Abstract
microRNAs (miRNAs) and small interfering RNAs (siRNAs) in plants bear a methyl group on the ribose of the 3′ terminal nucleotide. We showed previously that the methylation of miRNAs and siRNAs requires the protein HEN1 in vivo and that purified HEN1 protein methylates miRNA/miRNA* duplexes in vitro. In this study, we show that HEN1 methylates both miRNA/miRNA* and siRNA/siRNA* duplexes in vitro with a preference for 21–24 nt RNA duplexes with 2 nt overhangs. We also demonstrate that HEN1 deposits the methyl group on to the 2′ OH of the 3′ terminal nucleotide. Among various modifications that can occur on the ribose of the terminal nucleotide, such as 2′-deoxy, 3′-deoxy, 2′-O-methyl and 3′-O-methyl, only 2′-O-methyl on a small RNA inhibits the activity of yeast poly(A) polymerase (PAP). These findings indicate that HEN1 specifically methylates miRNAs and siRNAs and implicate the importance of the 2′-O-methyl group in the biology of RNA silencing.
INTRODUCTION
Small interfering RNAs (siRNAs) are 21–24 nt sequence-specific mediators of RNA silencing. siRNAs are processed from long double-stranded RNAs through the action of an RNase III enzyme Dicer [reviewed in (1)]. The products of Dicer-mediated cleavage are duplexes of siRNAs with 2 nt overhangs in each strand. Usually one strand of the siRNA duplex is selectively incorporated into an RNA silencing effector complex known as the RISC while the antisense strand is degraded. The RISC loaded with the siRNA targets RNAs with sequence complementarity to the siRNA for cleavage. siRNAs are also known to target chromatin modifications, such as histone methylation and DNA methylation (2–4). In addition to siRNAs from viruses and transgenes, plants appear to be rich in endogenous siRNAs, such as siRNAs from repeat sequences, transposons, as well as a class of trans-acting siRNAs (5–12). Plant siRNAs belong to two major size classes, 21–22 nt and 24 nt. The 21–22 nt class mediates RNA cleavage while the 24 nt class is believed to cause chromatin modification (5).
microRNAs (miRNAs) are 21–24 nt RNA products of non-protein coding genes. In animals, the biogenesis of miRNAs from their primary precursors involves at least two RNase III enzymes [reviewed in (13)]. A Drosha-containing protein complex processes the pri-miRNA into a hairpin pre-miRNA, in which the miRNA is located within the stem of the hairpin. The pre-miRNA is further processed by Dicer to a duplex of the miRNA and its antisense strand miRNA*. The miRNA/miRNA* duplex resembles a siRNA/siRNA* duplex in that each strand has a 2 nt overhang, but differs from a siRNA/siRNA* duplex in that it usually contains mis-matches in the two strands. The miRNA strand is selectively loaded on to RISC, where the miRNA mediates sequence-specific regulation of target mRNAs.
In plants, the biogenesis of miRNAs and all types of siRNAs involves an additional step, methylation. Arabidopsis miRNAs and siRNAs carry a methyl group on the ribose of the 3′ terminal nucleotide (14–16). Methylation of the small RNAs requires the protein HEN1. In hen1 mutants, small RNAs lack methylation and have additional nucleotides, primarily uridines, on their 3′ ends, suggesting that one function of small RNA methylation is to protect the 3′ ends of the small RNAs from an as yet unidentified enzymatic activity in vivo (15). In vitro, HEN1 was found to act on two miRNA/miRNA* duplexes with different sequences but not DNA duplexes, single-stranded miRNA, pre-miRNA or tRNA, suggesting that HEN1 prefers the structure of miRNA/miRNA* duplexes (16). Furthermore, we showed that the 2′ OH and 3′ OH of the 3′ terminal nucleotide in miRNA/miRNA* duplexes are both required for the activity of HEN1 (16).
In the previous study, we found that HEN1 was unable to methylate two 21 nt siRNA/siRNA* duplexes in vitro (16), which raised the question of whether HEN1 can recognize and methylate siRNA duplexes on its own. In addition, the position of the methyl group on the ribose of the 3′ terminal nucleotide was not determined. In this study, we characterized the substrate specificity of HEN1, determined the position of the methyl group, and tested the effect of the methylation on other enzymes. We showed that HEN1 prefers miRNA/miRNA* duplexes of 21–24 nt and that HEN1 also works on siRNA/siRNA* duplexes between 21 and 24 nt but it prefers 23–24 nt siRNA/siRNA* duplexes. We showed that HEN1 exclusively methylates the 2′ OH of the 3′ terminal nucleotide and that this modification in a small RNA affects the ability of the small RNA to serve as a substrate for two enzymes tested.
MATERIALS AND METHODS
Expression and purification of HEN1 protein from Escherichia coli
Glutathione S-transferase (GST) and GST-HEN1 were expressed and purified as described previously (16).
