Abstract
The outermost epidermal cell wall is specialized to withstand pathogens and natural stresses, and lipid-based cuticular polymers are the major barrier against incursions. The Arabidopsis thaliana mutant bodyguard (bdg), which exhibits defects characteristic of the loss of cuticle structure not attributable to a lack of typical cutin monomers, unexpectedly accumulates significantly more cell wall–bound lipids and epicuticular waxes than wild-type plants. Pleiotropic effects of the bdg mutation on growth, viability, and cell differentiation are also observed. BDG encodes a member of the α/β-hydrolase fold protein superfamily and is expressed exclusively in epidermal cells. Using Strep-tag epitope-tagged BDG for mutant complementation and immunolocalization, we show that BDG is a polarly localized protein that accumulates in the outermost cell wall in the epidermis. With regard to the appearance and structure of the cuticle, the phenotype conferred by bdg is reminiscent of that of transgenic Arabidopsis plants that express an extracellular fungal cutinase, suggesting that bdg may be incapable of completing the polymerization of carboxylic esters in the cuticular layer of the cell wall or the cuticle proper. We propose that BDG codes for an extracellular synthase responsible for the formation of cuticle. The alternative hypothesis proposes that BDG controls the proliferation/differentiation status of the epidermis via an unknown mechanism.
INTRODUCTION
In plants, the outermost epidermal cell wall is specially fortified and consists of hydrophilic and hydrophobic components. Cell wall carbohydrates and glycoproteins provide an underlying hydrophilic framework, whereas extracellular lipids constitute the impermeable hydrophobic outer portion of the cell wall, called cuticle. Some extracellular lipids (e.g., waxes) are soluble and can be extracted with organic solvents, whereas others (e.g., cutin) are interconnected by ester bonds, and their composition can only be studied after hydrolysis of the polyesters. Moreover, a fraction of the lipid polymers (e.g., cutan) is resistant to hydrolysis. Structurally, the outermost layer of the cuticle is composed of wax, whereas the insoluble polymers constituting the cuticle proper are deposited beneath it and are seen in electron micrographs as a thin lamellate layer of darkly staining material (Kolattukudy, 2001a). As an exterior protective and supporting structure, the cell wall determines cell shape and is involved in adhesion, defense, differentiation, and growth processes. Not surprisingly, therefore, many mutants associated with cell wall biogenesis and function (Arioli et al., 1998; Ellis et al., 2002; Nishimura et al., 2003) show pleiotropic phenotypes.
The cuticular layer of the cell wall is distinguished by the presence of lipid molecules and comprises the part of the cell wall where carbohydrate and lipid molecules are intimately interconnected. Its exact composition is not known, because it is not possible to physically separate the various constituents. However, the composition of the lipid polyesters that make up the structural component of the cuticle proper, cutin, has been precisely determined in many plant species that have a relatively thick cuticular membrane. Typically, it is characterized by the presence of C16 and C18 fatty acids bearing ω- and mid-chain hydroxyl groups (Kolattukudy, 2001a). However, α,ω-dicarboxylic fatty acids (40%) and 2-hydroxy fatty acids (14%) were identified as major depolymerization products in Arabidopsis thaliana cutin, although cutin-characteristic ω-hydroxy and mid-chain hydroxylated fatty acids were detected as well (7 and 8%, respectively) (Franke et al., 2005). Besides the unusual chemical composition, the cuticle proper is very thin in Arabidopsis compared with most plants studied (Nawrath, 2003).
Biochemical analysis revealed a striking similarity between Arabidopsis cutin and root suberin, which is a cell wall–linked polyester found in cells that provide a barrier to apoplastic water transport and that have a defensive function. It can be enzymatically detached and isolated with the use of polysaccharide hydrolases. Contrary to cutin, the aliphatic composition of suberin in Arabidopsis is very similar to that described previously for other plants (Kolattukudy, 2001b; Bernards, 2002). It is characterized by high levels of very-long-chain ω-hydroxy (43%) and α,ω-dicarboxylic (24%) fatty acids. Carboxylic fatty acids, fatty alcohols, and 2-hydroxy fatty acids are minor components (Franke et al., 2005).
Based on biochemical studies conducted in the 1970s and 1980s, many of the steps in the biosynthetic pathway for cuticular polymers have been deduced (Kolattukudy, 2001a; Lequeu et al., 2003). However, the failure to identify mutants in this pathway has made it difficult to verify these proposals. It now seems likely that a number of Arabidopsis mutants that exhibit adhesion responses (organ fusions), cell death, and retarded cell differentiation and growth (Lolle et al., 1998; Yephremov et al., 1999; Pruitt et al., 2000; Tanaka et al., 2001, 2004; Wellesen et al., 2001; Chen et al., 2003; Kurata et al., 2003; Schnurr et al., 2004; Kurdyukov et al., 2006) may, in fact, be associated with defects in the cuticular layer of the cell wall or in the cuticle proper. In turn, molecular cloning of several genes apparently involved in the formation of cuticle in Arabidopsis has raised further questions concerning the interrelationships between various components of the epidermal cell wall and the mechanisms by which these genes function in a diverse set of signaling pathways (Yephremov and Schreiber, 2005).
Lipid polymers constitute the major barrier against invasion by pathogens and restrict cell–cell interactions. To allow pollen–pistil interactions to occur and to enable pathogens to invade the plant, lipid cell wall polymers have to be weakened or degraded. Cutinases capable of doing this have been isolated from phytopathogenic fungi (Soliday et al., 1984), and the presence of enzymes with a similar activity has been reported in plants (Hiscock et al., 1994, 2002; Lavithis and Bhalla, 1995). However, in contrast with the case in fungi, corresponding plant genes have not been molecularly identified. Also, the postulated synthases involved in the last steps in the biosynthesis of cutin and suberin, and in attaching lipids to other components of the cell wall, have not yet been identified. The synthesis and hydrolysis of extracellular polymers have to be coordinated with extension of the cell surface during growth, but how this is achieved also remains to be explained.
Here, we report the molecular cloning of the BODYGUARD (BDG) gene, which is required for the formation of the cuticle in Arabidopsis. BDG is an epidermis-specific extracellular α/β-hydrolase fold–containing protein. Structural analysis showed that cell wall and cuticular materials are largely misplaced in the mutant, and a continuous functional cuticle proper is not formed, thus explaining its various phenotypes. These results are consistent with the idea that BDG may function in the cuticular layer of the cell wall or cuticle proper by serving as a synthase, but paradoxically, the loss of its function in the bdg mutant results in an overall increase in the level of cutin and wax constituents.
RESULTS
Morphological and Cuticular Defects Associated with the Loss of the BDG Function
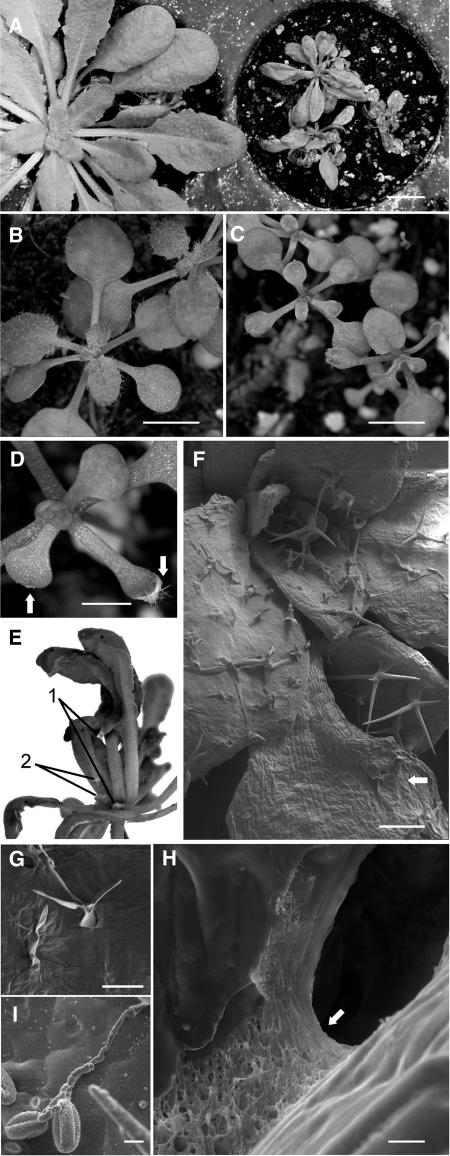
During regular mutant screens of an Arabidopsis population mutagenized with the maize (Zea mays) transposon Enhancer/Suppressor-mutator (En/Spm), two mutant families, 6W61 and 6W32, were selected for study because they both segregated dwarfish, bushy plants. Closer examination revealed that these mutants often display leaf fusions, which can be quite prominent, especially if the plants are grown first under short-day conditions. At early stages of development, both 6W61 and 6W32 (Figure 1C) were clearly distinguishable from wild-type plants (Figure 1B), because they were smaller, often produced deformed leaves, and showed reduced pubescence. At the age of several weeks, mutant plants exhibited strong growth retardation and abnormal leaf morphology (Figure 1A); however, the severity of these phenotypes varied. Generally, mutants were characterized by having curved, more elongated leaves with slightly serrated edges. The leaf deformations were observed on both mutants independently of the occurrence of leaf fusions. Some leaf fusions were so strong, it resulted in tearing of laminae and petioles during growth (Figure 1E), similar to what occurred in lcr (Wellesen et al., 2001). Despite a general growth deficiency in both mutants, they produced normal-looking fertile flowers and gave rise to seeds. It was noted also that spraying with fungicides, which were well tolerated by wild-type plants, was deleterious to these mutants and resulted in necrotic lesions and ultimately in death (data not shown). Complementation tests on the recessive mutants W32 and W61 showed that they were allelic. Because the phenotypes of these mutants suggested that they were defective in the formation of the cuticle, the mutant locus was called bdg, and the mutant alleles W32 and W61 were designated bdg-1 and bdg-2, respectively.
Figure 1.
Phenotype of the bdg Mutant of Arabidopsis.
The panels show photographs ([A] to [E]) and scanning electron micrographs ([F] to [I]). Bars = 10 mm for (A), 5 mm for (B) and (C), 2 mm for (D), 200 μm for (F), 100 μm for (G), and 10 μm for (H) and (I).
(A) When grown naturally in short daylengths (8 h of light), 6-week-old mutants (right) have smaller, often deformed leaves and may stunt their growth. Compare with the wild-type plant at left.
(B) and (C) Wild-type (B) and bdg (C) plants can be easily distinguished at the age of 3 weeks. Note the small size of bdg leaf blades and the reduced numbers of trichomes on them.
(D) Typical necrotic lesions affecting both the epidermis and the mesophyll at the tips of leaves in bdg (arrows).
(E) Deformation of a bdg plant caused by graft-like fusions of rosette leaves. Note the rupture of two petioles (lines 1 and 2 show upper and lower rips) caused by extremely strong fusions between growing organs.
(F) General view of the leaf surface showing defects in the epidermis in bdg. Some trichomes are fully developed, but many are misshapen and arrested at earlier stages. In many cases, trichomes become flattened, bent, and eventually adhere to the leaf surface (arrow).
(G) Collapse of mature trichomes.
(H) A suture zone (arrow) showing no signs of cuticle proper.
(I) Germination of wild-type pollen induced by contact with bdg leaf epidermis.
Scanning electron microscopy of plants grown in the greenhouse showed that epidermal cells of bdg are shrunken. Although bdg plants were able to develop mature trichomes, these frequently appeared to be flattened or otherwise misshapen (Figures 1F and 1G). Shrinkage probably could account for most of these deformations, because trichomes later collapsed and appeared dehydrated, having a flat shape and adhering to pavement epidermal cells (Figure 1G); however, the involvement of mechanisms specifically controlling cell death cannot be excluded. The epidermis seemed to be the primary target of the mutation; however, parenchymatous cells became affected as cell death spread from the leaf tips (Figure 1D).
Examination of suture zones in bdg revealed a network of fibrous material between normally separate parts of the plant (Figure 1H), similar to that found in suture zones of lcr mutants (Wellesen et al., 2001). These findings suggested that the cell walls are in direct contact with each other in such zones.
