Abstract
Ribonucleotide reductase (RNR), comprising two large (R1) and two small (R2) subunits, catalyzes a rate-limiting step in the production of deoxyribonucleotides needed for DNA replication and repair. Previous studies in yeast and mammals indicated that defective RNR often led to cell cycle arrest, growth retardation, and p53-dependent apoptosis, whereas abnormally increased RNR activities led to higher mutation rates. Because plants are constantly exposed to environmental mutagens and plant cells are totipotent, an understanding of RNR function in plants is important. We isolated and characterized mutations in all three R2 genes (TSO2, RNR2A, and RNR2B) in Arabidopsis thaliana. tso2 mutants had reduced deoxyribonucleoside triphosphate (dNTP) levels and exhibited developmental defects, including callus-like floral organs and fasciated shoot apical meristems. tso2 single and tso2 rnr2a double mutants were more sensitive to UV-C light, and tso2 rnr2a seedlings exhibited increased DNA damage, massive programmed cell death, and release of transcriptional gene silencing. Analyses of single and double r2 mutants demonstrated that a normal dNTP pool and RNR function are critical for the plant response to mutagens and proper plant development. The correlation between DNA damage accumulation and the subsequent occurrence of apoptotic nuclei in tso2 rnr2a double mutants suggests that perhaps plants, like animals, can initiate programmed cell death upon sensing DNA damage.
INTRODUCTION
Plants are distinct from animals in their lifestyle and their development. A sedentary lifestyle subjects plants to constant exposure to environmental mutagens, including UV irradiation and reactive oxygen species. Accordingly, plants must have evolved active surveillance mechanisms to protect their genomic integrity. A key element for genome surveillance is DNA repair. This essential function of plants must be tightly regulated by the surveillance mechanisms known as DNA damage checkpoint pathways.
Ribonucleotide reductase (RNR) is an important target of the DNA damage checkpoint pathways in yeast, mammals, and possibly higher plants (Huang et al., 1998). RNR catalyzes the reduction of all four ribonucleotide diphosphates (NDPs) into their corresponding deoxyribonucleosides (dNDPs), a rate-limiting step in DNA precursor synthesis (Elledge et al., 1992; Kolberg et al., 2004). RNR consists of two large subunits (R1) and two small subunits (R2). The R2 subunit houses the di-iron tyrosyl radical cofactor essential for the reduction of NDP to dNDP. The well-known chemotherapeutic agent hydroxyurea is a specific but reversible inhibitor of the R2 subunit, acting as a scavenger of the tyrosyl free radical. The R1 subunit binds the nucleoside diphosphate substrates and allosteric effectors to ensure the production of a balanced deoxyribonucleoside triphosphate (dNTP) pool. R1 is the target of feedback regulation, which ensures that dNTPs are not overproduced and that enough NDPs are left for RNA synthesis.
Genetic analyses of rnr mutants in yeast and mammals indicated that defective RNR often led to cell cycle arrest, growth retardation, and p53-dependent apoptosis. For example, failure to maintain a sufficient and balanced dNTP pool can lead to misincorporation of dNTPs into DNA, resulting in genetic abnormalities and cell death, as shown in the murine hemopoietic cell line (Oliver et al., 1996). Defective feedback regulation of R1 in budding yeast led to increased dNTP levels, resulting in increased mutation rates but enhanced resistance to mutagens (Chabes et al., 2003a). Loss-of-function rnr1 and rnr2 mutants of budding yeast showed depleted dNTP pools and displayed a cdc terminal phenotype, arresting at S/G2-phase as a large budded cell (Elledge et al., 1992). In humans, p53R2, an RNR small subunit, is a direct regulatory target of the tumor-suppressor protein p53 (Tanaka et al., 2000). It was suggested that p53R2 supplies dNDPs to the DNA damage-repair system outside the S-phase of the cell cycle. Most of the p53R2 knockout mice died from severe renal failure by the age of 14 weeks, and a greater number of apoptotic cells in kidneys were detected (Kimura et al., 2003). These genetic studies in mammals and fungi demonstrated that RNR plays a key role in maintaining genomic stability.
Considering the critical role of RNR in DNA synthesis and repair, it is not surprising that RNR expression and activity are under exquisite control both transcriptionally and posttranscriptionally, as shown by studies of yeast and mammalian cells. These include controlled degradation of RNR protein by Anaphase-Promoting Complex-Cdh1–mediated proteolysis in mouse (Chabes et al., 2003b), the redistribution of RNR2 and RNR4 proteins from nucleus to cytoplasm in response to DNA damage in yeast (Liu et al., 2003; Yao et al., 2003), the control of RNR mRNA stability by the yeast cytoplasmic Constitutive Invertase Derepression13 poly(A) polymerase (Saitoh et al., 2002), the inhibition of RNR activity by the Suppressor of mec1 Lethality1 protein in yeast (Chabes et al., 1999; Zhao et al., 2000), and the repression of RNR transcription by Constitutive RNR Transcription1, a yeast homolog of the mammalian RFX family of DNA binding proteins (Huang et al., 1998). The existence of multiple regulatory mechanisms for RNR activities underscores the importance of a proper dNTP pool for the fitness and survival of an organism.
In contrast with yeast and mammals, our knowledge of RNR function and regulation in plants is limited. R1 and R2 genes have been isolated from tobacco (Nicotiana tabacum) and Arabidopsis thaliana (Philipps et al., 1995; Chaboute et al., 1998), and their S-phase–specific expression was shown to depend on the E2F-like motifs in their promoters (Chaboute et al., 2000, 2002). Transient translocation of a green fluorescent protein–R1 protein from cytoplasm to nucleus in response to UV irradiation was reported recently (Lincker et al., 2004). However, no mutation in any of the plant RNR genes has been reported, and the function of RNR in maintaining plant genomic stability and genetic variability has not yet been demonstrated.
Here, we report the functional dissection of RNR in higher plants. Mutations in TSO2 were isolated in ethyl methanesulfonate (EMS) mutagenesis screens that caused defects in shoot apical meristem (SAM), flowers, leaves, and fertility. We show that TSO2 encodes one of the three R2 genes in Arabidopsis, revealing a direct role of TSO2 in DNA replication. Subsequent identification and characterization of mutations in all three R2 genes in the Arabidopsis genome provided insights into RNR function for plant growth and development and the response to environmental mutagens. More significantly, extensive programmed cell death (PCD) was found in tso2 rnr2a double mutants, which correlated with accumulating DNA damage in this double mutant. One possible explanation for this correlation between DNA damage and PCD is that perhaps plants, like animals, can initiate PCD upon DNA damage, despite the absence of a p53 homolog in the Arabidopsis genome. Finally, release of transcriptional gene silencing (TGS) was observed in tso2 rnr2a double mutants, demonstrating an interesting link between DNA replication/chromosome integrity and epigenetic inheritance.
RESULTS
tso2 Mutants Exhibit Various Developmental Defects
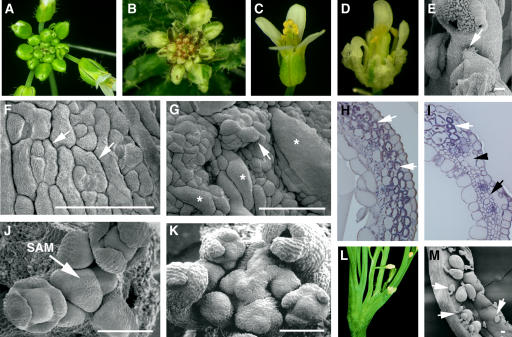
Four independent tso2 mutants (tso2-1, -2, -3, and -4) were isolated from EMS mutagenesis screens in the Arabidopsis Landsberg erecta (Ler) background (see Methods). These mutants were named tso2 because of their resemblance to tso1 mutants, with TSO meaning “ugly” in Chinese (Liu et al., 1997). All four tso2 alleles are recessive and phenotypically similar to each other. Therefore, tso2-1 was chosen for further analysis. At the seedling stage, tso2-1 mutants were similar to the wild type, with normal roots, cotyledons, and young leaves. Abnormalities first appeared in the fifth rosette leaf and persisted in all subsequent leaves, floral organs, and siliques. The abnormalities included white sectors in green organs (Figures 1A and 1B), uneven thickness, rough surfaces, and irregular margins of leaves or floral organs (Figures 1C and 1D). Stamens occasionally exhibited carpel characteristics indicating homeotic transformation (Figure 1E). The rough and uneven sepals apparently resulted from abnormally enlarged cells as well as cell clusters projecting above the epidermis (Figures 1F and 1G). The white sectors appeared to result from an absence of chloroplasts in patches of mesophyll cells and the formation of large air spaces or tiny cells in subepidermal cell layers (Figures 1H and 1I). Occasionally, tso2 mutants exhibited fasciated stems on which a single SAM, a group of self-renewing cells at the shoot tip, was enlarged and split into multiple SAMs (Figures 1J to 1L). Finally, tso2 mutants exhibited reduced fertility, with ∼29% aborted seeds (Figure 1M).
Figure 1.
Phenotypic Analyses of Wild-Type and tso2-1 Plants.
(A) A wild-type (Ler) inflorescence.
(B) A tso2-1 inflorescence showing white sectors on leaves and sepals.
(C) A wild-type flower.
(D) A tso2-1 flower showing sepals with white sectors and rough surfaces.
(E) Partial homeotic transformation in a tso2-1 floral organ, in which stigmatic tissues characteristic of carpels are found on top of an anther (arrow).
(F) Scanning electron micrograph of a wild-type sepal epidermis. Immature stomata (arrows) are interspersed with rectangular cells.
(G) A tso2-1 sepal epidermis. Clusters of epidermal cells are projected above the epidermal surface, an example of which is indicated by the arrow. Some epidermal cells are significantly large and are marked with asterisks.
(H) Cross section of a wild-type carpel valve. Chloroplasts that encircle and outline the photosynthetic mesophyll cells are stained purple and are indicated by arrows.
(I) Cross section of a tso2-1/tso2-1; rnr2b-1/+ carpel valve. Although some mesophyll cells possess chloroplasts (white arrow), others completely lack chloroplasts (black arrow) and are small. Abnormal air spaces (black arrowhead) are present beneath the epidermis.
(J) Scanning electron micrograph of a wild-type SAM showing spiral arrangements of young floral primordia.
(K) A tso2-1 SAM that is disorganized and is undergoing fasciation.
(L) A fasciated tso2-1 plant showing multiple splits of the shoot.
(M) An open tso2-1 silique showing mature seeds interspersed with aborted seeds or unfertilized ovules (arrows).
Bars = 100 μm.
