Abstract
Rho family small GTPases are signaling switches controlling many eukaryotic cellular processes. Conversion from the GDP- to GTP-bound form is catalyzed by guanine nucleotide exchange factors (GEFs). Rho GEFs in animals fall into two structurally distinct classes containing DH and DOCKER catalytic domains. Using a plant Rho GTPase (ROP1) as bait in yeast two-hybrid screens, we identified a family of Rho GEFs, named RopGEFs. The Arabidopsis thaliana RopGEF family of 14 members contains a conserved central domain, the domain of unknown function 315 (DUF315), and variable N- and C-terminal regions. In vitro GEF assays show that DUF315 but not the full-length version of RopGEF1 has high GEF activity toward ROP1. Our data suggest that the variable regions of RopGEF1 are involved in regulation of RopGEF through an autoinhibitory mechanism. RopGEF1 overexpression in pollen tubes produced growth depolarization, as does a constitutively active ROP1 mutant. The RopGEF1 overexpression phenotype was suppressed by expression of a dominant-negative mutant of ROP1, probably by trapping RopGEF1. Deletion mutant analysis suggested a requirement of RopGEF activity for the function of RopGEF1 in polar growth. Green fluorescent protein–tagged RopGEF1 was localized to the tip of pollen tubes where ROP1 is activated. These results provide strong evidence that RopGEF1 activates ROP1 in control of polar growth in pollen tubes.
INTRODUCTION
ROP GTPase, the sole family of a Rho-related GTPase from plants, has emerged as an important signaling molecule. ROP GTPase signaling regulates many diverse processes, including pollen tip growth, leaf cell morphogenesis, root hair development, H2O2 production, hormone responses, disease resistance, and cellulose synthesis (Li et al., 1998, 1999; Kawasaki et al., 1999; Fu et al., 2001, 2002; Lemichez et al., 2001; Molendijk et al., 2001; Ono et al., 2001; Baxter-Burrell et al., 2002; Jones et al., 2002; Nakanomyo et al., 2002; Tao et al., 2002; Zheng et al., 2002). As a molecular switch, ROP GTPase is presumably activated by GDP-GTP exchange factors (GEFs) and inactivated by GTPase-activating proteins (GAPs) and guanine nucleotide dissociation inhibitors. Both ROP GAPs and guanine nucleotide dissociation inhibitors have been identified ( Zarsky et al., 1997; Bischoff et al., 2000; Wu et al., 2000), but GEFs for ROPs in plants remained elusive until a recent report describing a novel family of Rho GEFs from Arabidopsis thaliana, termed RopGEFs (Berken et al., 2005).
Conventional Rho GEFs from animals and fungi contain diffuse B-cell lymphoma homology (DH) and pleckstrin homology domains (Cerione and Zheng, 1996). The DH domain, consisting of ∼150 amino acids, is a catalytic domain of Rho GEF (Hoffman and Cerione, 2002). BLAST searches using the DH domain from mammalian Rho GEF revealed no DH domain–containing proteins in the Arabidopsis and rice (Oryza sativa) genomes. Recently identified unconventional Rho GEFs contain Dock-homology region-1 (DHR1) and DHR2 with no sequence homology to DH domains (Brugnera et al., 2002; Cote and Vuori, 2002). DHR2 can mediate GTP loading toward Rac1 in vitro (Cote and Vuori, 2002). Another well-documented unconventional Rho GEF is the Salmonella typhimurium SopE protein. SopE protein activates Cdc42 in a Db1-like fashion despite its lack of sequence homology to DH domains (Buchwald et al., 2002).
A single Dock180 homolog in Arabidopsis, SPIKE1, was identified based on its mutations that caused trichome branching defect (Qiu et al., 2002). SPIKE1 mutations have pavement cell morphogenesis defect similar to that induced by the loss of function of ROP2 and ROP4 (Fu et al., 2002, 2005; Qiu et al., 2002). However, it remains to be determined whether SPIKE1 has ROP GEF activity. Arabidopsis RopGEFs were identified using ROP4 (D121N) as bait in a yeast two-hybrid screen (Berken et al., 2005). They contain a plant-specific Rop nucleotide exchanger (PRONE) domain with no sequence homology to DH or Docker domains, yet the PRONE domain was shown to contain GEF activity toward several Arabidopsis ROPs. Interestingly, the McCormick group has recently identified a tomato (Lycopersicon esculentum) homolog RopGEF, kinase partner protein (KPP), based on its interaction with the intracellular kinase domain of tomato receptor-like protein kinases (RLKs) PRK1 and PRK2 (Kaothien et al., 2005). Overexpression of this tomato RopGEF/KPP induced depolarized pollen tube growth similar to that induced by ROP1 overexpression (Kaothien et al., 2005). However, the functional significance of RopGEFs in the in vivo regulation of ROPs has not been studied.
ROP1 and its closely related members (likely ROP3 and ROP5) control polarized tip growth in pollen tubes (Lin and Yang, 1997; Kost et al., 1999; Li et al., 1999; Gu et al., 2004). ROP1 signaling controls two branch pathways that are required for efficient tip growth. These two pathways are controlled by two ROP1 target proteins: RIC3 and RIC4. RIC4 promotes F-actin assembly, whereas RIC3 activates Ca2+ signaling that leads to F-actin disassembly (Gu et al., 2005). ROP1 is localized to the tip of the plasma membrane (PM) in pollen tubes and is activated at the tip, forming an apical cap of active ROP1 (Lin and Yang, 1997; Li et al., 1999; Hwang et al., 2005). ROP1 activity oscillates in the same frequency as tip growth oscillation (Hwang et al., 2005). ROP1 activation at the tip temporally precedes a new growth burst and spatially predicts a new growth direction (Hwang et al., 2005). However, the molecular mechanism for the regulation of ROP1 activity in pollen tubes remains obscure.
In our efforts to identify ROP activators, we performed a yeast two-hybrid screen using ROP1 (D121A) as bait. Although our screens were independent of that performed by Berken et al. (2005), we identified the same family of RopGEFs. In agreement with the results of Berken et al. (2005), we independently show that the catalytic activity of RopGEF resides in the conserved plant-specific domain of unknown function 315 (DUF315) (similar to the PRONE domain). Interestingly, our data show that the variable regions of RopGEF1 participate in the regulation of RopGEF via an autoinhibitory mechanism. Significantly, our studies show that RopGEF1 functionally acts as an activator of ROP1 in the control of polarized pollen tube growth.
RESULTS
Identification of a Family of Plant-Specific Proteins That Interacts with the Dominant-Negative ROP1
In general, GEFs preferentially bind nucleotide-free and GDP-bound forms of GTPases. To identify potential RopGEFs, we conducted a yeast two-hybrid screen using a dominant-negative mutant of ROP1 as bait. To generate the bait construct, site-directed mutagenesis was used to create a dominant-negative form of ROP1 (DN-rop1) containing a D121A mutation (Li et al., 1999). A C188S mutation within the C-terminal isoprenylation motif was introduced into DN-rop1 to prevent its association with the PM (Wu et al., 2000). A total of 72 putative positive colonies were selected by their ability to grow on selective plates (SC-Trp-Leu-His) and for β-galactosidase activity using a filter-lift assay from a total of 3 million transformants screened. We recovered and sequenced 41 constructs from this collection. Among those sequenced, 23 were partial cDNAs encoding two families of plant-specific proteins. In this study, we focused on a family of 14 members containing a highly conserved central region with 315 amino acid residues, annotated as DUF315. During the preparation of this report, Berken et al. (2005) reported the same family of DUF315-containing proteins and named Arabidopsis DUF315 members RopGEF1 through RopGEF14. For consistency and brevity, we adopted Berken et al.'s nomenclature. Fourteen of the 23 cDNA clones corresponded to three distinct RopGEF proteins, RopGEF1, RopGEF12, and RopGEF14. To confirm the interaction between RopGEF proteins and DN-rop1, partial cDNA clones were subcloned into maltose binding protein (MBP) fusion vectors. In vitro pull-down assays confirmed the interaction between these three RopGEF proteins and DN-rop1 (data not shown).
Amino acid sequence alignment of the 14 RopGEFs from Arabidopsis showed that the N- and C-terminal regions outside of the DUF315 domain are highly variable (Figure 1). The DUF315 domain can be divided into three highly conserved subdomains (S1, S2, and S3) separated by two short stretches of variable amino acid residues (Figure 1). Database searches reveal that the RopGEFs are present in various plant species. The rice genome encodes at least 11 RopGEFs. Database searches have failed to identify any homologs of RopGEFs from nonplant organisms, suggesting that RopGEF is a plant-specific gene family.
Figure 1.
Comparison of Predicted Amino Acid Sequences among 14 RopGEFs.
To identify additional genes encoding DUF315 domain–containing proteins, the amino acid sequence of DUF315 domain was used as a query for BLASTP searches of the National Center for Biotechnology Information (NCBI) and The Arabidopsis Information Resource (TAIR) databases. Sequences that were identical among 14 RopGEFs are highlighted in black, and conserved consensus sequences are highlighted in gray. The conserved DUF315 domain is composed of three subdomains (S1, S2, and S3) and is indicated by dotted lines.
