. Cloning and biochemical analysis of the Tetrahymena origin binding protein TIF1—Competitive DNA binding in vitro and in vivo to critical rDNA replication determinants. J. Biol. Chem. 2001;276:45417–45426. doi: 10.1074/jbc.M106162200. [DOI] [PubMed] [Google Scholar]
- 22.Larson D.D., Blackburn E.H., Yaeger P.C., Orias E. Control of rDNA replication in Tetrahymena involves a cis-acting upstream repeat of a promoter element. Cell. 1986;47:229–240. doi: 10.1016/0092-8674(86)90445-9. [DOI] [PubMed] [Google Scholar]
- 23.Shang Y.H., Song X.Y., Bowen J., Corstanje R., Gao Y., Gaertig J., Gorovsky M.A. A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc. Natl Acad. Sci. USA. 2002;99:3734–3739. doi: 10.1073/pnas.052016199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orias E., Bruns P.J. Induction and isolation of mutants in Tetrahymena. Methods Cell Biol. 1976;13:247–282. [PubMed] [Google Scholar]
- 25.Bruns P.J., Cassidy-Hanley D. Biolistic transformation of macro- and micronuclei. Methods in Cell Biology. 2000;62:501–512. doi: 10.1016/s0091-679x(08)61553-8. [DOI] [PubMed] [Google Scholar]
- 26.Cassidy-Hanley D., Bowen J., Lee J.H., Cole E., VerPlank L.A., Gaertig J., Gorovsky M.A., Bruns P.J. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics. 1997;146:135–147. doi: 10.1093/genetics/146.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allen S.L. Genomic exclusion—a rapid means for inducing homozygous diploid lines in Tetrahymena pyriformis syngen I. Science. 1967;155:575–577. doi: 10.1126/science.155.3762.575. [DOI] [PubMed] [Google Scholar]
- 28.Engberg J., Din N., Saiga H., Higashinakagawa T. Nucleic Acids Res. Vol. 12. 1984. Nucleotide sequence of the 5′-terminal coding region for pre-rRNA and mature 17S rRNA in Tetrahymena thermophila rDNA; pp. 959–972. PubMed accession no. X00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Engberg J. The ribosomal-RNA genes of Tetrahymena—structure and function. Eur. J. Cell Biol. 1985;36:133–151. [PubMed] [Google Scholar]
- 30.Yao M.C., Zhu S.G., Yao C.H. Gene amplification in Tetrahymena thermophila—formation of extrachromosomal palindromic genes coding for ribosomal RNA. Mol. Cell Biol. 1985;5:1260–1267. doi: 10.1128/mcb.5.6.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao M.C., Zhu S.G., Yao C.H. Mol. Cell. Biol. Vol. 5. 1985. Gene amplification in Tetrahymena thermophila: formation of extrachromosomal palindromic genes coding for rRNA; pp. 1260–1267. PubMed accession no. M11155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cassidy-Hanley D., Bisharyan Y., Fridman V., Gerber J., Lin C., Orias E., Orias J.D., Ryder H., Vong L., Hamilton E.P. Genome-wide characterization of Tetrahymena thermophila chromosome breakage sites. II. Physical and genetic mapping. Genetics. 2005;170:1623–1631. doi: 10.1534/genetics.104.031435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamilton E.P., Bruns P.J., Lin C., Merriam V., Orias E., Vong L., Cassidy-Hanley D. Genome-wide characterization of Tetrahymena thermophila chromosome breakage sites. I. Cloning and identification of functional sites. Genetics. 2005;170:1611–1621. doi: 10.1534/genetics.104.031401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morrison T.L., Yakisich J.S., Cassidy-Hanley D., Kapler G.M. TIF1 represses rDNA replication initiation, but promotes normal S phase progression and chromosome transmission in Tetrahymena. Mol. Biol. Cell. 2005;16:2624–2635. doi: 10.1091/mbc.E05-02-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mpoke S.S., Wolfe J. Differential staining of apoptotic nuclei in living cells: Application to macronuclear elimination in Tetrahymena. J. Histochem. Cytochem. 1997;45:675–683. doi: 10.1177/002215549704500505. [DOI] [PubMed] [Google Scholar]
- 36.Yakisich J.S., Kapler G.M. The effect of phosphoinositide 3-kinase inhibitors on programmed nuclear degradation in Tetrahymena and fate of surviving nuclei. Cell Death Differ. 2004;11:1146–1149. doi: 10.1038/sj.cdd.4401473. [DOI] [PubMed] [Google Scholar]
- 37.Pan W.C., Blackburn E.H. Single extrachromosomal ribosomal-RNA gene copies are synthesized during amplification of the rDNA in Tetrahymena. Cell. 1981;23:459–466. doi: 10.1016/0092-8674(81)90141-0. [DOI] [PubMed] [Google Scholar]
