Abstract
Polar auxin transport (PAT) plays a crucial role in the regulation of many aspects of plant growth and development. We report the characterization of a semidominant Arabidopsis thaliana bushy and dwarf1 (bud1) mutant. Molecular genetic analysis indicated that the bud1 phenotype is a result of increased expression of Arabidopsis MAP KINASE KINASE7 (MKK7), a member of plant mitogen-activated protein kinase kinase group D. We showed that BUD1/MKK7 is a functional kinase and that the kinase activity is essential for its biological functions. Compared with the wild type, the bud1 plants develop significantly fewer lateral roots, simpler venation patterns, and a quicker and greater curvature in the gravitropism assay. In addition, the bud1 plants have shorter hypocotyls at high temperature (29°C) under light, which is a characteristic feature of defective auxin action. Determination of tritium-labeled indole-3-acetic acid transport showed that the increased expression of MKK7 in bud1 or the repressed expression in MKK7 antisense transgenic plants causes deficiency or enhancement in auxin transport, indicating that MKK7 negatively regulates PAT. This conclusion was further substantiated by genetic and phenotypic analyses of double mutants generated from crosses between bud1 and the auxin-related mutants axr3-3, tir1-1, doc1-1, and atmdr1-1.
INTRODUCTION
The plant hormone auxin, mainly indole-3-acetic acid (IAA), plays a crucial role in a wide variety of developmental processes, including embryogenesis, apical dominance, lateral root formation, and vascular differentiation (Benkova et al., 2003; Leyser, 2003; Reinhardt et al., 2003; Morris et al., 2004). IAA is biosynthesized mainly in the shoot apex and young leaves, and its transport is quite complex both in shoots and in roots. The basipetal polar auxin transport (PAT) in shoots is important for lateral shoot inhibition because either blocking PAT or removing the shoot apex promotes shoot branching, although auxin does not enter the lateral bud directly, suggesting the existence of secondary messengers in mediating auxin action (Gil et al., 2001; Booker et al., 2003, 2004, 2005; Leyser, 2003; Sorefan et al., 2003). Auxin can also be acropetally transported into the meristem through the epidermis to induce organ formation (Reinhardt et al., 2003). In roots, IAA moves acropetally (toward the root apex) through the central vasculature and basipetally (from the root apex toward the base) through the epidermis (Muday and DeLong, 2001; Kepinski and Leyser, 2005). Inhibition of acropetal auxin transport from shoots into roots or disruption of auxin signaling significantly inhibits lateral root development (Reed et al., 1998; Casimiro et al., 2001; Rogg et al., 2001; Fukaki et al., 2002). Appropriate distribution of auxin is also necessary for normal vascular differentiation in leaves and stems. Seedlings grown in media containing auxin transport inhibitor show altered leaf vascular patterns (Galweiler et al., 1998; Mattsson et al., 1999; Steynen and Schultz, 2003), and mutations in auxin transport or signaling components confer simpler leaf venation (Hardtke and Berleth, 1998; Steynen and Schultz, 2003). In inflorescence stems, differentiation of interfascicular fibers is influenced by inhibitor treatment and altered in auxin transport mutants (Mattsson et al., 1999; Zhong and Ye, 1999).
PAT requires asymmetrically localized influx and efflux carriers on the plasma membrane (Marchant et al., 1999; Steinmann et al., 1999; Swarup et al., 2001). The permease-like AUXIN RESISTANCE1 protein, which is asymmetrically localized on the plasma membrane of root protophloem, facilitates IAA entry into cells (Marchant et al., 1999; Swarup et al., 2001). On the other hand, IAA moving out of plant cells requires efflux facilitators (Palme and Galweiler, 1999; Muday and DeLong, 2001; Friml et al., 2002). PIN-FORMED1 (PIN1) encodes a membrane protein that is detectable at the basal end of auxin transport–competent cells in vascular tissues (Galweiler et al., 1998). Similarly, immunolocalization studies have shown that EIR1/AGR1/PIN2 that mediates gravitropism and PAT in roots is also asymmetrically localized in the carrier cells (Chen et al., 1998; Luschnig et al., 1998; Muller et al., 1998). Besides IAA influx and efflux facilitators, several other proteins identified in genetic studies in Arabidopsis thaliana have also been found to participate in the modulation of PAT. For example, mutation in chalcone synthase, the first enzyme in the flavonoid biosynthetic pathway, was demonstrated to enhance the auxin transport, reinforcing the conception that flavonoids may act as endogenous inhibitors in controlling auxin distribution in vivo (Brown et al., 2001; Peer et al., 2004). The BIG gene, encoding a calossin-like protein, was also found to play a role in auxin transport because disruption of BIG in the tir3-1 (doc1-1) mutant displays PAT disruption phenotypes, such as decreased inflorescence height and reduced apical dominance (Gil et al., 2001). Recently, ATP binding cassette proteins were also shown to be involved in auxin transport regulation. The double mutant atmdr1-1 atpgp1-1 was decreased in auxin transport, showing pleiotropic phenotypes such as reduced apical dominance, decreased fertility, wrinkled and curled leaves, and stunted stature (Noh et al., 2001; Luschnig, 2002). Further studies have shown that the atmdr1-1 mutant plant displays faster and greater gravitropism (Noh et al., 2003).
Reversible protein phosphorylation may also control the activity of auxin transport proteins. The broad-spectrum kinase inhibitors staurosporine and K252a rapidly reduce auxin efflux, suggesting that protein phosphorylation may be essential to sustain the activity of the efflux carrier (Delbarre et al., 1998). Treating with auxin transport inhibitor napthylphthalamic acid (NPA) and Tyr kinase inhibitors, Bernasconi (1996) found that Tyr phosphorylation is able to reduce the regulation of auxin efflux. Genetic studies have provided further evidence that protein phosphorylation is involved in auxin transport regulation. For instance, disruption of the RCN1 gene, which encodes the regulatory subunit of protein phosphatase 2A, leads to an increase in root basipetal auxin transport and a reduction in root gravity response (Garbers et al., 1996; Deruere et al., 1999). PINOID, a gene encoding a protein-Ser/Thr kinase, has been found to direct auxin transport by acting as a binary switch in apical-basal PIN polar targeting (Friml et al., 2004). Although these studies have provided useful clues, additional mutants and regulatory components are needed for a better understanding of the regulation of auxin transport.
Mitogen-activated protein kinase (MAPK) phosphorylation cascades are conserved in all eukaryotes and play essential roles in transmitting diverse stimuli, including mitogen, developmental cues, and various stress signals (Bogre et al., 2000; Ichimura, 2002; Jonak et al., 2002). A MAPK cascade consists of three classes of enzymes, MAPK, MAPK kinase (MAPKK), and MAPKK kinase (MAPKKK). MAPK is activated by dual phosphorylation of Thr and Tyr residues in a TXY motif by its upstream MAPKK, which is in turn activated by the phosphorylation of Ser and Thr residues by its upstream MAPKKK. MAPKKs with Asp at the third residue in the S/TXXXXXS/T consensus motif may also be autoactive (Kiegerl et al., 2000; Cardinale et al., 2002). Besides participating in response to various extracellular biotic and abiotic stimuli (Bogre et al., 2000; Petersen et al., 2000; Asai et al., 2002; Menke et al., 2004), MAPK cascades have been shown to be involved in developmental process and hormonal responses (Nishihama et al., 2001; Krysan et al., 2002; Lu et al., 2002; Soyano et al., 2003; Bergmann et al., 2004; Liu and Zhang, 2004).
Several lines of evidence have implicated the association of MAPK cascades with auxin action. It has been reported that extracts from 2,4-D–treated tobacco (Nicotiana tabacum) cells displayed a kinase activity on recombinant MPK2 protein threefold to fourfold more effective than that of the auxin-starved cells, suggesting that auxin may function as an activator of plant MAPK (Mizoguchi et al., 1994). Using an Arabidopsis leaf protoplast transient expression system, Kovtun and colleagues (2000) have shown that an oxidative stress MAPK cascade could negatively regulate early auxin response. In addition, auxin was shown to activate an unknown MAPK in Arabidopsis seedling roots (Mockaitis and Howell, 2000). However, these experiments were mainly performed in cultured cells and all lack genetic evidence. In this article, we report that Arabidopsis MAP KINASE KINASE7 (MKK7) negatively regulates PAT, which in turn affects the plant architecture.
RESULTS
Isolation and Morphological Characterization of bud1 Plants
A bushy and dwarf 1 (bud1) mutant was identified from a T1 population of transgenic plants generated by a sense/antisense RNA expression system (Mou et al., 2002). In the T2 progeny, self-pollinated bud1 plants segregated into three distinctive phenotypes, namely, severely bushy and dwarfed phenotype, an intermediate phenotype, and wild-type-like phenotype (Figures 1A, 1B, and 1E). Genetic analysis showed a segregation ratio of 95:214:84 for these three phenotypes, fitting the expected ratio of 1:2:1 (P > 0.05). These results suggested that the initial T1 plant was heterozygote with a semidominant mutation in a single nuclear gene, and the T2 segregants were homozygous, heterozygous, or lacking the mutation, respectively. To confirm this, T3 progeny derived from the self-pollinated wild-type-like plants and plants showing the intermediate phenotype were analyzed. As expected, all T3 progeny derived from wild-type-like T2 plants produced only wild-type plants, whereas those derived from the intermediate-phenotyped plants segregated into the three types with a ratio similar to the original T1 plant. Plants homozygous for bud1 were highly sterile due to their shorter stamens relative to gynoecium (Figures 1C and 1D), and hand-pollination with its own pollen can restore mutant fertility.
