Abstract
Plant cells frequently undergo endoreduplication, a process in which chromosomal DNA is successively duplicated in the absence of mitosis. It has been proposed that endoreduplication is regulated at its entry by mitotic cyclin-dependent kinase activity. However, the regulatory mechanisms for its termination remain unclear, although plants tightly control the ploidy level in each cell type. In the process of searching for regulatory factors of endoreduplication, the promoter of an Arabidopsis thaliana cyclin A gene, CYCA2;3, was revealed to be active in developing trichomes during the termination period of endoreduplication as well as in proliferating tissues. Taking advantage of the situation that plants encode highly redundant cyclin A genes, we were able to perform functional dissection of CYCA2;3 using null mutant alleles. Null mutations of CYCA2;3 semidominantly promoted endocycles and increased the ploidy levels achieved in mature organs, but they did not significantly affect the proportion of cells that underwent endoreduplication. Consistent with this result, expression of the CYCA2;3–green fluorescent protein fusion protein restrained endocycles in a dose-dependent manner. Moreover, a mutation in the destruction box of CYCA2;3 stabilized the fusion protein in the nuclei and enhanced the restraint. We conclude that CYCA2;3 negatively regulates endocycles and acts as a key regulator of ploidy levels in Arabidopsis endoreduplication.
INTRODUCTION
Eukaryotic cells generally proliferate through the mitotic cell cycle, which allows cells to maintain their DNA content at the 2C level after each cell division (1C is the DNA content of a haploid genome). However, certain cells undergoing differentiation increase their DNA contents to 4C or higher as a result of endoreduplication (for reviews, see Edgar and Orr-Weaver, 2001; Larkins et al., 2001; Kondorosi and Kondorosi, 2004; Lilly and Duronio, 2005). Although the phenomenon of endoreduplication has a number of variations, for the most part the process lacks all vestiges of mitosis, and the resulting ploidy numbers are powers of two. Therefore, endoreduplication is thought to be a process in which chromosomal DNA is successively duplicated in the absence of mitosis (Edgar and Orr-Weaver, 2001).
Plants exhibit endoreduplication more frequently than animals (for reviews, see Joubes and Chevalier, 2000; Larkins et al., 2001; Sugimoto-Shirasu and Roberts, 2003; Kondorosi and Kondorosi, 2004). Endoreduplication often occurs during the differentiation of cells that are highly specialized in their morphology or metabolism. An Arabidopsis thaliana trichome, a large branched cell on the surface of aerial organs (Figure 1R), generally has a DNA content of 32C (Hulskamp et al., 1994). Maize (Zea mays) endosperm cells, which accumulate starch and storage proteins, usually undergo four to five successive endocycles during seed development (Kowles and Phillips, 1985). Other cells, such as those in leaves and roots, also exhibit high ploidy. Arabidopsis cotyledons and leaf pavement cells have been observed to have ploidy levels from 2C to 32C and from 2C to 16C, respectively (Galbraith et al., 1991; Melaragno et al., 1993). Moreover, the ploidy levels of Arabidopsis hypocotyls vary depending on growth conditions, with levels of 2C to 8C under normal light conditions and 2C to 16C in darkness (Gendreau et al., 1997).
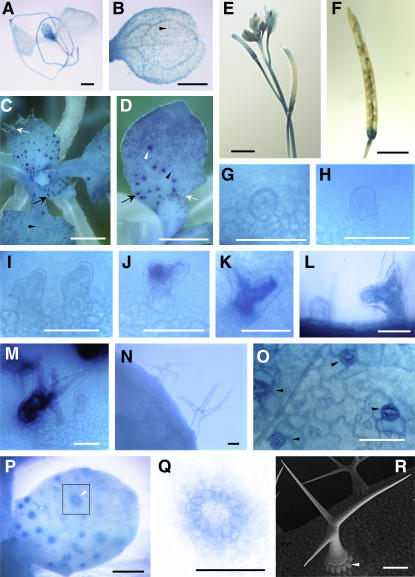
Figure 1.
Histochemical Analysis of CYCA2;3 Promoter Activity.
CYCA2;3 promoter activity was histochemically analyzed using a CYCA2;3 promoter–GUS reporter gene that was introduced into wild-type plants.
(A) Young seedling at 7 d after germination (DAG).
(B) Cotyledons at 5 DAG.
(C) Shoot apex.
(D) and (P) Young leaves.
(E) Inflorescence.
(F) Seedpod.
(G) to (M) Developing trichomes at stage 2 ([G], [H], and left part of [L]), early stage 3 (I), late stage 3 ([J] and [K]), and stage 4 (right part of [L] and [M]).
(N) Mature trichomes.
(O) Leaf surface.
(Q) Trichome socket cells.
(R) Scanning electron microscopic image of a mature trichome and trichome socket cells on the wild-type leaf surface.
Black and white arrows in (C) and (D) indicate developing and mature trichomes, respectively. Black and white arrowheads in (B) to (D), (O), (P), and (R) indicate stomata guard cells and trichome socket cells, respectively. A magnification of the region indicated by the square in (P) is shown in (Q). Bars = 1 mm in (A) to (D), 3 mm in (E) and (F), 0.05 mm in (G) to (O), and 0.1 mm in (P) to (R).
Genetic studies have shown that several Arabidopsis mutants exhibit trichomes with altered ploidy phenotypes (for reviews, see Marks, 1997; Hulskamp, 2004). Trichomes of one class of mutants, including triptychon and kaktus, have a DNA content of 64C (Hulskamp et al., 1994), whereas those of the glabra3-1 and glabra3-sst mutants have DNA contents lower and higher than the wild type, respectively (Hulskamp et al., 1994; Esch et al., 2003). These phenotypes suggest that the succession of endocycles is regulated both positively and negatively to terminate endoreduplication at 32C in trichomes. Furthermore, the trichome initial cells on leaves of the siamese mutant do not proceed with endoreduplication but instead display promoted mitotic cell cycles that result in multicellular trichomes (Walker et al., 2000), implying that a mechanism exists to trigger a switch from mitosis to endocycles. These genetic studies suggest that plant endoreduplication is actively regulated in both the entry and the endocycle succession.
For the entry into endoreduplication, the activity of cyclin-dependent kinase (CDK) associated with mitotic cyclins has been proposed as a key negative regulator (Edgar and Orr-Weaver, 2001; Larkins et al., 2001). In plants, reverse genetic studies of various cell cycle–related proteins have provided evidence to support this idea. Ectopic expression of the Arabidopsis mitotic cyclin CYCB1;2 in developing trichomes can switch from endocycles to mitotic cycles, resulting in multicellular trichomes (Schnittger et al., 2002b). A nonmitotic cyclin, CYCD3;1, has also been postulated to regulate mitotic CDK activity directly or indirectly, because its ectopic expression in developing trichomes induces not only DNA replication but also cell division (Schnittger et al., 2002a). Overexpression of a fusion protein between the Nicotiana tabacum mitotic cyclin CYCA3;2 and green fluorescent protein (GFP) causes an inhibitory effect on endoreduplication in Arabidopsis leaves (Yu et al., 2003). In Medicago truncatula, a reduction in ploidy levels is caused by antisense suppression of the cell cycle switch 52A (ccs52A) gene, which encodes a substrate-specific activator of the anaphase-promoting complex (APC) and mediates the degradation of mitotic cyclins (Cebolla et al., 1999). The WEE1 kinase, a negative regulator of CDK activity, is upregulated during endoreduplication in maize endosperm cells, suggesting its regulatory role in the entry into endoreduplication (Sun et al., 1999). Recently, moderate overexpression of the Arabidopsis CDK inhibitors Kip-related protein 1 (KRP1) and KRP2 was shown to promote the entry into endoreduplication (Verkest et al., 2005; Weinl et al., 2005). From results of analyses using transgenic plants overexpressing a dominant negative CDKB1;1 and KRP2 (Boudolf et al., 2004; Verkest et al., 2005), a regulatory cascade composed of the tandem negative switches CDKB1;1, KRP2, and CDKA1;1 has been proposed for the mitosis-to-endocycle transition (Verkest et al., 2005).
Little is known about the regulatory mechanisms of the termination of endoreduplication (i.e., the termination of endocycle succession at the appropriate ploidy level for each cell). In endocycles, much of the same G1/S regulatory machinery seen in the mitotic cell cycle is thought to be used (Edgar and Orr-Weaver, 2001). The transcription factor E2 promoter binding factor a-dimerization partner a (E2Fa-DPa), which activates genes for the G1/S machinery, and the transcriptional repressor DP-E2F-like 1, which antagonizes the E2Fa-DPa pathway (Mariconti et al., 2002), have been shown to be positive and negative regulatory factors of endoreduplication, respectively (De Veylder et al., 2002; Vlieghe et al., 2005). In the G1/S transition of the mitotic cell cycle, the chromosomal DNA is licensed to replicate only once by the assembly of the prereplication complex (pre-RC) (for reviews, see Diffley, 2004; Blow and Dutta, 2005). The Arabidopsis pre-RC components cell division cycle 6a (CDC6a) and CDC 10-dependent transcript 1a (CDT1a) cause increases in ploidy levels when ectopically overexpressed (Castellano et al., 2001; del Mar Castellano et al., 2004). The CDK activity suppresses the reassembly of the pre-RC (Diffley, 2004), which dissociates after the initiation of DNA replication. Thus, CDKs associated with mitotic cyclins may negatively regulate the succession of endocycles as well as the mitosis-to-endocycle transition. In turn, APC downregulates the CDK activity to allow reassembly of the pre-RC in early G1 by promoting the degradation of mitotic cyclins (Harper et al., 2002; Peters, 2002), suggesting that APC may also be involved in regulating the succession of endocycles. On the other hand, CDK activity is likely to be required for endoreduplication at a low level. It has been reported that expression of a dominant negative CDKA;1 reduces the ploidy levels of maize endosperm cells (Leiva-Neto et al., 2004) and that highly overexpressed CDK inhibitors reduce the ploidy levels in Arabidopsis leaves and trichomes (De Veylder et al., 2001; Jasinski et al., 2002; Zhou et al., 2003; Schnittger et al., 2003; Verkest et al., 2005; Weinl et al., 2005).