Preparation of miRNA/miRNA* and siRNA/siRNA* duplexes
Single-stranded RNA oligonucleotides were purchased from Integrated DNA Technologies or Dharmacon RNA Technologies in high-performance liquid chromatography (HPLC) purified forms. The RNAs were resuspended in the annealing buffer [50 mM Tris–HCl (pH 7.6), 100 mM KCl and 2.5 mM MgCl2], and the concentrations of the RNAs were verified by spectrometry. Annealing of the complementary strands was performed by mixing equal amounts of RNAs, heating the mixture to 95°C for 5 min, cooling to 37°C and incubating at 37°C for 2 h followed by room temperature incubation for 1 h.
miRNA and siRNA methyltransferase assay
The methyltransferase assay was performed as described previously (16). Briefly, a 100 µl of reaction mixture containing 50 mM Tris–HCl (pH 8.0), 100 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 2 mM DTT, 5% glycerol, 2 µl RNasin (Promega), 0.5 µCi S-adenosyl-L-[methyl-14C]methionine ([14C]SAM) (58.0 mCi/mmole; Amersham Biosciences), 5 µg purified protein and 1 nmol RNA substrate was incubated at 37°C for 2 h. The reaction was extracted with phenol/chloroform. Small RNAs were ethanol precipitated and analyzed in a 15% polyacrylamide urea gel. The gel was treated with an autoradiography enhancer (En3hance from PerkinElmer) and exposed to X-ray film at −80°C.
For the methylation of miR173/miR173*C (duplex 2 in Table 1) for use in the HPLC analysis, about 200 µg of the annealed miR173/miR173*C duplex was incubated with purified GST-HEN1 or GST in the presence of 50 mM SAM at 37°C for 2 h. The miRNAs were extracted with phenol/chloroform followed by ethanol precipitation. The extracted miRNAs were annealed and methylated again with GST-HEN1 or GST.
Table 1.
Sequences and potential structures of small RNA duplexes
aThe antisense strands are marked by an asterisk. The length of each strand is indicated by the numbers in the parentheses.
bThe sense strands and antisense strands are in red and blue, respectively. The sequences (5′ – 3′) are displayed from left to right for the sense strands and right to left for the antisense strands. ‘-’ indicates the absence of a nucleotide at a position. Letters in black represent nucleotides not present in the original miR173/miR173*. Duplexes 18–22 contain a 2′ deoxyribose on the 3′ terminal nucleotide of miR173*.
Reverse-phase HPLC analysis of methylated miR173 nucleosides
The miR173/miR173*C RNAs (duplex 2 in Table 1) from the GST or GST-HEN1 reactions were dissolved in a 45 µl volume of 20 mM sodium acetate buffer (pH 5.3) with 5 mM ZnCl2 and 50 mM NaCl, and then digested with 5 U of nuclease P1 (USB; 1000 U/ml) for 60 min at 37°C. After digestion, 10 µl of 1 M Tris–HCl (pH 8.0) and 1 U of calf intestine alkaline phosphatase (Roche) were added, and incubation was carried out for 30 min at 37°C. The miR173/miR173*C hydrolysate was subjected to reverse-phase HPLC with a Phenomenex Luna C18 (250 × 4.60 mm) column at a flow rate of 0.8 ml/min. The mobile phase was 50 mM triethylamine acetate [TEAA (pH 7.6)] and 2% acetonitrile (ACN). A gradient was used where the concentration of ACN was gradually increased at 15 min. The running program was as follows: 0–15 min: 2% ACN; 15–20 min: linear increase to 100% ACN.
miRNA 3′ end adenylation with yeast poly(A) polymerase (PAP)
A 25 µl mixture containing 0.5 µl yeast PAP (USB; 600 U/µl), 0.5 µl or 2.5 µl [α-32P]ATP (10 µCi/µl;PerkinElmer) and 80 nM miR173 was incubated at 37°C for 10 min. The radiolabeled RNAs were analyzed in a 15% polyacrylamide urea gel. The radioactive signals were visualized with a PhosphorImager.
Ligation with T4 RNA ligase
Synthesized miR173 RNA standard and miR173 with a methyl group on either the 2′ or 3′ OH (miR173-2′OMe or miR173-3′OMe) were treated with calf intestine alkaline phosphatase (New England Biolabs) to remove the 5′P to prevent self-ligation. Ligation of the miR173 RNAs to an RNA linker (5′-pUAUGAAGCC), in which the 3′ OH is blocked by a C-3 spacer (CH2CH2CH2OH), was performed at 16°C for 16 h in a 10 µl mixture containing 20 U T4 RNA ligase (Amersham Pharmacia), 1 ng of dephosphorylated miR173 and 1 µg RNA linker. Free miR173 and miR173 ligated to the RNA linker were resolved by gel electrophoresis and detected by RNA filter hybridization using a probe complementary to miR173.