RopGEFs Display Differential Interactions with and GEF Activity toward ROP1
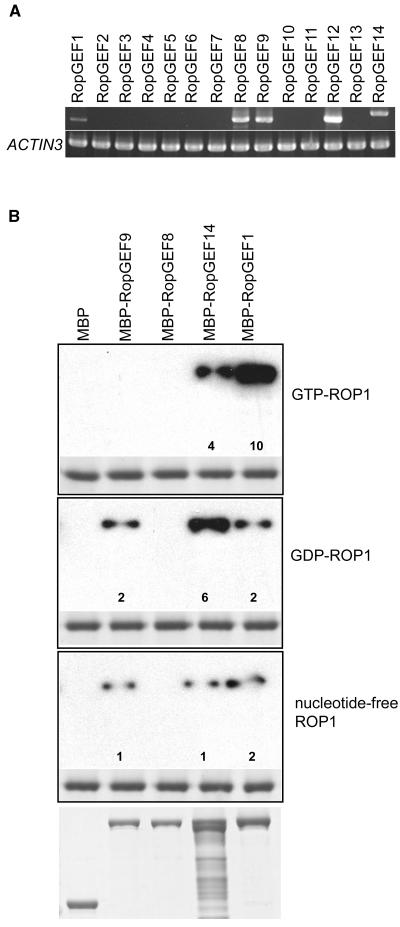
As a first step in testing the functional relationship between ROP1 and RopGEFs, we examined whether any of these RopGEFs are expressed in mature pollen because ROP1 is specifically expressed in pollen and is known to control pollen tube growth (Li et al., 1998). RT-PCR analysis showed that five RopGEF genes were expressed in mature pollen (Figure 2A). We then investigated which of these RopGEFs interacted with ROP1 using an in vitro pull-down assay. The predicted coding sequences of these five RopGEF genes were fused to the C terminus of MBP. Purified fusion proteins were pulled down with GTP-bound, GDP-bound, or nucleotide-free forms of the GST-ROP1 fusion protein and detected using an anti-MBP antibody (Wu et al., 2000). Despite numerous attempts, we were unable to express a RopGEF12 fusion protein in Escherichia coli; therefore, we only include four RopGEFs in our interaction assays. As shown in Figure 2, RopGEF fusion proteins exhibited differential interactions with different forms of ROP1 in vitro. RopGEF1 preferentially interacted with GTP-ROP1. RopGEF14 preferentially interacted with GDP-ROP1. Interestingly, RopGEF9 only interacted with GDP-bound and nucleotide-free forms of ROP1. RopGEF8 did not interact with any form of ROP1, although RopGEF8 and RopGEF9 share 79% identity at the amino acid sequence level.
Figure 2.
Differential Interactions between ROP1 and RopGEFs Expressed in Pollen.
(A) RT-PCR analysis of RopGEF genes in Arabidopsis pollen. Pollen RNA was isolated from mature pollen as described previously (Li et al., 1998) and was used for RT-PCR analysis as described in Methods. ACTIN3 was used as a positive control. Primers used for cDNA synthesis are listed in Table 1.
(B) GTP- or GDP-bound or nucleotide-free GST-ROP1 and MBP-RopGEF fusion proteins expressed in E. coli were purified by affinity chromatography and used for pull-down assays as described in Methods. Only four of the five pollen-expressed RopGEFs were used for this assay because we were unable to express RopGEF12 in E. coli. The top of each panel shows the binding of MBP-RopGEF fusion proteins to ROP1 in the indicated form, whereas the bottom of each panel shows GST-ROP1 loading control. Loading control for MBP-RopGEF fusion proteins is shown at the very bottom of the three panels. Numbers indicate relative signal intensity standardized with the loading control (see Methods).
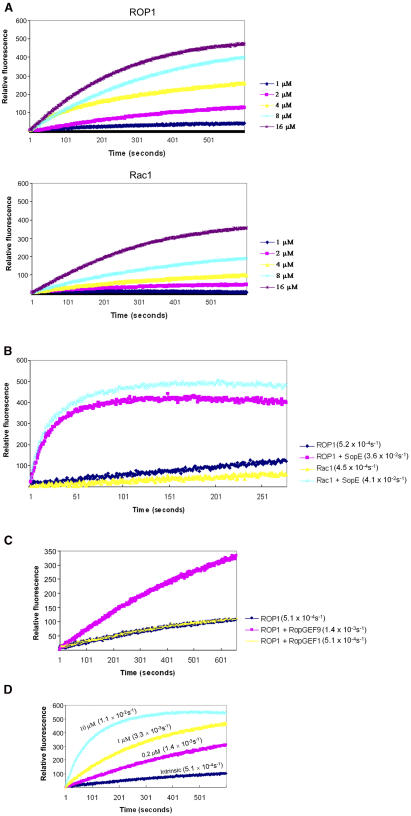
Since RopGEF9 differentially interacted with the GDP-bound and nucleotide-free forms of ROP1, we suspected that the RopGEF gene family might encode unconventional Rho GEFs for ROPs. To assay GEF activity, we used a fluorescence spectroscopy–based procedure for measuring nucleotide replacement on or dissociation from GTPase proteins (Leonard et al., 1994). GTPase protein was preloaded with unlabeled GDP, and the GEF reaction was started by the addition of GEF proteins and fluorescently labeled N-methylanthraniloyl (mant)-GDP. The incorporation rate for this nucleotide analog indicates GEF activity. For a positive control, we used SopE, an unconventional Rho GEF from the bacterial enteropathogen S. typhimurium (Rudolph et al., 1999). The intrinsic nucleotide replacement rate for GDP-ROP1 was faster than that of human GDP-Rac1 (Figure 3A). SopE was active on both human Rac1 and Arabidopsis ROP1 (Figure 3B).
Figure 3.
Differential in Vitro GEF Activities of RopGEF1 and RopGEF9 toward ROP1.
(A) Time course of Rac1 or ROP1 intrinsic nucleotide exchange. Assays contained 200 nM mant-GDP and the indicated concentration of GDP-Rac1 or GDP-ROP1.
(B) Bacterially expressed SopE (200 nM) stimulated the replacement of GDP by mant-GDP (200 nM) on 1 μM of Rac1 or ROP1 recombinant protein. Results are a representative of three independent assays that gave similar results. Rate constants (Kobs) are shown in parentheses. Kobs values were obtained by monophasic exponential fits using PeakFit v4.01.
(C) Differential GEF activity of RopGEFs toward ROP1. Assays contained 200 nM mant-GDP, 1 μM GDP-ROP1, and 200 nM RopGEF1 or RopGEF9. Results are a representative of three independent assays that gave similar results. Rate constants (Kobs) are shown in parentheses. Kobs values were obtained by monophasic exponential fits using PeakFit v4.01.
(D) Concentration-dependent GEF activity of RopGEF9 toward ROP1. Assays contained 200 nM mant-GDP, 1 μM GDP-ROP1, and indicated concentrations of RopGEF9. Rate constants (Kobs) are shown in parentheses. Kobs values were obtained by monophasic exponential fits using PeakFit v4.01.
We chose RopGEF1 and RopGEF9 for the in vitro GEF assay since they had differential interaction patterns with ROP1. The full-length RopGEF1 and RopGEF9 were purified as recombinant MBP fusion proteins and then cleaved by Factor Xa to remove MBP. Interestingly, the full-length RopGEF1 displayed no GEF activity, whereas RopGEF9 showed low but consistent GEF activity toward ROP1 (Figure 3C). RopGEF9 activity toward ROP1 showed measurable increase upon increasing concentration of RopGEF9 (Figure 3D). The ability of RopGEF9 to promote nucleotide exchange (loading) is consistent with its preferential interaction with GDP-bound and nucleotide-free forms of ROP1.
The Conserved DUF315 Domain Contains RopGEF Catalytic Activity
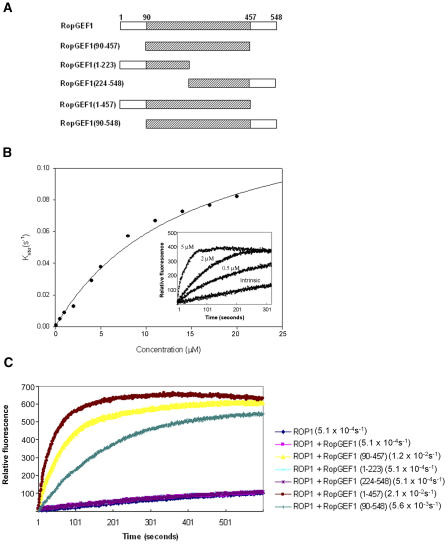
We next tested whether the DUF315 domain, which is highly conserved in all RopGEF members from different plant species, possesses the catalytic function of RopGEF. We suspected that the lack of RopGEF activity for RopGEF1 might be due to the presence of RopGEF inhibitory elements in the variable regions. Thus, we generated truncated RopGEF1 containing only the DUF315 domain (residues 90 to 457) (Figure 4A) and assayed its GEF activity. Indeed, this DUF315 domain had a high RopGEF activity toward ROP1 (Figure 4B). The apparent rate constant Kobs was determined by fitting data to a single exponential function using PeakFit. The rate constant increased with DUF315 domain concentrations in an apparently hyperbolic manner. The data obtained could be fitted to a hyperbolic function, and Kmax was determined to be 1.38 × 10−1 s−1. The DUF315 domain of RopGEF1 stimulates the nucleotide dissociation from ROP1 ∼380-fold. These results indicate that the DUF315 domain is sufficient for the catalysis of nucleotide exchange on ROP1.