Although the involvement of various cell cycle–related proteins in endoreduplication has been revealed, it is still unclear which proteins play key regulatory roles in endoreduplication, especially in the process of terminating endocycle succession at the appropriate ploidy levels that are determined by the genetic program and growth conditions. To identify key regulators of endoreduplication in plants, we searched for cell cycle–related genes expressed during Arabidopsis trichome development, in which endoreduplication occurs instead of the mitotic cell cycle. CDKA;1 has been revealed to be expressed during trichome development (Imajuku et al., 2001). In the process of identifying cyclins that are involved in endoreduplication, we found that the promoter of a cyclin A gene, CYCA2;3, is active not only in proliferating tissues but also in developing trichomes during the termination period of endoreduplication. Taking advantage of the situation that plants encode highly redundant cyclin A genes, we obtained CYCA2;3 null mutant lines and found that the defect semidominantly promoted endocycles and increased the ploidy levels achieved in mature organs, but it did not significantly affect the proportion of cells that underwent endoreduplication. Based on the results of our forward and reverse genetic approaches, we conclude that CYCA2;3 acts as a key regulator of ploidy levels in Arabidopsis endoreduplication.
RESULTS
The CYCA2;3 Promoter Is Active during the Termination Period of Endoreduplication in Trichomes
In the process of identifying cyclins that are involved in endoreduplication, we found that the promoter of CYCA2;3 is active in trichomes. Histochemical analysis using the Escherichia coli β-glucuronidase (GUS) reporter gene showed that a 2.4-kb DNA region upstream of the CYCA2;3 initiation codon exhibits strong promoter activity, preferentially in developing trichomes, trichome socket cells, root and shoot meristems, vascular systems, and stomata guard cells, and weak activity in many other tissues, including developing leaf epidermis (Figures 1A to 1D and 1O to 1Q). In reproductive organs, the styles, pedicels, and bases and vascular systems of seedpods showed strong GUS activity (Figures 1E and 1F). In developing trichomes, relatively stronger GUS activity than in the surrounding cells was not observed at stage 2 (Figures 1G, 1H, and the left half of 1L; stalk emergence and expansion [the stage definitions are according to Szymanski et al., 1999]) but became evident during the progression of stage 3 (Figures 1I to 1K; formation of branch structures). GUS activity was evident also at stage 4 (the right half of Figures 1L and 1M; expansion of the stalk and branches, which have blunt tips) and then gradually disappeared as the trichome matured (Figure 1N). This temporal expression pattern during trichome development is similar to that of CDKA;1 (Imajuku et al., 2001). Assuming that CYCA2;3 is involved in regulating the endoreduplication of trichome cells, it is likely to function in the termination, but not the initiation, of endoreduplication, as developing Arabidopsis trichomes undergo three of four endocycles before stage 2 and complete the final one at stage 3 (Hulskamp et al., 1994).
The Loss of CYCA2;3 Function Promotes Endocycles and Increases Ploidy Levels
To investigate the function of CYCA2;3 in plant developmental processes, we identified loss-of-function mutants of the gene. The lines SALK_086463 and SALK_092515 (ecotype Columbia) were obtained from the ABRC. PCR using genomic DNA as a template revealed that SALK_086463 and SALK_092515 have T-DNA insertions at positions ∼100 and 400 bp downstream of the CYCA2;3 initiation codon in the first and second exons, respectively (Figure 2A). RT-PCR using total RNA prepared from homozygous mutant plants amplified no transcripts corresponding to an internal region of the CYCA2;3 protein in either line (Figure 2C), indicating that both lines are null alleles of CYCA2;3. Therefore, we further analyzed SALK_086463 as a representative loss-of-function mutant unless indicated otherwise.
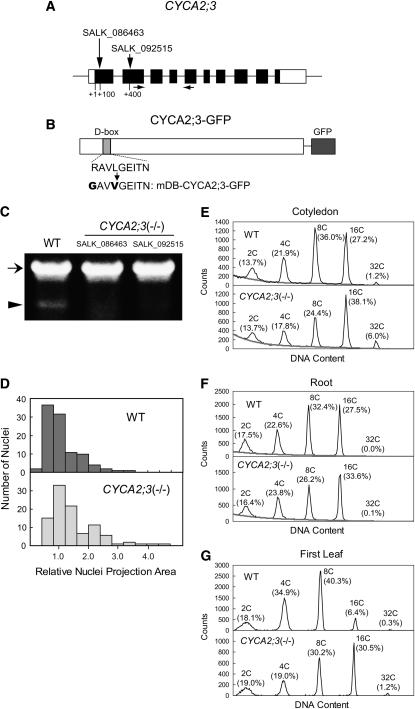
Figure 2.
Analysis of Loss-of-Function Mutations of CYCA2;3.
(A) The exon (boxes) and intron (lines) structure of CYCA2;3. Coding and noncoding regions are shown as black and white boxes, respectively. Vertical arrows indicate the sites of the T-DNA insertions in the lines SALK_086463 and SALK_092515. Horizontal arrows indicate the positions of the primers used in RT-PCR analysis.
(B) Structure of the fusion protein CYCA2;3-GFP. The amino acid sequence of the D-box and the altered sequence in mDB-CYCA2;3-GFP are shown below.
(C) RT-PCR analysis of CYCA2;3 expression in wild-type and CYCA2;3(−/−) plants of the SALK_086463 and SALK_092515 lines. RT-PCR was performed using total RNA prepared from 2-week-old plants. The positions of the bands corresponding to the ACT2 (a positive control; At3g18780) and CYCA2;3 transcripts are indicated by an arrow and an arrowhead, respectively.
(D) Comparison of the sizes of nuclei in trichomes of wild-type (top) and CYCA2;3(−/−) (bottom) plants. The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI), and the projection area was measured microscopically (see Methods). To calculate the relative value, the mean nuclear projection area from wild-type trichomes was arbitrarily set to 1.
(E) to (G) Ploidy distribution patterns in cotyledons (E), roots (F), and first leaves (G) of wild-type (top panels) and CYCA2;3(−/−) (bottom panels) plants. Seedlings grown under normal light conditions for 4 DAG ([E] and [F]) or 14 DAG (G) were dissected and subjected to flow cytometry. Ploidy levels and the proportion of cells with those levels are indicated above each peak. The gray lines in (E) and (F) are the calculated baselines (see Methods).
Homozygous mutant plants [CYCA2;3(−/−)] did not show any phenotype with morphology or growth rates obviously different from those of the wild type. However, the nuclei in trichomes of the mutant plants were larger than those of wild-type trichomes (Figure 2D). The proportion of nuclei with large projection areas was higher in the mutant than in the wild type (e.g., 23.0% of mutant nuclei and 8.0% of wild-type nuclei showed relative projection areas > 2.0). Moreover, the distribution pattern of mutant nuclei exhibited an additional peak at a projection area position larger than the major peak. Because it is thought that the major and additional peaks correspond to trichomes with ploidy levels of 32C and 64C, respectively, this result strongly suggests that the mutant contains a higher proportion of 64C trichomes than the wild type. In addition to the enlarged nuclei, the proportion of trichomes with four branches was significantly higher and that of three-branch trichomes was significantly lower in the mutant than in the wild type (Table 1). This observation supports the idea that the mutant trichomes have higher ploidy levels than wild-type trichomes, as the branch number of Arabidopsis trichomes is thought to be positively correlated with the ploidy level (Hulskamp, 2004).
Table 1.
Branch Numbers of Wild-Type and CYC2;3(−/−) Trichomes
| Number of Trichomes |
Proportion of Trichomes (%) |
||||
|---|---|---|---|---|---|
| Branch Number | Wild Type | CYC2;3(−/−) | Wild Type | CYC2;3(−/−) | Pa |
| 2 | 11 | 15 | 1.1 | 1.4 | 0.530 |
| 3 | 875 | 846 | 86.0 | 78.1 | <0.001 |
| 4 | 130 | 216 | 12.8 | 20.0 | <0.001 |
| 5 | 1 | 5 | 0.1 | 0.5 | 0.107 |
| Total | 1017 | 1082 | 100.0 | 100.0 | |
Trichomes with different numbers of branches on the adaxial surface of first leaves of 3-week-old wild-type and CYCA2;3(−/−) plants were counted.
Probability that the indicated proportions of trichomes would be observed if there were no differences in the proportions between the wild type and CYCA2;3(−/−).
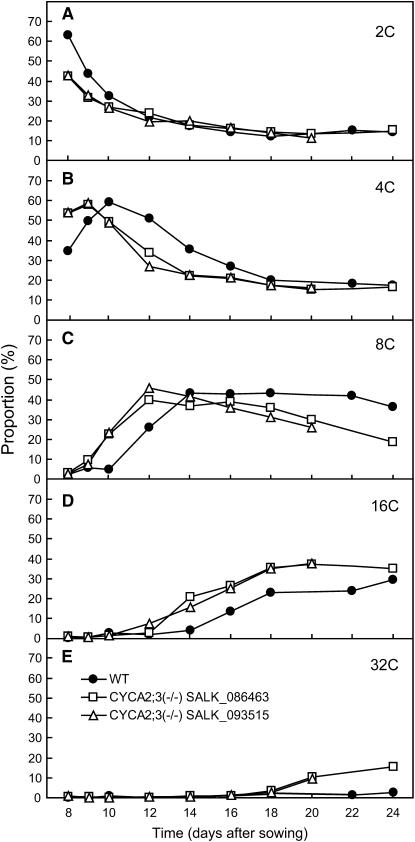
To further investigate the effect of this mutation on ploidy, we analyzed the ploidy distribution in cotyledons (4 DAG), roots (4 DAG), and first leaves (14 DAG) using flow cytometry (Figures 2E to 2G). The proportion of 2C cells was not significantly affected by the mutation in any examined organs. However, the mutation significantly increased the proportion of cells with a ploidy levels of 16C or higher (i.e., the proportions for the wild type and the mutant were 28.4 and 44.1% in cotyledons, 27.5 and 33.7% in roots, and 6.7 and 31.7% in first leaves, respectively). This ploidy phenotype was analyzed in more detail in a time-course experiment with developing first leaves (Figure 3). The proportions of 2C and 4C cells were lower and higher, respectively, in the mutant than in the wild type during a relatively early period of leaf development (8 and 9 d after sowing), when cells with a ploidy level of 8C or higher were not evident. During leaf development, 8C, 16C, and 32C cells became evident and increased in proportion earlier in the mutant than in the wild type, with the higher ploidy levels showing greater time differences between the mutant and the wild type. The proportion of 4C cells decreased earlier in the mutant. In late stages (≥20 d after sowing), the mutant leaves showed almost the same proportions of 2C and 4C cells, a lower proportion of 8C cells, and higher proportions of 16C and 32C cells compared with wild-type leaves. These results indicate that the first endocycle occurs earlier and that subsequent endocycles succeed faster in mutant leaves than in wild-type leaves, although the proportion of cells that underwent endoreduplication during leaf development was the same.