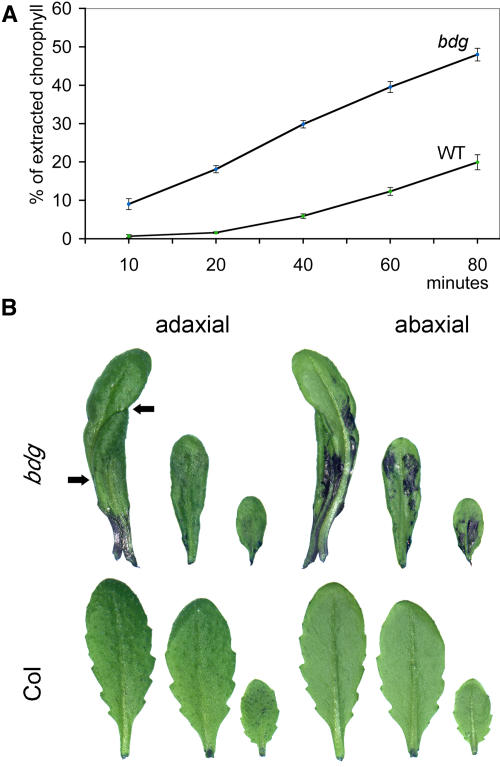
A reduction in the isolating properties of the outermost epidermal cell wall affected in the mutant also accounts for the germination of wild-type pollen grains on the leaf surface in bdg plants (Figure 1I), the enhancement of chlorophyll leaching in 80% ethanol (Lolle et al., 1997) (Figure 2A), and the increased staining with aqueous solutions of the cationic dye toluidine blue (Tanaka et al., 2004) (Figure 2B). In these respects, bdg is similar to most mutants that show organ fusions (Lolle et al., 1998).
Figure 2.
Defects in the Epidermal Cell Surface of bdg Leaves Revealed by Leaching of Chlorophyll and Staining with a Cationic Dye.
(A) Intact rosette leaves of bdg release chlorophyll faster than do wild-type leaves when immersed in 80% ethanol, probably because both the solvent and solute more easily penetrate the leaf surface. Each value is an average of six replicates. Bars indicate se. Differences between Columbia (Col) and bdg were significant at P < 0.01.
(B) Representative rosette leaves of bdg (top row) and Col (bottom row) are shown from adaxial and abaxial sides after 2 min of staining with an aqueous solution (0.05%) of toluidine blue, which is able to bind selectively to free anionic groups such as carboxylate, phosphate, and sulfate radicals (Tanaka et al., 2004). When stained, leaves of bdg show a patchy coloration pattern, in contrast with wild-type leaves, which remain virtually unstained because the intact cuticle excludes the dye. Notice that the dye penetrates more rapidly through the cuticle on the abaxial side. The two fused leaves of bdg shown at left (arrows) are reminiscent of staining to unfused leaves. This figure also illustrates changes in leaf morphology. In bdg, the leaves are elongated and narrow with a smooth margin, compared with the broad serrated leaves of the wild type.
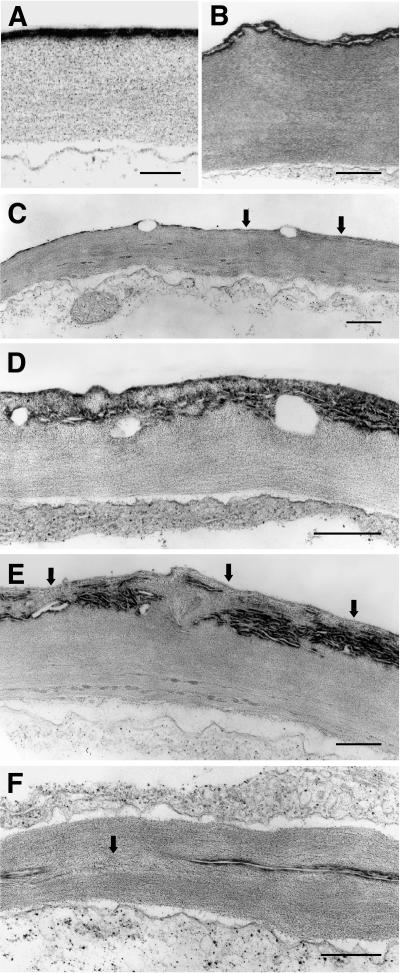
To prove that the cuticle is indeed impaired in bdg, we inspected the outermost cell wall of the epidermis by transmission electron microscopy. In electron micrographs, the cuticle of the wild-type leaf of Arabidopsis is seen as a continuous and regular layered structure: an outer electron-dense membrane of 20 to 30 nm corresponding to the cuticle proper is deposited over a less dense layer corresponding to the cell wall (Figure 3A). The structure of the cuticle in the bdg mutant, on the other hand, is characterized by a series of irregular layers in which electron-dense and less dense materials of the cell wall are intermingled with empty spaces (Figures 3B to 3E). In many cases (Figure 3C), the outermost cell wall boundary is protected by only a minimal amount of the electron-dense overlay, which is composed mainly of extracellular lipid polymers (e.g., cutin) (Kolattukudy, 2001a). The formation of multilayered structures is correlated with an increase in the thickness of the cuticular layer (Figures 3D and 3E). Irregular deposition of electron-opaque material occurs not only at the outermost surface but also within the deeper layers of the cell wall. Empty spaces are occasionally found, giving this complex structure a cavernous appearance (Figures 3C to 3E). In places, large amounts of electron-opaque material are deposited within the cell wall, whereas the outer boundary itself is hardly protected at all (Figure 3E).
Figure 3.
Ultrastructure of the Cuticle of the Leaf Epidermis.
The images were obtained with a transmission electron microscope. Bars = 500 nm.
(A) Regular electron-dense cuticle proper (membrane) formed by wild-type plants. Compare with the deformed cuticles in bdg ([B] to [F]).
(B) The mutant cell wall exhibiting a paste-like appearance with irregular, misshapen boundaries. The cuticular membrane is much thinner than in the wild type and is puckered, probably as a result of pressure from the underlying material associated with the cell wall.
(C) In places (arrows), the electron-opaque cuticle proper shows large-scale disruption and may be practically absent altogether.
(D) Pockets or caverns in the cuticular zone (see also [C]).
(E) A series of irregular layers formed by bulging of the less opaque cell wall material. The cuticular membrane is hardly detectable along the surface of the protuberance (arrows).
(F) A fusion zone (arrow) devoid of intervening cuticular membrane between two fused leaves.
The cavernous structure of the cuticle in bdg suggests that underlying cell wall constituents may bulge out continuously as a result of the fragility of the outer coat. Interestingly, the cuticular layer in the mutant is thicker overall, and the amount of extracellular lipid polymers does not appear to be reduced. As expected, no cuticle proper is formed between the walls of the epidermal cells in the suture zones (Figure 3F).
Thus, the outermost layer of the cell wall is mainly affected in bdg mutants, making it likely that BDG is involved in the pathway responsible for the formation of either the cuticular layer of the cell wall or the cuticle proper.
Phenotype of bdg under in Vitro Culture Conditions
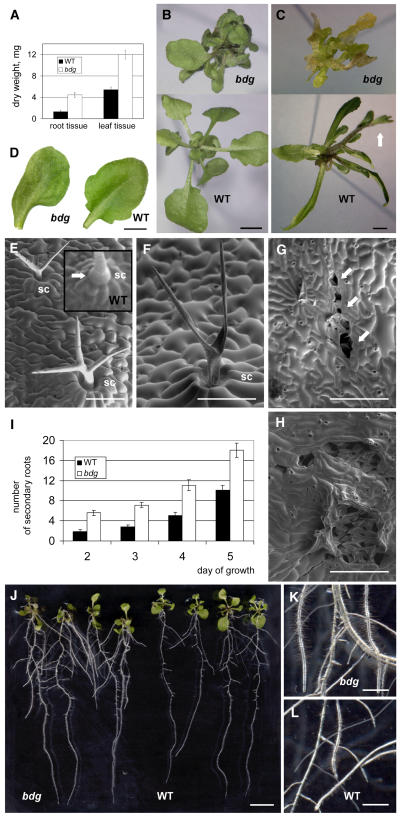
Based on the phenotypic performance of bdg maintained in the greenhouse, it is conceivable that the inability to form the cuticle makes bdg plants vulnerable to natural environmental conditions that do not adversely affect the growth of wild-type plants. To test this notion, bdg plants were grown on hormone-free Murashige and Skoog (MS) medium supplemented with agar under conditions of 100% humidity to avoid dehydration stress. Indeed, under these conditions, the mutants did not exhibit strong growth retardation during the first 2 weeks. On the contrary, bdg plants had two times greater dry weight yields of leaf and root tissue than wild-type plants (Figure 4A). Mutant plants, however, exhibited characteristic morphological features of bdg. They developed deformed leaves (Figures 4B and 4D) that had thick, short petioles, and occasional leaf fusions. Degenerative changes in bdg were first characterized by the progressive deterioration of trichomes and the appearance of ruptures in the leaf epidermis that were observed in severely deformed leaves and leaves that appeared almost wild type. In contrast with wild-type trichomes, which are characterized by a bulbous stalk dominating longitudinally expanded subsidiary cells (Figure 4E), bdg trichomes often lacked a bulbous expansion at the base, which appeared to be located in the depression area (Figure 4F), and later collapsed. Under scanning electron microscopy, small tears in the epidermis were seen (Figure 4G), suggesting that cell–cell adhesion is weakened in the epidermis of bdg. As the tears enlarged, they were transformed into concave lesions that lacked the epidermis (Figure 4H).
Figure 4.
In Vitro Culture of the bdg Mutant.
The panels show photographs ([B] to [D] and [J] to [L]) and scanning electron micrographs ([E] to [H]). Bars = 3 mm for (B) to (D), 100 μm for (E) and (F), 300 μm for (G) and (H), 10 mm for (J), and 250 μm for (K) and (L).
(A) Graph showing the mean dry weight of root and leaf tissue in bdg compared with the wild type. The plants from germinated seeds were grown for 7 d on vertical agar plates. Dry weight refers to the weight of tissue after it has been dried at 60°C. Results are presented as means ± se for 16 bdg and 32 wild-type plants.
(B) Twenty-four-day-old bdg and wild-type plants. Rosette leaves in bdg are severely misshapen (see also [D]) and have short, thick petioles, deformed laminae, and, occasionally, leaf fusions.
(C) At the age of 4 weeks, wild-type plants produced inflorescences, whereas mutant plants became vitrified and developed a calli-like appearance.
(D) A typical bdg leaf at the age of 24 d that did not exhibit any fusions with other leaves. It can be distinguished by misshapen lamina compared with the wild-type leaf.
(E) Branched trichomes on the abaxial surface of a mutant rosette leaf such as the one shown in (D). Note that bdg trichomes do not form bulbous structures at their bases as wild-type trichomes, shown in the inset for comparison, do (arrow). Subsidiary cells (sc) forming a ring around the trichome cell appear unaffected.
(F) Typical collapsed trichome in bdg. It appears similar to the trichomes shown in Figure 1G on the rosette leaves of bdg plants grown in a greenhouse.
(G) Epidermal ruptures (arrows) in the bdg rosette leaf such as the one shown in (D).
(H) Degenerative changes in the bdg epidermis resulting from the progression of tears.
(I) Graph showing the mean number of secondary roots in bdg and wild-type plants grown on vertical agar plates. Results are presented as means ± se for 16 bdg and 32 wild-type plants.
(J) Four bdg and four wild-type plants grown on a representative vertical plate for 1 week.
(K) and (L) Higher magnification of bdg roots (K) compared with wild-type roots (L). Mutant roots produce more and longer root hairs.
No difference was found in the length of the main root between bdg and the wild type (data not shown), although the growth of secondary roots appeared to be initiated earlier in bdg, and total root length and root dry weight were higher for bdg compared with the wild type (Figures 4I and 4J). Increased root branching in bdg was also observed when plants were grown on medium containing diverse polyethylene glycol concentrations (data not shown). The root hairs of bdg plants were found to be twice as long as wild-type hairs, contributing to the hairy appearance of excessively branched bdg roots (Figure 4K). Roots of the bdg mutant showed a normal response to gravitropism (data not shown).
During the first 10 d, bdg and wild-type plants developed slightly translucent leaves; however, wild-type plants gradually adapted to the tissue culture environment, recovered, and bolted within 4 weeks, whereas bdg plants continued to produce deformed vitrified leaves that occasionally developed tumor-like outgrowths along petioles. The mutant plants gradually became bushy as a result of the excessive proliferation of meristems. The bdg leaves failed to expand and exhibited rough, thickened lamina and disorganized mesophyll and epidermal layers (Figure 4C). Calli-like structures were observed in the apical zone in 30 of 33 mutants (∼90%) after 4 weeks of development. At the areas associated with proliferation and in appendages, leaves showed under scanning electron microscopy examination a complete lack of epidermal layer.
These observations support the idea that epidermal cells are the primary target of the bdg mutation. However, it appears that the phenotype conferred by bdg is the result of the complex interplay between genetic and environmental factors and that other factors besides the level of humidity are important in determining morphogenesis and cell viability in bdg.