TSO2 Encodes a Small Subunit of RNR
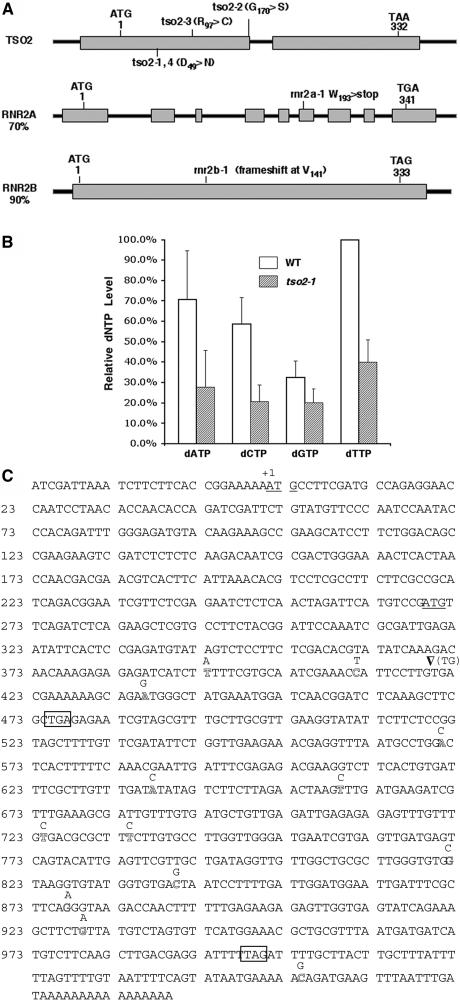
Using a map-based cloning approach, we isolated the TSO2 gene and showed that TSO2 encodes the small subunit (R2; At3g27060) of RNR (see Methods). Alignment of R2 proteins from different species revealed 16 amino acids that are absolutely conserved from bacteria and plants to human. These invariant residues are distributed throughout the protein. tso2-1, tso2-2, tso2-3, and tso2-4 are all caused by missense mutations affecting either invariant or highly conserved amino acids (Figure 2A). tso2-1 and tso2-4 affect the same invariant Asp-49 residue known to be involved in binding to the R1 large subunit (Philipps et al., 1995). tso2-2 and tso2-3 affect Gly-170 and Arg-97, respectively, which reside in areas important for iron binding and for tyrosyl radical environment (Philipps et al., 1995).
Figure 2.
Functional Analyses of Three Arabidopsis R2 Genes.
(A) Scheme of three Arabidopsis R2 genes. Rectangular boxes represent exons. The start and stop codons and the position of each r2 mutation are indicated. tso2-1 and tso2-4 are identical. Numbers indicate amino acid residues. Percentage identities between TSO2 and RNR2A or RNR2B are indicated.
(B) Relative dNTP levels in wild-type (Ler) and tso2-1 floral tissues. Results from three different experiments were averaged and normalized to the wild-type dTTP level. Error bars represent sd.
(C) Full-length RNR2B cDNA sequence from Col (BT004167). Two start codons are underlined, and two stop codons are boxed. The Ler RNR2B sequence differs from the Col sequence at 14 positions, which are indicated immediately above the corresponding Col positions. An arrowhead indicates a 2-bp (TG) deletion from Col resulting in premature translational termination at the first stop codon. A second start codon could potentially direct the translational initiation of a second peptide. Single base pair changes at positions 659, 734, 822, 840, and 929 are missense, but the remaining single base pair changes are silent. The RNR2B sequence from the Ws accession is identical to that of Ler.
Do the tso2 mutations cause abnormal dNTP levels? We measured dNTP levels in tso2-1 mutants using a polymerase-based method (Roy et al., 1999). We found that in wild-type Arabidopsis, the relative levels of the four dNTPs with respect to each other were similar to those of other organisms, with dTTP the most abundant and dGTP the least abundant (Figure 2B). tso2-1 floral tissues were found to contain significantly reduced levels of all four dNTPs (Figure 2B). Although dGTP level was reduced to 60% of the wild-type level, dCTP was reduced to 35% of the wild-type level. These differential reductions in dNTPs could lead to an imbalanced dNTP pool in tso2-1 floral tissues. Therefore, a reduced and possibly imbalanced dNTP pool may underlie the various phenotypes observed in tso2-1 mutants.
Functional Redundancy among the Three RNR Genes
Because tso2 mutants are all viable and fertile, redundant genes in the Arabidopsis genome may compensate for defective tso2. Examination of the Arabidopsis genome revealed two additional R2 genes named RNR2A (At3g23580) and RNR2B (At5g40942 and AY178109), the protein products of which are 70 and 90% identical, respectively, to TSO2. RNR2B is highly similar to TSO2 even at the nucleic acid level, with 82% identity. TSO2 and RNR2B also possess similar gene structures: TSO2 has one small intron and RNR2B has no introns (Figure 2A). By contrast, RNR2A has a distinct gene structure, with eight introns and nine exons, and exhibits almost no sequence similarity to TSO2 at the nucleic acid level, indicating that RNR2A probably has a distinct evolutionary origin from TSO2 and RNR2B.
Interestingly, RNR2B from the Columbia (Col) accession possesses a 2-bp deletion resulting in a frameshift at Val-141 and a subsequent stop, deleting more than half of the protein, including 11 of the 16 invariant residues (Figure 2C). A second start codon could potentially direct the translational initiation of a second peptide homologous with TSO2 at residues 141 to 332 (Figure 2C). This putative second peptide does not have the N-terminal half of the protein containing 5 of the 16 invariant residues. Given the severity of the frameshift mutation in Col RNR2B, it is likely that it represents a loss-of-function or reduction-of-function allele. We thus named Col RNR2B rnr2b-1. Nevertheless, Col plants, which carry rnr2b-1, do not exhibit any obvious phenotype.
By contrast, RNR2B from Ler and Wassilewskija (Ws) accessions (which are identical to each other) do not harbor the 2-bp deletion and differ from Col in 13 additional nucleotides (Figure 2C), resulting in an intact protein of 333 amino acid residues (AAO62422). To test whether Ler RNR2B is a functional protein, RNR2B genomic DNA from Ler (RNR2B-L) was fused to the strong and constitutive promoter 35S and transformed into the tso2-1 mutant. This 35S:RNR2B-L transgene was able to rescue all 36 tso2-1 plants, indicating that the RNR2B gene from Ler or Ws performs a function similar to TSO2.
Previously, a cDNA encoding RNR2A was reported and its mRNA was shown to specifically accumulate during the S-phase of the cell cycle in synchronized tobacco BY2 cells (Philipps et al., 1995). We tested RNR2A function by fusing the RNR2A cDNA to the 35S promoter and transforming the construct into tso2-1 mutants. The 35S:RNR2A transgene was able to rescue the tso2-1 phenotype in all 40 transgenic plants, indicating that RNR2A can substitute for TSO2 when it is expressed from the strong and constitutive promoter. Using a reverse-genetics method known as TILLING (for targeting induced local lesions in genomes) (McCallum et al., 2000), we isolated an EMS-induced rnr2a mutation (rnr2a-1) in the Col accession. rnr2a-1 is a nonsense mutation that deletes one-third of the RNR2A protein (Figure 2A). In the Col background, rnr2a-1 is actually an rnr2a-1 rnr2b-1 double mutant and is phenotypically wild-type (Figure 3A), indicating that TSO2 alone provides sufficient RNR activity for normal development.
Figure 3.
Phenotypes of r2 Single and Double Mutants.
(A) An rnr2a-1 rnr2b-1 seedling showing no obvious phenotype.
(B) A tso2-1 seedling. White tissues are forming in the emerging leaves.
(C) A typical tso2-1 rnr2a-1 seedling, which will not develop further.
(D) The SAM of a tso2-1 rnr2a-1 seedling that terminates with callus-like cells.
(E) A wild-type leaf.
(F) A tso2-1 rnr2a-1 leaf showing a severely disrupted leaf surface.
(G) A wild-type Ler plant and a tso2-1 plant.
(H) Six-week old tso2-1/tso2-1 plants singly or doubly heterozygous for rnr2a-1 and/or rnr2b-1: tso2-1/tso2-1 (1); tso2-1/tso2-1; rnr2b-1/+ (2); tso2-1/tso2-1; rnr2a-1/+ (3); and tso2-1/tso2-1; rnr2a-1/+; rnr2b-1/+ (4).
(I) tso2-1 siliques showing small white sectors in the green carpel valve.
(J) tso2-1/tso2-1; rnr2b-1/+ siliques showing larger white sectors and smaller siliques.
Bars in (A) to (F) = 1 mm.
Aided by PCR-based markers specific to each mutant allele, we analyzed the F2 progeny of a cross between tso2-1 (Ler) and rnr2a-1 rnr2b-1 (Col). tso2-1 rnr2a-1 and tso2-1 rnr2b-1 double mutants were identified, which exhibited a much stronger phenotype than either parent. First, tso2-1 rnr2a-1 seedlings did not develop beyond the two- to four-leaf stage (Figure 3C). Their SAMs were terminated with callus-like cells (Figure 3D), and their leaves exhibited massively disorganized surfaces (Figures 3E and 3F). Second, tso2-1 rnr2b-1 was embryo-lethal. PCR-based genotyping detected tso2-1 rnr2b-1 only among aborted seeds, and not among viable plants from tso2-1/tso2-1; rnr2b-1/+ parents (Table 1). This finding strongly suggests that Col RNR2B (i.e., rnr2b-1) does not have any or does not have the same level of activity as the Ler RNR2B. Finally, tso2-1/tso2-1 plants heterozygous for rnr2a-1, rnr2b-1, or rnr2a-1 rnr2b-1 were viable but exhibited a much stronger phenotype than tso2-1/tso2-1, as shown by their smaller stature, more severely reduced fertility, more frequent stem fasciation, and larger white sectors (Figures 3G to 3J). These results indicate that the functions of RNR2A and RNR2B are essential in the tso2-1 background.
Table 1.
Results of Genotyping among the Progeny of tso2-1/tso2-1; rnr2b-1/+ Parents
| rnr2b-1/rnr2b-1 | rnr2b-1/+ | +/+ | Total | |
|---|---|---|---|---|
| Number of plants | ||||
| Expected | 9 | 18 | 9 | 36 |
| Observed | 0 | 22 | 14 | 36 |
| Number of seeds | ||||
| Green (normal) | 0 | 13 | 5 | 18 |
| White (abnormal) | 2 | 33 | 3 | 38 |
Only the genotype at RNR2B is shown, as all plants are also tso2-1/tso2-1.
mRNA Expression Pattern of TSO2, RNR2A, and RNR2B
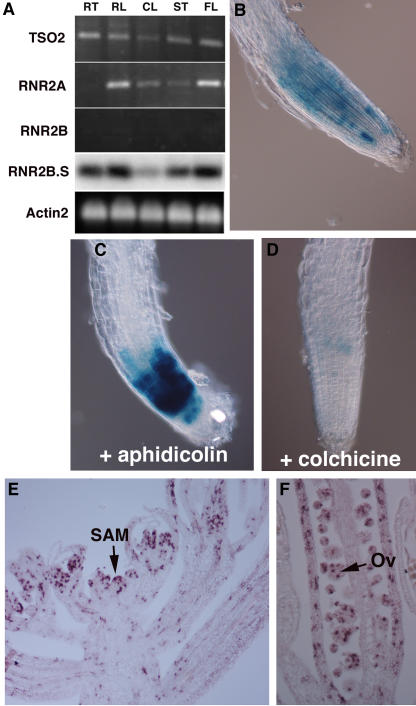
To test whether the R2 genes exhibit different tissue- or stage-specific expression patterns, semiquantitative RT-PCR was performed for all three R2 genes (Figure 4A). TSO2 transcripts were found in roots, rosette and cauline leaves, stems, and flowers. Although RNR2A transcripts were not detected in roots, they were detected in rosette and cauline leaves, stems, and flowers. By contrast, RNR2B transcripts were detected only on DNA gel blots of RT-PCR products and were present in all tissues tested (Figure 4A, RNR2B.S). Thus, all three genes, in most cases, are expressed widely, with RNR2B expression at a much lower level than TSO2 and RNR2A.