Figure 3.
Time-Course Ploidy Distribution Analysis of Mutant First Leaves in Leaf Development.
Proportions of cells with ploidy levels of 2C (A), 4C (B), 8C (C), 16C (D), and 32C (E) in wild-type (closed circles) and mutant (SALK_086463, open squares; SALK_092515, open triangles) first leaves harvested at the indicated times were determined by flow cytometry and plotted.
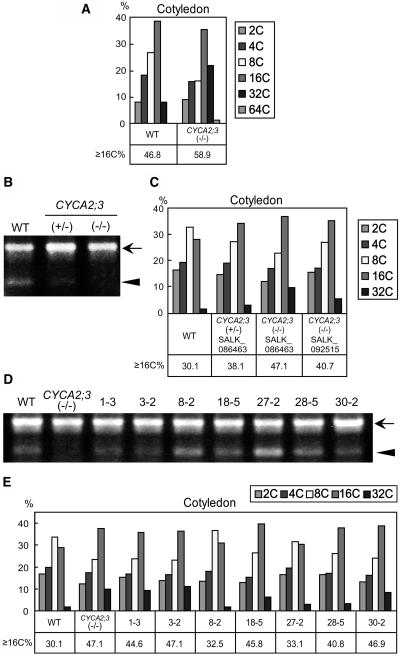
In fully expanded mutant cotyledons (7 DAG), the ploidy phenotype was enhanced and 64C cells, which are not detected in wild-type cotyledons, were produced (Figure 4A). This extra endocycle in cotyledons, together with the evidently increased proportion of 32C cells in mature first leaves, indicates that the mutation also affects the ploidy levels finally achieved in these organs. Homozygous mutant plants of the other line, SALK_092515, showed almost the same ploidy distribution patterns during leaf development (Figure 3) and in cotyledons (4 DAG; Figure 4C) as that of SALK_086463, confirming that the loss of function of CYCA2;3 is responsible for this ploidy phenotype.
Figure 4.
Ploidy Distribution Analyses of Cotyledons of Mutant and Complementation Plants.
(A) Ploidy distribution analysis of fully expanded cotyledon cells at 7 DAG in wild-type and CYCA2;3(−/−) plants.
(B) and (C) Quantitative RT-PCR analysis of the CYCA2;3 transcript (B) and ploidy distribution analysis of cotyledon cells at 4 DAG (C) in wild-type, heterozygous [CYCA2;3(+/−) SALK_086463 line], and homozygous [CYCA2;3(−/−) SALK_086463 and SALK_092515 lines] mutant plants.
(D) and (E) Quantitative RT-PCR analysis of CYCA2;3 (or CYCA2;3-GFP) transcripts (D) and ploidy distribution analysis of cotyledon cells at 4 DAG (E) in the wild type, CYCA2;3(−/−), and the complementation lines (1-3, 3-2, 8-2, 18-5, 27-2, 28-5, and 30-2) containing the CYCA2;3-GFP gene driven by the CYCA2;3 promoter. Quantitative RT-PCR was performed using total RNA from 2-week-old plants. The positions of the bands corresponding to the ACT2 (a positive control) and CYCA2;3 (or CYCA2;3-GFP) transcripts are indicated by the arrow and arrowhead, respectively. Ploidy distribution analysis was performed by flow cytometry of cells from cotyledons. The proportions of cells with the indicated ploidy levels were calculated (see Methods). ≥16C%, the proportion (%) of cells with a ploidy level of 16C or higher.
Expression of the Endogenous CYCA2;3 Gene and the CYCA2;3-GFP Transgenes Restrains Endocycles in a Dose-Dependent Manner
To investigate the functional manner of CYCA2;3, the ploidy levels of plants heterozygous for the SALK_086463 mutation were compared with those of wild-type and homozygous mutant plants. Quantitative RT-PCR analysis showed that CYCA2;3 transcripts were less abundant in the heterozygote than in the wild type (Figure 4B). Flow cytometry analysis of cotyledon cells of the heterozygote revealed a ploidy distribution pattern intermediate between those of the wild type and the homozygote (Figure 4B). These results indicate that the mutation is semidominant and that the expression level of CYCA2;3 negatively correlates with the ploidy level.
To examine the effect of overexpression of the CYCA2;3 function on endoreduplication, the CYCA2;3-GFP fusion protein (Figure 2B) was expressed. First, to determine whether the fusion protein has the same function as the authentic protein, functional complementation analysis was performed in CYCA2;3(−/−) plants under the control of the CYCA2;3 promoter, which was used in GUS histochemical analysis. Although no fluorescence resulting from the GFP moiety was observed in the transgenic plants (data not shown), the fusion gene transcript was detected at various levels in seven transgenic lines through quantitative RT-PCR (Figure 4D). The ploidy phenotype in cotyledons was complemented almost completely in lines 8-2 and 27-2, which expressed the fusion gene at relatively high levels, and was complemented partially (lines 18-5 and 28-5) or barely (lines 1-3, 3-2, and 30-2) in lines expressing the fusion gene at relatively low levels (Figures 4D and 4E). This result indicates that the fusion protein performs the authentic function of CYCA2;3 in the regulation of endoreduplication and that its expression also negatively correlates with the ploidy level.
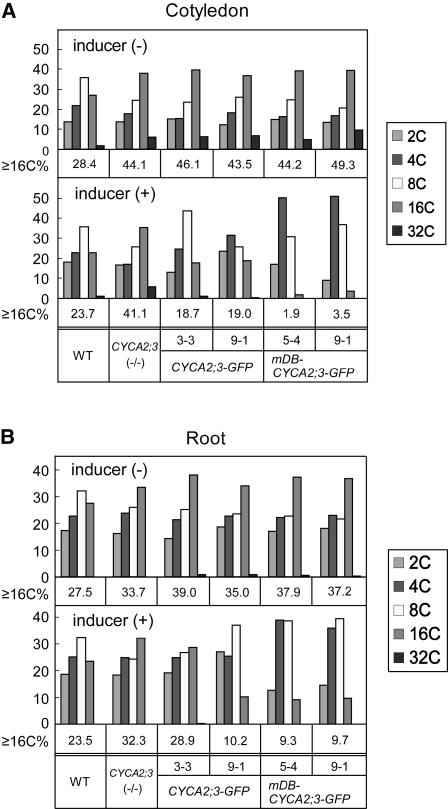
Next, the fusion protein was expressed ectopically using an estrogen-inducible gene expression system (Zuo et al., 2000). To quantitatively analyze the effect of the fusion protein in the absence of the endogenous protein, the transgene was introduced into CYCA2;3(−/−) plants. The transgenic plants showed no macroscopic changes, either in the absence or the presence of the inducer (Figure 5A). Induced GFP fluorescence was observed in nuclei at various levels in different lines (data not shown). Figure 5B shows the fluorescence of representative lines with low (line 3-3) and high (line 9-1) expression levels in roots. The ploidy distributions in cotyledons and roots of these two lines were the same as in the transgenic parental line CYCA2;3(−/−) in the absence of the inducer (Figure 6). When the fusion protein was expressed, the proportion of high-ploidy cells (16C or higher) was reduced to different levels depending on the levels of the fusion protein (Figure 6). In roots, the proportion was significantly lower in line 9-1 (10.2%) than in line 3-3 (28.9%). In cotyledons, although the proportion was almost the same in the two lines (18.7% in line 3-3 and 19.0% in line 9-1), the proportion of 8C cells was significantly lower in line 9-1 (25.8%) than in line 3-3 (43.7%). These results indicate that expression of the CYCA2;3 gene and the CYCA2;3-GFP transgenes quantitatively restrains endocycles in a dose-dependent manner.
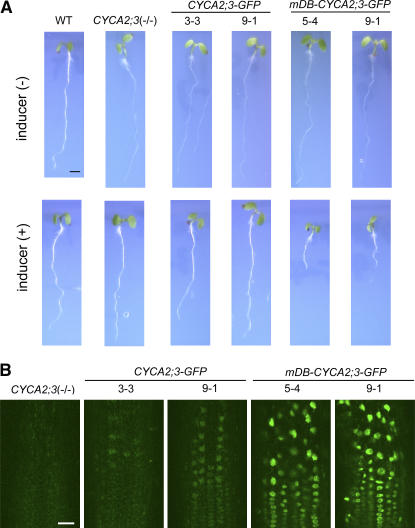
Figure 5.
Macroscopic Phenotypes of Mutant and Transgenic Seedlings, and Subcellular Localization Patterns of the GFP Fusion Proteins.
(A) Seedlings of the wild type, CYCA2;3(−/−), and transgenic lines containing the inducible CYCA2;3-GFP (lines 3-3 and 9-1) or mDB-CYCA2;3-GFP (lines 5-4 and 9-1) gene in the CYCA2;3(−/−) background. Seedlings grown for 4 DAG on agar medium lacking (top row) or containing (bottom row) 10 μM β-estradiol are shown. Bar = 2 mm.
(B) Fluorescence in inducer-treated CYCA2;3(−/−) roots and roots inducibly expressing CYCA2;3-GFP (lines 3-3 and 9-1) or mDB-CYCA2;3-GFP (lines 5-4 and 9-1) in the CYCA2;3(−/−) background. Seedlings were grown for 7 DAG on agar medium lacking β-estradiol and then for 3 d on agar medium containing 10 μM β-estradiol. The fluorescence from the GFP fusion proteins was observed using confocal laser-scanning microscopy. As a negative control for GFP fluorescence, CYCA2;3(−/−) roots were observed under the same conditions used for the CYCA2;3-GFP lines. Bar = 0.02 mm.
Figure 6.
Ploidy Distribution Analysis of Transgenic Plants Ectopically Expressing CYCA2;3-GFP or mDB-CYCA2;3-GFP.
Ploidy distributions in cotyledons (A) and roots (B) of the wild type, CYCA2;3(−/−), and transgenic lines expressing CYCA2;3-GFP (lines 3-3 and 9-1) or mDB-CYCA2;3-GFP (lines 5-4 and 9-1) in the CYCA2;3(−/−) background are shown. Cotyledons and roots from seedlings grown for 4 DAG on agar medium lacking (top panels) or containing (bottom panels) 10 μM β-estradiol were subjected to flow cytometry, and the proportions of cells with the indicated ploidy levels were calculated (see Methods). ≥16C%, the proportion (%) of cells with a ploidy level of 16C or higher.