RESULTS
HEN1 has a strict size requirement for miRNA/miRNA* duplexes
Our previous studies (16) showed that miRNA/miRNA* duplexes, but not pre-miRNA, DNA duplexes, or single-stranded miRNAs, are preferred substrates of HEN1 in vitro. We also showed that a miRNA/miRNA* duplex with blunt ends did not serve as a good substrate indicating that the 2 nt overhang is an important feature in the substrate. Furthermore, we demonstrated that the 2′ and 3′ hydroxyl groups (OH) on the 3′ terminal nucleotide are required in cis for methylation. Other features of the miRNA/miRNA* duplex, such as the sequence, the length of the duplex, the number and the sequence of the overhanging nucleotides and the presence of unpaired nucleotides, or bulges, in the duplex, may also influence or determine whether the duplex can serve as a substrate of HEN1 but were not examined.
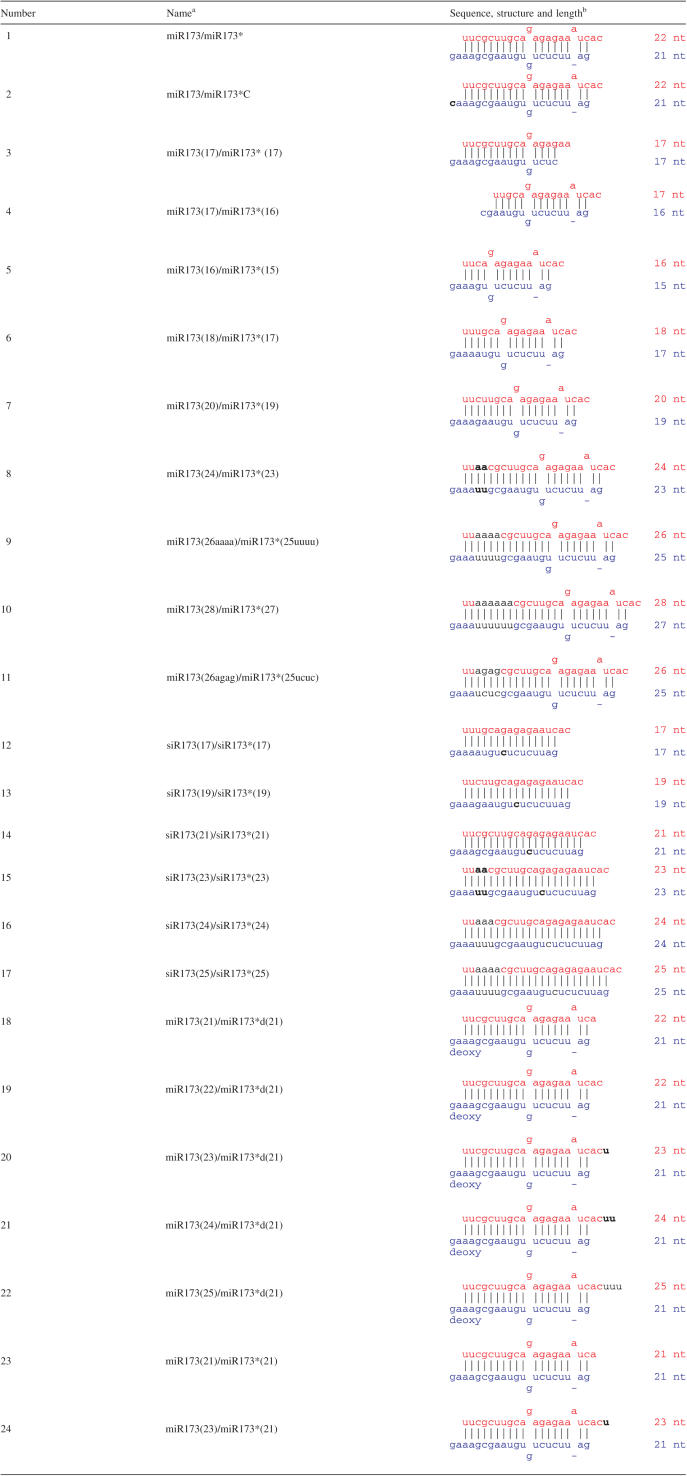
We decided to test the importance of these features by making various modifications of miR173/miR173* (duplex 1 in Table 1), which we previously showed to be a substrate of HEN1 (16). We first tested truncated versions of miR173/miR173* (duplexes 3 and 4 in Table 1) for their ability to serve as substrates for HEN1 in an in vitro methylation reaction. Duplex 3 represented a 5 nt truncation from the 3′ end of miR173 and a 4 nt truncation from the 5′ end of miR173* such that the length of the resulting duplex, the sequence of the overhanging nucleotides in the miR173 strand, and the number of bulges were different from the original miR173/miR173*. Duplex 4 represented a 5 nt truncation from the 5′ end of miR173 and the 3′ end of miR173* such that the length of the duplex and the sequence of the overhanging nucleotides in the miR173* strand were different from the original miR173/miR173*. While the miR173/miR173* duplex was methylated by HEN1 in vitro [(16), Figure 1a, lane 1], both truncated duplexes failed to be methylated (Figure 1a, lanes 3 and 4). In fact, both strands in the two duplexes failed to be methylated, although the overhanging sequences were not altered in one of the two strands. Since our previous work showed that both strands of the miR173/miR173* duplex are methylated in vitro, the fact that the sense strand of duplex 4 and the antisense strand of duplex 3, in which the sequences of the overhanging nucleotides were not affected, failed to be methylated suggests that the length of a duplex or the presence of bulges is an important feature in the substrate. In duplex 4, the number of bulges and the position of the bulges relative to the 3′ end of the sense strand are the same as that in miR173/miR173* and yet duplex 4 could not be methylated by HEN1. This suggests that the length of the RNA duplex is a critical factor in substrate recognition.