Figure 4.
mRNA Expression of TSO2, RNR2A, and RNR2B.
(A) Expression of TSO2, RNR2A, and RNR2B mRNA measured by 20 cycles of RT-PCR with gene-specific primers. RT, root; RL, rosette leaves; CL, cauline leaves; ST, stems; FL, flowers. The RT-PCR products of RNR2B were detected only by DNA gel blot hybridization with a RNR2B-specific probe (lane RNR2B.S). Actin2 was the loading control.
(B) pTSO2:GUS expression in a 3-d-old wild-type root. Note the sporadic GUS activity.
(C) pTSO2:GUS expression in a 3-d-old root treated with 5 μg/mL aphidicolin, revealing an increased number of GUS-expressing cells and an increased level of GUS expression.
(D) pTSO2:GUS expression in a 3-d-old root treated with 0.5% colchicine. Note the significantly reduced GUS activity.
(E) In situ hybridization showing TSO2 mRNA expression in a wild-type Ler SAM. Note the sporadic TSO2 expression throughout the young floral primordia.
(F) In situ hybridization showing sporadic TSO2 expression in developing ovules (Ov) and carpel valves.
To examine TSO2 transcription during the cell cycle, the β-glucuronidase (GUS) reporter gene was fused to the TSO2 1.2-kb promoter. Transgenic seedlings harboring pTSO2:GUS were treated with 5 μg/mL aphidicolin or 0.5% colchicine, which arrest cells at S- or M-phase, respectively. GUS expression was detected in more cells and at higher levels when the seedlings were arrested at S-phase (Figures 4B and 4C). By contrast, GUS expression was reduced dramatically when the seedlings were arrested at M-phase (Figure 4D). Hence, TSO2 transcription occurs predominantly at the S-phase of the cell cycle.
TSO2 mRNA distribution during reproductive development was examined by in situ hybridization. The sporadic rather than uniform pattern of TSO2 mRNA expression in developing floral tissues (Figures 4E and 4F) is characteristic of cell cycle phase–specific expression (Fobert et al., 1994). A large number of cells in the young floral meristem and developing floral organ primordia show a higher level of TSO2 mRNA expression (Figure 4E). Cells in the developing ovules and carpel valves also express TSO2 mRNA at a high level (Figure 4F). Therefore, the TSO2 mRNA expression pattern is consistent with its role in dNDP biosynthesis during DNA replication in actively dividing cells.
tso2-1 Exhibits Defects in Cell Cycle Progression
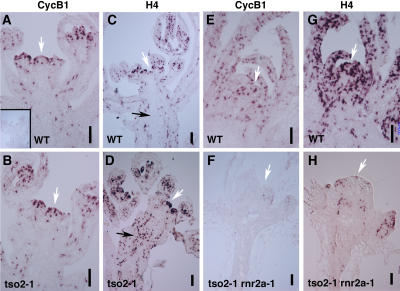
Because yeast rnr mutants display a cdc phenotype, we tested whether cell cycle progression is affected in tso2-1 mutants. In situ hybridization examining the expression of cell cycle phase–specific markers was performed. G2/M-specific Cyclin B1 (CycB1) and S-specific Histone4 (H4) expression was examined in wild-type and tso2-1 floral tissues. CycB1 and H4 were both expressed strongly in a sporadic manner in wild-type as well as tso2-1 tissues (Figures 5A to 5D). Although the CycB1 expression pattern was similar in the wild type and tso2-1, H4 expression was significantly different between the wild type and tso2-1. Many more cells in tso2-1 inflorescences expressed H4 than in the wild type, suggesting a prolonged S-phase in these tso2-1 cells. We also examined CycB1 and H4 expression in tso2-1 rnr2a-1 seedlings (Figures 5E to 5H). Although wild-type seedlings expressed both CycB1 and H4 strongly in the SAM and developing leaves, tso2-1 rnr2a-1 seedlings possessed little or no CycB1- or H4-expressing cells, perhaps because of a complete absence of cell division activity in tso2-1 rnr2a-1 seedlings.
Figure 5.
In Situ Hybridization Examining the Expression of Cell Cycle Phase–Specific Marker Genes.
(A) CycB1 mRNA expression in wild-type SAM and developing young flowers. The white arrow indicates the SAM. The inset shows a wild-type SAM hybridized with the sense CycB1 RNA probe as a negative control.
(B) CycB1 expression in tso2-1 SAM and young flowers.
(C) H4 expression in a wild-type inflorescence. The black arrow indicates a lack of H4-expressing cells in the stem.
(D) H4 expression in a tso2-1 inflorescence. A dramatic increase in H4-expressing cells in flowers and in the stem (black arrow) is evident.
(E) CycB1 RNA expression in a wild-type 3-week-old seedling.
(F) CycB1 RNA expression in a 3-week-old tso2-1 rnr2a-1 seedling.
(G) H4 expression in a wild-type 3-week-old seedling.
(H) H4 expression in a 3-week-old tso2-1 rnr2a-1 seedling.
Bars = 10 μm.
DNA Damage Accumulation in tso2-1 rnr2a-1
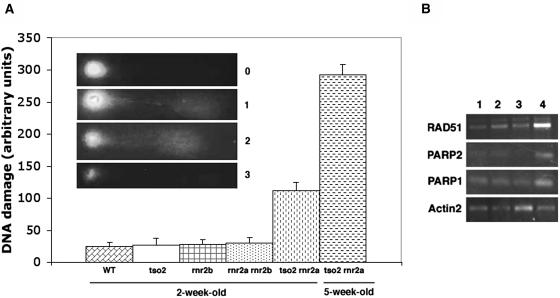
Are the reduced dNTP levels in tso2-1 sufficient to impede DNA replication fork progression and induce DNA strand breaks? Using the comet assay (Angelis et al., 1999), we measured DNA damage levels in 2-week-old seedlings of tso2 single and double mutants. Although tso2-1 or rnr2a-1 rnr2b-1 did not exhibit increased DNA damage, tso2-1 rnr2a-1 exhibited significant increases in DNA damage (Figure 6A). Furthermore, the DNA damage level in 5-week-old tso2-1 rnr2a-1 seedlings was much higher than that in 2-week-old tso2-1 rnr2a-1 seedlings.
Figure 6.
Increased DNA Damage in tso2-1 rnr2a-1 Seedlings.
(A) Results from the comet assay indicating the relative amount of DNA damage in different genotypes. Although tso2-1, rnr2b-1, or rnr2a-1 rnr2b-1 mutants exhibited levels of DNA damage similar to those in the wild type (Ler), tso2-1 rnr2a-1 double mutants exhibited an increased level of DNA damage even at 2 weeks old. The DNA damage level was increased further in 5-week-old tso2-1 rnr2a-1 plants. The extent of DNA damage in each nucleus is indicated by the units 0, 1, 2, 3, or 4. An increased unit correlated with a larger comet tail and a smaller comet head, as illustrated in the inset. The DNA damage units per genotype were derived by summing the units from 100 nuclei on each slide. Error bars represent sd values averaged from four slides.
(B) Twenty-seven to 30 cycles of RT-PCR were used to detect the induction of ATPARP1, ATPARP2, and ATRAD51 in 3-week-old seedlings. Lanes 1, 2, 3, and 4 correspond to the wild type (Ler), tso2-1, rnr2a-1 rnr2b-1, and tso2-1 rnr2a-1, respectively.
Consistent with the comet assay, only tso2-1 rnr2a-1 seedlings were found to induce the expression of molecular markers associated with DNA damage repair (Figure 6B). These markers included poly(ADP-ribose) polymerase1 (ATPARP1), ATPARP2, and ATRAD51 (Doutriaux et al., 1998; Doucet-Chabeaud et al., 2001). Both ATPARP1 and ATPARP2 are transcriptionally induced by increased levels of double-stranded DNA breaks. However, ATPARP2 can also be induced by oxidative stresses. ATRAD51 is involved in meiotic recombination and in homologous recombination repair and is induced by increased double-stranded DNA breaks (Osakabe et al., 2002). The induction of ATPARP1, ATPARP2, and ATRAD51 contrasts sharply with the nearly complete shutdown of CycB1 and H4 expression in tso2-1 rnr2a seedlings (Figures 5F and 5H), suggesting that tso2-1 rnr2a-1 double mutants did not simply shut down transcription machinery altogether but were able to selectively induce DNA repair genes.
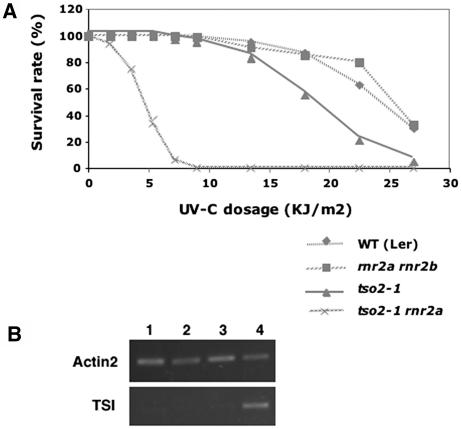
Increased Sensitivity to UV-C Light in tso2-1 and tso2-1 rnr2a-1 Seedlings
Because RNR also provides dNTPs for DNA damage repair, the reduced dNTP pool sizes in tso2-1 might lead to hypersensitivity to DNA-damaging agents. We tested the sensitivity of wild-type (Ler), tso2-1, rnr2a-1 rnr2b-1, and tso2-1 rnr2a-1 seedlings to UV-C light (Figure 7A). tso2-1 rnr2a-1 exhibited significantly increased sensitivity to UV-C light. tso2-1 exhibited slightly increased sensitivity to UV-C light, but only at high UV-C light levels. rnr2a-1 rnr2b-1 exhibited a level of UV-C light sensitivity similar to the wild type (Ler), indicating that TSO2 alone is sufficient to provide wild-type levels of protection against UV-C light.
Figure 7.
Increased Sensitivity to UV-C Light and Epigenetic Misregulation in r2 Mutants.
(A) Graph illustrating the survival rate of wild-type (Ler), tso2-1, rnr2a-1 rnr2b-1, and tso2-1 rnr2a-1 seedlings under increasing UV-C light levels.