Figure 1.
Morphological Comparison between Wild-Type and bud1 Mutant Plants.
(A) The 35-d-old wild-type plant grown on soil. Bar = 2 cm.
(B) The 35-d-old bud1 heterozygous (left) and homozygous (right) plants grown on soil. Bar = 2 cm.
(C) Flowers of wild-type (left), bud1 heterozygous (middle) and homozygous (right) plants.
(D) Flowers of wild-type (left) and bud1 homozygous (right) plants, showing differences in pistils and stamens.
(E) The 50-d-old bud1 heterozygous (left) and homozygous (right) plants grown on soil. Bar = 2 cm.
(F) The third rosette leaves of wild-type (left), bud1 heterozygous (middle), and homozygous (right) plants.
(G) to (I) Scanning electron microscopy of the third internodes of wild-type (G) and bud1 heterozygous (H) and homozygous (I) plants. Bars = 150 μm.
(J) to (L) Scanning electron microscopy of the fifth rosette leaves of wild-type (J) and bud1 heterozygous (K) and homozygous (L) plants. Bars = 150 μm.
Characteristic cells are marked in blue in (G) to (L).
One of the most characteristic features of the bud1 plant is its loss of apical dominance, producing significantly more branches than the wild type at the late developmental stage (Figure 1E, Table 1). Although the bud1 plant is strongly branched, only one lateral inflorescence emerges from each leaf axil (Figures 1B and 1E), suggesting that the extreme branching phenotype of the bud1 plant is formed due to the continuous formation of higher order branches by releasing dormant buds from axils of rosette and cauline leaves.
Table 1.
Morphological Abnormalities of the bud1 Plant
| Measurements | BUD1/BUD1 | BUD1/bud1 | bud1 |
|---|---|---|---|
| Height (cm) | 23.94 ± 0.76 | 10.11 ± 0.41 | 4.09 ± 0.28 |
| Internode length (cm) | 1.56 ± 0.05 | 0.82 ± 0.02 | 0.44 ± 0.03 |
| Leaf blade length (cm)a | 1.44 ± 0.05 | 0.63 ± 0.03 | 0.48 ± 0.03 |
| Leaf blade width (cm)a | 0.67 ± 0.02 | 0.46 ± 0.02 | 0.38 ± 0.03 |
| No. inflorescences (>5 mm) | 2.63 ± 1.73 | 6.04 ± 0.35 | 10.15 ± 0.68 |
| No. sterile siliques | 0 | 10.50 ± 0.59 | 17.50 ± 1.91 |
| No. plants measured | 35 | 28 | 33 |
| No. lateral roots per rootb | 11.88 ± 0.52 | – | 4.64 ± 0.46 |
| Hypocotyl length (cm)b | 0.57 ± 0.02 | – | 0.23 ± 0.01 |
Data shown are means ± se. The 50-d-old plants were used for the measurement unless indicated. The differences among wild-type (BUD1/BUD1), heterozygous (BUD1/bud1), and homozygous bud1 plants were determined with the least significant difference t test. All the data sets were significantly different among the three genotypes at P = 0.01.
The third-paired rosette leaves.
The 15-d-old seedlings grown on 0.5× MS plates were used for analysis (n = 30).
The bud1 mutant plant is significantly smaller and the inflorescence stem is more slender than the wild type (Figure 1, Table 1). When grown in soil under continuous white light, 50-d-old homozygous mutant plants are <5.0 cm in height. By contrast, the wild-type plants are taller than 24.0 cm at the same age (Table 1). The bud1 flowers also show an altered morphology, being less elongated both in sepals and petals than those of the wild type (Figures 1C and 1D). In addition, the bud1 petioles are shorter and the leaves are curled and smaller than those of the wild type (Figure 1F, Table 1). Scanning electron microscopic analysis further revealed defective cell expansion in bud1 stems and leaves (Figures 1G to 1L).
Abnormalities in Hypocotyl Elongation Induced by High Temperature and Formation of Lateral Roots and Vascular System in bud1 Plants
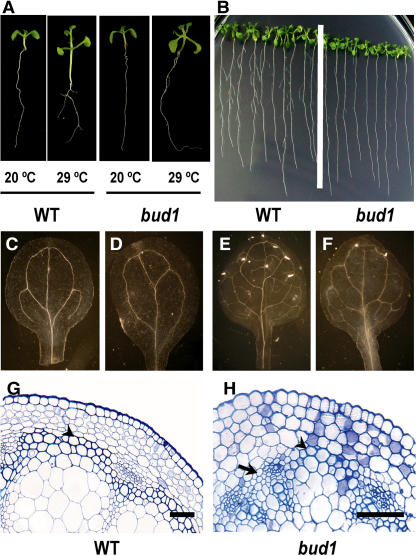
The loss of apical dominance and other morphological changes of bud1 plants suggest that the mutant might be defective in auxin action. Based on the previous findings that the deficiency in auxin biosynthesis, transporting, and/or signaling would specifically affect the hypocotyl elongation induced by high temperature (Gray et al., 1998), we compared the hypocotyl elongation at high temperature between the bud1 and wild-type plants. The hypocotyls of 9-d-old wild-type seedlings grown at 29°C were significantly longer than those at 20°C, whereas the hypocotyls of the bud1 seedlings were slightly elongated (Figure 2A), indicating that the hypocotyl elongation at high temperature is impaired in bud1 mutant plants.
Figure 2.
The bud1 Mutant Phenotypes Seem Auxin Related.
(A) Induction of hypocotyl elongation by high temperature. Wild-type and bud1 mutant seedlings were grown on 0.5× Murashige and Skoog (MS) plates at 20 and 29°C for 9 d and photographed with the same magnification.
(B) Comparison of lateral roots between bud1 and wild-type plants, which were grown on 0.5× MS plates for 14 d and photographed.
(C) to (F) Vascular systems of cleared specimens of wild-type ([C] and [E]) and bud1 ([D] and [F]) plants were viewed with dark-field optics. Photographs showing venation patterns of cotyledons ([C] and [D]) and leaves ([E] and [F]) of the seedlings grown on 0.5× MS plates for 12 d were taken with the same magnification.
(G) and (H) Microscopic comparison of the interfascicular fiber differentiation among wild-type (G) and homozygous bud1 (H) stems, showing the presence of three layers of interfascicular fibers in the wild type and one to two layers of sclerified interfascicular cells in homozygous bud1 (arrowhead) or no interfascicular fibers (arrow). Bars = 5 μm.
Reduction of lateral roots has also been regarded as a criterion of the PAT disruption from shoots into roots or the disruption of auxin signaling (Reed et al., 1998; Xie et al., 2000; Rogg et al., 2001; Fukaki et al., 2002; Gray et al., 2003). As shown in Figure 2B and Table 1, 2-week-old bud1 seedlings developed significantly less lateral roots than the wild type, suggesting that auxin polar transport or signaling may be altered in the mutant plants.
Plants exhibit characteristic vascular patterns in stems and leaves. Auxin has long been known to play a vital role in vascular patterning (Sachs, 2000; Fukuda, 2004). We therefore compared the vascular systems between wild-type and mutant plants. A simpler venation pattern was found in both cotyledon (Figures 2C and 2D) and true leaves of 12-d-old bud1 seedlings (Figures 2E and 2F), which is similar to the venation pattern of the auxin signaling mutant mp or auxin transport defective mutant pin1 (Hardtke and Berleth, 1998; Steynen and Schultz, 2003) and consistent with our previous observation that the β-glucuronidase (GUS) expression driven by the DR5 promoter was altered in the leaf vascular of bud1 mutant plants (Dai et al., 2003). Furthermore, transverse section of bud1 mutant stems also showed less interfascicular fibers (Figures 2G and 2H).
Cloning and Molecular Characterization of BUD1
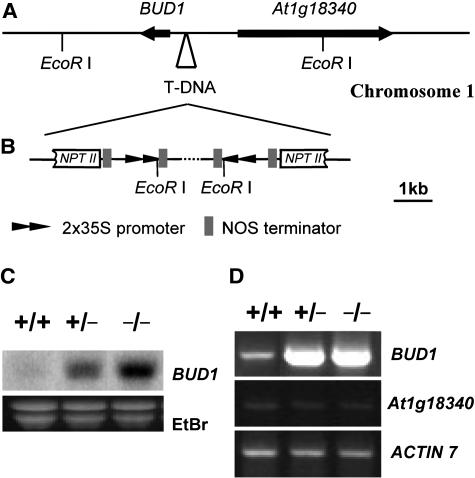
Genetic and molecular analysis indicated that the bud1 mutant resulted from a single T-DNA insertion event. Genomic sequences flanking the T-DNA were recovered by screening the bud1 genomic sublibrary using the cauliflower mosaic virus (CaMV) 35S enhancer as a probe, and the T-DNA insertion site was identified (Figures 3A and 3B). A BLASTN search of the Arabidopsis genomic database revealed that the T-DNA was inserted in Chromosome 1 without disrupting any known genes. However, careful analysis revealed that the T-DNA is inserted 513 bp upstream of the start codon of a putative MKK (At1g18350) and 1438 bp upstream of a putative basic transcription factor subunit (At1g18340) (Figure 3A). Sequence analysis of the recovered insertion locus showed that the T-DNA insertion fragment is composed of two inverted repeats, each of which contains a 2× CaMV 35S enhancer without any cDNA insertion (Figure 3B). Surprisingly, the NPTII gene in the T-DNA was truncated, probably due to DNA rearrangement that might occur in the process producing the T2 generation plants because the T1 plant of bud1 was resistant to kanamycin, but its T2 generation plants became sensitive to kanamycin. The rearrangement of the T-DNA did not affect its flanking plant DNA sequence.