Mutation of the Destruction Box Stabilizes the Fusion Protein and Enhances the Restraint of Endocycles
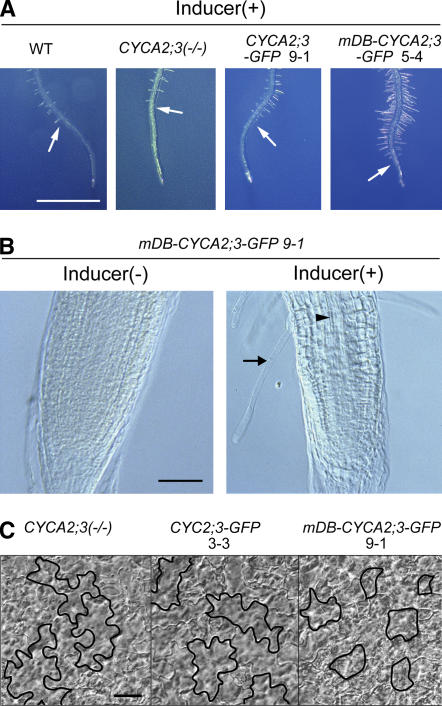
A-type cyclins contain destruction box (D-box) sequences, which have been reported to mediate protein degradation (Glotzer et al., 1991). To express the CYCA2;3-GFP fusion protein at even higher levels, we changed its D-box sequence to a nonfunctional one (from RAVLGEITN to GAVVGEITN; see Figure 2B) (Genschik et al., 1998) and inducibly expressed the protein, mDB-CYCA2;3-GFP, in CYCA2;3(−/−) plants. In the absence of the inducer, the transgenic plants showed no changes in either macroscopic morphology (Figure 5A) or ploidy distribution patterns (Figure 6) compared with the parental line CYCA2;3(−/−). In the presence of the inducer, the nuclei of these plants showed much stronger GFP fluorescence than those expressing CYCA2;3-GFP (Figure 5B). This observation suggests that authentic CYCA2;3 is downregulated in planta by D-box–mediated protein degradation. Expression of this modified cyclin resulted in dwarfism but not in the complete growth arrest of plants (Figure 5A). In the root apical region of the inducer-treated plants, root hair differentiation and vascular differentiation occurred closer to the root tip than normal (Figures 7A and 7B). This phenotype suggests that the abnormal accumulation of the mutant protein retards the mitotic cell cycle in proliferating tissues, because a similar phenotype has been reported as an effect of cell cycle arrest (Umeda et al., 2000). In cotyledon pavement cells, reductions in both size and shape complexity were observed (Figure 7C). These results suggest that reduced levels of both cell proliferation and cell expansion caused the dwarfism.
Figure 7.
Phenotypes of Roots and Cotyledon Epidermal Cells in Plants Expressing CYCA2;3-GFP or mDB-CYCA2;3-GFP.
(A) Phenotypes of root apical regions in the wild type, CYCA2;3(−/−), and transgenic lines containing the inducible CYCA2;3-GFP (line 9-1) or mDB-CYCA2;3-GFP (line 5-4) gene in the CYCA2;3(−/−) background. Root apical regions of seedlings grown for 4 DAG on agar medium containing 10 μM β-estradiol are shown. Arrows indicate the positions of root hair initiation. Bar = 2 mm.
(B) Internal structure of root tips of a transgenic line containing the inducible mDB-CYCA2;3-GFP gene in the CYCA2;3(−/−) background (line 9-1). Root tips of seedlings grown for 4 DAG on agar medium lacking (left) or containing (right) 10 μM β-estradiol were observed with Nomarski differential interference contrast (DIC) microscopy. Arrows and arrowheads indicate a root hair cell and a tracheary element, respectively. Bar = 0.1 mm.
(C) Adaxial cotyledon epidermal cells of CYCA2;3(−/−) and transgenic lines containing the inducible CYCA2;3-GFP (line 3-3) or mDB-CYCA2;3-GFP (line 9-1) gene in the CYCA2;3(−/−) background were observed with Nomarski DIC microscopy. Cotyledons of seedlings grown for 4 DAG on agar medium lacking or containing 10 μM β-estradiol were observed. The shapes of some pavement cells are traced with boldface lines for clarity. Bar = 0.02 mm.
Cotyledons and roots expressing mDB-CYCA2;3-GFP showed relatively severe decreases in the proportion of high-ploidy cells (16C or higher) compared with those expressing CYCA2;3-GFP (Figure 6). This result is consistent with the idea that CYC2;3 quantitatively restrains endocycles depending on its expression level. It is also notable that a significant increase in the proportion of 4C cells was observed specifically in mDB-CYCA2;3-GFP–expressing plants in both cotyledons and roots (see Discussion).
CYCA2;3 and CDKA;1 Physically Interact with Each Other and Possibly Coexist in Nuclei of Developing Trichomes during the Termination Period of Endoreduplication
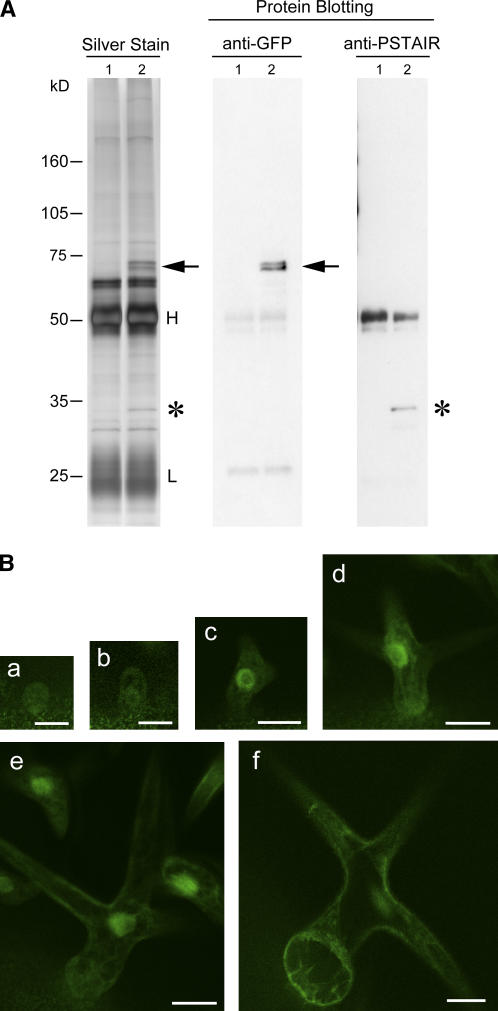
The similar patterns of promoter activities of CYCA2;3 and CDKA;1 in developing trichomes suggest that CYCA2;3 and CDKA;1 form a complex that regulates endoreduplication. We performed an immunoprecipitation experiment to test whether CYCA2;3 has the ability to bind to CDKA;1 in planta. Protein complexes containing mDB-CYCA2;3-GFP in inducer-treated roots (line 9-1) were immunoprecipitated with an anti-GFP antibody. After fractionation by SDS-PAGE, the immunoprecipitates showed evident bands specific to mDB-CYCA2;3-GFP–expressing plants at positions around 34 and 73 kD (Figure 8A). The immunoprecipitates were examined for the presence of CDKA;1 by protein gel blot analysis with an antibody specific to the peptide sequence PSTAIR, which CDKA;1 only contains in Arabidopsis. A signal band was detected at a position corresponding to the 34-kD band (Figure 8A), indicating that mDB-CYCA2;3-GFP binds to CDKA;1 in planta. The bands that were detected with the anti-GFP antibody around 73 kD are thought to correspond to mDB-CYCA2;3-GFP and its derivatives.
Figure 8.
Physical Interaction between mDB-CYCA2;3-GFP and CDKA;1 in Planta, and Intracellular Localization of CDKA;1-GFP in Developing Trichomes.
(A) Detection of CDKA;1 in protein complexes containing mDB-CYCA2;3-GFP. Seedlings of CYCA2;3(−/−) (control; lane 1) and mDB-CYCA2;3-GFP (line 9-1; lane 2) were grown for 7 DAG on agar medium lacking β-estradiol and then for 3 d on agar medium containing 10 μM β-estradiol. Protein extracts prepared from their roots were used in an immunoprecipitation experiment with an anti-GFP antibody. Immunoprecipitates were fractionated by SDS-PAGE and then visualized by silver staining (left) or subjected to protein gel blot analysis with the anti-GFP antibody (center) or an antibody specific to the peptide sequence PSTAIR (right). Arrows and asterisks indicate signal bands at the expected positions of mDB-CYCA2;3-GFP and CDKA;1, respectively. H and L indicate the positions of the heavy and light chains, respectively, of the antibody used in immunoprecipitation. The positions of molecular mass standards are indicated at left.
(B) Intracellular localization analysis of CDKA;1-GFP in developing trichomes. The fusion protein CDKA;1-GFP was expressed using the truncated CDKA;1 promoter, which contains developing trichome specific activity (Imajuku et al., 2001). The fluorescence from the GFP moiety was observed using confocal laser scanning microscopy. Developing trichomes at stage 2 ([a] and [b]), stage 3 (c), stage 4 (d), early stage 5 (e), and late stage 5 (f) are shown. Bars = 0.025 mm in (a) and (b) and 0.05 mm in (c) to (f).
CYCA2;3-GFP was observed to be localized in nuclei (Figure 4B). To examine the intracellular localization of CDKA;1, a CDKA;1-GFP fusion protein was expressed using the truncated CDKA;1 promoter, which shows activity specific to developing trichomes (Imajuku et al., 2001). The fusion protein was localized evidently in nuclei of developing trichomes in the period from stage 3 to early stage 5 (Figure 8B), whereas its cytosolic localization was observed throughout the period examined, from stage 2 to late stage 5. The period of nuclear localization coincides with that of CYCA2;3 promoter activity and includes the termination period of trichome endoreduplication (for comparison, see Supplemental Figure 1 online). The possible coexistence of CDKA;1 and CYCA2;3 in the nuclei and their ability to physically interact strongly suggest that the proteins form a complex in the nuclei of developing trichomes during the period of endoreduplication termination.