(B) Twenty-five cycles of RT-PCR were used to detect TSI transcripts in 3-week-old seedlings. Lanes 1, 2, 3, and 4 correspond to the wild type (Ler), tso2-1, rnr2a-1 rnr2b-1, and tso2-1 rnr2a-1, respectively.
Release of TGS in tso2-1 rnr2a-1
The phenotype of tso2-1 associated with meristem fasciation and increased sensitivity to DNA-damaging agents resembles that of several Arabidopsis mutants defective in DNA/chromatin replication and assembly, including mutants of BRU1, FAS1, FAS2, ATCAP-E1, ATCAP-E2, and ATMRE11 (Kaya et al., 2001; Bundock and Hooykaas, 2002; Siddiqui et al., 2003; Takeda et al., 2004). All of these mutants were found to release TGS (Kaya et al., 2001; Takeda et al., 2004) at the pericentromeric repeats called Transcriptional Silent Information (TSI) (Steimer et al., 2000). We found that tso2-1 rnr2a-1 seedlings also released silencing at TSI but tso2-1 seedlings did not (Figure 7B). Because tso2-1 plants exhibit more severe mutant phenotypes at reproductive stages, we tested tso2-1 inflorescence tissues for the release of TSI. No TSI expression was observed in tso2-1 inflorescence tissues (data not shown).
PCD in tso2-1 rnr2a-1
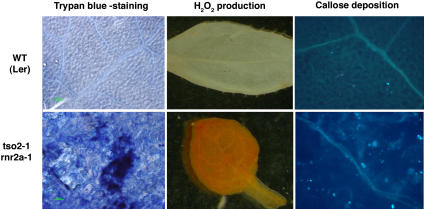
In animals, genomic instability often induces PCD in a p53-dependent manner (Chernova et al., 1995; Vogelstein et al., 2000). Furthermore, apoptosis was reported to occur in p53R2 knockout mice (Kimura et al., 2003). Therefore, we tested whether the accumulating DNA damage in tso2-1 rnr2a-1 seedlings described above could lead to PCD using histochemical markers. Trypan blue was used to stain dead cells (Rate et al., 1999). Large patches of trypan blue–stained cells were observed in the leaves of 10-d-old tso2-1 rnr2a-1 seedlings but not in wild-type or tso2-1 seedlings (Figure 8; data not shown). Furthermore, a high level of H2O2 and a large number of callose depositions were detected in the leaves of 10-d-old tso2-1 rnr2a-1 seedlings (Figure 8). Both H2O2 production and callose deposition are indicators of plant cells undergoing hypersensitive PCD during incompatible plant–pathogen interactions (Dietrich et al., 1994; Brodersen et al., 2002), suggesting that the cell death observed in tso2 rnr2a is likely programmed.
Figure 8.
Expression of Cell Death Markers in tso2-1 rnr2a-1 Mutants.
Rosette leaves of 10-d-old wild-type and tso2-1 rnr2a-1 plants were examined for PCD using three different histochemical markers: trypan blue staining, H2O2 production, and callose deposition. Dark blue patches stained by trypan blue, reddish-brown deposits (reaction products between 3,3-diaminobenzidine and H2O2), and callose deposition revealed by aniline blue staining indicated PCD in tso2-1 rnr2a-1 mutants.
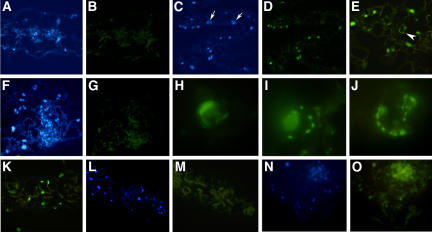
To further characterize tso2 rnr2a–mediated PCD, the first two leaves of 3-week-old wild-type (Ler) and tso2-1 rnr2a-1 plants were fixed, sectioned, and processed for terminal deoxyribonucleotidyl transferase–mediated dUTP nick-end labeling (TUNEL). TUNEL can sensitively detect DNA fragmentation, one of the hallmarks of PCD, by labeling exposed 3′ hydroxyl ends of DNA fragments with fluorescein-dUTP. The same leaf sections were simultaneously stained with 4′,6-diamidino-2-phenylindole (DAPI) to reveal all nuclei in each section (Figures 9A, 9C, and 9F). None of the nuclei in wild-type sections was TUNEL-positive (Figure 9B). By contrast, some but not all nuclei in tso2-1 rnr2a-1 leaf sections were TUNEL-positive (Figures 9C and 9D), indicating that not all cells in tso2-1 rnr2a-1 leaves had undergone PCD. Some TUNEL-positive nuclei in tso2-1 rnr2a-1 leaves exhibited nuclear morphologies characteristic of apoptotic nuclei, including marginalization of chromatin on the nuclear membrane, fragmentation of the nucleus into small bodies, and peripheral nuclear crescents (Figures 9E and 9H to 9J). The apoptotic nuclear morphology provides the most convincing evidence for the occurrence of PCD in the tso2 rnr2a mutants, as DNase I–treated nuclei in wild-type leaf sections fail to show apoptotic nuclear morphology even though they are TUNEL-positive (Figure 9K). This observation is consistent with other published work (Kressel and Groscurth, 1994), indicating that TUNEL labeling can only be considered specific for PCD when it is associated with the characteristic nuclear morphology.
Figure 9.
PCD in tso2-1 rnr2a-1 Double Mutants Detected by the TUNEL Assay.
Images shown in (A) to (K) are from 3-week-old seedlings. Images shown in (L) and (M) and in (N) and (O) are from 6- and 11-d-old seedlings, respectively. Images in (A) to (G) and (K) to (O) are magnified ×88. Images in (H) to (J) are magnified ×366.
(A) DAPI staining of nuclei in a wild-type (Ler) leaf section.
(B) The same wild-type leaf section shown in (A) is TUNEL-negative, showing no green fluorescent signal.
(C) DAPI staining of nuclei in a tso2-1 rnr2a-1 leaf section.
(D) Many TUNEL-positive (green fluorescent) nuclei are found in the same tso2-1 rnr2a-1 leaf section shown in (C). Some nuclei (indicated by arrows in [C]) are not fluorescent, indicating that some cells have not undergone PCD.
(E) TUNEL-positive nuclei in a tso2-1 rnr2a-1 leaf section. Nuclear morphologies characteristic of different stages of PCD are shown. The arrowhead indicates a cell enlarged in (J).
(F) DAPI staining of nuclei in a tso2-1 rnr2a-1 leaf section.
(G) A negative control for the TUNEL. The same leaf section shown in (F) was not provided with terminal deoxyribonucleotidyl transferase during TUNEL labeling.
(H) A TUNEL-positive nucleus in a tso2-1 rnr2a-1 leaf section. The crescent-shaped nuclear morphology was previously described for UV-C light–treated protoplast cells undergoing PCD (Danon and Gallois, 1998).
(I) A TUNEL-positive nucleus of tso2-1 rnr2a-1 exhibiting marginalization of chromatin on the nuclear membrane and fragmentation of the nucleus into small bodies.
(J) An enlargement of the TUNEL-positive nucleus shown in (E) showing fragmented DNA bodies at the nuclear peripheral.
(K) A wild-type (Ler) leaf section treated with DNase I before detection by the TUNEL. The DNA nicks created by DNase I give positive TUNEL signals. However, none of the TUNEL-positive nuclei exhibit apoptotic characteristics, as shown in (H) to (J).
(L) DAPI staining of nuclei in a cotyledon section of a 6-d-old tso2-1 rnr2a-1 seedling.
(M) An absence of TUNEL-positive nuclei in the same cotyledon section shown in (L). Chloroplasts are visible as a result of their autofluorescence.
(N) DAPI staining of nuclei in a cotyledon section of an 11-d-old tso2-1 rnr2a-1 seedling.
(O) TUNEL-positive nuclei in the same cotyledon section shown in (N).
Does the DNA damage occur before the initiation of the apoptosis-like PCD in these tso2-1 rnr2a-1 seedlings? DNA damage was already shown to occur in 2- and 5-week-old tso2-1 rnr2a-1 seedlings (Figure 6A). Subsequently, 6-d-old seedlings were also tested for DNA damage using the comet assay. The 6-d-old tso2-1 rnr2a-1 seedlings showed 52% (±10% se) more relative DNA damage units than the corresponding wild-type seedlings, indicating DNA damage accumulation in the 6-d-old tso2-1 rnr2a-1 seedlings. The TUNEL assay was then used to determine the time course of cell death in tso2-1 rnr2a-1 at 6, 11, and 16 d after germination. Although none of the 8-μm sections of 6-, 11-, and 16-d-old wild-type seedlings (including cotyledon and hypocotyls) was TUNEL-positive (data not shown), the tso2-1 rnr2a-1 seedling sections showed increased TUNEL signals with age. Specifically, the majority of the 6-d-old tso2-1 rnr2a-1 seedling sections was negative for the TUNEL signal (Figures 9L and 9M). By examining 200 sections, only an average of fewer than one (0.62; n = 200) TUNEL-positive nucleus per section was observed. At 11 d, an increased average number (1.8; n = 160) of TUNEL-positive nuclei per section was observed (Figures 9N and 9O). At 16 d, an average of 6.7 (n = 180) TUNEL-positive nuclei per section was observed. However, none of the TUNEL-positive nuclei in 6- and 11-d-old seedlings exhibited the apoptotic nuclear morphology (Figures 9L to 9O). Apoptotic nuclear morphology was observed starting in 16-d-old tso2 rnr2a seedlings (data not shown) and continuing in 3-week-old tso2 rnr2a seedlings (Figures 9E and 9H to 9J). Therefore, DNA damage was detected at least 10 d before the appearance of nuclei exhibiting characteristic apoptotic morphology in the tso2-1 rnr2a-1 seedlings.