Figure 3.
Cloning of BUD1.
(A) Physical map of the T-DNA insertion site. The T-DNA with two inverted repeats was inserted 513 bp upstream of BUD1 and 1438 bp upstream of At1g18340, respectively. The arrows indicate the transcription orientations of the putative open reading frames flanking the integrated T-DNA.
(B) Structure of the integrated T-DNA in the bud1 genome.
(C) RNA gel blot analysis of the total RNA prepared from 30-d-old soil-grown wild-type (+/+), bud1 heterozygous (+/−), and bud1 homozygous (−/−) plants. Ethidium bromide (EtBr)-stained total RNA was used as a loading control.
(D) RT-PCR analysis of the BUD1 and At1g18340 expression in 30-d-old soil-grown wild-type (+/+), bud1 heterozygous (+/−), and bud1 homozygous (−/−) plants. ACTIN7 was used as an internal control.
The T-DNA insertion between two putative genes suggests that the mutant phenotype may result from altered expression of At1g18340 and/or At1g18350. Therefore, the expression levels of both genes in the mutant and wild-type plants at the mature stage were investigated by RNA gel blot analysis. The expression level of At1g18350 was remarkably increased in mutant plants (Figure 3C), whereas the expression of At1g18340 was undetectable both in mutant and wild-type plants (data not shown), probably due to its extremely low expression level. We therefore employed a more sensitive method, RT-PCR, to monitor expression of these two genes. No difference of the At1g18340 expression between mutant and wild-type plants could be found, but a dramatic increase in the At1g18350 expression was observed (Figure 3D), as revealed by RNA gel blot hybridization. These results indicate that the bud1 phenotype is very likely caused by an increased expression of At1g18350, and accordingly we designated the At1g18350 gene as BUD1.
Confirmation by Transgenic Studies
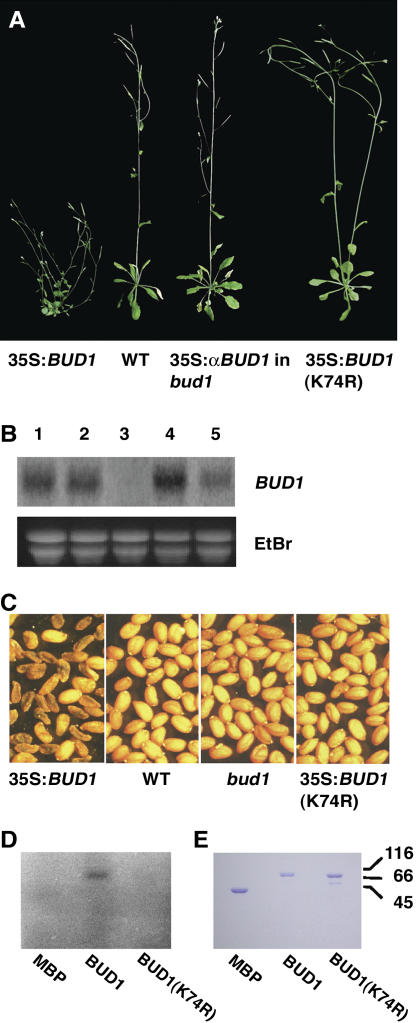
To verify whether At1g18350 represents BUD1, we performed two experiments. First, we placed an At1g18350 cDNA under the control of the CaMV 35S promoter in the binary vector pBI121 and then transformed wild-type plants. Among 77 primary T1 transformants, 6 showed a bud1-like mutant phenotype, 56 died at different stages, and 15 were wild-type-like plants (Figure 4A). RNA gel blot analysis demonstrated that the bud1-like transgenic plants had an increased expression level of BUD1, whereas the wild-type-like transgenic plants showed no apparent increase in the BUD1 expression level compared with that of the wild type (Figure 4B). These observations suggested that overexpression of At1g18350 is able to recapture the bud1 mutant phenotype. The phenotype of the surviving bud1-like transgenic lines includes small and curled leaves, reduced apical dominance, and reduced length and diameter of internodes (Figure 4A). Moreover, most of the seeds produced from these transgenic lines were black and shrunken (Figure 4C), lacking the ability to germinate and grow on the plates with or without supplement of kanamycin, indicating that they were embryo lethal. The seeds with normal appearance were able to geminate and grow on normal MS plates but died with supplement of kanamycin, indicating that they do not contain the transgene. These results indicated that the small number of surviving T1 transgenic lines is heterozygous for the transgene, which was confirmed by DNA gel blot analysis (data not shown), suggesting that both homozygote and heterozygote were completely lethal in the T2 generation, probably due to the ectopic expression of MKK7 driven by 35S promoter. Second, we overexpressed an antisense cDNA construct of At1g18350 driven by the CaMV 35S promoter in the heterozygous bud1 background. Out of the 23 transgenic lines, 17 displayed wild-type-like phenotype (Figure 4A), of which five lines were homozygous at the bud1 locus (data not shown). Taken together, we conclude that At1g18350 represents BUD1 that is overexpressed in the bud1 mutant plant.
Figure 4.
Confirmation and Molecular Characterization of the BUD1 Gene.
(A) Phenotypes of transgenic plants. The transgenic plants generated from the wild-type plant transformed with the 35S:BUD1 construct mimicked the bud1 phenotypes but not the plant transformed with 35S:BUD1(K74R). The bud1 mutant phenotype was suppressed when antisense BUD1 (35S:αBUD1) construct was introduced into the bud1 mutant plant.
(B) RNA gel blot analysis. Total RNA prepared from transgenic plants containing the 35S:BUD1 transgene and showing the bud1-like phenotype (lane 1), from transgenic plants containing the 35S:BUD1(K74R) transgene and showing the wild-type phenotype (lane 2), from transgenic plants containing the 35S:BUD1 transgene and showing the wild-type phenotype (lane 3), from bud1 homozygous plants (lane 4), and from bud1 heterozygous plants (lane 5). Ethidium bromide (EtBr)-stained total RNA was used as a loading control.
(C) T2 seeds of the bud1, wild-type, and transgenic plants.
(D) In vitro kinase activity assay. Autoradiogram showed the in vitro autophosphorylation of purified MBP-BUD1 or MBP-BUD1(K74R) separated by SDS-PAGE. Purified MBP was used as a negative control.
(E) Photograph of purified MBP, MBP-BUD1, and MBP-BUD1(K74R) separated by SDS-PAGE and stained with Coomassie blue.
BUD1 Encodes Arabidopsis MKK7
A BUD1 cDNA was cloned by rapid amplification of cDNA ends (RACE)-PCR from the bud1 total RNA by taking advantage of its overexpression feature. Sequence comparison between the BUD1 cDNA and the genomic DNA revealed that BUD1 contains an open reading frame of 924 bp without any intron, a 35-bp 5′ untranslated region (UTR), and a 129-bp 3′ UTR (see Supplemental Figure 1 online).
BLAST analysis suggested that BUD1 encodes the MKK7, a member of the Arabidopsis MAPK cascades. In the Arabidopsis genome, 10 putative MAPKKs have been identified and classified into four groups (Ichimura, 2002). Phylogenetic analysis revealed that BUD1 is one of the four members in Group D, whose functions have not been identified and studied in any detail (Ichimura, 2002; Jonak et al., 2002). BUD1 contains 11 conserved kinase domains and a highly conserved motif SLDYCNS (see Supplemental Figure 1 online), in which the Ser residues are thought to be phosphorylated (Ichimura et al., 1998).
The MKK7 expression pattern was studied using quantitative PCR. As shown in Supplemental Figure 2 online, MKK7 was expressed in all tissues examined, with a relatively higher level in leaves and lower level in roots and flowers. In the bud1 mutant plants, the expression pattern was similar, although the expression level was much higher. The ubiquitous expression pattern of MKK7 was consistent with the pleiotropic mutant phenotypes.
The BUD1 Kinase Activity Is Essential for Its Biological Function
To determine whether BUD1/MKK7 has a kinase activity, we expressed and purified a maltose binding protein (MBP)-BUD1 recombinant protein from Escherichia coli cells. The purified MBP-BUD1 displayed an autophosphorylation activity (Figures 4D and 4E). However, no kinase activity could be detected in MBP-BUD1(K74R), in which a conserved Lys residue (K) at the position 74 of the ATP binding site in the kinase domain II was replaced with an Arg residue (R). These data indicate that MKK7 is indeed a functional kinase and its ATP binding site is indispensable.