DISCUSSION
A2-Type Cyclins in Arabidopsis
Plants contain a number of cyclin genes whose functional overlap and differentiation remain largely unclear. Arabidopsis encodes 10 A-type cyclins that have been categorized into the subclasses A1, A2, and A3 based on their primary structures (Vandepoele et al., 2002). Because the genes for A-type cyclins show similar expression profiles within each subclass during the mitotic cell cycle, their functions in cell cycle progression are thought to be conserved in each subclass (Dewitte and Murray, 2003). A gene expression analysis with microarrays has shown that the transcript levels of A2-type cyclin genes have a distinct peak at early mitosis in synchronized Arabidopsis cell cultures (Menges et al., 2005). In a synchronized Medicago cell culture, the transcript levels of an A2-type gene increased steadily from late G1 to G2/M phase, whereas the protein levels increased during the S phase and remained at high levels from G2 to mid M phase (Roudier et al., 2000).
The Arabidopsis A2 subclass contains four members, CYCA2;1 to CYCA2;4. The CYCA2;1 promoter has been reported to be active in shoot and root meristems, vascular systems, and stomata guard cells but not in trichomes (Burssens et al., 2000). The 2.5-kb DNA region upstream of the CYCA2;4 initiation codon showed a promoter activity pattern similar to that of the CYCA2;1 promoter, except in stomata guard cells (see Supplemental Figure 2 online). The 2.0-kb DNA region upstream of the CYCA2;2 initiation codon showed a promoter activity pattern similar to that of the CYCA2;3 promoter, including that in trichome development (see Supplemental Figure 2 online). These observations suggest that the functions of Arabidopsis A2-type cyclin genes are highly redundant. This type of redundancy often becomes a major hindrance in genetic analysis. In our case, however, the redundancy allowed us to analyze the loss-of-function effect of CYCA2;3, which would have led to lethality if the gene were unique and indispensable, as is the case for animal cyclin A genes (Murphy et al., 1997; Murphy, 1999). We could clearly detect the ploidy phenotype of the null mutant plants, possibly because endocycles are quantitatively regulated by the overall level of A2-type cyclins, to which CYCA2;3 contributes significantly. Unlike in cotyledons and first leaves, the ploidy level in etiolated hypocotyls was not affected significantly by the mutation (see Supplemental Figure 3 online). CYCA2;3 promoter activity was not detected in the epidermis and cortex (see Supplemental Figure 3 online), which undergo endoreduplication most actively in this organ (Gendreau et al., 1998). Consistently, a real-time quantitative RT-PCR experiment showed that, compared with levels of other A2-type cyclin genes, the relative transcript levels of CYCA2;3 were high in first leaves and cotyledons but not in etiolated hypocotyls (see Supplemental Figure 4 online). Although the cyclin protein levels in these organs are unknown, the contribution of CYCA2;3 to the regulation of ploidy levels might be low in etiolated hypocotyls.
A Regulatory Role of CYCA2;3 in Endoreduplication and the Mitotic Cell Cycle
Altered expression and/or functions of cell cycle–related proteins often cause pleiotropic phenotypes in both endoreduplication and the mitotic cell cycles. It is noteworthy in our study that the phenotype of CYCA2;3(−/−) mutants was not pleiotropic but was specific to the change of ploidy levels. In first leaves, the first endocycle occurred earlier and subsequent endocycles succeeded faster in the mutant than in the wild type, whereas the proportion of cells that underwent endoreduplication during leaf development was the same. Therefore, CYCA2;3 is thought to negatively regulate every endocycle of cells undergoing endoreduplication but is not likely to be related to the mitosis–endocycle cell fate. In addition, cells with abnormally high ploidy levels were observed in the fully expanded mutant cotyledons, and the proportion of cells with the highest ploidy level (32C) was significantly larger in mutant mature first leaves than in those of the wild type. Thus, CYCA2;3 is thought to function in terminating endocycle succession at appropriate ploidy levels, consistent with the strong CYCA2;3 promoter activity in the period of the termination of trichome endoreduplication. The endogenous CYCA2;3 gene and CYCA2;3-GFP transgenes quantitatively reduced the ploidy levels depending on their expression levels. Moreover, stabilizing the fusion protein through the mutation in the D-box sequence further enhanced this reduction. These findings suggest that the amount of CYCA2;3 in a cell acts as a quantitative factor in restraining endocycles and that strong expression of the protein results in the termination of endocycle succession.
The effect on ploidy levels of mature leaves and cotyledons in the CYCA2;3(−/−) mutants is significant but not severe. Although it is unclear how many extra endocycles occurred in each cell, the effect is explainable by assuming that a limited proportion of cells undergo only one extra endocycle. Although the functional redundancy of other A2-type cyclin genes possibly reduces the mutant effect, negative regulatory pathways other than that involving A2-type cyclins might exist for endocycles. An endocycle might contain multiple check points for its progression, as does the mitotic cell cycle, and a defect in only one check point could allow a limited number of extra endocycles.
In proliferating tissues, in which promoters of CYCA2;3 and the other A2-type genes are active, A2-type cyclins are supposed to have some of the canonical cyclin A functions known in the mitotic cell cycle of animals (for review, see Sherr and Roberts, 2004). Strong CYCA2;3 promoter activity was also observed in mature stomata guard cells, in which neither the mitotic cell cycles nor endocycles occur. CYCA2;3 might prevent mature stomata guard cells from reentering the S phase, which leads to an extra mitotic cycle or endocycle through the repression of DNA replication licensing (see below). One of the Medicago A2-type cyclin genes, Medsa;cycA2;2, has been shown to be expressed in proliferating tissues but not in endoreduplicating tissues (Roudier et al., 2003), suggesting that some A2-type cyclin genes are specialized to the mitotic cell cycle.
A remarkable increase in the proportion of 4C cells was observed specifically in plants expressing mDB-CYCA2;3-GFP. This might be caused by retardation of the mitotic cell cycle in the M phase, in which the mutant protein escaping from D-box–mediated protein degradation is expected to accumulate abnormally. The fact that human cyclin A mutants lacking a D-box cause cell cycle arrest in anaphase (Geley et al., 2001) supports this idea. It is likely that mDB-CYCA2;3-GFP increases the 4C cell population through a totally different pathway from that for the termination of endoreduplication. Multicellular trichomes were not observed in either CYCA2;3-GFP– or mDB-CYCA2;3-GFP–overexpressing plants (data not shown), suggesting that CYCA2;3 does not contribute to the CDK activity that triggers cell divisions, unlike CYCB1;2 and CYCD3;1 (Schnittger et al., 2002a, 2002b).
Molecular Mechanism for the Negative Regulation of Endocycles
Studies of endoreduplication have proposed a regulatory mechanism for the mitosis-to-endocycle transition, in which mitotic CDK activity is a parameter that sustains mitotic cell cycles (Edgar and Orr-Weaver, 2001; Larkins et al., 2001; Lilly and Duronio, 2005). Various regulatory proteins of the mitotic CDK activity have been suggested as key regulators for the entry of endoreduplication in plants. However, the mechanisms for the termination of endocycle succession at appropriate ploidy levels remain unclear. Our study presents evidence that a particular cyclin acts as a key regulator of ploidy levels in plant endoreduplication.
The endocycle regulatory mechanism is thought to be common to the G1/S transition of the mitotic cell cycle (Edgar and Orr-Weaver, 2001). In the animal mitotic cell cycle, cyclin A/CDKs have been studied with respect to the promotion of S-phase entry and the downregulation of APC activity (Sherr and Roberts, 2004). Recently, it was revealed that animal cyclin A/CDKs function also in downregulating the components of pre-RC at the posttranslational level (Petersen et al., 1999; Coverley et al., 2000, 2002; Nishitani et al., 2004; Sugimoto et al., 2004). From this point of view, CYCA2;3 may negatively regulate endocycle succession through the downregulation of pre-RC function. The physical interaction between the recombinant CDT1a and CYCA2;2 proteins in an in vitro pull-down experiment (del Mar Castellano et al., 2004) supports this possibility. Animal cyclin A/CDKs also negatively regulate G1/S-related transcription factors, including E2F-DP (Dynlacht et al., 1994; Krek et al., 1994; Xu et al., 1994; Hayashi and Yamaguchi, 1999). In a transcription analysis using real-time quantitative PCR, however, significant differences in the transcript level of CDC6a, which is regulated by E2Fa-DPa (De Veylder et al., 2002), could not be detected among the wild type, the mutant, and the CYCA2;3-GFP and mDB-CYCA2;3-GFP overexpressors (see Supplemental Figure 5 online), suggesting that CYCA2;3 is not involved in transcriptional regulation.
The D-box mutation stabilized the CYCA2;3 fusion protein and enhanced the restraint of endocycles. This finding strongly suggests that the CYCA2;3 function in endocycles is negatively regulated by APC, which promotes D-box–mediated protein degradation. In Medicago nodules, a decreased level of the APC activator CCS52A causes a reduction in the population of endoreduplicated cells, especially those with a ploidy level of 16C or higher (Vinardell et al., 2003). CCS52 may be involved in the regulation of ploidy levels through the negative regulation of A2-type cyclins.
As for the CDK responsible for the negative regulation of endocycles, CDKA;1 is most likely to be the counterpart of CYCA2;3 based on our results (i.e., their possible coexistence and ability of physical interaction). Consistent with this, a Medicago A2-type cyclin, Medsa;CYCA2;2, specifically interacts with the PSTAIRE-type CDK in the yeast two-hybrid system (Roudier et al., 2000). In Arabidopsis, it has been proposed that CDKA;1 activity is downregulated at the entry of endoreduplication and remains at a low level for the progression of endoreduplication (Verkest et al., 2005). In this context, the upregulation of CDKA;1 activity could be one of the determinants of the termination of endoreduplication, and CYCA2;3 may play a pivotal role in regulating not only the activation but also the substrate specificity of CDKA;1.