Analysis of Bound Lipids in bdg
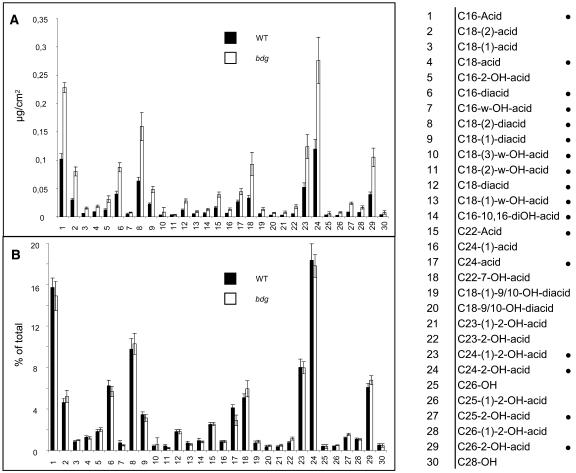
The composition of the lipid polyesters that constitute the structural component of the cuticle proper has been precisely determined in many plant species that have relatively thick cuticular membranes. However, isolation of the cuticle proper from wild-type Arabidopsis plants is difficult because of its extreme thinness; furthermore, the cuticle proper cannot be isolated as a continuous membrane from bdg mutants because it is highly deformed (Figure 3). Therefore, we compared the composition of residual bound lipids in leaves of bdg and wild-type plants grown under the same conditions. Residual bound lipids (i.e., those remaining after thorough extraction with methanol/chloroform) represent a fraction of the lipid polymers, which include cutin and other covalently bound lipids attached to the cell wall (Bonaventure et al., 2004; Franke et al., 2005; Kurdyukov et al., 2006). Of the 30 compounds identified in this fraction by gas chromatography–mass spectrometry (GC-MS), 17 were monomers of Arabidopsis cutin (Franke et al., 2005) (Figure 5). They constituted ∼83% of residual bound lipids. Compounds that are not found in cutin made up the remaining 17% of the total bound lipids in the samples. The comparison revealed a significant difference between bdg and the wild type in the quantity of the residual bound lipids per unit of leaf surface area (Figure 5A). Structural defects in the cuticle suggest that bdg may be incapable of synthesizing one or more essential cutin monomers. Strikingly, relative to leaf surface area, the amount of lipid (expressed in micrograms per square centimeter) found in bdg is 1.2 to 3 times that in the wild type, and this holds for all monomers. This increase correlates well with the deposition of more polymer material in the cell wall of bdg seen by transmission electron microscopy (Figures 3D and 3E). All of the lipid compounds analyzed, including cutin monomers, actually accumulate in higher amounts in the mutant than in the wild type, whereas the quantitative relationships between the different compounds (percentage composition) are very similar in bdg and the wild type (Figure 5B). This result suggests that BDG may be required not for the synthesis but for the cross-linking of molecules that form part of the cuticular layer of the cell wall or the cuticle proper.
Figure 5.
GC-MS Analysis of Residual Bound Lipids in bdg and Wild-Type Leaves.
Graphs (A) and (B) show the amount of each compound (expressed in micrograms per square centimeter and as a percentage of the total, respectively). Each value is the mean ± se of six replicates. The numbers correspond to the order of the peaks in the GC-MS scan. Monomers found in Arabidopsis cutin in an independent experiment (Franke et al., 2005) are indicated by black circles; see Franke et al. (2005) and Kurdyukov et al. (2006) for details of the method.
Wax Accumulation Is Increased in bdg through the Activation of the Decarbonylation Pathway
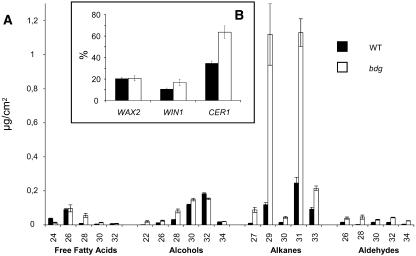
Like most mutants that show organ fusions (Yephremov et al., 1999), bdg plants do not exhibit any obvious reduction in epicuticular wax. On the contrary, analysis shows a 3.5-fold increase in wax load per square centimeter in the mutant compared with the wild type (Figure 6A). The amounts of C29, C31, and C33 alkanes, which represent the major constituents of the wax in Arabidopsis, were increased 9.4, 4.6, and 2.3 times, respectively, in bdg. Similar ratios were obtained for many aldehydes, alcohols, and acids (see Supplemental Table 2 online).
Figure 6.
Wax Biosynthesis in bdg.
(A) Analysis of the composition of epicuticular wax in bdg and wild-type leaves. Compound classes and carbon chain lengths are indicated on the x axis. Each bar shows the relative amount of a specific constituent as the mean ± se of five replicates.
(B) Increased expression levels of genes in the decarbonylation pathway. Quantification of mRNA was performed by RT-PCR. Transcript abundance is expressed relative to the amount of product amplified using primers for the control transcript, actin. The SHN1/WIN1 gene codes for an ethylene response factor–type transcription factor, which can activate several genes involved in wax biosynthesis in Arabidopsis, including CER1. CER1 encodes a putative fatty aldehyde decarbonylase, or a transmembrane protein that regulates it, or a protein involved in the secretion of alkanes (Aarts et al., 1995; Broun et al., 2004). WAX2/YRE has high sequence homology with CER1, but its function in the decarbonylation pathway is even less clear; it may encode an aldehyde-generating enzyme in the metabolic step preceding that catalyzed by CER1 (Chen et al., 2003; Kurata et al., 2003). The number of repetitions was 3 for WAX2/YRE and 12 for SHN1/WIN1 and CER1. Bars indicate means ± se. P values calculated by a nonparametric Wilcoxon-Mann-Whitney test (Statistica 6.0 package from StatSoft) are depicted above the graph bars if they are ≤0.05.
Taking the cuticular phenotype of bdg into consideration, we propose that the enhanced wax accumulation in bdg represents a compensatory reaction to the loss of cell wall and cuticle integrity.
In general, the alkanes with odd numbers of carbons contribute disproportionately to this increase, at the expense of other constituents; they make up ∼76% of the leaf wax in bdg compared with ∼46% in the wild type. This finding suggests that the major differences in wax biosynthesis in bdg can be attributed to activation of the decarbonylation pathway, which includes the reduction of even-carbon-number (C2n) fatty acids to aldehydes followed by the resultant aldehyde decarbonylase-catalyzed formation of alkanes with odd-numbered (C2n–1) chain lengths (Cheesbrough and Kolattukudy, 1984).
However, quantitative RT-PCR analyses of genes in the decarbonylation pathway suggest that BDG is not involved directly in wax biosynthesis: bdg cells accumulate higher levels of CER1 (∼184%) and SHN1/WIN1 (∼158%) transcripts compared with wild-type plants (Figure 6B). It remains to be determined whether transcriptional upregulation of these genes or enhanced stability of their transcripts is responsible for these changes. However, activation of the decarbonylation pathway in wax biosynthesis is likely to be one reason for the increased accumulation of alkanes in bdg.
Cloning of BDG by Transposon Tagging
Genetic analysis demonstrated that bdg-2 was a stable allele, whereas bdg-1 regularly reverted to the wild type (at rates of up to 11% per generation), suggesting that the latter mutation is caused by an En/Spm insertion. Segregation analysis of transposon flanking sequences was performed by transposon insertion display (Yephremov and Saedler, 2000) using progeny of heterozygous and homozygous mutants and wild-type plants containing up to 10 En/Spm insertions. One flanking fragment that cosegregated with the bdg-1 allele was identified and purified by HPLC (Transgenomic's WAVE system). Sequencing revealed that it constitutes a part of the first exon of the predicted gene At1g64670. The instability of bdg-1 made it difficult to prove by DNA gel blot hybridization that all bdg-1 mutants are homozygotes with regard to the En/Spm insertion at the At1g64670 locus, because a weak band corresponding to the wild-type product could often be seen. Therefore, we took a reverse genetic approach to obtain knockout mutants for the At1g64670 gene.
Reverse Genetics at the BDG Locus
First, we sequenced the At1g64670 gene in bdg-2 and found that it bears a 7-bp deletion at the site of the insertion in bdg-1 (see Supplemental Figure 1 online), suggesting that bdg-2 is a footprint allele derived from bdg-1.
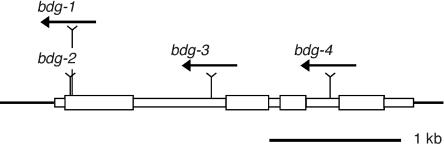
Second, using high-density filters containing transposon flanking sequences amplified from individual plants that had been mutagenized with En/Spm (Steiner-Lange et al., 2001), we identified families carrying other transposon insertions in the predicted At1g64670 gene (Figure 7). Progeny of these plants segregated mutants that were allelic to bdg-1 and indistinguishable from it with regard to the postgenital organ fusion phenotype. This finding confirmed that At1g64670 corresponds to the BDG gene.
Figure 7.
Mutant Alleles of the BDG Gene.
The location and orientation of En/Spm transposons in various mutant alleles of BDG (At1g64670, GenBank accession number AJ781319) are shown by the arrows. The bdg-2 allele is a derivative of bdg-1 and contains a transposon footprint (7-bp deletion) (see Supplemental Figure 1 online). Exons, introns, and 5′ and 3′ untranslated regions are boxed; exons are shown as thicker boxes.
Transgenic Complementation of bdg
Bearing in mind the facts that bdg mutants display a number of features, including changes in plant growth and architecture, and that unrelated mutations could potentially contribute to this pleiotropic phenotype, we attempted to complement the mutant by transformation with a 3.5-kb genomic fragment comprising the putative 0.9-kb promoter and the complete coding sequence of BDG. Plants with wild-type morphology were obtained in seven independent transgenic families, suggesting that this fragment can fully complement the phenotype conferred by the bdg mutant, and hence that all features of the bdg mutant result from a mutation in this single gene.
BDG Transcripts
RNA gel blot hybridization revealed that whereas BDG is expressed in various organs, including roots, the strongest expression is detected in flowers and floral buds (see Supplemental Figure 2 online). Cauline leaves show higher levels of BDG transcripts than do rosette leaves. Also, higher levels of BDG transcripts were observed in upper stem internodes than in bottom stem internodes. These results suggest that the expression of BDG is markedly higher in younger tissue toward the top of the stem, although this remains to be substantiated. The tissue expression patterns of BDG and ACE/HTH (Krolikowski et al., 2003; Kurdyukov et al., 2006) detected by RNA gel blot hybridization are quite similar when using the same filter after rehybridization (see Supplemental Figure 2 online).
The estimated size of BDG transcript was ∼1.7 kb, in good agreement with one of the two versions of At1g64670 that have been found in databases. To define the 5′ and 3′ untranslated regions, we mapped the actual beginning and the end of the transcript using rapid amplification of cDNA ends (RACE) techniques (see Supplemental Figure 1 online). The position of the transcription start was found to be very similar to that suggested by the sequencing of apparently full-length cDNA clones. Downstream of the previously reported polyadenylation site (GenBank accession number AK119137), 3′ RACE revealed the presence of an additional site (see Supplemental Figure 1 online). These results allowed us to conclude that BDG has four exons and that it encodes a protein of 469 amino acids.
BDG Defines a New Family of Plant-Specific α/β-Hydrolase Fold–Containing Proteins
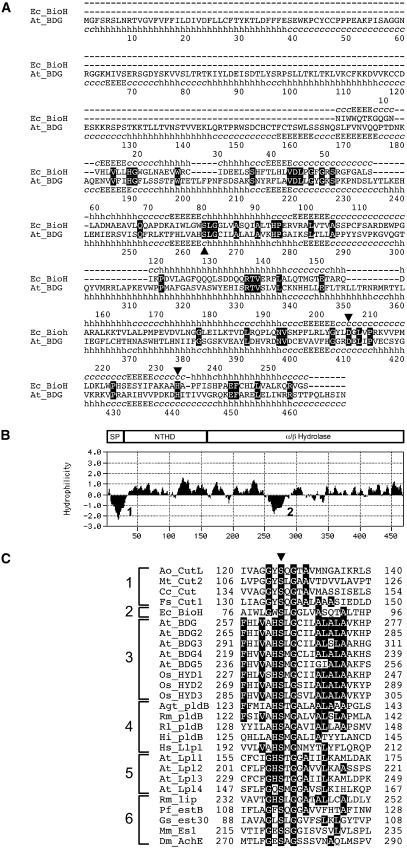
According to the annotations in the databases, BDG belongs to the α/β-hydrolase fold superfamily of proteins and contains the predicted hydrolase/acyltransferase domain (KOG1454). Members of this superfamily (hereafter referred to as α/β-hydrolases) show little amino acid sequence similarity, with the exception of the conserved catalytic triad composed of a base (His), a nucleophile (Ser), and an acid (Asp/Glu). However, studies of their three-dimensional structures have revealed that α/β-hydrolases diverged from a common ancestor and exhibit conservation of topology, having a characteristic α/β/α sandwich formed by a β-sheet core of five to eight strands connected by α-helices (Ollis et al., 1992; Hotelier et al., 2004). Therefore, to probe the sequence similarity of BDG to α/β-hydrolases further, we used the fold recognition programs GenTHREADER and 3D-PSSM (Kelley et al., 2000; McGuffin and Jones, 2003), which combine structural alignments to known three-dimensional structures with amino acid sequence alignments. With this method, we found that the BDG protein has a predicted α/β-hydrolase fold with seven strands connected by α-helices and a typical α/β-hydrolase catalytic triad comprising Ser-263, His-440, and Asp-410 (Figure 8A).