To investigate whether the kinase activity of BUD1 is required for its biological functions in vivo, we constructed a plant transformation plasmid containing the mutated BUD1 gene, BUD1(K74R), and transformed the wild-type plants. All of the 103 T1 transgenic lines containing the highly expressed BUD1(K74R) transgene showed the same phenotype as the wild type, whereas the transgenic plants containing the highly expressed normal BUD1 transgene mimic the bud1 phenotype (Figures 4A to 4C), suggesting that BUD1 kinase activity is essential for its biological activity.
Deficiency of PAT in bud1
Based on the finding that the bud1 phenotype results from its deficiency in auxin action, we performed experiments to find out whether the biosynthesis, transport, and/or signaling of auxin is altered in the mutant plants. The IAA levels in 15-d-old plants grown on MS plates were measured, and no significant difference was found between the homozygous bud1 and wild-type plants (data not shown). We then determined the sensitivity of bud1 plants to exogenous auxin by examining the root growth inhibition by 2,4-D. As shown in Figure 5A, both bud1 and wild-type roots showed similar sensitivity to 2,4-D, suggesting that auxin signaling in bud1 may not be affected. Expression of two auxin-responsive genes, IAA1 and IAA5, were induced to a similar degree in the roots treated with 2,4-D (Figure 5B). This is consistent with our previous result that the DR5 promoter–driven GUS expression induced by IAA treatment was similar in bud1 and wild-type plants (Dai et al., 2003). These results suggest that the bud1 phenotype may result from a deficiency in PAT, rather than the auxin biosynthesis or signaling.
Figure 5.
Auxin Transport and Gene Expression Analysis.
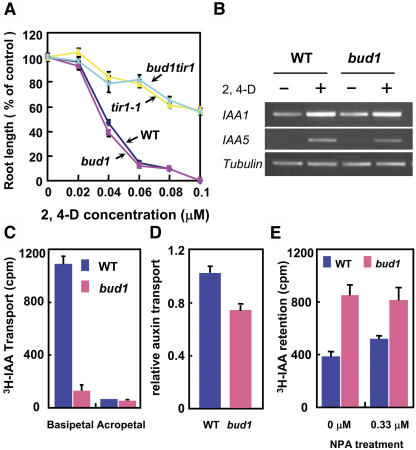
(A) Inhibition of root elongation by synthetic auxin 2,4-D. Each value represents the average of >10 seedlings. Bars represent ± se.
(B) Induction of auxin-responsive genes by 2,4-D (0.04 μM) treatment in roots.
(C) PAT of inflorescence stems. Each assay used 10 wild-type and bud1 homozygous seedlings. Values shown are means ± se.
(D) PAT of the bud1 and wild-type seedlings. Values are means ± se of three independent experiments. The difference between the wild type and bud1 is significant (Student's t test, P < 0.01).
(E) IAA efflux of dark-grown hypocotyl segments. Ten hypocotyl segments were used in each assay, and values are given as means ± se of three to five independent assays. The difference between the wild type and bud1 is significant (Student's t test, P < 0.05).
We therefore systematically compared the PAT between bud1 and wild-type plants. The tritium-labeled IAA (3H-IAA) basipetal transport in inflorescence stems excised from bud1 plants was reduced to one-tenth of that in the wild type (Figure 5C), as previously reported (Dai et al., 2003). To avoid the inaccuracy resulting from the slender bud1 inflorescence stem segments used in the assay, we further measured the auxin transport in the hypocotyl segments of both light- and dark-grown seedlings because the hypocotyl segments of bud1 and wild-type plants are morphologically similar under these conditions. To measure basipetal movement of 3H-IAA, a single microdroplet was applied to the apex of 5-d-old light-grown seedlings, and the auxin transport in the homozygous bud1 hypocotyl segments was reduced to 72% of that of the wild type (Figure 5D). The efflux rates of auxin transport in the etiolated hypocotyls were also determined by measuring the amount of auxin retained in the hypocotyl segments after sequential incubation in the auxin-containing solution and in auxin-free buffer. As shown in Figure 5E, the 3H-IAA retained in dark-grown homozygous bud1 hypocotyls was twice as much as that of the wild type and was less sensitive to the auxin transport inhibitor NPA than that in the wild type. Taken together, all these results indicated that auxin transport in bud1 mutant plants is deficient.
Enhanced PAT in MKK7 Knockdown Plants
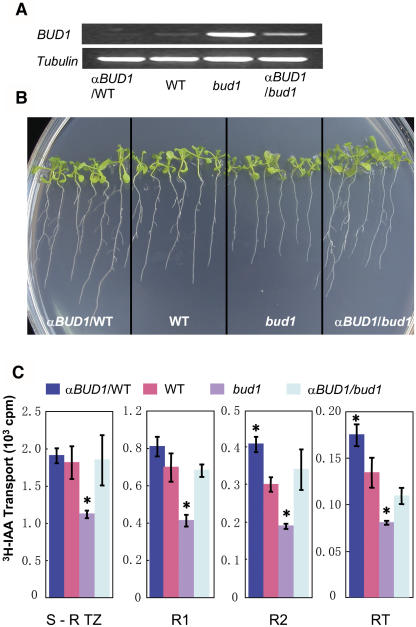
To investigate whether repressed MKK7 expression enhances auxin transport, we generated the BUD1 antisense transgenic lines both in the bud1 and wild-type backgrounds. Antisense BUD1 RNA significantly suppressed the BUD1 expression in the bud1 background (Figure 6A) and thereby reversed the mutant phenotype to wild type (Figures 6B and 4A) and resumed the auxin transport ability of the transgenic plant to the wild-type level (Figure 6C).
Figure 6.
Analysis of the BUD1 RNA Antisense Transgenic Plants.
(A) RT-PCR analysis of the BUD1 expression level in the wild type, bud1, antisense in the wild-type background (αBUD1/WT), and antisense in the bud1 background (αBUD1/bud1).
(B) Comparison of lateral roots. Plants were grown on 0.5× MS plates for 10 d and photographed.
(C) Auxin transport analysis of antisense BUD1 transgenic lines. Auxin transport from the shoot apex to the shoot–root transition zone (S-R TZ), its subsequent two 4-mm root segments below S-R TZ (R1 and R2), and the 2- to 3-mm root tip (RT) were determined using 4.5-d-old light-grown seedlings. Ten seedlings were used in each assay. Values shown were means ± se of five independent assays. Significance compared with the wild type (asterisks) was determined by the least significant difference t test at P < 0.05.
In the BUD1 antisense transgenic plants with wild-type background, the BUD1 expression level also decreased slightly compared with that of the wild type (Figure 6A). As the BUD1 expression decreased, the lateral roots of antisense transgenic plants appeared 1 d earlier and more lateral roots developed (15.91 ± 0.94) than that of the wild-type plants (11.89 ± 0.52, n = 30, P < 0.01) (Figure 6B). Consistent with the concept that acropetal auxin transport in roots affects the lateral root development (Reed et al., 1998; Casimiro et al., 2001), the auxin transport from the shoot apex into the R2 segment and the root tip was significantly enhanced, although no significant enhancement in S-R TZ and R1 segments was detected (Figure 6C).
Taken together, the finding that the auxin transport was disrupted by the increased expression of MKK7 in bud1 and enhanced by repressed expression of MKK7 in antisense transgenic plants suggests that MKK7 may function as a negative regulator of PAT.
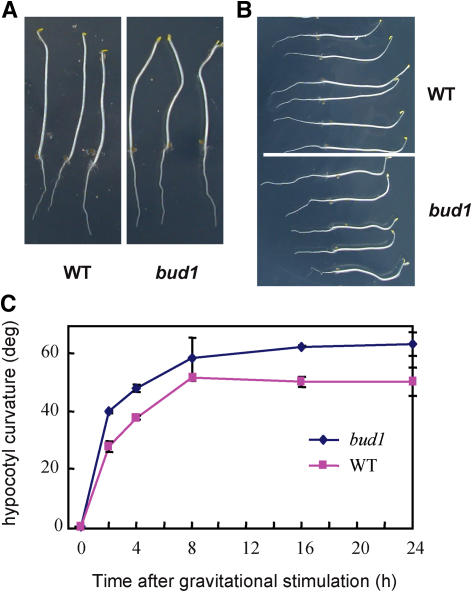
Altered Gravitropism Response in bud1 Plants
In Arabidopsis, PAT is required for hypocotyl elongation of light-grown seedlings but not for the dark-grown ones (Jensen et al., 1998). Consistent with this view, bud1 showed a normal hypocotyl elongation in the dark (Figure 7A) but a shorter hypocotyl under light (Figure 2A, Table 1). We also found the absence of the apical hook of dark-grown bud1 plants (Figure 7A), a phenotype that may be related to the auxin transport defect (Li et al., 2004; De Grauwe et al., 2005). Since the gravitropism of seedling hypocotyls is related to PAT, we further performed a gravitropism assay using etiolated seedlings. Compared with wild-type plants, the mutant hypocotyls bent to a greater degree after reorienting 90° for 18 h (Figure 7B). Kinetic analysis of the curvature development showed that the bud1 hypocotyls bent more quickly than the wild type within the beginning 2 h after reorientation, and the bending rate tended to be the same in a prolonged period (Figure 7C). The enhanced gravitropism response of bud1 plants was quite similar to that of atmdr1-1, a mutant that is defective in PAT (Noh et al., 2003).