Figure 1.
In vitro methyltransferase reactions by GST-HEN1 on miRNA/miRNA* duplexes. Various RNA duplexes were methylated (see Materials and Methods) in the presence of [14C]-SAM. The RNAs were then resolved on polyacrylamide gels, which were exposed to X-ray films to obtain the autoradiograms. The numbers of the duplexes above the lanes correspond to the ones in Table 1. Since the activity of GST-HEN1 varies between protein preparations, assays performed with the same preparation of GST-HEN1 are on the same autoradiogram and are comparable to one another. Signals from (a) cannot be compared to those in (b).
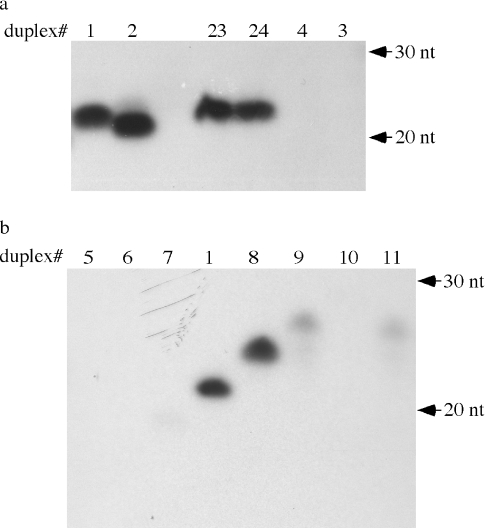
We further tested the length requirement by maintaining the 3′ overhanging nucleotides on both strands and introducing deletions or insertions of nucleotides in the fully complementary region encompassing the 5′ half of miR173 in the duplex. The deletions or insertions were introduced into position 3 from the 5′ end of miR173 so that the two bulges in the 3′ portion of the molecule were unlikely to be affected (Table 1). Duplexes 5–7 represented deletions of 6, 4 and 2 nt (starting from nucleotide 3 of the miR173 strand), respectively (Table 1). All three failed to be methylated by HEN1 in vitro (Figure 1b, lanes 5–7). Addition of two A-U base pairs (duplex 8 in Table 1) so that the two strands were 24 nt and 23 nt did not affect methylation efficiency (Figure 1b, lane 8). Addition of 4 A-U base pairs (duplex 9, Table 1) greatly reduced methylation efficiency (Figure 1b, lane 9) while addition of 6 A-U base pairs (duplex 10, Table 1) abolished methylation (Figure 1b, lane 10). To ensure that the effect of the additional nucleotides was not due to the sequence of the additional nucleotides, we tested another duplex (#11) with four additional nucleotides of different sequences from duplex 9 (Table 1). Duplex 11 behaved similarly to duplex 9 (Figure 1b, lanes 9 and 11). These data indicate that the most preferable length of miRNAs for HEN1-mediated methylation is 21–24 nt.
HEN1 methylates 21–24 nt siRNA/siRNA* duplexes in vitro
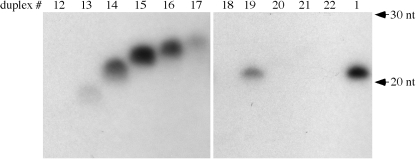
We previously reported that HEN1 was unable to methylate two 21 nt siRNA/siRNA* duplexes in vitro (16). Later we found that weak methylation signals could be observed upon prolonged exposure of the autoradiogram (data not shown), suggesting that HEN1 can act on siRNAs, perhaps at reduced efficiency. To further examine the ability of the enzyme to use siRNA/siRNA* duplexes as substrates as compared to miRNA/miRNA* duplexes, we designed siR173/siR173* (duplex 14 in Table 1) by eliminating the unpaired A in miR173 and by changing nucleotide 9 of miR173* from G to C such that the mismatch at this position is eliminated. Duplex 14 was tested together with miR173/miR173* under identical conditions and was found to be less well methylated as compared to miR173/miR173* (Figure 2, compare lane 14 to lane 1). This suggests that the mis-matches or bulges in the duplexes may be features recognized by HEN1. Alternatively, HEN1 may have a different size requirement for siRNA/siRNA* duplexes such that 21 nt is not an optimal length for siRNA/siRNA* duplexes.
Figure 2.