Figure 8.
Structure and Sequence Alignment of BDG with α/β-Hydrolases.
The amino acids that form the catalytic triad in α/β-hydrolases are indicated by triangles.
(A) Alignment of BDG with the representative, relatively high-scoring BioH protein from GenTHREADER searches (McGuffin and Jones, 2003). The known secondary structure sequences of BioH and predicted secondary structure sequences of BDG, represented by sequences of helixes (h), extended strands (E), and random coils (c), are correlated with the corresponding amino acid sequences.
(B) Proposed domain structure of the predicted BDG protein shown relative to a Kyte–Doolittle hydropathy plot generated by the MacVector sequence analysis package (Accelrys). In the plot, hydrophilic portions are above 0, and hydrophobic areas are below 0. Peaks with scores of >1.8 using a window size of 19 correspond to a possible N-terminal signal peptide (1) and a hydrophobic region in the α/β-hydrolase domain (2). NTHD, N-terminal hydrophilic domain; SP, signal peptide.
(C) Active-site Ser regions of various α/β-hydrolases. Shading highlights amino acid identities found in >35% of cases. Shown at left are functionally and structurally related gene families: (1) cutinases and cutinase-like proteins; (2) BioH family; (3) BDG family; (4) monoglyceridelipase and lysophospholipases; (5) putative lysophospholipases of Arabidopsis; and (6) various lipases and esterases.
With respect to α/β-hydrolases with known three-dimensional structures, BDG appeared to show no more than 12 to 16% sequence identity in its α/β-hydrolase domain. The region of homology occurs in the predicted C-terminal half of BDG, whereas for the 167–amino acid portion at the N terminus, no similar sequences could be identified in databases. In spite of the low sequence similarity, the predicted secondary structure of the C-terminal portion of BDG is very similar to that of BioH, a carboxylesterase involved in biotin biosynthesis in Escherichia coli (Figure 8A). The N-terminal portion of BDG is predicted to contain a 24– to 28–amino acid N-terminal signal sequence and a Leu zipper motif located at amino acids 80 to 101 in the hydrophilic domain (Figure 8B). The amino acid sequence of BDG seems to lack transmembrane-spanning domains, suggesting that the protein might be secreted out of the cells.
Whereas in the majority of α/β-hydrolases the GxSxG motif is conserved around the nucleophilic active site Ser, the predicted active site Ser-263 in members of the BDG family is situated in the (A,S)HSxG motif (Figure 8C). This pentapeptide is found in some bacterial lipases (e.g., lysophospholipases), in bacterial α/β-hydrolases whose function is not yet known, and in mammalian lysophospholipases (Taniyama et al., 1999). It has been demonstrated that substitutions at the first and last positions of the GxSxG motif do not always eliminate the enzymatic activity (Haruki et al., 1999); therefore, the importance of this active site architecture for protein function remains to be determined.
Strikingly, with regard to the amino acid residues surrounding the active site Ser-263, BDG is more reminiscent of lysophospholipases than is any of the four Arabidopsis proteins to which this function has hitherto been attributed on the basis of overall sequence similarity (Figure 8C), supporting the notion that BDG represents a distinct group of proteins.
Sequence homology searches with BLAST database search tools (http://www.ncbi.nlm.nih.gov/BLAST/) revealed highly related plant sequences, corresponding to predicted α/β-hydrolases with unknown functions. The Arabidopsis genome contains four sequences homologous with BDG (which show 82, 64, 45, and 40% amino acid identity to BDG in the α/β-hydrolase domain), whereas three similar sequences were found among predicted rice (Oryza sativa) proteins (with 64, 64, and 47% identity). All of these BDG-like proteins are also characterized by conservation of amino acid sequences at the active site Ser, suggesting that they evolved from a common ancestor and that their functions may be conserved in dicotyledonous and monocotyledonous plants.
Expression of BDG Is Specific to the Epidermis
Several genes associated with the organ fusions in plants are expressed specifically in the epidermis (Yephremov et al., 1999; Kurata et al., 2003; Kurdyukov et al., 2006). To study whether this is also true for BDG, we examined the expression of reporter genes in transgenic plants bearing promoter fusions with β-glucuronidase (GUS) (data not shown) and green fluorescent protein (GFP) (Figures 9A to 9E). In all organs of the BDG-GFP plants, GFP fluorescence was detected exclusively in protodermal and epidermal cells. In floral buds, the BDG promoter was ubiquitously active in all organs, but activity decreased as the flowers aged. In the pistil, which is the most rapidly growing part of the mature flower, GFP fluorescence was quite strong in the abaxial epidermal cells, whereas in the septum and the inner ovary wall, it was hardly detectable.
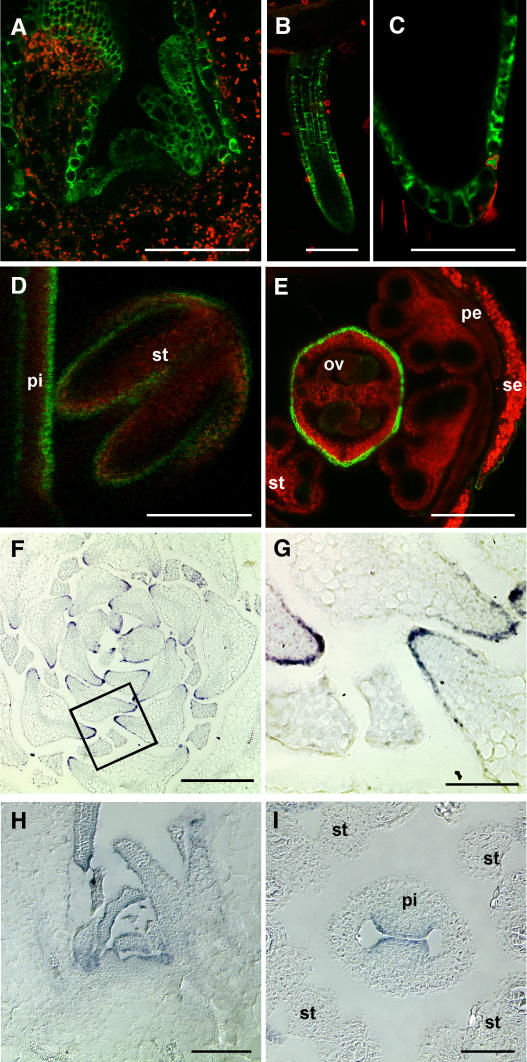
Figure 9.
Tissue-Specific Expression of BDG.
Series of images showing GFP fluorescence in transgenic Arabidopsis plants expressing GFP under the control of the BDG promoter ([A] to [E]) and in situ mRNA expression patterns of BDG ([F] to [I]) in wild-type plants. The GFP images were acquired using a confocal laser scanning microscope. GFP fluorescence is green (channel 520 to 540 nm), and autofluorescence is red (channel >590 nm). For mRNA expression analysis, tissue sections were hybridized with a digoxigenin-labeled riboprobe antisense to BDG cDNA. After immunodetection, using an alkaline phosphatase–conjugated anti-digoxigenin antibody, the sections were inspected with a light microscope. Bars = 100 μm for (A) to (C), 200 μm for (D) to (F), and 50 μm for (G) to (I).
(A) Longitudinal section through a vegetative apex.
(B) Lateral (secondary) root primordium.
(C) Optical section through the lateral root tip.
(D) Longitudinal section through a young floral bud showing the pistil (pi) and the stamen (st).
(E) Cross section through a floral bud at a later stage showing that strong GFP fluorescence is retained at the abaxial (external) side of the pistil. A weaker GFP signal is also visible in ovules (ov), petals (pe), and sepals (se).
(F) Cross section through a vegetative shoot apex of a short-day-grown wild-type plant. Note the particularly strong expression of BDG mRNA in the epidermis on the flanks of leaf primordia.
(G) The part of (F) inside the rectangle is magnified and displayed.
(H) Longitudinal section through a vegetative shoot apex of a long-day-grown wild-type plant. BDG expression is detected in the epidermis.
(I) Cross section through a floral bud showing BDG expression in the adaxial epidermis in prefusion carpels. At this stage, BDG is hardly detectable in other flower organs.
Interestingly, BDG appeared to be active in dermal cells of young lateral roots (Figures 9B and 9C), which lack a visible cuticle-like layer on the surface.
To validate the results of GUS and GPF reporter gene analysis, we examined gene expression by in situ hybridization. Tissue sections prepared from vegetative meristems and reproductive organs of wild-type Arabidopsis plants were hybridized with riboprobes directed against BDG mRNA. Compared with control probes such as epidermis-specific ACE/HTH (Krolikowski et al., 2003; Kurdyukov et al., 2006), the BDG probes yielded weaker hybridization signals. However, in all organs analyzed (leaf primordia, immature floral buds, sepals, petals, and anthers), expression was found to be limited to epidermal cells (Figures 9F to 9I; data not shown). The negative control sense probe produced no such signals (data not shown). Leaf primordia exhibited higher levels of BDG expression in lateral regions (Figures 9F and 9G).
Several features of the expression pattern suggested that BDG might be posttranscriptionally regulated, as we observed differences in expression level between in situ hybridization and GFP expression in the promoter-GFP marker lines. For example, prefused carpels showed in the adaxial epidermis a high in situ hybridization signal, whereas the strongest GFP signal was present in abaxial epidermal cells. However, this finding is preliminary and requires further study.
Thus, the epidermis-specific expression of BDG was confirmed in these experiments, suggesting that the 886-bp portion upstream from the coding sequence, used for reporter gene fusions, contains cis-regulatory elements for correct expression of the gene.
Immunolocalization of BDG
Given the high level of amino acid sequence conservation, anti-BDG antibodies would likely recognize other members of the BDG family. Therefore we used an epitope tagging–based approach to determine the localization of the BDG protein. To this end, bdg plants were transformed with a complementation construct carrying BDG fused in frame at the C terminus to the Strep-tag II epitope tag under the control of the native BDG promoter. Forty-three transgenic plants (T1 generation) were resistant to BASTA selection, and 38 of them (in nine independent families) appeared to be wild type. This finding demonstrated that full complementation of bdg was achieved by the epitope-tagged allele of BDG. The complementation of the phenotype conferred by bdg was maintained in the T2 and T3 BASTA-resistant progeny. The expression of the BDG–Strep-tag II fusion was also confirmed by RT-PCR and sequencing (data not shown); however, several attempts have failed to detect the Strep-tag II epitope with anti–Strep-tag II monoclonal antibody by immunolocalization and protein gel blot analysis, suggesting that BDG may be present at low levels. To enable ultrasensitive immunodetection, we used the tyramide signal amplification method based on the peroxidation of Alexa Fluor 488 tyramide. In the course of the peroxidation reaction, the highly reactive fluorescent radicals covalently couple to nucleophilic residues near the binding sites of a horseradish peroxidase antibody conjugate used as a secondary antibody (Haugland, 2002).
With this method, the Alexa Fluor 488–specific fluorescent signals were detected in the epidermis of transgenic bdg BDG–Strep-tag plants (Figures 10B to 10G) but not in the bdg mutant (Figure 10A) or wild-type plants (data not shown). Although present throughout the epidermis of incipient and developing leaf primordia, BDG accumulated to higher levels on the adaxial side of organs (Figures 10B and 10C). In contrast with mRNA in situ hybridization (Figures 9F and 9G), a stronger signal intensity was not detected in the lateral regions of leaf primordia.