Figure 4.
The DUF315 Domain of RopGEF1 Is Required and Sufficient for Nucleotide Exchange Activity toward ROP1.
(A) Diagram of truncated fragments of RopGEF1 used in GEF assays with ROP1. Hatched boxes indicate the conserved DUF315 domain.
(B) Kinetic analysis of the RopGEF activity of the DUF315 domain from RopGEF1. Kobs values at different concentrations of the DUF315 domain of RopGEF1 are plotted and fitted with a hyperbolic function using SigmaPlot. Kobs values were obtained by monophasic exponential fits using PeakFit v4.01. Inset shows the GEF activity of the DUF315 domain at three representative concentrations. Kmax value was obtained using the concentration curve fitted to a hyperbolic function.
(C) GEF activities of truncated fragments of RopGEF1 (100 nM) toward ROP1. The reaction contained 200 nM RopGEF1, 1 μM GDP-ROP1, and 200 nM mant-GDP. Rate constants (Kobs) are shown in parentheses. Kobs values were obtained by monophasic exponential fits using PeakFit v4.01.
To test whether the complete DUF315 domain is required for its GEF activity, we constructed two truncated fragments of RopGEF1, and both fragments lack part of the DUF315 domain. Both fragments have completely lost GEF activity toward ROP1 (Figure 4C). Based on these results, we conclude that the RopGEFs belong to a novel family of plant-specific Rho GEFs and that the conserved DUF315 domain is most likely a RopGEF catalytic domain. Our results are consistent with those of Berken et al. (2005) who used a slightly larger fragment of the DUF315 domain (residues 76 to 460) for GEF activity assays.
The Variable Terminal Regions of RopGEF1 Regulate GEF Activity
The full-length RopGEF1 had no in vitro GEF activity, whereas the conserved DUF315 domain of this RopGEF had a high RopGEF activity toward ROP1, suggesting that the variable N- and/or C-terminal regions contain regulatory motifs/sequences that are inhibitory to the RopGEF catalytic domain. To assess which regions contain the putative regulatory elements, we generated two truncated RopGEF1s that lack the N-terminal (residues 1 to 89) and C-terminal (residues 458 to 548) regions (Figure 4A), respectively. As shown in Figure 4C, deletion of the C-terminal region induced RopGEF activity to the degree similar to or slightly stronger than that of the DUF315 domain, whereas removing the N-terminal region partially activated GEF for ROP1. These results suggest that the inhibitory regulatory element(s) primarily resides in the C-terminal region, while the N-terminal region may assist in the inhibition of RopGEF activity mediated by the C-terminal region.
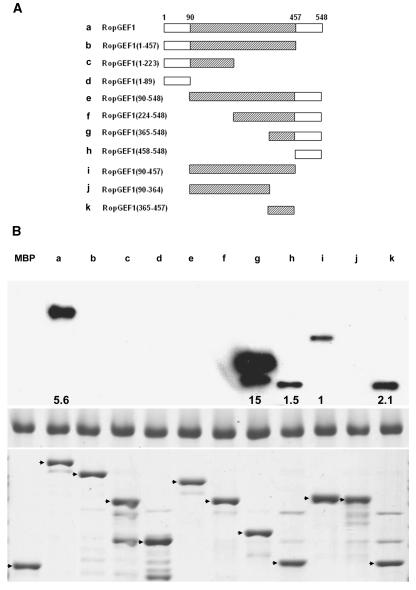
To test whether the C-terminal region of RopGEF1 confers the inhibitory effect by interacting with part of the GEF catalytic domain, we tested the ability of the C-terminal variable region (residues 458 to 548; fragment h, Figure 5A) to bind various truncated fragments of RopGEF1 in vitro. The DUF315 domain can be divided into three subdomains separated by short stretches of variable sequences (Figure 1), which was the basis of our design for the truncated fragment of RopGEF1. As shown in Figure 5, the C-terminal region interacts with RopGEF1 at its truncated fragments in a complex manner. First, it strongly interacted with the full-length RopGEF1 as well as the C-terminal fragment g (residues 365 to 548) that contains the C-terminal part of the DUF315 domain and the C-terminal variable region. Second, it weakly interacted with the following truncated fragments: fragment h (residues 458 to 548), fragment i (residues 90 to 457), and fragment k (residues 365 to 457). Third, it did not bind to any of the following truncated fragments: fragment b through fragment f and fragment j (Figure 5). These results demonstrate that the C-terminal region is capable of strong direct binding to a domain or motif located in the C-terminal part of RopGEF1 (residues 365 to 548; i.e., fragment g) but does not bind to subdomains S1 and S2 of the DUF315 domain (fragment j). The C-terminal interactive domain/motif seems to cover part of subdomain S3 of the DUF315 domain (fragment k) and part of the C-terminal variable region itself (fragment h) because either one alone bound weakly to the C-terminal variable region. The strong interaction is inhibited by the presence of subdomains S1 and/or S2 because fragment e (residues 90 to 548) and fragment f (residues 224 to 548) had no interaction with the C-terminal variable region. Moreover, this inhibition is released by the N-terminal variable region based on the fact that the full-length RopGEF1 is able to interact. These data are consistent with the observation (Figure 4C) that the removal of the C-terminal variable region completely releases the inhibition of RopGEF1 activity, whereas the deletion of the N-terminal variable region partially releases the inhibition (i.e., the N-terminal region only assists the inhibition mediated by the C-terminal region).
Figure 5.
In Vitro Interactions between the C-Terminal Variable Region and Various RopGEF1 Deletion Mutants.
(A) Diagram of truncated fragments of RopGEF1 used in the pull-down assays. Hatched boxes indicate the conserved DUF315 domain.
(B) In vitro binding assay for the interaction of the C-terminal variable region (amino acids 458 to 548) with various truncated fragments of RopGEF1. The top panel shows the binding of MBP fusion proteins of various RopGEF deletion mutants to the His-tagged C-terminal variable region of RopGEF1. The numbers at the bottom of each band indicate relative signal intensity standardized with the loading control (see Methods) for the His-tagged C-terminal region of RopGEF1, which is shown in the middle panel. The bottom panel shows purified MBP fusion proteins for various truncated fragments of RopGEF1 used for the pull-down assay. Arrows indicate protein bands of various truncated fragments of RopGEF1.
To test whether the binding of the C-terminal variable region to the DUF315 domain physically blocks GEF activity, we mixed the DUF315 domain with fragment g, k, or h. Neither fragment had a significant effect on the GEF activity of the DUF315 domain (data not shown). This suggests that the autoinhibitory mechanism most likely requires conformational changes that could result from an intramolecular interaction between the C-terminal region and the DUF315 domain.
Induction of Pollen Tube Growth Depolarization by RopGEF1 Overexpression Is Dependent upon ROP1 Activation
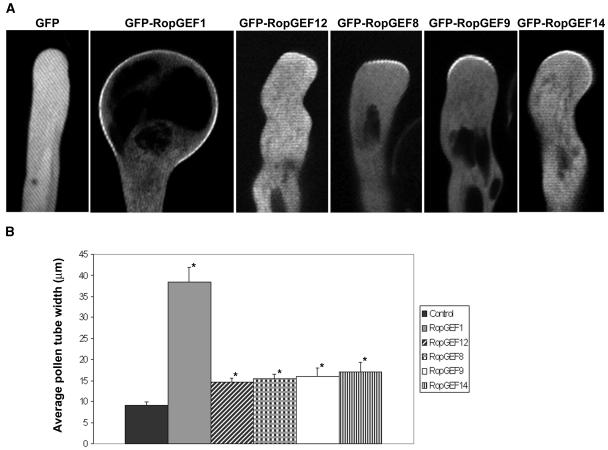
We next investigated whether RopGEF1 functionally activates ROP1 in the control of polarized growth in pollen tubes. Available single T-DNA insertional knockout mutants for RopGEF1, RopGEF9, RopGEF12, or RopGEF14 did not show any measurable defects in the growth of cultured pollen tubes or in pollination, suggesting that two or more of these RopGEFs expressed in pollen are functionally redundant. Thus, we resorted to a gain-of-function approach by overexpressing RopGEFs in tobacco (Nicotiana tabacum) pollen tubes. Previously, we found that expression of a constitutively active rop1 mutant (CA-rop1) caused severe depolarization of pollen tube growth, leading to the formation of bulbous tubes, while overexpression of wild-type ROP1 induced less severe depolarization of growth in pollen tubes (tip swelling) both in tobacco and Arabidopsis (Li et al., 1999; Gu et al., 2003). To facilitate monitoring of RopGEF expression and RopGEF subcellular localization, we tagged RopGEFs with green fluorescent protein (GFP). As shown in Figure 6, transient overexpression of each of the five RopGEFs induced growth depolarization in tobacco pollen tubes, with RopGEF1 causing the most severe phenotype. RopGEF1 overexpression produced a bulbous pollen tube, a phenotype similar to that induced by CA-rop1 expression. The RopGEF1 overexpression phenotype is similar to that of a tomato RopGEF/KPP, which interacts with the kinase domain of Le PRK1 (Kaothien et al., 2005). For every RopGEF examined, the GFP-RopGEF fusion gene and the RopGEF gene alone produced identical or nearly identical phenotypes when overexpressed under the pollen-specific LAT52 promoter, suggesting that GFP-RopGEF fusion genes are fully functional (data not shown).