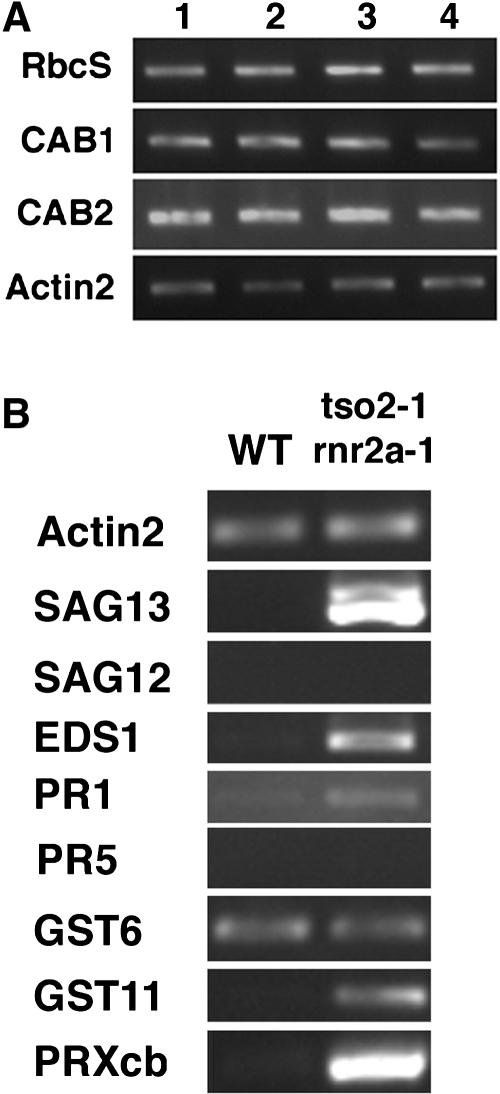
To date, PCD in plants has been studied extensively in the context of the hypersensitive response (HR) during incompatible plant–pathogen interactions and senescence (Greenberg, 1996; Kuriyama and Fukuda, 2002; Greenberg and Yao, 2004). PCD during senescence is often accompanied by a rapid loss of chlorophyll and reduced expression of photosynthesis-associated genes (PAGs), such as chlorophyll a/b binding protein (CAB1 and CAB2) and ribulose-1,5-bis-phosphate carboxylase/oxygenase small subunit (RbcS) (Miller et al., 1999). To test whether tso2 rnr2a–mediated PCD is caused by early senescence, we examined the expression of the PAGs CAB1, CAB2, and RbcS in 3-week-old seedlings of the wild type (Ler), tso2-1, rnr2a-1 rnr2b-1, and tso2-1 rnr2a-1 by semiquantitative RT-PCR (Figure 10A). All genotypes were similar to the wild type in expressing the three PAGs, in contrast with what would have been expected if tso2 rnr2a had undergone early senescence. In addition, we tested the expression of genes previously shown to be associated with HR-mediated PCD in plants (Brodersen et al., 2002), including Senescence-Associated Gene13 (SAG13), Peroxidase Cb (PRXcb), Glutathionine-S-Transferase11 (GST11), Pathogenesis-Related1 (PR1), and Enhanced Disease Susceptibility1 (EDS1). Additionally, SAG12, a Cys protease specific to senescence-induced PCD (Pontier et al., 1999), was tested. Using RT-PCR, we compared the expression of these genes in the wild type (Ler) and tso2-1 rnr2a-1 (Figure 10B). Although the expression of SAG13, GST11, and EDS1 was induced, the expression of SAG12 was not detected under the RT-PCR conditions used. Overall, tso2 rnr2a–mediated PCD appears similar to HR-mediated PCD in plants.
Figure 10.
Expression of Molecular Markers Associated with Senescence and HR-Mediated PCD.
(A) RT-PCR was used to examine photosynthesis-associated genes. Twenty, 27, and 17 PCR cycles were used to examine RbcS, CAB1, and CAB2, respectively. Actin2 was the loading control. Lanes 1, 2, 3, and 4 correspond to 3-week-old seedlings of the wild type (Ler), tso2-1, rnr2a-1 rnr2b-1, and tso2-1 rnr2a-1, respectively.
(B) Twenty-five cycles of RT-PCR were used to detect molecular markers associated with PCD in 3-week-old seedlings. SAG13, EDS1, PR-1, GST11, and PRXcb were induced to various levels in tso2-1 rnr2a-1 mutants. GST6 remained the same in both the wild type and tso2-1 rnr2a-1. SAG12 and PR-5 were not induced. However, SAG12 could be detected using 30 cycles of PCR (data not shown). A doublet was amplified with the SAG13 primers as a result of a nearly identical gene (At2g29350) encoding tropinone reductase. SAG13 corresponds to the upper band of the doublet.
DISCUSSION
Our molecular genetic studies of tso2 led to the identification and characterization of all three RNR2 genes in the Arabidopsis genome. The analyses of single and double rnr2 mutants demonstrated that normal dNTP pool and RNR function are critical for the plant response to mutagens and for proper plant development. Interestingly, although all four tso2 missense alleles exhibited morphological defects, single rnr2a-1 and rnr2b-1 and double rnr2a-1 rnr2b-1 mutants did not. It is likely that TSO2 normally plays a more predominant role than RNR2A and RNR2B. This is supported by the Affymetrix gene expression profiling study at AtGenExpress (Schmid et al., 2005), which shows that TSO2 is more abundantly and widely expressed than RNR2A, and by a recent transcriptome profiling study showing a 10-fold induction of TSO2 transcripts upon treatment with the genotoxins bleomycin plus mitomycin C (Chen et al., 2003). It is also possible that the four tso2 missense alleles are recessive antimorphic alleles. Recessive antimorphism is possible if the mutant TSO2 proteins interact with R1 or other R2 subunits, inactivating R1 or other R2 subunits. But this negative effect is masked by the wild-type TSO2 protein.
Although tso2 single mutants are viable, the tso2 rnr2a and tso2 rnr2b double mutants are seedling- and embryo-lethal, respectively, suggesting that both RNR2A and RNR2B function are essential in the tso2 mutant background. Interestingly, tso2 rnr2a seedlings exhibited a range of phenotypes not observed in tso2 single or rnr2a rnr2b double mutants, including the loss of CycB1 and H4 expression (Figures 5F and 5H), DNA damage accumulation (Figure 6), a high degree of UV-C light sensitivity (Figure 7A), release of TGS (Figure 7B), and PCD (Figures 8 and 9). It is likely that a greater degree of reduced or imbalanced dNTP pool in tso2 rnr2a could lead to greater DNA replication block and DNA damage, triggering subsequent phenotypes such as the release of TGS and PCD. However, the tso2 rnr2a double mutants did not just deregulate all genes nonspecifically. For example, the photosynthesis-associated genes CAB1, CAB2, and RbcS are expressed at a level similar to that in the wild type (Figure 10A). In addition, although the cell cycle genes CycB1 and H4 are turned off (Figures 5F and 5H), the DNA damage repair genes ATPARP1, ATPARP2, and ATRAD51 are specifically induced in tso2 rnr2a seedlings (Figure 6B). The identification and construction of single and double r2 mutants offer the opportunity to study the diverse functions of RNR in plant development.
Decreased dNTP Levels Affect Organelle Replication
One unique feature of tso2 mutants is the formation of white sectors on green organs. We observed a lack of chloroplasts in the photosynthetic mesophyll cells, unusual air spaces beneath the epidermis, and abnormally small mesophyll cells in tso2 tissues (Figures 1H and 1I). In yeast, lower RNR activity or reduced dNTP level was shown to increase the formation of mitochondrial DNA–deficient cells (petite cells) (Zhao et al., 1998, 2001), suggesting that organelle DNA replication is highly sensitive to reduced dNTP levels. Therefore, stochastic depletions of chloroplasts and/or mitochondria may underlie the white sectors and small cells in tso2 mutants. The development of white sectors occurring in the fifth leaf and in later-arising organs of tso2 could result from a gradual dilution of dNTP, which may be highly enriched in embryos/seedlings via salvage pathways (Reichard, 1988; Saada et al., 2001).
Defects in TGS May Underlie the Developmental Defects of tso2
Previous research has suggested that stochastic release of TGS in bru1 and fas mutants caused meristem fasciation as a result of the ectopic expression of WUSCHEL (WUS), a key meristem regulator (Mayer et al., 1998; Kaya et al., 2001; Takeda et al., 2004). Here, we showed that tso2-1 rnr2a-1 exhibited defects in epigenetic inheritance by releasing silencing at the TSI locus, suggesting that the fasciated SAM and homeotic transformation of floral organs observed in tso2-1 could be caused by stochastic release of silencing at loci including WUS and AGAMOUS. Because we failed to detect TSI in tso2-1 single mutants, it is possible that stochastic release of TSI or other regulatory genes is rare and difficult to detect in tso2-1. Future in situ hybridization experiments will be necessary to determine whether tso2-1 single mutants also exhibit defects in TGS. To date, there are no reports of defects in TGS in rnr mutants of yeast or mammals. It remains to be seen whether release of TGS could be the underlying basis of the growth abnormalities in rnr mutants of fungi or animals.
In addition to meristem fasciation and homeotic transformation of floral organs, tso2-1 mutants exhibited additional developmental defects, such as disorganized leaf cell layers (Figure 1I), aborted seed development (Figure 1M), unfused carpels (Figure 1E), and the formation of multiple carpels (data not shown). These developmental defects may be caused by additional mechanisms other than epigenetic misregulation. For example, the delayed cell division cycle shown by the significant increase in H4-expressing cells in tso2-1 (Figure 5) could lead to altered cell-to-cell communication, which is essential for proper organ fusion or proper cell layer organization. Alternatively, RNR may have additional functions not limited to the simple production of DNA building blocks (Chabes and Thelander, 2003). Downes et al. (2000) suggested that the normally high levels of intracellular purine dNDPs at S-phase act as an intracellular signal for the S-phase, retarding the progression toward M-phase. This may explain why RNR is more involved in oncogenic transformation than is expected for a simple metabolic enzyme (Downes et al., 2000).
tso2-1 rnr2a-1 Undergoes PCD
We demonstrated that cells in tso2-1 rnr2a-1 seedlings underwent PCD. Histochemical markers, molecular markers, and the TUNEL assay were used to analyze the cell death that occurred in tso2-1 rnr2a-1 double mutants. The nuclear morphology combined with the positive TUNEL signal indicative of DNA fragmentation provided strong support for the occurrence of apoptosis-like PCD in tso2 rnr2a mutants. Our data also revealed that the PCD in tso2 rnr2a plants resembles HR-mediated PCD with regard to histochemical markers and gene expression, suggesting that although the triggers of PCD may differ in tso2 rnr2a, many of the downstream effector genes of PCD may be shared among different PCD pathways in higher plants.
Possible Mechanisms Triggering PCD in tso2 rnr2a
What could be the trigger for PCD in tso2 rnr2a double mutants? In mammalian cells, DNA damage switches on the tumor-suppressor protein p53, which induces cell cycle arrest and PCD (apoptosis) (Chernova et al., 1995; Vogelstein et al., 2000). Whether PCD occurs in response to DNA damage as a defense mechanism in plants remains unresolved. A p53 homolog has not been identified in plants, even though the entire genome of Arabidopsis has been sequenced. Moreover, PCD was not detected in telomerase-deficient lines (Riha et al., 2001), and many mutants defective in DNA repair, such as mutants of the single-copy Arabidopsis gene KU80, are viable and wild type–like under normal growth conditions (West et al., 2002). However, fragmented nuclei resembling apoptotic nuclei have been mentioned in Arabidopsis mutants of ATTOP6B, which encodes a homolog of archaebacterial topoisomerase subunit B (Hartung et al., 2002). Nuclear degradation in aphidicolin-treated ataxia telangiectasia-mutated and rad3-related mutant cells was also reported (Culligan et al., 2004). A more in-depth characterization of UV light–induced PCD was performed using Arabidopsis protoplast cells (Danon and Gallois, 1998). The UV-C light–induced PCD was shown to be mediated by a caspase-like activity, as it can be suppressed by the caspase inhibitor p35 and Defender Against apoptotic Death (Danon et al., 2004). Because the UV irradiation could cause damage to other cellular components, whether DNA damage could directly induce PCD in plant cells is still debatable. If a p53-related pathway exists in plants, as suggested by Whittle et al. (2001), the p53-like gene might have little sequence homology with its mammalian counterpart.