Figure 7.
Comparison of Gravitropic Responses between Wild-Type and bud1 Plants.
(A) and (B) Morphologies of dark-grown seedlings and their gravitropic responses. Seedlings were grown in the dark for 4 d (A) and then the plates were reoriented by 90° and photographed 18 h thereafter (B). The bud1 seedlings showed greater gravitropic response than the wild type.
(C) Kinetics of hypocotyl gravitropism. Seedlings were grown in the dark for 4 d in 0.5× MS plates and reoriented by 90°. The gravitropic curvatures were measured at the time intervals as indicated. Values shown are means ± se of five independent assays.
Genetic Analyses of bud1 Double Mutants
To further investigate the function of BUD1, we generated double mutants of bud1 with either auxin-responsive or transport mutants. The auxin-resistant mutant axr3-3 shows an enhanced apical dominance, reduced root elongation, and increased adventitious rooting and lacks root gravitropism (Leyser et al., 1996). In the bud1 axr3-3 double mutant, the growth of axillary buds was significantly inhibited and the number of lateral branches was largely reduced compared with bud1 mutant plants (Figures 8B to 8D), but the length of inflorescence stems was only slightly increased and the size of leaves showed no apparent alteration. This indicates that the bud1 bushy phenotype was partially suppressed due to the hypersensitivity of axr3-3 to auxin, suggesting that BUD1 may act upstream of AXR3. Moreover, the root growth of the double mutant bud1 tir1-1 exhibited a similar strong resistance to auxin as tir1-1 (Figure 5A), a mutant defective in auxin signaling. These results suggested that BUD1 may act upstream of IAA signaling.
Figure 8.
Phenotypes of bud1 axr3-3, bud1 doc1-1, and bud1 atmdr1-1 Double Mutants.
Photographs were taken at 35 d after germination. (A) The wild type, (B) axr3-3, (C) bud1 axr3-3, (D) bud1, (E) doc1-1, (F) bud1 doc1-1, and (G) bud1 atmdr1-1. Bars = 2 cm.
Arabidopsis mutant doc1-1 is involved in both auxin transport and light-related pathways and displays reduced apical dominance (Gil et al., 2001). To test whether BUD1 functions in the same or different PAT pathways, we made a bud1 doc1-1 double mutant and compared its phenotype with its parents. As shown in Figures 8D to 8F, the double mutant bud1 doc1-1 exhibited an extreme dwarf phenotype, and the internode elongation between adjacent flowers was inhibited, suggesting that BUD1 and DOC1 might regulate auxin transport through different pathways. Since the pleiotropic phenotypes of bud1 are strikingly similar to atmdr1-1 and atmdr1-1 atpgp1-1 double mutants, we further crossed atmdr1-1 with bud1. The double mutant (Figure 8G) showed a very similar phenotype to bud1 in both height and lateral branching, suggesting that they may participate in the same PAT regulatory pathway.
DISCUSSION
In Arabidopsis, 10 members of MAPKKs have been identified based on the annotation of the genome sequence and have been divided into four groups phylogenetically (Ichimura, 2002). Most of the members in Groups A, B, and C have been shown to play important roles in biotic and abiotic responses (Matsuoka et al., 2002; Ren et al., 2002; Teige et al., 2004). However, the function of Group D members still remains to be elucidated. In this article, we report the isolation of the bud1 mutant and molecular genetic characterization of MKK7. We also present direct evidence that the bud1 phenotype results from the enhanced expression of MKK7, which negatively regulates PAT, thus affecting the formation of plant architecture.
Enhanced Expression of MKK7 Results in Reduced Apical Dominance
Activation tagging provides an effective approach to identify genes whose expression levels play a crucial role in normal plant growth and development. The bud1 mutant, isolated based on its abnormalities in plant architecture, may result from overexpression of MKK7 rather than from its ectopic or constitutive expression. First, as revealed by RNA gel blot and quantitative PCR analyses, the expression of MKK7 in bud1 plants is strongly increased compared with the wild type, and its relative expression pattern is similar to wild-type plants. However, it may have a threshold of the MKK7 transcripts to cause an apparent morphological phenotype because transgenic plants with weakly activated or inhibited MKK7 showed subtle phenotypes. Overexpression of MKK7 may result from the 2× 35S enhancer in the insertion. As previously reported, the CaMV 35S enhancer can lead to just an enhancement of the endogenous gene expression, and the phenotype resulting from such an enhancement, as opposed to true constitutive and ectopic activation, would be more likely to reflect the normal role of the activated gene (Weigel et al., 2000). Second, constitutive expression of MKK7 in wild-type plants leads to lethality of the transgenic plants. The transgenic study of the ectopic and constitutive expression of MKK7 driven by the CaMV 35S promoter demonstrated that most of the transgenic plants are lethal at different developmental stages, except that only a small percentage of the transgenic plants can grow like bud1 plants but set aborted seeds. The lethality of the transgenic plants is very likely due to the ectopic and constitutive expression of MKK7 rather than enhanced expression because RNA gel blot analysis showed that the MKK7 expression level in the transgenic plants is much weaker than that in the bud1 plants. Third, formation of reduced apical dominance requires the overexpression of the MKK7 active form. Overexpression of the mutated MKK7 form, MKK7(K74R), in which the kinase activity is abolished by a single amino acid substitution in the ATP binding site, failed to mimic the bud1 phenotype, demonstrating that the MAPK cascade is involved in the formation of normal plant architecture of Arabidopsis.
Generally, the MAPKK need to be phosphorylated by its upstream MAPKKKs. However, there is a kind of MAPKK in plants that contains an Asp residue in position 3 of the consensus sequence that forms S/TXDXXXS/T located between subdomains VII and VIII. These kinds of MAPKKs, such as SALT STRESS–INDUCED MKK and other MAPKKs of the PMKK2 subfamily, may constitute an autoactive class of MAPKKs (Kiegerl et al., 2000; Cardinale et al., 2002). BUD1 falls into this kind of MAPKK. Therefore, it is possible that enhanced expression of BUD1 could cause such a phenotype without upstream activation.
MKK7 Functions in Regulating PAT
The bud1 morphological phenotype and its defect in the induction of hypocotyl elongation by high temperature suggest its deficiency in IAA metabolism, signaling, or transport. The findings that there are no significant differences in IAA content or in response to auxin treatment between wild-type and bud1 plants ruled out the possibilities that IAA biosynthesis or signaling is involved in the formation of the bud1 phenotypes. Therefore, the research focused on the auxin transport. Systematic and in-depth studies indicate that the bud1 phenotypes result from the deficiency in polar auxin transport.
Direct evidence comes from the three different IAA transport assays, which all showed that the 3H-IAA transport in bud1 homozygous seedlings was significantly decreased. This was further strengthened by the finding that the auxin transport of bud1 plants is less sensitive to the auxin transport inhibitor NPA than that of the wild type because of the already reduced auxin transport in the bud1 plants. Moreover, we also showed supporting evidence that the BUD1 expression level is consistent with the auxin transporting ability, as manifested by auxin transport and phenotypic analyses of BUD1 antisense transgenic plants. The finding that the BUD1 antisense transgene was more effective in bud1 than in wild-type backgrounds may be explained by the extremely low basal expression of BUD1 or the existence of highly homologous subgroup D genes, for example, MKK9, which is ∼81% identical to BUD1.
The conclusion is also supported by the results obtained from the comparison of the hypocotyl elongation, gravitropism response, lateral root formation, lack of apical hook, and vascular patterns between the bud1 and wild-type plants. Unlike auxin signaling mutants such as shy2 and axr3, which cause photomorphogenesis in dark-grown seedlings, the dark-grown bud1 plants showed no apparent alterations in seedling morphology or hypocotyl elongation. However, the light-grown bud1 plants showed decreased hypocotyl elongation. This is consistent with previous studies that PAT is important for hypocotyl elongation in the light but not in the dark in Arabidopsis (Jensen et al., 1998). Like most PAT-deficient mutants, the bud1 mutant also produces less lateral roots, which is a characteristic feature showing deficiency in auxin-regulated root development (Reed et al., 1998). The apical hook formation of dark-grown seedlings was controlled by interactions of auxin, ethylene, and gibberellins. Recent studies have shown that auxin and its transport may play a crucial role in this process, which is consistent with the hookless phenotype of bud1 plants (Li et al., 2004; De Grauwe et al., 2005). Furthermore, the observation that the bud1 plant has abnormal vascular patterns in leaves and stems could partially be explained by deficiency in auxin action, although some auxin transport inhibition phenotype was not observed (Zhong and Ye, 1999; Steynen and Schultz, 2003). This is also consistent with our previous observations that the GUS expression pattern guided by DR5 promoter was altered in bud1 mutant plants but had a similar response to exogenous auxin treatment as the wild type (Dai et al., 2003).