Figure 6.
Subcellular Localization of GFP-RopGEFs and Their Overexpression Phenotypes in Tobacco Pollen Tubes.
To assess the function of five Arabidopsis RopGEFs expressed in pollen, they were tagged with GFP and transiently expressed under the LAT52 promoter in tobacco pollen grains. The transformed tubes indicated by GFP expression were visualized using confocal microscopy as described in Methods.
(A) Pollen tubes expressing different LAT52:GFP-RopGEFs. LAT52:GFP was used as control. All images are midplane confocal optical sections.
(B) Quantitative analysis of the phenotype of pollen tubes expressing different LAT52:GFP-RopGEFs. The degree of depolarized growth was determined by measuring the diameter of the widest region of the tube. Asterisks indicate a significant difference from control at the same data point (P < 0.05; t test). Approxiamately 25 to 30 pollen tubes were used for the analysis. Error bars indicate sd.
To assess subcellular localization of RopGEFs, tobacco pollen tubes expressing GFP-RopGEF fusions were examined using confocal microscopy. As shown in Figure 6, GFP alone was distributed evenly in the pollen tube cytoplasm. GFP-RopGEF1 was highly restricted to the apical region of the PM, a pattern similar to the localization of activated ROP1 (Hwang et al., 2005). However, GFP-RopGEF12 was barely detected in the pollen tube PM. Interestingly, GFP-RopGEF8, GFP-RopGEF9, and GFP-RopGEF14 were localized to the apical region of the pollen tube PM. These results indicate that different RopGEFs are localized differentially in pollen tubes and that ROP1 signaling may be regulated by multiple RopGEFs in pollen tubes. Furthermore, we selectively overexpressed RopGEF1 in Arabidopsis pollen, and multiple transgenic lines of RopGEF1 overexpression produced bulbous pollen tubes similar to that of ROP1 overexpression (data not shown). The overexpression phenotype and localization pattern of these RopGEFs resemble those of ROP1, indicating that they may be the activators of ROP1 in the control of polarized growth.
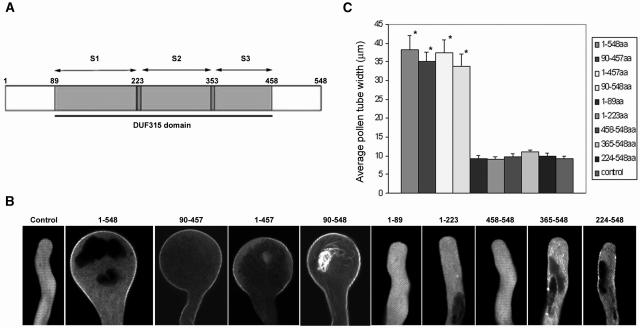
Overexpression of RopGEF1 induced the most severe depolarization of pollen tubes, and our biochemical analysis of RopGEFs mostly came from the use of RopGEF1; therefore, we focused on RopGEF1 for testing its functional relationship with ROP1. We assessed whether there was a tight correlation between RopGEF activity and the ability to cause depolarized growth among various truncated fragments of RopGEF1. Various truncated fragments of RopGEF1 were fused with GFP and transiently expressed in tobacco pollen tubes. As shown in Figure 7, all fragments retaining the intact DUF315 domain and RopGEF activity caused growth depolarization when overexpressed. The severity of growth depolarization is similar to that of full-length RopGEF1 or CA-rop1. By contrast, none of the fragments that lack the intact DUF315 domain and/or RopGEF activity induced depolarization. These results strongly suggest that the RopGEF activity of RopGEF1 is required for the function of RopGEF1 in the regulation of pollen tube growth, implying that ROP1 activation by RopGEF1 is critical for polarized growth in pollen tubes.
Figure 7.
Subcellular Localization and Overexpression Phenotype for GFP-Tagged Truncated Fragments of RopGEF1 in Tobacco Pollen Tubes.
Various GFP-tagged truncated fragments of RopGEF1 shown in (A) were transiently expressed under the LAT52 promoter in tobacco pollen grains as described in Figure 6. Approximately 5 h after bombardment, GFP localization in transformed pollen tubes was analyzed under a confocal microscope.
(A) Diagram of truncated fragments of RopGEF1 used in the analysis of subcellular localization. Shaded boxes indicate the conserved DUF315 domain. Numbers above shaded boxes indicate amino acid residues. The conserved subdomains within the DUF315 domain are indicated by arrows.
(B) Representative pollen tubes expressing different GFP-tagged truncated fragments of RopGEF1 under the control of the LAT52 promoter. LAT52:GFP was used as control. All images are midplane confocal optical sections.
(C) Quantitative analysis of the phenotype of pollen tubes expressing various GFP-tagged truncated fragments of RopGEF1. Asterisks indicate a significant difference from control at the same data point (P < 0.05; t test). LAT52:GFP was used as control. Approximately 25 to 30 pollen tubes were used for the analysis. Error bars indicate sd. aa, amino acids.
To assess whether there was a correlation between RopGEF activity and localization pattern among various truncated fragments of RopGEF1, we examined the localization of GFP-tagged RopGEF1 fragments. As shown in Figure 7B, various RopGEF1 fragments showed differential localization patterns. First, the fragment (residues 1 to 457) with the C-terminal variable region (residues 458 to 548) deleted was the only one that showed the exact same tip PM localization pattern as the full-length GFP-RopGEF1. Second, both the DUF315 domain and the fragment (residues 90 to 548) with the N-terminal variable region (residues 1 to 89) deleted expanded their PM localization all over the pollen tube. Third, all fragments that lack the intact DUF315 domain displayed similar localization as the GFP control except that the fragment (residues 224 to 548) showed PM localization on the side of pollen tubes but not at the extreme tip. Thus, the N-terminal variable region seems to contain a regulatory element that restricts RopGEF1 localization to the exact tip region of the PM. The DUF315 domain is not only required for its activation of ROP1 but also is essential for the proper targeting of RopGEF1 to the PM where ROP1 is located.
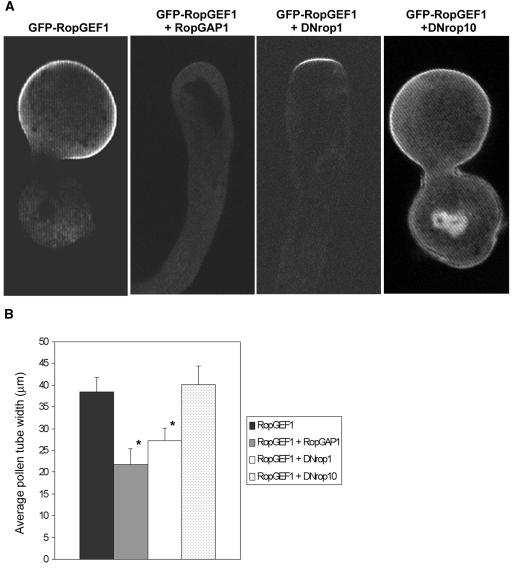
To further test whether RopGEF1 activates ROP1 in vivo, we examined the effect of RopGAP1 and DN-rop1 on the pollen tube phenotype induced by RopGEF1 overexpression. We reasoned that if RopGEF1 acts as a ROP1 activator, the RopGEF1-induced growth depolarization phenotype should be suppressed by RopGAP1 or DN-rop1, which inhibits ROP1 activation by promoting GTP hydrolysis or by trapping ROP1 activator, respectively. RopGAP1 coexpression suppressed GFP-RopGEF1–induced growth depolarization by reducing the width of tubes (from 38.4 to 21.8 μm; n = 22). Furthermore, RopGAP1 dramatically reduced GFP-RopGEF1 localization to the PM (81%, n = 22; Figure 8). DN-rop1 coexpression also reduced the PM localization of GFP-RopGEF1 and the tube width from 38.4 to 27.2 μm (n = 19), although DN-rop1 was not as effective as RopGAP1. To test whether the effect is specific for ROP1, we coexpressed RopGEF1 with a dominant-negative mutant of ROP10, which has been shown to be functionally distinct from ROP1 (Gu et al., 2005). Indeed coexpression of DN-rop10 had no effect on RopGEF1 overexpression phenotype or localization (87%, n = 25). Taken together, our data strongly suggest that RopGEF1 acts as a ROP1 activator by stimulating its GDP-GTP exchange.
Figure 8.
RopGEF1 Localization and Overexpression Phenotype Are Dependent on ROP1 Activation.
(A) The localization of GFP-RopGEF1 to the PM in pollen tubes was inhibited by coexpression of RopGAP1 or DN-rop1. Control shows typical GFP-RopGEF1 localization and tip morphology in tobacco pollen tubes transiently expressing LAT52:GFP-RopGEF1. DN-rop1, RopGAP1, or DN-rop10 shows tubes coexpressing LAT52:GFP-RopGEF1 with LAT52:DN-rop1, LAT52:RopGAP1, or LAT52:DN-rop10, respectively. All images are midplane confocal optical sections.