Could the DNA damage accumulated in tso2 rnr2a double mutants directly trigger PCD? Among all r2 single and double mutants examined, only the tso2 rnr2a double mutants accumulated DNA damage and only tso2 rnr2a exhibited PCD. The correlation between DNA damage and PCD in tso2 rnr2a suggests that the DNA damage in tso2 rnr2a may trigger these cells to undergo PCD to eliminate damaged cells. This suggestion is further supported by the time course analyses, which revealed DNA damage accumulation occurring at least 10 d before the appearance of apoptotic nuclei in the tso2 rnr2a seedlings (Figure 9). By examining whether PCD occurs in other plant mutants defective in genome integrity, one may identify the DNA/cellular lesions required to switch on PCD.
Although we favor the hypothesis that the DNA damage in tso2 rnr2a triggers PCD, alternative mechanisms could not be excluded. For example, a reduction of dNTP may directly serve as a signal to trigger PCD. Perturbation of dNTP pools has been suggested to trigger DNA fragmentation and cell death in mammalian cells (Oliver et al., 1996). Another possible trigger for PCD may be the formation of defective chloroplasts or mitochondria in these tso2 rnr2a mutants. The malfunction of these organelles may lead to the release of reactive oxygen species or cytochrome c, which may subsequently trigger PCD in the tso2 rnr2a double mutants. Nevertheless, the leaves of tso2 rnr2a seedlings, which showed massive PCD, did not exhibit the white sectors that are hallmarks of tso2-1 single mutants. A lack of correlation between white sectors in leaves and PCD suggests that chloroplast defects are unlikely to underlie PCD in tso2 rnr2a. Future experiments are necessary to further test hypotheses regarding the trigger for PCD in tso2 rnr2a. The availability of single and double mutations in all three R2 genes provides a unique opportunity to identify suppressor mutations in genes that act upstream and downstream of RNR, providing insights into DNA damage checkpoint pathways and DNA damage–induced PCD in higher plants.
METHODS
Plant Growth, Mutant Strains, and Materials
Arabidopsis thaliana plants were grown under a 16-h-light/8-h-dark cycle at 20°C. tso2-1, -2, -3, and -4 alleles were isolated in two separate EMS mutagenesis screens (Liu and Meyerowitz, 1995; Levin et al., 1998), and all four alleles were generated in the Ler background. rnr2a-1 was identified by the TILLING facility (McCallum et al., 2000). Specifically, the primer pair 5′-TTTGCTGTGAGGCTGGTCGCTTTT-3′ and 5′-CTTCCAGATTCGATGGCGGATTCA-3′ was used to amplify ∼1 kb of RNR2A genomic DNA from EMS-mutagenized Col-er M2 plants. HPLC-based detection of DNA heteroduplex led to the identification of rnr2a-1. Finally, primer pair 5′-CGATTAAATCTTCTTCACCGGA-3′ and 5′-GGCTCCAATCCTTTTTGGAT-3′ was used to PCR-amplify the entire RNR2B genomic DNA from three different Arabidopsis accessions: Col, Ler, and Ws. Sequence analyses revealed 14 differences between Col and Ler/Ws (Figure 2C). TSO2 was provided by the Kazusa DNA Research Institute.
Microscopy
For scanning electron microscopy, inflorescences were fixed, coated, dissected, and photographed as described previously (Bowman et al., 1989). Images were directly captured with the semicaps software and the AMRAY 1000A scanning electron microscope. Whole-mount floral photomicrographs were taken through a Zeiss Stemi SV6 dissecting microscope. Slides containing longitudinal sections of inflorescence from in situ hybridization experiments were examined and photographed with a Nikon ECL1PSE E600W microscope with Nomarski optics equipped with a DXM1200 digital still camera. Images from the comet assay and the aniline blue staining were captured with the Nikon Labophot-2 microscope equipped with fluorescein isothiocyanate and UV light filters using ×20 objectives and a Nikon digital camera.
Map-Based Cloning of TSO2
tso2-1 (Ler) was crossed to sup-2 (Col). A total of 358 F2 tso2-1 plants were assayed individually for linkage to various CAPS and dCAPS markers (see Supplemental Table 1 online), which mapped TSO2 to a 20-kb region within the P1 clone MOJ10. A cosmid library was constructed from MOJ10 using binary vector pCLD04541. Two overlapped cosmids, D and G, were transformed into tso2-1 mutants, and all eight transgenic plants harboring cosmid D but none of the five transgenic plants harboring cosmid G were wild type in phenotype, suggesting that cosmid D, not G, contains the TSO2 gene. Sequencing of the four candidate genes At3g27040, At3g27050, At3g27060, and At3g27070 in cosmid D identified only a mutation in At3g27060 in tso2-1.
Molecular Analyses of TSO2, RNR2A, and RNR2B
p35S:RNR2B-L was constructed by PCR-amplifying the open reading frame using Ler genomic DNA as a template and primer pair 5′-GCTCTAGACGATTAAATCTTCTTCACCGGA-3′ and 5′-CGGGATCCGGCTCCAATCCTTTTTGGAT-3′. The PCR fragment was inserted into the pBI121 vector (Clontech) using XbaI and BamHI. To construct p35S:RNR2A, mRNA isolated from wild-type (Col) inflorescences was reversed-transcribed into cDNA with oligo(dT) primer and SuperScript reverse transcriptase (Invitrogen). The first-strand cDNA served as the template for PCR with Expand high-fidelity DNA polymerase (Roche) using primer pair 5′-GCTCTAGAGAATTCGAGATAATGGGTTCG-3′ and 5′-CGGGATCCCAATGGAGAAGGGACAAGTGA-3′. The PCR products of RNR2A cDNA were digested with XbaI and BamHI and inserted into the pBI121 vector. The pBI121-RNR2A clones were sequenced to verify that the clones contained no mutation.
For pTSO2:GUS reporter constructs, a 2.1-kb BamHI/EcoRI fragment containing the GUS open reading frame and 3′ Nos was excised from pBI121 and inserted into the BamHI/EcoRI site of pBIN20 (Hennegan and Danna, 1998) to create the pBIN20GUS vector. A 1.2-kb promoter sequence of TSO2 was PCR-amplified with primer pair 5′-GCTCTAGAATAAGGCCCTGTTCGTTTCC-3′ and 5′-CGGGATCCGAATCTGTCTGGGGTTGGTG-3′. PCR products were digested with BamHI/XbaI and inserted into BamHI/XbaI-digested pBIN20GUS vector. All PCRs were performed with high-fidelity DNA polymerase Pwo or Tgo (Roche).
For semiquantitative RT-PCR, total RNA was isolated with Tri-Reagent (Sigma-Aldrich) from the wild type (Ler), including 1-week-old roots and seedlings, 2-week-old rosette leaves, and 4-week-old cauline leaves, stems, and inflorescences. Two micrograms of total RNA was used to synthesize cDNA with oligo(dT) and SuperScript II reverse transcriptase (Invitrogen). PCR was conducted with 1 μL of diluted RT reaction at 94°C for 20 s, 57°C for 20 s, and 72°C for 40 s for 20 to 25 cycles. The PCR products were DNA gel-blotted and hybridized with a RNR2B-specific probe PCR amplified using primers 5′-CAAGCTTGACGAGGATTTTT-3′ and 5′-GTGCTCCCTCTGCCAATAAA-3′. To assay the induction of cell death– and DNA damage–induced markers and the induction of TSI, RNA was isolated from 3-week-old seedlings using protocols and conditions similar to those described above. Primer sequences for all RT-PCRs are listed in Supplemental Table 2 online.
In situ hybridization was essentially as described previously (Liu et al., 2000) except that the RNA probes were synthesized using the DIG RNA labeling kit (SP6/T7) (Roche) and were not hydrolyzed. The TSO2 EST clone (RZL13g10F) was linearized with HindIII and served as a template for the TSO2 antisense probe using the T7 promoter. For the in situ hybridization shown in Figure 5, a cDNA clone of CycB1 (pCYC1At) (Hemerly et al., 1992) was obtained from Dirk Inze. pCYC1At was linearized with ClaI and served as the template for transcription to generate antisense and sense CycB1 RNA probes from T7 and SP6 promoters, respectively. A H4 EST clone (249N16) served as the template for PCR with primer pair 5′-TGGAAAGGGAGGAAAAGGTT-3′ and 5′-AACCCAGAAAACACAAACGC-3′. The 339-bp PCR product was cloned into the pCRII-TOPO vector (Invitrogen). Two clones containing inserts in opposite orientations relative to the T7 promoter were linearized with HindIII and served as templates for in vitro transcription from the T7 promoter to generate antisense and sense H4 RNA probes.
Genetic Analyses
tso2-1 (Ler) was crossed with rnr2a-1 rnr2b-1 (Col). The F1 progeny heterozygous for all three mutations are wild type. dCAPS and CAPS markers (see Supplemental Table 1 online) were used to screen tso2-like F2 plants at RNR2A and RNR2B loci to identify the plants of various genotypes shown in Figure 3H. PCR-based genotyping was performed for individual seeds from tso2-1/tso2-1; rnr2b-1/+ parents. tso2-1 rnr2b-1 double mutants were detected only among aborted seeds (Table 1). Finally, the rnr2a-1/rnr2a-1; tso2-1/+ plants, which are phenotypically normal, were identified from the same cross described above by PCR-based genotyping of normal-looking plants in F2. tso2-1 rnr2a-1 double mutants were obtained from the self progeny of a rnr2a-1/rnr2a-1; tso2-1/+ parent.
Drug Treatment of pTSO2:GUS Transgenic Plants
Seeds of pTSO2:GUS transgenic T2 plants of Ler ecotype were germinated on wet filter paper. One- to 2-d-old seedlings were then transferred to a 24-well plate containing 1 mL of water (nontreatment control), 0.5% colchicine, or 5 μg/mL aphidicolin. After 26 h, the seedlings were rinsed twice with water, stained overnight with 5-bromo-4-chloro-3-indolyl-β-glucuronic acid based on a previously described protocol (Parcy et al., 1998), and then photographed. Five different pTSO2:GUS transgenic lines and 15 to 20 T2 seedlings of each transgenic line were analyzed. Three T2 lines exhibited the same reported responses to different drug treatments.