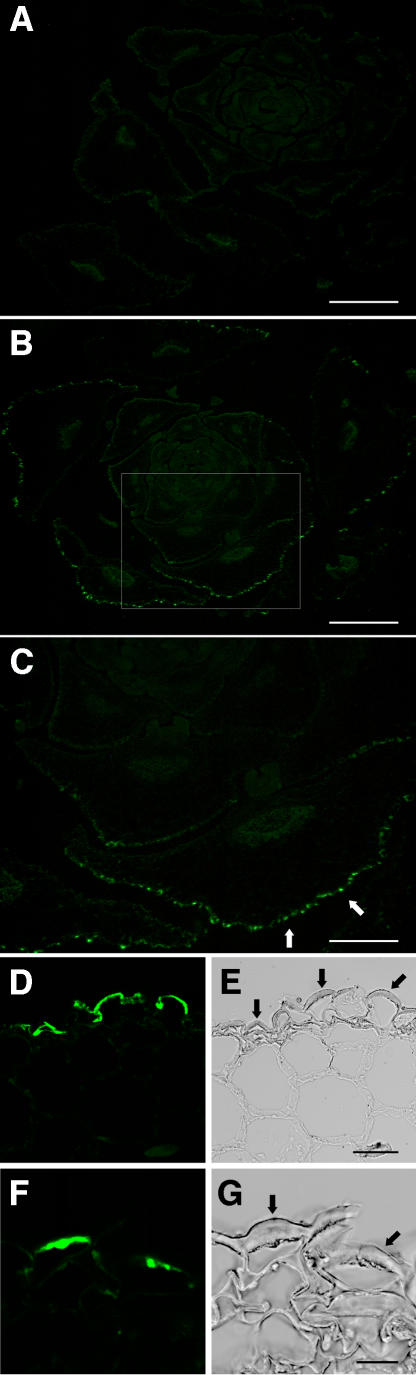
Figure 10.
Subcellular Immunolocalization of the Strep-Tag Epitope-Tagged BDG Protein.
Cross sections were cut from the paraffin-embedded vegetative shoot apexes of short-day-grown plants. After incubation of the sections with an epitope-specific monoclonal antibody, the fluorescent tyramide signal amplification method was used to detect BDG–Strep-tag. Images were taken with a fluorescence microscope ([A] to [C]) or a confocal laser scanning microscope ([D] to [G]). Bars = 500 μm for (A) and (B), 250 μm for (C), 40 μm for (D) and (E), and 15 μm for (F) and (G).
(A) A vegetative shoot apex in bdg used as a negative control together with that of a wild-type plant (data not shown).
(B) to (G) A vegetative shoot apex in bdg complemented with a construct carrying the BDG protein C-terminally tagged with the Strep-tag epitope sequence under the control of the native BDG promoter. Note that deformation in the shape of the leaf primordia occurs in bdg (A) but not in the complemented mutant (B). Specific fluorescent signals in the epidermis (arrows) were detected only in bdg BDG–Strep-tag plants ([B] to [G]) but not in negative controls. A magnified view of the leaf primordia boxed in (B) is shown in (C).
(D) and (F) Transverse sections corresponding to the light microscope images in (E) and (G), respectively. The Strep-tag–specific fluorescent signal is found localized in the outermost epidermal cell walls (arrows).
A polarly localized fluorescence pattern was observed in epidermal cells, suggesting that BDG contains a motif required for polar protein sorting (Figures 10D to 10G). In the epidermis, the epitope-specific signal was localized in the outermost cell wall layer. No pattern of Alexa Fluor 488–specific fluorescence was detected that could correspond to the nucleus, cytoplasm, vacuoles, or other cell compartments. In particular, no fluorescence that localized to a characteristic reticular network was observed, suggesting that BDG does not reside in the endoplasmic reticulum and Golgi apparatus. However, because of the limiting resolution of the method, it cannot be excluded that peripherally localized parts of these organelles contribute to the signal in the vicinity of the outermost cell wall. The fluorescence signal was observed in the inner but not the outer portion of the outermost epidermal cell wall hardened by cuticle secretion (Figures 10F and 10G). Cutinization of the outer portion of the cell wall is expected to cause a steric hindrance to the access of antibodies to antigenic sites. Therefore, it remains to be determined whether BDG was not detectable in these areas of the cell wall because of antigen masking or lower protein levels.
DISCUSSION
Sequence Comparisons Suggest a Novel Function for BDG
It is particularly difficult to predict the function of a member of the α/β-hydrolase superfamily, such as BDG, solely on the basis of its amino acid sequence, because sequence similarities are generally low, and because members of the superfamily catalyze the hydrolysis of a wide variety of substrates and include lipases, esterases, epoxide hydrolases, acyltransferases, and Ser proteases (Nardini and Dijkstra, 1999). Synthases belonging to this superfamily were also described (Belisle et al., 1997; Tonge, 2000). Nevertheless, families of proteins can be recognized on the basis of similarities in three-dimensional structures and functions. More than 30 such families have been defined in the α/β-hydrolase superfamily (Hotelier et al., 2004). However, the BDG family, comprising several genes in Arabidopsis and rice (Figure 8C), does not appear to be closely related to any of them. It follows that the function of BDG is novel and likely to be specific to plants.
BDG Gene Function in the Formation of the Epidermal Cell Wall and Cuticle
As described in detail above, mutations in BDG result in the severe deformation of epidermal cell walls and irregular deposition of electron-opaque material, which represents cutin. Whereas in many cases only a thin electron-dense membrane covers the lightly stained areas of the outermost cell wall boundary, deposition of extracellular lipid polymers within the deeper layers of the cell wall results in the formation of multilayered structures. The cuticular phenotype of bdg, which includes the structurally defective cuticle proper, suggests that the mutant may be unable to synthesize or secrete sufficient quantities of one or more cutin monomers. However, in correspondence with the overall enhanced deposition of electron-dense polymeric material, the amounts of insoluble lipids, representing typical cutin monomers, associated with cell walls are increased by twofold to threefold in bdg plants. Together, these results imply that biosynthesis of cutin precursors is probably not a direct target of the bdg mutation and distinguish bdg from the cuticular mutants of Arabidopsis isolated in previous studies. For example, it was shown that the att1 mutant forms a less osmiophilic cuticle, but more than twice as thick than that of the wild type, and exhibits a significant decrease in the levels of all cutin monomers, except for octadecadien-1,18-dioic acid. The ATT1 gene encodes CYP86A2, a cytochrome P450 monooxygenase catalyzing fatty acid ω-oxidation (Xiao et al., 2004). LACS2 encodes a long-chain acyl-CoA synthase that catalyzes the synthesis of ω-hydroxy fatty acyl-CoA. The thickness of the cutin layer on the abaxial surface of lacs2 leaves is reduced by 30% compared with that of wild-type plants, but its composition was not determined (Schnurr et al., 2004). The ace/hth mutant, deficient in fatty acid ω-alcohol dehydrogenase activity, shows in leaf polyester–specific reduction of the levels (∼70 to 80% of wild type) of α,ω-dicarboxylic fatty acids, which are the major constituents of cuticular polyesters (Bonaventure et al., 2004; Yephremov et al., 2004) and cutin (Franke et al., 2005) in Arabidopsis. The ace/hth mutant is the only mutant described to date in which the changes in the structure of the cuticle are reminiscent of those exhibited by bdg. In both mutants, the cuticle may be discontinuous or multilayered; however, overall enhanced deposition of electron-dense polymeric material was not observed in ace/hth, and the total amount of covalently bound lipids is insignificantly decreased in ace/hth (Kurdyukov et al., 2006). Interestingly, ace/hth and bdg not only both display a disrupted cuticle proper but also exhibit organ fusion phenotypes, whereas lacs2 and att1/CYP86A2 do not show organ fusions.
The bdg mutant accumulates approximately three times as much wax monomers as the wild type, and the alkanes with odd numbers of carbons contribute most to this increase. RT-PCR analysis of bdg shows the transcriptional activation of genes involved in the decarbonylation pathway, which catalyzes the formation of alkanes with odd-numbered chain lengths. Thus, in terms of genetics, BDG is a repressor of wax biosynthesis, although this does not mean necessarily that BDG is a direct repressor. It is more likely that the loss of crucial structural characteristics of cell wall polymers and the cuticle proper in bdg triggers wax biosynthesis via the upregulation of SHN1/WIN1 and CER1. This response, however, is not able to fully compensate for the structural defects in the cuticle and the reduced epidermal barrier function in bdg. It was reported that lacs2 shows a 1.5 times increase in the total wax load compared with wild-type plants (Schnurr et al., 2004). This is twice less than for bdg. Furthermore, the amounts of the major constituents of the wax, C29, C31, and C33 alkanes, were increased 9.4, 4.6, and 2.3 times in bdg, respectively, whereas in lacs2, C29 and C31 alkanes were increased 1.7 and 1.3 times, respectively, but the level of the C33 alkane was not changed (Schnurr et al., 2004). Therefore, it seems likely that either distinct mechanisms account for the increase of the wax accumulation in lacs2 and bdg or that the latter shows a much stronger compensatory response.
With regard to the function of the BDG gene, two possible scenarios could explain the observed cuticular defects. First, to explain the deformation of cell walls seen in bdg, one can assume that BDG is specifically involved in the postulated cross-linking of the outermost layer of epidermal cell walls that involves α,ω-dicarboxylic fatty acids (Kurdyukov et al., 2006) and/or the polymerization of cutin polyesters. In this scenario, cross-linking provides mechanical stability to the outermost layer of the cell wall, which normally prevents the underlying material from bulging outward. The increase in the amounts of lipids bound to cell walls in bdg could be accounted for if, under these conditions, more sites are available at which the polymerization of cutin monomers can be initiated. Indeed, the finding that BDG is expressed in the root dermal cells suggests that BDG may have a more general role in the biogenesis of the outermost cell walls than in cutin biosynthesis in aerial tissues. This finding is supported by reports showing the expression of several genes, such as LACS2 (Schnurr et al., 2004), FDH (Yephremov et al., 1999; Pruitt et al., 2000), LCR (Wellesen et al., 2001), WAX2/YRE (Kurata et al., 2003), and ACE/HTH (Krolikowski et al., 2003; Kurdyukov et al., 2006), that are required for cuticle development in roots. Subaerial tissues lack a visible cuticle-like layer on the surface, and transmission electron microscopy and histochemical staining reveal deposition of suberin in the cell walls in the endodermis and periderm but not in the epidermal cell walls (Franke et al., 2005). Nevertheless, the expression of ACE/HTH, which is probably directly involved in the biosynthesis of α,ω-dicarboxylic fatty acids, in the root epidermis suggests that these molecules contribute to covalently bound lipids associated with cell walls. Although the aliphatic composition of the root epidermal cell walls is not known, the presence of aromatic compounds is easily detected by fluorescence microscopy (Franke et al., 2005). Therefore, it seems likely that long-chain dicarboxylates are present as their esters to monolignols (p-coumaryl, coniferyl, and sinapyl alcohols) or glycerol, similar to what has been proposed for suberin (Bernards, 2002).
Although a direct involvement of BDG in the biosynthesis of the cuticular layer of the cell wall or cutin is currently one of the most appealing hypotheses for the function of this protein, the alternative scenario should be considered: that BDG plays no part in the biosynthesis of cuticle but rather contributes to the control of cell proliferation and differentiation. In this scenario, it is primarily the influence of the cell status that limits the ability to form the cuticle, whereas the accumulation of cutin monomers is preserved or enhanced. Based on the mutant phenotype under tissue culture conditions, BDG could be considered a negative regulator of cell proliferation. In support of this view, the phenotypes of bdg and pasticcino2/pepino mutants partially overlap, because mutations in PASTICCINO2/PEPINO (Bellec et al., 2002; Haberer et al., 2002) lead not only to uncoordinated cell divisions, uncontrolled growth, cell proliferation, and callus formation but also to organ fusions, strikingly similar to those in bdg, fdh, and lcr. Thus, BDG may code for an extracellular α/β-hydrolase fold–containing protein (e.g., lysophospholipase) the function of which is not directly related to cuticle.
Could BDG Be a Synthase Capable of Catalyzing Reactions in the Cuticular Layer of the Cell Wall?