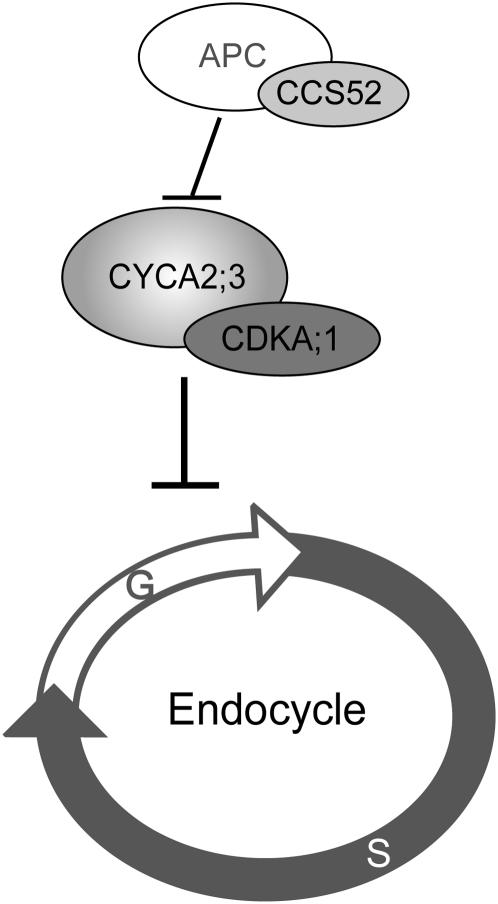
Figure 9 illustrates a working hypothesis of the signal transduction cascade in which CYCA2;3 acts as a negative regulator of endocycles. In this model, the rate of endocycles depends on the rate of entry into each DNA synthesis phase, and CYCA2;3/CDKA;1 negatively regulates the entry rate. When the endocycle succession is to be terminated, the activity of CYCA2;3/CDKA;1 is upregulated to prevent the entry into the next DNA synthesis phase. Upstream, CYCA2;3 is downregulated through protein degradation mediated by APC, the activation of which possibly involves the function of CCS52.
Figure 9.
Model for the Signal Cascade Negatively Regulating Endocycles through the Function of CYCA2;3.
A hypothesis for a signal cascade that negatively regulates endocycles is illustrated. CYCA2;3/CDKA;1 negatively regulates the rate of entry into the DNA synthesis phase of each endocycle. CYCA2;3 is downregulated through protein degradation mediated by APC, the activation of which possibly involves the function of CCS52. Other A2-type cyclins may have the same function as CYCA2;3. T bars indicate negative regulation.
Conclusion and Perspectives
CYCA2;3 negatively regulates endocycles and acts as a key regulator of ploidy levels in Arabidopsis endoreduplication. We propose that the level of CYCA2;3 protein, which is regulated in both tissue-specific and developmental stage–specific manners, is a parameter terminating endocycle succession. To further understand the regulatory mechanism for the termination of endocycle succession in plant cells, the interaction of the CYCA2;3/CDKA;1 complex with other regulatory proteins, including components of APC and pre-RC, should be investigated.
METHODS
Plant Materials and Growth Conditions
All Arabidopsis thaliana lines used were in the Columbia background. Columbia was used as the wild type. The T-DNA insertion lines SALK_086463 and SALK_092515 were identified in the collection of the Salk Institute Genomic Analysis Laboratory and obtained from the ABRC. Arabidopsis plants were grown on Murashige and Skoog medium containing 0.8% agar, B5 vitamins, and 1% sucrose under daylength conditions of 16 h of light/8 h of dark and at 22°C, unless noted otherwise.
Transgene Constructs and Arabidopsis Transformation
DNA fragments upstream of the CYCA2;2, CYCA2;3, and CYCA2;4 initiation codons were amplified by PCR using Arabidopsis genomic DNA and the following primer sets: 5′-GTCGACTTTCTGCCGCCGTTTAT-3′ and 5′-CCCGGGAATTTAGTCTGGAAAAAGGCC-3 for CYCA2;2, 5′-AAAGCCATGGTTACTATTAT-3′ and 5′-CCCCGGATCCGTTTAAAAATCCCACGATAAA-3′ for CYCA2;3, and 5′-GTCGACTTTGAAAACAAAGATAATTCTGAATAAG-3′ and 5′-CCCGGGGATTGAAACCCTTTGACACG-3′ for CYCA2;4 (the underlined sequences are the restriction sites used for the cloning of PCR fragments). To construct the CYCA2;2 promoter–GUS and CYCA2;4 promoter–GUS genes, the corresponding PCR fragments were digested with SalI and SmaI and substituted for the 35S promoter region in the binary vector pBI121 using the SalI and SmaI sites of the vector. To construct the CYCA2;3 promoter–GUS gene, the corresponding PCR fragment was digested with NcoI and BamHI and substituted for the 35S promoter region in a pBI121-derived vector containing a hygromycin resistance gene in the T-DNA region, using the HindIII and BamHI sites of the vector. The NcoI end of the fragment and the HindIII end of the vector were filled in before ligation. The DNA fragments encoding CYCA2;3 and the engineered GFP with Ser at position 65 (sGFP) (Niwa et al., 1999) were fused in-frame to construct the CYCA2;3-GFP gene. The junction sequence between the two coding sequences was 5′-CTATTCCTCGAGGTCGACGGTATCGCGATAAGCTTGATTGCGGCCGCCATG-3′ (the sequences encoding the C-terminal amino acids of CYCA2;3 and the initiation codon of sGFP are underlined). To construct the CYCA2;3 promoter–driven CYCA2;3-GFP gene, the fragment encoding the fusion protein was substituted for the GUS coding region in the CYCA2;3 promoter–GUS gene using the BamHI and SacI sites preceding and following the GUS coding region, respectively. The sequences of the BamHI and SacI junctions were 5′-GGATCCATGGGG-3′ and 5′-AAGTAAGAGCTC-3′, respectively (the initiation and termination codons of CYCA2;3-GFP are underlined). To construct the estrogen-inducible CYCA2;3-GFP gene, the DNA fragment encoding the CYCA2;3-GFP protein was fused between the XhoI and SpeI sites of pER-8 (Zuo et al., 2000). The sequences of the XhoI and SpeI junctions were 5′-CTCGAGATTTAAATATGGGG-3′ and 5′-AAGTAAACTAGT-3′, respectively (the initiation and termination codons of CYCA2;3-GFP are underlined). In the mDB-CYCA2;3-GFP gene, in vitro mutagenesis was used to change the DNA sequence encoding the D-box from 5′-CGAGCGGTTCTAGGGGAGATCACAAAT-3′ to 5′-GGGGCGGTTGTTGGGGAGATCACAAAT-3′ (the sequences encoding the mutated amino acids are underlined). To construct the CDKA;1-GFP gene, the DNA fragments encoding CDKA;1 and sGFP were fused in an in-frame manner and substituted for the GUS-coding fragment in the construction of P(−591/+4):GUS (Imajuku et al., 2001). The junction sequence between CDKA;1 and sGFP was 5′-GCATGCCCCCTCGAGGTGGTGGTGGCGCGGTGAGC-3′ (the sequences encoding the C-terminal amino acids of CDKA;1 and the N-terminal amino acids of sGFP are underlined). Strains of Agrobacterium tumefaciens LBA4404 carrying each construct were used to transform Arabidopsis by vacuum infiltration. The resulting transgenic plants were self-pollinated, and T3 plants homozygous for the transgene were used in subsequent experiments, unless noted otherwise.
RT-PCR Analysis
To detect the CYCA2;3 transcript, first-strand cDNA was synthesized and amplified with the SuperScript One-Step RT-PCR kit containing Platium Taq (Invitrogen Life Technologies) using total RNA and the CYCA2;3-specific primers 5′-AGGCACAGATAACACAGCTG-3′ and 5′-TGAGGTAGAGAGTGTCAGATGC-3′. As an internal control, primers specific to the ACT2 gene (At3g18780), 5′-TGGTGTCATGGTTGGGATG-3′ and 5′-CACCACTGAGCACAATGTTAC-3′, were used in the same reaction mixture. For quantitative RT-PCR analysis, first-strand cDNA was synthesized with the First-Strand cDNA Synthesis System for Quantitative RT-PCR (Marligen Biosciences) with total RNA. The cDNA was then subjected to PCR with the same primers. The PCR products (a 296-bp fragment for CYCA2;3 and a 782-bp fragment for ACT2) were fractionated by agarose gel electrophoresis and stained with ethidium bromide.
Real-Time Quantitative PCR
First-strand cDNA was synthesized from a total RNA fraction treated with DNase I and subjected to real-time quantitative PCR. Expression analysis of cyclin A2 genes was performed with the SYBR Green method using the DyNAmo HS SYBR Green qPCR kit (Finnzymes) and Rotor-Gene 3000A (Corbett Research). ACT2 transcripts were also quantified to normalize the amount of total transcripts in each sample. Primers used for the quantitative analysis were as follows: ACT2, 5′-AACCCAAAGGCCAACAGAGA-3′ and 5′-CCATCACCAGAATCCAGCAC-3′; CYCA2;1, 5′-AAAGACGAGCGGTGCTCAA-3′ and 5′-CGCCATCCACATCTACCAAA-3′; CYCA2;2, 5′-CGAAAGGGAGGCAACATCA-3′ and 5′-CATGGCACTGTTAGCACCTTC-3′; CYCA2;3, 5′-AGAGCCACTGGACCCAACA-3′ and 5′-ATGACCGCGTCCTTTCTTTATC-3′; CYCA2;4, 5′-TCTTGAAGAAATTTCTGGAGCGTA-3′ and 5′-GTAGCAGCCACTTCAGAAGATGTTA-3′. Standard cDNA templates for quantity calibration were produced by RT-PCR from a total RNA fraction and purified by fractionation with agarose gel electrophoresis. Primers used for this RT-PCR were as follows: ACT2, 5′-ATGGCTGAGGCTGATGA-3′ and 5′-TCAATTTGCATGTAAGAGAATTC-3′; CYCA2;1, 5′-ATGCATAGAGCTTCTTCTAAGCA-3′ and 5′-TCATCTTGAGAAGAGTGTGTTCA-3′; CYCA2;2, 5′-ATGTATTGCTCTTCTTCGATG-3′ and 5′-TCATCTTGAGAATAGTGATGTGAC-3′; CYCA2;3, 5′-ATCTCGATCTGTGTCCAAAGC-3′ and 5′-TCAGAATAGCGTGTCAAGTAGC-3′; CYCA2;4, 5′-GAGCTCATTTCCCCAAATTA-3′ and 5′-TTAGGAGATGAATAGCTTGTCC-3′.