Genetic analysis with bud1 double mutants also supports the conclusion that bud1 is an auxin transport mutant resulting from enhanced expression of MKK7. The phenotype of the double mutant bud1 axr3-3, which was generated from the cross of bud1 with auxin hypersensitive response mutant axr3-3 (Leyser et al., 1996), is much less severe than that of bud1 plants, indicating that the lose of apical dominance in bud1 can be partially suppressed by elevating its sensitivity to auxin. However, when crossed with the auxin transport decreased mutant doc1-1 (Gil et al., 2001), the double mutant bud1 doc1-1 showed an aggravated loss of apical dominance, which is much more severe than any parental mutant. Mutation of the BIG gene in doc1-1 not only causes an auxin transport disruption phenotype but also affects the expression of light-related genes, for example, CAB2 (Gil et al., 2001). However, the expression of CAB2 in bud1 plants was not affected (H. Wang, unpublished data). The extreme phenotype in bud1 doc1-1 and the different expression patterns of CAB2 in bud1 and doc1-1 plants may suggest that MKK7 and BIG regulate PAT through different pathways. However, the finding that bud1 and its double mutant bud1 atmdr1-1 had very similar morphological phenotypes, including curled leaves, increased lateral branching, decreased plant height, and reduced fertility, may suggest the involvement of BUD1 and MDR1 in the same PAT regulatory pathway in Arabidopsis.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia (Col-0) wild-type and mutant plants were grown on vermiculite saturated with 0.3× B5 medium under continuous illumination (80 to 120 μE m−2 s−1) at 23°C. For plants grown on 0.5× MS plates (Gibco BRL), seeds were surface sterilized. For temperature treatment, plants were germinated and grown on 0.5× MS in separate versatile environmental test chambers (Sanyo) under continuous illumination (70 μE m−2 s−1) at 20 and 29°C, respectively. The root auxin sensitivity was assayed as follows: 4-d-old light-grown seedlings were transferred to MS plates containing appropriate concentrations of synthetic auxin 2,4-D, and the root length was measured after 3 d of treatment.
Isolation of the T-DNA–Tagged Locus
Arabidopsis genomic DNA was digested completely with EcoRI, separated on agarose gel, blotted onto a Hybond N+ membrane (Amersham), and probed with CaMV 2× 35S promoter. The 7-kb DNA fragment that cosegregated with the mutant phenotype was recovered and ligated into the EcoRI-predigested λZAP II vector (Stratagene) to construct a sublibrary. The T-DNA–tagged locus was cloned by screening the sublibrary with 2× 35S enhancer as the probe and sequenced with a DNA Sequencer 3700 (Applied Biosystems).
RNA Gel Blot Analysis, RT-PCR, and RACE-PCR
Total RNA was prepared by a guanidine thiocyanate extraction method, and RNA gel blot analysis was performed as previously described (Hu et al., 2000). RNA (20 μg per lane) was separated in an agarose gel containing 10% formaldehyde, blotted onto a Hybond N+ membrane (Amersham), and probed with the PCR-amplified DNA fragments using the following primers: BamHISTART (5′-GGATCCTCTCTCTTCTATTTCCATGGC-3′) and SacIEND (5′-GAGCTCACAAGCAGTCGGATCTAAAG-3′) for BUD1/MKK7; TFF (5′-AGGAGATGTCTTCCGCTGATG-3′) and TFR (5′-AAGTCTTCATCGGATTCGTGTG-3′) for At1g18340.
For RT-PCR analysis, 2 μg of total RNA treated with DNase I (Gibco BRL) was added in a 20-μL reverse transcription reaction using the Superscript first-strand synthesis system for RT-PCR (Gibco BRL), and an appropriate amount of the products was further applied for a 20-μL PCR amplification reaction using gene-specific primers, including BamHISTART and SacIEND for BUD1/MKK7, TFF and TFR for At1g18340, and ACTIN7F (5′-TGGAATGGTGAAGGCTGGTTT-3′) and ACTIN7R (5′-CTGTTGGAAGGTGCTGAGGGA-3′) for ACTIN7.
The 5′ and 3′ UTR sequences of BUD1 were obtained by RACE-PCR with a 5′/3′ RACE kit (Roche). The specific primers used in RACE-PCR were SP1 (5′-GAGCTCAAGTATCATCACTCCG-3′), SP2 (5′-GCTAGTTGTTTCTCCGTAACGG-3′), SP3 (5′-CTGCTTCCTCTTCCGAGAAG-3′), and SP4 (5′-CCGTTACGGAGAAACAACTAGC-3′).
Plasmid Construction and Plant Transformation
To construct the plant transformation plasmids 35S:BUD1 and 35S:BUD1(K74R) in which the Lys residue (K) at the position of 74 was point mutated into Arg (R), the DNA fragment containing the entire CDS of BUD1 or BUD1(K74R) was double digested with BamHI/SacI and ligated into the pBI121 vector (Clontech). BUD1 was amplified with primers BamHISTART and SacIEND. BUD1(K74R) was generated by site-directed mutagenesis PCR (Ho et al., 1989) using primer pairs of BamHISTART/BUD1muR (5′-CGTTGACTGATCTCAGAGCG-3′) and BUD1muF (5′-CGCTCTGAGATCAGTCAACG-3′)/SacIEND (the mutated base pair is underlined).
To construct 35S:antisense BUD1 plant transformation plasmid, a 710-bp partial BUD1 fragment truncated at the 3′ end by SacI was cloned into pBI121 in an antisense orientation.
Plant transformation plasmids were introduced into Agrobacterium tumefaciens strain GV3101 (pMP90RK) (Koncz and Schell, 1986) by electroporation, and Arabidopsis plants were transformed via vacuum infiltration (Bechtold et al., 1993). Transformants (T1) were selected on plates containing 50 mg/L kanamycin and transferred to soil to allow self-pollination. Genetic and phenotypic analyses were performed mainly in T2 or T3 generations.
To generate 35S antisense BUD1 transgenic plants in the bud1 homozygous background, the antisense plasmid was introduced into BUD1/bud1 heterozygous plants, and the transformants were identified by PCR analysis using the following primers: P1 (5′-TCTAGAAGCCATGGAAATAGAAGAGAG-3′), P2 (5′-CTGCAGCCGTAGGGTCATGTGTGACTG-3′), P3 (5′-ACCACGTCTTCAAAGCAAGTG-3′), and P4 (5′-TATGATAATCATCGCAAGACCG-3′).
Protein Expression and Kinase Assay
To express and purify BUD1 and BUD1(K74R) proteins for kinase assay, we amplified the coding sequences from the above-described BUD1 and BUD1(K74R) constructs with primers BUD1EcoRIF and BUD1SalIR (5′-AGAATTCATGGCTCTTGTTCGTAAACGC-3′ and 5′-AGTCGACCTAAAGACTTTCACGGAGAAAAGG-3′). The PCR products were cloned into the Escherichia coli expression vector pMAL-c2 (New England Biolabs) and verified by sequencing. The fusion proteins were expressed and purified by amylose-affinity chromatography (New England Biolabs) and quantified by Bio-Rad protein assay reagent.
The autophosphorylation assay mixture (20 μL) containing 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, and 10 mM MnCl2, 10 μCi of γ-32P-ATP, and 1 to 2 μg MBP, MBP-BUD1, or MBP-BUD1(K74R) was incubated at 30°C for 30 min, and the reaction was terminated by adding 4 μL 6× SDS-PAGE sample buffer. The reaction mixture was heated at 95°C for 5 min and then separated on a 10% (w/v) SDS-PAGE. The gel was stained with Coomassie Brilliant Blue, and the 32P-labeled protein bands were visualized by the Variable Mode Imager Typhoon 8600 (Amersham Pharmacia Biotech).
Generation of bud1 Double Mutants
The double mutant bud1 axr3-3, bud1 doc1-1, bud1 atmdr1-1, or bud1 tir1-1 was generated from the cross of homozygous bud1 with axr3-3, doc1-1, atmdr1-1, or tir1-1 (Leyser et al., 1996) and identified from the F2 progeny grown on nutrient plates by comparing with its parental phenotypes and through PCR-based molecular analyses (Noh et al., 2001; Zenser et al., 2001).
IAA Measurement and Transport Assay
IAA content was measured mainly by the method described previously (Ouyang et al., 2000). Inflorescence stems of 6-week-old plants were used for auxin polar transport assay as described (Okada et al., 1991). Stem segments (2.5 cm in length) were placed in a 1.5-mL microcentrifuge tube with one end submerged in 30 μL of MES buffer (5 mM MES and 1% [w/v] sucrose, pH 5.5) containing 1.45 μM total IAA with 100 nM of 3H-IAA at room temperature in darkness for 24 h. Based on the orientation of the inflorescence segment within the tube, basipetal or acropetal auxin transport was measured. After incubation, the segment was removed and the last 5 mm of the nonsubmerged end was excised and placed into 2.5 mL of scintillation fluid. The samples were allowed to sit for at least 18 h before being counted in a liquid scintillation counter.
IAA transport of dark-grown hypocotyls was assayed as described previously (Garbers et al., 1996). In each assay, 10 apical hypocotyl segments each of 2.0-mm length were put in 30 μL of transporting buffer (5 mM sodium phosphate, pH 6.0, and 1% sucrose) containing 1.0 μM IAA plus 5.0 nM 3H-IAA and incubated at room temperature for 2 h with shaking at 100 rpm. The segments were washed twice and then incubated in 30 μL of buffer with shaking at 100 rpm for 2 h to allow the IAA to be transported out. After washing twice, the segments were incubated in 2.5-mL universal scintillation fluid for >18 h, and the radioactivity was counted by a liquid scintillation counter (1450 MicroBeta TriLux; Perkin-Elmer).