- 38.Fan Q.C., Yao M.C. A long stringent sequence signal for programmed chromosome breakage in Tetrahymena thermophila. Nucleic Acids Res. 2000;28:895–900. doi: 10.1093/nar/28.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brunk C.F., Conover R.K. Elimination of micronuclear specific DNA sequences early in anlagen development. Mol. Cell Biol. 1985;5:93–98. doi: 10.1128/mcb.5.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yokoyama R.W., Yao M.C. Elimination of DNA sequences during macronuclear differentiation in Tetrahymena thermophila, as detected by in situ hybridization. Chromosoma. 1982;85:11–22. doi: 10.1007/BF00344591. [DOI] [PubMed] [Google Scholar]
- 41.Coyne R.S., Yao M.C. Evolutionary conservation of sequences directing chromosome breakage and rDNA palindrome formation in tetrahymenine ciliates. Genetics. 1996;144:1479–1487. doi: 10.1093/genetics/144.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reischmann K.P., Zhang Z., Kapler G.M. Long range cooperative interactions regulate the initiation of replication in the Tetrahymena thermophila rDNA minichromosome. Nucleic Acids Res. 1999;27:3079–3089. doi: 10.1093/nar/27.15.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gorovsky M.A., Glover C., Johmann C.A., Keevert J.B., Mathis D.J., Samuelson M. Histones and chromatin structure in Tetrahymena macronuclei and micronuclei. Cold Spring Harb. Symp. Quant. Biol. 1977;42:493–503. doi: 10.1101/sqb.1978.042.01.052. [DOI] [PubMed] [Google Scholar]
- 44.Jenuewin T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 45.Wu M., Allis C.D., Sweet M.T., Cook R.G., Thatcher T.H., Gorovsky M.A. Four distinct and unusual linker proteins in a mitotically dividing nucleus are derived from a 71-kilodalton polyprotein, lack p34(Cdc2) sites, and contain protein kinase α sites. Mol. Cell Biol. 1994;14:10–20. doi: 10.1128/mcb.14.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stargell L.A., Bowen J., Dadd C.A., Dedon P.C., Davis M., Cook R.G., Allis C.D., Gorovsky M.A. Temporal and spatial association of histone-H2a variant Hv1 with transcriptionally competent chromatin during nuclear development in Tetrahymena thermophila. Genes Dev. 1993;7:2641–2651. doi: 10.1101/gad.7.12b.2641. [DOI] [PubMed] [Google Scholar]
- 47.Yu L.L., Gorovsky M.A. Constitutive expression, not a particular primary sequence, is the important feature of the H3 replacement variant hv2 in Tetrahymena thermophila. Mol. Cell Biol. 1997;17:6303–6310. doi: 10.1128/mcb.17.11.6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sutherland G.R., Baker E., Richards R.I. Fragile sites still breaking. Trends Genet. 1998;14:501–506. doi: 10.1016/s0168-9525(98)01628-x. [DOI] [PubMed] [Google Scholar]
- 49.Dudas A., Chovanec M. DNA double-strand break repair by homologous recombination. Mutat Res. 2004;566:131–167. doi: 10.1016/j.mrrev.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 50.Sabatier L., Ricoul M., Pottier G., Murnane J.P. The loss of a single telomere can result in instability of multiple chromosomes in a human tumor cell line. Mol. Cancer Res. 2005;3:139–150. doi: 10.1158/1541-7786.MCR-04-0194. [DOI] [PubMed] [Google Scholar]
- 51.Romero D.P., Blackburn E.H. Circular rDNA replicons persist in Tetrahymena thermophila transformants synthesizing GGGGTC telomeric repeats. J. Eukaryot. Microbiol. 1995;42:32–43. doi: 10.1111/j.1550-7408.1995.tb01537.x. [DOI] [PubMed] [Google Scholar]
- 52.Yu G.L., Blackburn E.H. Developmentally programmed healing of chromosomes by telomerase in Tetrahymena. Cell. 1991;67:823–832. doi: 10.1016/0092-8674(91)90077-c. [DOI] [PubMed] [Google Scholar]
- 53.Kirk K.E., Harmon B.P., Reichardt I.K., Sedat J.W., Blackburn E.H. Block in anaphase chromosome separation caused by a telomerase template mutation. Science. 1997;275:1478–1481. doi: 10.1126/science.275.5305.1478. [DOI] [PubMed] [Google Scholar]
- 54.Fan Q.C., Yao M.C. New telomere formation coupled with site-specific chromosome breakage in Tetrahymena thermophila. Mol. Cell Biol. 1996;16:1267–1274. doi: 10.1128/mcb.16.3.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]