(B) Quantitative analysis of the phenotype of pollen tubes coexpressing LAT52:GFP-RopGEF1 with LAT52:DN-rop1, LAT52:RopGAP1, or LAT52:DN-rop10. Asterisks indicate a significant difference from control at the same data point (P < 0.05; t test). Approximately 25 pollen tubes were analyzed for each treatment. Error bars indicate sd.
DISCUSSION
In our search for new regulators of ROP GTPases, we have identified a plant-specific novel family of Rho GEFs, termed RopGEFs, confirming a recent report independently showing that the catalytic activity of RopGEFs resides in the conserved DUF315/PRONE domain (Berken et al., 2005). Importantly, our work has provided experimental evidence that a member of the RopGEF family, RopGEF1, acts as an activator of ROP1 in the control of polarized growth in pollen tubes. Furthermore, our data have revealed an autoinhibitory mechanism by which the variable regions of RopGEF1 regulate its RopGEF activity. Our results suggest that this autoinhibition of RopGEF1 is released in vivo probably by another upstream signaling molecule that controls the pollen tube growth. These findings are particularly interesting, given a recent report showing that a tomato homolog of RopGEF/KPP induces a similar pollen tube phenotype as RopGEF1 when overexpressed and directly interacts with a pollen-specific tomato RLK (Kaothien et al., 2005).
RopGEFs: A Novel Family of Plant-Specific Rho GEFs
In this study, we report both in vitro biochemical studies and in vivo functional analysis showing that RopGEFs belong to a novel class of Rho GEFs that activate the plant-specific subfamily of Rho GTPases, ROPs. Our in vitro biochemical studies are consistent with those of Berken et al. (2005). RopGEFs share no sequence homology with any Rho GEFs identified to date and appear to be absent from fungi and animals. Thus, Berken et al. named the DUF315 domain PRONE. Berken's work and this report together have established a plant-specific novel family of Rho GEFs. The demonstration of a fourth class of structurally distinct Rho GEFs is in line with the fact that Rho family GTPases are versatile signaling switches that are capable of coupling to a variety of upstream signaling mechanisms. It would not be surprising if future studies reveal additional classes of Rho GEFs in plants or other organisms.
What is the significance of a new family of Rho GEFs that specifically function in plants? In fungi and animals, the Rho family is divided into several subfamilies, including Rho, Cdc42, and Rac. Interestingly, only a single Rho subfamily, Rop, has been found in plants, and Rop appears to have evolved prior to the splitting off of Rho Cdc42/Rac. SPIKE1, a homolog of the Docker family Rho GEFs, may have been the ancestral plant RopGEFs that could also activate ROPs (Qiu et al., 2002), although its GEF activity has yet to be demonstrated. The novel RopGEFs might have evolved specifically in plants to adapt to some specific structural features unique to the Rop subfamily of Rho GTPases. This is supported by the observation that the DUF315/PRONE domain of RopGEF1 appears to have no GEF activity toward mammalian Rac1 (Berken et al., 2005).
Another possible reason for the creation of a plant-specific subfamily of Rho GEFs may be the need for a plant-specific signaling mechanism upstream of RopGEFs. In mammals, Rho GEFs can be activated by G-protein-coupled receptors (GPCRs) and Tyr kinase receptors (Kjoller and Hall, 1999). GPCRs constitute the largest family of cell surface receptors, with >1000 members, and they perceive a diverse array of extracellular signals through their N-terminal ligand recognition and binding (Ellis and Miles, 2001). Lack of Tyr kinase receptors and likely scarcity of GPCRs in plants may explain the lack of conventional Rho GEFs. Instead, plants evolved a larger number of RLKs, and many of these are known to control various developmental processes and to perceive hormonal and environmental signals. It is tantalizing to speculate that ROPs act downstream of these cell surface receptors that are predominant in plants (see below).
RopGEF1 Acts as a ROP1 Activator in the Control of Pollen Tube Growth
We have provided evidence that RopGEF1 functionally acts upstream of ROP1 in the control of polar growth in pollen tubes. First, overexpression of either full-length or DUF315 domain of Arabidopsis RopGEF1 induced growth depolarization in tobacco pollen tubes as did CA-rop1. This finding is crucial for confirming a functional relationship between RopGEF1 and ROP1 because the full-length RopGEF1 has no GEF activity for ROP1 in vitro. Thus, the activation of RopGEF1 in vivo is important for its function. Second, deletion mutant analysis shows that the ability of the RopGEF1 fragments to induce depolarized growth is tightly correlated with the presence of the complete DUF315 domain (or RopGEF activity). Third, the physiological effect of RopGEF1 (i.e., the RopGEF1 overexpression phenotype) is dependent on the activation of ROP1 in vivo. Coexpression of RopGAP1 or DN-rop1 partially suppressed the depolarization of pollen tube growth caused by overexpression of RopGEF1. Finally, RopGEF1 and several other RopGEFs expressed in pollen are localized to the apical region of the pollen tube PM, similar to the localization of ROP1 and activated ROP1. These in vivo functional studies, together with the biochemical analysis showing that both RopGEF1 and RopGEF9 act as ROP1 GEFs, suggest that RopGEF1 and probably other functionally redundant RopGEFs directly activate ROP1 in the control of pollen tube tip growth. Nonetheless, analysis of multiple loss-of-function mutants for the RopGEFs expressed in pollen will be necessary to confirm this conclusion.
RopGEFs Are Regulated by an Autoinhibitory Regulatory Mechanism
In this report, we demonstrate an autoinhibitory regulation mechanism for RopGEF1. First, the survey of GEF activity for various truncated fragments of RopGEF1 reveals an inhibitory regulatory element(s) present in both C-terminal and N-terminal variable regions outside of the conserved central catalytic domain of RopGEF1. Second, the C-terminal variable region of RopGEF1 interacts strongly with a truncated RopGEF1 containing the C-terminal region and the subdomain S3 of the DUF315 domain. We propose that the C-terminal part of RopGEF1 can physically interact with the DUF315 domain, and this interaction blocks its GEF activity. Third, overexpression of full-length RopGEF1in tobacco pollen tubes induced cell depolarization similar to that induced by the DUF315 domain of RopGEF1. This indicates the existence of an unknown mechanism to release this autoinhibition toward RopGEF1 in pollen tubes, and this unknown mechanism is expected to be an upstream signaling molecule that activates RopGEF1 in pollen tubes.
The autoinhibition appears to involve an intramolecular inhibition mechanism. We have shown that the C-terminal regulatory region can physically associate with the central DUF315 domain. Thus, this interaction could physically block the access of RopGEF1 to ROP1, leading to the inhibition of the nucleotide exchange activity. Alternatively, the intramolecular interaction may produce an inactive RopGEF1 confirmation. To distinguish these two possibilities, we tested whether adding the inhibitory C-terminal region could affect the GEF activity of the DUF315 domain. The GEF activity of DUF315 domain was not poisoned by adding fragment g (residues 365 to 548), fragment k (residues 365 to 457), or fragment h (residues 458 to 548) (data not shown). Thus, the autoinhibition is not simply due to the physical blockage of the catalytic site by the regulatory region but likely requires the intramolecular interaction to generate a three-dimensional state that is inactive. In this case, an unknown activator of RopGEF1 would be required to alter this autoinhibitory state. Since RopGEF1 interacts with GTP-bound ROP1 in vitro, it is possible that the interaction between RopGEF1 and GTP-ROP1 (or other ROPs) can release this intramolecular autoinhibition of RopGEF1. This mechanism would be similar to that regulating the Ras GEF Sos. It has been demonstrated that Ras itself, when activated by the Ras GEF Sos and bound to GTP, can enhance the GEF activity of Sos by binding to a distinct site on the Sos molecule (Margarit et al., 2003). As discussed below, RLKs are other potential regulators of RopGEFs that could release the autoinhibition mediated by the C-terminal regulatory region.
RopGEFs May Link RLKs to Intracellular Signaling
RLKs are predominant transmembrane receptors in plants. They constitute a large superfamily (>600 RLKs in Arabidopsis alone), and many RLKs have been shown to be critical for plant growth, development, and hormone responses (Shiu and Bleecker, 2001a, 2001b; Dievart and Clark, 2003; Morris and Walker, 2003). Although several RLK-interacting proteins have been identified (Fujita et al., 2003; Hattan et al., 2004; Tarutani et al., 2004), the molecular basis for RLK-mediated intracellular signaling remains elusive. ROPs seem to form complexes with CLAVATA1-KAPP in Arabidopsis and Le PRK-KAPP in tomato (Trotochaud et al., 1999; Wengier et al., 2003). Interestingly, a tomato homolog of RopGEF/KPP was recently shown to interact with the kinase domain of tomato pollen–specific RLKs (Le PRK1 and Le PRK2) (Kaothien et al., 2005). KPP–Le PRK2 in pollen has been demonstrated by in vivo coimmunoprecipitation experiments (Kaothien et al., 2005). Furthermore, RopGEF/KPP overexpression in tobacco and tomato pollen induces pollen tube growth depolarization, as does Arabidopsis RopGEF1, suggesting that the tomato RopGEF/KPP may be a functional ortholog of Arabidopsis RopGEF1. The Arabidopsis genome encodes six homologs of Le PRK1 that are pollen specific (Kim et al., 2002; Becker et al., 2003; Honys and Twell, 2003). Future studies will determine whether these pollen RLKs indeed act upstream of RopGEF1 to activate ROP1 GTPase signaling to polarized pollen tube growth. Nonetheless, these observations raise an intriguing possibility that the RopGEF family members may be the elusive link between receptor kinases and intracellular signaling in plants.