Auxin transport of young seedlings of wild-type and homozygous bud1 mutant plants was assayed as previously described (Murphy et al., 2000). Plants were grown on filter paper saturated in 0.25× MS medium at 21°C for 5 d under continuous light. To assay IAA transport activity, a small drop (0.2 μL) of 10 nM 3H-IAA in ethanol (50 nCi μL−1) was applied onto each apical tip of 10 seedlings. After a 4-h transport period, the seedlings were rinsed with 0.25× MS medium. By removing the upper hypocotyls and cotyledons, the radioactivity in 2-mm sections excised above the transition zone was determined by a scintillation counter.
Auxin transport from shoot apex into roots was assayed using intact light-grown seedlings as described previously (Peer et al., 2004) with some modifications. Before assay, 10 seedlings grown 4.5 d after germination were transferred to vertically discontinuous filter paper strips saturated in one-quarter MS medium and allowed to equilibrate for 2 h. Auxin solutions were made up in 0.25% (w/v) agarose containing 25 mM MES, pH 5.2. A 0.2-μL microdroplet containing 500 nM unlabeled IAA and 500 nM 3H-IAA (specific activity 25 Ci/mmol) was placed on the shoot apical tip of seedlings. Seedlings were then incubated in the dark for 5 h. After incubation, the hypocotyls and cotyledons were removed. A 2-mm section of filter paper, upon which the S-R TZ was centered, was harvested along with the 2-mm segment of tissue containing the S-R TZ, and the subsequent three 4-mm filter paper strip sections containing the indicated root tissue or the 2- to 3-mm root tip were also collected. The samples were allowed to sit in 2 mL of universal scintillation fluid for at least 18 h before being counted in a liquid scintillation counter.
Accession Number
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number DQ185389.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. BUD1 cDNA Sequence.
Supplemental Figure 2. BUD1 Expression in Different Organs.
Supplementary Material
Acknowledgments
We thank Jianru Zuo and Qi Xie (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) and reviewers for critical comments on the manuscript, Huajun Chen (Beijing Forestry University, Beijing, China) for IAA measurements, Jiayi Xie (Institute of Microbiology, Chinese Academy of Sciences) for scanning electron microscopy examination, Ottoline Leyser (University of York, UK) for providing axr3-3 seeds, Joanne Chory (Salk Institute for Biological Studies, La Jolla, CA) for doc1-1 seeds, and Edgar P. Spalding (University of Wisconsin, Madison, WI) for atmdr1-1 and atmdr1-1 atpgp1-1 seeds. This work was supported by grants from the National Natural Science Foundation of China (30221002 and 30330040) and the Ministry of Sciences and Technology of China (J02-A-001).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jiayang Li (jyli@genetics.ac.cn).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.037846.
References
- Asai, T., Tena, G., Plotnikova, J., Willmann, M.R., Chiu, W.L., Gomez-Gomez, L., Boller, T., Ausubel, F.M., and Sheen, J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415 977–983. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of Arabidopsis thaliana plants. C. R. Acad. Sci. (Paris) 316 1194–1199. [Google Scholar]
- Benkova, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertova, D., Jurgens, G., and Friml, J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 591–602. [DOI] [PubMed] [Google Scholar]
- Bergmann, D.C., Lukowitz, W., and Somerville, C.R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304 1494–1497. [DOI] [PubMed] [Google Scholar]
- Bernasconi, P. (1996). Effect of synthetic and natural protein tyrosine kinase inhibitors on auxin efflux in zucchini (Cucurbita pepo) hypocotyls. Physiol. Plant 96 205–210. [Google Scholar]
- Bogre, L., Meskiene, I., Heberle-Bors, E., and Hirt, H. (2000). Stressing the role of MAP kinases in mitogenic stimulation. Plant Mol. Biol. 43 705–718. [DOI] [PubMed] [Google Scholar]
- Booker, J., Auldridge, M., Wills, S., McCarty, D., Klee, H., and Leyser, O. (2004). MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr. Biol. 14 1232–1238. [DOI] [PubMed] [Google Scholar]
- Booker, J., Chatfield, S., and Leyser, O. (2003). Auxin acts in xylem-associated or medullary cells to mediate apical dominance. Plant Cell 15 495–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker, J., Sieberer, T., Wright, W., Williamson, L., Willett, B., Stirnberg, P., Turnbull, C., Srinivasan, M., Goddard, P., and Leyser, O. (2005). MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev. Cell 8 443–449. [DOI] [PubMed] [Google Scholar]
- Brown, D.E., Rashotte, A.M., Murphy, A.S., Normanly, J., Tague, B.W., Peer, W.A., Taiz, L., and Muday, G.K. (2001). Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol. 126 524–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale, F., Meskiene, I., Ouaked, F., and Hirt, H. (2002). Convergence and divergence of stress-induced mitogen-activated protein kinase signaling pathways at the level of two distinct mitogen-activated protein kinase kinases. Plant Cell 14 703–711. [PMC free article] [PubMed] [Google Scholar]
- Casimiro, I., Marchant, A., Bhalerao, R.P., Beeckman, T., Dhooge, S., Swarup, R., Graham, N., Inze, D., Sandberg, G., Casero, P.J., and Bennett, M. (2001). Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R., Hilson, P., Sedbrook, J., Rosen, E., Caspar, T., and Masson, P.H. (1998). The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc. Natl. Acad. Sci. USA 95 15112–15117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, Y., Fu, Z., and Li, J. (2003). Isolation and characterization of an Arabidopsis bush and dwarf mutant. Acta Bot. Sin. 45 621–625. [Google Scholar]
- De Grauwe, L., Vandenbussche, F., Tietz, O., Palme, K., and Van Der Straeten, D. (2005). Auxin, ethylene and brassinosteroids: Tripartite control of growth in the Arabidopsis hypocotyl. Plant Cell Physiol. 46 827–836. [DOI] [PubMed] [Google Scholar]
- Delbarre, A., Muller, P., and Guern, J. (1998). Short-lived and phosphorylated proteins contribute to carrier-mediated efflux, but not to influx, of auxin in suspension-cultured tobacco cells. Plant Physiol. 116 833–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deruere, J., Jackson, K., Garbers, C., Soll, D., and Delong, A. (1999). The RCN1-encoded A subunit of protein phosphatase 2A increases phosphatase activity in vivo. Plant J. 20 389–399. [DOI] [PubMed] [Google Scholar]
- Friml, J., Benkova, E., Blilou, I., Wisniewska, J., Hamann, T., Ljung, K., Woody, S., Sandberg, G., Scheres, B., Jurgens, G., and Palme, K. (2002). AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108 661–673. [DOI] [PubMed] [Google Scholar]
- Friml, J., et al. (2004). A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306 862–865. [DOI] [PubMed] [Google Scholar]
- Fukaki, H., Tameda, S., Masuda, H., and Tasaka, M. (2002). Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 29 153–168. [DOI] [PubMed] [Google Scholar]
- Fukuda, H. (2004). Signals that control plant vascular cell differentiation. Nat. Rev. Mol. Cell Biol. 5 379–391. [DOI] [PubMed] [Google Scholar]
- Galweiler, L., Guan, C., Muller, A., Wisman, E., Mendgen, K., Yephremov, A., and Palme, K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282 2226–2230. [DOI] [PubMed] [Google Scholar]
- Garbers, C., DeLong, A., Deruere, J., Bernasconi, P., and Soll, D. (1996). A mutation in protein phosphatase 2A regulatory subunit A affects auxin transport in Arabidopsis. EMBO J. 15 2115–2124. [PMC free article] [PubMed] [Google Scholar]
- Gil, P., Dewey, E., Friml, J., Zhao, Y., Snowden, K.C., Putterill, J., Palme, K., Estelle, M., and Chory, J. (2001). BIG: A calossin-like protein required for polar auxin transport in Arabidopsis. Genes Dev. 15 1985–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., Muskett, P.R., Chuang, H.W., and Parker, J.E. (2003). Arabidopsis SGT1b is required for SCF(TIR1)-mediated auxin response. Plant Cell 15 1310–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., Ostin, A., Sandberg, G., Romano, C.P., and Estelle, M. (1998). High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. USA 95 7197–7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke, C.S., and Berleth, T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, S.N., Hunt, H.D., Horton, R.M., Pullen, J.K., and Pease, L.R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77 51–59. [DOI] [PubMed] [Google Scholar]
- Hu, Y., Bao, F., and Li, J. (2000). Promotive effect of brassinosteroids on cell division involves a distinct CycD3-induction pathway in Arabidopsis. Plant J. 24 693–701. [DOI] [PubMed] [Google Scholar]
- Ichimura, K. (2002). Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 7 301–308. [DOI] [PubMed] [Google Scholar]
- Ichimura, K., Mizoguchi, T., Hayashida, N., Seki, M., and Shinozaki, K. (1998). Molecular cloning and characterization of three cDNAs encoding putative mitogen-activated protein kinase kinases (MAPKKs) in Arabidopsis thaliana. DNA Res. 5 341–348. [DOI] [PubMed] [Google Scholar]