Despite the fact that BDG may be involved in distinct molecular mechanisms necessary for cuticle formation, it is reasonable to assume that BDG acts directly as an extracellular synthase. There are three lines of evidence that support this hypothesis. First, immunolocalization analysis (Figure 10) revealed that BDG is polarly localized in the cell and accumulates in the outermost cell wall layer of the epidermis, at the side where cuticular lipids are deposited. This result argues against the indirect involvement of BDG in cuticle formation. Computerized analysis of the BDG protein sequence revealed the presence at the N terminus of a putative signal peptide, followed by a hydrophilic domain and an α/β-hydrolase domain, supporting the immunolocalization data and the idea that BDG is secreted to the extracellular space. Second, the cell wall deformations seen in bdg, characterized by the stratification of the cuticular material, interrupted by unfilled pockets, are very similar to those seen in transgenic Arabidopsis plants that express a cutinase from the pathogenic fungus Fusarium solani pisi (Sieber et al., 2000). In particular, the zones of the outermost cell wall of bdg, at which the cuticle proper is largely absent, are reminiscent of the outermost cell walls of leaves of cutinase-expressing plants (Figure 3C) (C. Nawrath, unpublished data). Although cutinases were named for their unparalleled ability to hydrolyze cutin (Soliday et al., 1984), they can metabolize a large variety of carboxylic esters (Kolattukudy, 1984; Lauwereys et al., 1991). Therefore, defects in the cuticular layer of the cell wall of these transgenic plants may be explained by the broad hydrolytic activity of the cutinase. Similar defects in bdg can be attributed to the loss of an extracellular synthase involved in the formation of ester bonds in cutin and/or the cutinized layer of the cell wall (e.g., between α,ω-dicarboxylic fatty acids and monolignols or between α,ω-dicarboxylic fatty acids and ω-chain and mid-chain hydroxyls of fatty acids). Third, BDG and cutinases, which are small extracellular carboxylic ester hydrolases of phytopathogenic fungi with molecular masses of ∼22 to 25 kD, both belong to the superfamily of enzymes that contain the α/β-hydrolase fold and are likely to share the same catalytic triad. Genes encoding secreted proteins very similar to cutinases have been discovered in the genomes of Mycobacterium species, which are the causative agents of tuberculosis and leprosy in humans. Based on structural and amino acid sequence similarities, these proteins belong to the cutinase family (Hotelier et al., 2004), but obviously they are not cutinases. It was recently found that they act as mycolyltransferases during cell wall biosynthesis, catalyzing the esterification of trehalose (two d-glucose residues joined by their anomeric carbons) to two mycolic acid residues, which are very-long-chain α-alkyl,β-hydroxy fatty acids (Belisle et al., 1997; Tonge, 2000).
Many α/β-hydrolases show catalytic flexibility: whereas these enzymes catalyze condensations in organic solvents containing only traces of water, they catalyze hydrolytic reactions in aqueous media (Bornscheuer and Kazlauskas, 1999). A nonpolar milieu containing some water is presumably characteristic of the cuticular layer of the growing cell wall and seems to provide an adequate environment for both reaction directions required to maintain cuticle integrity. Therefore, it is not unlikely that BDG may prove to exhibit both synthase and hydrolase activities in the cell wall.
Despite the similarity in cuticle structure between the cutinase-expressing and bdg plants, their phenotypes are not identical. Thus, the large amounts of cuticular material that accumulate within the cell walls of the bdg mutants have never been seen in leaves of cutinase-expressing plants, implying that, unlike cutinase-expressing plants, bdg is possibly more capable of cross-linking cutin-like polymers. The enlarged cuticular layer observed in the leaves of bdg, however, may have a certain similarity to the cavernous structure of mixed composition of the cuticles of stems in cutinase-expressing plants (Sieber et al., 2000). Thus, BDG is an attractive candidate gene for further exploring the hypothesis that plant α/β-hydrolases are capable of catalyzing condensation reactions in the cuticular layer of the cell wall.
Cell Wall Integrity, Cell Death, and Morphogenesis in bdg
Loss of the BDG function has a dramatic effect on the morphology of the outermost epidermal cell wall. It appears that the proper strengthening of the primary cell wall ensures its regular deposition and should be in concert with the expansion of the cell surface. Microscopic tears in the epidermis of bdg seem to result from disruptions of junctions between lateral cell walls, indicating that extracellular lipid polymers are required for maintaining tissue integrity. The failure of the bdg mutant to preserve intercellular junctions between epidermal cells may account for uncoordinated morphological changes and result in the deformation of organs. Furthermore, it is possible that the compression of the inner tissue by the epidermis is weakened in bdg, thereby resulting in greater extensibility of the cortical tissues and promoting both the deformation and growth of organs. Under tissue culture conditions that reduce stress injury, this effect is seen in bdg during the initial 10-d period; however, bdg plants grown in the greenhouse exhibit stunted growth from the first days and often die, probably as a result of a combination of dehydration and stress.
The deleterious effect of the bdg mutation on cell viability may be illustrated in bdg by the death of mature trichome cells that occurs as leaves unfold. In the course of this process, bdg trichomes undergo morphological changes that are noticeably reminiscent of those observed in cyclin-dependent kinase inhibitor ICK1/KRP1-misexpressing trichomes (Schnittger et al., 2003). Strikingly, the trichome death can only be delayed but not fully avoided by growing bdg under tissue culture conditions. Trichome differentiation is also delayed and reduced in bdg, facilitating the identification of bdg at the three- to four-leaf stage, before most features of the phenotype are visible. However, whether this is a direct effect (e.g., related to cell wall integrity) or an indirect effect (e.g., related to a stress response of the plant) of the mutation remains to be investigated. The wax2/yre mutation strongly reduces (to one-sixth of that in the wild type) the amount of epicuticular wax and somewhat influences the morphogenesis of trichomes (Kurata et al., 2003). The unbranched stem trichomes were severely stunted and deformed in the double yre cer1 mutant but not in the corresponding single mutants. Because CER1 is also known to be involved in wax biosynthesis, this indicated to the authors that epicuticular wax is required for the normal growth of trichomes (Kurata et al., 2003). However, accumulation of epicuticular wax is greatly enhanced in bdg; therefore, another mechanism must be responsible for the observed trichome defects. The decrease in numbers of trichomes and changes in trichome branching patterns were observed in transgenic plants overexpressing the SHN1/WIN1 gene (Aharoni et al., 2004), which appeared to be activated as well as CER1 in bdg. SHN1/WIN1 itself is able to upregulate CER1 and several other wax biosynthesis pathway genes (Broun et al., 2004). Therefore, it is likely that SHN1/WIN1 accounts for similar trichome defects in bdg. SHN1/WIN1, which encodes an ethylene response factor–type transcription factor, probably plays a role in the control of cellular processes that go distinctly beyond wax biosynthesis. It is worth noting that several cuticular mutants, including fdh (Yephremov et al., 1999), lcr (Wellesen et al., 2001), and cer10 (Zheng et al., 2005), exhibit trichome phenotypes, indicating that the deformation of cuticle initiates a cascade of events ultimately affecting the fate of cells.
In this respect, the regulation of cuticle formation is particularly interesting in the septum of the pistil, because this tissue participates in epidermal cell–cell interactions and responds to the contact. Resembling that in a number of cuticular mutants (Lolle et al., 1998), pollen germination could be induced on the leaf epidermis of bdg (Figure 1I). Results of expression studies using the GFP reporter gene indicated that the downregulation of BDG in the inner ovary wall of the rapidly growing pistil (Figure 9E) may contribute to cell–cell interactions between pollen and the epidermis.
The Role of BDG in Cell Signaling Responses
It appears that BDG is an epidermis-specific extracellular α/β-hydrolase fold–containing protein required for cuticle formation; however, its exact role in this process remains unknown. Neither BDG mRNA nor the BDG protein traffics from the epidermis to subepidermal cells. Therefore, whether BDG is an enzyme that is capable of catalyzing condensation reactions in the cuticular lipid polyesters or anchoring the lipid polymers to the carbohydrate constituents of the cell walls, an interesting question is how it can modulate meristem proliferation, changes in morphology, and cell differentiation. Also, there is no simple explanation for the striking increase in the production of the epicuticular wax in bdg, even if it is a response to the loss of cell wall and cuticle integrity.
The involvement of lipid-derived signals has been proposed to explain the pleiotropic phenotype of the lcr organ-fusion mutant, which includes altered cell differentiation in the epidermis, reduced apical dominance, delayed senescence, and irregular shape of leaves (Wellesen et al., 2001). Further evidence in support of the idea that cuticle may be a source of signals for plants is given by the report that cutinase-expressing plants (Sieber et al., 2000) that are very similar to bdg with regard to deformations of the cell wall and cuticle show strong resistance to the necrotrophic fungus Botrytis cinerea (Chassot et al., 2004). Interestingly, bdg also possesses a certain level of resistance to this fungus, whereas trichomes are severely affected in cutinase-expressing plants as well as in bdg (C. Nawrath, unpublished data).
In any case, the nature of the signals that induce metabolic changes and a cell differentiation response in the epidermis and diffuse from the epidermis throughout the meristem to trigger cell proliferation is intriguing.
METHODS
Growth Conditions
Arabidopsis thaliana plants were grown in a greenhouse at 22 to 23°C under either an 8-h photoperiod (short day) or a 16-h photoperiod (long day) at 50 to 60% RH.
For in vitro experiments, seeds were surface-sterilized by applying a 70% ethanol solution for 10 min and a 10% bleach solution for 15 min. Seeds were then washed under sterile conditions, kept for synchronization in 0.1% agar for a few days in the dark at 4°C, and plated on agar containing half-strength MS medium and 3% sucrose. Finally, plants were transferred after germination onto agar-solid half-strength MS medium in glass pots or plates and grown under long-day conditions in a climate chamber (16 h of light/8 h of dark, 21 to 22°C).
Molecular Isolation of the bdg-1 Transposon Insertion Allele and Reverse Genetics
Leaf material and seeds were collected from 30 mutant and wild-type plants segregating for the reverting W32 mutant, and the seeds were sown for progeny testing. Then, three heterozygous and three homozygous W32 mutant plants were used for transposon insertion display with the 3′ end transposon primers as described previously (Yephremov and Saedler, 2000; Steiner-Lange et al., 2001). A single 800-bp fragment common to all six plants was detected, but initial attempts to reamplify DNA sequences cut from a silver-stained denaturing polyacrylamide gel failed. To circumvent this difficulty, separation of transposon insertion display products was conducted with Transgenomic's WAVE system (equipped with a fraction collector) run in nondenaturing mode at 50°C.
The reverse genetics screen for En/Spm insertions was performed essentially as described previously (Steiner-Lange et al., 2001) using a 2-kb gene probe prepared from genomic DNA by PCR.
Characterization of the 5′ and 3′ Termini of BDG mRNA
The 5′ and 3′ ends of the RNA were determined by RACE using a 5′/3′ RACE kit (Roche Applied Science). Modifications of the protocol included replacing dATP by dGTP for tailing.
The 5′ and 3′ RACE products were analyzed and purified by agarose gel electrophoresis and cloned into the pGEM-T vector. Twenty clones were size-selected, and the five longest were sequenced.
Transgene Constructs and Generation of Transgenic Plants
For transgenic complementation of the bdg-2 mutant, the BDG gene (with its native promoter) was amplified from genomic DNA of ecotype Col with the primers KS3F (5′-CGAAGCTTATGTATGTCAGTCGTCGTTGACCTCTGC-3′) and KS4R (5′-CTGAGCTCCGAAATTGGACGCACATTGCAAACCACT-3′), which introduced HindIII and SacI cloning sites (underlined), respectively, at the 5′ and 3′ ends. The PCR fragments, digested with HindIII and SacI, were ligated into the corresponding cloning sites of the binary vector pBHS (Wellesen et al., 2001). A colony-pooling strategy, which allowed us to avoid sequencing before the transformation of plants, was applied as follows. Independent colonies of Escherichia coli DH10B (eight clones) containing inserts were combined for plasmid DNA isolation. Agrobacterium tumefaciens GV3101 was transformed with the plasmid DNA, and positive colonies (16 clones) were combined again for in planta transformation of the mutant as described previously (Wellesen et al., 2001). Complementation was indicated by the appearance of plants with wild-type morphology in several (seven in this case) independent transgenic families.
For expression studies, the putative promoter of the BDG gene was amplified as an 886-bp fragment using PbdgHindIII (5′-GAGTCGGAAGCTTGATGCCACGCACACGTCCTTG-3′) and PbdgXba (5′-CGGTCTAGACAAAATGCGTGAGAGAGAGAA-3′) as primers, subcloned into pGEM-T (Promega), and verified by sequencing. The promoter was then inserted into pBHS-based reporter binary vectors containing a GUS or GFP mgfp5-ER (Haseloff and Siemering, 1998) gene as a reporter.
To generate a complementation construct in which the BDG protein was fused to the Strep-tag II epitope tag under the control of the native BDG promoter, the 3.5-kb genomic copy of BDG was amplified using PbdgHindIII and PbdgXma (5′-GCTCTGGGCCCATTTATTGAATGAAGTTGAGGAG-3′) as primers, digested with HindIII and XmaI, and cloned into the pBHS-based binary vector pB-ctST containing one copy of the Strep-tag II sequence (IBA). In the resulting construct, the BDG protein sequence was followed by the sequence PGSAWSHPQFEK, which contained the four–amino acid peptide linker and the Strep-tag II epitope tag (underlined).