METHODS
Yeast Two-Hybrid Screen
The dominant-negative ROP1 (D121A) and isoprenylation-defective mutations (C188S) were generated by site-directed mutagenesis as described previously (Li et al., 1999). The D121A/C188S double mutant (DN2S) was fused to the GAL4 DNA binding domain in bait vector pAS2. pAS2-DN2S was introduced into Y190 by electroporation. One transformant was designated as Y190-pAS2-DN2S and used for the transformation with 10 μg of the Arabidopsis thaliana cDNA library. All transformants were plated on SC-Trp-Leu-His + 100 mM 3-aminotriazole (Sigma-Aldrich) plates. Approximately 3 million transformants were obtained 1 week after transformation at 30°C. A filter assay was performed to test for the β-galactosidase activity on all transformants. Putative positives were traced back to plates using filter papers and grown in a fresh selective medium (SC-Trp-Leu-His) for 3 d at 30°C.
A total of 72 putative positives were recovered and confirmed by their ability to grow on SC-Trp-Leu-His and for β-galactosidase activity using filter assays for a second time. These putative positives were grown on SC-Leu containing 2.5 μg/mL cycloheximide to select for cells only containing prey vector. Yeast DNA was isolated from yeast cells according to yeast DNA preparation protocols (Hoffman, 1997) and transformed into Escherichia coli DH5α. Only 41 of the 72 putative clones were recovered. All plasmid DNAs were sequenced using the T7 primer.
Database Search, Sequence Alignment, and Phylogenetic Analysis
DNA sequences obtained from sequencing were used to identify Arabidopsis genes using BLAST-N against both NCBI (www.ncbi.nlm.nih.gov/blast) and TAIR databases (www.arabidopsis.org/Blast). Predicted amino acid sequences of RopGEFs were aligned using ClustalW (www.ebi.ac.uk/clustalw). Aligned sequences were processed using Boxshade (www.ch.embnet.org/software/BOX_form.html) in a fraction of 0.5 and output as new RTF format. Data generated from alignment was loaded to the Phylodendron Phylogenetic tree printer (iubio.bio.indiana.edu/treeapp) to reconstruct the rooted neighbor-joining phylogenetic tree by the distance-based method.
DNA Manipulation and Plasmid Construction
cDNAs for RopGEF1, RopGEF8, RopGEF9, RopGEF12, and RopGEF14 were amplified using PCR primers covering the predicted RopGEF coding sequences. All plasmids used for transient expression in pollen were constructed in the pLAT52:GFP vector or in the pLAT52 vector as described previously (Wu et al., 2001). GST-Rac1 and His-SopE were gifts from Michael Rosen (UT Southwestern Medical School, Dallas, TX). Various deletion mutants of RopGEF1 were amplified using PCR primers covering indicated amino acid residues in Figure7 and subcloned into pGEM-Teasy vector (Qiagen). The resulting constructs were then sequence confirmed and subcloned into either pLAT52:GFP or pMALC2 (MBP) or pET-30a (Novagen) fusion vectors.
Reverse Transcription and PCR Analysis of RopGEF Transcripts
Total RNA was isolated from Arabidopsis mature pollen using the RNeasy plant mini kit (Qiagen). Reverse transcription and PCR amplification were performed as described previously (Li et al., 1998). For all RopGEFs genes, 35 cycles of PCR amplification (94°C for 30 s, 54°C for 30 s, and 72°C for 1 min) were performed using the primers listed in Table 1. ACTIN3 was used as PCR amplification and loading controls. Ten microliters of each PCR product was loaded onto a 0.8% agarose gel to visualize the amplified cDNAs.
Table 1.
Primers Used for RT-PCR Analysis of RopGEF Expression in Mature Arabidopsis Pollen
| Gene | Forward Primer (5′/3′) | Reverse Primer (5′/3′) |
|---|---|---|
| RopGEF1 | GAAGATCTATGGGGAGCTTATCTTCT | CGGGTACCTTAATCTCTTTCCGGCGT |
| RopGEF2 | GGATCCCGATGGAGAATTTGCCAAATCACG | GGAATTCTCATTCTTCTCCTCTCAT |
| RopGEF3 | GAATTCATCCCTAGATGTCAGAATC | TTCACTACCTCTCATGG |
| RopGEF4 | ATGGAGAGTTCTTCGAATTCCGACC | AATCTCTACCACCACCACCCG |
| RopGEF5 | GGATCCCGATGGAGAATTTAGTGAAGAGC | GGAATTCTTAAGAGACAGTGTACTT |
| RopGEF6 | ATGGAGGATAATAGCTGTATCGGG | CCAATTATCTCCGGGGTTGA |
| RopGEF7 | GGATCCCGATGGATGGTTCGTCGGAA | GGAATTCGTCAAATCCCAGGATCAA |
| RopGEF8 | GGATCCATGGTTGCAGCGTTGGAA | GGTACCCTGCAGTTAATGCCTATCTTTGGG |
| RopGEF9 | GGATCCATGGTTCCATCGTTGGAA | GGTACCTCAATGCCTATCTTTAGG |
| RopGEF10 | ATGTTCGATGGTCGGAACTCT | TCAGTGTCTGTCACTAGGGC |
| RopGEF11 | ATGGAGCAAGAACAAGAGACT | GGTACCTCAGGAGTATCTTGCGGT |
| RopGEF12 | GAAGATCTATGGTTCGTGCTTCGGAA | GGAATTCTCAATGCCGTGCCGTTGG |
| RopGEF13 | ATGGTGAAAGCGAGTGAGAAAG | AAGGAATGTTGGAGACAAGATC |
| RopGEF14 | GGATCCCGATGATGCTGATGAGAAGA | CGATATCTCAAGGAGAAGTATCAGAAG |
Protein Expression and Purification
For protein expression in E. coli, RopGEFs were fused with MBP, and ROP1 was fused with glutathione S-transferase (GST) as described previously (Wu et al., 2001). Fusion proteins were expressed at 30°C for 4 h after induction with 1 mM isopropyl-β-d-thiogalactopyranoside. Cell cultures were spinned down at 5000 rpm for 10 min. For GST fusion proteins and MBP fusion proteins, cell pellets were resuspended in a binding buffer (20 mM Tris, pH 7.4, 200 mM NaCl, and 1 mM EDTA) and sonicated using 15-s pulses eight times. The supernatant was obtained by centrifugation at 8500 rpm for 30 min. The supernatant was mixed with glutathione-agarose beads (Sigma-Aldrich) or amylose beads (Biolabs) for GST fusion proteins and MBP fusion proteins, respectively. After 2 h of incubation, beads were washed with the binding buffer and eluted using maltose (10 mM) or glutathione (30 mM), respectively. For His-tagged proteins, cell pellets were resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole, pH 8.0). After sonication and centrifugation as described above, the supernatant was mixed with Ni-NTA beads (Qiagen) for 2 h. The mixture was then washed with a wash buffer (50 mM NaH2PO4, 300 mM NaCl, and 20 mM imidazole, pH 8.0). Purified His-tagged proteins were eluted using an elution buffer (50 mM NaH2PO4, 300 mM NaCl, and 250 mM imidazole, pH 8.0). MBP was cleaved using Factor Xa kits (Novagen), and His-tag was removed using Thrombin kits (Novagen) following the manufacturer's procedures. Proteins for GEF assays were dialyzed against the GEF assay buffer (see below).
Guanine Nucleotide Exchange Assay
Rac1 or ROP1 was preloaded with GDP as described (Nomanbhoy et al., 1996). Briefly, a 5-mL overnight culture of E. coli expressing GST-Rac1 or GST-ROP1 was used to inoculate 250 mL of YTA medium (16 g/L tryptone, 10g/L yeast extract, and 5 g/L NaCl, pH 7.0). Protein expression was induced at 30°C for 4 h with 1 mM isopropyl-β-d-thiogalactopyranoside after 2 h of growth at 37°C. Cells were harvested and resuspended in buffer A (20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM MgCl2, and 100 μM GDP). After sonication using 15-s pulses eight times, the supernatant was obtained by centrifugation at 8500 rpm for 30 min. The supernatant was mixed with glutathione-agarose beads for 2 h. The mixture was applied to a column and washed with 10 mL of buffer B (20 mM Tris, pH 8.0, 100 mM NaCl, 5 mM MgCl2, 2.5 mM CaCl2, and 40 μM GDP). To cleave the GST tag, 5 μL of 1 unit/μL thrombin was added to the mixture of beads. GDP-bound Rac1 or ROP1 was dialyzed against a nucleotide exchange buffer (20 mM Tris/HCl, pH 7.4, 50 mM NaCl, 2 mM MgCl2, and 1 mM DTT). Nucelotide exchange was monitored as the increase in relative fluorescence of a fluorescent GDP analog (mant-GDP) upon binding to GTPase proteins. Before the addition of 100 nM SopE or RopGEFs, 1 μM of GDP-ROP1 or Rac1 was incubated in the guanine nucleotide exchange buffer containing 200 nM mant-GDP (Molecular Probes). After equilibration, fluorescence was measured every second for 600 s using a spectrofluorometer with λexcitation at 360 nm and λemission at 440 nm. Spectral resolution was 5 nm for both excitation and emission paths. The Kobs values (pseudo-first-order rate constant) were obtained by monophasic-exponential fits using PeakFit v4.01 (Jandel Scientific Software).