- Jensen, P.J., Hangarter, R.P., and Estelle, M. (1998). Auxin transport is required for hypocotyl elongation in light-grown but not dark-grown Arabidopsis. Plant Physiol. 116 455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak, C., Okresz, L., Bogre, L., and Hirt, H. (2002). Complexity, cross talk and integration of plant MAP kinase signalling. Curr. Opin. Plant Biol. 5 415–424. [DOI] [PubMed] [Google Scholar]
- Kepinski, S., and Leyser, O. (2005). Plant development: Auxin in loops. Curr. Biol. 15 R208–R210. [DOI] [PubMed] [Google Scholar]
- Kiegerl, S., Cardinale, F., Siligan, C., Gross, A., Baudouin, E., Liwosz, A., Eklof, S., Till, S., Bogre, L., Hirt, H., and Meskiene, I. (2000). SIMKK, a mitogen-activated protein kinase (MAPK) kinase, is a specific activator of the salt stress-induced MAPK, SIMK. Plant Cell 12 2247–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204 383–396. [Google Scholar]
- Kovtun, Y., Chiu, W.L., Tena, G., and Sheen, J. (2000). Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA 97 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan, P.J., Jester, P.J., Gottwald, J.R., and Sussman, M.R. (2002). An Arabidopsis mitogen-activated protein kinase kinase kinase gene family encodes essential positive regulators of cytokinesis. Plant Cell 14 1109–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser, H.M., Pickett, F.B., Dharmasiri, S., and Estelle, M. (1996). Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 10 403–413. [DOI] [PubMed] [Google Scholar]
- Leyser, O. (2003). Regulation of shoot branching by auxin. Trends Plant Sci. 8 541–545. [DOI] [PubMed] [Google Scholar]
- Li, H., Johnson, P., Stepanova, A., Alonso, J.M., and Ecker, J.R. (2004). Convergence of signaling pathways in the control of differential cell growth in Arabidopsis. Dev. Cell 7 193–204. [DOI] [PubMed] [Google Scholar]
- Liu, Y., and Zhang, S. (2004). Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16 3386–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, C., Han, M.H., Guevara-Garcia, A., and Fedoroff, N.V. (2002). Mitogen-activated protein kinase signaling in postgermination arrest of development by abscisic acid. Proc. Natl. Acad. Sci. USA 99 15812–15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig, C. (2002). Auxin transport: ABC proteins join the club. Trends Plant Sci. 7 329–332. [DOI] [PubMed] [Google Scholar]
- Luschnig, C., Gaxiola, R.A., Grisafi, P., and Fink, G.R. (1998). EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 12 2175–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant, A., Kargul, J., May, S.T., Muller, P., Delbarre, A., Perrot-Rechenmann, C., and Bennett, M.J. (1999). AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J. 18 2066–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka, D., Nanmori, T., Sato, K., Fukami, Y., Kikkawa, U., and Yasuda, T. (2002). Activation of AtMEK1, an Arabidopsis mitogen-activated protein kinase kinase, in vitro and in vivo: Analysis of active mutants expressed in E. coli and generation of the active form in stress response in seedlings. Plant J. 29 637–647. [DOI] [PubMed] [Google Scholar]
- Mattsson, J., Sung, Z.R., and Berleth, T. (1999). Responses of plant vascular systems to auxin transport inhibition. Development 126 2979–2991. [DOI] [PubMed] [Google Scholar]
- Menke, F.L., van Pelt, J.A., Pieterse, C.M., and Klessig, D.F. (2004). Silencing of the mitogen-activated protein kinase MPK6 compromises disease resistance in Arabidopsis. Plant Cell 16 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi, T., Gotoh, Y., Nishida, E., Yamaguchi-Shinozaki, K., Hayashida, N., Iwasaki, T., Kamada, H., and Shinozaki, K. (1994). Characterization of two cDNAs that encode MAP kinase homologues in Arabidopsis thaliana and analysis of the possible role of auxin in activating such kinase activities in cultured cells. Plant J. 5 111–122. [DOI] [PubMed] [Google Scholar]
- Mockaitis, K., and Howell, S.H. (2000). Auxin induces mitogenic activated protein kinase (MAPK) activation in roots of Arabidopsis seedlings. Plant J. 24 785–796. [DOI] [PubMed] [Google Scholar]
- Morris, A.D., Friml, J., and Zazimalova, E. (2004). Auxin transport. In Plant Hormones: Biosynthesis, Signal Transduction, Action! P.J. Davies, ed (Dordrech, The Netherlands: Kluwer Academic Publishers), pp. 437–470.
- Mou, Z., Wang, X., Fu, Z., Dai, Y., Han, C., Ouyang, J., Bao, F., Hu, Y., and Li, J. (2002). Silencing of phosphoethanolamine N-methyltransferase results in temperature-sensitive male sterility and salt hypersensitivity in Arabidopsis. Plant Cell 14 2031–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muday, G.K., and DeLong, A. (2001). Polar auxin transport: Controlling where and how much. Trends Plant Sci. 6 535–542. [DOI] [PubMed] [Google Scholar]
- Muller, A., Guan, C., Galweiler, L., Tanzler, P., Huijser, P., Marchant, A., Parry, G., Bennett, M., Wisman, E., and Palme, K. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17 6903–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, A., Peer, W.A., and Taiz, L. (2000). Regulation of auxin transport by aminopeptidases and endogenous flavonoids. Planta 211 315–324. [DOI] [PubMed] [Google Scholar]
- Nishihama, R., Ishikawa, M., Araki, S., Soyano, T., Asada, T., and Machida, Y. (2001). The NPK1 mitogen-activated protein kinase kinase kinase is a regulator of cell-plate formation in plant cytokinesis. Genes Dev. 15 352–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh, B., Bandyopadhyay, A., Peer, W.A., Spalding, E.P., and Murphy, A.S. (2003). Enhanced gravi- and phototropism in plant mdr mutants mislocalizing the auxin efflux protein PIN1. Nature 423 999–1002. [DOI] [PubMed] [Google Scholar]
- Noh, B., Murphy, A.S., and Spalding, E.P. (2001). Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13 2441–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, K., Ueda, J., Komaki, M.K., Bell, C.J., and Shimura, Y. (1991). Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang, J., Shao, X., and Li, J.Y. (2000). Indole-3-glycerol phosphate, a branchpoint of indole-3-acetic acid biosynthesis from the tryptophan biosynthetic pathway in Arabidopsis thaliana. Plant J. 24 327–333. [DOI] [PubMed] [Google Scholar]
- Palme, K., and Galweiler, L. (1999). PIN-pointing the molecular basis of auxin transport. Curr. Opin. Plant Biol. 2 375–381. [DOI] [PubMed] [Google Scholar]
- Peer, W.A., Bandyopadhyay, A., Blakeslee, J.J., Makam, S.N., Chen, R.J., Masson, P.H., and Murphy, A.S. (2004). Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell 16 1898–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, M., et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103 1111–1120. [DOI] [PubMed] [Google Scholar]
- Reed, R.C., Brady, S.R., and Muday, G.K. (1998). Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiol. 118 1369–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt, D., Pesce, E.R., Stieger, P., Mandel, T., Baltensperger, K., Bennett, M., Traas, J., Friml, J., and Kuhlemeier, C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426 255–260. [DOI] [PubMed] [Google Scholar]
- Ren, D., Yang, H., and Zhang, S. (2002). Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J. Biol. Chem. 277 559–565. [DOI] [PubMed] [Google Scholar]
- Rogg, L.E., Lasswell, J., and Bartel, B. (2001). A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13 465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs, T. (2000). Integrating cellular and organismic aspects of vascular differentiation. Plant Cell Physiol. 41 649–656. [DOI] [PubMed] [Google Scholar]
- Sorefan, K., Booker, J., Haurogne, K., Goussot, M., Bainbridge, K., Foo, E., Chatfield, S., Ward, S., Beveridge, C., Rameau, C., and Leyser, O. (2003). MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 17 1469–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano, T., Nishihama, R., Morikiyo, K., Ishikawa, M., and Machida, Y. (2003). NQK1/NtMEK1 is a MAPKK that acts in the NPK1 MAPKKK-mediated MAPK cascade and is required for plant cytokinesis. Genes Dev. 17 1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann, T., Geldner, N., Grebe, M., Mangold, S., Jackson, C.L., Paris, S., Galweiler, L., Palme, K., and Jurgens, G. (1999). Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286 316–318. [DOI] [PubMed] [Google Scholar]
- Steynen, Q.J., and Schultz, E.A. (2003). The FORKED genes are essential for distal vein meeting in Arabidopsis. Development 130 4695–4708. [DOI] [PubMed] [Google Scholar]
- Swarup, R., Friml, J., Marchant, A., Ljung, K., Sandberg, G., Palme, K., and Bennett, M. (2001). Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev. 15 2648–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teige, M., Scheikl, E., Eulgem, T., Doczi, R., Ichimura, K., Shinozaki, K., Dangl, J.L., and Hirt, H. (2004). The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell 15 141–152. [DOI] [PubMed] [Google Scholar]
- Weigel, D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Q., Frugis, G., Colgan, D., and Chua, N.H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14 3024–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenser, N., Ellsmore, A., Leasure, C., and Callis, J. (2001). Auxin modulates the degradation rate of Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 98 11795–11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.H. (1999). IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 11 2139–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.