In vitro methyltransferase reaction by GST-HEN1 on siRNA/siRNA* duplexes and miRNA/miRNA* duplexes with 1–5 3′ overhangs. All duplexes in this figure were methylated with the same GST-HEN1 preparation and are therefore comparable to one another. The numbers above the lanes correspond to those in Table 1.
We tested the size requirement for siRNA duplexes. Starting from the 21 nt duplex 14, we deleted from position 3 of the siR173 strand 2 and 4 nt to result in a 19 nt duplex 13 and a 17 nt duplex 12, respectively. Duplex 13 was poorly methylated while duplex 12 was not detectably methylated by HEN1 (Figure 2, lanes 12 and 13). We also inserted 2, 3 or 4 A-U base pairs into position 3 of the siR173 strand of duplex 14. The 23 nt duplex 15 (Table 1) was much better methylated than the 21 nt duplex 14 (Figure 2, lanes 14 and 15). In fact, duplex 15 was methylated at a level comparable to that of miR173/miR173* (Figure 2, lanes 15 and 1). The 24 nt siRNA duplex 16 was less well methylated than the 23 nt duplex 15 (Figure 2, lanes 15 and 16). The 25 nt siRNA duplex 17 was barely methylated (Figure 2, lane 17). Therefore, HEN1 prefers 21–24 nt siRNA duplexes with 23 nt being the optimal length at least in the sequence context of siR173/siR173*.
HEN1 recognizes the 2 nt 3′ overhang
The 2 nt 3′ overhang is a feature specific to products of RNase III including miRNA and siRNA duplexes generated by Dicer (17). We determined whether the 2 nt 3′ overhang is an important feature in substrates of HEN1. We showed previously that a 2′ deoxy modification on the ribose of the 3′ terminal nucleotide prevented the methylation of the cis but not the trans strand of the duplex (16). Therefore, we employed miR173*d(21), which was identical to miR173* in sequence but which contained a 2′ deoxyribose on the 3′ terminal nucleotide, to allow us to monitor the methylation of only the miR173 strand in the duplex. We annealed miR173 oligos of various lengths to miR173*d(21) to generate duplexes 18–22 (Table 1) that have 1, 2, 3, 4 and 5 nt overhangs, respectively in the miR173 strand. As previously observed, duplex 19 with a 2 nt overhang in the miR173 strand was methylated (Figure 2, lane 19). All other duplexes, which had 1, 3, 4 or 5 nt overhangs in the miR173 strand, failed to be methylated (Figure 2, lanes 18, 20–22). This indicates a strict requirement for the length of the overhang for HEN1 activity.
We next asked whether the two overhangs on the two ends of the duplex were both required for the methylation of either end. We tested the ability of miR173* to be methylated when it was in duplexes 23 and 24 (Table 1) such that only the miR173* strand had a 2 nt overhang. The miR173 strands in duplexes 23 and 24 were the same as those in duplexes 18 and 20, respectively (Table 1). miR173* was efficiently methylated in these duplexes (Figure 1a, lanes 23 and 24). These results indicate that the 2 nt overhang acts in cis to allow methylation to occur.
HEN1 deposits a methyl group exclusively on to the 2′ OH of the 3′ terminal nucleotide
Chemical reactions showed that at least one of the two OH groups on the 3′ terminal nucleotide of small RNAs is blocked after methylation, suggesting that the methyl group is on either the 2′ OH, the 3′ OH or both positions. Chemical reactions done on miRNAs and siRNAs isolated from Arabidopsis showed that the small RNAs are blocked at the 2′ OH, 3′ OH or both positions in vivo. A recent study claimed that the methyl group is on the 2′ OH based on the fact that small RNAs, like 2′-O-Methyl (2′OMe) RNA oligonucleotides, showed reduced efficiency of ligation to an RNA linker by T4 RNA ligase (14). However, we found that a 3′OMe RNA oligonucleotide was also ligated to an RNA linker at a reduced efficiency as compared to an unmodified RNA (see below). Furthermore, the enzymatic reaction could not distinguish between 100% methylation at a single position versus a mixture of 2′ methylation and 3′ methylation in a population of molecules.