For the expression analysis of CDC6, E2Fa, and DPa genes, the TaqMan method (Roche Molecular Systems) was used with Rotor-Gene 3000A (Corbett Research). Primers and florescent probes used are listed as follows. ACT2 primers, 5′-TGTGCCAATCTACGAGGGTTTC-3′ and 5′-TTTCCCGCTCTGCTGTTGTG-3′; ACT2 probe, 5′-TET-ATGCCATCCTCCGTCTTGACCTTGCTG-TAMRA-3′; CDC6a primers, 5′-AAGTTAGAGGATCAATAGATCAAGAACG-3′ and 5′-TCTTGGACAATGCAGCTATCATATG-3′; CDC6a probe, 5′-FAM-TGACCACTTGACACTCTGGAACTGGACCTT-TAMRA-3′; DPa primers, 5′-GCAGAGAAGCCTTTGAATGAAAATG-3′ and 5′-ATATCCAACGCCATGAACACATTG-3′; DPa probe, 5′-FAM-CGCATCGTAGACTCTCCGCCTTATGTTCTT-TAMRA-3′; E2Fa primers, 5′-AGCCTTTCAAGAATCGAATACTTTGG-3′ and 5′-TGTAATACAGATACGTCAGCATCCTC-3′; E2Fa probe, 5′-FAM-TCGCCAGGACACGCATCAACTCCCT-TAMRA-3′ (TET, TAMRA, and FAM are tetrachlorofluorescein, 6-carboxytetramethylrhodamine, and 6-carboxyfluorescein, respectively).
Measurement of the Sizes of Trichome Nuclei
Trichomes were isolated from rosette leaves of 3-week-old plants as described (Zhang and Oppenheimer, 2004). The trichomes were incubated for 3 h in PBST (50 mM NaPO4, 150 mM NaCl, and 0.05% Triton X-100, pH 7.2) containing 0.5 μg/mL DAPI at room temperature and then washed three times with PBST. The DAPI-stained nuclei were photographed by fluorescence microscopy (AxioPhot2; Carl Zeiss), and the projection area of each nucleus was measured using the imaging software Image Ana-LITE (Omron).
Flow Cytometric Analysis
Plant ploidy was analyzed using the Ploidy Analyzer PA flow cytometer (Partec), according to the manufacturer's instructions. Baselines were calculated using the polynomial trendline function of Microsoft Excel (example baselines are the gray lines shown in Figures 2E and 2F). Before calculating the total count in each peak, the value at each position on the baseline was subtracted from the corresponding value of raw data.
Immunoprecipitation and Protein Gel Blotting
Plant samples were harvested, ground into powder in liquid nitrogen, and suspended in lysis buffer (Cockcroft et al., 2000) supplemented with protease inhibitor cocktail for plant cell and tissue extracts (Sigma-Aldrich). After centrifugation, the cleared lysate was incubated for 4 h at 4°C with BD Living Colors full-length A.v. polyclonal antibody (BD Bioscience) and protein A Sepharose CL-4B (Amersham Biosciences). Immunoprecipitate was washed with wash buffer (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 5 mM EGTA, and 1 mM DTT). The proteins in the precipitate were fractionated by SDS-PAGE and transferred onto polyvinylidene difluoride filter paper (Amersham Biosciences). Protein gel blotting was performed with mouse monoclonal antibodies specific to the PSTAIR peptide sequence (P7962; Sigma-Aldrich) and GFP (11E5; Molecular Probes) as primary antibodies and anti-mouse IgG antibody conjugated with peroxidase (A4416; Sigma-Aldrich) as a secondary antibody. Blotting signals was detected with the enhanced chemiluminescence protein gel blot detection system (Amersham Biosciences).
Other Methods
Histochemical GUS analysis was performed as described previously (Imajuku et al., 2001). The surfaces of wild-type leaves were observed with the S-3500N scanning electron microscope (Hitachi) in variable-pressure mode. GFP fluorescence was observed with the LSM510 confocal laser-scanning microscope (Carl Zeiss). For analyses of the inner structures of root tips and cotyledon epidermis cell shapes, plant tissues were fixed and cleared essentially as described (Umeda et al., 2000) and observed with Nomarski DIC microscopy using Axioplan 2 (Carl Zeiss).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers Z31589 (CYCA2;1), Z31402 (CYCA2;2), NM_101426 (CYCA2;3), NM_106686 (CYCA2;4), and NM_114734 (CDKA;1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Patterns of CYCA2;3 Promoter Activity and the Nuclear Localization of the CDKA;1-GFP Fusion Protein in Trichome Development.
Supplemental Figure 2. Histochemical Analysis of the CYCA2;2 and CYCA2;4 Promoters.
Supplemental Figure 3. Histochemical Analysis of CYCA2;3 Promoter Activity and Ploidy Distribution Analysis in Etiolated Hypocotyls.
Supplemental Figure 4. Expression Analysis of Arabidopsis A2-Type Cyclin Genes by Real-Time Quantitative PCR.
Supplemental Figure 5. Expression Analysis of G1/S-Related Genes by Real-Time Quantitative PCR.
Supplementary Material
Acknowledgments
We thank K. Yasuda for technical assistance; T. Wada, R. Tsugeki, and M. Ueda for technical advice on the microscopic observations; F. Toyoshima-Morimoto and Y. Miyata for technical advice on immunoprecipitation and protein gel blotting; Y. He, S. Hata, A. Sugiyama, and K. Yazaki for technical advice on real-time PCR; Y. Niwa for providing the GFP coding fragment; and N.-H. Chua for providing the estrogen-inducible gene expression system. This work was supported by Grants-in-Aid for Scientific Research (B) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan, to T.A. (Grant 16370023) and A.O. (Grant 16370022).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Takashi Aoyama (aoyama@scl.kyoto-u.ac.jp).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.037309.
References
- Blow, J.J., and Dutta, A. (2005). Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 6 476–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf, V., Vlieghe, K., Beemster, G.T.S., Magyar, Z., Torres Acosta, J.A., Maes, S., Van Der Scheren, E., Inzé, D., and De Veylder, L. (2004). The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16 2683–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burssens, S., de Almeida Engler, J., Beeckman, T., Richard, C., Shaul, O., Ferreira, P., Van Montagu, M., and Inzé, D. (2000). Developmental expression of the Arabidopsis thaliana CycA2;1 gene. Planta 211 623–631. [DOI] [PubMed] [Google Scholar]
- Castellano, M.M., del Pozo, J.C., Ramirez-Parra, E., Brown, S., and Gutierrez, C. (2001). Expression and stability of Arabidopsis CDC6 are associated with endoreplication. Plant Cell 13 2671–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebolla, A., Vinardell, J.M., Kiss, E., Olah, B., Roudier, F., Kondorosi, A., and Kondorosi, E. (1999). The mitotic inhibitor ccs52 is required for endoreduplication and ploidy-dependent cell enlargement in plants. EMBO J. 18 4476–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockcroft, C.E., den Boer, B.G., Healy, J.M., and Murray, J.A. (2000). Cyclin D control of growth rate in plants. Nature 405 575–579. [DOI] [PubMed] [Google Scholar]
- Coverley, D., Laman, H., and Laskey, R.A. (2002). Distinct roles for cyclins E and A during DNA replication complex assembly and activation. Nat. Cell Biol. 4 523–528. [DOI] [PubMed] [Google Scholar]
- Coverley, D., Pelizon, C., Trewick, S., and Laskey, R.A. (2000). Chromatin-bound Cdc6 persists in S and G2 phases in human cells, while soluble Cdc6 is destroyed in a cyclin A-cdk2 dependent process. J. Cell Sci. 113 1929–1938. [DOI] [PubMed] [Google Scholar]
- del Mar Castellano, M., Boniotti, M.B., Caro, E., Schnittger, A., and Gutierrez, C. (2004). DNA replication licensing affects cell proliferation or endoreplication in a cell type-specific manner. Plant Cell 16 2380–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., Beemster, G.T., Krols, L., Terras, F., Landrieu, I., van der Schueren, E., Maes, S., Naudts, M., and Inzé, D. (2001). Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13 1653–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., Beemster, G.T.S., de Almeida Engler, J., Ormenese, S., Maes, S., Naudts, M., Van Der Schueren, E., Jacqmard, A., Engler, G., and Inzé, D. (2002). Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J. 21 1360–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte, W., and Murray, J.A.H. (2003). The plant cell cycle. Annu. Rev. Plant Biol. 54 235–264. [DOI] [PubMed] [Google Scholar]
- Diffley, J.F.X. (2004). Regulation of early events in chromosome replication. Curr. Biol. 14 R778–R786. [DOI] [PubMed] [Google Scholar]
- Dynlacht, B.D., Flores, O., Lees, J.A., and Harlow, E. (1994). Differential regulation of E2F transactivation by cyclin/cdk2 complexes. Genes Dev. 8 1772–1786. [DOI] [PubMed] [Google Scholar]
- Edgar, B.A., and Orr-Weaver, T.L. (2001). Endoreplication cell cycles: More for less. Cell 105 297–306. [DOI] [PubMed] [Google Scholar]
- Esch, J.J., Chen, M., Sanders, M., Hillestad, M., Ndkiun, S., Idelkope, B., Neizer, J., and Marks, M.D. (2003). A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development 130 5885–5894. [DOI] [PubMed] [Google Scholar]
- Galbraith, D.W., Harkins, K.R., and Knapp, S. (1991). Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiol. 96 985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geley, S., Kramer, E., Gieffers, C., Gannon, J., Peters, J.M., and Hunt, T. (2001). Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J. Cell Biol. 153 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau, E., Hofte, H., Grandjean, O., Brown, S., and Traa, J. (1998). Phytochrome controls the number of endoreduplication cycles in the Arabidopsis thaliana hypocotyl. Plant J. 13 221–230. [DOI] [PubMed] [Google Scholar]
- Gendreau, E., Traas, J., Desnos, T., Grandjean, O., Caboche, M., and Hofte, H. (1997). Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 114 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik, P., Criqui, M.C., Parmentier, Y., Derevier, A., and Fleck, J. (1998). Cell cycle-dependent proteolysis in plants: Identification of the destruction box pathway and metaphase arrest produced by the proteasome inhibitor MG132. Plant Cell 10 2063–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer, M., Murray, A.W., and Krischner, M.W. (1991). Cyclin is degraded by the ubiquitin pathway. Nature 349 132–138. [DOI] [PubMed] [Google Scholar]
- Harper, J.W., Burton, J.L., and Solomon, M.J. (2002). The anaphase-promoting complex: It's not just for mitosis any more. Genes Dev. 16 2179–2206. [DOI] [PubMed] [Google Scholar]