In Vitro Protein–Protein Interaction Assays
To demonstrate direct interaction between RopGEFs and ROP1, we used MBP-RopGEFs and GST-ROP1 fusion proteins fused with GST for pull-down assays as described previously (Wu et al., 2000). GST-ROP1 was preloaded with GDP or GTP in a nucleotide loading buffer containing 3 mM corresponding nucleotide, 25 mM Tris-HCl, pH 7.5, 1 mM DTT, 10 mg/mL BSA, and 5 mM EDTA. Approximately 10 μg of GST-ROP1 fusion proteins were bound to glutathione-conjugated agarose beads, and similar amounts of MBP-RopGEF fusion proteins were used in each assay. Beads containing GST-ROP1 fusion proteins were incubated with MBP-RopGEF fusion proteins in an interaction buffer containing 20 mM HEPES, pH 7.4, 5 mM MgCl2, 1 mM DTT, 0.1% Triton X-100, and 1 mM EDTA for 2 h. After binding, beads containing GST-ROP1 and bound MBP-RopGEF were washed extensively to remove unbound proteins. The MBP fusion proteins were detected using a polyclonal antibody against MBP (New England Biolabs) and the BM chemiluminescence protein gel blot kit (Boehringer Mannheim). To standardize the interaction signal, films were scanned and the total signal intensity was measured using MetaMorph software (version 4.5; Universal Imaging). The relative intensity was calculated as a ratio of intensity from the protein gel blot signals to protein amount from the standardized GST-ROP1 loading control. To standardize the GST-ROP1 loading, protein gels were scanned and the total signal intensity was measured using MetaMorph software. The lowest amount of GST-ROP1 was arbitrary designated as one, and the rest of loading was calculated as a ratio versus the lowest amount of GST-ROP1.
Particle Bombardment–Mediated Transient Expression in Tobacco Pollen
Mature pollen grains collected from tobacco (Nicotiana tabacum) plants were used for transient expression using a particle bombardment procedure as described previously (Fu et al., 2001). Routinely, 0.5 mg of gold particles were coated with 0.5 μg of LAT52:GFP-RopGEF DNA or a mixture of 0.5 μg of LAT52:GFP-RopGEFs DNA with 0.5 μg of other fusion constructs. The pollen grains were incubated for 5 h before observation under an inverted microscope (model TE300; Nikon) as described previously (Fu et al., 2001) or a confocal microscope as described in the section following.
Analyses of RopGEFs Localization and Overexpression Phenotype
Approximately 5 h after bombardment, tubes expressing LAT52:GFP-RopGEFs were visualized and analyzed using laser scanning confocal microscopy as described previously (Fu et al., 2001). The degree of depolarized growth was determined by measuring the diameter of the widest region of the tube. To determine subcellular localization, tubes expressing GFP fusion proteins were analyzed using laser scanning confocal microscopy under a Nikon OPTIPHOT upright microscope equipped with a Bio-Rad MRC 600 confocal laser scanning device. One-micrometer optical sections were scanned and captured using Comos software. Confocal images were analyzed using Metamorph 4.5 software and processed using Photoshop 5.5 (Adobe Systems).
Generation of RopGEF1-Overexpressing Lines and Isolation of T-DNA Insertional Mutants
To generate RopGEF1-overexpressing binary constructs, the full-length RopGEF1 cDNA sequence was cloned into a pC1300LAT52 vector derived from pCAMBIA1300 (Cambia). The construct was introduced into Agrobacterium tumefaciens GV3101 by electroporation and transformed into Arabidopsis ecotype Columbia. T2 homozygous plants were selected for analysis. The screen for RopGEF T-DNA knockout lines from SIGNAL collection (http://signal.salk.edu/cgi_bin/tdnaexpress/) was based on a combination of database searches and PCR amplification of T-DNA flanking regions. For T-DNA lines identified from the SIGNAL collection, seeds were obtained from the Arabidopsis Knockout Facility (www.biotech.wisc.edu/arabidopsis/). PCR reactions were performed to identify single plants for the T-DNA insertion.
Accession Numbers
GenBank accession numbers for various annotated Arabidopsis RopGEFs are as follows: RopGEF1, NP_195556; RopGEF2, NP_171676; RopGEF3, NP_191956; RopGEF4, NP_182113; RopGEF5, NP_196213; RopGEF6, AAV84500; RopGEF7, NP_195821; RopGEF8, NP_189105; RopGEF9, CAB78366; RopGEF10, NP_197457; RopGEF11, NP_175634; RopGEF12, NP_178104; RopGEF13, NP_188234; RopGEF14, BAD94650.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Title Comparison of Predicted Amino Acid Sequences of Plant RopGEFs.
Supplemental Figure 2. Phylogenetic Tree of Plant RopGEFs.
Supplementary Material
Acknowledgments
We thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants, the Ohio State University Arabidopsis Biological Resources Center for seeds, Dimitri Niks and Amarasinghe Gaya for assistance with the GEF assay, and Michael Rosen for providing His-SopE and GST-Rac1 constructs. This work was supported by National Science Foundation grants to Z.Y. (MCB0111082 and IBN-0115078).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Zhenbiao Yang (zhenbiao.yang@ucr.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.036434.
References
- Baxter-Burrell, A., Yang, Z., Springer, P.S., and Bailey-Serres, J. (2002). RopGAP4-dependent Rop GTPase rheostat control of Arabidopsis oxygen deprivation tolerance. Science 296 2026–2028. [DOI] [PubMed] [Google Scholar]
- Becker, J.D., Boavida, L.C., Carneiro, J., Haury, M., and Feijó, J.A. (2003). Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol. 133 713–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berken, A., Thomas, C., and Wittinghofer, A. (2005). A new family of RhoGEFs activates the Rop molecular switch in plants. Nature 436 1176–1180. [DOI] [PubMed] [Google Scholar]
- Bischoff, F., Vahlkamp, L., Molendijk, A., and Palme, K. (2000). Localization of AtROP4 and AtROP6 and interaction with the guanine nucleotide dissociation inhibitor AtRhoGDI1 from Arabidopsis. Plant Mol. Biol. 42 515–530. [DOI] [PubMed] [Google Scholar]
- Brugnera, E., Haney, L., Grimsley, C., Lu, M., Walk, S.F., Tosello-Trampont, A.C., Macara, I.G., Madhani, H., Fink, G.R., and Ravichandran, K.S. (2002). Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat. Cell Biol. 4 574–582. [DOI] [PubMed] [Google Scholar]
- Buchwald, G., Friebel, A., Galan, J.E., Hardt, W.D., Wittinghofer, A., and Scheffzek, K. (2002). Structural basis for the reversible activation of a Rho protein by the bacterial toxin SopE. EMBO J. 21 3286–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerione, R.A., and Zheng, Y. (1996). The Dbl family of oncogenes. Curr. Opin. Cell Biol. 8 216–222. [DOI] [PubMed] [Google Scholar]
- Cote, J.F., and Vuori, K. (2002). Identification of an evolutionarily conserved superfamily of DOCK180-related proteins with guanine nucleotide exchange activity. J. Cell Sci. 115 4901–4913. [DOI] [PubMed] [Google Scholar]
- Dievart, A., and Clark, S.E. (2003). Using mutant alleles to determine the structure and function of leucine-rich repeat receptor-like kinases. Curr. Opin. Plant Biol. 6 507–516. [DOI] [PubMed] [Google Scholar]
- Ellis, B.E., and Miles, G.P. (2001). Plant biology. One for all? Science 292 2022–2023. [DOI] [PubMed] [Google Scholar]
- Fu, Y., Gu, Y., Zheng, Z., Wasteneys, G., and Yang, Z. (2005). Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120 687–700. [DOI] [PubMed] [Google Scholar]
- Fu, Y., Li, H., and Yang, Z. (2002). The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell 14 777–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y., Wu, G., and Yang, Z. (2001). Rop GTPase-dependent dynamics of tip-localized F-actin controls tip growth in pollen tubes. J. Cell Biol. 152 1019–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, H., Takemura, M., Tani, E., Nemoto, K., Yokota, A., and Kohchi, T. (2003). An Arabidopsis MADS-box protein, AGL24, is specifically bound to and phosphorylated by meristematic receptor-like kinase (MRLK). Plant Cell Physiol. 44 735–742. [DOI] [PubMed] [Google Scholar]
- Gu, Y., Fu, Y., Dowd, P., Li, S., Vernoud, V., Gilroy, S., and Yang, Z. (2005). A Rho family GTPase controls actin dynamics and tip growth via two counteracting downstream pathways in pollen tubes. J. Cell Biol. 169 127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Y., Vernoud, V., Fu, Y., and Yang, Z. (2003). ROP GTPase regulation of pollen tube growth through the dynamics of tip-localized F-actin. J. Exp. Bot. 54 93–101. [DOI] [PubMed] [Google Scholar]
- Gu, Y., Wang, Z., and Yang, Z. (2004). ROP/RAC GTPase: An old new master regulator for plant signaling. Curr. Opin. Plant Biol. 7 527–536. [DOI] [PubMed] [Google Scholar]
- Hattan, J., Kanamoto, H., Takemura, M., Yokota, A., and Kohchi, T. (2004). Molecular characterization of the cytoplasmic interacting protein of the receptor kinase IRK expressed in the inflorescence and root apices of Arabidopsis. Biosci. Biotechnol. Biochem. 68 2598–2606. [DOI] [PubMed] [Google Scholar]
- Hoffman, C.S. (1997). Saccharomyces cerevisiae. In Current Protocols in Molecular Biology, F.M. Ausubel, R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl, eds (New York: John Wiley & Sons), pp. 13.7.5–13.7.7, 13.11.1–13.11.3.