The promoter–reporter fusions and the epitope-tagged protein construct were then transformed into wild-type Arabidopsis (Col) via Agrobacterium as described above.
Staining with Toluidine Blue and Chlorophyll Leaching
Plants were grown in a greenhouse under short-day conditions for 7 to 8 weeks before experiments.
The staining method was adapted from Tanaka et al. (2004). Rosette leaves from Col and bdg mutant plants were stained at room temperature for 2 min without shaking, using a freshly prepared 0.05% solution of toluidine blue. Leaves were rinsed with tap water before being photographed.
The chlorophyll-leaching method was adapted from Lolle et al. (1997) with minor modifications. Six samples were prepared, each comprising 15 to 17 rosette leaves (∼0.5 g) taken from distinct plants. The samples were weighed before the addition of ethanol (80%). The extraction was conduced at 22°C in a rotating (18 rpm) water bath. Aliquots were taken after 10, 20, 40, 60, and 80 min. To determine the total amount of chlorophyll, the leaf samples were placed at 90°C for 15 min and cooled on ice before the final aliquot was taken. Measurements were performed using a UVIKON 810/820 spectrophotometer (Kontron), and calculations were done as described previously (Lolle et al., 1997).
Microscopic Techniques and in Situ Hybridization
Cryoscanning electron microscopy and transmission electron microscopy were performed as described by Yephremov et al. (1999) and Sieber et al. (2000), respectively. For confocal microscopy, plants or plant parts (transgenic plants expressing GFP, and Col plants as a control) were embedded in 5% low-melt agarose (Biozym, Hess) containing 0.005% Silwet L-77 (OSi Specialities). Sections (100 to 150 μm thick) were made with a Leitz 1512 Vibratome and viewed with a confocal laser scanning microscope (Leica TCS 4D) (Efremova et al., 2004).
For in situ hybridization probe synthesis, a 1.6-kb fragment was amplified from a BDG cDNA clone using primers (BDG-SP6, 5′-CTCGAGTTTAGGTGACACTATAGAACTGGAGGAAACCCTGCTACTGCTGTCCG-3′; BDG-T7, 5′-CTCGAGTAATACGACTCACTATAGGGAGATCA-GACCATCTTGGTGTTCTTTGC-3′) that were designed to include T7 and SP6 RNA polymerase promoter sites (BDG-specific sequences are underlined) on the flanking ends. The T7 polymerase transcribed the antisense digoxigenin-labeled probe used for the detection of BDG transcripts in sectioned tissue as described previously (Perbal et al., 1996). The sense probe to serve as a negative control was obtained using T7 RNA polymerase. Other controls included epidermis-specific probes (data not shown).
Immunocytochemistry Using Tyramide Signal Amplification
Vegetative apexes of 30-d-old plants grown under short-day conditions were fixed in methanol:acetic acid (3:1) at −20°C overnight. After fixation, chlorophyll was removed in 70% ethanol at −20°C. The tissues were dehydrated in a graded ethanol series and embedded in Paraplast Plus (Tyco Healthcare). Sections (6 μm) were cut with a microtome, placed onto Superfrost Plus GOLD microscope adhesive slides (Menzel-Glaser), air-dried for 24 h at 42°C, deparaffinized by washing in Histoclear, and rehydrated in an ethanol series. Heat-induced epitope retrieval was performed with 0.01 M sodium citrate, pH 9.0, at 80°C for 30 min. The sections were rinsed in Tris-buffered saline (TBS; 0.1 M Tris, pH 7.5, and 0.15 NaCl) containing 0.1% Tween 20 (TBST), incubated in 2% H2O2 in TBS for 1 h, blocked with 5 and 1% BSA in TBST, and then incubated overnight at 4°C with the mouse IgG monoclonal antibody (final dilution, 1.2 μg/mL in 1% BSA in TBST) against Strep-tag II peptide (IBA). The sections were washed in TBST and incubated for 1 h at room temperature with the tyramide signal amplification kit horseradish peroxidase conjugate of goat anti-mouse IgG antibody (Molecular Probes) diluted 1:100 in 1% BSA in TBST. To detect secondary antibody, slides were washed extensively with TBST (three times for 30 min at room temperature), rinsed with TBS, and incubated with Alexa Fluor 488 tyramide (Molecular Probes) for 10 to 15 min according to the manufacturer's specifications. After final washings with TBST (three times for 30 min at room temperature and overnight at 4°C), the cover slips were mounted on microscope slides using a 50% solution of glycerol. The samples were examined with a Leica MZ FIII microscope using a GFP filter and a Leica TCS SP2 confocal laser microscope. In the confocal system, fluorescence was excited at 488 nm and emission was read in the 495- to 540-nm interval.
Analysis of Residual Bound Lipids
The details of this procedure have been described elsewhere (Franke et al., 2005; Kurdyukov et al., 2006). Briefly, wild-type and mutant plants were grown until flowering. Mature leaves were harvested and their areas measured by scanning. Soluble lipids were removed from samples by extensive steeping in a methanol:chloroform mixture. This delipidation treatment allowed the extraction of most of the characteristic membrane lipids, as no traces of the highly unsaturated fatty acids 16:3 and 18:3 were detected (Kurdyukov et al., 2006). After drying, the leaf extract was weighed and stored or used directly for GC-MS analysis.
Wax Analysis
Rosette leaves (10 to 20) of 5-week-old plants were cut and immediately immersed in chloroform for 10 s at room temperature. The resulting solution of cuticular waxes was spiked with 10 μg of tetracosane (Fluka) as an internal standard. The solvent was evaporated under a stream of nitrogen, and compounds containing free hydroxyl and carboxyl groups were converted into their trimethylsilyl ethers and esters, respectively, with bis-(N,N-trimethylsilyl)-trifluoroacetamide (Machery-Nagel) in pyridine for 40 min at 70°C before GC-MS analysis. Wax constituents were identified by their electron-impact MS spectra (70 eV, m/z 50 to 700) after capillary GC (DB-1, 30 m × 0.32 mm, 0.1 μm [J&W]) on an Agilent 6890N gas chromatograph combined with a quadrupole mass-selective detector 5973N (Agilent Technologies). Samples were injected onto the column at 50°C, held at 50°C for 2 min, and then desorbed by increasing the temperature according to the following profile: 40°C/min to 200°C, 2 min at 200°C, 3°C/min to 310°C, and 30 min at 310°C. The flow rate of He carrier gas was 2 mL/min. Quantitative determination of wax components was performed with an identical GC system equipped with a flame-ionization detector. The extracted leaf area was determined based on the pixel number of digitized photocopies of the leaves.
Quantitative RT-PCR
The plants taken for RT-PCR experiments were the same plants used for toluidine blue staining and chlorophyll leaching (see above). Three independent samples each representing the wild type and bdg were compared. For each sample, ten 0.5- to 1-cm leaves taken from different plants were combined, and total RNA was extracted with the RNeasy plant mini kit (Qiagen). Each reaction contained 500 ng of total RNA, gene-specific primers, and components from the One Step RT-PCR kit (Qiagen). Primers for actin2 (At3g18780) and SHN1/WIN1(At1g15360) were described previously (Aharoni et al., 2004; Broun et al., 2004); WAX2/YRE (At5g57800) was amplified with the forward primer 5′-AAGCATCCTGACCTTAGAGTTCGTGTGGTTCAT-3′ and the reverse primer 5′-TAAGACCATACTTCATGGCTGCTTCCCACA-3′; CER1 (At1g02205) was amplified with the forward primer 5′-CAGGAACGGAGAGGTGTATATCCACAACCAT-3′ and the reverse primer 5′-GTATCTATCATACCACACCAGCATTGATAG-3′.
All RT-PCR targets were amplified within the linear range. Optimal cycle number was determined for each gene individually: it was 36 for SHN1/WIN1 and 24 for all other genes. Amplifications were repeated up to four times; PCR products were separated on a 1.5% agarose gel containing 0.25 μg/mL ethidium bromide and quantified with a Typhoon 8600 fluorescence scanner (Amersham Biosciences).
Accession Numbers
The accession numbers for the genes discussed in this article are as follows: At1g02205 (CER1), At1g15360 (SHN1/WIN1), At1g64670 (BDG), At1g72970 (ACE/HTH), At1g49430 (LACS2), At2g26250 (FDH), At2g45970 (LCR), At5g57800 (WAX2/YRE), At4g00360 (ATT1), At3g55360 (CER10), and At5g10480 (PASTICCINO2/PEPINO). Proteins in the alignment shown in Figure 8 are as follows (GenBank accession numbers are indicated in parentheses): Ao_CutL, Aspergillus oryzae cutinase (D38311); Mt_Cut2, Mycobacterium tuberculosis mycolyltransferase (Z77163); Cc_Cut, Colletotrichum capsici cutinase (M18033); Fs_Cut, Fusarium solani cutinase (AAA33334); Ec_BioH, Escherichia coli carboxylesterase (X15587); At_BDG, Arabidopsis thaliana BDG gene At1g64670; At_BDG2, Arabidopsis BDG2 gene At5g41900; At_BDG3, Arabidopsis BDG3 gene At4g24140; At_BDG4, Arabidopsis BDG4 gene At5g17780; At_BDG5, Arabidopsis BDG5 gene At5g17720; Os_HYD1, Oryza sativa putative hydrolase (AP003818); Os_HYD2, O. sativa putative hydrolase (AC092389); Os_HYD3, O. sativa putative hydrolase (AAO33147); Agt_pldB, Agrobacterium tumefaciens lysophospholipase (AE009319); Rm_pldB, Rhizobium meliloti lysophospholipase (AL591791); Rl_pldB, Rhizobium loti lysophospholipase L2 (AL591791); Hi_pldB, Haemophilus influenzae lysophospholipase L2 (U32747); Hs_Llpl, Homo sapiens lysophospholipase (AB017494); At_Lpl1, Arabidopsis lysophospholipase At1g18360 (AY070740); At_Lpl2, Arabidopsis lysophospholipase At5g11650 (AAM62693); At_Lpl3, Arabidopsis lysophospholipase At1g73480 (AY045929); At_Lpl4, Arabidopsis lysophospholipase At5g16120 (AY051019); Rm_lip, Rhizomucor miehei triacylglycerol lipase (P19515); Gs_est30, Geobacillus stearothermophilus esterase (AY186197); Dm_AchE, Drosophila melanogaster acetylcholinesterase (P07140); Pf_estB, Pseudomonas fluorescens carboxylesterase (S79600); and Mm_Es1 Mus musculus carboxylesterase (AAC04708).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Composition Analysis of Residual Bound Lipids in bdg and Wild-Type Leaves.
Supplemental Table 2. Composition Analysis of Epicuticular Wax in bdg and Wild-Type Leaves.
Supplemental Figure 1. Genomic Organization of BDG.
Supplemental Figure 2. RNA Gel Blot Hybridization of 10 μg of Total RNA.
Supplementary Material
Acknowledgments
We thank colleagues from the Zentrum zur Identifikation von Genfunktionen durch Instertionsmutagenese bei Arabidopsis thaliana project, in particular Christiane Horst, for helping us with the reverse genetics screen, Martine Schorderet, Ulrich Ryser, and Elmon Schmelzer for helping us with microscopy, Aldona Ratajek-Kuhn for taking care of our plants, and RIKEN for providing cDNA clones for our study. We especially appreciate the criticisms of the manuscript made by Paul Hardy. This work was supported by a Max-Planck-Gesellschaft fellowship to S.K., Marie Curie Host Fellowships for early-stage researcher training to D.V., a Deutsche Forschungsgemeinshaft research grant to L.S, and Research Grant NF 3100A0-104224/1 from the Swiss National Science Foundation to J.-P.M.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Alexander Yephremov (efremov@mpiz-koeln.mpg.de).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.036079.
References
- Aarts, M.G.M., Keijzer, C.J., Stiekema, W.J., and Pereira, A. (1995). Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell 7 2115–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni, A., Dixit, S., Jetter, R., Thoenes, E., van Arkel, G., and Pereira, A. (2004). The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16 2463–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arioli, T., et al. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279 717–720. [DOI] [PubMed] [Google Scholar]
- Belisle, J.T., Vissa, V.D., Sievert, T., Takayama, K., Brennan, P.J., and Besra, G.S. (1997). Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science 276 1420–1422. [DOI] [PubMed] [Google Scholar]
- Bellec, Y., Harrar, Y., Butaeye, C., Darnet, S., Bellini, C., and Faure, J.-D. (2002). PASTICCINO2 is a protein tyrosine phosphatase-like involved in cell proliferation and differentiation in Arabidopsis. Plant J. 32 713–722. [DOI] [PubMed] [Google Scholar]
- Bernards, M.A. (2002). Demystifying suberin. Can. J. Bot. 80 227–240. [Google Scholar]
- Bonaventure, G., Beisson, F., Ohlrogge, J., and Pollard, M. (2004). Analysis of the aliphatic monomer composition of polyesters associated with Arabidopsis epidermis: Occurrence of octadeca-cis-6,cis-9-diene-1,18-dioate as the major component. Plant J. 40 920–930. [DOI] [PubMed] [Google Scholar]
- Bornscheuer, U.T., and Kazlauskas, R.J. (1999). Hydrolases in Organic Synthesis: Regio- and Stereoselective Biotransformations (Weinheim, Germany: Wiley-VCH).