- Hayashi, S., and Yamaguchi, M. (1999). Kinase-independent activity of Cdc2/cyclin A prevents the S phase in the Drosophila cell cycle. Genes Cells 4 111–122. [DOI] [PubMed] [Google Scholar]
- Hulskamp, M. (2004). Plant trichomes: A model for cell differentiation. Nat. Rev. Mol. Cell Biol. 5 471–480. [DOI] [PubMed] [Google Scholar]
- Hulskamp, M., Misra, S., and Jurgens, G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76 555–566. [DOI] [PubMed] [Google Scholar]
- Imajuku, Y., Ohashi, Y., Aoyama, T., Goto, K., and Oka, A. (2001). An upstream region of the Arabidopsis thaliana CDKA;1 (CDC2aAt) gene directs transcription during trichome development. Plant Mol. Biol. 46 205–213. [DOI] [PubMed] [Google Scholar]
- Jasinski, S., Riou-Khamlichi, C., Roche, O., Perennes, C., Bergounioux, C., and Glab, N. (2002). The CDK inhibitor NtKIS1a is involved in plant development, endoreduplication and restores normal development of cyclin D3;1-overexpressing plants. J. Cell Sci. 115 973–982. [DOI] [PubMed] [Google Scholar]
- Joubes, J., and Chevalier, C. (2000). Endoreduplication in higher plants. Plant Mol. Biol. 43 735–745. [DOI] [PubMed] [Google Scholar]
- Kondorosi, E., and Kondorosi, A. (2004). Endoreduplication and activation of the anaphase-promoting complex during symbiotic cell development. FEBS Lett. 567 152–157. [DOI] [PubMed] [Google Scholar]
- Kowles, R.V., and Phillips, R.L. (1985). DNA amplification patterns in maize endosperm nuclei during kernel development. Proc. Natl. Acad. Sci. USA 82 7010–7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek, W., Ewen, M.E., Shirodkar, S., Arany, Z., Kaelin, W.G., Jr., and Livingston, D.M. (1994). Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin A-dependent protein kinase. Cell 78 161–172. [DOI] [PubMed] [Google Scholar]
- Larkins, B.A., Dilkes, B.P., Dante, R.A., Coelho, C.M., Woo, Y.-M., and Liu, Y. (2001). Investigating the hows and whys of DNA endoreduplication. J. Exp. Bot. 52 183–192. [PubMed] [Google Scholar]
- Leiva-Neto, J.T., Grafi, G., Sabelli, P.A., Dante, R.A., Woo, Y.M., Maddock, S., Gordon-Kamm, W.J., and Larkins, B.A. (2004). A dominant negative mutant of cyclin-dependent kinase A reduces endoreduplication but not cell size or gene expression in maize endosperm. Plant Cell 16 1854–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly, M.A., and Duronio, R.J. (2005). New insights into cell cycle control from the Drosophila endocycle. Oncogene 24 2765–2775. [DOI] [PubMed] [Google Scholar]
- Mariconti, L., Pellegrini, B., Cantoni, R., Stevens, R., Bergounioux, C., Cella, R., and Albani, D. (2002). The E2F family of transcription factors from Arabidopsis thaliana. Novel and conserved components of the retinoblastoma/E2F pathway in plants. J. Biol. Chem. 277 9911–9919. [DOI] [PubMed] [Google Scholar]
- Marks, M.D. (1997). Molecular genetic analysis of trichome development in Arabidopsis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 137–163. [DOI] [PubMed] [Google Scholar]
- Melaragno, J.E., Mehrotra, B., and Coleman, A.W. (1993). Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5 1661–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges, M., de Jager, S.M., Gruissem, W., and Murray, J.A.H. (2005). Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 41 546–566. [DOI] [PubMed] [Google Scholar]
- Murphy, M. (1999). Correction. Delayed early embryonic lethality following disruption of the murine cyclin A2 gene. Nat. Genet. 23 481. [DOI] [PubMed] [Google Scholar]
- Murphy, M., Stinnakre, M.G., Senamaud-Beaufort, C., Winston, N.J., Sweeney, C., Kubelka, M., Carrington, M., Brechot, C., and Sobczak-Thepot, J. (1997). Delayed early embryonic lethality following disruption of the murine cyclin A2 gene. Nat. Genet. 15 83–86. [DOI] [PubMed] [Google Scholar]
- Nishitani, H., Lygerou, Z., and Nishimoto, T. (2004). Proteolysis of DNA replication licensing factor Cdt1 in S-phase is performed independently of geminin through its N-terminal region. J. Biol. Chem. 279 30807–30816. [DOI] [PubMed] [Google Scholar]
- Niwa, Y., Hirano, T., Yoshimoto, K., Shimizu, M., and Kobayashi, H. (1999). Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J. 18 455–463. [DOI] [PubMed] [Google Scholar]
- Peters, J.M. (2002). The anaphase-promoting complex: Proteolysis in mitosis and beyond. Mol. Cell 9 931–943. [DOI] [PubMed] [Google Scholar]
- Petersen, B.O., Lukas, J., Sorensen, C.S., Bartek, J., and Helin, K. (1999). Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J. 18 396–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier, F., Fedorova, E., Gyorgyey, J., Feher, A., Brown, S., Kondorosi, A., and Kondorosi, E. (2000). Cell cycle function of a Medicago sativa A2-type cyclin interacting with a PSTAIRE-type cyclin-dependent kinase and a retinoblastoma protein. Plant J. 23 73–83. [DOI] [PubMed] [Google Scholar]
- Roudier, F., Fedorova, E., Lebris, M., Lecomte, P., Gyorgyey, J., Vaubert, D., Horvath, G., Abad, P., Kondorosi, A., and Kondorosi, E. (2003). The Medicago species A2-type cyclin is auxin regulated and involved in meristem formation but dispensable for endoreduplication-associated developmental programs. Plant Physiol. 131 1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger, A., Schobinger, U., Bouyer, D., Weinl, C., Stierhof, Y.-D., and Hulskamp, M. (2002. b). Ectopic D-type cyclin expression induces not only DNA replication but also cell division in Arabidopsis trichomes. Proc. Natl. Acad. Sci. USA 99 6410–6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger, A., Schobinger, U., Stierhof, Y.-D., and Hulskamp, M. (2002. a). Ectopic B-type cyclin expression induces mitotic cycles in endoreduplicating Arabidopsis trichomes. Curr. Biol. 12 415–420. [DOI] [PubMed] [Google Scholar]
- Schnittger, A., Weinl, C., Bouyer, D., Schobinger, U., and Hulskamp, M. (2003). Misexpression of the cyclin-dependent kinase inhibitor ICK1/KRP1 in single-celled Arabidopsis trichomes reduces endoreduplication and cell size and induces cell death. Plant Cell 15 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr, C.J., and Roberts, J.M. (2004). Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 18 2699–2711. [DOI] [PubMed] [Google Scholar]
- Sugimoto, N., Tatsumi, Y., Tsurumi, T., Matsukage, A., Kiyono, T., Nishitani, H., and Fujita, M. (2004). Cdt1 phosphorylation by cyclin A-dependent kinases negatively regulates its function without affecting geminin binding. J. Biol. Chem. 279 19691–19697. [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu, K., and Roberts, K. (2003). “Big it up”: Endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 6 544–553. [DOI] [PubMed] [Google Scholar]
- Sun, Y., Dilkes, B.P., Zhang, C., Dant, R.A., Carneiro, N.P., Lowe, K.S., Jung, R., Gordon-Kamm, W.J., and Larkins, B.A. (1999). Characterization of maize (Zea mays L.) Wee1 and its activity in developing endosperm. Proc. Natl. Acad. Sci. USA 96 4180–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski, D.B., Marks, M.D., and Wick, S.M. (1999). Organized F-actin is essential for normal trichome morphogenesis in Arabidopsis. Plant Cell 11 2331–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda, M., Umeda-Hara, C., and Uchimiya, H. (2000). A cyclin-dependent kinase-activating kinase regulates defferentiation of root initial cells in Arabidopsis. Proc. Natl. Acad. Sci. USA 97 13396–13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele, K., Raes, J., De Veylder, L., Rouze, P., Rombauts, S., and Inzé, D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkest, A., Manes, C.-L., Vercruysse, S., Maes, S., Van Der Schueren, E., Beeckman, T., Genschik, P., Kuiper, M., Inzé, D., and De Veylder, L. (2005). The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. Plant Cell 17 1723–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinardell, J.M., Fedorova, E., Cebolla, A., Kevei, Z., Horvath, G., Kelemen, Z., Tarayre, S., Roudier, F., Mergaert, P., Kondorosi, A., and Kondorosi, E. (2003). Endoreduplication mediated by the anaphase-promoting complex activator CCS52A is required for symbiotic cell differentiation in Medicago truncatula nodules. Plant Cell 15 2093–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe, K., Boudolf, V., Beemster, G.T.S., Mares, S., Magyar, Z., Atanassova, A., de Almeida Engler, J., De Groodt, R., Inzé, D., and De Veylder, L. (2005). The DP-E2F-like gene DEL1 controls the endocycle in Arabidopsis thaliana. Curr. Biol. 15 59–63. [DOI] [PubMed] [Google Scholar]
- Walker, J.D., Oppenheimer, D.G., Concienne, J., and Larkin, J.C. (2000). SIAMESE, a gene controlling the endoreduplication cell cycle in Arabidopsis thaliana trichomes. Development 127 3931–3940. [DOI] [PubMed] [Google Scholar]
- Weinl, C., Marquardt, S., Kuijt, S.J., Nowack, M.K., Jakoby, M.J., Hulskamp, M., and Schnittger, A. (2005). Novel functions of plant cyclin-dependent kinase inhibitors, ICK1/KRP1, can act non-cell-autonomously and inhibit entry into mitosis. Plant Cell 17 1704–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, M., Sheppard, K.-A., Peng, C.-Y., Yee, A.S., and Piwnica-Worms, H. (1994). Cyclin A/CDK2 binds directly to E2F-1 and inhibits the DNA-binding activity of E2F-1/DP-1 by phosphorylation. Mol. Cell. Biol. 14 8420–8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y., Steinmetz, A., Meyer, D., Brown, S., and Shen, W.-H. (2003). The tobacco A-type cyclin, Nicta;CYCA3;2, at the nexus of cell division and differentiation. Plant Cell 15 2763–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., and Oppenheimer, D.G. (2004). A simple and efficient method for isolating trichomes for downstream analyses. Plant Cell Physiol. 45 221–224. [DOI] [PubMed] [Google Scholar]
- Zhou, Y., Wang, H., Gilmer, S., Whitwill, S., and Fowke, L.C. (2003). Effects of co-overexpressing the plant CDK inhibitor ICK1 and D-type cyclin genes on plant growth, cell size and ploidy in Arabidopsis thaliana. Planta 216 604–613. [DOI] [PubMed] [Google Scholar]
- Zuo, J., Niu, Q.-W., and Chua, N.-H. (2000). An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24 265–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.