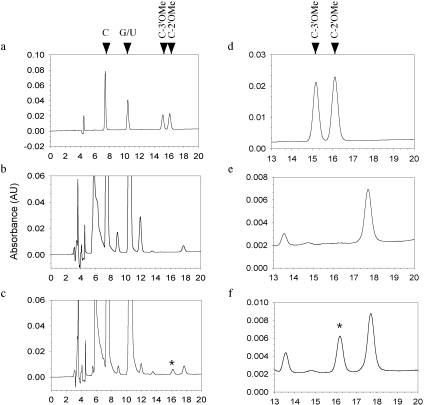
One way to definitively determine the position of the methyl group is to analyze the modified terminal nucleoside by HPLC. We first tested whether 3′OMe cytidine could be distinguished from 2′OMe cytidine by HPLC. We examined the elution profiles of 3′OMe cytidine, 2′OMe cytidine, cytidine, guanosine, uridine and adenosine on a C18 column. Indeed, 3′OMe cytidine and 2′OMe cytidine could be easily distinguished from each other and from other nucleosides (Figure 3a and d). Next, we designed an RNA oligonucleotide, miR173*C, by changing the last nucleotide of miR173* from G to C. We annealed miR173 to miR173*C (duplex 2 in Table 1) and performed the HEN1 methylation reaction. Since both strands would be methylated, the G to C change ensured that only methylated cytidine be present after the reaction. We digested the RNA oligonucleotides with nuclease P1 and calf intestine phosphatase to release the nucleosides, which were then loaded on to the HPLC column. The control reaction (with GST alone) was also similarly treated and examined by HPLC. By comparing the elution profile of the GST-HEN1 reaction (Figure 3c and f) to that of the GST reaction (Figure 3b and e), one extra peak was found in the GST-HEN1 profile (asterisk in Figure 3c and f). The elution time of the peak coincided with the 2′OMe cytidine standard (Figure 3a and d). The maximum absorbance of this peak is at 270 nm, characteristic of cytidine and methyl cytidine but distinct from other nucleosides (data not shown). This indicates that the extra peak represented 2′OMe. No peak corresponding to the 3′OMe cytidine standard was present. This result clearly demonstrated that HEN1 deposits a methyl group exclusively on to the 2′ OH of the ribose of the terminal nucleotide.
Figure 3.
Determination of the position of the methyl group by HPLC. (a) An elution profile of nucleoside standards. A, C, G, U, C-2′OMe and C-3′OMe were mixed and applied to the column. The retention times of C, G and U were determined by running each standard individually under identical conditions. The retention time of A was much longer than 20 min and is not shown. The retention times of C-3′OMe and C-2′OMe were determined by spiking the mix with more of either nucleoside in a separate run and noting which peak increased in amount. (b) An elution profile of the control methylation reaction with GST alone. (c) An elution profile of the methylation reaction with GST-HEN1. See ‘Materials and Methods’ for the treatment of the RNA duplexes before HPLC. The peak marked by an asterisk is the methyl-C generated by GST-HEN1. (d–f) Magnified versions of (a–c) in the region of 13–20 min.
2′-O-Methyl specifically affects yeast PAP
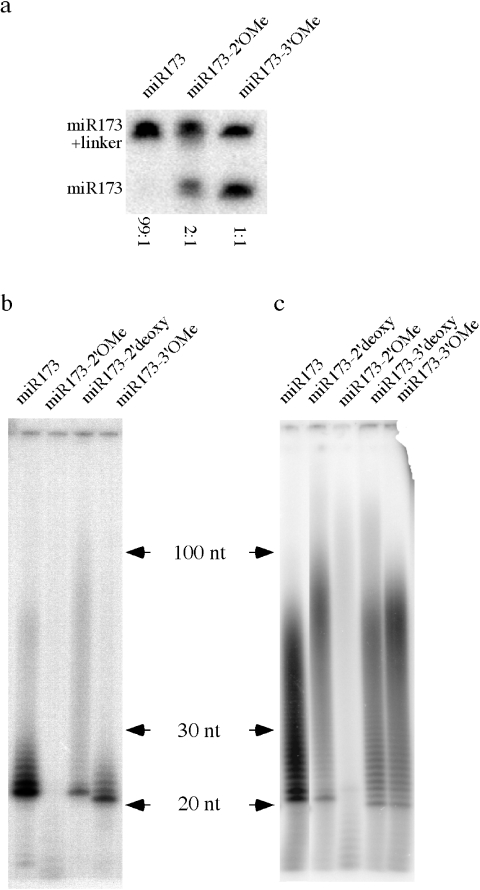
Having pinpointed the position of the methyl group allowed us to begin to determine the function of the modification on small RNAs biochemically. We tested the effect of a 2′OMe on the activities of two commercially available enzymes that act on RNAs, T4 RNA ligase and yeast PAP. We carried out ligation reactions of miR173, miR173 with a 2′OMe (miR173-2′OMe) and miR173 with a 3′OMe (miR173-3′OMe) to a linker molecule blocked at its 3′ end. After ligation, the small RNAs were resolved by electrophoresis, blotted to a membrane and detected by hybridization using a probe complementary to miR173. The ligated and free miR173 forms could be distinguished by their differential mobility (Figure 4a). In order to quantitatively compare the ligation efficiency of the three miR173 forms, we used an excess amount of the linker so that the miR173 forms would be limiting. Unmodified miR173 was nearly completely ligated to the linker. The 2′OMe group reduced the ligation efficiency by ∼30%. Intriguingly, HPLC purified miR173-3′OMe could also be ligated to the linker with an efficiency about 50% of the unmodified miR173. This indicates that T4 RNA ligase does not have an absolute requirement for a free 3′ OH. Consistent with this, miR173 with a 3′ deoxyribose on the 3′ terminal nucleotide could also be ligated to the linker under our experimental conditions (data not shown). Therefore, a methyl group on the 2′ OH of the terminal nucleotide of an RNA reduces the efficiency of T4 RNA ligase catalyzed ligation to the 3′ OH of the RNA. However, since a methyl group on the 3′ OH of the terminal nucleotide does not abolish T4 RNA ligase catalyzed ligation, this assay cannot be used to distinguish the position of the methyl group in small RNAs.