- Broun, P., Poindexter, P., Osborne, E., Jiang, C.-Z., and Riechmann, J.L. (2004). WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 101 4706–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassot, C., Nawrath, C., and Métraux, J.-P. (2004). Sensing of cuticular defects leads to pathogen defense. In X Cell Wall Meeting. Aug 29–Sep 3, Sorrento, Italy. (Neapel, Italy: University of Neapel), p. 49.
- Cheesbrough, T.M., and Kolattukudy, P.E. (1984). Alkane biosynthesis by decarbonylation of aldehydes catalyzed by a particulate preparation from Pisum sativum. Proc. Natl. Acad. Sci. USA 81 6613–6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., Goodwin, S.M., Boroff, V.L., Liu, X., and Jenks, M.A. (2003). Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 15 1170–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremova, N., Schreiber, L., Bär, S., Heidmann, I., Huijser, P., Wellesen, K., Schwarz-Sommer, Z., Saedler, H., and Yephremov, A. (2004). Functional conservation and maintenance of expression pattern of FIDDLEHEAD-like genes in Arabidopsis and Antirrhinum. Plant Mol. Biol. 56 821–837. [DOI] [PubMed] [Google Scholar]
- Ellis, C., Karafyllidis, I., Wasternack, C., and Turner, J.G. (2002). The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke, R., Briesen, I., Wojciechowski, T., Faust, A., Yephremov, A., Nawrath, C., and Schreiber, L. (2005). Apoplastic polyesters in Arabidopsis surface tissues—A typical suberin and a particular cutin. Phytochemistry 66 2643–2658. [DOI] [PubMed] [Google Scholar]
- Haberer, G., Erschadi, S., and Torres-Ruiz, R.A. (2002). The Arabidopsis gene PEPINO/PASTICCINO2 is required for proliferation control of meristematic and non-meristematic cells and encodes a putative anti-phosphatase. Dev. Genes Evol. 212 542–550. [DOI] [PubMed] [Google Scholar]
- Haruki, M., Oohashi, Y., Mizuguchi, S., Matsuo, Y., Morikawa, M., and Kanaya, S. (1999). Identification of catalytically essential residues in Escherichia coli esterase by site-directed mutagenesis. FEBS Lett. 454 262–266. [DOI] [PubMed] [Google Scholar]
- Haseloff, J., and Siemering, K.R. (1998). The uses of green fluorescent protein in plants. In Green Fluorescent Protein: Properties, Applications, and Protocols, M. Chalfie and S. Kain, eds (Chichester, UK: John Wiley & Sons), pp. 191–220.
- Haugland, R.P. (2002). Handbook of Fluorescent Probes and Research Products. (Eugene, OR: Molecular Probes).
- Hiscock, S.J., Bown, D., Gurr, S.J., and Dickinson, H.G. (2002). Serine esterases are required for pollen tube penetration of the stigma in Brassica. Sex. Plant Reprod. 15 65–74. [Google Scholar]
- Hiscock, S.J., Dewey, F.M., Coleman, J.O.D., and Dickinson, H.G. (1994). Identification and localization of an active cutinase in the pollen of Brassica napus L. Planta 193 377–384. [Google Scholar]
- Hotelier, T., Renault, L., Cousin, X., Negre, V., Marchot, P., and Chatonnet, A. (2004). ESTHER, the database of the alpha/beta-hydrolase fold superfamily of proteins. Nucleic Acids Res. 32 D145–D147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, L.A., MacCallum, R.M., and Sternberg, M.J. (2000). Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299 499–520. [DOI] [PubMed] [Google Scholar]
- Kolattukudy, P.E. (1984). Cutinases from fungi and pollen. In Lipases, B. Borgström and H.L. Brockman, eds (Amsterdam: Elsevier), pp. 471–504.
- Kolattukudy, P.E. (2001. a). Polyesters in higher plants. In Advances in Biochemical Engineering Biotechnology: Biopolyesters, W. Babel and A. Steinbuechel, eds (Berlin: Springer-Verlag), pp. 1–49. [DOI] [PubMed]
- Kolattukudy, P.E. (2001. b). Suberin from plants. In Biopolymers: Polyesters I. Biological Systems and Biotechnological Production, Y. Doi and A. Steinbuechel, eds (Muenster, Germany: Wiley-VCH), pp. 41–68.
- Krolikowski, K.A., Victor, J.L., Wagler, T.N., Lolle, S.J., and Pruitt, R.E. (2003). Isolation and characterization of the Arabidopsis organ fusion gene HOTHEAD. Plant J. 35 501–511. [DOI] [PubMed] [Google Scholar]
- Kurata, T., Kawabata-Awai, C., Sakuradani, E., Shimizu, S., Okada, K., and Wada, T. (2003). The YORE-YORE gene regulates multiple aspects of epidermal cell differentiation in Arabidopsis. Plant J. 36 55–66. [DOI] [PubMed] [Google Scholar]
- Kurdyukov, S., Faust, A., Trenkamp, S., Bär, S.T., Franke, B., Efremova, N., Tietjen, K., Schreiber, L., Saedler, H., and Yephremov, A. (2006). Genetic and biochemical evidence for involvement of HOTHEAD (HTH) in the biosynthesis of long chain α-,ω-dicarboxylic fatty acids and formation of extracellular matrix. Planta, in press. [DOI] [PubMed]
- Lauwereys, M., De Geus, P., De Meutter, J., Stanssens, P., and Matthyssens, G. (1991). Cloning, expression and characterization of cutinase, a fungal lipolytic enzyme. In Lipases—Structure, Mechanism and Genetic Engineering, L. Alberghina, R.D. Schmid, and R. Verger, eds (Weinheim, Germany: VCH), pp. 243–251.
- Lavithis, M., and Bhalla, P.L. (1995). Esterases in pollen and stigma of Brassica. Sex. Plant Reprod. 8 289–298. [Google Scholar]
- Lequeu, J., Fauconnier, M.-L., Chammai, A., Bronner, R., and Blee, E. (2003). Formation of plant cuticle: Evidence for the occurrence of the peroxygenase pathway. Plant J. 36 155–164. [DOI] [PubMed] [Google Scholar]
- Lolle, S.J., Berlyn, G.P., Engstrom, E.M., Krolikowski, K.A., Reiter, W.D., and Pruitt, R.E. (1997). Developmental regulation of cell interactions in the Arabidopsis fiddlehead-1 mutant: A role for the epidermal cell wall and cuticle. Dev. Biol. 189 311–321. [DOI] [PubMed] [Google Scholar]
- Lolle, S.J., Hsu, W., and Pruitt, R.E. (1998). Genetic analysis of organ fusion in Arabidopsis thaliana. Genetics 149 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin, L.J., and Jones, D.T. (2003). Improvement of the GenTHREADER method for genomic fold recognition. Bioinformatics 19 874–881. [DOI] [PubMed] [Google Scholar]
- Nardini, M., and Dijkstra, B.W. (1999). alpha/betaHydrolase fold enzymes: The family keeps growing. Curr. Opin. Struct. Biol. 9 732–737. [DOI] [PubMed] [Google Scholar]
- Nawrath, C. (September 1, 2003). The biopolymers cutin and suberin. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/10.1199/tab.0099, http://www.aspb.org/publications/arabidopsis/. [DOI] [PMC free article] [PubMed]
- Nishimura, M.T., Stein, M., Hou, B.-H., Vogel, J.P., Edwards, H., and Somerville, S.C. (2003). Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301 969–972. [DOI] [PubMed] [Google Scholar]
- Ollis, D.L., et al. (1992). The alpha/beta hydrolase fold. Protein Eng. 5 197–211. [DOI] [PubMed] [Google Scholar]
- Perbal, M.C., Haughn, G., Saedler, H., and Schwarz Sommer, Z. (1996). Non-cell-autonomous function of the Antirrhinum floral homeotic proteins DEFICIENS and GLOBOSA is exerted by their polar cell-to-cell trafficking. Development 122 3433–3441. [DOI] [PubMed] [Google Scholar]
- Pruitt, R.E., Vielle-Calzada, J.P., Ploense, S.E., Grossniklaus, U., and Lolle, S.J. (2000). FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proc. Natl. Acad. Sci. USA 97 1311–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger, A., Weinl, C., Bouyer, D., Schobinger, U., and Hulskamp, M. (2003). Misexpression of the cyclin-dependent kinase inhibitor ICK1/KRP1 in single-celled Arabidopsis trichomes reduces endoreduplication and cell size and induces cell death. Plant Cell 15 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnurr, J., Shockey, J., and Browse, J. (2004). The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell 16 629–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber, P., Schorderet, M., Ryser, U., Buchala, A., Kolattukudy, P., Métraux, J.-P., and Nawrath, C. (2000). Transgenic Arabidopsis plants expressing a fungal cutinase show alterations in the structure and properties of the cuticle and postgenital organ fusions. Plant Cell 12 721–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliday, C.L., Flurkey, W.H., Okita, T.W., and Kolattukudy, P.E. (1984). Cloning and structure determination of complementary DNA for cutinase: An enzyme involved in fungal penetration for plants. Proc. Natl. Acad. Sci. USA 81 3939–3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner-Lange, S., Gremse, M., Kuckenberg, M., Nissing, E., Schaechtele, D., Spenrath, N., Wolff, M., Saedler, H., and Dekker, K. (2001). Efficient identification of Arabidopsis knock-out mutants using DNA-arrays of transposon flanking sequences. Plant Biol. 3 391–397. [Google Scholar]
- Tanaka, H., Onouchi, H., Kondo, M., Hara-Nishimura, I., Nishimura, M., Machida, C., and Machida, Y. (2001). A subtilisin-like serine protease is required for epidermal surface formation in Arabidopsis embryos and juvenile plants. Development 128 4681–4689. [DOI] [PubMed] [Google Scholar]
- Tanaka, T., Tanaka, H., Machida, C., Watanabe, M., and Machida, Y. (2004). A new method for rapid visualization of defects in leaf cuticle reveals five intrinsic patterns of surface defects in Arabidopsis. Plant J. 37 139–146. [DOI] [PubMed] [Google Scholar]
- Taniyama, Y., Shibata, S., Kita, S., Horikoshi, K., Fuse, H., Shirafuji, H., Sumino, Y., and Fujino, M. (1999). Cloning and expression of a novel lysophospholipase which structurally resembles lecithin cholesterol acyltransferase. Biochem. Biophys. Res. Commun. 257 50–56. [DOI] [PubMed] [Google Scholar]
- Tonge, P.J. (2000). Another brick in the wall. Nat. Struct. Biol. 7 94–96. [DOI] [PubMed] [Google Scholar]
- Wellesen, K., Durst, F., Pinot, F., Benveniste, I., Nettesheim, K., Wisman, E., Steiner-Lange, S., Saedler, H., and Yephremov, A. (2001). Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different roles of fatty acid ω-hydroxylation in development. Proc. Natl. Acad. Sci. USA 98 9694–9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, F., Goodwin, M.S., Xiao, Y., Sun, Z., Baker, D., Tang, X., Jenks, M.A., and Zhou, J.-M. (2004). Arabidopsis CYP86A2 represses Pseudomonas syringae type III genes and is required for cuticle development. EMBO J. 23 2903–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yephremov, A., et al. (2004). Safeguarding the cuticular wall. In X Cell Wall Meeting. Aug 29–Sep 3, Sorrento, Italy. (Neapel, Italy: University of Neapel), p. 99.
- Yephremov, A., and Saedler, H. (2000). Display and isolation of transposon-flanking sequences starting from genomic DNA or RNA. Plant J. 21 495–505. [DOI] [PubMed] [Google Scholar]
- Yephremov, A., and Schreiber, L. (2005). The dark side of the cell wall: Molecular genetics of plant cuticle. Plant Biosyst. 139 74–79. [Google Scholar]
- Yephremov, A., Wisman, E., Huijser, P., Huijser, C., Wellesen, K., and Saedler, H. (1999). Characterization of the FIDDLEHEAD gene reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell 11 2187–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, H., Rowland, O., and Kunst, L. (2005). Disruptions of the Arabidopsis enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell 17 1467–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.