Measuring dNTP Levels
The dNTP pool was measured by a polymerase-based assay (Roy et al., 1999). Inflorescences with unopened flower buds were harvested in liquid nitrogen, ground to a fine powder, weighed, and extracted with 60% cold methanol by vigorous vortexing. The extracts were heated at 95°C for 5 min and centrifuged at 17,000g for 15 min. The supernatants were dried in a Speedvac, resuspended in sterile distilled water, and stored at −20°C. One microliter of each sample was used for the polymerase assay. Commercial dNTPs were tested in parallel to establish a linear calibration curve. As the internal control, a portion of the ground tissues was assayed for alcohol dehydrogenase activity (Russell et al., 1990). The alcohol dehydrogenase activity per gram of sample was then used to normalize the dNTP level for each gram of ground tissues.
Comet Assay for DNA Damage
The CometAssay kit from Trevigen (Gaithersburg, MD) was used with minor modifications. Seedlings were chopped with a razor in a Petri dish kept on ice and containing 500 μL of 1× PBS plus 20 mM EDTA. The resulting mixture was filtered through a 60-μm nylon mesh. Forty microliters of nuclei was mixed with 400 μL of 1% low-melting-point agarose (prewarmed at 37°C) and placed onto Trevigen-precoated slides. After incubating in lysis solution (2.5 M NaCl, 100 mM EDTA, pH 10, 10 mM Tris, 1% sodium lauryl sarcosinate, and 1% Triton X-100) for 1 h at 4°C, the nuclei on slides were unwound in alkaline solution (0.3 n NaOH and 5 mM EDTA) for 40 min and neutralized two to three times in 1× TBE (Tris-borate/EDTA) for 5 min. The slides were run at 1 V/cm for 10 min in 1× TBE and then dipped in 70% ethanol for 5 min. After air-drying, the slides were stained with a 1:10,000 dilution of SYBR green. Antifade solution (1% p-phenylenediamine dihydrochloride in 0.1× PBS and 90% glycerol) was added to slides that were examined by epifluorescence microscopy. The percentage of DNA in each comet tail (T DNA%) was evaluated with the Comet Score software (http://www.autocomet.com) and assigned a number (0 to 4), with a higher number corresponding to a higher T DNA% (Collins et al., 1997). DNA damage units for each genotype were derived by averaging the data from four slides. One hundred comets were scored per slide.
UV-C Light Treatment
Ten-day-old seedlings were irradiated at various doses of UV-C light using Stratalinker 1800 (Stratagene). Because the UV light detector of Stratalinker is situated 15 cm from the UV lamps, the UV-C light dosage presented in Figure 7A was calculated to reflect the 5-cm distance from seedlings to the UV lamps based on the formula that the UV-C dosage is a function of the square of the distance. After UV-C irradiation, seedlings were grown under F40GO gold fluorescent lights for 5 d to avoid photoreactivation. Plants were returned to normal lighting for 1 to 2 weeks before survival rate was scored. To test tso2-1 rnr2a-1, a large number of F2 seeds of tso2-1/+; rnr2a-1/rnr2a-1 were planted. One week after germination, tso2-1 rnr2a-1 double mutant seedlings were easily distinguished from siblings of tso2-1/+; rnr2a-1/rnr2a-1 or +/+; rnr2a-1/rnr2a-1 genotype. These siblings were pulled from the soil. Approximately 30 tso2-1 rnr2a-1 seedlings were left in each pot for UV-C light treatment.
Cell Death Assays
For trypan blue assay (Rate et al., 1999), 10-d-old rosette leaves were boiled in lactophenol containing 10 mg of trypan blue for 1 min, cleared in alcoholic lactophenol (95% ethanol:lactophenol, 2:1) for 2 min, washed in 50% ethanol, and stored in water. For aniline blue staining of callose (Rate et al., 1999), 10-d-old rosette leaves were boiled for 2 min in alcoholic lactophenol, rinsed in 50% ethanol, and then rinsed in water. Cleared and rinsed leaves were stained for 1 h at room temperature in a solution of 0.05% aniline blue in 0.15 M K2HPO4. Stained leaves were examined under UV epifluorescence. The 3,3′-diaminobenzidine uptake method (Thordal-Christensen et al., 1997) was used to detect H2O2. Ten-day-old seedlings were placed in 1 mg/mL 3,3′-diaminobenzidine in 10 mM ascorbic acid for 2 h. The seedlings were then boiled in 96% ethanol for 10 min and stored in 96% ethanol. H2O2 production is visualized as reddish-brown coloration.
The TUNEL assay was performed using the In Situ Cell Death Detection Kit-Fluorescein (Roche Diagnostics). Before detection, wild-type (Ler) and tso2-1 rnr2a-1 seedlings were fixed in 4% paraformaldehyde in 1× PBS at 4°C overnight and embedded in paraplasts. Eight-micrometer sections on glass slides were dewaxed in xylene, rehydrated, and then pretreated with 20 μg/mL proteinase K in 10 mM Tris-Cl, pH 7.5, for 20 min at room temperature. Two slides, treated with 1500 units/mL DNase I in 50 mM Tris-Cl, pH 7.5, 1 mM MgSO4, and 1 mg/mL BSA for 20 min at room temperature, served as positive controls. Two slides, labeled in the absence of the terminal deoxyribonucleotidyl transferase enzyme, served as negative controls. Vectashield mounting medium (Vector Laboratories) and DAPI (1 μg/mL) were used to mount the slides before they were viewed and photographed with a Zeiss Axiophot microscope.
Accession Numbers
The RNR2B sequence from Ler was submitted to GenBank with the accession number AY178109. The locus identifier numbers for TSO2, RNR2A, and RNR2B are At3g27060, At3g23580, and At5g40942, respectively. The accession number for TSO2 cDNA is RZL13g10F (AV546418).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. PCR-Based Molecular Markers Developed for This Study.
Supplemental Table 2. Primer Sequences Used in Semiquantitative RT-PCR.
Supplementary Material
Acknowledgments
We thank the ABRC and the Kazusa DNA Research Institute for DNA clones and seed stocks, Joshua Levin for tso2 alleles, Dirk Inze for the pCYC1At clone, Arabidopsis TILLING for identifying rnr2a-1, and Roy Beatrice for advice and protocol in dNTP measurement. We thank Eric Baehrecke, Caren Chang, Robert Franks, Steve Mount, Shin Takeda, Lee Zou, and members of the Liu laboratory for comments on the manuscript. We thank Tim Mougel for microscopy assistance (contribution no. 97 of the Laboratory for Biological Ultrastructure, University of Maryland, College Park). This work was supported by U.S. Department of Energy Grant 02-00ER20281 to Z.L.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Zhongchi Liu (zliu@umd.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.037044.
References
- Angelis, K.J., Dusinska, M., and Collins, A.R. (1999). Single cell gel electrophoresis: Detection of DNA damage at different levels of sensitivity. Electrophoresis 20 2133–2138. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen, P., Petersen, M., Pike, H.M., Olszak, B., Skov, S., Odum, N., Jorgensen, L.B., Brown, R.E., and Mundy, J. (2002). Knockout of Arabidopsis accelerated-cell-death11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev. 16 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundock, P., and Hooykaas, P. (2002). Severe developmental defects, hypersensitivity to DNA-damaging agents, and lengthened telomeres in Arabidopsis MRE11 mutants. Plant Cell 14 2451–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabes, A., Domkin, V., and Thelander, L. (1999). Yeast Sml1, a protein inhibitor of ribonucleotide reductase. J. Biol. Chem. 274 36679–36683. [DOI] [PubMed] [Google Scholar]
- Chabes, A., Georgieva, B., Domkin, V., Zhao, X., Rothstein, R., and Thelander, L. (2003. a). Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell 112 391–401. [DOI] [PubMed] [Google Scholar]
- Chabes, A., and Thelander, L. (2003). DNA building blocks at the foundation of better survival. Cell Cycle 2 171–173. [DOI] [PubMed] [Google Scholar]
- Chabes, A.L., Pfleger, C.M., Kirschner, M.W., and Thelander, L. (2003. b). Mouse ribonucleotide reductase R2 protein: A new target for anaphase-promoting complex-Cdh1-mediated proteolysis. Proc. Natl. Acad. Sci. USA 100 3925–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaboute, M.E., Clement, B., and Philipps, G. (2002). S phase and meristem-specific expression of the tobacco RNR1b gene is mediated by an E2F element located in the 5′ leader sequence. J. Biol. Chem. 277 17845–17851. [DOI] [PubMed] [Google Scholar]
- Chaboute, M.E., Clement, B., Sekine, M., Philipps, G., and Chaubet-Gigot, N. (2000). Cell cycle regulation of the tobacco ribonucleotide reductase small subunit gene is mediated by E2F-like elements. Plant Cell 12 1987–2000. [PMC free article] [PubMed] [Google Scholar]
- Chaboute, M.E., Combettes, B., Clement, B., Gigot, C., and Philipps, G. (1998). Molecular characterization of tobacco ribonucleotide reductase RNR1 and RNR2 cDNAs and cell cycle-regulated expression in synchronized plant cells. Plant Mol. Biol. 38 797–806. [DOI] [PubMed] [Google Scholar]
- Chen, I.P., Haehnel, U., Altschmied, L., Schubert, I., and Puchta, H. (2003). The transcriptional response of Arabidopsis to genotoxic stress—A high-density colony array study (HDCA). Plant J. 35 771–786. [DOI] [PubMed] [Google Scholar]
- Chernova, O.B., Chernov, M.V., Agarwal, M.L., Taylor, W.R., and Stark, G.R. (1995). The role of p53 in regulating genomic stability when DNA and RNA synthesis are inhibited. Trends Biochem. Sci. 20 431–434. [DOI] [PubMed] [Google Scholar]
- Collins, A., Dusinska, M., Franklin, M., Somorovska, M., Petrovska, H., Duthie, S., Fillion, L., Panayiotidis, M., Raslova, K., and Vaughan, N. (1997). Comet assay in human biomonitoring studies: Reliability, validation, and applications. Environ. Mol. Mutagen. 30 139–146. [DOI] [PubMed] [Google Scholar]
- Culligan, K., Tissier, A., and Britt, A. (2004). ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 16 1091–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon, A., and Gallois, P. (1998). UV-C radiation induces apoptotic-like changes in Arabidopsis thaliana. FEBS Lett. 437 131–136. [DOI] [PubMed] [Google Scholar]
- Danon, A., Rotari, V.I., Gordon, A., Mailhac, N., and Gallois, P. (2004). Ultraviolet-C overexposure induces programmed cell death in Arabidopsis, which is mediated by caspase-like activities and which can be suppressed by caspase inhibitors, p35 and Defender against Apoptotic Death. J. Biol. Chem. 279 779–787. [DOI] [PubMed] [Google Scholar]
- Dietrich, R.A., Delaney, T.P., Uknes, S.J., Ward, E.R., Ryals, J.A., and Dangl, J.L. (1994). Arabidopsis mutants simulating disease resistance response. Cell 77 565–577. [DOI] [PubMed] [Google Scholar]