Figure 4.
Effect of various modifications on the 3′ terminal nucleotide of a small RNA on T4 RNA ligase- and yeast PAP-catalyzed reactions. (a) T4 RNA ligase-mediated ligation of various miR173 forms to an RNA linker. (b) Activity of yeast PAP on various forms of miR173 in the presence of 2 pmol [α-32P]-ATP. The ladders or smears represent products of PAP-catalyzed reaction. (c) Activity of yeast PAP on various forms of miR173 in the presence of 10 pmol [α-32P]-ATP.
We next evaluated the effect of 2′OMe on yeast PAP activity. miR173 and miR173-2′OMe were incubated with [α-32P]ATP in the presence of yeast PAP. When 2 pmol of [α-32P]ATP was present, a small number of A residues were added to miR173 but not miR173-2′OMe (Figure 4b). A similar observation was also made by Ebhardt et al. (14). We considered two possibilities that could account for this observation. One, the 2′ OH is necessary for substrate recognition or catalysis by PAP. Two, PAP does not require the 2′ OH but a 2′OMe prevents substrate recognition or catalysis. To determine whether the presence of a 2′ OH was an absolute requirement for PAP activity, we tested whether miR173 with a 2′ deoxyribose on the 3′ terminal nucleotide (miR173-2′deoxy) could be extended by PAP. A residues were added to miR173-2′deoxy under identical conditions, although the amount of the products was lower (Figure 4b). This indicates that a 2′ OH is not an absolute requirement for PAP activity but rather a 2′OMe inhibits PAP activity. We also tested whether a 3′OMe group affected PAP activity. Although PAP is supposed to require the presence of 3′ OH, we found that miR173-3′OMe could be extended, at a reduced efficiency as compared to miR173, by PAP (Figure 4b). This suggests that yeast PAP could form 2′–5′ phosphodiester bonds. In fact, other members of the polymerase beta-like nucleotidyltransferase superfamily, to which PAP belongs, are known to make 2′–5′ phosphodiester bonds (18). Alternatively, a contaminating exonuclease might have first released the 3′ terminal nucleotide of miR173-3′OMe and the remaining oligonucleotide was extended by PAP. Consistent with the exonuclease hypothesis, the labeled RNAs in the miR173-3′OMe reaction appeared ∼1 nt shorter than those in the other reactions. However, if this was the case, miR173-2′OMe was not subjected to the same activity. We also tested the effect of various terminal modifications on PAP in the presence of 10 pmol of [α-32P]ATP. More A residues were incorporated into the products under this higher ATP concentration (Figure 4c). 2′OMe did not eliminate but greatly reduced ATP incorporation as compared to miR173 (Figure 4c). The 2′ deoxy, 3′ deoxy and 3′OMe reduced the amount of ATP incorporation but did not have an adverse effect as severe as 2′OMe (Figure 4c).
DISCUSSION
The present and a previous study together define the features of a small RNA duplex that allow recognition by HEN1 for methylation in vitro: free 2′ and 3′ OH on the 3′ terminal nucleotide, 2 nt 3′ overhang, and a length of 21–24 nt. Although a full mechanistic understanding of HEN1 catalyzed small RNA methylation awaits the structure of HEN1 together with its substrate, we can begin to deduce how HEN1 recognizes its substrates and catalyzes the reaction. We envision that both OH groups on the ribose of the 3′ terminal nucleotide are necessary for the positioning of the 2′ OH in the active site of the enzyme to ensure that only the 2′ OH of the terminal nucleotide (but not internal nucleotides) is methylated. The fact that the 2 nt overhang acts in cis suggests that the two ends of the molecules are independently recognized and methylated. However, both ends of the substrate must be contacted for the methylation of one end since HEN1 measures the length of the strands. Perhaps, HEN1 recognizes the substrate as a dimer with each monomer having one active site (two active sites being present in a HEN1 monomer is unlikely due to the lack of any sequence repeats). The two monomers each binds to the 3′ overhang on one strand and catalyzes the methylation of that strand. This model, however, is based on the assumption that both strands in a single RNA duplex are methylated, which has not been proven. The observation that both strands were methylated [(16) and data not shown]was made on a population of molecules. It is possible that the methylated duplexes were random mixtures of ones with either strand methylated. Alternatively, HEN1 measures the length of the substrate as a monomer and methylates only one end at a time.
HEN1 measures the length of the small RNA. This, together with the requirement for a 2 nt overhang and the presence of 2′ and 3′ OH on the terminal nucleotide, probably ensures that only miRNAs and siRNAs are methylated. A viral RNA silencing suppressor p19 also measures the length of a siRNA duplex for binding. It does so as a homodimer, in which each monomer has two tryptophan residues that interact with the 5′ and 3′ ends of each RNA strand (19,20). p19 does not require the 3′ overhang in siRNA duplexes for binding. While HEN1 prefers the 2 nt 3′ overhang in RNA duplexes, it can also act on RNAs with blunt ends with reduced efficiency (16).