- Hoffman, G.R., and Cerione, R.A. (2002). Signaling to the Rho GTPases: Networking with the DH domain. FEBS Lett. 513 85–91. [DOI] [PubMed] [Google Scholar]
- Honys, D., and Twell, D. (2003). Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol. 132 640–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, J.U., Gu, Y., Lee, Y.J., and Yang Z. (2005). Oscillatory ROP GTPase activation leads the oscillatory polarized growth of pollen tubes. Mol. Biol. Cell 16 5385–5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, M.A., Shen, J.J., Fu, Y., Li, H., Yang, Z., and Grierson, C.S. (2002). The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell 14 763–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaothien, P., Ok, S.H., Shuai, B., Wengier, D., Cotter, R., Kelley, D., Kiriakopolos, S., Muschietti, J., and McCormick, S. (2005). Kinase partner protein interacts with the LePRK1 and LePRK2 receptor kinases and plays a role in polarized pollen tube growth. Plant J. 42 492–503. [DOI] [PubMed] [Google Scholar]
- Kawasaki, T., Henmi, K., Ono, E., Hatakeyama, S., Iwano, M., Satoh, H., and Shimamoto, K. (1999). The small GTP-binding protein rac is a regulator of cell death in plants. Proc. Natl. Acad. Sci. USA 96 10922–10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.U., Cotter, R., Johnson, S., Senda, M., Dodds, P., Kulikauskas, R., Tang, W., Ezcurra, I., Herzmark, P., and McCormick, S. (2002). New pollen-specific receptor kinases identified in tomato, maize and Arabidopsis: The tomato kinases show overlapping but distinct localization patterns on pollen tubes. Plant Mol. Biol. 50 1–16. [DOI] [PubMed] [Google Scholar]
- Kjoller, L., and Hall, A. (1999). Signaling to Rho GTPases. Exp. Cell Res. 253 166–179. [DOI] [PubMed] [Google Scholar]
- Kost, B., Lemichez, E., Spielhofer, P., Hong, Y., Tolias, K., Carpenter, C., and Chua, N.H. (1999). Rac homologues and compartmentalized phosphatidylinositol 4, 5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J. Cell Biol. 145 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemichez, E., Wu, Y., Sanchez, J.P., Mettouchi, A., Mathur, J., and Chua, N.H. (2001). Inactivation of AtRac1 by abscisic acid is essential for stomatal closure. Genes Dev. 15 1808–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard, D.A., Evans, T., Hart, M., Cerione, R.A., and Manor, D. (1994). Investigation of the GTP-binding/GTPase cycle of Cdc42Hs using fluorescence spectroscopy. Biochemistry 33 12323–12328. [DOI] [PubMed] [Google Scholar]
- Li, H., Lin, Y., Heath, R.M., Zhu, M.X., and Yang, Z. (1999). Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to the tip-localized calcium influx. Plant Cell 11 1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Wu, G., Ware, D., Davis, K.R., and Yang, Z. (1998). Arabidopsis Rho-related GTPases: Differential gene expression in pollen and polar localization in fission yeast. Plant Physiol. 118 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y., and Yang, Z. (1997). Inhibition of pollen tube elongation by microinjected anti-Rop1Ps antibodies suggests a crucial role for Rho-type GTPases in the control of tip growth. Plant Cell 9 1647–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margarit, S.M., Sondermann, H., Hall, B.E., Nagar, B., Hoelz, A., Pirruccello, M., Bar-Sagi, D., and Kuriyan, J. (2003). Structural evidence for feedback activation by Ras.GTP of the Ras-specific nucleotide exchange factor SOS. Cell 112 685–695. [DOI] [PubMed] [Google Scholar]
- Molendijk, A.J., Bischoff, F., Rajendrakumar, C.S., Friml, J., Braun, M., Gilroy, S., and Palme, K. (2001). Arabidopsis thaliana Rop GTPases are localized to tips of root hairs and control polar growth. EMBO J. 20 2779–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, E.R., and Walker, J.C. (2003). Receptor-like protein kinases: The keys to response. Curr. Opin. Plant Biol. 6 339–342. [DOI] [PubMed] [Google Scholar]
- Nakanomyo, I., Kost, B., Chua, N.H., and Fukuda, H. (2002). Preferential and asymmetrical accumulation of a Rac small GTPase mRNA in differentiating xylem cells of Zinnia elegans. Plant Cell Physiol. 43 1484–1492. [DOI] [PubMed] [Google Scholar]
- Nomanbhoy, T.K., Leonard, D.A., Manor, D., and Cerione, R.A. (1996). Investigation of the GTP-binding/GTPase cycle of Cdc42Hs using extrinsic reporter group fluorescence. Biochemistry 35 4602–4608. [DOI] [PubMed] [Google Scholar]
- Ono, E., Wong, H.L., Kawasaki, T., Hasegawa, M., Kodama, O., and Shimamoto, K. (2001). Essential role of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. USA 98 759–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, J.L., Jilk, R., Marks, M.D., and Szymanski, D.B. (2002). The Arabidopsis SPIKE1 gene is required for normal cell shape control and tissue development. Plant Cell 14 101–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph, M.G., Weise, C., Mirold, S., Hillenbrand, B., Bader, B., Wittinghofer, A., and Hardt, W.D. (1999). Biochemical analysis of SopE from Salmonella typhimurium, a highly efficient guanosine nucleotide exchange factor for RhoGTPases. J. Biol. Chem. 274 30501–30509. [DOI] [PubMed] [Google Scholar]
- Shiu, S.H., and Bleecker, A.B. (2001. a). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 98 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, S.H., and Bleecker, A.B. (2001. b). Plant receptor-like kinase gene family: Diversity, function, and signaling. Sci. STKE 113, RE22. [DOI] [PubMed]
- Tao, L.Z., Cheung, A.Y., and Wu, H.M. (2002). Plant Rac-like GTPases are activated by auxin and mediate auxin-responsive gene expression. Plant Cell 14 2745–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarutani, Y., Sasaki, A., Yasuda, M., Nakashita, H., Yoshida, S., Yamaguchi, I., and Suzuki, Y. (2004). Identification of three clones which commonly interact with the kinase domains of highly homologous two receptor-like kinases, RLK902 and RKL1. Biosci. Biotechnol. Biochem. 68 2581–2587. [DOI] [PubMed] [Google Scholar]
- Trotochaud, A.E., Hao, T., Wu, G., Yang, Z., and Clark, S.E. (1999). The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell 11 393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengier, D., Valsecchi, I., Cabanas, M.L., Tang, W.H., McCormick, S., and Muschietti, J. (2003). The receptor kinases LePRK1 and LePRK2 associate in pollen and when expressed in yeast, but dissociate in the presence of style extract. Proc. Natl. Acad. Sci. USA 100 6860–6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, G., Gu, Y., Li, S., and Yang, Z. (2001). A genome-wide analysis of Arabidopsis Rop-interactive CRIB motif-containing proteins that act as Rop GTPase targets. Plant Cell 13 2841–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, G., Li, H., and Yang, Z. (2000). Arabidopsis RopGAPs are a novel family of Rho GTPase-activating proteins that require the Cdc42/Rac-interactive binding motif for Rop-specific GTPase stimulation. Plant Physiol. 124 1625–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarsky, V., Cvrckova, F., Bischoff, F., and Palme, K. (1997). At-GDI1 from Arabidopsis thaliana encodes a rab-specific GDP dissociation inhibitor that complements the sec19 mutation of Saccharomyces cerevisiae. FEBS Lett. 403 303–308. [DOI] [PubMed] [Google Scholar]
- Zheng, Z.L., Nafisi, M., Tam, A., Li, H., Crowell, D.N., Chary, S.N., Schroeder, J.I., Shen, J., and Yang, Z. (2002). Plasma membrane-associated ROP10 small GTPase is a specific negative regulator of abscisic acid responses in Arabidopsis. Plant Cell 14 2787–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.