- Doucet-Chabeaud, G., Godon, C., Brutesco, C., de Murcia, G., and Kazmaier, M. (2001). Ionising radiation induces the expression of PARP-1 and PARP-2 genes in Arabidopsis. Mol. Genet. Genomics 265 954–963. [DOI] [PubMed] [Google Scholar]
- Doutriaux, M.P., Couteau, F., Bergounioux, C., and White, C. (1998). Isolation and characterisation of the RAD51 and DMC1 homologs from Arabidopsis thaliana. Mol. Gen. Genet. 257 283–291. [DOI] [PubMed] [Google Scholar]
- Downes, C.S., Bachrati, C.Z., Devlin, S.J., Tommasino, M., Cutts, T.J., Watson, J.V., Rasko, I., and Johnson, R.T. (2000). Mammalian S-phase checkpoint integrity is dependent on transformation status and purine deoxyribonucleosides. J. Cell Sci. 113 1089–1096. [DOI] [PubMed] [Google Scholar]
- Elledge, S.J., Zhou, Z., and Allen, J.B. (1992). Ribonucleotide reductase: Regulation, regulation, regulation. Trends Biochem. Sci. 17 119–123. [DOI] [PubMed] [Google Scholar]
- Fobert, P.R., Coen, E.S., Murphy, G.J., and Doonan, J.H. (1994). Patterns of cell division revealed by transcriptional regulation of genes during the cell cycle in plants. EMBO J. 13 616–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg, J.T. (1996). Programmed cell death: A way of life for plants. Proc. Natl. Acad. Sci. USA 93 12094–12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg, J.T., and Yao, N. (2004). The role and regulation of programmed cell death in plant-pathogen interactions. Cell. Microbiol. 6 201–211. [DOI] [PubMed] [Google Scholar]
- Hartung, F., Angelis, K.J., Meister, A., Schubert, I., Melzer, M., and Puchta, H. (2002). An archaebacterial topoisomerase homolog not present in other eukaryotes is indispensable for cell proliferation of plants. Curr. Biol. 12 1787–1791. [DOI] [PubMed] [Google Scholar]
- Hemerly, A., Bergounioux, C., Van Montagu, M., Inze, D., and Ferreira, P. (1992). Genes regulating the plant cell cycle: Isolation of a mitotic-like cyclin from Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 89 3295–3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennegan, K.P., and Danna, K.J. (1998). pBIN20: an improved binary vector for Agrobacterium-mediated transformation. Plant Mol. Biol. Rep. 16 129–131. [Google Scholar]
- Huang, M., Zhou, Z., and Elledge, S.J. (1998). The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94 595–605. [DOI] [PubMed] [Google Scholar]
- Kaya, H., Shibahara, K.I., Taoka, K.I., Iwabuchi, M., Stillman, B., and Araki, T. (2001). FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell 104 131–142. [DOI] [PubMed] [Google Scholar]
- Kimura, T., Takeda, S., Sagiya, Y., Gotoh, M., Nakamura, Y., and Arakawa, H. (2003). Impaired function of p53R2 in Rrm2b-null mice causes severe renal failure through attenuation of dNTP pools. Nat. Genet. 34 440–445. [DOI] [PubMed] [Google Scholar]
- Kolberg, M., Strand, K.R., Graff, P., and Andersson, K.K. (2004). Structure, function, and mechanism of ribonucleotide reductases. Biochim. Biophys. Acta 1699 1–34. [DOI] [PubMed] [Google Scholar]
- Kressel, M., and Groscurth, P. (1994). Distinction of apoptotic and necrotic cell death by in situ labelling of fragmented DNA. Cell Tissue Res. 278 549–556. [DOI] [PubMed] [Google Scholar]
- Kuriyama, H., and Fukuda, H. (2002). Developmental programmed cell death in plants. Curr. Opin. Plant Biol. 5 568–573. [DOI] [PubMed] [Google Scholar]
- Levin, J.Z., Fletcher, J.C., Chen, X., and Meyerowitz, E.M. (1998). A genetic screen for modifiers of UFO meristem activity identifies three novel FUSED FLORAL ORGANS genes required for early flower development in Arabidopsis. Genetics 149 579–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincker, F., Philipps, G., and Chaboute, M.E. (2004). UV-C response of the ribonucleotide reductase large subunit involves both E2F-mediated gene transcriptional regulation and protein subcellular relocalization in tobacco cells. Nucleic Acids Res. 32 1430–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C., Powell, K.A., Mundt, K., Wu, L., Carr, A.M., and Caspari, T. (2003). Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint-dependent and -independent mechanisms. Genes Dev. 17 1130–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., Franks, R.G., and Klink, V.P. (2000). Regulation of gynoecium marginal tissue formation by LEUNIG and AINTEGUMENTA. Plant Cell 12 1879–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., and Meyerowitz, E.M. (1995). LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development 121 975–991. [DOI] [PubMed] [Google Scholar]
- Liu, Z., Running, M.P., and Meyerowitz, E.M. (1997). TSO1 functions in cell division during Arabidopsis flower development. Development 124 665–672. [DOI] [PubMed] [Google Scholar]
- Mayer, K.F., Schoof, H., Haecker, A., Lenhard, M., Jurgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95 805–815. [DOI] [PubMed] [Google Scholar]
- McCallum, C.M., Comai, L., Greene, E.A., and Henikoff, S. (2000). Targeting induced local lesions in genomes (TILLING) for plant functional genomics. Plant Physiol. 123 439–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J.D., Arteca, R.N., and Pell, E.J. (1999). Senescence-associated gene expression during ozone-induced leaf senescence in Arabidopsis. Plant Physiol. 120 1015–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, F.J., Collins, M.K., and Lopez-Rivas, A. (1996). dNTP pools imbalance as a signal to initiate apoptosis. Experientia 52 995–1000. [DOI] [PubMed] [Google Scholar]
- Osakabe, K., Yoshioka, T., Ichikawa, H., and Toki, S. (2002). Molecular cloning and characterization of RAD51-like genes from Arabidopsis thaliana. Plant Mol. Biol. 50 71–81. [DOI] [PubMed] [Google Scholar]
- Parcy, F., Nilsson, O., Busch, M.A., Lee, I., and Weigel, D. (1998). A genetic framework for floral patterning. Nature 395 561–566. [DOI] [PubMed] [Google Scholar]
- Philipps, G., Clement, B., and Gigot, C. (1995). Molecular characterization and cell cycle-regulated expression of a cDNA clone from Arabidopsis thaliana homologous to the small subunit of ribonucleotide reductase. FEBS Lett. 358 67–70. [DOI] [PubMed] [Google Scholar]
- Pontier, D., Gan, S., Amasino, R.M., Roby, D., and Lam, E. (1999). Markers for hypersensitive response and senescence show distinct patterns of expression. Plant Mol. Biol. 39 1243–1255. [DOI] [PubMed] [Google Scholar]
- Rate, D.N., Cuenca, J.V., Bowman, G.R., Guttman, D.S., and Greenberg, J.T. (1999). The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell 11 1695–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichard, P. (1988). Interactions between deoxyribonucleotide and DNA synthesis. Annu. Rev. Biochem. 57 349–374. [DOI] [PubMed] [Google Scholar]
- Riha, K., McKnight, T.D., Griffing, L.R., and Shippen, D.E. (2001). Living with genome instability: Plant responses to telomere dysfunction. Science 291 1797–1800. [DOI] [PubMed] [Google Scholar]
- Roy, B., Beuneu, C., Roux, P., Buc, H., Lemaire, G., and Lepoivre, M. (1999). Simultaneous determination of pyrimidine or purine deoxyribonucleoside triphosphates using a polymerase assay. Anal. Biochem. 269 403–409. [DOI] [PubMed] [Google Scholar]
- Russell, D.A., Wong, D.M.L., and Sachs, M.M. (1990). The anaerobic response of soybean. Plant Physiol. 92 401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saada, A., Shaag, A., Mandel, H., Nevo, Y., Eriksson, S., and Elpeleg, O. (2001). Mutant mitochondrial thymidine kinase in mitochondrial DNA depletion myopathy. Nat. Genet. 29 342–344. [DOI] [PubMed] [Google Scholar]
- Saitoh, S., Chabes, A., McDonald, W.H., Thelander, L., Yates, J.R., and Russell, P. (2002). Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA. Cell 109 563–573. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Scholkopf, B., Weigel, D., and Lohmann, J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37 501–506. [DOI] [PubMed] [Google Scholar]
- Siddiqui, N.U., Stronghill, P.E., Dengler, R.E., Hasenkampf, C.A., and Riggs, C.D. (2003). Mutations in Arabidopsis condensin genes disrupt embryogenesis, meristem organization and segregation of homologous chromosomes during meiosis. Development 130 3283–3295. [DOI] [PubMed] [Google Scholar]
- Steimer, A., Amedeo, P., Afsar, K., Fransz, P., Scheid, O.M., and Paszkowski, J. (2000). Endogenous targets of transcriptional gene silencing in Arabidopsis. Plant Cell 12 1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, S., Tadele, Z., Hofmann, I., Probst, A.V., Angelis, K.J., Kaya, H., Araki, T., Mengiste, T., Scheid, O.M., Shibahara, K., Scheel, D., and Paszkowski, J. (2004). BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev. 18 782–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, H., Arakawa, H., Yamaguchi, T., Shiraishi, K., Fukuda, S., Matsui, K., Takei, Y., and Nakamura, Y. (2000). A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature 404 42–49. [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen, H., Zhang, Z., Wei, Y., and Collinge, D.B. (1997). Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11 1187–1194. [Google Scholar]
- Vogelstein, B., Lane, D., and Levine, A.J. (2000). Surfing the p53 network. Nature 408 307–310. [DOI] [PubMed] [Google Scholar]
- West, C.E., Waterworth, W.M., Story, G.W., Sunderland, P.A., Jiang, Q., and Bray, C.M. (2002). Disruption of the Arabidopsis AtKu80 gene demonstrates an essential role for AtKu80 protein in efficient repair of DNA double-strand breaks in vivo. Plant J. 31 517–528. [DOI] [PubMed] [Google Scholar]
- Whittle, C.A., Beardmore, T., and Johnston, M.O. (2001). Is G1 arrest in plant seeds induced by a p53-related pathway? Trends Plant Sci. 6 248–251. [DOI] [PubMed] [Google Scholar]
- Yao, R., Zhang, Z., An, X., Bucci, B., Perlstein, D.L., Stubbe, J., and Huang, M. (2003). Subcellular localization of yeast ribonucleotide reductase regulated by the DNA replication and damage checkpoint pathways. Proc. Natl. Acad. Sci. USA 100 6628–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., Chabes, A., Domkin, V., Thelander, L., and Rothstein, R. (2001). The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J. 20 3544–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., Georgieva, B., Chabes, A., Domkin, V., Ippel, J.H., Schleucher, J., Wijmenga, S., Thelander, L., and Rothstein, R. (2000). Mutational and structural analyses of the ribonucleotide reductase inhibitor Sml1 define its Rnr1 interaction domain whose inactivation allows suppression of mec1 and rad53 lethality. Mol. Cell. Biol. 20 9076–9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., Muller, E.G., and Rothstein, R. (1998). A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2 329–340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.