We demonstrated that methylation occurs exclusively on the 2′ OH of the 3′ terminal nucleotide. We showed that 2′OMe on the terminal nucleotide of an RNA oligonucleotide reduced the efficiency of ligation by T4 RNA ligase through the 3′ OH and greatly reduced the ability of yeast PAP to act on the 3′ OH. Therefore, 2′OMe can affect enzymatic activities that target the 3′ OH. Since small RNAs have additional U residues on their 3′ ends in the absence of methylation in vivo (15), the 2′OMe probably prevents an unknown activity from accessing the 3′ OH in vivo. RNAi can spread into sequences adjacent to those homologous to the initial trigger in the target in both Caenorhabditis elegans and in plants and the spreading requires RNA-dependent RNA polymerases (RdRPs) (21–23). It has been proposed that siRNAs serve as primers for RdRPs to generate secondary siRNAs (21). The presence of the 2′OMe in plant siRNAs may influence, positively or negatively, the ability of RdRPs to use siRNAs as primers.
Acknowledgments
The authors thank Richard H. Ebright for suggesting use of enzymatic digestion and HPLC analysis to define the methylation site. The authors thank Julien Curaba for critical reading of the manuscript. The work was supported by a National Science Foundation grant (MCB 0343480) to X.C. and by an NIH grant R01-GM41376 to Richard H. Ebright. Funding to pay the Open Access publication charges for this article was provided by National Science Foundation (MCB0343480).
Conflict of interest statement. None declared.
REFERENCES
- 1.Hammond S.M. Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- 2.Matzke M.A., Birchler J.A. RNAi-mediated pathways in the nucleus. Nature Rev. Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- 3.Mette M.F., Aufsatz W., van der Winden J., Matzke M.A., Matzke A.J. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 2000;19:5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morel J.B., Mourrain P., Beclin C., Vaucheret H. DNA methylation and chromatin structure affect transcriptional and post-transcriptional transgene silencing in Arabidopsis. Curr. Biol. 2000;10:1591–1594. doi: 10.1016/s0960-9822(00)00862-9. [DOI] [PubMed] [Google Scholar]
- 5.Hamilton A., Voinnet O., Chappell L., Baulcombe D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002;21:4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Llave C., Kasschau K.D., Rector M.A., Carrington J.C. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14:1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mette M.F., van der Winden J., Matzke M., Matzke A.J. Short RNAs can identify new candidate transposable element families in Arabidopsis. Plant Physiol. 2002;130:6–9. doi: 10.1104/pp.007047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peragine A., Yoshikawa M., Wu G., Albrecht H.L., Poethig R.S. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sunkar R., Girke T., Jain P.K., Zhu J.K. Cloning and characterization of microRNAs from rice. Plant Cell. 2005;17:1397–1411. doi: 10.1105/tpc.105.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sunkar R., Girke T., Zhu J.K. Identification and characterization of endogenous small interfering RNAs from rice. Nucleic Acids Res. 2005;33:4443–4454. doi: 10.1093/nar/gki758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sunkar R., Zhu J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vazquez F., Vaucheret H., Rajagopalan R., Lepers C., Gasciolli V., Mallory A.C., Hilbert J.L., Bartel D.P., Crete P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 13.Kim V.N. MicroRNA biogenesis: coordinated cropping and dicing. Nature Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 14.Ebhardt H.A., Thi E.P., Wang M.B., Unrau P.J. Extensive 3′ modification of plant small RNAs is modulated by helper component-proteinase expression. Proc. Natl Acad. Sci. USA. 2005;102:13398–13403. doi: 10.1073/pnas.0506597102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J., Yang Z., Yu B., Liu J., Chen X. Methylation protects miRNAs and siRNAs from a 3′ end uridylation activity in Arabidopsis. Curr. Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu B., Yang Z., Li J., Minakhina S., Yang M., Padgett R.W., Steward R., Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 18.Aravind L., Koonin E.V. DNA polymerase beta-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res. 1999;27:1609–1618. doi: 10.1093/nar/27.7.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye K., Malinina L., Patel D.J. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature. 2003;426:874–878. doi: 10.1038/nature02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vargason J.M., Szittya G., Burgyan J., Tanaka Hall T.M. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115:799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- 21.Sijen T., Fleenor J., Simmer F., Thijssen K.L., Parrish S., Timmons L., Plasterk R.H., Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 22.Vaistij F.E., Jones L., Baulcombe D.C. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell. 2002;14:857–867. doi: 10.1105/tpc.010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voinnet O., Vain P., Angell S., Baulcombe D.C. Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell. 1998;95:177–187. doi: 10.1016/s0092-8674(00)81749-3. [DOI] [PubMed] [Google Scholar]